Significance
Studies of the 1940s Victoria blight of oats epidemic discovered that some fungal pathogens secrete HSTs responsible for symptom development and specificity of the associated disease. The causal necrotrophic pathogen of Victoria blight, C. victoriae, secretes the peptide HST victorin, which was, subsequently, shown to constitute a novel class of effectors that exploit host immunity pathways aimed at repelling biotrophic pathogens. Although these discoveries have broadened our mechanistic understanding of plant-pathogen interactions, the genetic and biochemical origins of victorin have remained elusive. Here, we solve this decades-old mystery by demonstrating that victorin is produced ribosomally, not, as assumed, by nonribosomal peptide synthetase. Furthermore, we identify a CAO enzyme responsible for converting victorin to its active form.
Keywords: RiPP, host-selective toxin, victorin, copper amine oxidase, Cochliobolus
Abstract
The necrotrophic fungal pathogen Cochliobolus victoriae produces victorin, a host-selective toxin (HST) essential for pathogenicity to certain oat cultivars with resistance against crown rust. Victorin is a mixture of highly modified heterodetic cyclic hexapeptides, previously assumed to be synthesized by a nonribosomal peptide synthetase. Herein, we demonstrate that victorin is a member of the ribosomally synthesized and posttranslationally modified peptide (RiPP) family of natural products. Analysis of a newly generated long-read assembly of the C. victoriae genome revealed three copies of precursor peptide genes (vicA1–3) with variable numbers of “GLKLAF” core peptide repeats corresponding to the victorin peptide backbone. vicA1–3 are located in repeat-rich gene-sparse regions of the genome and are loosely clustered with putative victorin biosynthetic genes, which are supported by the discovery of compact gene clusters harboring corresponding homologs in two distantly related plant-associated Sordariomycete fungi. Deletion of at least one copy of vicA resulted in strongly diminished victorin production. Deletion of a gene encoding a DUF3328 protein (VicYb) abolished the production altogether, supporting its predicted role in oxidative cyclization of the core peptide. In addition, we uncovered a copper amine oxidase (CAO) encoded by vicK, in which its deletion led to the accumulation of new glycine-containing victorin derivatives. The role of VicK in oxidative deamination of the N-terminal glycyl moiety of the hexapeptides to the active glyoxylate forms was confirmed in vitro. This study finally unraveled the genetic and molecular bases for biosynthesis of one of the first discovered HSTs and expanded our understanding of underexplored fungal RiPPs.
In the 1940s, the widespread planting across the United States of oat varieties possessing “Victoria-type” resistance to the biotrophic crown rust fungus Puccinia coronata provoked an epidemic of Victoria blight, a new oat disease caused by the necrotrophic pathogen C. victoriae (1). In 1947, based on their studies of Victoria blight, Meehan and Murphy reported the discovery and first demonstration that certain fungal plant pathogens can secrete secondary metabolites as HSTs that function as pathogenicity determinants at low concentrations (2). For a susceptible interaction, a corresponding, specific molecular target must be present in a host line or cultivar (3, 4). Indeed, pathogenicity of C. victoriae depends on the production of the HST victorin by the fungus, and on the host side, an oat cultivar carrying the Victoria blight susceptibility-conferring Vb gene. Remarkably, Vb is genetically inseparable from the Pc-2 gene conferring resistance against the biotrophic pathogen P. coronata (5, 6).
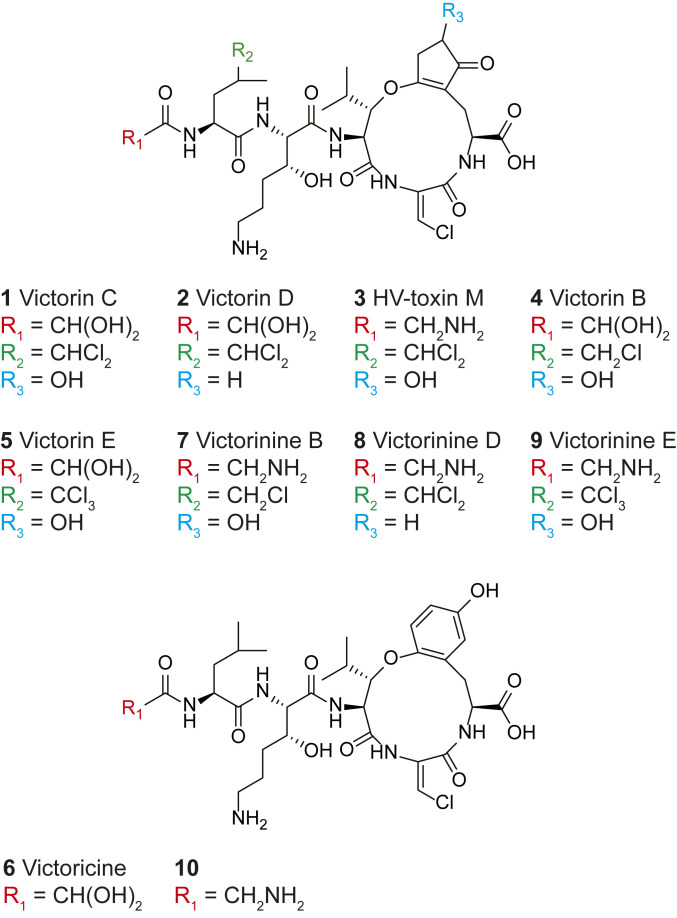
Victorin was isolated in the 1980s and shown to consist of a mixture of heterodetic cyclic hexapeptides with victorin B (4), C (1), D (2), E (5), victoricine (6), and HV-toxin M (3) as known members (Fig. 1) (7–9). All derivatives contain five peptide bonds, a 12-membered ring featuring an ether bond, and different degrees of chlorination with victorin C as the predominant form. Structural differences among victorin derivatives were shown to have little effect on bioactivity with the exception of 3 and 6, which displayed diminished host-selective toxicity (7, 10). Derivative 3 features a glycyl instead of a glyoxylate residue, while 6 has a phenyl moiety in place of a cyclopentenyl within its macrocycle (Fig. 1).
Fig. 1.
Structures of known victorin derivatives (1–6) and proposed structures of related biosynthesis intermediates (7–10).
In a 2007 landmark paper, Wolpert and colleagues identified the LOV1 gene in the victorin-sensitive model host Arabidopsis thaliana and showed that it encoded a nucleotide-binding-site leucine-rich repeat protein commonly involved in disease resistance against biotrophic pathogens (11). Subsequently, Wolpert and colleagues demonstrated that victorin binds to thioredoxin TRX-h5 involved in regulating plant immunity (12). The binding of victorin to TRX-h5 activates LOV1, which elicits a hypersensitive response-like host cell death typically deployed to halt infection by a biotroph but, in this case, is exploited by necrotrophic C. victoriae for host infection (12). The discovery that necrotrophic fungi can hijack components of plant defense signaling pathways as targets for disease susceptibility has added a new dimension to our knowledge of offensive arsenals deployed by necrotrophs (13). Despite these breakthroughs in understanding plant-pathogen interactions on the host side, on the pathogen side, the genetic and molecular bases for biosynthesis of victorin remained an enigma over seven decades after its initial discovery.
Fungi produce a huge variety of cyclic peptides through either nonribosomal or ribosomal pathways (14). Compared to the better known nonribosomal peptides biosynthesized by nonribosomal peptide synthetases (NRPSs), e.g., the immunosuppressive agent cyclosporine, the antifungal echinocandins, and the HST HC-toxin produced by Cochliobolus carbonum (15), the RiPPs from fungi have been discovered relatively recently. While bacterial- and plant-produced RiPPs have been known for decades (16), the first report of RiPPs in fungi was in 2007 with the discovery of the ribosomal origin of amatoxins and phallotoxins (17). Subsequently, ustiloxins (18, 19), epichloëcyclins (20), asperipin-2a (21), phomopsins (22), and, most recently, borosins (23, 24) were demonstrated to be produced ribosomally and classified as RiPPs.
The core peptides that form the backbones of several of the recently discovered RiPPs have been found to be encoded as multiple perfect or imperfect repeats within a single precursor peptide gene (16). After being excised from the precursor peptide by kexin proteases, the core peptides are cyclized and modified posttranslationally by enzymes encoded within the corresponding gene clusters. Based on some of these initial discoveries, an abundance of putative RiPP gene clusters has been identified in genomes of filamentous fungi, hinting at the untapped wealth of this class of compounds in fungi (21, 22). However, the lack of knowledge of fungal RiPP processing impedes the prediction of the mature products of those clusters. At the same time, extensive posttranslational modifications (PTMs) of the core peptide can mask the ribosomal origin of known cyclic peptides as was the case with ustiloxins (18).
Here, we reexamine the biosynthetic origin of victorin, which, like ustiloxins and phomopsins, are heterodetic peptides that include an ether bond as part of the macrocycle. A draft genome for the victorin-producing C. victoriae strain FI3 was completed in 2013 (25). Given that victorin consists of multiple nonproteinogenic amino acids and that C. victoriae is closely related to C. carbonum, which biosynthesizes the cyclic tetrapeptide HC-toxin via an NRPS, victorin was assumed to be produced by an NRPS as well. However, all C. victoriae mutants deleted for candidate NRPSs still produce victorin (25, 26). In the present study, we demonstrate that victorin is, in fact, synthesized ribosomally and is a member of the emerging family of kexin-processed fungal RiPPs. We further uncovered a CAO that converts the inactive forms of victorin to the potent HST.
Results
The Structure of Victorin Guided the Discovery of Its Precursor Peptide Gene.
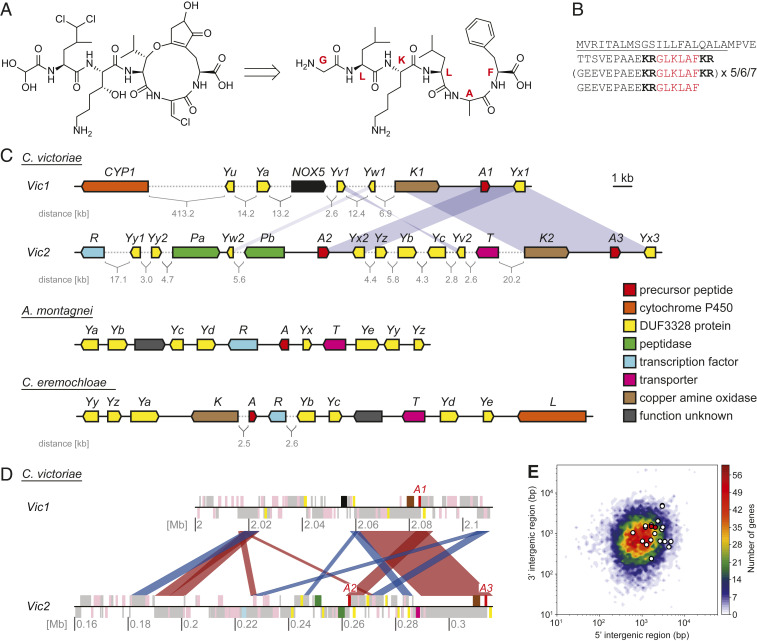
Retrobiosynthetic analysis of the chemical structure of victorin suggested that the peptide backbone is likely made up of the core peptide sequence "GLKLAF" if synthesized by a ribosomal pathway (Fig. 2A). Thus, we queried the C. victoriae FI3 genome on the JGI Mycocosm database (25) with this amino acid sequence. Among the BLASTp hits, one of the putative genes on a 1,240-bp scaffold (scaffold 582, NW_014574810) was predicted to encode a protein (ID 32336) with an N-terminal signal peptide and two repeats of the query sequence "GLKLAF," both flanked with “KR” corresponding to kexin protease cut sites. The predicted gene coding sequence was truncated 248 bp after the start codon. Absence of a stop codon as well as the fact that kexin-processed fungal RiPP precursor peptide genes usually contain a higher number of core peptide repeats led us to believe that a major part of the gene was missing.
Fig. 2.
Analysis of victorin structure, biosynthesis, and genetic background. (A) Deduction of the unmodified peptide backbone sequence of victorin C from its structure. Red letters indicate the individual amino acids that make up the peptide repeat. (B) Precursor peptide amino acid sequence. The number of sequence repeats varies among the three gene copies (Note: not all repeats encoded in vicA3 are in frame, thus, the predicted numbers of core peptides in VicA3 vary with different intron–exon structure predictions). The signal peptide is underlined, the kexin protease recognition sites are shown in bold, and the core peptide is shown in red. (C) Putative victorin biosynthesis genes (Top) and putative biosynthetic gene clusters of vicA homologs in Apiospora montagnei and Colletotrichum eremochloae. Blue shade represents duplication of the highlighted gene or region. Numbers following a gene name indicate duplicates. Gene name identity between species does not necessarily imply homology (SI Appendix, Table S2). (D) Schematic of the genomic regions harboring the victorin precursor peptide genes (annotated here as A1–A3). Each chromosome is shown as a black line with the position in megabases (Mb) indicated. Gray blocks show locations where transposable elements have been annotated, and pink blocks show annotated FI3 genes. Blocks appearing above the chromosome line are encoded in the forward direction, and blocks below the line in the reverse. Other colors indicate the locations of genes with putative victorin biosynthetic genes with the same color code as in C. The red and blue ribbons connecting the two contigs show regions >2 kb with >80% identity between the two chromosomes. Red ribbons indicate alignments in the same direction, and blue ribbons indicate inversions. (E) Gene density heat map of the C. victoriae genome. Genes were binned based on the lengths of their 5′ and 3′ intergenic regions. Putative victorin biosynthesis genes are overlaid as white dots, and precursor peptide genes are overlaid as red dots.
Long-Read Sequencing Revealed Putative Victorin Biosynthesis Genes at Two Loci.
To obtain the missing 3′ end of the precursor peptide gene as well as other victorin biosynthesis genes expected to be clustered in proximity, we performed long-read sequencing and assembled the strain FI3 genome de novo into 22 contigs (27). The assembly was polished using the short-read Illumina data available on the JGI Mycocosm database (28). Query of this assembly for the core peptide sequence GLKLAF resulted in identification of three victorin precursor peptide encoding genes (vicA1–3) with seven–nine core peptide repeats (Fig. 2B). The first putative precursor peptide gene vicA1 was on contig 7 (named the Vic1 locus), while two additional copies (vicA2 and A3) were located about 51 kb apart on contig 16 (named the Vic2 locus) (Fig. 2C). Alignment of the two contigs revealed 14 regions with over 70% sequence identity that were longer than 1 kb, 11 of which were within 100 kb of the three vicA gene copies (Fig. 2D). The aligned regions included many predicted transposable elements.
Compared with previously characterized fungal RiPPs gene clusters (21, 22), few candidate biosynthetic genes were within close proximity of the vicA genes. Analysis of intergenic regions in the FI3 genome showed that the three copies of vicA were in genomic regions of relatively low gene density (Fig. 2 C and E). Expanding the search region to within about 50 kb on both flanks of each precursor peptide gene, we identified candidate genes encoding putative RiPP biosynthesis-related proteins (designated according to fungal RiPP gene nomenclature where appropriate). These included genes encoding a putative CAO (two copies, vicK1/2), one NADPH oxidase (NOX5), three DUF3328 proteins (vicYa–c), two peptidases (vicPa/b), a transporter (vicT), and a transcription factor (vicR). Genes encoding DUF3328 proteins (given the letter Y in fungal RiPP gene nomenclature) have been a common feature found in all kexin-processed RiPP gene clusters reported so far with several of these genes observed within some RiPP gene clusters (29). It has recently been proposed that DUF3328 proteins are a novel family of oxidative cyclization enzymes (30). The DUF3328 domain possesses two “HXXHC” motifs, which are assumed to form the active sites (21). Although only vicYa–c were predicted by NCBI CD search to encode DUF3328 proteins, we found six additional genes at the Vic loci encoding hypothetical proteins containing this characteristic double motif with some of them appearing as duplicates. We tentatively included these genes as candidate victorin biosynthesis genes and assigned them as vicYu–z (Fig. 2C).
Identification of Homologous Gene Clusters Supported Our Prediction of Victorin Biosynthesis Genes.
One strategy to identify missing genes in fragmented gene clusters or support the involvement of unclustered biosynthetic genes in a biosynthetic pathway is to search for homologs, as the biosynthetic genes could be tightly clustered in the genome of another organism (31, 32). Thus, we searched for vicA homologs in all Ascomycota genomes in NCBI and JGI databases and identified a single homolog in the genomes of two Sordariomycetes, A. montagnei and C. eremochloae, which share remarkable similarity with VicA in terms of overall protein sequence and the core peptides contained therein (“QLKFNF” and “LFKFNF,” respectively) (SI Appendix, Fig. S1). Examination of genomic regions around these two vicA homologs revealed clustered genes encoding putative PTM enzymes with varying amino acid sequence similarity to proteins encoded by putative vic genes (Fig. 2C and SI Appendix, Table S2). Multiple hypothetical proteins featuring the double "HXXHC" motifs from both clusters shared up to 58% protein identity with putative VicY proteins not predicted to contain the DUF3328 domain. Additionally, within the C. eremochloae cluster was a cytochrome P450 (CYP) gene for which a corresponding homolog (CYP1) with 44% protein identity could be found on contig 7 of the C. victoriae genome long-read assembly but located at about 400 kb from other putative vic genes (Fig. 2C). These shared gene homologs and their tight clustering in A. montagnei and C. eremochloae genomes support our predictions that the scattered candidate vic genes in C. victoriae are encoding proteins involved in victorin biosynthesis.
Gene Deletions Resulted in Loss of Victorin Production and Host-Specific Toxicity of Culture Filtrates.
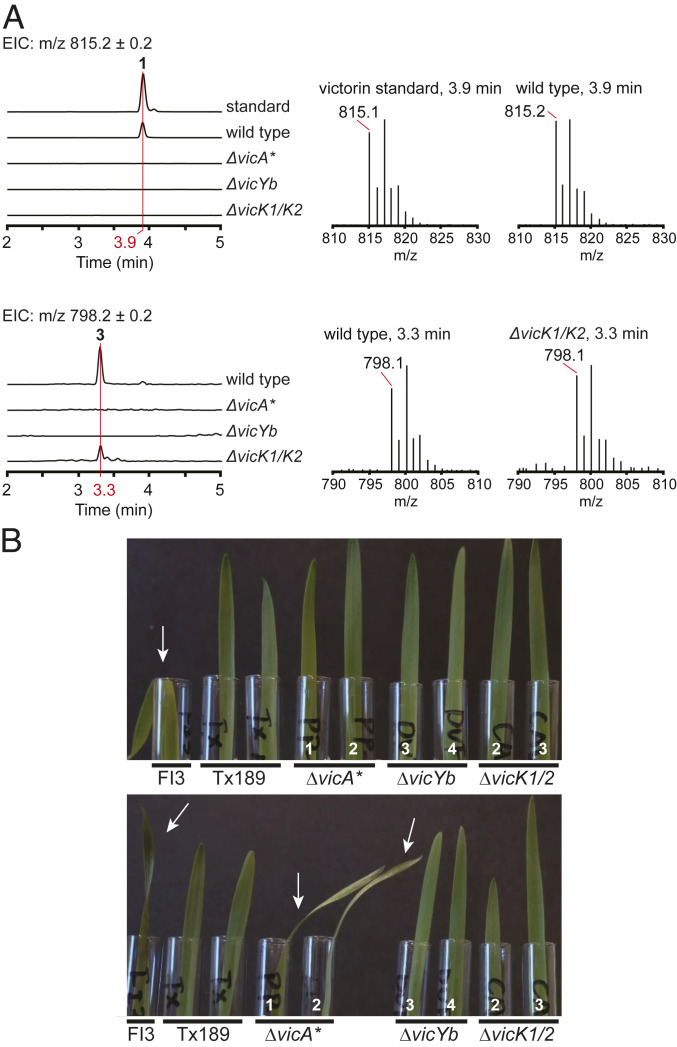
C. victoriae strain FI3 was cultured as previously reported in ref. 9. The production of victorin C, the major form of victorin, was verified by liquid chromatography–mass spectrometry (LC–MS) in comparison to a standard characterized in previous studies (8, 9) (Fig. 3A). Other previously reported victorin derivatives could also be detected by LC–MS (SI Appendix, Fig. S2), and all of them were verified by LC–MS/MS (SI Appendix, Fig. S3).
Fig. 3.
Victorin production analysis in deletion mutants. (A) LC–MS extracted ion chromatograms (EICs) of victorin C standard and of culture filtrate extractions from different C. victoriae strains (Left) and spectra at indicated elution times (Right). m/z of 815.2 (Top) corresponds to victorin C (1), m/z of 798.2 (Bottom) corresponds to HV-toxin M (3). (B) Two replicates (Top/Bottom) of victorin toxicity bioassay on susceptible oat cultivar (Fulgrain) leaves with undiluted culture filtrate from different C. victoriae strains. Arrows indicate leaf wilting. Numbers in white indicate the independent transformants (SI Appendix, Table S5). Note only culture filtrate from WT strain FI3 caused the typical leaf wilting symptom in the first assay replicate (Top), whereas in the second assay replicate (Bottom), the leaf wilting symptom was also observed with culture filtrate from the ΔvicA* mutant strain.
To determine if the candidate genes were involved in victorin biosynthesis, C. victoriae mutants with deletions of the precursor peptide gene vicA (ΔvicA* and ΔvicA**) and selected putative PTM genes encoding DUF3328 protein VicYb (ΔvicYb), CAO VicK (ΔvicK1/2), and NADPH oxidase NOX5 (ΔNOX5) were generated (SI Appendix, Table S5). For mutant ΔvicA*, diagnostic PCRs confirmed the deletion of at least one copy of vicA and the presence of at least one remaining copy (SI Appendix, Fig. S4). Subsequently, this mutant was subjected to a successful second deletion experiment with a different selectable marker. The resulting mutant ΔvicA** still had one copy of vicA in the genome, indicating that ΔvicA* was a single copy deletion mutant (SI Appendix, Fig. S4). Because the flanking DNA sequences of the three copies of vicA are identical, we do not know which copies were deleted. Examination of vicK mutants confirmed that a single targeted deletion experiment successfully deleted both copies of vicK. Targeted deletions of single copy genes vicYb and NOX5 were also confirmed by diagnostic PCR.
The culture filtrate of each mutant was analyzed by LC–MS (Fig. 3A and SI Appendix, Fig. S2) to detect changes in victorin production in comparison to the wild type (WT) FI3 strain. The culture filtrates were also tested with detached leaves of susceptible and resistant oat cultivars for host-specific toxicity (Fig. 3B and SI Appendix, Fig. S5) (10). LC–MS analysis of ΔvicA* mutant culture filtrate showed complete loss of production for all victorin derivatives (Fig. 3A and SI Appendix, Fig. S2). This indicated that vicA was indeed the precursor peptide gene for victorin. However, considering that two copies of the three precursor peptide genes remained in this strain, the complete absence of victorin production was surprising. Culture filtrate of ΔvicA* mutants elicited leaf wilting symptoms in some replicates of the toxicity test but not consistently (Fig. 3B and SI Appendix, Fig. S5). In comparison, characteristic wilting symptoms were observed for every replicate of WT strain FI3 culture filtrates (positive control on susceptible oats) and for none of the replicates with victorin-minus mutant Tx189 culture filtrates (negative control on susceptible oats). This host-specific toxicity assay is known to be highly sensitive, with victorin concentrations as low as 10 pM reported to induce diseaselike symptoms (33). Thus, this indicated that the deletion of at least one of three vicA copies did not completely eliminate but strongly diminished the ability of the mutant strain to produce victorin. The culture filtrate of ΔvicA** double deletion mutants appeared to exhibit reduced host-specific toxicity compared to that of ΔvicA* (SI Appendix, Fig. S5).
The deletion of vicYb, vicK, and NOX5 were expected to result in the accumulation of victorin pathway intermediates if they were involved in later stages of victorin biosynthesis. For the ΔvicYb mutant, no victorin could be detected in the culture filtrate (Fig. 3A and SI Appendix, Fig. S2). The absence of any victorin derivative or intermediate indicated VicYb is essential and involved in early steps of victorin biosynthesis. Consistent with this result, the culture filtrate bioassay showed no sign of toxicity, indicating a complete absence of victorin (Fig. 3B and SI Appendix, Fig. S5). In contrast, the ΔNOX5 mutant showed undiminished victorin production (SI Appendix, Figs. S5 and S6). Therefore, the NADPH oxidase encoded by NOX5 did not appear to be involved in victorin biosynthesis. Interestingly, the culture filtrate of the ΔvicK1/2 mutant contained 3, but no other previously known victorin derivative. Like the ΔvicYb mutant, the ΔvicK1/2 mutant lost host-specific toxicity, which was not unexpected as 3 was reported to exhibit significantly reduced activity compared to 1 (7).
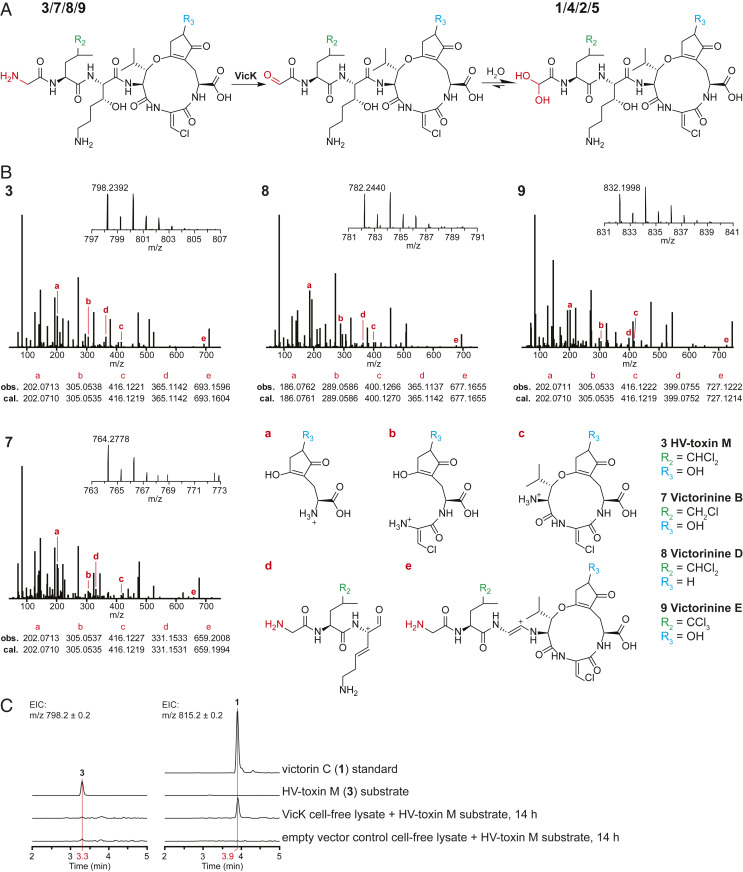
Comparative Metabolite Analysis Uncovered Novel Victorin Derivatives and the Role of CAO VicK.
The predominant victorin C (1) contains a glyoxylate residue. In contrast, HV-toxin M (3) possesses an unmodified N-terminal glycine instead of glyoxylate, thus, 3 is 17 Da less in mass compared to 1 (Fig. 1). The fact that mutant strain ΔvicK1/2 lost its ability to produce the latter but not the former led us to believe that VicK is involved in the conversion of 3 to 1 via oxidative deamination of the glycine residue to glyoxylate (Fig. 4A). Aside from 3, we detected three novel putative victorin derivatives by LC–MS analysis in the culture filtrate of strain ΔvicK1/2 featuring both the characteristic isotopic pattern expected of chlorinated compounds and a mass difference of about −17 Da compared to other known victorin derivatives. These compounds appeared to be lighter derivatives of victorin D (2), B (4), and E (5). Based on the same mass difference between 1 and 3, we proposed structures 7–9 for these novel derivatives and performed LC–MS/MS on the culture filtrate for confirmation. Select fragment masses matched masses of predicted fragment structures (Fig. 4B). When comparing MS/MS data of 7–9 against 4, 2, and 5 (SI Appendix, Fig. S3), respectively, fragments covering the N-terminal moiety showed the expected mass difference while fragments excluding the N terminus displayed the same mass for each respective pair as in the case when comparing 3 against 1. The MS/MS fragments of 7–9 could also be mapped to corresponding fragments observed for the 1 standard with the expected mass difference (SI Appendix, Fig. S7). Based on these results, we conclude VicK to be involved in oxidative deamination of 2–4 (Fig. 4A). We propose the names victorinine B (7), D (8), and E (9) for the novel structures.
Fig. 4.
Biosynthetic function of VicK. (A) Proposed reaction catalyzed by VicK. (B) MS1 and MS2 spectra of ions corresponding to masses of compounds 3, and 7–9 with proposed structures based on diagnostic fragment ions a–e in comparison to the corresponding glyoxylate analogs 1, 4, 2, and 5 (SI Appendix, Fig. S3). (C) In vitro oxidative deamination of 3 with cell-free lysate from A. nidulans expressing VicK. m/z of 798.2 corresponds to 3, m/z of 815.2 to 1. Chromatograms of samples containing cell-free lysates are from after 14 h of incubation with the substrate. Y axes of all seven chromatograms are scaled equally.
In Vitro Oxidative Deamination of HV-Toxin M Yields Victorin C.
To corroborate the function of VicK, we attempted to assay the VicK protein in vitro. Unfortunately, expression of VicK fused to maltose-binding protein in Escherichia coli was unsuccessful. Thus, we heterologously expressed VicK in Aspergillus nidulans host strain LO8030 and used its cell-free mycelium lysate to assay 3, a technique we previously used to characterize other fungal biosynthetic proteins (34, 35). Incubation of the cell-free lysate with purified 3 resulted in the accumulation of 1, whereas 1 was absent in the negative control (Fig. 4C). Discrepancies in peak size between 3 in the negative control and 1 after incubation with VicK-containing cell-free lysate were observed and investigated by boiling cell-free lysate from empty vector-carrying mycelium and incubating it with either 1 or 3 (SI Appendix, Fig. S8). Whereas 1 was largely unaffected, higher amounts of 3 in the boiled sample compared to the untreated cell lysate suggest selective degradation of 3 by endogenous A. nidulans enzymes. Collectively, these results confirmed VicK as a novel CAO responsible for oxidative deamination of the glycyl moiety of 3 to the predominant 1. Although 7–9 could not be isolated in sufficient quantities to perform the same assay, it is very likely that VicK acts on them in the same way, converting them to the glyoxylate-containing 2, 4 and 5.
Discussion
Studies on HST victorin, the molecular basis for Victoria blight of oats, has contributed significantly to our understanding of the interactions between necrotrophic pathogens and plant hosts. Over seven decades after its initial discovery (2) and three decades after its structures were elucidated (8, 9), the genetic basis for the biosynthesis of victorin has finally been solved. Contrary to previous assumptions of it being produced nonribosomally, here, we have conclusively demonstrated that the heterodetic victorin peptides are RiPPs. In contrast to known fungal RiPP biosynthetic gene clusters (21, 22), which consist of a single copy of the precursor peptide gene with most biosynthesis-related genes in close genomic proximity, the vic genes are loosely clustered at two different loci and include multiple copies of the precursor peptide gene. Although multiple gene duplicates are found between Vic1 and Vic2 loci, we are unable to determine at this stage whether either locus alone is sufficient for biosynthesis of victorin due to the presence of candidate genes that are unique to each locus, e.g., vicYa/Yu in Vic1 and vicPa/Pb in Vic2. Both loci are gene-sparse transposon-rich genomic regions. These Vic1 and Vic2 structural features are reminiscent of regions carrying biosynthetic genes for other well-known HSTs produced by Cochliobolus taxa, including HC-toxin (two diffuse gene clusters spanning >500 kb) from C. carbonum, a close relative of C. victoriae (36), and T-toxin from Cochliobolus heterostrophus (two loci on different chromosomes, spanning >1.2 Mb) (37). Thus, the genomic context of Vic1 and Vic2 loci is congruent with their role in biosynthesis of a virulence determinant. Additionally, in virulence loci (e.g., encoding effector proteins) of some filamentous plant pathogens, similar structural features have been implicated as characteristics of genomic regions that evolve at an accelerated rate compared to the rest of the genome (38).
Interestingly, there are no vic homologs found in any of the dothideomycete genomes in the NCBI (233 species) and JGI Mycocosm (172 species) databases. We are reminded of parallel findings with the C. heterostrophus T-toxin biosynthetic genes, which have been discovered in unrelated Dothideomycetes and a few Eurotiomycetes but not in any other of the more than 50 Cochliobolus species (32). However, surprisingly, we found two biosynthetic gene clusters containing multiple vic homologs, including precursor peptide gene vicA homologs, in two distantly related plant-associating sordariomycete fungi. The two “vic-like” RiPP gene clusters in the sordariomycetes A. montagnei and C. eremochloae featured eight genes each where their predicted proteins showed over 30% sequence identity to predicted proteins in the Vic1 and Vic2 loci, supporting their involvement in victorin biosynthesis either as PTM enzymes or in other roles (e.g., regulation, transport, and resistance). Additionally, the cytochrome P450 gene apparently associated with the C. eremochlae RiPP cluster led us to discover a homolog on contig 7 of the C. victoriae genome. Despite the long genomic distance to other victorin candidate genes, the homology between the two genes could indicate involvement of CYP1 in the biosynthesis of victorin. The fact that NOX5 was shown to not be necessary for victorin production, despite being relatively close to putative vicY genes, corresponds to the lack of a NOX5 homolog in the two Sordariomycete vic-like gene clusters.
In RiPP biosynthesis, the precursor peptide gene is directly transcribed and translated into the precursor substrate that feeds into the pathway. Thus, the transcript dosage of the precursor peptide gene has a proportionate effect on the yield of the RiPP product, a trend that has been observed with the heterologous expression of bacterial RiPP cyanobactins (39). This is reflected in the significant drop in victorin production in the ΔvicA* strain, even though two copies of vicA remain. By contrast, the deletion of DUF3328 gene vicYb abolished victorin production completely. DUF3328 proteins have been shown to be involved in the biosynthesis of the first cyclic ustiloxin intermediate (40) and in two macrocyclization reactions in asperipin-2a (30). We reasoned that the absence of any victorin derivative in the culture filtrate of the ΔvicYb strain supports that VicYb is required for the formation of the macrocycle in victorin, as cyclization of peptides has been shown to improve metabolic stability (41).
The aldehyde functional group on the glyoxylate moiety of victorin was previously shown to be essential for both its host-selective toxicity (10) and binding to the thioredoxin TRX-h5 via covalent association (12). Here, we identified the CAO VicK to be responsible for converting victorin to the active form by oxidizing the N-terminal glycyl residue in the peptides to glyoxylate. The deletion of vicK in C. victoriae abolished production of glyoxylate-containing forms of victorin and yielded three novel glycyl derivatives of victorin B, D, and E alongside HV-toxin M. This observation, along with the presence of multiple victorin derivatives in WT, suggest relaxed substrate specificity of enzymes in the victorin biosynthetic pathway, resulting in a metabolic grid that produces a set of analogs (42). The involvement of VicK in oxidative deamination of the glycyl residue to glycoxylate was further verified in vitro. CAOs are known to be involved in primary amine metabolism (43), but the participation of CAOs in secondary metabolite biosynthesis is rare. In fungi, a CAO has been discovered to be involved in the early step biosynthesis of the anti-Alzheimer’s alkaloid huperzine A (44). However, the involvement of CAO in RiPP biosynthesis is unprecedented and, thus, expands the known RiPP PTM enzymes.
In conclusion, this paper sets the foundation for further study of victorin biosynthesis, in which many intriguing questions remain. The chlorination on unactivated carbon centers in victorin is a novelty in fungal RiPPs that likely requires a different type of enzyme than the one responsible for chlorination of phomopsins (45). Presumably, they are also different from bacterial nonheme iron/α-ketoglutarate-dependent halogenases that can chlorinate unactivated carbon (46), as no homolog was found in the C. victoriae genome. Furthermore, the multitude of DUF3328 and DUF3328-like vicY genes suggest that some of them likely fulfill biosynthetic tasks not yet associated with them, supported by the presence of multiple DUF3328 genes in some RiPP clusters beyond the number of macrocycles in the associated RiPP products (e.g., phomopsins, ref. 22). Importantly, the discovery of homologous vic-like gene clusters in two distantly related plant-associating sordariomycete fungi and their absence in related dothideomycetes hint at lateral gene transfer as the evolutionary origin of vic genes in C. victoriae, which belongs to Pleosporales, an order proposed to have a propensity for acquiring genes laterally (47). The discovery also opened up questions about the biological function of the sordariomycete victorin-like RiPPs especially whether they are involved in interactions with plant hosts. Given that victorin binds and inhibits a thioredoxin (12), a promising target for anticancer drug development (48), the chemistry and biology of this subgroup of victorin-like RiPPs warrants further investigation.
Materials and Methods
Fungal Strains and Growth Conditions.
All C. victoriae mutant strains created in this study (SI Appendix, Table S5) were constructed by deleting genes from WT strain C. victoriae field isolate FI3 (victorin+, MAT1-2, unknown geographical origin) (25). Victorin-null mutant strain Tx189, previously created using restriction enzyme mediated integration (REMI) mutagenesis (49), was used as a negative control for victorin production.
All C. victoriae strains were stored in liquid complete medium (CM) (50) containing 25% glycerol at −80 °C (37). For each experiment, an aliquot was recovered from glycerol and grown on plates containing CM with xylose (CMX) (51) at 22–24 °C under a 16:8-h light:dark cycle as previously described in ref. 37. For media compositions, see SI Appendix, Supplementary Material and Methods.
Victorin Bioassays.
For victorin bioassays, 10 2 × 2-mm plugs were cut from CMX plates and transferred to 20 mL modified liquid Fries medium (52) for 12 d in the dark. Liquid was collected from below the floating mat of mycelium, and 1-mL aliquots of undiluted or 1:10 dilutions of culture filtrate placed in 15-mL glass tubes. Replicate cut leaves of 1-wk-old susceptible (Fulgrain) and resistant (Red Rustproof) oats were placed in the filtrates. Leaves were monitored for wilting daily for 3 d.
Victorin Detection by MS.
WT strain FI3 and mutants were grown on CMX plates and transferred to liquid still culture as described above. Liquid cultures contained 50 mL of modified liquid Fries medium supplemented with 1.25 g of oats in a 250-mL Erlenmeyer flask and were incubated at 25 °C under a 16:8-h light:dark (wavelength 365-nm) cycle for 14 d. Approximately 1 g of Amberlite XAD-16 resin was added to the culture fluid, incubated for 1 h at room temperature and 160 rpm, filled into a column, washed with 5% acetonitrile, and extracted with three volumes of 1.5 mL of acetonitrile water mixtures (20:80, 40:60, and 60:40). Acetonitrile was evaporated from the collected fractions, and the remaining aqueous solution analyzed with MS.
In Vitro Oxidative Deamination of HV-Toxin M.
Roughly 3 µg of HV-toxin M was incubated with 1-mM phenylmethylsulfonyl fluoride and 1.5 mL of cell-free lysate from A. nidulans mycelium carrying either pYFAC-pyrG-vicK1/2 or pYFAC-pyrG for 14 h at 25 °C and 400 rpm. After centrifuging, the supernatant was passed through a Bond Elut C18 column (100 mg, Agilent, Part no.: 12,102,001). The column was washed with 5% acetonitrile and eluted with 2 mL of 75% acetonitrile. Acetonitrile was evaporated from the collected elution, and the remaining aqueous solution analyzed by LC–MS.
Supplementary Material
Acknowledgments
We thank Professor Thomas Wolpert (Oregon State University) for the victorin C standard. This work was supported by Australian Research Council (ARC) Discovery Projects (DP210102180 and DP200101880, to Y.-H.C. and P.S.S.). Y.-H.C. is supported by an ARC Future Fellowship (FT160100233). S.C.K. was supported by ARC and The University of Western Australia scholarships. C. victoriae strain FI3 was originally sequenced under JGI Proposal 336/300660 (awarded to B.G.T.). X.Z. was partially supported by the China Scholarship Council. M.C.M. was supported by the Australian National University, Grains and Research Development Corporation and NSW Department of Primary Industries co-investment (DAN00203).
Footnotes
This article is a PNAS Direct Submission.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2010573117/-/DCSupplemental.
Data Availability.
Nanopore reads used for assembly and genome sequence have been deposited in NCBI GenBank (accession no.: SRR10412887). The updated FI3 assembly at NCBI is accession no. AMCY00000000. The genome assembly, gene annotations transferred from the original FI3 assembly, transposon annotations, and manual vic gene annotations are available at GitHub, https://github.com/megancamilla/Cvictoriae_FI3_assembly_annotations. All other study data are included in the article and supporting information.
References
- 1.Meehan F., Murphy H. C., A new Helminthosporium blight of oats. Science 104, 413–414 (1946). [DOI] [PubMed] [Google Scholar]
- 2.Meehan F., Murphy H. C., Differential phytotoxicity of metabolic by-products of Helminthosporium victoriae. Science 106, 270–271 (1947). [DOI] [PubMed] [Google Scholar]
- 3.Wolpert T. J., Dunkle L. D., Ciuffetti L. M., Host-selective toxins and avirulence determinants: What’s in a name? Annu. Rev. Phytopathol. 40, 251–285 (2002). [DOI] [PubMed] [Google Scholar]
- 4.Petrov V., Qureshi M. K., Hille J., Gechev T., Occurrence, biochemistry and biological effects of host-selective plant mycotoxins. Food Chem. Toxicol. 112, 251–264 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Wolpert T. J., Lorang J. M., Victoria Blight, defense turned upside down. Physiol. Mol. Plant Pathol. 95, 8–13 (2016). [Google Scholar]
- 6.Mayama S., Bordin A. P. A., Morikawa T., Tanpo H., Kato H., Association of avenalumin accumulation with co-segregation of victorin sensitivity and crown rust resistance in oat lines carrying the Pc-2 gene. Physiol. Mol. Plant Pathol. 46, 263–274 (1995). [Google Scholar]
- 7.Kono Y., Kinoshita T., Takeuchi S., Daly J., Structure of amino acids isolated from hydrolyzed HV-toxin M, a host-specific toxin-related compound produced by Helminthosporium victoriae. Agric. Biol. Chem. 53, 505–511 (1989). [Google Scholar]
- 8.Wolpert T. J., Macko V., Acklin W., Jaun B., Arigoni D., Structure of minor host-selective toxins from Cochliobolus victoriae. Experientia 42, 1296–1299 (1986). [Google Scholar]
- 9.Macko V. et al., Characterization of victorin C, the major host-selective toxin from Cochliobolus victoriae: Structure of degradation products. Experientia 41, 1366–1370 (1985). [Google Scholar]
- 10.Wolpert T. J., Macko V., Acklin W., Arigoni D., Molecular features affecting the biological activity of the host-selective toxins from Cochliobolus victoriae. Plant Physiol. 88, 37–41 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lorang J. M., Sweat T. A., Wolpert T. J., Plant disease susceptibility conferred by a “resistance” gene. Proc. Natl. Acad. Sci. U.S.A. 104, 14861–14866 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lorang J. et al., Tricking the guard: Exploiting plant defense for disease susceptibility. Science 338, 659–662 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hammond-Kosack K. E., Rudd J. J., Plant resistance signalling hijacked by a necrotrophic fungal pathogen. Plant Signal. Behav. 3, 993–995 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang X., Lin M., Xu D., Lai D., Zhou L., Structural diversity and biological activities of fungal cyclic peptides, excluding cyclodipeptides. Molecules 22, 2069 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bills G. et al., New insights into the echinocandins and other fungal non-ribosomal peptides and peptaibiotics. Nat. Prod. Rep. 31, 1348–1375 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Arnison P. G. et al., Ribosomally synthesized and post-translationally modified peptide natural products: Overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 30, 108–160 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hallen H. E., Luo H., Scott-Craig J. S., Walton J. D., Gene family encoding the major toxins of lethal Amanita mushrooms. Proc. Natl. Acad. Sci. U.S.A. 104, 19097–19101 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Umemura M. et al., Characterization of the biosynthetic gene cluster for the ribosomally synthesized cyclic peptide ustiloxin B in Aspergillus flavus. Fungal Genet. Biol. 68, 23–30 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Umemura M. et al., MIDDAS-M: Motif-independent de novo detection of secondary metabolite gene clusters through the integration of genome sequencing and transcriptome data. PLoS One 8, e84028 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson R. D. et al., A novel family of cyclic oligopeptides derived from ribosomal peptide synthesis of an in planta-induced gene, gigA, in Epichloë endophytes of grasses. Fungal Genet. Biol. 85, 14–24 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Nagano N. et al., Class of cyclic ribosomal peptide synthetic genes in filamentous fungi. Fungal Genet. Biol. 86, 58–70 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Ding W. et al., Biosynthetic investigation of phomopsins reveals a widespread pathway for ribosomal natural products in Ascomycetes. Proc. Natl. Acad. Sci. U.S.A. 113, 3521–3526 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van der Velden N. S. et al., Autocatalytic backbone N-methylation in a family of ribosomal peptide natural products. Nat. Chem. Biol. 13, 833–835 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Ramm S. et al., A self-sacrificing N-methyltransferase is the precursor of the fungal natural product omphalotin. Angew. Chem. Int. Ed. Engl. 56, 9994–9997 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Condon B. J. et al., Comparative genome structure, secondary metabolite, and effector coding capacity across Cochliobolus pathogens. PLoS Genet. 9, e1003233 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zainudin N. A. I. M. et al., Virulence, host-selective toxin production, and development of three Cochliobolus phytopathogens lacking the Sfp-type 4 ’-phosphopantetheinyl transferase Ppt1. Mol. Plant Microbe Interact. 28, 1130–1141 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Kessler S. C., et al. , SRR10412887: Fastq file containing raw unprocessed Oxford Nanopore Reads for Cochliobolus victoriae FI3. NCBI SRA. https://www.ncbi.nlm.nih.gov/sra/SRR10412887. Deposited 18 January 2020.
- 28.Kessler S. C., et al. , AMCY00000000.1: Genome assembly for Cochliobolus victoriae FI3 constructed with Nanopore reads. NCBI GenBank. https://www.ncbi.nlm.nih.gov/nuccore/578057235. Deposited 22 April 2020.
- 29.Vogt E., Künzler M., Discovery of novel fungal RiPP biosynthetic pathways and their application for the development of peptide therapeutics. Appl. Microbiol. Biotechnol. 103, 5567–5581 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Ye Y. et al., Heterologous production of asperipin-2a: Proposal for sequential oxidative macrocyclization by a fungi-specific DUF3328 oxidase. Org. Biomol. Chem. 17, 39–43 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Cacho R. A., Tang Y., Chooi Y. H., Next-generation sequencing approach for connecting secondary metabolites to biosynthetic gene clusters in fungi. Front. Microbiol. 5, 774 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Condon B. J. et al., Clues to an evolutionary mystery: The genes for T-toxin, enabler of the devastating 1970 Southern corn leaf blight epidemic, are present in ancestral species, suggesting an ancient origin. Mol. Plant Microbe Interact. 31, 1154–1165 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Walton J. D., Host-selective toxins: Agents of compatibility. Plant Cell 8, 1723–1733 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu J., Li H., Chooi Y. H., Fungal dirigent protein controls the stereoselectivity of multicopper oxidase-catalyzed phenol coupling in viriditoxin biosynthesis. J. Am. Chem. Soc. 141, 8068–8072 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Li H. et al., Biosynthesis of a tricyclo[6.2.2.0(2,7)]dodecane system by a berberine bridge enzyme-like aldolase. Chemistry 25, 15062–15066 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Ahn J. H., Cheng Y. Q., Walton J. D., An extended physical map of the TOX2 locus of Cochliobolus carbonum required for biosynthesis of HC-toxin. Fungal Genet. Biol. 35, 31–38 (2002). [DOI] [PubMed] [Google Scholar]
- 37.Inderbitzin P., Asvarak T., Turgeon B. G., Six new genes required for production of T-toxin, a polyketide determinant of high virulence of Cochliobolus heterostrophus to maize. Mol. Plant Microbe Interact. 23, 458–472 (2010). [DOI] [PubMed] [Google Scholar]
- 38.Dong S., Raffaele S., Kamoun S., The two-speed genomes of filamentous pathogens: Waltz with plants. Curr. Opin. Genet. Dev. 35, 57–65 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Gu W., Sardar D., Pierce E., Schmidt E. W., Roads to Rome: Role of multiple cassettes in cyanobactin RiPP biosynthesis. J. Am. Chem. Soc. 140, 16213–16221 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ye Y. et al., Unveiling the biosynthetic pathway of the ribosomally synthesized and post-translationally modified peptide ustiloxin B in filamentous fungi. Angew. Chem. Int. Ed. Engl. 55, 8072–8075 (2016). [DOI] [PubMed] [Google Scholar]
- 41.Driggers E. M., Hale S. P., Lee J., Terrett N. K., The exploration of macrocycles for drug discovery–An underexploited structural class. Nat. Rev. Drug Discov. 7, 608–624 (2008). [DOI] [PubMed] [Google Scholar]
- 42.Tianero M. D. et al., Metabolic model for diversity-generating biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 113, 1772–1777 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brazeau B. J., Johnson B. J., Wilmot C. M., Copper-containing amine oxidases. Biogenesis and catalysis; a structural perspective. Arch. Biochem. Biophys. 428, 22–31 (2004). [DOI] [PubMed] [Google Scholar]
- 44.Kang X. et al., Genomic characterization provides new insights into the biosynthesis of the secondary metabolite huperzine a in the endophyte Colletotrichum gloeosporioides Cg01. Front. Microbiol. 9, 3237 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Neumann C. S., Fujimori D. G., Walsh C. T., Halogenation strategies in natural product biosynthesis. Chem. Biol. 15, 99–109 (2008). [DOI] [PubMed] [Google Scholar]
- 46.Galonić D. P., Vaillancourt F. H., Walsh C. T., Halogenation of unactivated carbon centers in natural product biosynthesis: Trichlorination of leucine during barbamide biosynthesis. J. Am. Chem. Soc. 128, 3900–3901 (2006). [DOI] [PubMed] [Google Scholar]
- 47.Friesen T. L., Faris J. D., Solomon P. S., Oliver R. P., Host-specific toxins: Effectors of necrotrophic pathogenicity. Cell. Microbiol. 10, 1421–1428 (2008). [DOI] [PubMed] [Google Scholar]
- 48.Zhang J., Li X., Han X., Liu R., Fang J., Targeting the thioredoxin system for cancer therapy. Trends Pharmacol. Sci. 38, 794–808 (2017). [DOI] [PubMed] [Google Scholar]
- 49.Schiestl R. H., Petes T. D., Integration of DNA fragments by illegitimate recombination in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 88, 7585–7589 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leach J., Lang B. R., Yoder O. C., Methods for selection of mutants and in vitro culture of Cochliobolus heterostrophus. J. Gen. Microbiol. 128, 1719–1729 (1982). [Google Scholar]
- 51.Tzeng T. H., Lyngholm L. K., Ford C. F., Bronson C. R., A restriction fragment length polymorphism map and electrophoretic karyotype of the fungal maize pathogen Cochliobolus heterostrophus. Genetics 130, 81–96 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pringle R. B., Scheffer R. P., Purification of selective toxin of Periconia circinata. Phytopathology 53, 785–787 (1963). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Nanopore reads used for assembly and genome sequence have been deposited in NCBI GenBank (accession no.: SRR10412887). The updated FI3 assembly at NCBI is accession no. AMCY00000000. The genome assembly, gene annotations transferred from the original FI3 assembly, transposon annotations, and manual vic gene annotations are available at GitHub, https://github.com/megancamilla/Cvictoriae_FI3_assembly_annotations. All other study data are included in the article and supporting information.