Abstract
Background and Aims
Intestinal biopsy sampling during IBD trials represents a valuable adjunct strategy for understanding drug responses at the tissue level. Given the length and distinctive embryonic origins of the proximal and distal colon, we investigated whether inherent regional differences of immune cell composition could introduce confounders when sampling different disease stages, or pre/post drug administration. Here, we capitalise on novel mass cytometry technology to perform deep immunophenotyping of distinct healthy colonic segments, using the limited numbers of biopsies that can be harvested from patients.
Methods
Biopsies [2.8 mm] were collected from the caecum, transverse colon, descending colon, and rectum of normal volunteers. Intestinal leukocytes were isolated, stained with a panel of 37 antibodies, and mass cytometry data acquired.
Results
Site-specific patterns of leukocyte localisation were observed. The proximal colon featured increased CD8+ T cells [particularly resident memory], monocytes, and CD19+ B cells. Conversely, the distal colon and rectum tissues exhibited enrichment for CD4+ T cells and antibody-secreting cells. The transverse colon displayed increased abundance of both γδ T cells and NK cells. Subsets of leukocyte lineages also displayed gradients of expression along the colon length.
Conclusions
Our results show an inherent regional immune cell variation within colonic segments, indicating that regional mucosal signatures must be considered when assessing disease stages or the prospective effects of trial drugs on leukocyte subsets. Precise protocols for intestinal sampling must be implemented to allow for the proper interpretation of potential differences observed within leukocyte lineages present in the colonic lamina propria.
Keywords: mass cytometry, lamina propria, randomised controlled trials
Introduction
The inflammatory bowel diseases, Crohn’s disease [CD] and ulcerative colitis [UC], are a group of complex and diverse intestinal conditions, which develop due to a combination of genetic, immune, environmental, and microbial factors, recently described as the interactome.1 Their incidence, in the Western and developing world, is on the rise.2 A number of novel therapeutics have been developed over recent decades, targeting multiple components of the immune response, including proinflammatory cytokines, signalling molecules, and adhesion molecules that allow for cell traffic to the intestine [e.g., vedolizumab, ozanimod].3–5
A major hurdle for the clinical testing of novel IBD therapies has been the reliance on imprecise, subjective clinical endpoints, such as the Crohn’s Disease Activity Index [CDAI], despite the recent inclusion of blinded endoscopic and histological assessments.6–8 The holy grail of efficacy readouts for therapeutics that target components of the immune response are predictive biomarkers, whether peripheral or tissue-specific. The ideal biomarker should specifically predict whether a novel therapeutic is having the desired effect, and aid in identifying patients who may or may not respond to a specific therapy in the intended manner. The emergence of novel, high-parameter techniques, such as mass cytometry and transcriptomics, has allowed for robust characterisation of the immune response during IBD, using minimal tissue amounts that can be obtained from intestinal biopsies or peripheral blood.9–13
This revolution in scientific methodology represents a valuable tool for the clinical assessment and mechanistic understanding of ongoing and future IBD therapies. Despite this, the complexity and variability of the composition of the human intestinal mucosa represents a huge hurdle for the application of tissue-based biological readouts for treatment efficacy or mode of action studies.14 Sampling methods vary among clinicians, with no clear consensus on an ideal biopsy sampling methodology for IBD studies.10,15 Alongside this fact is that the knowledge of the immune composition of the human intestine remains limited, with much insight based on studies in mice. Examination of the immune composition of the human intestine would be beneficial, by identifying not only the steady state conditions that are perturbed in IBD, but also drug responses at the tissue level. A robust biopsy sampling methodology that can be uniformly implemented during future IBD studies, to measure disease pathogenesis, drug mode of action, and therapeutic efficacy, is needed.14
Ulcerative colitis is restricted to the large bowel, and Crohn’s disease may also present in the colon in a substantial proportion of patients.2 The colon itself can be divided into multiple segments; the ascending, transverse, descending, and sigmoid colon, followed by the rectum, all of which can be described by fundamental differences in intra-abdominal location, embryology, and genetic, and immune factors. The embryological development of the colon occurs in two distinct stages; the mid-gut develops first, from which the ascending colon, the proximal half of the transverse, the small intestine and caecum/appendix are derived. The hind-gut derivatives include the distal half of the transverse, as well as the descending and sigmoid colon, and the rectum.16 It has been known for decades that there is segmental cell heterogeneity, as antimicrobial-producing Paneth cells are observed in the proximal but not the distal colon of healthy individuals.17 Furthermore, the vascularisation and lymphatic drainage of the colon highlights this division; blood supply to the proximal colon depends on the superior mesenteric artery, whereas the distal part of the transverse colon onwards is supplied by the inferior mesenteric artery.18 This observed heterogeneity results in significant alterations between the two major embryologically distinct segments in terms of numbers and phenotype of antibody-secreting cells.18 Other studies have revealed large variations in gene expression between the ascending and descending colon in adults,19 and dendritic cell subsets in the ascending colon have been reported to display a more mature phenotype as compared with their distal counterparts.20 Further studies of DNA methylation,21 bacterial abundance, composition and activity,22,23 and the immune composition of the murine colon24,25 further highlight the biological, functional, and immunological differences between segments of the human colon.
Given the multiple factors that vary between the different colonic segments, it is logical to predict that the immune composition of each segment may also vary substantially. However, to our knowledge this have never been previously addressed systematically, in part due to technical limitations related to obtaining enough material from a patient, and the challenges related to developing an inclusive antibody panel for standard flow cytometry, given spectral overlap issues.26 With the potential for significant immune population variation along the colonic mucosa, we attempted to characterise the immune composition of distinct colonic sites, with the aim of examining whether the biopsy sampling site could introduce confounders for mechanism of action studies or clinical trials. We used mass cytometry and a panel of 37 common immune markers to identify most of diverse immune cell lineages within the colon. These analyses highlighted the diversity of each colonic segment across a range of immune lineages, with potential implications for study design and the need for highly defined standardisation of intestinal biopsy sampling during randomised controlled trials or mechanism of action studies.
Methods
Human samples
Adult patients [ages 30–70 years] without history of gastrointestinal disease, undergoing ileocolonoscopy for evaluation of abdominal symptoms or colorectal cancer screening, were recruited after written informed consent, as per SDVAMC and UCSD Institutional Review Board approved protocols [H130266 and 161756].9 Six standard biopsies were obtained from four different colonic sites; caecal base, transverse colon [70 cm from anal verge], descending colon [40 cm from anal verge], and rectum using standard [2.8 mm] intestinal biopsy forceps, deposited in 50-mL conical tubes containing 15 mL Hanks Balanced Salt Solution [ThermoFisher Scientific, San Diego, CA].
Isolation of immune cells from intestinal biopsy samples
All biopsies were frozen within 2 h of harvest, as per our own and others’ previously published protocols.9,12 In brief: biopsies were transferred to cryovials containing freezing buffer [50% complete RPMI, 40% FBS, 10% DMSO] and slow-frozen in a Mr Frosty freezing container [Sigma-Aldrich]. For cell isolation, biopsies were thawed rapidly at 37°C and washed immediately in 10 ml complete RPMI (RPMI supplemented with 10% fetal bovine serum [FBS], 50 mg/mL penicillin and streptomycin, and 2 mM sodium pyruvate [all ThermoFisher Scientific, San Diego, CA]). Biopsies were subsequently transferred to weighing boats containing 0.5 ml complete RPMI and finely minced using surgical scissors. Minced tissue was next incubated in a digestion solution, comprising complete RPMI with 1.5 mg/mL collagenase VIII [Sigma Aldrich, Carlsbad, CA] and 50 µg/mL DNase I [Roche, Pleasanton, CA], and incubated for 20 min at 37°C on a rocker. Following digestion, remaining tissue debris was removed using a 70-µm cell strainer. Cell suspensions were washed with complete RPMI and labelled with mass cytometry antibodies.
Antibody staining
Cell suspensions were prepared for mass cytometry acquisition as previously described,9 but with additional barcoding steps. In brief: isolated cells were resuspended in Maxpar cell staining buffer [CSB; Fluidigm, San Francisco, CA] and subsequently stained as follows:
viability staining using Cisplatin-195Pt [Fluidigm] for 5 min;
initial fixation step using Maxpar Fix I buffer [Fluidigm] for 20 min;
permeabilization and barcode labelling of each sample using the Cell-ID 20-Plex Pd Barcoding Kit [Fluidigm]. Following barcoding, cells from up to 20 different samples were combined into a single tube for further antibody labelling;
FC receptor blockade using Human TruStain FcX [BioLegend, San Diego, CA] for 10 min;
surface antibody staining [Fluidigm; BioLegend, San Diego, CA, full antibody list Supplementary Table 1, available as Supplementary data at ECCO-JCC online] for 30 min;
second fixation step using 1.6% formaldehyde [methanol-free, Thermo Scientific] for 10 min; and
DNA-Intercalator labelling using Maxpar Fix & Perm Buffer [Fluidigm] and Cell-ID Intercalator-Ir [Fluidigm], incubated at 4°C overnight.
Following overnight intercalator staining, samples were washed twice with cell staining buffer and stored at -80°C for 2 weeks, in a suspension comprising 90% FBS, 10% DMSO.9,27 Where indicated, purified antibodies were conjugated with metal isotopes in house, using antibody labelling kits [Fluidigm]. As barcoding reagents are limited to 20 samples per barcode set, two identical barcode sets were used for mass cytometry sample staining. Equal numbers of samples from each sample group [n = 5 caecum/transverse colon/descending colon/rectum] were combined together into each barcode set, to ensure comparable data acquisition and to minimise batch effects in the downstream data analysis. Supplementary Figure 1 [available as Supplementary data at ECCO-JCC online] displays staining intensity of several subset markers across the two barcode sets.
Sample acquisition and data processing
Before acquisition, cells were washed twice with Milli-Q water and resuspended in a 1:10 dilution of EQ Four Element Calibration Beads [Fluidigm] to a concentration of 0.5 × 106 cells/mL. Samples were acquired using a CyTOF Helios [Fluidigm], according to the manufacturer’s directions. Data were normalised to mass bead signal using the Nolan lab Matlab software28 [Github: https://github.com/nolanlab]. Barcode sets 1 and 2 were acquired on sequential days, and each set was acquired within 1 day of run time.
Data analysis
Following normalisation, barcoded samples were de-barcoded using the Nolan lab single-cell de-barcoder tool [Github; https://github.com/nolanlab]. Mass cytometry data were analysed using a number of online analysis platforms: Cytobank [Cytobank Inc., Santa Clara, CA] for biaxial gating and t-SNE [vi-SNE] analysis29; OMIQ [Omiq, Inc.] for biaxial gating and opt-SNE30 analysis31; and Astrolabe [Astrolabe Diagnostics, Inc., NJ, USA] for automated cell subset determination and quantification, as previously described.32 Before t-distributed Stochastic Neighbor Embedding [t-SNE] analysis, mass cytometry data were gated on nucleated, live, CD45+ events, then gated on the indicated populations of interest. Unless otherwise stated, vi-SNE and opt-SNE analyses were conducted using 100 000 total events proportionally drawn from samples, with 1000 iterations and a perplexity value of 30. Heatmaps were generated using the OMIQ heatmap algorithm. Summary graphs were produced using GraphPad Prism version 8 software [GraphPad Software Inc., La Jolla, California]. t-SNE plot lineage overlays were produced using Inkscape software. t-SNE and downstream analyses were performed using the markers shown in Supplementary Table 1.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 7 software [GraphPad Software Inc.]. Column statistics tests were used to assess parametric distribution of datasets. For comparison of two groups, the Wilcoxon matched-pairs signed rank test was used, or the Mann-Whitney test for unpaired samples. For comparison of multiple groups, analysis of variance [ANOVA] was utilised for parametric datasets, followed by Tukey’s multiple comparison test, and the Kruskal-Wallis test was used for non-parametric datasets, followed by the Dunn’s multiple comparison test. Descriptive statistics are displayed as mean ± standard deviation in all figures. Significance is defined as p-values of <0.05, and resulting statistical significances of difference are indicated in figures as p <0.05, p <0.01, p <0.001, p <0.0001.
Results
The immune cell composition differs between colonic segments
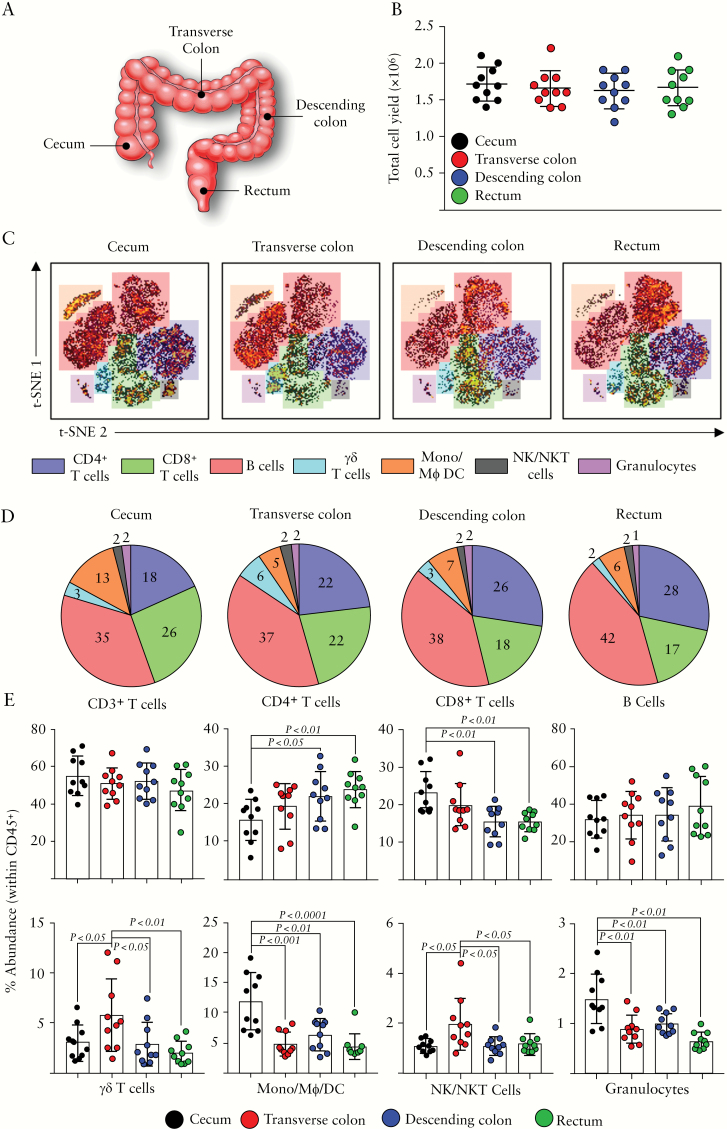
We assessed the variability of immune cell composition of the colon by examining the representation of the major immune lineages within each colonic segment. The four colonic segments examined included the caecum, transverse colon, descending colon, and rectum [Figure 1A]. Cellular yields were consistent between segments, with an average of 1.8 million cells obtained from each set of six colonic biopsies [Figure 1B]. t-SNE analysis was conducted on nucleated, live, CD45+ immune cell events from the four colonic segments, and the phenotype of cell clusters was determined using expression of markers denoting the main immune lineages [Figure 1C]. Discrete populations belonging to the following lineages were observed on the t-SNE map; CD4+ T cells, CD8+ T cells, B cells, γδ T cells, monocytes/macrophages/dendritic cells [Mono/Mϕ/DC], NK/NKT cells, and granulocytes, similar to those previously described.9 Average representation of each lineage observed [Figure 1D] and individual donor expression [Figure 1E] provide an insight into how these populations are represented within each colonic segment examined. Whereas there was no significant difference between the segments regarding overall CD3+ T cell representation, both CD4+ and CD8+ T cell subsets displayed marked differences throughout the colonic mucosa. CD4+ T cells were least abundant in the caecum, increasing in abundance towards the distal colon, whereas CD8+ T cells followed the opposite pattern, being more densely represented in the proximal colon. There appeared to be minimal variation in the abundance of the B cell lineage throughout the colon. The transverse colon featured heightened representation of both the γδ T cell and NK/NKT cell lineages, as compared with the remainder of the colonic mucosa. Finally, the caecum contained larger populations of cells which fell into the categories of monocyte/macrophage/dendritic cells, and granulocytes. Overall, the dominant populations throughout the colon were lymphocytes of the T and B cell lineages; the proximal colon appears enriched with CD8+ T cells and monocytic cells, the transverse colon with γδ T cells and NK cells, and the distal colon with CD4+ T cells.
Figure 1.
Representation of the major immune lineages along the colonic tract. [A] Diagram displaying the colonic sites of biopsy sampling for this study. [B] Total cellular yields derived from six pinch biopsies, taken from the indicated sites. [C] vi-SNE analysis of CD45+ immune cells derived from the indicated intestinal sites and acquired by mass cytometry. Colour overlays depict representation of the immune lineages described. [D] Representation of each immune lineage identified in [C] across all four sampled intestinal sites. Values indicate mean percentage abundance of immune lineages identified. [E] Percentage abundances of the indicated lineages across each intestinal site, determined by biaxial gating. Each data point represents a single donor, n = 10 total. Error bars represent standard deviation of mean. Statistical significance determined using analysis of variance [ANOVA], followed by Tukey’s multiple comparison test (for colour figure refer online version).
CD8+ T cells along the colonic tract
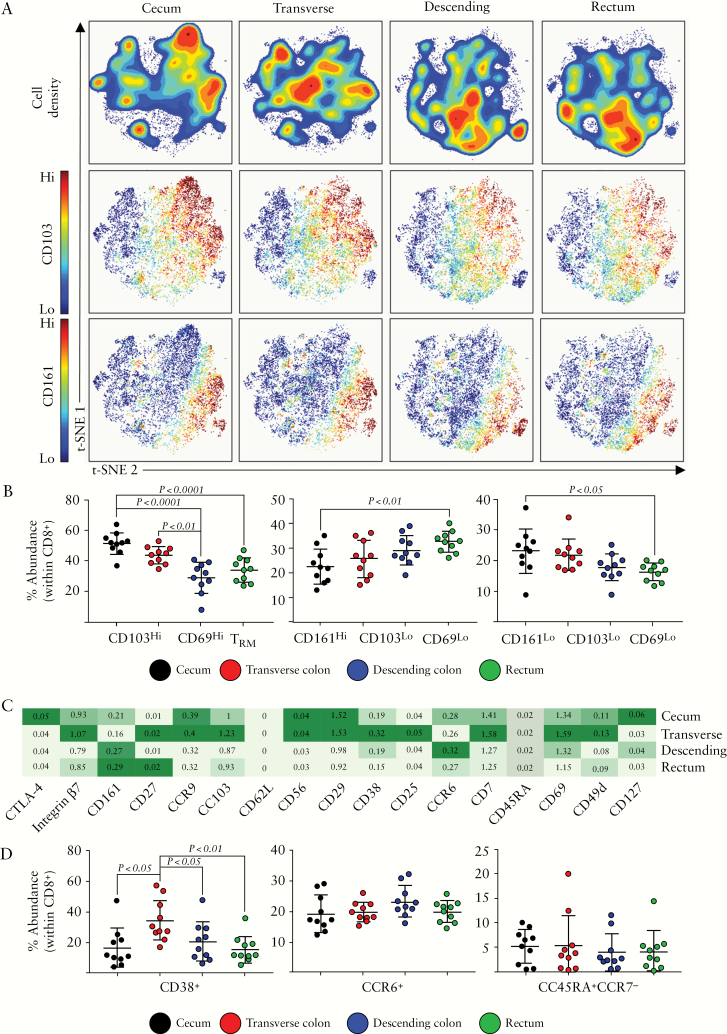
The abundance of CD8+ T cells in the colon was highest proximally, decreasing towards the rectum [Figure 1]. The dominant population of CD8+ T cells in the human intestine are tissue-resident memory cells [TRM] which form part of the intraepithelial cell compartment.33 These cells can be defined by an expression of CD103 and CD69, and represent a long-lived T cell population which provides a first response to infections at the mucosal surface.34 Indeed, we observed a prominent population of CD8+CD103+ cells [Figure 2A], which also co-expressed CD69 and integrin β7 [data not shown]. TRM were most abundant in the caecum, and displayed decreased abundance towards the distal colon and rectum [Figure 2B]. This abundance correlated with increased abundances of CD8+ T cells in general in the proximal colon. CD8+ T cells could also be sub-divided based on their expression of CD161 [Figure 2A], the C-type lectin-like receptor. We observed a population of CD8+CD161+ cells which expressed low levels of the TRM markers CD103 and CD69, described previously as a highly functional memory CD8+ T cell subset within the intestinal lamina propria,35 but may also include the mucosal-associated invariant T [MAIT] cell population as well.36 This population displayed an increased abundance towards the distal colonic segments [Figure 2B]. Subsequently, the remaining CD8+ T cells which expressed neither TRM markers nor CD161 displayed a moderately increased abundance in the caecum as compared with rectal tissue.
Figure 2.
CD8+ T cell subsets along the colonic tract. [A] t-SNE analysis of CD8+ cellular events isolated from intestinal biopsies derived from the caecum, transverse, or descending colon, or rectum. Density plots are displayed, alongside plots displaying CD103 and CD161 expression, generated using the OMIQ opt-SNE algorithm. [B] Proportions of the indicated CD8+ cellular subsets within each colonic compartment, determined using the Astrolabe profiling step,32 by limiting clusters to CD8+ events. [C] A heatmap displaying the relative expression of the cellular markers indicated, within the CD8+ T cell population, across the four colonic compartments. [D] Proportions of the indicated CD8+ T cell subsets within each colonic segment. Each data point represents a single donor, n = 10 total. Error bars represent standard deviation of mean. Statistical significance determined using analysis of variance [ANOVA], followed by Tukey’s multiple comparison test.
Further delineation of the CD8+ T cell populations revealed additional CD8+ T cell sub-populations, based on expression of CD38, CCR6, another marker of MAIT cells,35,36 and the memory markers CD45RA/CCR7 [Figure 2C/D]. CD38+CD8+ T cells were higher in abundance in the transverse colon as compared with the remaining colonic compartments. Expression of the chemokine receptor CCR6 identified a sub-population of CD8+ T cells, whose abundance did not differ throughout the colon. Finally, terminally differentiated [TEMRA] cells, defined as CD45RA+CCR7-, represented a small and consistent population throughout [Figure 2D], with the remaining CD8+ T cells exhibiting a CD45RO+CCR7- T effector memory [TEM] phenotype.
CD4+ T cells along the colonic tract
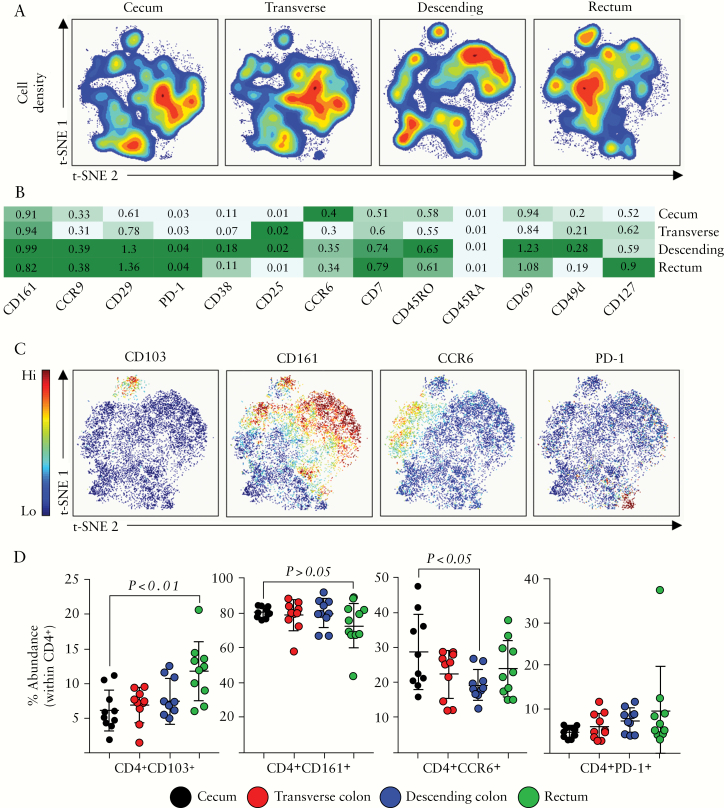
In contrast to CD8+ T cells, which were more abundant in the proximal colon, CD4+ T cells were most abundant in the distal colon [Figure 1]. t-SNE analysis of the CD4+ T cell pool across the colonic segments revealed the variation within this immune subset at each colonic site [Figure 3A]. Expression of a number of markers relevant to the CD4+ T cell population revealed several sub-populations which were detected within the colonic mucosa [Figure 3B]. Similar to CD8+ T cells, there is evidence that CD4+ T cells are capable of becoming a resident memory population in the intestine and at other mucosal sites, and this population can be defined by the expression of CD103.37 Indeed, we detected a small but significant population of CD4+CD103+TRM cells at each colonic site [Figure 3C], which were highest in abundance among CD4+ T cells in the rectum of healthy individuals [Figure 3D]. In contrast, there was no significant difference between the caecum, transverse, and descending colonic sites in terms of TRM abundance. Alongside CD103, this CD4+TRM population also expressed integrin β7, and CD69 [data not shown].
Figure 3.
CD4+ T cell subsets along the colonic tract. [A] opt-SNE analysis of CD4+ cellular events isolated from intestinal biopsies derived from the caecum, transverse, or descending colon, or rectum. [B] A heatmap displaying the relative expression of the cellular markers indicated, within the CD4+ T cell population, across the four colonic compartments. [C] Representative opt-SNE plots displaying expression of CD103, CD161, CCR6, and PD-1 among CD4+ cellular events. opt-SNE plots derived from the transverse colon from a single representative donor. [D] Summarised data of the expression of each marker in [C] displayed, determined using the Astrolabe profiling step,32 by limiting clusters to CD4+ events. Each data point represents a single donor, n = 10 total. Error bars represent standard deviation of mean. Statistical significance determined using analysis of variance [ANOVA], followed by Tukey’s multiple comparison test.
CD161, an important antigen in intestinal immunity, has also been shown to be expressed by CD4+ T cells.38 We identified a prominent population of CD161+CD4+ T cells within all segments [Figure 3C, D], and this population remained relatively constant throughout. Additional subsets of CD4+ T cells may be defined by the presence or absence of the chemokine receptor CCR6, previously described to be expressed by a number of subsets, including Th17 cells and T regulatory cells.39–41 Significantly higher abundance of CCR6+CD4+ T cells was observed in the caecum compared with the descending colon [Figure 3C, D]. Finally, expression of the programmed cell death-1 receptor [PD-1] was able to define a small but robust population of colonic CD4+ T cells [Figure 3C, D] which were consistent in abundance and phenotype along the colonic tract.
B cells along the colonic tract
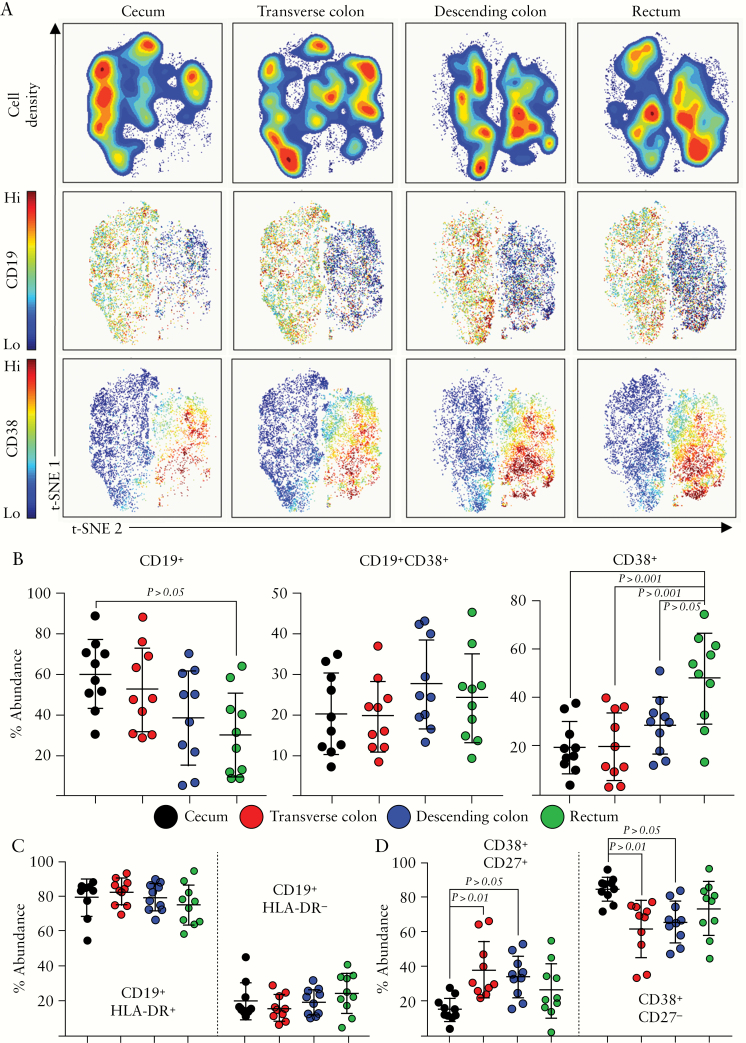
The role of B cells in IBD is unclear, whether they play an active role in pathogenesis or their frequency is simply modulated by inflammation.42–48 Despite the fact that B cell abundance did not differ between colonic segments [Figure 1], we examined whether specific B cell subsets may display altered abundance. Indeed, t-SNE analysis [Figure 1] identified two major B cell sub-populations which appeared to differ along the colonic tract. Gating on these populations and performing downstream t-SNE analysis revealed the identity of these two groups, and expression of CD19 versus CD38 was able to differentiate these two clusters of cells [Figure 4A]. CD19+ cells were defined as B cells, whereas CD38+ cells lacking all other lineage markers [Lineage-, Lin-] were defined as antibody-secreting cells [ASC]. In terms of the different colonic mucosal sites, there was a significantly higher abundance of CD19+ B cells in the proximal colon, which decreased towards the rectum [Figure 4B]. Conversely, CD38+ ASC were markedly increased in the rectum as compared with the rest of the colonic mucosa. Within the B cell population, two further sub-populations were described by differentiation based on the expression of HLA-DR [Figure 4C]. Despite this observation, there were no significant differences in the abundance of these sub-sets [CD19+HLA-DR+/CD19+HLA-DR-] between colonic segments. ASC were also able to be differentiated based on the expression of CD27 [Figure 4D]; the caecum showed increased and decreased abundance of CD38+CD27- and CD38+CD27+ subsets, respectively. In summary, although the total representation of the B cell lineage along the colonic tract remained unchanged, we observed a marked shift from B cells to ASC towards the distal portion of the colonic mucosa.
Figure 4.
B cell subsets along the colonic tract. [A] Representative opt-SNE analysis of CD19+/Lin-CD38+ cellular events isolated from intestinal biopsies, derived from the caecum, transverse, or descending colon, or rectum. Density plots are displayed, alongside expression plots displaying the expression of CD19 and CD38. [B] Proportions of the indicated B cell lineage subsets within each colonic compartment, determined using the Astrolabe profiling step,32 by limiting clusters to CD19+/Lin-CD38+ events. [C] Summary data displaying abundances of CD19+HLA-DR+ and CD19+ HLA-DR- subsets within each colonic segment. [D] Summary data displaying abundances of CD38+CD27+ and CD38+CD27- subsets within each colonic segment. Each data point represents a single donor, n = 10 total. Error bars represent standard deviation of mean. Statistical significance determined using analysis of variance [ANOVA], followed by Tukey’s multiple comparison test.
γδ T cells along the colonic tract
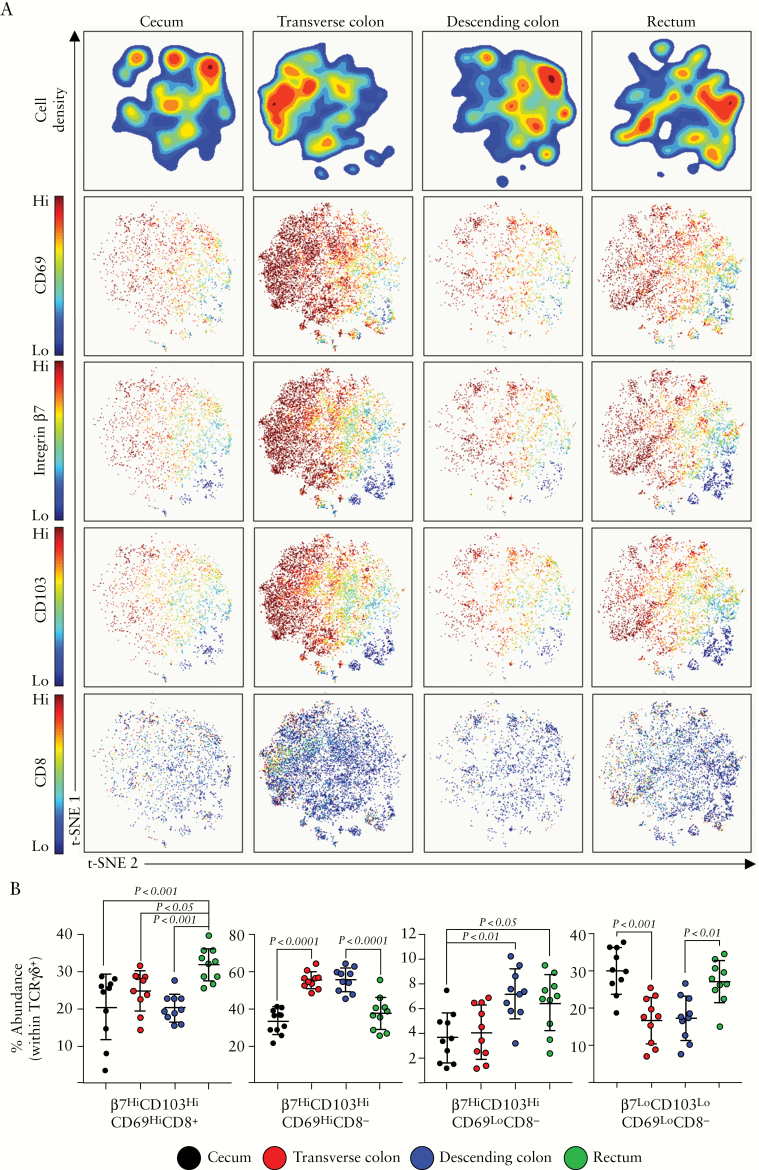
As γδ T cell abundance peaked in the transverse colon [Figure 1], we then examined whether this peak was associated with a particular subset within this lineage. Human γδ T cells subsets are defined based on the composition of their T cell receptor. γδ T cells expressing a Vδ2 subunit of the TCR predominate in peripheral blood, whereas tissue-resident γδ T cells mainly express the Vδ1 subunit and are mainly found at mucosal sites.49,50 Indeed, the vast majority of TCRγδ + events identified in the colonic mucosa of all segments were TCRVδ2-, indicating that these cells are mainly tissue-resident Vδ1+ cells [data not shown]. t-SNE analysis of the four compartments revealed shifting phenotypes of γδ T cells within each segment [Figure 5A]. We used the online analysis platform Astrolabe to automatically identify γδ T cell sub-populations, which revealed four prominent subsets of colonic-resident γδ T cells, based on their expression of four markers: integrin β7, CD103, CD69, and CD8. Co-expression of integrin β7, CD103, and CD69 is typical of tissue residency,51 reinforcing the observation that the majority of cells were Vδ1+. Indeed, expression of each of these markers revealed sub-populations of γδ T cells on the t-SNE map [Figure 5A]. A subset of γδ T cells have been shown to express CD8 in the intestine, and here we observed a similar population. These TCRγδ +β7HiCD103HiCD69HiCD8+ cells were most prominent in rectum, whereas most other γδ T cell subsets were most abundant in transverse colon [Figure 5B]. In contrast, TCRγδ +CD8- cells peaked in the transverse and descending segments, and cells featuring a non-tissue resident phenotype [β7Lo/CD103Lo/CD69Lo] were higher in abundance in the caecum and rectum. Overall, γδ T cells in the colon appeared to largely represent a homogeneous, tissue-resident phenotype, but there are significant alterations in these populations throughout the colon.
Figure 5.
TCRγδ + T cell subsets along the colonic tract. [A] opt-SNE analysis of TCRγδ + cellular events isolated from intestinal biopsies derived from the caecum, transverse, or descending colon, or rectum. Density plots are displayed, alongside plots displaying the expression of CD69, integrin β7, CD103, and CD8. [B] Proportions of the indicated TCRγδ + cellular subsets within each colonic compartment, determined using the Astrolabe profiling step,32 by limiting clusters to TCRγδ + events. Each data point represents a single donor, n = 10 total. Error bars represent standard deviation of mean. Statistical significance determined using analysis of variance [ANOVA], followed by Tukey’s multiple comparison test.
NK and NKT cells along the colonic tract
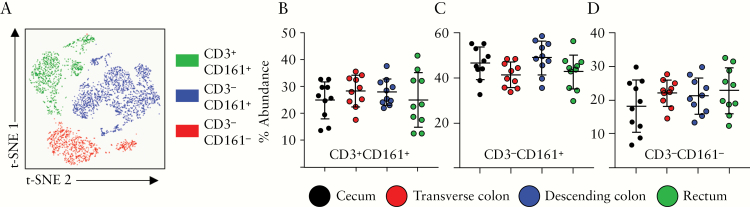
Similar to γδ T cells, the NK cell lineage displayed a peak abundance in the transverse colon, compared with all other segments [Figure 1]. Cells within this lineage [CD56+NKp46+CD16+] could further be divided into three subsets: CD3+CD161+ NKT cells, CD3-CD161+ NK cells, and CD3-CD161- NK cells [Figure 6A]. NKT cells in humans can be described by a co-expression of CD3 alongside CD56, NKp46, and CD16, and CD161 has been used to describe subsets of proinflammatory NK cells.52 Despite the differences in abundance of the NK lineage in general in the colonic mucosa, there were no significant differences in abundance between the three NK subsets identified, across the four colonic segments [Figure 6B]. As such the numbers of NK and NKT cells may peak in the transverse colon, but the composition of this lineage was similar within the remaining segments.
Figure 6.
NK and NKT cells along the colonic tract. [A] Representative t-SNE plot displaying events grouped into one of three sub-populations: CD3+CD161+, CD3-CD161+, and CD3-CD161-, pre-gated on Lin-CD56+ cellular events. In all, 25 000 cellular events were used for generation of t-SNE plots. [B] Abundance of CD3+CD161+ NKT cells, [C] CD3-CD161+ NK cells, and [D] CD3-CD161- NK cells in the four colonic compartments examined, determined using the Astrolabe profiling step,32 by limiting clusters to Lin-CD56+ events. Each data point represents a single donor, n = 10 total. Error bars represent standard deviation of mean. Statistical significance determined using analysis of variance [ANOVA], followed by Tukey’s multiple comparison test.
Discussion
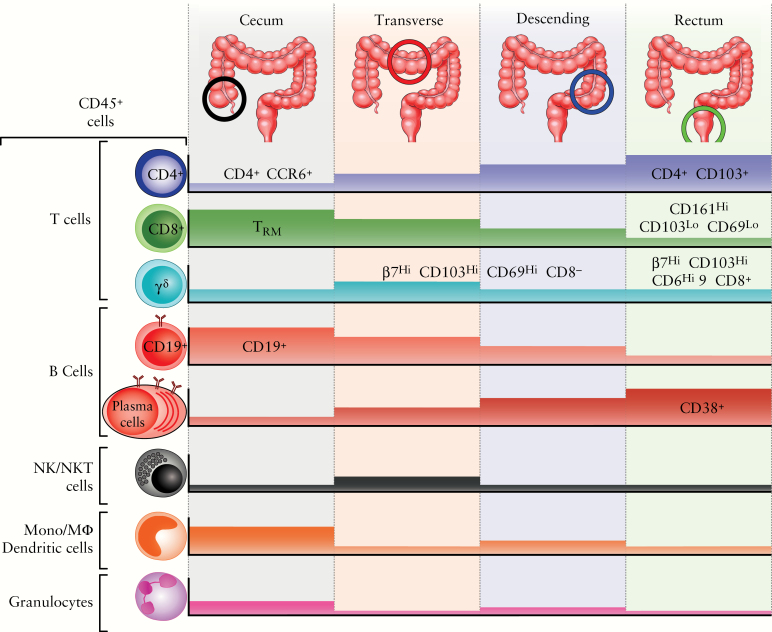
The major finding of the present study is the detection of regional variation in the relative abundance of immunocyte populations along the human colon [Figure 7]. Previous studies have reported variable frequency of immune cell subsets in separate parts of the gastrointestinal [GI] tract [i.e. upper GI vs small intestine vs large intestine], predominantly stemming from mouse data.24 Our results show that compartmentalisation of mucosal immunophenotypes takes place within what are often considered uniform GI segments, such as the large intestine. This highlights the importance of defined protocols for intestinal tissue harvest. Employing mass cytometry and analysing separately these segments, we observe site-specific patterns of leukocyte subset localisation. Given that examined tissue specimens were derived from healthy individuals, this variability appears to represent an inherent tissue property and not reactive recruitment or differentiation due to a local disease process. Nonetheless, our findings clearly indicate that regional mucosal signatures should be considered and controlled for during immune system-related clinical or translational studies for IBD or any other inflammatory condition of the large bowel. For example, in UC there is a continuous pattern of colonic involvement, upon both endoscopic and histological examination; thus, it is easy to assume that the location of intestinal biopsies is of minor importance for mucosal profiling. Similarly, in colonic CD, patchy distribution of the lesions may render optimal localisation problematic. In both cases however, as our present study shows, baseline immunoprofiling in control samples highly depends on the colonic segment from which biopsies are obtained. Thus, accurate spatial matching of intestinal sampling between control and disease-related groups, or prospectively before and after trial drug administration, is of paramount importance for the proper interpretation of differences observed in immunological expression patterns.
Figure 7.
Regional compartmentalisation of immune cells along the human colonic tract. Diagramatic representation of the distribution of major leukocyte lineages from the caecum to the rectum. Coloured blocks represent relative abundance of each lineage displayed (for colour figure refer online version).
In our CyTOF analysis, all types of immunocytes examined were present throughout the colon. Nevertheless, significant gradients of specific cell categories were detected between the examined segments. Such differentiation became more prominent when we compared proximal and distal colonic segments. In particular, the right colon appears to be enriched for CD8+ T cells, CD19+ B cells, and innate immune populations, such as granulocytes and cells of the monocytic lineage. On the other hand, the distal colon harbours increased numbers of CD4+ cells and antibody-secreting cells. Such dissociation most likely denotes the merging of distinct embryological origins, varied ecological pressure potentially by the microbiota, and discrete functional specialisation between the two areas.19,53 The proximal colon [caecum, ascending, and initial two-thirds of transverse] develop from the midgut, whereas the distal [terminal one-third of transverse, descending, and sigmoid] from the hindgut.16
This embryological dissimilarity may contribute to variation later on, as it may also explain why the transverse colon demonstrated some unique characteristics. It also results in disparate lymphatic drainage along the length of the colonic mucosa.54 In rats, lymph from the caecum and ascending colon drain to mesenteric lymph nodes [MLNs], and from the descending colon and rectum to the caudal lymph node, whereas the transverse colon drains separately into intrapancreatic nodes.55 As regional differences in bacterial flora composition and dietary components may exist, this disparate lymphoid uptake may lead to distinctive antigen delivery to corresponding lymph nodes and discrete local immunological signatures.18 Added to this embryonic dissociation is a differentiated genetic programming between left and right colon, which is much more prominent during adult compared with fetal life.19 This indicates that such events take place during postnatal development. Along the same line, genome-wide methylation analysis of normal DNA, extracted from colonic biopsies, detected more than 8000 methylated loci that distinguished right from left colon.21 The aforementioned studies point to significant embryological, genetic, and epigenetic differences between proximal and distal human colon, which may also underline the relatively distinct segmental immunological microenvironments observed.
We found prominent regional differences in the distribution of certain sub-populations of immune cells in the human colon. One such population consists of tissue-resident memory cells [TRM]. TRM can be either CD4+ or CD8+ and display combinations of surface proteins [CD103, CD49a, CD69] which facilitate binding to local ligands, thus allowing for their retention within peripheral tissues and preventing recirculation.56 Conflicting results have been reported in IBD samples, where CD103+ T-cell numbers were found to be decreased, unaltered, or increased in various studies, depending also on mucosal positioning [intraepithelial vs lamina propria].57–59 Our results demonstrate proximal predilection for CD8+CD103HiCD69Hi TRM and distal for CD4+CD103+ cells, and may explain these contradictory results, emphasising the need for rigorous sampling localisation for accurate interpretation of distribution patterns. The functional importance of such differences remains to be defined during homeostatic and inflammatory states. A primary regulatory, tissue-protective function of CD8+CD103Hi cells has been proposed.60 Those cells were found to be less prominent in the distal colon, whereas decreased numbers of CD8+CD103Hi cells were recently reported in UC.61 Taken together, we can speculate that, in the distal colon of patients with active UC, the number of mucosal CD8+ TRM falls below a critical threshold and this may lead to defective regulatory control of effector responses, contributing to the inflammatory process.
Cells of the γδ T cell lineage displayed marked heterogeneity along the colonic tract. Based on the markers included in this panel, we observed a peak of TCRγδ + cells within the transverse colon and a robust CD8+ resident memory-like population of γδ T cells in the rectum. γδ T cells have been implicated in IBD pathogenesis62 and its resolution, in particular the observed CD8+ population.63 Ulcerative colitis begins in the rectum and proceeds to affect the proximal colon in later stages. As such, immune populations specific for rectal tissue may provide valuable insight into potential initial triggers of UC pathogenesis. The abundance of CD4+ T cells towards the distal portion of the colon may also have implications for HIV transmission.64,65
Our study, similar to others that involve acquisition of large amounts of data, has limitations that need to be considered for the appropriate interpretation of results. First, on the purely technical level, there is a need for unified collection and cell isolation protocols, as procedural differences may lead to unbalanced enzymatic cleavage of surface markers and erroneous estimates of cell subset localisation. In addition, CyTOF is not widely available, which lessens the generalised applicability of deep mucosal immunoprofiling at the moment. CyTOF may be envisioned as a discovery tool, from which standard flow cytometry marker panels may be developed, focused on the most relevant immune subsets. Although our study group consisted of healthy individuals, inter-individual variability remains a problem. Nevertheless, for the majority of markers we observed acceptable standard deviations for their expression. Third, the antibody panel used for cell characterisation has its own limitations and it is always possible that relevant cell subsets are insufficiently accounted for. A representative but important example is the lack of adequate characterisation of innate lymphoid cells within the current protocol. This creates the need for additional markers that will detect ILCs and discriminate between their various fractions in the colonic segments. Indeed, studies of T helper subsets and regulatory T cell populations within the intestinal segments would be a valuable adjunct, given the emergence of therapies targeting distinct cytokine pathways [IL-12/IL-23].66 Fourth, since our starting material is endoscopically obtained intestinal tissue, it does not discriminate between the intraepithelial and lamina propria compartments. Finally, regional characterisation of the colon should be expanded to other parts of the GI tract as well. This is of particular significance for the terminal ileum, which is a common site of involvement in over half of patients with CD but may also be affected by the so-called ‘backwash ileitis’ in UC. Thus, comparison between the immunological composition of terminal ileum vs colonic segments will be important for understanding both common and diverse inflammatory pathways in these two locations. We are currently exploring such extra-colonic immune cell distribution patterns, and aim to validate findings and dissect biomarker applicability in cohorts of IBD patients.
Since our current findings indicate that immune composition of the human intestinal tract varies significantly between anatomical sites, the question arises as to how sampling should be performed during studies of CD and UC, in order to ensure robust and reproducible measurements that may be related to disease pathogenesis and drug efficacy. Here we propose several recommendations to facilitate this process. When comparing different clinical groups [e.g., UC patients vs controls] care should be taken in obtaining samples from identical sites across individuals. For example, if comparing healthy individuals with patients with active UC, sampling should be performed consistently at a defined distance from the anal verge using the colonoscope centimetre markers [e.g., 40 cm from anal verge] in both patient cohorts. If comparisons are to be made between patients with active disease, care should be taken to sample both visually inflamed and non-inflamed areas that can be matched across patient cohorts. For example in patients with distal ulcerative colitis, defined areas of descending colon could be sampled, whereas a defined area of the transverse colon may represent an uninvolved segment across cohorts.
However, this internal uninflamed control strategy will not be an option for patients with pancolitis. A logistical challenge may be to compare cell composition in patients with variable disease severity, which may then be defined ideally by both endoscopic and histological severity. Crohn’s disease represents an additional challenge for reproducible biopsy collection, given its segmental involvement. As with UC, care should be taken to sample sites that are as close as possible across subjects. Where patchy inflammation occurs, biopsies should be obtained from adjacent inflamed and non-inflamed sites at a defined distance from the anal verge. For longitudinal studies, where the response to therapeutics is to be assessed over time [e.g., pre- and post-therapy], tattooing of initial biopsy sites for subsequent sampling may improve consistency. Multiple intestinal sites could be sampled in this way in order to build a picture of the overall intestinal response to therapy. For mass cytometric analyses, we9 and others12 have determined that six standard-sized biopsies obtained with 2.8-mm forceps are sufficient to identify minor immune populations within the intestinal mucosa. Once clinically relevant markers are identified, focused standard flow or spectral panels can be developed including intracellular cytokines and transcription factors. CyTOF-derived discoveries may then be more readily accessible to the greater IBD research community.
Our study demonstrates potential challenges for the proper interpretation of mass cytometry and mRNA expression data and the need for appropriate validation controls across different laboratories and between separate trial sites. It also establishes an experimental setup that could be incorporated as a secondary endpoint in clinical trials with new drugs for IBD [e.g., patterns of phosphorylated STAT expression after a JAK inhibitor]. This is in line with the current trend of exploring treatment outcomes that go beyond the traditional Selecting Therapeutic taRgets in Inflammatory bowel DiseasE [STRIDE]-proposed combinations of clinical plus endoscopic evaluations.67,68 Recently, histological parameters have been also examined and reported, as they may have better long-term prognostic value. Based on the present findings, we propose that cellular and molecular analyses of biopsy tissue at baseline and their responses during treatment may also be included as secondary endpoints in IBD clinical trials. Immune profiling may be performed at baseline, but also prospectively at defined time points during treatment. Tattooing the biopsy site may be a practical solution to achieve consistency during prospective mechanistic studies. Newer technologies,69 including spectral flow cytometry,70 may make this approach more accessible to the wider community. Such an approach will be of paramount importance for the discovery of molecular biomarkers with prognostic significance, for the natural history of the disease and, importantly, for confirmation of local response to a specific therapy. This type of information may lead to a better stratification of candidate study subjects and lay a foundation for a personalised approach to IBD therapy.
Supplementary Material
Acknowledgements
We thank Ms Gelareh Gangi for assistance with maintenance of the IRB protocol, and Michael Solga of the University of Virginia Flow Cytometry Core for assistance with mass cytometry event acquisition.
Funding
This work was supported by grants from: National Institutes of Health [DK108670, DK118927]; VA Merit BLRD-I01 BX003436; Takeda Pharmaceuticals U.S.A; IISR-2017–102050 grant to JRN; San Diego Digestive Diseases Research Center [P30 DK120515]; Shared Instrumentation Grant [SIG] Program [S10]: CyTOF Mass Cytometer S10 OD018499-01 to LJI; support from Chiba University-UC San Diego Program in Mucosal Immunology, Allergy and Vaccines; and PSD is supported by an AGA Research Scholar Award.
Conflict of Interest
PSD has received consulting and research support from Takeda, Abbvie, Janssen, Pfizer.
Author Contributions
CJT, JR-N for study concept and design; JR-N, TP-J, AY, BB, PSD, DP for sample collection; CJT, LRL, SY for sample processing and data acquisition; CJT, MG for analysis and interpretation of data; CJT, GB, JR-N for drafting of manuscript; LRL, SY, MG, TP-J, AY, BB, PSD, GB, JR-N for critical revision of manuscript; CJT for statistical analysis.
References
- 1. de Souza HSP, Fiocchi C, Iliopoulos D. The IBD interactome: an integrated view of aetiology, pathogenesis and therapy. Nat Rev Gastroenterol Hepatol 2017;14:739–49. [DOI] [PubMed] [Google Scholar]
- 2. Abraham C, Cho JH. Inflammatory bowel disease. N Engl J Med 2009;361:2066–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pagnini C, Pizarro TT, Cominelli F. Novel pharmacological therapy in inflammatory bowel diseases: beyond anti-tumor necrosis factor. Front Pharmacol 2019;10:671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pérez-Jeldres T, Tyler CJ, Boyer JD, et al. . Cell trafficking interference in inflammatory bowel disease: therapeutic interventions based on basic pathogenesis concepts. Inflamm Bowel Dis 2019;25:270–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pérez-Jeldres T, Tyler CJ, Boyer JD, et al. . Targeting cytokine signaling and lymphocyte traffic via small molecules in inflammatory bowel disease: JAK inhibitors and S1PR agonists. Front Pharmacol 2019;10:212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sandborn WJ, Feagan BG, Hanauer SB, et al. . A review of activity indices and efficacy endpoints for clinical trials of medical therapy in adults with Crohn’s disease. Gastroenterology 2002;122:512–30. [DOI] [PubMed] [Google Scholar]
- 7. D’Haens G, Sandborn WJ, Feagan BG, et al. . A review of activity indices and efficacy end points for clinical trials of medical therapy in adults with ulcerative colitis. Gastroenterology 2007;132:763–86. [DOI] [PubMed] [Google Scholar]
- 8. Levesque BG, Sandborn WJ, Ruel J, Feagan BG, Sands BE, Colombel JF. Converging goals of treatment of inflammatory bowel disease from clinical trials and practice. Gastroenterology 2015;148:37–51.e1. [DOI] [PubMed] [Google Scholar]
- 9. Tyler CJ, Pérez-Jeldres T, Ehinger E, et al. . Implementation of mass cytometry as a tool for mechanism of action studies in inflammatory bowel disease. Inflamm Bowel Dis 2018;24:2366–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rubin SJS, Bai L, Haileselassie Y, et al. . Mass cytometry reveals systemic and local immune signatures that distinguish inflammatory bowel diseases. Nat Commun 2019;10:2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chan SN, Low END, Raja Ali RA, Mokhtar NM. Delineating inflammatory bowel disease through transcriptomic studies: current review of progress and evidence. Intest Res 2018;16:374–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Konnikova L, Boschetti G, Rahman A, et al. . High-dimensional immune phenotyping and transcriptional analyses reveal robust recovery of viable human immune and epithelial cells from frozen gastrointestinal tissue. Mucosal Immunol 2018;11:1684–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. van Unen V, Li N, Molendijk I, et al. . Mass cytometry of the human mucosal immune system identifies tissue- and disease-associated immune subsets. Immunity 2016;44:1227–39. [DOI] [PubMed] [Google Scholar]
- 14. Agace WW, McCoy KD. Regionalized development and maintenance of the intestinal adaptive immune landscape. Immunity 2017;46:532–48. [DOI] [PubMed] [Google Scholar]
- 15. Danese S, Sandborn WJ, Colombel JF, et al. . Endoscopic, radiologic, and histologic healing with vedolizumab in patients with active Crohn’s disease. Gastroenterology 2019;157:1007–18.e7. [DOI] [PubMed] [Google Scholar]
- 16. Morton DA, Foreman KB, Albertine KH. Midgut and hindgut. In: The Big Picture: Gross Anatomy. New York, NY: McGraw-Hill Companies; 2011. [Google Scholar]
- 17. Simmonds N, Furman M, Karanika E, Phillips A, Bates AW. Paneth cell metaplasia in newly diagnosed inflammatory bowel disease in children. BMC Gastroenterol 2014;14:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Eriksson K, Quiding-Järbrink M, Osek J, et al. . Anatomic segmentation of the intestinal immune response in nonhuman primates: differential distribution of B cells after oral and rectal immunizations to sites defined by their source of vascularization. Infect Immun 1999;67:6210–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Glebov OK, Rodriguez LM, Nakahara K, et al. . Distinguishing right from left colon by the pattern of gene expression. Cancer Epidemiol Biomarkers Prev 2003;12:755–62. [PubMed] [Google Scholar]
- 20. Bernardo D, Durant L, Mann ER, et al. . Chemokine [C-C Motif] receptor 2 mediates dendritic cell recruitment to the human colon but is not responsible for differences observed in dendritic cell subsets, phenotype, and function between the proximal and distal colon. Cell Mol Gastroenterol Hepatol 2016;2:22–39.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kaz AM, Wong CJ, Dzieciatkowski S, Luo Y, Schoen RE, Grady WM. Patterns of DNA methylation in the normal colon vary by anatomical location, gender, and age. Epigenetics 2014;9:492–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cummings JH, Macfarlane GT. The control and consequences of bacterial fermentation in the human colon. J Appl Bacteriol 1991;70:443–59. [DOI] [PubMed] [Google Scholar]
- 23. Hevia A, Bernardo D, Montalvillo E, et al. . Human colon-derived soluble factors modulate gut microbiota composition. Front Oncol 2015;5:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mowat AM, Agace WW. Regional specialization within the intestinal immune system. Nat Rev Immunol 2014;14:667–85. [DOI] [PubMed] [Google Scholar]
- 25. Suzuki H. Differences in intraepithelial lymphocytes in the proximal, middle, distal parts of small intestine, caecum, and colon of mice. Immunol Invest 2009;38:780–96. [DOI] [PubMed] [Google Scholar]
- 26. Bagwell CB, Adams EG. Fluorescence spectral overlap compensation for any number of flow cytometry parameters. Ann N Y Acad Sci 1993;677:167–84. [DOI] [PubMed] [Google Scholar]
- 27. Sumatoh HR, Teng KW, Cheng Y, Newell EW. Optimization of mass cytometry sample cryopreservation after staining. Cytometry A 2017;91:48–61. [DOI] [PubMed] [Google Scholar]
- 28. Finck R, Simonds EF, Jager A, et al. . Normalization of mass cytometry data with bead standards. Cytometry A 2013;83:483–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Amir el-AD, Davis KL, Tadmor MD, et al. . Visne enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat Biotechnol 2013;31:545–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Belkina AC, Ciccolella CO, Anno R, et al. . Automated optimized parameters for T-distributed stochastic neighbor embedding improve visualization and analysis of large datasets. Nat Commun 2019; 10: 5415. doi:10.1038/s41467-019-13055-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen H, Lau MC, Wong MT, Newell EW, Poidinger M, Chen J. Cytofkit: a bioconductor package for an integrated mass cytometry data analysis pipeline. PLoS Comput Biol 2016;12:e1005112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Amir ED, Lee B, Badoual P, et al. . Development of a comprehensive antibody staining database using a standardized analytics pipeline. Front Immunol 2019;10:1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Konjar Š, Ferreira C, Blankenhaus B, Veldhoen M. Intestinal barrier interactions with specialized CD8 T cells. Front Immunol 2017;8:1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schenkel JM, Masopust D. Tissue-resident memory T cells. Immunity 2014;41:886–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fergusson JR, Hühn MH, Swadling L, et al. . CD161[int]CD8+ T cells: a novel population of highly functional, memory CD8+ T cells enriched within the gut. Mucosal Immunol 2016;9:401–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Godfrey DI, Koay HF, McCluskey J, Gherardin NA. The biology and functional importance of MAIT cells. Nat Immunol 2019;20:1110–28. [DOI] [PubMed] [Google Scholar]
- 37. Nguyen QP, Deng TZ, Witherden DA, Goldrath AW. Origins of CD4+ circulating and tissue-resident memory T-cells. Immunology 2019;157:3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kleinschek MA, Boniface K, Sadekova S, et al. . Circulating and gut-resident human Th17 cells express CD161 and promote intestinal inflammation. J Exp Med 2009;206:525–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lee AY, Eri R, Lyons AB, Grimm MC, Korner H. CC chemokine ligand 20 and its cognate receptor CCR6 in mucosal T cell immunology and inflammatory bowel disease: odd couple or axis of evil? Front Immunol 2013;4:194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kitamura K, Farber JM, Kelsall BL. CCR6 marks regulatory T cells as a colon-tropic, IL-10-producing phenotype. J Immunol 2010;185:3295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Godefroy E, Alameddine J, Montassier E, et al. . Expression of CCR6 and CXCR6 by gut-derived CD4+/CD8α+ T-regulatory cells, which are decreased in blood samples from patients with inflammatory bowel diseases. Gastroenterology 2018;155:1205–17. [DOI] [PubMed] [Google Scholar]
- 42. Jinno Y, Ohtani H, Nakamura S, et al. . Infiltration of CD19+ plasma cells with frequent labeling of Ki-67 in corticosteroid-resistant active ulcerative colitis. Virchows Arch 2006;448:412–21. [DOI] [PubMed] [Google Scholar]
- 43. Hosomi S, Oshitani N, Kamata N, et al. . Increased numbers of immature plasma cells in peripheral blood specifically overexpress chemokine receptor CXCR3 and CXCR4 in patients with ulcerative colitis. Clin Exp Immunol 2011;163:215–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tarlton NJ, Green CM, Lazarus NH, et al. . Plasmablast frequency and trafficking receptor expression are altered in pediatric ulcerative colitis. Inflamm Bowel Dis 2012;18:2381–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. MacDermott RP, Nash GS, Bertovich MJ, Seiden MV, Bragdon MJ, Beale MG. Alterations of IgM, IgG, and IgA synthesis and secretion by peripheral blood and intestinal mononuclear cells from patients with ulcerative colitis and Crohn’s disease. Gastroenterology 1981;81:844–52. [PubMed] [Google Scholar]
- 46. Wang X, Jiang Y, Zhu Y, et al. . Circulating memory B cells and plasmablasts are associated with the levels of serum immunoglobulin in patients with ulcerative colitis. J Cell Mol Med 2016;20:804–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang Z, Zhu M, Luo C, et al. . High level of IgG4 as a biomarker for a new subset of inflammatory bowel disease. Sci Rep 2018;8:10018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Uo M, Hisamatsu T, Miyoshi J, et al. . Mucosal CXCR4+ IgG plasma cells contribute to the pathogenesis of human ulcerative colitis through FcγR-mediated CD14 macrophage activation. Gut 2013;62:1734–44. [DOI] [PubMed] [Google Scholar]
- 49. Tyler CJ, Doherty DG, Moser B, Eberl M. Human Vγ9/Vδ2 T cells: Innate adaptors of the immune system. Cell Immunol 2015;296:10–21. [DOI] [PubMed] [Google Scholar]
- 50. Lawand M, Déchanet-Merville J, Dieu-Nosjean MC. Key features of gamma-delta T-cell subsets in human diseases and their immunotherapeutic implications. Front Immunol 2017;8:761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kumar BV, Ma W, Miron M, et al. . Human tissue-resident memory T cells are defined by core transcriptional and functional signatures in lymphoid and mucosal sites. Cell Rep 2017;20:2921–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kurioka A, Cosgrove C, Simoni Y, et al. ; Swiss HIV Cohort Study; Oxford IBD Cohort Investigators CD161 defines a functionally distinct subset of pro-inflammatory natural killer cells. Front Immunol 2018;9:486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Distler P, Holt PR. Are right- and left-sided colon neoplasms distinct tumors? Dig Dis 1997;15:302–11. [DOI] [PubMed] [Google Scholar]
- 54. Van den Broeck W, Derore A, Simoens P. Anatomy and nomenclature of murine lymph nodes: descriptive study and nomenclatory standardization in BALB/cAnNCrl mice. J Immunol Methods 2006;312:12–9. [DOI] [PubMed] [Google Scholar]
- 55. Tilney NL. Patterns of lymphatic drainage in the adult laboratory rat. J Anat 1971;109:369–83. [PMC free article] [PubMed] [Google Scholar]
- 56. Mueller SN, Mackay LK. Tissue-resident memory T cells: local specialists in immune defence. Nat Rev Immunol 2016;16:79–89. [DOI] [PubMed] [Google Scholar]
- 57. Elewaut D, Van Damme N, De Keyser F, et al. . Altered expression of alpha E beta 7 integrin on intra-epithelial and lamina propria lymphocytes in patients with Crohn’s disease. Acta Gastroenterol Belg 1998;61:288–94. [PubMed] [Google Scholar]
- 58. Oshitani N, Watanabe K, Maeda K, et al. . Differential expression of homing receptor CD103 on lamina propria lymphocytes and association of CD103 with epithelial adhesion molecules in inflammatory bowel disease. Int J Mol Med 2003;12:715–9. [PubMed] [Google Scholar]
- 59. Ichikawa R, Lamb CA, Eastham-Anderson J, et al. . AlphaE integrin expression is increased in the ileum relative to the colon and unaffected by inflammation. J Crohns Colitis 2018;12:1191–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lamb CA, Mansfield JC, Tew GW, et al. . αEβ7 integrin identifies subsets of pro-inflammatory colonic CD4+ T lymphocytes in ulcerative colitis. J Crohns Colitis 2017;11:610–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Roosenboom B, Wahab PJ, Smids C, et al. . Intestinal CD103+CD4+ and CD103+CD8+ T-cell subsets in the gut of inflammatory bowel disease patients at diagnosis and during follow-up. Inflamm Bowel Dis 2019;25:1497–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Catalan-Serra I, Sandvik AK, Bruland T, Andreu-Ballester JC. Gammadelta T cells in Crohn’s disease: a new player in the disease pathogenesis? J Crohns Colitis 2017;11:1135–45. [DOI] [PubMed] [Google Scholar]
- 63. Kadivar M, Petersson J, Svensson L, Marsal J. CD8αβ+ γδ T cells: a novel T cell subset with a potential role in inflammatory bowel disease. J Immunol 2016;197:4584–92. [DOI] [PubMed] [Google Scholar]
- 64. McElrath MJ, Smythe K, Randolph-Habecker J, et al. ; NIAID HIV Vaccine Trials Network Comprehensive assessment of HIV target cells in the distal human gut suggests increasing HIV susceptibility toward the anus. J Acquir Immune Defic Syndr 2013;63:263–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Mylvaganam GH, Velu V, Hong JJ, et al. . Diminished viral control during simian immunodeficiency virus infection is associated with aberrant PD-1hi CD4 T cell enrichment in the lymphoid follicles of the rectal mucosa. J Immunol 2014;193:4527–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Weaver KN, Gregory M, Syal G, et al. . Ustekinumab is effective for the treatment of Crohn’s disease of the pouch in a multicenter cohort. Inflamm Bowel Dis 2019;25:767–74. [DOI] [PubMed] [Google Scholar]
- 67. Pouillon L, Peyrin-Biroulet L. It is time to revise the STRIDE guidelines determining therapeutic goals for treat-to-target in inflammatory bowel disease. J Crohns Colitis 2018;12:509. [DOI] [PubMed] [Google Scholar]
- 68. Peyrin-Biroulet L, Sandborn W, Sands BE, et al. . Selecting Therapeutic Targets in Inflammatory Bowel Disease [STRIDE]: determining therapeutic goals for treat-to-target. Am J Gastroenterol 2015;110:1324–38. [DOI] [PubMed] [Google Scholar]
- 69. Chattopadhyay PK, Gierahn TM, Roederer M, Love JC. Single-cell technologies for monitoring immune systems. Nat Immunol 2014;15:128–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Schmutz S, Valente M, Cumano A, Novault S. Spectral cytometry has unique properties allowing multicolor analysis of cell suspensions isolated from solid tissues. PLoS One 2016;11:e0159961. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.