Abstract
The gasdermins, family of pore‐forming proteins, are emerging key regulators of infection, autoinflammation and antitumor immunity. Multiple studies have recently characterised their crucial roles in driving pyroptosis, a lytic pro‐inflammatory type of cell death. Additionally, gasdermins also act as key effectors of NETosis, secondary necrosis and apoptosis. In this review, we will address current understanding of the mechanisms of gasdermin activation and further describe the protective and detrimental roles of gasdermins in host defence and autoinflammatory diseases. These data suggest that gasdermins play a prominent role in innate immunity and autoinflammatory disorders, thereby providing potential new therapeutic avenues for the treatment of infection and autoimmune disease.
Keywords: Gasdermins, inflammatory bowel disease, multiple sclerosis, NETosis, pyroptosis, rheumatoid arthritis
This review summarises the latest advances in several aspects of mode of activation of gasdermins, which finally drives pyroptosis, NETosis, secondary necrotic death and apoptosis. Here, we also highlight the current understanding of gasdermin involvement in infections and autoimmunity disorders, providing a promising translational approach to the treatment of infectious and autoinflammatory diseases.

Introduction
The gasdermins constitute a protein superfamily classified by the gasdermin domain and include six members [gasdermin A (GSDMA), gasdermin B (GSDMB), GSDMC, GSDMD, GSDME and Pejvakin (PJVK)]. The name gasdermin (gastro + dermato) is derived from the relative unique expression pattern of GSDMA in the upper gastrointestinal (GI) tract and the cutaneous epithelium. 1 Besides the gasdermin domain, all gasdermin family members share approximately 45% overall sequence homology. At the structural level, gasdermins consist of two functional domains, the gasdermin‐N domain and gasdermin‐C domain. The N‐terminal domain (GSDM‐NT) of all gasdermin proteins, except PJVK, can oligomerise and therefore form membrane‐spanning pores in the plasma membrane leading to the release of inflammatory molecules, disruption of ionic gradients, osmotic cellular swelling and cytolysis, ultimately resulting in cell death. In healthy cells, however, the cytotoxic effects mediated through the N‐terminal domain can be suppressed by the autoinhibitory C‐terminal domain. 2 , 3 , 4
In this review, we highlight recent advances in understanding the activation mechanism of the gasdermin. Moreover, we also focus on their roles in host immunity and autoinflammation diseases.
The members of gasdermin family
GSDMD
GSDMD, located at chromosome 8q24.2, is the best‐studied member of the gasdermin family. Initially, GSDMD was found to be expressed in the epithelial cells of the oesophagus and stomach. 5 Recent studies revealed that GSDMD is also highly expressed in macrophages, neutrophils, monocytes, T cells and B cells, 6 , 7 , 8 , 9 where it acts as a key executor of inflammatory cell death.
GSDMD consists of a N‐terminal pore‐forming domain and a C‐terminal repressor domain, with a range of diverse cleavage sites for different caspases or granzymes within the linker region (Table 1). The crystal structure of GSDMD provides further understanding of the connection between these two domains. Liu and co‐workers demonstrated that two aromatic residues in β1‐β2 loop of N‐terminal domain (NTD; F50 and W51 in mouse GSDMD or F49 and W50 in human GSDMD) insert into a hydrophobic pocket consisting of L292, E295, Y376, A380, S470 and A474 in the C‐terminal domain. This hydrophobic pocket at the NTD‐CTD interface is conserved among members of the gasdermin family. Functional assays further indicated that this domain interface is critical for the autoinhibition of GSDMD. 4
Table 1.
Key cleaved residues of the gasdermin family
| Key caspases/proteases | Gasdermin family | Key residues | Biofunctions | Reference |
|---|---|---|---|---|
| Caspase‐1 | GSDMD |
D275 in human being D276 in mouse |
Cleavage by caspase‐1 leads to pyroptosis | 6 |
|
Caspase‐4/5 (caspase‐11 in mouse) |
GSDMD |
D275 in human being D276 in mouse |
Cleavage by caspase‐4,5 contributes to pyroptosis | 10 |
| Caspase‐8 | GSDMD |
D275 in human being D276 in mouse |
Cleavage by caspase‐8 promotes pyroptosis | 11, 12, 13, 14, 15 |
| Caspase‐3 | GSDMD |
D87 in human being D88 in mouse |
Cleavage by caspase‐3 counteracts pyroptosis | 14, 15 |
| Neutrophil elastase (NE) | GSDMD |
C268 in human being V251 in mouse |
Cleavage by NE results in NETosis | 7, 16 |
|
Caspase‐4/5 (caspase‐11 in mouse) |
GSDMD |
D275 in human being D276 in mouse |
Cleavage by caspase‐4,5 causes NETosis in neutrophils | 58 |
| Cathepsin G | GSDMD | L274 in mouse | Activation of GSDMD by cathepsin G drives inflammation in neutrophils | 8 |
|
Granzyme A (GzmA) |
GSDMB | Lys244 | Cleavage by GzmA triggers pyroptosis | 17 |
| Granzyme B (GzmB) | GSDME | D270 in human being and mouse | Cleavage by GzmB induces pyroptosis | 18 |
| Caspase‐3 | GSDME | D270 in human being and mouse | Cleavage by caspase‐3 initiates pyroptosis/secondary necrosis/apoptosis |
Previous studies found that GSDMD can be cleaved by active caspase‐1 downstream of canonical inflammasome complexes, including NLRC4 inflammasome sensing of bacterial flagellin, NLRP3 inflammasome sensing of reactive oxygen species (ROS) and K+ and AIM2 inflammasome sensing of viral DNA. 6 Additionally, GSDMD can also be cleaved by active caspase‐11 (caspase‐4/5 in humans) downstream of lipopolysaccharide (LPS)‐activated non‐canonical inflammasome complexes. 10 Recently, the identification of GSDMD cleavage by caspase‐3/8, 11 , 12 , 13 , 14 , 15 neutrophil‐specific elastase (ELANE), 7 , 16 cathepsin G 8 and granzymes 17 , 18 has advanced our understanding of GSDMD activation and increased the number of possible activation mechanisms.
GSDMA
Firstly, GSDMA, located at chromosome 17q21, was identified through its elevated expression in cells of GI tract and skin. Secondly, GSDMA expression appears silenced in primary gastric cancers and gastric cancer cell lines. 1 These results suggest that GSDMA may play a critical role in suppressing carcinogenesis of gastric tissue. Later research indicated that transforming growth factor‐β (TGF‐β) indirectly upregulates GSDMA expression via induction of LIM domain only 1 (LMO1) in the gastric epithelial cell lines, which finally induces gastric epithelium cell apoptosis. 19
By virtue of its expression in the epithelium of the skin, several studies have addressed the role of GSDMA in cutaneous biology. In mice, there exist three isoforms of GSDMA encoded by Gsdma1, Gsdma2 and Gsdma3. Mice with spontaneous or chemically induced mutations in Gsdma3 exhibited epidermal hyperplasia, hyperkeratosis and a hair‐loss phenotype in the skin. 20 , 21 , 22 However, GSDMA3−/− mice had no visible developmental skin abnormalities, 23 suggesting that these mutations confer a gain of function. These hair‐loss‐related mutations were shown to abrogate the interaction between the C‐ and N‐terminal domains of GSDMA3, and in a later study, the N‐terminal domain of GSDMA3 was shown to induce pyroptosis. 6 Together, the physiological function of GSDMA in the skin appears to be associated with the regulation of proliferation/ differentiation of epidermal stem cells and the hair follicle cycle.
GSDMB
The human GSDMB gene, also located at chromosome 17q21, consists of 12 exons and has at least four alternatively spliced transcripts ranging in length from 1578 to 1646bp. However, there is no counterpart of the human GSDMB gene in the mouse genome.
GSDMB is highly expressed in certain tissues, particularly lung, oesophageal and gastric tissues. 5 , 24 The high expression of GSDMB in airway epithelial cells contributes to the pathology of asthma. 24 Additionally, unlike the reduced expression of GSDMA, GSDMC and GSDMD in gastric and oesophageal cancers, GSDMB is overexpressed in gastric, uterine cervix and breast cancers, indicating that GSDMB may function as a potential oncogene. 25 , 26 , 27 Given the high expression and potential tumorigenesis effects of GSDMB, targeting GSDMB might be a therapeutic strategy for gastric, uterine cervix and breast cancers. Moreno‐Bueno and co‐workers provided evidence that intracellular delivery of an antibody targeting GSDMB reduces aggressiveness of HER2‐positive breast cancer. 28 Recently, Shao and colleagues indicated that cytotoxic lymphocytes, such as cytotoxic T cells (CTLs) and natural killer (NK) cells, can cause tumor cell death through GSDMB‐dependent pyroptosis, suggesting that GSDMB might be a good target to increase effectiveness of cancer immunotherapy. 17
GSDMC
GSDMC, located at chromosome 8q24.2, is also known as melanoma‐derived leucine zipper (MLZE) since its high expression in metastatic melanoma cells. 2 However, it was first shown to function as an oncogene in colorectal cancer. 29 GSDMC is expressed in the epithelium of the skin, and recently, it was shown that ultraviolet light (UV) can induce GSDMC expression in the skin via TRPV1/calcium/calcineurin/NFATc1 signalling. 30 GSDMC in its turn upregulates MMP‐1, an important mediator of tissue damage by UV irradiation, through activating ERK and JNK pathways. 31 However, in general, the functions of GSDMC remain poorly understood.
GSDME
GSDME, located at chromosome 7p15, was originally identified as DFNA5 (deafness, autosomal dominant 5) since it is found mutated in familial ageing‐related hearing loss. 32 Genetic mutations within intron 7 of human GSDME cause skipping of exon 8 and truncation of the autoinhibitory C‐terminus at residue 315, finally leading to hearing loss. 32 , 33 These observations suggest that the N‐terminal domain of GSDME, like the GSDMD N‐terminal domain, may possess spontaneous pore formation ability and therefore induce pyroptosis in cochlear cells.
GSDME expression is silenced in most cancer cells but expressed in many normal tissues. Identification of promoter hypermethylation and silencing of GSDME expression in colorectal cancer (CRC) suggests GSDME may function as a potential tumor suppressor for colon tumorigenesis. 34 Consistent with its tumor suppressive role in CRC, GSDME deficiency accelerates melanoma growth. 35 In addition, reduced GSDME levels are associated with worse 5‐year survival and increased metastases from breast cancers. 36 Chemotherapeutics, such as cisplatin, lobaplatin and doxorubicin, were shown to trigger pyroptosis in cancer cell through caspase‐3‐dependent cleavage of GSDME. 37 , 38 Recently, GSDME has been found to exert a marked effect on the tumor immune microenvironment via pyroptosis. 18 , 39 Zhang and his group indicated that cytotoxic lymphocytes, including CD8+ T and NK cells, suppress tumor growth via GSDME. Mechanistically, granzyme B (GzmB) from killer cytotoxic lymphocytes induces GSDME‐dependent pyroptosis in tumor cells, by both directly cleaving GSDME and indirectly activating caspase‐3 to cleave GSDME. 18 Additionally, Erkes and colleagues found that combined BRAF inhibitor and MEK inhibitor treatment causes GSDME‐mediated pyroptosis in BRAF V600E/K ‐mutant melanoma via active caspase‐3, ultimately resulting in HMGB‐1 release to activate antitumor T‐cell responses. 39 Collectively, these studies define a new functional intersection between GSDME‐dependent pyroptosis and T‐cell/NK cell responses to tumor cells.
Pejvakin
Firstly, pejvakin (PJVK or DFNB59), located at chromosome 2q31.1–q31.3, was identified in patients with auditory neuropathy. 40 , 41 Unlike other gasdermin members, PJVK has a truncated C‐terminal domain and lacks therefore the autoinhibitory function of this domain. Secondly, PJVK has not been shown to induce pore formation, yet has been shown to drive pexophagy, a selective form of autophagy that targets damaged peroxisomes. 42 PJVK is accordingly broadly expressed in the membrane of peroxisomes in inner hair cells, can function as a ROS sensor and can recruit the autophagy machinery to selectively degrade peroxisomes (pexophagy) during exposure to loud noises. These results indicate that PJVK‐mediated pexophagy plays a critical role in reducing peroxisome proliferation, ultimately protecting auditory hair cells against noise‐induced oxidative stress damage.
Gasdermin family members are involved in a diverse variety of mechanisms of programmed cell death
Pyroptosis, NETosis, apoptosis and secondary necrosis are four well‐studied modes of programmed cell death (PCD) that initially were considered to be independent of one another, but emerging evidence indicates that there is extensive crosstalk between these forms of PCD and that all four pathways can be activated by the same cell death effectors – the pore‐forming gasdermin proteins.
Pyroptosis induced by GSDMDNT via inflammatory caspases
Pyroptosis is a form of PCD that was originally described in 2000, 43 , 44 and is characterised by pore formations in the plasma membrane, swelling, rupture of the cell and release of cytosolic contents such as interleukin‐1 (IL‐1β), interleukin‐18 (IL‐18) and high mobility group box 1 (HMGB1). It is a form of necrotic cell death that has emerged as an important innate immune mechanism against intracellular pathogens, including Escherichia coli, Salmonella typhimurium, Shigella flexneri and Burkholderia thailandensis. 7 , 10 , 45 , 46
Despite the important biological function of pyroptosis, the activation mechanism for pyroptosis remained unclear until recently. In 2015, three research groups independently and simultaneously, using different techniques (CRISPR/Cas‐9 nuclease screening, ENU‐forward mouse genetic screening and high‐sensitive quantitative mass spectrometry), demonstrated that GSDMD is a novel substrate of inflammatory caspase‐1 and caspase‐11 (caspase‐4/5 in humans), which cleave GSDMD between the N‐terminal domain (pore‐forming, pyroptotic‐inducing domain) and C‐terminal domain (autoinhibitory domain) downstream of canonical or non‐canonical inflammasome activation 6 , 10 , 47 (Figure 1a and b). Recent structural insight into caspase‐1/4/11–GSDMD complex formation reveals that caspase‐1/4/11, which are autoprocessed after inflammasome activation, recognise and insert into a hydrophobic groove formed by Leu306, Leu310, Val367 and Leu370 of GSDMD‐C domain, ultimately cleaving GSDMD. 48
Figure 1.
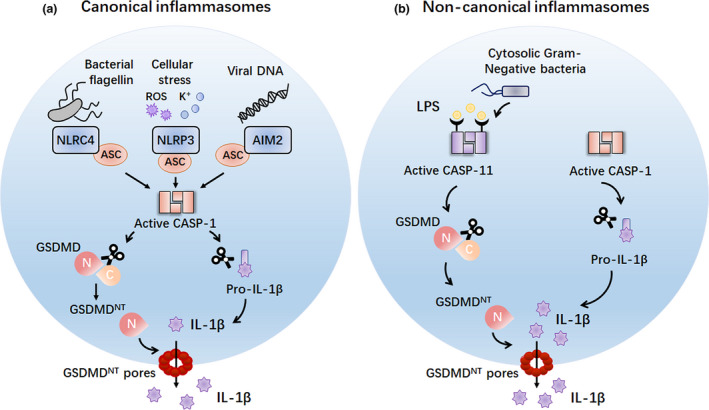
Inflammatory caspases induce GSDMDNT‐dependent pyroptosis. (a) The canonical inflammasome sensors, including NLRC4, NLRP3 and AIM2, detect diverse microbial signals and activate caspase‐1 through the ASC in macrophages. Active caspase‐1 can cleave GSDMD to liberate the pyroptotic GSDMDNT domain to form pores in the plasma membrane, ultimately resulting in pyroptosis. In addition, active caspase‐1 also processes the pro‐inflammatory cytokine pro‐IL‐1β to generate mature IL‐1β, which is presumably released by cell lysis during pyroptosis. (b) The non‐canonical inflammasome detects cytosolic Gram‐negative bacteria or LPS molecules and activates caspase‐11 (or caspase‐4/5 in humans) in macrophages. Active caspase‐11 can cleave GSDMD to release the cytotoxic GSDMDNT fragment to drive pyroptosis. ACS, apoptosis‐associated speck‐like protein containing a CARD; AIM2, absent in melanoma 2; CASP‐1, caspase‐1; CASP‐11, caspase‐11; LPS, lipopolysaccharide; NLRC4, NLR caspase activation and recruitment domain (CARD) domain‐containing 4; NLRP3, Nod‐like receptor (NLR) pyrin domain‐containing 3; ROS, reactive oxygen species.
Intriguingly, it has been shown that the N‐terminal domain of GSDMD (GSDMDNT), the cytotoxic fragment with approximate molecular weight of 30Kd (p30), which is released after caspase‐1/4/11 processing, selectively binds to phosphatidylinositol phosphates (PIPs), phosphatidylserine (PS) and phosphoinositide (PI; presence of which is restricted to the inner leaflet of the mammalian cell membrane) and cardiolipin (present in the inner and outer leaflets of bacterial membranes). 45 , 49 , 50 Once GSDMD is cleaved, GSDMDNT can bind to these lipids, oligomerise and form ring‐like pores (with an estimated inner diameter of about 10–16 nm) in the plasma membrane. 11 , 45 , 49 , 50 , 51 , 52 Using high‐resolution atomic force microscopy (AFM), Mulvihill et al. further revealed the mechanism of membrane pore formation by GSDMDNT. The time‐lapse AFM images indicated that GSDMDNT can assemble arc‐, slit‐ and ring‐shaped transmembrane oligomers that insert into the PI‐, PIP‐ or PS‐containing membrane. Over time, these arc‐, slit‐ and ring‐shaped transmembrane oligomers can incorporate additional oligomers, thereby forming large and stable ring‐shaped oligomers. Gradual loss of membrane lipids in the inner side of these ring‐shaped structures ultimately generates complete transmembrane pores. 53 Additionally, they observed no significant changes in the height of GSDMDNT oligomers during GSDMDNT pore formation, which demonstrates that the pore‐forming process of GSDMDNT is initiated directly in lipid membranes without prepore‐to‐pore transitions. 53 , 54 Recent structural research further indicates that the lipid‐binding ability is dependent on the α1 helix and β1‐β2 loop of GSDMDNT. 4 Additionally, three interfaces, including interface I (reverse‐parallel β3 and β8 strands), interface II (interface between α3 helix and β2 strand from one subunit and α2 helix and β11 strand from neighbouring subunit) and interface III (interface between α1 and α1′ helix from one subunit and α1 helix and β3 strand from adjacent subunit), are critical for the oligomerisation of the GSDMDNT fragment. 4 GSDMD‐mediated pore formation in the cellular plasma membrane finally results in the perturbation of electrochemical ion gradients and therefore causes cell swelling, rupture of plasma membrane and secretion of the cytosolic content, including proteins such as IL‐1β, IL‐18 and HMGB1. This feature of pore formation and release of cytosolic content is shared with GSDMANT and GSDMENT. These data identify gasdermin protein family members, GSDMA, GSDMD and GSDME, as the direct executors of pyroptotic cell death downstream of inflammatory caspase‐1 and caspase‐11.
Pyroptosis triggered by GSDMDNT and GSDMENT via pro‐apoptotic caspases
Besides the inflammatory caspases, growing evidence suggests that pro‐apoptotic caspases, such as caspase‐8 and caspase‐3, also play a key role in regulating GSDMD activity in macrophages. Three independent studies revealed that caspase‐8 drives pyroptosis by inducing GSDMD cleavage. 12 , 13 , 14 Sarhan et al., Orning et al. and Chen et al. observed caspase‐8, which is activated downstream of Yersinia infection or LPS/TNF‐α/TAK1 inhibitor co‐stimulation, cleaves GSDMD to generate active p30 fragments in macrophages, ultimately leading to pyroptosis (Figure 2a).
Figure 2.
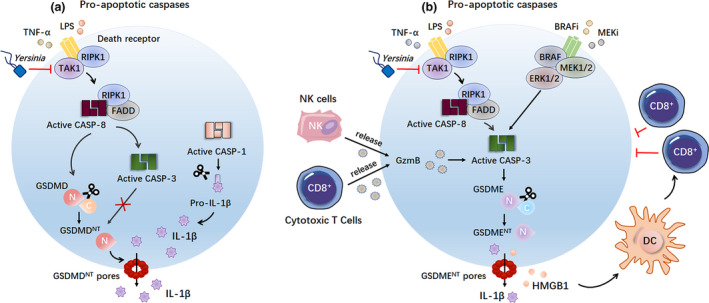
Pro‐apoptotic caspases trigger GSDMDNT‐ and GSDMENT‐mediated pyroptosis. (a) Caspase‐8, activated downstream of Yersinia infection or LPS/TNF‐α/TAK1 inhibitor co‐stimulation, cleaves GSDMD to generate the pyroptotic GSDMDNT domain to trigger pyroptosis. Interestingly, caspase‐3, which is active downstream of caspase‐8 activation, inhibits GSDMDNT‐mediated pyroptosis by counteracting the cleavage of GSDMD. (b) Caspase‐3, which is active downstream of caspase‐8 activation upon Yersinia infection or LPS/TNF‐α/TAK1 inhibitor co‐stimulation, induces GSDME cleavage to trigger GSDME‐dependent pyroptosis in macrophages. In addition, both GzmB from killer cytotoxic lymphocytes and treatment with BRAF inhibitors and MEK inhibitors can induce caspase‐3 activation in cancer cells, finally leading to caspase‐3‐mediated GSDME‐dependent pyroptosis. BRAFi, BRAF kinase inhibitors; CASP‐1, caspase‐1; CASP‐3, caspase‐3; CASP‐8, caspase‐8; DC, dendritic cells; FADD, Fas‐associated protein with death domain; GzmB, granzyme B; HMGB1, high mobility group box 1; MEKi, MEK kinase inhibitors; RIPK1, receptor‐interacting protein kinase 1; TAK1, TGF‐β‐activated kinase 1.
Interestingly, in addition to the active p30 fragment, it is also observed that GSDMD was cleaved into a p43 and p20 fragment upon Yersinia infection or LPS/TNF‐α/TAK1 inhibitor co‐stimulation. Chen et al. 12 and Sarhan et al. 14 suggested that caspase‐3, which is activated downstream of caspase‐8, counteracts the GSDMD‐dependent cell death by further processing the pyroptotic p30 fragment and full‐length GSDMD into the inactive p20 and p43 fragments, respectively (Figure 2a). Similarly, Taabazuing et al. provide evidence that caspase‐3 and caspase‐7 can also inactivate GSDMD by cleaving GSDMD at position D87 resulting into the p20 and p43 fragments. 15 These results suggest that there is a balance between caspase‐8‐mediated GSDMD activation and caspase‐3‐mediated GSDMD inactivation. Future work should further explore how to regulate this balance upon infection or inflammation.
In contrast to GSDMD, GSDME can be cleaved via caspase‐3 in macrophages during Yersinia infection or LPS/TNF‐α/TAK1 inhibitor co‐stimulation. Additionally, as already mentioned, both GzmB from killer cytotoxic lymphocytes 18 or combined BRAF inhibitor and MEK inhibitor treatment 39 can trigger GSDME‐dependent pyroptosis in cancer cells downstream of caspase‐3 activation (Figure 2b). Interestingly, the cytokines released from pyroptotic cancer cells have a markedly promoting effect on the cytotoxic lymphocytes, giving a novel insight into the antitumor immunity.
Pyroptosis caused by GSDMBNT and GSDMENT via granzyme
In addition to caspases, gasdermins can be directly cleaved by granzyme. Indeed, Zhang et al. 18 show that GzmB can directly cleave GSDME (Figure 3). This direct GzmB‐triggered pyroptosis provides a simple mechanism to induce pyroptosis without canonical or non‐canonical inflammasome activation.
Figure 3.
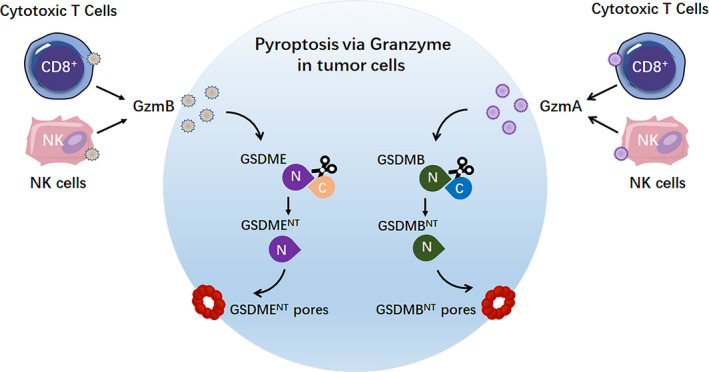
Granzymes initiate GSDMBNT‐ and GSDMENT‐induced pyroptosis. GzmA or GzmB, which is released from killer cytotoxic lymphocytes, induces GSDMB‐ or GSDME‐dependent pyroptosis in tumor cells, respectively. GzmA, granzyme A; GzmB, granzyme B; NK cells, natural killer cells.
Shao et al. extended this model by showing that granzyme A (GzmA) from NK cells and cytotoxic T lymphocytes, similar to GzmB, cleaves and activates GSDMB to induce cancer cell pyroptosis 17 (Figure 3). These findings reinforce the view that pyroptosis can also be initiated in a less complex manner independent of caspases. Additionally, these data deepen our understanding of critical functions of gasdermin proteins in cytotoxic lymphocyte‐mediated tumor clearance.
NETosis induced by GSDMDNT
Neutrophils, a specific type of white blood cell, function as an essential player of innate immunity directed against fungal and bacterial pathogens. NETosis, which is defined to be programmed neutrophil cell death, is an important antipathogen strategy employed by neutrophils. During NETosis, neutrophils can release chromatin, granular and cytoplasmic proteins and associated proteases to form an extracellular web‐like matrix, named neutrophil extracellular traps (NETs). 55 , 56 NETs can immobilise, catch and kill pathogens through their web‐like structures that consist of various antimicrobial proteins, including neutrophil elastase (NE), cathepsin G and histones. 57 Given the key effects of NETosis on releasing antimicrobial NETs, NETosis is now recognised as a vital contributor to immune defence. However, the precise mechanisms of NETosis are not yet fully clear.
Recently, Chen et al. 58 found that cytosolic Gram‐negative pathogen or LPS triggers GSDMD‐dependent NETosis via a non‐canonical (caspase‐4/11) inflammasome signalling pathway (Figure 4a). However, neutrophils resist pyroptosis upon canonical inflammasome activation. 59 , 60 Interestingly, Sollberger and Hiroto Kambara found that GSDMD can be cleaved through a caspase‐independent way in the formation process of NET 7 , 16 (Figure 4b).
Figure 4.
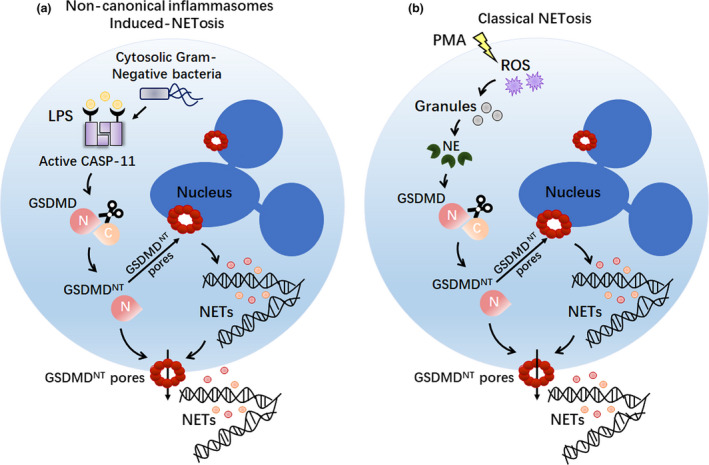
NETosis induced by pore‐forming GSDMDNT fragment. (a) Upon cytoplasmic Gram‐negative bacterial infection or LPS stimulation, active caspase‐11 triggers robust GSDMD cleavage to generate GSDMDNT domain. Then, the GSDMDNT fragments form pores in nuclear and plasma membranes to NETs in neutrophils. (b) Upon treatment with classical NETosis activators (such as PMA), ROS induces NE release from the granules, leading to NE‐mediated GSDMD cleavage and activation. Cleaved GSDMDNT fragments rupture nuclear and plasma membranes of neutrophil cells, resulting in NET extrusion. CASP‐11, caspase‐11; LPS, lipopolysaccharide; NE, neutrophil elastase; NETs, neutrophil extracellular traps; PMA, phorbol 12‐myristate 13‐acetate; ROS, reactive oxygen species.
Taken together, these results provide a new avenue of understanding as to how GSDMD contributes to NET formation following diverse stimuli, including Gram‐negative pathogens, LPS, ROS or phorbol 12‐myristate 13‐acetate (PMA).
GSDMENT fragment switches apoptosis to secondary necrosis/pyroptosis
Apoptosis is an immunologically silent form of PCD driven by caspase‐3/7/8/9 that typically results in rapidly engulfing dead cells by nearby phagocytes. However, in some instance, the scavenging of apoptotic cells is inhibited or insufficient in a timely manner, which causes these dying cells into a process called secondary necrosis. Secondary necrosis refers to a terminal phase at the end of the apoptotic programme characterised by plasma membrane permeabilisation, swelling, lysis and the release of intracellular pro‐inflammatory molecules including activated caspase‐3 and HMGB1. 61 However, the molecular mechanism of secondary necrosis remains poorly understood. Emerging evidence indicates that secondary necrosis is orchestrated by the activity of apoptotic caspase‐3, which directly cleaves GSDME to produce a necrotic GSDMENT fragment that targets and permeabilises the plasma membrane of macrophages 62 (Figure 5a).
Figure 5.
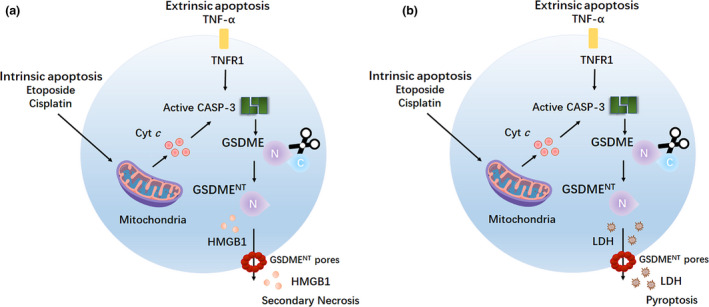
GSDMENT switches apoptosis to secondary necrosis/pyroptosis. (a) Caspase‐3, which is activated by intrinsic or extrinsic apoptotic pathway, cleaves GSDME to generate the pyroptotic GSDMENT fragment, which permeabilises the plasma membrane and releases pro‐inflammatory DAMP molecules such as HMGB1, finally leading to secondary necrosis in macrophages. (b) Caspase‐3, which is activated by intrinsic or extrinsic apoptotic pathway, mediates GSDME cleavage to generate the pyroptotic GSDMENT fragment, which permeabilises the plasma membrane and releases LDH, finally leading to pyroptosis in cancer cells. CASP‐3, caspase‐3; Cyt c, cytochrome c; DAMP, danger‐associated molecular pattern; HMGB1, high mobility group box 1; LDH, lactate dehydrogenase.
In addition to inducing secondary necrosis, GSDME can also switch caspase‐3‐mediated apoptosis to pyroptosis 18 , 38 (Figure 5b). As cancer cells express little GSDME, reversal of GSDME silencing can potentially sensitise cancer cells to chemotherapy drugs. Of caution here is that GSDME is highly expressed in various normal tissues, these GSDME‐mediated pyroptotic effects contribute to the extensive normal tissue damage and inflammation that occur in patients undergoing chemotherapy, and thus, lowering GSDME may be an alternative approach and a beneficial target for alleviating such adverse effects of chemotherapy drugs.
Taken together, these findings also extend our understanding of apoptosis. The expression level of GSDME may determine the form of cell death, including secondary necrosis or pyroptosis, upon ‘apoptotic stimulations’ in caspase‐3‐activated cells.
Gasdermins augment the mitochondrial apoptotic pathway
Interestingly, and complementary to the above, new evidence demonstrated that members of the gasdermin family also play additional roles in the apoptotic programme (Figure 6).
Figure 6.
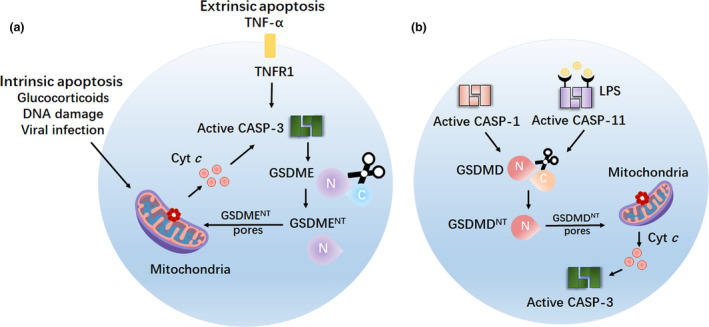
Gasdermins augment the mitochondrial apoptotic pathway. (a) Caspase‐3, which is activated by intrinsic or extrinsic apoptotic pathway, cleaves GSDME to generate the pyroptotic GSDMENT fragment, which permeabilises the mitochondria membrane, releases cytochrome c (Cyt c) and activates the apoptosome in cancer cells and immortalised bone marrow‐derived macrophages (iBMDMs). Cyt c released through GSDMENT mitochondrial pores, in turn, drives caspase‐3 activation and GSDME cleavage. (b) Canonical or non‐canonical inflammasome‐induced GSDMDNT fragment can permeabilise the mitochondria and release Cyt c, finally enhancing the mitochondrial apoptotic pathway in cancer cells and iBMDMs. TNFR1, tumor necrosis factor receptor type 1; Cyt c, cytochrome c; CASP‐3, caspase‐3; CASP‐1, caspase‐1; CASP‐11, caspase‐11.
GSDMENT, which is cleaved by active caspase‐3, can also form pores in the mitochondrial membrane resulting in the release of pro‐apoptotic molecules, such as cytochrome c (Cyt c) and high temperature requirement protein A2 (HtrA2/Omi). This event creates a positive feedback loop that promotes caspase‐3 activation and further GSDME cleavage, ultimately augmenting the apoptotic programme 35 (Figure 6a). The above mechanism might explain how the human deafness‐associated GSDME mutant induces cochlear cell death. Indeed, the absence of the gasdermin‐C domain in the GSDME hearing‐loss mutant leads to form pores in the mitochondria, release Cyt c and activate caspase‐3, finally resulting in cochlear hair cell apoptosis and sensorineural hearing loss. Furthermore, this function appears to be conserved within the superfamily, as other members such as GSDMANT and GSDMDNT also permeabilise the mitochondria to release pro‐apoptotic factors 35 (Figure 6b).
Roles of Gasdermin family members in the host defence against microbial infection
In normal physiology, gasdermin family members play an important role in antimicrobial innate immune defence through pyroptosis, NETosis and direct killing of bacteria.
As mentioned above, active GSDMD, which is cleaved by caspase‐1 downstream of canonical inflammasome complexes, can restrict Legionella pneumophila, Rotavirus and Francisella novicida infections through pyroptosis in bone marrow‐derived macrophages and epithelial cells. 63 , 64 , 65 Additionally, GSDMD‐dependent pyroptosis in bone marrow‐derived macrophages and monocytes, which is triggered through mouse caspase‐11 or human caspase‐4, is another critical host response against Gram‐negative bacteria, such as E. coli, S. typhimurium, S. flexneri and B. thailandensis. 10 , 47 Finally, these infective and pyroptotic macrophages functionally trap the bacteria, which results in pore‐induced intracellular trap‐mediated efferocytic clearance of bacteria by neighbouring macrophages and neutrophils in vitro and in vivo. 66
In addition to pyroptosis, GSDMD can also exert host protective effects by promoting NETosis in neutrophils. GSDMD‐dependent neutrophil death, which is triggered by non‐canonical (caspase‐4/11) inflammasome downstream of cytosolic Gram‐negative bacteria, induces antimicrobial NET extrusion via forming pores in the nuclear and plasma membrane of neutrophils. 58
Previous studies demonstrated that bacteria, such as E. coli, Mycobacterium and δ‐proteobacterium Bdellovibrio bacteriovorus, accumulate cardiolipin, PI and PS in the bacterial membrane. 67 , 68 , 69 Given the selective lipid‐binding properties of the GSDMDNT fragment, GSDMDNT can bind to bacteria, which are enriched with cardiolipin, PI and PS in the membrane. Then, GSDMDNT can directly and efficiently kill intracellular and extracellular bacteria by forming transmembrane pores and ultimately increasing permeabilities in the bacterial membrane. 3 , 45 , 46 , 50 Lipid‐specific binding may explain selective killing by GSDMDNT, yet different bacterial classes also differ in the composition and structure of their cell wall. The bacterial cell wall imposes a structural barrier for passage of gasdermin fragments. Whether and how, for example, GSDMDNT passes the bacterial wall remains elusive, and thus, the apparent differential sensitivity of bacteria to gasdermin‐mediated killing is likely a combination of both cell wall composition and membrane lipid composition.
Role of the gasdermin family in inflammatory diseases
GSDMD activation may in fact be a double‐edged sword in host responses. Excessive inflammatory cell deaths by gasdermin activations may play a detrimental role in host defence. In agreement, several studies demonstrated that gasdermin family members contribute to septic shock and autoinflammatory diseases, including multiple sclerosis (MS), systemic sclerosis (SSc), inflammatory bowel disease (IBD), rheumatoid arthritis (RA), familial Mediterranean fever (FMF) and type 1 diabetes (T1D; Table 2).
Table 2.
The role of gasdermins in autoimmunity diseases
| Autoimmunity diseases | Gasdermins | Study type | Biofunctions | Reference |
|---|---|---|---|---|
| Multiple sclerosis (MS) | GSDMD | Experimental evidence | GSDMD‐mediated pyroptosis inducing neuroinflammation via promoting and recruiting active T‐cell infiltration into the central nervous system (CNS) | 78 |
| Systemic sclerosis (SSc) | GSDMA | GWAS | The variant rs3894194, a missense mutation of GSDMA, is a novel susceptibility locus for SSc | 79 |
| Inflammatory bowel disease (IBD) |
GSDMA GSDMB GSDMD |
GWAS Experimental evidence |
GSDMA and GSDMB were proved to be IBD susceptibility genes; GSDMD‐mediated pyroptosis contributes to IBD | 80, 81, 82, 83 |
| Rheumatoid arthritis (RA) |
GSDMB GSDMD |
GWAS Experimental evidence |
GSDMB locus is associated with the risk of RA; GSDMD‐conferred monocyte pyroptosis is involved in RA | 84, 85, 86 |
| Familial Mediterranean fever (FMF) | GSDMD | Experimental evidence | GSDMD‐mediated pyroptosis is critical for autoinflammatory pathology in FMF | 89 |
|
Type 1 diabetes (T1D) |
GSDMB | GWAS | A risk allele of rs2290400 in GSDMB probably determines the risk of T1D predominantly in early childhood | 90, 91, 92, 93 |
GWAS, genome‐wide association studies.
Septic shock
Sepsis, which is defined as life‐threatening organ dysfunctions caused by a host's dysregulation following infection, is still a major challenge for healthcare systems. In general, pyroptosis protects multicellular host organisms against invasive pathogenic microbial infections; however, excessive pyroptosis might lead to an overwhelming inflammatory response, finally resulting in sepsis and septic shock. Emerging evidence suggests that GSDMD‐dependent pyroptosis induced by caspase‐1/11 inflammasome activation drives sepsis downstream of LPS exposure/TMEM173 activation/lipid peroxidation. 70 , 71 , 72 , 73 In addition, magnesium, cAMP and vitamin E can protect against sepsis by blocking GSDMD‐induced pyroptosis 70 , 74 , 75 These results provide novel insight into the role of GSDMD‐triggered pyroptosis in sepsis, thereby revealing some potential targets for therapeutic intervention for lethal infection.
Multiple sclerosis
Multiple sclerosis, a chronic inflammatory disease of the central nervous system (CNS), is characterised by inflammatory demyelination, chronic axonal damage and neurodegeneration. Previously, NLRP3 inflammasome, caspase‐1 and IL‐1β all have been implicated in the pathogenesis of MS. 76 , 77 In a recent study, Li et al. 78 identified GSDMD is a driver of experimental autoimmune encephalomyelitis (EAE) in mice, an animal model of MS. Mechanistically, GSDMD‐mediated pyroptosis in peripheral myeloid cells promotes the priming, differentiation and activation of T cells in the secondary lymphoid organs and then recruits T‐cell infiltration into the CNS, ultimately inducing neuroinflammation in EAE. Collectively, this finding represents the first indication of a possibly essential role of GSDMD in the pathogenesis of MS. Consequently, targeting GSDMD‐mediated pyroptosis might be a potential therapeutic strategy for MS treatment.
Systemic sclerosis
Systemic sclerosis is an autoimmune rheumatic disease characterised by excessive production and accumulation of fibrosis in the skin and internal organs and by injuries to small arteries. Although genetic underlying of SSc is suspected, key susceptibility gene of SSc remains unknown. Using transethnic meta‐analysis of genome‐wide association studies (GWASs) in the Japanese and European populations and two replication studies, Terao et al. 79 identified rs3894194, a missense mutation of GSDMA, as a novel susceptibility locus for SSc. Interestingly, they found the signal of rs3894194 is enriched in keratinocytes and fibroblasts. These findings may suggest the potential pathogenic role of keratinocytes and fibroblasts in SSc. Further functional annotation of the rs3894194 allele in SSc is needed in the future.
Inflammatory bowel disease
Inflammatory bowel disease entails diseases characterised by chronic inflammation of the GI tract, including Crohn's disease and ulcerative colitis. Genetic factors, dysbiosis of the microbiome, dysregulation of the immune system and environment may all contribute to the pathogenesis of IBD. However, the exact cause of IBD is largely unknown. Many studies aim to identify genetic risk factors for IBD. Intriguingly, GSDMA and GSDMB are identified to be IBD susceptibility genes. 80 , 81 , 82 However, the mechanism by which GSDMA and GSDMB affect IBD onset and/or progression still requires further experimental investigation.
Recently, it was shown that GSDMD‐dependent pyroptosis in intestinal epithelial cells, which is triggered downstream of never in mitosis gene A (NIMA)‐related kinase 7 (NEK7)‐mediated NLRP3 inflammasome activation, is implicated in the pathogenesis of IBD in vitro and in vivo. 83 This study provides the first experimental evidence that may help in understanding how GSDMD‐mediated pyroptosis may contribute to IBD.
Rheumatoid arthritis
Rheumatoid arthritis, a disabling autoimmune disorder, is characterised by chronic inflammation, stiffness and destruction of the synovial joints. While the cause of RA is not clear, again genetic factors are believed to be involved in the aetiology of RA. To unravel the complex genetic architecture of RA, a multiethnic GWAS and bioinformatic analysis were used to identify the most likely causal variants and genes in RA. These researches identified that GSDMB locus is associated with risk of RA. 84 Further studies showed that CD4+ and B lymphocytes, the important inflammatory executors in RA, exhibit overlapping expression quantitative trait locus at GSDMB locus. 85 These data extend our understanding of the landscape of GSDMB in early RA, especially in several critical lymphocyte populations of early RA.
A recent study from Zhang and his team demonstrated that complement C1q synergises with pentraxin 3 (PTX3) in promoting NLRP3 overactivation, ultimately resulting in GSDMD‐mediated monocyte pyroptosis and excessive inflammatory cytokine release in RA. Additionally, the release of inflammatory cytokines, such as IL‐6, in turn, drives PTX3 plus C1q‐induced GSDMD‐dependent monocyte pyroptosis in a positive feedback. 86 These findings further explore the pathogenic effects of gasdermin family on RA, suggesting a potential novel therapeutic strategy by targeting pyroptosis in RA.
Familial Mediterranean fever
Familial Mediterranean fever, the most common monogenic autoinflammatory disease worldwide, is characterised by recurrent fever, abdominal pain, headache, rash, serositis, arthritis and dermal manifestations. It is caused by missense mutations in MEFV gene, which results in exaggeratedly activating the pyrin inflammasome. 87 , 88 However, the pathophysiologic mechanisms driven by excessive activation of pyrin inflammasome in FMF are incompletely understood. Recently, Kanneganti et al. demonstrated that GSDMD‐dependent pyroptosis, which is activated downstream of pyrin inflammasome, contributes to the autoinflammation‐associated growth retardation, anaemia, neutrophilia, cytokine production and tissue damage in Mefv V726A/V726A FMF mouse model. Importantly, deletion of GSDMD in vivo fully rescued these autoinflammation‐associated features. 89 These findings identify GSDMD‐mediated pyroptosis is critical for autoinflammatory pathology in FMF, providing a potential therapeutic target for the treatment of FMF.
Type 1 diabetes
Type 1 diabetes, which is characterised by insulin deficiency, is a common autoimmune disorder that arises from the action of multiple genetic and environmental risk factors. Chromosome 17q21, containing a cluster of five genes [GSDMA, GSDMB, ORM1‐like 3 (ORMDL3), IKAROS family zinc finger 3 (IKZF3) and zona pellucida‐binding protein 2 (ZPBP2)], was first linked to susceptibility to T1D in 2009. 90 , 91 , 92 Then, Ayabe et al. 93 further clarified the variant rs2290400, which is a non‐coding SNP located in intron 3 of GSDMB, functions as a T1D susceptibility allele in Japanese children. Importantly, this risk allele of rs2290400 in GSDMB probably determines the risk of T1D predominantly in early childhood. These studies may potentially shed light on the pathogenesis of T1D, especially the early childhood T1D. Further function study is needed to explore how the variant rs2290400 in GSDMB contributes to the early‐onset T1D in children.
Therapeutic targeting of GSDMD
As gasdermin‐induced pore formation plays a pivotal role in sepsis and numerous autoinflammatory diseases, there is increasing interest to develop small molecule inhibitors targeting GSDMD and other gasdermin family members.
Since Cys191/192 (human/mouse) of GSDMD is the critical residue for oligomerisation during pyroptosis‐related pore formation, 45 targeting Cys191/192 might be an attractive pharmacological strategy to block GSDMD‐induced pyroptosis. Rathkey and his group identified necrosulphonamide (NSA) to bind directly to GSDMD via Cys191 and therefore to inhibit GSDMD‐mediated pyroptosis downstream of inflammasome activation. In addition, NSA treatment significantly increased survival in LPS‐induced sepsis in vivo, suggesting that GSDMD inhibitors may show clinically efficacy in treating sepsis. 94
In parallel, Hu et al. 95 discovered disulphiram (a drug used to treat alcohol addiction) and Bay 11‐7082 (a previously identified NF‐κB inhibitor) to potently inhibit GSDMD pore formation in liposomes and inflammasome‐mediated pyroptosis via covalently modifying Cys191/192 (human/mouse) of GSDMD. In line with Hu's 95 study, mice administered with disulphiram are protected from LPS‐induced septic shock and MOG35–55 peptide‐triggered EAE 78 in vivo. These results also demonstrated that GSDMD might be an alternative target for the treatment of sepsis, MS and potentially additional inflammatory diseases.
In addition, using a chemical screen of 182 710 small molecules and proteomics, a small molecule LDC7559 was identified to specifically block PMA‐induced GSDMD‐dependent NET formation. 16 Mechanistically, LDC7559 strongly suppressed the release of myeloperoxidase and NE from granules, ultimately inhibiting NE‐cleaved GSDMD to execute NETosis upon PMA stimulation. This compound, or its derivatives, might be attractive lead compounds for targeting GSDMD activity.
Conclusions and future perspectives
Understanding of pyroptotic cell death has rapidly progressed since the identification of GSDMD as a key mediator of pyroptosis. Initially, pyroptosis has been defined as a form of lytic cell death downstream of inflammatory caspase activation. Recent studies revealed that the active pro‐apoptotic caspases or granzymes are also involved in the progression of pyroptosis. 12 , 13 , 14 Moreover, besides pyroptosis, gasdermins also participate in NETosis, secondary necrotic death and apoptosis. 7 , 58 , 61 Further studies are needed to explore in detail the mode of activation of gasdermins and their precise role(s) in these diverse modes of cell death.
Accumulating evidence demonstrates that gasdermin‐mediated pyroptosis contributes to the pathology of several autoinflammatory disorders, including MS, SSc, IBD and RA. 96 , 97 , 98 , 99 However, the potential pathogenic role of pyroptosis in other autoimmune diseases, such as psoriasis, systemic lupus erythematosus and ankylosing spondylitis, remains unknown. It would be of great interest to further investigate whether gasdermin‐triggered pyroptosis contributes to these and other autoinflammatory diseases.
Recently, GSDMB, GSDMD and GSDME are described to be cleaved and activated by inflammatory/pro‐apoptotic caspases or granzymes. Nevertheless, we still know very little about GSDMA and GSDMC. GSDMA and GSDMC also contain the pyroptotic N‐terminal domain; however, the physiological or pathological signals that mediate GSDMA or GSDMC cleavage remain unclear. In addition, the role of GSDMA‐ and GSDMC‐mediated pore formation in biology and diseases is also still poorly understood.
The activation of GSDMD or GSDME downstream of active caspase‐3 has opposing consequences. 12 , 14 , 15 It is therefore important to determine the structural effect(s) of active caspase‐3 on the distinct gasdermin proteins and the biological consequences hereof. Thus, future research should explore the balance of caspase‐3 in inhibiting and activating pyroptosis and apoptosis, respectively, and how the crosstalk between these pathways regulates the immune response to microbial infection in vivo.
As previously mentioned, the GSDMDNT fragment can directly and efficiently kill intracellular and extracellular bacteria by binding to specific lipid classes present in bacterial membranes and therefore can form transmembrane pores in these bacterial membranes. 45 , 50 Given the efficient killing bacteria properties, the pyroptotic GSDMDNT fragment may be considered a novel antimicrobial agent to treat E. coli, Mycobacterium and δ‐proteobacterium Bdellovibrio bacteriovorans infections. In addition, this gasdermin‐derived pyroptotic fragment might be considered relatively safe since its ability to form pores is restricted by the lipid‐binding properties and thus the inner leaflet of mammalian cytomembranes. However, distinct bacteria compose a large diversity of structural lipids in their bacterial membrane. 69 For example, the membrane of Campylobacter jejuni is enriched with phosphatidylethanolamine (PE) and phosphatidylglycerol (PG). 100 The major membrane lipids of Borrelia burgdorferi are PG, phosphatidylcholine (PC) and glycolipids (GLs). 101 , 102 Additionally, Porphyromonas gingivalis accumulates PE, PG and sphingolipids in the membrane. 103 Furthermore, the membrane of Clostridium perfringens is formed by PG, PE, alanyl‐PG (APG) and lysyl‐PG. 104 , 105 As a result of the diversity of bacterial membrane lipids and the selective lipid‐binding preferences of GSDMDNT fragment, it will be not come as a surprise that pathogens such as Campylobacter jejuni, B. burgdorferi, P. gingivalis and C. perfringens are resistant to GSDMD killing.
Finally, it should be noted that in order to become effective, the GSDMDNT fragment should be able to reach the membrane compartment where it can bind and form pores. As such, the bacterial wall may impose a physical barrier that will not be passed passively, and thus, the effective use of GSDMDNT fragment should be accompanied by treatments that enable the crossing of the GSDMDNT fragment over the bacterial wall.
Growing evidence indicates that distinct gasdermin proteins display a cell‐type and tissue‐specific expression pattern. Although mechanism of gasdermin activation and participation in the different modes of cell death display common features for all gasdermins, there are also clear differences as described here. Therefore, it will be very interesting to determine the cell‐type and tissue‐specific role(s) of the different gasdermins as this may help to explain their varying role in the different diseases. Future studies should also further characterise the function of different gasdermins in immune cells and non‐immune cells during infections and diseases. For example, GSDMB, which is highly expressed in oesophageal and gastric tissues, may be associated with IBD susceptibility. However, the detailed cellular mechanism by which GSDMB contributes to IBD progression remains largely unknown. Further studies are required to investigate whether GSDMB‐mediated pyroptosis contributes to IBD.
The field of microbe–host interplay has gained much attention in recent years. Increasing evidence suggests that microbial dysbiosis in skin or gut is oftentimes involved in diverse inflammatory diseases, including psoriasis, atopic dermatitis and IBD. 106 , 107 , 108 , 109 Given the high expression and the host‐defensive role of GSDMA, GSDMC and GSDME in the skin, it is therefore reasonable to speculate that GSDMA, GSDMC and GSDME may exert critical physiological functions in skin. Additionally, GSDMA, GSDMC and GSDME may act as a bridge between microbiome and skin inflammatory diseases.
Since GSDMD drives a variety of inflammatory diseases, the identification of drugs targeting GSDMD and other gasdermin family members is attractive. The newly found GSDMD inhibitors do open novel therapeutic avenues for sepsis, MS and other autoinflammatory diseases. However, NSA, disulphiram and Bay 11‐7082 likely lack sufficient specificity and will also inhibit other proteins in the pyroptotic pathway, 94 , 95 or display off‐target effects. Further research will be needed to identify novel GSDMD inhibitors with high specificity. The recent descriptions of the crystal structure of the various gasdermin proteins may offer a structural basis to design specific inhibitors of GSDMD in future. 3 , 4 , 110
Author Contributions
Lipeng Tang: Conceptualization; Writing‐original draft; Writing‐review & editing. Chuanjian Lu: Writing‐review & editing. Guangjuan Zheng: Writing‐review & editing. Boudewijn MT Burgering: Writing‐review & editing.
Conflict of Interest
The authors declare no conflict of interest.
Acknowledgments
This study was supported by Innovative and Enhancement Research Program of Guangdong Province (No. 2019KTSCX025); the Specific Research Fund for TCM Science and Technology of Guangdong Provincial Hospital of Chinese Medicine (YN2018QJ03); the Natural Science Foundation of Guangdong Province (No. 2017A030313639); and Medical Scientific Research Foundation of Guangdong Province (No. A2017277). We thank Tao Shi and Tianshu Gui for assistances in paper submission and modification.
Contributor Information
Chuanjian Lu, Email: lcj@gzucm.edu.cn.
Guangjuan Zheng, Email: zhengguangjuan@gzucm.edu.cn.
Boudewijn MT Burgering, Email: b.m.t.burgering@umcutrecht.nl.
References
- 1. Saeki N, Kuwahara Y, Sasaki H, Satoh H, Shiroishi T. Gasdermin (Gsdm) localizing to mouse Chromosome 11 is predominantly expressed in upper gastrointestinal tract but significantly suppressed in human gastric cancer cells. Mamm Genome 2000; 11: 718–724. [DOI] [PubMed] [Google Scholar]
- 2. Watabe K, Ito A, Asada H et al Structure, expression and chromosome mapping of MLZE, a novel gene which is preferentially expressed in metastatic melanoma cells. Jpn J Cancer Res 2001; 92: 140–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kuang S, Zheng J, Yang H et al Structure insight of GSDMD reveals the basis of GSDMD autoinhibition in cell pyroptosis. Proc Natl Acad Sci USA 2017; 114: 10642–10647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Liu Z, Wang C, Yang J et al Crystal structures of the full‐length murine and human Gasdermin D reveal mechanisms of autoinhibition, lipid binding, and oligomerization. Immunity 2019; 51: 43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Saeki N, Usui T, Aoyagi K et al Distinctive expression and function of four GSDM family genes (GSDMA‐D) in normal and malignant upper gastrointestinal epithelium. Genes Chromosomes Cancer 2009; 48: 261–271. [DOI] [PubMed] [Google Scholar]
- 6. Shi J, Zhao Y, Wang K et al Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 2015; 526: 660–665. [DOI] [PubMed] [Google Scholar]
- 7. Kambara H, Liu F, Zhang X et al Gasdermin D exerts anti‐inflammatory effects by promoting neutrophil death. Cell Rep 2018; 22: 2924–2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Burgener SS, Leborgne NGF, Snipas SJ, Salvesen GS, Bird PI, Benarafa C. Cathepsin G inhibition by Serpinb1 and Serpinb6 prevents programmed necrosis in neutrophils and monocytes and reduces GSDMD‐driven inflammation. Cell Rep 2019; 27: 3646–3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xi G, Gao J, Wan B et al GSDMD is required for effector CD8+ T cell responses to lung cancer cells. Int Immunopharmacol 2019; 74: 105713. [DOI] [PubMed] [Google Scholar]
- 10. Kayagaki N, Stowe IB, Lee BL et al Caspase‐11 cleaves gasdermin D for non‐canonical inflammasome signalling. Nature 2015; 526: 666–671. [DOI] [PubMed] [Google Scholar]
- 11. Chen X, He WT, Hu L et al Pyroptosis is driven by non‐selective gasdermin‐D pore and its morphology is different from MLKL channel‐mediated necroptosis. Cell Res 2016; 26: 1007–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen KW, Demarco B, Heilig R et al Extrinsic and intrinsic apoptosis activate pannexin‐1 to drive NLRP3 inflammasome assembly. EMBO J 2019; 38: 101638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Orning P, Weng D, Starheim K et al Pathogen blockade of TAK1 triggers caspase‐8‐dependent cleavage of gasdermin D and cell death. Science 2018; 362: 1064–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sarhan J, Liu BC, Muendlein HI et al Caspase‐8 induces cleavage of gasdermin D to elicit pyroptosis during Yersinia infection. Proc Natl Acad Sci USA 2018; 115: E10888–E10897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Taabazuing CY, Okondo MC, Bachovchin DA. Pyroptosis and apoptosis pathways engage in bidirectional crosstalk in monocytes and macrophages. Cell Chem Biol 2017; 24: 507–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sollberger G, Choidas A, Burn GL et al Gasdermin D plays a vital role in the generation of neutrophil extracellular traps. Sci Immunol 2018; 3: eaar6689. [DOI] [PubMed] [Google Scholar]
- 17. Zhou Z, He H, Wang K et al Granzyme A from cytotoxic lymphocytes cleaves GSDMB to trigger pyroptosis in target cells. Science 2020; 368: eaaz7548. [DOI] [PubMed] [Google Scholar]
- 18. Zhang Z, Zhang Y, Xia S et al Gasdermin E suppresses tumour growth by activating anti‐tumour immunity. Nature 2020; 579: 415–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Saeki N, Kim DH, Usui T et al GASDERMIN, suppressed frequently in gastric cancer, is a target of LMO1 in TGF‐β‐dependent apoptotic signalling. Oncogene 2007; 26: 6488–6498. [DOI] [PubMed] [Google Scholar]
- 20. Runkel F, Marquardt A, Stoeger C et al The dominant alopecia phenotypes Bareskin, Rex‐denuded, and Reduced Coat 2 are caused by mutations in gasdermin 3. Genomics 2004; 84: 824–835. [DOI] [PubMed] [Google Scholar]
- 21. Tanaka S, Tamura M, Aoki A et al A new Gsdma3 mutation affecting anagen phase of first hair cycle. Biochem Biophys Res Commun 2007; 359: 902–907. [DOI] [PubMed] [Google Scholar]
- 22. Zhou Y, Jiang X, Gu P, Chen W, Zeng X, Gao X. Gsdma3 mutation causes bulge stem cell depletion and alopecia mediated by skin inflammation. Am J Pathol 2012; 180: 763–774. [DOI] [PubMed] [Google Scholar]
- 23. Shi P, Tang A, Xian L et al Loss of conserved Gsdma3 self‐regulation causes autophagy and cell death. Biochem J 2015; 468: 325–336. [DOI] [PubMed] [Google Scholar]
- 24. Das S, Miller M, Beppu AK et al GSDMB induces an asthma phenotype characterized by increased airway responsiveness and remodeling without lung inflammation. Proc Natl Acad Sci USA 2016; 113: 13132–13137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Komiyama H, Aoki A, Tanaka S et al Alu‐derived cis‐element regulates tumorigenesis‐dependent gastric expression of GASDERMIN B (GSDMB). Genes Genet Syst 2010; 85: 75–83. [DOI] [PubMed] [Google Scholar]
- 26. Hergueta‐Redondo M, Sarrio D, Molina‐Crespo A et al Gasdermin B expression predicts poor clinical outcome in HER2‐positive breast cancer. Oncotarget 2016; 7: 56295–56308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hergueta‐Redondo M, Sarrio D, Molina‐Crespo A et al Gasdermin‐B promotes invasion and metastasis in breast cancer cells. PLoS One 2014; 9: e90099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Molina‐Crespo Á, Cadete A, Sarrio D et al Intracellular delivery of an antibody targeting gasdermin‐B reduces HER2 breast cancer aggressiveness. Clin Cancer Res 2019; 25: 4846–4858. [DOI] [PubMed] [Google Scholar]
- 29. Miguchi M, Hinoi T, Shimomura M et al Gasdermin C is upregulated by inactivation of transforming growth factor β receptor Type II in the presence of mutated Apc, promoting colorectal cancer proliferation. PLoS One 2016; 11: e0166422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kusumaningrum N, Lee DH, Yoon HS, Park CH, Chung JH. Ultraviolet light‐induced gasdermin C expression is mediated via TRPV1/calcium/calcineurin/NFATc1 signaling. Int J Mol Med 2018; 42: 2859–2866. [DOI] [PubMed] [Google Scholar]
- 31. Kusumaningrum N, Lee DH, Yoon HS, Kim YK, Park CH, Chung JH. Gasdermin C is induced by ultraviolet light and contributes to MMP‐1 expression via activation of ERK and JNK pathways. J Dermatol Sci 2018; 90: 180–189. [DOI] [PubMed] [Google Scholar]
- 32. Van Laer L, Huizing EH, Verstreken M et al Nonsyndromic hearing impairment is associated with a mutation in DFNA5. Nat Genet 1998; 20: 194–197. [DOI] [PubMed] [Google Scholar]
- 33. Booth KT, Azaiez H, Kahrizi K et al Exonic mutations and exon skipping: Lessons learned from DFNA5. Hum Mutat 2018; 39: 433–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kim MS, Chang X, Yamashita K et al Aberrant promoter methylation and tumor suppressive activity of the DFNA5 gene in colorectal carcinoma. Oncogene 2008; 27: 3624–3634. [DOI] [PubMed] [Google Scholar]
- 35. Rogers C, Erkes DA, Nardone A, Aplin AE, Fernandes‐Alnemri T, Alnemri ES. Gasdermin pores permeabilize mitochondria to augment caspase‐3 activation during apoptosis and inflammasome activation. Nat Commun 2019; 10: 1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Croes L, Beyens M, Fransen E et al Large‐scale analysis of DFNA5 methylation reveals its potential as biomarker for breast cancer. Clin Epigenetics 2018; 10: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yu J, Li S, Qi J et al Cleavage of GSDME by caspase‐3 determines lobaplatin‐induced pyroptosis in colon cancer cells. Cell Death Dis 2019; 10: 193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang Y, Gao W, Shi X et al Chemotherapy drugs induce pyroptosis through caspase‐3 cleavage of a gasdermin. Nature 2017; 547: 99–103. [DOI] [PubMed] [Google Scholar]
- 39. Erkes DA, Cai W, Sanchez IM et al Mutant BRAF and MEK inhibitors regulate the tumor immune microenvironment via pyroptosis. Cancer Discov 2020; 10: 254–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Delmaghani S, del Castillo FJ, Michel V et al Mutations in the gene encoding pejvakin, a newly identified protein of the afferent auditory pathway, cause DFNB59 auditory neuropathy. Nat Genet 2006; 38: 770–778. [DOI] [PubMed] [Google Scholar]
- 41. Schwander M, Sczaniecka A, Grillet N et al A forward genetics screen in mice identifies recessive deafness traits and reveals that pejvakin is essential for outer hair cell function. J Neurosci 2007; 27: 2163–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Defourny J, Aghaie A, Perfettini I, Avan P, Delmaghani S, Petit C. Pejvakin‐mediated pexophagy protects auditory hair cells against noise‐induced damage. Proc Natl Acad Sci USA 2019; 116: 8010–8017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Brennan MA, Cookson BT. Salmonella induces macrophage death by caspase‐1‐dependent necrosis. Mol Microbiol 2000; 38: 31–40. [DOI] [PubMed] [Google Scholar]
- 44. Miao EA, Leaf IA, Treuting PM et al Caspase‐1‐induced pyroptosis is an innate immune effector mechanism against intracellular bacteria. Nat Immunol 2010; 11: 1136–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liu X, Zhang Z, Ruan J et al Inflammasome‐activated gasdermin D causes pyroptosis by forming membrane pores. Nature 2016; 535: 153–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang J, Deobald K, Re F. Gasdermin D protects from melioidosis through pyroptosis and direct killing of bacteria. J Immunol 2019; 202: 3468–3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. He WT, Wan H, Hu L et al Gasdermin D is an executor of pyroptosis and required for interleukin‐1β secretion. Cell Res 2015; 25: 1285–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang K, Sun Q, Zhong X et al Structural mechanism for GSDMD targeting by autoprocessed caspases in pyroptosis. Cell 2020; 180: 941–955. [DOI] [PubMed] [Google Scholar]
- 49. Sborgi L, Ruhl S, Mulvihill E et al GSDMD membrane pore formation constitutes the mechanism of pyroptotic cell death. EMBO J 2016; 35: 1766–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ding J, Wang K, Liu W et al Pore‐forming activity and structural autoinhibition of the gasdermin family. Nature 2016; 535: 111–116. [DOI] [PubMed] [Google Scholar]
- 51. Aglietti RA, Estevez A, Gupta A et al GsdmD p30 elicited by caspase‐11 during pyroptosis forms pores in membranes. Proc Natl Acad Sci USA 2016; 113: 7858–7863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Russo HM, Rathkey J, Boyd‐Tressler A, Katsnelson MA, Abbott DW, Dubyak GR. Active caspase‐1 induces plasma membrane pores that precede pyroptotic lysis and are blocked by lanthanides. J Immunol 2016; 197: 1353–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mulvihill E, Sborgi L, Mari SA, Pfreundschuh M, Hiller S, Muller DJ. Mechanism of membrane pore formation by human gasdermin‐D. EMBO J 2018; 37: e98321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ding J, Shao F. Growing a gasdermin pore in membranes of pyroptotic cells. EMBO J 2018; 37: e100067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Jorch SK, Kubes P. An emerging role for neutrophil extracellular traps in noninfectious disease. Nat Med 2017; 23: 279–287. [DOI] [PubMed] [Google Scholar]
- 56. Papayannopoulos V. Neutrophil extracellular traps in immunity and disease. Nat Rev Immunol 2018; 18: 134–147. [DOI] [PubMed] [Google Scholar]
- 57. Thiam HR, Wong SL, Wagner DD, Waterman CM. Cellular mechanisms of NETosis. Annu Rev Cell Dev Biol 2020. (Online ahead of print). 10.1146/annurev-cellbio-020520-111016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chen KW, Monteleone M, Boucher D et al Noncanonical inflammasome signaling elicits gasdermin D‐dependent neutrophil extracellular traps. Sci Immunol 2018; 3: eaar6676. [DOI] [PubMed] [Google Scholar]
- 59. Karmakar M, Katsnelson M, Malak HA et al Neutrophil IL‐1β processing induced by pneumolysin is mediated by the NLRP3/ASC inflammasome and caspase‐1 activation and is dependent on K+ efflux. J Immunol 2015; 194: 1763–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chen KW, Gross CJ, Sotomayor FV et al The neutrophil NLRC4 inflammasome selectively promotes IL‐1β maturation without pyroptosis during acute Salmonella challenge. Cell Rep 2014; 8: 570–582. [DOI] [PubMed] [Google Scholar]
- 61. Silva MT. Secondary necrosis: the natural outcome of the complete apoptotic program. FEBS Lett 2010; 584: 4491–4499. [DOI] [PubMed] [Google Scholar]
- 62. Rogers C, Fernandes‐Alnemri T, Mayes L, Alnemri D, Cingolani G, Alnemri ES. Cleavage of DFNA5 by caspase‐3 during apoptosis mediates progression to secondary necrotic/pyroptotic cell death. Nat Commun 2017; 8: 14128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zhu Q, Zheng M, Balakrishnan A, Karki R, Kanneganti TD. Gasdermin D promotes AIM2 inflammasome activation and is required for host protection against Francisella novicida . J Immunol 2018; 201: 3662–3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Goncalves AV, Margolis SR, Quirino GFS et al Gasdermin‐D and Caspase‐7 are the key Caspase‐1/8 substrates downstream of the NAIP5/NLRC4 inflammasome required for restriction of Legionella pneumophila . PLoS Pathog 2019; 15: e1007886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhu S, Ding S, Wang P et al Nlrp9b inflammasome restricts rotavirus infection in intestinal epithelial cells. Nature 2017; 546: 667–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Jorgensen I, Zhang Y, Krantz BA, Miao EA. Pyroptosis triggers pore‐induced intracellular traps (PITs) that capture bacteria and lead to their clearance by efferocytosis. J Exp Med 2016; 213: 2113–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Jackson M, Crick DC, Brennan PJ. Phosphatidylinositol is an essential phospholipid of mycobacteria. J Biol Chem 2000; 275: 30092–30099. [DOI] [PubMed] [Google Scholar]
- 68. Nguyen NA, Sallans L, Kaneshiro ES. The major glycerophospholipids of the predatory and parasitic bacterium Bdellovibrio bacteriovorus HID5. Lipids 2008; 43: 1053–1063. [DOI] [PubMed] [Google Scholar]
- 69. Sohlenkamp C, Geiger O. Bacterial membrane lipids: diversity in structures and pathways. FEMS Microbiol Rev 2016; 40: 133–159. [DOI] [PubMed] [Google Scholar]
- 70. Kang R, Zeng L, Zhu S et al Lipid peroxidation drives Gasdermin D‐mediated pyroptosis in Lethal Polymicrobial sepsis. Cell Host Microbe 2018; 24: 97–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Zhang H, Zeng L, Xie M et al TMEM173 drives lethal coagulation in sepsis. Cell Host Microbe 2020; 27: 556–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wang D, Yu S, Zhang Y et al Caspase‐11‐GSDMD pathway is required for serum ferritin secretion in sepsis. Clin Immunol 2019; 205: 148–152. [DOI] [PubMed] [Google Scholar]
- 73. Hagar JA, Powell DA, Aachoui Y, Ernst RK, Miao EA. Cytoplasmic LPS activates caspase‐11: implications in TLR4‐independent endotoxic shock. Science 2013; 341: 1250–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Chen R, Zeng L, Zhu S et al cAMP metabolism controls caspase‐11 inflammasome activation and pyroptosis in sepsis. Sci Adv 2019; 5: eaav5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Wang D, Zheng J, Hu Q et al Magnesium protects against sepsis by blocking gasdermin D N‐terminal‐induced pyroptosis. Cell Death Differ 2020; 27: 466–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Olcum M, Tastan B, Kiser C, Genc S, Genc K. Microglial NLRP3 inflammasome activation in multiple sclerosis. Adv Protein Chem Struct Biol 2020; 119: 247–308. [DOI] [PubMed] [Google Scholar]
- 77. Lin CC, Edelson BT. New insights into the role of IL‐1β in experimental autoimmune encephalomyelitis and multiple sclerosis. J Immunol 2017; 198: 4553–4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Li S, Wu Y, Yang D et al Gasdermin D in peripheral myeloid cells drives neuroinflammation in experimental autoimmune encephalomyelitis. J Exp Med 2019; 216: 2562–2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Terao C, Kawaguchi T, Dieude P et al Transethnic meta‐analysis identifies GSDMA and PRDM1 as susceptibility genes to systemic sclerosis. Ann Rheum Dis 2017; 76: 1150–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Christodoulou K, Wiskin AE, Gibson J et al Next generation exome sequencing of paediatric inflammatory bowel disease patients identifies rare and novel variants in candidate genes. Gut 2013; 62: 977–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Soderman J, Berglind L, Almer S. Gene expression‐genotype analysis implicates GSDMA, GSDMB, and LRRC3C as contributors to inflammatory bowel disease susceptibility. Biomed Res Int 2015; 2015: 834805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. McGovern DP, Gardet A, Torkvist L et al Genome‐wide association identifies multiple ulcerative colitis susceptibility loci. Nat Genet 2010; 42: 332–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Chen X, Liu G, Yuan Y, Wu G, Wang S, Yuan L. NEK7 interacts with NLRP3 to modulate the pyroptosis in inflammatory bowel disease via NF‐κB signaling. Cell Death Dis 2019; 10: 906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Kurreeman FA, Stahl EA, Okada Y et al Use of a multiethnic approach to identify rheumatoid‐ arthritis‐susceptibility loci, 1p36 and 17q12. Am J Hum Genet 2012; 90: 524–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Thalayasingam N, Nair N, Skelton AJ et al CD4+ and B lymphocyte expression quantitative traits at rheumatoid arthritis risk loci in patients with untreated early arthritis: implications for causal gene identification. Arthritis Rheumatol 2018; 70: 361–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Wu XY, Li KT, Yang HX et al Complement C1q synergizes with PTX3 in promoting NLRP3 inflammasome over‐activation and pyroptosis in rheumatoid arthritis. J Autoimmun 2020; 106: 102336. [DOI] [PubMed] [Google Scholar]
- 87. Ozdogan H, Ugurlu S. Familial Mediterranean fever. Presse Med 2019; 48: e61–e76. [DOI] [PubMed] [Google Scholar]
- 88. Moghaddas F, Llamas R, De Nardo D et al A novel Pyrin‐associated autoinflammation with neutrophilic dermatosis mutation further defines 14‐3‐3 binding of pyrin and distinction to Familial Mediterranean Fever. Ann Rheum Dis 2017; 76: 2085–2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Kanneganti A, Malireddi RKS, Saavedra PHV et al GSDMD is critical for autoinflammatory pathology in a mouse model of Familial Mediterranean Fever. J Exp Med 2018; 215: 1519–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Verlaan DJ, Berlivet S, Hunninghake GM et al Allele‐specific chromatin remodeling in the ZPBP2/GSDMB/ORMDL3 locus associated with the risk of asthma and autoimmune disease. Am J Hum Genet 2009; 85: 377–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Grant SF, Qu HQ, Bradfield JP et al Follow‐up analysis of genome‐wide association data identifies novel loci for type 1 diabetes. Diabetes 2009; 58: 290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Barrett JC, Clayton DG, Concannon P et al Genome‐wide association study and meta‐analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet 2009; 41: 703–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Ayabe T, Fukami M, Ogata T et al Variants associated with autoimmune Type 1 diabetes in Japanese children: implications for age‐specific effects of cis‐regulatory haplotypes at 17q12‐q21. Diabet Med 2016; 33: 1717–1722. [DOI] [PubMed] [Google Scholar]
- 94. Rathkey JK, Zhao J, Liu Z et al Chemical disruption of the pyroptotic pore‐forming protein gasdermin D inhibits inflammatory cell death and sepsis. Sci Immunol 2018; 3: eaat2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Hu JJ, Liu X, Xia S et al FDA‐approved disulfiram inhibits pyroptosis by blocking gasdermin D pore formation. Nat Immunol 2020; 21: 736–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Spel L, Martinon F. Inflammasomes contributing to inflammation in arthritis. Immunol Rev 2020; 294: 48–62. [DOI] [PubMed] [Google Scholar]
- 97. Faliti CE, Gualtierotti R, Rottoli E et al P2X7 receptor restrains pathogenic Tfh cell generation in systemic lupus erythematosus. J Exp Med 2019; 216: 317–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Yi YS. Role of inflammasomes in inflammatory autoimmune rheumatic diseases. Korean J Physiol Pharmacol 2018; 22: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Guo Q, Wu Y, Hou Y et al Cytokine secretion and pyroptosis of thyroid follicular cells mediated by enhanced NLRP3, NLRP1, NLRC4, and AIM2 inflammasomes are associated with autoimmune thyroiditis. Front Immunol 2018; 9: 1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Leach S, Harvey P, Wali R. Changes with growth rate in the membrane lipid composition of and amino acid utilization by continuous cultures of Campylobacter jejuni. J Appl Microbiol 1997; 82: 631–640. [DOI] [PubMed] [Google Scholar]
- 101. Wang XG, Scagliotti JP, Hu LT. Phospholipid synthesis in Borrelia burgdorferi: BB0249 and BB0721 encode functional phosphatidylcholine synthase and phosphatidylglycerolphosphate synthase proteins. Microbiology 2004; 150: 391–397. [DOI] [PubMed] [Google Scholar]
- 102. Ostberg Y, Berg S, Comstedt P, Wieslander A, Bergstrom S. Functional analysis of a lipid galactosyltransferase synthesizing the major envelope lipid in the Lyme disease spirochete Borrelia burgdorferi . FEMS Microbiol Lett 2007; 272: 22–29. [DOI] [PubMed] [Google Scholar]
- 103. Nichols FC, Riep B, Mun J et al Structures and biological activities of novel phosphatidylethanolamine lipids of Porphyromonas gingivalis . J Lipid Res 2006; 47: 844–853. [DOI] [PubMed] [Google Scholar]
- 104. Roy H, Ibba M. RNA‐dependent lipid remodeling by bacterial multiple peptide resistance factors. Proc Natl Acad Sci USA 2008; 105: 4667–4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Johnston NC, Baker JK, Goldfine H. Phospholipids of Clostridium perfringens: a reexamination. FEMS Microbiol Lett 2004; 233: 65–68. [DOI] [PubMed] [Google Scholar]
- 106. Fyhrquist N, Muirhead G, Prast‐Nielsen S et al Microbe‐host interplay in atopic dermatitis and psoriasis. Nat Commun 2019; 10: 4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Halfvarson J, Brislawn CJ, Lamendella R et al Dynamics of the human gut microbiome in inflammatory bowel disease. Nat Microbiol 2017; 2: 17004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Zuo T, Ng SC. The Gut microbiota in the pathogenesis and therapeutics of inflammatory bowel disease. Front Microbiol 2018; 9: 2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Lee SY, Lee E, Park YM, Hong SJ. Microbiome in the Gut‐Skin Axis in Atopic Dermatitis. Allergy Asthma Immunol Res 2018; 10: 354–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Ruan J, Xia S, Liu X, Lieberman J, Wu H. Cryo‐EM structure of the gasdermin A3 membrane pore. Nature 2018; 557: 62–67. [DOI] [PMC free article] [PubMed] [Google Scholar]


