Abstract
Background
Feline calicivirus (FCV) is a major and highly infectious pathogen in cats worldwide. However, there have been limited studies about the status of FCV infections in Korea.
Objectives
To investigate the current status of FCV infections in stray cats in Korea.
Methods
A novel reverse transcription polymerase chain reaction (RT-PCR) assay was developed based on the conserved nucleotide sequences of reported FCV strains. Field swab samples were collected from 122 cats (2 hospital admitted cats and 120 stray cats) in 2016 and 2017. All the samples were tested by virus isolation and 2 different RT-PCRs, including the novel RT-PCR, for the detection of FCV.
Results
The novel RT-PCR assay showed no cross-reactivity to the nucleic acids of the other feline pathogens tested, and the limit of detection was calculated as 100 TCID50/mL based on an in vitro assessment. The novel RT-PCR assay detected 5 positive samples from the 122 field samples, which showed perfect agreement with the results of the virus isolation method. In contrast, another RT-PCR assay used in a previous study in Korea detected no positive samples. The prevalence of FCV infection in stray cats was 2.5% (3/120) based on the results of virus isolation and the novel RT-PCR assays.
Conclusions
The current study is the first report of the detection and prevalence of FCV in stray cats in Korea. The novel RT-PCR assay developed in this study showed high sensitivity and specificity, which indicates a useful diagnostic assay to identify FCV infection in cats.
Keywords: Feline calicivirus, reverse transcriptase PCR, prevalence
INTRODUCTION
Worldwide, feline calicivirus (FCV) is a major infectious pathogen widely distributed in cats. FCV infection generally leads to acute, mild to moderate, stomatitis, and upper respiratory tract disease (URTD). It can also cause arthritis and hemorrhagic-like fever, and more recently, virulent mutants of FCV have been identified as the cause of severe and acute virulent systemic disease [1,2,3].
It is difficult to differentiate FCV infection from respiratory infections caused by other respiratory pathogens, such as feline herpes virus type 1 (FHV-1) and Chlamydia psittaci, due to the similarity of clinical symptoms [4,5]. Thus, laboratory methods such as reverse transcription polymerase chain reaction (RT-PCR) assay, virus isolation, immunofluorescence tests, or serological tests are necessary for accurate diagnosis.
In general, virus isolation is considered to be a gold standard assay to detect FCV [6], and it is less sensitive to genomic variation which happens frequently in RNA viruses. However, this method is not easily applied for routine diagnosis. Virus isolation based on cell culture requires skillful techniques as well as increased labor and time [7]. Also, virus isolation might fail due to the presence of antibodies in extracellular fluids which inhibit viral replications in vitro [7,8].
Compared to virus isolation, the RT-PCR assay is a very useful method due to its easy, rapid, sensitive, and inexpensive features [5,9]. For the aforementioned reasons, an FCV specific RT-PCR assay has been developed and widely used for the detection of FCV infection. However, the high degree of gene variation in FCV due to RNA genome features can reduce the diagnostic sensitivity of FCV-specific molecular assays [10,11]. As gene mutations frequently occur, the nucleotide sequences of primer binding sites are likely to be changed. These changes may hamper the sensitivity of previously established RT-PCR assays [12].
In the present study, a novel RT-PCR assay was developed with a new primer set designed based on the sequences of ORF2 and ORF3 conserved regions. The new RT-PCR was applied to field samples, and the results were compared to those of a previously used RT-PCR and virus isolation methods to validate the accuracy.
MATERIALS AND METHODS
Sample collection
A total of 122 samples were obtained from the conjunctival sac, oropharynx, and nasal cavities of cats during the period from September 2016 to August 2017. Among them, 2 samples were collected from hospital-admitted cats with clinical signs; one with mild nasal discharge and the other had severe URTD (nasal bleeding, oral ulcer, and conjunctivitis). The remaining 120 samples were collected from the healthy stray cats captured for the purpose of trap-neuter-return. The swab samples collected from the conjunctival sac, oropharynx, and nose from a same cat were suspended together in 2 mL phosphate buffered saline (PBS). The suspension was centrifuged at 4,200 × g for 10 min at 4°C and the supernatant was collected. The supernatant was subjected to PCR assay and virus isolation.
Development of the novel RT-PCR
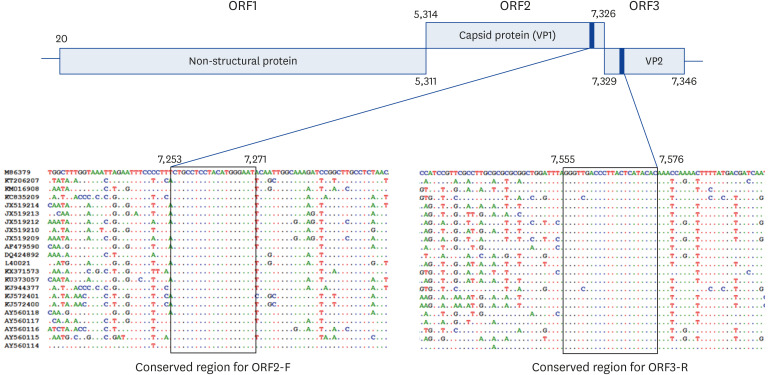
To design a novel primer set specific for FCV, the currently available whole genome sequences of FCV isolates were downloaded from GenBank (accession No. M86379, KT206207, KM016908, KC835209, JX519214, JX519213, JX519209, AF479590, DQ424892, L40021, KX371573, KU373057, KJ944377, KJ572401, KJ572400, AY560118, AY560114). After sequence alignment of the 17 global isolates, 2 conserved regions located on ORF2, encoding a major capsid protein VP1, and ORF3, encoding a minor capsid protein VP3, were identified and used for the novel PCR (Fig. 1). The forward primer, ORF2-F (5′-CTGCCTCCTACATGGGAAT-3′), binds on region F of ORF2, corresponding to the position of 7,253–7,271 bps of FCV strain F9 (GenBank accession No. M86379, Fig. 1). The binding site of reverse primer, ORF3-R (5′-GTGTATGAGTAAGGGTCRACCC-3′), is located on the starting region of ORF3, corresponding to the position 7,555–7,576 bps of strain F9. The expected size of the target amplicon was calculated as 324 bps.
Fig. 1. Genome map of FCV and the conserved regions. ORF2-F and ORF3-R primers were derived from the conserved regions from ORF2 and ORF3, respectively.
FCV, feline calicivirus.
RT-PCR conditions were optimized using total RNA extracted from the combined FVRC attenuated vaccine (PureVax feline4; Boehringer Ingelheim, Germany) which contains FHV-1 (F2 strain), FCV (F9 strain), feline parvovirus (FPV; Johnson Leopard Origin strain) and Chlamydophila felis (905 strain). Briefly, cDNA was synthesized from the extracted RNA using RNA to cDNA EcoDry Premix (Clontech, Japan, Cat No. 639546) following the manufacturer's recommendations. The PCR reaction was performed in 20 μL mixture containing 2 μL of cDNA solution, 1 μL of each primer (10 pmol), 2.5 mM of each dNTP, 10 mM Tris-HCL (pH 8.8), 1.5 mM MgCl2, 50 mM KCl, and 2.5 U of Taq polymerase. Thermal cycling conditions consisted of denaturation (95°C, 5 min), followed by 35 cycles of denaturation (95°C, 30 sec), primer annealing (57°C, 30 sec), and primer extension (72°C, 30 sec). A final extension was performed at 72°C (5 min).
To validate the RT-PCR assay, the amplified PCR products were purified and sequenced by Sanger's method [13]. A blast search of the obtained sequences was performed using the Blastn program of National Center for Biotechnology Information (NCBI) to identify the FCV-specific sequences. To determine the limit of detection (LOD), the FCV SU (the first FCV field isolate in this study by virus isolation as described below) were cultivated in Crandell-Rees feline kidney (CRFK) cells and the collected culture supernatant was adjusted to 106 TCID50/mL. The adjusted supernatant was serially diluted up to a concentration of 10−1 TCID50/mL by a 10-fold serial dilution method and these 8 diluents were subjected to the novel RT-PCR assay.
Detection of FCV in field samples by virus isolation
All of the supernatants prepared above were filtered through a 0.2-μm-pore-size filter and used as an inoculum for virus isolation. CRFK cells were used to cultivate and isolate FCV. In brief, CRFK cells were plated on 12-well plates at the concentration of 1 × 105 cells/mL in each well. After overnight incubation, the plates were washed 3 times with PBS, and then 100 μL of inoculum was added on the cell monolayer in each well. For absorption of the virus, the inoculated plates were incubated at 37°C with 5% CO2 for 2 h. After 2 h incubation, the plates were washed 3 times with PBS, and refilled with 300 μL of Dulbecco's Modified Eagle's medium containing 5% fetal bovine serum. Cytopathic effect (CPE) was observed for 3 days. If CPE developed, the cell culture was maintained until 50%–70% CPE developed, and then the cell culture supernatant was collected. Virus identification was performed by antigen detection using the RT-PCR assay as described above. If CPE were not observed after 5 passages, the virus isolation result was considered to be negative.
Detection of FCV in field samples by molecular methods
All of the 122 swab samples collected in this study were tested for the presence of FCV by 2 different RT-PCR methods; the first was a previously developed RT-PCR (Cal-RT-PCR) method widely used in many studies including a previous study in Korea [5,11,14,15,16], and the second was the RT-PCR developed in this study. RNA extraction and cDNA generation from all samples were conducted as described above. The 2 RT-PCR assays were performed as described previously [14] and above, respectively. If the RT-PCR results were positive with an expected amplicon size, the PCR products were sequenced for the further identification of FCV, as described above. If the RT-PCR results were false negative, compared to those of virus isolations, high-titer FCV field isolates (1 × 1010 TCID50/mL) were prepared and the LOD of the PCR assay was determined as described above.
Comparison of RT-PCR and virus isolation methods for the detection of FCV in field samples
Kappa statistics were used to compare the new PCR assay with virus isolation and Cohen's kappa coefficient (κ) was calculated from the following formula:
| κ = (Po − Pe)/(1 − Pe) |
| Po = (a + d)/(a + b + c + d), Pe = Pyes + Pno |
| Pyes = (a + b)/(a + b + c + d) ∙ (a + c)/(a + b + c + d), Pno = (c + d)/(a + b + c + d) ∙ (b + d)/(a + b + c + d) |
Where ‘a’ is the number of positives in both methods, ‘b’ is the number of positives in RT-PCR and negatives in virus isolation, ‘c’ is the number of negatives in RT-PCR and positives in virus isolation, ‘d’ is the number of negatives in both methods. The κ varies from 0 to 1 and it means that 0 = agreement equivalent to chance, 0.10–0.20 = slight agreement, 0.21–0.40 = fair agreement, 0.41–0.60 = moderate agreement, 0.61–0.80 = substantial agreement, 0.81–0.99 = near perfect agreement, and 1 = perfect agreement [17].
RESULTS
Validation of the new RT-PCR assay
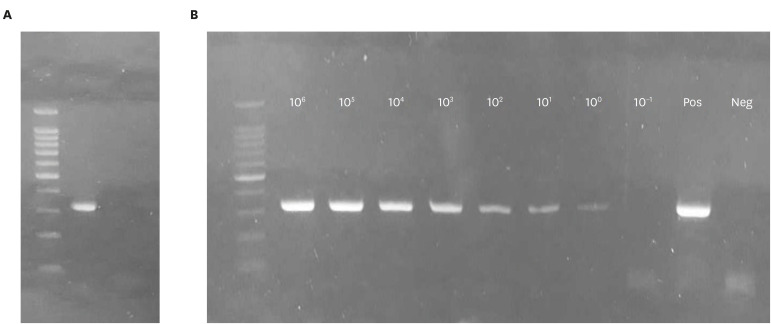
The specificity of a new primer set was first tested in silico, using Nucleotide-BLST and Primer-BLAST programs on NCBI website (https://blast.ncbi.nlm.nih.gov/Blast.cgi). No possible PCR product was detected for all known feline pathogens and feline genomes. Then, the RT-PCR was performed with the extracted nucleic acids from the combined FVRC attenuated vaccine containing feline pathogens (FCV, FHV-1, FPV, and C. felis). Only one band around 324 bp was visualized, which was the expected size of the FCV target amplicon and there was no cross-reactivity to the nucleic acids of the other feline pathogens used for specificity evaluation (Fig. 2A). Also, non-specific fragments were not observed in electrophoresis of the novel RT-PCR performed with 122 field samples (data not shown). The LOD was determined to be as low as 100 TCID50 for the novel RT-PCR developed in the current study (Fig. 2B).
Fig. 2. Validation of the novel RT-PCR. (A) The result of a new RT-PCR assay conducted with nucleic acids of feline pathogens (FCV, FHV-1, FPV, and Chlamydophila felis) extracted from the combined FVRC attenuated vaccine. Only one target amplicon (324 bp) was visualized and there was no cross-reactivity. Lane 1, DNA size marker; lane 2, RT-PCR with FVRC attenuated vaccine. (B) LOD of the new RT-PCR was determined with a cultured filed isolate (FCV 60). Lane 1, DNA size marker; lane 1–9, 10-fold serial diluents (106 to 10− 1 TCID50); lane 10–11, positive and negative controls, respectively. The target amplicons (312 bp) were identified from lanes 2 (106 TCID50) to 8 (100 TCID50). The LOD was determined as 100 TCID50/mL.
RT-PCR, reverse transcription polymerase chain reaction; FCV, feline calicivirus; FHV-1, feline herpes virus type 1; LOD, limit of detection.
Detection of FCV in field samples by virus isolation and RT-PCR assays
A total of 122 field samples were subjected to virus isolation and the 2 RT-PCR assays. In the virus isolation method, a total of 7 samples developed CPE in the cell culture after inoculation. Of these 7 samples, 5 samples (sample No. 60, 64, 88, 120, and SU) were confirmed as FCV positive by the novel PCR assay followed by sequence analyses, but 2 samples were FCV negative. The 2 FCV-negative samples that still showed CPE were concluded to be FHV-1 infections based on FHV-1 specific PCR positive reactions [16] (data not shown). Overall, 5 FCV isolates were recovered from the field samples. Of these 5 samples, 3 were from the healthy stray cats and the other 2 were from the cats with clinical symptoms (Table 1 and Fig. 3).
Table 1. Comparison of the virus isolation method and a novel RT-PCR assay for the detection of FCV in 122 field samples.
| Novel RT-PCR | Virus isolation | ||
|---|---|---|---|
| Positive | Negative | Total | |
| Positive | 5* | 0 | 5 |
| Negative | 0 | 117 | 117 |
| Total | 5 | 117 | 122 |
Cohen's kappa coefficient (κ) between the 2 assays was 1.
RT-PCR, reverse transcription polymerase chain reaction; FCV, feline calicivirus.
*The 5 positive samples by both 2 methods were from 2 hospital-admitted clinical cats (sample No. 121 and SU) and 3 healthy stray cats (sample No. 60, 64 and 88).
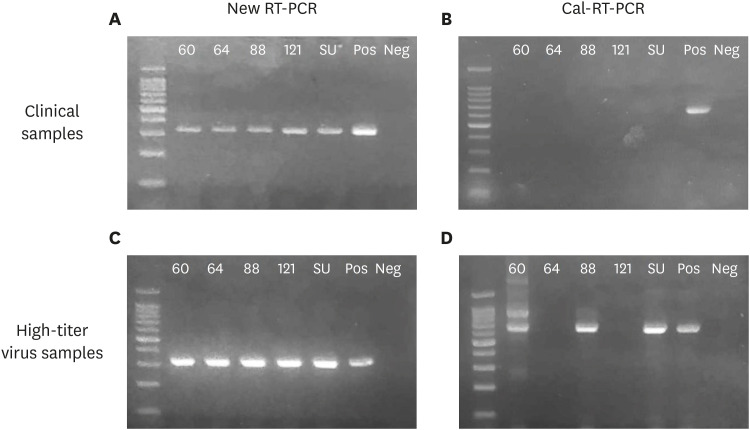
Fig. 3. Detection of FCV by 2 RT-PCR assays. The new RT-PCR assay developed in this study (left panel) and a RT-PCR assay developed by Seal [14] (Cal-RT-PCR, right panel) were performed for all 122 field samples to detect FCV infections. (A) The new RT-PCR assay amplified targeted amplicons (324 bp) from all 5 positive samples identified by virus isolation; however, (B) the old RT-PCR assay did not detect any FCV. (C, D) RT-PCR assays were performed for high virus titer samples of the 5 positive isolates (1010 TCID50). (C) The new RT-PCR assay amplified targeted amplicons (324 bp) from all of the samples, (D) but the old RT-PCR assay amplified the targeted amplicon (673 bp) in only 3 of the 5 positive samples with some non-specific bands. Lane 1, DNA size marker; lanes 2–4, FCV positive samples from healthy stray cats; lanes 5, FCV positive sample from a cat with nasal discharge; lane 6, FCV positive sample from a cat with severe URTD; lanes 7–8, a positive and a negative control, respectively.
FCV, feline calicivirus; RT-PCR, reverse transcription polymerase chain reaction; URTD, upper respiratory tract disease.
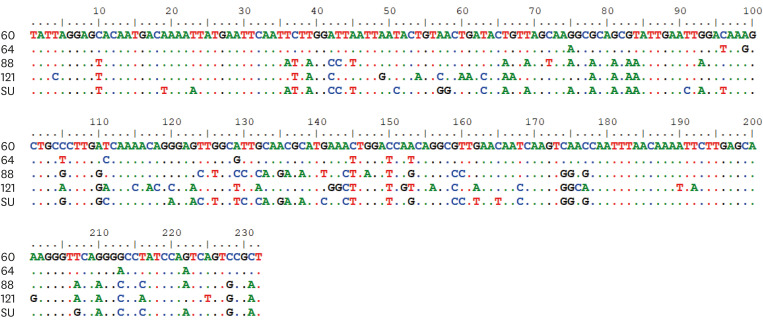
The 2 RT-PCR assays showed different levels of accuracy. The Cal-RT-PCR developed in a previous study [14] detected no positive sample at all (Fig. 3B). In contrast, the novel RT-PCR developed in this study showed 5 positive reactions from the 122 samples, which were further confirmed as true positives by sequence analyses (Fig. 3A). The sequence alignment results revealed that the similarity among the 5 isolates ranged from 75.4% to 95.2% each other (Table 2 and Fig. 4). The 5 RT-PCR positive samples were consistent with the virus isolation positive samples. The virus isolation assay is considered the gold standard, and the new RT-PCR showed 100% sensitivity and specificity. Taken together, the results of the novel RT-PCR assay showed a perfect agreement with the virus isolation method in the detection of FCV from 122 field samples (κ value = 1, Table 1).
Table 2. Nucleotide sequence similarity (%) matrix among PCR product sequences derived from 5 Korean FCV isolates.
| Isolate ID | 60 | 64 | 88 | 121 | SU |
|---|---|---|---|---|---|
| 60 | - | 95.2 | 80.6 | 77.5 | 75.4 |
| 64 | 95.2 | - | 81.0 | 77.1 | 77.5 |
| 88 | 80.6 | 81.0 | - | 78.8 | 91.8 |
| 121 | 77.5 | 77.1 | 78.8 | - | 78.0 |
| SU | 75.4 | 77.5 | 91.8 | 78.0 | - |
PCR, polymerase chain reaction; FCV, feline calicivirus.
Fig. 4. A comparison of nucleotide sequences of the 5 FCV isolates recovered in this study. The sequence alignment results of the 5 isolates for 232 bp on the PCR fragments are presented. They were all identified as FCVs by blast n search in NCBI database. The sequence similarity of the 5 FCV isolates ranged from 75.4% to 95.2% each other.
FCV, feline calicivirus; PCR, polymerase chain reaction; NCBI, National Center for Biotechnology Information.
Cal-RT-PCR was further tested with high-titer virus samples (1 × 1010 TCID50/mL) prepared with the cultured FCV field isolates from this study. Of these 5 isolates, the target band (673 bp) was amplified by Cal-RT-PCR only in the preparation of FCV isolates 60, 88, and SU. The nucleic acids of isolates 64 and 121 were not amplified even with the high-titer virus samples. Isolates 88 and 122 were used to determine the LOD of Cal-RT-PCR, since these 2 isolates only generated a specific target band without any nonspecific bands at high virus concentration (Fig. 3C and D). The LOD of Cal-RT-PCR was determined as 1 × 107 TCID50/mL (data not shown).
Prevalence of FCV in healthy stray cats in Korea
A total of 120 healthy stray cats were tested by virus isolation and the 2 different RT-PCR assays in this study. The result was considered FCV positive if any one of the tests was positive in this study. Virus isolation and the novel RT-PCR identified the same samples as positive (Table 1). Virus isolation or RT-PCR detection-based FCV apparent prevalence was calculated as 2.5% (3/120) in this study.
DISCUSSION
FCV is widely spread in the general cat population, including both clinically diseased and healthy cats worldwide [18]. The virus has even been detected and isolated from shelter cats and stray cats in other countries [9,19,20]. In contrast, there have been very limited studies about the prevalence of FCV in cat populations in Korea. Kang and Park [16] previously used the Cal-RT-PCR method and reported a 0% FCV prevalence in cats residing in an animal shelter. However, FCV has high genetic variability due to the RNA genome feature and this feature may lead to false negatives in the detection of FCV by RT-PCR methods [10,11,12]. The number of mutations on the primer binding sites significantly affects the efficiency of RT-PCR detection assays. It has been reported that 2 different RT-PCRs detected different portions of FCV in positive cats in the same tested samples due to nucleotide differences in the primer targeting sequences [21]. Recent studies in Europe reported that there were extensive variabilities in the nucleotide sequences of FCV isolated through European countries and there were no predominant strains that were internationally distributed. Rather, some strain clusters were restricted to a single country [20,22]. Therefore, the previous Korean study could have underestimated the prevalence of FCV if the molecular detection method (Cal-RT-PCR) was not appropriate to detect Korean isolates due to the nucleotide sequence differences as aforementioned. With this in mind, we developed a novel RT-PCR for FCV detection and used it to investigate the prevalence of FCV in stray cats, in comparison with virus isolation and Cal-RT-PCR methods.
The new primer set was designed based on the conserved sequences of 17 FCV strains available through the GenBank website. The condition of a novel RT-PCR was optimized using FVRC attenuated vaccine in which the FCV virus was included. Then, in the application with the 122 field samples, the novel RT-PCR detected 5 FCV positive samples; the analytical sensitivity of the new test was measured as low as 100 TCID50 with a Korean isolate [21], which indicates that the RT-PCR was very sensitive to detect FCV from feline samples. In contrast, Cal-RT-PCR used in the previous study in Korea [16] detected no positive samples although at least 5 cats were truly FCV positive (Table 1). With high-titer virus preparation of the 5 field isolates, Cal-RT-PCR only detected 3 of them (Fig. 3D). The LOD of Cal-RT-PCR measured with 2 field isolates in this study was 1 × 107 TCID50/mL, which suggested that this RT-PCR could not be used to detect FCV directly from swab samples without viral cultures in this study. The efficiency of Cal-RT-PCR to detect FCV was demonstrated to be comparable to that of virus isolation in a previous study in Australia [11]. The discrepancy of results between the previous study and this study might be attributed to the geographical distribution of different FCV strains. Taken together, the results of this study suggest that Cal-RT-PCR is not appropriate to investigate the prevalence of FCV infection in Korea, and the previous Korean study [16] might have underestimated the prevalence of FCV in Korean cats.
In general, sensitivity and specificity are the most important parameters in diagnostic assays. To determine the absolute sensitivity and specificity of a diagnostic test, enough numbers of true positive and negative samples are required. However, it is difficult, sometimes impossible, to obtain those numbers of samples in many kinds of infectious animal diseases. FCV is one of those cases in Korea. Alternatively, many new diagnostic tests were evaluated with considered reference tests. Virus isolation is a biological diagnostic method that is almost unaffected by genetic variation, as compared to molecular diagnostic methods [7]. Thus, virus isolation has been frequently used for the validation and comparison of sensitivity and specificity in development of RT-PCR for FCV [4,11,23,24]. Similarly, the new RT-PCR was further validated by comparison of the 122 field sample results with those of virus isolation in this study. Although limited numbers were tested, the results revealed perfect agreement (κ = 1) between the 2 FCV detection methods, which indicated both the specificity and sensitivity of the new RT-PCR at 100%. In addition to the identical results with those of viral isolation, the high specificity of the novel RT-PCR was further supported by the results with 122 field samples, in which no non-specific band was detected. The high sensitivity, especially to detect diverse FCV strains, was also supported by detecting 5 isolates showing high genetic diversity (Table 2 and Fig. 4). Taken together, the results indicate that the novel RT-PCR is applicable to field testing for feline FCV infection in Korea; also, it has several advantages of molecular detection over virus isolation methods.
This is the first report of FCV detection and isolation from stray cats in Korea. All of the tested stray cats were healthy without any recognized symptoms. From those animals, the detection and isolation-based prevalence was 2.5% (3 of 120). Several studies have reported the presence of FCV in healthy cats sampled from different locations, including households, veterinary hospitals, animal shelters, and breeding catteries in many countries [9,25,26,27]. FCV infection in cats is sometime prone to be asymptomatic, and the asymptomatic infected cats can be the major source of FCV spreading in the general cat populations [18]. Stray cats are unvaccinated and freely roam long distances in urban and suburban areas. Therefore, asymptomatic infected stray cats shedding the virus are expected to be good carriers, and thus allow FCV to circulate and evolve in cat populations and other members of the felidae family. Thus, appropriate surveillance measures in wild cats are important to control FCV in countries. A similar study recently conducted in Spain reported a slightly higher prevalence of FCV in stray cats (5.2%) [19].
FCV has been considered a major pathogen causing respiratory problems in cat for long time and high virulent strains have been constantly emerging recently [2,3]. However, there has been limited information on the situation of FCV infection in cat populations, especially in stray cats, in Korea. This is partly attributed to the lack of efficient and convenient diagnostic assays. In this study, we developed a novel RT-PCR for FCV detection in Korea and demonstrated that the novel detection method is a useful tool for rapid and accurate diagnosis of FCV infection. In addition, using the novel RT-PCR and virus isolation methods, we reported the infection and prevalence of FCV in stray cats in Korea for the first time. To establish a proper control program for FCV infection in cats, continuous surveillance is needed. The novel RT-PCR developed in this study may be useful to screen the FCV infection in different cat populations in Korea.
Footnotes
Funding: This work was supported by the 2019 Inje University Research Grant.
Conflict of Interest: The authors declare no conflicts of interest.
- Conceptualization: Kim JS, Park YH, Park KT.
- Data curation: Kim JS.
- Formal analysis: Kim JS.
- Funding acquisition: Park KT.
- Investigation: Kim JS.
- Methodology: Kim JS.
- Project administration: Park KT.
- Resources: Kim JS, Park YH.
- Software: Kim JS.
- Supervision: Park KT.
- Validation: Kim JS, Park KT.
- Visualization: Kim JS.
- Writing - original draft: Kim JS, Park KT.
- Writing - review & editing: Kim JS, Park YH, Park KT.
References
- 1.Cai Y, Fukushi H, Koyasu S, Kuroda E, Yamaguchi T, Hirai K. An etiological investigation of domestic cats with conjunctivitis and upper respiratory tract disease in Japan. J Vet Med Sci. 2002;64(3):215–219. doi: 10.1292/jvms.64.215. [DOI] [PubMed] [Google Scholar]
- 2.Hurley KE, Pesavento PA, Pedersen NC, Poland AM, Wilson E, Foley JE. An outbreak of virulent systemic feline calicivirus disease. J Am Vet Med Assoc. 2004;224(2):241–249. doi: 10.2460/javma.2004.224.241. [DOI] [PubMed] [Google Scholar]
- 3.Hurley KF, Sykes JE. Update on feline calicivirus: new trends. Vet Clin North Am Small Anim Pract. 2003;33(4):759–772. doi: 10.1016/s0195-5616(03)00025-1. [DOI] [PubMed] [Google Scholar]
- 4.Marsilio F, Di Martino B, Decaro N, Buonavoglia C. A novel nested PCR for the diagnosis of calicivirus infections in the cat. Vet Microbiol. 2005;105(1):1–7. doi: 10.1016/j.vetmic.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 5.Sykes JE, Allen JL, Studdert VP, Browning GF. Detection of feline calicivirus, feline herpesvirus 1 and Chlamydia psittaci mucosal swabs by multiplex RT-PCR/PCR. Vet Microbiol. 2001;81(2):95–108. doi: 10.1016/s0378-1135(01)00340-6. [DOI] [PubMed] [Google Scholar]
- 6.Murphy FA, Gibbs EP, Horzinek MC, Studdert MJ. Veterinary Virology. 3rd ed. Cambridge: Academic Press; 1999. Chapter 12: Laboratory diagnosis of viral diseases; pp. 193–224. [Google Scholar]
- 7.Radford AD, Addie D, Belák S, Boucraut-Baralon C, Egberink H, Frymus T, et al. Feline calicivirus infection. ABCD guidelines on prevention and management. J Feline Med Surg. 2009;11(7):556–564. doi: 10.1016/j.jfms.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaskell RM, Dennis PE, Goddard LE, Cocker FM, Wills JM. Isolation of felid herpesvirus I from the trigeminal ganglia of latently infected cats. J Gen Virol. 1985;66(Pt 2):391–394. doi: 10.1099/0022-1317-66-2-391. [DOI] [PubMed] [Google Scholar]
- 9.Berger A, Willi B, Meli ML, Boretti FS, Hartnack S, Dreyfus A, et al. Feline calicivirus and other respiratory pathogens in cats with feline calicivirus-related symptoms and in clinically healthy cats in Switzerland. BMC Vet Res. 2015;11:282. doi: 10.1186/s12917-015-0595-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chao L. Fitness of RNA virus decreased by Muller's ratchet. Nature. 1990;348(6300):454–455. doi: 10.1038/348454a0. [DOI] [PubMed] [Google Scholar]
- 11.Sykes JE, Studdert VP, Browning GF. Detection and strain differentiation of feline calicivirus in conjunctival swabs by RT-PCR of the hypervariable region of the capsid protein gene. Arch Virol. 1998;143(7):1321–1334. doi: 10.1007/s007050050378. [DOI] [PubMed] [Google Scholar]
- 12.Ohe K, Sakai S, Sunaga F, Murakami M, Kiuchi A, Fukuyama M, et al. Detection of feline calicivirus (FCV) from vaccinated cats and phylogenetic analysis of its capsid genes. Vet Res Commun. 2006;30(3):293–305. doi: 10.1007/s11259-006-3232-5. [DOI] [PubMed] [Google Scholar]
- 13.Griffin HG, Griffin AM. DNA sequencing. Recent innovations and future trends. Appl Biochem Biotechnol. 1993;38(1-2):147–159. doi: 10.1007/BF02916418. [DOI] [PubMed] [Google Scholar]
- 14.Seal BS. Analysis of capsid protein gene variation among divergent isolates of feline calicivirus. Virus Res. 1994;33(1):39–53. doi: 10.1016/0168-1702(94)90016-7. [DOI] [PubMed] [Google Scholar]
- 15.Coyne KP, Dawson S, Radford AD, Cripps PJ, Porter CJ, McCracken CM, et al. Long-term analysis of feline calicivirus prevalence and viral shedding patterns in naturally infected colonies of domestic cats. Vet Microbiol. 2006;118(1-2):12–25. doi: 10.1016/j.vetmic.2006.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang BT, Park HM. Prevalence of feline herpesvirus 1, feline calicivirus and Chlamydophila felis in clinically normal cats at a Korean animal shelter. J Vet Sci. 2008;9(2):207–209. doi: 10.4142/jvs.2008.9.2.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smeeton NC. Early history of the kappa statistic. Biometrics. 1985;41(3):795. [Google Scholar]
- 18.Radford AD, Coyne KP, Dawson S, Porter CJ, Gaskell RM. Feline calicivirus. Vet Res. 2007;38(2):319–335. doi: 10.1051/vetres:2006056. [DOI] [PubMed] [Google Scholar]
- 19.Ravicini S, Pastor J, Hawley J, Brewer M, Castro-López J, Beall M, et al. Prevalence of selected infectious disease agents in stray cats in Catalonia, Spain. JFMS Open Rep. 2016;2(1):2055116916634109. doi: 10.1177/2055116916634109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Afonso MM, Pinchbeck GL, Smith SL, Daly JM, Gaskell RM, Dawson S, et al. A multi-national European cross-sectional study of feline calicivirus epidemiology, diversity and vaccine cross-reactivity. Vaccine. 2017;35(20):2753–2760. doi: 10.1016/j.vaccine.2017.03.030. [DOI] [PubMed] [Google Scholar]
- 21.Meli ML, Berger A, Willi B, Spiri AM, Riond B, Hofmann-Lehmann R. Molecular detection of feline calicivirus in clinical samples: a study comparing its detection by RT-qPCR directly from swabs and after virus isolation. J Virol Methods. 2018;251:54–60. doi: 10.1016/j.jviromet.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 22.Hou J, Sánchez-Vizcaíno F, McGahie D, Lesbros C, Almeras T, Howarth D, et al. European molecular epidemiology and strain diversity of feline calicivirus. Vet Rec. 2016;178(5):114–115. doi: 10.1136/vr.103446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abd-Eldaim MM, Wilkes RP, Thomas KV, Kennedy MA. Development and validation of a TaqMan real-time reverse transcription-PCR for rapid detection of feline calicivirus. Arch Virol. 2009;154(4):555–560. doi: 10.1007/s00705-009-0337-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burgesser KM, Hotaling S, Schiebel A, Ashbaugh SE, Roberts SM, Collins JK. Comparison of PCR, virus isolation, and indirect fluorescent antibody staining in the detection of naturally occurring feline herpesvirus infections. J Vet Diagn Invest. 1999;11(2):122–126. doi: 10.1177/104063879901100203. [DOI] [PubMed] [Google Scholar]
- 25.Mochizuki M, Kawakami K, Hashimoto M, Ishida T. Recent epidemiological status of feline upper respiratory infections in Japan. J Vet Med Sci. 2000;62(7):801–803. doi: 10.1292/jvms.62.801. [DOI] [PubMed] [Google Scholar]
- 26.Henzel A, Brum MC, Lautert C, Martins M, Lovato LT, Weiblen R. Isolation and identification of feline calicivirus and feline herpesvirus in Southern Brazil. Braz J Microbiol. 2012;43(2):560–568. doi: 10.1590/S1517-83822012000200017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holst BS, Berndtsson LT, Englund L. Isolation of feline herpesvirus-1 and feline calicivirus from healthy cats in Swedish breeding catteries. J Feline Med Surg. 2005;7(6):325–331. doi: 10.1016/j.jfms.2005.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]