Significance
So far, no targeted therapy has been approved for KRAS mutant tumors. We report that combined genetic inactivation of CDK4 and RAF1 in advanced KRAS/p53 mutant lung tumors leads to effective tumor regression without inducing significant toxicities. In spite of this therapeutic response, CDK4/RAF1-resistant cells appeared. We have identified and pharmacologically validated two independent resistance mechanisms involving hypermethylation of tumor-suppressor genes and activation of the PI3K pathway.
Keywords: KRAS, CDK4/RAF1 inhibition, Tumor Regression, Lung Cancer, Resistance Mechanisms
Abstract
KRAS mutant lung adenocarcinomas remain intractable for targeted therapies. Genetic interrogation of KRAS downstream effectors, including the MAPK pathway and the interphase CDKs, identified CDK4 and RAF1 as the only targets whose genetic inactivation induces therapeutic responses without causing unacceptable toxicities. Concomitant CDK4 inactivation and RAF1 ablation prevented tumor progression and induced complete regression in 25% of KRAS/p53-driven advanced lung tumors, yet a significant percentage of those tumors that underwent partial regression retained a population of CDK4/RAF1-resistant cells. Characterization of these cells revealed two independent resistance mechanisms implicating hypermethylation of several tumor suppressors and increased PI3K activity. Importantly, these CDK4/RAF1-resistant cells can be pharmacologically controlled. These studies open the door to new therapeutic strategies to treat KRAS mutant lung cancer, including resistant tumors.
Despite intense research efforts, KRAS mutant tumors remain mostly intractable for targeted therapies. Clinical development of selective KRASG12C inhibitors show significant promise for lung tumors that carry this mutation (1, 2). However, the remaining KRAS oncoproteins are still undruggable (3). On the other hand, most KRAS effectors, including the members of the MAPK pathway, are druggable kinases for which there is a wide spectrum of selective inhibitors. Genetic studies using genetically engineered mice have revealed that ablation of the MEK and ERK families of kinases results in the rapid death of adult mice (4). These observations may explain why none of the MAPK inhibitors have been so far approved for the treatment of KRAS mutant tumors due to their unacceptable toxicities (5). Among them, MEK1/2 inhibitors have attracted the most attention, with some reaching phase III clinical trials. However, in the SELECT-1 trial, addition of selumetinib to docetaxel did not improve progression-free survival in advanced KRAS mutant lung cancer patients at tolerated doses (6). A second MEK1/2 inhibitor, trametinib, has been approved for B-RAFV600E mutant melanoma in combination with dabrafenib (7). However, in a randomized phase II study (GSK1120212) with KRAS mutant lung tumors, trametinib was not superior to docetaxel as a single agent (8). Moreover, grade 4 toxicity events only occurred in the trametinib arm (8). These observations raise the possibility that the potential benefit derived from tampering with MAPK pathway signaling in KRAS mutant tumors might be limited by toxicity in a manner not too different from that observed with classic cytotoxics known to block essential cellular functions. Although several ERK1/2 and RAF inhibitors have also entered clinical trials in recent years, none of them have so far advanced beyond phase II, either as single agents or in combination with other drugs (3).
Genetic interrogation of each of the individual members of the RAF family of kinases has established that they are not essential for adult homeostasis. More importantly, ablation of RAF1, but not B-RAF, in Kras/Trp53 tumor-bearing animals results in significant therapeutic activity (9). Surprisingly, RAF1 ablation does not affect MAPK signaling neither in normal or tumor tissue, a property likely to explain the absence of significant toxicities (4, 9), yet the therapeutic impact observed upon genetic ablation of RAF1 is limited. Although two-thirds of the Kras/Trp53-driven lung tumors underwent partial regression, only 10% of them underwent complete regression. Therefore, the potential clinical use of forthcoming RAF1-selective inhibitors will require combination therapies.
One of the main downstream effectors of MAPK signaling are the cyclin-dependent kinases (CDKs). Previous studies have illustrated that genetic ablation or pharmacological inhibition of CDK4 results in limited therapeutic activity against Kras-driven lung adenocarcinomas without inducing detectable toxicities (10). In this study, we have interrogated the therapeutic consequences of combining CDK4 inhibition with RAF1 ablation in advanced Kras/Trp53-driven lung adenocarcinomas.
Results
Genetic Inhibition of CDK4 Kinase Activity in Established KRAS-Driven Tumors Induce Limited Tumor Regression.
Genetic interrogation of the role of potential therapeutic targets in either tumor development or progression has been carried out using knockout strategies, yet target ablation does not mimic pharmacological inhibition. Hence, to evaluate the therapeutic effect of inhibiting CDK4, we designed a Cdk4 conditional knocked-in allele, Cdk4FxKD, which upon Cre-mediated recombination generates a Cdk4KD allele that expresses a kinase dead isoform, CDK4KD, in which the K35 residue responsible for ATP binding was substituted by a methionine residue (SI Appendix, Fig. S1A). The lack of catalytic activity of CDK4KD was confirmed using a baculovirus expression system in the presence of CYCLIN D1 (SI Appendix, Fig. S1B). Western blot analysis of mouse embryonic fibroblasts carrying the recombined Cdk4KD allele demonstrated that the levels of expression of the CDK4KD isoform are similar to those of the endogenous CDK4 protein (SI Appendix, Fig. S1C). Unfortunately, the unrecombined Cdk4FxKD allele does not express CDK4 (SI Appendix, Fig. S1C). Therefore, induction of the genetic switch that replaces the wild-type CDK4 protein by the CDK4KD kinase dead isoform required a double heterozygous strategy involving the Cdk4FxKD allele described above and Cdk4L, a conditional floxed allele (11). As illustrated in SI Appendix, Fig. S1A, Cre-mediated recombination of these alleles led to the generation of the Cdk4KD allele that encoded the CDK4KD isoform and a Cdk4– allele that eliminated expression of the wild-type CDK4 protein.
Cdk4FxKD and Cdk4L alleles were incorporated to a “therapeutic strain,” Kras+/FSFG12V;Trp53F/F;hUBC-CreERT2+/T, in which expression of KRASG12V and ablation of p53 is mediated by infection with adenoviral particles expressing the Flp(o) recombinase (9). Activation of the inducible CreERT2 recombinase, encoded by the hUBC-CreERT2 transgene (12), was achieved by exposing mice to a tamoxifen (TX)-containing diet. As indicated above, Cre-mediated recombination of Cdk4FxKD and Cdk4L into Cdk4KD and Cdk4– alleles resulted in the replacement of the wild-type CDK4 protein by the kinase dead CDK4KD isoform (SI Appendix, Fig. S1A). This genetic switch faithfully mimics a scenario in which a putative CDK4 inhibitor will block CDK4 kinase activity without affecting other CDKs or causing off-target effects. Tumor extracts were submitted to Western blot analysis to confirm proper recombination of the Cdk4FxKD and Cdk4L alleles (SI Appendix, Fig. S1D). The limited number of tumors that retained expression of the wild-type CDK4 protein were not included in the waterfall plot depicted in Fig. 1A.
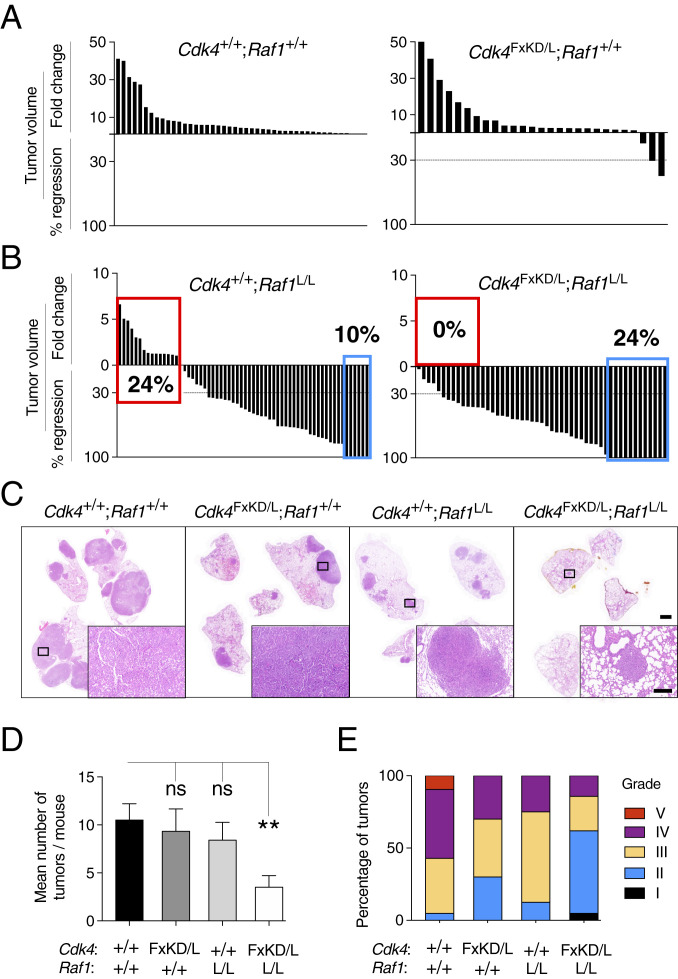
Fig. 1.
Concomitant inhibition of CDK4 and RAF1 ablation results in regression of all lung adenocarcinomas. (A) Waterfall plot representing the increase in tumor volume (fold-change) and the percentage of tumor regression as determined by CT scans performed at the beginning and end of the trial of individual lung tumors present in Kras+/FSFG12V;Trp53F/F;hUBC-CreERT2+/T;Cdk4+/+;Raf1+/+ (n = 14 mice/45 tumors) and Kras+/FSFG12V;Trp53F/F;hUBC-CreERT2+/T;Cdk4FxKD/L;Raf1+/+ (n = 16 mice/27 tumors) exposed to TX for 9 wk. (B) Waterfall plot representing the increase in tumor volume (fold-change) and the percentage of tumor regression as determined by CT scans performed at the beginning and end of the trial of individual lung tumors present in Kras+/FSFG12V;Trp53F/F;hUBC-CreERT2+/T;Cdk4+/+;Raf1L/L (n = 12 mice/62 tumors) and Kras+/FSFG12V;Trp53F/F;hUBC-CreERT2+/T;Cdk4FxKD/L;Raf1L/L (n = 19 mice/51 tumors) exposed to TX for 9 wk. (C) H&E staining of paraffin sections of whole lungs obtained from representative tumor-bearing Kras+/FSFG12V;Trp53F/F;hUBC-CreERT2+/T mice harboring the indicated alleles after 9 wk of TX exposure. (Scale bars, 2 [lower magnification] and 0.2 mm [inset].) (D) Quantification of the mean number of tumors per mouse as determined by postmortem analysis in Kras+/FSFG12V;Trp53F/F;hUBC-CreERT2+/T mice harboring Cdk4+/+;Raf1+/+ (solid bar; n = 4), Cdk4FxKD/L;Raf1+/+ (dark gray bar; n = 6), Cdk4+/+;Raf1L/L (clear gray bar; n = 10), and Cdk4FxKD/L;Raf1L/L (open bar; n = 8) alleles after 9 wk of TX exposure. Error bars indicate mean ± SEM. P values were calculated using the unpaired Student´s t test. **P < 0.01, ns, not significant. (E) Percentage of tumors of different histopathological grades (I to V) present in Kras+/FSFG12V;Trp53F/F;hUBC-CreERT2+/T mice harboring Cdk4+/+;Raf1+/+ (n = 4), Cdk4FxKD/L;Raf1+/+ (n = 3), Cdk4+/+;Raf1L/L (n = 10), and Cdk4FxKD/L;Raf1L/L (n = 8) alleles after 9 wk of TX exposure.
Exposure of tumor-bearing Kras+/FSFG12V;Trp53F/F;hUBC-CreERT2+/T;Cdk4FxKD/L mice carrying advanced lung adenocarcinomas to a continuous TX diet for 9 wk only induced measurable tumor regression (>30% decrease in tumor burden) in 2 of 27 tumors (Fig. 1A and SI Appendix, Table S1). Systemic expression of CDK4KD in a related mouse tumor model that retains a wild-type p53 protein resulted in increased, but still rather limited percentage of tumor regressions (4 of 44 tumors) (SI Appendix, Fig. S1E and Table S1) (10). As expected, no regressions were observed in control mice expressing wild-type CDK4 (Fig. 1A and SI Appendix, Table S1). These observations indicate that systemic inhibition of CDK4 kinase activity has rather limited therapeutic activity in KRAS-driven lung adenocarcinomas.
Concomitant Inhibition of CDK4 Kinase Activity and Ablation of RAF1 Expression Induces Acceptable Toxicities in Adult Mice.
Systemic replacement of the wild-type CDK4 kinase by CDK4KD was well tolerated even by 1-y-old mice (SI Appendix, Fig. S2A). These observations indicated that CDK4 inactivation could be combined with other therapeutic targets without increasing the toxicity commonly associated with combination therapies. Hence, we interrogated whether adult mice could tolerate the systemic inhibition of CDK4 along with RAF1 ablation, a step prior to test this therapeutic strategy in Kras/Trp53-driven lung adenocarcinomas (9). As illustrated in SI Appendix, Fig. S2B, 8-mo-old mice exposed to a TX diet for 20 wk also tolerated well the systemic inhibition of these targets, although in this case mice displayed a more pronounced weight loss than the control cohort. Mice were killed at 1 y of age and submitted to detailed histological analysis of representative tissues, such as lung, pancreas, intestine, and liver. As illustrated in SI Appendix, Fig. S2C, none of the sections analyzed revealed detectable abnormalities when compared with the same tissues from control hUBC-CreERT2+/T;Cdk4+/+;Raf1+/+ mice of the same age submitted to an identical protocol. Targeted allele excision in those tissues examined histologically confirmed complete recombination of the corresponding Cdk4 and Raf1 conditional alleles, resulting in CDK4KD expression as well as absence of RAF1 (SI Appendix, Fig. S2D). These results demonstrate that systemic inactivation of CDK4 and RAF1 does not affect normal homeostasis even in old mice.
CDK4 and RAF1 Are Essential for Progression of Kras/Trp53-Driven Lung Adenocarcinomas.
Next, we interrogated whether concomitant inactivation of CDK4 could cooperate with RAF1 ablation to induce regression of Kras/Trp53-driven lung adenocarcinomas (9). To this end, we used three cohorts of mice that carried either wild-type Cdk4+ and Raf1+ alleles, Raf1L alleles, or the combination of Cdk4FxKD/L and Raf1L alleles. Animals carrying tumors (average of five tumors per mouse) ranging from 0.3 to 76 mm3 as determined by computed-tomography (CT) imaging, were exposed to TX for 9 wk. As expected, no tumor regressions were observed in control mice carrying wild-type alleles (Fig. 1A). In agreement with our previous results, 6 of the 62 tumors (10%) initially present in the Kras+/FSFG12V;Trp53F/F;hUBC-CreERT2+/T;Raf1L/L cohort completely regressed (CR) (Fig. 1B and SI Appendix, Table S1) (9). Similarly, 34 tumors (55%) regressed by more than 30% of their initial volume, thus qualifying as partial regressions (PR, as per the response evaluation criteria in solid tumors) (Fig. 1B and SI Appendix, Table S1). In contrast, 15 tumors (25%) displayed progressive disease (PD). Finally, the remaining seven tumors (11%), either did not grow or regressed less than 30%, hence they were classified as stable disease (SD).
Combined inactivation of CDK4 kinase activity and ablation of RAF1 expression elicited significantly better results. As illustrated in Fig. 1B, 24% of the tumors (12 of 51) underwent CR, more than twice the number observed in mice in which we only targeted RAF1 (Fig. 1B and SI Appendix, Table S1). The number of PRs (34 of 51, 66%) were also more abundant than in the cohort that underwent RAF1 ablation. More importantly, in this combined therapeutic scenario, none of the 51 tumors monitored displayed PD (Fig. 1B and SI Appendix, Table S1). Tumor response was similar regardless of the initial size of the lesions as determined by CT analysis (SI Appendix, Fig. S3 A and B). Histological analysis revealed additional lesions not detected by CT in both cohorts, yet the number of these lesions was also significantly lower in the mice that underwent systemic inactivation of CDK4 and RAF1 (Fig. 1 C and D). Finally, histological analysis also revealed that tumors lacking CDK4 kinase activity and RAF1 expression were less aggressive, according to the grading criteria of Jackson et al. (13) (Fig. 1E).
Real-time PCR (RT-PCR) analysis of tumor cells isolated by laser-capture microdissection of tumors displaying PR or SD at the end of the 9-wk-long trial indicated significant excision of the conditional Cdk4 and Raf1 alleles in most tumors (SI Appendix, Table S2). Thus, suggesting that at least a percentage of those cells present in these tumors represent cells resistant to CDK4 and RAF1 inactivation. Next, we analyzed the survival of Kras+/FSFG12V;Trp53F/F;hUBC-CreERT2+/T, Kras+/FSFG12V;Trp53F/F;hUBC-CreERT2+/T;Raf1L/L, and Kras+/FSFG12V;Trp53F/F;hUBC-CreERT2+/T;Cdk4FxKD/L;Raf1L/L tumor-bearing mice that had not been used for other purposes after the 9-wk-long trial. Control mice expressing wild-type CDK4 and RAF1 died within 7 wk (median survival 2 wk). Mice in which RAF1 has been ablated died by 8 wk after the end of the trial (median survival 5 wk), whereas those in which both targets were inactivated survived considerably longer, up to 15 wk (median survival 5.5 wk) (SI Appendix, Fig. S3C). The similar median survival data for these two cohorts of mice is due to the limited number of surviving mice, and we do not consider it to be informative. In any case, the death of these mice could not be attributed to tumor burden, considering the limited size of these resistant tumors. Careful examination of various tissues, including intestine, kidney, and liver did not reveal significant abnormalities. Hence, the premature dead of those mice that underwent CDK4 and RAF1 inactivation is likely to result from a combination of factors, such as the continued exposure to TX, the lung damage caused by the advanced tumors prior to their therapeutic response, and the relatively old age of the mice.
Concomitant CDK4 and RAF1 Inactivation Reduced Cell Proliferation and Increased Cell Death.
Ki67 immunostaining revealed significantly fewer proliferating cells upon CDK4 and RAF1 inactivation than those expressing the corresponding wild-type proteins (Fig. 2 A and B). Moreover, the apoptotic effect induced by loss of RAF1 expression was significantly enhanced upon additional CDK4 kinase inhibition (Fig. 2 A and B). Combined inactivation of CDK4 and RAF1 also led to increased infiltration of CD3+ T lymphocytes, CD4+ T helper cells, and CD8+ cytotoxic T cells compared to wild-type controls (SI Appendix, Fig. S4A). Quantification of the GranzymeB+/CD8– cell population showed an increase of NK cells (SI Appendix, Fig. S4B). Infiltrating F4/80+ macrophages were also slightly reduced in the double targeted tumors (SI Appendix, Fig. S4B). To what extent these immune responses contribute to the enhanced therapeutic activity observed upon inactivation of CDK4 and RAF1 remains to be determined.
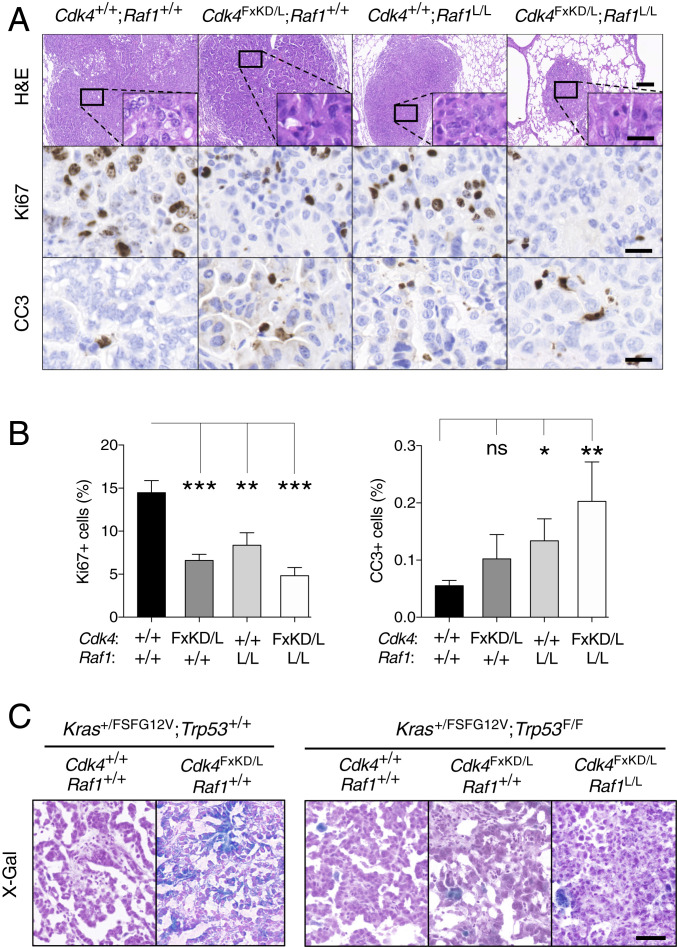
Fig. 2.
CDK4 inactivation and RAF1 ablation cooperate to reduce tumor proliferation and induce cell death. (A) Representative images of H&E, Ki67, and cleaved caspase 3 (CC3) stainings in paraffin-embedded sections of tumors from Kras+/FSFG12V;Trp53F/F;hUBC-CreERT2+/T mice harboring Cdk4+/+;Raf1+/+, Cdk4FxKD/L;Raf1+/+, Cdk4+/+;Raf1L/L, and Cdk4FxKD/L;Raf1L/L alleles after 9 wk of TX exposure. (Scale bars, 0.2 [H&E lower magnification] and 0.02 mm [H&E inset, Ki67 and CC3].) (B) Quantification of the percentage of Ki67+ and CC3+ cells present in sections of tumors from Kras+/FSFG12V;Trp53F/F;hUBC-CreERT2+/T mice harboring Cdk4+/+;Raf1+/+ (n = 3 mice/28 tumors) (solid bar), Cdk4FxKD/L;Raf1+/+ (n = 3 mice/28 tumors) (dark gray bar), Cdk4+/+;Raf1L/L (n = 3 mice/23 tumors) (light gray bar), and Cdk4FxKD/L;Raf1L/L (n = 8 mice/28 tumors) (open bar) alleles after 9 wk of TX exposure. Error bars indicate mean ± SEM. P values were calculated using the unpaired Student´s t test. *P < 0.05, **P < 0.01, and ***P < 0.001, ns, not significant. (C) Representative images of senescent cells identified by X-Gal staining in OCT sections of tumors from Kras+/FSFG12V;Trp53+/+;hUBC-CreERT2+/T mice harboring Cdk4+/+;Raf1+/+ and Cdk4FxKD/L;Raf1+/+ alleles and Kras+/FSFG12V;Trp53F/F;hUBC-CreERT2+/T mice harboring Cdk4+/+;Raf1+/+, Cdk4FxKD/L;Raf1+/+ and Cdk4FxKD/L;Raf1L/L alleles after 9 wk of TX exposure. (Scale bar, 0.05 mm.)
Previous studies have shown that ablation of Cdk4 in KrasG12V-driven tumors containing a functional p53 induced the appearance of senescent cells (10). In agreement with these observations, expression of a CDK4 kinase dead isoform in these tumors also induced senescence (Fig. 2C). However, elimination of CDK4 activity either by itself or in combination with RAF1 ablation in tumors lacking p53 failed to activate a senescence response, possibly due to the absence of this tumor suppressor (Fig. 2C) (10). Hence, tumor regression in the absence of CDK4 kinase activity and RAF1 expression appears to be primarily mediated by a decreased proliferative response combined with increased apoptosis.
To better define the molecular mechanisms responsible for tumor regression, we treated control Kras+/G12V;Trp53−/−;hUBC-CreERT2+/T lung tumor cell lines, as well as those carrying conditional Cdk4FxKD/L and Raf1L/L alleles, either separately or in combination, by infection with AdenoCre (AdCre) particles followed by exposure to 4-hydroxy-tamoxifen (4OHT) in order to achieve efficient recombination of these conditional alleles. Whereas independent inactivation of each target significantly reduced cell proliferation (70 to 80%), combined inactivation of both targets almost completely prevented the ability of these tumor cells to proliferate (SI Appendix, Fig. S5A). Expression of CDK4KD along with ablation of the wild-type CDK4 and RAF1 proteins did not affect the levels of phospho-ERK, indicating that combined CDK4 and RAF1 inactivation did not interfere with canonical MAPK signaling (Fig. 3A) (9). Similarly, the absence of CDK4 activity did not substantially alter phosphorylation of the RB tumor suppressor, possibly due to compensation by the related CDK2 or CDK6 kinases (Fig. 3A) (14).
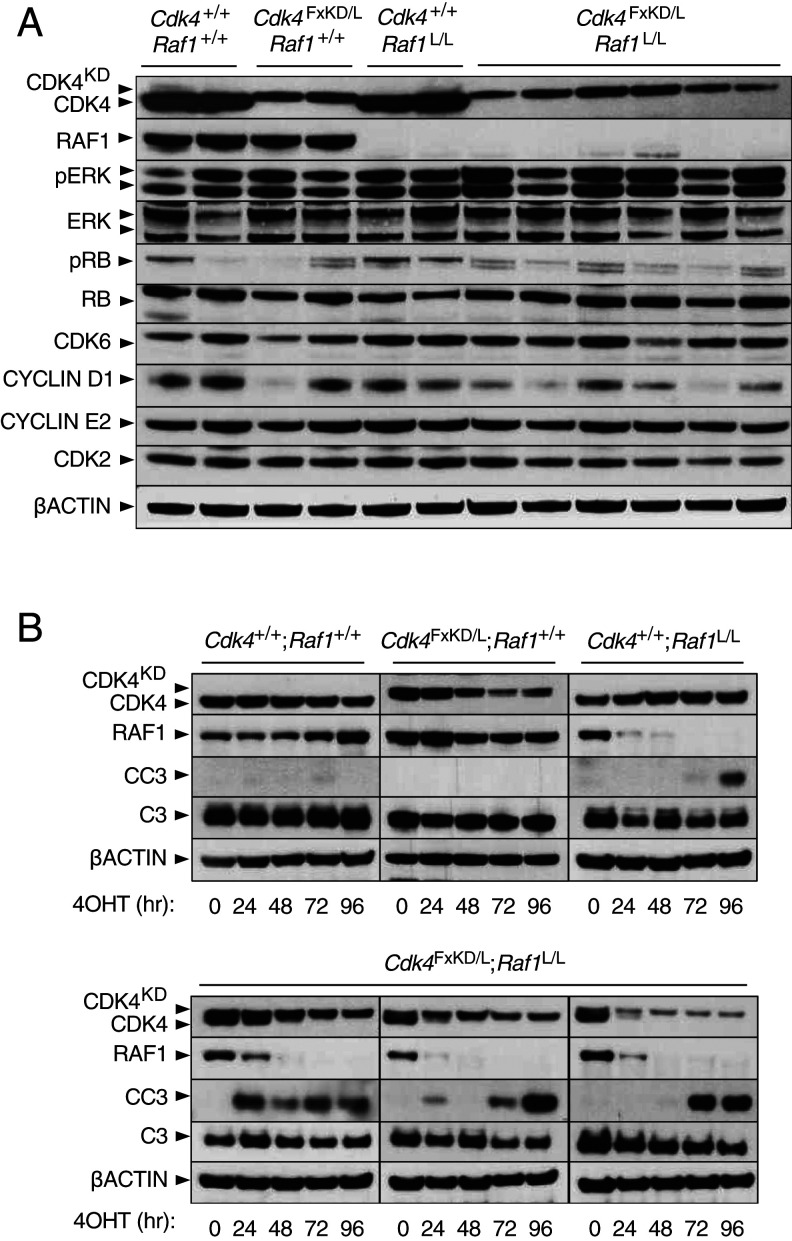
Fig. 3.
Concomitant CDK4 inactivation and RAF1 ablation boosts apoptosis. (A) Western blot analysis of CDK4, CDK4KD, RAF1, phospho-ERK (pERK), ERK1/2 (ERK), phospho-RB (pRB), RB, CDK6, CYCLIN D1, CYCLIN E2, and CDK2 expression in lysates obtained from Kras+/G12V;Trp53−/−;hUBC-CreERT2+/T lung tumor cell lines harboring Cdk4+/+;Raf1+/+, Cdk4FxKD/L;Raf1+/+, Cdk4+/+;Raf1L/L, and Cdk4FxKD/L;Raf1L/L alleles 96 h after 4OHT exposure. β-Actin was used as loading control. Migration of the above proteins is indicated by arrowheads. (B) Western blot analysis of CDK4, CDK4KD, RAF1, cleaved caspase-3 (CC3), caspase-3 (C3) expression in lysates obtained from Kras+/G12V;Trp53−/−;hUBC-CreERT2+/T lung tumor cell lines harboring Cdk4+/+;Raf1+/+, Cdk4FxKD/L;Raf1+/+, Cdk4+/+;Raf1L/L, and Cdk4FxKD/L;Raf1L/L alleles maintained in 4OHT containing media. Samples were harvested at the indicated times (hr, hours). β-Actin was used as loading control. Migration of the above proteins is indicated by arrowheads.
Combined inactivation of CDK4 and RAF1 led to significant levels of cell death as determined by TO-PRO-3/Hoechst assay in 3D cultures (SI Appendix, Fig. S5B). This effect appears to be mediated by an apoptotic mechanism since both CDK4 and RAF1 proteins synergistically cooperate to induce the rapid appearance of cleaved caspase-3 (CC3) within a 24- to 96-h time range (Fig. 3B). Analysis of β-galactosidase, a well-known senescence marker, failed to reveal significant levels of senescent cells. These results add further evidence indicating that apoptosis, rather than senescence, is the primary mechanism behind tumor regression upon combined inhibition of CDK4 and RAF1.
Transcriptional Consequences of Combined CDK4 and RAF1 Inactivation.
Next, we carried out RNA-sequencing (RNA-seq) analysis of KRAS/p53 mutant lung tumor cells before and after combined inactivation of CDK4 and RAF1 to identify those molecular events implicated in the antitumor response induced by these targets. Single-sample gene set enrichment analysis (ssGSEA) revealed down-regulation of pathways associated with cell cycle progression (RB pathway Biocarta). Similarly, we observed overexpression of pathways that contain genes known to be up-regulated upon G1 cell cycle arrest (ARF pathway Biocarta) (SI Appendix, Fig. S5C). Pathways linked to apoptosis are also up-regulated (hallmark of apoptosis or death pathway Biocarta) (SI Appendix, Fig. S5C). In fact, several proapoptotic and antiapoptotic-related genes, such as Bcl2l1, Bcl2, Bak1, Cdkn1a, Gadd45g, Sqstm1, and Fas appeared differentially expressed (SI Appendix, Fig. S5D). Overall, most oncogenic pathways—including those driven by KRAS, PI3K/AKT/mTOR, MYC, WNT, NOTCH, or ERBB—were down-regulated, thus supporting the concept that combined inhibition of CDK4 and RAF1 leads to a slowdown in signaling activity (SI Appendix, Fig. S5C).
Pharmacological Inhibition of CDK4 and RAF1 Does Not Phenocopy Genetic Inhibition.
The Food and Drug Administration has recently approved a series of CDK4/6 inhibitors that have shown a significant therapeutic effect in estrogen-positive breast cancer (15, 16). To pharmacologically validate our genetic studies, we compared the therapeutic effect of expressing CDK4KD with that of two independent CDK4/6 inhibitors, abemaciclib and palbociclib, in the context of RAF1 ablation. Kras+/FSFG12V;Trp53F/F tumor-bearing mice were treated with pharmacologically effective doses of abemaciclib (50 mg/kg; n = 10) or palbociclib (50 mg/kg; n = 5). As illustrated in Fig. 4A, the observed therapeutic benefit was significantly less effective than that observed upon genetic inactivation of CDK4. Indeed, neither inhibitor induced PRs, whereas expression of the CDK4KD isoform resulted in PR of 20% of the tumors. The differential effect between genetic vs. pharmacological inactivation of CDK4 was even more pronounced in the context of RAF1 ablation, since neither inhibitor phenocopied the cooperative effect observed upon genetic inactivation of CDK4 (Fig. 4A). These results indicate that the wider inhibitory spectrum of abemaciclib or palbociclib on CDK6 is irrelevant for KRAS-driven lung adenocarcinomas (10). Moreover, they indicate that in order to observe a therapeutic benefit for patients with KRAS mutant lung tumors, it will be necessary to generate more potent CDK4 inhibitors.
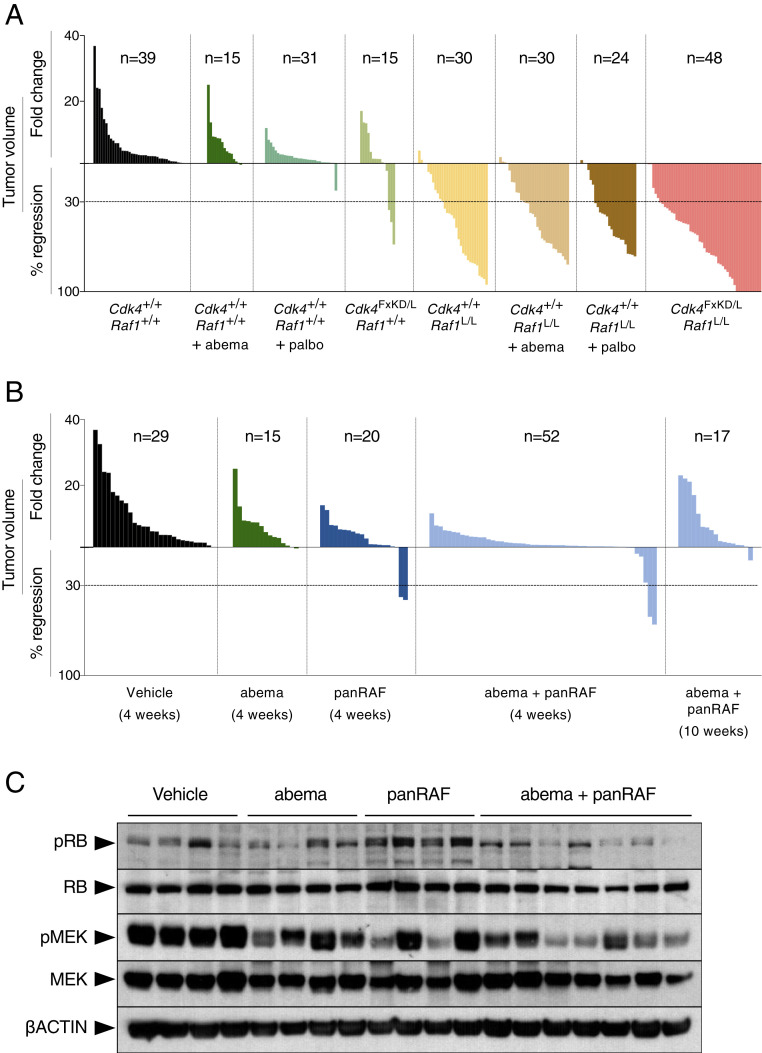
Fig. 4.
Pharmacological inhibition of CDK4/RAF1 does not phenocopy genetic inhibition. (A) Waterfall plot representing the increase in tumor volume (fold-change) and the percentage of tumor regression as determined by CT scans performed at the beginning and at the end of the trial of individual lung tumors present in Kras+/FSFG12V;Trp53F/F;hUBC-CreERT2+/T;Cdk4+/+;Raf1+/+ mice either untreated (solid bars) or treated with abemaciclib (abema; dark green bars) or treated with palbociclib (palbo; middle green bars), Kras+/FSFG12V;Trp53F/F;hUBC-CreERT2+/T;Cdk4FxKD/L;Raf1+/+ (light green bars), Kras+/FSFG12V;Trp53F/F;hUBC-CreERT2+/T;Cdk4+/+;Raf1L/L mice either untreated (yellow bars) or treated with abemaciclib (abema; brown bars) or treated with palbociclib (palbo; dark brown bars), and Kras+/FSFG12V;Trp53F/F;hUBC-CreERT2+/T;Cdk4FxKD/L;Raf1L/L mice (pink bars) exposed to TX for 4 wk. The number of tumors analyzed for each group is indicated. (B) Waterfall plot representing the increase in tumor volume (fold-change) and the percentage of tumor regression as determined by CT scans performed at the beginning and at the end of the trial of individual lung tumors present in Kras+/FSFG12V;Trp53F/F mice treated with vehicle (Vehicle; solid bars), abemaciclib (abema; dark green bars), panRAF inhibitor (panRAF; dark blue), and the combination of abemaciclib and panRAF for the indicated number of weeks (light blue bars). The number of tumors analyzed for each group is indicated. (C) Western blot analysis of phospho-RB (pRB), RB, phospho-MEK (pMEK), MEK1/2 (MEK), expression in lysates obtained from representative tumors described in B. β-Actin was used as loading control. Migration of the above proteins is indicated by arrowheads.
We also attempted to pharmacologically validate RAF1 by inhibiting its kinase activity. To this end, we used four independent RAF kinase inhibitors, including MLN2480 (Millennium Pharmaceuticals), a panRAF kinase inhibitor currently in clinical trials; GW5074 (GlaxoSmithKline), an RAF inhibitor with better (10-fold) inhibitory activity on RAF1 than on BRAF; PLX8394 (Plexicon), a paradox breaker also in clinical trials; and LSN3074753 (Eli Lilly), a surrogate of the panRAF inhibitor LY3009120 suitable for use in mice (17). These inhibitors were tested against cell lines derived from two independent KRAS mutant lung PDX models (PDX dc-1 and PDX dc-2). As illustrated in SI Appendix, Fig. S6, only LSN3074753 displayed a submicromolar GI50. Treatment of tumor-bearing Kras+/FSFG12V;Trp53F/F mice for 4 wk with either abemaciclib (50 mg/kg, once a day) or LSN3074753 (20 mg/kg, twice a day) alone induced a limited reduction in tumor growth (Fig. 4B). Combination of both inhibitors (50 mg/kg once a day and 20 mg/kg once a day, respectively) further reduced tumor growth but did not increase the number of PRs (5 of 52, 10%). Longer treatments (up to 10 wk) did not increase the therapeutic activity of this combination (Fig. 4B). To determine the relative contribution of either inhibitor to the observed reduction in tumor burden, we analyzed the levels of pRB and pMEK, the two main downstream targets of these inhibitors. As illustrated in Fig. 4C, both inhibitors reduced the phosphorylation of their respective downstream targets, therefore making it difficult to assess which pathway is more relevant for tumor progression. Yet the fact that the panRAF inhibitor induced partial regression of a limited number of tumors suggests a more prevalent role of the MAPK pathway than inhibition of the cell cycle. These results, taken together, indicate that pharmacological validation of the genetic results will require more potent and selective inhibitors.
Characterization of CDK4/RAF1-Resistant Tumor Cells.
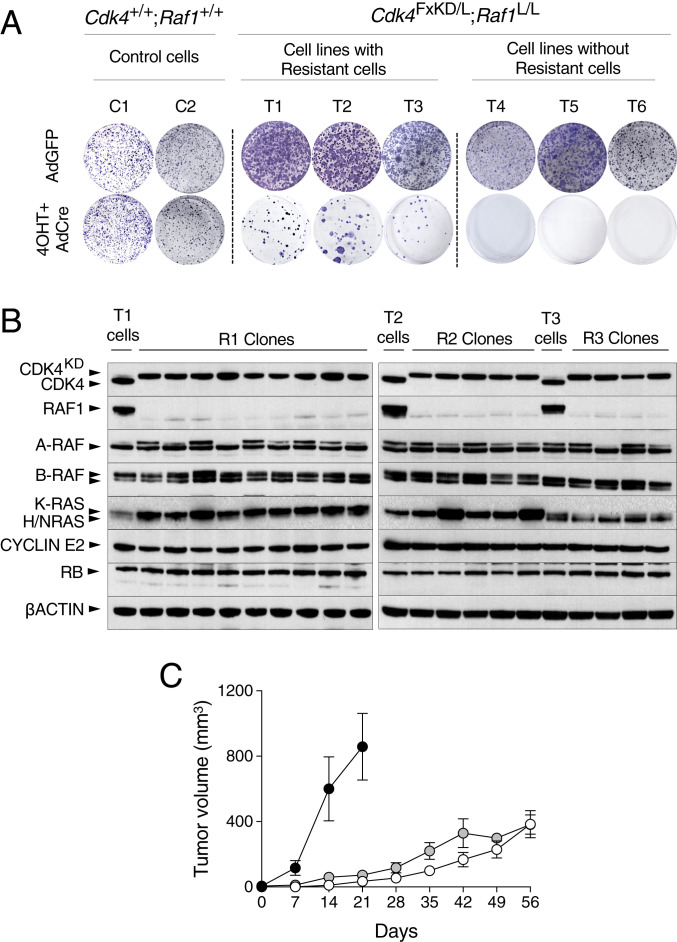
Despite the significant therapeutic response observed upon CDK4 and RAF1 inactivation, most tumors (66%) underwent PRs rather than CRs, thus suggesting the presence of resistant cells (Fig. 1B). To interrogate those mechanisms implicated in the lack of response to combined CDK4/RAF1 inactivation, we used cell lines from lung tumors obtained from control Kras+/FSFG12V;Trp53F/F;hUBC-CreERT2+/T and experimental Kras+/FSFG12V;Trp53F/F;hUBC-CreERT2+/T;Cdk4FxKD/L;Raf1L/L mice. These cells were treated with 4OHT followed by infection with AdCre particles to ensure efficient recombination of the Cdk4FxKD, Cdk4L, and Raf1L alleles and subsequently allowed to grow in standard colony formation assays. Whereas control cells grew efficiently, cell lines carrying Cdk4 and Raf1 conditional alleles underwent either partial or complete cell death, thus reproducing in vitro the PRs and CRs observed in vivo, respectively (Fig. 5A). Clones generated from three independent tumor cell lines were picked, expanded, and submitted to RT-PCR analysis to ensure the complete excision of the conditional Cdk4FxKD, Cdk4L, and Raf1L alleles in each of the clones. Correct excision of these alleles was also confirmed at the RNA level. Short-read alignments from the RNA-seq data were mapped to the mouse genome, confirming transcription of the Cdk4KD allele, by the presence of the K35M miscoding mutation in exon 2 (SI Appendix, Fig. S7 A, Left, red line), as well as complete deletion of the Raf1 exon 3 (SI Appendix, Fig. S7 A, Right). Next, we submitted these clones to Western blot analysis. As expected, all clones express the CDK4KD isoform instead of the wild-type protein. Similarly, none of the clones exhibited detectable levels of RAF1 (Fig. 5B). Absence of RAF1 expression was not compensated by either increased expression of the A-RAF and B-RAF isoforms or of members of the RAS protein family (Fig. 5B) Finally, these clones were able to proliferate in vitro and form tumors when implanted in immunodeficient mice (Fig. 5C), hence demonstrating that they are bona fide double CDK4/RAF1-resistant cells.
Fig. 5.
Generation of CDK4/RAF1 resistant tumor cell clones. (A) Kras+/G12V;Trp53−/−;hUBC-CreERT2+/T;Cdk4+/+;Raf1+/+ (C1 and C2) and Kras+/G12V;Trp53−/−;hUBC-CreERT2+/T;Cdk4FxKD/L;Raf1L/L (T1 to T6) lung tumor cell lines exposed to 4OHT and infected with AdCre to inactivate CDK4 and RAF1 were used in a colony formation assay to identify resistant clones. Adeno GFP (AdGFP) was used as negative control. (B) Western blot analysis of CDK4, CDK4KD, RAF1, A-RAF, B-RAF, K-RAS, H-RAS, N-RAS, CYCLIN E2, and RB expression levels in lysates of parental T1, T2, and T3 cell lines expressing CDK4 and RAF1 and of resistant clones (R1, R2, and R3 clones) derived from these cell lines upon CDK4 inactivation and RAF1 ablation. β-Actin was used as loading control. Migration of the above proteins is indicated by arrowheads. (C) Tumor growth of individual CDK4/RAF1 resistant clones R1.1, R1.2, and R1.3 (solid circles), R2.1, R2.2, and R2.3 (open circles), and R3.1, R3.2 and R3.3 (gray circles) after subcutaneous implantation in immunodeficient mice. Error bars indicate mean ± SEM.
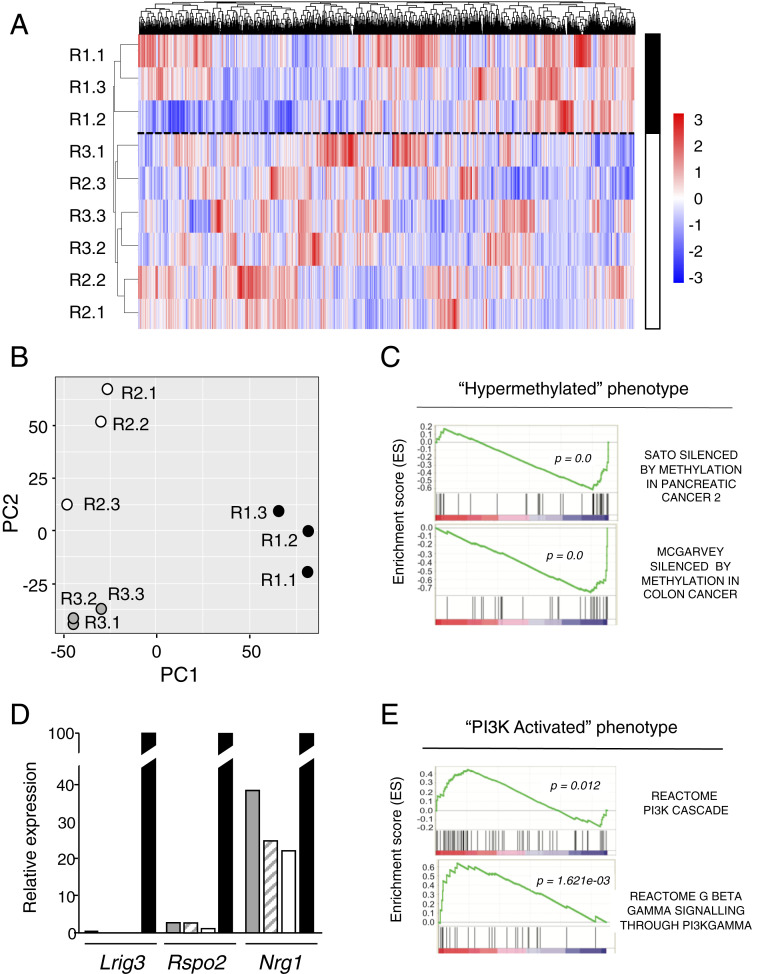
Three independent CDK4/RAF1-resistant clones derived from each of the three different tumor cell lines (R1.1, R1.2, R1.3, R2.1, R2.2, R2.3, R3.1, R3.2, and R3.3) along their respective parental cells (T1, T2, and T3) were submitted to RNA-seq analysis. ssGSEA of those differentially expressed genes (DEGs) indicated that whereas the parental T1, T2, and T3 cell lines that retained CDK4 and RAF1 expression displayed a similar transcriptional pattern, the CDK4/RAF1-resistant clones exhibited a significant degree of heterogeneity (SI Appendix, Fig. S7B). Yet, deeper analysis comparing these resistant clones among themselves, either by heatmap or principal component analysis (PCA), revealed two distinct transcriptional profiles predictive of different resistance mechanisms (Fig. 6 A and B). Comparison of the transcriptional profiles of resistant clones R1.1, R1.2, and R1.3 with that of their T1 parental cell line correlated with a “hypermethylated” phenotype, leading to a significant decrease in the expression of a series of tumor-suppressor genes, such as Lrig3 (log2 fold-change [log2FC] = −6.3), Rspo2 (log2FC = −4.3), and Nrg1 (log2FC = −1.5) (Fig. 6C). These observations were further validated by RT-PCR (Fig. 6D). On the other hand, resistant clones R2.1, R2.2, and R2.3, as well as the R3.1, R3.2, and R3.3 clones derived from the T2 and T3 tumor cell lines, respectively, have significantly up-regulated genes related to a “PI3K-activated” phenotype (Fig. 6E).
Fig. 6.
Resistance to CDK4/RAF1 inactivation is mediated by independent mechanisms. (A) Heatmap representing color-coded expression levels of differentially expressed genes among the indicated CDK4/RAF1 resistant tumor cell clones. The solid and open bars separated by a discontinuous line indicate the two main hierarchical clusters found among these resistant clones. (B) Distribution of the 9 CDK4/RAF1-resistant tumor cell clones in a 2D space determined by the first and second principal components generated by PCA. (C) GSEA for the "hypermethylated" phenotype comparing resistant clones R1.1, R1.2, and R1.3 with their parental T1 cell lines before CDK4/RAF1 inactivation. Adjusted P < 0.05. (D) Validation of the three main down-regulated genes Lrig3, Rspo2, and Nrg1, in CDK4/RAF1-resistant tumor cell clones exhibiting a "hypermethylated" phenotype. Expression levels were determined by RT-PCR in CDK4/RAF1 resistant tumor cell clones R1.1 (gray bars), R1.2 (striped bars), R1.3 (open bars) relative to those present in the parental T1 cells (solid bars). (E) GSEA for the "PI3K-activated" phenotype comparing resistant clones R2.1, R2.2, and R2.3 as well as R3.1, R3.2, and R3.3 with their parental cell lines T2 and T3, respectively, before CDK4/RAF1 inactivation. Adjusted P < 0.05.
Next, we interrogated whether the differential transcriptional profiles of these two classes of resistant clones had any consequences on their tumorigenic properties in vivo. As illustrated in Fig. 5C, those tumors exhibiting the "hypermethylated" phenotype (R1.1, R1.2, and R1.3) grew at a much faster pace than those whose resistance was due to a "PI3K-activated" phenotype (R2.1, R2.2, R2.3 and R3.1, R3.2, R3.3). We also investigated whether those resistant tumors with a "PI3K-activated" phenotype may have acquired neuroendocrine properties, as observed in a small percentage of EGFR mutated lung adenocarcinomas (18). As illustrated in SI Appendix, Fig. S7C, synaptophysin, a well-known neuroendocrine marker, was expressed in mastocytes or dendritic cells but not in tumor cells. In the case of a second marker, chromogranin A, we observed similar levels of expression in parental and resistant tumor cells, indicating that expression of this marker is not related to the acquisition of resistance via PI3K activation (SI Appendix, Fig. S7C).
Pharmacological Vulnerabilities to Overcome CDK4/RAF1 Resistance.
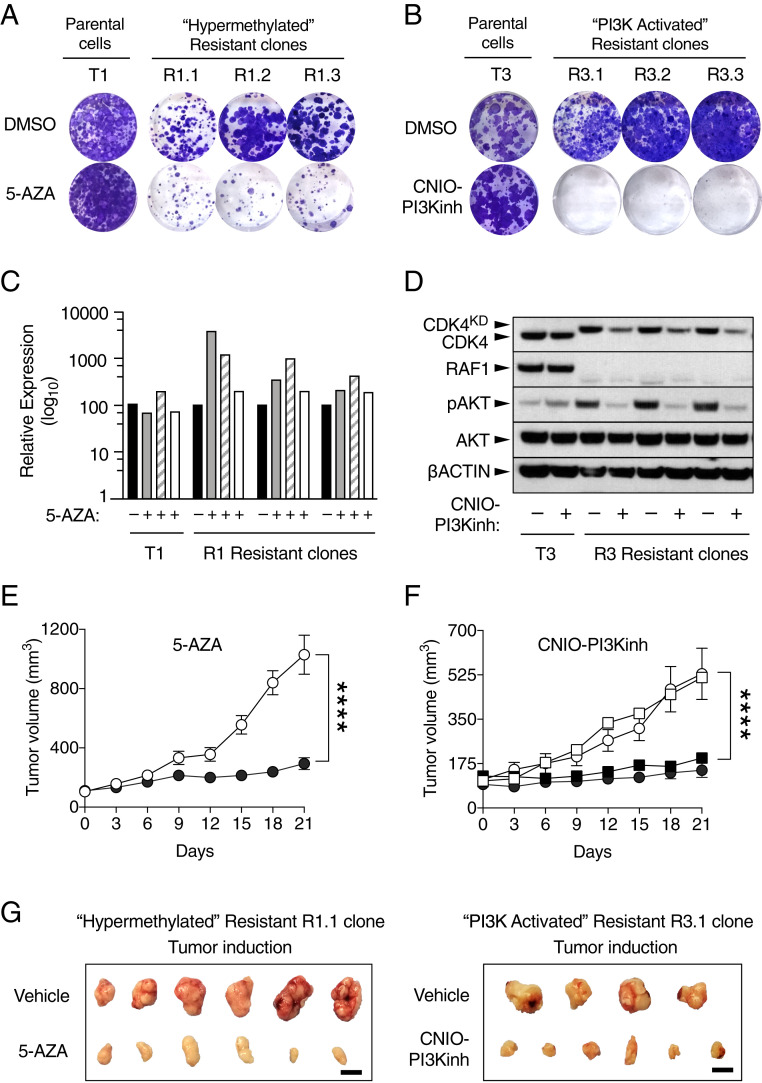
To pharmacologically validate the bioinformatic analysis described above, we exposed the CDK4/RAF1-resistant clones with the "hypermethylated" phenotype (R1.1, R1.2, and R1.3) to the demethylation agent 5-azacytidine (5-AZA). As illustrated in Fig. 7A, these resistant clones, but nor their parental cells, were exquisitely sensitive to this drug, demonstrating that the sensitivity to 5-AZA requires prior inactivation of CDK4 and RAF1. Similarly, exposure of the R2- and R3-resistant clones displaying the "PI3K-activated" phenotype, to the CNIO-PI3K inhibitor (19), revealed that their response to PI3K inhibitors required CDK4 and RAF1 inactivation (Fig. 7B).
Fig. 7.
Pharmacological validation of the hypermethylated and PI3K-activated resistance mechanisms. (A) Colony formation assay of the parental T1 cell line and the CDK4/RAF1 hypermethylated resistant clones R1.1, R1.2, and R1.3 treated with DMSO or with 2 µM 5-AZA for 9 d. (B) Colony formation assay of the parental T3 cell line and CDK4/RAF1 PI3K-activated resistant clones R3.1, R3.2, and R3.3 treated with DMSO or with 1 µM PI3K inhibitor (CNIO-PI3Kinh) for 9 d. (C) RT-PCR validation of the levels of expression of Lrig3 (gray bars), Rspo2 (striped bars), and Nrg1 (open bars) after 72 h in the absence or presence of 5-AZA relative to those in the DMSO treated cells (solid bars) in T1 parental cells and R1 resistant clones. (D) Western blot analysis of CDK4, CDK4KD, RAF1, phospho-AKT (pAKT), and AKT expression after 24 h in the absence or presence of the PI3K inhibitor (CNIO-PI3Kinh) in T3 parental cells and R3 resistant clones. β-Actin was used as loading control. Migration of the above proteins is indicated by arrowheads. (E) Tumor growth of CDK4/RAF1 resistant clones R1.1 and R1.2 treated with vehicle (empty circles; n = 15 tumors) or with 6 mg/kg of 5-AZA (solid circles; n = 15 tumors) for 21 d. Error bars indicate mean ± SEM. P values were calculated using the unpaired Student’s t test. ****P < 0.0001. (F) Tumor growth of CDK4/RAF1-resistant clones R2.1 (circles) and R3.1 (squares) treated with vehicle (open symbols) (R2.1, n = 8 tumors and R3.1, n = 9 tumors) or with 15 mg/kg of PI3K inhibitor (CNIO-PI3Kinh) (solid symbols) (R2.1, n = 9 tumors and R3.1, n = 8 tumors) for 21 d. Error bars indicate mean ± SEM P values were calculated using the unpaired Student’s t test. ****P < 0.0001. (G) Representative pictures of tumors derived from CDK4/RAF1-resistant clone R1.1 treated with vehicle or with 5-AZA and of tumors derived from CDK4/RAF1-resistant clone R3.1 treated with vehicle or with the PI3K inhibitor (CNIO-PI3Kinh). (Scale bar, 1 cm.)
Interestingly, 5-AZA treatment of the R1-resistant clones rescued the expression of Lrig3, Rspo2, and Nrg1 genes (Fig. 7C). Therefore, those tumor suppressors could be used as biomarkers for those resistant cells exhibiting a "hypermethylated" phenotype (20–22). Similarly, Western blot analysis of the R3-resistant clones revealed the presence of increased levels of phospho-AKT compared to their parental cell line T3. As expected, these phosphorylation levels decreased significantly upon exposure to the CNIO-PI3K inhibitor (Fig. 7D).
We also searched for potential vulnerabilities in both classes of CDK4/RAF1-resistant cells by exposing them to a single concentration (5 µM) of a collection of 114 inhibitors known to act in most oncogene-related pathways (SI Appendix, Table S3). None of these compounds selectively blocked those resistant cells with the "hypermethylated" phenotype (SI Appendix, Fig. S8A). In contrast, we identified 24 compounds that selectively reduced cell proliferation (>75%) in those cells with the activated PI3K pathway (SI Appendix, Fig. S8A). Interestingly, 4 of these 24 compounds were inhibitors of the various PI3K isoforms that significantly inhibited the resistant cells displaying a "PI3K-activated" phenotype but not those with a "hypermethylated" phenotype (SI Appendix, Fig. S8B).
To validate these resistance mechanisms in vivo, we subcutaneously implanted resistant cells displaying both phenotypes in immunodeficient mice. Once the tumors reached a volume averaging between 100 and 150 mm3, mice were treated either with vehicle or with the corresponding inhibitor, 5-AZA, for those mice bearing tumors with the "hypermethylated" phenotype and CNIO-PI3Kinh for those carrying tumors showing a "PI3K-activated" phenotype. As illustrated in Fig. 7 E and G, treatment with 5-AZA for 3 wk significantly halted tumor progression when compared to those exposed to vehicle. Similarly, those tumors with an activated PI3K pathway also showed a significant therapeutic benefit when compared to the controls upon treatment with CNIO-PI3Kinh (Fig. 7 F and G). Taken together, these results illustrate that the resistance of KRAS/p53 mutant lung tumors to a combined CDK4/RAF1 therapeutic treatment can be mediated by at least two independent mechanisms involving either epigenetic modifications or activation of the PI3K pathway.
Discussion
Attempts to block KRAS mutant tumors using inhibitors against KRAS downstream effectors have not yielded positive results in the clinic, mainly due to lack of significant antitumor activity at tolerated doses (3). Previous studies have illustrated that expression of the MEK1/2 or ERK1/2 kinase families is essential to maintain adult homeostasis (4), hence providing a plausible explanation for the clinical results. Indeed, genetic analysis of each of the KRAS effectors within the MAPK pathway has revealed that only targeting RAF1 elicits a significant therapeutic response without causing unacceptable toxicities (4, 9). However, RAF1 ablation only induced complete responses in a small percentage of tumors, indicating the need to identify combination partners. Previous studies have indicated that ablation of CDK4 or exposure to the CDK4/6 inhibitor palbociclib had limited therapeutic activity against KRAS mutant lung tumors (10, 23). As illustrated here, replacement of the wild-type CDK4 protein by a kinase dead isoform also resulted in tumor inhibition, albeit with limited efficacy. However, concomitant inactivation of CDK4 kinase activity and loss of RAF1 expression significantly increased the therapeutic effect observed upon RAF1 ablation, leading to complete regression of a quarter of all lung tumors. Perhaps more importantly, this therapeutic strategy completely halted tumor progression.
Previous studies have shown that combined treatment of similar Kras/Trp53-driven lung tumors with a combination of the MEK inhibitor trametinib and palbociclib drives tumor cells toward senescence followed by their clearance executed by activated NK cells (23). We have also observed that either ablation of CDK4 or expression of a kinase dead isoform in lung tumors exclusively driven by KrasG12V induced a senescence response that compromised tumor development (10). This effect is likely to be mediated by p53 activation, since we have not observed such senescent phenotype upon CDK4 inactivation in Kras/Trp53 mutant lung tumors. Instead, genetic inactivation of CDK4 and RAF1 induced tumor regression by a combination of increased apoptosis and reduced proliferative capabilities without a significant increase in inflammatory responses. Whether the senescence phenotype observed by Ruscetti et al. (23) could be induced by the drug treatment remains to be determined (24, 25).
Surprisingly, exposure of tumor-bearing mice to the CDK4/6 inhibitors abemaciclib or palbociclib did not improve the therapeutic effect mediated by RAF1 ablation. Thus, suggesting the need to generate more potent CDK4/6 inhibitors that could translate the results obtained upon genetic inhibition of CDK4 activity to treat KRAS mutant lung cancer patients. Similarly, treatment with a panRAF inhibitor was significantly less effective than ablating RAF1 expression. Moreover, combination of this panRAF inhibitor with abemaciclib did not result in a significant increase in therapeutic efficacy. These results, along with the fact that trametinib as well as panRAF inhibitors have already failed in clinical trials against KRAS mutant lung tumors, makes it necessary to identify more potent and less toxic drugs before these results can be translated to the clinic.
Despite the significant levels of tumor regression observed upon combined CDK4 and RAF1 inhibition, a significant number of tumor cells remain resistant to this therapeutic approach and may serve as a reservoir for the development of resistant tumors (26, 27). Transcriptome analysis of these CDK4/RAF1-resistant cells did not identify a unique pathway or gene signature, suggesting the existence of multiple resistance mechanisms. However, bioinformatic analysis revealed that a limited subset of resistant cells displayed up-regulated epigenetic changes leading to a "hypermethylated" phenotype that inhibits a series of tumor suppressors likely to be responsible for the observed tumor resistance. Indeed, tumors rising from these resistant cells significantly slowed down their growth capacity upon treatment with 5-AZA, a DNA methylation inhibitor. Functional validation of each of these methylated tumor suppressors should provide relevant information regarding those molecular pathways involved in the acquisition of resistance upon CDK4/RAF1 inhibition, eventually opening new avenues for the identification of additional therapeutic targets. Other resistant cells displayed an activated PI3K signaling cascade and responded well to PI3K inhibitors. Interestingly, Kras/Trp53 mutant lung tumor cells were not sensitive to either 5-AZA or PI3K inhibitors unless CDK4 and RAF1 were previously inactivated, thus demonstrating that their observed pharmacological vulnerabilities to methylation or PI3K inhibitors represent bona fide resistance mechanisms derived from CDK4/RAF1 inhibition. Yet, the high toxicities displayed by 5-AZA and PI3K inhibitors in the clinic underscores the need for better compounds to combat the resistance of these KRAS mutant lung tumor cells to CDK4/RAF1 inhibition. Hopefully, the design of more potent and selective inhibitors against those targets and signaling pathways described here will allow the translation of these results to the clinic in a not too distant future.
Materials and Methods
Mice.
The strains Kras+/FSFG12V (9), Trp53F/F (28), hUBC-CreERT2+/T (12), Raf1L/L (29), and Cdk4L/L (11) have been described previously. Generation of Cdk4+/FxKD mice is described below. All animal experiments were approved by the Ethical Committees of the Spanish National Cancer Research Centre (CNIO), the Carlos III Health Institute, and the Autonomous Community of Madrid (PROEX 81/16), and were performed in accordance with the guidelines stated in the International Guiding Principles for Biomedical Research Involving Animals, developed by the Council for International Organizations of Medical Sciences. Mice were housed in specific-pathogen-free conditions at CNIO’s Animal Facility (Association for Assessment and Accreditation of Laboratory Animal Care, JRS: dpR 001659). Female and male mice were used for the experiments. All mice were genotyped at the CNIO’s Genomic Unit.
Generation of Cdk4+/FxKD and Cdk4FxKD/L Mice.
To generate the Cdk4FxKD conditional allele, we designed a targeting vector based on the pFLEX strategy (30) that contained the entire Cdk4 locus (exons 1 to 8) along with an inverted exon 2 (2KD) that carried an amino-terminal Flag sequence as well as a mutation in the ATP-binding sequences that replaced the lysine located at position 35 by a methionine residue (K35M). This targeting vector also carried an frt-PGKneo-frt cassette located between the mutant exon 2 and exon 3 as well as a PGK-TK cassette at its 3′ end for selection purposes. The vector was electroporated into mouse V6.4 (C57BL/6J × 129S4/SvJae) F1 embryonic stem cells and recombinant clones were selected in the presence of G418 and ganciclovir. Southern blot analysis of 112 G418/Gan-resistant clones identified two homologous recombinants. These clones were aggregated with eight cell-stage CD-1 embryos. Male chimeras derived from two of the clones transmitted the targeted allele to their progeny. These chimeras were crossed with pCAG-Flpe transgenic mice (31) to excise the PGKneo cassette resulting in the conditional Cdk4FxKD allele (SI Appendix, Fig. S1A).
Tumor Induction and Target Inactivation.
The Cdk4FxKD allele along with the floxed Cdk4L and Raf1L alleles were subsequently incorporated to the therapeutic tumor model, Kras+/FSFG12V;Trp53F/F;hUBC-CreERT2+/T, in which expression of KRASG12V and ablation of p53 was carried out by intranasal instillation of a single dose of 106 pfu per mouse of adenoviral particles encoding the Flp (Ad-Flp) recombinase in anesthetized (ketamine 75 mg/kg, xylazine 12 mg/kg) 8- to 10-wk-old mice. Adenoviral preparations were purchased from Iowa University (Iowa City, IA). Target inactivation was carried out by activation of the inducible CreERT2 recombinase encoded by the hUBC-CreERT2 transgene by exposing the corresponding mice to a TX-containing diet (Teklad CRD TAM400 diet, Harlan) during the duration of the experiment.
Kinase Assays.
CDK4, CDK4KD, and CYCLIN D1 proteins were expressed in a baculovirus system (32), purified by affinity chromatography, and incubated in kinase assay buffer (33) containing 1 mM ATP, 10 μCi [γ-32P] ATP, and 2 μg of RB recombinant protein (Millipore, 12-439) for 30 min at 30 °C. Reactions were stopped by adding 4× loading buffer and 10× reducing agent (Nupage, Invitrogen) followed by boiling for 5 min. Proteins were electrophoresed and checked for incorporated radioactive ATP before being transferred to nitrocellulose membranes. Anti-CDK4 (Santa Cruz, Sc-260) and anti CYCLIN D1 antibodies (Neomarkers, MS-210-P1; 1:500 dilution) were revealed with HRP-conjugated goat anti-rabbit antibody (Dako, P0448) and goat anti-mouse antibody (Dako, P0447), respectively.
Micro-CT Imaging.
Mice were anesthetized with a continuous flow of 1 to 3% isoflurane/oxygen mixture (0.5 L/min). The chest area was imaged by 3D micro-CT performed with a CompaCT scanner (SEDECAL, GE). Data were acquired with 720 projections by 360° scan, integration time of 100 ms, with three frames, photon energy of 50 KeV, and current of 100 μA. Tumor measurements were obtained with GE MicroView software v2.2. Tumor volume was calculated according to the following formula: Short axis × short axis × long axis/2.
Laser-Capture Microdissection.
When residual tumors were too small to perform biochemical analysis, samples were microdissected with the help of a laser-capture microdissector in order to check for target excision. Ten-thousand cells were microdissected using a PALM microbeam Zeiss Axio Observer (Carls Zeiss) from 4-μm thickness paraffin sections. DNA was isolated from the captured samples and resuspended in 25 μL of lysis buffer (1× Taq Platinum Buffer supplemented with both 1% TX 100 and 5 mg/mL of proteinase K dissolved in 1 mM CaCl2).
In Vivo Pharmacological Treatments in Genetically Engineered Mice Tumor Models.
Drug treatments were carried out with tumor-bearing Kras+/FSFG12V;Trp53F/F and Kras+/FSFG12V;Trp53F/F;hUBC-CreERT2+/T;Raf1L/L mice. The latter were exposed to a TX-containing diet during 4 wk to induce efficient RAF1 ablation. All mice harbored at least one tumor larger than 3 mm3 as determined by CT analysis. When used as single agents, abemaciclib was dosed at 50 mg/kg (once a day, by mouth) in 1% hidroxyethylcellulose, 25 mM phosphate buffer; palbociclib was dosed at 50 mg/kg (once a day, by mouth) in 0.5% methylcellulose and the panRAF inhibitor (LSN3074753) was dosed at 20 mg/kg (twice a day, intraperitoneally) in 25 mM phosphate buffer containing 20% hidroxypropyl β cyclodextrin. When used in combination with the panRAF inhibitor dosing was reduced to 20 mg/kg, once a day. Drug efficacy was monitored by CT analysis.
In Vivo Pharmacological Treatments in Allograft Tumor Models.
Drug treatments were carried out in immunodeficient Foxn1nu/nu mice. Tumors were induced by injecting a suspension of 2.5 × 104 cells diluted in 1:1 PBS:Matrigel (Corning, 354234) in a final volume of 100 μL per flank. Once the tumor reached 100 to 150 mm3 in size as determined by caliper measurements, mice were daily treated with either 6 mg/kg of 5-AZA (once a day, intraperitoneally) in sterile saline solution or 15 mg/kg CNIO-PI3Kinh (once a day, by mouth) in 10% N-Methyl-2-pyrrolidone (NMP; Fluka 69118)/90% PEG300 (vol/vol). A minimum of eight tumors per group were followed-up. Tumor measurements were performed every 3 d over 21 d. Tumor growth over 1,200 mm3 was considered humane endpoint criteria.
Histopathology and Immunohistochemistry.
For routine histological analysis, lung lobes were fixed in 10%-buffered formalin (Sigma) and embedded in paraffin. H&E-stained sections were used to quantify the number of tumors and classify them according to standard histopathological grading (13). Antibodies used for immunostaining included those raised against Ki67 (Master Diagnostica, 0003110QD), CC3 (Cell Signaling, 9661), CD3ε (Santa Cruz, sc-1127), CD4 (Cell Signaling, 25229), CD8 (CNIO, AM-OTO94A) GranzymeB (Cell Signaling, 46890), F4/80 (ABD Serotec, MCA497), Chromogranin A (Abcam, ab15160), and Synaptophysin (Master Diagnostica, 000313QD). Tissue slides were scanned using the AxioScan Z1 scanner (Zeiss). Immunostainings were quantified using the Zen Lite software (Zeiss).
Cell Proliferation and Colony Formation Assays.
Kras+/G12V;Trp53−/−;hUBC-CreERT2+/T lung tumor cell lines harboring Cdk4L, Cdk4FxKD, and Raf1L conditional alleles were infected with AdCre (multiplicity of infection = 10) and maintained in DMEM supplemented with 10% FBS and 600 nM of 4OHT (Sigma, H70904) for 4 d to ensure proper cleavage of the corresponding alleles. For proliferation assays, cells were seeded in 96-well plates, exposed to AdCre+4OHT treatment and their proliferation rate measured by using the MTT assay (Roche, 11465007001). For colony formation assays, 5,000 cells were seeded in 10-cm dishes, exposed to AdCre+4OHT treatment and maintained in DMEM supplemented with 10% FBS for 10 d. Plates were either fixed during 5 min at room temperature in 1% glutaraldehyde and stained in 0.2% Crystal violet overnight or used to isolate individual colonies. Excision of the conditional alleles in these clonal cells was indirectly assessed by Western blot analysis.
Spheroids Formation and Cell Death Assays.
Cells (1,000 cells per well) were dispensed in ULA 96-well round-bottomed plates (Costar, 7007) using a multichannel pipette. After seeding, plates were centrifuged (1,600 rpm × 10 min) and incubated for 4 d at 37 °C until spheroids reach 380- to 400-μm diameter. Once the spheroids were formed, TO-PRO-3 (1 μM) and Hoechst (0.1 μM) were added. Images were obtained using a confocal ultraspectral microscope (Leica TCS-SP5) or the Opera High Content Screening system (Perkin-Elmer). Quantifications were performed using the ImageJ software.
Western Blot Analysis.
Protein extraction was performed in protein lysis buffer (50 mM Tris⋅HCl pH 7.4, 150 mM NaCl, 0.5% Nonidet P-40) supplemented with a mixture of protease and phosphatase inhibitors (cOmplete Mini, Roche, 11836153001; Phosphatase Inhibitor Mixture 2 and 3, Sigma, P5726 and P0044). To quantify the amount of protein obtained the Bradford (Bio-Rad) method was used. Next, 25 μg of protein extracts obtained from tumor tissues were separated on NUPAGE 4 to 12% Bis-Tris Midi gels (Invitrogen), transferred to a nitrocellulose membrane (GE Healthcare) and blotted with antibodies raised against: CDK4 (Santa Cruz 260), RAF1 (BD Biosciences, 610151), B-RAF (Santa Cruz 5284), A-RAF (Cell Signaling 4432), panRAS (Calbiochem OP40), ERK1 (BD Pharmingen, 554100), ERK2 (BD Biosciences, 610103), phospho-ERK (Cell Signaling, 9101), MEK1 (Santa Cruz 219), pMEK1/MEK2 (Cell Signaling, 9154), AKT (Cell Signaling, 9272), phospho-AKT (Cell Signaling, 4060), caspase-3 (C3) (Cell Signaling, 9662), cleaved caspase-3 (CC3) (Cell Signaling, 9661), phosphor-RB (Cell Signaling 9308), RB (Santa Cruz 50), CYCLIN D1 (Neomarkers MS-210), CDK6 (Neomarkers MS-451), CDK2 (Abcam 32147), CYCLIN E2 (Santa Cruz 28351), and β-actin (Sigma, A5441). Primary antibodies were detected as previously described (9).
Apoptosis Assays.
Kras+/G12V;Trp53−/−;hUBC-CreERT2+/T tumor cells harboring Cdk4L, Cdk4FxKD, and/or Raf1L conditional alleles were maintained at 37 °C in DMEM (Gibco) containing 10% FBS (Gibco) and 600 nM 4OHT for 4 d. Cells were harvested at 0, 24, 48, 72, and 96 h postexposure to 4OHT. Cell lysates were analyzed by Western blot for the expression of CDK4, CDK4KD, RAF1, cleaved caspase-3 (CC3), caspase-3, and β-actin.
Senescence-Associated β-Galactosidase (SA-β-gal) Expression.
To identify senescent cells in vivo, lung tissues were harvested during necropsy and embedded in optimal cutting temperature compound (OCT) and frozen at −80 °C. Then, 10-μm-thick sections were cut using a cryostat. Tissues were fixed for 5 min at room temperature with 2% formaldehyde/0.2% glutaraldehyde solution (Sigma Aldrich). After fixation, samples were incubated at 37 °C in 5-bromo-4-chloro-3-indolyl β-d-galactopyranoside (X-Gal) staining solution (1× citric acid/sodium phosphate buffer pH6.0; 500 mM potassium ferricyanide [Prolab]; 500 mM potassium ferrocyanide [Prolab]; 150 mM NaCl; 2 mM MgCl2, and 1 mg/mL of the substrate X-Gal [Applichem] dissolved in N-N-dimethylformamide). After 12 h, samples were washed in PBS and representative pictures were taken under bright-field microscopy.
In Vitro Drug Treatments.
Lung tumor cell lines were plated at 3,000 cells per well in triplicates in 96-well plates and grown for 24 h. Doses were separated by threefold and spanned from 1 nM to 20 μM. Control cells were incubated with media containing DMSO. To calculate the GI50, values were plotted against the inhibitor concentrations and fit to a sigmoid dose–response curve using GraphPad Prism (v7.01) software. Cell viability was assessed using the Cell Titer Glo Luminescent Cell Viability Kit (Promega, G7571). Luminescence counts were read in a Victor plate reader (Perkin-Elmer). For colony formation assays, 500 cells per well were seeded in 96-well plates; 2 µM 5-AZA (Sigma, A2385) was added on days 0, 1, 2, 3, 6, and 9. Then, 1 µM CNIO-PI3Kinh was added on days 0, 3, 6, and 9. For compound screenings, a library of 114 drugs was used at a unique 5-µM dose during 72 h. The compounds included in this library are indicated in SI Appendix, Table S3.
Quantitative RT-PCR.
Total RNA was extracted using RNeasy Micro Kit (Qiagen) following the manufacturer’s protocol. DnaseI digestion was performed to eliminate residual genomic DNA. cDNA synthesis was performed using the SuperScript II Reverse Transcription Kit (Thermo Fisher Scientific) following the manufacturer’s protocol. Primers used included: Lrig3 forward: 5′ CTGACTGTGCTAGAGACGCC 3′; Lrig3 reverse: 5′ GGGCTGTCATCTTTCGTCCA 3′; Rspo2 forward: 5′ GCCGCTGCTTTGATGAATGT 3′; Rspo2 reverse: 5′ ACAATCTGCCGTGTTCTGGT 3′; Nrg1 forward: 5′ TCTGTGTGAATGGAGGCGAG 3′; Nrg1 reverse: 5′ CACTTGCACAAGTATCTTGAGGG 3′; β-actin forward: 5′ GGCACCACACCTTCTACAATG 3′ and β-actin reverse: 5′ GTGGTGGTGAAGCTGTAGCC 3′. Quantitative RT-PCR assays were performed with the AB 7900 Fast Real Time PCR System using Power SYBR Green PCR Master Mix (Applied Biosystems). The relative mRNA expression was calculated using the comparative cycle threshold (Ct), where ΔCt is the Ct value for any sample normalized to the endogenous housekeeping gene (β-actin).
RNA-Seq and Data Analysis.
The quality of the RNA-seq reads was evaluated with FastQC (www.bioinformatics.babraham.ac.uk/projects/fastqc/). The RNA-seq reads were mapped onto the reference mouse genome GRCm38/mm10 using STAR (v2.4.0j) (34). The abundance of each gene was quantified as transcripts per million value and evaluated by a statistical method RNA-seq by expectation maximization. DESeq2 analysis was used to call the DEGs between two conditions compared (35). The DEGs were called using adjusted P < 0.05 as the cutoff. To identify the heterogeneity of the different CDK4/RAF1 resistant clones, we compared the parental cell lines (T1, T2, and T3) with their corresponding clones, R1.1, R1.2, R1.3, R2.1, R2.2, R2.3, R3.1, R3.2, and R3.3, respectively. In addition, all of the CDK4/RAF1 resistant clones were compared among each other. We used the DEGs among the nine resistant clones to generate a hierarchical clustering and heatmaps with the “pheatmap” package in R software. PCA, an unsupervised learning technique, was used to generate first and second principal components and the resistant clones were clustered based on these principal components. GSEA (36) was employed to determine the gene sets, including Kyoto Encyclopedia of Genes and Genomes, gene ontology, cancer hallmarks, and chemical and genetic perturbations databases, enriched by a preranked list of all genes, which were sorted by the statistical significance of differential expression defined by DESeq2 analysis. Chemical and genetic perturbations database includes 3,433 gene sets, representing expression signatures of genetic and chemical perturbations. Since GSEA gene sets were annotated with human genes, we converted the corresponding mouse genes to their human orthologs for this analysis. ssGSEA (37) was used to generate an activity profile of pathways that were significantly enriched by DEGs (false-discovery rate adjusted P < 0.05).
Statistical Analysis.
All values are expressed as mean ± SEM. P values were calculated with the unpaired Student’s t test using the GraphPad Prism (v7.01) software. A P value ≤ 0.05 is considered statistically significant. Significant differences between experimental groups were: *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Supplementary Material
Acknowledgments
We thank S. Ortega for the generation of the Cdk4FxKD mouse model; and M. San Román, R. Villar, M. C. González, A. López, N. Cabrera, P. Villanueva, J. Condo, J. Klett, A. Cebriá, A. Otero, O. Domínguez, G. Luengo, G. Garaulet, F. Mulero, and D. Megías for excellent technical support. This work was supported by European Research Council Grant ERC-2015-AdG/695566, THERACAN, Spanish Ministry of Science, Innovation, and Universities Grant RTC-2017-6576-1, and the Autonomous Community of Madrid Grant B2017/BMD-3884 iLUNG-CM (to M.B.); Spanish Ministry of Science, Innovation, and Universities Grant RTI2018-094664-B-I00 (to M.B. and M.M.); and National Natural Science Foundation of China Grant 31771469 (to H.W.). M.B. is a recipient of an Endowed Chair from the AXA Research Fund. L.E.-B. is the recipient of an FPI fellowship from the Spanish Ministry of Economy and Competitiveness. F.F.-G., M.S., and P.N. were supported by an FPU fellowships from the Spanish Ministry of Education.
Footnotes
Competing interest statement: S.-B.P. is an employee at the Oncology Research at Eli Lilly and Company. M.B. reports a research contract with Oncology Research, Eli Lilly and Company.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2002520117/-/DCSupplemental.
Data Availability.
All data are available in the main text and SI Appendix.
References
- 1.Canon J. et al., The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature 575, 217–223 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Ostrem J. M., Peters U., Sos M. L., Wells J. A., Shokat K. M., K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature 503, 548–551 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drosten M., Barbacid M., Targeting the MAPK pathway in KRAS-driven tumors. Cancer Cell 37, 543–550 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Blasco R. B. et al., c-Raf, but not B-Raf, is essential for development of K-Ras oncogene-driven non-small cell lung carcinoma. Cancer Cell 19, 652–663 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ryan M. B., Corcoran R. B., Therapeutic strategies to target RAS-mutant cancers. Nat. Rev. Clin. Oncol. 15, 709–720 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Jänne P. A. et al., Selumetinib plus docetaxel compared with docetaxel alone and progression-free survival in patients with KRAS-mutant advanced non-small cell lung cancer: The SELECT-1 randomized clinical trial. JAMA 317, 1844–1853 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flaherty K. T. et al.; METRIC Study Group , Improved survival with MEK inhibition in BRAF-mutated melanoma. N. Engl. J. Med. 367, 107–114 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Blumenschein G. R. Jr. et al., A randomized phase II study of the MEK1/MEK2 inhibitor trametinib (GSK1120212) compared with docetaxel in KRAS-mutant advanced non-small-cell lung cancer (NSCLC). Ann. Oncol. 26, 894–901 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanclemente M. et al., c-RAF ablation induces regression of advanced Kras/Trp53 mutant lung adenocarcinomas by a mechanism independent of MAPK signaling. Cancer Cell 33, 217–228.e4 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Puyol M. et al., A synthetic lethal interaction between K-Ras oncogenes and Cdk4 unveils a therapeutic strategy for non-small cell lung carcinoma. Cancer Cell 18, 63–73 (2010). [DOI] [PubMed] [Google Scholar]
- 11.Lagarrigue S. et al., CDK4 is an essential insulin effector in adipocytes. J. Clin. Invest. 126, 335–348 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Ruzankina Y. et al., Deletion of the developmentally essential gene ATR in adult mice leads to age-related phenotypes and stem cell loss. Cell Stem Cell 1, 113–126 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jackson E. L. et al., The differential effects of mutant p53 alleles on advanced murine lung cancer. Cancer Res. 65, 10280–10288 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Barrière C. et al., Mice thrive without Cdk4 and Cdk2. Mol. Oncol. 1, 72–83 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beaver J. A. et al., FDA approval: Palbociclib for the treatment of postmenopausal patients with estrogen receptor-positive, HER2-negative metastatic breast cancer. Clin. Cancer Res. 21, 4760–4766 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Corona S. P., Generali D., Abemaciclib: A CDK4/6 inhibitor for the treatment of HR+/HER2- advanced breast cancer. Drug Des. Devel. Ther. 12, 321–330 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peng S. B. et al., Inhibition of RAF isoforms and active dimers by LY3009120 leads to anti-tumor activities in RAS or BRAF mutant cancers. Cancer Cell 28, 384–398 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Oser M. G., Niederst M. J., Sequist L. V., Engelman J. A., Transformation from non-small-cell lung cancer to small-cell lung cancer: Molecular drivers and cells of origin. Lancet Oncol. 16, e165–e172 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ortega-Molina A. et al., Pharmacological inhibition of PI3K reduces adiposity and metabolic syndrome in obese mice and rhesus monkeys. Cell Metab. 21, 558–570 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jonna S. et al., Detection of NRG1 gene fusions in solid tumors. Clin. Cancer Res. 25, 4966–4972 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kvarnbrink S. et al., LRIG1 is a prognostic biomarker in non-small cell lung cancer. Acta Oncol. 54, 1113–1119 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Wu L. et al., R-spondin family members as novel biomarkers and prognostic factors in lung cancer. Oncol. Lett. 18, 4008–4015 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ruscetti M. et al., NK cell-mediated cytotoxicity contributes to tumor control by a cytostatic drug combination. Science 362, 1416–1422 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schick U. et al., Trametinib radiosensitises RAS- and BRAF-mutated melanoma by perturbing cell cycle and inducing senescence. Radiother. Oncol. 117, 364–375 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Wang Y. et al., Mutant LKB1 confers enhanced radiosensitization in combination with trametinib in KRAS-mutant non-small cell lung cancer. Clin. Cancer Res. 24, 5744–5756 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Hangauer M. J. et al., Drug-tolerant persister cancer cells are vulnerable to GPX4 inhibition. Nature 551, 247–250 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramirez M. et al., Diverse drug-resistance mechanisms can emerge from drug-tolerant cancer persister cells. Nat. Commun. 7, 10690 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee C. L. et al., Generation of primary tumors with Flp recombinase in FRT-flanked p53 mice. Dis. Model. Mech. 5, 397–402 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jesenberger V. et al., Protective role of Raf-1 in Salmonella-induced macrophage apoptosis. J. Exp. Med. 193, 353–364 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schnütgen F. et al., A directional strategy for monitoring Cre-mediated recombination at the cellular level in the mouse. Nat. Biotechnol. 21, 562–565 (2003). [DOI] [PubMed] [Google Scholar]
- 31.Rodríguez C. I. et al., High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat. Genet. 25, 139–140 (2000). [DOI] [PubMed] [Google Scholar]
- 32.Harashima H., Sekine M., Measurement of plant cyclin-dependent kinase activity using immunoprecipitation-coupled and affinity purification-based kinase assays and the baculovirus expression system. Methods Mol. Biol. 779, 65–78 (2011). [DOI] [PubMed] [Google Scholar]
- 33.Matsushime H. et al., D-type cyclin-dependent kinase activity in mammalian cells. Mol. Cell. Biol. 14, 2066–2076 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dobin A. et al., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Subramanian A. et al., Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barbie D. A. et al., Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 462, 108–112 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the main text and SI Appendix.