Significance
Understanding the evolution of sex determination mechanisms and sex chromosomes is of fundamental importance in biology. Here we have reconstructed the evolution of the sex-determining region in the Atlantic herring. The region is small and contains only three intact genes. The candidate sex-determining factor BMPR1BBY is an evolutionary innovation in the herring lineage. It encodes a truncated form of a BMP type I receptor, which originated by gene duplication and underwent rapid protein evolution. The receptor has maintained its kinase activity and has the potential to induce development of testis. The other two genes in the sex-determining region, CATSPERG and CATSPER3Y, are male beneficial genes because they encode proteins predicted to be essential for sperm to fertilize the egg.
Keywords: sex determination, BMPR1, CatSper, gene duplication, molecular evolution
Abstract
The mechanisms underlying sex determination are astonishingly plastic. Particularly the triggers for the molecular machinery, which recalls either the male or female developmental program, are highly variable and have evolved independently and repeatedly. Fish show a huge variety of sex determination systems, including both genetic and environmental triggers. The advent of sex chromosomes is assumed to stabilize genetic sex determination. However, because sex chromosomes are notoriously cluttered with repetitive DNA and pseudogenes, the study of their evolution is hampered. Here we reconstruct the birth of a Y chromosome present in the Atlantic herring. The region is tiny (230 kb) and contains only three intact genes. The candidate male-determining gene BMPR1BBY encodes a truncated form of a BMP1B receptor, which originated by gene duplication and translocation and underwent rapid protein evolution. BMPR1BBY phosphorylates SMADs in the absence of ligand and thus has the potential to induce testis formation. The Y region also contains two genes encoding subunits of the sperm-specific Ca2+ channel CatSper required for male fertility. The herring Y chromosome conforms with a characteristic feature of many sex chromosomes, namely, suppressed recombination between a sex-determining factor and genes that are beneficial for the given sex. However, the herring Y differs from other sex chromosomes in that suppression of recombination is restricted to an ∼500-kb region harboring the male-specific and sex-associated regions. As a consequence, any degeneration on the herring Y chromosome is restricted to those genes located in the small region affected by suppressed recombination.
The mechanisms underlying sex determination are astonishingly plastic. Particularly the triggers for the molecular machinery, which recalls either the male or female program, are highly variable and have evolved independently and repeatedly (1–8). Fish show a rich diversity of sex determination systems, including both genetic and environmental triggers (1, 9, 10). Adaptive hypotheses prevail in explaining the evolution of the various sex-determining triggers that guarantee a stable sex ratio of the population. Genetic sex determination is generally explained to be stabilized through the evolution of sex chromosomes, when sexually antagonistic selection links the genetic sex determiner to a locus that is beneficial to the same sex or even antagonistic to the opposite sex (11–15).
The Atlantic herring (Clupea harengus) is one of the most abundant vertebrates on Earth. Neither its karyotype (16) nor the male and female linkage map (17) indicated the presence of sex chromosomes. Unexpectedly, our analysis of the Atlantic herring genome revealed a Y chromosome having a minute male-specific region (∼230 kb), a small sex-associated region (∼300 kb), and a large pseudoautosomal region (∼30 Mb) corresponding to chromosome 8 in the reference assembly (17). Here we reconstruct the birth of this chromosome involving two gene duplications followed by rapid protein evolution of the candidate male-determining gene BMPR1BBY.
Results
Identification of the Sex-Determining Region.
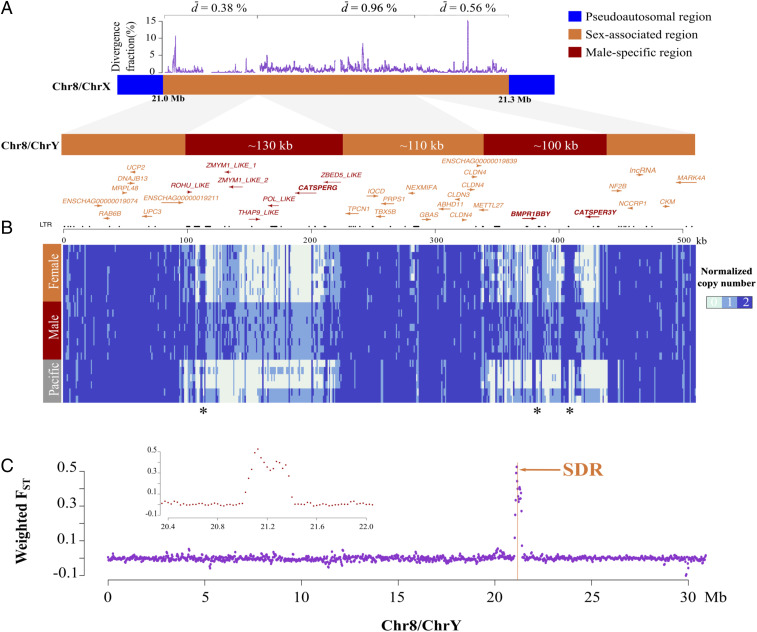
The sex-associated region was identified by genome-wide association analysis using whole-genome sequencing data aligned to the reference genome (17). We compared SNP (single nucleotide polymorphism) allele frequencies between eight males and eight females by χ2 analysis (Dataset S1); 765 SNPs spanning ∼160 kb on chromosome 8 showed significant allele frequency differences (P ≤ 1.0 × 10−10) (SI Appendix, Fig. S1A). Analysis of SNP genotypes across the sex-associated region for the 16 fish revealed that males were heterozygous (XY), whereas females were homozygous (XX) for most sex-associated SNPs (SI Appendix, Fig. S1B). Thus, males are the heterogametic sex. An additional 17 Atlantic herring of unknown sex followed this pattern perfectly with individuals either being heterozygous or homozygous across the sex-associated region. The sequence of this region in the current assembly (17) represents the female haplotype (SI Appendix, Fig. S1B). In conclusion, herring chromosome 8 can now be split into chromosome (Chr) X and Y.
To characterize the male-specific region on ChrY, we examined the sequence of all unplaced scaffolds from the current assembly (Methods) (17). Two unplaced scaffolds, 18 and 229, showed regions of high identity with the female-specific sequence on ChrX (SI Appendix, Fig. S1 C and D). These regions were interrupted by male-specific sequences that are present in only one copy in males (Fig. 1 A and B). We also analyzed data from six individually sequenced Pacific herring of unknown sex (18) and found that four had no coverage in the male-specific regions, whereas two had a single copy of the male-specific regions (Fig. 1B). The results provide evidence that the male-specific region evolved prior to the divergence of the Atlantic and Pacific herring that separated ∼2 My before present (19).
Fig. 1.
Assembly of chromosome Y in Atlantic herring. (A) Assembly and annotation of male-specific region (red), sex-associated region (orange), and flanking pseudoautosomal region (blue). Some genes (annotated as “_like”) within the male-specific region show partial similarity with known proteins. LTR track, long terminal repeats. d, nucleotide sequence divergence between ChrX and Y in 500-bp windows. (B) Normalized copy number of sequenced individuals (eight female and eight male Atlantic herring and six Pacific herring of unknown sex) across the male-specific and sex-associated regions. Regions with high coverage marked with an asterisk overlap repetitive sequences. (C) Genetic differentiation (FST) in 50-kb overlapping windows between males (n = 8) and females (n = 8) across ChrY. The SDR is indicated by a vertical line. The male-specific region was not included in this analysis, since it is missing on ChrX. (Inset) FST values in the near vicinity of the SDR.
We used the Xdrop method (20) for targeted resequencing of long DNA molecules (up to ∼100 kb) from the borders between the male-specific and the flanking region. The alignment of these reads to our ChrY assembly confirmed a correct orientation and did not reveal any gaps in the alignment (SI Appendix, Fig. S1D).
Our assembly of the ∼230-kb male-specific region of ChrY shows that the male-specific sequence is split into two parts (Fig. 1A, in red) separated by an ∼110-kb sex-associated region (Fig. 1B, in orange) that is well assembled and annotated (17). It contains 13 genes that are present on both X and Y, as indicated by equal sequence depth in males and females (Fig. 1B). The region showing strong genetic differentiation between males and females is remarkably sharp and only extends for ∼300 kb from ∼21.0 to ∼21.3 Mb on ChrX (Fig. 1C), and the male-specific region together with the sex-associated region extends for ∼500 kb on ChrY (SI Appendix, Fig. S1). These results imply that there is no meiotic recombination in the male-specific regions (red) and no or at least suppressed recombination in the sex-associated regions (orange) (Fig. 1A). The genes located in the region showing strong genetic differentiation between X and Y are listed in SI Appendix, Table S1. The nucleotide distance between X and Y in the sex-associated region located between the male-specific regions (d = 0.96%; Fig. 1A) is only 2.5-fold higher than the estimated average nucleotide distance between Atlantic and Pacific herring (∼0.4%), implying either a divergence time of ∼5 My or ongoing genetic exchange in the sex-associated region.
The male-specific region contains only three intact genes—CATSPERG, CATSPER3Y, and BMPR1BBY—that all are of high interest for male function (Fig. 1A). In addition, we identified other genes annotated ab initio, but these were not well supported by RNA-seq data from gonadal tissue; one of these, POL-like, is of retroviral origin. CATSPERG and CATSPER3Y encode subunits of the sperm-specific Ca2+ channel CatSper that is required for sperm function and male fertility in mouse, humans, and sea urchin (21–25). A phylogenetic tree analysis shows that the substitution rates of CATSPERG in Atlantic herring and its orthologs in other teleost are similar, consistent with a maintained function as a CatSper subunit (SI Appendix, Fig. S2). CATSPER3 exists in two copies, CATSPER3Y in the male-specific region and a pseudoautosomal copy, CATSPER3A, ∼1 Mb apart from the Y copy (Chr8, 22,259,380 to 22,268,493 bp). We estimate that CATSPER3Y arose by duplication about 15 Mya (SI Appendix, Fig. S3). Although the two CATSPER3 genes in herring are closely related, CATSPER3Y is more well conserved compared with paralogs in other species (SI Appendix, Fig. S3), suggesting that its function is maintained. By contrast, two findings suggest that CATSPER3A is nonfunctional: its exon–intron organization is not intact, and the dN/dS ratio of nonsynonymous (dN) to synonymous (dS) substitutions is close to 1.0, which is characteristic for pseudogenes.
The third gene in the male-specific region BMPR1BBY encodes a severely truncated form of BMPR1B (bone morphogenetic protein receptor 1B), a transmembrane protein that hosts an intracellular Ser/Thr kinase domain and is a member of the TGFβ receptor family. Components of TGFβ signaling control the development of male gonads (4). Sequence comparison of BMPR1BBY with the paralogs BMPR1BA and BMPR1BB on chromosomes 7 and 21, respectively, demonstrated that BMPR1BBY is more closely related to BMPR1BB (SI Appendix, Fig. S4). Transcriptome analysis of testis RNA revealed two isoforms with different 5′ start sites (S- and L-BMPR1BBY) containing only coding sequences corresponding to exon 7 to 11 of the autosomal paralogs (SI Appendix, Fig. S4). In summary, BMPR1BBY is a truncated copy of BMPR1BB that has been translocated to chromosome 8; it is a strong candidate for being the male sex-determining factor.
We genotyped 200 individuals with known sex for the presence of BMPR1BBY. The high consistency between the X/Y genotype and sex phenotype (94%; SI Appendix, Table S2) demonstrates strong association to sex determination. Eleven out of 12 discrepancies between genotype and phenotype concerned XY females carrying BMPR1BBY, suggesting that the herring Y may not show full penetrance. Such incomplete penetrance of a young genetic sex determination system has also been observed in the northern pike (26).
Tissue Expression of Genes from the Male-Specific Region.
We compared the tissue expression of BMPR1BBY, CATSPER3Y, CATSPERG, and the autosomal copies BMPR1BB and CATSPER3A, as well as two other TGFβ signaling genes, the anti-Müllerian hormone (AMH) and its cognate receptor (AMHR2), involved in vertebrate sexual development. No expression of CATSPER3A was detected. The comparison revealed that the three Y-specific genes were only expressed in males as expected (SI Appendix, Fig. S5). BMPR1BBY and, surprisingly, also CATSPER3Y were expressed in both testis and brain, whereas CATSPERG showed testis-specific expression. The autosomal copy of BMPR1BB was more broadly expressed with the highest level in gonads; female gonads showed higher transcript levels than male gonads (SI Appendix, Fig. S5B). As expected, AMH and AMHR2 were highly expressed in male and female gonads (SI Appendix, Fig. S5 D and F), consistent with their important roles in sex determination and gonad differentiation.
Rapid Protein Evolution of BMPR1BBY.
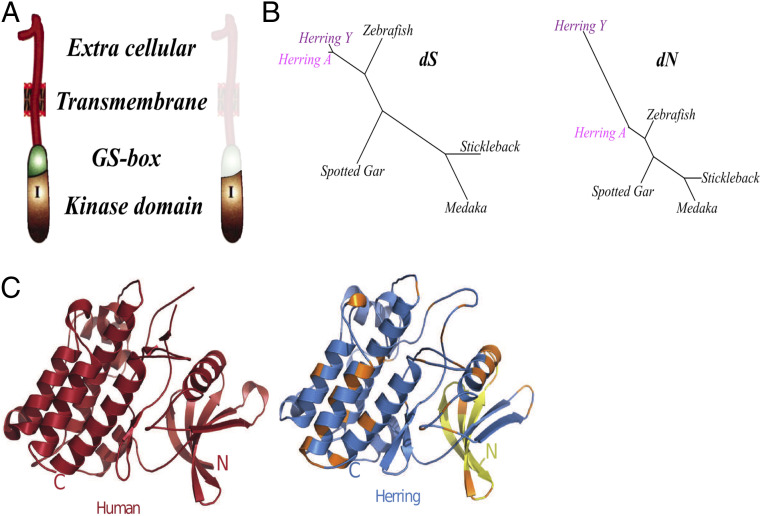
The predicted structure of the autosomal herring BMPR1BB and the canonical vertebrate BMPR1 receptor is similar (Fig. 2A) (27). The BMPR1BBY transcripts are lacking exons 1 to 6 of the autosomal BMPR1BB (SI Appendix, Fig. S4) that encode the extracellular, transmembrane, and part of the intracellular domain (Fig. 2A). These domains mediate ligand binding, receptor activation, and membrane localization and entail the phosphorylation sites of the GS loop. Amino acid alignment of BMPR1BBY and BMPR1BB from herring and other fish reveals more than 40 changes that have occurred since BMPR1BBY arose (SI Appendix, Fig. S4C). The rate of dS and dN substitutions between BMPR1BBY and BMPR1BB is 0.12 and 0.07, respectively, yielding a dN/dS ratio of 0.58, a dramatic and highly significant (P < 10−21) increase compared with dN/dS ratios of 0.01 to 0.02 in other branches of the tree (Fig. 2B and SI Appendix, Fig. S6). We estimate that BMPR1BBY arose by duplication about 20 Mya (SI Appendix, Fig. S6).
Fig. 2.
Evolution of the ChrY-specific BMPR1BBY gene and protein. (A) Schematic representation of herring BMPR1BB and BMPR1BBY proteins showing the architecture of BMP receptors and highlighting the absence of the extracellular, transmembrane and GS-box domains in the truncated BMPR1BBY. (B) dS and dN phylogenetic trees of BMPR1BB (Herring A) and BMPR1BBY (Herring Y) compared to BMPR1BB in other species. (C) Crystal structure of the intracellular domain of human BMPR1B (PDB 3MDY, amino acids 206 to 500, red) and a model of herring BMPR1BBY (blue). Yellow color represents the N-terminal extension of L-BMPR1BBY compared to S-BMPR1BBY. Orange color indicates amino acid substitutions between BMPR1BB and BMPR1BBY. N, N terminus; C, C terminus.
We built a structural model of the intracellular domain of herring BMPR1BBY and BMPR1BB based on human BMPR1B (Fig. 2C). Both herring BMPR1BB protein folds are essentially identical to that of the human paralog (Cα root-mean-square deviation of 0.008, 0.077, and 0.073 Å for BMPR1BB, L-BMPR1BBY, and S-BMPR1BBY, respectively; SI Appendix, Fig. S7 and Table S5), suggesting that the truncated form may still display Ser/Thr kinase activity.
To conclude, BMPR1BBY shows an accelerated protein evolution typical for sex-determination genes (28) but appears to maintain the BMPR1B protein fold.
Physiological Activity of BMPR1BBY.
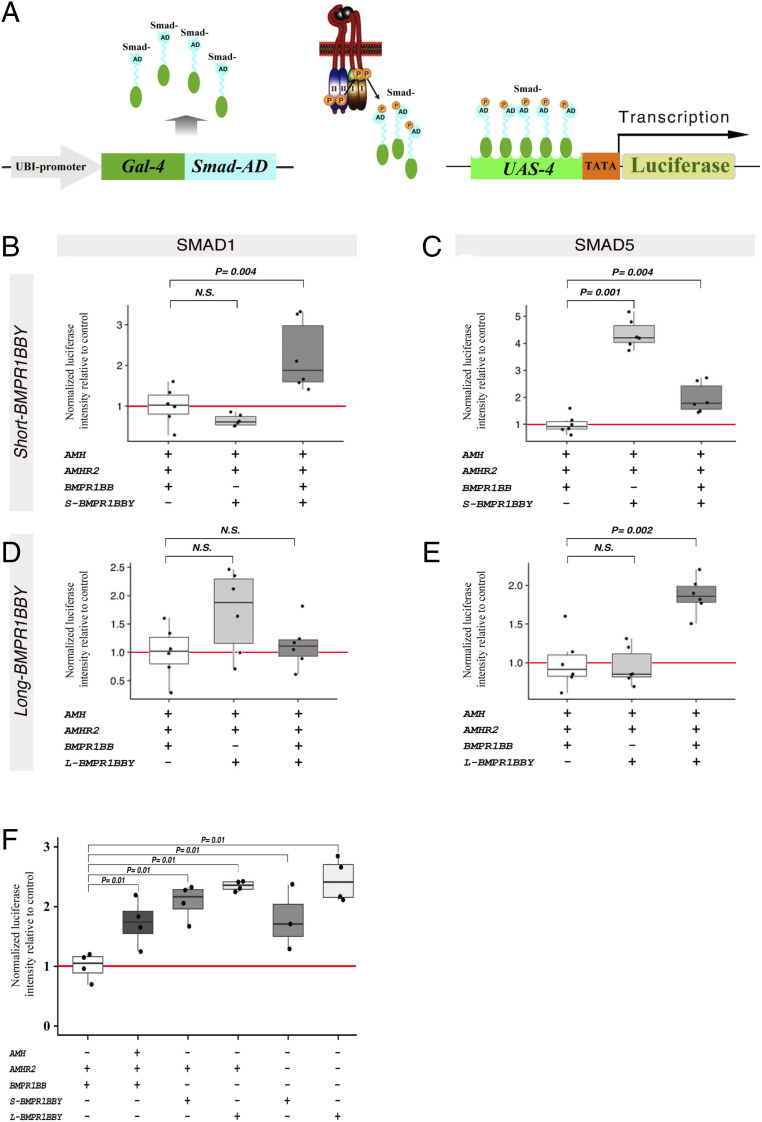
TGFβ signaling by the anti-Müllerian hormone (AMH) system is critically involved in sex determination (29). BMPR1BB, a type-I TGFβ receptor, is activated by ligand-bound type-II receptor and, in turn, phosphorylates the transcription factor SMAD. AMH-induced signaling occurs primarily via SMAD5 phosphorylation and to some extent via SMAD1 and SMAD8 (30). To probe the kinase activity of truncated BMPRBBY, we established an in vitro reporter system for SMAD activation (Fig. 3A and Methods). The system consists of a Gal4 transcriptional activator coupled to the transactivation domains of SMAD1 and SMAD5, which are specific for AMH signaling (31), and a UAS-Luc reporter driving luciferase expression. We tested three different scenarios: 1) the canonical AMH-signaling machinery including herring AMH, AMHR2, and BMPR1BB; 2) the canonical AMH machinery for which BMPR1BB has been replaced by either S-BMPR1BBY or L-BMPR1BBY; and 3) the canonical AMH machinery supplemented by either S-BMPR1BBY or L-BMPR1BBY (Fig. 3 B–E). Phosphorylation levels of SMAD1 did not differ significantly when BMPR1BB was replaced by either S-BMPR1BBY or L-BMPR1BBY (Fig. 3 B and D). By contrast, S-BMPR1BBY is fourfold more active than BMPR1BB in SMAD5 phosphorylation (Fig. 3C). Both SMAD1 and SMAD5 activity were enhanced when both BMPR1BB and S-BMPR1BBY were present (Fig. 3 B and C). No significant increase of SMAD1 and only low increase of SMAD5 phosphorylation were recorded after addition of L-BMPR1BBY (Fig. 3 D and E). Thus, S- and L-BMPR1BBY can substitute for BMPR1BB in activating SMAD1 and SMAD5.
Fig. 3.
Analysis of BMPR1BBY signaling. (A) Schematic of the UAS-Gal4-Smad-AD reporter assay for quantifying the degree of SMAD1 and SMAD5 phosphorylation upon application of different combinations of TGFβ ligands and receptors (see Methods for detailed information). In absence of AMHR2 or BMPR1BBY-induced stimulations, the Gal4-Smad-AD fusion protein remains in the cytoplasm and therefore does not activate transcription of the UAS-4 promoter driving expression of the luciferase. In this situation, only background luciferase expression levels are recorded and set a blank value. In presence of an active type I receptor, the Gal4-Smad-AD fusion proteins are selectively phosphorylated by the receptor through the Smad-AD domain and then translocate to the nucleus. Here they bind and transactivate the UAS-4 promoter through its Gal4 DNA binding domain, which leads to transcription of the luciferase gene. The resulting luciferase activity thus strictly correlates to the degree of activation (phosphorylation) of the Smads, which in turn is a measure of the specific upstream signals elicited by the various combinations of ligands and receptors. AD, specific transactivation domains of the different SMADS. (B–E) Normalized luciferase intensities relative to controls after transfection of different combinations of herring AMH, AMHR2, BMPR1BB, and S- and L-BMPR1BBY. (F) Normalized luciferase intensities relative to controls using the SMAD1 reporter after transfection of different combinations of herring AMH, AMHR2, BMPR1BB, and S- and L-BMPR1BBY.
Next, we examined whether S- and L-BMPR1BBY synergize with canonical AMH signaling. To this end, AMHR2, BMPR1BB, and S- or L-BMPR1BBY were tested either in the presence or absence of the AMH ligand. While canonical AMH signaling (AMH, AMHR2, and BMPR1BB) was clearly stimulated by AMH (SI Appendix, Fig. S8A), AMH did not modulate S- or L-BMPR1BBY-mediated signaling when associated with AMHR2 (SI Appendix, Fig. S8 B and C). Moreover, S- or L-BMPR1BBY–mediated signaling via SMAD1 phosphorylation was unaffected when AMH was expressed without its cognate receptor AMHR2 (SI Appendix, Fig. S8 D and E), indicating that BMPR1BBY-mediated signaling is not further modulated by the AMH ligand. Finally, in the absence of AMH, the presence or absence of AMHR2 did not affect S- or L-BMPR1BBY–induced signaling (Fig. 3F), while differential synergistic effects were observed for BMPR1BBYs when expressed together with the whole AMH machinery (AMH ligand, AMHR2, and BMPR1BB; Fig. 3 B–E).
In summary, BMPR1BBY has maintained its enzymatic activity despite the severe truncation and the accelerated evolution at the protein level. S- and L-BMPR1BBY–mediated signaling requires neither the presence of the AMH ligand nor the AMHR2 receptor or the autosomal BMPR1BB, yet is able to enhance canonical AMH signaling. Thus, the Y-linked receptor has the enzymatic features to act as an upstream regulator of male gonad development.
CatSper3Y and CatSperG Are Essential Components of the Herring Sperm Proteome.
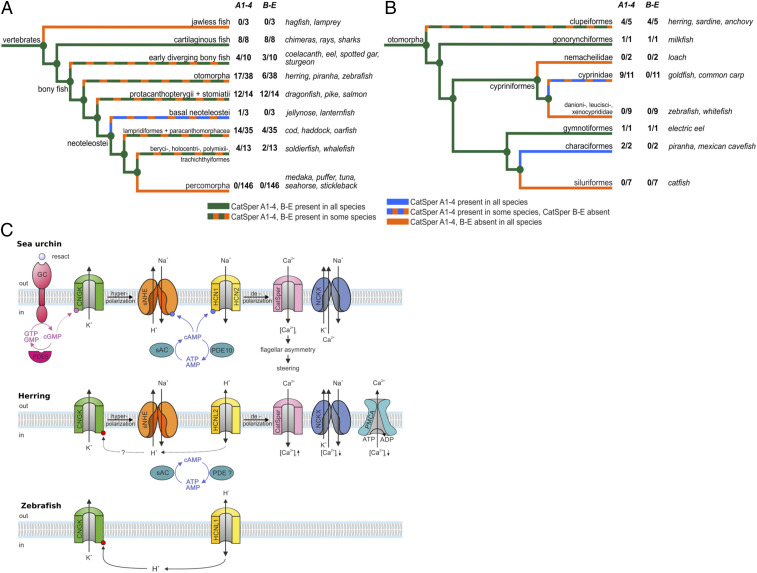
The location of two CatSper genes on the herring Y chromosome is intriguing because CatSper has been thought to be lost in several metazoan lineages including fish (32). We revisited this issue and identify here CatSper genes in many but not all fish genomes (Fig. 4 A and B and Dataset S2). CatSper is a uniquely complex Ca2+ channel that consists of at least nine different subunits CatSper1–CatSper4, CatSperB, CatSperG, CatSperD, CatSperE, and CatSperZ. The Atlantic herring genome contains all CatSper genes except CatSperZ (33) (Dataset S2). Using mass spectrometry (MS), seven CatSper subunits including CatSper3 and CatSperG were identified in mature herring sperm (Datasets S3 and S4). Signaling pathways control CatSper and, thereby, sperm motility, navigation, and fertilization from marine invertebrates to mammals (34–36). The CatSper channel is gated open by membrane voltage and alkaline pHi. Three signaling events are key to CatSper activation in sperm of the sea urchin Arbacia punctulata: hyperpolarization of the membrane potential Vm by a K+ channel CNGK (37, 38) followed by repolarization via hyperpolarization-activated and cyclic nucleotide-gated (HCN) channels, so-called pacemaker channels (39, 40), and alkalization by a sperm-specific voltage-gated sodium/proton exchanger (sNHE or SLC9C1) (41). The activity of the SLC9C1 exchanger and HCN channel is modulated by cAMP, which is synthesized by a soluble adenylate cyclase (sAC or adcy10 gene). Remarkably, all these signaling proteins upstream of CatSper are present in herring sperm, suggesting that CatSper and Ca2+ also control motility of herring sperm (Fig. 4C and Dataset S4). Two functionally important variations are striking. First, herring sperm carry an HCN-like (HCNL) channel that is distinct from classic HCN channels and falls into a new gene subfamily (42). HCNL1 in zebrafish sperm, unlike classic HCN channels, is proton-selective and not cAMP-sensitive (42). Although herring sperm carry the HCNL2 isoform that has not yet been characterized functionally, structural features underlying proton permeation are conserved between HCNL1 and HCNL2 (34). Second, the zebrafish CNGK channel is also gated not by cyclic nucleotides but by alkaline pHi, indicating that the cellular messengers in fish sperm are protons rather than cAMP or cGMP (42, 43). These results suggest that herring sperm employ a signaling pathway for fertilization that combines signaling proteins and mechanisms from marine invertebrates, zebrafish, and mammals. As more signaling molecules are identified, the herring data strengthen an emerging picture of evolutionary significance: sperm preferentially adopt unique, sperm- and species-specific signaling molecules and pathways that are tuned to their respective habitat of reproduction (35).
Fig. 4.
Characterization of CatSper genes and the Ca2+ signaling pathway in sperm. (A and B) Multiple independent gene losses of CatSper1 to CatSper4 and CatSperB, CatSperG, CatSperD, and CatSperE in fish. Cladogram of major phylogenetic groups based on Betancur et al. (70). Colored branches indicate the presence of at least three of each the CatSper1 to CatSper4 and CatSperB, CatSperG, CatSperD, and CatSperE in all (green for both and blue for CatSper1 to CatSper4 only), none (orange), or some (green and orange for both, blue and orange for CatSper1 to CatSper4 only) species of the clade. Number of species with CatSper genes/all species investigated. Typical members of the respective clade are shown on the right. (A) Full tree including all fish species with searchable genomes. Clade groupings are simplified: early diverging bony fish comprise several groups—Cladistia, Chondrostei, coelacanths, eels, and Holostei. Note the near-complete retention of CatSper in cartilaginous fish and salmon-related fish (Protacanthopterygii and Stomiatii). (B) Presence and absence of CatSper in the Otomorpha clade. Note the almost complete retention of CatSper1 to CatSper4 in Cyprinidae, whereas all three sister clades have lost CatSper1 to CatSper4. Within Clupeiformes, four of five species have retained CatSper. (C) Comparison of signaling pathways in sperm of the sea urchin (Arbacia punctulata), Atlantic herring (C. harengus), and zebrafish (D. rerio). GC, chemoreceptor guanylate cyclase; CNGK, K+-selective cyclic nucleotide-gated channel; sNHE, sperm specific Na+/H+ exchanger SLC9C1; HCN1 and HCN2, hyperpolarization activated and cyclic nucleotide-gated channels; CatSper, sperm-specific Ca2+ channel; NCKX, Na+/Ca2+/K+ exchanger; PMCA, plasma membrane Ca2+-ATPase; HCNL1, HCN-like 1 channel; HCNL2, HCN-like 2 channel.
In summary, we conclude that the herring Y-specific region contains genes encoding two proteins, CatSperG and CatSper3Y, expected to be essential for sperm function based on the functional conservation of the CatSper channel in both invertebrates and vertebrates.
Discussion
BMPR1BBY is a very strong candidate sex-determining factor in the Atlantic herring. It is clearly the best candidate out of the only three genes in the male-specific region because of the well-established role for TGF/BMPR signaling in sexual development (29). Its presence is strongly correlated with the male phenotype, it shows an accelerated protein evolution, and it can replace the AMH/BMPR1B/AMHR pathway for SMAD phosphorylation. The dN/dS ratio of 0.58 for BMPR1BBY is dramatically higher than the dN/dS = 0.01 to 0.02 for the autosomal paralogs (SI Appendix, Fig. S6). However, a dN/dS ratio of 0.58, well below 1.0, may be explained by relaxed purifying selection or a combination of positive selection at some sites and purifying selection at other sites. Our findings that the structural modeling predicts a conserved protein structure as well as its conserved kinase activity strongly suggest that the latter is the correct explanation. An important topic for future studies will be to further characterize the expression pattern of BMPR1BBY and in particular explore if it is expressed during early testis development at the sex determination stage. However, such experiments are challenging in a pelagic fish that does not reproduce in captivity.
The herring Y chromosome provides a unique opportunity to reconstruct the birth of a sex chromosome due to the relatively small size of the male-specific region that is not overloaded with repetitive DNA and other junk material like most other sex chromosomes. The evolutionary process must have started with the emergence of a truncated copy of BMPR1BB that was translocated to chromosome 8 (Fig. 5). The truncation assigned a new protein function to the gene. At the same time, because of the loss of its 5′ end including the promoter and intron 1, BMPR1BBY must have lost the expression regulation of the autosomal gene and most likely acquired a new spatial and temporal expression pattern required to trigger male gonad development. Such transcriptional rewiring of emerging sex-determining genes, in particular if derived from gene duplication of a downstream gonadal factor, has been postulated to be a hallmark of sex chromosome evolution (4, 44).
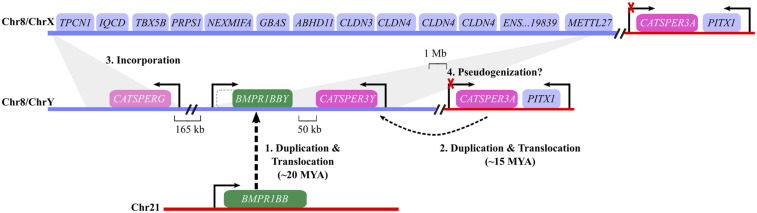
Fig. 5.
Reconstruction of the evolution of sex chromosomes in Atlantic herring. The evolution of ChrY occurred in four steps, but the exact order of steps 2 to 4 is not yet known. 1) Duplication and translocation of BMPR1BB from Chr21 to Chr8. 2) Duplication and translocation of CATSPER3 within Chr8/ChrY. 3) Incorporation of CATSPERG in ChrY and loss from ChrX. 4) CATSPER3A becomes pseudogenized or evolved a new function. MYA, million years ago.
The insertion of the duplicated fragment from chromosome 21 into chromosome 8 created a region of nonhomology that cannot recombine in meiosis. Thus, a proto Y was formed and maintained by suppressed recombination ab initio. It is likely that CATSPER3 and CATSPERG were already colocalized on the ancestral chromosome 8 when BMPR1BBY arrived on the scene because they are located on the same chromosome in many teleosts (SI Appendix, Table S3). In goldfish, the two genes are <1 Mb apart. Possibly the next step was duplication of CATSPER3 and its translocation near BMPR1BBY during or after the duplication event (Fig. 5). The synteny in that region is highly conserved across vertebrates, including CATSPER3 and the homeobox gene PITX1, although CatSper genes have been lost in birds and many teleosts (SI Appendix, Fig. S9, and Fig. 4). Notably, this region is more rearranged in the herring than in other vertebrates (SI Appendix, Fig. S9), suggesting that chromosomal rearrangements contributed to the evolution of the Y-specific region. A third event was the incorporation of CATSPERG into the Y-specific region and its loss from a pseudoautosomal location. Finally, CATSPER3A underwent the fate of most duplicated genes, namely, degeneration, because it was redundant and CATSPER3Y benefited from its tighter linkage to the male sex-determining gene. By this process, a minimal sex-determining region (SDR) has evolved in the Atlantic herring; the region does not recombine with chromosome X and contains only three functional genes, a candidate male-determining factor and two genes essential for sperm function.
It is possible that the SDR in herring as present today is much younger than the emergence of the BMPR1BBY gene about 20 Mya (SI Appendix, Fig. S6), as indicated by the modest sequence divergence in the sex-associated region which rather suggests a divergence time of a few million years (Fig. 1A). However, it is possible that there is ongoing genetic exchange (recombination or gene conversion) between X and Y in this region. In fact, one of the Pacific herring chromosomes included in Fig. 1B may represent a recombinant haplotype because it appears X-like over the BMPR1BBY-CATSPER3Y region but Y-like over CATSPERG. It will be of considerable interest to further explore the evolution of this SDR region in a closely related clupeid like the sprat (Sprattus sprattus).
The association of BMPR1BBY with CATSPER3Y and CATSPERG in the herring SDR is consistent with one of the most characteristic features of sex chromosomes, namely, suppressed recombination between a sex-determining factor and genes beneficial for that sex (11–15). However, the Atlantic herring Y differs from most previously described sex chromosomes as this suppressed recombination has not spread along the chromosome. The region showing strong genetic differentiation between males and females is remarkably small and extends only over ∼300 kb despite the existence of the herring Y for at least a few million years. As a comparison, the Y chromosome in the threespine stickleback, which is less than 26 My old, has a nonrecombining region of 17.5 Mb that shows extensive degeneration (45). Unlike the stickleback Y chromosome, the herring Y does not lack any gene present on the X chromosome; it only carries three additional genes that are likely essential for male development and function. Any degeneration that may have occurred is expected to be restricted to the genes located in the sex-associated region showing sequence divergence between X and Y (SI Appendix, Table S1).
Methods
Whole-Genome Sequence Analyses.
We analyzed whole-genome sequence (WGS) data of 16 individuals with known sex, collected from Atlantic and Baltic with on average ∼12X sequence coverage (19). The reads were aligned to two assemblies of the Atlantic herring genome (17, 19), and high confidence SNPs were called as described (17, 19). A genome-wide association analysis for the detection of sex-associated regions was done by comparing allele frequency between males and females with a χ2 test using read counts extracted from sites called in the older assembly (19). The depth of coverage across the chromosome-based genome assembly (17) was extracted using GATK:DepthOfCoverage (version 3.5.0) (46). The data were converted to normalized depth 1-kb windows, and we compared the depth between males and females. No Animal Care and Use Committee approval was required because the herring were not harvested specifically for research purposes.
Assembly of the Chromosome Y Region.
The assembly of the Y region is based on two unassigned scaffolds (18 and 229) and parts from an earlier version of our current PacBio-assembly of the herring genome (17), which is based on a male Atlantic herring, but only the ChrX region was included in the previously reported assembly (17). The Y assembly presented here was validated by investigating uniformity of depth of coverage of individual PacBio reads across the assembled sequence.
We used eight males and eight females to estimate the genetic differentiation between males and females by Weir–Cockerham FST (47) using vcftools v0.1.16 (48) in 50-kb overlapping windows across chromosome Y.
Annotation of the Male-Specific Region.
We identified male-unique regions on unassigned scaffolds from the current PacBio-based herring assembly which was not annotated by the Ensembl annotation pipeline (https://www.ensembl.org) because the male-unique region was not included in this assembly (17). Thus, we annotated these regions by the annotation pipeline implemented by National Bioinformatics Infrastructure Sweden based on the Maker package (version 3.01.02) (49) (https://github.com/NBISweden/GAAS, version 2019-07-11). We first generated an evidence-built annotation based on protein similarity using Uniprot (50) (downloaded August 11, 2016). We also generated an ab initio built annotation by first training Augustus (version 2.7) (51) using 1,213 genes annotated by Ensembl (17). Then we combined the two annotations to generate the final annotation. We manually curated BMPR1BBY by comparing it with the autosomal copy on chromosome 21 and generated the final gene structure. In addition, we used GenScan (52) to do another round of ab initio annotation which was merged with the Maker annotation.
To validate the structure of the gene model we conducted 3′ RACE and Nanopore whole-transcriptome and gene-specific cDNA sequencing. Total RNA was prepared from the testis of an adult Atlantic herring using the RNeasy Mini Kit (Qiagen). The 3′ RACE reaction was performed with 1 μg RNA using the FirstChoiceTM RLM-RACE Kit (Thermo Fisher Scientific). Nested RACE PCRs were performed in a 25-μL reaction containing KAPA HiFi Fidelity Buffer with MgCl2, 0.32 mM dNTPs, 0.4 μM each of the forward and reverse primer, 0.8 U KAPA HiFi DNA Polymerase (Kapa Biosystems) and 1 μL of the cDNA or Outer RACE PCR product (1:5 dilution) as PCR template. Amplification was carried out following the program: 95 °C for 5 min, 35 cycles of 98 °C for 20 s, 58 °C for 30 s and 72 °C for 1 min, and a final extension of 10 min at 72 °C.
Targeted Resequencing Using Xdrop.
To confirm the orientation of the male-specific region in relation to the flanking pseudoautosomal region we used the Xdrop method for targeted sequencing of long DNA molecules (20). Primer pairs for the amplification of fragments from the male-specific region in the near vicinity of the border to the pseudoautosomal region were designed. The following primer pairs were used to amplify a fragment in the vicinity of CATSPER3Y: for dPCR, Fw-TGCACTAGTGGGTGACTACCAA and R- TGTAAGACACCAGAATTTCCCCAC (product size 173 bp, Tm = 76.25 ± 0.08 °C), and for qPCR, Fw-TACTCAGATGGCCGACGAAATCTTTA and R-GCGTCAAAGACGTTATATCCGC (product size 111 bp, Tm = 77.88 ± 0.11 °C). For the amplicon from the scaffold18 region, the following primers were used: for dPCR, Fw-ACCGGCTTTATCCATCCGTC and R- AGTGACGAAAACGAAACCGTC (product length 165 bp, Tm = 76.97 ± 0.08 °C), and for qPCR, Fw:CTTCGGATCTGCACGATTCAC and R-TCAGAAGACCGGAGACTGGA (product length 127, Tm = 80.98 ± 0.11 °C). The reactions were run in a final volume of 10 μL, of which 5 μL were of dPCR mix, 0.5 μL of qPCR dye, 0.4 μL of 10 μM primer pair, and 0.1 μL of water. The enrichment and Nanopore sequencing of the genomic regions was performed by Samplix services (https://samplix.com/).
Genotyping Sex-Specific Markers.
We genotyped 200 individuals for the presence/absence of BMPR1BBY. These samples had accurate sex records and were collected from two localities, Björköfjärden, Kattegat and Brofjorden, Skagerrak (19). The following primer pair was designed using primer3 with default settings, bby-fw CCAGACTGACTTGTGGTTCC and bby-R CTTCCCAGCAACAAGATGG, giving a 132-bp product with a melting temperature of 85.89 ± 0.07 °C (average and SD, respectively). The PCR reactions were composed of 5 μL SYBR Green PCR Master Mix (Applied Biosystems by Thermo Fisher Scientific), 1 μL of primer pair working solution at a concentration of 4 μM, and 4 μL of the DNA samples at a final concentration of 1 ng per reaction. Control reactions with no template were included at all points to spot any primer dimer formation. PCR was done using a QuantStudio 6 Flex instrument (Applied Biosystems by Thermo Fisher Scientific) with the thermal profile 2 min at 50 °C and 2 min at 95 °C, followed by 40 cycles of 15 s at 95 °C, 30 s at 57 °C, and 30 s at 72 °C. Melting curves were generated with incubation for 15 s at 95 °C, 1 min at 60 °C, and 15 s at 95 °C. The ramp rate between the two last steps was 0.05 °C/s. Primer sequences are also in SI Appendix, Table S4.
Nanopore Sequencing.
Total RNA (500 ng) from testis was converted into cDNA following the Oxford Nanopore Technologies (ONT) protocol for rapid cDNA sequencing with the SuperScript IV reverse transcriptase (Invitrogen). To improve yield of full-length cDNA the incubation times were extended to 50 min at 50 °C and 20 min at 42 °C. The cDNA was amplified with LongAmp polymerase (NEB) and click primers (cPRM, ONT) for 14 cycles with 6 min elongation. Sequencing was done as partial runs on R9.4 and R9.5 flow cells as 1D reads. BMPR1BBY and CATSPERG were amplified from testis cDNA with LongAmp and internal primers and in combination with the ONT forward and reverse adaptor primers in order to capture the full transcript. PCR products were sequenced using the LSK309 1d2 ligation sequencing protocol (ONT).
Genomic Alignments.
Similarity between CATSPER3Y and BMPR1BBY and their presumed homologous autosomal loci was assessed by performing local alignments of the genomic nucleotide sequence, including flanking regions, of the two Y-specific gene copies against genomic sequence fragments, extracted from assembly version Ch_v2.0.2 (GCA_900700415.1) that covered the potentially orthologous autosomal genes. The targets were identified from the Ensembl-provided annotation of the herring genome (Database version 99.202). The alignments were performed using Clustal Omega version 1.2.3 (53) using default parameters and analyzed in R (54) using the package “ape” version 5.3 (55).
We compared the gene content in the sex-associated region on X and Y using Clustal Omega version 1.2.3 (53) implemented in “ape” R package version 5.3 (55) as described above.
Phylogenetic Analysis by Maximum Likelihood (PAML).
First, we collected a reference set of CATSPER3 and BMPR1BB genes from the ENA database. After manual inspection of the possible transcripts, we retained the option for each species that most closely resembled the herring Y copy. Second, in-frame nucleotide alignments spanning the extent of the Y-specific copies were generated for both genes. For BMPR1BB, which is highly conserved, the nucleotide alignments were already in frame, as verified by the “trans” function from “ape” and subsequent comparison with the reference sequences. For CATSPER3, which is more variable, PAL2NAL (56) was used to translate the amino acid alignment to a nucleotide equivalent using the reference cDNA sequences. In both cases, ambiguous positions were discarded during the rate analysis. Next, we extracted the taxa represented in each alignment from the Fish Tree of Life (57) to generate unrooted guide trees for the PAML analysis. These trees were edited to split the herring entry into a Y-specific and autosomal copy, and for CATSPER3, the coelacanth was added as an additional unresolved branch. Finally, the alignments and guide trees were fed into the “codeml” module of PAML version 4.9 (58) and run under three different models: 1) All branches evolve at the same rate (NULL model). 2) Only the “herring Y-” (for BMPR1BB) or the “herring A-” (for CATSPER3) branch has a unique rate (two-rates model). 3) Both the “herring Y” and “herring A” branches have a unique rate (three-rates model).
The models were then compared using a likelihood ratio test statistic, which was evaluated against a χ2-distribution with degrees of freedom equal to the difference in estimated parameters between the compared models. The PAML control files, guide trees, and alignments used are found at https://github.com/nimarafati/Atlantic_herring_SDR.
We used dS values from the PAML analysis to estimate divergence times by extracting fossil-calibrated LCA estimates from the Fish Tree of Life (57) and dividing the time interval proportionally to the lengths of the terminal and internal branches.
Structure Modeling.
Structural prediction of the intracellular domain of BMPR1BB, full-length L-BMPR1BBY, and full-length S-BMPR1BBY was performed using the RaptorX server (59), which gave the best models using Protein Data Bank 3MDY (human BMPR1B) (60) as the template. Structure alignments and protein representations were generated using the PyMOL Molecular Graphics System (Version 1.3, Schrödinger, LLC).
Preparation and Mass Spectrometric Analysis of Flagella from Herring Sperm.
The preparation of flagella from C. harengus sperm was as described (39) using Herring Ringer’s (HR) solution instead of artificial seawater. Briefly, sperm were washed two times with HR (in mM: 206 NaCl, 7.2 KCl, 2.1 CaCl2, 3.1 MgCl2, 10 Hepes/NaOH, pH 7.6) (61). Hepes was used for buffering instead of NaHCO3. Sperm suspension was sheared 20 times by centrifugation for 30 s at 75 × g and 4 °C through a 40-µm mesh of a cell strainer (BD Biosciences). Flagella were washed in HR and stored as pellet.
Flagella protein (20 µg) was subjected to GeLCMS as described (62) with the following modifications: excision of 10 gel slices, use of Proteome Discoverer Rev. 2.2, C. harengus protein sequences from National Center for Biotechnology (NCBI) release GCF_900700415.1 (date April 16, 2019) supplemented with additional sequences (Dataset S4), recalibration of precursor MS spectra, precursor mass error tolerance 8 ppm, MS/MS mass error tolerance 0.6 Da, oxidation of Met, phosphorylation at Ser, Thr, Tyr, and N-terminal pyro-Glu as variable, Cys-Propionamide as static modification.
Bioinformatic Analysis of CatSper Genes.
All fish genomes available from NCBI (https://www.ncbi.nlm.nih.gov/genome/) as of March 11, 2020, were analyzed for the presence of highly conserved β actin as positive control. The whole-genome shotgun databases (WGS) for these genomes were searched with TBlastN (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=tblastn) for β actin, using the Danio rerio (zebrafish) sequence as query. Genomes which were unsearchable or yielded poor hits were excluded. The research resulted in 271 genomes, which were evaluated for the presence of CatSper1 to CatSper4 and CatSperB, CatSperG, CatSperD, and CatSperE. We then performed TBlastN searches in the WGS databases of fish genomes using previously annotated protein sequences for CatSper1 to CatSper4 and CatSperB, CatSperG, CatSperD, and CatSperE of Lepisosteus occulatus (spotted gar), Latimeria chalumnae (coelacanth), Callorhinchus milii (elephant shark), Salmo salar (Atlantic salmon), Esox lucius (northern pike), and Carassius auratus (goldfish). The search was recursive, whenever more closely related sequences were identified. Candidates were evaluated according to e value until outgroup genes were found; a rigid cutoff by e value was ineffective. Candidate genes were predicted by genewise (https://www.ebi.ac.uk/Tools/psa/genewise/) (63) using closely related validated sequences as template. To validate candidates, we employed phylogenetic tree analysis. Trees were constructed from the candidate genes, the query sequences and other validated CatSper genes (e.g., Mus musculus [mouse], for full list, see Dataset S2). For CatSper1 to CatSper4, representative members from related ion channel families were used as outgroup (CACNA and SCN channels of mouse and zebrafish, TPCN and TRP channels of zebrafish). CatSperB, CatSperG, CatSperD, and CatSperE served as each other’s respective outgroup. Sequences were aligned using the MAFFT (multiple alignment using fast Fourier transform) tool version 7 (https://mafft.cbrc.jp/alignment/server/) (64, 65) with the iterative refinement strategy called E-INS-I strategy and otherwise default parameters. Gap Strip Squeeze (https://hcv.lanl.gov/content/sequence/GAPSTREEZE/gap.html) was used to remove regions of the multiple sequence alignments which contained gaps in more than 90% of the sequences. Trees were subsequently calculated using the PhyML online tool, which is a phylogenetic tool that employs a maximum likelihood (ML) algorithm (http://www.atgc-montpellier.fr/phyml/) (66). Tree improvement was set to SPR (subtree pruning and regrafting), and branch support was set to χ2-based aLRT (approximate likelihood ratio test). Otherwise the default settings were unchanged. Branch support for single CatSper1 to CatSper4 clades was always maximal; thus, CatSper1 to CatSper4 candidates that fell into these clades were considered validated. Branch support for outgroup clades (CACNA, SCN, and TRP) was maximal, and candidates which were sorted to the outgroup genes were discarded. For CatSperB, CatSperG, CatSperD, and CatSperE, branch support was always close to maximal; thus, candidates falling into these groups were considered validated.
RNA Preparation and Expression Analysis.
Total RNA was extracted from the gonad, heart, spleen, kidney, gills, intestine, hypothalamus and saccus vasculosus (BSH), and brain without BSH (brain) of three adult male and three female Atlantic herring using the RNeasy Mini Kit (Qiagen). RNA was reverse transcribed into cDNA with a High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific). Real-Time PCR with SYBR Green chemistry was performed for BMPR1BB, CATSPER3Y, CATSPERG, AMH, and AMHR2 in 10 μL reactions of SYBR Green PCR Master Mix (Thermo Fisher Scientific), 0.3 μM primers, and cDNA template, with a program of an initial denaturation of 10 min at 95 °C followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. TaqMan Gene Expression assay (Thermo Fisher Scientific) containing 0.30 μM primers and 0.25 μM BMPR1BBY TaqMan probe (Integrated DNA Technologies) was performed to compare the relative expression levels of BMPR1BBY. ACTIN was used as the housekeeping gene in all expression assays. All of the primers and probes are listed in SI Appendix, Table S4.
The expression of CATSPER3A in gonads and brain was investigated by RT-PCR. Briefly, cDNA was produced with the High Capacity cDNA Synthesis kit (Thermo Fisher Scientific), without RNase inhibitor. As this kit does not contain any oligo-dT primer, that was added to the reaction master mix to a final concentration of 5 mM [Oligo(dT)18 primer; Thermo Fisher Scientific]. Primer pairs were designed with the NCBI tool (SI Appendix, Table S4). RT-PCRs were run with the AmpliTaq Gold DNA Polymerase with Buffer II and MgCl2 (Thermo Fisher Scientific) together with dNTPs (dNTP Mix, 5 mM each) at a final concentration of 200 mM each. Primer pairs were added to a final concentration of 0.4 mM each, and the working concentration of DNA polymerase was 2 U/reaction. The total sample amount was 40 ng in a final volume of 10 μL. The reactions were incubated according to the following profile: 95 °C for 10 min, 35 cycles of 95 °C for 15 s, 57 °C for 30 s, 72 °C for 1 min, followed by 72 °C for 5 min. All reactions were checked by gel electrophoresis. No detectable amplification was observed for either primer pair.
Transfection Experiments.
Full-length clones for AMH, AMHR2, and autosomal BMPR1BB as well as L-BMPR1BBY and S-BMPR1BBY clones were designed including a fish consensus Kozak sequence (67), synthesized and cloned (GenScript) into the expression vector pcDNA3.1 (-) (Thermo Fisher Scientific).
Medaka spermatogonial stem cells (Sg3) were cultured as previously described (68). For transfection, cells were grown to 80% confluency in six-well plates and subsequently transfected with 5 µg expression vectors using the FuGene (Roche) reagent as described by the manufacturer.
For monitoring differential activation of Smads upon selective AMH, AMHR2, BMPR1B, or S- or L-BMPR1BBY expression, Sg3 cells were seeded in six-well plates and cotransfected with a combination of four kinds of plasmids (Fig. 3A): 1) an expression plasmid which encodes a fusion protein (31, 69) (Smad1-AD-Gal4-DBD or Smad5-AD-Gal4-DBD; 300 ng per well) that will translocate to the nucleus upon phosphorylation by the different TGFßs and in return transactivate the UAS-4 promoter through its Gal-4 DNA binding domain; 2) a reporter plasmid, which codes for luciferase under the control of a minimal promoter, which contains UAS sequences (UAS-luc, Firefly luciferase; 300 ng per well); 3) plasmids coding for the different ligands and receptors to be tested for signaling activity [either pcDNA3.1(-)-AMH, pcDNA3.1(-)-AMHR2, pcDNA3.1(-)-BMPR1BB, pcDNA3.1(-)-S-BMPR1BY, pcDNA3.1(-)-L-BMPR1B or a control plasmid (empty pcDNA3.1(-)); 400 ng per well]; and 4) a Gaussia luciferase expression plasmid for normalization (pCMV-GLuc; 5 ng per well). After 24 h, cells were washed with 2× PBS solution and lysed with 75 µL of passive lysis buffer (Dual Luciferase Reporter Kit Assay; Promega) and then subjected to luciferase assay. Firefly luciferase activity (UAS-luc reporter constructs) was quantified using the Dual Luciferase Reporter Assay System (Promega) and normalized against cotransfected Gaussia luciferase expressing plasmid. Datasets are the results of at least four independent cell transfections and luciferase measurements. Statistical significance was assessed by means of the Wilcoxon–Mann–Whitney test (n = 4 or 8). Raw data are in Dataset S5.
Supplementary Material
Acknowledgments
We thank Carl-Henrik Heldin and Aristidis Moustakas for valuable comments and Chunheng Mo for technical assistance. The study was supported by the Knut and Alice Wallenberg Foundation (to L.A.), Vetenskapsrådet (to L.A.), Research Council of Norway project 254774, GENSINC (to A.F., F.B. and L.A.), and AquaCRISPR (ANR-16-COFA-0004-01) (to A.H.). The National Genomics Infrastructure (NGI)/Uppsala Genome Center and UPPMAX (Uppsala Multidisciplinary Center for Advanced Computational Science) provided service in massive parallel sequencing and computational infrastructure. Work performed at NGI/Uppsala Genome Center has been funded by RFI/VR (Council for Research Infrastructure/Vetenskapsrådet) and Science for Life Laboratory, Sweden.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2009925117/-/DCSupplemental.
Data Availability.
The reference genome used in this study is deposited in European Nucleotide Archive (ENA) (PRJEB31270) and the sequence of the SDR together with its annotation as well as other sequences and codes used in this study are available on Github: https://github.com/nimarafati/Atlantic_herring_SDR. Variant calls used in the χ2-analysis were from Ref. 19 deposited in Dryad (5r774). Also, Samplix and cDNA data generated for this study are deposited in ENA (PRJEB38031)
References
- 1.Bachtrog D. et al.; Tree of Sex Consortium , Sex determination: Why so many ways of doing it? PLoS Biol. 12, e1001899 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Charlesworth D., Mank J. E., The birds and the bees and the flowers and the trees: Lessons from genetic mapping of sex determination in plants and animals. Genetics 186, 9–31 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Doorn G. S., Evolutionary transitions between sex-determining mechanisms: A review of theory. Sex Dev. 8, 7–19 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Herpin A., Schartl M., Plasticity of gene-regulatory networks controlling sex determination: Of masters, slaves, usual suspects, newcomers, and usurpators. EMBO Rep. 16, 1260–1274 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schartl M., Galiana-Arnoux D., Schultheis C., Böhne A., Volff J.-N., . “A primer of sex determination in poeciliids” in Ecology and Evolution of Poeciliid Fishes, Pilastro A., Evans J., Schlupp I., Eds. (The University of Chicago Press, 2010), pp. 264–275. [Google Scholar]
- 6.Matsuda M., Sakaizumi M., Evolution of the sex-determining gene in the teleostean genus Oryzias. Gen. Comp. Endocrinol. 239, 80–88 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Gammerdinger W. J., Kocher T. D., Unusual diversity of sex chromosomes in African cichlid fishes. Genes (Basel) 9, 480 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pennell M. W., Mank J. E., Peichel C. L., Transitions in sex determination and sex chromosomes across vertebrate species. Mol. Ecol. 27, 3950–3963 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Devlin R. H., Nagahama Y., Sex determination and sex differentiation in fish: An overview of genetic, physiological, and environmental influences. Aquaculture 208, 191–364 (2002). [Google Scholar]
- 10.Schartl M., Herpin A., “Sex determination in vertebrates” in Encyclopedia of Reproduction, Skinner M. K., Ed., (Elsevier, 2018), pp. 159–167. [Google Scholar]
- 11.Fisher R. A., The evolution of dominance. Biol. Rev. Camb. Philos. Soc. 6, 345–368 (1931). [Google Scholar]
- 12.Bull J. J., Evolution of Sex Determining Mechanisms, (Benjamin/Cummings, Menlo Park, CA, 1983). [Google Scholar]
- 13.Rice W. R., The accumulation of sexually antagonistic genes as a selective agent promoting the evolution of reduced recombination between primitive sex-chromosomes. Evolution 41, 911–914 (1987). [DOI] [PubMed] [Google Scholar]
- 14.Charlesworth D., Charlesworth B., Marais G., Steps in the evolution of heteromorphic sex chromosomes. Heredity 95, 118–128 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Kirkpatrick M., The evolution of genome structure by natural and sexual selection. J. Hered. 108, 3–11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ida H., Oka N., Hayashigaki K., Karyotypes and cellular DNA contents of three species of the subfamily Clupeinae. J. Ichthyol. 38, 289–294 (1991). [Google Scholar]
- 17.Pettersson M. E. et al., A chromosome-level assembly of the Atlantic herring genome-detection of a supergene and other signals of selection. Genome Res. 29, 1919–1928 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lamichhaney S. et al., Parallel adaptive evolution of geographically distant herring populations on both sides of the North Atlantic Ocean. Proc. Natl. Acad. Sci. U.S.A. 114, E3452–E3461 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martinez Barrio A. et al., The genetic basis for ecological adaptation of the Atlantic herring revealed by genome sequencing. eLife 5, e12081 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Madsen E. B., Kvist T., Höijer I., Ameur A., Mikkelsen M. J., Xdrop: Targeted sequencing of long DNA molecules from low input samples using droplet sorting bioRxiv: 10.1101/409086 (6 September 2018). [DOI] [PMC free article] [PubMed]
- 21.Ren D. et al., A sperm ion channel required for sperm motility and male fertility. Nature 413, 603–609 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seifert R. et al., The CatSper channel controls chemosensation in sea urchin sperm. EMBO J. 34, 379–392 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brenker C. et al., Action of steroids and plant triterpenoids on CatSper Ca2+ channels in human sperm. Proc. Natl. Acad. Sci. U.S.A. 115, E344–E346 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith J. F. et al., Disruption of the principal, progesterone-activated sperm Ca2+ channel in a CatSper2-deficient infertile patient. Proc. Natl. Acad. Sci. U.S.A. 110, 6823–6828 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hildebrand M. S. et al., Genetic male infertility and mutation of CATSPER ion channels. Eur. J. Hum. Genet. 18, 1178–1184 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pan Q. et al., Identification of the master sex determining gene in Northern pike (Esox lucius) reveals restricted sex chromosome differentiation. PLoS Genet. 15, e1008013 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herpin A., Lelong C., Favrel P., Transforming growth factor-β-related proteins: An ancestral and widespread superfamily of cytokines in metazoans. Dev. Comp. Immunol. 28, 461–485 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Mawaribuchi S., Yoshimoto S., Ohashi S., Takamatsu N., Ito M., Molecular evolution of vertebrate sex-determining genes. Chromosome Res. 20, 139–151 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adolfi M. C., Nakajima R. T., Nóbrega R. H., Schartl M., Intersex, hermaphroditism, and gonadal plasticity in vertebrates: Evolution of the Müllerian duct and Amh/Amhr2 signaling. Annu. Rev. Anim. Biosci. 7, 149–172 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Visser J. A., AMH signaling: From receptor to target gene. Mol. Cell. Endocrinol. 211, 65–73 (2003). [DOI] [PubMed] [Google Scholar]
- 31.Sèdes L. et al., Anti-Müllerian hormone recruits BMPR-IA in immature granulosa cells. PLoS One 8, e81551 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cai X., Clapham D. E., Evolutionary genomics reveals lineage-specific gene loss and rapid evolution of a sperm-specific ion channel complex: CatSpers and CatSperbeta. PLoS One 3, e3569 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chung J.-J. et al., CatSperζ regulates the structural continuity of sperm Ca2+ signaling domains and is required for normal fertility. eLife 6, e23082 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wachten D., Jikeli J. F., Kaupp U. B., Sperm sensory signaling. Cold Spring Harb. Perspect. Biol. 9, a028225 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaupp U. B., Strünker T., Signaling in sperm: More different than similar. Trends Cell Biol. 27, 101–109 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Lishko P. V. et al., The control of male fertility by spermatozoan ion channels. Annu. Rev. Physiol. 74, 453–475 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bönigk W. et al., An atypical CNG channel activated by a single cGMP molecule controls sperm chemotaxis. Sci. Signal. 2, ra68 (2009). [DOI] [PubMed] [Google Scholar]
- 38.Strünker T. et al., A K+-selective cGMP-gated ion channel controls chemosensation of sperm. Nat. Cell Biol. 8, 1149–1154 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Trötschel C. et al., Absolute proteomic quantification reveals design principles of sperm flagellar chemosensation. EMBO J. 39, e102723 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gauss R., Seifert R., Kaupp U. B., Molecular identification of a hyperpolarization-activated channel in sea urchin sperm. Nature 393, 583–587 (1998). [DOI] [PubMed] [Google Scholar]
- 41.Windler F. et al., The solute carrier SLC9C1 is a Na+/H+-exchanger gated by an S4-type voltage-sensor and cyclic-nucleotide binding. Nat. Commun. 9, 2809 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wobig L. et al., A family of hyperpolarization-activated channels selective for protons. Proc. Natl. Acad. Sci. U.S.A. 117, 13783–13791 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fechner S. et al., A K(+)-selective CNG channel orchestrates Ca(2+) signalling in zebrafish sperm. eLife 4, e07624 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Herpin A. et al., Transcriptional rewiring of the sex determining dmrt1 gene duplicate by transposable elements. PLoS Genet. 6, e1000844 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peichel C. L. et al., Assembly of the threespine stickleback Y chromosome reveals convergent signatures of sex chromosome evolution. Genome Biol. 21, 177 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McKenna A. et al., The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weir B. S., Cockerham C. C., Estimating F-statistics for the analysis of population structure. Evolution 38, 1358–1370 (1984). [DOI] [PubMed] [Google Scholar]
- 48.Danecek P. et al.; 1000 Genomes Project Analysis Group , The variant call format and VCFtools. Bioinformatics 27, 2156–2158 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cantarel B. L. et al., MAKER: An easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 18, 188–196 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Magrane M.; UniProt Consortium , UniProt knowledgebase: A hub of integrated protein data. Database (Oxford) 2011, bar009 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stanke M., Diekhans M., Baertsch R., Haussler D., Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 24, 637–644 (2008). [DOI] [PubMed] [Google Scholar]
- 52.Burge C., Karlin S., Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268, 78–94 (1997). [DOI] [PubMed] [Google Scholar]
- 53.Sievers F. et al., Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Team RC , R: A Language and Environment for Statistical Computing, (R Foundation for Statistical Computing, Vienna, Austria, 2019). [Google Scholar]
- 55.Paradis E., Schliep K., ape 5.0: An environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 35, 526–528 (2019). [DOI] [PubMed] [Google Scholar]
- 56.Suyama M., Torrents D., Bork P., PAL2NAL: Robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34, W609–W612 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rabosky D. L. et al., An inverse latitudinal gradient in speciation rate for marine fishes. Nature 559, 392–395 (2018). [DOI] [PubMed] [Google Scholar]
- 58.Yang Z., PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591 (2007). [DOI] [PubMed] [Google Scholar]
- 59.Källberg M. et al., Template-based protein structure modeling using the RaptorX web server. Nat. Protoc. 7, 1511–1522 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chaikuad A., et al. , Crystal structure of the cytoplasmic domain of the bone morphogenetic protein receptor type-1B (BMPR1B) in complex with FKBP12 and LDN-193189. Protein Data Bank. https://www.rcsb.org/structure/3MDY (accessed 3 February 2020).
- 61.Vines C. A. et al., Motility initiation in herring sperm is regulated by reverse sodium-calcium exchange. Proc. Natl. Acad. Sci. U.S.A. 99, 2026–2031 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Raju D. et al., Accumulation of glucosylceramide in the absence of the beta-glucosidase GBA2 alters cytoskeletal dynamics. PLoS Genet. 11, e1005063 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Birney E., Clamp M., Durbin R., GeneWise and Genomewise. Genome Res. 14, 988–995 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Katoh K., Misawa K., Kuma K., Miyata T., MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Katoh K., Standley D. M., MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Guindon S. et al., New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321 (2010). [DOI] [PubMed] [Google Scholar]
- 67.Grzegorski S. J., Chiari E. F., Robbins A., Kish P. E., Kahana A., Natural variability of Kozak sequences correlates with function in a zebrafish model. PLoS One 9, e108475 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hong Y. et al., Establishment of a normal medakafish spermatogonial cell line capable of sperm production in vitro. Proc. Natl. Acad. Sci. U.S.A. 101, 8011–8016 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu F. et al., A human Mad protein acting as a BMP-regulated transcriptional activator. Nature 381, 620–623 (1996). [DOI] [PubMed] [Google Scholar]
- 70.Betancur-R R. et al., The Tree of Life and a New Classification of Bony Fishes, (PLOS Currents Tree of Life, ed. 1, 2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The reference genome used in this study is deposited in European Nucleotide Archive (ENA) (PRJEB31270) and the sequence of the SDR together with its annotation as well as other sequences and codes used in this study are available on Github: https://github.com/nimarafati/Atlantic_herring_SDR. Variant calls used in the χ2-analysis were from Ref. 19 deposited in Dryad (5r774). Also, Samplix and cDNA data generated for this study are deposited in ENA (PRJEB38031)