Abstract
The RNA quality of tissue biobank is crucial for translational research; however, the effects of the ex vivo ischemia time on RNA integrity and expression of genes related to hypoxia, stress, apoptosis and autophagy remains elusive. A total of 18 carcinoma tissues were stored at room temperature for 15 min, 30 min, 1, 2, 4, 8 and 24 h. The integrity and purity of isolated RNA were analyzed. Furthermore, the gene expression of mTOR, hypoxia-inducible factor 1α, phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit β isoform (PI3KCB), threonine kinase 1 (AKT1), NF-κB, protein kinase AMP-activated catalytic subunit α1 (AMPKα1), caspase 8 (CASP8), unc-51 like autophagy activating kinase 1 and Fas cell surface death receptor were analyzed using reverse transcription-quantitative PCR. The results demonstrated that RNA integrity numbers (RINs) remained stable in carcinoma tissues following ex vivo ischemia for 2 h at room temperature and that degradation began at 4 h (P<0.001). Additionally, the expression of PI3KCB, AKT1, AMPKα1 and CASP8 decreased at time points 8–24 h following ex vivo ischemia and delayed processing (P<0.001). In conclusion, >2 h of ex vivo ischemia and delayed processing induced RNA degradation and a decrease in RIN, and the gene expressions of PI3KCB, AKT1, AMPKα1 and CASP8 may be considered as markers to evaluate tissue quality at the gene expression level, providing a method for the standard processing and assessment of tissue specimen.
Keywords: ex vivo ischemia time, delayed processing time, tissue biobank, pre-analysis variable, RNA integrity number
Introduction
Tissue biobanks support the collection, analysis, storage and distribution of biospecimens for basic, translational and clinical research. With the advent of proteomics, transcriptomics and metabolomics, as well as the development of bioinformatics, molecular imaging and drug discovery, the demand for high-quality tissue biospecimens continues to increase (1,2), large-scale and high-flux research requirements for high quantity and quality biospecimens have allowed tissue biobanks to develop from undeveloped, small-scale storages to complex and multifaceted large-scale storages (3).
Quality assurance and quality control are critical for tissue biobanks to ensure that biospecimens distributed to researchers are comparable and produce accurate results; as a rough estimate, the number of stored tissue biospecimens was >300 million in the US around 2000, with this number increasing at the rate of 20 million/year (4). However, according to a survey taken by >700 cancer researchers, 47% of them were of the opinion that sample quality is difficult to control and 60% questioned the quality of samples (4).
There are various factors influencing the quality of biospecimens, and pre-analytical variables are crucial (5,6). Pre-analytical variables are divided into the pre-acquisition and post-acquisition phases. In the pre-acquisition phase, factors such as anesthesia (7), surgical trauma (8,9) and in vivo ischemia (10) might affect the cell integrity and gene expression profiles of the biospecimen, but they cannot be controlled very well because of the uncertainty of clinical treat requirements (11). Rather, factors such as ex vivo ischemia time, delayed processing time and processing temperature, which are crucial for bio-specimen quality (12), can be controlled and standardized effectively.
To the best of our knowledge, little research has been conducted of the impact of ex vivo ischemia time and delayed processing of biospecimens on hypoxic, stress, apoptotic and autophagic pathways. Therefore, the current study systematically assessed the impact of ex vivo ischemia time and delayed processing time on fresh tissue specimens in terms of the RNA quality and relative mRNA expression of genes involved in hypoxia, stress, apoptosis and autophagy to provide a theoretical basis for the standard processing and assessment of tissue biospecimens.
Materials and methods
Procurement of tissues
A total of 18 carcinoma tissue specimens were collected from patients who underwent surgery at Peking Union Medical College Hospital, Beijing, China from April 2017 to June 2017. The clinical characteristics of 18 patients are presented in Table I. The median age of the patients at diagnosis was 49.67±12.76 years (range, 29–79 years), of which 16 patients were female (88.9%) and 2 patients were male (11.1%). The collected tissues included specimens from 10 patients with thyroid cancer (55.5%), 3 with ovarian cancer (16.6%), 2 with breast cancer (11.1%), 1 with pancreatic cancer (5.6%), 1 with polymorphous adenocarcinoma (5.6%) and 1 with lung cancer (5.6%). These 18 specimens and 6 types of cancer were included due to their prevalence and commonality in the clinical and tissue biobank. All patients provided signed informed consent for the donation of the specimens. The current study was approved by the Ethics Committee of the Institutional Review Board of Peking Union Medical College Hospital.
Table I.
Clinical characteristics of the patients (n=18).
| Clinical characteristic | Number of patients, n (%) |
|---|---|
| Sex | |
| Male | 2 (11.1) |
| Female | 16 (88.9) |
| Age, years | |
| ≤50 | 8 (44.4) |
| >50 | 10 (55.6) |
| Type of carcinoma | |
| Thyroid | 10 (55.5) |
| Breast | 3 (16.6) |
| Ovary | 2 (11.1) |
| Pancreas | 1 (5.6) |
| Polymorphous adenocarcinoma | 1 (5.6) |
| Lung | 1 (5.6) |
Following gross examination, the tissues were resected and washed several times with PBS (pH 7.2) to remove cell debris and remaining blood. Tissues were then dissected into two parts, one part of the specimen (1 cm3) was used for clinical pathology diagnosis, while the other part was divided to 7 smaller specimens (volume, 100 mm3). One of the 7 small specimens was immediately snap-frozen in liquid nitrogen, thus the time from tissue resection to storage in liquid nitrogen was ~15 min, which was set as the first point of ex vivo ischemia time. The other 6 specimens were stored at room temperature for 15, 45, 105, 225, 465 and 1,425 min, and then were stored in liquid nitrogen, so the ex vivo ischemia time was 30 min, 1, 2, 4, 8 or 24 h, respectively. Following storage, the specimens were cryopreserved for the subsequent RNA extraction, gene expression analysis and morphological features assessment, which were reviewed by two experienced pathologists to confirm final diagnoses.
RNA extraction and integrity control
Total RNA was extracted from all tissue samples; ~30 mg of 30 mm3 tissue specimen was homogenized using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) with a homogenizer (IKA Werke GmbH & Co. KG) for RNA extraction, according to the manufacturers' protocols. RNA concentration was quantified using NanoDrop™ One (Thermo Fisher Scientific, Inc.) by measuring the extinction coefficient at 260 nm. Additionally, the ratio of optical density (OD) 260/230 and OD 260/280, which indicates RNA purity, was tested using the NanoDrop One. An Agilent 2100 Bioanalyzer and an Agilent RNA 6000 Nano kit (Agilent Technologies, Inc.) were used to determine RNA quality, according to the manufacturers' protocol. RNA integrity numbers (RINs) were calculated using Agilent Technologies 2100 Bioanalyzer expert software (version B.02.0×; Agilent Technologies, Inc.), and were presented as a value (1–10) to indicate the degree of degradation.
Reverse transcription
Total RNA (1 µg) was diluted to a final volume of 25 µl. A reverse transcription master mix was prepared as follows to reverse transcribe RNA into cDNA: 5 µl 5X reaction buffer, 1 µl random primers (cat. no. C1181; Promega Corporation), 1.25 µl dNTPs (cat. no. U1330; Promega Corporation), 1 µl M-MLV reverse transcriptase (cat. no. M1701; Promega Corporation), 0.5 µl RNase inhibitors (Takara Bio, Inc.) and 6.25 µl nuclease-free water (cat. no. AM9937; Invitrogen; Thermo Fisher Scientific, Inc.). Following reverse transcription, all samples were diluted to a final volume of 100 µl.
Reverse transcription-quantitative PCR (RT-qPCR)
All PCR procedures were set up using a QIAgility robotic system (Qiagen, Inc.). Mixes were prepared with 5 µl Fast SYBR1 Green Master mix (Applied Biosystems; Thermo Fisher Scientific, Inc.; cat. no. 4385612) and 2 µl primer mix (1.67 µM; Table II). Then, 2 µl cDNA (10 ng/µl) were added for a final volume of 10 µl/well. Gene expression quantification was performed via RT-qPCR using an Applied Biosystems® 7500 Real-Time PCR system (Thermo Fisher Scientific, Inc.). The following thermocycling conditions were used: 95°C for 20 sec; followed by 40 cycles of 95°C for 3 sec and 60°C for 30 sec. The expression of the housekeeping gene GAPDH was used for normalization and mRNA levels were quantified using the 2−ΔΔCq method (13,14). The primers used in the analysis of gene expression were associated with hypoxia, stress, apoptosis and autophagy: Caspase 8 (CASP8), Fas cell surface death receptor (FAS), hypoxia-inducible factor 1α (HIF1α), unc-51 like autophagy activating kinase 1 (ULK1), mTOR, NF-κB, phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit β isoform (PI3KCB), AKT serine/threonine kinase 1 (AKT1), protein kinase AMP-activated catalytic subunit α1 (AMPKα1) and (Table II).
Table II.
Primer sequences used for reverse transcription-quantitative PCR.
| Gene | Primer sequences (5′→3′) | Fragment size, bp |
|---|---|---|
| CASP8 | F: GCCCCCATCTATGAGCTGAC | 100 |
| R: TATCCCCCTGACAAGCCTGA | ||
| FAS | F: GTGGACCCGCTCAGTACG | 121 |
| R: GGACGATAATCTAGCAACAGACG | ||
| HIF1α | F:@ TTGGCAACCTTGGATTGGATG | 190 |
| R: AAATCTCCGTCCCTCAACCTC | ||
| ULK1 | F: GTCACACGCCACATAACAG | 90 |
| R: TCTTCTAAGTCCAAGCACAGT | ||
| mTOR | F: GCTTAGAGGACAGCGGGGAA | 120 |
| R: AAGCATCTTGCCCTGAGGTT | ||
| NF-κB | F: CCACAAGACAGAAGCTGAAG | 149 |
| R: AGATACTATCTGTAAGTGAACC | ||
| PI3KCB | F: TGGAACTTGGCACTGGAACT | 114 |
| R: GCTGGGAAACAAGGGGAGAA | ||
| AKT1 | F: CTGGTCCTGTCTTCCTCATGTT | 190 |
| R: AGGCAGCCCCTTTGACTTCT | ||
| AMPKα1 | F: TGAAAGAAAAGTGTGTGCTGT | 64 |
| R: TGGGTGAGAAGATGAGGGAAAGA | ||
| GAPDH | F: GAAGGTGAAGGTCGGAGTC | 226 |
| R: GAAGATGGTGATGGGATTTC |
bp, base pair; CASP8, caspase 8; FAS, Fas cell surface death receptor; HIF1α, hypoxia-inducible factor 1α; ULK1, unc-51 like autophagy activating kinase 1; PI3KCB, phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit β isoform; AKT1, threonine kinase 1; STAT3, signal transduced and activator of transcription.
Histological assessment
The other part of all the fresh tissues specimen were used for the pathological clinical diagnosis. In addition, after the RNA isolation, the frozen thyroid tissues in liquid nitrogen with different ex vivo ischemia times were fixed in 10% neutral buffered formalin at room temperature for 24 h. Specimens were then embedded in paraffin. Sections (thickness, 5 µm) were microtomed and placed on slides. Slides were baked for 1 h at 60°C in an oven and stained with hematoxylin and eosin, both at room temperature for 1 h with an autostainer XL (ST5010; Leica Microsystems GmbH). Histological evaluations were completed by two experienced pathologists on a light microscope (magnification, ×100 or ×200; Axio Scope.A1; Zeiss GmbH).
Statistical analysis
Statistical analysis was performed using SPSS (version 19.0; IBM Corp.) and Prism software (version 5; GraphPad Software, Inc.). One-way ANOVA followed by Tukey's post hoc test was used to analyse differences between multiple groups. Pearson's correlation coefficient was used for correlation analysis. P<0.05 was considered to indicate a statistically significant difference.
Results
Effect of ex-vivo ischemia time on RNA integrity
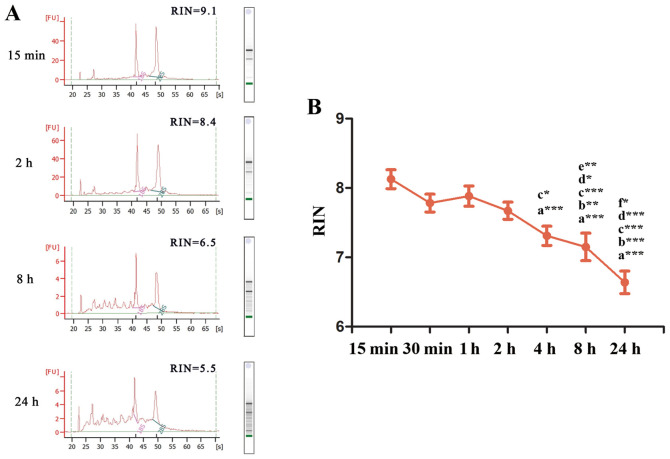
To evaluate the influence of tissue ex vivo ischemia time points on RNA integrity under controlled conditions, 18 fresh carcinoma tissues from different organs were collected, separated and stored at room temperature for various ex-vivo ischemia times prior to being snap-frozen in liquid nitrogen. The RNA quality was assessed according to integrity and purity. The RNA integrity data of tissues ex vivo ischemia at 15 min, 2, 8 and 24 h are presented in Fig. 1A. The distinct ribosomal peaks of 18S and 28S indicated good RNA integrity at 15 min and 2 h, but not at 8 and 24 h. The RINs of each group were 8.12±0.576, 7.78±0.546, 7.88±0.621, 7.67±0.523, 7.31±0.589, 7.15±0.844 and 6.63±0.688 for ex vivo ischemia time points 15, 30 min, 1, 2, 4, 8 and 24 h, respectively (Fig. 1B). SPSS analysis demonstrated that the RINs at 4, 8 and 24 h were significantly decreased compared with 15 min (P<0.001). Furthermore, compared with 30 min, RINs at 8 h (P<0.01) and 24 h (P<0.001) were significantly decreased. Similarly, compared with 1 h, the RINs at 4 h (P<0.05), 8 h (P<0.001) and 24 h (P<0.001) were decreased, and the RINs at 8 h (P<0.05) and 24 h (P<0.001) were significantly declined compared with 2 h. In addition, the RINs at 8 h were decreased compared with 4 h (P<0.01), and similarly, the RINs at 24 h were significantly decreased compared with 8 h (P<0.05). The results indicated that RNA remained intact following 2 h of ex vivo ischemia at room temperature and integrity began to gradually decline after 4 h.
Figure 1.
RNA integrity of tissues for various ex vivo ischemia time points. (A) mRNA RINs at 15 min, 2, 8 and 24 h of ex vivo ischemia. (B) Changes in RNA integrity at increasing ex vivo ischemia time points at room temperature. a, vs. 15 min; b, vs. 30 min; c, vs. 1 h; d, vs. 2 h; e, vs. 4 h; f, vs. 8 h. *P<0.05, **P<0.01, ***P<0.001. RINs, RNA integrity numbers.
The means of RNA OD260/280 and OD260/230 ratios of 7 time points were 2.05±0.1 and 1.78±0.43, respectively (n=126). The OD260/280 ratios of each group were 2.028±0.099, 2.101±0.144, 2.043±0.091, 2.051±0.085, 2.031±0.045, 2.011±0.076 and 2.085±0.128 for ex vivo ischemia time points 15, 30 min, 1, 2, 4, 8 and 24 h, respectively (Fig. 2). The OD260/230 ratios were 1.713±0.398, 1.960±0.330, 1.916±0.367, 1.675±0.561, 1.839±0.391, 1.550±0.500 and 1.799±0.481 for ex vivo ischemia time points 15, 30 min, 1, 2, 4, 8 and 24 h, respectively. The ratios of OD260/280 and OD260/230 of all groups indicated that the degradation of RNA integrity does not affect RNA purity reflected by absorbance under ultraviolet light.
Figure 2.
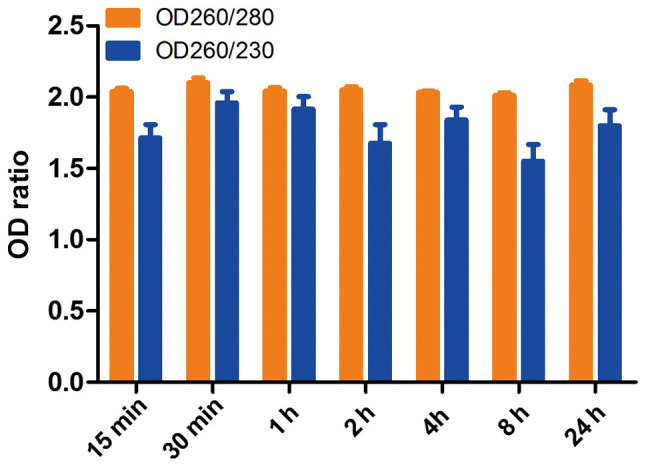
OD260/280 and OD260/230 of isolated RNA for various ex vivo ischemia time points. OD, optical density.
Effect of ex vivo ischemia time on gene expression
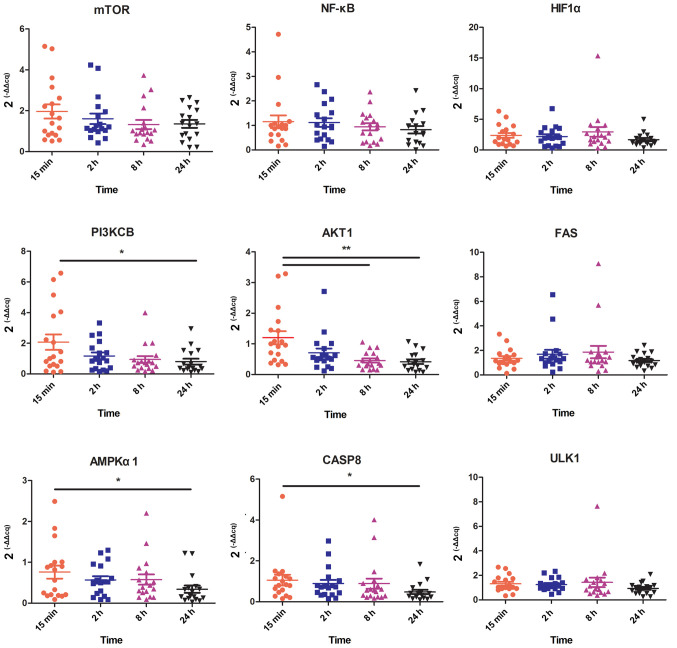
To explore the influence of ex vivo ischemia time points on cell functions involving hypoxia, stress, apoptosis and autophagy, the gene expression of mTOR, HIF1α, PI3KCB, AKT1, NF-κB, AMPKα1, CASP8, ULK1 and FAS were evaluated using RT-qPCR. The RINs of the 7 different ex vivo ischemia times indicated that RNA remained intact following 2 h of ex vivo ischemia at room temperature and integrity began to gradually decline after 4 h. Thus, the present study mainly focused on ex vivo ischemia times of 15 min, 2, 8 and 24 h, and more times will be explored in further study. The results demonstrated that AKT1 was significantly decreased at 8 h compared with 15 min (P<0.01) and that PI3KCB, AMPKα1 and CASP8 were significantly decreased at 24 h compared with 15 min (P<0.05; Fig. 3). However, the expression of mTOR, HIF1α, NF-κB, ULK1 and FAS were not significantly different between time points.
Figure 3.
Gene expression of genes associated with hypoxic, stress, apoptotic and autophagic pathways for various ex vivo ischemia time points. HIF1α, hypoxia-inducible factor 1α; PI3KCB, phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit β isoform; AKT1, threonine kinase 1; FAS, Fas cell surface death receptor; AMPKα1, protein kinase AMP-activated catalytic subunit α1; CASP8, caspase 8; ULK1, unc-51 like autophagy activating kinase 1. *P<0.05, **P<0.01.
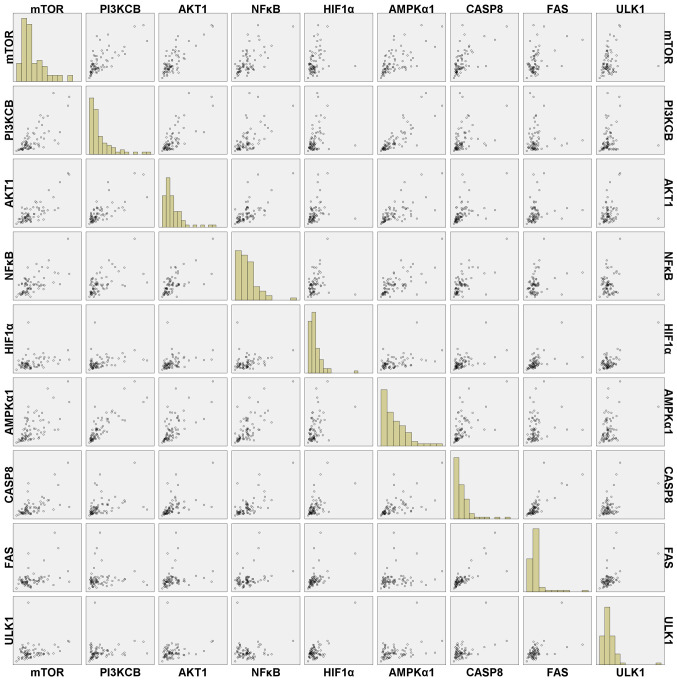
Further analysis indicated that there was no significant correlation between RIN and gene expression (Table III). In addition, the interclass Pearson correlation coefficients among the gene expression levels were strong, such as rmTOR/PI3KCB=0.721, rmTOR/AKT1=0.792, rmTOR/NF-κB=0.640, rmTOR/AMPKα1=0.630, rPI3KCB/AKT1=0.741, rPI3KCB/NF-κB=0.572, rPI3KCB/AMPKα1=0.835, rAKT1/NF-κB=0.646 and rAKT1/AMPKα1=0.666, rAKT1/CASP8=0.508, rCASP8/FAS=0.762, rCASP8/mTOR=0.607, rHIF1α/ULK1=0.728 (P<0.001) (Table IV). In addition, HIF1α had slight positive correlation with mTOR, PI3KCB and AKT1 (r=0.281, r=0.293 and r=0.298, respectively; P<0.05), and FAS has correlation with NF-κB (r=0.281; P<0.05). In addition, there were no correlations observed between some gene expression (P>0.05), such as AKT1 vs. FAS, ULK1 vs. AKT1, AMPKα1 vs. FAS, ULK1 vs. mTOR, ULK1 vs. PI3KCB, ULK1 vs. NF-κB and FAS vs. PI3KCB. A scatter plot of gene expression, including the correlation between the expression of these genes is presented in Fig. 4; the scatterplot trend was consistent with the correlation coefficients of the gene expression presented in Table IV.
Table III.
Correlation coefficients between RIN and gene expression.
| RIN | ||
|---|---|---|
| Gene | Pearson correlation coefficient | Significance (bilateral) |
| mTOR | −0.065 | 0.590 |
| PI3KCB | −0.051 | 0.668 |
| AKT1 | 0.172 | 0.150 |
| NF-κB | 0.102 | 0.394 |
| HIF1α | 0.018 | 0.882 |
| AMPKα1 | −0.040 | 0.741 |
| CASP8 | 0.133 | 0.267 |
| FAS | 0.066 | 0.582 |
| ULK1 | 0.024 | 0.840 |
RIN, RNA integrity number; PI3KCB, phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit β isoform; AKT1, threonine kinase 1; HIF1α, hypoxia-inducible factor 1α; AMPKα1, protein kinase AMP-activated catalytic subunit α1; CASP8, caspase 8; FAS, Fas cell surface death receptor; ULK1, unc-51 like autophagy activating kinase 1.
Table IV.
Pearson's correlation coefficients of gene expression.
| Gene expression | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Variable | mTOR | PI3KCB | AKT1 | NF-κB | HIF1α | AMPKα1 | CASP8 | FAS | ULK1 |
| mTOR | |||||||||
| r-value | 1 | 0.721c | 0.792c | 0.640c | 0.281a | 0.630c | 0.607c | 0.409c | 0.221 |
| Significance (bilateral) | N/A | 0.000 | 0.000 | 0.000 | 0.017 | 0.000 | 0.000 | 0.000 | 0.063 |
| N | 72 | 72 | 72 | 72 | 72 | 72 | 72 | 72 | 72 |
| PI3KCB | |||||||||
| r-value | 0.721c | 1 | 0.741c | 0.572c | 0.293a | 0.835c | 0.372b | 0.019 | 0.150 |
| Significance (bilateral) | 0.000 | N/A | 0.000 | 0.000 | 0.013 | 0.000 | 0.001 | 0.876 | 0.208 |
| N | 72 | 72 | 72 | 72 | 72 | 72 | 72 | 72 | 72 |
| AKT1 | |||||||||
| r-value | 0.792c | 0.741c | 1 | 0.646c | 0.298a | 0.666c | 0.508c | 0.181 | 0.189 |
| Significance (bilateral) | 0.000 | 0.000 | N/A | 0.000 | 0.011 | 0.000 | 0.000 | 0.127 | 0.111 |
| N | 72 | 72 | 72 | 72 | 72 | 72 | 72 | 72 | 72 |
| NF-κB | |||||||||
| r-value | 0.640c | 0.572c | 0.646c | 1 | 0.226 | 0.674c | 0.658c | 0.281a | 0.021 |
| Significance (bilateral) | 0.000 | 0.000 | 0.000 | N/A | 0.056 | 0.000 | 0.000 | 0.017 | 0.861 |
| N | 72 | 72 | 72 | 72 | 72 | 72 | 72 | 72 | 72 |
| HIF1α | |||||||||
| r-value | 0.281a | 0.293a | 0.298a | 0.226 | 1 | 0.433c | 0.525c | 0.482c | 0.728c |
| Significance (bilateral) | 0.017 | 0.013 | 0.011 | 0.056 | N/A | 0.000 | 0.000 | 0.000 | 0.000 |
| N | 72 | 72 | 72 | 72 | 72 | 72 | 72 | 72 | 72 |
| AMPKα1 | |||||||||
| r-value | 0.630c | 0.835c | 0.666c | 0.674c | 0.433c | 1 | 0.557c | 0.137 | 0.342c |
| Significance (bilateral) | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | N/A | 0.000 | 0.250 | 0.003 |
| N | 72 | 72 | 72 | 72 | 72 | 72 | 72 | 72 | 72 |
| CASP8 | |||||||||
| r-value | 0.607c | 0.372c | 0.508c | 0.658c | 0.525c | 0.557c | 1 | 0.762c | 0.467c |
| Significance (bilateral) | 0.000 | 0.001 | 0.000 | 0.000 | 0.000 | 0.000 | N/A | 0.000 | 0.000 |
| N | 72 | 72 | 72 | 72 | 72 | 72 | 72 | 72 | 72 |
| FAS | |||||||||
| r-value | 0.409c | 0.019 | 0.181 | 0.281a | 0.482c | 0.137 | 0.762c | 1 | 0.501c |
| Significance (bilateral) | 0.000 | 0.876 | 0.127 | 0.017 | 0.000 | 0.250 | 0.000 | N/A | 0.000 |
| N | 72 | 72 | 72 | 72 | 72 | 72 | 72 | 72 | 72 |
| ULK1 | |||||||||
| r-value | 0.221 | 0.150 | 0.189 | 0.021 | 0.728c | 0.342b | 0.467c | 0.501c | 1 |
| Significance (bilateral) | 0.063 | 0.208 | 0.111 | 0.861 | 0.000 | 0.003 | 0.000 | 0.000 | N/A |
| N | 72 | 72 | 72 | 72 | 72 | 72 | 72 | 72 | 72 |
P<0.05
P<0.01
P<0.001. PI3KCB, phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit β isoform; AKT1, threonine kinase 1; HIF1α, hypoxia-inducible factor 1α; AMPKα1, protein kinase AMP-activated catalytic subunit α1; CASP8, caspase 8; FAS, Fas cell surface death receptor; ULK1, unc-51 like autophagy activating kinase 1; N/A, not applicable.
Figure 4.
Scatter plots of the expression of genes associated with hypoxic, stress, apoptotic and autophagic pathways for various ex vivo ischemia time points. PI3KCB, phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit β isoform; AKT1, threonine kinase 1; HIF1α, hypoxia-inducible factor 1α; AMPKα1, protein kinase AMP-activated catalytic subunit α1; CASP8, caspase 8; FAS, Fas cell surface death receptor; ULK1, unc-51 like autophagy activating kinase 1.
Histological morphology
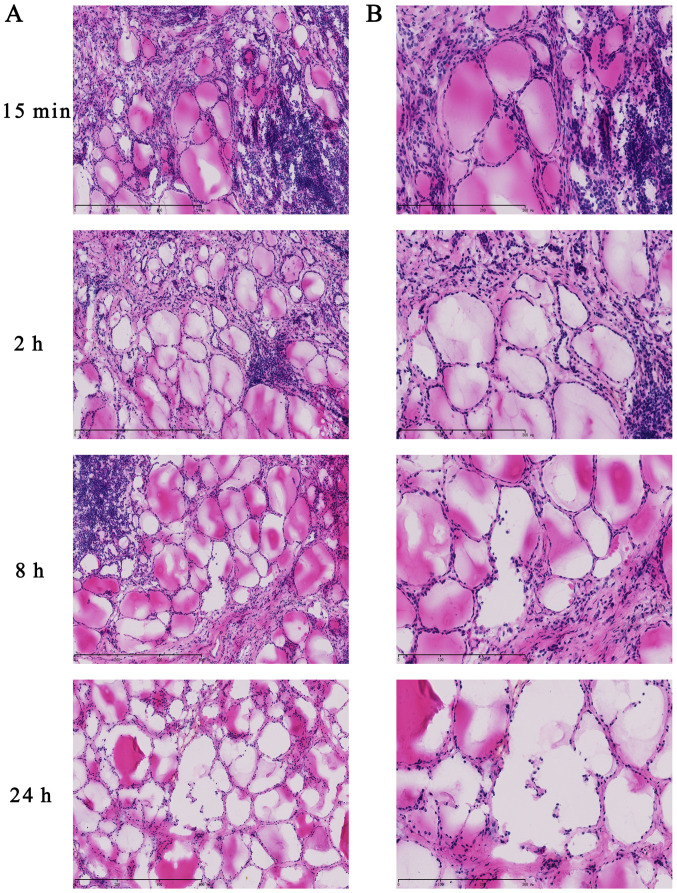
To evaluate histomorphology at various ex vivo ischemia time points, frozen tissue specimens were fixed, sectioned and stained with H&E. The histological morphology demonstrated that follicular epithelial cells were arranged closely and maintained integrity at 15 min and 2 h in thyroid tissues (Fig. 5). However, the microstructure of the thyroid follicles was destroyed at 8 h and cells displayed looseness and follicle disintegration at 24 h. These results demonstrated that ex vivo ischemia and delayed processing induced morphological changes in thyroid carcinoma, which was accompanied by RNA degradation.
Figure 5.
Haematoxylin and eosin-stained thyroid tissues at various ex vivo ischemia time points. Thyroid tissues at ex vivo ischemia time points of 15 min, 2, 8 and 24 h at (A) magnification ×100 and (B) magnification ×200.
Discussion
RNA degradation is one of the major issues when storing tissues in biobanks, and previous studies have investigated the effect of tissue type, storage buffers and ex vivo ischemia times on RNA quality (15,16). Among these factors, ex vivo ischemia and delayed processing time are crucial pre-analysis challenges for RNA quality (6,12). The results of the current study demonstrated that RNA integrity decreased significantly following ex vivo ischemia for 4 h, indicating that RNA remained intact for 2 h at room temperature.
Previous research reported that the effects of ex vivo ischemia time on RNA integrity varied in different types of tumors. Bao et al (17) demonstrated that RNA remained stable both at room temperature and on ice for ≤4 h in colon cancer tissues. Additionally, retained ileum mucosa was revealed to exhibit high RNA integrity for 1.5 h at room temperature and 6 h at 4°C (18). In kidney renal cell carcinoma tissues, RNA degradation was observed at room temperature for 1 h and degradation was not detected following storage on ice for 4 h (19,20). Similarly, RIN gradually decreased in tumor and adjacent normal tissues at 24°C at ischemia time points 15, 30, 60 and 120 min in hepatocellular carcinoma tissues (21). Even though our study did not verify whether the RNA integrity from different tumor tissues (such as thyroid, ovary, breast, pancreas, jaw and lung) had different sensitivities to ex vivo ischemia time points, it could be verified by further studies with more carcinoma specimens.
In addition, a previous study demonstrated that the RIN of non-small cell lung cancer tissue was significantly lower (OR=0.08; P=0.01) in samples that were preserved for >3 h prior to cryopreservation, and prolonged in vivo (>2 h) and ex vivo ischemia (>10 h) times were associated with lower patient-derived xenograft engraftment rates (22). Furthermore, long term ex vivo ischemia of human brain tumor biopsy tissues affected ribosomal RNA integrity and increased RNA degradation (23). When snap freezing and multiple sampling of biopsies were combined, the percentage of specimens with degraded RNA was reduced by two-fold (23). However, another previous study on RNA integrity and gene expression in human myocardial tissue reported that there was no significant correlation between RIN and a post-mortem interval of 5–24 h at 4°C (24). Furthermore, RNA integrity was not correlated with 1 h ex vivo ischemia time points in different types of tumors, including breast, colon, stomach, oesophageal, gall bladder, ovarian, kidney, liver, lung, pancreatic, small intestinal, spleen, common bile duct, retroperitoneum, testis and urinary bladder cancer (24,25).
Additionally, the duration of ex vivo ischemia affects the related gene expression profile (20). The results of the current study demonstrated that the gene expression of PI3KCB, AKT1, AMPKα1 and CASP8 were significantly decreased following ex vivo ischemia at time points 8–24 h compared with 15 min. PI3KCB, AKT1, AMPKα1 and CASP8 serve important roles in the hypoxic, stress, apoptotic and autophagic signaling pathways (26,27). AMPKα1 is critical for hypoglycemia, hypoxia, ischemia and heat shock (28), and regulates growth, metabolism reprogramming, glucose and fatty acid metabolism and mitochondrial function (29). Oncogenes AKT1 and PI3KCB are major factors in the regulation of various cellular functions, including metabolism, growth, proliferation, survival, transcription and protein synthesis (30,31). Furthermore, AMPKα1 stimulates autophagy by inhibiting mTOR, whereas PI3KCB, Ras and AKT1 inhibit autophagy by activating mTOR (32). Additionally, CASP8 is induced by endoplasmic reticulum stress and proteotoxic stress (33), which participate in the apoptosis signal pathway (34).
An increased number of studies have demonstrated that when the blood supply is cut off, tissues undergo oxygen deprivation and mammalian cells undergo hypoxemia, autophagy and endoplasmic reticulum stress response (35). Then, hypoxemia, autophagy, apoptosis and endoplasmic reticulum stress occur and coordinate (36,37). Furthermore, a previous study has reported that exposure to ex vivo ischemia significantly altered gene expression of G-protein signaling 1 and eukaryotic translation elongation factor 1 α 1 in normal and colorectal cancer tissue samples (38).
Dr Carolyn Compton, a former Office of Biorepository and Biospecimen Research executive, proposed that when biological samples are excised, cut off from a blood supply and exposed to abrupt changes in temperature, cellular behavior becomes difficult to predict (4). Gene expression and protein phosphorylation fluctuate extensively and cellular self-destruct pathways may be activated (4). The American Society of Clinical Oncology, College of American Pathologists (2014), Biorepositories and Biospecimen Research Branch, National Cancer Institute (2016), Clinical Laboratory Standards Institute (2011), Clinical Laboratory Standards Institute (2005), International Standards Organization/Technical Committee (2016) and European Committee for Standardization/Technical Committee (2015) demonstrated that ex vivo ischemia times in tissues should be as short as possible (optimally <20 min and not >1 h) for the analysis of biomarkers, molecular analysis, and RNA, DNA and protein isolation (6).
Although the results of the current study revealed that the expression of PI3KCB, AKT1, AMPKα1 and CASP8 decreased with time, there was no correlation between RIN and gene expression, indicating that the decrease in gene expression was not induced by RNA degradation. Instead, the decrease in gene expression was associated with hypoxia, stress, apoptosis and autophagy induced by ex vivo ischemia and delayed processing.
Furthermore, the results demonstrated positive correlations between the expression of mTOR, PI3KCB, AKT1, NF-κB and AMPKα1, indicating that their expression levels may be coordinated during ex vivo ischemia and delayed processing. Certain stress responsive transcription factors, including NF-κB, serve a major role in autophagic response (39) and orchestrate numerous transcriptional responses, including autophagy and execution of cell death (40). Wang et al (26) reported that endoplasmic reticulum stress and mitochondrial injury on myocardial ischemia and reperfusion upregulated phosphorylated PI3KCB and AKT1. Hypoxia and reperfusion can evoke autophagy and apoptosis via the AKT1 and mTOR signaling pathways in microglia cells and the expression of mTOR and AKT1 was not altered under hypoxic conditions for 6 h and reperfusion injury at 24; however, the levels of p-AKT1 and mTOR were altered (27). Thus, the results of the current study indicated that while mRNA expression was altered due to ex vivo ischemia and delayed processing, whether the expression of phosphorylated proteins, which is important in cell signal transduction, may be altered requires further study.
Furthermore, the results demonstrated that there were fewer positive correlations between HIF1α and mTOR, PI3KCB and AKT1. A prior study revealed that HIF1α promotes the expression of hundreds of genes involved in autonomous and non-autonomous cellular adaptations to hypoxia, which are upregulated at the protein level via mTOR or at the mRNA level via STAT3 and NF-κB signaling (41). Although the current study did not report ULK and/or FAS downregulation at the mRNA level or an association with the other factors, the activity of ULK is required for autophagy (42). ULK-mediated Beclin-1 phosphorylation in vivo is crucial for the function of Beclin-1 in autophagy (42).
Moreover, in addition to RNA degradation, the results of the present study showed that ex vivo ischemia and delayed processing induced morphological changes in thyroid carcinoma tissues, which is another factor to be evaluated following delayed processing.
In addition to the processing variables, biological and environmental factors, including donor sex, age (43), drug use (44) and diet, exercise and lifestyle habits, such as smoking, could affect the quality of biospecimens (45). Furthermore, various acquisition methods, such as collecting during the surgical operation (in situ) or after resection (ex vivo) (9), and surgical approaches, such as laparoscopic colectomy or open colectomy (8) could influence gene expression analysis.
There were certain limitations of the present study. Firstly, the current study was a preliminary study that could not verify whether RNA integrity from various tumor tissues had different sensitivities to ex vivo ischemia time due to the small sample size. Future studies should include larger sample sizes and more tumor specimens to comprehensively examine the effects of various ex vivo ischemia time points and delayed processing on RNA integrity and tissue quality in different carcinoma tissues. Secondly, tissue biobanks are the basic infrastructure that provide biospecimens to various institutions for basic, translational and clinical research, and their procedures for the collection, analysis, storage and distribution of biospecimens may affect the results of downstream studies. Furthermore, tissue banks should set up individual procedures according to the requirements by researchers to accommodate research institutions that require tissues for specific purposes. Therefore, a future study should investigate the commonality of more tissues in biobanks, such as breast, colon, stomach, oesophagus, gall bladder, ovary, kidney, liver, lung and pancreas, which could be used to provide research institutions with standardized processing procedures for specimens.
In conclusion, ex vivo ischemia time and delayed processing induced RNA degradation and altered gene expression in various carcinoma tissues. Fresh tissues should be processed within 2 h to avoid RNA degradation to acquire high quality biospecimens. Furthermore, the gene expression of PI3KCB, AKT1, AMPKα1 and CASP8 may be considered as markers to evaluate tissue quality at the gene expression level, providing a method for the standard processing and assessment of tissue specimens.
Acknowledgements
Not applicable.
Funding
The current study was supported by the Chinese Academy of Medical Sciences Initiative for Innovative Medicine (grant nos. 2017-I2M-1-001 and 2017-I2M-2-001) and The National Key Research and Development Program of China: The Cluster Construction of Human Genetic Resource Biobank in North China (grant no. 2016YFC1201703).
Availability of data and materials
All data generated or analyzed during the current study are included in this published article.
Authors' contributions
AW, SZ and DC processed the tissue specimens, performed the RNA extraction and qPCR. DG and TX designed the study, contributed to the analysis of the data and wrote the manuscript. JS performed the histological analysis and interpreted the patient data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All patients provided signed informed consent for the donation of the specimens. The current study was approved by the Ethics Committee of the Institutional Review Board of Peking Union Medical College Hospital.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Watson PH, Wilson-McManus JE, Barnes RO, Barnes, Giesz SC, Png A, Hegele RG, Brinkman JN, Mackenzie IR, Huntsman DG, Junker A, et al. Evolutionary concepts in biobanking-the BC BioLibrary. J Transl Med. 2009;7:95. doi: 10.1186/1479-5876-7-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nishihara H. Clinical biobank: A novel system to support cancer clinical sequencing in Japan. Rinsho Byori. 2017;65:167–172. (In Japanese) [PubMed] [Google Scholar]
- 3.Ginsburg GS, Burke TW, Febbo P. Centralized biorepositories for genetic and genomic research. JAMA. 2008;299:1359–1361. doi: 10.1001/jama.299.11.1359. [DOI] [PubMed] [Google Scholar]
- 4.Baker M. Biorepositories: Building better biobanks. Nature. 2012;486:141–146. doi: 10.1038/486141a. [DOI] [PubMed] [Google Scholar]
- 5.Carithers LJ, Agarwal R, Guan P, Odeh H, Sachs MC, Engel KB, Greytak SR, Barcus M, Soria C, Jason Lih CJ, et al. The biospecimen preanalytical variables program: A multiassay comparison of effects of delay to fixation and fixation duration on nucleic acid quality. Arch Pathol Lab Med. 2019;143:1106–1118. doi: 10.5858/arpa.2018-0172-OA. [DOI] [PubMed] [Google Scholar]
- 6.Compton CC, Robb JA, Anderson MW, Berry AB, Birdsong GG, Bloom KJ, Branton PA, Crothers JW, Cushman-Vokoun AM, Hicks DG, et al. Preanalytics and precision pathology: Pathology practices to ensure molecular integrity of cancer patient biospecimens for precision medicine. Arch Pathol Lab Med. 2019;143:1346–1363. doi: 10.5858/arpa.2019-0009-SA. [DOI] [PubMed] [Google Scholar]
- 7.Kahn N, Riedlinger J, Roessler M, Rabe C, Lindner M, Koch I, Schott-Hildebrand S, Herth FJ, Schneider MA, Meister M, Muley TR. Blood-sampling collection prior to surgery may have a significant influence upon biomarker concentrations measured. Clin Proteomics. 2015;12:19. doi: 10.1186/s12014-015-9093-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galissier T, Schneider C, Nasri S, Kanagaratnam L, Fichel C, Coquelet C, Diebold MD, Kianmanesh R, Bellon G, Dedieu S, et al. Biobanking of fresh-frozen human adenocarcinomatous and normal colon tissues: Which parameters influence RNA quality? PLoS One. 2016;11:e0154326. doi: 10.1371/journal.pone.0154326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou JH, Sahin AA, Myers JN. Biobanking in genomic medicine. Arch Pathol Lab Med. 2015;139:812–818. doi: 10.5858/arpa.2014-0261-RA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olsen J, Kirkeby LT, Eiholm S, Jess P, Troelsen JT, Gögenür I, Olsen J. Impact of in vivo ischemic time on RNA quality-experiences from a colon cancer biobank. Biopreserv Biobank. 2015;13:255–262. doi: 10.1089/bio.2015.0009. [DOI] [PubMed] [Google Scholar]
- 11.Zheng XH, Zhang SD, Zhang PF, Li XZ, Hu YZ, Tian T, Zhu L, Wang RZ, Jia WH. Tumor cell content and RNA integrity of surgical tissues from different types of tumors and its correlation with ex vivo and in vivo ischemia. Ann Surg Oncol. 2018;25:3764–3770. doi: 10.1245/s10434-018-6723-z. [DOI] [PubMed] [Google Scholar]
- 12.Betsou F, Lehmann S, Ashton G, Barnes M, Benson EE, Coppola D, DeSouza Y, Eliason J, Glazer B, Guadagni F, et al. Standard preanalytical coding for biospecimens: Defining the sample PREanalytical code. Cancer Epidemiol Biomarkers Prev. 2010;19:1004–1011. doi: 10.1158/1055-9965.EPI-09-1268. [DOI] [PubMed] [Google Scholar]
- 13.Mazzei M, Vascellari M, Zanardello C, Melchiotti E, Vannini S, Forzan M, Marchetti V, Albanese F, Abramo F. Quantitative real time polymerase chain reaction (qRT-PCR) and RNAscope in situ hybridization (RNA-ISH) as effective tools to diagnose feline herpesvirus-1-associated dermatitis. Vet Dermatol. 2019;30:491–e147. doi: 10.1111/vde.12787. [DOI] [PubMed] [Google Scholar]
- 14.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 15.Micke P, Ohshima M, Tahmasebpoor S, Ren ZP, Ostman A, Pontén F, Botling J. Biobanking of fresh frozen tissue: RNA is stable in nonfixed surgical specimens. Lab Invest. 2006;86:202–211. doi: 10.1038/labinvest.3700372. [DOI] [PubMed] [Google Scholar]
- 16.Rudloff U, Bhanot U, Gerald W, Klimstra DS, Jarnagin WR, Brennan MF, Allen PJ. Biobanking of human pancreas cancer tissue: Impact of ex-vivo procurement times on RNA quality. Ann Surg Oncol. 2010;17:2229–2236. doi: 10.1245/s10434-010-0959-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bao WG, Zhang X, Zhang JG, Zhou WJ, Bi TN, Wang JC, Yan WH, Lin A. Biobanking of fresh-frozen human colon tissues: Impact of tissue ex-vivo ischemia times and storage periods on RNA quality. Ann Surg Oncol. 2013;20:1737–1744. doi: 10.1245/s10434-012-2440-1. [DOI] [PubMed] [Google Scholar]
- 18.Lee SM, Schelcher C, Thasler R, Schiergens TS, Thasler WE. Pre-analytical determination of the effect of extended warm or cold ischaemia on RNA stability in the human ileum mucosa. PLoS One. 2015;10:e0138214. doi: 10.1371/journal.pone.0138214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun H, Sun R, Hao M, Wang Y, Zhang X, Liu Y, Cong X. Effect of duration of ex vivo ischemia time and storage period on RNA quality in biobanked human renal cell carcinoma tissue. Ann Surg Oncol. 2016;23:297–304. doi: 10.1245/s10434-014-4327-9. [DOI] [PubMed] [Google Scholar]
- 21.Zheng H, Tao YP, Chen FQ, Li HF, Zhang ZD, Zhou XX, Yang Y, Zhou WP. Temporary ischemia time before snap freezing is important for maintaining high-integrity RNA in hepatocellular carcinoma tissues. Biopreserv Biobank. 2019;17:425–432. doi: 10.1089/bio.2019.0003. [DOI] [PubMed] [Google Scholar]
- 22.Guerrera F, Tabbo F, Bessone L, Maletta F, Gaudiano M, Ercole E, Annaratone L, Todaro M, Boita M, Filosso PL, et al. The influence of tissue ischemia time on RNA integrity and patient-derived xenografts (PDX) engraftment rate in a non-small cell lung cancer (NSCLC) biobank. PLoS One. 2016;11:e0145100. doi: 10.1371/journal.pone.0145100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Castells Domingo X, Ferrer-Font L, Davila M, Candiota AP, Simões RV, Fernández-Coello A, Gabarrós A, Boluda S, Barceló A, Ariño J, Arús C. Improving ribosomal RNA integrity in surgically resected human brain tumor biopsies. Biopreserv Biobank. 2016;14:156–164. doi: 10.1089/bio.2015.0086. [DOI] [PubMed] [Google Scholar]
- 24.Gonzalez-Herrera L, Valenzuela A, Marchal JA, Lorente JA, Villanueva E. Studies on RNA integrity and gene expression in human myocardial tissue, pericardial fluid and blood, and its postmortem stability. Forensic Sci Int. 2013;232:218–228. doi: 10.1016/j.forsciint.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 25.Song SY, Jun J, Park M, Park SK, Choi W, Park K, Jang KT, Lee M. Biobanking of fresh-frozen cancer tissue: RNA is stable independent of tissue type with less than 1 hour of cold ischemia. Biopreserv Biobank. 2018;16:28–35. doi: 10.1089/bio.2017.0062. [DOI] [PubMed] [Google Scholar]
- 26.Wang Z, Wang Y, Ye J, Lu X, Cheng Y, Xiang L, Chen L, Feng W, Shi H, Yu X, et al. bFGF attenuates endoplasmic reticulum stress and mitochondrial injury on myocardial ischaemia/reperfusion via activation of PI3K/Akt/ERK1/2 pathway. J Cell Mol Med. 2015;19:595–607. doi: 10.1111/jcmm.12346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen CM, Wu CT, Yang TH, Chang YA, Sheu ML, Liu SH. Green tea catechin prevents hypoxia/reperfusion-evoked oxidative stress-regulated autophagy-activated apoptosis and cell death in microglial cells. J Agric Food Chem. 2016;64:4078–4085. doi: 10.1021/acs.jafc.6b01513. [DOI] [PubMed] [Google Scholar]
- 28.Mihaylova MM, Shaw RJ. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat Cell Biol. 2011;13:1016–1023. doi: 10.1038/ncb2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qi D, Young LH. AMPK: Energy sensor and survival mechanism in the ischemic heart. Trends Endocrinol Metab. 2015;26:422–429. doi: 10.1016/j.tem.2015.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hers I, Vincent EE, Tavare JM. Akt signalling in health and disease. Cell Signal. 2011;23:1515–1527. doi: 10.1016/j.cellsig.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 31.Manning BD, Cantley LC. AKT/PKB signaling: Navigating downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Avalos Y, Canales J, Bravo-Sagua R, Criollo A, Lavandero S, Quest AF. Tumor suppression and promotion by autophagy. Biomed Res Int. 2014;2014:603980. doi: 10.1155/2014/603980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iurlaro R, Munoz-Pinedo C. Cell death induced by endoplasmic reticulum stress. FEBS J. 2016;283:2640–2652. doi: 10.1111/febs.13598. [DOI] [PubMed] [Google Scholar]
- 34.Saggioro FP, Neder L, Stavale JN, Paixão-Becker ANP, Malheiros SMF, Soares FA, Pittella JEH, Matias CCMS, Colli BO, Carlotti CG, Jr, Franco M. Fas, FasL, and cleaved caspases 8 and 3 in glioblastomas: A tissue microarray-based study. Pathol Res Pract. 2014;210:267–273. doi: 10.1016/j.prp.2013.12.012. [DOI] [PubMed] [Google Scholar]
- 35.Schito L, Semenza GL. Hypoxia-inducible factors: Master regulators of cancer progression. Trends Cancer. 2016;2:758–770. doi: 10.1016/j.trecan.2016.10.016. [DOI] [PubMed] [Google Scholar]
- 36.LaGory EL, Giaccia AJ. The ever-expanding role of HIF in tumour and stromal biology. Nat Cell Biol. 2016;18:356–365. doi: 10.1038/ncb3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kurokawa M, Kornbluth S. Caspases and kinases in a death grip. Cell. 2009;138:838–854. doi: 10.1016/j.cell.2009.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lange N, Unger FT, Schoppler M, Pursche K, Juhl H, David KA. Identification and validation of a potential marker of tissue quality using gene expression analysis of human colorectal tissue. PLoS One. 2015;10:e0133987. doi: 10.1371/journal.pone.0133987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pietrocola F, Izzo V, Niso-Santano M, Vacchelli E, Galluzzi L, Maiuri MC, Kroemer G. Regulation of autophagy by stress-responsive transcription factors. Semin Cancer Biol. 2013;23:310–322. doi: 10.1016/j.semcancer.2013.05.008. [DOI] [PubMed] [Google Scholar]
- 40.Verzella D, Pescatore A, Capece D, Vecchiotti D, Ursini MV, Franzoso G, Alesse E, Zazzeroni F. Life, death, and autophagy in cancer: NF-κB turns up everywhere. Cell Death Dis. 2020;11:210. doi: 10.1038/s41419-020-2399-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karar J, Maity A. PI3K/AKT/mTOR pathway in angiogenesis. Front Mol Neurosci. 2011;4:51. doi: 10.3389/fnmol.2011.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nazarko VY, Zhong Q. ULK1 targets beclin-1 in autophagy. Nat Cell Biol. 2013;15:727–728. doi: 10.1038/ncb2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim SJ, Dix DJ, Thompson KE, Murrell RN, Schmid JE, Gallagher JE, Rockett JC. Effects of storage, RNA extraction, genechip type, and donor sex on gene expression profiling of human whole blood. Clin Chem. 2007;53:1038–1045. doi: 10.1373/clinchem.2006.078436. [DOI] [PubMed] [Google Scholar]
- 44.Becker N, Lockwood CM. Pre-analytical variables in miRNA analysis. Clin Biochem. 2013;46:861–868. doi: 10.1016/j.clinbiochem.2013.02.015. [DOI] [PubMed] [Google Scholar]
- 45.Willemse EA, Teunissen CE. Biobanking of cerebrospinal fluid for biomarker analysis in neurological diseases. Adv Exp Med Biol. 2015;864:79–93. doi: 10.1007/978-3-319-20579-3_7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during the current study are included in this published article.