Abstract
Mechanical ventilation (MV) and lipopolysaccharide (LPS) infection are common causes of acute lung injury. The aim of the present study was to identify the key genes and potential mechanisms involved in mechanical ventilation (MV) and lipopolysaccharide (LPS)-induced acute lung injury (ALI). Gene expression data of adult C57BL/6 mice with ALI induced by inhaling LPS, MV and LPS + MV were downloaded from the Gene Expression Omnibus database. Differentially expressed genes (DEGs) associated with MV, LPS and LPS + MV were screened, followed by functional enrichment analysis, protein-protein interaction network construction, and prediction of transcription factors and small molecule drugs. Finally, the expression of key genes was verified in vivo using reverse transcription-quantitative PCR. A total of 63, 538 and 1,635 DEGs were associated with MV, LPS and LPS + MV, respectively. MV-associated genes were significantly enriched in the ‘purine ribonucleotide metabolic process’. LPS and LPS + MV-associated genes were significantly enriched in ‘cellular response to cytokine stimulus’ and ‘cell chemotaxis’. All three conditions were enriched in ‘TNF signaling pathway’ and ‘IL-17 signaling pathway’. Expression levels of C-X-C motif chemokine ligand (CXCL)2, CXCL3 and CXCL10 were upregulated in the LPS and LPS + MV groups. Adenosine A2b receptor, zinc finger and BTB domain-containing 16 and hydroxycarboxylic acid receptor 2 were identified as DEGs in the MV group. Compared with the control group, Early growth response 1 and activating TF 3 was upregulated in all three groups. Similarities and differences were observed among the MV- and LPS-induced ALI, and MV may enhance the effects of LPS on gene expression. MV may affect urine ribonucleotide metabolic-related processes, whereas LPS may cause cell chemotaxis and cytokine stimulus responses in ALI progression. The inflammatory response was shared by MV and LPS. The results of the present study may provide insight into a theoretical basis for the study and treatment of ALI.
Keywords: acute lung injury, mechanical ventilation, lipopolysaccharide, chemokines, inflammatory response
Introduction
Acute lung injury (ALI) and acute respiratory distress syndrome, which is an intricate cascade process that develops from ALI, are multifactorial clinical disorders with a high incidence that frequently cause acute respiratory failure and subsequent death (1,2). The major characteristic of ALI is an exaggerated inflammatory response, which may lead to alveolar capillary destruction, respiratory failure and even multiple organ failure (3,4). ALI can be caused by several factors; for example, aspiration of low-pH gastric fluid (acid aspiration) can induce an inflammatory pulmonary reaction associated with neutrophilic infiltration and alveolar epithelial damage, termed aspiration pneumonitis, which may be followed by infection (5). In addition, sepsis, transfusion, trauma and other factors may lead to ALI (6). Several animal models, such as sepsis or mechanical ventilation-induced acute lung injury, have been established to explore the mechanism and treatment of ALI (7,8).
LPS is a component of the cell walls of gram-negative bacteria (9). LPS causes microvascular lung injury, increases macrophage and neutrophil presence, and induces pulmonary inflammation in animals and humans (10). The feasibility of analyzing the pathogenesis of ALI with an in vitro ALI model induced by LPS has been verified in previous studies (11,12). Zhang et al (13) have reported that amygdalin treatment decreases inflammatory cell infiltration and the release of inflammatory cytokines, and represses the activation of nuclear factor-κB (NF-κB) and NLR family pyrin domain containing 3 in LPS-induced ALI mice, exerting a protective effect. Pedrazza et al (14) have demonstrated that decreases in inflammation, oxidative damage and the formation of neutrophil extracellular traps are observed in an LPS-induced ALI model following treatment with mesenchymal stem cells, as well as an increased survival curve. Mechanical ventilation (MV) is a necessary treatment for patients with respiratory dysfunction (15,16). However, the ventilation effect and oxygenation provided by MV and the negative side effects should be considered when using MV (17). Lung injury caused by MV has attracted increasing attention from the medical community (18). The occurrence of such lung injury is not only associated with high airway pressure, but also with ventilation capacity, ventilation mode and inhaled oxygen concentration (19,20). Chian et al (21) have demonstrated that an inflammatory reaction and NF-κB activation are implicated in ventilator-induced lung injury, and treatment with an inhibitory peptide of NF-κB (SN50) reduces such lung injury. Despite previous advances, the specific mechanisms underlying LPS-induced and ventilator-induced ALI remain unclear.
In clinical practice, a number of patients with sepsis-related lung injury often require the support of a ventilator, and MV may further aggravate lung injury, resulting in a potential exacerbation of the condition or an additional condition in the patients (22). However, the differences between LPS-induced lung injury and MV-induced lung injury, as well as the effects of the combination of the two means of inducing lung injury have not been studied previously. To assess this, the gene expression dataset GSE18341 of ALI in adult C57BL/6 mice was used to analyze the underlying mechanisms in the present study. The genes that were differentially expressed in response to LPS, MV and LPS + MV treatment were screened, followed by functional enrichment analysis, construction of interaction networks, and prediction of transcription factors (TFs) and small molecule drugs. The results of the present study may provide novel insight into a theoretical basis for the study and treatment of ALI.
Materials and methods
Dataset
The gene expression dataset GSE18341 of ALI in adult C57BL/6 mice caused by inhaling LPS, MV and LPS combined with MV were downloaded from the Gene Expression Omnibus (GEO; http://ncbi.nlm.nih.gov/geo/) (23). A total of 4 groups of data in the dataset were selected to analyze the possible mechanisms and key genes of ALI induced by mechanical ventilation and LPS, including differential, Venn diagram, enrichment, protein-protein interaction (PPI) network and transcription factor (TF) prediction analysis, as well as drug target gene interaction network construction, in order to verify the associated genes and signaling pathways.
Data source and preprocessing
To clarify the differences and relationship between LPS- and MV-induced lung injury, as well as the effects of the combination of the two factors on lung tissue, a total of 16 samples were divided into 4 groups (n=4 samples/group): i) MV group (treated with MV for 2 h); ii) LPS group (treated with inhaled LPS, then spontaneous breathing for 2 h); iii) MV + LPS group (treated with inhaled LPS, then MV for 2 h); and iv) control group (untreated, spontaneously breathing). The data were collected on the GPL1261 [Mouse430_2] Affymetrix Mouse Genome 430 2.0 Array. The raw data were preprocessed using the RMA method in the affy package (Version 1.50.0; http://bioconductor.org/packages/release/bioc/html/affy.html) used with R (version 3.3.3) (24), including background correction, normalization and expression calculation. The probes were matched to gene symbols based on the annotations file, and probes without matched gene symbols were removed, whereas the mean expression value was selected as the final expression value when multiple probes matched to one gene symbol. The data were selected to analyze the possible mechanism and key genes of ALI induced by MV and LPS, and the identified genes were verified using reverse transcription-quantitative (RT-q)PCR.
Differential expression and Venn analysis
Differential expression analysis of MV vs. control, LPS vs. control, MV + LPS vs. control, MV + LPS vs. MV and MV + LPS vs. LPS was performed using the limma package (version 3.10.3; www.bioconductor.org/packages/2.9/bioc/html/limma.html) to obtain the corresponding P-value and log fold change (FC); then, the adjusted P-value (adj.P) was obtained using the Benjamini and Hochberg test. The differentially expressed genes (DEGs) were screened with a threshold of adj.P<0.05 and |log FC|>0.585. The DEGs in each compared group were used to perform the Venn analysis. Specifically, the overlapping genes in the MV vs. control, MV + LPS vs. Control and MV + LPS vs. LPS groups were considered to be genes associated with MV. Similarly, the overlapping genes in LPS vs. control, MV + LPS vs. control and MV + LPS vs. MV were considered to be genes associated with LPS. Notably, the DEGs in MV + LPS vs. control were considered to comprise the gene response to LPS combined with MV. The Venn diagrams were plotted using an online tool (http://bioinformatics.psb.ugent.be/webtools/Venn/).
Functional enrichment analysis
ClusterProfiler (version 3.8.1; bioconductor.org/packages/release/bioc/html/clusterProfiler.html) in R was used to perform functional enrichment analysis of the genes associated with MV, LPS or the gene response to LPS combined with MV, including identification of the biological processes in Gene Ontology (GO_BP) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. The terms with P<0.05 and an enriched gene count ≥2 were selected as significantly enriched results.
PPI network
The interactions between coding proteins of DEGs associated with MV, LPS or the gene response to LPS combined with MV were predicted using Search Tool for the Retrieval of Interacting Genes/Proteins (version 10.0; string-db.org/) with the following parameters: Species, Mus musculus; and PPI score, 0.9, highest confidence. The PPI networks associated with MV, LPS and MV + LPS were visualized using Cytoscape (version 3.4.0, chianti.ucsd.edu/cytoscape-3.4.0/). Subsequently, CytoNCA (version 2.1.6, apps.cytoscape.org/apps/cytonca) was used to analyze the topological properties of the nodes in the PPI networks, and the parameter was set to ‘without weight’.
Prediction of TFs
The TFs of the DEGs were predicted using TRRUST (version 2, www.grnpedia.org/trrust/). The TF-gene interaction pairs with P-values <0.05 were selected to construct the regulatory network using Cytoscape.
Drug-gene interaction prediction
Drug targets of the DEGs were predicted using The Drug Gene Interaction database (dgidb.org/search_interactions) with the parameter ‘FDA approved’. Drug-gene interaction pairs with references and known interaction type were selected, and drug-gene interaction networks were visualized using Cytoscape.
Experimental animals
A total of 28 male C57BL/6 mice (age, 7–9 weeks; weight, 24–28 g) were purchased from Shanghai SLAC Laboratory Animal Co., Ltd. Mice were randomly divided into four groups. 28 mice were randomly divided into four groups. Mice were housed in an antigen- and virus-free room at 25°C with 40–70% humidity and ad libitum access to food and water under a natural day/night cycle. All animal experiments were approved by the Ethics Committee of Experimental Animals of Shanghai Jiaotong University School of Medicine. Based on the results of the bioinformatics analysis, the expression levels of the identified genes associated with MV, LPS or MV + LPS response were verified in mice in four different groups. For the MV group, after being anesthetized with intraperitoneal injection of ketamine (75 mg/kg) and xylazine (10 mg/kg), the mice were orotracheally intubated with 20 G arterial cannule (BD Pharmingen, San Jose, CA), and mechanically ventilated (Inspira, Harvard Apparatus, Boston, MA) with 20 ml/kg at 70 breaths per minute for 2 h. For the LPS group, after anesthesia, mice were administered a single intratracheal dose of purified LPS extracted from the membrane of Escherichia coli 0111:B4 (Sig-maeAldrich, St. Louis, MO, USA) at 5 mg/kg in a total volume of 50 µl for 2 h. For MV + LPS group, after anesthesia, mice were treated with 5 mg/kg LPS and then were ventilated with 20 ml/kg tidal volume for 2 h. Control mice underwent incubation but breathed spontaneously. Lungs were harvested at the time points indicated.
RT-qPCR verification
Total RNA was isolated from the lung tissues using TRIzol® reagent (cat. no. 9109; Takara Bio, Inc.), and the RNA concentration and quality were determined by detecting absorbance at a wavelength of 260 nm. Subsequently, reverse transcription (25°C for 10 min, 42°C for 1 h, 85°C for 10 min, 4°C hold) of total RNA was performed using PrimeScript™ RT Master mix (cat. no. RR036A; Takara Bio, Inc.), followed by qPCR using Power SYBR® Green PCR Master mix (Roche Diagnostics) with the following thermocycling conditions: 50°C for 2 min; 95°C for 10 min; followed by 40 cycles of 95°C for 10 sec and 58–62°C for 30 sec. The melting curve was analyzed between 60 and 95°C at an incremental rate of 0.5°C/10 sec. The primer sequences are presented in Table SI. β-actin was measured as an internal control. The relative expression levels of genes were calculated using the 2−ΔΔCq method (25). RT-qPCR was repeated 6 times to obtain statistical values.
ELISA verification
The left upper lung tissues were homogenized in PBS, centrifuged for 10 min at 4,000 × g at 4°C and sonicated in 1 ml PBS containing protease inhibitors (2 mM phenylmethylsulfonyl fluoride and 1 µg/ml each antipain, leupeptin, and pepstatin A). IL-6 (cat. no. F10830), C-X-C motif chemokine ligand (CXCL)2 (cat. no. F11170), CXCL3 (cat. no. F10244), CXCL10 (cat. no. F10933) and TNF-α (cat. no. F11630) protein levels in lung tissue homogenates were measured using commercially available ELISA kits (Shanghai Westang Bio-Tech Co., Ltd.) according to the manufacturer's protocol. ELISAs were repeated 7 times to obtain statistical values.
Lung histopathological examination
Lung tissues were fixed using 10% formalin for 24 h at room temperature and embedded in paraffin for histopathological analysis; 4-µm sections were cut and stained with hematoxylin and eosin (H&E). The total staining of each slide was scored by two blinded expert lung pathologists. The criteria for scoring lung inflammation were set as previously described (26): 0, normal tissue; 1, minimal inflammatory change; 2, mild to moderate inflammatory changes (no obvious damage to the lung architecture); 3, moderate inflammatory injury (thickening of the alveolar septae); 4, moderate to severe inflammatory injury (formation of nodules or areas of pneumonitis that distorted the normal architecture); and 5, severe inflammatory injury with total obliteration of the field. Histopathological examination was repeated 7 times.
Statistical analysis
Data are presented as the mean ± standard error of the mean. Statistical significance was calculated using a Student's t-test or one-way ANOVA followed by the Bonferroni's post hoc test. P<0.05 was considered to indicate a statistically significant difference. Statistical analyses were performed using SPSS software (version 16.0.1; SPSS, Inc.).
Results
Data preprocessing
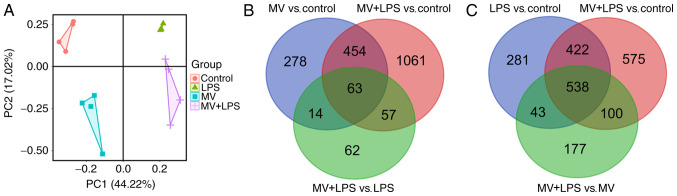
A 21,499-gene expression matrix was obtained according to the method described above, and principal component analysis (PCA) was performed based on this gene expression matrix. As presented in Fig. 1A, the samples from each group were clustered, suggesting that LPS, MV and MV + LPS had notable effects on lung injury.
Figure 1.
Principal component and Venn diagram analysis. (A) Principal component analysis was performed based on the gene expression matrix. (B) Venn diagram analysis identified 63 overlapping DEGs related to MV. (C) Venn diagram analysis identified a total of 538 overlapping DEGs related to LPS. DEG, differentially expressed gene; MV, mechanical ventilation; LPS, lipopolysaccharide; PC1, principal component 1; PC2, principal component 2.
Differential expression and Venn diagram analysis
A total of 809, 1,284, 1,635, 858 and 196 DEGs were identified between MV vs. control, LPS vs. control, MV + LPS vs. control, MV + LPS vs. MV and MV+LPS vs. LPS, respectively (Table I). The 1,635 genes that were differentially expressed in samples with MV + LPS treatment compared with the control were considered to be the genes responsive to LPS combined with MV. Following Venn diagram analysis, 63 overlapping DEGs among MV vs. control, MV + LPS vs. control and MV + LPS vs. LPS were selected as the genes associated with MV (Fig. 1B). Similarly, the 538 overlapping DEGs among LPS vs. control, MV + LPS vs. control and MV + LPS vs. MV were considered to be the genes associated with LPS (Fig. 1C).
Table I.
Results of DEG analysis.
| DEGs | LPS vs. control | MV vs. control | MV + LPS vs. control | MV + LPS vs. LPS | MV + LPS vs. MV |
|---|---|---|---|---|---|
| Upregulated | 688 | 187 | 824 | 121 | 721 |
| Downregulated | 596 | 622 | 811 | 75 | 137 |
| Total | 1284 | 809 | 1635 | 196 | 858 |
DEG, differentially expressed gene; MV, mechanical ventilation; LPS, lipopolysaccharide.
Functional enrichment analysis
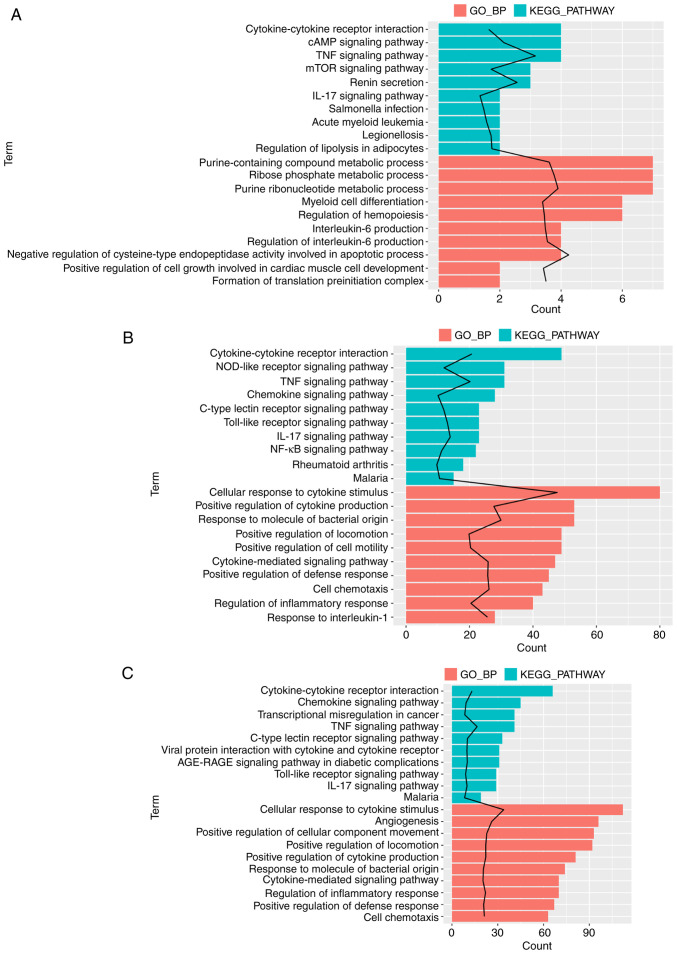
Enrichment analysis was performed for the aforementioned DEGs associated with MV, LPS or the gene response to LPS combined with MV to determine their functions. The gene set associated with MV was significantly enriched in genes associated with 110 GO_BP terms and 12 KEGG pathways; among these, the GO_BP term ‘negative regulation of cysteine-type endopeptidase activity involved in apoptotic process’ was the most significant (Fig. 2A). The gene sets associated with LPS were significantly enriched in genes associated with 268 GO_BP terms and 70 KEGG pathways (Fig. 2B), and the MV + LPS gene set was significantly enriched in genes associated with 339 GO_BP terms and 90 KEGG pathways (Fig. 2C). The term ‘cellular response to cytokine stimulus’ was the most significant in the DEGs associated with LPS and MV + LPS. Fig. 2 demonstrates the top 10 significantly enriched GO_BP terms and KEGG pathways arranged by P-value. These DEG sets were all significantly enriched in genes associated with the GO_BP terms and KEGG pathways involved in the inflammatory response, including the ‘TNF signaling pathway’, ‘IL-17 signaling pathway’, ‘cellular response to cytokine stimulus’ and ‘regulation of the inflammatory response’, amongst others (Fig. 2).
Figure 2.
Top 10 significantly enriched GO_BP terms and KEGG pathways arranged by P-value. (A) The top 10 significantly enriched GO_BP terms and KEGG pathways associated with MV. (B) The top 10 significantly enriched GO_BP terms and KEGG pathways associated with LPS. (C) The top 10 significantly enriched GO_BP terms and KEGG pathways associated with the response to LPS +MV. Black line indicates -log10 (P-value). GO, Gene Ontology; BP, biological process; KEGG, Kyoto Encyclopedia of Genes and Genomes; MV, mechanical ventilation; LPS, lipopolysaccharide.
Construction of the PPI network
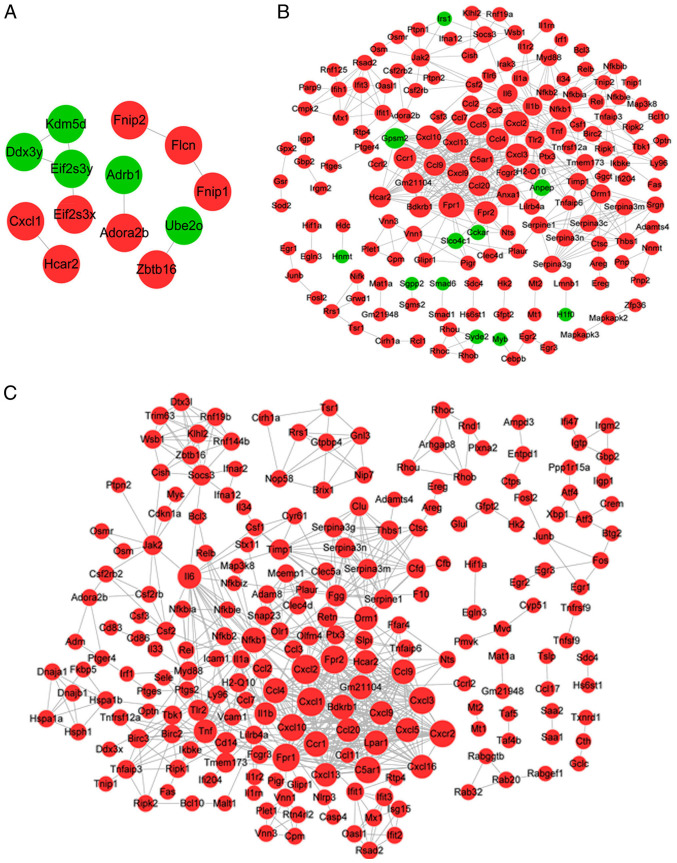
For the genes associated with MV, the PPI network contained 13 genes and 9 interactions, among which 8 genes were upregulated and 5 were downregulated (Fig. 3A). For example, adenosine A2b receptor (ADORA2B), zinc finger and BTB domain containing 16 (ZBTB16) and hydroxycarboxylic acid receptor 2 (HCAR2) were upregulated.
Figure 3.
PPI networks. (A) The PPI network for genes related to MV. (B) The PPI network for genes related to LPS. (C) The PPI network for genes related to LPS + MV. Red indicates upregulation, green indicates downregulation, black lines indicate PPI, and node size indicates connectivity. PPI, protein-protein interaction; MV, mechanical ventilation; LPS, lipopolysaccharide.
For the genes associated with LPS, the PPI network comprised 166 genes, including 11 downregulated and 155 upregulated genes, and 472 interactions (Fig. 3B). The topological properties of the nodes (top 20, arranged by score) are listed in Table II. Formyl peptide receptor (FPR)1, FPR2, CXCL2, CXCL3, CXCL10 and nuclear factor κB subunit 1 (NFKB1), tumor necrosis factor (TNF) and interleukin 6 (IL6) were identified as the hub nodes in the PPI network as their degree, betweenness and closeness scores were all in the top 20.
Table II.
Top 20 genes in the PPI network of genes associated with lipopolysaccharide treatment.
| A, Top 20 genes based on degree | |
|---|---|
| Gene | Degree |
| Fpr1a | 27 |
| Cxcl2a | 26 |
| Cxcl10a | 24 |
| C5ar1 | 23 |
| Fpr2a | 22 |
| Cxcl3a | 21 |
| Ccr1 | 20 |
| Ccl4 | 20 |
| Bdkrb1 | 19 |
| Ccl9 | 19 |
| Anxa1 | 19 |
| Ccl5 | 19 |
| Nfkb1a | 19 |
| Gm21104 | 18 |
| Gpsm2 | 17 |
| Hcar2 | 17 |
| Cxcl13 | 17 |
| Ccl20 | 17 |
| Il6a | 15 |
| Tnfa | 15 |
| B, Top 20 genes based on betweenness | |
| Gene | Betweenness |
| Il6a | 3,750.5457 |
| Cxcl10a | 2,844.6838 |
| Nfkb1a | 2,505.5789 |
| Tnfa | 2,152.0164 |
| Ifit1 | 1,969.5000 |
| Fpr1a | 1,809.1663 |
| Cxcl2a | 1,700.6371 |
| Orm1 | 1,651.6079 |
| Jak2 | 1,479.5116 |
| Tlr2 | 1,104.2716 |
| Socs3 | 1,070.4786 |
| Csf2 | 1,046.3145 |
| Cxcl3a | 813.3837 |
| Il1b | 801.5933 |
| Vnn1 | 690.0000 |
| Timp1 | 662.3790 |
| Tnfaip3 | 609.1126 |
| Fpr2a | 561.2064 |
| Tmem173 | 525.8387 |
| Tbk1 | 519.1886 |
| C, Top 20 genes based on closeness | |
| Gene | Closeness |
| Cxcl2a | 0.02045 |
| Il6a | 0.02043 |
| Cxcl10a | 0.02042 |
| Tnfa | 0.02040 |
| Nfkb1a | 0.02039 |
| Ccl5 | 0.02037 |
| Cxcl3a | 0.02036 |
| Tlr2 | 0.02035 |
| Il1b | 0.02035 |
| Ccl4 | 0.02035 |
| Fpr1a | 0.02034 |
| Ccl2 | 0.02032 |
| C5ar1 | 0.02032 |
| Ccr1 | 0.02031 |
| Fpr2a | 0.02030 |
| Bdkrb1 | 0.02029 |
| Ccl9 | 0.02029 |
| Anxa1 | 0.02029 |
| Gm21104 | 0.02028 |
| Orm1 | 0.02028 |
In the top 20 by degree, betweenness and closeness values.
For the gene response to LPS combined with MV, the PPI network contained 216 genes and 679 interactions, and all the genes were upregulated (Fig. 3C). Table III displays the top 20 nodes; CXCL2, CXCL3, CXCL10, CXCL11, FPR1, FPR2, TNF, IL6, NFKB1 and orosomucoid 1 (ORM1) were the hub nodes in the PPI network as their degree, betweenness and closeness scores were all in the top 20. Notably, most of the hub genes in the LPS and MV + LPS PPI networks were consistent, with the exception of CXCL11 and ORM1.
Table III.
Top 20 genes in the PPI network of genes associated with lipopolysaccharide treatment and mechanical ventilation.
| A, Top 20 genes based on degree | |
|---|---|
| Gene | Degree |
| Cxcl2a | 31 |
| Cxcl1a | 30 |
| Fpr1a | 28 |
| Fpr2a | 28 |
| Cxcl3a | 26 |
| Cxcr2 | 26 |
| Cxcl10a | 26 |
| C5ar1 | 25 |
| Ccr1 | 23 |
| Ccl4 | 22 |
| Nfkb1a | 22 |
| Bdkrb1 | 21 |
| Lpar1 | 21 |
| Ccl9 | 21 |
| Gm21104 | 20 |
| Cxcl5 | 20 |
| Il6a | 20 |
| Orm1a | 19 |
| Tnfa | 19 |
| Cxcl16 | 19 |
| B, Top 20 genes based on betweenness | |
| Gene | Betweenness |
| Il6a | 6,930.810 |
| Cxcl10a | 3,350.056 |
| Nfkb1a | 3,270.180 |
| Tlr2 | 2,781.188 |
| Tnfa | 2,772.925 |
| Socs3 | 2,672.574 |
| Fpr1a | 2,494.769 |
| Il1b | 2,181.053 |
| Cxcl2a | 2,124.261 |
| Ifit1 | 2,034.400 |
| Jak2 | 2,021.862 |
| Fpr2a | 1,942.306 |
| Cxcl1a | 1,859.115 |
| Csf2 | 1,650.456 |
| Orm1a | 1,555.060 |
| Hspa1b | 1,473.000 |
| Fgg | 1,185.236 |
| Vnn1 | 1,184.000 |
| Tbk1 | 801.417 |
| Cxcl3a | 750.258 |
| C, Top 20 genes based on closeness | |
| Gene | Closeness |
| Il6a | 0.01541 |
| Cxcl2a | 0.01541 |
| Cxcl1a | 0.01539 |
| Cxcl10a | 0.01538 |
| Nfkb1a | 0.01538 |
| Tnfa | 0.01538 |
| Tlr2 | 0.01536 |
| Il1b | 0.01536 |
| Cxcl3a | 0.01535 |
| Ccl4 | 0.01535 |
| Fpr1a | 0.01534 |
| Ccl2 | 0.01534 |
| Cxcr2 | 0.01534 |
| C5ar1 | 0.01534 |
| Ccl3 | 0.01533 |
| Cxcl5 | 0.01532 |
| Ccr1 | 0.01532 |
| Fpr2a | 0.01532 |
| Orm1a | 0.01531 |
| Bdkrb1 | 0.01531 |
In the top 20 by degree, betweenness and closeness values.
Prediction of TFs
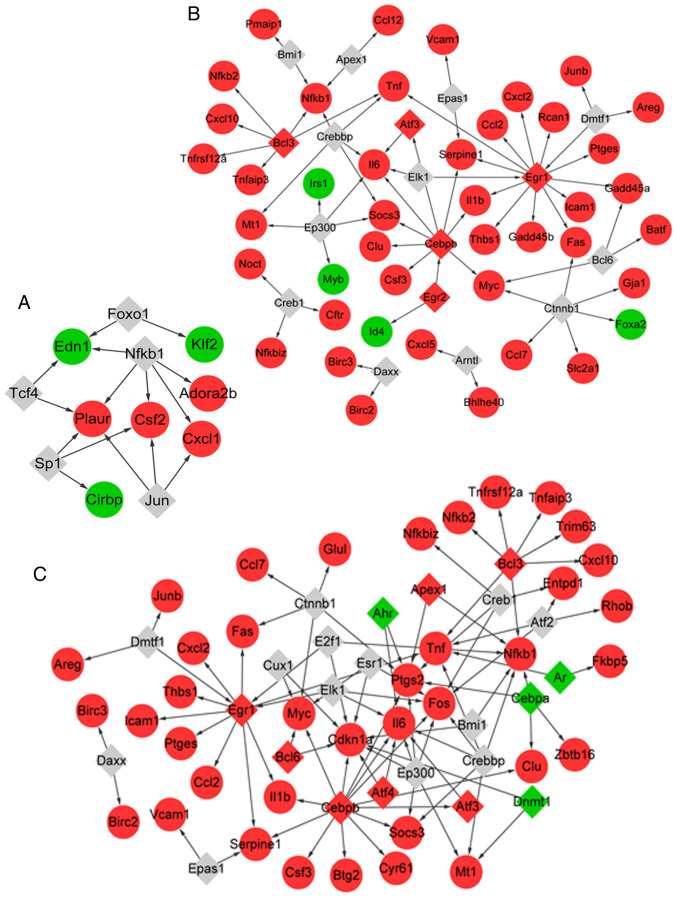
The TFs of the genes associated with MV were predicted. In total, 15 TF-target pairs were obtained (Fig. 4A), among which five TFs (such as NFKB1) were predicted to target seven genes (such as ADORA2B). Similarly, 17 TFs were predicted to target 46 genes associated with LPS, comprising 67 TF-target pairs (Fig. 4B). A total of five out of the 17 TFs were differentially expressed in samples with LPS treatment vs. control, including early growth response 1 (EGR1), activating TF 3 (ATF3) and BCL3 transcription coactivator (BCL3).
Figure 4.
TF-target interaction networks. (A) The TF-target interaction network for genes related to MV. (B) The TF-target interaction network for genes related to LPS. (C) The TF-target interaction network for genes related to LPS + MV. Gray diamonds indicate predicted TFs, red diamond indicate upregulated TFs, red circles indicate upregulated target genes, green circles indicate downregulated target genes, and black arrows indicate target genes regulated by the TFs. TF, transcription factor; MV, mechanical ventilation; LPS, lipopolysaccharide.
In addition, a total of 24 TFs were predicted to target 40 genes that responded to LPS combined with MV, which comprised 91 TF-target pairs (Fig. 4C). In total, 11 out of the 24 TFs were differentially expressed in samples from animals treated with MV + LPS, including aryl hydrocarbon receptor (AHR) and androgen receptor (AR).
Prediction of drug-gene interactions
For the genes associated with MV, 27 drugs were predicted to target 5 genes, including 4 upregulated and 1 downregulated gene (Fig. 5A). A total of 23 drugs were identified to target adrenoceptor β1 (ADRB1), and most of these were agonists of ADRB1 such as norepinephrine (Table SII).
Figure 5.
Drug-gene interaction networks. (A) The drug-gene interaction network for genes related to M. (B) The drug-gene interaction network for genes related to LPS. (C) The drug-gene interaction network for genes related to LPS+MV. Red circles indicate upregulated target genes, green circles indicate downregulated target genes, gray squares indicate predicted drugs. MV, mechanical ventilation; LPS, lipopolysaccharide.
Of the genes associated with LPS, 40 genes (36 upregulated and 4 downregulated genes) were targeted by 65 drugs, and the drug-gene network consisted of 74 drug-gene interactions (Fig. 5B). For example, glucosamine and aspirin were identified to be antagonists of NFKB2. Thalidomide was an antagonist of NFKB1, and theophylline was an antagonist of ADORA2B (Table SII).
A total of 218 drugs were predicted to target 78 genes that responded to LPS combined with MV, and the drug-gene network comprised 248 drug-gene interactions (Fig. 5C). For example, niacin was predicted to be an agonist of HCAR2, whereas siltuximab was identified to be an inhibitor, antibody and antagonist of IL6 (Table SII).
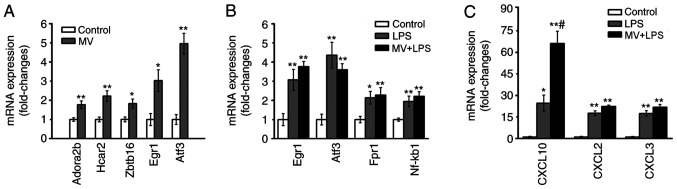
Expression of the verified genes
According to the results of the bioinformatics analyses, the genes ADORA2B, HCAR2 and ZBTB16, and the TFs EGR1 and ATF3 were selected for further verification as they were significantly upregulated in the mechanical ventilation group. The RT-qPCR analysis demonstrated that expression levels of ADORA2B, HCAR2, ZBTB16, EGR1 and ATF3 were upregulated in the lung tissues obtained from mice in the MV group compared with those in the control group (P<0.05 or P<0.01; Fig. 6A). In the LPS and MV + LPS groups, CXCL10, CXCL2, CXCL3, FPR1, IL-6 and NFKB1 were identified as the key nodes in the PPI network, and EGR1, ATF3 and BCl3 were identified as the key TFs; thus, a number of their encoding genes were chosen for verification according to the results of the bioinformatics analysis. The expression levels of EGR1, ATF3, FPR1, NFKB1 CXCL10, CXCL2 and CXCL3 were increased in the lung tissues of mice in the LPS and MV + LPS groups compared with those in the control group (P<0.05 or P<0.01; Fig. 6B and C).
Figure 6.
Verification of differentially expressed genes in vivo. mRNA expression levels in mice lung tissue were determined by reverse transcription-quantitative PCR (n=6). (A) mRNA expression of the verified genes between the control group and MV group. (B and C) mRNA expression of the verified genes between the control, LPS and MV + LPS groups. *P<0.05, **P<0.01 vs. control; #P<0.05 vs. MV. MV, mechanical ventilation; LPS, lipopolysaccharide.
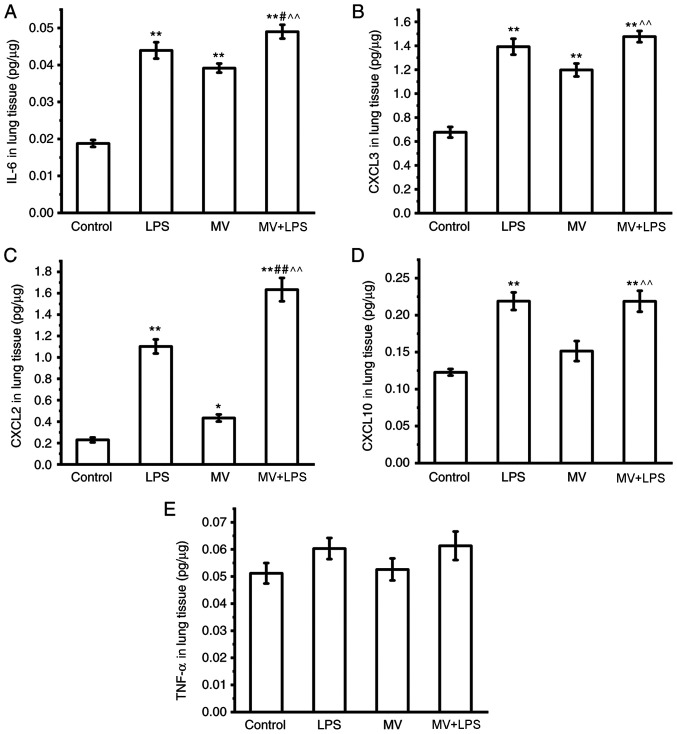
ELISA was used to further confirm the changes at the protein expression level of the selected genes. As presented in Fig. 7, the protein levels of IL-6, CXCL3 and CXCL2 in mouse lung tissues were increased in the MV, LPS and MV + LPS groups compared with those in the control group (P<0.05 or P<0.01; Fig. 7A-C). The protein levels of CXCL10 were increased in the LPS and MV + LPS group, but not in the MV group compared with those in the control group (P<0.01; Fig. 7D). No significant changes were observed in the levels of TNF-α in each group (Fig. 7E).
Figure 7.
Protein levels of the verified genes. (A) IL-6, (B) CXCL3, (C) CXCL2, (D) CXCL10 and (E) TNF-α protein expression levels in mouse lung tissues were determined by ELISA (n=7). *P<0.05, **P<0.01 vs. control; #P<0.05 vs. LPS; ##P<0.01 vs. LPS; ^^P<0.01 vs. MV. MV, mechanical ventilation; LPS, lipopolysaccharide; CXCL, C-X-C motif chemokine ligand.
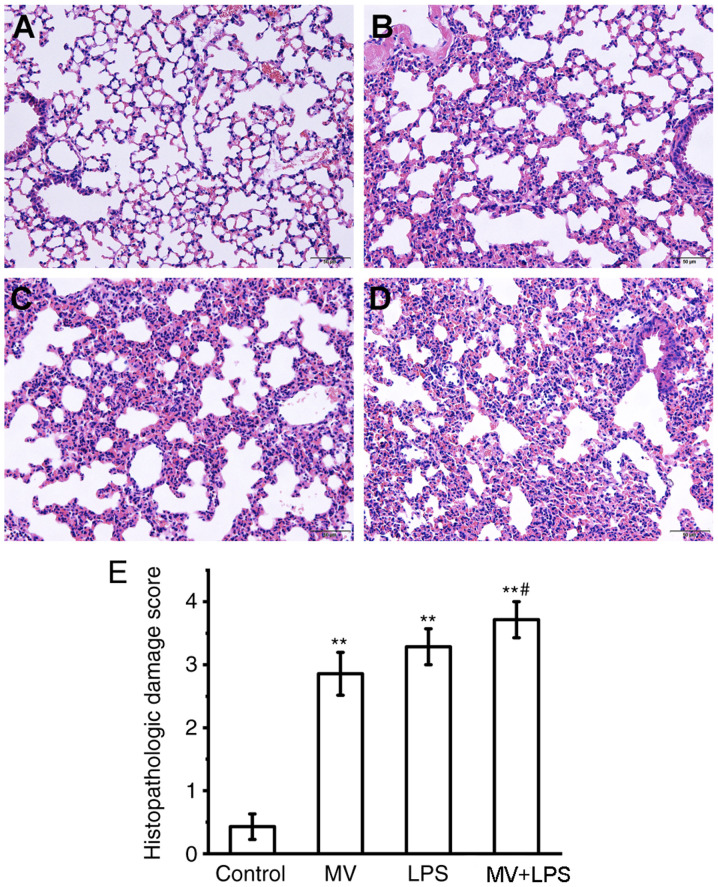
Histological evaluation of lung tissue. Sections of mouse lung tissue were stained with H&E and scored by histopathological analysis. Histological analysis of lung tissue sections revealed that MV, LPS or LPS combined with MV all induced diffuse interstitial edema, alveolar thickening and marked decreases in alveolar air space, as well as lung recruitment of leukocyte and a high histopathological damage score compared with those in the control group (P<0.01; Fig. 8). Additionally, compared with the MV group, MV + LPS treatment further aggravated lung injury (P<0.05 vs. MV group; Fig. 8E).
Figure 8.
Lung histopathological changes in mice subjected to MV, LPS or MV + LPS. Left lower lungs were removed for histopathological analysis using hematoxylin and eosin staining. (A) Control; (B) MV; (C) LPS treatment; and (D) MV + LPS. Magnification, ×200. Scale bar, 100 µm. (E) The severity of lung injury was scored in the stained tissues (n=7). **P<0.01 vs. control; #P<0.05 vs. MV. MV, mechanical ventilation; LPS, lipopolysaccharide.
Discussion
In the present study, a total of 63, 538 and 1,635 DEGs were identified as associated with MV, LPS, and MV + LPS, respectively. The MV DEG set was significantly enriched in genes associated with ‘negative regulation of cysteine-type endopeptidase activity involved in apoptotic process’ and ‘purine ribonucleotide metabolic process’. The LPS and MV + LPS DEG sets were significantly enriched in genes associated with ‘cellular response to cytokine stimulus’, ‘response to molecule of bacterial origin’ and ‘cell chemotaxis’. Notably, these three groups of genes were significantly enriched in the ‘TNF signaling pathway’ and ‘IL-17 signaling pathway’. Li et al (27) have reported that LPS-induced ALI may be attenuated by IL-17 via inhibition of the expression of extracellular signal-regulated kinase 1/2 and NF-κB. Patel et al (28) have indicated that TNF induces dysfunction of the alveolar epithelia through induction of death signaling, and blocking such signaling results in favorable effects in ALI. These findings suggest that treatment of the inflammatory response is shared amongst the different treatments, although certain differences are present between MV and LPS treatment. In the present study, the MV + LPS group exhibited the highest number of DEGs, which was greater than the sum of each group alone, indicating that MV may enhance the effects of LPS on gene expression. Similar views were proposed in a previous study by Chen et al (11). Chen et al (11) reported that the MV + LPS group generated the most DEGs, suggesting that MV is able to augment the influence of LPS on gene expression. In addition, in the present study, the chemokines of the CXC subfamily (including CXCL2, CXCL3 and CXCL10), EGR1 and ATF3 were upregulated in the LPS and MV + LPS groups compared with the control group, whereas ADORA2B, ZBTB16, HCAR2, EGR1 and ATF3 were differentially expressed in the MV group both in the bioinformatics analysis and in vivo.
Chemokines are typical basic heparin-binding proteins with a molecular weight of 7–10 kDa that perform crucial roles in the migration, recruitment and recirculation of leukocytes (29). Chemokines are secreted through stimulation of the resident lung cells by inflammatory mediators and bacterial products, and are retained at inflammatory sites by matrix heparin sulfate proteoglycans, forming a gradient of chemokines toward inflammatory lesions (30). In ALI, based on the chemokine gradients, inflammatory cells are recruited to the lung, including neutrophils, macrophages and mononuclear cells, which together with chemokines have been identified to serve important roles in the pathogenesis of ALI (30). The levels of chemokines are increased in the lungs of ALI animal models, and the severity of lung injury can be reduced by neutralizing chemokines or their corresponding receptors (31,32). For instance, Wang et al (31) have demonstrated that influenza A-induced ALI can be attenuated by treatment with a monoclonal antibody against CXCL10. Chen et al (10) have suggested that rutin exhibits a protective effect against LPS-induced ALI by decreasing macrophage inflammatory protein 2α (also termed CXCL2) levels and inactivating matrix metalloproteinase 9. Yang et al (33) have demonstrated that tephrosin exerts a favorable effect against sepsis-induced ALI by downregulating the expression of intracellular adhesion molecule 1 and CXCL2. C-C motif chemokine ligand 2 (CCL2), also termed MCP1, was also significantly upregulated in the LPS and MV + LPS groups in the present study. A previous study has reported high levels of CCL2 in H7N9 virus-induced ALI in mice and in infected lung tissues, and that the injury is attenuated in CCL2-deficient mice (34). The results of the present study were consistent with the aforementioned previous reports. In the drug-gene prediction, danazol was predicted to be an inhibitor of CCL2. Although there are no studies that focus on the effect of danazol on CCL2 in ALI, it has been reported that danazol can directly inhibit the expression of CCL2 in endometrial epithelial cells in a dose-dependent manner (35,36). This may provide novel ideas and a theoretical basis for the study and treatment of ALI.
ADORA2B encodes the adenosine A2b receptor, also termed A2BAR, which is a G protein-coupled receptor (37). The expression of ADORA2B was upregulated in the MV group compared with that in the control group in the present study. Eckle et al (38) have previously demonstrated that the pulmonary adenosine level is increased in mice treated with MV compared with the control group, and myeloid and pulmonary A2BAR signaling mediates the inflammatory response of ventilator-induced lung injury; however, pulmonary A2BARs weaken the alveolar-capillary barrier and improve alveolar fluid transport, suggesting that A2BAR is a potential therapeutic target in ALI. A2BAR may serve roles in the inflammatory response and airway wall remodeling in asthma, suggesting that A2BAR antagonists may be a potential new therapeutic approach (39). Theophylline has been predicted to be an antagonist of A2BAR in previous studies (39,40). The mechanisms behind the protective effects of A2BAR remain unclear and require further investigation.
In conclusion, the similarities and differences between ALI induced by different treatments were analyzed in the present study. The gene response to MV was significantly enriched in urine ribonucleotide metabolism-related processes, whereas the gene response to LPS and LPS+MV was significantly enriched in ‘cellular response to cytokine stimulus’ and ‘cell chemotaxis’. The involvement in the inflammatory response was shared between the DEGs identified in the MV and LPS-induced ALI groups. In addition, MV may enhance the effect of LPS on gene expression. The results of the present study provide novel insight and a theoretical basis for the study and treatment of ALI.
Supplementary Material
Acknowledgements
Not applicable.
Funding
This work was supported by grants from The Hospital Foundation of Xin Hua Hospital (grant nos. 15YJ04 and 15YJ14) and The National Natural Science Foundation of China (grant no. 81901991).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
WD designed the study and analyzed the bioinformatics data. ZF conducted the verification experiment. YZ was responsible for data acquisition and data analysis, and participated in the animal modeling experiment. ZR conducted statistical analysis. LJ designed the study, and made substantial contributions in drafting the manuscript and revising it critically for important intellectual content. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All animal experiments were approved by the Ethics Committee of Experimental Animals of Shanghai Jiaotong University School of Medicine.
Patient consent for publication
Not applicable.
Competing interests
The authors confirm that they have no competing interests.
References
- 1.Wang C, Zeng L, Zhang T, Liu J, Wang W. Casticin inhibits lipopolysaccharide-induced acute lung injury in mice. Eur J Pharmacol. 2016;789:172–178. doi: 10.1016/j.ejphar.2016.07.035. [DOI] [PubMed] [Google Scholar]
- 2.Zeng M, Sang W, Chen S, Chen R, Zhang H, Xue F, Li Z, Liu Y, Gong Y, Zhang H, Kong X. 4-PBA inhibits LPS-induced inflammation through regulating ER stress and autophagy in acute lung injury models. Toxicol Lett. 2017;271:26–37. doi: 10.1016/j.toxlet.2017.02.023. [DOI] [PubMed] [Google Scholar]
- 3.Niu X, Liu F, Li W, Zhi W, Zhang H, Wang X, He Z. Cavidine ameliorates lipopolysaccharide-induced acute lung injury via NF-κB signaling pathway in vivo and in vitro. Inflammation. 2017;40:1111–1122. doi: 10.1007/s10753-017-0553-1. [DOI] [PubMed] [Google Scholar]
- 4.Modrykamien AM, Gupta P. The acute respiratory distress syndrome. Proc (Bayl Univ Med Cent) 2015;28:163–171. doi: 10.1080/08998280.2015.11929219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gramatté J, Pietzsch J, Bergmann R, Richter T. Causative treatment of acid aspiration induced acute lung injury-recent trends from animal experiments and critical perspective. Clin Hemorheol Microcirc. 2018;69:187–195. doi: 10.3233/CH-189113. [DOI] [PubMed] [Google Scholar]
- 6.Fanelli V, Ranieri VM. Mechanisms and clinical consequences of acute lung injury. Ann Am Thorac Soc. 2015;12(Suppl 1):S3–S8. doi: 10.1513/AnnalsATS.201407-340MG. [DOI] [PubMed] [Google Scholar]
- 7.Zhao H, Zhao M, Wang Y, Li F, Zhang Z. Glycyrrhizic acid prevents sepsis-induced acute lung injury and mortality in rats. J Histochem Cytochem. 2016;64:125–137. doi: 10.1369/0022155415610168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang W, Luo F, Lu Q, Liu J, Li P, Wang X, Fu Y, Hao K, Yan T, Ding X. The protective effect of Trillin LPS-induced acute lung injury by the regulations of inflammation and oxidative state. Chem Biol Interact. 2016;243:127–134. doi: 10.1016/j.cbi.2015.09.010. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Y, Zhao C, He W, Wang Z, Fang Q, Xiao B, Liu Z, Liang G, Yang S. Discovery and evaluation of asymmetrical monocarbonyl analogs of curcumin as anti-inflammatory agents. Drug Des Devel Ther. 2014;8:373–382. doi: 10.2147/DDDT.S58168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen WY, Huang YC, Yang ML, Lee CY, Chen CJ, Yeh CH, Pan PH, Horng CT, Kuo WH, Kuan YH. Protective effect of rutin on LPS-induced acute lung injury via down-regulation of MIP-2 expression and MMP-9 activation through inhibition of Akt phosphorylation. Int Immunopharmacol. 2014;22:409–413. doi: 10.1016/j.intimp.2014.07.026. [DOI] [PubMed] [Google Scholar]
- 11.Chen Y, Zhou X, Rong L. Analysis of mechanical ventilation and lipopolysaccharide induced acute lung injury using DNA microarray analysis. Mol Med Rep. 2015;11:4239–4245. doi: 10.3892/mmr.2015.3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen H, Bai C, Wang X. The value of the lipopolysaccharide-induced acute lung injury model in respiratory medicine. Expert Rev Respir Med. 2010;4:773–783. doi: 10.1586/ers.10.71. [DOI] [PubMed] [Google Scholar]
- 13.Zhang A, Pan W, Lv J, Wu H. Protective effect of Amygdalin on LPS-induced acute lung injury by inhibiting NF-κB and NLRP3 signaling pathways. Inflammation. 2017;40:745–751. doi: 10.1007/s10753-017-0518-4. [DOI] [PubMed] [Google Scholar]
- 14.Pedrazza L, Cunha AA, Luft C, Nunes NK, Schimitz F, Gassen RB, Breda RV, Donadio MV, de Souza Wyse AT, Pitrez PMC, et al. Mesenchymal stem cells improves survival in LPS-induced acute lung injury acting through inhibition of NETs formation. J Cell Physiol. 2017;232:3552–3564. doi: 10.1002/jcp.25816. [DOI] [PubMed] [Google Scholar]
- 15.Bouferrache K, Vieillard-Baron A. Acute respiratory distress syndrome, mechanical ventilation, and right ventricular function. Curr Opin Crit Care. 2011;17:30–35. doi: 10.1097/MCC.0b013e328342722b. [DOI] [PubMed] [Google Scholar]
- 16.Gordo-Vidal F, Enciso-Calderón V. Acute respiratory distress syndrome, mechanical ventilation and right ventricular function. Med Intensiva. 2012;36:138–142. doi: 10.1016/j.medin.2011.08.012. (In Spanish) [DOI] [PubMed] [Google Scholar]
- 17.Nieman GF, Gatto LA, Bates JH, Habashi NM. Mechanical ventilation as a therapeutic tool to reduce ARDS incidence. Chest. 2015;148:1396–1404. doi: 10.1378/chest.15-0990. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Mekontso Dessap A, Boissier F, Charron C, Bégot E, Repessé X, Legras A, Brun-Buisson C, Vignon P, Vieillard-Baron A. Acute cor pulmonale during protective ventilation for acute respiratory distress syndrome: Prevalence, predictors, and clinical impact. Intensive Care Med. 2016;42:862–870. doi: 10.1007/s00134-015-4141-2. [DOI] [PubMed] [Google Scholar]
- 19.Brochard L, Slutsky A, Pesenti A. Mechanical ventilation to minimize progression of lung injury in acute respiratory failure. Am J Respir Crit Care Med. 2017;195:438–442. doi: 10.1164/rccm.201605-1081CP. [DOI] [PubMed] [Google Scholar]
- 20.Sadowitz B, Jain S, Kollisch-Singule M, Satalin J, Andrews P, Habashi N, Gatto LA, Nieman G. Preemptive mechanical ventilation can block progressive acute lung injury. World J Crit Care Med. 2016;5:74–82. doi: 10.5492/wjccm.v5.i1.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chian CF, Chiang CH, Chuang CH, Liu SL, Tsai AC. SN50, a cell-permeable-inhibitor of nuclear factor-κB, attenuates ventilator-induced lung injury in an isolated and perfused rat lung model. Shock. 2016;46:194–201. doi: 10.1097/SHK.0000000000000563. [DOI] [PubMed] [Google Scholar]
- 22.Slutsky AS, Ranieri VM. Ventilator-induced lung injury. N Engl J Med. 2013;369:2126–2136. doi: 10.1056/NEJMra1208707. [DOI] [PubMed] [Google Scholar]
- 23.Smith LS, Gharib SA, Frevert CW, Martin TR. Effects of age on the synergistic interactions between lipopolysaccharide and mechanical ventilation in mice. Am J Respir Cell Mol Biol. 2010;43:475–486. doi: 10.1165/rcmb.2009-0039OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.The R Development Core Team, corp-author. R: A language and environment for statistical computing. 2014 [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Dong WW, Liu YJ, Lv Z, Mao YF, Wang YW, Zhu XY, Jiang L. Lung endothelial barrier protection by resveratrol involves inhibition of HMGB1 release and HMGB1-induced mitochondrial oxidative damage via an Nrf2-dependent mechanism. Free Radic Biol Med. 2015;88:404–416. doi: 10.1016/j.freeradbiomed.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 27.Li TJ, Zhao LL, Qiu J, Zhang HY, Bai GX, Chen L. Interleukin-17 antagonist attenuates lung inflammation through inhibition of the ERK1/2 and NF-κB pathway in LPS-induced acute lung injury. Mol Med Rep. 2017;16:2225–2232. doi: 10.3892/mmr.2017.6837. [DOI] [PubMed] [Google Scholar]
- 28.Patel BV, Wilson MR, O'Dea KP, Takata M. TNF-induced death signaling triggers alveolar epithelial dysfunction in acute lung injury. J Immunol. 2013;190:4274–4282. doi: 10.4049/jimmunol.1202437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Puneet P, Moochhala S, Bhatia M. Chemokines in acute respiratory distress syndrome. Am J Physiol Lung Cell Mol Physiol. 2005;288:L3–L15. doi: 10.1152/ajplung.00405.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bhatia M, Zemans RL, Jeyaseelan S. Role of chemokines in the pathogenesis of acute lung injury. Am J Respir Cell Mol Biol. 2012;46:566–572. doi: 10.1165/rcmb.2011-0392TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang W, Yang P, Zhong Y, Zhao Z, Xing L, Zhao Y, Zou Z, Zhang Y, Li C, Li T, et al. Monoclonal antibody against CXCL-10/IP-10 ameliorates influenza A (H1N1) virus induced acute lung injury. Cell Res. 2013;23:577–580. doi: 10.1038/cr.2013.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bao Z, Ye Q, Gong W, Xiang Y, Wan H. Humanized monoclonal antibody against the chemokine CXCL-8 (IL-8) effectively prevents acute lung injury. Int Immunopharmacol. 2010;10:259–263. doi: 10.1016/j.intimp.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 33.Yang J, Tian H, Huang X. Tephrosin attenuates sepsis induced acute lung injury in rats by impeding expression of ICAM-1 and MIP-2. Microb Pathog. 2018;117:93–99. doi: 10.1016/j.micpath.2018.02.017. [DOI] [PubMed] [Google Scholar]
- 34.Lai C, Wang K, Zhao Z, Zhang L, Gu H, Yang P, Wang X. C-C motif chemokine ligand 2 (CCL2) mediates acute lung injury induced by lethal influenza H7N9 virus. Front Microbiol. 2017;8:587. doi: 10.3389/fmicb.2017.00587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boucher A, Lemay A, Akoum A. Effect of hormonal agents on monocyte chemotactic protein-1 expression by endometrial epithelial cells of women with endometriosis. Fertil Steril. 2000;74:969–975. doi: 10.1016/S0015-0282(00)01540-5. [DOI] [PubMed] [Google Scholar]
- 36.Jolicoeur C, Lemay A, Akoum A. Comparative effect of danazol and a GnRH agonist on monocyte chemotactic protein-1 expression by endometriotic cells. Am J Reprod Immunol. 2015;45:86–93. doi: 10.1111/j.8755-8920.2001.450204.x. [DOI] [PubMed] [Google Scholar]
- 37.Vecchio EA, White PJ, May LT. The adenosine A2B G protein-coupled receptor: Recent advances and therapeutic implications. Pharmacol Ther. 2019;198:20–33. doi: 10.1016/j.pharmthera.2019.01.003. [DOI] [PubMed] [Google Scholar]
- 38.Eckle T, Grenz A, Laucher S, Eltzschig HK. A2B adenosine receptor signaling attenuates acute lung injury by enhancing alveolar fluid clearance in mice. J Clin Invest. 2008;118:3301–3315. doi: 10.1172/JCI34203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Holgate ST. The Quintiles Prize Lecture 2004. The identification of the adenosine A2B receptor as a novel therapeutic target in asthma. Br J Pharmacol. 2005;145:1009–1015. doi: 10.1038/sj.bjp.0706272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Philipp S, Yang XM, Cui L, Davis AM, Downey JM, Cohen MV. Postconditioning protects rabbit hearts through a protein kinase C-adenosine A2b receptor cascade. Cardiovasc Res. 2006;70:308–314. doi: 10.1016/j.cardiores.2006.02.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.