Abstract
Cancer is one of the most leading causes of death and a major public health problem, universally. According to accumulated data, annually, approximately 8.5 million people died because of the lethality of cancer. Recently, a novel RNA domain-containing endonuclease-based genome engineering technology, namely the clustered regularly interspaced short palindromic repeat (CRISPR)-associated protein-9 (Cas9) have been proved as a powerful technique in the treatment of cancer cells due to its multifunctional properties including high specificity, accuracy, time reducing and cost-effective strategies with minimum off-target effects. The present review investigates the overview of recent studies on the newly developed genome-editing strategy, CRISPR/Cas9, as an excellent pre-clinical therapeutic option in the reduction and identification of new tumor target genes in the solid tumors. Based on accumulated data, we revealed that CRISPR/Cas9 significantly inhibited the robust tumor cell growth (breast, lung, liver, colorectal, and prostate) by targeting the oncogenes, tumor-suppressive genes, genes associated to therapies by inhibitors, genes associated to chemotherapies drug resistance, and suggested that CRISPR/Cas9 could be a potential therapeutic target in inhibiting the tumor cell growth by suppressing the cell-proliferation, metastasis, invasion and inducing the apoptosis during the treatment of malignancies in the near future. The present review also discussed the current challenges and barriers, and proposed future recommendations for a better understanding.
Keywords: CRISPR/Cas9, Genome editing, RNA editing, Solid tumor, Off-target, Viral delivery
Graphical abstract
Highlights
-
S•
CRISPR/Cas9 is a pioneering efficient and less time-consuming genome editing technology.
-
•
CRISPR/Cas9 tool followed the specific PAM sequence (2–6 bp) on target DNA for genome editing.
-
•
CRISPR/Cas9 efficiently repressed the expression of different oncogenes in solid tumors.
-
•
Cas9 should update its application to the stomach and cervix to treat more cancer.
-
•
Chemical agents should be used in the Cas9 synthesis to reduce its off-target effects.
1. Introduction
Globally cancer is considered one of the major leading causes of death. In 2018, approximately 9.6 million people had died because of cancer. Cancer is characterized by the heterogeneity and accumulation of different genetic mutations and alternations, including modification in epigenetic factors (DNMT1), variation in genes like multidrug resistance genes (MDR1/Pgp and ABC), activation in tumor suppressor (TP53) and oncogenes (RAS) [1]. The tumor heterogeneity is categorized into intratumoral and intertumoral heterogeneity. Intratumoral heterogeneity mainly occurs due to the unequal circulation of genetically distinct tumor subpopulations across single or different disease sites. In contrast, intertumoral heterogeneity is associated with patients' specific factors such as patients-somatic profile and germline alternations. Intertumoral heterogeneity also circulates between patients harboring tumors of the same histological type [2]. However, genetic instability generated several critical substrates for tumor heterogeneity that are maintained by specific therapeutic processes. Resultingly, this heterogeneity triggers the site-specific response and ultimately provides the seeds for the development of cancer resistance [3].
During the past few decades, different therapies, including immunotherapy, targeted antibodies, hormone therapy, chemotherapy, targeted drug therapy, stem cell transplant, and surgery have been introduced into the medical field for the betterment of prognosis in cancer patients, which significantly eradicate or condense the cancerous cells and upsurges the maximum lifespan of about only five years [4,5]. Even after these therapies, cancer continues to evolve and ultimately give rise to molecular heterogeneity attributed to various molecular events like epigenetic, genetic, phenotypic, and transcriptomic alternations. However, due to their several limitations, including toxicity, high cost, and adverse effects (affect the healthy cells and caused the partial or complete loss of organ functions) these therapies did not improve as much survival of patients. Because of the multi-step existence of cancer, the prevention and treatment of cancer are difficult. So, it is prerequisite to study its prognosis, pathogenesis, etiology, and phenotypes to improve the existing therapies and develop a new treatment [6].
In recent years, a genome editing engineering technology has been proposed for the cancer treatment that can target any gene in the affected area and make knock-in and knock-out alternations. DNA domain binding conventional techniques, including transcriptional activator-like effector nuclease (TALENs), and zinc finger nuclease (ZFNs) have greatly influenced in molecular biology for engendering animal, cellular cancer models, and therapy investigations. But because of their complexity and time-consuming strategies, their widespread use has been restricted [1,7,8]. However, RNA interference (RNAi) is considered a great establish for gene expression in cancer therapy research, but due to its temporary knockdown effects, it needs to be continuously administrated to gain the acceptance level of knockdown [9]. In the present days, a new RNA domain-containing endonuclease-based genome engineering tool, namely the clustered regularly interspaced short palindromic repeat (CRISPR)-associated protein-9 (Cas9) have been developed for cancer treatment that gained a significant amendment in the gene expression and gene therapy due to its simplicity, and less time-consuming target abilities with high accuracy and efficiency [10].
Solid tumors (see glossary; Supplementary material) including breast, lung, liver, prostate, and colorectal cancer are the most common types of tumors, that showed less progress toward their treatment by gene therapies as compared to non-solid tumors like leukemia. Thus, with the development of CRISPR/Cas-9, this situation rapidly changed. Previously published data suggested that CRISPR/Cas-9 can effectively target the cancer cells and suppress tumor growth by inducing apoptosis and inhibiting cell proliferation and metastasis [11,12]. The single guide-RNA (sgRNA), endonuclease enzyme Cas-9 are the two fundamental components of the CRISPR system that guided the CRISPR technology to treat the cancer cells. Cas-9 protein induced double-stranded DNA breaks (DSBs) at target sites. It initiates the DNA repair process by non-homologous end joining (NHEJ) repair mechanism of insertion and deletion of a small part of a sequence. However, Cas-9 can also practice another way for repair mechanism, including homology-directed repair (HDR) using template DNA. CRISPR/Cas-9 technique was first applied in animal and mammalian cells six years ago. It was used as the most ingenious and versatile strategy against cancer exhibiting and treatment explorations [13,14].
The principal objective of the present review is to investigate the potential therapeutic properties of CRISPR/Cas9 against solid tumors, including breast, lung, liver, colorectal, and prostate cancer. The present review also investigates the structure, mechanism, current challenges and future prospective, and delivery of CRISPR/Cas9 to the target site. Furthermore, in the present comprehensive review, many genetic mutations in genes regarding cancer treatment by CRISPR/Cas9 are discussed to gain the focus of the researcher for the translation of laboratory-research to clinics.
2. Advantages of genome editing CRISPR/Cas9 technology
In the molecular biology field, the genome editing is usually carried out by binding the sequence of specific DNA binding domain to a non-specific binding domain which allows precise and efficient alternations of a gene of interest by DNA double-strand breaks (DSBs) via stimulating the DNA repair mechanism [15]. The emerging evidence suggested that the efficiency of genome editing have been improved by programmable nucleases that used to correct the mutations in several diseases such as sickle cell disease, several combined immunodeficiency (SCID), cancer, and hemophilia [16]. Conventionally, two different genome editing techniques such as transcription activator-like effector nucleases (TALENs) and zinc-finger nucleases (ZFNs) have been extensively used for DNA repair by targeting the gene of interest. The efficiency of these techniques entirely depends on specificity and affinity of nucleases [17]. Traditionally, TALENs and ZFNs editing technologies were used to target several oncogenes like human Papillomavirus oncogenes. A study showed that Compo-Zr ZFNs significantly targeted the E6 gene in HPV-16 cell lines with the editing activity of 50% but showed less activity in CaSki and SiHa cervical cell lines. Similarly, TALENs effectively edited the E7 gene in SiHa cells but did not show the desirable activity in other cell lines [16]. Recently, a study revealed that ZFNs significantly removed the HIV-1 pro-viral DNA and gave a new hope to target the other viral genomes in single or combine form (TALEN/ZFN) [18]. Although these genome editing techniques showed somehow better results, but these both techniques exert off-target effects and also very time taking and less efficient techniques. So, there was a need to develop a highly efficient and accurate technique to compensate the old ones [19].
Based on the emerging achievement of CRISPR/Cas9 in the molecular field, this technique has superiority over the other conventional genome editing techniques such as transcriptional activator-like effector nuclease (TALENs), and zinc finger nuclease (ZFNs) due to its limited off-target effects and less time-consuming target ability. The CRISPR-Cas9 only required the short complementary sequence of sg-RNA to target the DNA, which is a relatively cost-effective and much easier method than conventional tools (TALENs, ZFNs, and RNAi) to edit desired part of DNA [20,21]. According to the accumulated data, the CRISPR/Cas9 system was initially identified by the prokaryotic (Escherichia coli) adaptive immune system, which is considered good anti-phages and antiviruses system [22]. CRISPR/Cas9 comprises three distinct prime components, including CRISPR RNA (crRNA), trans-activating CRISPR RNA (tracrRNA), and endonuclease Cas-9 [23].
CRISPR/Cas9 possess several distinct variants (dCas9, CRISPR-X, CRISPR-a, and CRISPR-i), which can perform various functions under different circumstances, for instance, deactivated Cas-9 (d-Cas9) can be used to target the epigenome (see glossary) by suppressing the enzymatic activities of HNH domains without disrupting the sequence [1]. Similarly, another study reported that CRISPR-I significantly blocked the endogenous genes, including CXCR-4 and CD-71 in the HeLa cells [24]. The literature reported that the CRISPR/Cas9 has the highest genome editing proficiency than all conventional techniques, including ZFNs and TALEN, and more than 79% effectiveness of CRISPR/Cas9 is recorded to edit the human pluripotent stem cells (PSCs; see glossary) [25]. The emerging evidence revealed that CRISPR/Cas9 tool effectively targeted at the DNA level and resulted in the temporary or permanent inactivation of genes, which make it best genome editing technique with less off-target effects as compared to the RNAi technique [26,27].
Based on the multi-step transmutation progression of cancer, there is a need to develop a new multifunctional therapeutic drug/tool. However, count on the ability of CRISPR/Cas9 to edit the multiple genes at a time, this technique is considered as a direct and parallel target and recognized as an efficient test center tool against cancer-associated genomic mutations both in vivo and in vitro studies [26,27].
3. Structure and mechanism of CRISPR/Cas9 system
The accumulated data suggested that CRISPR/Cas9 system comprises three main components including Cas9 protein with DNA endonuclease activity, a single guided RNA (sgRNA), and a tracrRNA, which makes attraction with Cas9 (see Fig. 1 ) [28]. The sgRNA contains a crucial function in the specificity, efficiency, and accuracy of the CRISPR/Cas9 tool. The first 10–12 nucleotides of sgRNA at 3ˊ end next to a proto-spacer adjacent motif (PAM; see glossary) known as seed sequence directed the Cas9 protein in the genome editing performance by binding to the target sites [29]. The recent evidence also suggested that the truncated sgRNA with less than 20 nucleotides effectively minimized the off-target effects of about 5000 folds without losing efficiency [22]. In addition, another study also reported that the development of about 5 base pairs in sgRNA duplex excellently enhanced the knockout proficiency of CRISPR/Cas9 [23]. Cas9 is a CRISPR class 2 isolated (Streptococcus pyogenes) protein guided by RNA-guided nuclease and responsible for DNA double-strand destruction [11]. Cas9 protein contains two lobes, including alpha and nuclease lobes. The nuclease lobe consists of two extra domains, including RuvC and HNH domain. The RuvC (retrovirus integrase protein) cuts the non-targeted DNA double strands, whereas the HNH cleaved the specifically targeted strand of DNA. Both of these domains usually formed a DSB (double-strand break), but due to mutation at D10A and H840A in RuvC and HNH domains respectively, results in deactivation of Cas9 (dCas9) [31,32].
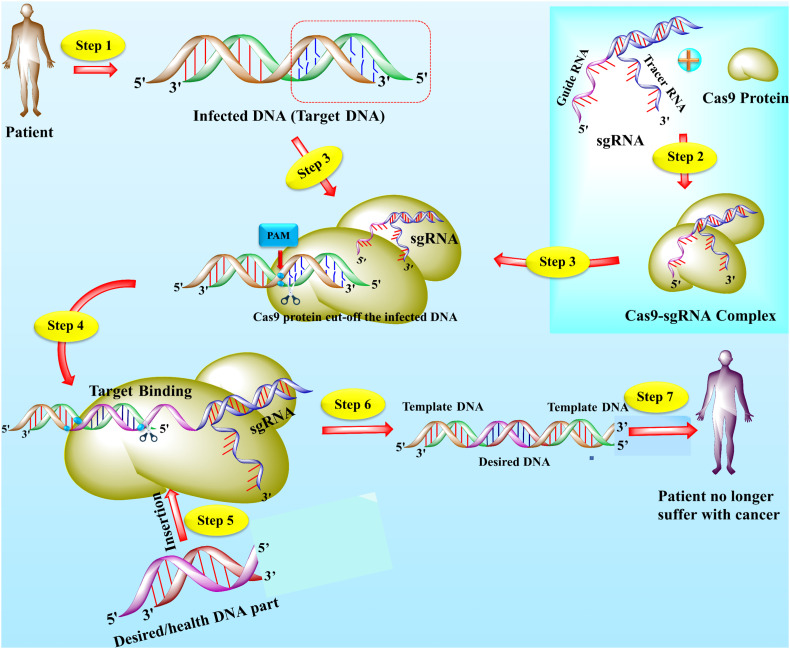
Fig. 1.
The structure and mechanism of action of CRISPR/Cas9 technology. The CRISPR/Cas9 is an emerging gene-editing technology that is used by scientists to edit the gene to treat lethal diseases like cancer. Usually, CRISPR/Cas9 technology is consists of two main parts including single-guided RNA (sgRNA) and Cas9 protein. The sgRNA (contain a sequence complementary to target DNA) directed the Cas9 protein toward targets and Cas9 is an enzyme that cut off the DNA. The specific part of DNA that is affected by a disease like cancer, acts as a target or template DNA (step 1). The sgRNA and Cas9 stick together and seek out the target DNA downstream to the specific protospacer adjacent motif (PAM) (step 2). The Cas9-sgRNA complex recognized the PAM of the effected genome and bound to target DNA (step 3). The Cas9 unzipped the double-strand of target DNA at 3 or 4 nucleotides upstream of the PAM sequence and guided RNA matches the complementary sequence. If the complementary sequence matched, the Cas9 used the molecular scissors to cut the affected part of the target DNA and insert the healthy or desired part of DNA (step 4–5). The double-strand break (DSB) is repaired by enzymes in two ways, either by homologous recombination (HR) or non-homologous end-joining (NHEJ). In the present figure, the DNA is repaired by the HR method (step 6). Finally, if all the process succeeds, cancer (but not limited to cancer) patient become healthy (step 7) [8,113,114].
4. The potential therapeutic target effects of CRISPR/Cas9 in solid tumors
In the past few years, due to the specificity, efficiency, and accuracy of CRISPR/Cas9, this genome editing tool is excessively used in the research laboratories, which helped the researchers to detect the role of different oncogenes in the cancer cells [33]. The tumor-suppressor genes (ETS1, CPEB2, BRCA1, PGC1a, TP53, MiR-1205, and SOX15) comprise their significant role in tumorigenesis, which can suppress cell mitigation, cell proliferation, enhance cell differentiation, and downregulate the cancer progression [34,35]. CRISPR/Cas9 technology targeted these tumor-suppressor genes to inhibit or reduce the tumorigenesis by restoring the activities of tumor-suppressor genes [36]. CRISPR/Cas9 barcoding technology is used to identify the cancer heterogeneity and its therapeutic targets against different cancer cells [37]. Herein, we discussed the novel molecular therapeutic effects of CRISPR/Cas9 technology in the treatment of most accruing solid cancers, including breast, lung, colorectal, prostate, and liver cancers.
4.1. Breast cancer
Breast cancer is a common leading cause of death among women, comprising 30% of all new diagnosis tumors, globally. The molecular profile of breast cancer reveals the heterogeneity and existence of several cancer subtypes with clinical consequences [38]. One of the major challenges of the breast tumor is the complexity of cancer, as cancer is not the form of a single cell; instead, it is composed of diverse cells including the appearances of progenitor or stem cells [39]. Breast epithelium is composed of four different subtypes based on the estrogen receptor (ER) expression, including luminal A, luminal B, Her2-enriched, and triple-negative breast cancer (TNBC) [40]. The luminal subtypes (ER-positive) are the most lethal and common forms of breast cancer (70%) with 30% resistance to endocrine therapies in infected patients [41]. However, a reduction of breast cancer is crucial, particularly in re-occurrence. Recently, CRISPR/Cas9 has proved a new, revolutionary, and an effective therapeutic target in the treatment of breast cancer cells (see Fig. 2 ).
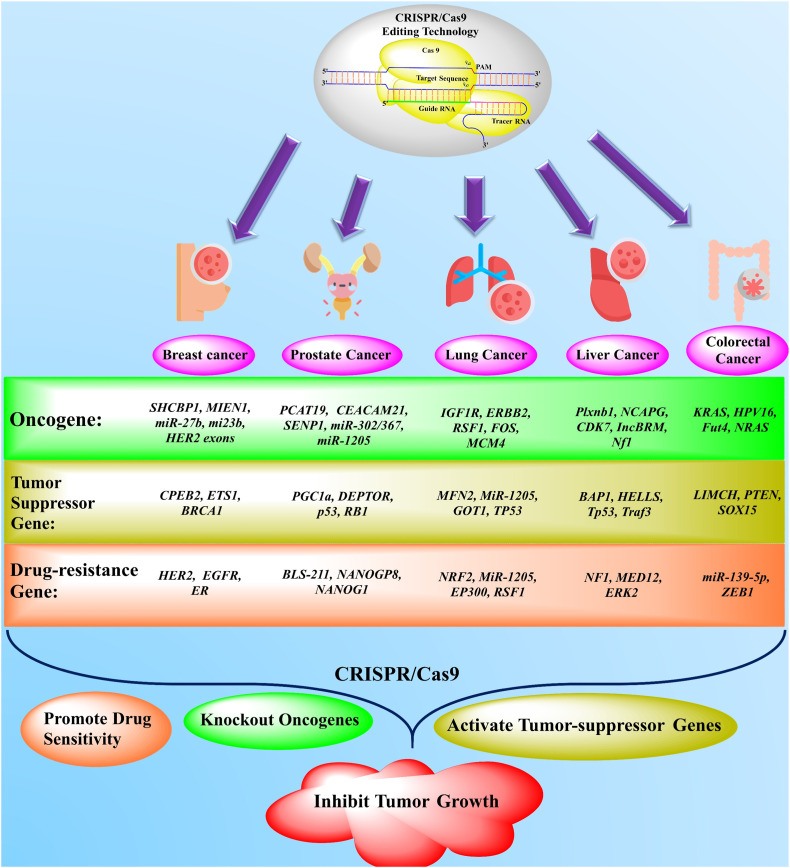
Fig. 2.
The application of newly emerged gene-editing technology (CRISPR/Cas9) in the treatment of different solid cancers. The novel gene-editing technology, namely CRISPR/Cas9, effectively targeted the oncogenes, drug-resistance, and tumor-suppressor genes in the treatment of various solid tumors, including breast, lung, liver, prostate, and colorectal cancers from the past couple of years. Based on accumulated data, the CRISPR/Cas9 has been used in knockout/delete the various oncogenes in different cancer treatments as present in a green box. The brown box represents the activation of different tumor suppressor genes. The orange box characterizes the inhibition/suppression of drug-resistance genes by CRISPR/Cas9 in solid tumors [31,35,45,70,74,[115], [116], [117], [118], [119], [120], [121], [122], [123], [124], [125], [126], [127], [128]]. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Mintz et al. [42] performed an experiment to treat the breast tumor (BRCA1m and PARP1m) and check the therapeutic proficiency of different chemotherapies synergy with CRISPR/Cas9, both in 2D and 3D tumor-chip models. They reported that both BRCA1m and PARP1m, triple-negative breast cancer (TNBC), cell lines were more sensitive with three different chemotherapeutic drugs, including docetaxel, gemcitabine, and doxorubicin in the 2D tumor culture model as compared to 3D tumor-chip model. Similarly, another study revealed that the non-viral delivery of sgRNA (a component of CRISPR) into HCC1806 and MCF10A cell lines significantly knockout both alleles of APOBEC3G genes in the treatment of breast cancer via blocking the conversion of the G1 to S phase in the cell cycle and inhibiting the cell proliferation [43]. Yang and his research group exposed that CRISPR/Cas9 technology successfully deleted the individual and co-knockout of CXCR7 and CXCR4 genes (both alleles) in the TNBC cell line (MDA-MB-231) during the treatment of breast cancer by suppressing the tumor cell proliferation, invasion, tumor growth, and invasion [44].
Hannafon and co-workers reported that the lentiviral delivery of CRISPR/Cas9 (sgRNA) into the xenografted nude mice (see glossary) significantly knockout both alleles of miRNA (miR-27b and miR-23b genes on chromosome 9) in the MCF7 mammary tumor cells to reduce tumor growth by inducing the cell-proliferation and mitigation at the endpoint by using a ki-67 marker (cell cycle marker) [45]. Similarly, another study exposed that in vivo lentiviral delivery of CRISPR/Cas-mediated somatic genome in mice successfully reduced the breast tumor susceptibility in triple-negative breast cancer (TNBC; see glossary) cell lines of p53 and K14Cre [46]. Alvares-Fernández et al. [47] stated that during in vivo study, the CRISPR-based interruption of MASTL kinase effectively condensed the cell-proliferation in humans mammary tumor cell lines and showed its highest expression in cancer cells as a contrast to normal cells. However, the suppression of MASTL could be an excellent target option in the treatment of breast cancer. Hence, the preclinical studies of breast cancer with CRISPR/Cas9 lead to identifying many new proteins and genes that could bring significant treatment outcomes in breast tumor suppression.
4.2. Liver cancer
Liver cancer is aligned as the fourth leading cause of deaths among both women and men of all cancer-related mortalities, globally. Unlike BRAF mutations in melanoma and EFGR mutation in lung tumors, the significant mutations in a liver tumor are still undruggable. Different therapies, including inhibitors (sorafenib and THZ1) are recently used in the treatment of liver cancer, but they are limited to drug resistance and molecular targets [48]. Currently, the new emerging genome editing technique, namely CRISPR/Cas9, is showing admirable results in the identification of new genes (CDK7, CD44, and Nf1) as therapeutic targets in the treatment of liver tumor [49]. The effect of CRISPR/Cas9 in the different liver tumor's oncogenes, tumor-suppressor genes, and drug resistance genes are presented in Fig. 2.
Wang et al. [50] reported that an in vitro lentiviral delivery of sgRNA (a component of the CRISPR system) into two cell lines of liver, including Huh7 and Hep38, identified a cyclin-dependent kinase namely CDK7 (see glossary) as hits during the screening by next-generation sequence. They also revealed that CRISPR/Cas9-mediated CDK7 significantly affected the cell-proliferation activity of Huh7 and Hep38 cell lines in both long-term colony formation assays and short-term IncuCyte cell-proliferation assay. Hence, CDK7 is identified as an important therapeutic target in the treatment of hepatocellular carcinoma by the CRISPR/Cas9 genome-editing tool. Similarly, Han and co-workers [51] stated that lentiviral delivery (in vivo) of sgRNA and Cas9 into C3A-iCSCs stem cell effectively knockout the CD44 gene (transcriptional regulatory gene) in the liver tumor by promoting the differentiation and cancer stem cells assessed by ChIP-qPCR.
Wang et al. [52] performed both in vitro and in vivo studies with CRISPR/Cas9 to investigate the limitations with the synergistic effects of selumetinib and sorafenib (see glossary) drugs during the liver tumor treatment. They exposed that inhibition of ERK2 (MAPK1) alerted several liver tumor cell lines to sorafenib and suggested that patients with high basal p-ERK (>30% of all liver tumors) are most likely to assistance from such synergistic treatment. Therefore, the inhibition of kinases, including ERK and MEK could be a potent therapeutic target in the treatment of liver carcinoma. In addition, Song and his research group potently revealed that Plxnb1, B9d1, Flrt 2, and Nf1 genes are the liver tumor-suppressor genes using CRISPR/Cas9 technology. They exposed that the lentiviral delivery of sgRNA and Cas9 into p53 and MYC-Cas9 liver cells successfully knockout the NF1 gene in the mouse liver model. They also reported that the inactivation or suppression of these genes (previously thought they are not related to a liver tumor) by CRISPR/Cas9 leads to the acceleration of liver tumors in mice and upregulates the liver progenitor cell markers namely SOX9 and HMGA2. However, for the first time, they founded that Plxnb1, B9d1, Flrt 2, and Nf1 are the liver tumor-suppressive genes by using CRISPR/Cas9 technology [53]. However, the simultaneous inhibition of SOX9 and HMGA2 might be a potent therapeutic target in the reduction of liver carcinoma. Similarly, Zhu et al. [54] deleted the IncBRM gene in the Huh7 and Hep3B cell lines of a liver tumor by using CRISPR/Cas9 technology and reported that knockout of IncBRM gene impaired the serial sphere formation without affecting the expression of neighboring genes, which suggested that IncBRM gene perform its role in trans. They suggested that the IncBRM gene could also serve as a biomarker (see glossary) for the analysis and drug discovery against liver cancer.
4.3. Lung cancer
Lung tumor is the most leading cause of cancer-related deaths, both in women and men with frequent metastasis and recurrence [55]. According to emerging evidence, several genes, including JUN, FOS, ROS1, ERBB2, RAS, RAF1, and MYC are considered as lung-tumor-associated proto-oncogenes. Similarly, APC, MCC, RB, CDKN2A, TP53, and NM23 are known as cancer-suppressor genes in the lung tumor (see Fig. 2). A recent study reported that both oncogene overexpression and gene mutations are involved in lung tumor growth. However, the utilization of recently developed gene-editing technology (CRISPR/Cas9) could significantly remove lung cancer by targeting the tumor-suppressive genes, genes associated with therapies by inhibitors, oncogenes, and genes associated with chemotherapy drug resistance [35,56].
Most recently, Lu and co-workers performed a clinical trial-based experiment (NCT02793856) to investigate the feasibility and safety of CRISPR/Cas9-mediated T-cell therapy in the treatment of lung cancer by targeting the PD-1 gene. They revealed that the median overall survival rate was 42.6 weeks (CI: 95%, 10.3–74.9 weeks), with 0.05% off-target effects in 18 patients accessed by next-generation sequence. They also claimed that the lifespan of gene-editing T-cell was short, which suggested limited off-target effects, and proved that CRISPR/Cas9 genome editing technology is clinically feasible for lung tumor treatment [57]. However, in the future, more clinical trials should be performed to treat lung cancer. Similarly, Perumal et al. [58] experimented on Slug/Snail non-small lung cancer cells (NSLC; see glossary) by CRISPR/Cas9 genome editing tool. They reported that non-viral delivery (plasmid) of Cas9 into NCI-H460 and A549 cell lines of lung carcinoma effectively knockout the PTEN gene using Epithelial-mesenchymal transition (EMT) marker. They revealed that the knockout of the PTEN gene showed high invasion, metastasis, and cancer cell growth than a PTEN wild type by promoting the expression of phosphorylation of GSK-3β and AKT pathway while suppressing β-catenin. Hence, the inactivation of the PTEN gene could be contributed to the EMT marker by regulating the translocation of β-catenin (see glossary) in the lung carcinoma treatment.
A study revealed that knockout of AMPK (α1 and α2) signaling pathway by CRISPR/Cas9 technology in murine lung tumor (KRAS gene) significantly reduced the size of lung carcinoma cells [59]. Similarly, Cheung et al. [60] experimented using CRISPR/Cas9 genome editing technology to target the point mutation in lung tumors (EGFR). They reported that CRISPR/Cas9 involved in specific genome targets in L858R mutant cells and suggested that this novel strategy could be used to target the specific area of lung cancer (15 to 35%) with T > G, A > G, and C > G point mutations. Moreover, the lentiviral delivery of Cas9 into A549 cell lines effectively knock out the MYC gene, which resultingly revealed that the silencing of long non-coding RNA EPIC1 (lnc-EPICI) by CRISPR/Cas9 that significantly reduced the cell-proliferation, tumor size, and cancer cell survival [61]. Another study showed that the knockout of p107 and closely related p130 genes by CRISPR/Cas9 strategy effectively enhance the cancer progression (see glossary). They also suggested that this novel strategy could be feasible and safe to tumor suppressor genes in small cell lung cancer (SCLC; type of lung cancer), which might be anticipated in the development of a novel potential drug against the progression of lung carcinomas [62].
4.4. Colorectal cancer
Because of high-prevalence, genetic physiognomies (MSI, BRAF, NRAS, and KRAS), and clinic-pathogenesis of colorectal cancer, it has attained the much attention during the past couple of decades, which imitated the preliminary step toward precision medicine [63,64]. Conventionally, several anti-tumor agents, including anti-VEGF and anti-EGFR have been used against colorectal cancer, but they are limited to restrained efficiency and metastatic settings [65]. However, most recently, the CRISPR/Cas9 has proven as a revolutionary technique in the molecular biology and oncology research field due to its multifunctional strategies, including high accuracy in genome editing and high specificity [66]. The effect of CRISPR/Cas9 in the different colorectal oncogenes, tumor-suppressor genes, and drug resistance genes are presented in Fig. 2.
Blanas et al. [67] activates the transcriptional expression of Fut4 and Fut9 genes in a murine colorectal tumor cell line (MC38), by using the de-novo gene (see glossary) expression system as CRISPR/dCas9-VPR. They reported that introduction of these genes (Fut4 and Fut9) into murine cell lines lead to the neo-expression of functional Lewis-antigen on the cell surface and effected the expressions of sialylation, core-fucosylation, and antennary of N-glycans in murine cell line (MC38) glycovariants. Hence, CRISPR/dCas9 could be a promising strategy for gene transcription during the treatment of colorectal cancer and glycobiology in the near future. Similarly, Hsu and his research group grow the HPV16 + (anal cancer-derived gene) into immunodeficient mice to check could CRISPR/Cas9 suppress anal cancer growth by targeting the HPV16 (E6 and E7) genes or not. They revealed that the adeno-associated virus (AAV) delivery of Cas9/sgRNA (a component of CRISPR) into mice effectively reduced the anal tumor size by targeting the HPV16 (E6 and E7) genes and suggested that CRISPR/Cas9 might be a potential option in the treatment of HPV-induced cancers in humans [68].
Li et al. [69] demonstrated that the lentiviral delivery of Cas9/sgRNA into CaCO-2 cell line successfully knock out the Par3L protein in colorectal tumor cells by inhibiting the cell-proliferation, inducing apoptosis, and activating the expression of signaling cascade-3. They observed that Par3L proteins are more sensitive to anti-tumor chemotherapies and suggested that the inactivation of Par3L might be a potential target by CRISPR/Cas9 technology in colorectal cancer cell survival by suppressing the AMPK signaling transduction (see glossary). Wan and co-workers revealed that hyaluronic acid (HA)-associated CP/Ad-SS-GD/RNP nano-complexes delivery in cancer effective mice efficiently inhibited the colorectal cancer cell growth and metastasis by targeting the KRAS mutant gene [70].
4.5. Prostate cancer
Prostate cancer is ranked as the second most leading cause of cancer-related deaths among men, a commonly diagnosed tumor in European men. The development of tumor size leads to many genomic alternations, including a mutation in function and structure of the genome, and these alternations depend on the tumor size and cancer-cell types [71]. Conventionally different strategies, including chemotherapies, have been used against these alternations, but they remain poorly characterized [72]. However, the CRISPR/Cas9 proves as a potential target against prostate cancer due to its limited off-target effects and high accuracy [10].
Most recently, Fenner and his research group [73] demonstrated that the knockout of ER-β gene by CRISPR/Cas9 in mice genome effectively involved in regulating the prostate tumor size and act as a tumor-suppressor gene, that suggested its (ER-β gene) role in the prostate cancer which was previously not reported. Hence, the study claimed the role of ER-β gene as a repressor gene in the prostate tumor androgen receptor, which could be led to the discovery of novel and better drugs against the lethality of prostate tumors. Batir et al. [74] revealed that lentiviral delivery of sgRNA (a component of CRISPR/Cas9) into human prostate tumor cell line (PC-3), successfully repaired the TP53 414delC mutation, caused by mutant tumor protein, p53, at the rate of 26% with ssODN2 by inhibiting the cell-proliferation of PC-3 cell. However, the utilization of CRISPR/Cas9 in repairing the mutations carried by alternations in the genome during prostate cancer could be a potential target soon. Yoshikawa and his research group observed the knockout of PTEN (Phosphatase and tensin homolog) from murine prostate tumor cells by CRISPR/Cas9 that leading to the activation of cyclin D1 expression, and RAC-alpha serine/threonine-protein kinase phosphorylation, which suggested that the knockout of PTEN in prostate tumor mobilize many critical genes for cancer cell survival [75].
The emerging evidence suggested that an in vitro lentiviral delivery of sgRNA (a component of CRISPR/Cas9) effectively targets the androgen receptor (AR) in a prostate cancer cell line. They revealed that the inactivation of AR by CRISPR/Cas9 technology successfully inhibited the prostate tumor cell size by inhibiting cell proliferation and promoting apoptosis. Hence, it is suggested that CRISPR/Cas9 could be a revolutionary and potential technique against prostate tumors in the upcoming time [76]. Similarly, Ye et al. [77] demonstrated that the lentiviral delivery of CRISPR/Cas9 into the human xenograft mouse model (PC-3) significantly inactivated the GPRC6A receptor. They suggested that the knockout of the GPRC6A receptor by CRISPR/Cas9 might be resulted in reducing the prostate cancer cell size by inhibiting the mitigation and cell proliferation.
The applications of CRISPR/Cas9 in the treatment of different solid cancer cells, including breast, lung, liver, colorectal, and prostate are presented in Table 1 .
Table 1.
The in vitro and in vivo applications of CRISPR/Cas9 in the treatment of different solid tumors during the last five years.
| Cancer type | Target choice | Cell line/gene | Study type | Vector | Screening/verification | CRISPR effect | Mechanism | Reference |
|---|---|---|---|---|---|---|---|---|
| Breast cancer | TNBC | MDA-MB-231 | In vitro | Lentiviral | Western blotting, PCR | Knockout of the CXCR4 or CXCR7 gene | Delay the conversion of the G1/S cycle, inhibit cell proliferation, invasion, and mitigation | [44] |
| BRCA1 wild-type, BRCA1m | MDA-MB-231, MDA-MB-436 | In vitro | Plasmid | (Sanger sequencing) PCR and T7EI assay | Knockout PARP1 inhibitors | Apoptosis | [42] | |
| PP2A-B55 | Cal-51 | In vivo | Lentiviral | Flow cytometry | Knock-out of both alleles | Inhibit cell proliferation and tumor suppression | [47] | |
| Nutlin-3a (organoid) | 16 PM0462, 16 PM0408, 8 PM0050 | In vivo | Lentiviral | Western blot analysis | Knock-out of PTEN, NF1, P53, and RB1 | Induction of cellular senescence and cell proliferation | [108] | |
| miR-23b, miR27b | MCF7 cells | In vitro | Lentiviral | DNA sequencing, qRT-PCR, MTS growth assay | Knockout of miR-23b and miR-27b | Inhibit cell proliferation and promote migration in miR-27b depleted cells | [45] | |
| APOBEC3G | MCF10A, HCC1806 | In vitro | Plasmid | Sanger sequencing. | Knock-out of both alleles of APOBEC3 | Inhibit cell proliferation | [43] | |
| Sox2, Sox9 | MDA-MB-231, MCF7TamR | In vivo | Cas9n vector | Immunofluorescence, ALDEFLUOR assays | Knockout | Promote Wnt signaling and reduce invasion | [38] | |
| Lung cancer | PD-1 | T-cells | Phase 1 clinical trial | Plasmid | Next-generation sequencing | Knockout the PD-1 | Effectively targeted exon 2 of PD-1 gene to reduce lung cancer | [57] |
| PTEN | A549, NCI-H460 | In vitro | Plasmid | RT-PCR, Immunocytochemistry | Knockout of both alleles of PTEN | CRISPR/Cas9 KO the PTEN in slug/snail lung tumor cells, which contributes to EMT by nuclear translocation of β-catenin | [58] | |
| EGFR | NCI-H1975, NCI-H1650 | In vitro | Lentiviral | T7 Endonuclease I assay, Sanger sequencing | Knockout of EGFR | Inhibit cell proliferation and EGFR expression in lung cancer | [60] | |
| p107 and p130 | Trp53, Rb1, Rosa26 | In vivo | Adenoviral | Flow cytometry | Knockout of genes | Loss of tumor-suppressor genes (p107 and p130) induce metastasis, promote cell proliferation | [62] | |
| Liver cancer | β-catenin | PTEN and p53 (TP53 and Trp53) | In vivo | Plasmid | PCR | Knockout of both alleles | Akt phosphorylation | [109] |
| C57-HBV | PTEN, p53 | In vivo | Plasmid | PCR, Immunohistochemical (IHC) analysis | Loss-of-function alterations of p53 and Pten genes in H2.35 cells | Akt phosphorylation that resulted in corresponding somatic dysfunction, and lipid accumulation | [49] | |
| HMGA2 (Nf1, Plxnb1, Flrt2, and B9d1) |
p53 | In vivo | Lentiviral | RNA sequencing, bioinformatics analysis | Knockout of both alleles | Reduce the mitogen-activated protein kinase | [53] | |
| CD44 | CD44 − C3A-iCSCs D6 clone and CD44− C3A-iCSCs C10 clone | In vitro | px458 vector | Immunofluorescence, Western blotting | Knockout of CD44 | Lower cell proliferation | [51] | |
| Factor IX (F9) | AAV9-HS-CRM8-TTRmin-Cas9, AAV9-U6-mF9-Exon1-gRNA | In vivo | AAV | PCR, bioinformatics analysis | Loss of FIX activity | Induce a dsDNA break in a DNA-sequence-specific manner | [110] | |
| Colorectal cancer | KRAS | SW-480 CRC cells | In vitro | Polymer | Flow cytometry | Knockout of both alleles | Induce apoptosis, suppress cell proliferation and promote tumor cell death | [70] |
| Par3L | CaCO-2 | In vitro | Plasmid | Western blot assay, flow cytometry | Knockout of both alleles | Induce apoptosis and inhibit cell proliferation | [69] | |
| Fut4 and Fut9 | MC38 | In vitro | pX330 vector | qRT-PCR analysis, flow cytometry | Inhibit glycosylation | CRISPR-dCas9-VPR system activates the Fut4 and Fut9 genes to induce the neo-expression | [111] | |
| Anal cancer | HPV16 | 293 T | In vivo | AVV | PCR | Knockout oncogenes (E6 and E7) | Inhibit the expression of HPV16 E6 and E7 genes to reduce anal tumor | [68] |
| Prostate cancer | ERRα, PGC1α | PC3, DU145, 293FT | In vivo | Lentiviral | Chromatin immunoprecipitation, bioinformatic analysis | Knockout of ERRα | Inhibit MYC levels to suppress metastasis and invasion of lung cancer growth | [72] |
| NANOG,NANOGP8 | DU145 | In vivo | pX330 vector | Western blot analysis | Knockout of NANOG and NANOGP8 | Inhibit cell proliferation, and cell migration capacity, promote drug sensitivity | [112] | |
| GPRC6A | PC-3, LNCap, DU145, 22Rv1 | In vitro | Lentiviral | RT-PCR, Western blot | Activation of GPRC6A | Block ERK, mTOR, and AKT phosphorylation, and inhibit promoted cell proliferation | [77] | |
| AR | LNCaP | In vitro | Lentiviral | PCR | Inhibition of AR | Induce apoptosis and inhibit cell proliferation | [76] | |
| PTEN | ΔPTEN, 2924 V, PSA | In vivo | Plasmid | DNA sequencing, western blotting | Knockout of PTEN | CRISPR/Ca9 KO the PTEN gene in ΔPTEN cell line to check the role of PTEN in prostate cancer | [75] | |
| TP53 | PC-3 (CRL-1435 and ATCC) | In vitro | Plasmid | Flow cytometry, Immuno-fluorescence analysis, qPCR | Knockout of both alleles of TP53 | Induce apoptosis, inhibit cell proliferation | [74] |
TNBC: triple-negative breast cancer; KO: knockout; AVV: adeno-associated vector.
5. Other Cas family members
Besides to Cas9 protein, researchers are also constantly trying to develop the new members of CRISPR system. Recently some CRISPR-associated (Cas) proteins such as Cas12a, Cas12b, Cas13a, and Cas13b proteins have been developed and studied to minimize the off-target effects and enhance genome editing efficiency of CRISPR system. Cas12a is a simpler and smaller RNA-guided endonuclease protein as compare to Cas9 [19]. The type V of CRISPR/Cas system (Cas12a and Cas12b) represent some unique characteristics that make them distinguish from Cas9. The type V proteins members possess the RuvC nuclease domain instead HNH domain to identify the site-specific T-rich PAM (5′) sequence just upstream to target region. In addition, Cas12 nucleases built staggered DSBs distal to PAM sequence unlike to Cas9 that helped to generate the unique knock-in strategies for genome editing through HNEJ mechanism [78].
A study revealed that Cas12a (also known as Cpf1) showed less off-target effects and could be used in gene therapy soon to target the gene of interest. Most recently, Gier et al. [79] observed that lentiviral delivery of Acidaminococcus Cas12a (AsCas12a) in RN2c12 cell line successfully showed the double knockout screening against epigenetic regulators. They also revealed that this screen showed the tight interactions between Kat6a and Jmjd6, Brd9 and Jmjd6, and Brpf1 and Jmjd6 genes in leukemia cells. Similarly, Yoon et al. [80] demonstrated that intratumoral delivery of oAd/Cas12a/crEGFR effectively showed the highly accurate and efficient editing activity in a tumor-specific gene by targeting epidermal growth factor receptor (EGFR) gene with fewer off-target effects via inducing the apoptosis and inhibiting the tumor cell proliferation. In addition to Cas12s, Cas12b or C2c1 is 1129 aa (3.4 kb) in size. Unlike to Cas12a, Cas12b requisites both trancrRNA and crRNA to form a complex for genome editing. The emerging evidence revealed that due to small size of Cas12b, it could easily be delivered to the target site by viral vector and Cas12b produced a long sticky end after double-strand DNA editing which could be expected to enhance the efficiency of ligation after DNA break [81]. It is also noteworthy that Cas12b is very sensitive and effective to single-base mismatch of about 20 nucleotides that make it highly specific CRISPR/Cas12b system [82].
Like to type V CRISPR/Cas system, the type VI also possesses two important Cas nucleases such as Cas13a and Cas13b. These two proteins also carried some unique features, unlike Cas9, these proteins cleaved the ssRNA instead of DNA because they have no catalytical DNA domain. Recently, studies stated that Cas13 proteins possess two identified highly conserved prokaryotic and eukaryotic nucleotide-binding domains containing specific RNA cleavage site [78,83]. Zhao et al. [84] stated that bacterial Cas13a and CRISPR-RNA (crRNA) showed the successful knockout effects in mutant KRAS mRNA expression with the knockout efficiency of about 94%. They also observed that in vitro delivery of CRISPR/Cas13a effectively knock down the KRAS-G12D mRNA expression (70%) via inducing the apoptosis and inhibiting the tumor cell proliferation. Fan et al. [85] reported that liposomes delivery of CRISPR/Cas13a into BCa cells significantly inhibited the bladder tumor genes and observed that codon-optimized LwaCas13a plays an important role in the target site regulation. Similarly, Li and co-workers [86] designed the dm6ACRISPR by fusing the ALKBH5 to C or N-terminal of inactive Cas13b with the help of six amino acids sequence such as GSGGGG to target the RNA demethylation. They found that dm6ACRISPR successfully targeted the N 6-methyladenosine demethylation in cytochrome b 5 form A (CYB5A) mRNA which was responsible for coding the oncoproteins including EGFR and suggested that dm6ACRISPR could be a potential target to inhibit the tumor cell proliferation. Although some recently emerged Cas protein systems such as Cas12 and Cas13 showed the remarkable results in the tumor field, but still extensive research is required to properly understand the mechanism of action, efficiency and off-target effects of these nucleases.
6. Delivery of CRISPR/Cas9
After the discovery of the CRISPR/Cas9 genome editing technique, the delivery of CRISPR-components remains a major challenge. According to emerging evidence, the viral vector has been used over the decades in reverse genetic engineering (see glossary) to deliver the oncogene to the tissue to induce tumors. Initially, for this purpose, different viral-vectors have been used, but among all, the recombinant retroviral vectors got considerable attention due to their minimum off-target effects, high specificity, and accuracy to target cell genome. Since the repurposing of the CRISPR system into biotechnology, the viral vectors were rapidly developed for the delivery of sgRNAs or Cas9 to the target cell genome [87]. Initially, the lentiviral libraries were pooled to deliver the CRISPR components due to the substantial size of the Cas9 gene (4.1 kb), which could efficiently fit into the lentivirus [88,89]. Nevertheless, several major concerns, including mutagenesis and immunogenicity are associated with the use of adenoviral and lentiviral vectors as the Cas9 ribonucleoproteins delivery method (see Fig. 3 ) [90].
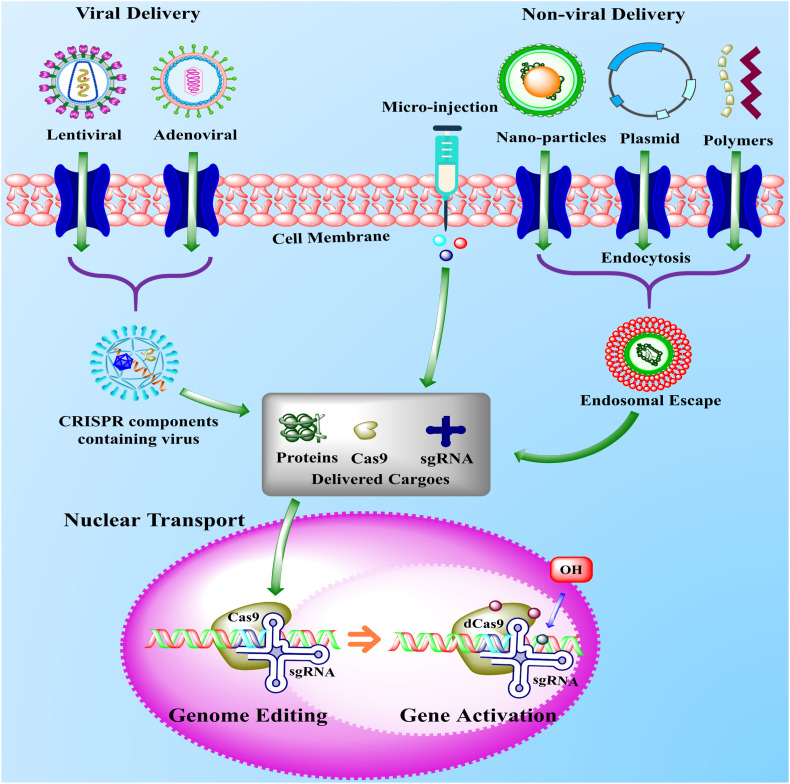
Fig. 3.
The schematic representation of viral and non-viral delivery systems of CRISPR-components inside the cell for genome editing. The delivery of CRISPR-components is one of the most important steps in the genome editing strategy to minimize the off-target effects. The CRISPR-components, including sgRNA, ribonucleoproteins, and Cas9 are delivered to the target site in two common ways, either by viral vectors (lentiviral and adenoviral) or non-viral delivery system including DNA plasmids, nanoparticles, polymers, and micro-injection. In the non-viral delivery system, the DNA plasmids, nanoparticles, and polymers containing CRISPR-components cross the cell membrane (light red) either by endocytosis or pore formation and formed the endosomal escape (red and green) inside the cell. The endosomal escape released the CRISPR-components inside the cell (grey box), which enter into the nucleus (pink) and recognized the target site and finally activated the tumor-suppressor genes or suppressed the oncogenes. Similar to non-viral delivery, the viral delivery performed the same function, but viral deliveries are associated with the use of adenoviral and lentiviral vectors as a Cas9 ribonucleoprotein delivery method [129,130]. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Besides to viral vectors, the plasmid DNA is known as another alternative method for the delivery of CRISPR components as sgRNA and Cas9 for the genome editing approach in vitro and in vivo studies. In recent years the standard molecular cloning techniques have been used to develop the plasmid, including cell-type-specific transcriptional targeting elements, diverse Cas9 structures, and different sgRNAs [91,92]. But the emerging evidence associated with the several problems with the plasmid DNA delivery system, including plasmid DNA, enhances the perseverance time of CRISPR RNPs inside the cell, which results in improve off-target ability. Besides, the plasmid DNA persuades the mutagenesis effects when active promoter elements implanted into the host genome [93,94].
Although the viral vectors have been a significant option in the delivery of CRISPR component, there is a need to develop a vector with both clinical and pre-clinical studies delivery strategies and restricted payload size. One of the major concerns with adeno-associated viruses (AAV) is the packing of Cas9 into separate viral particles, which resultingly reduced the accuracy [95]. However, to tackle this problem, the scientist has discovered another effective delivery method of CRISPR components known as non-viral delivery to target the genome, as presented in Fig. 3. The emerging evidence processed that the non-viral delivery method has superiority than viral-vectors and plasmid DNA methods due to high packing ability, fewer adverse-effects, high accuracy, and spatiotemporal specificity. Different types of materials, including graphene oxide, liposome, metal framework, gold nanoparticles, and cationic polymers, have shown promising results in the delivery of CRISPR/Cas9 components [96,97]. Recently, the cationic polymers (see glossary) have received considerable attention due to the effective delivery method during the treatment of cancer cells by the CRISPR/Cas9 tool. Wan et al. [70] efficiently prepared the supramolecular polymer delivery system (in vitro) by binding the disulfide-bridge gunidyl adamantine (Ad-SS-GD) with beta-cyclodextrin polyethyleneimine (CP). They reported that this nano-complex Ad-SS-GD/CP/RNP effectively released the Cas9 ribonucleoproteins (Cas9 RNPs) to the intracellular environment by breaking the disulfide bonds during the treatment of colorectal cancer and 293 T cells. However, the delivery of sgRNA and Cas9 as a ribonucleoprotein complex (RNP) is still challenging, especially in vivo with the count of affluence of denaturation and encapsulation difficulties [90].
7. Current challenges and future directions
Cancer is one of the leading causes of death and the most studied field during the past few decades due to its complexity, heterogeneity of the disease, and high epidemiology among humans. The multi-mutated and multigenic characteristics of cancer give them a unique heterogeneity ability that could exist differently in different cancer-affected patients with the same cancer type. However, this unique characteristic of heterogeneity triggered the major challenge for clinical rehabilitation [98]. The specific, accurate, and simple genome editing ability of CRISPR/Cas9 attained the special attention of the scientific community around the globe, especially during the last five years in the cancer biology field. The discovery of Cas9 nuclease has broadened the era of molecular biology, specifically to treat the multifunctional genes of cancer. The modern researchers have been using the CRISPR/Cas9 technology, particularly in suppressing the oncogenes in mice models in terms of cost-effective, high specificity, accuracy, and time length without applying multifunctional colonies of mice [99].
No doubt, the CRISPR/Cas9 proved as a revolutionary tool and showed remarkable results in the treatment of cancer cells, but there are also still a few challenges that should be improved on urgent bases. One of the major challenges during the synthesis of the CRISPR system is to reduce the off-target effects of Cas9 nuclease [100]. The Cas9 nuclease is a promising factor to control the off-target characteristics and undesirable adverse effects of the CRISPR system. To regulate the off-target challenge of Cas9, the researchers should investigate the different physical or chemical agents, but not limited to tetracycline and doxycycline, as a promising approach to develop the desired expression of Cas9. In addition, the various online tools are available for the assistance of researchers to design sgRNA with minimum off-target effects to promote specificity toward the cell genome editing [101,102]. Recently few genome editing variants of the CRISPR system, including truncated sgRNA, Cas9 nickase, and interaction of Fok1 with dCas9, proved as a promising tool in the reduction of off-target effects [22,103].
In addition to minimizing the off-target effects, the in vivo delivery system of Cas9 should also extend its application to the cervix and stomach to locate and treat the more cancers. The enormous size of Cas9 is associated with packing problems, particularly in low immunogenic adeno-associated viral (AAV) vectors, which are excessively used in the in vitro and in vivo gene deliveries [104]. The most recent studies claim that the persistent binding of Cas9 nuclease with DSBs suppressing the entry of repair proteins to the target site, which resultingly plummeting the repair efficiency. This remains the rate-limited step in a Cas9-DSB complex in the genome editing in vivo [105]. However, the translocating RNA polymerase could be used to dislocate the template bounded Cas9, only after RNA polymerase locates the DSBs from an exact direction [104,106]. Moreover, there are still some oncogenic genomic variations, including chromothripsis, aneuploidy, and LINE-1-mediated genome mutation, that are difficult to treat with the current version of the CRISPR/Cas9 system. However, the mutagenic efficiency of CRISPR/Cas9 should be improved in the near future by designing the potent Cas9, develop efficient delivery methods with prevailing sgRNA [103,107].
8. Concluding remarks
In summary, the present review made a comprehensive discussion on the structure, function, delivery, current challenges and future direction of CRISPR/Cas9 to treat solid tumors, including breast, lung, liver, colorectal, and prostate. Because of the multi-step existence and epigenetic variations of cancer, the prevention and treatment of cancer cells are difficult. So, there is a need to develop a multifunctional target against cancer. Recently, RNA domain-containing endonuclease-based genome engineering technology, namely the clustered regularly interspaced short palindromic repeat (CRISPR)-associated protein-9 (Cas9) has led to the identification and reduction of tumor-associated genes (PTEN, CD44, IncBRM, ROS1, TP53, MCC, Fut4, HPV16 +, and APOBEC3G) due to its rapid immunological response, specificity, optimization of delivery systems, high efficiency, and less off-target ability. Based on the latest findings, the present review also observed that CRISPR/Cas9 successfully identified the new latent targets, including CDK7, Nf1, Plxnb1, B9d1, Flrt 2, PD-1, EGFR, lnc-EPICI, p107, Par3L, and ER-β genes during the tumor cell study. But some challenges, including reducing the off-target effects of Cas9 and delivery method of sgRNA to target cells, are still associated with the performance of CRISPR/Cas9 technology. However, it is recommended to improve the optimization of Cas9 and minimize the off-target effects to meet the prerequisite for potential therapeutic targets in the oncology research field. In addition, to minimizing the off-target effects, the in vivo delivery system of Cas9 should also extend its application to the cervix and stomach to locate and treat the more cancers.
9. Outstanding questions
-
1.
CRISPR/Cas9 is an efficient timesaving genome editing technology, but currently, genome editing results are based on in vivo and in vitro studies. So, more clinical trials should be carried on in the near future for a better understanding of its mechanism.
-
2.
One of the major concerns about CRISPR/Cas9 technology is its off-target effects. Some studies have raised the question of its off-target effects. It is suggested that more research should be conducted to fully aware of its off-target effects.
-
3.
Could polymers as deliver vector instead of lentiviral and adenoviral reduce the off-target effects of CRISPR/Cas9?
-
4.
Based on the accumulated data, we noticed that CRISPR/Cas9 effectively regulates the apoptosis pathway. Could CRISPR/Cas9 technology use as a potential therapeutic target in the treatment of age-related diseases, including diabetes and cardiovascular, by regulating the apoptosis and mitochondrial-derived peptide like humanin?
-
5.
Could the use of CRISPR/Cas9 as genome editing technology reduce the risk of COVID-19?
-
6.
Could the use of chemical agents like tetracycline and doxycycline prove as a potential agent in the reduction of off-target effects of the current version of CRISPR/Cas9 during its synthesis?
Acknowledgments
Acknowledgement
Hereby, we extend our gratitude to A.Q Research Group, Pakistan, for reviewing the article and providing helpful comments.
Funding statement
This research received no specific grant from any funding agency in public, commercial or not-for-profit sectors.
Informed consent
For this type of study, informed consent is not required.
CRediT authorship contribution statement
A. Hazafa, and M. Yameen: Conceived the presented data, Writing - original draft, Software, & Supervision. A. Mumtaz, S. Bilal, and M. F. Farooq: Developed the theory, Formal analysis, & Investigation. H. Naeem, M. O. Ullah, and M. S. Mukhtiar: Software. F. Zafar, S. N. Chaudhry, and M. Firdous: Revision.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.lfs.2020.118525.
Appendix A. Supplementary data
Supplementary material
References
- 1.Yi L., Li J. CRISPR-Cas9 therapeutics in cancer: promising strategies and present challenges. Biochimica et Biophysica Acta -Reviews on Cancer. 2016;1866(2):197–207. doi: 10.1016/j.bbcan.2016.09.002. [DOI] [PubMed] [Google Scholar]
- 2.Jordan E.J., Kim H.R., Arcila M.E., Barron D., Chakravarty D., Gao J., Chang M.T., Ni A., Kundra R., Jonsson P. Prospective comprehensive molecular characterization of lung adenocarcinomas for efficient patient matching to approved and emerging therapies. Cancer discovery. 2017;7(6):596–609. doi: 10.1158/2159-8290.CD-16-1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dagogo-Jack I., Shaw A.T. Tumour heterogeneity and resistance to cancer therapies. Nat. Rev. Clin. Oncol. 2018;15(2):81. doi: 10.1038/nrclinonc.2017.166. [DOI] [PubMed] [Google Scholar]
- 4.Yum S., Li M., Chen Z.J. Old dogs, new trick: classic cancer therapies activate cGAS. Cell Res. 2020:1–10. doi: 10.1038/s41422-020-0346-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jubeen F., Liaqat A., Sultan M., Iqbal S.Z., Sajid I., Sher F. Green synthesis and biological evaluation of novel 5-fluorouracil derivatives as potent anticancer agents. Saudi Pharmaceutical Journal. 2019;27(8):1164–1173. doi: 10.1016/j.jsps.2019.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maresso K.C., Tsai K.Y., Brown P.H., Szabo E., Lippman S., Hawk E.T. Molecular cancer prevention: current status and future directions. CA Cancer J. Clin. 2015;65(5):345–383. doi: 10.3322/caac.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen S., Sun H., Miao K., Deng C.-X. CRISPR-Cas9: from genome editing to cancer research. Int. J. Biol. Sci. 2016;12(12):1427. doi: 10.7150/ijbs.17421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaushik I., Ramachandran S., Srivastava S.K. CRISPR-Cas9: a multifaceted therapeutic strategy for cancer treatment. Semin. Cell Dev. Biol. 2019;96:4–12. doi: 10.1016/j.semcdb.2019.04.018. Elsevier. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Agrawal N., Dasaradhi P., Mohmmed A., Malhotra P., Bhatnagar R.K., Mukherjee S.K. RNA interference: biology, mechanism, and applications. Microbiol. Mol. Biol. Rev. 2003;67(4):657–685. doi: 10.1128/MMBR.67.4.657-685.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kregel S., Wang C., Han X., Xiao L., Fernandez-Salas E., Bawa P., McCollum B.L., Wilder-Romans K., Apel I.J., Cao X. Androgen receptor degraders overcome common resistance mechanisms developed during prostate cancer treatment. Neoplasia. 2020;22(2):111–119. doi: 10.1016/j.neo.2019.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu B., Saber A., Haisma H.J. CRISPR/Cas9: a powerful tool for identification of new targets for cancer treatment. Drug Discov. Today. 2019;24(4):955–970. doi: 10.1016/j.drudis.2019.02.011. [DOI] [PubMed] [Google Scholar]
- 12.Koo T., Yoon A.-R., Cho H.-Y., Bae S., Yun C.-O., Kim J.-S. Selective disruption of an oncogenic mutant allele by CRISPR/Cas9 induces efficient tumor regression. Nucleic Acids Res. 2017;45(13):7897–7908. doi: 10.1093/nar/gkx490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang F., Wen Y., Guo X. CRISPR/Cas9 for genome editing: progress, implications and challenges. Hum. Mol. Genet. 2014;23(R1):R40–R46. doi: 10.1093/hmg/ddu125. [DOI] [PubMed] [Google Scholar]
- 14.Sánchez-Rivera F.J., Jacks T. Applications of the CRISPR–Cas9 system in cancer biology. Nat. Rev. Cancer. 2015;15(7):387–395. doi: 10.1038/nrc3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wyman C., Kanaar R. DNA double-strand break repair: all's well that ends well. Annual Review Genetics. 2006;40:363–383. doi: 10.1146/annurev.genet.40.110405.090451. [DOI] [PubMed] [Google Scholar]
- 16.Pal A., Kundu R. Human papillomavirus E6 and E7: the cervical cancer hallmarks and targets for therapy. Front. Microbiol. 2020;10:3116. doi: 10.3389/fmicb.2019.03116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qu X., Wang P., Ding D., Li L., Wang H., Ma L., Zhou X., Liu S., Lin S., Wang X. Zinc-finger-nucleases mediate specific and efficient excision of HIV-1 proviral DNA from infected and latently infected human T cells. Nucleic Acids Res. 2013;41(16):7771–7782. doi: 10.1093/nar/gkt571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shankar S., Prasad D., Sanawar R., Das A.V., Pillai M.R. TALEN based HPV-E7 editing triggers necrotic cell death in cervical cancer cells. Sci. Rep. 2017;7(1):1–12. doi: 10.1038/s41598-017-05696-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen S., Yao Y., Zhang Y., Fan G. CRISPR system: discovery, development and off-target detection. Cell. Signal. 2020:109577. doi: 10.1016/j.cellsig.2020.109577. [DOI] [PubMed] [Google Scholar]
- 20.Wang H., Yang H., Shivalila C.S., Dawlaty M.M., Cheng A.W., Zhang F., Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153(4):910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cong á., Ran F., Cox D., Lin S., Barretto R., Habib N., Hsu P., Wu X., Jiang W., Marraffini L. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu Y., Sander J.D., Reyon D., Cascio V.M., Joung J.K. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 2014;32(3):279. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dang Y., Jia G., Choi J., Ma H., Anaya E., Ye C., Shankar P., Wu H. Optimizing sgRNA structure to improve CRISPR-Cas9 knockout efficiency. Genome Biol. 2015;16(1):280. doi: 10.1186/s13059-015-0846-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gilbert L.A., Larson M.H., Morsut L., Liu Z., Brar G.A., Torres S.E., Stern-Ginossar N., Brandman O., Whitehead E.H., Doudna J.A. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154(2):442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Z., Zhang Y., Gao F., Han S., Cheah K.S., Tse H.-F., Lian Q. CRISPR/Cas9 genome-editing system in human stem cells: current status and future prospects. Molecular Therapy-Nucleic Acids. 2017;9:230–241. doi: 10.1016/j.omtn.2017.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen S., Sanjana N.E., Zheng K., Shalem O., Lee K., Shi X., Scott D.A., Song J., Pan J.Q., Weissleder R. Genome-wide CRISPR screen in a mouse model of tumor growth and metastasis. Cell. 2015;160(6):1246–1260. doi: 10.1016/j.cell.2015.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sánchez-Rivera F.J., Papagiannakopoulos T., Romero R., Tammela T., Bauer M.R., Bhutkar A., Joshi N.S., Subbaraj L., Bronson R.T., Xue W. Rapid modelling of cooperating genetic events in cancer through somatic genome editing. Nature. 2014;516(7531):428–431. doi: 10.1038/nature13906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen X., Liu J., Janssen J.M., Gonçalves M.A. The chromatin structure differentially impacts high-specificity CRISPR-Cas9 nuclease strategies. Molecular Therapy-Nucleic Acids. 2017;8:558–563. doi: 10.1016/j.omtn.2017.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tian X., Gu T., Patel S., Bode A.M., Lee M.-H., Dong Z. CRISPR/Cas9–an evolving biological tool kit for cancer biology and oncology. NPJ precision oncology. 2019;3(1):1–8. doi: 10.1038/s41698-019-0080-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nishimasu H., Ran F.A., Hsu P.D., Konermann S., Shehata S.I., Dohmae N., Ishitani R., Zhang F., Nureki O. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156(5):935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu J., Sareddy G.R., Zhou M., Viswanadhapalli S., Li X., Lai Z., Tekmal R.R., Brenner A., Vadlamudi R.K. Differential effects of estrogen receptor β isoforms on glioblastoma progression. Cancer Res. 2018;78(12):3176–3189. doi: 10.1158/0008-5472.CAN-17-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun H., Huang D., Liu G., Jian F., Zhu J., Zhang L. SIRT4 acts as a tumor suppressor in gastric cancer by inhibiting cell proliferation, migration, and invasion. OncoTargets therapy. 2018;11:3959. doi: 10.2147/OTT.S156143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang C., Lin X., Zhao Z. Applications of CRISPR/Cas9 technology in the treatment of lung cancer. Trends Mol. Med. 2019;25(11):1039–1049. doi: 10.1016/j.molmed.2019.07.007. [DOI] [PubMed] [Google Scholar]
- 36.Xu K., Chen G., Li X., Wu X., Chang Z., Xu J., Zhu Y., Yin P., Liang X., Dong L. MFN2 suppresses cancer progression through inhibition of mTORC2/Akt signaling. Sci. Rep. 2017;7:41718. doi: 10.1038/srep41718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guernet A., Mungamuri S.K., Cartier D., Sachidanandam R., Jayaprakash A., Adriouch S., Vezain M., Charbonnier F., Rohkin G., Coutant S. CRISPR-barcoding for intratumor genetic heterogeneity modeling and functional analysis of oncogenic driver mutations. Mol. Cell. 2016;63(3):526–538. doi: 10.1016/j.molcel.2016.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Domenici G., Aurrekoetxea-Rodríguez I., Simões B.M., Rábano M., Lee S.Y., San Millán J., Comaills V., Oliemuller E., López-Ruiz J.A., Zabalza I. A Sox2–Sox9 signalling axis maintains human breast luminal progenitor and breast cancer stem cells. Oncogene. 2019;38(17):3151–3169. doi: 10.1038/s41388-018-0656-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Visvader J.E., Stingl J. Mammary stem cells and the differentiation hierarchy: current status and perspectives. Genes Dev. 2014;28(11):1143–1158. doi: 10.1101/gad.242511.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goldhirsch A., Winer E.P., Coates A., Gelber R., Piccart-Gebhart M., Thürlimann B., Senn H.-J., members P., Albain K.S., André F. Personalizing the treatment of women with early breast cancer: highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Ann. Oncol. 2013;24(9):2206–2223. doi: 10.1093/annonc/mdt303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Padua M.B., Bhat-Nakshatri P., Anjanappa M., Prasad M.S., Hao Y., Rao X., Liu S., Wan J., Liu Y., McElyea K. Dependence receptor UNC5A restricts luminal to basal breast cancer plasticity and metastasis. Breast Cancer Res. 2018;20(1):35. doi: 10.1186/s13058-018-0963-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mintz R.L., Lao Y.H., Chi C.W., He S., Li M., Quek C.H., Shao D., Chen B., Han J., Wang S. CRISPR/Cas9-mediated mutagenesis to validate the synergy between PARP1 inhibition and chemotherapy in BRCA1-mutated breast cancer cells. Bioengineering Translational Medicine. 2020:e10152. doi: 10.1002/btm2.10152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mendes de Almeida R., Bandarra S., Clara Ribeiro A., Mascarenhas P., Bekman E., Barahona I. Inactivation of APOBEC3G gene in breast cancer cells using the CRISPR/Cas9 system. Ann. Med. 2019;51(sup1):1–8. [Google Scholar]
- 44.Yang M., Zeng C., Li P., Qian L., Ding B., Huang L., Li G., Jiang H., Gong N., Wu W. Impact of CXCR4 and CXCR7 knockout by CRISPR/Cas9 on the function of triple-negative breast cancer cells. OncoTargets therapy. 2019;12:3849. doi: 10.2147/OTT.S195661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hannafon B.N., Cai A., Calloway C.L., Xu Y.-F., Zhang R., Fung K.-M., Ding W.-Q. miR-23b and miR-27b are oncogenic microRNAs in breast cancer: evidence from a CRISPR/Cas9 deletion study. BMC Cancer. 2019;19(1):642. doi: 10.1186/s12885-019-5839-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pulver E., Silva A.M. da, Bouwman P., Annunziato S., Jonkers J. PO-333 somatic engineering of mammary gland epithelial cells using CRISPR/Cas9 for rapid testing of breast cancer susceptibility genes in mouse models. BMJ Publishing Group Limited. 2018 doi: 10.1136/esmoopen-2018-EACR25.363. [DOI] [Google Scholar]
- 47.Álvarez-Fernández M., Sanz-Flores M., Sanz-Castillo B., Salazar-Roa M., Partida D., Zapatero-Solana E., Ali H.R., Manchado E., Lowe S., VanArsdale T. Therapeutic relevance of the PP2A-B55 inhibitory kinase MASTL/Greatwall in breast cancer. Cell Death Differentiation. 2017:1–13. doi: 10.1038/s41418-017-0024-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kieckhaefer J.E., Maina F., Wells R.G., Wangensteen K.J. Thieme Medical Publishers; 2019. Liver Cancer Gene Discovery Using Gene Targeting, Sleeping Beauty, and CRISPR/Cas9, Seminars in Liver Disease; pp. 261–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu Y., Qi X., Zeng Z., Wang L., Wang J., Zhang T., Xu Q., Shen C., Zhou G., Yang S. CRISPR/Cas9-mediated p53 and Pten dual mutation accelerates hepatocarcinogenesis in adult hepatitis B virus transgenic mice. Sci. Rep. 2017;7(1):1–11. doi: 10.1038/s41598-017-03070-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang C., Jin H., Gao D., Wang L., Evers B., Xue Z., Jin G., Lieftink C., Beijersbergen R.L., Qin W. A CRISPR screen identifies CDK7 as a therapeutic target in hepatocellular carcinoma. Cell Res. 2018;28(6):690–692. doi: 10.1038/s41422-018-0020-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Han S., Guo J., Liu Y., Zhang Z., He Q., Li P., Zhang M., Sun H., Li R., Li Y. Knock out CD44 in reprogrammed liver cancer cell C3A increases CSCs stemness and promotes differentiation. Oncotarget. 2015;6(42) doi: 10.18632/oncotarget.6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang C., Jin H., Gao D., Lieftink C., Evers B., Jin G., Xue Z., Wang L., Beijersbergen R.L., Qin W. Phospho-ERK is a biomarker of response to a synthetic lethal drug combination of sorafenib and MEK inhibition in liver cancer. J. Hepatol. 2018;69(5):1057–1065. doi: 10.1016/j.jhep.2018.07.004. [DOI] [PubMed] [Google Scholar]
- 53.Song C.-Q., Li Y., Mou H., Moore J., Park A., Pomyen Y., Hough S., Kennedy Z., Fischer A., Yin H. Genome-wide CRISPR screen identifies regulators of mitogen-activated protein kinase as suppressors of liver tumors in mice. Gastroenterology. 2017;152(5):1161–1173. doi: 10.1053/j.gastro.2016.12.002. (e1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhu P., Wang Y., Wu J., Huang G., Liu B., Ye B., Du Y., Gao G., Tian Y., He L. LncBRM initiates YAP1 signalling activation to drive self-renewal of liver cancer stem cells. Nat. Commun. 2016;7(1):1–13. doi: 10.1038/ncomms13608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hazafa A., Rehman K.-U., Jahan N., Jabeen Z. The role of polyphenol (flavonoids) compounds in the treatment of cancer cells. Nutrition cancer. 2020;72(3):386–397. doi: 10.1080/01635581.2019.1637006. [DOI] [PubMed] [Google Scholar]
- 56.Nair J., Nair A., Veerappan S., Sen D. Translatable gene therapy for lung cancer using Crispr CAS9—an exploratory review. Cancer Gene Ther. 2019:1–9. doi: 10.1038/s41417-019-0116-8. [DOI] [PubMed] [Google Scholar]
- 57.Lu Y., Xue J., Deng T., Zhou X., Yu K., Deng L., Huang M., Yi X., Liang M., Wang Y. Safety and feasibility of CRISPR-edited T cells in patients with refractory non-small-cell lung cancer. Nat. Med. 2020:1–9. doi: 10.1038/s41591-020-0840-5. [DOI] [PubMed] [Google Scholar]
- 58.Perumal E., Youn K.S., Sun S., Seung-Hyun J., Suji M., Jieying L., Yeun-Jun C. PTEN inactivation induces epithelial-mesenchymal transition and metastasis by intranuclear translocation of β-catenin and snail/slug in non-small cell lung carcinoma cells. Lung Cancer. 2019;130:25–34. doi: 10.1016/j.lungcan.2019.01.013. [DOI] [PubMed] [Google Scholar]
- 59.Eichner L.J., Brun S.N., Herzig S., Young N.P., Curtis S.D., Shackelford D.B., Shokhirev M.N., Leblanc M., Vera L.I., Hutchins A. Genetic analysis reveals AMPK is required to support tumor growth in murine Kras-dependent lung cancer models. Cell Metab. 2019;29(2):285–302. doi: 10.1016/j.cmet.2018.10.005. (e7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cheung A.H.-K., Chow C., Zhang J., Zhou Y., Huang T., Ng K.C.-K., Or T.C.-T., Yao Y.Y., Dong Y., Fung J.M.-W. Specific targeting of point mutations in EGFR L858R-positive lung cancer by CRISPR/Cas9. Lab. Investig. 2018;98(7):968–976. doi: 10.1038/s41374-018-0056-1. [DOI] [PubMed] [Google Scholar]
- 61.Zhang B., Lu H.-Y., Xia Y.-H., Jiang A.-G., Lv Y.-X. Long non-coding RNA EPIC1 promotes human lung cancer cell growth. Biochemical biophysical research communications. 2018;503(3):1342–1348. doi: 10.1016/j.bbrc.2018.07.046. [DOI] [PubMed] [Google Scholar]
- 62.Ng S.R., Rideout W.M., Akama-Garren E.H., Bhutkar A., Mercer K.L., Schenkel J.M., Bronson R.T., Jacks T. CRISPR-mediated modeling and functional validation of candidate tumor suppressor genes in small cell lung cancer. Proc. Natl. Acad. Sci. 2020;117(1):513–521. doi: 10.1073/pnas.1821893117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kyrochristos I.D., Roukos D.H. Comprehensive intra-individual genomic and transcriptional heterogeneity: evidence-based colorectal cancer precision medicine. Cancer Treat. Rev. 2019;101894 doi: 10.1016/j.ctrv.2019.101894. [DOI] [PubMed] [Google Scholar]
- 64.Jubeen F., Liaqat A., Amjad F., Sultan M., Iqbal S.Z., Sajid I., Khan Niazi M.B., Sher F. Synthesis of 5-fluorouracil cocrystals with novel organic acids as coformers and anticancer evaluation against HCT-116 colorectal cell lines. Crystal Growth Design. 2020;20(4):2406–2414. [Google Scholar]
- 65.Franko J., Shi Q., Meyers J.P., Maughan T.S., Adams R.A., Seymour M.T., Saltz L., Punt C.J., Koopman M., Tournigand C. Prognosis of patients with peritoneal metastatic colorectal cancer given systemic therapy: an analysis of individual patient data from prospective randomised trials from the analysis and research in cancers of the digestive system (ARCAD) database. The Lancet Oncology. 2016;17(12):1709–1719. doi: 10.1016/S1470-2045(16)30500-9. [DOI] [PubMed] [Google Scholar]
- 66.Matano M., Date S., Shimokawa M., Takano A., Fujii M., Ohta Y., Watanabe T., Kanai T., Sato T. Modeling colorectal cancer using CRISPR-Cas9–mediated engineering of human intestinal organoids. Nat. Med. 2015;21(3):256. doi: 10.1038/nm.3802. [DOI] [PubMed] [Google Scholar]
- 67.Blanas A., Cornelissen L.A., Kotsias M., van der Horst J.C., van de Vrugt H.J., Kalay H., Spencer D.I., Kozak R.P., van Vliet S. Transcriptional activation of fucosyltransferase (FUT) genes using the CRISPR-dCas9-VPR technology reveals potent N-glycome alterations in colorectal cancer cells. Glycobiology. 2019;29(2):137–150. doi: 10.1093/glycob/cwy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hsu D.S., Kornepati A.V., Glover W., Kennedy E.M., Cullen B.R. Targeting HPV16 DNA using CRISPR/Cas inhibits anal cancer growth in vivo. Futur. Virol. 2018;13(07):475–482. doi: 10.2217/fvl-2018-0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li T., Liu D., Lei X., Jiang Q. Par3L enhances colorectal cancer cell survival by inhibiting Lkb1/AMPK signaling pathway. Biochemical biophysical research communications. 2017;482(4):1037–1041. doi: 10.1016/j.bbrc.2016.11.154. [DOI] [PubMed] [Google Scholar]
- 70.Wan T., Chen Y., Pan Q., Xu X., Kang Y., Gao X., Huang F., Wu C., Ping Y. Genome editing of mutant KRAS through supramolecular polymer-mediated delivery of Cas9 ribonucleoprotein for colorectal cancer therapy. J. Control. Release. 2020;322:236–247. doi: 10.1016/j.jconrel.2020.03.015. [DOI] [PubMed] [Google Scholar]
- 71.Ferlay J., Colombet M., Soerjomataram I., Dyba T., Randi G., Bettio M., Gavin A., Visser O., Bray F. Cancer incidence and mortality patterns in Europe: estimates for 40 countries and 25 major cancers in 2018. Eur. J. Cancer. 2018;103:356–387. doi: 10.1016/j.ejca.2018.07.005. [DOI] [PubMed] [Google Scholar]
- 72.Valcarcel-Jimenez L., Macchia A., Crosas-Molist E., Schaub-Clerigué A., Camacho L., Martín-Martín N., Cicogna P., Viera-Bardón C., Fernández-Ruiz S., Rodriguez-Hernandez I. PGC1α suppresses prostate cancer cell invasion through ERRα transcriptional control. Cancer Res. 2019;79(24):6153–6165. doi: 10.1158/0008-5472.CAN-19-1231. [DOI] [PubMed] [Google Scholar]
- 73.Fenner A. CRISPR–Cas9 ERβ deletion reveals roles in prostate. Nature Reviews Urology. 2020;17(4):192–193. doi: 10.1038/s41585-020-0302-3. [DOI] [PubMed] [Google Scholar]
- 74.Batır M.B., Şahin E., Çam F.S. Evaluation of the CRISPR/Cas9 directed mutant TP53 gene repairing effect in human prostate cancer cell line PC-3. Mol. Biol. Rep. 2019;46(6):6471–6484. doi: 10.1007/s11033-019-05093-y. [DOI] [PubMed] [Google Scholar]
- 75.Takao A., Yoshikawa K., Karnan S., Ota A., Uemura H., De Velasco M.A., Kura Y., Suzuki S., Ueda R., Nishino T. Generation of PTEN-knockout (−/−) murine prostate cancer cells using the CRISPR/Cas9 system and comprehensive gene expression profiling. Oncol. Rep. 2018;40(5):2455–2466. doi: 10.3892/or.2018.6683. [DOI] [PubMed] [Google Scholar]
- 76.Wei C., Wang F., Liu W., Zhao W., Yang Y., Li K., Xiao L., Shen J. CRISPR/Cas9 targeting of the androgen receptor suppresses the growth of LNCaP human prostate cancer cells. Mol. Med. Rep. 2018;17(2):2901–2906. doi: 10.3892/mmr.2017.8257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ye R., Pi M., Cox J.V., Nishimoto S.K., Quarles L.D. CRISPR/Cas9 targeting of GPRC6A suppresses prostate cancer tumorigenesis in a human xenograft model. Journal of Experimental Clinical Cancer Research. 2017;36(1):90. doi: 10.1186/s13046-017-0561-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang J., Zhang C., Feng B. The rapidly advancing class 2 CRISPR-Cas technologies: a customizable toolbox for molecular manipulations. Journal of Cellular Molecular Medicine. 2020;24(6):3256–3270. doi: 10.1111/jcmm.15039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gier R.A., Budinich K.A., Evitt N.H., Cao Z., Freilich E.S., Chen Q., Qi J., Lan Y., Kohli R.M., Shi J. High-performance CRISPR-Cas12a genome editing for combinatorial genetic screening. Nat. Commun. 2020;11(1):1–9. doi: 10.1038/s41467-020-17209-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yoon A.-R., Jung B.-K., Choi E., Chung E., Hong J., Kim J.-S., Koo T., Yun C.-O. CRISPR-Cas12a with an oAd induces precise and cancer-specific genomic reprogramming of EGFR and efficient tumor regression. Mol. Ther. 2020;28:2286–2296. doi: 10.1016/j.ymthe.2020.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yang H., Gao P., Rajashankar K.R., Patel D.J. PAM-dependent target DNA recognition and cleavage by C2c1 CRISPR-Cas endonuclease. Cell. 2016;167(7):1814–1828. doi: 10.1016/j.cell.2016.11.053. (e12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu L., Chen P., Wang M., Li X., Wang J., Yin M., Wang Y. C2c1-sgRNA complex structure reveals RNA-guided DNA cleavage mechanism. Mol. Cell. 2017;65(2):310–322. doi: 10.1016/j.molcel.2016.11.040. [DOI] [PubMed] [Google Scholar]
- 83.Abudayyeh O.O., Gootenberg J.S., Konermann S., Joung J., Slaymaker I.M., Cox D.B., Shmakov S., Makarova K.S., Semenova E., Minakhin L. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016;353(6299) doi: 10.1126/science.aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhao X., Liu L., Lang J., Cheng K., Wang Y., Li X., Shi J., Wang Y., Nie G. A CRISPR-Cas13a system for efficient and specific therapeutic targeting of mutant KRAS for pancreatic cancer treatment. Cancer Lett. 2018;431:171–181. doi: 10.1016/j.canlet.2018.05.042. [DOI] [PubMed] [Google Scholar]
- 85.Fan J., Liu Y., Liu L., Huang Y., Li X., Huang W. A multifunction lipid-based CRISPR-Cas13a genetic circuit delivery system for bladder cancer gene therapy. ACS Synth. Biol. 2019;9(2):343–355. doi: 10.1021/acssynbio.9b00349. [DOI] [PubMed] [Google Scholar]
- 86.Li J., Chen Z., Chen F., Xie G., Ling Y., Peng Y., Lin Y., Luo N., Chiang C.-M., Wang H. Targeted mRNA demethylation using an engineered dCas13b-ALKBH5 fusion protein. Nucleic Acids Res. 2020;48(10):5684–5694. doi: 10.1093/nar/gkaa269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xiao-Jie G.E.L., Li-Juan J., Zuckermann M., Kawauchi D., Gronych J. Applications of the CRISPR/Cas9 system in murine cancer modeling. Briefings in functional genomics. 2017;16(1):25–33. doi: 10.1093/bfgp/elw021. [DOI] [PubMed] [Google Scholar]
- 88.Zhou Y., Zhu S., Cai C., Yuan P., Li C., Huang Y., Wei W. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature. 2014;509(7501):487–491. doi: 10.1038/nature13166. [DOI] [PubMed] [Google Scholar]
- 89.Shalem O., Sanjana N.E., Hartenian E., Shi X., Scott D.A., Mikkelsen T.S., Heckl D., Ebert B.L., Root D.E., Doench J.G. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343(6166):84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li M., Xie H., Liu Y., Xia C., Cun X., Long Y., Chen X., Deng M., Guo R., Zhang Z. Knockdown of hypoxia-inducible factor-1 alpha by tumor targeted delivery of CRISPR/Cas9 system suppressed the metastasis of pancreatic cancer. J. Control. Release. 2019;304:204–215. doi: 10.1016/j.jconrel.2019.05.019. [DOI] [PubMed] [Google Scholar]
- 91.Shen B., Zhang W., Zhang J., Zhou J., Wang J., Chen L., Wang L., Hodgkins A., Iyer V., Huang X. Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects. Nat. Methods. 2014;11(4):399. doi: 10.1038/nmeth.2857. [DOI] [PubMed] [Google Scholar]
- 92.Konermann S., Brigham M.D., Trevino A.E., Joung J., Abudayyeh O.O., Barcena C., Hsu P.D., Habib N., Gootenberg J.S., Nishimasu H. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517(7536):583. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yin H., Kanasty R.L., Eltoukhy A.A., Vegas A.J., Dorkin J.R., Anderson D.G. Non-viral vectors for gene-based therapy. Nat. Rev. Genet. 2014;15(8):541–555. doi: 10.1038/nrg3763. [DOI] [PubMed] [Google Scholar]
- 94.Rui Y., Varanasi M., Mendes S., Yamagata H.M., Wilson D.R., Green J.J. Poly (beta-amino ester) nanoparticles enable non-viral delivery of CRISPR/Cas9 plasmids for gene knockout and gene deletion. Mol. Ther.–Nucleic Acids. 2020;20:661–672. doi: 10.1016/j.omtn.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yu W., Mookherjee S., Chaitankar V., Hiriyanna S., Kim J.-W., Brooks M., Ataeijannati Y., Sun X., Dong L., Li T. Nrl knockdown by AAV-delivered CRISPR/Cas9 prevents retinal degeneration in mice. Nat. Commun. 2017;8(1):1–15. doi: 10.1038/ncomms14716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen X., Chen Y., Xin H., Wan T., Ping Y. Near-infrared optogenetic engineering of photothermal nanoCRISPR for programmable genome editing. Proc. Natl. Acad. Sci. 2020;117(5):2395–2405. doi: 10.1073/pnas.1912220117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wan T., Niu D., Wu C., Xu F.-J., Church G., Ping Y. Material solutions for delivery of CRISPR/Cas-based genome editing tools: current status and future outlook. Mater. Today. 2019;26:40–66. [Google Scholar]
- 98.Zhang J., Fujimoto J., Zhang J., Wedge D.C., Song X., Zhang J., Seth S., Chow C.-W., Cao Y., Gumbs C. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science. 2014;346(6206):256–259. doi: 10.1126/science.1256930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chira S., Gulei D., Hajitou A., Zimta A.-A., Cordelier P., Berindan-Neagoe I. CRISPR/Cas9: transcending the reality of genome editing. Molecular Therapy-Nucleic Acids. 2017;7:211–222. doi: 10.1016/j.omtn.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Xing H., Meng L.-h. CRISPR-cas9: a powerful tool towards precision medicine in cancer treatment. Acta Pharmacol. Sin. 2019:1–5. doi: 10.1038/s41401-019-0322-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dow L.E., Fisher J., O’rourke K.P., Muley A., Kastenhuber E.R., Livshits G., Tschaharganeh D.F., Socci N.D., Lowe S.W. Inducible in vivo genome editing with CRISPR-Cas9. Nat. Biotechnol. 2015;33(4):390–394. doi: 10.1038/nbt.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Richardson C.D., Ray G.J., DeWitt M.A., Curie G.L., Corn J.E. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat. Biotechnol. 2016;34(3):339–344. doi: 10.1038/nbt.3481. [DOI] [PubMed] [Google Scholar]
- 103.Lu X.-J., Qi X., Zheng D.-H., Ji L.-J. Modeling cancer processes with CRISPR-Cas9. Trends Biotechnol. 2015;33(6):317–319. doi: 10.1016/j.tibtech.2015.03.007. [DOI] [PubMed] [Google Scholar]
- 104.Ghosh D., Venkataramani P., Nandi S., Bhattacharjee S. CRISPR–Cas9 a boon or bane: the bumpy road ahead to cancer therapeutics. Cancer Cell Int. 2019;19(1):12. doi: 10.1186/s12935-019-0726-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Clarke R., Heler R., MacDougall M.S., Yeo N.C., Chavez A., Regan M., Hanakahi L., Church G.M., Marraffini L.A., Merrill B. Enhanced bacterial immunity and mammalian genome editing via RNA-polymerase-mediated dislodging of Cas9 from double-strand DNA breaks. Mol. Cell. 2018;71(1):42–55. doi: 10.1016/j.molcel.2018.06.005. (e8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Su S., Hu B., Shao J., Shen B., Du J., Du Y., Zhou J., Yu L., Zhang L., Chen F. CRISPR-Cas9 mediated efficient PD-1 disruption on human primary T cells from cancer patients. Sci. Rep. 2016;6:20070. doi: 10.1038/srep20070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Huang C.-H., Lee K.-C., Doudna J.A. Applications of CRISPR-Cas enzymes in cancer therapeutics and detection. Trends in cancer. 2018;4(7):499–512. doi: 10.1016/j.trecan.2018.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Dekkers J.F., Whittle J.R., Vaillant F., Chen H.-R., Dawson C., Liu K., Geurts M.H., Herold M.J., Clevers H., Lindeman G.J. Modeling breast cancer using CRISPR-Cas9–mediated engineering of human breast organoids. JNCI: Journal of the National Cancer Institute. 2020;112(5):540–544. doi: 10.1093/jnci/djz196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Xue W., Chen S., Yin H., Tammela T., Papagiannakopoulos T., Joshi N.S., Cai W., Yang G., Bronson R., Crowley D.G. CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature. 2014;514(7522):380–384. doi: 10.1038/nature13589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Singh K., Evens H., Nair N., Rincón M.Y., Sarcar S., Samara-Kuko E., Chuah M.K., VandenDriessche T. Efficient in vivo liver-directed gene editing using CRISPR/Cas9. Mol. Ther. 2018;26(5):1241–1254. doi: 10.1016/j.ymthe.2018.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Blanas A., Cornelissen L.A., Kotsias M., van der Horst J.C., van de Vrugt H.J., Kalay H., Spencer D.I., Kozak R.P., van Vliet S.J. Transcriptional activation of fucosyltransferase (FUT) genes using the CRISPR-dCas9-VPR technology reveals potent N-glycome alterations in colorectal cancer cells. Glycobiology. 2019;29(2):137–150. doi: 10.1093/glycob/cwy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kawamura N., Nimura K., Nagano H., Yamaguchi S., Nonomura N., Kaneda Y. CRISPR/Cas9-mediated gene knockout of NANOG and NANOGP8 decreases the malignant potential of prostate cancer cells. Oncotarget. 2015;6(26) doi: 10.18632/oncotarget.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Marino N.D., Pinilla-Redondo R., Csörgő B., Bondy-Denomy J. Anti-CRISPR protein applications: natural brakes for CRISPR-Cas technologies. Nat. Methods. 2020:1–9. doi: 10.1038/s41592-020-0771-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Esposito R., Bosch N., Lanzós A., Polidori T., Pulido-Quetglas C., Johnson R. Hacking the cancer genome: profiling therapeutically actionable long non-coding RNAs using CRISPR-Cas9 screening. Cancer Cell. 2019;35(4):545–557. doi: 10.1016/j.ccell.2019.01.019. [DOI] [PubMed] [Google Scholar]
- 115.Jägle S., Dertmann A., Schrempp M., Hecht A. ZEB1 is neither sufficient nor required for epithelial-mesenchymal transition in LS174T colorectal cancer cells. Biochemical biophysical research communications. 2017;482(4):1226–1232. doi: 10.1016/j.bbrc.2016.12.017. [DOI] [PubMed] [Google Scholar]
- 116.Chen Y., Zhang Y. Application of the CRISPR/Cas9 system to drug resistance in breast cancer. Advanced Science. 2018;5(6) doi: 10.1002/advs.201700964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hu W., Guo G., Chi Y., Li F. Construction of Traf3 knockout liver cancer cell line using CRISPR/Cas9 system. J. Cell. Biochem. 2019;120(9):14908–14915. doi: 10.1002/jcb.28753. [DOI] [PubMed] [Google Scholar]
- 118.Hua J.T., Chen S., He H.H. Landscape of noncoding RNA in prostate cancer. Trends Genet. 2019;35(11):840–851. doi: 10.1016/j.tig.2019.08.004. [DOI] [PubMed] [Google Scholar]
- 119.Kweon J., Jang A.-H., Shin H.R., See J.-E., Lee W., Lee J.W., Chang S., Kim K., Kim Y. A CRISPR-based base-editing screen for the functional assessment of BRCA1 variants. Oncogene. 2020;39(1):30–35. doi: 10.1038/s41388-019-0968-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lee Y.-R., Chen M., Pandolfi P.P. The functions and regulation of the PTEN tumour suppressor: new modes and prospects. Nat. Rev. Mol. Cell Biol. 2018;19(9):547–562. doi: 10.1038/s41580-018-0015-0. [DOI] [PubMed] [Google Scholar]
- 121.Mou H., Kennedy Z., Anderson D.G., Yin H., Xue W. Precision cancer mouse models through genome editing with CRISPR-Cas9. Genome medicine. 2015;7(1):53. doi: 10.1186/s13073-015-0178-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Parolia A., Cieslik M., Chu S.-C., Xiao L., Ouchi T., Zhang Y., Wang X., Vats P., Cao X., Pitchiaya S. Distinct structural classes of activating FOXA1 alterations in advanced prostate cancer. Nature. 2019;571(7765):413–418. doi: 10.1038/s41586-019-1347-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Pereira M.A., Lima M.K., Couto P.G.P., Penna M.G., Alvim L.B., Nani T.F., Freire M.C.M., Araújo L.H. Springer; 2020. Cancer Genomics in Precision Oncology: Applications, Challenges, and Prospects, 'Essentials of Cancer Genomic, Computational Approaches and Precision Medicine; pp. 453–499. [Google Scholar]
- 124.Tordjman J., Majumder M., Amiri M., Hasan A., Hess D., Lala P.K. Tumor suppressor role of cytoplasmic polyadenylation element binding protein 2 (CPEB2) in human mammary epithelial cells. BMC Cancer. 2019;19(1):561. doi: 10.1186/s12885-019-5771-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wang S., Yang H., Chen X., Jiang Z. Effects of SOX15 on the colorectal cancer cells via downregulation of the Wnt/β-catenin signaling pathway. Future Oncol. 2018;14(19):1921–1932. doi: 10.2217/fon-2017-0688. [DOI] [PubMed] [Google Scholar]
- 126.Wang Y., Gao B., Tan P.Y., Handoko Y.A., Sekar K., Deivasigamani A., Seshachalam V.P., OuYang H.-Y., Shi M., Xie C. Genome-wide CRISPR knockout screens identify NCAPG as an essential oncogene for hepatocellular carcinoma tumor growth. FASEB J. 2019;33(8):8759–8770. doi: 10.1096/fj.201802213RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wang Y., Li X., Liu W., Li B., Chen D., Hu F., Wang L., Liu X.M., Cui R., Liu R. MicroRNA-1205, encoded on chromosome 8q24, targets EGLN3 to induce cell growth and contributes to risk of castration-resistant prostate cancer. Oncogene. 2019;38(24):4820–4834. doi: 10.1038/s41388-019-0760-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Artegiani B., van Voorthuijsen L., Lindeboom R.G., Seinstra D., Heo I., Tapia P., López-Iglesias C., Postrach D., Dayton T., Oka R. Probing the tumor suppressor function of bap1 in crispr-engineered human liver organoids. Cell Stem Cell. 2019;24(6):927–943. doi: 10.1016/j.stem.2019.04.017. (e6) [DOI] [PubMed] [Google Scholar]
- 129.Liu C., Zhang L., Liu H., Cheng K. Delivery strategies of the CRISPR-Cas9 gene-editing system for therapeutic applications. J. Control. Release. 2017;266:17–26. doi: 10.1016/j.jconrel.2017.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Glass Z., Lee M., Li Y., Xu Q. Engineering the delivery system for CRISPR-based genome editing. Trends Biotechnol. 2018;36(2):173–185. doi: 10.1016/j.tibtech.2017.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material