Abstract
Infection by the severe acute respiratory syndrome (SARS) coronavirus-2 (SARS-CoV-2) is the cause of the new viral infectious disease (coronavirus disease 2019; COVID-19). Emerging evidence indicates that COVID-19 may be associated with a wide spectrum of neurological symptoms and complications with central nervous system (CNS) involvement. It is now well-established that entry of SARS-CoV-2 into host cells is facilitated by its spike proteins mainly through binding to the angiotensin-converting enzyme 2 (ACE-2). Preclinical studies have suggested that neuropilin-1 (NRP1), which is a transmembrane receptor that lacks a cytosolic protein kinase domain and exhibits high expression in the respiratory and olfactory epithelium, may also be implicated in COVID-19 by enhancing the entry of SARS-CoV-2 into the brain through the olfactory epithelium. In the present study, we expand on these findings and demonstrate that the NRP1 is also expressed in the CNS, including olfactory-related regions such as the olfactory tubercles and paraolfactory gyri. This furthers supports the potential role of NRP1 as an additional SARS-CoV-2 infection mediator implicated in the neurologic manifestations of COVID-19. Accordingly, the neurotropism of SARS-CoV-2 via NRP1-expressing cells in the CNS merits further investigation.
Keywords: neuropilin-1, SARS-CoV-2, COVID-19, brain, central nervous system, neurotropism, neurologic symptoms
Introduction
Infection by the severe acute respiratory syndrome (SARS) coronavirus-2 (SARS-CoV-2) is the cause of a new viral infectious disease, named as coronavirus disease 2019 (COVID-19) (1). Following the initial outbreak of COVID-19 cases at the end of 2019, COVID-19 has reached pandemic status within a few months with rapidly increasing rates of infection and mortality. Thus, as of August 21st, 2020, over 21 million confirmed cases and over 760,000 deaths have been reported worldwide (2). Although COVID-19 is asymptomatic or mild in most cases and manifests primarily as a lower respiratory tract infection which is transmitted mainly via air droplets (3), emerging evidence indicates that SARS-CoV-2 can also invade and attack the central nervous system (CNS) leading to a wide repertoire of neurological symptoms and complications (4–7). As such, increasing research focus has been placed on identifying the mediators that facilitate the SARS-CoV-2 infection in different human organs/tissues.
It is now well-established that entry of SARS-CoV-2 into cells is facilitated by its spike proteins mainly through binding to the angiotensin-converting enzyme 2 (ACE-2) (8,9). Moreover, the SARS-CoV-2 spike viral proteins are primed/activated by the transmembrane protease serine 2 (TMPRSS2), which appears to also play a key role in this viral infection (8,10,11). Accordingly, there is now increasing interest in identifying additional molecular mediators that may also facilitate the SARS-CoV-2 infection of host cells and COVID-19 symptoms, such as the receptor for advanced glycation end products (RAGE), angiotensin II receptor type 2 (AGTR2), CD147 and olfactory receptors (12–15). As such, recent data suggest that another protein/receptor termed neuropilin-1 (NRP1) may also be implicated in the SARS-CoV-2 infection (16–18). NRP1 is a transmembrane receptor which, due to the lack of a cytosolic protein kinase domain, acts primarily as a co-receptor for various ligands and, thus, induces a multitude of effects, including cell proliferation, angiogenesis and axon control (19,20).
As aforementioned, two preclinical studies have indicated that NRP1 may play a role as a new mediator of the SARS-CoV-2 infection (17,18). Indeed, in the first of these studies, NRP1 was identified as a factor for SARS-CoV-2 infection, since, in addition to ACE-2, the spike (S1) proteins of this coronavirus can also bind to NRP1 in vitro (18). Similar results were reported by a second study in which neuropathological analysis of COVID-19 autopsies further revealed that SARS-CoV-2 infected NRP1-positive cells in the olfactory epithelium and bulb (17). Based on these findings, we hypothesise that NRP1 may be further implicated in the CNS involvement and neurological features noted in patients with COVID-19. To this aim, we explored in detail the expression of NRP1 in the human brain.
Materials and methods
To examine the detailed expression of NRP1 in the human brain, we employed a database which measured gene expression through single cell RNA sequencing (RNA-Seq) and, thus, allowed to identify expression in cell types reported as fragments per kilobase of transcript per million mapped (FPKM) reads (21). A second RNA-Seq dataset was also used to examine brain region specific gene expression using Cap Analysis of Gene expression (CAGE) generated by the FANTOM5 project and reported as tags per million (22–24). Human protein expression of NRP1 in the brain was also extracted from the protein atlas database (proteinatlas.org) (23,24). Visualisation of NRP1 expression in the brain was performed using the Allen brain atlas with six human donors as assessed by microarray and presented as a heat map (25).
Results
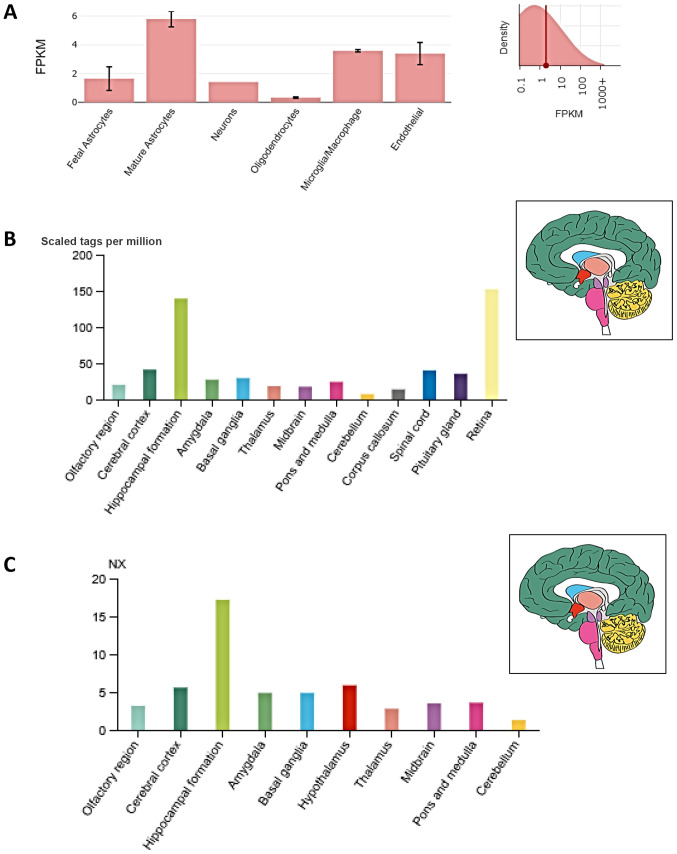
Fig. 1A presents the single cell expression of NRP1 RNA in specific cell types in the human brain. These data reveal that NRP1 expression is highest in mature astrocytes (5.8 FPKM), whilst also showing expression in endothelial cells (3.4 FPKM). Moreover, FANTOM5 data analysis revealed NRP1 expression in the olfactory region (20.4 tags per million), which is higher than in other regions of the brain such as the cerebellum and thalamus (Fig. 1B). Of note, highest expression of NRP1 was noted at both gene and protein level in the hippocampal formation (i.e., the region located in the temporal lobe of each cerebral cortex, medial to the inferior horn of the lateral ventricle) (Fig. 1C).
Figure 1.
NRP1 expression in the human brain. (A) Regarding cell types, based on RNA-Seq data, NRP1 is highly expressed in mature astrocytes (mature astrocytes have multiple functions, including controlling the permeability of the blood brain barrier and maintaining extracellular homeostasis), microglia/macrophages, and endothelial cells. Insert on panel A; FPKM reads. (B) Regarding brain regions, NRP1 exhibits widespread expression in the human brain (including the olfactory regions) based on data from both RNA-Seq expression using CAGE on FANTOM5 and (C) RNA expression (NX) of normalised NRP1 expression levels from 10 brain regions. The insert panel of brain anatomy in panels B and C indicates in colour the different anatomical structures. NRP1, neuropilin-1; RNA-Seq, RNA sequencing; FPKM, fragments per kilobase of transcript per million mapped; CAGE, Cap Analysis of Gene expression; FANTOM5, functional annotation of mammalian genome 5.
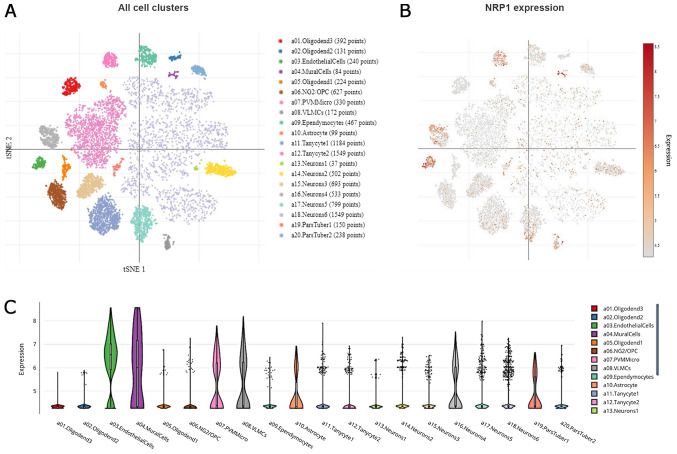
To gain better insight, we further expanded our observations on single cell analysis of the hypothalamic arcuate and median eminence (Arc-ME) complex, using the Single Cell Portal (26). Using T-distributed Stochastic Neighbor Embedding (tSNE), a machine learning algorithm for visualization, distinct cell sub-populations appear to express NRP1 (Fig. 2A and B). More specifically, high expression was noted in endothelial cells, mural cells (vascular smooth muscle cells and pericytes), a specific cluster of neurons, perivascular macrophages (PVM) and microglia, as well as in vascular and leptomeningeal cells (VLMCs), as presented in Fig. 2C.
Figure 2.
Single cell analysis of the hypothalamic Arc-ME complex, using the Single Cell Portal, revealed distinct expression of NRP1. (A) Arc-ME single-cell transcriptomics data are displayed as a spectral tSNE plot of 20,921 cells, coloured per density clustering and annotated according to known cell types. (B) Sub-populations of cell types which are enriched for NRP1 expression, with expression intensity demonstrated by a heat map. (C) Single cell analysis of the hypothalamic Arc-ME complex, using the Single Cell Portal, enriched for population subtypes, represented as violin plots. Arc-ME, arcuate and median eminence; tSNE, T-distributed Stochastic Neighbor Embedding.
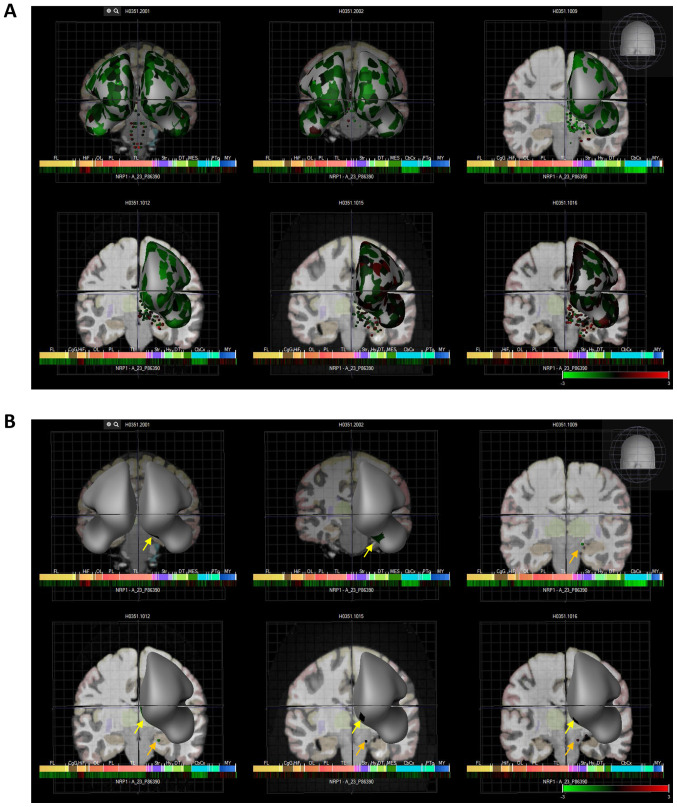
Finally, NRP1 expression is also noted in the paraolfactory gyri of the human brain. Indeed, six patient brains with global NRP1 expression are demonstrated in Fig. 3, as well as the olfactory specific regions of the brain; in humans the olfactory bulb inputs to the olfactory tubercle which in turn outputs to the limbic system (27,28). These data are displayed as heat maps with Log2 expression values presented in Fig. 3 and Table I, and show that NRP1 is expressed in the olfactory tubercles and paraolfactory gyri at reasonable levels in most brains.
Figure 3.
Six human donors and heat map expression of NRP1 as assessed by microarray (Allen brain atlas). (A) Indicates global brain expression of NRP1 as detected by microarray probes, with expression of cerebral areas indicated on the hemispheres and expression in brain nuclei indicated as dots. Heat maps of expression are displayed below individual brains and organised by anterior to posterior regions with the FL, HiF, OL, PL, TL, Str, DT, CbCx and MY marked on the heat map. (B) Shows expression in olfactory accessory tissues with yellow arrows indicating the paraolfactory gyri and orange arrows indicating the olfactory tubercle. NRP1, neuropilin-1; FL, frontal lobe; HiF, hippocampal formation; OL, occipital lobe; PL, parietal lobe; TL, temporal lobe; Str, striatum; DT, dorsal thalamus; CbCx, cerebral cortex, MY, myelencephalon.
Table I.
Log2 expression of NRP1 in the brains of six human donors showing that NRP1 is expressed in the olfactory tubercles and paraolfactory gyri at reasonable levels in most of these brains.
| Brain region | Donor ID | Log2 expression |
|---|---|---|
| Olfactory tubercle | H0351.2001 | Not available |
| H0351.2002 | Not available | |
| H0351.1009 | 6.8 | |
| H0351.1012 | 7.2 | |
| H0351.1015 | 8.9 | |
| H0351.1016 | 9.2 | |
| Paraolfactory gyri | H0351.2001 | 7.8 |
| H0351.2002 | 9.4 | |
| H0351.1009 | Not available | |
| H0351.1012 | 7.0 | |
| H0351.1015 | 10.9 | |
| H0351.1016 | 8.9 |
NRP1, neuropilin-1.
Discussion
This study presents a detailed in silico analysis of the expression of NRP1 in the human brain, highlighting the potential role of NRP1 as an additional SARS-CoV-2 infection mediator in the CNS via NRP1-expressing cells. Our present findings further support the data of recent preclinical studies (16–18), suggesting that NRP1 may be implicated in the neurologic features and CNS involvement of COVID-19.
It is known that human coronaviruses can lead to neurological sequelae, including vision alterations, seizures, motor coordination disturbances and a plethora of other neurological symptoms which may be evidenced with various imaging findings, including encephalopathy, oedema, haemorrhage and ischaemia (29–32). In accord with this, an increasing body of evidence now consistently indicates that SARS-CoV-2 infection may also be associated to a wide spectrum of neurological symptoms and complications from the CNS (3–5,7). Indeed, prominent neurologic features, including encephalopathy, agitation and confusion, as well as corticospinal tract signs, were present in a cohort of 58 consecutive patients (median age: 63 years) admitted to Strasbourg University Hospital with COVID-19 related acute respiratory distress syndrome (ARDS) (33). In addition, in this study single acute ischemic strokes were noted in two of the 13 patients who underwent magnetic resonance imaging (MRI) of the brain. Furthermore, in a retrospective case series study including 214 consecutive hospitalized patients (women/men: 127/87; mean age: 52.7 years) with laboratory-confirmed COVID-19 from Wuhan, China (the first epicentre of COVID-19), almost half (45.5%) of the patients with severe COVID-19 experienced impaired consciousness and other neurologic manifestations (e.g., acute cerebrovascular disease) (5). Overall, in this study neurologic symptoms were noted in more than one third (36.4%) of these 214 hospitalized patients with severe and non-severe COVID-19. This observation is further supported by the findings of a more recent UK study which reported that patients with COVID-19 may exhibit a wide spectrum of neurological syndromes, including encephalopathies with delirium/psychosis, CNS inflammatory syndromes (e.g., encephalitis and encephalomyelitis) and stroke (34). Furthermore, a study from Germany that reported the autopsy findings in six patients who died from COVID-19 with SARS-CoV-2 pneumonia (women/men: 2/4, age range: 58–82 years) noted pronounced CNS involvement with meningitis, panencephalitis and brainstem neuronal cell damage in all these cases (35). Of note, disruption to micro-structural and functional brain integrity in the recovery stages of COVID-19 further suggests longer-term CNS consequences/complications of SARS-CoV-2 infection (36).
So far, it has been hypothesised that SARS-CoV-2 can enter into the CNS through neural or haematogenous propagation, including through the olfactory epithelium of the nasal cavity and the olfactory bulb via retrograde transport along axons of olfactory sensory neurons, as well as via interaction with ACE-2 of the capillary endothelium causing impairment of the blood-brain-barrier (BBB), or due to BBB disruption induced in the context of an underlying cytokine storm (5,37). In addition to these potential mechanisms, Cantuti-Castelvetri et al (17) recently showed that NRP1, which is highly expressed in the respiratory and olfactory epithelium, enhances SARS-CoV-2 entry into the brain, and that this can be inhibited by blocking NRP1 with a monoclonal antibody. Interestingly, NRP1 has been shown to also play a role in mediating cell entry of other viruses, such as the Epstein-Barr virus (EBV) (38). Indeed, NRP1 interacts directly with the EBV glycoprotein B, whilst EBV infection is suppressed upon NRP1 knockdown (38).
Given this newly identified role of NRP1 in enhancing SARS-CoV-2 entry into the CNS, characterizing the precise expression of NRP1 in the human brain becomes important in the context of the neurologic involvement of COVID-19. Our present findings collectively show that NRP1 expression is present in the human brain, including olfactory regions, which corroborates the previously described data showing that NRP1 may mediate entry of SARS-CoV-2 into the brain through the olfactory epithelium/bulb (17). This further highlights the potential importance of the olfactory tubercle for SARS-CoV-2 entry into the brain since in humans the olfactory bulb inputs to the olfactory tubercle which in turn outputs to the limbic system (27,28). Finally, the parolfactory gyri which receive inputs from the olfactory bulb and provide input to the limbic system, also exhibit NRP1 expression, and so their potential involvement in the SARS-CoV-2 infection of the CNS merits further research.
In conclusion, in the present study we demonstrate that the NRP1, which is highly expressed in the respiratory and olfactory epithelium, is also expressed in the CNS, including olfactory related regions such as the olfactory tubercles and paraolfactory gyri. Given the data by Cantuti-Castelvetri et al (17) showing that NRP1 may mediate the entry of SARS-CoV-2 into the brain through the olfactory epithelium (17), our findings warrant further investigation in order to elucidate the potential role of NRP1 as an additional SARS-CoV-2 infection mediator implicated in the neurologic manifestations of COVID-19. Indeed, the neurotropism of SARS-CoV-2 via NRP1-expressing cells in the CNS should be investigated in preclinical and clinical studies, since clarifying how SARS-CoV-2 enters and spreads in the CNS is vital for developing effective approaches to prevent this viral dissemination and its consequent complications.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analysed during this study are included in this published article.
Authors' contributions
JD, EK, IK consulted the literature, produced the figures and manuscript; MH and KC contributed to critical revision of the article; JD, HSR, EK, IK and DAS contributed to the writing of the manuscript and final edits. IK and EK contributed equally to the conception of the work, data and literature analysis as well as interpretation. All authors read and approved the final manuscript. JD is an alumnus of Brunel University London. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
DAS is the Editor-in-Chief for the journal, but had no personal involvement in the reviewing process, or any influence in terms of adjudicating on the final decision, for this article. The other authors declare that they have no competing interests.
References
- 1.Domingo P, Mur I, Pomar V, Corominas H, Casademont J, de Benito N. EBioMedicine. Vol. 58. Elsevier B.V.; 2020. (2020). The four horsemen of a viral Apocalypse: The pathogenesis of SARS-CoV-2 infection (COVID-19) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organisation (WHO), corp-author Coronavirus disease 2019 (COVID-19): Weekly Epidemiological Update. 21 August 2020. www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports. Last accessed August 22, 2020. [Google Scholar]
- 3.Gavriatopoulou M, Korompoki E, Fotiou D, Ntanasis- Stathopoulos I, Psaltopoulou T, Kastritis E, Terpos E, Dimopoulos MA. Organ-specific manifestations of COVID-19 infection. https://doi.org/10.1007/s10238-020-00648-x. Clin Exp Med. doi: 10.1007/s10238-020-00648-x. [DOI] [PMC free article] [PubMed]
- 4.De Felice FG, Tovar-Moll F, Moll J, Munoz DP, Ferreira ST. Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) and the Central Nervous System. Trends Neurosci. 2020;43:355–357. doi: 10.1016/j.tins.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mao L, Jin H, Wang M, Hu Y, Chen S, He Q, Chang J, Hong C, Zhou Y, Wang D, et al. Neurologic manifestations of hospitalized patients with Coronavirus Disease 2019 in Wuhan, China. JAMA Neurol. 2020;77:683. doi: 10.1001/jamaneurol.2020.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mao XY, Jin WL. The COVID-19 Pandemic: Consideration for brain infection. Neuroscience. 2020;437:130–131. doi: 10.1016/j.neuroscience.2020.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Needham EJ, Chou SHY, Coles AJ, Menon DK. Neurological implications of COVID-19 infections. Neurocrit Care. 2020;32:667–671. doi: 10.1007/s12028-020-00978-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu N-H, Nitsche A, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shang J, Wan Y, Luo C, Ye G, Geng Q, Auerbach A, Li F. Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci USA. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iwata-Yoshikawa N, Okamura T, Shimizu Y, Hasegawa H, Takeda M, Nagata N. TMPRSS2 contributes to virus spread and immunopathology in the airways of murine models after coronavirus infection. J Virol. 2019;93:e01815–18. doi: 10.1128/JVI.01815-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Katopodis P, Anikin V, Randeva HS, Spandidos DA, Chatha K, Kyrou I, Karteris E. Pan-cancer analysis of transmembrane protease serine 2 and cathepsin L that mediate cellular SARS-CoV-2 infection leading to COVID-19. Int J Oncol. 2020;57:533–539. doi: 10.3892/ijo.2020.5071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cui C, Huang C, Zhou W, Ji X, Zhang F, Wang L, Zhou Y, Cui Q. AGTR2, one possible novel key gene for the entry of SARS-CoV-2 into human cells. IEEE/ACM Trans Comput Biol Bioinformatics. 2020 Jul 14; doi: 10.1109/TCBB.2020.3009099. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 13.Kerkeni M, Gharbi J. RAGE receptor: May be a potential inflammatory mediator for SARS-COV-2 infection? Med Hypotheses. 2020;144:109950. doi: 10.1016/j.mehy.2020.109950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kerslake R, Hall M, Randeva HS, Spandidos DA, Chatha K, Kyrou I, Karteris E. Co-expression of peripheral olfactory receptors with SARS-CoV-2 infection mediators: Potential implications beyond loss of smell as a COVID-19 symptom. Int J Mol Med. 2020;46:949–956. doi: 10.3892/ijmm.2020.4646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xia P, Dubrovska A. Tumor markers as an entry for SARS CoV 2 infection? FEBS J. 2020 Aug 1; doi: 10.1111/febs.15499. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bittmann S, Weissenstein A, Moschüring-Alieva E, Villalon G, Villalon G. Neuropilin-1 in transmission process of COVID-19. J Regen Biol Med. 2020;2:1–2. [Google Scholar]
- 17.Cantuti-Castelvetri L, Ojha R, Pedro L, Djannatian M, Franz J, Kuivanen S, Kallio K, Kaya T, Anastasina M, Smura T, et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and provides a possible pathway into the central nervous system. https://doi.org/10.1101/2020.06.07.137802. bioRxiv. doi: 10.1126/science.abd2985. [DOI] [PMC free article] [PubMed]
- 18.Daly JL, Simonetti B, Antón-Plágaro C, Kavanagh Williamson M, Shoemark DK, Simón-Gracia L, Klein K, Bauer M, Hollandi R, Greber UF, et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. https://doi.org/10.1101/2020.06.05.134114. bioRxiv. doi: 10.1126/science.abd3072. [DOI] [PMC free article] [PubMed]
- 19.Guo HF, Vander Kooi CW. Neuropilin Functions as an Essential Cell Surface Receptor. J Biol Chem. 2015;290:29120–29126. doi: 10.1074/jbc.R115.687327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roy S, Bag AK, Singh RK, Talmadge JE, Batra SK, Datta K. Multifaceted role of neuropilins in the immune system: potential targets for immunotherapy. Front Immunol. 2017;8:1228. doi: 10.3389/fimmu.2017.01228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y, Sloan SA, Clarke LE, Caneda C, Plaza CA, Blumenthal PD, Vogel H, Steinberg GK, Edwards MSB, Li G, et al. Purification and characterization of progenitor and mature human astrocytes reveals transcriptional and functional differences with mouse. Neuron. 2016;89:37–53. doi: 10.1016/j.neuron.2015.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Forrest AR, Kawaji H, Rehli M, Baillie JK, de Hoon MJ, Haberle V, Lassmann T, Kulakovskiy IV, Lizio M, Itoh M, et al. FANTOM Consortium and the RIKEN PMI and CLST (DGT) A promoter-level mammalian expression atlas. Nature. 2014;507:462–470. doi: 10.1038/nature13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thul PJ, Åkesson L, Wiking M, Mahdessian D, Geladaki A, Ait Blal H, Alm T, Asplund A, Björk L, Breckels LM, et al. A subcellular map of the human proteome. Science. 2017;356:eaal3321. doi: 10.1126/science.aal3321. [DOI] [PubMed] [Google Scholar]
- 24.Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419–1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 25.Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, Shen EH, Ng L, Miller JA, van de Lagemaat LN, Smith KA, Ebbert A, Riley ZL, et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489:391–399. doi: 10.1038/nature11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Campbell JN, Macosko EZ, Fenselau H, Pers TH, Lyubetskaya A, Tenen D, Goldman M, Verstegen AMJ, Resch JM, McCarroll SA, et al. A molecular census of arcuate hypothalamus and median eminence cell types. Nat Neurosci. 2017;20:484–496. doi: 10.1038/nn.4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ikemoto S. Dopamine reward circuitry: Two projection systems from the ventral midbrain to the nucleus accumbens - olfactory tubercle complex. Brain Res Brain Res Rev. 2007;56:27–78. doi: 10.1016/j.brainresrev.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wesson DW, Wilson DA. Sniffing out the contributions of the olfactory tubercle to the sense of smell: Hedonics, sensory integration, and more? Neurosci Biobehav Rev. 2011;35:655–668. doi: 10.1016/j.neubiorev.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arabi YM, Harthi A, Hussein J, Bouchama A, Johani S, Hajeer AH, Saeed BT, Wahbi A, Saedy A, Al Dabbagh T, et al. Severe neurologic syndrome associated with Middle East respiratory syndrome corona virus (MERS-CoV) Infection. 2015;43:495–501. doi: 10.1007/s15010-015-0720-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hung ECW, Chim SSC, Chan PKS, Tong YK, Ng EKO, Chiu RWK, Leung C-B, Sung JJY, Tam JS, Lo YMD. Detection of SARS coronavirus RNA in the cerebrospinal fluid of a patient with severe acute respiratory syndrome. Clin Chem. 2003;49:2108–2109. doi: 10.1373/clinchem.2003.025437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lau KK, Yu WC, Chu CM, Lau ST, Sheng B, Yuen KY. Possible central nervous system infection by SARS coronavirus. Emerg Infect Dis. 2004;10:342–344. doi: 10.3201/eid1002.030638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li K, Wohlford-Lenane C, Perlman S, Zhao J, Jewell AK, Reznikov LR, Gibson-Corley KN, Meyerholz DK, McCray PB., Jr Middle East respiratory syndrome coronavirus causes multiple organ damage and lethal disease in mice transgenic for human dipeptidyl peptidase 4. J Infect Dis. 2016;213:712–722. doi: 10.1093/infdis/jiv499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Helms J, Kremer S, Merdji H, Clere-Jehl R, Schenck M, Kummerlen C, Collange O, Boulay C, Fafi-Kremer S, Ohana M, et al. Neurologic features in severe SARS-CoV-2 infection. N Engl J Med. 2020;382:2268–2270. doi: 10.1056/NEJMc2008597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paterson RW, Brown RL, Benjamin L, Nortley R, Wiethoff S, Bharucha T, Jayaseelan DL, Kumar G, Raftopoulos RE, Zambreanu L, et al. UCL Queen Square National Hospital for Neurology and Neurosurgery COVID-19 Study Group The emerging spectrum of COVID-19 neurology: Clinical, radiological and laboratory findings. Brain. 2020 Jul 8; doi: 10.1093/brain/awaa240. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.von Weyhern CH, Kaufmann I, Neff F, Kremer M. Early evidence of pronounced brain involvement in fatal COVID-19 outcomes. Lancet. 2020;395:e109. doi: 10.1016/S0140-6736(20)31282-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu Y, Li X, Geng D, Mei N, Wu P-Y, Huang C-C, Jia T, Zhao Y, Wang D, Xiao A, et al. Cerebral micro-structural changes in COVID-19 patients - an MRI-based 3-month follow-up study. EClinicalMedicine. 2020;25:100484. doi: 10.1016/j.eclinm.2020.100484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Conde Cardona G, Quintana Pájaro LD, Quintero Marzola ID, Ramos Villegas Y, Moscote Salazar LR. Neurotropism of SARS-CoV 2: Mechanisms and manifestations. J Neurol Sci. 2020;412:116824. doi: 10.1016/j.jns.2020.116824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang HB, Zhang H, Zhang JP, Li Y, Zhao B, Feng GK, Du Y, Xiong D, Zhong Q, Liu WL, et al. Neuropilin 1 is an entry factor that promotes EBV infection of nasopharyngeal epithelial cells. Nat Commun. 2015;6:6240. doi: 10.1038/ncomms7240. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analysed during this study are included in this published article.