Significance
Human ALIX is involved in many essential cellular processes. The carboxy (C)-terminal proline-rich domain (PRD) of ALIX, which interacts with multiple partners, has eluded analysis owing to the presence of polyproline stretches that hinder recombinant protein production. By dividing it into two fragments, the amino (N)- and the C-terminal portions, we carry out a detailed biophysical investigation of ALIX-PRD. The disordered N-terminal fragment comprises three proline-rich motifs that compete for binding to its partner, TSG101-UEV. In contrast, the C-terminal fragment forms reversible amyloids modulated by tyrosine phosphorylation, the first known example where posttranslational modifications dictate the dissolution of amyloid fibrils. This study illustrates the mechanistic interdependence of multivalency brought out by polymerization of ALIX-PRD and its effect on ALIX function.
Keywords: intrinsically disordered protein, posttranslational modifications, amyloids, NMR, signal transduction
Abstract
Proline-rich domains (PRDs) are among the most prevalent signaling modules of eukaryotes but often unexplored by biophysical techniques as their heterologous recombinant expression poses significant difficulties. Using a “divide-and-conquer” approach, we present a detailed investigation of a PRD (166 residues; ∼30% prolines) belonging to a human protein ALIX, a versatile adaptor protein involved in essential cellular processes including ESCRT-mediated membrane remodeling, cell adhesion, and apoptosis. In solution, the N-terminal fragment of ALIX-PRD is dynamically disordered. It contains three tandem sequentially similar proline-rich motifs that compete for a single binding site on its signaling partner, TSG101-UEV, as evidenced by heteronuclear NMR spectroscopy. Global fitting of relaxation dispersion data, measured as a function of TSG101-UEV concentration, allowed precise quantitation of these interactions. In contrast to the soluble N-terminal portion, the C-terminal tyrosine-rich fragment of ALIX-PRD forms amyloid fibrils and viscous gels validated using dye-binding assays with amyloid-specific probes, congo red and thioflavin T (ThT), and visualized by transmission electron microscopy. Remarkably, fibrils dissolve at low temperatures (2 to 6 °C) or upon hyperphosphorylation with Src kinase. Aggregation kinetics monitored by ThT fluorescence shows that charge repulsion dictates phosphorylation-mediated fibril dissolution and that the hydrophobic effect drives fibril formation. These data illuminate the mechanistic interplay between interactions of ALIX-PRD with TSG101-UEV and polymerization of ALIX-PRD and its central role in regulating ALIX function. This study also demonstrates the broad functional repertoires of PRDs and uncovers the impact of posttranslational modifications in the modulation of reversible amyloids.
Proline-rich domains (PRDs) are ubiquitous in the eukaryotic proteome and function as docking sites for a multitude of signaling protein modules (1–4). Prolines are also favored in intrinsically disordered proteins (IDPs), which lack a well-defined structure (5, 6). The relative abundance of proline is primarily due to its unusual structural properties. The proline sidechain is fused to its backbone amide nitrogen, which imposes unique conformational constraints and provides a distinct entropic advantage for binding to its signaling partners. The most common structure formed by consecutive proline residues is a left-handed helix, termed polyproline type II (PPII), with an extended rigid conformation that favors entropy-driven protein–protein interactions (7–9). A PPII structure can tolerate various combinations of proline and nonproline residues without compromising its structural integrity. PRDs are predicted to be disordered (10) and contain strategically placed short segments comprising proline residues and polyproline stretches. These contiguous functional segments, termed linear motifs (11), maintain the structural plasticity of PRDs where the presence of prolines results in extended conformations, rendering the linear motifs accessible for interactions with their signaling partners. A typical PRD contains sequentially similar linear epitopes, and this multivalency results in an increase in the avidity of protein–protein interactions and solution-to-gel phase transitions (12). Such interactions have been less studied in the context of intracellular signaling networks as compared with the interactions between extracellular ligands and their receptors, namely antibody–receptor and carbohydrate–lectin systems (13–15). To make use of ubiquitous PRDs, eukaryotes employ an array of proline-recognition domains (16, 17), which comprise several well-known motifs, including Src homology 3 (SH3), WW, GYF, and ubiquitin E2 variant (UEV), as well as many others. The interactions between proline-recognition domains and PRDs create dynamic signaling networks where a PRD can rapidly switch between binding partners and form multivalent complexes that dictate cell growth, transcription, cell signaling, and other essential cellular functions. Despite their importance, however, PRDs are usually removed from the protein constructs used for X-ray crystallography and NMR studies, due to the problems associated with their recombinant expression and their lack of structure. Since prolines lack amide protons, they are not amenable to conventional 1H-detected NMR methods used to study IDPs at atomic resolution (18). As a result, there are clear gaps in our current understanding of how PRDs regulate cellular functions.
The apoptosis-linked gene-2 interacting protein X (ALIX), a human cellular protein, functions within the endosomal-sorting complexes required for transport (ESCRT) pathway (19, 20). The ESCRT pathway comprises a collection of proteins that form polymeric filaments and mediates membrane scission to facilitate cytokinetic abscission, biogenesis of multivesicular bodies, plasma membrane repair, and budding of enveloped viruses such as HIV-1 and Ebola. The ESCRT machinery consists of five functionally distinct protein complexes, ESCRT-0, -I, -II, and -III and AAA ATPase VPS4. ESCRT-0, -I, and -II are involved in sorting ubiquitinated proteins to endosomes. ESCRT-III filaments comprising polymeric assemblies of charged multivesicular body proteins (CHMPs) and ATPase VPS4 are involved in membrane scission. ALIX, also known as programmed cell death 6 interacting protein (PDCD6IP), interacts with ESCRT-I protein tumor-susceptibility gene 101 (TSG101) and recruits ESCRT-III proteins to carry out membrane scission. In addition to ESCRT-mediated membrane remodeling, ALIX is involved in apoptosis, cell adhesion, and endocytosis (21). ALIX consists of an amino (N)-terminal Bro1 domain, a central V-domain, and a carboxy (C)-terminal PRD containing ∼30% prolines (Fig. 1A). The Bro1 and V domains have been extensively studied by structural methods (22, 23). Relatively little is known about the PRD, despite its functional importance. ALIX-PRD is highly conserved among vertebrates (SI Appendix, Fig. S1) and is known to interact with multiple binding partners (21), including the UEV domain of TSG101 (TSG101-UEV) (24, 25), tyrosine kinases such as Src (26) and Hck (27), endocytic proteins endophilins (28), and many others. The interactions between ALIX-PRD and TSG101-UEV are mediated by the N-terminal portion of PRD, which is also implicated in the autoinhibition of ALIX (29). The C-terminal tyrosine-rich portion of PRD is implicated in ALIX multimerization (30). The biological importance of ALIX-PRD and the lack of prior biophysical studies make it a prototypical system to study the mechanistic role of PRDs in cell signaling.
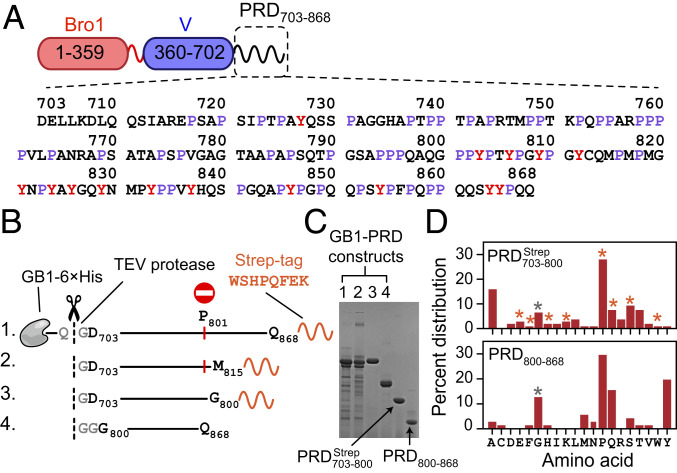
Fig. 1.
ALIX domain organization and summary of ALIX-PRD constructs used in the current work. (A) Schematic of ALIX organization. Primary sequence of PRD is shown with prolines (∼30%) and tyrosines (∼9%) labeled in purple and red, respectively. (B) Recombinant PRD constructs, namely GB1– GB1– GB1– and GB1–PRD800–868. The positions of purification tags, 6×His and strep, are marked (primary sequence of strep tag is shown). TEV protease cutting sites are shown in gray and marked with dashed lines and scissors. Recombinant expression of GB1– and GB1– resulted in truncated fragments because of ribosomal stalling induced by polyproline stretches, especially at residue P801, marked by a red circle and vertical red line. (C) SDS-PAGE analysis of purified PRD constructs [16% wt/vol tris(hydroxymethyl)aminomethane–glycine gel]; the order of GB1–PRD fusion constructs is the same as the one depicted in B. TEV-cleaved products, namely and PRD800–868, are marked with arrows. (D) Amino acid composition of (Top) and PRD800–868 (Bottom). Vertical bars marked with orange and gray asterisks denote contributions from nonnative strep tag and remnant-glycine residues of TEV cleavage sites, respectively.
By dividing it into two recombinant constructs, representing the N- and the C-terminal portions of ALIX-PRD, we carry out a detailed investigation using a range of biophysical methods, including heteronuclear NMR spectroscopy, fluorescence spectroscopy, analytical ultracentrifugation (AUC), negative-stain electron microscopy (EM), and mass spectrometry (MS). Note that poor bacterial expression (31) and the overall size of the full-length protein (∼96 kDa) do not allow for a similar biophysical investigation of PRD in the context of full-length ALIX. We show that the three tandem proline-rich motifs of the N-terminal portion of ALIX-PRD compete for binding with TSG101-UEV, suggesting that a PRD-mediated multimerization of ALIX will result in enhanced functional affinity for its binding partner, TSG101-UEV. We demonstrate that the C-terminal tyrosine-rich portion of ALIX-PRD forms amyloid fibrils and viscous gels under near-physiological conditions, as evidenced by dye-binding assays with amyloid-specific probes, congo red (CR) and thioflavin T (ThT), and visualized by negative-stain EM. Remarkably, the fibril formation is reversible as the fibrils dissolve at low temperatures (2 to 6 °C) or upon Src kinase-mediated hyperphosphorylation of ALIX-PRD. This reversible polymerization is strikingly different from those exhibited by pathological amyloids, including the ones associated with Alzheimer’s and Parkinson’s diseases that resist disassembly. We show that amyloid formation is driven by the hydrophobic effect, likely mediated by intra- and intermolecular interactions between tyrosine and proline residues, whereas phosphorylation-mediated dissolution is governed by charge repulsion.
Results and Discussion
Recombinant Constructs.
The current work made use of two codon-optimized PRD constructs, namely the N-terminal fragment, and the C-terminal fragment, PRD800–868 (Fig. 1 B–D). This “divide-and-conquer” strategy was employed since we were unable to express intact full-length PRD as well as its shorter truncated counterpart in Escherichia coli and respectively). Sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE) (Fig. 1C) and liquid chromatography–electrospray ionization–time-of-flight mass spectrometry (LC-ESI-TOFMS) (SI Appendix, Fig. S2) analyses of the expression patterns of the latter two constructs revealed ribosomal stalling and subsequent translational arrest induced by polyproline stretches that are ubiquitous in ALIX-PRD, with regions 758PPPP761 and 801PPYP804 being particularly problematic. Essentially no intact proteins were produced as both and did not express beyond residue P801. These results agree with prior reports that show that translation of consecutive proline residues leads to ribosomal stalling (32–34). Each PRD construct carried a cleavable N-terminal fusion tag comprising B1 domain of protein G, GB1 (35), followed by a polyhistidine affinity tag (6×His) and a tobacco etch virus (TEV) protease cleavage site; GB1–6×His–TEV, hereafter referred to as GB1. In the case of a noncleavable strep affinity tag (36) was engineered at the C terminus (801WSHPQFEK808), which served two purposes. It allowed a ready separation of intact protein from its truncated fragments using affinity chromatography and facilitated precise measurements of protein concentrations using ultraviolet absorbance; extinction coefficient at 280 nm (37): 6,970 versus 1,280 M−1cm−1, with and without the C-terminal strep tag, respectively (see Fig. 1D for the amino acid composition of the two PRD constructs used in current work). We also made use of recombinant human full-length Src kinase and TSG101-UEV (see SI Appendix, Fig. S3 for LC-ESI-TOFMS characterization of the constructs used in current work).
NMR Analysis of
yielded a high-quality 1H-15N transverse relaxation-optimized spectroscopy–heteronuclear single quantum coherence spectroscopy (TROSY-HSQC) spectrum (Fig. 2A). The chemical shifts of the backbone amide proton resonances were clustered in a narrow 1 part per million (ppm) window (∼7.7 to ∼8.7 ppm), a hallmark of random-coil conformation. To further explore the structural propensity of in solution, we recorded NMR-backbone chemical shifts, three bond 3JHNHα couplings, and 15N-longitudinal and transverse relaxation rates (R1 and R2, respectively). Nearly complete (∼97%) backbone resonance assignments were achieved using a combination of three-dimensional conventional 1H-detected and modern 13C-detected NMR methods (18) (Fig. 2B). Differences between observed chemical shifts and the corresponding random-coil values, referred to as secondary chemical shifts (Δδ), are sensitive indicators of local secondary structure. By using the random-coil values corrected for pH, ionic strength, and temperature (38, 39), rms values of 0.19, 0.35, 0.29, and 0.64 ppm were obtained for secondary chemical shifts of 13Cα, 13Cβ, 13C′, and 15N, respectively (SI Appendix, Fig. S4A). Note that the chemical shifts of the last C-terminal residue, K808, were not considered for rms calculations. Δδ(13Cα) values, the best reporters of local secondary structure, were evenly distributed around 0 ppm, indicative of a random-coil conformation. The absence of structural ordering was confirmed by analysis of backbone chemical shifts using the program δ2D (40), which yielded low values of the average secondary-structure propensities, ∼2% of α-helix and ∼11% of β-sheet (SI Appendix, Fig. S4B). For nearly all residues of the population of PPII fell in the ∼20 to 30% range. A few notable exceptions were regions 752PQPPAR757 and 782AAP784, which showed elevated PPII propensities (∼30 to 40%). Finally, nothing definitive can be said about the conformation of the two polyproline stretches, namely 758PPPP761 and 794PPP796, due to the lack of δ2D predictions for these regions. These results are in excellent agreement with experimental CheZOD Z-scores (41), which indicated that nearly all residues of are disordered with Z-scores of <3 (SI Appendix, Fig. S4C). Note that the 734GHAPTP739 motif showed slightly elevated Z-scores (∼3 to 6), indicating a possible presence of a transient ordered structure in that region. 3JHNHα couplings of were highly correlated with the random-coil 3JHNHα values predicted using nearest‐neighbor effects (42), with a Pearson’s correlation coefficient of ∼0.8 (SI Appendix, Fig. S4D). Analysis of 15N-relaxation rates, measured at 800 MHz at 30 °C, revealed average R1 and R2 values of ∼1.2 and ∼2.1 s−1, respectively, indicating rapid backbone dynamics in the pico- to nanosecond regime (SI Appendix, Fig. S4E). Taken together, NMR results establish that is disordered in solution with the lack of dominant PPII population(s).
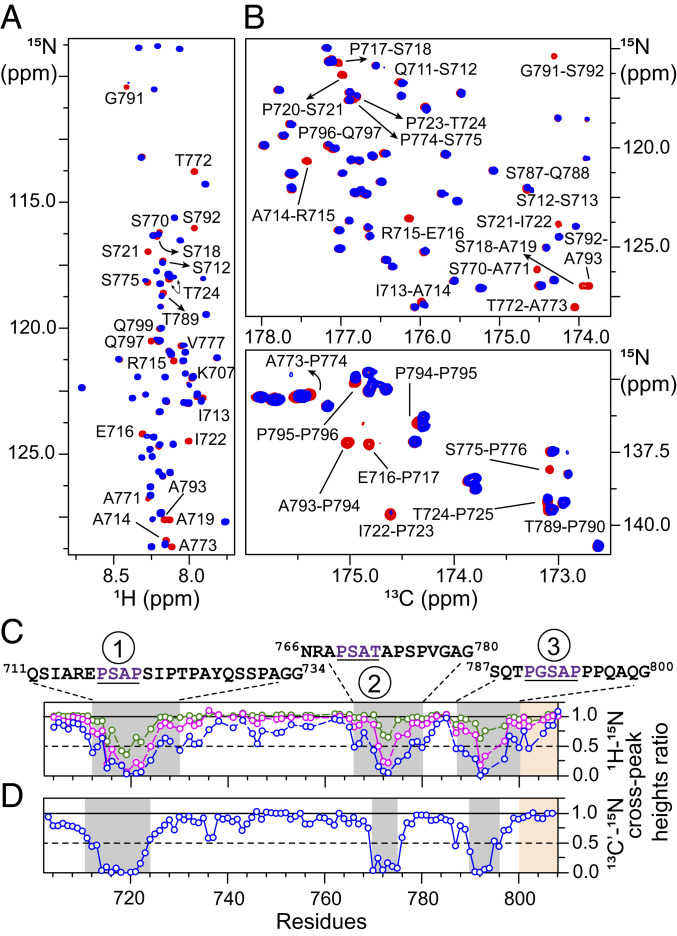
Fig. 2.
NMR analyses of interactions of with TSG101-UEV. (A and B) Overlay of expanded regions of the 1H-15N TROSY (A) and 13C-15N CO-N (B) correlation spectra of 100 µM in the absence (red) and presence (blue) of TSG101-UEV (molar ratio: 1:3). Some isolated cross-peaks that exhibit significant reduction in intensities on addition of TSG101-UEV are labeled. (C and D) The reduction in 1H-15N (C) and 13C′-15N (D) cross-peak intensities of on addition of TSG101-UEV is indicative of intermediate exchange on the chemical-shift time scale. Color scheme is as follows: + TSG101-UEV molar ratio: green, 1:0.25; magenta, 1:1.5; blue, 1:3. Affected regions are highlighted with semitransparent gray rectangles; primary sequences of each interacting PRD site (1, 2, and 3) are shown above the graphs, with recurring PTAP-like motifs underlined and labeled in dark purple. The position of the C-terminal strep tag (residues 801 to 808) is denoted by semitransparent orange rectangles.
Interactions of NMR-Visible with Unlabeled TSG101-UEV.
TSG101-UEV binds to P(T/S)AP motifs found in both cellular and viral proteins, including the p6 domain of HIV-1 Gag polyprotein (43). Direct interactions between the 717PSAP720 motif of ALIX-PRD and TSG101-UEV were reported using yeast-two hybrid and surface plasmon resonance (SPR) measurements (25). The latter were carried out using recombinant TSG101-UEV and an 11-residue peptide analog of ALIX-PRD, PRD714–723, and yielded an equilibrium dissociation constant, KD, of 142 ± 0.5 μM. To explore interactions between and TSG101-UEV, we made use of NMR-titration experiments, where an increasing concentration of unlabeled TSG101-UEV was added to NMR-visible (Fig. 2 A and B and SI Appendix, Fig. S5). Significant reductions in 1HN/15N and 13C′/15N cross-peak intensities were observed for residues surrounding the 717PSAP720 motif on the addition of unlabeled TSG101-UEV, a manifestation of intermediate-exchange regime on the chemical-shift time scale. The affected PRD region encompassed residues 711 to 734. As expected for an intermediate-exchange regime, only a few residues exhibited small but detectable 1HN/15N and 13C′/15N chemical-shift perturbations (SI Appendix, Fig. S5). Surprisingly, two additional regions (residues 766 to 780 and 787 to 800) exhibited significant reductions in cross-peak intensities on the addition of TSG101-UEV, indicative of secondary interactions. Examination of primary sequences of all three affected PRD regions, namely residues 711 to 734, 766 to 780, and 787 to 800 (hereafter referred to as sites 1, 2, and 3, respectively), revealed a plausible explanation for these interactions. All three sites comprise sequentially similar motifs, namely 717PSAP720, 769PSAT772, and 790PGSAP794 (Fig. 2C), which interact with TSG101-UEV. 15N-relaxation rates of were measured as a function of TSG101-UEV concentration (SI Appendix, Fig. S6). Interactions between and TSG101-UEV are localized around the sites mentioned above as a significant increase in 15N-R2 rates on TSG101-UEV addition was found only at these three interaction sites (SI Appendix, Fig. S6 A and B). These results indicate that the TSG101-UEV–bound state of is dynamically disordered, with local ordering around the three interaction sites. These observations are in excellent agreement with the X-ray structure of the complex between TSG101-UEV and HIV-1 p6 analog comprising the PTAP motif (44) as the entire nine-residue p6 analog (PEPTAPPEE) was visualized in an extended conformation in the binding groove of TSG101-UEV. Sedimentation studies of and TSG101-UEV demonstrated that both proteins were monodisperse and monomeric (SI Appendix, Fig. S7). Sedimentation velocity experiments on returned a best-fit frictional ratio of ∼2.0, indicative of an IDP. In contrast, the frictional ratio for TSG101-UEV was ∼1.3, a value typical for globular proteins. We also carried out sedimentation-equilibrium experiments on (SI Appendix, Fig. S8) and TSG101-UEV (SI Appendix, Fig. S9), which showed that both proteins remain monodisperse at high concentrations with no indication of self-association. These results corroborate and confirm NMR-titration (cf., Fig. 2) and -relaxation data (cf., SI Appendix, Fig. S6) and establish that the reduction in cross-peak intensities of observed on the addition of unlabeled TSG101-UEV is due to the association of the two proteins.
Interactions of NMR-Visible TSG101-UEV with Unlabeled
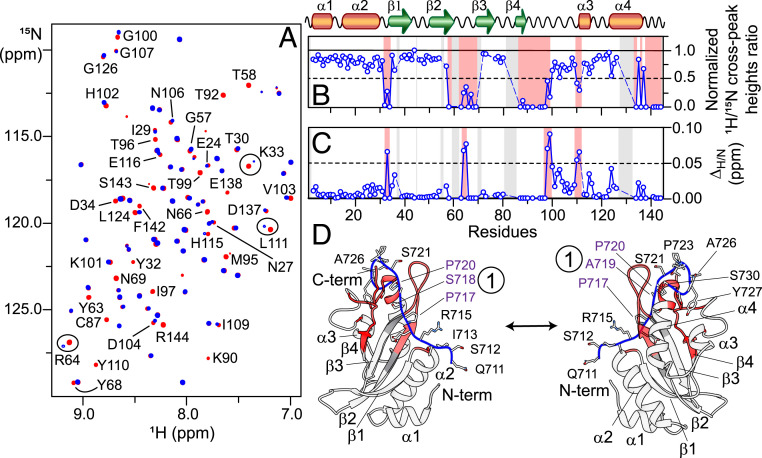
To further explore TSG101-UEV + interactions, NMR-titration experiments were carried out on NMR-visible TSG101-UEV in the absence and presence of unlabeled (Fig. 3A). The solution conformation of TSG101-UEV in the absence of was assessed using residual dipolar couplings (RDCs), which provide information on orientations of bond vectors (45–47). Excellent agreement was observed between the backbone amide (1DNH) RDCs, measured in polyethyleneglycol/hexanol alignment medium (48), and those calculated from X-ray coordinates of TSG101-UEV (Protein Data Bank [PDB] entry 3OBS) (44), indicating that the structure of TSG101-UEV remains unaltered in solution (SI Appendix, Fig. S10). A significant reduction in 1HN/15N cross-peak intensities and small 1HN/15N chemical-shift perturbations (∼0.05 to 0.1 ppm) were observed for the following TSG101-UEV regions on the addition of 4 molar equivalents of (Fig. 3 B and C): residues 32 to 34, 58 to 69, 87 to 103, 110 to 111, and 134 to 144. Comparison of these results against X-ray/NMR complexes of TSG101-UEV + HIV-1 p6 analogs (43, 44) revealed that occupies the same binding pocket of TSG101-UEV as that of the HIV-1 p6 peptide. We, therefore, carried out docking calculations using the X-ray structure of TSG101-UEV + HIV-1 p6 peptide complex as a template (PDB entry 3OBU) (44), supplemented by distance restraints derived from NMR-titration data in Xplor-NIH (49). To simplify calculations, only a large fragment of one of the interacting sites of site 1, was used as a PRD analog (residues 711 to 730). The lowest-energy structure from these calculations is shown in Fig. 3D. This structure confirms that TSG101-UEV carries a single binding site for a PTAP-like motif. Therefore, among the three PTAP-like motifs of namely 717PSAP720, 769PSAT772, and 790PGSAP794, only one can interact with one TSG101-UEV molecule at any given time.
Fig. 3.
NMR and structural analyses of interactions of TSG101-UEV with (A) Overlay of expanded region of the 1H-15N TROSY correlation spectra of 15N/2H-labeled 100 µM TSG101-UEV in the absence (red) and presence (blue) of (molar ratio: 1:4). Some isolated cross-peaks that exhibit significant reduction in intensities upon addition of are labeled. Cross-peaks that undergo chemical-shift changes on addition of are marked by circles. (B and C) The reduction in 1H-15N cross-peak intensities (B) and the perturbations in 1HN/15N chemical shifts (C) of TSG101-UEV on addition of Affected regions are highlighted in semitransparent red rectangles (secondary structure elements are indicated above the graphs). Semitransparent gray rectangles and dashed blue lines indicate the residues that could not be assigned unambiguously. (D) A ribbon diagram of model of TSG101-UEV + PRD711–730 complex. TSG101-UEV and PRD711–730 are colored in white and blue, respectively. Regions marked in red represent residues of TSG101-UEV that are most affected on addition of Gray ribbons indicate residues around the binding site that could not be assigned unambiguously. For PRD711–730, the side chains of individual residues are also shown; 717PSAP720 motif (site 1) is marked with dark purple labels.
Quantitative Characterization of Interactions of with TSG101-UEV.
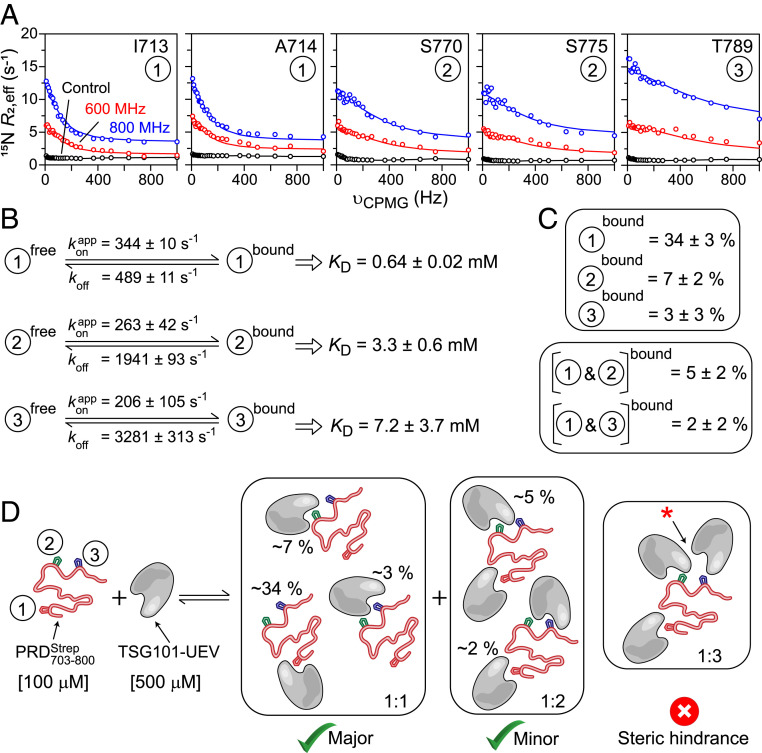
To quantitate + TSG101-UEV interactions, we collected Carr−Purcell−Meiboom−Gill (CPMG) relaxation dispersion experiments, which probe exchange between free and bound states on a time scale ranging from ∼0.1 to 10 ms (50–52). Analysis of CPMG profiles via propagation of the Bloch–McConnell equations (53) yields the kinetic rate constants for the exchange process(es) and population(s) of the bound state(s). Fig. 4A shows representative 15N-CPMG relaxation dispersion profiles observed for each of the interaction site of NMR-visible in the presence of 5-molar excess of unlabeled TSG101-UEV; the corresponding profiles in the absence of TSG101-UEV are flat. Consistent with 1HN/15N chemical-shift perturbation data (SI Appendix, Fig. S5), only a few residues exhibited significant dispersions. Therefore, to aid the quantitative analysis, 15N-CPMG measurements were recorded on NMR-visible in the presence of 2- and 3-molar excess of unlabeled TSG101-UEV (one at a time; SI Appendix, Fig. S11). Because only one site of can interact with one TSG101-UEV at any given time, dispersion profiles for each interaction site were fit independently to a two-state exchange model, A ↔ B, comprising a free (State A) and a bound (State B). For each PRD site, all of the CPMG data collected as a function of increasing TSG101-UEV concentration were fit simultaneously. The results of the CPMG fits are shown in Fig. 4A and SI Appendix, Fig. S11. The kinetic parameters extracted from the fits are shown in Fig. 4B (also see SI Appendix, Table S1). The values of fitted dissociation rate constants, koff, were 489 ± 11 s−1 (site 1) versus 1,941 ± 93 s−1 (site 2), and 3,281 ± 313 s−1 (site 3). Under the conditions of the experiment (100 μM + 500 μM TSG101-UEV), the values were 344 ± 10 s−1 (site 1), 263 ± 42 s−1 (site 2), and 206 ± 105 s−1 (site 3), where is the fitted pseudo first-order association rate constant. The exchange rate constants, between free and bound were ∼830 s−1 (site 1), ∼2,200 s−1 (site 2), and ∼3,500 s−1 (site 3), indicating that the exchange at site 1 is ∼3 and ∼4 times slower than at sites 2 and 3, respectively. This is largely because the koff rates for each PRD site were significantly different even though the corresponding values were comparable. The similarity of values is consistent with the observations that each PRD site is composed of sequentially similar PTAP-like motifs (cf., Fig. 2C), whereas differences in the corresponding koff rates can be attributed to the fact that, although similar, sites 1, 2, and 3 are not identical to one another. The total amount of unbound TSG101-UEV was calculated to be 453 μM under the conditions of the experiment (100 μM + 500 μM TSG101-UEV; see SI Appendix, SI Materials and Methods for additional details). Because only one PRD site can bind to one TSG101-UEV at any given time, second-order association rate constants (kon) were readily computed to be 76 ± 2, 58 ± 9, and 45 ± 23 (× 104 M−1⋅s−1) for sites 1, 2, and 3, respectively; , where [L] is the concentration of unbound TSG101-UEV. The resultant equilibrium dissociation constant, KD = koff/kon, values were 0.64 ± 0.02 mM (site 1), 3.3 ± 0.6 mM (site 2), and 7.2 ± 3.7 mM (site 3), implying that site 1 of is the primary site of interaction, whereas sites 2 and 3 are secondary. Note that the KD reported here for site 1 (∼0.64 mM; measured at 30 °C) is approximately five times weaker than that reported in a previous study (25) carried out using a peptide analog of ALIX-PRD (∼0.14 mM; measured using SPR at 20 °C). The discrepancy in binding affinity observed between intact (this work) versus its peptide analog (PRD714–723) (25) can be attributed to differences in constructs and experimental conditions, specifically the temperature. These findings are consistent with our earlier investigations of interactions between HIV-1 Gag and protease, which demonstrated that native substrates often interact weakly with their binding partners compared with the corresponding peptide analogs (51, 54).
Fig. 4.
Quantitative analyses of interactions of with TSG101-UEV. (A) Representative backbone 15N-CPMG relaxation dispersion profiles observed for 100 µM on addition of TSG101-UEV (molar ratio: 1:5); dispersions were recorded at 600 MHz (red) and 800 MHz (blue). The experimental data are displayed as circles, and the solid lines represent the global best fits to a two-state exchange model. Control relaxation dispersions at 800 MHz obtained in the absence of TSG101-UEV are shown in black. For the control data, black lines are used to guide the eye. (B) Summary of kinetic parameters obtained upon globally best fitting all CPMG data to a two-site exchange mode. Each interacting site (1, 2, and 3) is fit individually. is an apparent pseudo–first-order association rate constant that pertains to 100 μM and 500 μM TSG101-UEV used in the CPMG experiments. koff is the dissociation rate constant. The equilibrium dissociation constant, KD, for each individual site is given by koff/kon, where and [L] is the concentration of unbound TSG101-UEV. (C) Populations of 1:1 and 1:2 complexes formed between and TSG101-UEV, calculated using KD values for each individual PRD site and mass-action law. Note that sites 2 and 3 are mutually exclusive as they cannot simultaneously interact with two TSG101-UEV molecules due to steric hindrance (cf., Fig. 2C). (D) Scheme depicting potential modes of interactions of with TSG101-UEV; populations of bound sites are labeled.
In addition to a 1:1 stoichiometry where one site is occupied by one TSG101-UEV molecule, there exists a possibility that sites 1 and 2, which are ∼40 residues apart (cf., Fig. 2 C and D), can be occupied by two TSG101-UEV molecules at any given time: a 1:2 stoichiometry. Similarly, sites 1 and 3 are ∼50 residues apart and can thus form a 1:2 complex. Sites 2 and 3, however, are mutually exclusive as they are separated by ∼10 residues and, therefore, cannot simultaneously interact with two TSG101-UEV molecules due to steric hindrance. The total percentage occupancy of each site under the conditions of the experiment (100 μM + 500 μM TSG101-UEV) was calculated using corresponding KD values for each site and mass-action law (Fig. 4 C and D). The bound populations for the 1:1 complex were 34 ± 3% (site 1), 7 ± 2% (site 2), and 3 ± 3% (site 3), and for the 1:2 complex were 5 ± 2% (site 1 + site 2) and 2 ± 2% (site 1 + site 3). The above observations establish that contains three PTAP-like motifs that compete for binding to TSG101-UEV, where site 1 is the primary site of interaction, and sites 2 and 3 are secondary. The C-terminal portion of PRD is implicated in multimerization of ALIX (30) (see Amyloids and Gels of PRD800-868), indicating that in the event of ALIX multimerization, the sites mentioned above are likely to increase the functional affinity of ALIX-PRD for its interactions with TSG101.
Amyloids and Gels of PRD800–868.
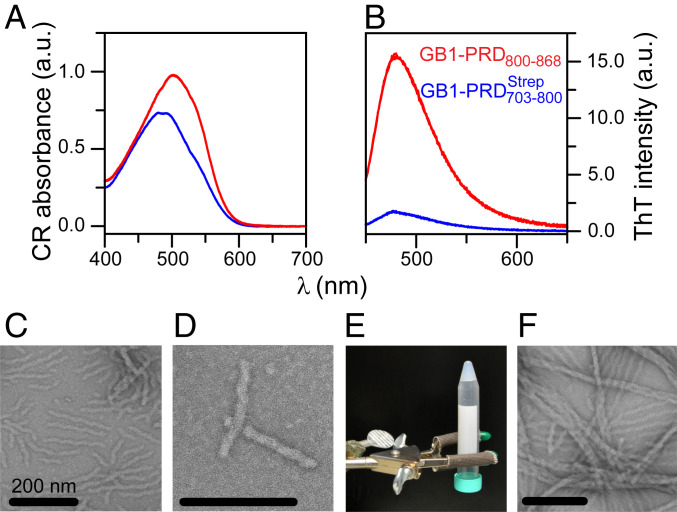
In contrast to which exhibited high solubility (soluble up to ∼2 mM), the C-terminal portion, PRD800–868, was poorly soluble under aqueous conditions (<30 μM; pH 4.5 to 7). The presence of an N-terminal GB1 fusion tag, often employed to overcome protein solubility and stability issues (55, 56), greatly improved the solubility of PRD800–868 (soluble up to ∼0.5 mM). Due to its favorable properties, including small size, high solubility, and extreme thermal stability, and because it usually does not influence structural characteristics of the tagged partner, the GB1 tag is often considered to be an ideal solubility-enhancement tag. The use of GB1 fusion construct allowed us to collect highly reproducible data and characterize aggregation properties of PRD800–868 (a serendipitous observation during purification of recombinant PRD800–868 revealed its aggregation-prone behavior). Spectral-shift assays were carried out using an amyloid-specific dye, CR, to explore the possibility that GB1–PRD800–868 aggregates were amyloidogenic (57). Clear shifts toward 540 nm were observed for aggregates of GB1–PRD800–868 in contrast to its N-terminal soluble counterpart, GB1– indicating the presence of amyloid structures for GB1–PRD800–868 aggregates (Fig. 5A). Emission assays carried out using ThT (58), another amyloid-specific dye that is thought to form ordered arrays along the lengths of fibrils, leading to increase in its fluorescence, indicated the presence of cross–β-sheet conformations (59) for GB1–PRD800–868 aggregates, corroborating their amyloidogenic nature (Fig. 5B). Analysis of GB1–PRD800–868 aggregates by negative-stain EM elucidated that these aggregates consist of unbranched filaments, characteristic of amyloid fibrils (Fig. 5 C and D). After ∼3 d at room temperature under nonagitated (quiescent) conditions, GB1–PRD800–868 aggregates coalesced to form a dense gel. Upon inverting the tube, the GB1–PRD800–868 solution remained at the bottom of the tube instead of falling into the cap, indicating a significant increase in viscosity (Fig. 5E). Analysis of GB1–PRD800–868 gel by negative-stain EM revealed a dense network of intertwined fibrils (Fig. 5F). Note that the gel-forming behavior of GB1–PRD800–868 is consistent with a wide variety of amyloid-forming polypeptides that have been shown to form hydrogels (60).
Fig. 5.
Amyloid fibrils and gel formed by GB1–PRD800–868. (A and B) Absorbance spectra of CR (A) and emission spectra of ThT (B) for aggregates formed by GB1–PRD800–868 (red). GB1– samples (blue) were used as controls. Raw data of three replicates (n = 3) are plotted against wavelength (nm). (C and D) Negatively stained EM images of GB1–PRD800–868 amyloid fibrils. (Scale bars, 200 nm.) (E) Formation of a gel by GB1–PRD800–868. To initiate the gel formation, a 0.1 mM GB1–PRD800–868 sample was kept at room temperature under quiescent conditions for ∼3 d. (F) EM images of GB1–PRD800–868 gel.
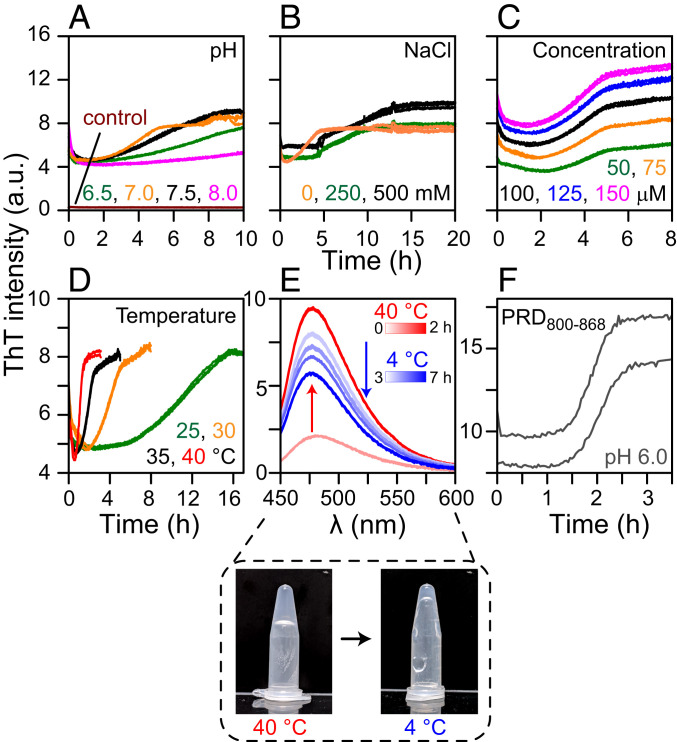
To validate and gain a mechanistic understanding of GB1–PRD800–868 amyloid formation, we monitored aggregation kinetics using ThT fluorescence. The effects of the following experimental conditions on aggregation kinetics were monitored (one at a time): pH, ionic strength, protein concentration, and temperature (Fig. 6). GB1–PRD800–868 exhibited sigmoidal aggregation profiles, ThT signal against time, a hallmark of amyloid formation (61). Most profiles displayed an initial dip in ThT signals, followed by a small lag phase, a robust growth phase, and a final plateau. High-quality data were obtained with continuous linear shaking with essentially no variations between replicates (raw data of three replicates from one plate are shown, n = 3; Fig. 6 A–D). To rule out the possible contributions from the GB1 fusion tag in PRD800–868 aggregation, identical experiments were performed on GB1– and on the GB1 tag itself, which showed no obvious ThT signals (Fig. 6A). ThT signals of GB1–PRD800–868 samples at zero time point (t0) were considerably higher than the corresponding controls, indicating the presence of substantial GB1–PRD800–868 aggregates at t0. The lyophilized GB1–PRD800–868 samples were treated with ammonium hydroxide to produce an aggregate-free solution (62); however, this method was not fully effective and did not result in complete solubilization and disaggregation of GB1–PRD800–868 samples. Faster aggregation was observed at pH 7.0 versus pH 6.5, 7.5 and 8.0 (Fig. 6A) and with 0 mM NaCl versus 250 and 500 mM NaCl (Fig. 6B). Changes in the concentration of GB1–PRD800–868 did not affect its aggregation rates (Fig. 6C; also see SI Appendix, Fig. S12 for measurements carried out under quiescent conditions, which show a similar trend). A significant increase in rates was observed upon an increase in temperature (Fig. 6D). At 40 °C, a half-maximal signal was reached within ∼1 h (t1/2 = 1 h), whereas the corresponding t1/2 values for 25, 30, and 35 °C were ∼11, ∼4, and ∼2 h, respectively. To further assess the impact of temperature on aggregation, ThT emission spectra were recorded (Fig. 6E; n = 3). GB1–PRD800–868 samples were incubated at 40 °C until the maximum ThT signal was reached (∼2 h), whereupon the temperature was reduced to 4 °C. Surprisingly, ThT signals of GB1–PRD800–868 amyloid fibrils decreased as a function of time at 4 °C (incubation time: ∼4 h), indicating that PRD800–868 forms reversible amyloids. Similar measurements carried out at 2, 6, 8, and 10 °C showed a slightly faster dissolution rate at 2 °C compared with 4 °C (incubation time: ∼3 h) and essentially no decrease in ThT signals at 8 and 10 °C, indicating that the PRD800–868 filaments are stable at and above 8 °C (SI Appendix, Fig. S13). A similar behavior was seen with GB1–PRD800–868 gel, which became fragile and liquid-like at 4 °C (Fig. 6 E, Lower). ThT measurements were also carried out on PRD800–868 (without the GB1 tag), which displayed characteristic sigmoidal profiles (Fig. 6F). Poor solubility of PRD800–868 resulted in significant variations between replicates and, therefore, precluded us from carrying out a thorough kinetic analysis.
Fig. 6.
Formation and dissolution of GB1–PRD800–868 amyloid fibrils. (A–D) ThT fluorescence was monitored to determine the effects of pH (A), ionic strength (B), concentration (C), and temperature (D) on the aggregation kinetics of GB1–PRD800–868 (raw data of three replicates [n = 3] are plotted against time). Control curves (dark red) (A) are collected using GB1– Control experiments were also carried out on GB1 fusion tag and showed no ThT signal. For A–C, the measurements were carried out at 30 °C. (E) Dissolution of GB1–PRD800–868 amyloid fibrils at 4 °C. ThT emission spectra of GB1–PRD800–868 (n = 3). Samples were incubated at 40 °C for ∼2 h (0 and 2-h time points are shown in light and dark red, respectively). Temperature was dropped to 4 °C, and fluorescence was measured until the reading was stabilized (3 to 7 h; light-to-dark blue gradient). The blue and red gradient color bars denote incubation time at 4 and 40 °C, respectively. The gel formed by GB1–PRD800–868 at 40 °C became fragile at 4 °C (E, Lower). ThT assays were also carried out on PRD800–868. Poor solubility of PRD800–868 resulted in large variations in kinetic curves (n = 2) (F). a.u., arbitrary units.
Src-Mediated Dissolution of GB1–PRD800–868 Amyloid Fibrils.
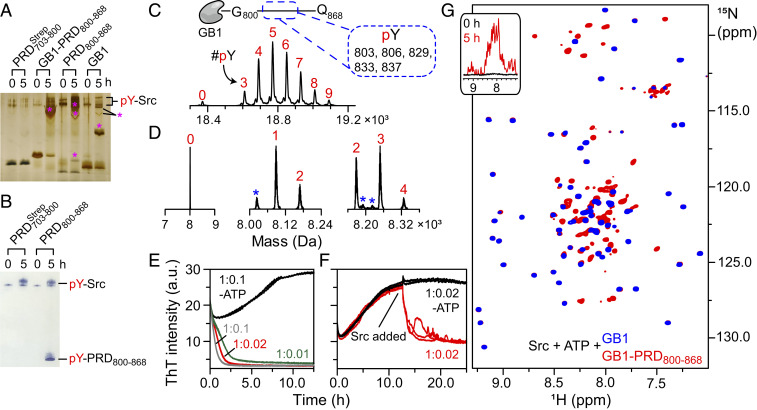
Src kinase was shown to hyperphosphorylate ALIX-PRD, culminating in a cellular redistribution of ALIX and initiating the formation of ESCRT-mediated intraluminal vesicles (26, 63). To study Src–PRD interactions, we carried out in vitro phosphorylation reactions using full-length Src. Recombinant full-length Src activity was measured using ADP-Glo assays (SI Appendix, Fig. S14) and was found to be consistent with previous reports (64). PRD constructs, namely and PRD800–868, were incubated with Src in the presence of adenosine triphosphate (ATP) (30 °C; incubation time: 5 h), and the products were monitored using Phos-tag SDS-PAGE (65). The latter is a modified version of SDS-PAGE that carries a phosphate-binding tag, which attenuates the migration of phosphorylated proteins, leading to a distinct separation of nonphosphorylated and phosphorylated species. Mobility shifts of phosphorylated PRD800–868 products were readily visible (Fig. 7A). No such shifts were observed for implying lack of phosphorylation and PRD800–868 contain 1 versus 14 tyrosines, respectively; cf., Fig. 1 A and D). To circumvent the poor solubility of PRD800–868, a kinase reaction was carried out on GB1–PRD800–868, and the corresponding mobility shifts indicated its phosphorylation. However, a control reaction carried out on the GB1 fusion tag revealed that the tag itself, comprising four tyrosines, undergoes residual phosphorylation in the presence of Src (Fig. 7A and SI Appendix, Table S2). Src-mediated phosphorylation of PRD800–868 and the lack of phosphorylation of were unambiguously confirmed by Western blotting and MS (Fig. 7 B–D, respectively). LC-ESI-TOFMS analyses of in vitro kinase reactions revealed the formation of hyperphosphorylated PRD800–868, elucidating phosphorylation of 9 (out of 18) and 4 (out of 14) tyrosine residues of GB1–PRD800–868 and PRD800–868, respectively (Fig. 7 C and D and SI Appendix, Table S2). LC–tandem MS (LC-MS/MS) sequencing of chymotrypsin-digested GB1–PRD800–868 revealed phosphorylation of the following PRD800–868 tyrosine residues upon Src treatment: 803, 806, 829, 833, and 837 (Fig. 7 C, Upper; also see SI Appendix, Fig. S15). Finally, we note that nothing definitive can be said about the phosphorylation status of remaining tyrosine residues of PRD800–868, namely 809, 812, 821, 824, 826, 846, 854, 864, and 865, because of sequencing coverage gaps arising from problems associated with proteolytic digestion of PRD800–868, which carries no native lysine and arginine residues (cf., Fig. 1D) and, thus, is not amenable to traditional trypsin-based proteomics (66).
Fig. 7.
Dissolution of GB1–PRD800–868 amyloids upon Src-mediated tyrosine phosphorylation. (A–D) Characterization of in vitro phosphorylation of PRD constructs using Phos-tag SDS-PAGE (A), Western blotting (B), and MS (C and D). For Phos-tag gel, the following constructs, namely GB1–PRD800–868, PRD800–868, and the GB1 tag, were incubated with Src (substrate to kinase molar ratio: 1:0.01). Phosphorylated products were visualized by silver staining and are marked by pink asterisks. (B) In vitro phosphorylation of and PRD800–868 by Western blotting. (C and D) LC-ESI-TOFMS and LC-MS/MS analyses of in vitro phosphorylation reactions revealed hyperphosphorylated states of GB1–PRD800–868 (C) and PRD800–868 (D). A schematic representation of GB1–PRD800–868 along with phosphorylated tyrosines (dashed rectangle) are shown above the graph in C. The numbers in red represent the number of phosphorylated tyrosines, labeled as pY (peaks marked with blue asterisks represent sodium/iron adducts; SI Appendix, Table S2). (E–G) The impact of tyrosine phosphorylation on aggregation kinetics of GB1–PRD800–868 was assessed using ThT assays (E and F) and NMR spectroscopy (G). For ThT assays (n = 3) (E), 150 µM GB1–PRD800–868 samples were incubated at 30 °C with 2 mM ATP and varying concentrations of Src (molar ratios: 1:0.1 [gray], 1:0.02 [red], and 1:0.01 [green]). a.u., arbitrary units. Control experiments were carried out on a GB1–PRD800–868 + Src mixture in the absence of ATP (black; molar ratio: 1:0.1). (F) Samples of 150 µM GB1–PRD800–868 were incubated at 30 °C without Src (n = 5) for ∼11 h. Src + ATP were then added to three samples (red), whereas the remaining two (black) received only Src. G shows the overlay of expanded regions of the 1H-15N TROSY-HSQC spectra of phosphorylated GB1–PRD800–868 (red) and the phosphorylated GB1 tag (blue). Both were incubated with Src (molar ratio: 1:0.1) in the presence of 2 mM ATP for 5 h at 30 °C. (G, Inset) Corresponding one-dimensional profiles of 15N-labeled GB1–PRD800–868 recorded at 0 and 5 h (black and red, respectively) after addition of unlabeled Src and ATP.
A previous report suggested direct interactions between the SH3 domain of Src and the 752PQPPAR757 motif of ALIX-PRD (26). To investigate + Src interactions, NMR-titration experiments were carried out using NMR-visible and unlabeled Src (SI Appendix, Fig. S16). With a minor exception of residues P754 and P759 (13C′/15N cross-peak intensity ratios of ∼0.55 and ∼0.63, respectively), no discernible 1HN/15N and 13C′/15N cross-peak intensity changes and chemical-shift perturbations were observed for on the addition of 3 molar equivalents of Src, indicating that full-length Src did not interact with The apparent discrepancy between these two results can be attributed to the use of full-length Src (this work) versus the isolated SH3 domain of Src (26). Recombinant Src is phosphorylated at multiple sites (SI Appendix, Fig. S3C) and, thus, likely exists in a clamped autoinhibited state (67). This closed conformation is expected to limit the access of the SH3 domain of Src to leading to no noticeable in vitro interactions between the two proteins. In line with these observations, the study mentioned above demonstrated that a constitutively active form of full-length Src that exists in an open conformation could interact with ALIX in Src−/−/Yes−/−/Fyn−/− (SYF) mouse embryo fibroblasts (26). Our results are in agreement with a recently proposed model of Src–ALIX interactions in exosomes (63), in which active Src associated with ALIX-PRD via its SH3 domain and these interactions were precluded by the clamped conformation of autoinhibited Src.
The impact of phosphorylation on the aggregation of GB1–PRD800–868 was assessed using fluorescence and NMR spectroscopy. For ThT assays (Fig. 7E), 150 μM GB1–PRD800–868 samples were incubated with varying amounts of Src (1.5, 3, and 15 μM) and 2 mM ATP (pH 7.5). As seen above (cf., Fig. 6 A–D), ThT signals at t0 were considerably higher than the baseline levels, likely due to the presence of preexisting GB1–PRD800–868 aggregates. However, a complete loss of ThT signals was observed as phosphorylation proceeded in real time. The dissolution rates were proportional to the amount of Src and, therefore, to the rate of phosphorylation. Half-lives of GB1–PRD800–868 aggregates were as follows: ∼1.5, ∼1.0, and ∼0.6 h at 30 °C for samples containing 1.5, 3, and 15 μM Src, respectively. Corresponding control experiments carried out on GB1–PRD800–868 + Src samples (without ATP) displayed characteristic sigmoidal profiles, indicating that the presence of Src per se does not hinder GB1–PRD800–868 aggregation. In a separate experiment, 3 μM Src + 2 mM ATP was added to 150 μM GB1–PRD800–868 samples upon reaching a stationary phase, which resulted in a significant loss of ThT signal with the corresponding half-life of ∼1 h (Fig. 7F). These observations indicate that both preformed GB1–PRD800–868 aggregates at t0 as well as aggregates formed in the plateau phase are susceptible to Src-mediated dissolution and establish that Src-mediated phosphorylation results in the dissolution of GB1–PRD800–868 fibrils.
Src-mediated dissolution of GB1–PRD800–868 assemblies was further validated by NMR spectroscopy (Fig. 7G). Two 15N-1H TROSY-HSQC spectra were recorded on 15N-labeled GB1–PRD800–868 in the presence of Src and ATP. The spectrum recorded at t0 exhibited spectroscopic signs of aggregation, i.e., resonances were broadened beyond the limits of detection (Fig. 7 G, Inset), confirming the presence of very high-molecular-weight GB1–PRD800–868 aggregates at t0. The reaction was allowed to proceed for 5 h, whereupon the same sample yielded a well-dispersed, high-quality spectrum (Fig. 7G). These observations established that phosphorylation dissolves GB1–PRD800–868 amyloids into NMR-amenable species that are likely monomeric. Control experiments were carried out on the GB1 fusion tag in the presence of Src and ATP under identical conditions. A near-perfect superimposition of spectra of these two proteins, phosphorylated GB1–PRD800–868 against phosphorylated GB1 tag, allowed a ready identification of PRD800–868 resonances. A narrow backbone amide proton dispersion of PRD800–868 cross-peaks, ∼7.3 to ∼8.7 ppm, revealed that (phosphorylated) PRD800–868, like its N-terminal counterpart, is also disordered in solution. Taken together, the above observations corroborate and confirm that GB1–PRD800–868 forms reversible amyloid fibrils.
Concluding Remarks
In summary, we made use of two recombinant constructs and PRD800–868) to carry out a thorough characterization of ALIX-PRD in solution (Fig. 1). We performed a detailed NMR characterization of the conformational plasticity of (a 98-residue polypeptide representing the N-terminal portion of ALIX-PRD) and carried out quantitative analyses of its interactions with TSG101-UEV. In addition, we uncovered remarkable aggregation properties of PRD800–868 (a 69-residue polypeptide representing the tyrosine-rich C-terminal portion of ALIX-PRD), which forms amyloids under near-physiological conditions that dissolve upon posttranslational modifications (Src-mediated tyrosine phosphorylation) or at low temperatures (2 to 6 °C).
Interactions between ALIX-PRD and TSG101-UEV.
NMR-backbone chemical shifts, 3JHNHα couplings, and 15N-relaxation rates indicate that is disordered in solution with no indication of any significant secondary-structure propensity (SI Appendix, Fig. S4). NMR-titration experiments demonstrate that contains three PTAP-like motifs, namely 717PSAP720, 769PSAT772, and 790PGSAP794, which compete for a single binding site on TSG101-UEV (Figs. 2 and 3 and SI Appendix, Fig. S5). 15N-relaxation measurements carried out on in the presence of TSG101-UEV show a lack of global ordering upon complex formation but point to local ordering around the interaction sites of (SI Appendix, Fig. S6). Quantitative analyses of 15N-CPMG dispersion data (Fig. 4) indicate low-affinity interactions for each of the three PTAP-like motifs (∼0.6 to ∼7 mM) and elucidate that the N-terminal 717PSAP720 motif is the primary site of interaction, whereas the other two sites, 769PSAT772 and 790PGSAP794, are secondary. These observations are in excellent agreement with a previous in vivo study (30), which indicated that mutations in the 717PSAP720 motif did not abolish ALIX–TSG101 interactions. They are also consistent with the fact that among the three PTAP-like motifs of ALIX-PRD, the 717PSAP720 motif is completely conserved among vertebrates (SI Appendix, Fig. S1). The analyses of CPMG dispersions allowed us to quantitate the amount of complex formed between each of the three PTAP-like motifs in the context of and TSG101-UEV, as well as populations of higher-order complexes in which two PTAP-like motifs are simultaneously occupied by two TSG101-UEV molecules (steric hindrance between 769PSAT772 and 790PGSAP794 motifs is likely to prevent a 1:3 stoichiometry between and TSG101-UEV). We hypothesize that the conformational plasticity of disordered will provide plausible opportunities for its unbound PTAP-like motifs to (re)bind before they diffuse out of proximity of TSG101-UEV. Since the C-terminal PRD forms amyloidogenic aggregates, in the event of PRD-mediated oligomerization of ALIX, we predict that ALIX oligomers will exhibit a markedly increased binding affinity to TSG101-UEV. These observations agree with a prior in vivo study (30), which showed that the multimerization-deficient ALIX constructs carrying the C-terminal PRD deletions and mutations were unable to interact with TSG101 despite comprising all three PTAP-like motifs mentioned above. Moreover, restoration of ALIX multimerization via a chimeric attachment of a GCN4 leucine zipper was shown to reinstate binding to TSG101. The lack of in vivo interactions between monomeric ALIX and TSG101 confirms the ultraweak nature of these interactions. The additive effect of multivalency is likely responsible for generating detectable in vivo interactions between the two proteins upon ALIX oligomerization. In addition to the UEV domain, full-length TSG101 comprises a coiled-coil domain that is shown to form tetrameric assemblies in solution (68). Based on these observations, we predict a dramatic increase in avidity for interactions between ALIX oligomers and TSG101 tetramers in vivo.
Amyloid Fibrils of ALIX-PRD.
The amyloid fibrils and gel of PRD800–868, visualized by negative-stain EM, revealed rope-like unbranched assemblies and a dense phase comprising an intertwined fibrous network, respectively (Fig. 5). In addition, fibril formation yielded a characteristic ThT-fluorescence enhancement (Fig. 6). Aggregation kinetics of PRD800–868, monitored by ThT fluorescence, show that aggregation is influenced by pH and ionic strength (faster aggregation at pH 7.0 and zero ionic strength) as well as by temperature, with high temperatures promoting aggregation (∼10-fold increase in aggregation at 40 versus 25 °C) and low temperatures (2 to 6 °C), promoting dissolution of amyloid fibrils. The discovery that temperature affects aggregation kinetics highlights the potential role of the hydrophobic effect in the formation of PRD800–868 fibrils. It is well known that the clustering of hydrophobic groups in polar solvents gives rise to a temperature-dependent hydrophobic effect that peaks between 30 and 80 °C and becomes weaker at lower and higher temperatures (69). Since the initial protein concentration did not affect the aggregation rates (Fig. 6C), we hypothesize that aggregation of PRD800–868 is primarily governed by a monomer-independent secondary-nucleation step in the form of fibril fragmentation (61). Further, we speculate that the presence of ∼20% tyrosines in PRD800–868 (Fig. 1D) drives its fibril formation through extensive stacking of aromatic rings of tyrosine residues accompanied by hydrophobic CH/π tyrosine–proline interactions (70). The remarkable conservation of PRD800–868 among vertebrates, with a near-complete conservation of tyrosine and proline residues (SI Appendix, Fig. S1), supports this hypothesis. It is further supported by the phosphorylation-mediated dissolution of amyloid fibrils (Fig. 7), where the introduction of negatively charged phosphate groups creates charge repulsion. Surprisingly, aggregates formed at the stationary phase are also susceptible to phosphorylation-mediated dissolution (Fig. 7F), which provides important clues about their structure. We hypothesize that a few tyrosine residues are solvent-exposed in PRD800–868 aggregates and, therefore, accessible for Src-mediated phosphorylation. Charge–repulsion created by phosphorylation of these tyrosine residues will likely initiate the disassembly. We hypothesize that both soluble and insoluble aggregates are present at the stationary phase and that the soluble aggregates are amenable to Src-mediated dissolution, whereas insoluble fibrils are inaccessible to Src. This hypothesis is supported by the fact that the addition of an Src + ATP mixture to PRD800–868 samples at the stationary phase does not culminate in a complete loss of ThT signal, indicating the presence of residual fibrils in the PRD800–868 samples.
ALIX-PRD Polymerization and ALIX Function.
The ESCRT-III proteins, CHMPs, polymerize into filamentous structures and are the main drivers of ESCRT-mediated membrane remodeling (19, 20). Note that the CHMP4 paralogues (CHMP4A and -B; Snf7 in yeast) are the most abundant components of ESCRT-III. The CHMP4 proteins exist as soluble monomers in solution and, therefore, need to be activated. This is achieved by nucleation factors, which trigger the polymerization of CHMP4. ALIX is a well-known nucleator factor of CHMP4 proteins (71). The N-terminal Bro1 domain of ALIX directly binds to and nucleates CHMP4 (72). The Bro1 domain interacts with lysobisphosphatidic acid, a phospholipid found in late endosomes, which allows ALIX to mediate the highly selective recruitment of CHMP4 paralogs to late endosomes (73). Based on our results, we predict that the oligomerization of ALIX mediated by the amyloidogenic assemblies of the C-terminal PRD will likely result in a multiplicative increase in binding affinity between the Bro1 domain and CHMP4 proteins, resulting in nucleation and polymerization of CHMP4. ALIX colocalizes with active Src in endosomal membranes (63). This ultimately drives the relocation of ALIX to the cytoplasm and reduces its association with its binding partners (26). Our results are in excellent agreement with this in vivo study as we show that hyperphosphorylation of PRD800–868 by Src shifts the equilibrium toward soluble monomeric species. Finally, we note that the N-terminal portion of ALIX-PRD is implicated in interdomain interactions with the N-terminal Bro1 domain, resulting in a closed conformation of ALIX (29). The preliminary NMR-titration experiments carried out using NMR-visible and unlabeled recombinant Bro1 domain did not yield any noticeable in vitro interactions between these two domains. Therefore, we cannot say anything definitive about these interdomain interactions of ALIX.
PRDs are often viewed as a collection of proline-rich motifs that serve as points of interactions for their signaling partners. Data presented here provide detailed quantitative analyses of interactions of a PRD with its binding partner. Our data also show that a PRD can form reversible amyloids and viscous gels. An array of “functional” amyloids that play physiological roles in humans have recently been identified, and a few of them have been shown to dissolve under physiological conditions (74). Further investigation is needed to determine whether amyloid formation of ALIX occurs in vivo and to uncover the exact cellular function(s) of these aggregates. From an application perspective, however, a reversible amyloid is of great significance for the development of novel biopolymers (75) that can be used for targeted drug delivery, tissue engineering, development of biosensors, and others.
Experimental Procedures
Full details relating to expression and purification of recombinant proteins; NMR, AUC, fluorescence spectroscopy, MS, and EM measurements; global fitting of relaxation dispersion data; X-ray and NMR-based docking calculations; in vitro kinase assays are provided in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank V. Tugarinov, Z. Jiang, A. Szabo, D. H. Appella, M. Wälti, A. Ceccon, Y. Shen, J. Ying, P. A. Jennings, B. Ramaraju, A. Shihora, I. Budin, and J. A. McCammon for useful discussions and X. Huang (University of California, San Diego [UCSD] Biomolecular NMR Facility), J. Yu (NMR Facility, Sanford Burnham Prebys Medical Discovery Institute), and L. Gross and Y. Su (UCSD Molecular Mass Spectrometry Facility) for technical support. We thank A. van der Vliet (University of Vermont), D. S. Waugh (National Cancer Institute, NIH), and N. Tjandra (National Heart, Lung and Blood Institute, NIH) for Src kinase, TEV protease, and TSG101-UEV constructs, respectively. This work was funded, in part, by a 2018 Development Award from the San Diego Center for AIDS Research (an NIH-funded program; NIH Grant P30 AI036214) (to L.D.); by Collaborative Development Awards from the Cellular Host Elements in Egress, Trafficking, and Assembly of HIV Center (University of Utah; an NIH-funded program; Grant P50 AI150464) and the HIV Interactions in Viral Evolution (HIVE) Center (The Scripps Research Institute; an NIH-funded program; NIH Grant U54 AI150472) (to L.D.); NSF Graduate Research Fellowship Grant DGE-1650112 (to R.D.E.); and NIH Grant R01 GM031749 (to W.M.). V.S.R. was supported by HIVE Center Grant U54 AI150472. C.D.S. was supported by the Center for Information Technology Intramural Research Program at the NIH. R.G. was supported by the National Institute of Diabetes and Digestive and Kidney Diseases Intramural Research Program at the NIH.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2010635117/-/DCSupplemental.
Data Availability.
The ALIX-PRD plasmids, namely and PRD800–868, have been deposited in the Addgene Repository, https://www.addgene.org/ (accession nos. 141344 and 141345 [PRD800–868]). The chemical-shift assignments of have been deposited in the Biological Magnetic Resonance Bank, http://www.bmrb.wisc.edu/ (accession no. 28111) (76). The coordinates of the docking model have been deposited in the Protein Model DataBase, http://srv00.recas.ba.infn.it/PMDB/ (accession no. PM0083242) (77).
References
- 1.MacArthur M. W., Thornton J. M., Influence of proline residues on protein conformation. J. Mol. Biol. 218, 397–412 (1991). [DOI] [PubMed] [Google Scholar]
- 2.Williamson M. P., The structure and function of proline-rich regions in proteins. Biochem. J. 297, 249–260 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rubin G. M., et al. , Comparative genomics of the eukaryotes. Science 287, 2204–2215 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morgan A. A., Rubenstein E., Proline: The distribution, frequency, positioning, and common functional roles of proline and polyproline sequences in the human proteome. PLoS One 8, e53785 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Theillet F. X., et al. , The alphabet of intrinsic disorder: I. Act like a Pro: On the abundance and roles of proline residues in intrinsically disordered proteins. Intrinsically Disord. Proteins 1, e24360 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wright P. E., Dyson H. J., Intrinsically disordered proteins in cellular signalling and regulation. Nat. Rev. Mol. Cell Biol. 16, 18–29 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adzhubei A. A., Sternberg M. J. E., Makarov A. A., Polyproline-II helix in proteins: Structure and function. J. Mol. Biol. 425, 2100–2132 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Tossavainen H., Aitio O., Hellman M., Saksela K., Permi P., Structural basis of the high affinity interaction between the alphavirus nonstructural protein-3 (nsP3) and the SH3 domain of Amphiphysin-2. J. Biol. Chem. 291, 16307–16317 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liao T.-J., Jang H., Nussinov R., Fushman D., High-affinity interactions of the nSH3/cSH3 domains of Grb2 with the C-terminal proline-rich domain of SOS1. J. Am. Chem. Soc. 142, 3401–3411 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Linding R., Russell R. B., Neduva V., Gibson T. J., GlobPlot: Exploring protein sequences for globularity and disorder. Nucleic Acids Res. 31, 3701–3708 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van der Lee R., et al. , Classification of intrinsically disordered regions and proteins. Chem. Rev. 114, 6589–6631 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li P., et al. , Phase transitions in the assembly of multivalent signalling proteins. Nature 483, 336–340 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee Y. C., Lee R. T., Carbohydrate-protein interactions: Basis of glycobiology. Acc. Chem. Res. 28, 321–327 (1995). [Google Scholar]
- 14.Mammen M., Choi S. K., Whitesides G. M., Polyvalent interactions in biological systems: Implications for design and use of multivalent ligands and inhibitors. Angew. Chem. Int. Ed. Engl. 37, 2754–2794 (1998). [DOI] [PubMed] [Google Scholar]
- 15.Kiessling L. L., Lamanna A. C., “Multivalency in biological systems” in Chemical Probes in Biology, Schneider M. P., Ed. (Springer Netherlands, 2003), pp. 345–357. [Google Scholar]
- 16.Kay B. K., Williamson M. P., Sudol M., The importance of being proline: The interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J. 14, 231–241 (2000). [PubMed] [Google Scholar]
- 17.Zarrinpar A., Bhattacharyya R. P., Lim W. A., The structure and function of proline recognition domains. Sci. STKE 2003, RE8 (2003). [DOI] [PubMed] [Google Scholar]
- 18.Bastidas M., Gibbs E. B., Sahu D., Showalter S. A., A primer for carbon-detected NMR applications to intrinsically disordered proteins in solution. Concepts Magnet. Resonance A 44, 54–66 (2015). [Google Scholar]
- 19.Schöneberg J., Lee I.-H., Iwasa J. H., Hurley J. H., Reverse-topology membrane scission by the ESCRT proteins. Nat. Rev. Mol. Cell Biol. 18, 5–17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vietri M., Radulovic M., Stenmark H., The many functions of ESCRTs. Nat. Rev. Mol. Cell Biol. 21, 25–42 (2020). [DOI] [PubMed] [Google Scholar]
- 21.Ren X., Hurley J. H., Proline-rich regions and motifs in trafficking: From ESCRT interaction to viral exploitation. Traffic 12, 1282–1290 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fisher R. D., et al. , Structural and biochemical studies of ALIX/AIP1 and its role in retrovirus budding. Cell 128, 841–852 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Zhai Q., et al. , Structural and functional studies of ALIX interactions with YPX(n)L late domains of HIV-1 and EIAV. Nat. Struct. Mol. Biol. 15, 43–49 (2008). [DOI] [PubMed] [Google Scholar]
- 24.Strack B., Calistri A., Craig S., Popova E., Göttlinger H. G., AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell 114, 689–699 (2003). [DOI] [PubMed] [Google Scholar]
- 25.von Schwedler U. K., et al. , The protein network of HIV budding. Cell 114, 701–713 (2003). [DOI] [PubMed] [Google Scholar]
- 26.Schmidt M. H. H., Dikic I., Bögler O., Src phosphorylation of Alix/AIP1 modulates its interaction with binding partners and antagonizes its activities. J. Biol. Chem. 280, 3414–3425 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi X., et al. , Identification and biophysical assessment of the molecular recognition mechanisms between the human haemopoietic cell kinase Src homology domain 3 and ALG-2-interacting protein X. Biochem. J. 431, 93–102 (2010). [DOI] [PubMed] [Google Scholar]
- 28.Chatellard-Causse C., et al. , Alix (ALG-2-interacting protein X), a protein involved in apoptosis, binds to endophilins and induces cytoplasmic vacuolization. J. Biol. Chem. 277, 29108–29115 (2002). [DOI] [PubMed] [Google Scholar]
- 29.Zhou X., Si J., Corvera J., Gallick G. E., Kuang J., Decoding the intrinsic mechanism that prohibits ALIX interaction with ESCRT and viral proteins. Biochem. J. 432, 525–534 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carlton J. G., Agromayor M., Martin-Serrano J., Differential requirements for Alix and ESCRT-III in cytokinesis and HIV-1 release. Proc. Natl. Acad. Sci. U.S.A. 105, 10541–10546 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhai Q., et al. , Activation of the retroviral budding factor ALIX. J. Virol. 85, 9222–9226 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doerfel L. K., et al. , EF-P is essential for rapid synthesis of proteins containing consecutive proline residues. Science 339, 85–88 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Ude S., et al. , Translation elongation factor EF-P alleviates ribosome stalling at polyproline stretches. Science 339, 82–85 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Woolstenhulme C. J., et al. , Nascent peptides that block protein synthesis in bacteria. Proc. Natl. Acad. Sci. U.S.A. 110, E878–E887 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huth J. R., et al. , Design of an expression system for detecting folded protein domains and mapping macromolecular interactions by NMR. Protein Sci. 6, 2359–2364 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmidt T. G., Skerra A., The Strep-tag system for one-step purification and high-affinity detection or capturing of proteins. Nat. Protoc. 2, 1528–1535 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Gill S. C., von Hippel P. H., Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 182, 319–326 (1989). [DOI] [PubMed] [Google Scholar]
- 38.Kjaergaard M., Brander S., Poulsen F. M., Random coil chemical shift for intrinsically disordered proteins: Effects of temperature and pH. J. Biomol. NMR 49, 139–149 (2011). [DOI] [PubMed] [Google Scholar]
- 39.Kjaergaard M., Poulsen F. M., Sequence correction of random coil chemical shifts: Correlation between neighbor correction factors and changes in the Ramachandran distribution. J. Biomol. NMR 50, 157–165 (2011). [DOI] [PubMed] [Google Scholar]
- 40.Camilloni C., De Simone A., Vranken W. F., Vendruscolo M., Determination of secondary structure populations in disordered states of proteins using nuclear magnetic resonance chemical shifts. Biochemistry 51, 2224–2231 (2012). [DOI] [PubMed] [Google Scholar]
- 41.Nielsen J. T., Mulder F. A. A., Quality and bias of protein disorder predictors. Sci. Rep. 9, 5137 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shen Y., Roche J., Grishaev A., Bax A., Prediction of nearest neighbor effects on backbone torsion angles and NMR scalar coupling constants in disordered proteins. Protein Sci. 27, 146–158 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pornillos O., Alam S. L., Davis D. R., Sundquist W. I., Structure of the Tsg101 UEV domain in complex with the PTAP motif of the HIV-1 p6 protein. Nat. Struct. Biol. 9, 812–817 (2002). [DOI] [PubMed] [Google Scholar]
- 44.Im Y. J., et al. , Crystallographic and functional analysis of the ESCRT-I/HIV-1 Gag PTAP interaction. Structure 18, 1536–1547 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bax A., Grishaev A., Weak alignment NMR: A hawk-eyed view of biomolecular structure. Curr. Opin. Struct. Biol. 15, 563–570 (2005). [DOI] [PubMed] [Google Scholar]
- 46.Deshmukh L., et al. , Structure and dynamics of full-length HIV-1 capsid protein in solution. J. Am. Chem. Soc. 135, 16133–16147 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deshmukh L., Ghirlando R., Clore G. M., Conformation and dynamics of the Gag polyprotein of the human immunodeficiency virus 1 studied by NMR spectroscopy. Proc. Natl. Acad. Sci. U.S.A. 112, 3374–3379 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rückert M., Otting G., Alignment of bological macromolecules in novel nonionic liquid crystalline media for NMR experiments. J. Am. Chem. Soc. 122, 7793–7797 (2000). [Google Scholar]
- 49.Schwieters C. D., Kuszewski J. J., Tjandra N., Clore G. M., The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 160, 65–73 (2003). [DOI] [PubMed] [Google Scholar]
- 50.Palmer A. G., 3rd, Chemical exchange in biomacromolecules: Past, present, and future. J. Magn. Reson. 241, 3–17 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Deshmukh L., Tugarinov V., Louis J. M., Clore G. M., Binding kinetics and substrate selectivity in HIV-1 protease-Gag interactions probed at atomic resolution by chemical exchange NMR. Proc. Natl. Acad. Sci. U.S.A. 114, E9855–E9862 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Deshmukh L., Tugarinov V., Appella D. H., Clore G. M., Targeting a dark excited state of HIV-1 nucleocapsid by antiretroviral thioesters revealed by NMR spectroscopy. Angew. Chem. Int. Ed. Engl. 57, 2687–2691 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McConnell H. M., Reaction rates by nuclear magnetic resonance. J. Chem. Phys. 28, 430–431 (1958). [Google Scholar]
- 54.Deshmukh L., Louis J. M., Ghirlando R., Clore G. M., Transient HIV-1 Gag-protease interactions revealed by paramagnetic NMR suggest origins of compensatory drug resistance mutations. Proc. Natl. Acad. Sci. U.S.A. 113, 12456–12461 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cheng Y., Patel D. J., An efficient system for small protein expression and refolding. Biochem. Biophys. Res. Commun. 317, 401–405 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou P., Wagner G., Overcoming the solubility limit with solubility-enhancement tags: Successful applications in biomolecular NMR studies. J. Biomol. NMR 46, 23–31 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Klunk W. E., Jacob R. F., Mason R. P., Quantifying amyloid by congo red spectral shift assay. Methods Enzymol. 309, 285–305 (1999). [DOI] [PubMed] [Google Scholar]
- 58.Aliyan A., Cook N. P., Martí A. A., Interrogating amyloid aggregates using fluorescent probes. Chem. Rev. 119, 11819–11856 (2019). [DOI] [PubMed] [Google Scholar]
- 59.Tycko R., Amyloid polymorphism: Structural basis and neurobiological relevance. Neuron 86, 632–645 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jean L., Foley A. C., Vaux D. J. T., The physiological and pathological implications of the formation of hydrogels, with a specific focus on amyloid polypeptides. Biomolecules 7, E70 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Michaels T. C. T., et al. , Chemical kinetics for bridging molecular mechanisms and macroscopic measurements of amyloid fibril formation. Annu. Rev. Phys. Chem. 69, 273–298 (2018). [DOI] [PubMed] [Google Scholar]
- 62.Ryan T. M., et al. , Ammonium hydroxide treatment of Aβ produces an aggregate free solution suitable for biophysical and cell culture characterization. PeerJ 1, e73 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hikita T., Kuwahara A., Watanabe R., Miyata M., Oneyama C., Src in endosomal membranes promotes exosome secretion and tumor progression. Sci. Rep. 9, 3265 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Heppner D. E., et al. , Direct cysteine sulfenylation drives activation of the Src kinase. Nat. Commun. 9, 4522 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kinoshita E., Kinoshita-Kikuta E., Koike T., Separation and detection of large phosphoproteins using Phos-tag SDS-PAGE. Nat. Protoc. 4, 1513–1521 (2009). [DOI] [PubMed] [Google Scholar]
- 66.Tsiatsiani L., Heck A. J., Proteomics beyond trypsin. FEBS J. 282, 2612–2626 (2015). [DOI] [PubMed] [Google Scholar]
- 67.Boggon T. J., Eck M. J., Structure and regulation of Src family kinases. Oncogene 23, 7918–7927 (2004). [DOI] [PubMed] [Google Scholar]
- 68.White J. T., Toptygin D., Cohen R., Murphy N., Hilser V. J., Structural stability of the coiled-coil domain of tumor susceptibility gene (TSG)-101. Biochemistry 56, 4646–4655 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huang D. M., Chandler D., Temperature and length scale dependence of hydrophobic effects and their possible implications for protein folding. Proc. Natl. Acad. Sci. U.S.A. 97, 8324–8327 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zondlo N. J., Aromatic-proline interactions: Electronically tunable CH/π interactions. Acc. Chem. Res. 46, 1039–1049 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Christ L., et al. , ALIX and ESCRT-I/II function as parallel ESCRT-III recruiters in cytokinetic abscission. J. Cell Biol. 212, 499–513 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McCullough J., Fisher R. D., Whitby F. G., Sundquist W. I., Hill C. P., ALIX-CHMP4 interactions in the human ESCRT pathway. Proc. Natl. Acad. Sci. U.S.A. 105, 7687–7691 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Larios J., Mercier V., Roux A., Gruenberg J., ALIX- and ESCRT-III-dependent sorting of tetraspanins to exosomes. J. Cell Biol. 219, e201904113 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jackson M. P., Hewitt E. W., Why are functional amyloids non-toxic in humans? Biomolecules 7, E71 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mankar S., Anoop A., Sen S., Maji S. K., Nanomaterials: Amyloids reflect their brighter side. Nano Rev. 2, 6032 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Elias R. D., Deshmukh L., Backbone 1H, 13C and 15N chemical shift assignments of the N-terminal portion of ALIX-PRD. Biological Magnetic Resonance Data Bank. http://www.bmrb.wisc.edu/data_library/summary/index.php?bmrbId=28111. Deposited 27 March 2020.
- 77.Elias R. D., Deshmukh L., Docking model of the complex between ALIX-PRD and TSG101-UEV. Protein Model DataBase, ID PM0083242. http://srv00.recas.ba.infn.it/PMDB/. Deposited 26 June 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The ALIX-PRD plasmids, namely and PRD800–868, have been deposited in the Addgene Repository, https://www.addgene.org/ (accession nos. 141344 and 141345 [PRD800–868]). The chemical-shift assignments of have been deposited in the Biological Magnetic Resonance Bank, http://www.bmrb.wisc.edu/ (accession no. 28111) (76). The coordinates of the docking model have been deposited in the Protein Model DataBase, http://srv00.recas.ba.infn.it/PMDB/ (accession no. PM0083242) (77).