Significance
Despite its enormous evolutionary success (it is the carboxylating enzyme of all photosynthetic pathways from microorganisms to higher plants), Rubisco is rather inefficient due to wasteful competitive inhibition by molecular oxygen. Quite critically, the intimate mechanism of O2 addition is unknown. We show here that isotope effects (13C, 18O, 25Mg, or 2H) and high level computations of redox potential are all consistent with oxygen acting as an oxidant in a redox reaction generating superoxide which then attacks the substrate. Our results explain why the elimination of oxygenation by enzymatic bioengineering is so difficult, because it would require a drastic change in electrostatic and/or redox potential of the substrate, and this would alter carboxylation activity.
Keywords: Rubisco, oxygenation, mechanism, photosynthesis, isotope effect
Abstract
Ribulose 1,5-bisphosphate carboxylase/oxygenase (Rubisco) is the cornerstone of atmospheric CO2 fixation by the biosphere. It catalyzes the addition of CO2 onto enolized ribulose 1,5-bisphosphate (RuBP), producing 3-phosphoglycerate which is then converted to sugars. The major problem of this reaction is competitive O2 addition, which forms a phosphorylated product (2-phosphoglycolate) that must be recycled by a series of biochemical reactions (photorespiratory metabolism). However, the way the enzyme activates O2 is still unknown. Here, we used isotope effects (with 2H, 25Mg, and 18O) to monitor O2 activation and assess the influence of outer sphere atoms, in two Rubisco forms of contrasted O2/CO2 selectivity. Neither the Rubisco form nor the use of solvent D2O and deuterated RuBP changed the 16O/18O isotope effect of O2 addition, in clear contrast with the 12C/13C isotope effect of CO2 addition. Furthermore, substitution of light magnesium (24Mg) by heavy, nuclear magnetic 25Mg had no effect on O2 addition. Therefore, outer sphere protons have no influence on the reaction and direct radical chemistry (intersystem crossing with triplet O2) does not seem to be involved in O2 activation. Computations indicate that the reduction potential of enolized RuBP (near 0.49 V) is compatible with superoxide (O2•−) production, must be insensitive to deuteration, and yields a predicted 16O/18O isotope effect and energy barrier close to observed values. Overall, O2 undergoes single electron transfer to form short-lived superoxide, which then recombines to form a peroxide intermediate.
Rubisco is the most abundant enzyme of the biosphere (1) and catalyzes the fixation of about 120 gigatons (10 petamol) of carbon from atmospheric CO2 each year. Rubisco-catalyzed carboxylation is in fact the first step of autotrophic metabolism in the vast majority of photo- and chemosynthetic organisms. However, Rubisco is also responsible for O2 fixation. About 100 gigatons (3.1 petamol) of O2 are assimilated each year by terrestrial vegetation. This huge flux participates in defining the 16O/18O isotope signature of atmospheric oxygen and the so-called Dole effect (O2 naturally 18O-enriched by 23.5‰ compared to mean ocean water). Furthermore, it leads to a loss of carbon (in the form of CO2) of about 20 gigatons per year in photorespiratory metabolism. Rubisco-catalyzed oxygenation is thus considered as an energy and carbon loss for plant production, and its suppression is assumed to have the potential to increase crop yield by up to 55% (2). Although this assumption is probably too simplistic because photorespiration is involved in nitrogen metabolism, sulfur assimilation, and pathogen defense (3–6), it is certainly desirable to minimize oxygenation to improve photosynthesis. However, methods to suppress oxygenation are currently limited by our poor understanding of its chemical mechanism, in contrast to carboxylation (mechanism summarized in Fig. 1 and detailed in SI Appendix, Fig. S1). Also, manipulating Rubisco in crops to improve its specificity faces other important challenges such as molecular interaction with proteins (e.g., Rubisco activase and chaperones) (7) or a persistent trade-off between specificity and carboxylation velocity (8).
Fig. 1.
Simplified mechanism of Rubisco-catalyzed oxygenation and carboxylation showing the presumed fate of protons. For clarity, C3 and O3 protons are labeled in blue and red, respectively. This figure only shows Lys residues (numbering in higher plants) directly involved in the mechanism. Enolization forms a cycle that eventually leads to the loss of the C3 proton in solvent water when ribulose 1,5-bisphosphate (RuBP) is regenerated. This figure assumes that gas addition and hydration are concerted events and accompanied by a proton relay. A larger and more detailed scheme is provided in SI Appendix, Fig. S1. The involvement of active site residues is further explained in SI Appendix, Fig. S2. The notation “Lys201C” stands for carbamylated Lys-201.
Rubisco is the first cofactor-less oxygenase that has been discovered (9), and oxygenation consists of O2 addition to the enolized form of RuBP (enolate), forming a peroxide (Fig. 1). In its ground state, O2 is a biradical triplet (3Σg−) and its addition to singlet enolate is spin forbidden. To overcome this problem, most oxygenases involve a metal or reducing cofactors (10). Rubisco-catalyzed oxygenation is believed to stem from intrinsic reactivity of the enolate toward O2 and has thus been assumed to be inevitable (11, 12). However, reactivity with O2 is not systematic in enolate-forming enzymes (and more generally, carbanion-forming enzymes, involving enolates, enaminates, or quinonoid intermediates) since they may or may not have an oxygenase activity; furthermore, some oxygenases can catalyze O2 addition onto aromatic substrates without cofactor or metal ion (13). In other words, the reactivity of the enolate with O2 depends on subtle stereochemical and electrostatic constraints in the enzyme active site and is not a fixed, inevitable feature. It is also possible that in the case of Rubisco, the geometry of the active site allowing CO2 fixation adventitiously leads to O2 binding. In fact, in higher plants, the apparent Michaelis constant (Km) for O2 is much larger than 1) that for CO2 and 2) cellular dissolved O2 concentration, suggesting that perhaps, O2 binding is an intrinsic property of the chemical environment created by the active site and thus cannot be easily suppressed.
Two main mechanisms can be considered that enable O2 to overcome the “spin problem” inherent in cofactor-less oxygenases like Rubisco (10, 14, 15): 1) single electron transfer (SET) where the RuBP enolate reduces O2, yielding radical intermediates that recombine to form a RuBP peroxide; or 2) direct O2 attack (electrophilic O2 addition) whereby one or another of the reactants undergoes intersystem crossing (ISC) so as to permit the formation of the peroxide.
A SET-based mechanism has been suggested early on (16) but there is presently little experimental evidence. Unfortunately, direct detection of superoxide (O2•−) during the reaction using electron paramagnetic resonance (EPR) remains problematic, not only because of superoxide’s short lifetime, but also because of the presence of multiple signals of the enzyme itself (without reaction) at the same magnetic strength as superoxide (g ∼ 2) (17). However, H2O2 has been shown to be generated during oxygenation (about one molecule per 200 catalytic cycles) (18), suggesting that a reactive oxygen species might be involved in the mechanism (or that the peroxide can be broken down, yielding hydrogen peroxide). Ab initio calculations with density functional theory (DFT) using energy expression based on hybrid energy functionals B3LYP have suggested that SET is plausible, since the minimum-energy structure with bound oxygen is found to correspond to a biradical complex with partial electron transfer to O2 (19). It has been suggested that most cofactor-less enzymes such as glucose oxidase activate O2 by SET followed by spin transition via spin orbit coupling (SOC) (14, 20), and Rubisco, perhaps, involves a similar mechanism.
Three variants of the mechanism based on ISC can be considered:
-
1)
O2 ISC. If a direct O2 attack occurs, ground-state O2 could be excited to a singlet form (1Δg) which could then react with the enolate. This assumption has found some support from experiments where Mg2+ was substituted by Mn2+, thereby causing chemiluminescence during O2 fixation (21). However, the spectrum of emitted light is not compatible with singlet O2 deexcitation (22), and furthermore, singlet O2 deexcitation should be far too rapid to allow reaction with the enolate (reviewed in ref. 23).
-
2)
RuBP enolate ISC. Computations have suggested that a direct attack is possible because the C2–C3 torsion in enzyme-bound RuBP likely lowers the energy gap of the singlet-to-triplet transition of the enolate (24, 25). The major problem with the direct O2 attack is that it must be sensitive to slight alterations in the hydrogen bonds network as well as other effects of the Mg2+ coordination sphere, which all affect both the force constant between C2 and attacking O2, and how the forming partial charge of O2 is stabilized. Thus, in principle, significant differences in kinetic 16O/18O isotope effect among Rubisco forms are expected, for which evidence is to the contrary (26, 27).
-
3)
Paramagnetic enhanced ISC. It is well known that the presence of paramagnetic species can enhance ISC when reactions involve radical pairs (RPs) (28). Substitution of the physiologically relevant divalent metal ion Mg2+ with other paramagnetic divalent ions (Mn2+, Co2+, or Ni2+) is known to favor the oxygenase reaction over the carboxylation reaction (29), a phenomenon that might be attributable to paramagnetic enhanced ISC. Also, paramagnetic transition metal ions (like Fe2+, Cu2+, or Mn2+) that facilitate spin-forbidden reactions are commonly found in oxygenases. The Mg2+ ion is seldom considered as a magnetic species. However, bulk Mg is a mixture of three isotopes, 24Mg (∼79%), 25Mg (∼10%), and 26Mg (∼11%). Isotope 25Mg is commercially available in almost isotopically pure form. Although the 25Mg2+ ion does not have unpaired electrons and thus is not paramagnetic, it has a nuclear magnetic moment (30). The possible role of magnetic susceptibility of 25Mg2+ causing mass-independent isotope fractionation phenomena is debated for several biological reactions (31). We therefore considered the possibility that the oxygenase reaction catalyzed by Mg2+-activated Rubisco might be the consequence of magnetically enhanced ISC.
Altogether, the O2 attack hypothesis implies ISC of the enolate without charge separation and then reaction of a noncharged RP (triplet enolate + biradical O2) while SET directly forms a charged RP (RuBP enolate•+ + O2•‒) that undergoes reaction. Despite this obvious chemical difference, efforts devoted to Rubisco’s chemistry in the past 30 y have not provided definitive experimental evidence and thus explained O2 addition. Here, we took advantage of multiple isotopic substitutions and looked at their impact on oxygenation kinetics. We tested whether the 16O/18O kinetic isotope effect, which gives direct information on O2 chemistry, varies when enzyme CO2/O2 specificity changes, the reaction is slowed down with deuterium, or the active site metal is changed. By studying how these conditions affected O2 fixation, and by comparing with carboxylation, we present a catalytic scenario for Rubisco-catalyzed O2 activation.
Results
Proton Exchange Capacity in Rubisco Forms.
Before carrying out isotopic substitution (with deuterium, 2H) to measure isotope effects, the ability to carry out proton exchange and thus wash out the H3 proton of RuBP (32) was assessed. In fact, enolization is reversible and may result in H exchange whereby the H3 of RuBP is lost in the solvent and replaced by a proton (deuteron) from the solvent (Fig. 1).
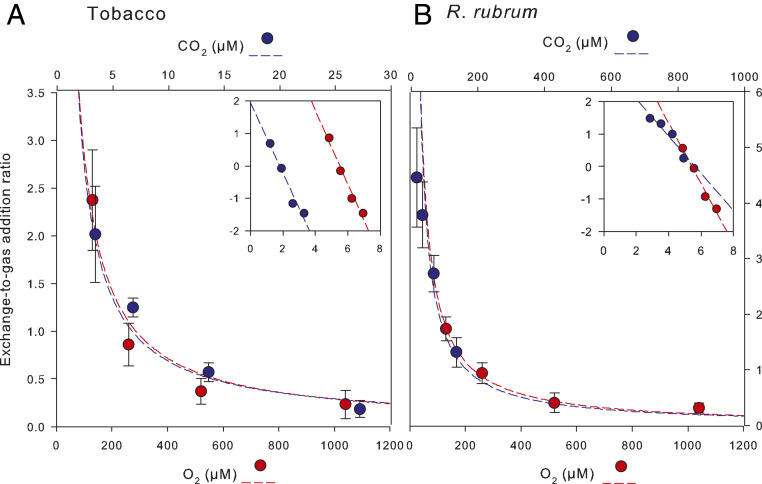
Two Rubisco forms (multimeric L8S8 higher plant form, tobacco; and dimeric L2 prokaryotic form, Rhodospirillum rubrum) of contrasted specificity (about 80 and 9, respectively) were used. The 1H/2H isotope composition of RuBP at H3 (along with the amount of catalysis products) was monitored by NMR when enzymes were assayed in 2H2O with natural RuBP (h-RuBP) at saturating concentration (Fig. 2). The appearance of 2H in RuBP at H3 was expressed as the exchange-to-catalysis ratio (mathematics explained in SI Appendix, Notes S1). As expected (33), there was a clear hyperbolic decrease of exchange rate, reflecting the higher commitment to catalysis as dissolved gas concentration increased. At low dissolved gas concentration, the rate of exchange with the prokaryotic enzyme was much higher than in the plant enzyme (about double). Regressions show that the reverse commitment to catalysis (deenolization-to-gas addition ratio of rate constants) was 7 ± 1 (CO2) and 500 ± 40 (O2) µM in tobacco and 25 ± 3 and 180 ± 25 (O2) µM in R. rubrum, demonstrating a much higher propensity to achieve H exchange in the prokaryotic enzyme during carboxylation catalysis. Nevertheless, the prokaryotic enzyme is much less CO2 specific than the tobacco enzyme and therefore, oxygenation is relatively faster, giving less opportunity for H exchange when assayed with O2.
Fig. 2.
Exchange to gas-addition kinetic partitioning in the carboxylase (blue) or oxygenase (red) reaction versus dissolved CO2 or O2 concentration, respectively, using Rubisco from tobacco (A) or R. rubrum (B). Dashed lines represent hyperbolic regressions. (Insets) Log-log relationship with linear regressions. Data are mean ± SE of n = 4 replicates. Exchange was monitored using the appearance of deuterium at the H3 position when the enzyme was assayed with ordinary ribulose 1,5-bisphosphate (deuterium at natural abundance) in 96% D2O as a solvent.
Deuteration Alters CO2- but Not O2-Addition Chemistry.
Gas addition rates, CO2/O2 specificity (kinetic partitioning), and 12C/13C and 16O/18O isotope effects were then measured with natural or heavy water (2H2O) as a solvent, using natural or 2H-3-RuBP (denoted as d-RuBP hereafter) as a substrate. Since the reaction starts with RuBP enolization via abstraction of the H3 proton of RuBP, a significant 1H/2H isotope effect was expected on both CO2 and O2 fixation rates, as observed before (34, 35). In fact, in both Rubisco forms, assays with solvent H2O and d-RuBP slowed down CO2 fixation significantly (Fig. 3). However, the tobacco enzyme was much more affected by d-RuBP (in H2O) than the prokaryotic enzyme (isotope effect of 2.7, compared to 1.5), showing little loss of 2H via deenolization. When 2H2O was used as a solvent with natural RuBP (h-RuBP), there was a moderate solvent isotope effect in tobacco (1.7, also reflecting isotope effects on steps other than enolization) while the effect was large in R. rubrum and could not be increased further with d-RuBP as a substrate.
Fig. 3.
Rubisco catalysis rate for carboxylation (vc, blue) and oxygenation (vo, red) under different conditions of deuteration, using the enzyme from tobacco (A and B) or R. rubrum (C and D). Assays were carried out using ordinary water (H2O) or deuterated water (D2O, 96%) as a solvent, with ordinary (i.e., deuterium at natural abundance) or deuterated (99% D at the H3 atom position) ribulose 1,5-bisphosphate (RuBP). Carboxylation was assayed at 45 µM (tobacco) and 150 µM (R. rubrum) dissolved CO2, and oxygenation at 200 µM dissolved O2. Data are mean and SD of n = 4 replicates. Letters stand for statistical classes: when two bars harbor distinct letters, it indicates that the values are statistically different (ANOVA, P < 0.01). Numbers above bars indicate the average H/D isotope effect (with reference to H2O + h-RuBP).
Quite critically, the isotope effect (with either 2H2O or d-RuBP) on the oxygenation rate was significantly lower than that on carboxylation (Fig. 3 B and D), with a value of 1.8 only in tobacco and a very small isotope effect (1.3) in R. rubrum with 2H2O + d-RuBP (and no isotope effect at all under other conditions). In other words, even when the enzyme was forced to use deuterons to either reform RuBP or process the oxygenation intermediate, the isotope effect remained small (1.3 to 1.8) in contrast to carboxylation (2.9 to 3.6), demonstrating that O2 addition itself is unlikely to depend on H bonds with attacking O2 or to involve protonation of oxygen atoms.
The small effect of deuteration on oxygenation rate was not due to a dramatic decrease in specificity making oxygenation much less rate limiting. In fact, there was a small increase in specificity with 2H2O + d-RuBP in tobacco (Fig. 4A). In the prokaryotic enzyme, the use of 2H2O decreased specificity including when natural RuBP was the substrate, while the isotope effect on oxygenation rate was unity, simply reflecting the fact that deuteration affected carboxylation much more than oxygenation (Fig. 3 C and D). Furthermore, there was no change at all in the V/K 16O/18O kinetic isotope effect [denoted as 18(V/K)] associated with oxygenation, in clear contrast with the 12C/13C isotope effect [13(V/K)], which was considerably lower in 2H2O, by up to 15‰ (Fig. 4). This effect was not caused by proton exchange and the formation of d-RuBP during deenolization, simply because the use of d-RuBP as a substrate induced an increase—not a decrease—in the 12C/13C isotope effect.* This implies that CO2 addition appears to be quite sensitive to the H bond network of the active site or to acid/base properties. That is, chemical events concerted with CO2 addition, such as C3 hydration or O2/O3 reprotonation are essential to define the force constant between C2 and attacking CO2 in the transition state. We observe that 2H2O decreased 13(V/K) more in R. rubrum than in the tobacco enzyme (Fig. 4), probably reflecting a higher dependence of CO2 addition on C3 hydration and/or O2/O3 reprotonation in the prokaryotic enzyme. By contrast, O2 addition is independent of such chemical events in both the prokaryotic and plant enzyme.
Fig. 4.
Rubisco CO2/O2 specificity (Sc/o, gray), carbon and oxygen kinetic isotope effects (13(V/K), blue, and 18(V/K), red, respectively) under different conditions of deuteration, using the enzyme from tobacco (A, C, and E) or R. rubrum (B, D, and F). Assays were carried out using ordinary water (H2O) or deuterated water (D2O, 96%) as a solvent, with ordinary (i.e., deuterium at natural abundance) or deuterated (99% D at the H3 atom position) ribulose 1,5-bisphosphate (RuBP). Data are mean and SD of n = 4 replicates. Letters stand for statistical classes as in Fig. 3 (P < 0.01). The asterisk in A stands for near significance (P = 0.08) for the difference between solvent H2O and solvent D2O with ordinary ribulose 1,5-bisphosphate as the substrate. NS, no significant difference across assay conditions.
Proton Substitution also Concerns O3.
Of particular significance is the small but significant increase in 13(V/K) when the enzyme is assayed with d-RuBP (Fig. 4 C and D). In fact, it demonstrates that the H3 proton has not yet been exchanged with the solvent when gas addition takes place, and therefore, participates in the H bond network or protonation events concerted with CO2 addition. This result is consistent with the involvement of a proton relay between residues (including the carbamate, and water) whereby the H3 proton can be redistributed to another site and eventually lost in the solvent during or after CO2/O2 addition. Presumably, such a proton relay must render proton exchange in the enolate at O2 and O3 easier, with either the H3 proton or another H atom that comes from water. RuBP isotopologues formed during assays with natural RuBP (substrate) + 2H2O (solvent) were monitored with accurate mass liquid chromatography-mass spectrometry (LC-MS), in order to detect the possible appearance of bideuterated molecules (Fig. 5). Samples obtained after 60-s reaction time were frozen and vacuum dried to remove the solvent (2H2O) and then resuspended in a H2O-based buffer for LC-MS analysis. The H3 proton is not exchangeable, and the spontaneous proton exchange at O3 with the solvent is relatively slow (37). That is, the amount of 2H in O3 obtained in this analysis is certainly underestimated because of the loss of 2H during sample preparation for LC-MS. Still, RuBP molecules carrying two 2H atoms could be easily detected, in addition to monodeuterated RuBP. Here, the appearance of a second 2H atom in RuBP is interpreted as being due to O3 deuteration. We recognize that an isotopic exchange could also have occurred at other positions, such as H atoms attached to C1. However, this contribution must be very small considering their high pKa and the very slow rate of H+ dissociation, even under alkaline conditions (38, 39). In addition, there was no doubly labeled RuBP formed without the enzyme (Fig. 5), demonstrating that the incorporation of both 2H atoms was not spontaneous. Calculation of % 2H at each site from observed isotopologue abundance showed that the O3 proton was more abundant at low commitment for catalysis, like the H3 proton. Accordingly, the highest value of % 2H at O3 was observed at nonsaturating dissolved O2 concentration (Fig. 5 E and F). We thus conclude that the lack of 2H/1H isotope effect on 18(V/K) (Fig. 4) shows that O2 addition was independent of H exchange not only at H3 but also at O3.
Fig. 5.
Isotopic species of ribulose 1,5-bisphosphate (RuBP) after incomplete reaction run (5% reaction) in D2O as a solvent with ordinary RuBP (deuterium at natural abundance) as a substrate and Rubisco from tobacco. Assays were carried out with CO2 (carboxylase, blue) or O2 (oxygenase, red) at the concentration (in µM) shown on x axes. A control with no reaction (NR) is also shown. Isotopic species were monitored using accurate mass high resolution LC-MS analysis. Protiated [i.e., nondeuterated (A), monodeuterated (B), and dideuterated (C)] RuBP in % of total nonconsumed RuBP. (D and E) Proportions of deuterium at the two sites of exchange (presumably, C3 and O3) calculated from B and C. (F) Relationship between % D at the two sites. Letters stand for statistical classes (P < 0.05, ANOVA). Other nondeuterated isotopic species that did not vary or were negligible (18O, 13C, and 13C2) are not shown. ND, value hardly visible on this graph (0.004%).
Reduction Potential of the Enolate and Expected 18(V/K).
The reduction potential of the enolate was estimated by quantum-chemical calculations, with or without isotopic substitution to deuterium at O3 and when applicable (fully protonated species), also at O2 (Fig. 6). The reduction potential (vs. standard hydrogen electrode [SHE]) of the noncharged species (enol) was found to be +0.49 V, which is higher than that of O2 → O2•− (−0.18 V at normal dissolved O2 concentration). Importantly, this value is not substantially affected by deuterium substitution (isotope effects on Keq of 1.00 to 1.08) meaning that SET from the enolate to O2 should not be affected by 2H, as observed. Similarly, there is little isotope effect on the oxidation potential of the charged species. However, in its dissociated (enolate) form, the reduction potential is much more negative, at about −0.37 V. Consistently, when the negative charge at O2 is accommodated by a point charge, the reduction potential has an intermediate value which depends on the distance between O2 and such a point charge (SI Appendix, Fig. S3). That is, the calculated reduction potential is quite sensitive to the charge carried by O2. However, in Rubisco’s active site, it is unlikely that both the O2 and O3 atoms are fully deprotonated or that the charge is not accommodated for, since there are H bonds stabilizing the structure with possible H+ shuttling back and forth with amino acid residues (His or Lys) (as suggested by both quantum mechanics/molecular mechanics (40) and the crystal structure (SI Appendix, Fig. S2) which suggests O2 is only ∼3 Å away from Lys residues) and furthermore, O2 and O3 participate in the Mg2+ coordination sphere. Fully deprotonated O2 and O3 is also unlikely in the context of a proton relay where deuteration at O3 (Fig. 5) may take place. Under the assumption that the potential is +0.49 V effectively, the Marcus theory of electron transfer provides an estimate of the kinetic 16O/18O isotope effect and the energy barrier, of 1.021 and 23 kcal mol−1, for a reorganization energy of about 55 kcal mol−1 (SI Appendix, Fig. S4). These values are consistent with both the observed isotope effect 18(V/K) (Fig. 3), and the presumed energy barrier of about 20 kcal mol−1 (23). As expected, the reorganization energy is larger than the average of self-reorganization energy in oxidoreduction of O2 (to superoxide) (46 kcal mol−1) and enols (∼20 kcal mol−1), that is, 33 kcal mol−1 as this does not account for distorting bonds and angles in reactants.
Fig. 6.
Estimated reduction potential of enediol(ate) forms of ribulose 1,5-bisphosphate (vs. SHE). For simplicity, R and R′ represent C1 (CH2O–PO42−) and C4–C5 groups (CHOH–CH2O–PO42−). The thermodynamic (equilibrium) H/D isotope effect associated with the redox half reaction (HK/DK) is shown in blue (positions of isotopic substitution recalled below; M stand for the nonsubstituted [monoisotopic] form). Here, calculations were carried out assuming that phosphate groups were deprotonated. Note that the form deprotonated in C2 (C2–O−) is less oxidant than oxygen while the form protonated in C2 is more oxidizing than oxygen. Reduction potentials of couples involving O2 as an oxidant (62) are indicated in red. Further calculations with variations in the point charge accommodating O2 are provided in SI Appendix, Fig. S3.
Isotopic Magnesium and Other Metals Do Not Change Oxygenation Kinetics.
Rubisco’s active site contains Mg2+ as an essential component for its structure and reactivity of the enolate since O2 and O3 participate in the Mg2+ coordination sphere. Although Mg2+ does not form a covalent O–Mg bond and is not reduced to Mg+ during catalysis, changing the metal ion is relevant here. In fact, the rate of ISC followed by RP recombination is low in most organic reactions, but may increase with magnetic fields and/or paramagnetic species via so-called “spin chemistry” or paramagnetic enhanced ISC (41, 42). As stated in the Introduction, when assayed with paramagnetic metal ions (such as Mn2+ or Co2+) instead of Mg2+, oxygenation is favored over carboxylation (29). However, such a change in specificity is not due to a large increase in the rate of oxygenation but a suppression of carboxylation. Furthermore, oxygenation with Co2+ or Mn2+ could involve a mechanism different from that with Mg2+ (22). Here, we used 25Mg (nuclear magnetic isotope) and assayed the enzyme under a small magnetic field (∼1.7 mT). In principle, if the reaction involves a weakly coupled radical pair (here, molecular oxygen and triplet enolate), magnetic properties must alter the probability of ISC via hyperfine coupling (28). However, we found no change in either specificity, 18(V/K), or oxygenation rate with either 25Mg or pure 24Mg, and there was no mass-independent isotope fractionation between 17O and 18O (SI Appendix, Figs. S5 and S6 and Table S1). In addition, we found no change in 18(V/K) with Mn2+ or Co2+, which promote oxygenation over carboxylation (SI Appendix, Fig. S6 and Table S1). Since the Mg2+ coordination sphere directly involves the enolate (Mg2+ being at a close distance from O2 and O3, SI Appendix, Fig. S2), the lack of effect of 25Mg/24Mg substitution or use of paramagnetic metals on 18(V/K) indicates that O2 addition is independent of the local magnetic field created by the metal and, therefore, paramagnetic enhanced ISC is unlikely.
Discussion
To solve the catalytic mechanism of Rubisco-catalyzed oxygenation, we studied processes occurring during O2 addition, using 16O/18O kinetic isotope effects, which provide direct information on oxygen chemistry. In fact, 18(V/K) in Rubisco catalysis practically equals 16ko/18ko, where ko is the rate constant of O2 addition (mathematics given in SI Appendix, Notes S1).
Our results show that 18(V/K) is 1.021 and invariant, not only in different Rubisco forms, but also with solvent 2H2O or d-RuBP (Fig. 4 E and F), although we show that the deuteron from d-RuBP remains in the active site during gas addition (as revealed by the large 2H effect on 13(V/K), Fig. 4 C and D). It demonstrates that O2 chemistry is independent of bond with protons and charge stabilization of oxygen atoms by H+ and thus rules out O2 attack at C2 with H+ stabilization of a nascent charge, HOO• production, or point charge stabilization of superoxide. A similar situation has been found in glucose oxidase, where the invariance of the isotope effect revealed a SET mechanism whereby O2 is reduced to superoxide which then attacks the reactant (43, 44). DFT calculations also indicate that in glucose oxidase, an electron is transferred from the cofactor FADH2 to O2 (where SET is facilitated by the low ionization potential of FADH2 and attraction of superoxide to a protonated histidine residue of the active site) and after triplet radical pair formation, the triplet-to-singlet transition involves SOC (reviewed in ref. 20). Similarly, such a two-step mechanism with superoxide formation occurs in many cofactor-less or cofactor-dependent oxygenases (10, 15).
The observed isotope effect of Rubisco-catalyzed oxygenation (1.021) is lower than the equilibrium isotope effect for O2 → O2•− half reaction (1.033) (44) and in agreement with the kinetic isotope effect calculated with the Marcus theory of redox reactions (when superoxide is formed with the enolate at a potential E0 of 0.49 V, Fig. 6 and SI Appendix, Fig. S4). However, this calculation neglects possible O atoms motion, while O atoms could be nonfrozen (Franck–Condon factor) and the O–O distance could change during superoxide production (45). In that case, the 16O/18O isotope effect is expected to be very sensitive to the O–O bond order, and is equal to 1.021 when the assumed O–O distance is increased by 0.05 Å (SI Appendix, Fig. S4). Under the assumption of an O2 attack on triplet enolate at C2 (instead of SET), computations of the isotope effect (Bigeleisen–Wolfsberg theory) also predicts an isotope effect of 1.021 when the O–O bond order decreases a lot (SI Appendix, Fig. S4A). However, in this scenario, the nascent charge on the distal O atom would be stabilized by Mg2+ and/or charged (protonated) residues and thus must depend on active site geometry and deuteration. In fact, substitution of a proton by a deuteron in charge stabilization should, in principle, change 18(V/K) by about 3 to 4‰ (SI Appendix, Fig. S4B), in contrast to experimental evidence. We also show that oxygenation is independent of the permanent magnetic environment created by the metal, which also rules out ISC of a (weakly coupled) radical pair such as O2 (biradical) + enolate triplet (28, 46). Instead, formation of the peroxide intermediate from superoxide might involve a triplet-to-singlet transition via electron orbital transition, which creates a transient magnetic field and spin flip (i.e., SOC).
The chemistry of O2 addition is thus considerably different from that of carboxylation, since the transition state of CO2 addition has a variable geometry among Rubisco forms [as shown by 13(V/K)], and carboxylation is sensitive to 2H substitution in both the solvent and substrate RuBP (Figs. 3 and 4). With 2H2O, the C2–COO bond order of the nascent carboxylate group appears to be lower leading to lower stretching wavenumber νCC (which predominates numerically in the kinetic isotope effect) and thus smaller 13(V/K). In other words, this secondary deuterium effect likely comes from charge destabilization of the nascent carboxylate, or O2 or O3 atoms. However, there are at least two sites of exchangeable H with contrasted effects, since d-RuBP increases 13(V/K), in contrast to 2H2O. The increase in 13(V/K) with d-RuBP does not come from carboxylation being more rate-limiting since 1) enolization is by far the most impacted step (intrinsic isotope effect near 9, ref. 35) and takes place prior to CO2 addition (33) (Fig. 1); and 2) 13(V/K) practically equals 12kc/13kc (where kc is the rate constant of CO2 addition) and thus can hardly be influenced by the rate constant of (de)enolization. It is also unlikely that the effect of deuterium is related to considerable changes in active site structure itself or Mg2+ coordination since substitution to Mn2+ does not change 13(V/K) (SI Appendix, Table S2). Rather, it shows that the H3 proton of RuBP participates somehow in the geometry of the transition state associated with CO2 addition. It perhaps participates in protonation of His-294/287 (residue numbering in higher plants/R. rubrum, respectively), Lys-175/166, O2, or O3, providing some support to the proton relay hypothesis from the carbamate (Lys-201/191) to other residues of the active site (such as His-294/287) (47) (SI Appendix, Fig. S2). Of course, the H3 proton is eventually lost (in the solvent) as shown using RuBP tritiated at H3 (48).
Altogether, we show that Rubisco catalyzes oxygenation via SET, likely facilitated by the proper dielectric environment of the active site. This piece of information is capital to understand the determinants of Rubisco specificity. Of course, facilitating O2 reduction was not a driving force of enzyme evolution since the oxygenase reaction is detrimental to cellular metabolism. Still, adapting the active site to faster carboxylation or higher specificity to CO2 did not eliminate the propensity for oxygen to react. Our results suggest that active site evolution has not been associated with a better charge stabilization of O2•− and O2, and thereby avoided undesirable increase in the driving force (−ΔG0) of O2 reduction. Variations in oxygenation rate between Rubisco forms are more likely linked to small changes in E0 and/or reorganization energy but little change in bound O2/O2•− zero-point energy, explaining 18(V/K) invariance. The present study also explains why eliminating oxygenation by bioengineering of the enzyme has proved unsuccessful so far, because it would require changing the dielectric environment of the active site or increasing E0 of the enolate, without affecting enolate geometry and H+ redistribution during CO2 addition. Obtaining such subtle rearrangements of the active site might be out of reach with current molecular technologies.
Materials and Methods
Chemicals and Protein Preparation.
All chemicals were purchased from Sigma-Aldrich, unless otherwise stated. Ninety-six percent 25MgO and 99% 24MgO were purchased from Trace Sciences International and converted to MgCl2 by the addition of a 10% HCl solution. Ninety-nine percent enriched D-[3-2H]ribose was purchased from Omicron Biochemicals. Purifed ribokinase was a gift from Kwaku Dayie, University of Maryland, College Park, MD. Tobacco (Nicotiana tabacum var. Wisconsin) Rubisco was purified as described in ref. 49. Briefly, Rubisco was precipitated from tobacco leaf extracts using 12% polyethylene glycol and then allowed to crystalize in a Tris buffer (25 mM) at pH 7.2. Rubisco crystals were collected and washed and finally redissolved (in a Tris buffer 25 mM at pH 7.6 containing 20% glycerol) for storage at −80 °C. R. rubrum Rubisco (N-terminal His4 tagged) was expressed in Escherichia coli (Bl21) using a pet28a(+) vector. Transformed E. coli were grown at 37 °C in a 1-L glass culture container in LB-ampicillin medium up to an OD of 0.5 to 0.7. Rubisco expression was induced overnight with 0.5 mM IPTG at 19 °C. Harvested cells were resuspended in 10 mL buffer (Tris 50 mM, NaCl 300 mM, imidazole 10 mM, pH 8.0) and lysed with an EmulsiFlex-B15 cell disruptor (Avestin) at a homogenizing pressure of about 15,000 psi. After centrifugation (20,000 × g, 5 min), the Rubisco protein was purified with a His-trap column on an ATKA system using an imidazole gradient (10 mM to 250 mM over 10 min). Purified Rubisco was precipitated with 50% ammonium sulfate, redissolved in 25 mM Tris (with EDTA 1 mM, pH 7.8) and desalted on a PD10 column. Glycerol (10% final content) was added for storage at −80 °C. Phosphoribulokinase (PRK) from Synechococcus was prepared similarly.
RuBP Synthesis.
RuBP was prepared enzymatically from ribose 5-phosphate by the sequential action of ribose 5-phosphate isomerase and PRK (50) in a reaction medium containing 10 mM ribose 5-phosphate, 20 mM ATP and 10 mM MgCl2 (adjusted to pH 7.4 with NaHCO3). Reaction completion was checked with 31P-NMR. Activated charcoal was then added to remove nucleotides, before adding a fivefold molar excess of barium acetate and ethanol to precipitate RuBP. The precipitate was then redissolved in water, and Ba2+ ions were removed with washed Dowex 50 H+. The product was frozen dried for storage at −80 °C. Purity was checked with 1H- and 31P-NMR. The same protocol was used to synthesize 2H-3-RuBP, using H3 deuterated ribose 5-phosphate (produced with D-[3-2H]ribose and ribokinase) as a starting material. The absence of signal of the H3 proton in the product was checked by 1H-NMR.
Specificity and Catalytic Rates.
Rubisco specificity factor (Sc/o) was measured with 31P-NMR according to ref. 51. Reactions were carried out at 25 °C in septum-capped 2-mL conical vials, filled with 300 µL of buffer (Hepes 100 mM, 20 mM MgCl2, pH 8). The solution was equilibrated for 1 h with a gas mixture 800 ppm CO2/40% O2 (in N2) produced with high precision mass-flow controllers (FC260, Tylan Inc.). CO2 and O2 mole fractions were continuously monitored with an IRGA (Li-6251, Li-Cor Inc) and an oxygen sensor (MAX-250, Maxtec). A total of 10 µL of RuBP was then injected to get a final concentration of 3 mM. The reaction was started with 10 µL of Rubisco extract that had been activated for 20 min with 20 mM NaHCO3 and 15 mM MgCl2. The reaction ran for 30 min with constant gas bubbling and was quenched with 240 µL EDTA at 100 mM. After centrifugation, the supernatant was adjusted to pH 6.5 and mixed with 40 µL D2O for 31P-NMR analysis (example given in SI Appendix, Fig. S7B). NMR analyses were conducted on a Bruker Advance 700 MHz NMR spectrometer (Bruker Biospin) in 5-mm NMR tubes, using a proton-decoupled (Waltz 16 sequence) pulse program (zgpg). The acquisition time and relaxation delay were set to 1.5 s and 2 s, respectively, and 2,000 scans were accumulated. Absolute carboxylation and oxygenation velocities (vc and vo, in mol mol−1 site s−1) were determined using CO2 and O2 consumption rates measured by mass spectrometry (see below, 12C/13C Isotope Effect and 16O/18O Isotope Effect). Carboxylase assays were performed without dissolved O2, and oxygenation assays were performed without added dissolved CO2 (except for residual CO2 used for activation during assay preparation). The number of active sites was determined using 14C-CABP binding (52). For all kinetic parameters (specificity, exchange, and isotope effects) shown in figures, we used separate assay replicates.
Proton Exchange.
Proton exchange (Fig. 2) was measured by 1H-NMR (example given in SI Appendix, Fig. S7A), using samples from assays carried out as above. After quenching, samples were instant frozen with liquid nitrogen, lyophilized, and redissolved in ultrapure D2O (99.96% D2O, Sigma-Aldrich). NMR analyses were performed at 11 °C using 1D acquisition (zg30). Isotopologue composition of RuBP (Fig. 5) was assessed using high-resolution MS (Orbitrap, Thermo Scientific) (monitored m/z is shown in SI Appendix, Fig. S8). Lyophilized samples were redissolved in water, ions were removed with washed Dowex 50H+, and the eluate containing RuBP was instant frozen, lyophilized, and redissolved in 100 µL water. Samples were injected directly (infusion). MS analysis was operated in negative polarity in the full MS scan mode (mass scan range 50 to 750 m/z) with the following source settings: source voltage 3,500 V, resolution 70,000, automatic gain control target 1·106, mass scan range 60 to 600 m/z, sheath gas 40, auxiliary gas 10, sweep gas 1.5, probe temperature 300 °C, capillary temperature 250 °C, and S-lens radiofrequency level 50.
12C/13C Isotope Effect.
The carbon isotope effect associated with CO2 addition was measured according to ref. 34. Briefly, CO2 uptake was monitored by a membrane-inlet (MI) system comprising a 700-µL temperature-controlled cuvette (25 °C) connected to the vacuum line of the isotope ratio mass spectrometer (IRMS Isochrom, Elementar). For each assay, five injections of a bicarbonate solution just before starting the reaction were used to correct the baseline and check linearity. The Craig correction was applied to the whole dataset to convert 45/44 and 46/44 ratios into 13C/12C ratios. True δ13C and δ18O values for bicarbonate (measured independently by IRMS), corrected for the equilibrium isotope effect of acid-base dissociation, were used as a reference to compute the true 13C/12C isotope ratio of the reference gas (vs. Vienna Pee Dee Belemnite). All data were then corrected against the reference gas to obtain absolute 13C/12C ratios. The Rubisco isotope effect 13(V/K), thereafter denoted as α, was expressed vs. dissolved CO2 and calculated with the slope of the isotope ratio given by the Rayleigh equation: ln R = ln R0 + (1/α − 1)·ln f, where R is the 13C/12C isotope ratio in CO2, and f is the fraction of unreacted CO2.
16O/18O Isotope Effect.
The oxygen isotope effect was measured by MI-IRMS, according to ref. 53 (illustrated in SI Appendix, Fig. S5). A specific semipermeable membrane was used (purpose-built multilayered membrane made of Teflon, paraffin-polyolefin, and Teflon) through which O2 could permeate. The paraffin-polyolefin layer was chosen here for its low permeability for O2, so as to avoid excessive O2 consumption through the membrane. As for 13C, injection of water equilibrated with increasing % O2 was used for baseline correction and checking linearity. The isotope effect 18(V/K) was calculated vs. dissolved O2 using the slope in the Rayleigh equation.
Computations.
Predicted isotope effects (SI Appendix, Fig. S2) were calculated according to refs. 23 and 45. The reduction potential of RuBP enolate was calculated via the Nernst equation and referenced against the SHE in aqueous solution (ESHE = 4.28 V). All of the geometries of species investigated in the study were optimized at the M06-2X/6–31+G(d) level of theory (54) and frequencies were also calculated at this level. Geometries were verified as local minima. Entropies, thermal corrections, and zero-point vibrational energies were scaled by recommended scale factors (55). Single-point energies were calculated using the high-level composite ab initio method G3(MP2,CC) to improve the accuracy (56). For all species investigated, conformational searching was performed with the energy-directed tree search (EDTS) algorithm (57) to identify conformations with lowest Gibbs free energy. Gibbs free energies in the gas phase were calculated using standard ideal gas partition functions under the harmonic oscillator approximation. Gibbs free energies in solution were obtained via a thermocycle, in which Gibbs free energies in the gas phase were combined with Gibbs free energies of solvation and the necessary phase change correction term (58). The SMD solvent model (59) was used to correct for implicit solvent effects in water. For this purpose, geometries were fully optimized in solution at the M06-2X/6-31+G(d) level. All standard ab initio molecular orbital theory and DFT calculations were carried out using the Gaussian 09 (60) and Molpro 2015 (61) software packages.
Supplementary Material
Acknowledgments
C.B. and G.D.F. acknowledge funding by the Australian Government through the Australian Research Council Centre of Excellence for Translational Photosynthesis (Project CE140100015), and G.T. thanks the Australian Research Council for its support via a fellowship under contract FT140100645. We thank Dr. Hilary Stuart-Williams for membrane-inlet IRMS measurements and Dr. Spencer Whitney for providing material to produce R. rubrum Rubisco.
Footnotes
The authors declare no competing interest.
*Our result on the effect of d-RuBP on 13(V/K) is somewhat different from that found in ref. 36, where deuteration tended to decrease the isotope effect by several per mil (with a large SE). This difference is likely explained by the method used. In ref. 36, there was a slight residual carboxylation activity during the oxygenase assay, which affected the measurement of the positional isotope effect in C3 and thus the calculation of 13(V/K) from the δ13C value of products (3-phosphoglycerate).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2008824117/-/DCSupplemental.
Data Availability.
All study data are included in the article and SI Appendix.
References
- 1.Bar-On Y. M., Milo R., The global mass and average rate of rubisco. Proc. Natl. Acad. Sci. U.S.A. 116, 4738–4743 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walker B. J., VanLoocke A., Bernacchi C. J., Ort D. R., The costs of photorespiration to food production now and in the future. Annu. Rev. Plant Biol. 67, 107–129 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Kangasjärvi S., Neukermans J., Li S., Aro E.-M., Noctor G., Photosynthesis, photorespiration, and light signalling in defence responses. J. Exp. Bot. 63, 1619–1636 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Mignolet-Spruyt L. et al., Spreading the news: Subcellular and organellar reactive oxygen species production and signalling. J. Exp. Bot. 67, 3831–3844 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Abadie C., Carroll A., Tcherkez G., “Interactions between day respiration, photorespiration, and N and S assimilation in leaves” in Plant Respiration: Metabolic Fluxes and Carbon Balance, Tcherkez G., Ghashghaie J., Eds. (Springer, Amsterdam, 2017), pp. 1–18. [Google Scholar]
- 6.Abadie C., Tcherkez G., Plant sulphur metabolism is stimulated by photorespiration. Commun. Biol. 2, 379 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilson R. H., Whitney S. M., “Improving CO2 fixation by enhancing Rubisco performance” in Directed Enzyme Evolution: Advances and Applications, Alcalde M., Ed. (Springer International Publishing, Cham, 2017), pp. 101–126. [Google Scholar]
- 8.Flamholz A. I. et al., Revisiting trade-offs between Rubisco kinetic parameters. Biochemistry 58, 3365–3376 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schloss J. V., Oxygen toxicity from plants to people. Planta 216, 38–43 (2002). [DOI] [PubMed] [Google Scholar]
- 10.Romero E., Gómez Castellanos J. R., Gadda G., Fraaije M. W., Mattevi A., Same substrate, many reactions: Oxygen activation in flavoenzymes. Chem. Rev. 118, 1742–1769 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Lorimer G., Andrews T., Plant photorespiration—An inevitable consequence of the existence of atmospheric oxygen. Nature 243, 359–360 (1973). [Google Scholar]
- 12.Andrews T., Photorespiration—still unavoidable? FEBS Lett. 90, 1–9 (1978). [Google Scholar]
- 13.Bathellier C., Tcherkez G., Lorimer G. H., Farquhar G. D., Rubisco is not really so bad. Plant Cell Environ. 41, 705–716 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Silva P. J., Refining the reaction mechanism of O2 towards its co-substrate in cofactor-free dioxygenases. PeerJ 4, e2805 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mattevi A., To be or not to be an oxidase: Challenging the oxygen reactivity of flavoenzymes. Trends Biochem. Sci. 31, 276–283 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Lorimer G. H., The carboxylation and oxygenation of ribulose 1, 5-bisphosphate: The primary events in photosynthesis and photorespiration. Annu. Rev. Plant Physiol. 32, 349–382 (1981). [Google Scholar]
- 17.Styring S., Brändén R., Co2+-and Cu2+-incubated ribulose-1,5-bisphosphate carboxylase/oxygenase from Rhodospirillum rubrum studied with electron paramagnetic resonance spectroscopy. Biochim. Biophys. Acta Protein Struct. Molec. Enzym. 832, 113–118 (1985). [Google Scholar]
- 18.Kim K., Portis A. R. Jr., Oxygen-dependent H2O2 production by Rubisco. FEBS Lett. 571, 124–128 (2004). [DOI] [PubMed] [Google Scholar]
- 19.Kannappan B., Cummins P. L., Gready J. E., Mechanism of oxygenase-pathway reactions catalyzed by Rubisco from large-scale Kohn–Sham density functional calculations. J. Phys. Chem. B 123, 2833–2843 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Minaev B. F., Ågren H., Minaeva V., “Spin-orbit coupling in enzymatic reactions and the role of spin in biochemistry” in Handbook of Computational Chemistry, Leszczynski J., Ed. (Springer, Dordrecht, 2016), pp. 1–31. [Google Scholar]
- 21.Mogel S. N., McFadden B. A., Chemiluminescence of the Mn2(+)-activated ribulose-1,5-bisphosphate oxygenase reaction: Evidence for singlet oxygen production. Biochemistry 29, 8333–8337 (1990). [DOI] [PubMed] [Google Scholar]
- 22.Lilley R. M., Wang X., Krausz E., Andrews T. J., Complete spectra of the far-red chemiluminescence of the oxygenase reaction of Mn2+-activated ribulose-bisphosphate carboxylase/oxygenase establish excited Mn2+ as the source. J. Biol. Chem. 278, 16488–16493 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Tcherkez G., The mechanism of Rubisco-catalysed oxygenation. Plant Cell Environ. 39, 983–997 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Andrés J., Safont V., Tapia O., Straining the double bond in 1, 2-dihydroxyethylene. A simple theoretical model for the enediol moiety in Rubisco’s substrate and analogs. Chem. Phys. Lett. 198, 515–520 (1992). [Google Scholar]
- 25.Oliva M., Safont V. S., Andrés J., Tapia O., Transition structures for D-ribulose-1, 5-bisphosphate carboxylase/oxygenase-catalyzed oxygenation chemistry: Role of carbamylated lysine in a model magnesium coordination sphere. J. Phys. Chem. A 105, 4726–4736 (2001). [Google Scholar]
- 26.Tcherkez G. G., Farquhar G. D., Andrews T. J., Despite slow catalysis and confused substrate specificity, all ribulose bisphosphate carboxylases may be nearly perfectly optimized. Proc. Natl. Acad. Sci. U.S.A. 103, 7246–7251 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guy R. D., Fogel M. L., Berry J. A., Photosynthetic fractionation of the stable isotopes of oxygen and carbon. Plant Physiol. 101, 37–47 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grissom C. B., Magnetic field effects in biology: A survey of possible mechanisms with emphasis on radical-pair recombination. Chem. Rev. 95, 3–24 (1995). [Google Scholar]
- 29.Christeller J. T., The effects of bivalent cations on ribulose bisphosphate carboxylase/oxygenase. Biochem. J. 193, 839–844 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Forsén S., Lindman B., Calcium and magnesium NMR in chemistry and biology. Annu. Rep. NMR Spectrosc. 11, 183–226 (1981). [Google Scholar]
- 31.Hore P. J., Are biochemical reactions affected by weak magnetic fields? Proc. Natl. Acad. Sci. U.S.A. 109, 1357–1358 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saver B. G., Knowles J. R., Ribulose-1,5-bisphosphate carboxylase: Enzyme-catalyzed appearance of solvent tritium at carbon 3 of ribulose 1,5-bisphosphate reisolated after partial reaction. Biochemistry 21, 5398–5403 (1982). [DOI] [PubMed] [Google Scholar]
- 33.Pierce J., Lorimer G. H., Reddy G. S., Kinetic mechanism of ribulose bisphosphate carboxylase: Evidence for an ordered, sequential reaction. Biochemistry 25, 1636–1644 (1986). [Google Scholar]
- 34.Tcherkez G. G. et al., D2O solvent isotope effects suggest uniform energy barriers in ribulose-1,5-bisphosphate carboxylase/oxygenase catalysis. Biochemistry 52, 869–877 (2013). [DOI] [PubMed] [Google Scholar]
- 35.Van Dyk D. E., Schloss J. V., Deuterium isotope effects in the carboxylase reaction of ribulose 1,5-bisphosphate carboxylase/oxygenase. Biochemistry 25, 5145–5156 (1986). [Google Scholar]
- 36.Roeske C. A., O’Leary M. H., Carbon isotope effects on enzyme-catalyzed carboxylation of ribulose bisphosphate. Biochem. J. 23, 6275–6284 (1984). [DOI] [PubMed] [Google Scholar]
- 37.Wu J., Serianni A. S., Vuorinen T., Furanose ring anomerization: Kinetic and thermodynamic studies of the D-2-pentuloses by 13C-n.m.r. spectroscopy. Carbohydr. Res. 206, 1–12 (1990). [DOI] [PubMed] [Google Scholar]
- 38.Richard J. P., Acid-base catalysis of the elimination and isomerization reactions of triose phosphates. J. Am. Chem. Soc. 106, 4926–4936 (1984). [Google Scholar]
- 39.Isbell H. S., Frush H. L., Wade C. W. R., Hunter C. E., Transformations of sugars in alkaline solutions. Carbohydr. Res. 9, 163–175 (1969). [Google Scholar]
- 40.Kannappan B., Gready J. E., Redefinition of rubisco carboxylase reaction reveals origin of water for hydration and new roles for active-site residues. J. Am. Chem. Soc. 130, 15063–15080 (2008). [DOI] [PubMed] [Google Scholar]
- 41.Jones A. R., Magnetic field effects in proteins. Mol. Phys. 114, 1691–1702 (2016). [Google Scholar]
- 42.Woodward J., Radical pairs in solution. Prog. React. Kinet. Mech. 27, 165–207 (2002). [Google Scholar]
- 43.Roth J. P., Klinman J. P., Catalysis of electron transfer during activation of O2 by the flavoprotein glucose oxidase. Proc. Natl. Acad. Sci. U.S.A. 100, 62–67 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Su Q., Klinman J. P., Nature of oxygen activation in glucose oxidase from Aspergillus niger: The importance of electrostatic stabilization in superoxide formation. Biochemistry 38, 8572–8581 (1999). [DOI] [PubMed] [Google Scholar]
- 45.Roth J. P. et al., Oxygen isotope effects on electron transfer to O2 probed using chemically modified flavins bound to glucose oxidase. J. Am. Chem. Soc. 126, 15120–15131 (2004). [DOI] [PubMed] [Google Scholar]
- 46.Cohen A. E., Nanomagnetic control of intersystem crossing. J. Phys. Chem. A 113, 11084–11092 (2009). [DOI] [PubMed] [Google Scholar]
- 47.Cleland W. W., Andrews T. J., Gutteridge S., Hartman F. C., Lorimer G. H., Mechanism of Rubisco: The carbamate as general base. Chem. Rev. 98, 549–562 (1998). [DOI] [PubMed] [Google Scholar]
- 48.Fiedler F., Müllhofer G., Trebst A., Rose I. A., Mechanism of ribulose-diphosphate carboxydismutase reaction. Eur. J. Biochem. 1, 395–399 (1967). [DOI] [PubMed] [Google Scholar]
- 49.Servaites J. C., Crystalline ribulose bisphosphate carboxylase/oxygenase of high integrity and catalytic activity from Nicotiana tabacum. Arch. Biochem. Biophys. 238, 154–160 (1985). [DOI] [PubMed] [Google Scholar]
- 50.Horecker B., Ribulose diphosphate. Biochem. Prep. 6, 83–90 (1958). [Google Scholar]
- 51.Wang Z.-Y., Luo S., Sato K., Kobayashi M., Nozawa T., Measurements of the CO2/O2 specificity of ribulose 1,5-bisphosphate carboxylase/oxygenase by 31P-and 1H-NMR. Photosynth. Res. 58, 103–109 (1998). [Google Scholar]
- 52.Ruuska S. et al., The interplay between limiting processes in C3 photosynthesis studied by rapid-response gas exchange using transgenic tobacco impaired in photosynthesis. Funct. Plant Biol. 25, 859–870 (1998). [Google Scholar]
- 53.Cheah M. H. et al., Online oxygen kinetic isotope effects using membrane inlet mass spectrometry can differentiate between oxidases for mechanistic studies and calculation of their contributions to oxygen consumption in whole tissues. Anal. Chem. 86, 5171–5178 (2014). [DOI] [PubMed] [Google Scholar]
- 54.Zhao Y., Truhlar D. G., The M06 suite of density functionals for main group thermochemistry, thermochemical kinetics, noncovalent interactions, excited states, and transition elements: Two new functionals and systematic testing of four M06-class functionals and 12 other functionals. Theor. Chem. Acc. 120, 215–241 (2008). [Google Scholar]
- 55.Alecu I. M., Zheng J., Zhao Y., Truhlar D. G., Computational thermochemistry: Scale factor databases and scale factors for vibrational frequencies obtained from electronic model chemistries. J. Chem. Theory Comput. 6, 2872–2887 (2010). [DOI] [PubMed] [Google Scholar]
- 56.Curtiss L. A., Raghavachari K., Redfern P. C., Baboul A. G., Pople J. A., Gaussian-3 theory using coupled cluster energies. Chem. Phys. Lett. 314, 101–107 (1999). [Google Scholar]
- 57.Izgorodina E. I., Lin C. Y., Coote M. L., Energy-directed tree search: An efficient systematic algorithm for finding the lowest energy conformation of molecules. Phys. Chem. Chem. Phys. 9, 2507–2516 (2007). [DOI] [PubMed] [Google Scholar]
- 58.Ho J., Klamt A., Coote M. L., Comment on the correct use of continuum solvent models. J. Phys. Chem. A 114, 13442–13444 (2010). [DOI] [PubMed] [Google Scholar]
- 59.Marenich A. V., Cramer C. J., Truhlar D. G., Universal solvation model based on solute electron density and on a continuum model of the solvent defined by the bulk dielectric constant and atomic surface tensions. J. Phys. Chem. B 113, 6378–6396 (2009). [DOI] [PubMed] [Google Scholar]
- 60.Frisch M., et al. , Gaussian 09 Revision E. 01, 2009 (Gaussian Inc, Wallingford, CT, 2009).
- 61.Werner H., et al. , MOLPRO, Version 2015.1, A Package of Ab Initio Programs (University of Cardiff, Cardiff, UK, 2015).
- 62.Koppenol W. H., Stanbury D. M., Bounds P. L., Electrode potentials of partially reduced oxygen species, from dioxygen to water. Free Radic. Biol. Med. 49, 317–322 (2010). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and SI Appendix.