Abstract
Milk serves as a mode of protection to neonate through transferring the host defense proteins from mother to offspring. It also guards the mammary gland against various types of infections. Along with the presence of six vital proteins, bovine milk (whey) contains a massive class of minor proteins, not all of which have been comprehensively reported. In this study, we performed an LC-MS/MS-based ultra-deep identification of the milk whey proteome of Indian zebu (Sahiwal) cattle. Three independent search engines that are Comet, Tandem, and Mascot-based analysis resulted in the discovery of over 6,210 non-redundant proteins commonly identified. Genome-wise mapping revealed that chromosome 1 showed a minimum expression of 14 proteins, whereas chromosome 19 expressed 250 maximum proteins in milk whey. These results demonstrate that milk proteome in Sahiwal cattle is quite complicated, and minor milk fractions play a significant role in host defense.
Keywords: host defense, Indian zebu, mass spectrometer, milk whey, proteomics, Sahiwal
Introduction
Milk synthesis is an absolute feature that defines the mammalian class. It has received considerable interest because of its nutritional and functional properties to newborn and young offspring (1). It majorly acts as a vital source for the transfer of defense molecules against pathogens from mother to child. Worldwide, humans consume bovine milk in their diet, and a regular diet accounts for 80–90% of total bovine milk intake and acts as an essential source of nutrition to them. Milk acts as a medium for the transfer of host defense proteins, yet its repertoire of minor proteins has been only partly characterized (2). In addition, bovine milk is exceedingly being utilized for dairy products, including yogurt, curd, cheese, butter, and, to some extent, as bioactive peptides (3).
Milk from different species was previously studied as a substitute for human milk in infant food. Several studies were mainly focused on major milk components (4). The low abundant proteins in milk could be explored with the development of analytical methods. The comprehensive understanding of milk of different species could be beneficial in promoting the utilization of milk as a source of nutrition. Attaining a complete knowledge of the milk whey proteome could serve as a significant foundation for the production of functional foods and infant products.
More recently, proteomic approaches based on mass spectrometry have been used for farm animal milk proteome research (5). Various proteomic approaches have been identified for exploring the molecular pathways and cellular functions of the complex milk proteins in caseins, whey protein, and milk-fat globular membrane (6). Proteomic studies have been conducted for exploring the milk whey proteome in humans and bovine for the identification of proteins related to host defense and immune system (7).
Sahiwal (Bos indicus) cattle breed is the dominant milch breed of Indian origin having native tract in North-Western region of India but a much broader breeding tract in the country. In terms of milk production, it is a high milk-producing breed (8); it is known to be genetically more thermotolerant and less disease susceptible (9). In the tropics, Indicine cattle have lesser disease susceptibility for reproductive and productive disorders and parasitic infections compared to taurine cattle (10). Also, Indian native zebu cattle (Sahiwal) can survive comparatively well on low input production systems with moderate milk production but yield much more when maintained on the intensive system of production. Therefore, it is crucial to generate the proteome profile of Indian zebu cattle with the aim of identifying low abundant proteins involved in the host defense mechanism of Indicine cattle. The protein composition of milk varies with genetic and non-genetic factors (11, 12). In cow milk, the significant determinants of milk proteome include breed, feed conversion, stage of lactation, parity of production, and environmental determinants (13). The foremost fraction of milk includes caseins, and ~20% of milk composition is the whey proteome comprising alpha-lactalbumin and beta-lactoglobulin. Bioactive proteins and peptides in cow milk were reported to play a different role in cellular and physiological functions. It acts as a significant determinant in the development of immune response, protective functions against fungal, bacterial, and viral infections, and milk also plays a vital role in the maintenance of intestinal microbiota (14). Most of the reported proteins responsible for high biological activity are immunoglobulins, lactoferrin, α-lactalbumin, β-lactoglobulin, αS1-, αS2-, β-, k-caseins, and lactadherin (15). The presence of immunoglobulins in whey confers the initial line of passive defense to neonates, and in adults, it regulates a person's immune system (16). β-Lactoglobulin and lactoferrin fractions of milk proteome exhibit anticarcinogenic, immunomodulatory, antimicrobial, and antioxidant responses (17). Therefore, it is essential to perform the in-depth comparison of whey proteome of different species for unraveling the diversity in genetics and biological traits for a given species. Previously, a study identified the limited number of 211 proteins but demonstrated the uniqueness of a few proteins in the given species (1). Recent reports suggest the comparison of milk proteins within Holstein and Jersey breeds of dairy cattle (18). They reported differences in the low abundant proteins present in the skimmed milk fractions of Holstein and Jersey breeds fed on the same diet with similar management conditions. A report on comparison of different lactation stage proteins was conducted in Malanad Gidda (Bos indicus) cattle (19); however, to date, no such report is available in Indian zebu (Sahiwal) cattle. The present study was conducted to explore the milk whey proteome of Indian zebu cattle. As per our knowledge, it is the first report of milk whey proteome of Sahiwal cattle of Indian origin and can be used as a comprehensive reference database for cattle genetic resources of India.
Materials and Methods
Chemicals and Reagents
All chemicals, reagents, and organic solvents were purchased from Sigma-Aldrich (St. Louis, MO, USA) unless otherwise specified. Ultra-pure grade water was used throughout. All reagents used for this study were of molecular grade.
Selection of Animals for Milk Collection
Indian zebu (Bos indicus) cattle, Sahiwal breed (n = 10), in different significant lactation phases were used for the current study. The animals were reared in the animal herd under uniform feeding and breeding practices at the ICAR–National Dairy Research Institute, Karnal, India. Before the study, all animals were screened for the absence of mastitis by the California mastitis test. Only the healthy cows with somatic cell count ≤200 lakhs per cell without any sign of mammary gland infection were included in the study, and it was ascertained that the milk is free from bacteria. Before the collection of milk, the rectal examination for the absence of high body temperature was obtained twice a day, and cows having normal body temperature range were used for milk collection. All cows had free access to water and offered the same ad libitum diet.
The ICAR–National Dairy Research Institute, Karnal, India, animal care, and the committee approved all procedures used in this study. The milk samples were immediately transported to the laboratory under an icebox within 1 h and pooled before the experiment. The pooled samples aim to reduce the animal-to-animal variations.
Sample Preparation
Raw milk samples were defatted by centrifugation at 4,000 rpm for 15 min to obtain skimmed milk samples. Subsequently, the skimmed milk samples were ultracentrifuged at 65,000×g for 2 h at 4°C for separating casein, and the liquid portion was referred to as milk whey. Phenyl methyl sulfonate was added at a concentration of 0.01% to prevent proteolytic degradation of milk whey samples. The samples were stored at −80°C for further experimentation and analysis.
Optimization by Various Protein Extraction Methods
Various protein extraction procedures were followed and optimized, which includes (a) ultracentrifugation followed by acetone precipitation, (b) CaCl2 precipitation methods at different concentrations (60, 90, and 120 mM), and (c) TCA/acetone precipitation methods, respectively.
-
Ultracentrifugation followed by acetone precipitation
The milk was subjected to ultracentrifugation at 65,000×g for 2 h for whey preparation with minor modifications (7). Whey samples were further precipitated by adding chilled acetone and incubated at −20°C for 14–16 h followed by protein precipitation at 13,000 rpm at 4°C for 10 min (20). The supernatant was discarded, and the pellet was reconstituted with 1× PBS by adding 0.2% DEA (diethylamine) and 20% 0.5 M Tris of pH 6.8. Subsequently, the sample was dried in the vacuum-sealed concentrator and processed for tryptic digestion and mass spectrometric analysis.
-
CaCl2 precipitation
CaCl2 precipitation method was optimized at different concentrations (60, 90, and 120 mM) of CaCl2, respectively, for precipitating milk whey proteins from defatted milk samples followed by SDS-PAGE profiling of the whey precipitate.
-
TCA/acetone precipitation
TCA/acetone precipitation method was used for precipitating protein samples. Then 1.5 ml of 10% TCA/acetone was added to the whey sample and stored at −20°C overnight (21). The sample was centrifuged at 13,000 rpm for 10 min at 4°C, and the supernatant was discarded. The pellet was further re-suspended in 1× PBS and 2% DEA (diethylamine) for pellet dissolution.
Protein Quantitation
2D-Clean Up kit (GE Healthcare, USA) was used for removal of interfering substances from precipitated milk protein preparation, and total protein concentration was estimated using Bradford protein estimation kit (Bio-Rad) as per the manufacturer's instruction.
SDS-PAGE
Twenty-five micrograms (25 μg) of pooled Proteo-miner enriched milk whey sample (pooled from 10 animals of different lactation stages, n = 10), respectively, were subjected to 12% SDS-PAGE (10 × 10.5 cm) in a Mini VE complete gel electrophoresis system (GE Healthcare, USA). The gel was stained with colloidal Coomassie brilliant blue dye for in-gel digestion.
In-gel Tryptic Digestion
The gel lane with the enriched sample was cut into 12 equal pieces, which was further de-stained using 40% ACN (acetonitrile) and 40 mM NH4HCO3 (ammonium bicarbonate) at a ratio of 1:1 (v/v), respectively. The de-stained gel bands were reduced with 5 mM dithiothreitol (DTT) in 40 mM NH4HCO3, followed by alkylation with 20 mM iodoacetamide in 40 mM NH4HCO3. Overnight digestion of gel bands was carried out using 12.5 ng/μl trypsin (modified sequencing grade; Promega, USA) at 37°C. Further, peptide extraction was done from gel pieces, lyophilized, and desalted using zip tip (Millipore, Germany) following the manufacturer's instruction and stored at −80°C for peptide identification by LC-MS/MS.
In-solution Tryptic Digestion
For in-solution digestion, 500 μg of pooled protein samples (50 μg each from 10 animals pooled from different stages of lactation) from TCA/acetone precipitated protein samples were processed separately. Further, 45 mM DTT (dithiothreitol) was dissolved in 50 mM NH4HCO3 (ammonium bicarbonate) to reduce disulfide bonds followed by alkylation of cysteine residues using 10 mM IAA (iodoacetamide) in 50 mM NH4HCO3 (ammonium bicarbonate). Digestion was carried out overnight using trypsin (1:100) at 37°C. The reaction was subsequently stopped with 10% TFA (trifluoroacetic acid), and peptides were further vacuum dried. Samples were fractionated into 24 fractions, further desalted by zip tip, and further subjected for peptide identification by LC-MS/MS (22).
Electrospray Ionization Tandem Mass Spectrometry LC-MS/MS Analysis
The lyophilized peptide fractions were reconstituted in 0.1% formic acid in LC-MS-grade water and subjected to nano-LC (Nano-Advance; Bruker, Germany) followed by identification in captive ion source (Bruker Captive Spray tip) spray-in Maxis-HD qTOF (Bruker) mass spectrometer (MS) with high mass accuracy and sensitivity. The peptides were enriched in nano-trap column (Acclaim Pep Map, particle size 5 μm, pore size 100 Å; Thermo Scientific) and eluted on to nano-analytical column (Kaya Tech HIQ SIL C18HS/3, 0.1 × 150 mm, 3 μm particle size, and 200 Å pore size). The peptide elution was carried out using a linear gradient of 5–45% acetonitrile at 400 nl/min flow rate in a total run time of 135 keeping the solvent system as follows: solvent A, 100% water in 0.1% formic acid; and solvent B, 100% acetonitrile in 0.1% formic acid. Positive ions were generated by electrospray, and the q-TOF was operated in data-dependent acquisition mode to automatically switch between MS and MS/MS acquisition. Precursor ion TOF MS survey scan was acquired with a range of 300–1,800 m/z with resolution R = 75,000. Q1 sequentially selects six most intense precursor ions for fragmentation using collision-induced dissociation for MS-MS analysis with a fixed cycle time of 3 s along with 2 min of release for exclusion filter (Data acquisition otof software, version 24.8; Bruker Daltonics).
Data Processing and Analysis
The vendor-created.d file format was analyzed with three different search engines that are Mascot, Comet, and Tandem for high confidence identification of the profiled whey proteome. For Mascot (Matrix Science, UK, version 2.4.1), we used the ProteinScape platform (version 2.0). Initially, the peak lists were generated through Hyster (Bruker Daltonics) to make spectral data in the form of mgf format (Mascot Generic Files). Resultant mgf files were used for the identification of proteins using the UniProt database downloaded April 3, 2019 along with usual contaminant proteins for spectra examination. The parameter for MS/MS ion search contains tryptic digested sites with two missed cleavage allowed, flexible modification on amino acids—methionine for oxidation, N-terminal acetylation, Gln-pyro Glu, and Glu-pyro Glu, while static modification of cysteine as carboxy amidomethylation or propionamide. The peptide precursor ion tolerance was 20 ppm, with MS/MS tolerance of 0.01 Da. The “ion score cutoff” was set to 30, thereby eliminating the lowest quality matches and minimum peptide length as six amino acid residues. To increase the confidence and remove the false-positive identification, a 1% false discovery rate (FDR) was used at equal peptide and protein levels. The decoy reversed sequences database was included for the calculation of the FDR.
The data were also re-searched in Trans-Proteomic Pipeline (version 5.1.0) for Comet and Tandem search engines. The in-depth procedure for performing the analysis was reported in Suhail et al. (23). Briefly, in the first step, the raw data were transformed to open format mzML files. The converted files were searched using the TPP pipeline keeping the aforementioned common database and the search parameters. However, on a different note, TPP initially performs the peptide assignments using Tandem and Comet search engine. In addition, PeptideProphet and ProteinProphet algorithms were employed in the pipeline to calculate the probability values for both independently examined peptides and the corresponding proteins. The exact mass model for high probability in PeptideProphet was utilized to boost the confidence for peptide associations and also to promote the definite possibility of peptides. Extra protein validation level was performed applying both PeptideProphet and ProteinProphet scores, where the protein was confirmed if it includes at least two top-ranked peptides with each peptide probability score above 95% (Supplementary Table 1). iProphet algorithm validated and merged all search engine results. This method takes as the input of PeptideProphet spectrum-level results from multiple LC-MS/MS runs and then computes a unique probability at the level of a unique peptide sequence (or protein sequence). This structure provides for the incorporation of outcomes from various search engines. It brings into account additional promoting determinants, including the number of sibling experiments distinguishing the equivalent peptide ions, the number of replicate ion identifications, sibling ions, and sibling modification states. A model of iProphet performance with regard to the abundance of correct entries vs. error is provided in Supplementary Table 1. An iProphet probability of higher than 0.9999 was accepted as the cutoff for the conclusive identification of the protein. For protein quantitation, ≥2 unique peptides per protein were estimated to ensure high-quality quantitation.
Bioinformatics Analysis
All the graphical analyses were performed in the R environment using respective tools. Histograms, density scatter plots, and principal component analysis (PCA) were generated using the ggPlot2 package. All the identified proteins were mapped with UniProt bovine chromosomal proteome information and parsed to create files appropriately formatted for input to Circos. The Gene Ontology (GO) categories were analyzed using the DAVID bioinformatics resources, and only the genes with a p adjusted value (FDR) of <0.05 were included and subsequent GO term plotted.
Results
Assessment of Different Methods for the Quality of Aqueous Milk Whey Protein Extraction
The present study aims to determine the comprehensive profile of bovine milk whey proteome in Indian zebu (Sahiwal) cattle. The method optimization using three different procedures (ultracentrifugation-acetone precipitation, CaCl2 precipitation at different concentrations, and TCA/acetone precipitation methods) for the isolation of the aqueous whey fraction showed that TCA/acetone precipitation performed best and resulted in the identification of maximum peptides/proteins in quality check LC-MS/MS runs (data not shown). The SDS-PAGE profiles for different protein extraction procedures and method optimization for milk whey samples also showed a better profile of the TCA/acetone precipitation method in comparison with others (Supplementary Figures 1A,B). Before running the whole experiment, we performed the quality assurance test; we ran the 2D gel for the TCA/acetone precipitated whey and identified the clear sorted pattern of spots (Supplementary Figure 2), suggesting its suitability for further in-depth proteome analysis.
Protein Identification by LC-MS/MS
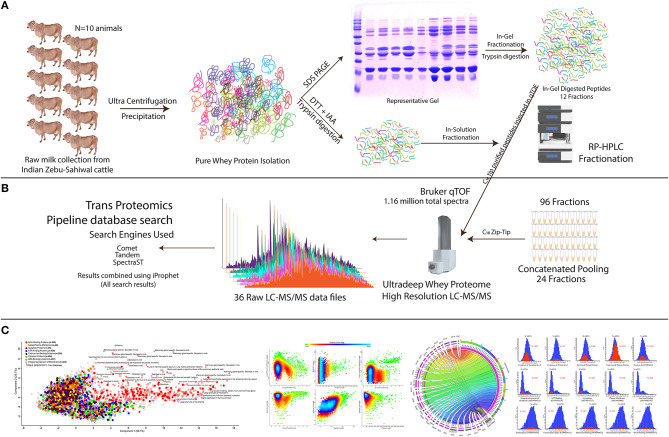
We employed the optimized bovine milk whey extraction workflow for the identification of ultra-deep proteome (Supplementary Figures 1, 2). We ran the SDS-PAGE whey proteome profile of 18 animals separately (Supplementary Figure 3) and selected 10 animals whose profile was matching almost similar for sample pooling. In this way, we can determine the whey proteome profile of the population and neglected the animal-to-animal variations. The isolated proteome of indigenous zebu (Sahiwal) milk whey samples were fractionated using a combined approach: (1) in-gel digestion LC-MS/MS methods and (2) in-solution digestion LC-MS/MS methods. Samples prepared from in-gel and in-solution tryptic digestion were subjected to high-resolution qTOF, nano-LC-MS/MS (Figure 1). An outstanding feature of analyzing this dataset is the conversion of open file format and its interchange data processing between modules (i.e., comet, tandem, SpectraST) under the umbrella of Trans Proteomics Pipeline (TPP). Finally, the total combined run of 36 fractions resulted in the identification of 1,154,123 spectra, 57,286 peptides, and 6210 proteins. The authenticity and confidence of the identified proteome were also confirmed through cross-mapping the proteome information using three independent proteomics software (TPP and Protein Scape) (Supplementary Tables 1, 2). Only the 6,210 common proteins identified in all the three platforms were selected for further analysis. All the proteins were selected at <1% FDR and having the protein prophet and iProphet value of 0.9999 and 0.95, respectively. Next, we chose the MS2-based Normalized Spectral Index for the quantification of individual proteins using the StPeter algorithm implemented in TPP for whey proteome quantitation. It allows the identification of individual proteins in the sample.
Figure 1.
Experimental setup and systemic proteomics workflow for the comprehensive discovery of bovine milk whey proteome. (A) From 10 individual animals and pooled milk samples, the milk whey proteome is isolated and purified using ultracentrifugation and TCA/acetone precipitation strategy. In-gel fractionation through SDS-PAGE and in-sol digested peptides were subjected to C18 chromatography for long gradient 96 fractionation (see Materials and Methods). (B) C18 separated fractions were concatenated, mixed, and analyzed using a high-resolution qTOF instrument. All the raw data were extracted, and the resulting peptides and proteins were analyzed using the TPP pipeline utilizing Comet, Tandem, and SpectraST search engines, and the obtained results were compiled using iProphet algorithm as described in Materials and Methods section. (C) Brief description of the data analysis; detailed information is provided in the Materials and Methods section. For determination of the high confidence bovine whey proteome, data were filtered using the stringent parameter. Evidence of the proteome at the different levels was determined along with the identification of the different Gene Ontology (GO) classes in it.
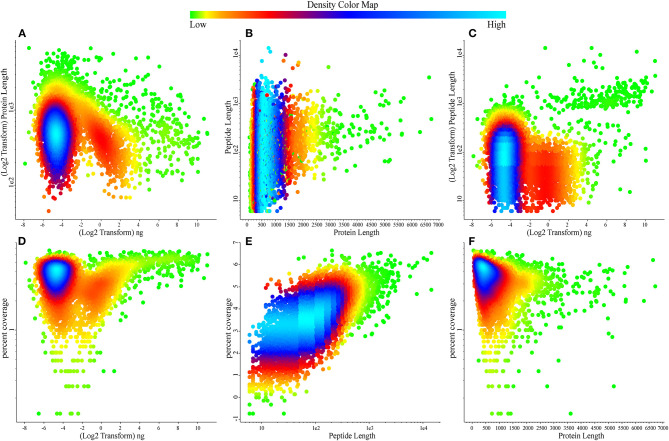
To date, this is the largest comprehensive whey proteome dataset. Our next aim was to determine the quality pattern of the proteins present in the whey. We plotted density plots to identify the information; most of the densely populated proteins were determined in the concentration range of 0.0156–0.0625 ng (Figures 2A,B) with the identified high-density percentage coverage of 20–65% (Figures 2C,D) in whey proteome. The protein length is ranging in between 500 and 800 amino acid residues (Figure 2F) with the identification of tryptic peptides ranging between 7 and 15 amino acid residues (Figure 2E).
Figure 2.
Scatter relationship plots describing the density of attributes over the bovine whey proteome data. (A) Protein lengths over quantification values. (B) Peptide length over protein length. (C) Peptide length over quantification values. (D) Protein percentage coverage over quantification values. (E) Protein percentage coverage over peptide length. (F) Protein percentage coverage over protein length. Color map reports the density of the values in the data, green reports the low, dense data points while sky blue reports highly dense data points.
Chromosomal Mapping of Bovine Whey Proteome
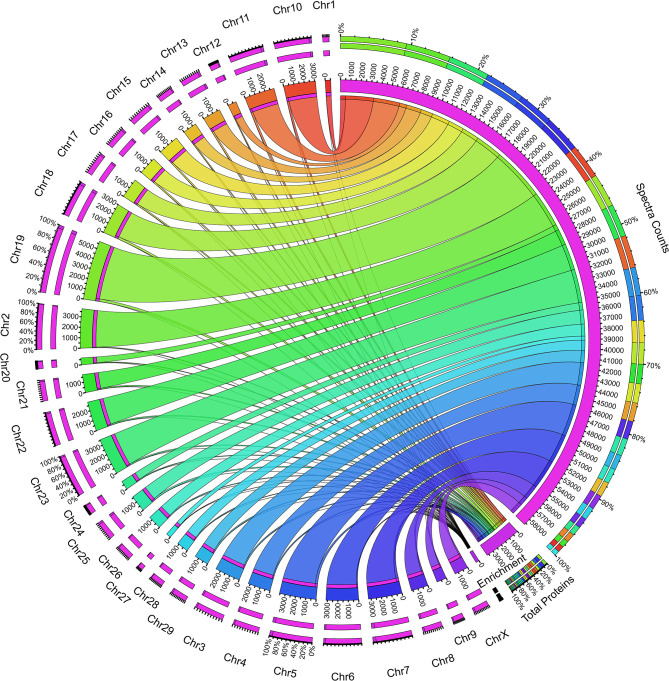
Next, we sought to determine the expression of the proteins from the individual chromosomes of the genome. We performed the chromosomal mapping analysis creating a Circos plot based on the UniProt data. Total spectra and protein information fetched from the search engine for identification of the proteins were mapped using the UniProt database. We found the contribution of all the 29 + X chromosomes in the whey proteome, but the analysis resulted in interesting facts such as the uneven distribution of the protein expression in the chromosome-wise manner. Chromosome 1 showed a minimum expression of 14 proteins, whereas chromosome 19 expressed 250 maximum proteins (Figure 3); the average expression number of proteins per chromosomes is 123. To make the analysis unbiased, we calculated the enrichment percentage of all the chromosomes which is the division of the total proteins assigned in the database to the actual number of proteins identified in the mapping analysis; however, the results are the same (Supplementary Table 3). The factors responsible for such high expression of chromosome 19 and very low expression of chromosome 1 contributing to the whey proteome are unknown. Nonetheless, further studies on genome-wide proteome analysis of mammary gland cells are required to answer these questions.
Figure 3.
Circos plot showing the whey protein expression mapped to individual chromosomes in the bovine genome. The right inner pink ring indicates the total spectra collected during the proteome analysis. The connecting set of ribbons describes the collection of spectra identified from individual chromosomes, and its width showed the number of spectra assigned. The higher the number spectra, the broader the ribbon width and vice versa. The bottom of the plot describes the total proteins assigned and the enrichment value based on the protein coded for the respective chromosome. The left outer ring describes the chromosomes and the number of protein assignment, which expresses bovine whey.
Functional Significance of Bovine Milk Whey Proteins
To understand the overall function of bovine whey protein/proteome, we characterize the subcellular localization using the program ngLOC utilizing the n-gram-based Naïve Bayesian classification model for the fixed-length peptide sequences density distributions over each distinct subcellular location (Supplementary Table 4). The results showed the identification of 11 different organelle classification while maximum proteins 46.13% (3,012 proteins) in whey are annotated to come from nucleus followed by cytoplasm 18.63% (1,220 proteins) and plasma membrane 17.20% (1,123 proteins). Further, we performed the ontological gene classification and found the majority of proteins in functional GO terms were nuclear proteins (46.13%) followed by proteins present in the cytoplasm (18.68%), placenta (17.2%), exosomes (7.08%), and Golgi bodies (1.18%). The cellular component GO enriched term reveals the identification of 18.92% proteins in the cytoplasm, 17.41% in the nucleus, 16.5% proteins were phosphoproteins, and 15.5% were present in extracellular exosomes. The majority of proteins had a functional role in metal ion binding (9.88%) and ATP binding (9.5%) (Supplementary Table 5).
Molecular functions in terms of protein percentage reveal the involvement of the majority of proteins in ATP binding (9.5%) followed by metal ion binding, RNA binding, zinc ion binding, DNA binding, and transcription factor activity. The proteins in biological process class were majorly identified in transcription, DNA template, oxidation–reduction process, intracellular signal transduction, and regulation of transcription. In similar terms, the majority of proteins in the KEGG pathway were present in metabolic pathways, PI3K, Akt, MAPK, and RAS signaling pathways. Also, several proteins have a role in pathways related to cancer, focal adhesion, regulation of actin cytoskeleton, and endocytosis. In the SMART term, the majority of proteins are SMW382: AAA. In Interpro term counts, the majority of protein percentage was found in P-loop containing NTP hydrolase, Protein kinases like domain, Homeodomain, and Serine/threonine–protein kinase active site. Clustering the proteins in terms of tissue specificity, milk proteins revealed expression in the mammary gland, milk, fetal skin and muscle, brain, and oviduct.
In Reactome term counts, 1.09% proteins were involved in platelet degranulation, followed by ubiquinilation and proteasome degradation, anchoring of the basal body to the plasma membrane, cell adhesion, in utero embryonic development, oxidation–reduction process, and positive regulation of gene expression (Supplementary Figure 4).
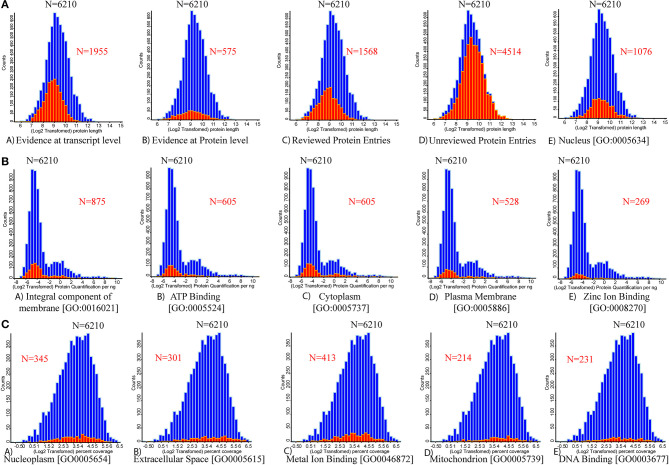
Bioactive proteins present in the whey fraction are involved in a wide range of physiological activities, including anti-inflammatory effects and protection against pathogen-induced intestinal inflammation (24). The relationship between counts vs. log2-transformed protein length reveals that out of 6,210 proteins, 1,955 proteins had evidence at transcript levels; however, 575 proteins have evidence at the protein level. Out of 6,210 entries, 1,568 protein entries are reviewed, and 4,514 entries are unreviewed (Figure 4). The relationship between counts vs. log2 protein quantification per ng reveals 875 proteins as integral components of the membrane (GO: 0016021). The GO terms reveal that 1076 proteins (GO: 0005634) were present in the nucleus, 605 in the cytoplasm (GO: 0005737), 528 in the plasma membrane (GO: 0005886), 345 proteins in nucleoplasm (GO: 0005654), 301 in extracellular space (GO: 0005615), and 214 in mitochondria (GO: 0005739). Some other proteins involved in different metabolic processes include 605 proteins in ATP binding (GO: 0005524), 269 proteins in zinc ion binding (GO: 0008270), 413 proteins in metal ion binding (GO: 0046872), and 231 proteins in DNA binding (GO: 0003677).
Figure 4.
Histograms for the distributions of the protein lengths, protein quantification, and percentage coverage are presented. The vertical axis of the histogram presents the number of each categorical protein in each bin. (A) Distribution of varying protein length (full length) counts identified through database mapping to the number of proteins. (A) Evidenced at transcript level. (B) Evidenced at protein level. (C) Reviewed UniProt database protein entries. (D) Un-reviewed UniProt database protein entries. (E) Protein associated with nucleus origin. (B) Mapping the number of protein quantification values for five high abundant UniProt-based GO terms. (A) Integral component of membrane. (B) ATP binding. (C) Cytoplasm. (D) Plasma membrane. (E) Zinc ion binding proteins. (C) Mapping the number of individual protein percentage coverage for the next five high abundant UniProt-based GO terms. (A) Nucleoplasm. (B) Extracellular space. (C) Metal ion binding. (D) Mitochondrion. (E) DNA binding. Red color overlapped histogram presents the respective mapped attribute over the whole bovine whey proteome data (n = 6,210).
PCA Score Plots for Milk Whey Proteome
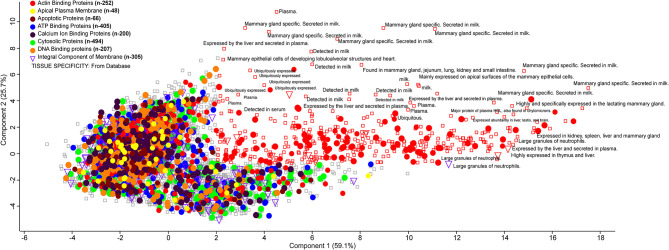
Milk whey proteins can be graphically represented using a PCA score plot where two components explain maximum variability. It is used as a data reduction tool where the score plot represents a dataset of 6,210 proteins. These were assigned as actin-binding proteins (n = 252), apical plasma membrane (n = 48), apoptotic protein (n = 66), ATP binding protein (n = 405), Ca ion binding protein (n = 200), cytosolic protein (n = 494), DNA binding protein (n = 207), and integral component of membrane (n = 305). The score plot classifies the whey proteins based on their involvement in several metabolic, biochemical, and cellular processes. Actin-binding proteins are maximally distributed, and the majority of proteins were mammary gland specific, secreted in milk. Some of the integral components are found in the large granules of neutrophils with their possible role in immunoregulation (Figure 5).
Figure 5.
PCA plot describing the orthogonal dispersion of the proteins reported in the deep whey proteome analysis. Color coding indicates the different class of major proteins identified in the whey proteome data. All the proteins identified to specific tissue from the UniProt database are reported with a rectangle.
Many of the milk bioactive proteins and peptides are also known to exhibit multifunctional physiological properties. In the present study, the majority of proteins with a high score as beta-lactoglobulin, lactoperoxidase, caseins, alpha-lactalbumin, alpha-2 macroglobulin, complement C3, lactotransferrin, alpha-S2 and S1 casein, xanthine oxidase, serotransferrin, and lactadherin were found to play a role in immune regulation and host defense. The top 60 lowest abundant proteins identified with high confidence levels and significant scores are shown in Table 1. The majority of the high score proteins have a role in immune status and host defense against invading microbes. However, the high abundant proteins like complement C3, lactoferrin, lactoperoxidase, and xanthine oxidase proteins exhibit immunomodulatory and anti-inflammatory properties. Polymeric immunoglobulin receptor (PIGR) and osteopontin proteins play a role in the innate immune system (25). Heat shock proteins (HSP 70) were found to be involved in pathways related to thermotolerance and help in maintaining homeostasis against adverse climatic conditions in livestock (26).
Table 1.
Top 60 low abundant proteins identified in whey proteome with respective database information, molecular function, cellular component, biological process, Prosite, Pfam, InterPro, SMART, and SUPFAM.
| Protein | Protein names | Gene symbol | ng | Molecular function | Cellular component | Biological process | Prosite | Pfam | InterPro | SMART | SUPFAM |
|---|---|---|---|---|---|---|---|---|---|---|---|
| F1MCK2 | AHNAK nucleoprotein | AHNAK | 0.074 | Costamere | Regulation of RNA splicing | PS50106 | IPR001478 | SM00228 | SSF50156 | ||
| E1BPD7 | Marker of proliferation Ki-67 | MKI67 | 0.066 | PS50006 | PF00498 | IPR000253 | SM00240 | SSF49879 | |||
| F1N415 | Piccolo presynaptic cytomatrix protein | PCLO | 0.075 | Metal ion binding | Presynaptic active zone | Synapse assembly | PS50004 | PF00168 | IPR000008 | SM00239 | SSF50156 |
| E1BGB0 | Kinesin family member 13B | KIF13B | 0.054 | 14-3-3 protein binding | Axon | Microtubule-based movement | PS00845 | PF01302 | IPR036859 | SM01052 | SSF49879 |
| F1MV51 | APC regulator of WNT signaling pathway | APC | 0.094 | Beta-catenin binding | Beta-catenin complex | Cell cycle arrest | PS50176 | PF05972 | IPR026836 | SM00185 | SSF48371 |
| E1BLA0 | Shugoshin 2 | SGOL2 | 0.057 | Centromeric region | Meiotic sister chromatid cohesion | IPR026706 | |||||
| E1BKT9 | Desmoplakin | DSP | 0.018 | Cell adhesion molecule binding | Plasma membrane | Adherens junction organization | PS50002 | PF00681 | IPR028462 | SM00250 | SSF75399 |
| E1BPB1 | Kinesin family member 24 | KIF24 | 0.093 | ATPase activity | kinesin complex | Microtubule-based movement | PS00411 | PF00225 | IPR027640 | SM00129 | SSF47769 |
| L8IEV2 | Dynein heavy chain 1, axonemal | M91_21125 | 0.078 | ATPase activity | Dynein complex | Cilium movement | PF12774 | IPR035699 | SSF52540 | ||
| E1BHP0 | Alpha-mannosidase | MAN2B2 | 0.095 | Alpha-mannosidase activity | Vacuolar membrane | Mannose metabolic process | PF09261 | IPR011013 | SM00872 | SSF74650 | |
| L8IV13 | Leucine-rich repeat-containing protein KIAA1731 | M91_18003 | 0.083 | Microtubule binding | Centriole | Positive regulation of centriole elongation | PF15309 | IPR029299 | |||
| L8HSL9 | WD repeat-containing protein 87 | M91_17430 | 0.085 | PS50082 | PF00400 | IPR015943 | SM00320 | SSF50978 | |||
| E1BKZ5 | Golgin B1 | GOLGB1 | 0.084 | cis-Golgi network | Protein localization to pericentriolar material | IPR026202 | |||||
| F1N2K7 | Phosphoinositide kinase, FYVE-type zinc finger containing | PIKFYVE | 0.033 | 1-Phosphatidylinositol-3-phosphate 5-kinase activity | Cell–cell junction | Intracellular signal transduction | PS50186 | PF00118 | IPR002423 | SM00049 | |
| E1BC24 | Midasin | MDN1 | 0.062 | ATPase activity | Cytosol | Ribosomal large subunit assembly | PS50234 | PF07728 | IPR003593 | SM00382 | SSF52540 |
| E1BKC4 | Nipped-B protein | NIPBL | 0.071 | Chromatin binding | Chromatin | Brain development | PF12765 | IPR011989 | SSF48371 | ||
| L8IT59 | Vacuolar protein sorting-associated protein 13A | M91_05527 | 0.081 | PF09333 | IPR015412 | ||||||
| E1BKI3 | Nitric oxide synthase (EC 1.14.13.39) | NOS1 | 0.065 | Calmodulin binding | Cytosol | Arginine catabolic process | PS51384 | PF00667 | IPR003097 | SM00228 | SSF50156 |
| E1B9N6 | Envoplakin | EVPL | 0.035 | Intermediate filament binding | Cornified envelope | Epidermis development | PF00681 | IPR041615 | SM00250 | SSF75399 | |
| E1BPX1 | Vacuolar protein sorting 13 homolog C | VPS13C | 0.103 | Cytosol | Mitochondrion organization | PF09333 | IPR015412 | ||||
| O46382 | Brefeldin A-inhibited guanine nucleotide-exchange protein 1 | ARFGEF1 | 0.091 | ARF guanyl-nucleotide exchange factor activity | Cytosol | Endomembrane system organization | PS50190 | PF16213 | IPR016024 | SM00222 | SSF48371 |
| F1MC51 | ATP binding cassette subfamily B member 5 | ABCB5 | 0.067 | ATPase activity | Plasma membrane | PS50929 | PF00664 | IPR003593 | SM00382 | SSF52540 | |
| E1BHT5 | Ubiquitin protein ligase E3 component n-recognin 4 | UBR4 | 0.065 | Ubiquitin-protein transferase activity | Centrosome | Ubiquitin-dependent protein catabolic process | PS51157 | PF13764 | IPR016024 | SM00396 | SSF48371 |
| E1BCI2 | E3 ubiquitin protein ligase (EC 2.3.2.27) | RNF40 | 0.061 | Metal ion binding | HULC complex | Histone H2B ubiquitination | PS00518 | IPR013956 | SM00184 | ||
| E1BL95 | Centromere protein J | CENPJ | 0.037 | Identical protein binding | Centriole | Microtubule polymerization | PF07202 | IPR033068 | |||
| E1B7W1 | Thyroid hormone receptor associated protein 3 | THRAP3 | 0.092 | RNA splicing | PF15440 | IPR026667 | |||||
| F1N137 | Inositol 1,4,5-trisphosphate receptor type 2 | ITPR2 | 0.094 | Inositol 1,4,5 trisphosphate binding | Endoplasmic reticulum | PS50919 | PF08709 | IPR014821 | SM00472 | SSF100909 | |
| F6RF21 | Structural maintenance of chromosomes flexible hinge domain containing 1 | SMCHD1 | 0.047 | ATPase activity | Barr body | Double-strand break repair | PF06470 | IPR036890 | SM00968 | SSF55874 | |
| F1MHT1 | Amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase | AGL | 0.047 | 4-Alpha-glucanotransferase activity | Cytoplasm | Glycogen biosynthetic process | PF06202 | IPR008928 | SSF48208 | ||
| L8J1D8 | Eukaryotic translation initiation factor 3 subunit B (eIF3b) | EIF3B | 0.072 | Translation initiation factor activity | 43S complex | Cytoplasmic translation initiation complex | PS50102 | PF08662 | IPR011400 | SM00360 | SSF54928 |
| F1MJ95 | Unc-80 homolog, NALCN channel complex subunit | UNC80 | 0.081 | Cation channel activity | Axon | Cation homeostasis | PF15778 | IPR031542 | |||
| E1B9N8 | Lysine methyltransferase 2D | KMT2D | 0.033 | Histone binding | MLL3/4 complex | RNA polymerase II | PS51805 | PF05965 | IPR034732 | SM00542 | SSF47095 |
| F1MWK8 | Protein tyrosine kinase 7 (inactive) | PTK7 | 0.089 | ATP binding | Cell–cell junction | Actin cytoskeleton reorganization | PS50835 | PF07679 | IPR007110 | SM00409 | SSF48726 |
| E1BCV4 | Nucleoporin 98 | NUP98 | 0.059 | mRNA binding | Kinetochore | Nuclear pore complex assembly | PS51434 | PF04096 | IPR037665 | SSF82215 | |
| A4IFK4 | Synaptopodin 2 | SYNPO2 | 0.076 | Actin binding | Cytoskeleton | Actin bundle assembly | PS50106 | PF00595 | IPR001478 | SM00228 | SSF50156 |
| A6QLI5 | Melanoma-associated antigen D4 (MAGE-D4 antigen) | MAGED4 | 0.094 | PS50838 | PF01454 | IPR037445 | SM01373 | ||||
| E1BLB6 | Protein kinase, DNA-activated, catalytic subunit | PRKDC | 0.086 | ATP binding | DNA-dependent kinase- ligase 4 complex | Activation of innate immune response | PS51189 | PF02259 | IPR016024 | SM01343 | SSF48371 |
| F1MGR3 | Arginine-glutamic acid dipeptide repeats | RERE | 0.100 | Chromatin binding | Histone deacetylase | Branching of a nerve | PS51038 | PF03154 | IPR002951 | SM00439 | SSF46689 |
| A6H737 | Lysyl oxidase homolog 2 (Lysyl oxidase-like protein 2) | LOXL2 | 0.080 | Calcium ion binding | Basement membrane | Cellular protein modification process | PS00926 | PF01186 | IPR001695 | SM00202 | SSF56487 |
| A6QQJ9 | GPRASP1 protein | GPRASP1 | 0.085 | PF04826 | IPR006911 | SSF48371 | |||||
| F1MZM7 | Synapse defective Rho GTPase homolog 2 | SYDE2 | 0.054 | Signal transduction | PS50238 | PF00620 | IPR035892 | SM00324 | SSF48350 | ||
| A0JN33 | Kin of IRRE like 3 | KIRREL3 | 0.083 | Plasma membrane | PS50835 | PF07679 | IPR007110 | SM00409 | SSF48726 | ||
| F1MNU5 | Phospholipase A2 | PLA2G4F | 0.078 | Calcium-dependent phospholipase A2 | Cytoplasm | Arachidonic acid secretion | PS50004 | PF00168 | IPR016035 | SM00239 | SSF52151 |
| E1B949 | FAT atypical cadherin 4 | FAT4 | 0.086 | Calcium ion binding | Apical part of cell | Branching involved in ureteric bud | PS00010 | PF00028 | IPR002126 | SM00112 | |
| F1MZU6 | Collagen alpha-3(IV) chain | COL4A3 | 0.048 | Extracellular matrix | Collagen | Apoptotic process | PS51403 | PF01413 | IPR008160 | SM00111 | SSF56436 |
| E1BEB3 | Histone acetyltransferase | KAT6A | 0.102 | DNA binding | Cytosol | Cellular senescence | PS51504 | PF01853 | IPR016181 | SM00526 | SSF46785 |
| E1BKZ9 | Sortilin | SORT1 | 0.070 | Cytosol | Endocytosis | PF15902 | IPR031777 | SM00602 | |||
| L8HZQ1 | Kielin/chordin-like protein | M91_00882 | 0.063 | PS01208 | PF00093 | IPR036084 | SM00832 | SSF57567 | |||
| F1MG90 | WDFY family member 4 | WDFY4 | 0.023 | PS50197 | PF02138 | IPR016024 | SM01026 | ||||
| L8J3H5 | Sperm-associated antigen 1 | M91_19629 | 0.096 | PS50005 | PF13877 | IPR025986 | SM00028 | SSF48452 | |||
| E1BHN4 | RING-type domain | RNF213 | 0.063 | ATPase activity | Cytoplasm | Wnt signaling pathway | PS00518 | PF00097 | IPR003593 | SM00382 | SSF52540 |
| F1MFU7 | Zinc finger protein 862 | ZNF862 | 0.069 | Nucleic acid binding | Transcription | PS50805 | PF05699 | IPR008906 | SM00349 | SSF10964 | |
| E1BNA3 | Adaptor related protein complex 4 subunit epsilon 1 | AP4E1 | 0.064 | AP-4 adaptor complex | Vesicle-mediated transport | PF01602 | IPR017109 | SM01356 | SSF48371 | ||
| P98167 | SCO-spondin | SSPO | 0.044 | Peptidase inhibitor | Extracellular | Cell adhesion | PS01225 | PF08742 | IPR006207 | SM00832 | KOG1215 |
| E1BEJ9 | NLR family apoptosis inhibitor | NAIP | 0.056 | ATP binding | Plasma membrane | Apoptotic process | PS50143 | PF00653 | IPR003593 | SM00382 | SSF52540 |
| F1MFH3 | FRAS1 related extracellular matrix protein 2 | FREM2 | 0.055 | Basement membrane | Cell communication | PS51854 | PF03160 | IPR038081 | SM00237 | SSF141072 | |
| E1B754 | Microtubule associated serine/threonine kinase 2 | MAST2 | 0.086 | ATP binding | Microtubule cytoskeleton | Cytoskeleton organization | PS51285 | PF08926 | IPR000961 | SM00228 | |
| E1BP05 | Usherin | USH2A | 0.092 | Collagen binding | Apical plasma membrane | Animal organ morphogenesis | PS01248 | PF00041 | IPR013320 | SM00180 | SSF49265 |
| A3KN18 | CEBPZ protein | CEBPZ | 0.067 | Ribosome biogenesis | PF03914 | IPR016024 | SSF48371 |
Discussion
To date, to the best of our knowledge, the current report is the most comprehensive characterization of minor bovine milk proteome. Bovine milk is a complex body fluid containing various secreted proteins and is an important source of human nutrition. However, the minor immune-related host defense proteins have not been wholly characterized and need to be explored exhaustively. Milk-derived bioactive peptides present in whey and casein have potential roles as health-promoting agents against cancer, diabetes, hypertension, osteoporosis, and several other disorders (27). Whey proteins can serve promising health benefits in terms of their ability to act as antioxidant, antihypertensive, anti-tumor, anti-viral, anti-bacterial, and hypo-lipidemic (28). Bioactive peptides present in milk exosomes, colostrum, milk fat globular membrane, skimmed milk, and whey provide information on generating large bovine milk proteome datasets.
The present study was designed with the aim to explore the in-depth milk whey proteome of Indian zebu (Sahiwal) cattle, which was not studied earlier. Indian zebu (Sahiwal) cattle have been acclaimed for the potential to disease resistance and adaptability for heat stress as compared with exotic Bos taurus cattle. For proteomics analysis purpose, different protocols for milk protein extraction were attempted including acetone precipitation of full cream milk (20), ultracentrifugation to pellet caseins (7), ammonium sulfate precipitation of caseins to isolate serum (29), and acetic acid removal of caseins to isolate whey proteins (30). Further, optimization of protein extraction methods in skimmed milk of Holstein and Jersey breeds was studied (21), where they compared three protein extraction procedures that were urea-based buffer, TCA/acetone, and methanol/chloroform. The results reported the methanol/chloroform method as the best-optimized method for peptide identification in cattle breeds. In our study, we optimized three methods for protein extraction, which includes ultracentrifugation followed by acetone precipitation, CaCl2 precipitation at different concentrations, and TCA/acetone precipitation method. Out of these three methods, TCA/acetone precipitation was found to be the best-optimized method with maximum peptide identification in comparison with other protein extraction methods.
In an attempt to generate the proteome map, Smolenski et al. used bovine skimmed milk, whey, and milk fat globular membrane during peak lactation, colostrum, and mastitis stages. The results identified only 53 minor milk host defense proteins with 2,903 peptides (2). The latest report on differentially expressed changes in lactation and parity stage-specific whey proteome determined 103 proteins on primiparous cows (31). According to earlier reports, milk proteins from exosomes, milk fat globular membrane, skimmed milk, and whey have a nutraceutical role (32). Putative biomarkers of negative energy balance from milk proteins of early lactation in dairy cows were identified (33). The study found six milk proteins out of 59 proteins linked to metabolism and mammary gland cell proliferation. Another study was conducted on the characterization of bovine milk whey proteome during early lactation in Holstein and Jersey breeds by Tacoma et al. (18). They identified 935 low abundant proteins in skimmed milk fractions of the two breeds and highlighted the breed difference between proteome of Jersey and Holstein Friesian cows. However, no reports are available to date on milk whey profiling in Indian zebu cattle. Thus, the present study focuses on the global proteomic profiling of Indian zebu (Sahiwal) cattle, an important milch breed of the Indian subcontinent, to generate the proteomic profiling data for the first time. The findings suggest the identification and role of pathways related to activated immune response and stress tolerance homeostasis mechanisms. However, comparative proteomic analysis across different lactation stages was conducted in Malnad Gidda cattle (19) and Jersey and Kashmiri cattle (34). A recent report compiled an online database with 3,100 proteins from milk whey, milk fat globular membrane, and exosomes (35). Similarly, another separate study reported 4,654 cow milk proteins in five lactation stages to create the database. The stages were colostrum, early lactation, mid-lactation, peak lactation, and milk fractions consisting of exosomes, skimmed milk, and milk fat globular membrane (36).
Altogether, small sets of bovine milk proteome have been studied earlier, but none of them was able to provide the exhaustive proteome profile. The current report is the first time identification of 6,210 proteins in milk whey of Indian zebu (Sahiwal) cattle. It provides an in-depth identification of milk whey proteome to understand the role in human health and its biological significance. We determined proteome with the role in metal ion binding, zinc binding, and many more. Notably, from 6,210 proteins, a total of 1,568 proteins have UniProt reviewed protein entries, but the large majority (4,514) are unreviewed protein entries that have been validated by our data and could be further explored. Our bioinformatics analysis found that identified proteins are involved in various activities as ATP binding, DNA binding, metal ion binding, and zinc ion binding. Our fuzzy clustering analysis identified the presence of metal ion binding proteins, clusters in two independent databases Pfam and Panther, suggesting the ample presence of these proteins in whey (Supplementary Figure 5). The PCA revealed that actin-binding proteins are maximally distributed, and the majority of proteins were mammary gland specific, secreted in milk. Some of the integral components are found in the large granules of neutrophils with their possible role in immunoregulation.
The cellular component–centered gene ontology analysis reveals the involvement of major identified proteins in the nucleus, cytoplasm, plasma membrane, nucleoplasm, and mitochondria. Based on the interactive studies of whey proteins with metal ions, our whey proteome dataset proposed that it has potential applications in nutraceuticals and food industry (37). The metal ion binding properties of whey proteins could be highly useful and provide a way for designing functional foods. Purification of whey proteins such as lactoferrin which is known for its anti-bacterial and antimicrobial role can be utilized as a nutritional and dietary supplement as well as synergetic treatment in patients with anemia, kidney disease (38), and also in cancer (39). The low abundant minor milk proteins have a potential role in host defense and immune regulation. Several studies reviewed the role of immunoglobulins in milk for health and immunity (40–42). The presence of antimicrobial proteins and peptides such as beta-defensin, cathelicidins, complement proteins, and several other minor milk whey proteins were reported (42–46). In our study, we identified several minor proteins as cathelicidin 1 and 2, gelsolin, interferon 12 subunit alpha, fatty acid-binding protein, nucleobindin 2, transthyretin, claudin, haptoglobin, MFGE8 protein, polymeric immunoglobulin receptor, ubiquilin 4, and collectin 46 with different roles and biological significance (Table 1).
The complete proteome advocates that milk acts as an excellent medium for microbial growth that is evident from the enormous diversity of different high concentration peptides and proteins. It is the best source of nitrogen content for microbial growth. It allows the natural growth and habitat of beneficial microbes. Recently, we identified the occupancy of probiotics in fermented milk products and found a constant abundance (47). The determination of different attributes from the isolated microorganism showed positive results to classify them in the probiotics category (48). Combined genomics and proteomics analysis provide the landmarks for several genes and proteins coded in the genome for the probiotic attributes (49, 50). Hence, it strongly suggests that the whole milk supplementation allows the nourishment and abundant growth of healthy gut microbiota.
Apart from the significant milk proteins with a role in the innate immune system and host defense, numerous minor proteins were found to be overwhelming and found to be involved in response to regulation of immunity and inflammation. Quantification of the protein amount in ng reveals that significant whey proteins as alpha-lactalbumin, beta-lactoglobulin, beta-casein, ribonuclease pancreatic, albumin, and immunoglobulin chain proteins were identified in higher protein concentrations. Several other low abundant proteins (Table 1) as gelsolin, tetranectin, haptoglobin, cathepsin z, lysozyme, brain ribonuclease, peptidoglycan recognition protein 1, cathelicidin 2, vimentin, and others have been identified with a possible role in biological and physiological mechanisms.
Conclusion
Milk proteome profiling was reported for the first time in Indian zebu (Sahiwal) cattle. Although studies were conducted in Holstein and Jersey (B. taurus) cattle, there is a lack of milk proteome profiling data in Indian zebu (Sahiwal) cattle. In this study, different protein extraction procedures as ultracentrifugation followed by acetone precipitation, CaCl2 precipitation at different concentrations, and TCA/acetone precipitation were optimized where TCA/acetone precipitation gave maximum peptide identification by LC-MS/MS. This study led to the identification of 6,210 proteins from in-gel digestion and in-sol digestion. All the generated proteomics raw files were analyzed with three different search engines, namely, Mascot, Comet, and Tandem, for high confidence identification of the profiled whey proteome. The majority of the proteins identified with a high score were found to play a role in immune regulation and host defense system implicating lesser disease susceptibility and better adaptability of Indian zebu (Sahiwal) cattle. The present study represents the first time reported Sahiwal milk proteome data in Indian zebu cattle, which could serve as a database for different cattle genetic resources of India.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
AC and SA equally designed, conducted data analysis, and wrote the article. AC, SA, SB, PR, and VV helped in designing and performing the experiments. SA performed the data analysis and created the illustrations for the article. SK and AM supervised the whole project and reviewed the article. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are thankful to DST-SERB for providing funding. SA is thankful to the Indian Council of Medical Research (ICMR) for the SRF fellowship. We would also like to thank the Director of the ICAR–National Dairy Research Institute (NDRI), India, for providing the necessary facilities to carry out research work.
Footnotes
Funding. Department of Science and Technology funded this work, DST-SERB under the National Post Doctoral Fellowship scheme File No. PDF/2015/000770.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnut.2020.00150/full#supplementary-material
(A) Comparison of SDS-PAGE patterns of milk whey samples for optimization of different extraction procedures. 1D image of milk whey protein extraction procedures followed for optimization. (a) Ultracentrifugation followed by acetone precipitation. (b) CaCl2 precipitation method at different concentrations: 60, 90, and 120 mM. (c) TCA/acetone precipitation method. (B) Comparison of different methods of preparation of bovine milk whey in Indian Sahiwal cattle. (a) Acid precipitation at different pH. (b) Ultracentrifugation at 65,000×g for 2 h. (c) Ultracentrifugation followed by acid precipitation.
Two-dimensional gel electrophoretogram of bovine milk whey in Indian Sahiwal cattle.
Animal-to-animal proteome SDS profile of 18 different animals shown on two individual gels (A,B). Ten best sharing proteome profiles were selected for the determination of deep whey proteome.
All the different Gene Ontological Information determined for the whey proteome data shown in (A–J). (A) ngLog mapped total proteome. (B) Functional annotation terms specified. (C) Complete cellular component counts. (D) The total molecular function counts. (E) The total biological function counts. (F) Full KEGG pathway annotation. (G) Total SMART term counts. (H) Total InterPro terms counts. (I) Tissue-specific proteins determined. (J) Total reactome term counts.
Fuzzy C-means clustering of proteins identified with the metal ion binding ability.
Final complete table for identifications of total whey proteins in Trans Proteomics Pipeline (TPP) using Comet and Tandem search engine through in-gel and in-solution digestion.
Final complete table for identifications of total whey proteins in ProteinScape using Mascot search engine through in-gel and in-solution digestion.
Total number of chromosome-associated spectra counts, total proteins mapped, and enrichment values.
ngLOC-based classification of whey proteome in respective cellular organelle classification.
DAVID-based classification of all the proteins mapped in whey proteome.
References
- 1.Yang Y, Bu D, Zhao X, Sun P, Wang J, Zhou L. Proteomic analysis of cow, yak, buffalo, goat, and camel milk whey proteins: quantitative differential expression patterns. J Proteome Res. (2013) 12:1660–7. 10.1021/pr301001m [DOI] [PubMed] [Google Scholar]
- 2.Smolenski G, Haines S, Kwan FYS, Bond J, Farr V, Davis SR, et al. Characterisation of host defence proteins in milk using a proteomic approach. J Proteome Res. (2007) 6:207–15. 10.1021/pr0603405 [DOI] [PubMed] [Google Scholar]
- 3.Reddi S, Kumar N, Vij R, Mada SB, Kapila S, Kapila R. Akt drives buffalo casein-derived novel peptide-mediated osteoblast differentiation. J Nutr Biochem. (2016) 38:134–44. 10.1016/j.jnutbio.2016.08.003 [DOI] [PubMed] [Google Scholar]
- 4.Spertino S, Cipriani V, De Angelis C, Giuffrida MG, Marsano F, Cavaletto M. Proteome profile and biological activity of caprine, bovine and human milk fat globules. Mol Biosyst. (2012) 8:967–74. 10.1039/C2MB05400K [DOI] [PubMed] [Google Scholar]
- 5.Roncada P, Piras C, Soggiu A, Turk R, Urbani A, Bonizzi L. Farm animal milk proteomics. J Proteomics. (2012) 75:4259–74. 10.1016/j.jprot.2012.05.028 [DOI] [PubMed] [Google Scholar]
- 6.Wada Y, Lönnerdal B. Bioactive peptides derived from human milk proteins—mechanisms of action. J Nutr Biochem. (2014) 25:503–14. 10.1016/j.jnutbio.2013.10.012 [DOI] [PubMed] [Google Scholar]
- 7.Hettinga K, Van Valenberg H, De Vries S, Boeren S, Van Hooijdonk T, Van Arendonk J, et al. The host defense proteome of human bovine milk. PloS ONE. (2011) 6:e19433. 10.1371/journal.pone.0019433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Joshi BK, Singh A, Gandhi RS. Performance evaluation, conservation and improvement of Sahiwal cattle in India. Anim Genet Resour. (2001) 31:43–54. 10.1017/S1014233900001474 [DOI] [Google Scholar]
- 9.Gaughan JB, Mader TL, Holt SM, Josey MJ, Rowan KJ. Heat tolerance of Boran and Tuli crossbred steers. J Anim Sci. (1999) 77:2398–405. 10.2527/1999.7792398x [DOI] [PubMed] [Google Scholar]
- 10.Glass EJ, Preston PM, Springbett A, Craigmile S, Kirvar E, Wilkie G, et al. Bos taurus and Bos indicus (Sahiwal) calves respond differently to infection with Theileria annulata and produce markedly different levels of acute phase proteins. Int J Parasitol. (2005) 35:337–47. 10.1016/j.ijpara.2004.12.006 [DOI] [PubMed] [Google Scholar]
- 11.Scarso S, McParland S, Visentin G, Berry DP, McDermott A, De Marchi Genetic M. and nongenetic factors associated with milk color in dairy cows. J Dairy Sci. (2017) 100:7345–61. 10.3168/jds.2016-11683 [DOI] [PubMed] [Google Scholar]
- 12.Sorathiya LM, Tyagi KK, Patel MD, Raval AP. Effect of non-genetic factors on milk compositions in Surti buffaloes. Indian Vet J. (2018) 95:43–45. [Google Scholar]
- 13.Vargas-Bello-Pérez E, Márquez-Hernández RI, Hernández-Castellano LE. Bioactive peptides from milk: animal determinants and their implications in human health. J Dairy Res. (2019) 86:136–44. 10.1017/S0022029919000384 [DOI] [PubMed] [Google Scholar]
- 14.Affolter M, Grass L, Vanrobaeys F, Casado B, Kussmann M. Qualitative, and quantitative profiling of the bovine milk fat globule membrane proteome. J Proteomics. (2010) 73:1079–88. 10.1016/j.jprot.2009.11.008 [DOI] [PubMed] [Google Scholar]
- 15.Pereira PC. Milk nutritional composition and its role in human health. Nutrition. (2014) 30:619–27. 10.1016/j.nut.2013.10.011 [DOI] [PubMed] [Google Scholar]
- 16.Hurley WL, Theil PK. Perspectives on immunoglobulins in colostrum and milk. Nutrients. (2011) 3:442–74. 10.3390/nu3040442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mills S, Ross RP, Hill C, Fitzgerald GF, Stanton C. Milk intelligence: mining milk for bioactive substances associated with human health. Int Dairy J. (2011) 21:377–401. 10.1016/j.idairyj.2010.12.011 [DOI] [Google Scholar]
- 18.Tacoma R, Fields J, Ebenstein DB, Lam YW, Greenwood SL. Characterization of the bovine milk proteome in early-lactation Holstein and Jersey breeds of dairy cows. J Proteomics. (2016) 130:200–10. 10.1016/j.jprot.2015.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mol P, Kannegundla U, Dey G, Gopalakrishnan L, Dammalli M, Kumar M, et al. Bovine milk comparative proteome analysis from early, mid, and late lactation in the cattle breed, Malnad Gidda (Bos indicus). Omics. (2018) 22:223–35. 10.1089/omi.2017.0162 [DOI] [PubMed] [Google Scholar]
- 20.Danielsen M, Codrea MC, Ingvartsen KL, Friggens NC, Bendixen E, Røntved CM. Quantitative milk proteomics–host responses to lipopolysaccharide-mediated inflammation of bovine mammary gland. Proteomics. (2010) 10:2240–9. 10.1002/pmic.200900771 [DOI] [PubMed] [Google Scholar]
- 21.Vincent D, Ezernieks V, Elkins A, Nguyen N, Moate PJ, Cocks BG, et al. Milk bottom-up proteomics: method optimization. Front Genet. (2016) 6:360. 10.3389/fgene.2015.00360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ali SA, Kaur G, Kaushik JK, Malakar D, Mohanty AK, Kumar S. Examination of pathways involved in leukemia inhibitory factor (LIF)-induced cell growth arrest using label-free proteomics approach. J Proteomics. (2017) 168:37–52. 10.1016/j.jprot.2017.07.014 [DOI] [PubMed] [Google Scholar]
- 23.Suhail A, Rizvi ZA, Mujagond P, Ali SA, Gaur P, Singh M, et al. DeSUMOylase SENP7-mediated epithelial signaling triggers intestinal inflammation via expansion of gamma-delta t cells. Cell Rep. (2019) 29:3522–38. 10.1016/j.celrep.2019.11.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lönnerdal B. Infant formula and infant nutrition: bioactive proteins of human milk and implications for composition of infant formulas. Am J Clin Nutr. (2014) 99:712–7. 10.3945/ajcn.113.071993 [DOI] [PubMed] [Google Scholar]
- 25.Johansen FE, Kaetzel CS. Regulation of the polymeric immunoglobulin receptor and IgA transport: new advances in environmental factors that stimulate pIgR expression and its role in mucosal immunity. Mucosal Immunol. (2011) 4:598–602. 10.1038/mi.2011.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mishra SR, Palai TK. Importance of heat shock protein 70 in livestock-at cellular level. J Mol Pathophysiol. (2014) 3:30–32. 10.5455/jmp.20141028023220 [DOI] [Google Scholar]
- 27.Mohanty DP, Mohapatra S, Misra S, Sahu PS. Milk derived bioactive peptides and their impact on human health–a review. Saudi J Biol Sci. (2016) 23:577–83. 10.1016/j.sjbs.2015.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keri Marshall ND. Therapeutic applications of whey protein. Altern Med Rev. (2004) 9:136–56. [PubMed] [Google Scholar]
- 29.Hogarth CJ, Fitzpatrick JL, Nolan AM, Young FJ, Pitt A, Eckersall PD. Differential protein composition of bovine whey: a comparison of whey from healthy animals and from those with clinical mastitis. Proteomics. (2004) 4:2094–100. 10.1002/pmic.200300723 [DOI] [PubMed] [Google Scholar]
- 30.Senda A, Fukuda K, Ishii T, Urashima T. Changes in the bovine whey proteome during the early lactation period. Anim Sci J. (2011) 82:698–706. 10.1111/j.1740-0929.2011.00886.x [DOI] [PubMed] [Google Scholar]
- 31.Yang Y, Zheng N, Zhao X, Yang J, Zhang Y, Han R, et al. Changes in whey proteome with lactation stage and parity in dairy cows using a label-free proteomics approach. Food Res Int. (2020) 128:108760. 10.1016/j.foodres.2019.108760 [DOI] [PubMed] [Google Scholar]
- 32.Verma A, Ambatipudi Challenges K. and opportunities of bovine milk analysis by mass spectrometry. Clin. Proteomics. (2016) 13:8. 10.1186/s12014-016-9110-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Delosière M, Pires J, Bernard L, Cassar-Malek I, Bonnet M. Milk proteome from in silico data aggregation allows the identification of putative biomarkers of negative energy balance in dairy cows. Sci Rep. (2019) 9:1–11. 10.1038/s41598-019-46142-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bhat SA, Ahmad SM, Ibeagha-Awemu EM, Mobashir M, Dar MA, Mumtaz PT, et al. Comparative milk proteome analysis of Kashmiri and Jersey cattle identifies differential expression of key proteins involved in immune system regulation and milk quality. BMC Genomics. (2020) 21:161. 10.1186/s12864-020-6574-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maity S, Bhat AH, Giri K, Ambatipudi K. BoMiProt: a database of bovine milk proteins. J. Proteomics. (2020) 215:103648. 10.1016/j.jprot.2020.103648 [DOI] [PubMed] [Google Scholar]
- 36.Delosière M, Pires J, Bernard L, Cassar-Malek I, Bonnet M. Dataset reporting 4654 cow milk proteins listed according to lactation stages and milk fractions. Data Brief . (2020) 2019:105105. 10.1016/j.dib.2019.105105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodzik A, Pomastowski P, Sagandykova GN, Buszewski B. Interactions of whey proteins with metal ions. Int J Mol Sci. (2020) 21:2156. 10.3390/ijms21062156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chan S, Au K, Francis RS, Mudge DW, Johnson DW, Pillans PI. Phosphate binders in patients with chronic kidney disease. Aust Prescr. (2017) 40:9–14. 10.18773/austprescr.2017.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Butler JS, Sadler PJ. Targeted delivery of platinum-based anticancer complexes. Curr Opin Chem Biol. (2013) 17:175–88. 10.1016/j.cbpa.2013.01.004 [DOI] [PubMed] [Google Scholar]
- 40.Sordillo LM, Shafer-Weaver K, DeRosa D. Immunobiology of the mammary gland. J Dairy Sci. (1997) 80:1851–65. 10.3168/jds.S0022-0302(97)76121-6 [DOI] [PubMed] [Google Scholar]
- 41.Korhonen H, Marnila P, Gill HS. Milk immunoglobulins and complement factors. Br J Nutr. (2000) 84:75–80. 10.1017/S0007114500002282 [DOI] [PubMed] [Google Scholar]
- 42.Wheeler TT, Smolenski GA, Harris DP, Gupta SK, Haigh BJ, Broadhurst MK, et al. Host-defence-related proteins in cows' milk. Animal. (2012) 6:415. 10.1017/S1751731111002151 [DOI] [PubMed] [Google Scholar]
- 43.Eckersall PD, Young FJ, McComb C, Hogarth CJ, Safi S, Fitzpatrick JL, et al. Acute phase proteins in serum and milk from dairy cows with clinical mastitis. Vet Rec. (2001) 148:35–41. 10.1136/vr.148.2.35 [DOI] [PubMed] [Google Scholar]
- 44.Rainard P. The complement in milk and defense of the bovine mammary gland against infections. Vet Res. (2003) 34:647–70. 10.1051/vetres:2003025 [DOI] [PubMed] [Google Scholar]
- 45.Swanson K, Gorodetsky S, Good L, Davis S, Musgrave D, Stelwagen K, et al. Expression of a β-defensin mRNA, lingual antimicrobial peptide, in bovine mammary epithelial tissue is induced by mastitis. Infect Immun. (2004) 72:7311–14. 10.1128/IAI.72.12.7311-7314.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murakami M, Dorschner RA, Stern LJ, Lin KH, Gallo RL. Expression and secretion of cathelicidin antimicrobial peptides in murine mammary glands and human milk. Pediatr Res. (2005) 57:10–15. 10.1203/01.PDR.0000148068.32201.50 [DOI] [PubMed] [Google Scholar]
- 47.Panicker AS, Ali SA, Anand S, Panjagari NR, Kumar S, Mohanty AK, et al. Evaluation of some in vitro probiotic properties of Lactobacillus fermentum strains. J. Food Sci Technol. (2018) 55:2801–7. 10.1007/s13197-018-3197-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parijat LV, Munjal K, Ali SA, Rawat P, Kumar S, Behare P, et al. Evaluation of stationary phase and bile stress related protein spots in Lactobacillus fermentum NCDC 400 by 2-DE method. Indian J Dairy Sci. (2016) 69:455–9. [Google Scholar]
- 49.Ali SA, Kumar S, Mohanty AK, Behare P. Draft genome sequence of Lactobacillus fermentum NCDC 400, isolated from a traditional Indian dairy product. Genome Announc. (2018) 6:e01492-17. 10.1128/genomeA.01492-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pragya P, Kaur G, Ali SA, Bhatla S, Rawat P, Lule V, et al. High-resolution mass spectrometry-based global proteomic analysis of probiotic strains Lactobacillus fermentum NCDC 400 and RS2. J Proteomics. (2017) 152:121–30. 10.1016/j.jprot.2016.10.016 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Comparison of SDS-PAGE patterns of milk whey samples for optimization of different extraction procedures. 1D image of milk whey protein extraction procedures followed for optimization. (a) Ultracentrifugation followed by acetone precipitation. (b) CaCl2 precipitation method at different concentrations: 60, 90, and 120 mM. (c) TCA/acetone precipitation method. (B) Comparison of different methods of preparation of bovine milk whey in Indian Sahiwal cattle. (a) Acid precipitation at different pH. (b) Ultracentrifugation at 65,000×g for 2 h. (c) Ultracentrifugation followed by acid precipitation.
Two-dimensional gel electrophoretogram of bovine milk whey in Indian Sahiwal cattle.
Animal-to-animal proteome SDS profile of 18 different animals shown on two individual gels (A,B). Ten best sharing proteome profiles were selected for the determination of deep whey proteome.
All the different Gene Ontological Information determined for the whey proteome data shown in (A–J). (A) ngLog mapped total proteome. (B) Functional annotation terms specified. (C) Complete cellular component counts. (D) The total molecular function counts. (E) The total biological function counts. (F) Full KEGG pathway annotation. (G) Total SMART term counts. (H) Total InterPro terms counts. (I) Tissue-specific proteins determined. (J) Total reactome term counts.
Fuzzy C-means clustering of proteins identified with the metal ion binding ability.
Final complete table for identifications of total whey proteins in Trans Proteomics Pipeline (TPP) using Comet and Tandem search engine through in-gel and in-solution digestion.
Final complete table for identifications of total whey proteins in ProteinScape using Mascot search engine through in-gel and in-solution digestion.
Total number of chromosome-associated spectra counts, total proteins mapped, and enrichment values.
ngLOC-based classification of whey proteome in respective cellular organelle classification.
DAVID-based classification of all the proteins mapped in whey proteome.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.