Significance
Current leukemia treatments focus on targeting leukemic cells and neglect the influence of the malignant BM environment, which is modified to nurture and protect LSCs. By abrogating K3-mediated integrin adhesion of LSCs to BM niches, either through genetics or an LSC-specific RNA aptamer-mediated K3-siRNA delivery, we impaired integrin-mediated niche retention of LSCs, slowed down the course of leukemia in vivo, and prolonged the survival of mice suffering from CML. Therefore, delivering K3-depleting compounds via tumor-specific surface receptors represents a strategy to abolish the function of multiple integrin classes on LSCs and interactions with numerous niche components at once.
Keywords: Kindlin-3, integrin adhesion, BM niche, microenvironment, leukemic stem cells
Abstract
Kindlin-3 (K3)–mediated integrin adhesion controls homing and bone marrow (BM) retention of normal hematopoietic cells. However, the role of K3 in leukemic stem cell (LSC) retention and growth in the remodeled tumor-promoting BM is unclear. We report that loss of K3 in a mouse model of chronic myeloid leukemia (CML) triggers the release of LSCs from the BM into the circulation and impairs their retention, proliferation, and survival in secondary organs, which curbs CML development, progression, and metastatic dissemination. We found de novo expression of cytotoxic T lymphocyte-associated antigen 4 (CTLA-4) on CML-LSCs but not normal hematopoietic stem cells and this enabled us to specifically deplete K3 with a CTLA-4–binding RNA aptamer linked to a K3-siRNA (small interfering RNA) in CTLA-4+ LSCs in vivo, which mobilized LSCs in the BM, induced disease remission, and prolonged survival of mice with CML. Thus, disrupting interactions of LSCs with the BM environment is a promising strategy to halt the disease-inducing and relapse potential of LSCs.
Chronic myeloid leukemia (CML) is a clonal, hematopoietic malignancy of the myeloid lineage caused by the transformation of a hematopoietic stem cell (HSC) into a disease-initiating leukemic stem cell (LSC). The course of CML typically progresses from a chronic phase (CP-CML) characterized by the accumulation of neutrophils and LSCs in the bone marrow (BM) and peripheral blood (PB) and the remodeling of the BM microenvironment into a malignant BM niche to a fatal blast crisis (BC-CML) characterized by the rapid expansion of differentiation-arrested myeloid or lymphoid precursor cells (1–8). In almost all cases of CML, the malignant transformation results from a characteristic reciprocal translocation between chromosomes 9 and 22 in HSCs, which generates the breakpoint cluster region–Abelson (BCR-ABL) fusion gene. Depending on the BCR breaking point, the chimeric gene is translated into the p190, p210 (most common), or p230 BCR-ABL oncoprotein (9, 10). The underlying mechanism(s) for the CP-CML–to–BC-CML transition is not known. It is surmised, however, that secondary genetic and/or epigenetic changes in leukemic cells lead to the formation and expansion of leukemic clones with increasingly malignant characteristics (2, 11).
CML is treated with ABL tyrosine kinase inhibitors (TKIs) such as imatinib, which block proliferation of leukemic cells and prevent the accumulation of neutrophils in the PB and BM (12) and the onset of a BC-CML (13–16). Although treatment of CML with TKIs often leads to complete remission and normal life expectancy, a drawback of the inhibitor treatment is the poor effect on a subpopulation of Bcr-Abl–positive self-renewing LSCs protected within the leukemic BM niche (4, 17–20), resulting in a lifelong treatment requirement. Moreover, around 16% of imatinib-treated patients develop an intolerance against ABL TKIs and around 10% acquire mutations in the BCR-ABL fusion gene, resulting in therapy resistance (16).
Hematopoietic stem and progenitor cells (HSPCs) receive biochemical and biophysical cues from the BM microenvironment that promote retention, self-renewal, regeneration, and differentiation (3, 4). The positioning and retention of HSPCs within the BM critically depend on integrin-mediated adhesion to extracellular matrix (ECM) proteins such as osteopontin, fibronectin (FN), collagens, laminins, and cell surface-presented ligands such as vascular cell adhesion protein 1 (VCAM-1) or intercellular adhesion molecule 1 (ICAM-1) (21–27). We have shown that both in mice and humans, BM retention and survival of HSPCs depend on Kindlin-3 (K3) (28), which is expressed in all hematopoietic cells (29). K3 is an essential integrin-binding and -activating adaptor protein (30), whose loss leads to dysfunctional erythrocytes, platelets, and immune effector cells collectively termed leukocyte adhesion deficiency type III (LAD-III) in humans (31–36).
Since LSCs remodel the cellular and ECM composition of BM niches, which in turn further fuel LSC malignancy (3–8), we investigated in a murine CML model if and how K3 loss influences the association of LSCs with the BM microenvironment and the lifespan of leukemic mice, and whether K3 depletion can be utilized as a novel treatment strategy for CML. Our findings on the involvement of K3 in CML induction, development, and intervention are discussed.
Results
Kindlin-3 Retains LSCs in the BM and Promotes CML Development.
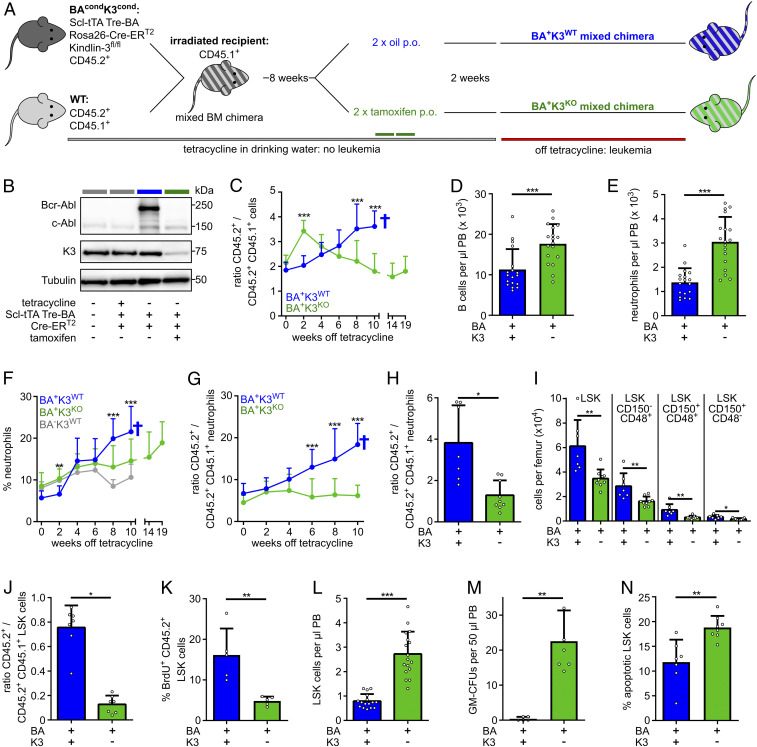
To test the function of K3 for LSCs, we established a mouse strain in which the expression of the Bcr-Abl oncogene can be conditionally induced and the floxed Fermt3 gene can be conditionally disrupted in HSPCs (28, 37). To obtain this mouse strain, we intercrossed mice carrying the following transgenes: the stem cell leukemia-driven tetracycline-controlled transactivator (Scl-tTA), a tetracycline-responsive element-driven Bcr-Abl (Tre-Bcr-Abl), a tamoxifen-regulated and ubiquitously expressed Cre (Rosa26-Cre-ERT2), and a floxed Fermt3 (K3fl/fl) on a C57BL/6 CD45.2+ background (termed BAcondK3cond). To avoid anemia, dysfunctional effector cells, and impaired BM retention of HSPCs in Fermt3-null mice (28, 31–33, 38), we generated mixed BM chimeras by transplanting CD45.2+ BAcondK3cond BM cells from tetracycline-treated donor animals and wild-type (WT) BM cells from CD45.2+CD45.1+ F1 donor animals in a 1:1 ratio into lethally irradiated WT congenic CD45.1+ C57BL/6.SJL recipient mice. The resulting mixed BM chimeras were further treated with tetracycline to suppress Bcr-Abl expression in BAcondK3cond (CD45.2+) cell populations. Once the mixed hematopoietic system was successfully established 8 wk post transplantation, we administered tamoxifen to knockout the Fermt3 gene (K3KO), or oil to retain expression of the floxed K3 gene (K3WT), followed by the withdrawal of tetracycline and the induction of Bcr-Abl expression 2 wk later. Using this strategy, we finally obtained mixed BM chimeric animals containing WT together with either BA+K3WT or BA+K3KO hematopoietic cells (Fig. 1A). To indicate that the BA+K3WT (CD45.2+) and the BA+K3KO (CD45.2+) chimeras were competitively transplanted with WT BM (CD45.2+CD45.1+) cells, we termed them BA+K3WT and BA+K3KO mixed chimeras, respectively. Western blots verified the functionality of our transgenes and confirmed K3 protein loss and induction of Bcr-Abl expression in CD45.2+ splenocytes from mixed chimeras upon tamoxifen treatment and withdrawal of tetracycline. CD45.2+ splenocytes isolated from BA+K3KO chimeras did not express Bcr-Abl oncoprotein, suggesting that they originate from nontransformed HSCs (Fig. 1B; see also below Fig. 5).
Fig. 1.
K3 loss mobilizes LSCs. (A) BA+K3WT and BA+K3KO mixed chimeras generated by transferring BM cells carrying a conditional, HSPC-inducible, and tetracycline-controlled Bcr-Abl (BA) transgene and a conditional, tamoxifen-inducible, Fermt3-null allele wild type mixed with WT BM cells into lethally irradiated recipient mice. The congenic CD45.1 and CD45.2 surface receptors are used to distinguish transgenic, WT, and host cells. Tamoxifen or oil were administered orally (p.o.). (B) Bcr-Abl, c-Abl, K3, and tubulin protein expression in CD45.2+ splenocytes. (C) Ratio between transgenic (CD45.2+) and WT (CD45.2+CD45.1+) leukocytes in the PB of BA+K3WT (n ≥ 8) and BA+K3KO (n ≥ 11) mixed chimeras. Statistics by multiple t test, two-sided. (D and E) Numbers of B220+ B cellc (D) and Gr1high/Mac1+ neutrophils (E) per microliter of PB determined 2 wk after leukemia induction (n = 18). Statistics by Mann–Whitney U test (D) and unpaired t test, two-sided (E). (F and G) Percentage of PB neutrophils (F) and ratio (G) between transgenic (CD45.2+) and WT (CD45.2+CD45.1+) PB neutrophils in BA+K3WT (n ≥ 8), BA+K3KO (n ≥ 11), and BA−K3WT (n = 7) chimeras at the indicated time points after tetracycline withdrawal. Statistics by multiple t test, two-sided. (H) Ratio between transgenic (CD45.2+) and WT (CD45.2+CD45.1+) BM neutrophils in BA+K3WT and BA+K3KO mixed chimeras 6 wk after tetracycline withdrawal (n ≥ 7). Statistics by unpaired t test, two-sided. (I) Numbers of LSK, LSK CD150−CD48+, LSK CD150+CD48+, and LSK CD150+CD48− cells per femur in BA+K3WT and BA+K3KO mixed chimeras 6 wk off tetracycline (n ≥ 7). Statistics by unpaired t test, two-sided. (J) Ratio between transgenic (CD45.2+) and WT (CD45.2+CD45.1+) LSK cells in the BM of BA+K3WT and BA+K3KO mixed chimeras 6 wk after tetracycline withdrawal (n ≥ 7). Statistics by unpaired t test, two-sided. (K) Percentage of BrdU+ (bromodeoxyuridine-positive) transgenic (CD45.2+) LSK cells in the BM of BA+K3WT and BA+K3KO mixed chimeras 6 wk off tetracycline (n = 5). Statistics by unpaired t test, two-sided. (L) Number of LSK cells per microliter of PB in BA+K3WT and BA+K3KO mixed chimeras 2 wk after tetracycline withdrawal (n ≥ 16). Statistics by unpaired t test, two-sided. (M) Frequency of GM-CFUs in 50 µL PB of BA+K3WT and BA+K3KO mixed chimeras 2 wk off tetracycline after 13 d of culture (n ≥ 5). Statistics by Mann–Whitney U test, two-sided. (N) Percentage of apoptotic Annexin-V+ PB LSK cells in BA+K3WT and BA+K3KO mixed chimeras 2 wk after tetracycline withdrawal (n ≥ 7). Statistics by unpaired t test, two-sided. Data are shown as mean ± SD. *P < 0.05, **P < 0.01, and ***P < 0.001.
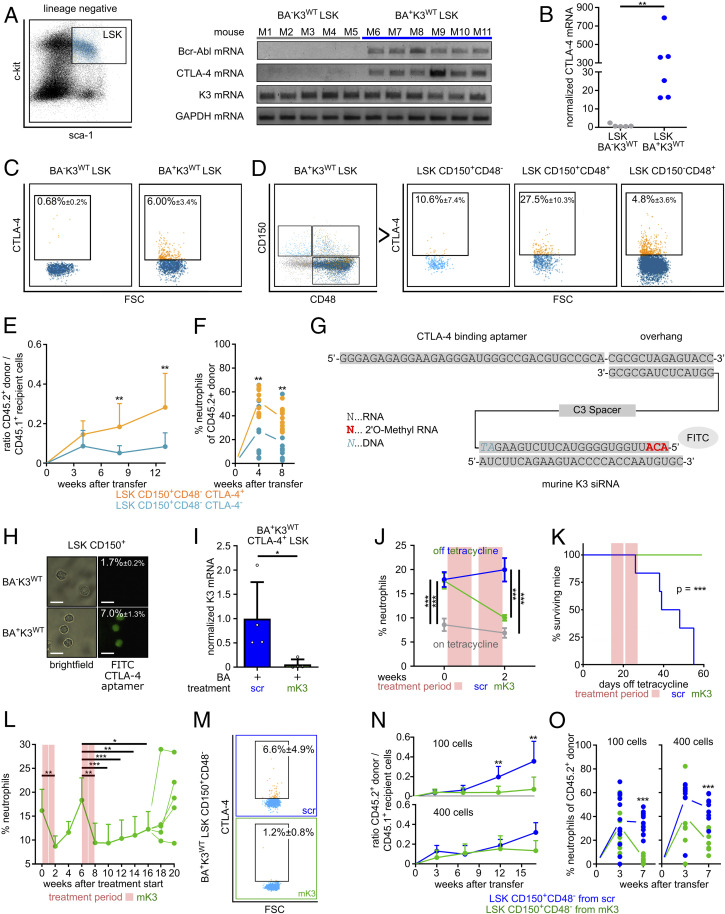
Fig. 5.
Depletion of K3 in LSCs with a CTLA-4 RNA aptamer-tagged K3-siRNA. (A) Bcr-Abl, CTLA-4, K3, and GAPDH mRNA levels in LSK cells isolated from the BM of individual wild-type (BA−K3WT) or leukemic (BA+K3WT) mice. (B) CTLA-4 mRNA levels measured by qPCR. Statistics by Mann–Whitney U test, two-sided. (C) CTLA-4 surface expression on LSK cells isolated from the BM of BA−K3WT or BA+K3WT mice. (D) CTLA-4 surface expression on HSPC subpopulations isolated from the BM of BA+K3WT mice. (E and F) Ratio between PB CD45.2+ donor and CD45.1+ host cells (E) and percentage of PB neutrophils of CD45.2+ donor cells (F) in sublethally irradiated CD45.1+ WT recipients transplanted with 100 CD45.2+ LSK CD150+CD48− CTLA4+ or 100 CD45.2+ LSK CD150+CD48− CTLA-4− cells isolated from BA+K3WT mice (n ≥ 9). Statistics by two-way ANOVA, Sidak’s multiple-comparison test (E), and multiple t test, two-sided (F). (G) Sequence of the CTLA-4–targeting RNA aptamer linked to stabilized mK3-siRNA labeled with fluorescein isothiocyanate (FITC). (H) Bright-field images of BM-derived LSK CD150+ cells from BA−K3WT or BA+K3WT mice stained with FITC-labeled CTLA-4 aptamer. (Scale bars, 10 µm.) Percentages indicate the amount of FITC+ LSK CD150+ cells determined by flow cytometry (n ≥ 5). (I) K3 mRNA levels of CTLA-4+ LSK cells isolated from CTLA-4 aptamer-tagged scrambled control (scr)- or mK3-siRNA–treated BA+K3WT mice (n = 4). Statistics by Mann–Whitney U test, two-sided. (J) PB neutrophils of BA+K3WT mice before and after treatment with CTLA-4 aptamer-tagged scrambled control- or mK3-siRNA (n ≥ 5). Statistics by two-way ANOVA, Sidak’s multiple-comparison test. Data are shown as mean ± SEM. (K) Kaplan–Meier survival curve of BA+K3WT mice treated with CTLA-4 aptamer-tagged scrambled control- or mK3-siRNA (n ≥ 5). Statistics by log-rank test. (L) PB neutrophil numbers of CTLA-4 aptamer-tagged scrambled control- or mK3-siRNA–treated BA+K3WT mice suffering from a CML relapse (n = 5). Statistics by unpaired t test, two-sided. (M) CTLA-4 surface expression on LSK CD150+CD48− cells isolated from the BM of BA+K3WT mice treated with CTLA-4 aptamer tagged-scrambled control- or mK3-siRNA (n ≥ 3). (N and O) Ratio between PB CD45.2+ donor and CD45.1+ host cells (N) and percentage of PB neutrophils of CD45.2+ donor cells (O) in sublethally irradiated CD45.1+ WT recipients receiving 100 or 400 CD45.2+ LSK CD150+CD48− cells derived from CTLA-4 aptamer-tagged scrambled control- or mK3-siRNA–treated BA+K3WT mice (n ≥ 8). Statistics by two-way ANOVA, Sidak’s multiple-comparison test (N), and multiple t test, two-sided (O). Data are shown as mean ± SD if not indicated otherwise. *P < 0.05, **P < 0.01, and ***P < 0.001.
Next, we monitored the disease onset by determining the ratio of BA+K3WT or BA+K3KO (CD45.2+) and WT (CD45.2+/CD45.1+) leukocytes in 2-wk intervals in the PB using flow cytometry. Although BAcondK3cond (CD45.2+) and WT (CD45.2+CD45.1+) BM cells were transplanted in a 1:1 ratio, we observed a 2:1 ratio of CD45.2+:CD45.2+CD45.1+ cells 10 wk after transplantation (time point 0) in BA+K3WT and BA+K3KO mixed chimeras. In BA+K3WT mixed chimeras the percentage of CD45.2+ BA+K3WT cells steadily increased and the CD45.2+CD45.1+ WT cells decreased in the PB, indicating that normal hematopoiesis is outcompeted by malignant hematopoiesis, which together with the death of the mice is a typical hallmark of CML progression (Fig. 1C and SI Appendix, Fig. S1A). In BA+K3KO mixed chimeras, the percentage of CD45.2+ BA+K3KO cells transiently increased in the PB 2 wk after tetracycline withdrawal (Fig. 1C), which was due to an elevated release of BA+K3KO B220+ B cells and Gr1hi/Mac1+ neutrophils, but not T cells, into the PB (Fig. 1 D and E and SI Appendix, Fig. S1 B and C). The BA+K3KO cell numbers began to decrease at around 4 wk after tetracycline withdrawal and reached a plateau 8 to 10 wk later (Fig. 1C), while CD45.2+CD45.1+ WT cell numbers rose and reached normal levels 10 wk after tetracycline withdrawal (SI Appendix, Fig. S1A). The CD45.2+ Gr1hi/Mac1+ neutrophils also rose 6 wk post tetracycline withdrawal in BA+K3WT mixed chimeras, whereas CD45.2+ Gr1hi/Mac1+ neutrophils remained at normal levels for more than 14 wk in BA+K3KO mixed chimeras and began slowly rising ∼19 wk after tetracycline withdrawal (Fig. 1 F and G).
The significantly lower numbers of CD45.2+ Gr1hi/Mac1+ neutrophils in the BM of BA+K3KO mixed chimeras (Fig. 1H) were associated with a reduction of lineage−Sca-1+c-kit+ (LSK) cells. To define the LSK subpopulation(s) affected by K3 loss, we used the SLAM family markers CD150 and CD48 to define which LSK subpopulations were affected by K3 loss. Flow cytometry revealed that the LSK CD150+CD48− population highly enriched with HSCs/LSCs (7, 39) and LSK CD150+CD48+ and LSK CD150−CD48+ populations enriched with hematopoietic progenitor cells (HPCs) (39) were significantly reduced in the BM of BA+K3KO mixed chimeras (Fig. 1I). The reduction of the different LSK populations in the BM of BA+K3KO mixed chimeras resulted in a decline of the CD45.2+ BA+K3KO LSK fraction (Fig. 1J), which was associated with a diminished proliferation (Fig. 1K) and release into the PB as early as 2 wk after tetracycline withdrawal (Fig. 1L). In line with the CD45.2+ BA+K3KO LSK cell release into the PB, PB samples from BA+K3KO mixed chimeras showed an elevated colony formation potential in colony-forming unit-granulocyte, monocyte (CFU-GM) assays (Fig. 1M). Short-term homing assays indicated that the BM entry of CD45.2+ BA+K3KO LSK CD150+CD48− cells was reduced (SI Appendix, Fig. S1D), whereas spleen infiltration was unaffected (SI Appendix, Fig. S1E). Interestingly, however, CD45.2+ BA+K3KO LSK CD150+CD48− cells did not expand in spleens of mixed BM chimeras for up to 6 wk post leukemia induction (SI Appendix, Fig. S1F). In contrast to the pronounced defects upon K3 loss, BA+K3WT mixed chimeras contained similar numbers of highly proliferative CD45.2+ BA+K3WT and CD45.2+CD45.1+ WT LSK cells in the BM and exhibited low LSK cell numbers in the PB, rare colony formation potential in CFU-GM assays, high BM homing rates, and increased spleen cellularity (Fig. 1 J–M and SI Appendix, Fig. S1 D–F). Furthermore, the few circulating CD45.2+ BA+K3WT LSK cells were prone to apoptosis, which was more pronounced in CD45.2+ BA+K3KO LSK cells (Fig. 1N). These findings indicate that K3 is essential for LSC retention in protective BM niches as well as secondary organs such as the spleen.
Kindlin-3 Loss Reduces CML Mortality.
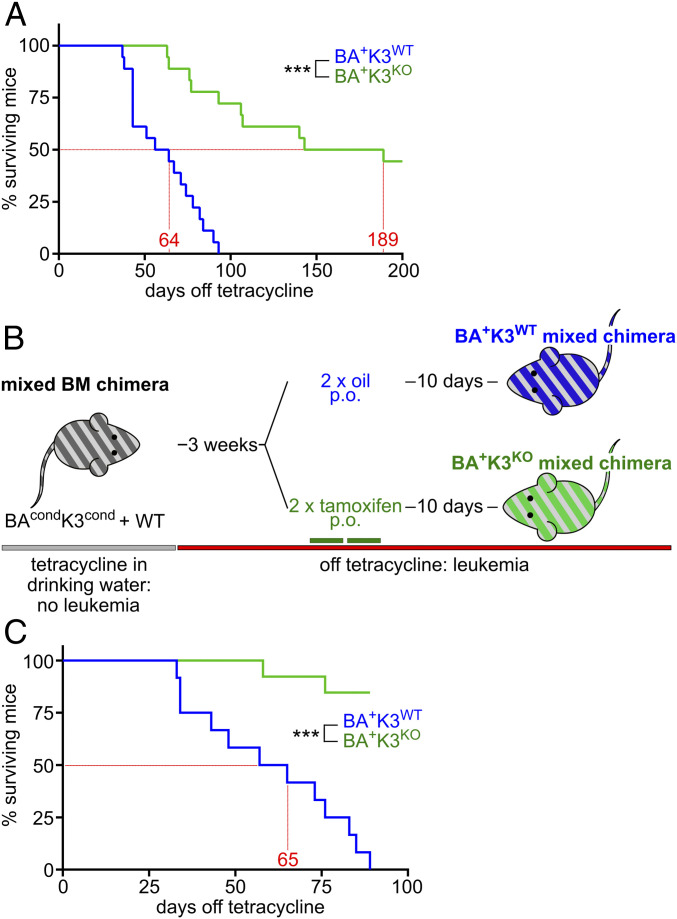
Based on our findings, K3 loss should have a distinct impact on the survival of BA+K3KO mixed chimeras. Indeed, Kaplan–Meier survival analysis revealed a profound lifespan extension of BA+K3KO mixed chimeras, which survived on average 189 d as compared with 64 d of BA+K3WT mixed chimeras (Fig. 2A). Moribund BA+K3WT mixed chimeras suffered from several characteristic end-stage hematological abnormalities (16) including anemia, hemorrhage, and leukemic cell infiltration in the lung and liver, while BA+K3KO mixed chimeras developed none of the symptoms, indicating that K3 enables CML development. Despite efficient Cre-mediated Fermt3 deletion, a few BA+K3KO mixed chimeras showed a shortened lifespan, which was not due to CML but rather caused by the poorer engraftment efficiency of CD45.2+CD45.1+ WT cells.
Fig. 2.
Deletion of Fermt3 prolongs lifespan. (A) Kaplan–Meier survival curve of BA+K3WT and BA+K3KO mixed chimeras treated orally (p.o.) with tamoxifen before tetracycline withdrawal (n = 18). (B) BAcondK3cond mixed chimeras first withdrawn from tetracycline (BA+) followed by tamoxifen (K3KO) or oil (K3WT) treatment. (C) Kaplan–Meier survival curve of BA+K3WT and BA+K3KO mixed chimeras treated with tamoxifen after tetracycline withdrawal (n ≥ 12). Statistics by log-rank test. ***P < 0.001.
To investigate whether loss of K3 is also able to stop or retard full-blown CML, we first withdrew tetracycline to generate BA+K3cond mixed chimeras and upon CML development (>20% PB neutrophils; termed leukemic mice) administered tamoxifen to delete the Fermt3 gene. Control cohorts received oil to retain K3 expression (Fig. 2B). Whereas oil-treated BA+K3WT mice had enlarged spleens, leukemic cell infiltrations into the spleen, and survived on average 65 d after tetracycline withdrawal, more than 75% of the tamoxifen-treated BA+K3KO mixed chimeras were still alive with normal spleen size and cellularity at 110 d post tetracycline withdrawal (Fig. 2C). These findings strongly suggest that loss of K3-mediated integrin adhesion delays CML development as well as progression in vivo.
Loss of Kindlin-3 Attenuates the Blast Crisis Induced by Bcr-Abl+ Myeloblasts.
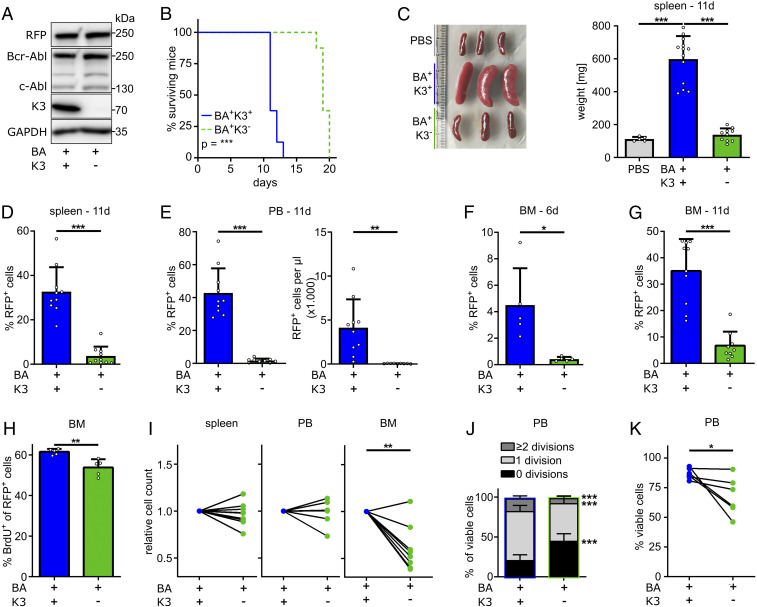
The profound expansion of mature myeloid cells and leukemic organ infiltration results in the death of Scl-tTA/Tre-Bcr-Abl mice before the onset of a myeloid blast crisis (7, 37). Therefore, we used our recently established 32D blast cell crisis model (40) to investigate whether K3 expression also influences the course of a blast-like CML in vivo. To this end, we generated red fluorescent protein (RFP)-tagged Bcr-Abl–expressing murine 32D myeloblast cells (BA+K3+), produced BA+K3− cells by disrupting the K3-encoding Fermt3 gene using CRISPR-Cas9 technology (Fig. 3A), injected RFP+ BA+K3+ or RFP+ BA+K3− cells into nonirradiated syngeneic C3H/HeOuJ mice, and monitored their survival. BA+K3+ cell-injected recipient mice died ∼12 d after injection, whereas BA+K3− cell-treated mice survived ∼20 d (Fig. 3B). At 11 d post injection, spleen size and weight in BA+K3+ cell-injected animals were increased and the splenic architecture was severely damaged, which was neither observed in BA+K3− cell- nor phosphate-buffered saline (PBS)-injected control mice (Fig. 3C and SI Appendix, Fig. S2A). Blast cell infiltration and expansion in the spleen, PB, and BM were determined by quantifying RFP+ BA+K3+ and BA+K3− cell numbers 6 and 11 d post injection. In line with normal spleen size, RFP+ tumor cells were barely detectable in the spleen and PB of BA+K3− cell-injected animals. The percentage of RFP+ cells was ∼40% in the PB of BA+K3+ cell-injected mice, which corresponds to an 83-fold higher absolute tumor load in BA+K3+ compared with BA+K3− cell-injected animals (Fig. 3 D and E). RFP+ BA+K3+ cells were readily detectable in the BM 6 d after injection and further increased 11 d post injection (Fig. 3 F and G). In contrast, BA+K3− cells were barely detected in the BM 6 d post injection (Fig. 3F) and increased 11 d after injection to a level that was fivefold lower compared with BA+K3+ cell-injected animals (Fig. 3G).
Fig. 3.
K3 promotes CML blast crisis. (A) Protein expression of RFP, Bcr-Abl, c-Abl, K3, and GAPDH by Bcr-Abl–expressing (BA+) 32D myeloblasts with or without a Fermt3 gene deletion (K3+ or K3−). (B) Kaplan–Meier survival curve of syngeneic C3H/HeOuJ mice transplanted with either RFP+ BA+K3+ or RFP+ BA+K3− cells (n = 8). Statistics by log-rank test. (C–G) Engraftment parameters including spleen weights 11 d after transplantation (n ≥ 5) (C), RFP+ cells in spleen (D), PB (E), and BM (F and G) at 11 d (D–F; n ≥ 9) or 6 d (G; n = 5) after transplantation. Statistics by one-way ANOVA, Tukey’s multiple-comparison test (C), or unpaired t test, two-sided (D–G). (H) Percentage of BrdU+ 32D cells in the BM at day 11 after transplantation (n = 5). Statistics by unpaired t test, two-sided. (I) Relative homing rates of CFSE- or CTV-labeled BA+K3+ or BA+K3− 32D cells in the spleen, PB, and BM 3 h after intracardial injection (n ≥ 7). Statistics by paired t test, two-sided. (J and K) PB 32D (J) cell divisions and (K) cell viability 3 h post injection (n ≥ 5). Statistics by paired t test, two-sided. Data are shown as mean ± SD. *P < 0.05, **P < 0.01, and ***P < 0.001.
Despite the low numbers, BA+K3− cell proliferation was only slightly reduced in the BM (Fig. 3H). To investigate whether impaired homing contributes to the low BA+K3− cell numbers in the BM, we mixed equal numbers of carboxyfluorescein succinimidyl ester (CFSE)-labeled BA+K3− and CellTrace violet (CTV)-labeled BA+K3+ cells or vice versa, injected them intracardially into syngeneic, nonirradiated C3H/HeOuJ mice, and quantified dye+ cells in the BM, PB, and spleen 3 h post injection. The experiments revealed significantly fewer BA+K3− cells in the BM but similar numbers of BA+K3+ and BA+K3− cells in the PB and spleen (Fig. 3I). In line with the decreased proliferation of BA+K3− cells in the BM (Fig. 3H), they also underwent fewer cell divisions in the PB (Fig. 3J) and were more prone to apoptosis (Fig. 3K). Of note, cell-cycle progression, proliferation, and viability of in vitro cultured BA+K3− and BA+K3+ cells were alike, indicating that K3 modulates 32D cell proliferation and survival in the in vivo environment but not when propagated in vitro (SI Appendix, Fig. S2 B–D).
Bcr-Abl Amplifies Integrin Activity in a K3-Dependent Manner.
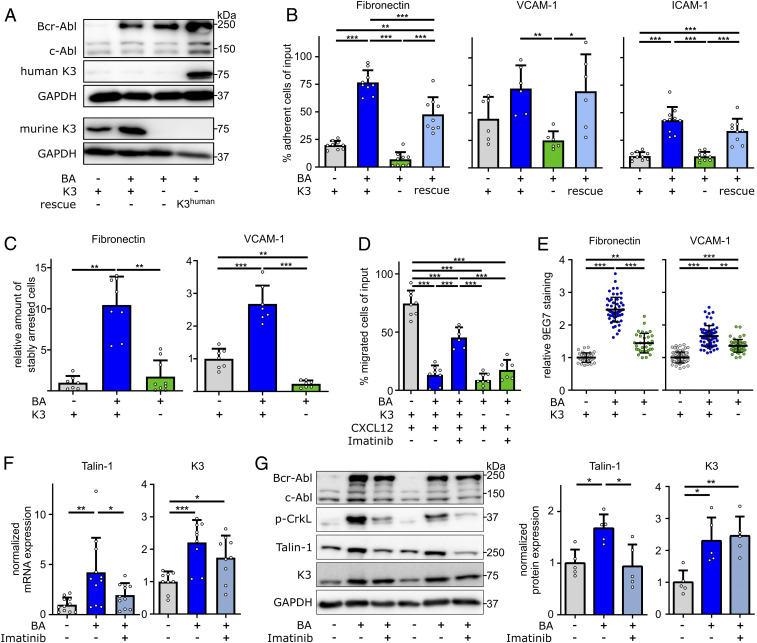
To investigate how K3 promotes the life-shortening blast phase of Bcr-Abl–expressing 32D cells, we compared adhesion and migration of parental Bcr-Abl–negative (BA−K3+), BA+K3+, and BA+K3− 32D cells (Fig. 4A). In plate and wash assays, BA+K3+ cells adhered significantly more strongly to FN, VCAM-1, and ICAM-1 than parental BA−K3+ cells. Similarly, as reported for erythroleukemia-type K562 cells (41), the enforced adhesion caused by Bcr-Abl was abolished upon loss of K3 expression in BA+K3− cells (Fig. 4B) and regained upon reexpression of the human K3-encoding complementary DNA (BA+K3rescue; Fig. 4 A and B). BA+K3+ cells also resisted shear forces of 2.5 dyn/cm2 significantly better than BA−K3+ and BA+K3− cells and arrested on FN and VCAM-1 in an α5- and α4-integrin–dependent manner (Fig. 4C and SI Appendix, Fig. S3A). The strong integrin-mediated adhesion of BA+K3+ cells as well as the absent adhesion of BA+K3− cells curbed migration toward a CXCL12 chemokine gradient to background level (Fig. 4D). Bcr-Abl kinase inhibition with imatinib partially restored the chemokine-induced migration of BA+K3+, suggesting that the Bcr-Abl kinase activity strengthens adhesion and diminishes migration (Fig. 4D).
Fig. 4.
Bcr-Abl elevates integrin-mediated cell adhesion in a K3-dependent manner. (A) Protein expression of Bcr-Abl, c-Abl, murine K3, human K3 (rescue), and GAPDH by 32D myeloblasts with or without Bcr-Abl expression (BA+ or BA−), with or without Fermt3 gene deletion (K3+ or K3−), and with or without human K3 reexpression (rescue). (B) 32D cell adhesion to FN (n = 9), VCAM-1 (n = 6), or ICAM-1 (n ≥ 9) under static conditions. (C) 32D cells stably adhering to FN or VCAM-1 under flow (n ≥ 7). (D) 32D cell chemotaxis toward CXCL12 with or without preincubation with imatinib overnight (n ≥ 6). (E) 9EG7 staining intensities in 32D cells plated on FN (n ≥ 33) or VCAM-1 (n ≥ 49). Statistics by Kruskal–Wallis test. (F and G) K3 and Talin-1 mRNA (F; n ≥ 9) and Bcr-Abl, c-Abl, p-CrkL, K3, Talin-1, and GAPDH protein (G; n = 5) expression in 32D cells with or without 24-h imatinib treatment. Data are shown as mean ± SD. Statistics by one-way ANOVA, Tukey’s multiple-comparison test if not indicated otherwise. *P < 0.05, **P < 0.01, and ***P < 0.001.
The enhanced adhesion of BA+K3+ to FN and VCAM-1 was not due to an altered integrin cell-surface profile, which was comparable to BA+K3− cells and only slightly changed when compared with BA−K3+ cells (SI Appendix, Fig. S3B). However, levels of β1-integrin activation-associated 9EG7 epitope were elevated on BA+K3+ cells and reduced on BA+K3− cells (Fig. 4E). Integrin activation requires binding of the adaptor proteins Talin-1 and K3 to the cytoplasmic domain of β-integrins (30), which were both up-regulated at the messenger RNA (mRNA) as well as protein level in BA+K3+ cells. Of note, K3 mRNA levels were also elevated in human CML cells (SI Appendix, Fig. S3C) (42, 43). Interestingly, inhibition of the constitutive Abl kinase activity of Bcr-Abl for 24 h with imatinib reduced phosphorylation of the Bcr-Abl substrate CrkL and normalized Talin-1 but not K3 levels, suggesting that Talin-1 expression is controlled by the Bcr-Abl kinase activity and K3 expression is controlled by the Bcr-Abl adaptor function (Fig. 4 F and G).
Therapeutic Strategy to Target Kindlin-3 Specifically in LSCs.
Our findings suggested that depletion of K3 in LSCs might be a novel strategy to combat CML. To deliver a K3-depleting agent, such as a K3-siRNA (small interfering RNA), into LSCs, we first searched for cell-surface proteins whose expression is absent on normal HSPCs but induced on disease-initiating LSCs. Since immune checkpoint molecules are frequently up-regulated or de novo expressed on transformed cells (44), we isolated LSK cells from leukemic CD45.2+ Scl-tTA/Tre-Bcr-Abl mice (15 to 20% neutrophils in the PB; termed BA+K3WT mice) and performed qRT-PCR measurements of programmed cell death protein 1 (PD-1), programmed cell death 1 ligand 1 (PDL1) and ligand 2 (PDL2), lymphocyte-activation gene 3 (LAG3), T cell immunoglobulin and mucin-domain containing 3 (TIM3), and cytotoxic T lymphocyte-associated antigen 4 (CTLA-4). None of the immune checkpoint molecules tested were expressed by wild-type (BA−K3WT) LSK cells, whereas only CTLA-4 was detectable in BA+K3WT LSK cells (Fig. 5 A and B and SI Appendix, Fig. S4A). Flow cytometry revealed CTLA-4 protein expression on ∼6% of the BA+K3WT LSK cells and on very few CTLA-4+ BA−K3WT LSK cells, which—in light of the absent CTLA-4 mRNA expression by BA−K3WT LSK cells (Fig. 5 A and B)—likely represents a nonspecific staining signal (Fig. 5C). A refined analysis identified CTLA-4+ leukemic cells in all HSPC subpopulations (Fig. 5D). To confirm that the disease-initiating LSCs are enriched in the CTLA-4+ HSC population, we engrafted 100 CD45.2+ LSK CD150+CD48− CTLA-4+ or 100 CD45.2+ LSK CD150+CD48− CTLA-4− cells from the BM of BA+K3WT mice into sublethally irradiated CD45.1+ WT recipients and monitored PB donor cell repopulation and CML development. Whereas the LSK CD150+CD48− CTLA-4+ cells rapidly engrafted and induced a leukemic neutrophil population in 10 out of 10 recipients, the LSK CD150+CD48− CTLA-4− cells showed reduced repopulation potential and increased neutrophil numbers in only 3 out of 11 recipients (Fig. 5 E and F), strongly suggesting that the LSCs are enriched in the CTLA-4+ HSC population.
To transfer these findings into an in vivo CML treatment protocol, we utilized the rapidly internalizing and recycling CTLA-4 surface protein (45) as an LSC-specific anchor to deliver a specific and highly stable murine K3 (mK3)-siRNA linked to a CTLA-4–binding RNA aptamer (46) (Fig. 5G and SI Appendix, Fig. S4 B–D). The high siRNA stability was achieved by methylating the first three ribonucleotides of the mK3-siRNA, which efficiently decreased K3 mRNA and protein in nucleofected BA+K3+ 32D cells (SI Appendix, Fig. S4 B–D). The treatment of CTLA-4–expressing BA+K3+ 32D cells with the CTLA-4 aptamer-tagged mK3-siRNA also reduced K3 mRNA more than 15-fold when compared with cells treated with CTLA-4 aptamer-tagged scrambled control-siRNA (scr; SI Appendix, Fig. S4E).
In line with the expression of CTLA-4 on ∼10% BA+K3WT LSK CD150+ cells (Fig. 5D), the CTLA-4 aptamer bound to 5 to 10% of BA+K3WT but was unable to bind to BA−K3WT LSK CD150+ cells in vitro (Fig. 5H). A daily short-term treatment of BA+K3WT mice with the CTLA-4 aptamer-tagged mK3-siRNA for 4 consecutive days efficiently decreased the K3 mRNA in CTLA-4+ LSK cells (Fig. 5I). Impressively, two cycles of five daily injections of the CTLA-4 aptamer-tagged mK3-siRNA over the course of 14 d normalized neutrophil counts in the PB and spleen size and dramatically extended the lifespan of BA+K3WT mice compared with animals treated with CTLA-4 aptamer-tagged scrambled control-siRNA (Fig. 5 J and K and SI Appendix, Fig. S4 F and G). Moreover, retreatment of BA+K3WT mice suffering from a relapse, caused by de novo Bcr-Abl expression and/or treatment-evading LSCs, resulted in prolonged remissions (Fig. 5L). Importantly, the clinical benefits of the CTLA-4 aptamer-tagged mK3-siRNA treatment were due to the K3-siRNA, as treatments with CTLA-4 aptamer-tagged scrambled control-siRNA or anti–CTLA-4 blocking antibodies did not improve elevated neutrophil counts in the PB, CML course or activity, and numbers of T effector cells in leukemic BA+K3WT mice (SI Appendix, Fig. S4 H–J).
The reduced CTLA-4+ LSK CD150+CD48− cell numbers in the BM (Fig. 5M) and the extended lifespan (Fig. 5K) of CTLA-4 aptamer-tagged mK3-siRNA–treated BA+K3WT mice strongly suggest that the treatment targets, mobilizes, and eventually eradicates disease-initiating LSCs. To test the hypothesis, we first engrafted 100 CD45.2+ LSK CD150+CD48− cells derived from the BM of CTLA-4 aptamer-tagged scrambled control- or mK3-siRNA–treated BA+K3WT mice into sublethally irradiated CD45.1+ WT recipients and monitored PB donor cell repopulation and CML development. Whereas the LSK CD150+CD48− cells from scrambled control-siRNA–treated BA+K3WT mice rapidly engrafted and developed a leukemic neutrophil population in 9 out of 12 recipient mice, the LSK CD150+CD48− cells from mK3-siRNA–treated BA+K3WT animals showed poor repopulation potential and increased neutrophil numbers in only 1 out of 12 recipient mice (Fig. 5 N and O). The engraftment potential of 400 CD45.2+ LSK CD150+CD48− cells from mK3-siRNA–treated BA+K3WT donor mice was comparable to that of 400 LSK CD150+CD48− cells from scrambled control-siRNA–treated BA+K3WT donor mice. However, the increase of CD45.2+ leukemic neutrophils in the PB was only observed when mice received LSK CD150+CD48− cells from scrambled control-siRNA–treated leukemic BA+K3WT donor mice (Fig. 5 N and O). The calculation of disease-initiating LSCs revealed that the BM of BA+K3WT mice treated with CTLA-4 aptamer-tagged scrambled control-siRNA harbored 1 LSC per 71 LSK CD150+CD48− cells, while the BM of BA+K3WT mice treated with CTLA-4 aptamer-tagged mK3-siRNA harbored 1 LSC per 927 LSK CD150+CD48− cells.
Taken together, our results demonstrate that the in vivo delivery of a K3-siRNA via LSC-specific anchors targets and displaces disease-initiating LSCs in the BM and halts CML progression in and extends lifespan of CML mice.
Discussion
A hallmark of hematological malignancies that initiate from LSCs is the profound remodeling and adaptation of the normal HSPC BM environment, which reinforces the activity of LSCs and in the end leads to the displacement of normal hematopoiesis (3, 4). Attempts to halt disease progression by releasing LSCs from their malignant BM microenvironment, however, have produced conflicting results (47–53). The major drawback of the approach is relocation of the leukemia from the BM to secondary organs such as the spleen or central nervous system (48, 54). Since most blood cells including normal HSPCs require integrins to extravasate and retain in the BM (28, 55, 56), we asked whether abrogating K3 expression and thereby abolishing the activation and function of most if not all integrin classes on LSCs is sufficient to induce LSC detachment from the BM microenvironment, prevent their settlement in distant organs, and protect from leukemia.
We used the inducible double-transgenic Scl-tTA/Tre-Bcr-Abl (BA+) murine CML model (37) to investigate K3 function(s) in LSCs and for the course of leukemia. Tamoxifen-induced deletion of K3 followed by inducing the expression of Bcr-Abl in BA+ mixed BM chimeras produced a rapid release of BA+K3− HSPCs into the circulation, a delayed increase of neutrophil counts in the PB, and a significant lifespan extension. The circulating BA+K3− HSPCs were unable to settle in secondary hematopoietic organs and induce an extramedullary leukemia. The marked lifespan extension of leukemic mice was apparent irrespective of whether Bcr-Abl expression was induced before or after the deletion of Fermt3, indicating that K3 loss curbs both the development and the progression of CML.
Our findings also indicate that K3 deficiency curbs the deadly blast-like crisis induced with p210 Bcr-Abl–expressing (BA+) 32D myeloid progenitor cells (40, 57) in syngeneic recipient mice. The lack of K3 expression reduced peripheral survival, proliferation, and BM homing of BA+ 32D cells and prolonged the survival of engrafted mice. K3 loss abolished the Bcr-Abl–mediated integrin hyperactivity of BA+ 32D cells caused by up-regulating Talin-1 and K3 mRNA transcription. Inducing integrin hyperactivity is a subtle and highly efficient strategy of the Bcr-Abl oncogene to induce remodeling of the BM environment and strong adhesion to and retention of LSCs in the protective microenvironment of the BM and secondary organs, which eventually results in metastatic leukemia.
The disruptive power of K3 loss on LSCs and the course of CML identified K3 depletion as a treatment strategy. The vital role of K3 for several hematopoietic cells including normal HSPCs (28), however, demanded a delivery system that transfers an anti-K3 compound such as K3-siRNA into LSCs and yet avoids normal HSPCs. In our search for such a delivery system, we discovered that the immune checkpoint receptor CTLA-4 is expressed on ∼10% of BA+ (LSK CD150+CD48−) HSCs but not normal (BA−) HSCs, and that the disease-initiating LSCs are enriched in the CTLA-4+ BA+ HSC population. To exploit CTLA-4 expression on LSCs for a targeted treatment of leukemic BA+K3WT mice, we fused a CTLA-4–binding RNA-based aptamer (46) to a stable K3-siRNA. This treatment approach efficiently depleted K3 in CTLA-4+ BA+ LSK cells, decreased the disease-initiating LSC numbers in the BM and neutrophil counts in the PB, prevented LSC dissemination into extramedullary organs, prolonged the survival of the CML mice, and, most impressively, induced long-lasting remissions in mice suffering from CML relapses. Importantly, the CTLA-4 RNA aptamer by itself showed no obvious side effects such as inflammation or T cell activation, which is probably due to the poor anti–CTLA-4 inhibitory activity of the monomeric form of the RNA aptamer (46) used in our study.
Transplantation experiments with BA+ LSK CD150+CD48− cells derived from the BM of CTLA-4 aptamer scrambled control-siRNA–treated BA+ leukemic mice indicate that the disease-initiating LSCs are rare and lowered by more than 100-fold upon CTLA-4 aptamer-tagged K3-siRNA treatment. Our calculations indicate that the leukemia-initiating capacity is restricted to ∼1% of the BA+ LSK CD150+CD48− cell population, which is in line with a published study (58), and to ∼10% of CTLA-4+ BA+ LSK CD150+CD48− cells, which suggests that the size of the CTLA-4+ BA+ LSK CD150+CD48− cell population exceeds the number of CTLA-4+ disease-initiating BA+ LSCs. It is conceivable, therefore, that BA+ LSK CD150+CD48− cells acquire mutations that induce the expression of CTLA-4 before a mutation(s) arises that cooperates with Bcr-Abl to generate disease-initiating LSCs. If so, dislodging the entire pool of CTLA-4+ BA+ LSK CD150+CD48− cells from the BM environment might eliminate not only disease-initiating LSCs but also their premalignant progenitors.
Inspection of the published literature revealed that also a CML patient-derived CD34+ BM cell population expresses CTLA-4 (59). Unfortunately, this CTLA-4+ cell population was not further characterized, and therefore it is not possible to deduce from these data whether the human disease-initiating LSCs are, like in the mouse, enriched in this CTLA-4+ BM cell population. Clearly, the CTLA-4 aptamer also binds to human CTLA-4 (60) and efficiently delivers a stable human K3-siRNA into activated T cells, decreasing K3 mRNA expression. These findings highlight the principal feasibility of our approach in the human system and call for trials in a humanized mouse model.
Materials and Methods
Detailed descriptions are provided in SI Appendix.
Experimental Models.
Mice.
CML was induced in mice carrying the Scl-tTA and Tre-Bcr-Abl (BAcond) transgenes and blocked by tetracycline administration via drinking water (0.5 g/L; bela-pharm) (37). K3flox/flox Rosa26Cre-ERT2 (K3cond) mice have been described (28). These mice were backcrossed and kept on a C57BL/6 background. C3H/HeOuJ mice were obtained from Charles River. Mice were kept under specific pathogen-free conditions in the animal facility of the Max Planck Institute of Biochemistry. The mouse experiments were approved by the Government of Upper Bavaria.
Cell lines.
All cell lines were cultured at 37 °C and 5% CO2. All cell lines were routinely screened for mycoplasma contamination by PCR-based screenings (PCR Mycoplasma Test Kit II; PanReac AppliChem) following the instructions of the manufacturer. Cell lines were routinely characterized by surface marker expression using flow cytometry.
32D cells (clone 3, mouse, ACC-411; DSMZ) were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium (Gibco) supplemented with 10% fetal calf serum (FCS), 10% preconditioned supernatant of Wehi-3B cell (mouse, ACC-26; DSMZ) cultures, glutamine, and antibiotics. Wehi-3B cells were cultured in RPMI-1640 supplemented with 10% FCS, glutamine, and antibiotics.
Quantification and Statistical Analysis.
Statistical analysis was performed with GraphPad Prism 6. For every reported dataset at least two independent experiments were performed. All datasets were tested for normal distribution. Statistical significance was assumed at P < 0.05. *P < 0.05, **P < 0.01, and ***P < 0.001. Statistical tests used for the experiments are described in the figure legends. Unless otherwise noted, all values are reported as mean ± SD.
Supplementary Material
Acknowledgments
We thank Alexander Felber and Dr. Johannes B. Jäger for expert technical assistance; Dr. Raphael Ruppert for help in setting up the mouse models; Drs. Eric So, Robert S. Welner, and Andreas Herrmann for inspiring discussions regarding LSCs, CTLA-4 expression, and CTLA-4 targeting; and Drs. Mercedes Costell, Peter Murray, and Roy Zent for critically commenting on the manuscript. The work was funded by the European Research Council (Proposal ID 810104) and the Max Planck Society.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. C.W. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2009078117/-/DCSupplemental.
Data Availability.
We confirm that the data supporting the findings of this study are available within the article and SI Appendix or in public resources (Microarray Innovations in Leukemia Study; National Center for Biotechnology Information Gene Expression Omnibus [GSE13159]).
References
- 1.Eisterer W. et al., Different subsets of primary chronic myeloid leukemia stem cells engraft immunodeficient mice and produce a model of the human disease. Leukemia 19, 435–441 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Calabretta B., Perrotti D., The biology of CML blast crisis. Blood 103, 4010–4022 (2004). [DOI] [PubMed] [Google Scholar]
- 3.Schepers K., Campbell T. B., Passegué E., Normal and leukemic stem cell niches: Insights and therapeutic opportunities. Cell Stem Cell 16, 254–267 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krause D. S., Scadden D. T., A hostel for the hostile: The bone marrow niche in hematologic neoplasms. Haematologica 100, 1376–1387 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lane S. W., Scadden D. T., Gilliland D. G., The leukemic stem cell niche: Current concepts and therapeutic opportunities. Blood 114, 1150–1157 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhatia R., McGlave P. B., Dewald G. W., Blazar B. R., Verfaillie C. M., Abnormal function of the bone marrow microenvironment in chronic myelogenous leukemia: Role of malignant stromal macrophages. Blood 85, 3636–3645 (1995). [PubMed] [Google Scholar]
- 7.Reynaud D. et al., IL-6 controls leukemic multipotent progenitor cell fate and contributes to chronic myelogenous leukemia development. Cancer Cell 20, 661–673 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Welner R. S. et al., Treatment of chronic myelogenous leukemia by blocking cytokine alterations found in normal stem and progenitor cells. Cancer Cell 27, 671–681 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rowley J. D., Letter: A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature 243, 290–293 (1973). [DOI] [PubMed] [Google Scholar]
- 10.Deininger M. W., Goldman J. M., Melo J. V., The molecular biology of chronic myeloid leukemia. Blood 96, 3343–3356 (2000). [PubMed] [Google Scholar]
- 11.Perrotti D., Jamieson C., Goldman J., Skorski T., Chronic myeloid leukemia: Mechanisms of blastic transformation. J. Clin. Invest. 120, 2254–2264 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kobayashi S. et al., BCR-ABL promotes neutrophil differentiation in the chronic phase of chronic myeloid leukemia by downregulating c-Jun expression. Leukemia 23, 1622–1627 (2009). [DOI] [PubMed] [Google Scholar]
- 13.O’Brien S. G. et al.; IRIS Investigators , Imatinib compared with interferon and low-dose cytarabine for newly diagnosed chronic-phase chronic myeloid leukemia. N. Engl. J. Med. 348, 994–1004 (2003). [DOI] [PubMed] [Google Scholar]
- 14.Copland M. et al., Dasatinib (BMS-354825) targets an earlier progenitor population than imatinib in primary CML but does not eliminate the quiescent fraction. Blood 107, 4532–4539 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Jørgensen H. G., Allan E. K., Jordanides N. E., Mountford J. C., Holyoake T. L., Nilotinib exerts equipotent antiproliferative effects to imatinib and does not induce apoptosis in CD34+ CML cells. Blood 109, 4016–4019 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Jabbour E., Kantarjian H., Chronic myeloid leukemia: 2018 update on diagnosis, therapy and monitoring. Am. J. Hematol. 93, 442–459 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Holtz M. S. et al., Imatinib mesylate (STI571) inhibits growth of primitive malignant progenitors in chronic myelogenous leukemia through reversal of abnormally increased proliferation. Blood 99, 3792–3800 (2002). [DOI] [PubMed] [Google Scholar]
- 18.Graham S. M. et al., Primitive, quiescent, Philadelphia-positive stem cells from patients with chronic myeloid leukemia are insensitive to STI571 in vitro. Blood 99, 319–325 (2002). [DOI] [PubMed] [Google Scholar]
- 19.Corbin A. S. et al., Human chronic myeloid leukemia stem cells are insensitive to imatinib despite inhibition of BCR-ABL activity. J. Clin. Invest. 121, 396–409 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hamilton A. et al., Chronic myeloid leukemia stem cells are not dependent on Bcr-Abl kinase activity for their survival. Blood 119, 1501–1510 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nilsson S. K. et al., Osteopontin, a key component of the hematopoietic stem cell niche and regulator of primitive hematopoietic progenitor cells. Blood 106, 1232–1239 (2005). [DOI] [PubMed] [Google Scholar]
- 22.Matsunaga T. et al., Interaction between leukemic-cell VLA-4 and stromal fibronectin is a decisive factor for minimal residual disease of acute myelogenous leukemia. Nat. Med. 9, 1158–1165 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Sahin A. O., Buitenhuis M., Molecular mechanisms underlying adhesion and migration of hematopoietic stem cells. Cell Adhes. Migr. 6, 39–48 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar R., Godavarthy P. S., Krause D. S., The bone marrow microenvironment in health and disease at a glance. J. Cell Sci. 131, jcs201707 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Bhatia R., Wayner E. A., McGlave P. B., Verfaillie C. M., Interferon-alpha restores normal adhesion of chronic myelogenous leukemia hematopoietic progenitors to bone marrow stroma by correcting impaired beta 1 integrin receptor function. J. Clin. Invest. 94, 384–391 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peled A. et al., The chemokine SDF-1 activates the integrins LFA-1, VLA-4, and VLA-5 on immature human CD34(+) cells: Role in transendothelial/stromal migration and engraftment of NOD/SCID mice. Blood 95, 3289–3296 (2000). [PubMed] [Google Scholar]
- 27.Gu Y. C. et al., Laminin isoform-specific promotion of adhesion and migration of human bone marrow progenitor cells. Blood 101, 877–885 (2003). [DOI] [PubMed] [Google Scholar]
- 28.Ruppert R. et al., Kindlin-3-mediated integrin adhesion is dispensable for quiescent but essential for activated hematopoietic stem cells. J. Exp. Med. 212, 1415–1432 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ussar S., Wang H. V., Linder S., Fässler R., Moser M., The Kindlins: Subcellular localization and expression during murine development. Exp. Cell Res. 312, 3142–3151 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Moser M., Legate K. R., Zent R., Fässler R., The tail of integrins, talin, and kindlins. Science 324, 895–899 (2009). [DOI] [PubMed] [Google Scholar]
- 31.Moser M., Nieswandt B., Ussar S., Pozgajova M., Fässler R., Kindlin-3 is essential for integrin activation and platelet aggregation. Nat. Med. 14, 325–330 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Moser M. et al., Kindlin-3 is required for beta2 integrin-mediated leukocyte adhesion to endothelial cells. Nat. Med. 15, 300–305 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Moretti F. A. et al., Kindlin-3 regulates integrin activation and adhesion reinforcement of effector T cells. Proc. Natl. Acad. Sci. U.S.A. 110, 17005–17010 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuijpers T. W. et al., LAD-1/variant syndrome is caused by mutations in FERMT3. Blood 113, 4740–4746 (2009). [DOI] [PubMed] [Google Scholar]
- 35.Malinin N. L. et al., A point mutation in KINDLIN3 ablates activation of three integrin subfamilies in humans. Nat. Med. 15, 313–318 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Svensson L. et al., Leukocyte adhesion deficiency-III is caused by mutations in KINDLIN3 affecting integrin activation. Nat. Med. 15, 306–312 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koschmieder S. et al., Inducible chronic phase of myeloid leukemia with expansion of hematopoietic stem cells in a transgenic model of BCR-ABL leukemogenesis. Blood 105, 324–334 (2005). [DOI] [PubMed] [Google Scholar]
- 38.Schmidt S. et al., Kindlin-3-mediated signaling from multiple integrin classes is required for osteoclast-mediated bone resorption. J. Cell Biol. 192, 883–897 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pietras E. M. et al., Functionally distinct subsets of lineage-biased multipotent progenitors control blood production in normal and regenerative conditions. Cell Stem Cell 17, 35–46 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schemionek M. et al., Mtss1 is a critical epigenetically regulated tumor suppressor in CML. Leukemia 30, 823–832 (2016). [DOI] [PubMed] [Google Scholar]
- 41.Qu J. et al., Kindlin-3 interacts with the ribosome and regulates c-Myc expression required for proliferation of chronic myeloid leukemia cells. Sci. Rep. 5, 18491 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haferlach T. et al., The clinical utility of microarray-based gene expression profiling in the diagnosis and sub-classification of leukemia: Final report on 3252 cases from the International MILE Study Group. Blood 112, 753 (2008). [Google Scholar]
- 43.Bagger F. O. et al., BloodSpot: A database of gene expression profiles and transcriptional programs for healthy and malignant haematopoiesis. Nucleic Acids Res. 44, D917–D924 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seidel J. A., Otsuka A., Kabashima K., Anti-PD-1 and anti-CTLA-4 therapies in cancer: Mechanisms of action, efficacy, and limitations. Front. Oncol. 8, 86 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qureshi O. S. et al., Constitutive clathrin-mediated endocytosis of CTLA-4 persists during T cell activation. J. Biol. Chem. 287, 9429–9440 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Santulli-Marotto S., Nair S. K., Rusconi C., Sullenger B., Gilboa E., Multivalent RNA aptamers that inhibit CTLA-4 and enhance tumor immunity. Cancer Res. 63, 7483–7489 (2003). [PubMed] [Google Scholar]
- 47.Weisberg E. et al., Inhibition of CXCR4 in CML cells disrupts their interaction with the bone marrow microenvironment and sensitizes them to nilotinib. Leukemia 26, 985–990 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Agarwal A. et al., Effects of plerixafor in combination with BCR-ABL kinase inhibition in a murine model of CML. Blood 120, 2658–2668 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nervi B. et al., Chemosensitization of acute myeloid leukemia (AML) following mobilization by the CXCR4 antagonist AMD3100. Blood 113, 6206–6214 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zeng Z. et al., Targeting the leukemia microenvironment by CXCR4 inhibition overcomes resistance to kinase inhibitors and chemotherapy in AML. Blood 113, 6215–6224 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Löwenberg B. et al.; Dutch-Belgian Hemato-Oncology Cooperative Group; Swiss Group for Clinical Cancer Research , Effect of priming with granulocyte colony-stimulating factor on the outcome of chemotherapy for acute myeloid leukemia. N. Engl. J. Med. 349, 743–752 (2003). [DOI] [PubMed] [Google Scholar]
- 52.Boyd A. L. et al., Niche displacement of human leukemic stem cells uniquely allows their competitive replacement with healthy HSPCs. J. Exp. Med. 211, 1925–1935 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zittoun R. et al., Granulocyte-macrophage colony-stimulating factor associated with induction treatment of acute myelogenous leukemia: A randomized trial by the European Organization for Research and Treatment of Cancer Leukemia Cooperative Group. J. Clin. Oncol. 14, 2150–2159 (1996). [DOI] [PubMed] [Google Scholar]
- 54.Fruehauf S., Zeller W. J., Calandra G., Novel Developments in Stem Cell Mobilization: Focus on CXCR4, (Springer, New York, 2012). [Google Scholar]
- 55.Hirsch E., Iglesias A., Potocnik A. J., Hartmann U., Fässler R., Impaired migration but not differentiation of haematopoietic stem cells in the absence of beta1 integrins. Nature 380, 171–175 (1996). [DOI] [PubMed] [Google Scholar]
- 56.Potocnik A. J., Brakebusch C., Fässler R., Fetal and adult hematopoietic stem cells require beta1 integrin function for colonizing fetal liver, spleen, and bone marrow. Immunity 12, 653–663 (2000). [DOI] [PubMed] [Google Scholar]
- 57.Neviani P. et al., FTY720, a new alternative for treating blast crisis chronic myelogenous leukemia and Philadelphia chromosome-positive acute lymphocytic leukemia. J. Clin. Invest. 117, 2408–2421 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang B. et al., Altered microenvironmental regulation of leukemic and normal stem cells in chronic myelogenous leukemia. Cancer Cell 21, 577–592 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pistillo M. P. et al., CTLA-4 is not restricted to the lymphoid cell lineage and can function as a target molecule for apoptosis induction of leukemic cells. Blood 101, 202–209 (2003). [DOI] [PubMed] [Google Scholar]
- 60.Herrmann A. et al., CTLA4 aptamer delivers STAT3 siRNA to tumor-associated and malignant T cells. J. Clin. Invest. 124, 2977–2987 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
We confirm that the data supporting the findings of this study are available within the article and SI Appendix or in public resources (Microarray Innovations in Leukemia Study; National Center for Biotechnology Information Gene Expression Omnibus [GSE13159]).