Abstract
The aim of the present study was to use the clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated (Cas) 9-mediated gene knockout technology for the rapid classification of the differential function of micro (mi)RNAs screened using miRNA expression profiling by microarray. The rational design of single guide RNAs for the CRISPR/Cas9 system was verified to function in human LNCaP cells with rapid and efficient target gene editing. miRNA (miR)-205, miR-221, miR-222, miR-30c, miR-224, miR-455-3p, miR-23b and miR-505 were downregulated in patients with prostate cancer (PCa) and were experimentally validated to function as tumor suppressors in prostate cancer cells, affecting tumor proliferation, invasion and aerobic glycolysis. In addition, the data of the present study suggested that miR-663a and mfiR-1225-5p were upregulated in prostate cancer tissues and cell proliferation of miR-663a and miR-1225-5p knockout PCa cells was significantly lower compared with miR-NC cells. Furthermore, knockout of miR-1225-5p and miR-663a significantly decreased the lactate production in LNCaP cells in vitro. In conclusion, the present study offered a simple and efficient method for rapidly classifying miRNA function by applying CRISPR/Cas9 in LNCaP cells. The present study suggested, for the first time to the best of the authors' knowledge, that the aberrant expression of miR-663a and miR-1225-5p may be involved with the progression of prostate cancer, implying their potential as candidate markers for this type of cancer. However, the precise role of miR-663a and miR-1225-5p in accelerating the development of prostate cancer and promoting tumor progression remains to be elucidated.
Keywords: prostate cancer, CRISPR/Cas9, microRNA-663a, microRNA-1225-5p
Introduction
Prostate cancer (PCa), which is a clinically heterogeneous-multifocal disease, is the second most frequently diagnosed cancer in males worldwide (1). The most recent US statistic reported that in 2017 the number of new cases and cases of mortality associated with PCa in the US were 161,360 and 26,730, respectively (1). microRNAs (miRNAs or miRs) are small noncoding RNAs (length ~22 nt) expressed in animal and plant cells (2,3). miRNAs are the smallest known carriers of gene-encoded, post-transcriptional regulatory information in plants and animals (4), and suppress a variety of gene expression at the post-transcriptional level by pairing with complementary nucleotide sequences in the 3′-untranslated region (UTR) of specific target genes (5). Accumulated evidence suggests that miRNAs serve as tumor suppressor or oncogenes depending on the target gene (6,7) and dysregulation of miRNAs may serve important roles in the initiation and progression of types of cancer of various tissue origins (8).
Similar to other malignancies, PCa has a distinctive miR expression profile, which has been the basis for the functional study of miRNA in PCa (7–9). Porkka et al (7) demonstrated that 37 miRNAs were downregulated and 14 upregulated in PCa tissues compared with benign tissues. A previous study obtained differential expression data of miRNAs in PCa determined by miRNA microarray analyses and reported that 11 miRNAs were upregulated and 17 miRNAs were downregulated in PCa (10). However, these previous gain-of-function studies did not include loss-of-function analyses using miRNA knockout and applying the clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated (Cas) 9 system, which can effectively, specifically and stably suppress gene expression in vitro and in vivo (11,12). CRISPR/Cas9 is a recently discovered genome editing system, which has markedly changed the way that researchers study genes and their functions in mammalian systems (12,13). It is derived from the CRISPR/Cas bacterial-acquired immune system and Cas9 is directed by guide (g)RNAs, which match the DNA targeted in cleavage to modify the respective gene (11,13).
To classify rapidly the differential functions of miRNAs, which were identified through miRNA expression profiling in PCa, the CRISPR/Cas9 system was used to knockout the expression of PCa-associated miRNAs, including miR-205, miR-221, miR-455-3p, miR-222, miR-224, miR-505, miR-23b, miR-30c, miR-1225-5p and miR-663a in LNCaP cells. The proliferation, invasion and metabolic changes were determined.
Materials and methods
Prostate cancer-associated miRNA Data
The prostate cancer-associated miRNA is the differentially expressed miRNA in the PCa tissue compared with the adjacent benign prostate gland tissues, which is from our miRNA expression profiling microarray data (GEO, http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE34932) and the Taylor database (GEO, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE21032; Table I) (10).
Table I.
Differentially expressed miRNAs in PCa (Fold change ≥1.5, P<0.05).
| miRNA microarray data | Taylor database | |||||
|---|---|---|---|---|---|---|
| Name | Regulation | Fold change | P-value | Fold change | P-value | Chromosomal location |
| hsa-miR-205 | Down | 58.96 | 0.009 | 3.19 | <0.001 | 1:209605478-209605587[+] |
| hsa-miR-221 | Down | 5.15 | 0.036 | 3.24 | <0.001 | X:45605585-45605694[-] |
| hsa-miR-455-3p | Down | 4.39 | 0.014 | 1.63 | 0.003 | 9:116971714-116971809[+] |
| hsa-miR-222 | Down | 3.94 | 0.036 | 3.55 | <0.001 | X:45606421-45606530[-] |
| hsa-miR-221 | Down | 3.50 | 0.050 | 1.59 | 0.008 | X:45605585-45605694[-] |
| hsa-miR-224 | Down | 3.32 | 0.011 | 2.56 | <0.001 | X:151127050-151127130[-] |
| hsa-miR-505 | Down | 2.72 | 0.038 | 1.52 | <0.001 | X:139006307-139006390[-] |
| hsa-miR-23b | Down | 2.70 | 0.033 | 1.86 | <0.001 | 9:97847490-97847586[+] |
| hsa-miR-30c | Down | 2.09 | 0.018 | 1.60 | <0.001 | 1:41222956-41223044[+] |
| hsa-miR-1225-5p | Up | 3.37 | 0.041 | 1.56 | 0.005 | 16:2140196-2140285[-] |
| hsa-miR-663a | Up | 2.71 | 0.044 | 2.64 | <0.001 | 20:26188822-26188914[-] |
miR, microRNA; PCa, prostate cancer; hsa, human.
Cell culture
The human PCa cell line, LNCaP was purchased from the American Type Culture Collection and cultured in RPMI-1640 medium (HyClone; Cytiva) supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.), 2 mM L-glutamine and 1% penicillin/streptomycin antibiotics (Thermo Fisher Scientific, Inc.). Cells were maintained at 37°C in a humidified chamber supplemented with 5% CO2.
Single guide (sg)RNA design and plasmid construction
Target sequences of miR-205, miR-221, miR-455-3p, miR-222, miR-224, miR-505, miR-23b, miR-30c, miR-1225-5p and miR-663a for CRISPR interference were designed. Taking miR-205 as an example (Gene ID406988; http://www.ncbi.nlm.nih.gov/gene/406988): The gene sequence 5′-AAAGATCCTCAGACAATCCATGTGCTTCTCTTGTCCTTCATTCCACCGGAGTCTGTCTCATACCCAACCAGATTTCAGTGGAGTGAAGTTCAGGAGGCATGGAGCTGACA-3′ was retrieved and downloaded from GeneBank and the sgRNA was designed by Zhang lab (https://zlab.bio/guide-design-resources) using Target Finder (version 2014, http://targetfinder.flycrispr.neuro.brown.edu/) and DNA 2.0 gRNA (https://www.dna20.com). A total of four optimal target sequences for each miRNA were selected and four scramble sequences were used as controls (Table SI). Subsequently, two complementary oligonucleotides with Bbsl restriction sites for gRNAs were synthesized and cloned into CRISPR/Cas9 lentiCRISPR-v2 vector (cat. no. 52961; Addgene, Inc.) by HYYMed Company using T4 DNA ligase (cat. no. D2011B; Takara Biotechnology Co., Ltd.) (14).
Cell line construction and transfection
LNCaP cells were seeded in 6-well plates (3×105), grown to ~70% confluence and transfected with the CRISPR/Cas9 lentiCRISPR-v2 vector plasmid construct (2 µg) using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocols. On the day following transfection, cells were treated with 10 mg/ml of puromycin (Beyotime Institute of Biotechnology) and maintained at 37°C in a humidified chamber supplemented with 5% CO2 for >2 days. Subsequently, the cells were harvested or cultured for further experiments.
T7 endonuclease I (T7E1) assays for knockout efficiency
T7E1 digestion assay was performed to analyze the gene knockout efficiency as previously described (15,16). Genomic DNA of transfected cells was extracted using HiPure Tissue DNA Mini kit (cat. no. D3121-03; Angen Biotech Co., Ltd.) according to the manufacturer's protocols. The target site of sgRNA was amplified by PCR and DNA fragments were subjected to digestion with the mismatch-sensitive T7E1 (cat. no. E3321; New England BioLabs, Inc.). For T7E1 digestion, amplified PCR products were denatured at 95°C for 5 min and slowly cooled to room temperature to allow formation of heteroduplex DNA; the annealed products were incubated with 0.5 µl T7E1 for 30 min at 37°C and the digested DNA was separated on 2% agarose gels. Based on relative band intensities, the small insertion and deletion (indel) percentage was calculated using the formula, 1-√ 1-(b+c)/(a+b+c) ×100, where a is the integrated intensity of the undigested PCR product, and b and c are the integrated intensities of each cleavage product (14).
Cell proliferation assay
For cell viability assays, cells were seeded in 96-well plates at 5,000 cells/200 µl per well and cultured for 24, 48 and 72 h. Cells were then incubated with 20 µl Cell Counting Kit-8 (CCK-8) solution (cat. no. C0038; Beyotime Institute of Biotechnology) for 2 h at 37°C according to the manufacturer's protocol. The absorbance was measured at 450 nm using a spectrophotometer.
Cell invasion assay
Transwell inserts (pore size, 8 µm) were filled with 50 µl of a mixture of serum-free RPMI-1640 medium and Matrigel (ratio, 1:10; BD Biosciences) for 30 min at 37°C. The inserts were placed in 24-well tissue culture plates (Transwell; Corning Inc.) containing 10% FBS-medium. Following solidification by incubation in 37°C for 4 h, 8×104 cells in 200 µl medium were placed in upper chambers. Following a 48-h incubation at 37°C with 5% CO2, culture medium with mitomycin to halt the mitosis was added and the membranes were fixed with 10% formalin for 5 min at room temperature and stained with 0.05% crystal violet for 10 min at room temperature. Migrated cells were assessed and the data were expressed as the mean ± standard deviation.
Detection of lactic acid
The culture medium was collected and the concentration of lactic acid was determined using the GEM Premier 3000 Blood Gas analyzer (Instrumentation Laboratory Inc.). Lactate was measured by amperometry based on the following principle: Lactate oxidase, immobilized on a lactate biochip sensor, selectively converted lactate into pyruvate and hydrogen peroxide (H2O2), the released H2O2 oxidizes on a platinum electrode and produces an electric current that is proportional to the lactate concentration in the blood sample.
Statistical analysis
Data are expressed as the mean ± standard deviation. Data analysis was performed by using one-way ANOVA followed by Tukey's post hoc test or independent samples t-test between control and experimental groups using SPSS 17.0 (SPSS, Inc.). P<0.05 was considered to indicate a statistically significant difference.
Results
Prostate cancer-associated miRNAs selection
The PCa differentially expressing miRNAs with fold change >1.5 were selected for miRNA expression profiling (Table I). The following differentially expressing miRNAs were selected: hsa-miR-205, hsa-miR-221, hsa-miR-455-3p, hsa-miR-222, hsa-miR-224, hsa-miR-505, hsa-miR-23b, hsa-miR-30c, hsa-miR-1225-5p and hsa-miR-663a (downregulated, 8; upregulated, 2; Table I) (10). To further study the above miRNAs function in prostate cancer cell, the use of CRISPR/Cas9-mediated gene knockout technology for the rapid classification of the function of differential miRNAs was assessed in the following.
miRNAs knockout by CRISPR/Cas9 system
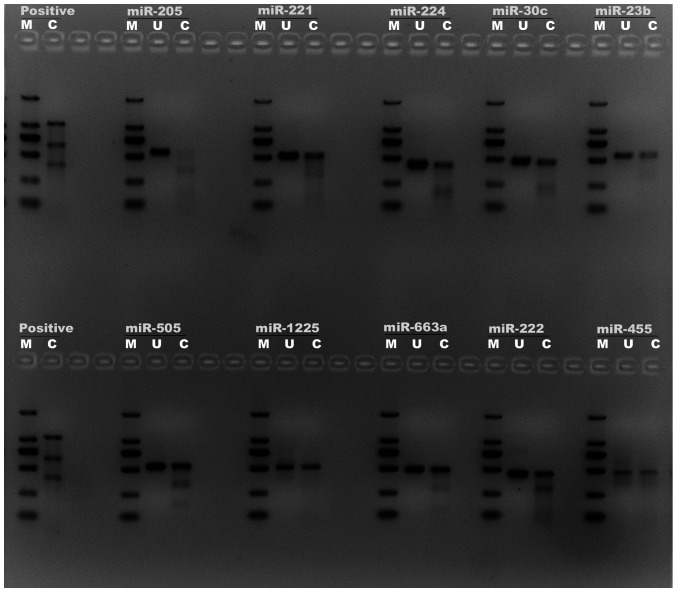
In the present study, a total of four optimal target sgRNA sequences for each miRNA were designed (Table SI), and every sgRNA position in genomic DNA was illustrated in Fig. SIA. The sgRNA sequence was synthesized (Table SII) and cloned into the CRISPR/Cas9 lentiCRISPR-v2 vector as demonstrated in Fig. S1B (14). To verify the knockout efficiency of miRNAs in human LNCaP cells, a mismatch-sensitive T7E1 assay was conducted to analyze the sgRNA target genomics profiling as previously described (15). The PCR primers for T7E1 assay of miRNAs were designed (Table SIII and Fig. S2). The T7E1 assay result is presented in Fig. 1. By comparing the bright lanes of ‘uncut’ (U) and ‘cut’ (C), it demonstrated that the mixture of the miR-205 sgRNA vectors demonstrated effective knockout of the miR-205 gene in LNCaP cells with >90% indel percentage and the other miRNAs also demonstrated a positive knockout effect.
Figure 1.
T7E1 assay for knockout efficiency. Each sample consists of three lanes in the agarose gel: The left lane is an DNA marker (M), the middle lane is ‘uncut’ (U), meaning no treatment, and the right lane is ‘cut’ (C), digested by the T7E1 endonuclease. The ‘cut’ lane demonstrated additional lower DNA bands and the decreased bright of the bands compare the ‘uncut’ lane. Diverse intensities of lower bands presumably reflect different degrees of mosaic mutations. PUC19 digested by the T7E1 is used as the positive control. T7E1, T7 endonuclease I.
miRNAs affect prostate cancer cell proliferation
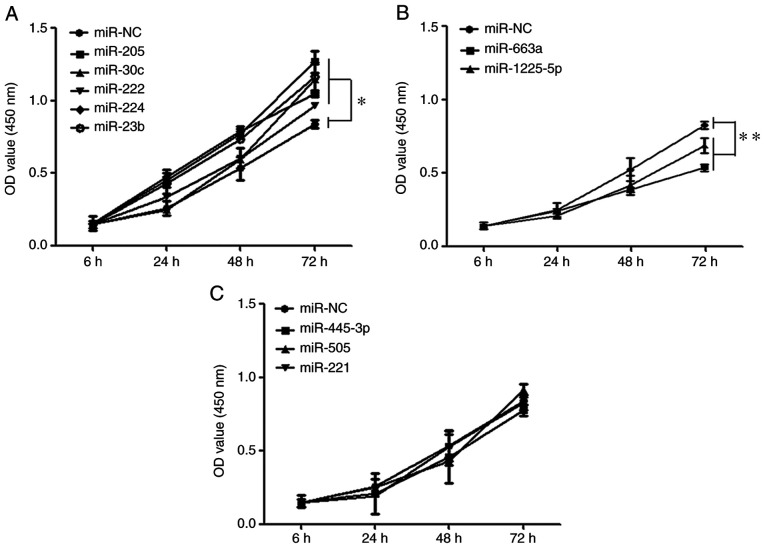
To study the influence of the differentially expressed miRNAs on PCa cells, a lentiviral vector knockout system for the miRNAs using the CRISPR/Cas9 system was applied and stable LNCaP cells were established following lentivector transfection. Proliferation was investigated by CCK-8 assay. The results indicated that the proliferative abilities of miR-222, miR-224, miR-23b, miR-205 and miR-30c knockout cells were significantly enhanced (P<0.05) compared with the blank vector groups in LNCaP cells lines at 72 h following transfection (Fig. 2A). In contrast, the proliferative abilities of miR-1225-5p and miR-663a knockout were significantly reduced (P<0.01; Fig. 2B). The other group of miR-221, miR-455-3p, miR-505 showed no function on the cell proliferation of prostate cancer (P>0.05; Fig. 2C).
Figure 2.
Cell proliferation assay. (A) The miRNAs group of enhancing the cell proliferation. (B) The miRNAs group of reducing the cell proliferation. (C) The miRNAs group of un-affecting the cell proliferation. *P<0.05 and **P<0.01. NC, negative control with scramble vector; miRNAs, micro RNAs.
miRNAs affect prostate cancer cell invasion
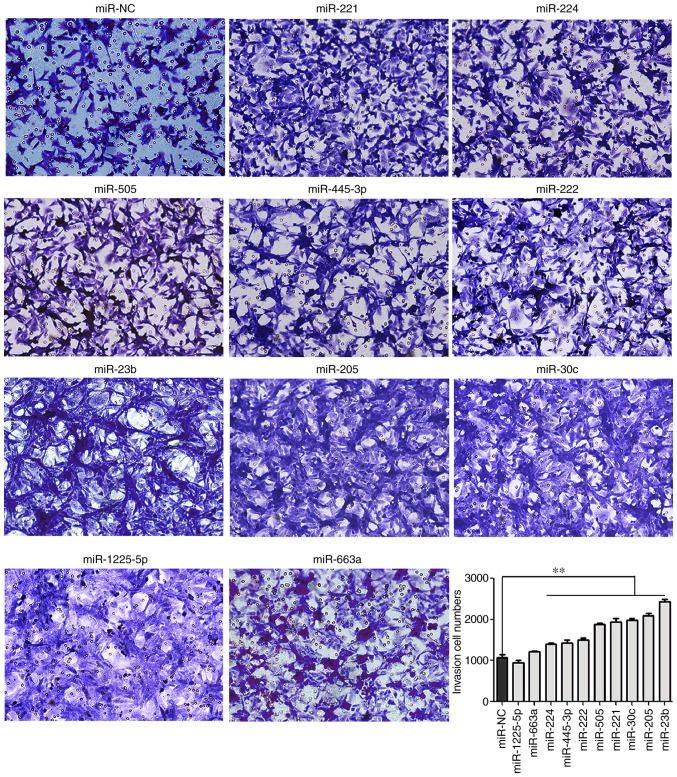
In the present study, Transwell assays were performed to study the effect of various miRNAs on PCa cell invasion. The results revealed that knockout of miR-205, miR-221, miR-455-3p, miR-222, miR-224, miR-505, miR-23b and miR-30c significantly increased invasive activities of LNCaP cells compared with the control cells (P<0.01; Fig. 3), which suggested that these miRNAs may act as tumor suppressors in PCa.
Figure 3.
Cell invasion assay. Cell invasion was detected by Transwell invasion assay. Cell numbers were counted in six independent symmetrical visual fields under a microscope at ×200 magnification. **P<0.01. NC, negative control with scramble vector.
miRNAs affect prostate cancer cell metabolism
In the present study, the lactic acid concentration in culture medium of miRNA-knockout LNCaP cells was detected in vitro to represent the cell metabolism. As presented in Fig. 4, knockout of miR-505 and miR-23b demonstrated increased lactate production in LNCaP cells in vitro (P<0.01), while knockout of miR-1225-5p, miR-663a, miR-205, miR-30c, miR-222 and miR-224 significantly decreased the lactic acid concentration (P<0.01).
Figure 4.
Measurement of lactic acid. Culture medium was collected and the concentration of lactic acid was determined using the GEM Premier 3000 Blood Gas analyzer. Data are presented as the mean ± standard deviation of three independent experiments. **P<0.01. NC, negative control with scramble vector.
Discussion
PCa is one of the most common malignancies in Europe and the US and poses a serious threat to the health of older males (1,17). miRNAs are small non-coding RNA molecules that regulate gene expression by base pairing with their target mRNAs and they contribute to cancer initiation, progression and metastasis by directly modulating oncogenic or tumor suppressor pathways (18). Previous studies demonstrated that miRNA expression patterns serve as phenotypic signatures of various types of cancer and may be used as diagnostic, prognostic and therapeutic tools (9,10,19). Previous studies have analyzed global miRNA expression profiles or the functional role of miRNAs in PCa; however, results are inconsistent (20,21).
In the present study, eight down- and two upregulated miRNAs in PCa tissues were selected based on previous miRNA expression profiling microarray data and the Taylor database. Downregulation of miR-205, miR-221, miR-455-3p, miR-222, miR-224, miR-505, miR-23b and miR-30c in PCa tissues led to the hypothesis that these miRNAs may function as tumor suppressors. CRISPR/Cas9-mediated gene knockout technology was applied to classify the differentially expressed miRNA functions. Proliferation and invasive abilities of the miRNAs were subsequently validated by CCK-8 and Transwell assays. According the Warburg effect, aggressive tumors frequently exhibit metabolic alteration and reveal an increasing dependence on the glycolytic pathway to generate ATP, even in the presence of oxygen (22). The increased glycolysis may be a response to the hypoxic conditions characterizing the microenvironment of malignant cells (22). The generation and accumulation of lactic acid leads to microenvironmental acidosis and facilitates tumor proliferation, invasion and metastasis (23). In the present study, the lactic acid concentration of the miRNA knockout LNCaP cells was detected in vitro.
miR-205 is reported to be downregulated in patients with PCa and acts as a tumor suppressor (24–26). Verdoodt et al (27) reported that miRNA-205 is a novel regulator of the anti-apoptotic protein B-cell lymphoma 2 and promotes apoptosis in PCa cells in response to DNA damage by cisplatin and doxorubicin treatment, and it further inhibits proliferation in PC3 and LNCaP cells. Recently, studies (28,29) suggested that miRNA-205 serves as a prognostic factor and suppresses proliferation and invasion in various types of cancers, which is consistent with the results of the present study. Downregulation of miR-221 and miR-222 are frequently observed in PCa samples and certain differentially expressed gene targets are associated with PCa (9,10,21,30). The results of the present study indicated that the downregulation of miR-30c promoted cell proliferation and invasion, which was consistent with previous studies (31,32). Other studies (33,34) suggest that miR-224 directly targets the Ras-association domain family and acts as a tumor promoter in cervical and gastric cancer progression. Liu et al (35) indicated that miR-224 inhibits proliferation and migration of breast cancer cells by downregulating Fizzled-5 expression. The present study demonstrated that miR-224 was downregulated in PCa and knockout of miR-224 promoted proliferation and invasion in LNCaP cells. Recently, miRNA-455-3p was described to be markedly downregulated in PCa cells and clinical tumor specimens and it functions as a tumor suppressor by targeting eukaryotic translation initiation factor 4E and by inhibiting proliferation of PCa cells (36). Notably, the present study revealed that knockout of miRNA-455-3p promoted invasion of LNCaP cells. It was reported that miR-505 functions as a tumor suppressor in endometrial cancer by targeting tumor growth factor α and miR-505 modulated cancer proliferation and migration in human non-small cell lung cancer through inverse regulation of FZD4 (37). However, the functions remain to be elucidated in human PCa.
The present study described that knockout of miR-505 efficiently enhanced PCa cell invasion. A downregulated expression of miR-23b was described for malignant PCa tissues. The miR-23b/-27b cluster functions as a metastasis-suppressor by decreasing Huntingtin-interacting protein 1-related protein levels in preclinical models of PCa (38), which is consistent with the results of the present study that miR-23b inhibited PCa proliferation and invasion. Knockout of miR-505 and miR-23b demonstrated increased lactate production in LNCaP cells in vitro, which indicated that miR-505 and miR-23b may be involved in the metabolic regulation. The difference in lactic acid levels between the control and miRNA-knockout groups was not as significant as anticipated, this may be a limitation in the detection method of the GEM Premier 3,000 Blood Gas analyzer; the lactic acid levels were detected in conditional medium but not in cells. A better method and devices, such as Seahorse XF Analyzers, can be applied to live-cell metabolic assays.
miRNA-663a has been revealed to be downregulated in non-small cell lung cancer and to suppress cell proliferation and invasion by targeting JunD (39) and miR-1225-5p was revealed to serve to constrain gastric carcinoma growth and the metastatic potential via inhibition of insulin receptor substrate-1 and β-catenin signaling (40). The role of miR-663a and miR-1225-5p in PCa has not been previously addressed. The data from the present study indicated that miR-663a and miR-1225-5p were upregulated in PCa tissues and cell proliferation in miR-663a and miR-1225-5p knockout PCa cells was significantly lower compared with miR-NC cells. In addition, knockout of miR-1225-5p and miR-663a significantly decreased the lactate production in the LNCaP cells in vitro. Collectively, the data demonstrated that miR-663a and miR-1225-5p functioned as tumor promoters in PCa progression. The results provided a starting point for future research into the function of miR-663a and miR-1225-5p and suggested that miR-663a and miR-1225-5p upregulation may be involved in the progression of PCa and this may promote the clinical application of miR-663a and miR-1225-5p as PCa biomarkers.
In conclusion, the present study offered a simple and efficient method for rapidly classifying miRNA function using the CRISPR/Cas9 system in LNCaP cells. miR-205, miR-221, miR-222, miR-30c, miR-224, miRNA-455-3p, miR-23b and miR-505 downregulation in patients with PCa were experimentally validated to function as tumor suppressors in PCa cells. The data from the present study, for the first time to the best of the authors' knowledge, suggested that the aberrant expression of miR-663a and miR-1225-5p may be involved with PCa progression, implying their potential as candidate markers for this type of cancer. However, the precise roles of miR-663a and miR-1225-5p in accelerating the development of PCa and promoting tumor progression require further clarification. Limitations of the present study were that only one cell line, LNCap, was investigated and that the CRISPR/Cas9 system has a potential off-target problem, which may cause the cell function to change by the knockout of another unexpected gene, thus the detailed function of miRNA requires further study by overexpression and knockout of miRNA in vitro and in vivo.
Supplementary Material
Acknowledgements
The authors would like to thank Professor Yanqiong Zhang (Institute of Chinese Materia Medica, China Academy of Chinese Medical Sciences) who provided guidance and revised the language.
Funding
The present study was supported by grants from National Natural Science Foundation of China (grant nos. 81660426 and 81873608), the Natural Science Foundation of Guangdong Province (grant nos. 2014A030304068, 2014A030310088 and 2014A020212471), the High-level Innovative Talent Project of Guizhou Province in 2018 [grant no. (2018)5639] and the Science and Technology Project of HuaDu District (grant no. HD15CXY005).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
FNJ, JGZ and WDZ designed the study and edited the manuscript. YXL, WW, CYZ, GXC, YPW, ZZL, YY and ZDH performed the experiments and analyzed the data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MiRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ayub SG, Kaul D, Ayub T. Microdissecting the role of miRNAs in the pathogenesis of prostate cancer. Cancer Genet. 2015;208:289–302. doi: 10.1016/j.cancergen.2015.02.010. [DOI] [PubMed] [Google Scholar]
- 4.Zhao Y, Cong L, Lukiw WJ. Plant and Animal microRNAs (miRNAs) and their potential for inter-kingdom communication. Cell Mol Neurobiol. 2018;38:133–140. doi: 10.1007/s10571-017-0547-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 6.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Porkka KP, Pfeiffer MJ, Waltering KK, Vessella RL, Tammela TL, Visakorpi T. MicroRNA expression profiling in prostate cancer. Cancer Res. 2007;167:6130–6135. doi: 10.1158/0008-5472.CAN-07-0533. [DOI] [PubMed] [Google Scholar]
- 8.Plaisier CL, Pan M, Baliga NS. A miRNA-regulatory network explains how dysregulated miRNAs perturb oncogenic processes across diverse cancers. Genome Res. 2012;22:2302–2314. doi: 10.1101/gr.133991.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song CJ, Chen H, Chen LZ, Ru GM, Guo JJ, Ding QN. The potential of microRNAs as human prostate cancer biomarkers: A meta-analysis of related studies. J Cell Biochem. 2018;119:2763–2786. doi: 10.1002/jcb.26445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He HC, Han ZD, Dai QS, Ling XH, Fu X, Lin ZY, Deng YH, Qin GQ, Cai C, Chen JH, et al. Global analysis of the differentially expressed miRNAs of prostate cancer in Chinese patients. BMC Genomics. 2013;14:757. doi: 10.1186/1471-2164-14-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 12.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mussolino C, Morbitzer R, Lütge F, Dannemann N, Lahaye T, Cathomen T. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. 2011;39:9283–9293. doi: 10.1093/nar/gkr597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim D, Bae S, Park J, Kim E, Kim S, Yu HR, Hwang J, Kim JI, Kim JS. Digenome-seq: Genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat Methods. 2015;12:237–243. doi: 10.1038/nmeth.3284. [DOI] [PubMed] [Google Scholar]
- 17.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 18.Baranwal S, Alahari SK. miRNA control of tumor cell invasion and metastasis. Int J Cancer. 2010;126:1283–1290. doi: 10.1002/ijc.25014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saini S, Majid S, Dahiya R. Diet, microRNAs and prostate cancer. Pharm Res. 2010;27:1014–1026. doi: 10.1007/s11095-010-0086-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szczyrba J, Löprich E, Wach S, Jung V, Unteregger G, Barth S, Grobholz R, Wieland W, Stöhr R, Hartmann A, et al. The microRNA profile of prostate carcinoma obtained by deep sequencing. Mol Cancer Res. 2010;8:529–538. doi: 10.1158/1541-7786.MCR-09-0443. [DOI] [PubMed] [Google Scholar]
- 21.Schaefer A, Jung M, Mollenkopf HJ, Wagner I, Stephan C, Jentzmik F, Miller K, Lein M, Kristiansen G, Jung K. Diagnostic and prognostic implications of microRNA profiling in prostate carcinoma. Int J Cancer. 2010;126:1166–1176. doi: 10.1002/ijc.24827. [DOI] [PubMed] [Google Scholar]
- 22.Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nat Rev Cancer. 2004;4:891–899. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- 23.Airley RE, Mobasheri A. Hypoxic regulation of glucose transport, anaerobic metabolism and angiogenesis in cancer: Novel pathways and targets for anticancer therapeutics. Chemotherapy. 2007;53:233–256. doi: 10.1159/000104457. [DOI] [PubMed] [Google Scholar]
- 24.Gandellini P, Folini M, Longoni N, Pennati M, Binda M, Colecchia M, Salvioni R, Supino R, Moretti R, Limonta P, et al. miR-205 exerts tumor-suppressive functions in human prostate through down-regulation of protein kinase Cepsilon. Cancer Res. 2009;69:2287–2295. doi: 10.1158/0008-5472.CAN-08-2894. [DOI] [PubMed] [Google Scholar]
- 25.Majid S, Dar AA, Saini S, Yamamura S, Hirata H, Tanaka Y, Deng G, Dahiya R. MicroRNA-205-directed transcriptional activation of tumor suppressor genes in prostate cancer. Cancer. 2010;116:5637–5649. doi: 10.1002/cncr.25488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang N, Li Q, Feng NH, Cheng G, Guan ZL, Wang Y, Qin C, Yin CJ, Hua LX. miR-205 is frequently downregulated in prostate cancer and acts as a tumor suppressor by inhibiting tumor growth. Asian J Androl. 2013;15:735–741. doi: 10.1038/aja.2013.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verdoodt B, Neid M, Vogt M, Kuhn V, Liffers ST, Palisaar RJ, Noldus J, Tannapfel A, Mirmohammadsadegh A. MicroRNA-205, a novel regulator of the anti-apoptotic protein Bcl2, is downregulated in prostate cancer. Int J Oncol. 2013;43:307–314. doi: 10.3892/ijo.2013.1915. [DOI] [PubMed] [Google Scholar]
- 28.Pang H, Yue X. MiR-205 serves as a prognostic factor and suppresses proliferation and invasion by targeting insulin-like growth factor receptor 1 in human cervical cancer. Tumour Biol. 2017;39:1010428317701308. doi: 10.1177/1010428317701308. [DOI] [PubMed] [Google Scholar]
- 29.Chen S, Jin L, Nie S, Han L, Lu N, Zhou Y. MiR-205 inhibits growth and invasion of neuroblastoma by targeting cAMP responsive element binding protein 1. Oncol Res. 2018;26:445–455. doi: 10.3727/096504017X14974834436195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tong AW, Fulgham P, Jay C, Chen P, Khalil I, Liu S, Senzer N, Eklund AC, Han J, Nemunaitis J. MicroRNA profile analysis of human prostate cancers. Cancer Gene Ther. 2009;16:206–216. doi: 10.1038/cgt.2008.77. [DOI] [PubMed] [Google Scholar]
- 31.Ling XH, Han ZD, Xia D, He HC, Jiang FN, Lin ZY, Fu X, Deng YH, Dai QS, Cai C, et al. MicroRNA-30c serves as an independent biochemical recurrence predictor and potential tumor suppressor for prostate cancer. Mol Biol Rep. 2014;41:2779–2788. doi: 10.1007/s11033-014-3132-7. [DOI] [PubMed] [Google Scholar]
- 32.Zhang J, Wang X, Wang Y, Peng R, Lin Z, Wang Y, Hu B, Wang J, Shi G. Low expression of microRNA-30c promotes prostate cancer cells invasion involved in downregulation of KRAS protein. Oncol Lett. 2017;14:363–368. doi: 10.3892/ol.2017.6163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang Y, Li Y, Wang FF, Lv W, Xie X, Cheng X. Over-expressed miR-224 promotes the progression of cervical cancer via targeting RASSF8. PLoS One. 2016;11:e0162378. doi: 10.1371/journal.pone.0162378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He C, Wang L, Zhang J, Xu H. Hypoxia-inducible microRNA-224 promotes the cell growth, migration and invasion by directly targeting RASSF8 in gastric cancer. Mol Cancer. 2017;16:35. doi: 10.1186/s12943-017-0603-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu F, Liu Y, Shen J, Zhang G, Han J. MicroRNA-224 inhibits proliferation and migration of breast cancer cells by down-regulating Fizzled 5 expression. Oncotarget. 2016;7:49130–49142. doi: 10.18632/oncotarget.9734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao Y, Yan M, Yun Y, Zhang J, Zhang R, Li Y, Wu X, Liu Q, Miao W, Jiang H. MicroRNA-455-3p functions as a tumor suppressor by targeting eIF4E in prostate cancer. Oncol Rep. 2017;37:2449–2458. doi: 10.3892/or.2017.5502. [DOI] [PubMed] [Google Scholar]
- 37.Negrete-Garcia MC, Ramírez-Rodriguez SL, Rangel-Escareño C, Muñoz-Montero S, Kelly-García J, Vázquez-Manríquez ME, Santillán P, Ramírez MM, Ramírez-Martínez G, Ramírez- Venegas A, Ortiz-Quintero B. Deregulated MicroRNAs in cancer-associated fibroblasts from front tumor tissues of lung adenocarcinoma as potential predictors of tumor promotion. Tohoku J Exp Med Oct. 2018;246:107–120. doi: 10.1620/tjem.246.107. [DOI] [PubMed] [Google Scholar]
- 38.Rice MA, Ishteiwy RA, Magani F, Udayakumar T, Reiner T, Yates TJ, Miller P, Perez-Stable C, Rai P, Verdun R, et al. The microRNA-23b/-27b cluster suppresses prostate cancer metastasis via Huntingtin-interacting protein 1-related. Oncogene. 2016;35:4752–4761. doi: 10.1038/onc.2016.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y, Xu X, Zhang M, Wang X, Bai X, Li H, Kan L, Zhou Y, Niu H, He P. MicroRNA-663a is downregulated in non-small cell lung cancer and inhibits proliferation and invasion by targeting JunD. BMC Cancer. 2016;16:315. doi: 10.1186/s12885-016-2350-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng H, Zhang F, Lin X, Huang C, Zhang Y, Li Y, Lin J, Chen W, Lin X. MicroRNA-1225-5p inhibits proliferation and metastasis of gastric carcinoma through repressing insulin receptor substrate-1 and activation of β-catenin signaling. Oncotarget. 2016;7:4647–4663. doi: 10.18632/oncotarget.6615. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article.