Significance
The plant hormone auxin controls many aspects of growth and development. It does so by changing the activity of AUXIN RESPONSE FACTOR (ARF) proteins that recognize specific DNA elements in plant genes and switch gene expression on or off. A major question in plant biology is how these ARF proteins bind unique DNA sequences and thus select which genes are regulated by auxin. Here, the authors systematically study the DNA architecture required for high-affinity binding of ARF proteins, and for auxin-dependent gene regulation in the plant Arabidopsis thaliana. This work shows that repeats of ARF recognition sites are critical for function, and reveals the molecular basis for the distinct activities of different repeat structures.
Keywords: auxin, transcriptional regulation, protein–DNA interaction, ARF transcription factors, plant biology
Abstract
The hormone auxin controls many aspects of the plant life cycle by regulating the expression of thousands of genes. The transcriptional output of the nuclear auxin signaling pathway is determined by the activity of AUXIN RESPONSE transcription FACTORs (ARFs), through their binding to cis-regulatory elements in auxin-responsive genes. Crystal structures, in vitro, and heterologous studies have fueled a model in which ARF dimers bind with high affinity to distinctly spaced repeats of canonical AuxRE motifs. However, the relevance of this "caliper" model, and the mechanisms underlying the binding affinities in vivo, have remained elusive. Here we biochemically and functionally interrogate modes of ARF–DNA interaction. We show that a single additional hydrogen bond in Arabidopsis ARF1 confers high-affinity binding to individual DNA sites. We demonstrate the importance of AuxRE cooperativity within repeats in the Arabidopsis TMO5 and IAA11 promoters in vivo. Meta-analysis of transcriptomes further reveals strong genome-wide association of auxin response with both inverted (IR) and direct (DR) AuxRE repeats, which we experimentally validated. The association of these elements with auxin-induced up-regulation (DR and IR) or down-regulation (IR) was correlated with differential binding affinities of A-class and B-class ARFs, respectively, suggesting a mechanistic basis for the distinct activity of these repeats. Our results support the relevance of high-affinity binding of ARF transcription factors to uniquely spaced DNA elements in vivo, and suggest that differential binding affinities of ARF subfamilies underlie diversity in cis-element function.
The hormone auxin is a central growth regulator that contributes to almost all aspects of the plant life cycle by promoting development from embryogenesis to maturity and mediating various environmental growth responses (1–6). Changes in cellular auxin concentrations trigger transcriptional responses of numerous genes, mediated by AUXIN RESPONSE FACTOR (ARF) transcription factors (7–9). Most ARFs have three conserved functional domains: an N-terminal DNA-binding domain (DBD), C-terminal PB1 for protein–protein interaction, and a middle region that defines the transcription-regulatory activity.
Under low-auxin conditions, members of the Auxin/INDOLE ACETIC ACID (Aux/IAA) protein family bind ARFs through the shared PB1 domain, and suppress the expression of genes near ARF-binding sites by recruiting TOPLESS corepressors (9–11). The small auxin molecule promotes the physical interaction between Aux/IAA proteins and TIR1/AFB F-box proteins leading to Aux/IAA protein ubiquitination and degradation in the 26S proteasome (12–16). Thus, auxin promotes the activity of ARFs by releasing these from Aux/IAA-mediated inhibitors. Liberated ARFs then regulate the genes that have cognate DNA target sequences - so-called auxin response cis-elements (AuxREs) in their promoters (17, 18). Auxin, therefore, triggers specific cellular responses by turning genetic switches composed of ARFs and AuxREs (19–22).
A combination of molecular, genetic, and biochemical studies on the promoter regions of early auxin-responsive genes from soybean identified the hexanucleotide sequence motif 5′-TGTCTC-3′ to act as an AuxRE, which not only enabled the prediction of auxin responsiveness of genes but also the creation of auxin-reporter systems (7, 23, 24) and the identification of the first ARF protein (25). Fluorophore- or enzyme-reporter genes driven by synthetic promoters including a tandem direct repeat of TGTCTC spaced at 5-bp intervals, termed DR5 have been widely used for visualizing the distribution pattern of auxin signal in many plant species (26–33).
Analyses using protein-binding microarrays, which estimates the binding preference of proteins to all possible short double-stranded (ds)DNA sequences, have proposed 5′-TGTCGG-3′ rather than the canonical AuxRE TGTCTC as the preferred sequence for Arabidopsis ARF1, ARF5, and ARF3 (34, 35). DNA affinity purification sequencing (DAP-seq) has also demonstrated that TGTCGG is a probable hexanucleotide-target motif of ARF2 and ARF5 (36). The activity of this newly identified motif as functional AuxRE has been confirmed in Arabidopsis. The meta-analysis using publicly available microarray datasets collected from auxin-treated plants revealed that, among 5′-TGTCNN-3′ variants, TGTCGG is significantly overrepresented/enriched in the regulatory region of auxin up-regulated genes in nine datasets and in down-regulated genes in two datasets. In comparison, TGTCTC is associated with up-regulation in five datasets (37). Furthermore, a synthetic promoter based on the TGTCGG element, demonstrated its increased auxin sensitivity compared to the version based on the TGTCTC element (38).
Besides the base sequence of each motif, the positional relationship between closely associated AuxREs is thought to define auxin response. Together with the atomic-resolution structural analyses of the DBD and PB1 domain of a few ARFs, in vitro biochemical assays have suggested that ARFs can dimerize through these domains, and the dimerization ability allows strong binding to dsDNA carrying a pair of AuxREs arranged in inverted repeat with a spacer of a specific length (34, 39). Seven- or 8 bp of spacing between the half-sites is required for such enhanced interaction with ARF1, whereas ARF5 accepts spacing from 5 to 9 bp (25, 34). In addition, a synthetic reporter based on a direct repeat of 7–9 AuxREs, each spaced by 5 bases (DR5), is highly responsive to auxin (26, 38), suggesting that this repeat constellation is biologically meaningful.
To date, there is limited information on how ARFs recognize AuxRE variants and how the preference of ARFs to composite AuxREs, which consist of cooperating DNA motifs, is reflected in their gene-regulatory function in vivo. In this study, we address the biochemical basis underlying the specificity in ARF–DNA interactions, and its biological relevance. We illuminate the mode of interaction between ARF-DBD with a high-affinity AuxRE, and show the critical contribution of inverted AuxREs repeats to gene regulation in vivo. We find strong genome-wide association between auxin regulation and the presence of direct or inverted AuxRE repeats with a short spacer, and propose a mechanism underlying the distinct activities of such repeats. Our work establishes the in vivo relevance of high-affinity ARF binding to unique DNA repeats, and provides a plausible mechanism for diversity in cis-element function.
Results
Structural Basis of ARF Binding to a High-Affinity Element.
The DNA sequence 5′-TGTC-3′ is critical for recruiting ARFs based on in vitro binding assays, as well as the effect of mutations in this element in promoters on the expression of some genes (25, 34, 40, 41). However, the two nucleotides (NN) following the core tetranucleotide play an important role in determining ARF binding affinity in vitro (25, 42), and there is strong conservation of several NN variants in auxin-responsive genes in many angiosperms (43). Importantly, protein-binding microarrays revealed large differences in apparent binding affinity to all TGTCNN hexamers (34, 35), and a variant with GG as final bases conferred a more potent transcriptional auxin response than a TC variant (38).
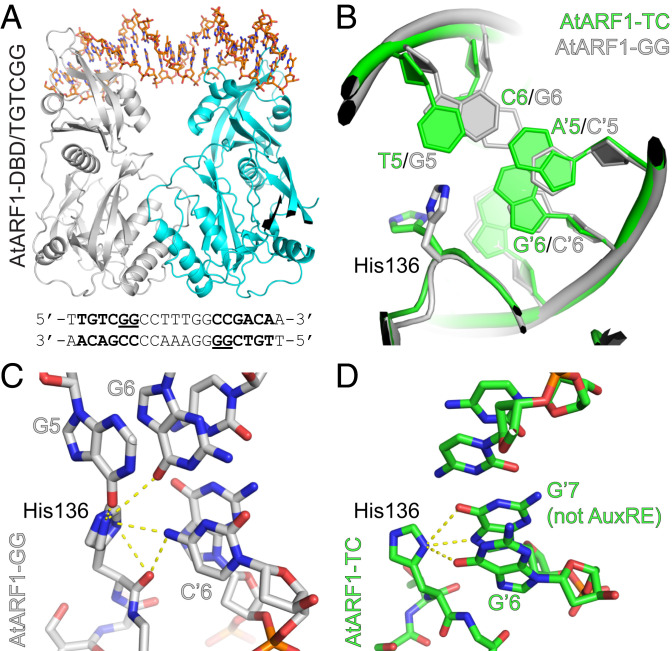
To investigate the mechanisms by which the last two nucleotides in the hexanucleotide motif 5′-TGTCNN-3′ influence ARF-DNA binding, we crystalized the DBD of Arabidopsis ARF1 (AtARF1-DBD) in complex with a double-stranded oligonucleotide carrying a 7-bp–spaced inverted repeat of the high-affinity TGTCGG hexanucleotide, analogous to the previous complex between AtARF1-DBD and a TGTCTC-containing repeat (34). The sequence of the spacer between the two inverted AuxRE sites was chosen based on its earlier use in DNA-binding assays (25) and in the ARF1-DBD-ER7 structure (34). The ARF1-TGTCGG structure was solved to a resolution of 1.65 Å (SI Appendix, Table S1), which facilitated the construction of loops Q228-P233 and E299-K306 that were not visible in the previously reported structures of AtARF-1DBD (Protein Data Bank ID code: 4LDX). The overall structure (Fig. 1A) is highly similar to that of the protein bound to the TGTCTC inverted repeat (34), with an rmsd of 0.78 Å. The B3 domain recognizes the DNA major groove, while the regions N- and C-terminal to the B3 domain form a single domain that enables the protein to dimerize (Fig. 1A). Superimposing the structures of the two ARF1-DBD-DNA complexes, we observed that the same residues contribute to DNA recognition in the two ARF1-DBD structures, but the conformation differs among residues H136 and G137, which contact the GG bases of the TGTCGG and the last TC bases of the TGTCTC hexanucleotide (Fig. 1B). In the TGTCGG-bound protein, the H136 sidechain flips and thereby comes into proximity of the two GG bases, which allows it to make hydrogen bonds to the O6 atom of either guanine, depending on the orientation of the imidazole ring (Fig. 1C). When bound to TGTCTC, this orientation of the H136 sidechain is not possible due to steric hindrance of the cytosine N4 atom and the guanidine O6 atom of the cytosine–guanidine base pair (Fig. 1 C and D). This extra contact of the TGTCGG sequence with G5/G6 displaces the loop of residues S134-L142 to 1.4–1.8 Å in superimposition with the ARF1-DBD-TGTCTC complex (Fig. 1B). Besides, the region encompassing bases C4-G5-G6-C7 is displaced away from the protein by about 1.9 Å, making room for H136 to penetrate deeper into the major groove (Fig. 1B). The displacement of the DNA is local, as further up- and downstream of the G5G6 bases, the DNA structures of TGTCTC-bound and TGTCGG-bound proteins coincide (SI Appendix, Fig. S1 A and B). A slightly higher curvature occurs in the TGTCGG-containing oligonucleotide (SI Appendix, Fig. S1B) resulting in a slight contraction of the protein structure along the direction of the helix axis of the DNA and bringing the two B3 domains about 1.2 Å closer to each other in comparison with the TGTCTC-bound structure (SI Appendix, Fig. S1 B and C).
Fig. 1.
Atomic basis for high-affinity ARF-DNA binding. (A) Crystal structure of an AtARF1-DBD dimer bound to dsDNA containing two inverted TGTCGG elements (sequence below, AuxREs in bold and high-affinity residues underlined). The two monomers are colored gray and cyan, respectively, and the DNA is shown as a stick representation in orange. (B) Overlay of the ARF1-DBD [H136-G137] region and interacting nucleotides in the structures of ARF1-DBD/TGTCGG complex (AtARF1-GG, gray) and ARF1-DBD/TGTCTC complex (AtARF1-TC, green). Note that in the AtARF1-GG structure, the DNA is displaced upwards and H136 enters deeper into the major groove, allow interactions with the G5 and G6 bases. (C and D) Hydrogen bonding of ARF1 His136 with AuxRE nucleotides. Interaction of H136-G137 with the G5G6 bases in the TGTCGG-containing oligonucleotide (C) and with G6G7 bases of the complementary strand in TGTCTC-containing oligonucleotide (D). Note that the G7 base is not part of the AuxRE. A backbone oxygen contributes to DNA interaction in the GG structure (C).
These results suggest that ARF dimers are adaptable to a range of AuxRE variants and adopt local conformational changes depending on the sequence of bases neighboring to the TGTC core. This structural analysis also shows that the increased affinity to the TGTCGG element is caused by subtle changes in the conformation of the protein loop and the two interacting guanidines, and through the formation of a single additional hydrogen bond.
Cooperatively Acting AuxREs Are Critical for Regulation of the TMO5 Gene.
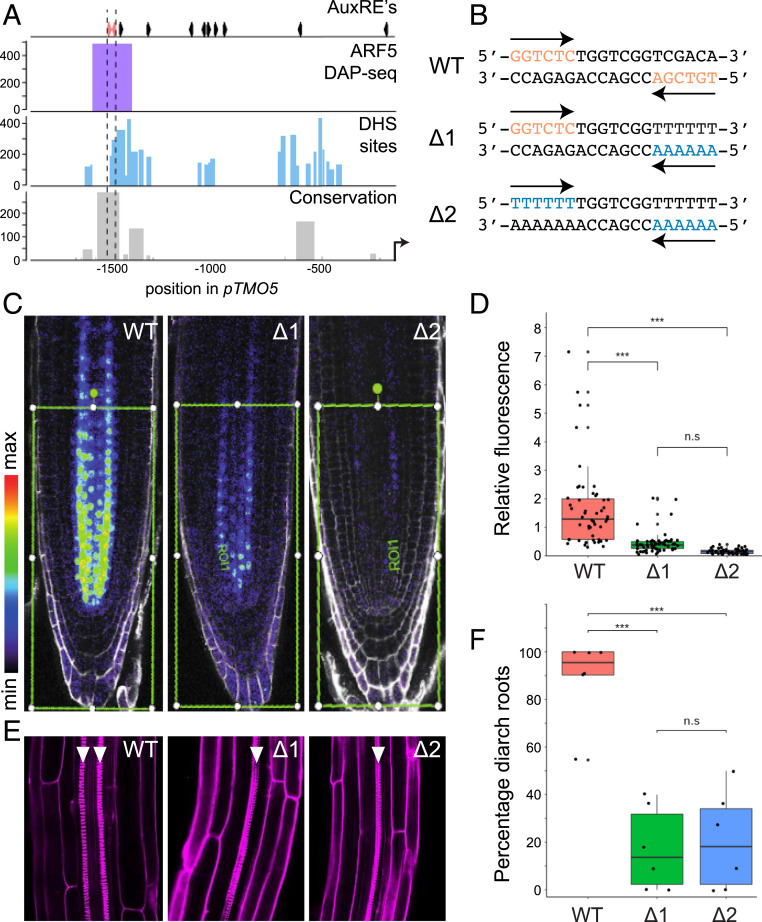
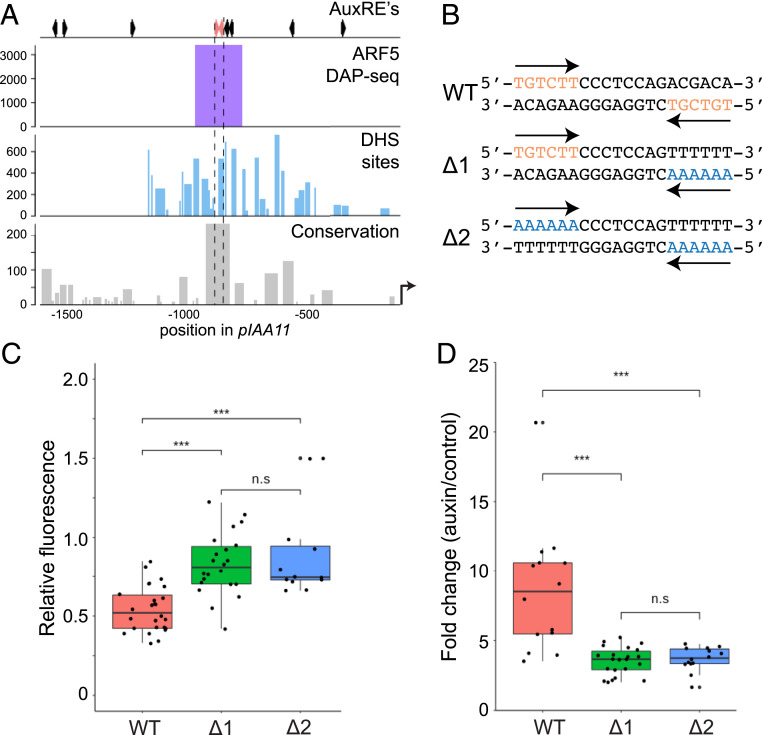
ARFs can bind with high affinity to a composite element composed of two inverted AuxREs, a property that requires dimerization of ARF DBDs (34, 42). While the significance of cooperative binding in vitro is clear, it is not known whether gene regulation in biological context depends on this phenomenon. Although the number of well-characterized genes directly targeted by ARFs with known biological functions is limited, previous work identified transcriptional targets for ARF5, including TARGET OF MONOPTEROS5 (TMO5) in Arabidopsis (44). Loss of TMO5 and its paralog TMO5-LIKE1 (T5L1) leads to strong defects in vascular tissue development (45), and this ARF5 target gene therefore offers a good model to study the relevance of cis-elements. Within the ∼2,3 kb promoter region that is sufficient to drive expression of a complementing TMO5 gene (45), there are 11 AuxRE-like motifs (Fig. 2A). Among these, only two are in an inverted repeat constellation with 7-bp spacing (Fig. 2A). TMO5 function appears deeply conserved across vascular plants (46), and we thus explored conservation of promoter regions in the plant kingdom [63 species of 7 lineages from the PlantRegMap database (47)]. At 1,589 bp upstream from the start codon in Arabidopsis, there is a highly conserved sequence which includes the two noncanonical AuxRE-like hexanucleotide motifs (GGTCTC; TGTCGA) in inverted repeat (Fig. 2A). Reanalysis of DAPseq ARF5 binding data and DNase I hypersensitivity (Fig. 2A) shows that this conserved sequence corresponds to an ARF5 binding region, and is located in accessible chromatin.
Fig. 2.
Cooperative action of two AuxRE motifs in an inverted repeat. (A) Genomic features of the promoter region of TMO5. (Top) Positions of AuxRE-like motifs across the promoter. The position of the two AuxRE-like motifs in an inverted repeat constellation with 7-bp spacing are indicated in red. (Bottom) Rows show AtARF5 DAP-seq peaks, DNase hypersensitive sites (DHSs), and sequence conservation among TMO5 homologs in 63 angiosperm species. (B) WT and mutated (∆1 and ∆2) sequence at 1,588–1,569 bp upstream of the start codon in the TMO5 promoter. AuxRE-like hexanucleotide motifs and the corresponding mutated nucleotides are indicated in red and blue, respectively. (C) Representative images of 5-d-old root tips that express TMO5-3xGFP driven by the WT TMO5 promoter, and ∆1 and ∆2 mutants. The roots were counterstained with propidium iodide (PI, gray). Green frames indicate the area in which GFP signals were quantified. (D) Boxplot showing the levels of fluorescent signals taken from TMO5-3xGFP driven by the WT and mutant promoter. Each dot represents the mean intensity measured from an individual root tip, and data were collected from multiple individual transgenics (SI Appendix, Fig. S3). (E) Representative images of 1-wk-old primary roots of tmo5 tmo5l1 seedlings that carry a transgene to express TMO5-tdT under the control of WT or mutant TMO5 promoter. Arrowheads indicate protoxylem strands. The roots were stained with PI. (F) Boxplots showing percentages of diarch seedlings in independent transgenic tmo5 tmo5l1 mutant seedlings carrying the transgene WT or mutated pTMO5. Each dot represents an individual transgenic line. The numbers of observed roots are shown in SI Appendix, Table S2. Asterisks in D and F indicate statistically significant differences (P value < 0.001) assessed by one-way ANOVA with post hoc Tukey test; n.s., not significant.
To investigate whether these adjacent AuxRE-like motifs are biologically relevant, and to determine if the two motifs in the repeat act redundantly, additively or cooperatively, we examined expression and biological activity of TMO5-3xGFP genomic fragments (44, 45) in which either one (proTMO5∆1) or both (proTMO5∆2) half-sites were mutated (Fig. 2B).
We first determined binding of ARF proteins to the wild-type and mutated inverted repeat by size-exclusion chromatography. ARF5 DBD interacted readily with the wild-type pTMO5 element (SI Appendix, Fig. S2), while no appreciable binding of ARF1-DBD could be detected. Mutation in either one or both AuxREs abrogated binding of ARF5-DBD in this assay (SI Appendix, Fig. S2), thus validating the prediction that the inverted repeat is a direct ARF5 binding site, and that the mutations interfere with binding in vitro.
Both single and double mutations in the half-sites strongly affected expression of the TMO5-3xGFP fusion protein, compared with the signal driven by the wild-type (WT) promoter, which is detected mainly in the young root vascular tissue (Fig. 2C). It is clear from our data that the loss of only one motif drastically impaired promoter activity (Fig. 2 C and D and SI Appendix, Fig. S3). Furthermore, neither proTMO5∆1 and proTMO5∆2 was ectopically expressed (Fig. 2C), suggesting that the base-pair substitutions did not create new, functional cis-elements to the promoter. Quantification of fluorescence signal intensity in root tips showed that the signal driven by the proTMO5∆1 was much lower than the value intermediate between those obtained from WT promoter and proTMO5∆2 (Fig. 2D), and in fact the single and double mutants were not significantly different from one another (one-way ANOVA with post hoc Tukey, P = 0.18). These results indicate that the region containing the composite element determines the expression level of TMO5 and the two half-sites function cooperatively and not redundantly or additively.
To determine whether cooperativity between the two AuxREs in the TMO5 promoter is required for biological function, we analyzed complementation of the tmo5 t5l1 double mutant (45) by the WT and mutated genomic fragments. WT Arabidopsis roots develop two xylem poles as well as two phloem poles, which establish a bisymmetric (diarch) pattern. Both single mutants in TMO5 or T5L1 are WT in appearance, but tmo5 tmo5l1 double-mutant roots show a single xylem and phloem pole (monarch) (45, 48, 49). When introduced into the tmo5 t5l1 mutant, a transgene encoding TMO5-tdTomato (TMO5-tdT) driven by the WT proTMO5 promoter fully complemented the monarch defect (Fig. 2 E and F and SI Appendix, Table S2). In contrast, neither proTMO5∆1 nor proTMO5∆2 could drive TMO5 protein levels sufficient for mutant complementation. Also here, mutation of a single half-site led to an effect indistinguishable from removing both half-sites (Fig. 2F). Thus, also with regards to biological activity, the cooperativity among two inverted AuxREs appears critical, which firmly supports a model where high-affinity binding of ARF dimers to repeats controls gene activity.
Genome-Wide Association of AuxRE Repeats with Auxin Response.
The occurrence and significance of the 7-bp–spaced bipartite inverted AuxREs in the TMO5 promoter confirms the critical prediction from the ARF–DNA structure and from in vitro binding assays that ARFs can act as dimers. However, it is unclear to what extent this mode of regulation reflects a general principle in auxin response. Given that TGTC-based hexamers are abundant in plant genomes, and cannot by themselves explain or help predicting auxin response confidently (37), it is likely that a composite element is involved in auxin-dependent gene regulation at genome-wide scale. Earlier enrichment of particular AuxRE compositions, including inverted repeats spaced by seven or eight bases, have been shown in the promoters of auxin-responsive genes and in ARF5 DAP-Seq regions (50).
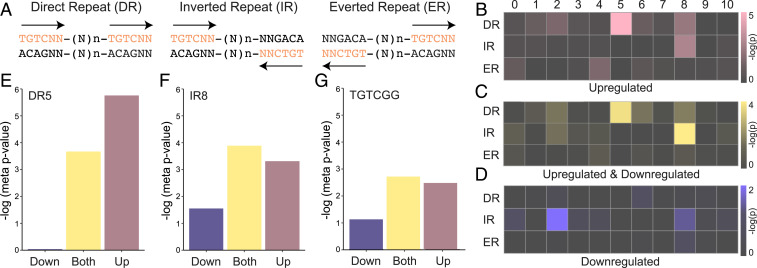
We systematically addressed if the occurrence of spaced repeats of AuxREs in gene upstream regions is associated with auxin responsiveness of gene expression in individual transcriptomes. First, we searched potential bipartite AuxREs: direct, inverted, and everted repeat (DR, IR, and ER) of TGTCNN hexamer with spacing between the half-sites ranging from 0 to 10 bp (Fig. 3A) in the 1,500-bp upstream of each gene in the Arabidopsis genome. Nomenclature of AuxRE repeats has not been uniform in the literature (25, 34, 36, 39, 50), and we here refer to these according to their actual physical orientation, given that each AuxRE has polarity (5′ to 3′) and can be considered a vector. Thus, when both vectors point in the same direction, this is a DR, when vectors point to each other, we refer to this as IR, and when they point away from each other, it is an ER (Fig. 3A). We adopt the definition of spacing from Pierre-Jerome et al. (39). Note that this is in contrast to our earlier use of ER (34) for the same repeats that we now name IR. We hope that this nomenclature will be adopted by the community to avoid further confusion. The −1,500 region was chosen as optimal for bioinformatics analysis on Arabidopsis (51), because its genome is compact, with many genes having only short distance to an upstream neighbor (52). The inter-AuxRE spacer range was based on the maximum distance that could reasonably be expected to allow protein–protein interaction when two ARFs are bound. To evaluate the association of these composite elements with auxin responsiveness, we conducted a meta-analysis of publicly available transcriptomic datasets (5 RNA-Seq and 10 microarray datasets) related to auxin (listed in SI Appendix, Table S3) using the metaRE pipeline (53). For each dataset, we estimated statistically (Fisher’s exact test) if any of the 33 AuxRE repeats (IR0-10, ER0-10, DR0-10) were enriched in upstream regions of auxin-dependent genes. In this analysis, we united the lists of genes that were up- or down-regulated after auxin treatment. We next combined P values for each repeat into meta p–values (under Fisher’s method) independently for RNA-Seq and microarray datasets. Following statistical analysis of association, we found only two composite motifs to be clearly associated with auxin-dependent gene regulation both in microarray and RNA-Seq datasets: DR5 and IR8 (Fig. 3C and SI Appendix, Fig. S4 and Tables S4 and S5). The DR5 and IR8 elements were distributed evenly in the −3,000-bp upstream region across the Arabidopsis genome (SI Appendix, Fig. S5). Strikingly, these are precisely the two motifs that have been studied functionally. Several other repeats showed weaker association and only in one of two independent analyses (Fig. 3C and SI Appendix, Tables S4 and S5).
Fig. 3.
Genome-wide association of AuxRE repeats with auxin responsiveness. (A) Definition of DRn, IRn, and ERn of TGTCNN hexanucleotide. N indicates A, C, G, or T. n indicates the number of nucleotides between two TGTCNN half-sites (0 ≤ n ≤10). (B–D) Association of DRn-, IRn-, or ERn variants present in the upstream regions (1,500 bp; 5′UTR) of the genes with auxin responsiveness. Color saturation visualizes the significance of overrepresentation for each composite element in −log10(meta p-value) units. Auxin-activated versus -nonactivated genes (B), both up- and down-regulated versus nonresponding genes (C), and down-regulated versus noninhibited genes (D). (E–G) Significance levels for association of DR5 (E), IR8 (F), and a single TGTCGG motif (G) with up-regulation, down-regulation, and both, in −log10(meta p-value) units.
We next performed an alternative analysis where up- and down-regulated genes were analyzed separately. This revealed that both the single TGTCGG element and the DR5 motif were more strongly associated with up-regulation than with down-regulation (Fig. 3 B, E, and G), whereas the IR8 motif was associated both with up- and down-regulation (Fig. 3 D and F). Weak association of IR8 with down-regulation (metap value < 0.05) was detected only in RNA-Seq datasets, while we also identified IR2 as an enriched motif for down-regulated genes. This analysis suggests that there are two major AuxRE repeats linked to the auxin transcriptional response: DR5 being prominently associated with gene activation, and IR8 with both gene activation and repression. Both repeats are more strongly associated with auxin-responsive gene expression than even the best single AuxRE (TGTCGG; Fig. 3G), suggesting a genome-wide role for composite elements in auxin response.
IR8 and DR5 Elements Predict Auxin-Responsive Transcription.
In addition to the previous in vitro demonstration of the physical interaction between ARFs and dsDNA containing a palindrome of TGTCTC spaced by eight nucleotides on a few model genes (25, 34), our current metaanalysis suggests that this motif may be commonly used in auxin-responsive genes. Furthermore, the association with the DR5 motif is consistent with substantial auxin responsiveness of synthetic promoters carrying a tandemly repeated TGTCTC or TGTCGG with 5-bp spacing, which was examined in plant cells, as well as in yeast (26, 38, 39, 54).
The meta-analysis detects a genome-wide association, but does not inform about individual genes. We therefore tested how predictive the presence of the motifs in gene promoters is of their auxin-dependence. We performed quantitative RT-PCR (qRT-PCR) expression analysis of genes harboring these elements in their upstream regions. In the Arabidopsis genome. We found 297 and 701 genes to contain (at least) one IR8 and DR5 element in their (−1,500-bp including 5′UTR) regulatory region, respectively. Among these genes, we randomly selected 21 genes containing IR8 and 18 genes that harbor DR5 and examined if and how each of these genes responds to exogenous auxin treatment in a time course on 5-d-old seedlings that includes both an early (15 min) and a late (6 h) time point. The selected genes are spread across different chromosomes, the IR8 and DR5 elements localize at different distances from the transcription start site (SI Appendix, Fig. S6), and the genes encode a range of activities such as signaling, transcription, and metabolism, which should minimize any bias (Datasets S1 and S2).
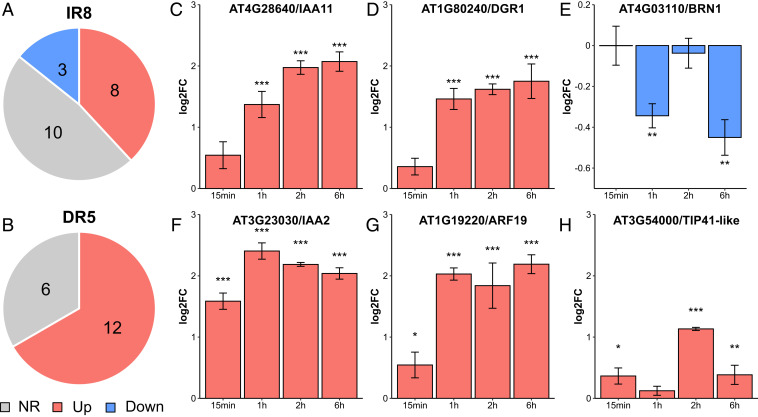
Genes were considered auxin-responsive in the time series under two-way ANOVA for “auxin treatment” factor (P < 0.05) and one-way ANOVA with post-hoc Tukey test for time points (P < 0.05). Out of the 21 IR8-containing genes, 11 changed their expression level in response to auxin: the transcript levels of 10 (e.g., IAA11, DGR1) increased, while of 3 (e.g., BRN1) decreased within 6 h of treatment with 1 µM IAA (Fig. 4 A and C–E and Dataset S1). On the other hand, the levels of 12 out of the 18 DR5-containing genes (e.g., IAA2, ARF19, TIP41-like) showed up-regulation in auxin-treated plants, and no genes displayed a significant decrease. This trend is consistent with the upregulation–specific overrepresentation detected by the meta-analysis (Fig. 4 B and F–H and Dataset S2).
Fig. 4.
IR8 and DR5 repeats accurately predict auxin response. (A and B) The proportion of genes that showed up-regulation (red) and down-regulation (blue) by qRT-PCR within 6 h of 2,4-D treatment among genes containing an IR8- (A) or DR5-element (B) in their upstream region. Auxin responsiveness was estimated by both two-way ANOVA for a time series (P < 0.05) with one-way ANOVA with Tukey post hoc test for a time point (P < 0.05). NR, not responsive to auxin. (C–H) Time courses of the auxin response of three representative genes downstream of IR8-element: AT4G28640/IAA11, AT1G80240/DGR1, and AT4G03110/BRN1 (C–E), and three downstream of DR5-element: AT3G23030/IAA2, AT1G19220/ARF19, and AT3G54000/TIP41-like (F–H). The ratio of expression levels in auxin-treated seedlings to that in mock sample at four time points: 15 min, 1 h, 2 h, 6 h are shown as mean ± SD. Asterisks indicate significant differences by one-way ANOVA with Tukey post hoc test (*<0.05, **<0.01, ***<0.001).
Given that the genes were randomly selected from among all IR8- or DR5-containing ones, and auxin response was tested only at a single stage of seedling development, we conclude that the presence of these elements predicts auxin responsiveness with more than 50% accuracy. This finding supports a central role for IR8 and DR5 elements in auxin-dependent gene regulation across a wide range of primary auxin-responsive genes.
Biological Relevance of the IR8 Element.
Much of the analysis of ARF-DNA binding and gene regulation, including the identification of ARF1, has been based on IR7 elements (25, 34, 39). Indeed, our analysis underlines the biological significance of the IR7 motif in TMO5 promoter (Fig. 2). However, meta-analysis did not identify IR7, but rather a repeat spaced by eight bases (IR8), as being strongly associated with auxin responses. To determine the in vivo relevance of this motif, and cooperativity of its half-sites, we chose the IAA11 gene given its substantial degree of up-regulation by auxin (Fig. 4C).
The IR8-AuxRE-like sequence (Fig. 5 A and B) in the 5′-upstream region of Arabidopsis IAA11 is contained within a region that is highly conserved among land plants [63 species of 7 lineages from ref. (47)]. Furthermore, this IR8 element is located in a region where peaks are detected in ARF5 and ARF2 DAP-seq, and also appears to be a DNase I-hypersensitive site, suggesting its accessibility to the transcriptional machinery (Fig. 5A).
Fig. 5.
An IR8 element in the IAA11 promoter functions as a composite AuxRE. (A) (Top) Positions of AuxRE-like motifs across the IAA11 promoter. The position of the two AuxRE-like motifs in an inverted repeat constellation with 7-bp spacing are indicated in red. (Bottom) Rows show AtARF5 DAP-seq peaks, DHSs, and sequence conservation among IAA11 homologs in 63 angiosperm species. (B) Sequence of the WT and mutant IR8 element in the IAA11 promoter (C) Boxplot compares the intensity levels from n3GFP driven by pIAA11WT (red), pIAA11∆1 (green), and pIAA11∆2 (blue) in vascular cells in the transition zone of primary roots grown without exogenous auxin (control condition). Each dot indicates the mean intensity over an area of same size measured for an individual root, and data were collected from multiple individual transgenics. (D) Auxin response of the WT and mutated IAA11 promoters based on the signals from n3GFP coupled to the promoters. The boxplot indicates the ratio between the signal levels in vascular cells in the elongation zone of roots treated with auxin for 6 h and that of roots under control condition. Each dot indicates the ratio estimated for individual root relative to the normalized control level for specific line. Asterisks in C and D indicate statistically significant differences (P < 0.001 by one-way ANOVA with post hoc Tukey test). n.s: not significant.
By observing lines in which nuclear-localized triple GFP (n3GFP) was driven from WT or mutated IAA11 promoters, we evaluated the effect of base substitutions in one or both of the half-sites composing the IR8 element (pIAA11∆1; pIAA11∆2 respectively; Fig. 5B). All reporter lines showed fluorescent signals in the vascular bundle within the transition zone of primary roots grown on hormone-free media, and the signal enhanced significantly (one-way ANOVA with post hoc Tukey test, P < 1 × 10−4) in the lines with mutations in one or both half-sites (Fig. 5C and SI Appendix, Fig. S7). This finding suggests that IR8 plays an important role in repressing IAA11 gene expression in the absence of external auxin.
Consistent with qPCR results, auxin treatment enhanced the intensity of the pIAA11WT::n3GFP signal (Fig. 5 C and D). Compared to pIAA11WT reporter lines, the intensity of GFP signal upon auxin treatment significantly weakened in both the pIAA11∆1::n3GFP and pIAA11∆2::n3GFP lines (Fig. 5D and SI Appendix, Fig. S6). This reduction in auxin responsiveness by mutation indicates that the IR8 element indeed functions as an AuxRE controlling the auxin-dependent induction of IAA11, and hence, is probably targeted by ARFs. Furthermore, the synergism of the adjacent TGTCNN motifs, which are consistent with interaction with dimerized/oligomerized ARFs, is supported by the observation that introducing the mutations at single half-site attenuated the degree of auxin response as much as the deletion of the two motifs did.
Differential ARF-DNA Affinities Underlie Distinct Repeat Activity.
Based on the meta-analysis and the measurements of gene transcripts, IR8 elements can mediate both auxin-dependent activation and repression. In contrast, DR5 elements seem specific to up-regulation, and indeed the widely used and highly auxin-responsive synthetic DR5 promoter contains 7–9 AuxREs spaced by five bases, a concatenation of DR5 elements (26, 38). Different interactions with ARF transcription factors presumably underlie the functional differences between IR8 and DR5 elements.
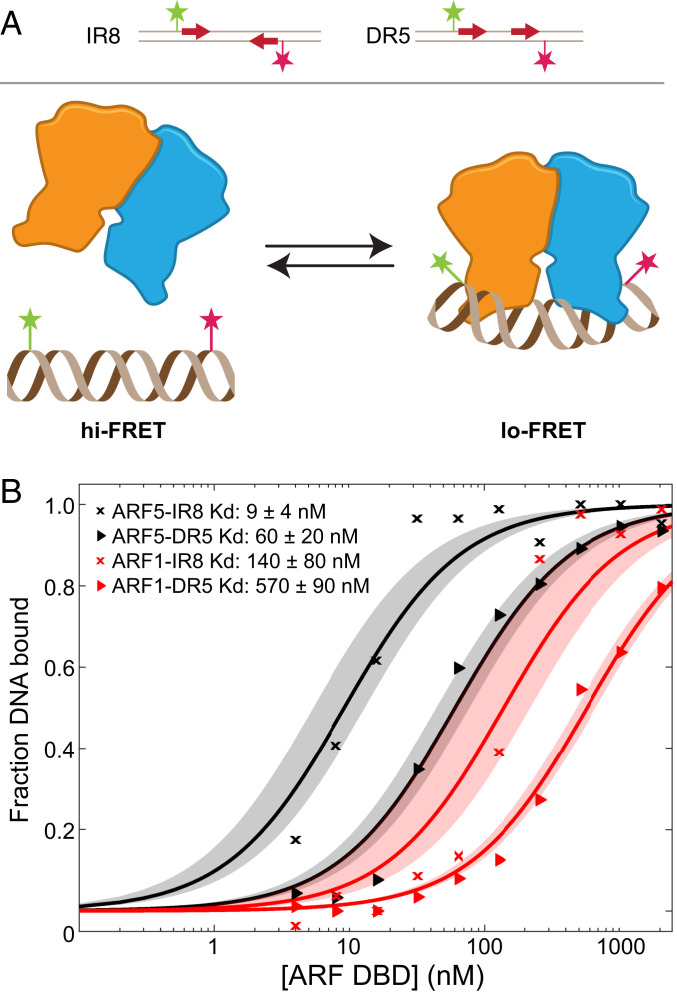
ARFs are phylogenetically divided into three major groups––class A, B, and C (22, 55). ARFs differ most prominently in the amino acid enrichment within their middle region, where class A ARFs are considered transcriptional activators and class B and C ARFs are thought to function as transcriptional repressors (9, 56). In the liverwort Marchantia polymorpha, A- and B-class ARFs appear to compete for binding sites, and the stoichiometry of activating A- and B-class ARF determines gene activity (56). To explore if differential interactions of DR5 and IR8 elements with ARF classes may underlie their differential activity, we quantitatively compared interactions of the DBDs of Arabidopsis ARF5 (A class) and ARF1 (B class) with IR8 and DR5 elements using a single-molecule Förster resonance energy transfer (FRET) assay (56). In this assay (Fig. 6A), the efficiency of energy transfer between two fluorescent dyes on an immobilized double-stranded oligonucleotide with ARF binding sites is modulated by ARF binding. As the titration of ARF proteins changes the fraction of occupied DNA molecules, a binding constant (Kd) can be derived. In this assay, AtARF5 displayed approximately sevenfold higher affinity to IR8 (9 nM) than to DR5 (60 nM; Fig. 6B and SI Appendix, Fig. S8), supporting that ARFs increase avidity through head-to-head dimerization of DBD upon binding to IR8 AuxREs. A similar trend, but with substantially weaker binding, was observed for AtARF1 (Fig. 6B). AtARF1 bound to IR8 with a Kd of 140 nM, a value 16-fold lower than the Kd derived from ARF5, and to DR5 with a Kd of 570 nM, a value 10-fold weaker (Fig. 6B).
Fig. 6.
Single-molecule FRET assays reveal differential ARF-DNA affinities. (A) Cartoon describing the single-molecule FRET assay. IR8- or DR5-containing (arrows) ds oligonucleotide carrying two FRET-compatible dyes (magenta and green stars) are immobilized on a coverslip. Arrows indicate position of TGTCGG sequences. Binding of ARF proteins (dimer indicated in orange/blue) to the oligo leads to bending of DNA bending and slight displacement of the dyes thereby decreasing FRET efficiency. (B) Titrations of ARF1-DBD and ARF5-DBD proteins (concentration in nM on x-axis) on surface-immobilized DR5 or IR8 oligonucleotides. The y-axis shows the fraction of DNA bound to protein derived from FRET efficiency distribution. Dissociation constants (Kd) are given in the legend, with their 95% intervals of confidence from the fit.
Thus, both ARF5 and ARF1 bind to IR8 with higher affinity than to DR5; furthermore, for both repeats, DNA binding affinity of ARF5 is higher than that of ARF1. Consequently, interaction of ARF1 with DR5 is far weaker compared to the other ARF-AuxRE combinations. While it should be noted that ARF1 and ARF5 may not represent the typical behavior of all class B- and A-ARFs, these results are in agreement with the notion that B-class ARFs in Arabidopsis may have too little interaction with DR5 composite elements to effectively regulate the expression level of genes, while IR8-AuxREs are likely to be accessible both to activators and repressors in the nucleus.
Discussion
How auxin regulates the transcription of specific gene sets leading to diverse, local cellular responses is a largely unanswered, yet fundamental question in plant biology.
Given that ARF transcription factors mediate most, if not all, of these diverse auxin responses, their interactions with DNA must contribute to the diversity of auxin responses.
Detailed analysis of promotors of auxin-regulated genes identified a minimal AuxRE element (23, 26, 57). Recent analysis showed that variation in the TGTCNN AuxRE contributes substantially to quantitative differences in the activity of the motif (38, 43). Furthermore, AuxREs can occur in repeats, and the first ARF protein was indeed identified based on its binding to an inverted AuxRE repeat (25). Structural analysis showed the biochemical basis for cooperative, high-affinity binding of ARF dimers to AuxRE repeats (34). Importantly, the binding affinity of two ARFs differed substantially depending on spacing between the inverted AuxRE repeats, which led to the formulation of a caliper model that helps ARFs discriminate binding sites. Importantly however, there are many open questions related to the requirements of AuxREs and their higher-order organization for ARF binding and auxin-dependent gene regulation in vivo. Here, we have addressed several outstanding questions.
Firstly, we provide a molecular basis for the sequence adaptability of ARFs by solving the high-resolution structure of ARF1-DBD complexed to a high-affinity AuxRE. When bound to TGTCGG, we find that His136 enters the major groove increasing the hydrophobic surface to interact with the DNA bases and allowing for subsidiary hydrogen bonding of the H136-G137 region with either guanine, thus increasing binding affinity. Based on homology modeling (22) and a crystal structure of the M. polymorpha ARF2-DBD in complex with DNA (56), all ARFs likely share a tertiary structure topology in their DBD, and share most residues in the DNA-contacting surface (22). Strikingly, the histidine residue corresponding to the His136 in AtARF1 is conserved among A- and B-class ARFs in land plants, suggesting the same adaptability of all these ARFs to AuxREs of varying affinities. The structural evidence presented here rationalizes the increased activity of the TGTCGG element in the DR5v2 promoter (38), and demonstrates how highly localized changes in hydrogen-bonding patterns and repositioning of structural elements can condition high-affinity ARF-DNA binding. It will be interesting to see how adaptive the protein–DNA interface is, and how it can accommodate the conserved variants at the NN positions (43).
Secondly, we tested the in vivo significance of the AuxRE repeats in the Arabidopsis IAA11 (IR8) and TMO5 (IR7) promoters. For both, we found that expression from the promoters is reduced by far more than 50% if a single AuxRE is mutated. This is perfectly consistent with the sites acting cooperatively, rather than redundantly or additively, and mirrors the earlier observation that an ARF5 protein whose DBD cannot dimerize, fails to complement the arf5/mp mutant (34). It should be noted that the IR7 AuxRE motif in both promoters lies in a well-conserved region. Apparently, despite the presence of numerous other AuxRE sites in each promoter, the IR7/8 motifs are the biologically relevant ones. Thus, as proposed based on in vitro data (25, 34, 42), cooperativity of two adjacent and spaced AuxRE elements underlies ARF binding and biological activity in at least these two promoters, which renders ARF dimers as the likely biologically active species.
Genome-wide association of AuxRE repeat motifs with auxin-dependent gene regulation strongly suggest the association of IR8 and DR5 motifs in gene promoters with transcriptional auxin response, supporting the insights gained over two decades by assays with synthetic model DNA sequences (25, 26, 34, 36). As a case in point, our validation of DR5 and IR8 associations using qPCR on randomly selected gene suggests that one can actually predict relevant transcription-factor binding sites and modes of DNA–protein interaction for hormone responses by integrating multiple transcriptomic and genomic datasets. Such associations could not have been inferred from individual transcriptome experiments, but follow from meta-analysis. Also, the positive validation rate (>50%) is very high, especially considering that transcriptomes used in the metaanalysis were derived from multiple genotypes, growth stages, and auxin treatment regimes, while validation was performed on a single genotype, growth stage, and auxin treatment regime. We expect that the presence of an IR8 or DR5 element in a gene upstream region will likely be a high-confidence predictor of auxin regulation. Considering that there are many more auxin-responsive genes (SI Appendix, Table S3) than genes with DR5 and IR8 repeats (998 in total), other AuxRE arrangements as well as single AuxREs might be sufficient to mediate transcriptional auxin response as well. Here, we detected the single AuxRE TGTCGG as being enriched in upstream regions of auxin-responsive genes, but with lower significance than the IR8 or DR5 repeat elements. It is possible that such single AuxREs are in fact accompanied by minimal adjacent AuxREs in which the critical residues are present, but that do not contain the TGTC core. Alternatively, it is possible that for such genes, ARFs indeed bind a single AuxRE. This was recently shown to be the case for AtARF19 in a yeast reconstitution assay (54). Especially because for some enriched motifs, such as IR2 (Fig. 3D), it would be difficult to envision protein–protein interaction among two interacting ARFs, there still remains an open question what the mechanistic basis for these enrichments is.
It should be noted that, in contrast to the predictions from ARF biochemical analysis in vitro (34), no significant association was found between inverted AuxRE repeats with spacing with more or fewer than eight base pairs, and auxin response. It could therefore be that the “caliper” model does not apply in vivo. However, the genome-wide association can only identify the major elements that generically associate with auxin regulation. Any motifs that drive auxin-dependent expression of a subset of genes, or in a specific context, will be missed in this analysis. Whether ARFs can “measure” spacing length in vivo therefore remains an open question.
Metatranscriptome analysis, followed by qPCR validation, illuminated functional differences between AuxRE repeats. While IR8 seems associated both with auxin-dependent up- and down-regulation, DR5 appears specific to up-regulated genes. Indeed, the pattern of the DR5-GFP reporter in roots can be derived from computational models that do not invoke any auxin-dependent gene repression (58). The three ARF classes have distinct activities, where A-class ARFs mostly seem to activate transcription and B/C-class ARF repress (9, 22, 55, 59). We recently explored the simple M. polymorpha auxin response system to derive a simple model in which A- and B-class ARFs compete for the same binding sites, and C-class ARFs are not part of the auxin response system. A-class ARFs act as repressors in the absence of auxin, and switch to activation upon auxin perception. B-class ARFs are nor auxin sensitive, but counterbalance A-ARF activity. Stoichiometry of A- and B-class ARFs thus defined auxin responsiveness (56). In this model, differential affinities of a DNA element to A- and B-class ARFs could explain association with up- or down-regulation. We found that the affinity of the A-class ARF5 to both IR8 and DR5 is higher than that of the B-class ARF1 to the same elements. Thus, upregulation-specific association of DR5 can be explained by a trade-off between affinity and specificity. In analogy, there is an inverse correlation between affinity and specificity of DNA sequences for Hox-Extradenticle complexes in SELEX-seq data. The Drosophila svb enhancer enables a precise expression pattern by exploiting a cluster of low-affinity binding sites that confer specificity for Ubx and AbdA over other Hox proteins (60, 61). It could likewise be that due to the fact that DR5 is an inherently poorer ARF binding site than IR8, coupled to the lower intrinsic DNA-binding affinity of B-class ARFs, there is no effective interaction of B-class ARFs to DR5 sites under physiological protein concentrations. Consequently, genomic DR5 elements may specifically interact with A-class ARFs, while IR8 elements, being better binding sites, can effectively recruit both A-and B-class ARFs. In addition to the protein-DNA binding affinity, the concentration of ARF proteins determines the degree of DNA interaction and the ability of B-class ARFs to compete with A-class ARFs. Systematic analysis of ARF gene expression showed that patterns in Arabidopsis are diverse, and there are certainly cells in which B-class ARFs are more abundant than A-class ARFs (62). It will be interesting to see how well these ARF accumulation patterns correlate with the sites of preferred expression of DR5- versus IR8-containting promoters. Given that only ARF5 and ARF1 were tested as representatives of the A- and B class, it is possible that there is diversity in DNA binding properties within the ARF subclasses, also with respect to the relative affinity to IR8 and DR5 elements. Obviously, a major question remains the biochemical and structural basis for eventual cooperative ARF binding to DR5 elements. Structural studies will have to reveal whether this is mediated by alternative dimerization interfaces.
Materials and Methods
A complete overview of the materials and methods used in this research can be found in SI Appendix. This includes information on crystallization, size-exclusion chromatography, bioinformatics, molecular cloning, plant growth, phenotypic and expression analysis, qPCR, and single-molecule FRET analysis.
Supplementary Material
Acknowledgments
This work was supported by Excellent Chemisch Onderzoek (ECHO; 711.011.002) and VICI (865.14.001) grants from the Netherlands Organization for Scientific research to D.W., by an Overseas Research Fellowship from the Japanese Society for the Promotion of Science to K.T., by the Ministry of Economy and Competitiveness of the Spanish Government (Grant BIO2016-77883-C2-2-P and Grant FIS2015-72574-EXP) (Agencia Estatal de Investigación; AEI/FEDER, EU) to D.R.B., a fellowship from the Graduate School Experimental Plant Sciences to J.H., the Russian Foundation for Basic Research (Grant RFBR 18-04-01130) to Y.S. and V.M.; Grant RFBR 18-29-13040 to V.L., and a Budget Project from the Russian Government (Project 0324-2019-0040-C-01) to V.M. and V.L.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2009554117/-/DCSupplemental.
Data Availability.
Protein structural coordinates are available at the Protein Data Bank (www.rcsb.org) under accession number 6YCQ. All other data are available in the main text or SI Appendix.
References
- 1.Woodward A. W., Bartel B., Auxin: Regulation, action, and interaction. Ann. Bot. 95, 707–735 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vanneste S., Friml J., Auxin: A trigger for change in plant development. Cell 136, 1005–1016 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Abel S., Theologis A., Odyssey of auxin. Cold Spring Harb. Perspect. Biol. 2, 1–14 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoshida S., Saiga S., Weijers D., Auxin regulation of embryonic root formation. Plant Cell Physiol. 54, 325–332 (2013). [DOI] [PubMed] [Google Scholar]
- 5.de Wit M., Lorrain S., Fankhauser C., Auxin-mediated plant architectural changes in response to shade and high temperature. Physiol. Plant. 151, 13–24 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Kato H., Nishihama R., Weijers D., Kohchi T., Evolution of nuclear auxin signaling: Lessons from genetic studies with basal land plants. J. Exp. Bot. 69, 291–301 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Guilfoyle T., Hagen G., Ulmasov T., Murfett J., How does auxin turn on genes? Plant Physiol. 118, 341–347 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guilfoyle T. J., Ulmasov T., Hagen G., The ARF family of transcription factors and their role in plant hormone-responsive transcription. Cell. Mol. Life Sci. 54, 619–627 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weijers D., Wagner D., Transcriptional responses to the auxin hormone. Annu. Rev. Plant Biol. 67, 539–574 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Long J. A., Ohno C., Smith Z. R., Meyerowitz E. M., TOPLESS regulates apical embryonic fate in Arabidopsis. Science 312, 1520–1523 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Szemenyei H., Hannon M., Long J. A., TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319, 1384–1386 (2008). [DOI] [PubMed] [Google Scholar]
- 12.Gray W. M., Kepinski S., Rouse D., Leyser O., Estelle M., Auxin regulates SCFTIR1-dependent degradation of AUX/IAA proteins. Nature 414, 271–276 (2001). [DOI] [PubMed] [Google Scholar]
- 13.Dharmasiri N., Dharmasiri S., Estelle M., The F-box protein TIR1 is an auxin receptor. Nature 435, 441–445 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Kepinski S., Leyser O., The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435, 446–451 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Tan X. et al., Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446, 640–645 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Calderón Villalobos L. I. A. et al., A combinatorial TIR1/AFB-Aux/IAA co-receptor system for differential sensing of auxin. Nat. Chem. Biol. 8, 477–485 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ballas N., Wong L. M., Theologis A., Identification of the auxin-responsive element, AuxRE, in the primary indoleacetic acid-inducible gene, PS-IAA4/5, of Pea (Pisum sativum). J. Mol. Biol. 233, 580–596 (1993). [DOI] [PubMed] [Google Scholar]
- 18.Guilfoyle T. J., Hagen G., Auxin response factors. Curr. Opin. Plant Biol. 10, 453–460 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Salehin M., Bagchi R., Estelle M., ScfTIR1/AFB-based auxin perception: Mechanism and role in plant growth and development. Plant Cell 27, 9–19 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hayashi K. I., The interaction and integration of auxin signaling components. Plant Cell Physiol. 53, 965–975 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Kato H. et al., Auxin-mediated transcriptional system with a minimal set of components is critical for morphogenesis through the life cycle in Marchantia polymorpha. PLoS Genet. 11, 1–26 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mutte S. K. et al., Origin and evolution of the nuclear auxin response system. eLife 7, e33399 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ulmasov T., Liu Z.-B., Hagen G., Guilfoyle T. J., Composite structure of auxin response elements. Plant Cell 7, 1611–1623 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hagen G., Guilfoyle T., Auxin-responsive gene expression: Genes, promoters and regulatory factors. Plant Mol. Biol. 49, 373–385 (2002). [PubMed] [Google Scholar]
- 25.Ulmasov T., Hagen G., Guilfoyle T. J., ARF1, a transcription factor that binds to auxin response elements. Science 276, 1865–1868 (1997). [DOI] [PubMed] [Google Scholar]
- 26.Ulmasov T., Murfett J., Hagen G., Guilfoyle T. J., Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9, 1963–1971 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sabatini S. et al., An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99, 463–472 (1999). [DOI] [PubMed] [Google Scholar]
- 28.Friml J. et al., Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426, 147–153 (2003). [DOI] [PubMed] [Google Scholar]
- 29.Gallavotti A., Yang Y., Schmidt R. J., Jackson D., The relationship between auxin transport and maize branching. Plant Physiol. 147, 1913–1923 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moreno-Risueno M. A. et al., Oscillating gene expression determines competence for periodic Arabidopsis root branching. Science 329, 1306–1311 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen Y., Yordanov Y. S., Ma C., Strauss S., Busov V. B., DR5 as a reporter system to study auxin response in Populus. Plant Cell Rep. 32, 453–463 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Yang J. et al., Dynamic regulation of auxin response during rice development revealed by newly established hormone biosensor markers. Front. Plant Sci. 8, 1–17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goldental-Cohen S., Israeli A., Ori N., Yasuor H., Auxin response dynamics during wild-type and entire flower development in tomato. Plant Cell Physiol. 58, 1661–1672 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Boer D. R. et al., Structural basis for DNA binding specificity by the auxin-dependent ARF transcription factors. Cell 156, 577–589 (2014). [DOI] [PubMed] [Google Scholar]
- 35.Franco-Zorrilla J. M. et al., DNA-binding specificities of plant transcription factors and their potential to define target genes. Proc. Natl. Acad. Sci. U.S.A. 111, 2367–2372 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O’Malley R. C. et al., Cistrome and epicistrome features shape the regulatory DNA landscape. Cell 165, 1280–1292 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zemlyanskaya E. V., Wiebe D. S., Omelyanchuk N. A., Levitsky V. G., Mironova V. V., Meta-analysis of transcriptome data identified TGTCNN motif variants associated with the response to plant hormone auxin in Arabidopsis thaliana L. J. Bioinform. Comput. Biol. 14, 1641009 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Liao C. Y. et al., Reporters for sensitive and quantitative measurement of auxin response. Nat. Methods 12, 207–210 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pierre-Jerome E., Moss B. L., Lanctot A., Hageman A., Nemhauser J. L., Functional analysis of molecular interactions in synthetic auxin response circuits. Proc. Natl. Acad. Sci. U.S.A. 113, 11354–11359 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamaguchi N. et al., A molecular framework for auxin-mediated initiation of flower primordia. Dev. Cell 24, 271–282 (2013). [DOI] [PubMed] [Google Scholar]
- 41.Konishi M., Donner T. J., Scarpella E., Yanagisawa S., MONOPTEROS directly activates the auxin-inducible promoter of the Dof5.8 transcription factor gene in Arabidopsis thaliana leaf provascular cells. J. Exp. Bot. 66, 283–291 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ulmasov T., Hagen G., Guilfoyle T. J., Dimerization and DNA binding of auxin response factors. Plant J. 19, 309–319 (1999). [DOI] [PubMed] [Google Scholar]
- 43.Lieberman-Lazarovich M., Yahav C., Israeli A., Efroni I., Deep conservation of cis-element variants regulating plant hormonal responses. Plant Cell 31, 2559–2572 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schlereth A. et al., MONOPTEROS controls embryonic root initiation by regulating a mobile transcription factor. Nature 464, 913–916 (2010). [DOI] [PubMed] [Google Scholar]
- 45.De Rybel B. et al., A bHLH complex controls embryonic vascular tissue establishment and indeterminate growth in Arabidopsis. Dev. Cell 24, 426–437 (2013). [DOI] [PubMed] [Google Scholar]
- 46.Lu K. J. et al., Evolution of vascular plants through redeployment of ancient developmental regulators. Proc. Natl. Acad. Sci. U.S.A. 117, 733–740 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tian F., Yang D. C., Meng Y. Q., Jin J., Gao G., PlantRegMap: Charting functional regulatory maps in plants. Nucleic Acids Res. 48, D1104–D1113 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.De Rybel B. et al., Plant development. Integration of growth and patterning during vascular tissue formation in Arabidopsis. Science 345, 1255215 (2014). [DOI] [PubMed] [Google Scholar]
- 49.Ohashi-Ito K. et al., A bHLH complex activates vascular cell division via cytokinin action in root apical meristem. Curr. Biol. 24, 2053–2058 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Stigliani A. et al., Capturing auxin response factors syntax using DNA binding models. Mol. Plant 12, 822–832 (2019). [DOI] [PubMed] [Google Scholar]
- 51.Mironova V. V., Omelyanchuk N. A., Wiebe D. S., Levitsky V. G., Computational analysis of auxin responsive elements in the Arabidopsis thaliana L. genome. BMC Genomics 15 (suppl. 12), S4 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goodman H. M., Ecker J. R., Dean C., The genome of Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 92, 10831–10835 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Novikova D. D., Cherenkov P. A., Sizentsova Y. G., Mironova V. V., metaRE R package for meta-analysis of transcriptome data to identify the cis-regulatory code behind the transcriptional reprogramming. Genes 11, 634 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lanctot A., Taylor-Teeples M., Oki E. A., Nemhauser J., Specificity in auxin responses is not explained by the promoter preferences of activator ARFs. Plant Physiol. 182, 1533–1536 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Finet C., Berne-Dedieu A., Scutt C. P., Marlétaz F., Evolution of the ARF gene family in land plants: Old domains, new tricks. Mol. Biol. Evol. 30, 45–56 (2013). [DOI] [PubMed] [Google Scholar]
- 56.Kato H. et al., Design principles of a minimal auxin response system. Nat. Plants 6, 473–482 (2020). [DOI] [PubMed] [Google Scholar]
- 57.Liu Z. B., Ulmasov T., Shi X., Hagen G., Guilfoyle T. J., Soybean GH3 promoter contains multiple auxin-inducible elements. Plant Cell 6, 645–657 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grieneisen V. A., Xu J., Marée A. F. M., Hogeweg P., Scheres B., Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature 449, 1008–1013 (2007). [DOI] [PubMed] [Google Scholar]
- 59.Ulmasov T., Hagen G., Guilfoyle T. J., Activation and repression of transcription by auxin-response factors. Proc. Natl. Acad. Sci. U.S.A. 96, 5844–5849 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Crocker J. et al., Low affinity binding site clusters confer HOX specificity and regulatory robustness. Cell 160, 191–203 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Crocker J., Preger-Ben Noon E., Stern D. L., “The soft touch: Low-affinity transcription factor binding sites in development and evolution” in Curr. Top. Dev. Biol., (2016), Vol. 117, pp. 455–469. [DOI] [PubMed] [Google Scholar]
- 62.Rademacher E. H. et al., A cellular expression map of the Arabidopsis AUXIN RESPONSE FACTOR gene family. Plant J. 68, 597–606 (2011). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Protein structural coordinates are available at the Protein Data Bank (www.rcsb.org) under accession number 6YCQ. All other data are available in the main text or SI Appendix.