Abstract
Tetralogy of Fallot (TOF) is the most common form of cyanotic congenital heart disease (CHD). Although a lower methylation level of whole genome has been demonstrated in TOF patients, little is known regarding the DNA methylation changes in specific gene and its associations with TOF development. NOTCH4 is a mediator of the Notch signalling pathway that plays an important role in normal cardiac development. However, the role of epigenetic regulation of the NOTCH4 gene in the pathogenesis of TOF remains unclear. Considering the NOTCH4 low mutation frequency and reduced expression in the TOF patients, we hypothesized that abnormal DNA methylation change of NOTCH4 gene may influence its expression and responsible for TOF development. In this study, we measured the promoter methylation status of NOTCH4 and was measured and its regulation mechanism was explored, which may be related to TOF disease. Additionally, the promoter methylation statuses of NOTCH4 was measured in order to further understand epigenetic mechanisms that may serve a role in the development of TOF. Immunohistochemical analysis was used to examine NOTCH4 expression in right ventricular outflow tract myocardial tissues in patients with TOF. Compared with healthy controls, patients with TOF displayed significantly reduced in NOTCH4 expression (P=0.0055). Moreover, bisulphite sequencing suggested that the methylation levels of CpG site 2 in the NOTCH4 promoter was significantly higher in the patients than in the controls (P=0.0459). NOTCH4 expression was negatively associated with CpG site 2 methylation levels (r=−0.51; P=0.01). ETS1 transcription factor can serve as transcriptional activators by binding to specific DNA sequences of target genes, such as DLL4 and NOTCH4, which serves an important role in normal heart development. Dual-luciferase reporter and electrophoretic mobility shift assays indicated that the ETS1 transcription factor could bind to the NOTCH4 promoter region. However, binding of ETS1 to the NOTCH4 promoter was abrogated by methylation at the putative ETS1 binding sites. These findings suggested that decreased NOTCH4 expression in patients with TOF may be associated with hypermethylation of CpG site 2 in the NOTCH4 promoter region, due to impaired binding of ETS1.
Keywords: NOTCH4, CpG methylation, ETS1 transcription factor, tetralogy of Fallot
Introduction
Congenital heart defects (CHDs), are the most common type of birth defect, affecting ~ eight in 1,000 livebirths (1,2). Among these congenital conditions, tetralogy of Fallot (TOF) is the most common cyanotic heart malformation and accounts for approximately 10% of all CHD cases (3). TOF results in anatomic defects, such as obstruction of the right ventricular outflow tract (RVOT), ventricular septal defect, aortic dextroposition and right ventricular hypertrophy (4). The pathogenesis of TOF is not completely understood. Previous studies have demonstrated that cardiac tissue-specific transcription factors, genes and associated signalling pathways, such as the Notch signalling pathway, play important roles in normal cardiac development, and that abnormal expression of such genes can contribute to TOF (5–7). In addition to DNA sequence variants, environmental factors, such as opioid exposure during early pregnancy and maternal pre-pregnancy obesity (8), may also be associated with the aetiology of TOF, as indicated by epidemiological data (9).
Notch is a highly conserved signalling pathway that regulates cell specification, differentiation, and organ formation and morphogenesis involved in development (10). Moreover, tissue-specific endocardial Notch signalling regulates cardiac morphogenesis through interactions with multiple myocardial, epicardial and neural crest-derived signals (11). Mutations in Notch signalling elements result in CHD in humans and mice, demonstrating its important role in normal cardiac development (12–14). NOTCH4 serves as a membrane-bound receptor that regulates cell fate (15). Notch1-deficient embryos display severe vascular developmental defects, which are exacerbated in Notch1/Notch4 double-mutant embryos (16). Constitutive activation of Notch4 in the embryonic vasculature also leads to defects in vascular remodelling (17,18). In addition, a previous study have demonstrated that Notch4 activation in endothelial cells causes trans-differentiation to a mesenchymal phenotype, suggesting that implicates Jagged1-Notch interactions promote epithelial-mesenchymal transition, which is required for normal endocardial cushion differentiation and vascular smooth muscle cell development (19). Furthermore, a previous study has demonstrated that the NOTCH4 gene was downregulated and had low frequency of genetic variants in the NOTCH4 coding region in patients with TOF (20).
DNA methylation is an epigenetic modification that is vital for embryonic development, and changes in methylation may be associated with the development of cardiovascular diseases (21). The interactions between DNA and transcription factors could be influenced directly or indirectly by DNA methylation, due to recruitment of methyl-CpG-binding proteins (22). Sheng et al (23,24) suggested that aberrant methylation levels at the promoter CpG island shore of the ZFPM2 and HAND1 genes may be responsible for gene transcription regulation in patients with TOF. Gong et al (25) demonstrated that demethylation in the TBX20 promoter region may be associated with overexpression in the cardiac tissue of patients with TOF. However, a recent study on severe anxiety has reported that a single CpG site located in the promoter of the Asb1 gene may be responsible for a methylation increase of 48.5% (26). Methylation at specific loci in genes, such as tissue factor F3, interleukin-6 and toll-like receptor 2, is influenced by exposure to air pollution, thus leading to several adverse health effects (27–29). Therefore it may be hypothesised that changes in DNA methylation contribute to downregulation of NOTCH4 in patients with TOF.
NOTCH4 has been demonstrated to be a ETS factor- regulated gene, and ETS may activate the expression of Notch signalling components to initiate Notch signalling in the early artery (30). The ETS1 oncogene belongs to a large family within the ETS domain family of transcription factors (31). It is widely expressed in the developing embryo, and could be detectable in the day-15 embryos of murine (32). Previous studies have demonstrated that ETS1 plays an important role in human heart development and was associated with the development of CHD (33,34). ETS proteins can function as transcriptional activators by binding to specific DNA sequences of target genes, thereby increasing (such as DLL4, NOTCH4 and NKp46) or decreasing (such as MMP1 and BCL2) gene transcription in response to various stimuli, including changes of genetics, histone modification or DNA methylation (30,35–37).
The aim of the present study was to examine the epigenetic mechanisms that regulate the NOTCH4 gene and their effect on NOTCH4 protein expression in patients with TOF. The findings of this study may provide insight into the aetiology of TOF.
Materials and methods
Clinical samples
The present study was approved by The Ethics Committee of The Children's Hospital of Fudan University (approval no. 2015) (26). Written informed consent was obtained from the parents or relatives of all study participants. Patients with TOF were recruited from the Children's Hospital of Fudan University between January 2016 to July 2018. TOF was diagnosed using an echocardiogram and confirmed by surgery. The control samples from autopsy specimens were provided by the Department of Forensic Medicine of Soochow University. In total, 24 patients with TOF were enrolled, including 14 (58.3%) males and 10 (41.7%) females. Patient age ranged from 1 month to14 years (mean ± SD, 2.54 ± 0.86 years). Control samples were obtained from five male subjects, aged 1 day to 7 months (mean ± SD, 0.35 ± 0.19 years). All samples were taken from RVOT myocardial tissue and immediately stored at −80°C in RNAlater solution (Ambion; Thermo Fisher Scientific, Inc.). Patient characteristics are summarized in Table I.
Table I.
Clinical characteristics of patients with TOF and healthy controls.
| Clinical variable | Patients with TOF | Controls | P-valuea |
|---|---|---|---|
| Age, years, mean ± SD | 2.54±0.86 | 0.35±0.19 | |
| Age category, years, n (%) | |||
| <1 | 15 (62.5) | 4 (80.0) | 0.796a |
| 1-2 | 4 (16.7) | 1 (20.0) | |
| >2 | 5 (20.8) | 0 (0) | |
| Sex, n (%) | |||
| Male | 14 (58.3) | 5 (100) | 0.134a |
| Female | 10 (41.7) | 0 (0) |
P-value for patients with TOF (n=24) vs. healthy controls (n=5); Fisher's exact test. TOF, tetralogy of Fallot.
Immunohistochemistry
RVOT myocardial tissues were fixed in 10% neutral buffered formalin at room temperature for 48 h and embedded in paraffin. The sections were then cut into 4-µm sections and dried overnight at 56°C. After deparaffination, hydration and antigen retrieval with citric acid buffer (0.01 mol/l; pH 6.0), endogenous peroxidase activity was blocked by incubating the sections with 3% hydrogen peroxide (H2O2) at room temperature for 20 min and blocking with 5% bovine serum (cat. no. 143183; Bio Forxx) for 1 h at room temperature. The slides were incubated with a rabbit anti-human monoclonal antibody against NOTCH4 (dilution 1:200, cat. no. ab184742; Abcam) overnight at 4°C. After primary antibody incubation, the slides were incubated with goat anti-rabbit/mouse IgG (cat. no. GK500710; Gene Tech Co., Ltd.) for 30 min at room temperature, then washed in TBST three times for 5 min each time. Finally, the slides were stained with 3,3-diaminobenzidine (DAB) for 40 sec and counterstained with haematoxylin for 1 min. After dehydration, the sections were mounted with mounting medium. For each sample, three visual fields were randomly chosen and examined under a light microscope at ×200 magnification. Images of cardiomyocytes were quantified using ImageJ (version 1.48; National Institutes of Health). To quantify the intensity of NOTCH4 protein expression, images were captured with a light microscope. Three randomly selected fields (×200 magnification) per tissue section were scanned and analyzed using ImageJ software (version 1.48). The integrated optical density (IOD) sum of each image was measured after the optical density was adjusted with the segmentation set at a level to allow for detection of positive immunostaining. For statistical analysis, the mean value of the total three counted fields was calculated.
DNA extraction and bisulphite sequencing PCR (BSP)
Genomic DNA was extracted from RVOT myocardial tissue samples from patients with TOF and control subjects using a QIAamp DNA Mini kit (Qiagen GmbH) according to the manufacturer's instructions. For each sample, bisulphite treatment of genomic DNA from RVOT myocardial tissues was performed using the EZ DNA Methylation-Gold kit (Zymo Research Corp.) according to the manufacturer's protocol. The bisulphite-treated DNA samples were then used as templates for BSP. The primers used to amplify the NOTCH4_R region (from position −240 to +113 bp, containing five CpG sites) were designed using Methyl Primer Express v1.0 software (Applied Biosystem; Thermo Fisher Scientific, Inc.). Primer sequences were as follows: NOTCH4-BSP-F, 5′-AATAGTAGGGTTGGGATTGTTTAGG-3′ and NOTCH4-BSP-R, 5′-ACAAAAACCACCTCCTCTACTCC-3′. For sequencing, the PCR products were purified using the AxyPrep DNA Gel Extraction kit (Axygen; Corning, Inc.) according to the manufacturer's protocol. The purified PCR products were then cloned into the pGEM-T easy vector system (Promega Corporation) and transformed into DH5α competent cells (Tiangen Biotech Co., Ltd.). After a 12-h incubation at 37°C, blue/white and ampicillin screening was carried out. For each p-GEM-T vector, 10 clones were determined the methylation status using Sanger sequencing The BIQ Analyser software (https://biq-analyzer.bioinf.mpi-inf.mpg.de/index.php) was used to analyse the sequencing results. The percent methylation of each CpG site in the samples was calculated as the number of methylated CpG sites relative to the total number of observed sequenced clones, which represents the overall methylation status of a specific region in a screened sample.
Reporter plasmid construction and in vitro methylation
pGL3-basic lacks eukaryotic promoter and enhancer sequences, and serves as a negative control. The region [position −179 to −53 bp, relative to the transcription start site (TSS)] of the human NOTCH4 gene was amplified with Premix PrimeSTAR HS DNA polymerase (cat. no. R040A; Takara Bio, Inc.), using human genomic DNA isolated from tissues of normal controls as a template. The PCR primers were synthesized by Generay Biotech Co., Ltd., as follows: NOTCH4-KpnI-F, 5′-ATAGGTACCAGATTCCTTCTCCCCTCCTA-3′ and NOTCH4-XhoI-R, 5′-GATCTCGAGGAGGAAGAGTGGAGGAACAC-3′. The amplified products were digested with KpnI–XhoI (cat. no. R3142V and R0146S; New England BioLabs, Inc.) and then inserted into the pGL3-promoter vector (Promega Corporation). The products were transformed into DH5α-competent cells (Tiangen Biotech Co., Ltd.), smeared on a lysogeny broth (LB) ampicillin (AMP) plate and cultured at 37°C overnight. An LB culture containing AMP was used to screen for the objective colonies. The fidelity of the plasmids was verified by Sanger sequencing. This construct is referred to as pGL3-NOTCH4_-179/-53 plasmid thereafter.
In addition, The Mut-pGL3-NOTCH4_-179/-53 plasmid was constructed by mutating a C base in the CpG site 2 to a T base to simulate its hypermethylation status which might abrogate binding of the target to NOTCH4_R. The Mut-pGL3-NOTCH4_-179/-53 plasmid was constructed using the KOD-plus-mutagenesis kit (cat. no. SMK-101; Toyobo Life Science) according to the manufacturer's protocol. The primers used for mutagenesis were synthesized by Generay Biotech Co., Ltd., as follows: Mut-NOTCH4-F, 5′-TGTCCTACTTCCCCCTACTTCCCCA-3′ and Mut-NOTCH4-R, 5′-GTGTGCCTGGAGGGCAGGTGATAGG-3′.
Lastly, the Me-pGL3-NOTCH4_-179/-53 plasmid was generated by incubating the pGL3-NOTCH4_-179/-53 plasmid with the M.SssI CpG methyltransferase (cat. no. M0226V; New England BioLabs) for 4 h at 37°C. The methylation status was confirmed by BSP, as aforementioned.
Transfections and dual luciferase reporter assays
HeLa and HL-1 cells were purchased from the Cell Bank of the Type Culture Collection of the Chinese Academy of Sciences, which were cultured in Dulbecco's modified Eagle's medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% foetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.) at 37°C with 5% CO2. For transfection, the cells were plated in 96-well plates 12 h before transfection at a density of 1–4×105 cells/well, then separately transfected with 100 ng of pGL3-basic (negative control), pGL3-promoter (empty vector), pGL3-NOTCH4_-179/-53 or Me-pGL3-NOTCH4_-179/-53 vector using Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc.).
In order to study the mechanism underlying the effect of the NOTCH4_R region on gene transcription activity, the NOTCH4_R region sequence was analysed by a TF search (http://www.cbrc.jp/research/db/TFSEARCH.html) and the JASPAR database (http://jaspar.binf.ku.dk/cgi-bin/jaspar_db.pl?-rm=browse&db=core&tax_group=vertebrates), and the results demonstrated the potential ETS1 transcription factor binding sites, and the CpG site 2 was within the ETS1 binding site. For dual luciferase reporter assays, 100 ng of pGL3-NOTCH4_-179/-53 (unmethylated), Mut-pGL3-NOTCH4_-179/-53 (mutated) and Me-pGL3-NOTCH4_-179/-53 (methylated) were co-transfected with 100 ng of ETS1 transcription factor expression vector which was constructed by cloning the entire human ETS1 cDNA (accession no. NM_001143820) into a pcDNA3.1 (+) expression vector (cat. no. G105592; YouBio). The pGL3-basic and pGL3-promoter vectors were also used as controls. The pRL-TK plasmid (Promega Corporation) was co-transfected with these plasmids to normalize luciferase activity 48 h following transient transfection. Three independent experiments were performed.
Electrophoretic mobility shift assay (EMSA) and shift western blotting
293T cells originally purchased from the Cell Bank of the Type Culture Collection of the Chinese Academy of Sciences. The cells were plated in 6-well plates 12 h before transfection at a density of 1–2×106 cells/well. The cells were transfected with 2.5 µg pcDNA3.1-ETS1 expression plasmid, and the nuclear extracts were obtained using cytoplasmic extraction reagent and nuclear extraction reagent (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. The protein concentration was measured using a bicinchoninic acid protein assay kit (Takara Biotechnology Co., Ltd.) according to the manufacturer's protocol. Subsequently, ETS1 overexpression was confirmed by western blotting. Protein samples (~10 µg) were separated by 10% SDS-PAGE gels, transferred to a nitrocellulose membrane (Whatman plc; GE Healthcare Life Sciences), and blocked with phosphate-buffered saline (PBS) with 1% Tween-20 (PBST) containing 5% BSA for 2 h at room temperature. Next, the membrane was probed with primary antibodies against ETS1 (1:1,000; cat. no. ab220361; Abcam) and GAPDH (1:3,000; cat. no. ab9482; Abcam) at 4°C overnight, then incubated with horseradish peroxidase (HRP)-conjugated anti-rabbit secondary antibody (1:3,000; cat. no. M21002; Abmart) and HRP-conjugated anti-mouse secondary antibody (1:3,000; cat. no. M21001; Abmart). The blots were detected by enhanced chemiluminescence (Thermo Fisher Scientific, Inc.), and were visualized using GE ImageQuant LAS4000 mini (GE Healthcare Life Sciences). GAPDH was used as a loading control.
Biotin-labelled oligonucleotide probes (Biotin-probe) specific for the ETS1-binding site of the NOTCH4 gene (5′-CGTCCT-3′; position-138 to −133) were synthesized by Generay Biotech Co., Ltd. and the oligonucleotide probes was annealed into double strands by heating it to 95°C, then cooling to room temperature. The mutant biotin-labelled oligonucleotide probes (Biotin-Mut-probe) were used to confirm the ETS1 binding specificity. An unlabelled oligonucleotide probe (Competitor-WT), a mutant unlabelled oligonucleotide probe (Competitor-Mut) and an methylated oligonucleotide probe (Competitor-Met) were used as competitors. The sequences of these oligonucleotide probes are listed in Table II.
Table II.
Sequences of oligonucleotide probes used for electrophoretic mobility shift assay.
| Probe | Sequence (5′-3′) |
|---|---|
| Biotin-probe-F | GCCCTCCAGGCACACCGTCCTACTTCCC |
| Biotin-probe-R | GGGAAGTAGGACGGTGTGCCTGGAGGGC |
| Biotin-Mut-probe-F | GCCCTCCAGGCACACATAACACTAGCCC |
| Biotin-Mut-probe-R | GGGCTAGTGTTATGTGTGCCTGGAGGGC |
| Competitor-WT-F | GCCCTCCAGGCACACCGTCCTACTTCCC |
| Competitor-WT-R | GGGAAGTAGGACGGTGTGCCTGGAGGGC |
| Competitor-Mut-F | GCCCTCCAGGCACACATAACACTAGCCC |
| Competitor-Mut-R | GGGCTAGTGTTATGTGTGCCTGGAGGGC |
| Competitor-Met-F | GCCCTCCAGGCACACCGTCCTACTTCCC |
| Competitor-Met-R | GGGAAGTAGGACGGTGTGCCTGGAGGGC |
Mut, mutant; me; methylated; F, forward; R, reverse.
The DNA-binding ability of ETS1 to the NOTCH4 gene was detected by EMSA using a LightShift™ EMSA kit (cat. no. 20148; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. Specifically, 10 µg nuclear extract were incubated with 20 fmol biotin-labelled probes in binding buffer at room temperature for 30 min, and a 200-fold excess of unlabelled/methylated probes was added to the reaction as competitors. The protein-DNA complexes were separated from the free probes in a 6% polyacrylamide gel at 100 V for 50 min, then transferred onto a nylon membrane at 380 mA for 30 min. Subsequently, the membrane was analysed using a Fujifilm Las3000 Luminescent Image Analyzer (FUJIFILM Wako Pure Chemical Corporation). Shift-western blotting was performed by transferring the protein-DNA complexes from the polyacrylamide gel to a PVDF membrane in 0.5X Tris-borate-EDTA (TBE) buffer for 30 min. For protein detection, the membrane was blocked with blocking buffer (PBS with 1% Tween-20 and 5% BSA) for 2 h at room temperature, then incubated with a primary antibody against ETS1 (dilution 1:1,000; cat. no. ab220361; Abcam) at 4°C overnight. Following primary antibody incubation, the membrane was incubated with a HRP-conjugated anti-rabbit secondary antibody (dilution 1:3,000; cat. no. M21002; Abmart) at room temperature for 2 h. The blots were visualized using a enhanced chemiluminescence (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol, and were visualized using GE ImageQuant LAS4000 mini (GE Healthcare Life Sciences).
Statistical analysis
All data are presented as the mean ± SD of three independent experiments. Statistical analysis was performed using SPSS software v20.0 (IBM Corp.). Differences between two groups were analysed by the Mann-Whitney test. Differences of luciferase activity assays between multiple groups were analysed using one-way ANOVA, followed by the Least Significant Difference post hoc test. Pearson's correlation analysis was performed to analyse the relationship between the immunohistochemistry data and bisulphite sequencing data. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of the NOTCH4 protein in patients with TOF and controls
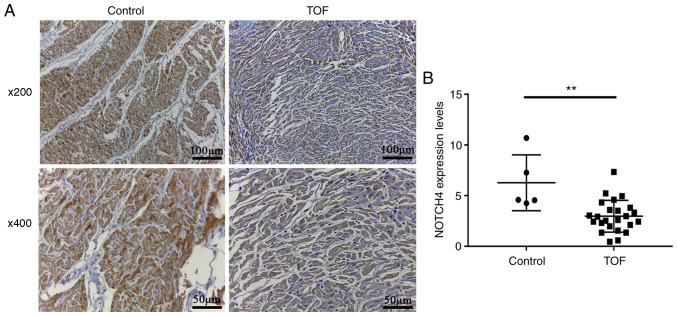
Immunohistochemistry was carried out to detect NOTCH4 protein expression in RVOT myocardial tissue from 24 patients with TOF and five controls. NOTCH4 protein was detectable by yellow or brown staining in the nucleus or cytoplasm. The staining intensity of cardiomyocytes from patients with TOF was weaker than that in the control subjects (Fig. 1A). Statistical analysis confirmed that NOTCH4 expression was significantly lower in the patients, compared with the controls (3.0 ± 0.3 vs. 6.3 ± 1.2; P=0.0055; Fig. 1B).
Figure 1.
NOTCH4 expression in myocardial tissue. (A) Immunohistochemical staining for NOTCH4 expression in ROVT myocardial tissue from patients with TOF (n=24) and healthy controls (n=5). NOTCH4 was expressed in both the nucleus and cytoplasm. The staining intensity of NOTCH4 was relatively weaker in patients with TOF. (B) Quantification of immunohistochemical staining. **P<0.01; Mann-Whitney test. TOF, tetralogy of Fallot; ROVT, right ventricular outflow tract.
NOTCH4 promoter methylation status is associated with protein expression
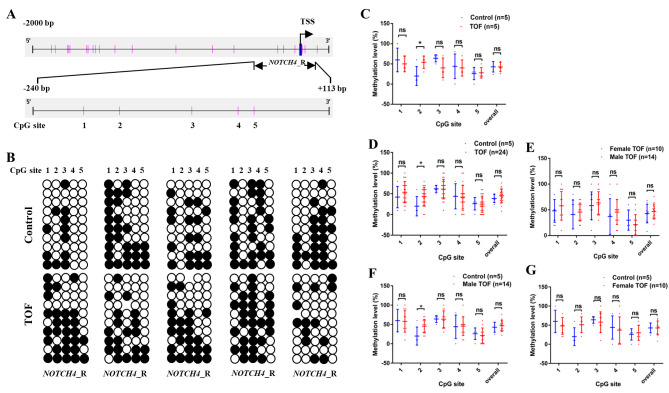
BSP was carried out in order to determine whether reduced NOTCH4 expression was caused by changes in its epigenetic regulation, BSP was performed on the NOTCH4 promoter using tissue samples collected from patients with TOF and controls. Considering that there is no CpG island in the promoter region (−2,000 to 200 bp), only the region that was targeted for sequencing (NOTCH4_R: −240 to +113 bp, containing 5 CpG sites; Fig. 2A) was studied.
Figure 2.
Methylation status of the NOTCH4 promoter region and its association with NOTCH4 protein expression. (A) Schematic representation of NOTCH4_R (−240 to +113 bp, relative to TSS) in the NOTCH4 promoter region (−2,000 to +200 bp, relative to TSS). CpG sites 1–5 within NOTCH4_R are displayed as vertical pink bars. (B) Bisulphite sequencing results for NOTCH4_R methylation in right ventricular outflow tract myocardial tissues from patients with TOF and healthy controls. White and black circles represent unmethylated and methylated CpG sites, respectively. n=5 in each group. (C) Initial screen: Methylation levels in patients with TOF (n=5) and healthy controls (n=5). (D) Second screen: Methylation levels patients with TOF (n=24) and healthy controls (n=5), (E) male (n=14) and female patients with TOF (n=10), and (F) male patients with TOF (n=14) and healthy controls (n=5). (G) Female patients with TOF (n=10) and healthy controls (n=5). Overall NOTCH4_R methylation levels are displayed, together with methylation levels at each CpG site. Data are presented as the mean ± SD. *P<0.05; Mann-Whitney test. TOF, tetralogy of Fallot; TSS, transcription start site.
In an initial screen, the methylation levels in NOTCH4_R were analysed in five patients with TOF and five control subjects (Fig. 2B). The overall methylation levels of NOTCH4_R (CpG sites 1–5) in patients with TOF did not significantly differ from the controls (42.4 ± 5.1 vs. 42.8 ± 5.8; Fig. 2C). Interestingly, only one CpG site (CpG site 2) in NOTCH4_R exhibited significantly higher methylation levels, compared with the controls (54.0 ± 6.8 vs. 20.0 ± 10.5; P=0.0476).
Following this initial screen, the methylation status of NOTCH4_R was determined in another 19 TOF patients and the data for the combined cohort was analysed (n=24; Fig. 2D). The methylation levels of CpG site 2 were significantly higher in 24 patients with TOF, compared with the controls (43.8 ± 4.5 vs. 20.0 ± 10.5; P=0.0459; Fig. 2D).
In addition, the methylation levels of all five CpG sites, as well as overall NOTCH4_R promoter methylation, were compared between male (n=14) and female (n=10) patients with TOF. None of the five CpG sites significantly differed between male and female patients with respect to methylation levels (Fig. 2E). Furthermore, the methylation levels of the five CpG sites were also compared between male patients with and normal controls. Consistent with the aforementioned results, only CpG site 2 in NOTCH4_R displayed significantly higher methylation levels in male patients with TOF, compared with controls (45.7 ± 4.7 vs. 20.0 ± 10.5; P=0.0263; Fig. 2F). In addition, the methylation levels of the five CpG sites were also compared between female patients with and normal controls. No significant difference was observed between female and normal controls (male) with respect to methylation levels (45.0 ± 9.2 vs. 20.0 ± 10.5; P=0.1758; Fig. 2G). After excluding two extreme values, CpG site 2 in NOTCH4_R showed higher methylation levels in female patients with TOF, compared with controls (56.2 ± 6.8 vs. 20.0 ± 10.5; P=0.0111). Thus, it can be concluded that no sex differences exist in methylation levels in the TOF group and the normal control group.
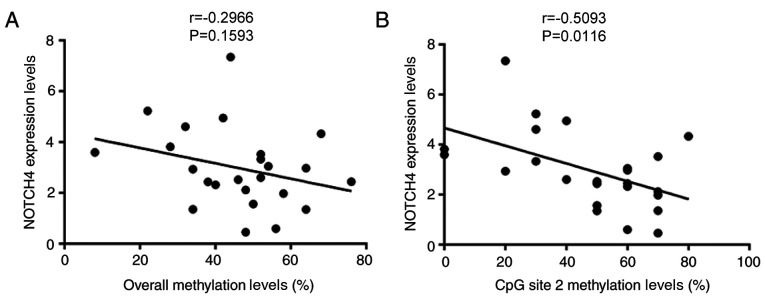
Pearson's correlation was carried out to determine whether NOTCH4_R methylation status was associated with NOTCH4 protein expression. There was no correlation between NOTCH4 protein expression and the overall methylation levels of NOTCH4_R (r=−0.2966; P=0.1593; n=24; Fig. 3A) in patients with TOF. However, a significant negative association was observed between NOTCH4 expression and the methylation status of the CpG site 2 (r=−0.5063; P=0.0116; n=24; Fig. 3B) in patients with TOF.
Figure 3.
Correlation analysis of NOTCH4 protein expression and DNA methylation levels. (A) Association between the overall methylation levels of NOTCH4_R (CpG sites 1–5) with NOTCH4 protein expression in patients with TOF. (B) Association between NOTCH4 CpG site 2 methylation levels with NOTCH4 protein expression in patients with TOF. n=24. TOF, tetralogy of Fallot.
Effect of NOTCH4 CpG site 2 methylation on gene transcription in vitro
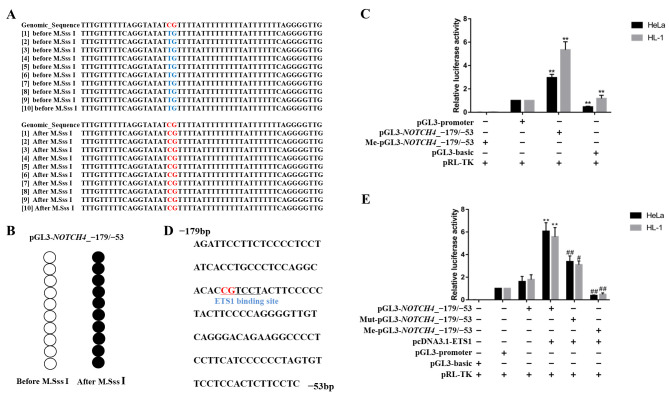
A dual-luciferase assay was carried out in HeLa and HL-1 cells to determine whether single CpG site 2 methylation could influence NOTCH4 gene transcription activity. The pGL3-NOTCH4_-179/-53 plasmid was constructed by cloning the NOTCH4 promoter region (−179 to −53 bp, containing only the CpG site 2) into the pGL3-promoter vector. In addition, a methylated plasmid was generated using M.SssI treatment. BSP was used to determine the methylation status of the unmethylated and methylated plasmids (Fig. 4A, B). As indicated in Fig. 4C, the luciferase activity of pGL3-NOTCH4_-179/-53 was significantly higher than that of pGL3-promoter (HeLa: P=0.0022; HL-1: P=0.0022). Moreover, the luciferase activity of me-pGL3-NOTCH4_-179/-53 decreased nearly six-fold, compared with pGL3-NOTCH4_-179/-53. These results indicated that methylation in the NOTCH4_R region regulated gene transcription and that a methylation of a single CpG site could inhibit transcription of the NOTCH4 gene.
Figure 4.
Effect of NOTCH4_R CpG site 2 methylation on gene transcription activity. (A and B) Bisulphite sequencing results for pGL3-NOTCH4_-179/-53. After M.SssI treatment, all CpG sites were methylated. (C) Luciferase assay for pGL3-basic, pGL3-promoter, pGL3-NOTCH4_-179/-53 and Me-pGL3-NOTCH4_-179/-53 in HeLa and HL-1 cell lines. (D) Sequence of the ETS1 transcription factor binding sites predicted in the NOTCH4 promoter (−179 to −53 bp, relative to TSS). The underlined sequence represents the ETS1 binding sites, containing only CpG site 2. (E) Luciferase assay for pGL3-basic, pGL3-promoter, pGL3-NOTCH4_-179/-53, Me-pGL3-NOTCH4_-179/-53 and Mut-pGL3-NOTCH4_-179/-53 co-transfected with pcDNA3.1-ETS1 in HeLa and HL-1 cell lines. Data are presented as the mean ± SD. **P<0.01 vs. pGL3-promoter; #P<0.05 and ##P<0.01 vs. pGL3 NOTCH4_-179/-53. One-way ANOVA and Least Significant Difference test. Me, methylated, Mut, mutant; TSS, transcription start site.
NOTCH4 expression is regulated by the ETS1 transcription factor binding to its promoter region
To clarify the mechanism through which NOTCH4_R methylation affects gene transcription, the NOTCH4_R sequence was analysed for potential binding sites for the ETS1 transcription factor using an online TF search and the JASPAR database (38,39) (Fig. 4D). To evaluate the effect of ETS1 on NOTCH4 transcriptional activity, we co-transfected ETS1-overexpressing plasmids with pGL3-NOTCH4_-179/-53, Mut-pGL3-NOTCH4_-179/-53 and Me-pGL3-NOTCH4_-179-/53, respectively, into HeLa and HL-1 cells (Fig. 4E). Following co-transfection with pGL3-NOTCH4_-179/-53, luciferase activity was significantly increased, compared with pGL3-basic. In addition, a significant increase in luciferase activity following co-transfection with pGL3-NOTCH4_-179/-53 and ETS1 transcription factor was observed. However, the luciferase gene activity driven by Mut-pGL3-NOTCH4_-179/-53 was significantly reduced in the presence of ETS1, compared with pGL3-NOTCH4_-179/-53 (HeLa: P=0.0022; HL-1: P=0.0159). Moreover, me-pGL3-NOTCH4_-179/-53 resulted in significantly reduced luciferase activity when co-transfected with the ETS1-overexpression plasmid (HeLa: P=0.0022; HL-1: P=0.0095).
Altogether, these findings demonstrated that the ETS1 transcription factor could bind to the NOTCH4 promoter region and promote gene expression. This interaction was inhibited by methylation changes in the ETS1 binding site.
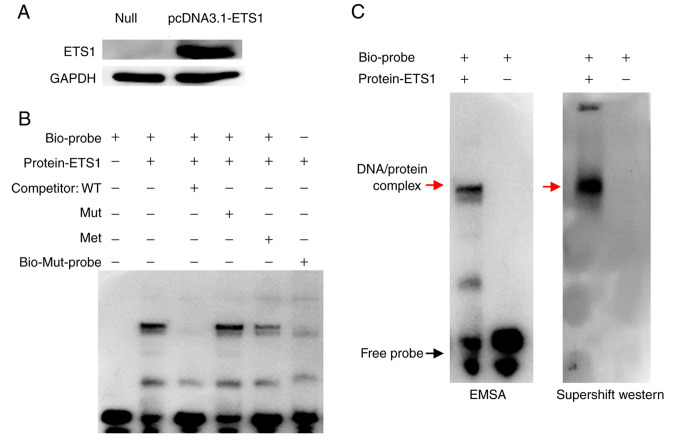
Impact of single CpG site 2 methylation on ETS1 binding affinity
The ETS1 transcription factor was overexpressed in the 293T cell line (Fig. 5A), and an EMSA was carried out to further confirm the effect of single CpG site 2 methylation on ETS1 binding affinity to the NOTCH4 promoter region. The biotinylated probe (Biotin-probe) could bind to the ETS1 transcription factor in the nuclear protein extract from transfected cells, forming a visible DNA/protein complex (Fig. 5B, lane 2). However, when the unlabelled probe (Competitor-WT) was added as a competitor, the band corresponding to the DNA/protein complex was not observed (Fig. 5B, lane 3). However, with the mutant unlabelled competition probe (Competitor-Mut), the binding of ETS1 to the biotinylated probe was not affected (Fig. 5B, lane 4). Moreover, with the addition of an unlabelled methylated probe (Competitor-Met), the band of the DNA/protein complex in lane 5 was lighter than that in lane 2, but heavier than that in lane 3 (Fig. 5B, lane 5). In addition, the complex was not detected when the mutated biotinylated probe (Biotin-Mut-probe) was added (Fig. 5B, lane 6). A super-shift western blot was conducted to confirm that the DNA/protein banding was caused by the presence of ETS1 protein. The specific DNA/protein complexes are indicated by red arrows. In addition, the results also demonstrated that the biotinylated probe could bind to the ETS1 transcription factor, and the DNA/protein complex was induced by ETS1 antibody, indicating that the complex consists of ETS1 (Fig. 5C).
Figure 5.
Effect of NOTCH4_R CpG site 2 methylation on ETS1 binding affinity. (A) ETS1 overexpression in 293T cell lines. Null, cells transfected with a pcDNA3.1 empty vector; pcDNA3.1-ETS1, cells transfected with an pcDNA3.1-ETS1 vector. (B) EMSA was performed to evaluate the effect of CpG site 2 methylation on binding of the ETS1 transcription factor to the NOTCH4_R region. (C) EMSA and super-shift western blot analysis confirmed ETS1 protein binding to the NOTCH4_R region. Specific DNA/protein complexes are indicated by a red arrow. Met, methylated, Mut, mutant; EMSA, Electrophoretic mobility shift assay.
Collectively, the present findings indicated that the ETS1 transcription factor could directly bind to the NOTCH4_R region and was affected by methylation of the CpG 2 site.
Discussion
The Notch signalling pathway is evolutionarily conserved and plays a critical role in the growth and development of diverse organisms, including in normal cardiac development (16–18). In vertebrates, Notch4 is distributed in the aorta, the endocardium and the endothelial cells of arteries, including pulmonary and cardiac vessels (40,41). In the present study, immunohistochemical staining demonstrated that NOTCH4 expression in RVOT myocardial tissues was significantly decreased in patients with TOF, compared with controls, suggesting that the developmental defects leading to TOF are associated with distinct changes in NOTCH4 expression. Considering the low frequency of genetic variants in the NOTCH4 coding region in patients with TOF (20), it was hypothesized that epigenetic changes might be at play in the abnormal expression of this gene.
Several studies have demonstrated that both genetic and epigenetic mechanisms control the expression of cardiac genes in a spatiotemporal manner during cardiac development (42,43). DNA methylation is one of the major epigenetic mechanisms controlling gene expression, which can increase or decrease the levels of gene transcription based on the methylation status of the target gene (44,45). Abnormal methylation can change the normal expression of genes and lead to different diseases. Abnormal DNA methylation has been associated with the pathogenesis of tetralogy of Fallot and can exacerbate defects, such as RVOT and ventricular septal defect, leading to pathogenic cardiac remodelling (46,47). Our previous study demonstrated that altered expression of NK2 homeobox 5, heart and neural crest derivatives expressed 1 and long interspersed nuclear element-1 may be associated with epigenetic regulation and involved in the development of TOF (22,48). Moreover, a single CpG site-based methylation change was also demonstrated to be responsible for changes in gene expression in different diseases (49,50). In the current study, the methylation status of the NOTCH4 promoter region and its association with gene expression was assessed. Considering that there is no CpG island in the NOTCH4 promoter, an important regulatory region near the TSS containing five CpG sites was selected for methylation analysis. Overall, no significant difference was observed in the overall methylation level between the patients with TOF and controls. However, when the individual methylation status of each CpG site was analysed separately, CpG site 2 exhibited significantly higher methylation levels in the cardiac tissue of patients with TOF, compared with the controls. Interestingly, in the patients, a significant negative correlation was also observed between NOTCH4 expression and the methylation levels at the CpG site 2. These findings indicated that CpG site 2 methylation changes may affect NOTCH4 expression.
To determine the effect of CpG site 2 methylation changes in the NOTCH4 promoter on gene expression and the underlying molecular mechanism, a dual-luciferase assay combined with an in vitro methylation assay was carried out. Following in vitro methylation, decreased transcriptional activity of pGL3-NOTCH4_-179/-53 was observed, suggesting that increased methylation of the CpG site 2 had a negative impact on transcriptional regulatory activity. Several studies have suggested that CpG site-specific regulation of DNA methylation could be mediated by transcription factors binding to specific gene promoter regions (51,52). Therefore, using online prediction software was used to identify a potential transcriptional factor that could represent a target for the NOTCH4 promoter region. A potential binding site for the ETS1 transcription factor was identified in in the NOTCH4 promoter that contained CpG site 2. The ETS1 transcription factor plays an important role in normal cardiac development (32), and can regulate transcriptional activity of target genes (such as DLL4, NOTCH4 and MMP1) by binding to specific sites (30,34–36).
In the present study, in vitro experiments demonstrated that NOTCH4 expression in patients with TOF was regulated by binding of ETS1 to NOTCH4_R. The present findings suggested that, although ETS1 could bind to the NOTCH4 promoter region and promote gene expression, binding affinity could be influenced when a single CpG site 2 was methylated, which might lead to a marked reduction of gene expression. However, CpG site 2 hypermethylation could result in reduced gene expression in vivo was not evaluated in this study and remains unclear. Thus, the direct function of single CpG site 2 methylation and its role in the regulation of NOTCH4 expression require further study.
Previous studies have demonstrated that DNA methylation occurs not only on CpG islands, but also on CpG island shores. Abnormal methylation in these regions may affect gene expression by altering the chromosomal structure (53,54). However, there is increasing evidence for the importance of the methylation status of individual CpG sites in the regulation of gene expression. For example, the methylation levels of a single CpG site was inversely correlated with oestrogen receptor α positivity in breast cancer specimens in a previous study (49). CpG site-specific methylation was demonstrated to alter the binding affinities of specific transcription factors that can activate or repress transcription (55,56). In the present study, CpG site 2 was differentially methylated in patients with TOF and controls, and the influence of sex and age was excluded through statistical analysis. In addition, abnormal methylation of CpG site 2 affected the binding affinity of the ETS1 transcription factor to the NOTCH4 gene and downregulate NOTCH4 expression. Furthermore, the causes of TOF development are complex, and involve many other factors, such as environmental, genetic and maternal factors such as age of the mother, radiation and drugs used by mother. Therefore, other potential interfering factors in the development of TOF cannot be excluded and should be evaluated in future studies.
A limitation of this study was the restricted sample size, due to difficulties in obtaining sufficient matched cardiac tissue from patients with TOF and controls. Further studies with a larger number of samples are required to confirm the present findings. In addition, it is uncertain whether the methylation patterns observed in the samples were effects or causes of TOF, since the onset of TOF preceded the measurement of methylation. The exact mechanism underlying NOTCH4 promoter methylation in the onset of TOF should also be explored in a large and prospective cohort, as well as animal models.
In conclusion, the present study suggested that single CpG site 2 in the NOTCH4 promoter region was hypermethylated in RVOT myocardial tissue from patients with TOF, which may lead to decreased NOTCH4 expression. Specifically, NOTCH4 expression may be regulated by an epigenetic mechanism, in which single CpG site methylation at the binding site of the ETS1 transcription factor in NOTCH4_R decreases ETS1 binding affinity and downregulates NOTCH4 expression. This, in turn, could contribute to the development of TOF.
Acknowledgements
Not applicable.
Funding
The current study was supported by The National Key Research and Development Program of China (grant no. 2016YFC1000500), The National Natural Science Foundation of China (grant nos. 81570282, 81570283, 81873482 and 81873483) and The Youth Program of National Natural Science Foundation of China (grant no. 81800282).
Availability of the data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
WS and GH made major contributions to the conception and design of this study. MY and HX collected samples and communicated with the patients' families. YZ performed the experiments and wrote the manuscript. RG, MC, XL, and XM helped collect and analyse the data. WS and GH supervised the study and edited the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of The Children's Hospital of Fudan University [approval no. 2015 (26)]. Written informed consent was obtained from the parents or relatives of all study participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.van der Linde D, Konings EE, Slager MA, Witsenburg M, Helbing WA, Takkenberg JJ, Roos-Hesselink JW. Birth prevalence of congenital heart disease worldwide: A systematic review and meta-analysis. J Am Coll Cardiol. 2011;58:2241–2247. doi: 10.1016/j.jacc.2011.08.025. [DOI] [PubMed] [Google Scholar]
- 2.Zhao QM, Ma XJ, Ge XL, Liu F, Yan WL, Wu L, Ye M, Liang XC, Zhang J, Gao Y, et al. Pulse oximetry with clinical assessment to screen for congenital heart disease in neonates in China: A prospective study. Lancet. 2014;384:747–754. doi: 10.1016/S0140-6736(14)60198-7. [DOI] [PubMed] [Google Scholar]
- 3.Bedard E, Mccarthy KP, Dimopoulos K, Giannakoulas G, Gatzoulis MA, Ho SY. Structural abnormalities of the pulmonary trunk in tetralogy of Fallot and potential clinical implications: A morphological study. J Am Coll Cardiol. 2009;54:1883–1890. doi: 10.1016/j.jacc.2009.06.040. [DOI] [PubMed] [Google Scholar]
- 4.Ho S, Mccarthy KP, Josen M, Rigby ML. Anatomic- echocardiographic correlates: An introduction to normal and congenitally malformed hearts. Heart. 2001;86(Suppl 2):II3–II11. doi: 10.1136/heart.86.suppl_2.ii3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kathiriya IS, Nora EP, Bruneau BG. Investigating the transcriptional control of cardiovascular development. Circ Res. 2015;116:700–714. doi: 10.1161/CIRCRESAHA.116.302832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaynak B, von Heydebreck A, Mebus S, Seelow D, Hennig S, Vogel J, Sperling HP, Pregla R, Alexi-Meskishvili V, Hetzer R, et al. Genome-wide array analysis of normal and malformed human hearts. Circulation. 2003;107:2467–2474. doi: 10.1161/01.CIR.0000066694.21510.E2. [DOI] [PubMed] [Google Scholar]
- 7.Nemer M. Genetic insights into normal and abnormal heart development. Cardiovasc Pathol. 2008;17:48–54. doi: 10.1016/j.carpath.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 8.Simeone RM, Tinker SC, Gilboa SM, Agopian AJ, Oster ME, Devine OJ, Honein MA, National Birth Defects Prevention Study Proportion of selected congenital heart defects attributable to recognized risk factors. Ann Epidemiol. 2016;26:838–845. doi: 10.1016/j.annepidem.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pierpont ME, Basson CT, Benson DW, Jr, Gelb BD, Giglia TM, Goldmuntz E, McGee G, Sable CA, Srivastava D, Webb CL, et al. Genetic basis for congenital heart defects: Current knowledge: A scientific statement from the American heart association congenital cardiac defects committee, council on cardiovascular disease in the young: Endorsed by the American academy of pediatrics. Circulation. 2007;115:3015–3038. doi: 10.1161/CIRCULATIONAHA.106.183056. [DOI] [PubMed] [Google Scholar]
- 10.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: Cell fate control and signal integration in developmen. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 11.Luxán G, D'amato G, Macgrogan D, de la Pompa JL. Endocardial Notch signaling in cardiac development and disease. Circ Res. 2016;118:e1–e18. doi: 10.1161/CIRCRESAHA.115.305350. [DOI] [PubMed] [Google Scholar]
- 12.Mccright B, Lozier J, Gridley T. A mouse model of Alagille syndrome: Notch2 as a genetic modifier of Jag1 haploinsufficiency. Development. 2002;129:1075–1082. doi: 10.1242/dev.129.4.1075. [DOI] [PubMed] [Google Scholar]
- 13.Digilio MC, Luca AD, Lepri F, Guida V, Ferese R, Dentici ML, Angioni A, Marino B, Dallapiccola B. JAG1 mutation in a patient with deletion 22q11.2 syndrome and tetralogy of Fallot. Am J Med Genet A 161A. 2013:3133–3136. doi: 10.1002/ajmg.a.36148. [DOI] [PubMed] [Google Scholar]
- 14.Garg V. Notch signaling in aortic valve development and disease. Etiology and morphogenesis of congenital heart disease: From gene function and cellular interaction to morphology [internet] In: Nakanishi T, Markwald RR, Baldwin HS, Keller BB, Srivastava D, Yamagishi H, editors. Tokyo: Springer; 2016. 2016. Chapter 53. Jun 25. [DOI] [PubMed] [Google Scholar]
- 15.Meester J, Verstraeten A, Alaerts M, Schepers D, Van Laer L, Loeys BL. Overlapping but distinct roles for NOTCH receptors in human cardiovascular disease. Clin Genet. 2019;95:85–94. doi: 10.1111/cge.13382. [DOI] [PubMed] [Google Scholar]
- 16.Krebs LT, Xue Y, Norton CR, Shutter JR, Maguire M, Sundberg JP, Gallahan D, Closson V, Kitajewski J, Callahan R, et al. Notch signaling is essential for vascular morphogenesis in mice. Genes Dev. 2000;14:1343–1352. [PMC free article] [PubMed] [Google Scholar]
- 17.Leong KG, Hu X, Li L, Noseda M, Larrivée B, Hull C, Hood L, Wong F, Karsan A. Activated Notch4 inhibits angiogenesis: Role of beta 1-integrin activation. Mol Cell Biol. 2002;22:2830–2841. doi: 10.1128/MCB.22.8.2830-2841.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uyttendaele H, Ho J, Rossant J, Kitajewski J. Vascular patterning defects associated with expression of activated Notch4 in embryonic endothelium. Proc Natl Acad Sci USA. 2001;98:5643–5648. doi: 10.1073/pnas.091584598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noseda M, Mclean G, Niessen K, Chang L, Pollet I, Montpetit R, Shahidi R, Dorovini-Zis K, Li L, Beckstead B, et al. Notch activation results in phenotypic and functional changes consistent with endothelial-to-mesenchymal transformation. Circ Res. 2004;94:910–917. doi: 10.1161/01.RES.0000124300.76171.C9. [DOI] [PubMed] [Google Scholar]
- 20.Page DJ, Miossec MJ, Williams SG, Monaghan RM, Fotiou E, Cordell H, Sutcliffe L, Topf A, Bourgey M, Bourque G, et al. Whole exome sequencing reveals the major genetic contributors to nonsyndromic tetralogy of Fallot. Circ Res. 2019;124:553–563. doi: 10.1161/CIRCRESAHA.118.313250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shames DS, Minna JD, Gazdar AF. DNA methylation in health, disease, and cancer. Curr Mol Med. 2007;7:85–102. doi: 10.2174/156652407779940413. [DOI] [PubMed] [Google Scholar]
- 22.Boyes J, Bird A. DNA methylation inhibits transcription indirectly via a methyl-CpG binding protein. Cell. 1991;64:1123–1134. doi: 10.1016/0092-8674(91)90267-3. [DOI] [PubMed] [Google Scholar]
- 23.Sheng W, Chen L, Wang H, Ma X, Ma D, Huang G. CpG island shore methylation of ZFPM2 is identified in tetralogy of Fallot samples. Pediatr Res. 2016;80:151–158. doi: 10.1038/pr.2016.42. [DOI] [PubMed] [Google Scholar]
- 24.Sheng W, Qian Y, Wang H, Ma X, Zhang P, Diao L, An Q, Chen L, Ma D, Huang G. DNA methylation status of NKX2-5, GATA4 and HAND1 in patients with tetralogy of Fallot. BMC Med Genomics. 2013;6:46. doi: 10.1186/1755-8794-6-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gong J, Sheng W, Ma D, Huang G, Liu F. DNA methylation status of TBX20 in patients with tetralogy of Fallot. BMC Med Genomics. 2019;12:75. doi: 10.1186/s12920-019-0534-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Emeny RT, Baumert J, Zannas AS, Kunze S, Wahl S, Iurato S, Arloth J, Erhardt A, Balsevich G, Schmidt MV, et al. Anxiety associated increased CpG methylation in the promoter of Asb1: A translational approach evidenced by epidemiological and clinical studies and a murine model. Neuropsychopharmacology. 2018;43:342–353. doi: 10.1038/npp.2017.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bind MA, Coull BA, Peters A, Baccarelli AA, Tarantini L, Cantone L, Schwartz JD, Vokonas PS, Koutrakis P, Schwartz JD. Beyond the mean: Quantile regression to explore the association of air pollution with gene-specific methylation in the normative aging study. Environ Health Perspect. 2015;123:759–765. doi: 10.1289/ehp.1307824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Somineni HK, Zhang X, Biagini MJ, Kovacic MB, Ulm A, Jurcak N, Ryan PH, Hershey GKK, Ji H. Ten-eleven translocation 1 (TET1) methylation is associated with childhood asthma and traffic-related air pollution. J Allergy Clin Immunol. 2016;137:797–805.e5. doi: 10.1016/j.jaci.2015.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Plusquin M, Guida F, Polidoro S, Vermeulen R, Raaschou-Nielsen O, Campanella G, Hoek G, Kyrtopoulos SA, Georgiadis P, Naccarati A, et al. DNA methylation and exposure to ambient air pollution in two prospective cohorts. Environ Int. 2017;108:127–136. doi: 10.1016/j.envint.2017.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wythe JD, Dang LT, Devine WP, Boudreau E, Artap ST, He D, Schachterle W, Stainier DYR, Oettgen P, Black BL, et al. ETS factors regulate Vegf-dependent arterial specification. Dev Cell. 2013;26:45–58. doi: 10.1016/j.devcel.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oikawa T, Yamada T. Molecular biology of the Ets family of transcription factors. Gene. 2003;303:11–34. doi: 10.1016/S0378-1119(02)01156-3. [DOI] [PubMed] [Google Scholar]
- 32.Kola I, Brookes S, Green AR, Garber R, Tymms M, Papas TS, Seth A. The Ets1 transcription factor is widely expressed during murine embryo development and is associated with mesodermal cells involved in morphogenetic processes such as organ formation. Proc Natl Acad Sci USA. 1993;90:7588–7592. doi: 10.1073/pnas.90.16.7588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ye M, Coldren C, Liang X, Mattina T, Goldmuntz E, Benson DW, Ivy D, Perryman MB, Garrett-Sinha LA, Grossfeld P. Deletion of ETS-1, a gene in the Jacobsen syndrome critical region, causes ventricular septal defects and abnormal ventricular morphology in mice. Hum Mol Genet. 2010;19:648–656. doi: 10.1093/hmg/ddp532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao Z, Kim GH, Mackinnon AC, Flagg AE, Bassett B, Earley JU, Svensson EC. Ets1 is required for proper migration and differentiation of the cardiac neural crest. Development. 2010;137:1543–1551. doi: 10.1242/dev.047696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramirez K, Chandler KJ, Spaulding C, Zandi S, Sigvardsson M, Graves BJ, Kee BL. Gene deregulation and chronic activation in natural killer cells deficient in the transcription factor ETS1. Immunity. 2012;36:921–932. doi: 10.1016/j.immuni.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li RZ, Pei HP, Watson DK, Papas TS. EAP1/Daxx interacts with ETS1 and represses transcriptional activation of ETS1 target genes. Oncogene. 2000;19:745–753. doi: 10.1038/sj.onc.1203385. [DOI] [PubMed] [Google Scholar]
- 37.Pham VN, Lawson ND, Mugford JW, Dye L, Castranova D, Lo B, Weinstein BM. Combinatorial function of ETS transcription factors in the developing vasculature. Dev Biol. 2007;303:772–783. doi: 10.1016/j.ydbio.2006.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.TFSEARCH, corp-author. http://www.cbrc.jp/research/db/TFSEARCH.html. [Jun 13;2020 ];
- 39.JASPAR database, corp-author. http://jaspar.binf.ku.dk/cgi-bin/jaspar_db.pl?rm=browse&db=core&tax_group=vertebrates. [Jun 13;2020 ];
- 40.Iso T, Hamamori Y, Kedes L. Notch signaling in vascular development. Arterioscler Thromb Vasc Biol. 2003;23:543–553. doi: 10.1161/01.ATV.0000060892.81529.8F. [DOI] [PubMed] [Google Scholar]
- 41.Villa N, Walker L, Lindsell CE, Gasson J, Iruela-Arispe ML, Weinmaster G. Vascular expression of Notch pathway receptors and ligands is restricted to arterial vessels. Mech Dev. 2001;108:161–164. doi: 10.1016/S0925-4773(01)00469-5. [DOI] [PubMed] [Google Scholar]
- 42.Wu B, Zhang Z, Lui W, Chen X, Wang Y, Chamberlain AA, Moreno-Rodriguez RA, Markwald RR, O'Rourke BP, Sharp DJ, et al. Endocardial cells form the coronary arteries by angiogenesis through myocardial-endocardial VEGF signaling. Cell. 2012;151:1083–1096. doi: 10.1016/j.cell.2012.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu B, Wang Y, Lui W, Langworthy M, Tompkins KL, Hatzopoulos AK, Baldwin HS, Zhou B. Nfatc1 coordinates valve endocardial cell lineage development required for heart valve formation. Circ Res. 2011;109:183–192. doi: 10.1161/CIRCRESAHA.111.245035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Borgel J, Guibert S, Li Y, Chiba H, Schübeler D, Sasaki H, Forné T, Weber M. Targets and dynamics of promoter DNA methylation during early mouse development. Nat Genet. 2010;42:1093–1100. doi: 10.1038/ng.708. [DOI] [PubMed] [Google Scholar]
- 45.Jones PA, Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293:1068–1070. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- 46.Chamberlain AA, Lin M, Lister RL, Maslov AA, Wang Y, Suzuki M, Wu B, Greally JM, Zheng D, Zhou B. DNA methylation is developmentally regulated for genes essential for cardiogenesis. J Am Heart Assoc. 2014;3:e976. doi: 10.1161/JAHA.114.000976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Feinberg AP. The key role of epigenetics in human disease prevention and mitigation. N Engl J Med. 2018;378:1323–1334. doi: 10.1056/NEJMra1402513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sheng W, Wang H, Ma X, Qian Y, Zhang P, Wu Y, Zheng F, Chen L, Huang G, Ma D. LINE-1 methylation status and its association with tetralogy of fallot in infants. BMC Med Genomics. 2012;5:20. doi: 10.1186/1755-8794-5-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsuboi K, Nagatomo T, Gohno T, Higuchi T, Sasaki S, Fujiki N, Kurosumi M, Takei H, Yamaguchi Y, Niwa T, Hayashi SI. Single CpG site methylation controls estrogen receptor gene transcription and correlates with hormone therapy resistance. J Steroid Biochem Mol Biol. 2017;171:209–217. doi: 10.1016/j.jsbmb.2017.04.001. [DOI] [PubMed] [Google Scholar]
- 50.Fürst RW, Kliem H, Meyer HH, Ulbrich SE. A differentially methylated single CpG-site is correlated with estrogen receptor alpha transcription. J Steroid Biochem Mol Biol. 2012;130:96–104. doi: 10.1016/j.jsbmb.2012.01.009. [DOI] [PubMed] [Google Scholar]
- 51.Bartels SJ, Spruijt CG, Brinkman AB, Jansen PW, Vermeulen M, Stunnenberg HG. A SILAC-based screen for Methyl-CpG binding proteins identifies RBP-J as a DNA methylation and sequence-specific binding protein. PLoS One. 2011;6:e25884. doi: 10.1371/journal.pone.0025884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mann IK, Chatterjee R, Zhao J, He X, Weirauch MT, Hughes TR, Vinson C. CG methylated microarrays identify a novel methylated sequence bound by the CEBPB|ATF4 heterodimer that is active in vivo. Genome Res. 2013;23:988–997. doi: 10.1101/gr.146654.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Irizarry RA, Ladd-Acosta C, Wen B, Wu ZJ, Montano C, Onyango P, Cui HM, Gabo K, Rongione M, Webster M, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Portela A, Esteller M. Epigenetic modifications and human disease. Nat Biotechnol. 2010;28:1057–1068. doi: 10.1038/nbt.1685. [DOI] [PubMed] [Google Scholar]
- 55.Claus R, Lucas DM, Stilgenbauer S, Ruppert AS, Yu L, Zucknick M, Mertens D, Bühler A, Oakes CC, Larson RA, et al. Quantitative DNA methylation analysis identifies a single CpG dinucleotide important for ZAP-70 expression and predictive of prognosis in chronic lymphocytic leukemia. J Clin Oncol. 2012;30:2483–2491. doi: 10.1200/JCO.2011.39.3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ceccarelli V, Racanicchi S, Martelli MP, Nocentini G, Fettucciari K, Riccardi C, Marconi P, Nardo PD, Grignani F, Binaglia L, Vecchini A. Eicosapentaenoic acid demethylates a single CpG that mediates expression of tumor suppressor CCAAT/enhancer-binding protein delta in U937 leukemia cells. J Biol Chem. 2011;286:27092–27102. doi: 10.1074/jbc.M111.253609. [DOI] [PMC free article] [PubMed] [Google Scholar]