Abstract
Background
Interleukin-12 (IL-12) is considered to be a risk factor for cancer; however, its role in hepatocellular carcinoma (HCC) remains unknown. This study aimed to explore the impacts of the IL-12 rs3212227 and rs568408 gene polymorphisms on HCC.
Methods
We searched PubMed, Embase, Web of Science, and Chinese Knowledge Infrastructure databases for studies on the associations between HCC and IL-12 rs568408 and rs3212227 polymorphisms published prior to 1 May 2020. The effects of the polymorphisms on HCC susceptibility were presented as odds ratios (ORs) and associated 95% confidence intervals.
Results
Seven studies were ultimately included, including 2375 cases and 3445 controls. The rs3212227 polymorphism was significantly associated with the risk of HCC in both the dominant model (CC+AC vs. AA, OR=1.22) and the allele model (C vs. A, OR=1.12). Combined analysis of rs568408 yielded a significant relative risk for HCC in the dominant (AA+AG vs. GG, OR=1.13), recessive (AA vs. AG+GG, OR=1.72), allele (A vs. G, OR=1.29), heterozygote (AG vs. GG, OR=1.27), and homozygote models (AA vs. GG, OR 1.17).
Conclusion
The IL-12 rs3212227 and rs568408 gene polymorphisms are associated with an increased risk of HCC.
Keywords: Interleukin-12, hepatocellular carcinoma, polymorphism, meta-analysis, cancer risk, rs3212227, rs568408
Introduction
Hepatocellular carcinoma (HCC) is a fatal disease that poses a major health threat worldwide. Hepatitis B virus (HBV) is a major risk factor for HCC,1 with an estimated 75%–85% of HCC cases attributable to persistent infection with HBV.2 Growing evidence has also highlighted the role of genetic polymorphisms in HCC, suggesting an important role for genetic factors in carcinogenesis.3,4 However, the potential value of genetic polymorphisms in HCC remains to be elucidated.
Cytokines are important immunoregulatory substances that produce inflammatory responses, thus influencing the occurrence of chronic inflammation, the tumor microenvironment,5 and the pathogenesis of HCC.6 Interleukin-12 (IL-12) comprises two subunits (p35 and p40) connected by a disulfide bond to produce an isoprene dimer cytokine with multiple biological effects.7 Gene polymorphisms have been identified in the promoter region, intron, and 3'-untranslated region of the portion of the IL-12 gene that codes for the p40 subunit.8 IL-12 plays a major role not only in inducing an appropriate immune responses against viral infections (including HBV), but also in the antitumor immune response.9 IL-12 has been shown to inhibit the proliferation and metastasis of various malignant tumors, in vitro and in vivo,10 and to inhibit the differentiation of natural killer and Th2 T helper cells, contributing to a strong anti-tumor effect.11 IL-12 is encoded by two genes, IL12A and IL12B, and numerous studies have demonstrated that the IL12A and IL12B genes include multiple functional polymorphic sites that might affect the occurrence and progression of non-Hodgkin lymphoma,12 tuberculosis,13 autoimmune disease,14 gastric carcinoma,15 lung carcinoma,16 and other malignancies.17 Tan et al. showed that the IL12A rs568408 variant may be a marker single-nucleotide polymorphism (SNP) associated with an increased risk of poor HBV clearance and HBV-related HCC development.9 Meanwhile, Ben-Selma et al.18 suggested that IL-12A rs568408 and gene–gene interactions between IL-12A rs568408 and IL-12B rs3212227 contributed to the outcome of chronic HBV infection, indicating their potential usefulness as predictive and diagnostic biomarkers of HCC. Additional evidence has shown that the IL-12 signaling pathway plays a pivotal role during HBV infection and may contribute to the pathogenesis of HCC.19,20 An accurate analysis of the distribution of IL-12 gene polymorphisms among human populations from various geographical regions may reveal a correlation between these polymorphisms and the occurrence of HCC.
Researchers have previously investigated the role of IL-12 in the development and progression of HCC;21 however, the specific association between IL-12 gene polymorphisms and susceptibility to HCC remains controversial. We conducted a meta-analysis of related case-control studies with large sample sizes to obtain additional evidence to allow the analysis of the association between IL-12 gene polymorphisms and susceptibility to HCC, to provide a scientific basis for the early diagnosis and treatment of HCC.
Methods and materials
Search strategy
Previous reports on the associations between the IL-12 rs568408 and rs3212227 polymorphisms and HCC were collected by searching the PubMed, Embase, Web of Science, Chinese Knowledge Infrastructure (CNKI), and Wanfang databases using the following key words: (“interleukin-12” OR “Interleukin-12” OR “IL-12” OR “IL12” OR “rs568408” OR “rs3212227”), and (“polymorphisms” OR “polymorphism” OR “SNP” OR “variation” OR “variant” OR “mutation” OR “genetic” OR “genotype”), and (“hepatocellular carcinoma” OR “HCC” OR “liver cancer”). According to the scope statements on Medline and Embase, other subject headings were included based on previous indexes and keywords that contained synonyms for the words included in the range description, to ensure that topic headings were used correctly, according to their definitions.
Only studies published before 1 May 2020 were included in the meta-analysis. We also screened the reference lists to identify related titles.
Selection criteria
Studies included in the meta-analysis had to fulfill the following inclusion criteria: 1) case-control study; 2) investigation of the association between IL-12 rs3212227 or rs568408 polymorphism with HCC; and 3) complete data, including numbers of patients in the case and control groups, distribution of genotype frequency, and related statistical indicators. If multiple publications referred to the same study sample, the study with the largest sample was selected for inclusion in the meta-analysis.
Data extraction
Cunqing Kong and Miao Chen independently screened the literature and extracted the following data: first author, year of publication, country, ethnicity, baseline characteristics of the control group, sample size of the case and control groups, genetic frequency, Hardy–Weinberg equilibrium (HWE) values for the control group, HCC risk; rs3212227 polymorphism; rs568408 polymorphism; rs3212227 gene mutation (A to C); and rs568408 gene mutation (C to A).
This was not a primary research study, and ethical approval and informed consent were therefore not required.
Statistical analysis
All statistical analyses were performed using Stata software, version 12.0 (StataCorp LP, College Station, TX, USA) and Review Manager 5.3 (Cochrane Collaboration). All binary variables were presented as summary risk ratios (RR) with 95% confidence intervals (CI), calculated using a fixed-effects model except in the event of high heterogeneity, in which case a random-effects model was used. The associations between IL-12 polymorphisms and HCC risk were evaluated using recessive, dominant, homozygote, heterozygote, and allelic models. Unadjusted odds ratios (ORs) and 95%CIs were used to describe the susceptibility of patients with rs3212227 or rs568408 polymorphisms to HCC. HWE values in the control group were determined by Fisher’s exact test and the effect of combining OR values on HCC was determined by the Z-test. Heterogeneity was evaluated using fixed-effect (I2<50%) and random-effect models (I2≥50%). Subgroup analysis was performed to determine the effect of ethnicity on the associations of rs3212227 and rs568408 polymorphism with HCC. We calculated the summary RR and 95%CI using a fixed-effect model, or a random-effect model in cases of high heterogeneity. The influence of study quality on the results was assessed by sensitivity analysis. The extent of heterogeneity was determined as the total percentage of variation between studies, measured with the I2 statistic. I2 values were categorized as low if I2 (0%–25%), moderate (25%–50%), and high (50%–90%). The Q-statistic was used to assess the presence of heterogeneity. A PQ statistic ≥0.05 was considered to indicate no significant heterogeneity among the included studies.
Results
Description of included studies
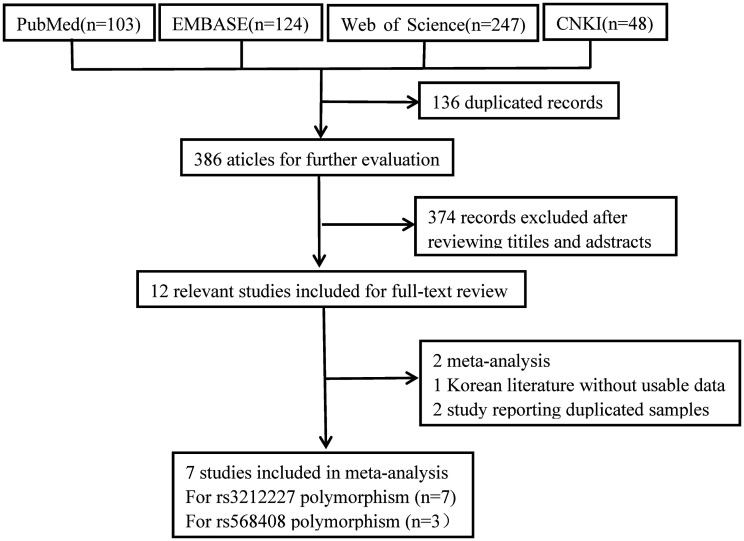
The search process and inclusion criteria for this study are shown in Figure 1. A total of 522 studies written in English or Chinese and published in the searched databases prior to 1 May 2020 were systematically screened (Figure 1). Seven studies9,21–26 were finally included in this meta-analysis (Table 1). These seven studies demonstrated the association between the rs3212227 polymorphism and HCC risk (2375 cases, 3445 controls) and three studies9,21,22 also evaluated the relationship between the rs568408 polymorphism and HCC risk (1342 cases, 2026 controls). The genetic distribution of the rs568408 polymorphism in all but two controls9,22 was consistent with the HWE (P>0.05).
Figure 1.
Flow diagram of literature research and selection process.
Table 1.
Characteristics of studies included in this meta-analysis.
| First author | Year | Country | Ethnicity | Genotyping method | P for HWE | Type of control group | Cases | Controls | NOS score |
|---|---|---|---|---|---|---|---|---|---|
| IL12A (rs568408) | |||||||||
| Liu22 | 2011 | China | Asian | PCR-RFLP | Disequilibrium | Cancer-free | 869 | 891 | 7 |
| Tan9 | 2015 | China | Asian | IMLDR | Disequilibrium | Healthy, CHB | 395 | 979 | 6 |
| Elsayed21 | 2016 | Egypt | African | PCR-RFLP | Equilibrium | Healthy, CHC | 78 | 156 | 5 |
| IL12B (rs3212227) | |||||||||
| Nieters23 | 2005 | China | Asian | PCR-RFLP | Equilibrium | Cancer-free | 249 | 250 | 7 |
| Ognjanovic24 | 2009 | America | North American | Taqman assays | No checking | Healthy | 117 | 223 | 5 |
| Liu22 | 2011 | China | Asian | PCR-RFLP | Equilibrium | Cancer-free | 869 | 891 | 9 |
| Yang4 | 2011 | China | Asian | Taqman assays | Equilibrium | Cancer-free | 608 | 612 | 8 |
| Saxena25 | 2014 | India | Asian | PCR-RFLP | Equilibrium | Healthy, CHB | 59 | 336 | 6 |
| Tan9 | 2015 | China | Asian | IMLDR | Equilibrium | Healthy, CHB | 395 | 977 | 7 |
| Elsayed21 | 2016 | Egypt | African | PCR-RFLP | Equilibrium | Healthy, CHC | 78 | 156 | 6 |
PCR, polymerase chain reaction; RFLP, restriction fragment length polymorphism; IMLDR, improved multiplex ligase detection reaction; HWE, Hardy–Weinberg equilibrium; CHB, chronic hepatitis B; CHC, chronic hepatitis C; NOS, Newcastle–Ottawa Scale.
Meta-analysis of association between rs3212227 polymorphism and HCC risk
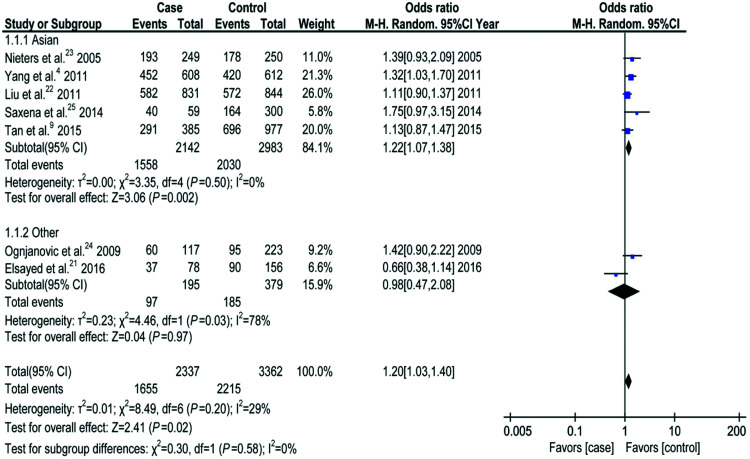
The associations between rs3212227 polymorphism and HCC risk for five models and various subgroups are shown in Table 2. All seven studies included in the meta-analysis (2,375 cases) reported an association between rs3212227 polymorphism and HCC. A fixed-effect analyses showed that rs3212227 was associated with increased risk for HCC in the dominant model (CC+AC vs. AA, OR 1.22, 95%CI 1.07–1.38, P = 0.003) (Figure 2) and the allele model (C vs. A, OR 1.12, 95%CI 1.03–1.21, P = 0.01) (Table 2). These results suggested that the C-allele polymorphism of rs3212227 may increase susceptibility to HCC. However, fixed-effect analyses using the other models showed no significant association between rs3212227 polymorphism and HCC risk (recessive model: CC vs. AG+AA, OR 1.15, 95%CI 0.99–1.33; heterozygote model: AC vs. AA, OR 0.96, 95%CI 0.86–1.08; homozygote model: CC vs. AA, OR 1.13, 95%CI 0.99–1.30).
Table 2.
Meta-analysis of association of rs3212227 polymorphisms with risk of hepatocellular carcinoma.
| Genotype comparison and genetic model | Group and subgroups |
Sample size (n) |
OR [95% CI] | P value | Analysis model |
Test of heterogeneity |
||
|---|---|---|---|---|---|---|---|---|
| Case | Control | I2 (%) | P value | |||||
| Dominant model | Overall | 2337 | 3362 | 1.22 [1.07–1.38] | 0.003 | Fixed | 0 | 0.40 |
| (CC+AC vs AA) | Asian | 2142 | 2983 | 1.22 [1.07–1.38] | 0.002 | Fixed | 0 | 0.50 |
| Other | 195 | 379 | 1.04 [0.74–1.47] | 0.82 | Random | 78 | 0.03 | |
| Recessive model | Overall | 1971 | 2889 | 1.15 [0.99–1.33] | 0.06 | Fixed | 0 | 0.75 |
| (CC vs AG+AA) | Asian | 1893 | 2733 | 1.15 [0.99–1.33] | 0.07 | Fixed | 0 | 0.60 |
| Other | 79 | 156 | 1.19 [0.59–2.40] | 0.63 | ||||
| Allele model | Overall | 3942 | 5778 | 1.12 [1.03–1.21] | 0.01 | Fixed | 25 | 0.26 |
| (C vs A) | Asian | 3786 | 5466 | 1.13 [1.04–1.23] | 0.005 | Fixed | 12 | 0.33 |
| Other | 156 | 312 | 0.84 [0.56–1.27] | 0.41 | ||||
| Heterozygote model | Overall | 1971 | 2889 | 0.96 [0.86–1.08] | 0.53 | Fixed | 47 | 0.11 |
| (AC VS AA) | Asian | 1893 | 2733 | 0.94 [0.83–1.06] | 0.29 | Fixed | 0 | 0.65 |
| Other | 78 | 156 | 1.77 [0.98–3.19] | 0.06 | ||||
| Homozygote model | Overall | 1554 | 2348 | 1.13 [0.98–1.30] | 0.88 | Fixed | 33 | 0.22 |
| (CC vs AA) | Asian | 1491 | 2218 | 1.18 [1.02–1.35] | 0.02 | Fixed | 0 | 0.54 |
| Other | 63 | 130 | 1.77 [0.98–3.19] | 0.06 | ||||
OR, odds ratio; 95%CI, 95% confidence interval.
Figure 2.
Association between rs3212227 polymorphism and hepatocellular carcinoma risk in the dominant model.
M-H, Mantel–Haenszel; CI, confidence interval.
Subgroup analysis according to ethnicity revealed significant differences between Asian and other ethnicities (2,180 cases, 3,066 controls; dominant model: CC+AC vs. AA, OR 1.22, 95%CI 1.07–1.38, P = 0.002; allele model: C vs. A, OR 1.13, 95%CI 1.04–1.23, P = 0.005; homozygote model: CC vs. AA, OR 1.18, 95%CI 1.02–1.35, P = 0.02). These results indicated a significant association between the IL-12 rs3212227 polymorphism and HCC risk in the Asian population (Table 2).
Meta-analysis of association between rs568408 polymorphism and HCC risk
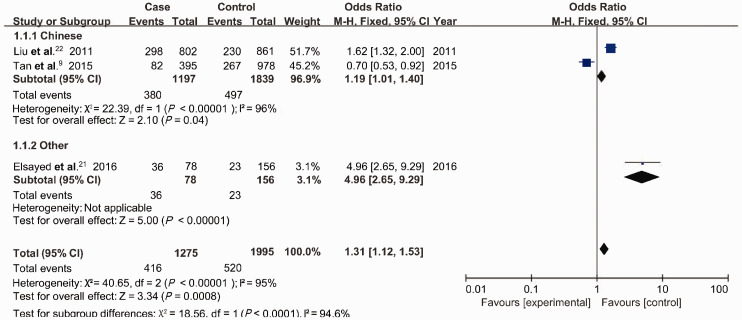
The associations between rs3212227 polymorphism and HCC risk for five models and various subgroups are shown in Table 3. The rs568408 polymorphism was significantly associated with increased risk of HCC in the overall study population (1,342 cases, 2,026 controls). The significant association between the rs568408 polymorphism and HCC risk was supported by ORs of 1.13 in the dominant model (AA+AG vs. GG: 95%CI 1.01–1.28, P<0.001) (Figure 3) and 1.72 in the recessive model (AA vs. AG+GG: 95%CI 1.07–2.78, P = 0.003). Similar trends were observed for the allele (A vs. G: OR = 1.29, 95%CI 1.12–1.48, P<0.001), heterozygote (AG vs. GG: OR = 1.27, 95%CI 1.08–1.49, P<0.001), and homozygote models (AA vs. GG: OR = 1.17, 95%CI 1.88–3.03, P = 0.009) (Table 3).
Table 3.
Meta-analysis of association of rs568408 polymorphisms with risk of hepatocellular carcinoma.
| Genotype comparison and genetic model | Group and subgroups |
Sample size (n) |
OR [95% CI] | P value | Analysis model |
Test of heterogeneity |
||
|---|---|---|---|---|---|---|---|---|
| Case | Control | I2 (%) | P value | |||||
| Dominant model | Overall | 1275 | 1995 | 1.13 [1.01–1.28] | 0.0008 | Random | 95 | <0.001 |
| (AA+AG vs GG) | Chinese | 1197 | 1839 | 3.13 [2.00–4.90] | 0.04 | Random | 95 | <0.001 |
| Other | 78 | 156 | 1.21 [1.08–1.36] | 0.001 | Random | |||
| Recessive model | Overall | 1197 | 1839 | 1.72 [1.07–2.78] | 0.003 | Random | 68 | 0.04 |
| (AA vs AG+GG) | Chinese | 1275 | 1995 | 1.40 [0.81–2.42] | 0.23 | Random | 74 | 0.05 |
| Other | 78 | 156 | 3.68 [1.28–10.53] | 0.02 | Random | |||
| Allele model | Overall | 2550 | 3990 | 1.29 [1.12–1.48] | 0.0003 | Random | 95 | <0.001 |
| (A vs G) | Chinese | 2394 | 3678 | 1.18 [1.02–1.36] | 0.03 | Random | 95 | <0.001 |
| Other | 156 | 312 | 4.08 [2.44–6.82] | <0.00001 | Random | |||
| Heterozygote model | Overall | 1238 | 1958 | 1.27 [1.08–1.49] | 0.0004 | Random | 94 | <0.001 |
| (AG vs GG) | Chinese | 1170 | 1808 | 1.18 [1.00–1.39] | 0.06 | Random | 95 | <0.001 |
| Other | 68 | 150 | 4.84 [2.40–9.78] | <0.00001 | Random | |||
| Homozygote model | Overall | 890 | 1512 | 1.88 [1.17–3.03] | 0.009 | Random | 79 | 0.009 |
| (AA vs GG) | Chinese | 844 | 1373 | 1.46 [0.85–2.51] | 0.17 | Random | 81 | 0.02 |
| Other | 52 139 | 5.28 [1.81–15.39] | 0.002 | Random | ||||
OR, odds ratio; 95%CI, 95% confidence interval.
Figure 3.
Association between rs568408 polymorphism and hepatocellular carcinoma risk in the dominant model.
M-H, Mantel–Haenszel.
Subgroup analysis of a study conducted in China that included 1264 cases and 1870 control showed that the rs568408 polymorphism was associated with HCC risk in the dominant (AA+AG vs. GG: OR 3.13, 95%CI 2.00–4.90, P = 0.04), allele (A vs. G: OR 1.18, 95%CI 1.02–1.36, P = 0.03), and heterozygote models (AG vs. GG: OR 1.18, 95%CI 1.00–1.39, P = 0.06) (Table 3). These results indicated that the A allele of rs568408 might be a genetic factor associated with increased risk of HCC in China.
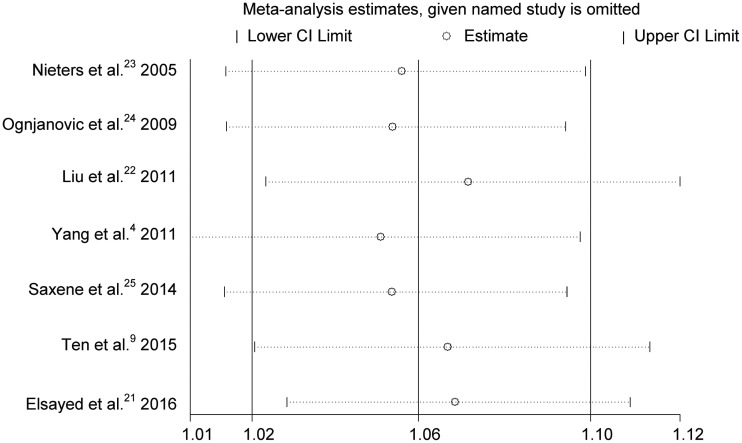
Sensitivity analysis
Sensitivity analysis was conducted by excluding one study at a time to assess the influence on the overall OR. Sensitivity analysis of the studies investigating the rs3212227 polymorphism showed that the results were credible (Figure 4).
Figure 4.
Sensitivity analysis for IL12 polymorphisms at rs3212227.
CI, confidence interval.
Discussion
Numerous studies have evaluated the associations between various polymorphisms of the IL-12 gene and HCC risk, but the results have been inconsistent. We therefore conducted a qualitative meta-analysis to explore the potential association between inherited variations in the IL-12 gene and HCC susceptibility in a large patient cohort. Our findings indicated that the rs3212227 and rs568408 polymorphisms were significantly correlated with HCC risk.
Elsayed et al.21 and Liu et al.22 previously showed that the IL-12 rs568408 A allele polymorphism was significantly correlated with HCC susceptibility. However, Tan et al.9 reported conflicting findings, and found no significant association between this polymorphism and an increased risk for HCC. Xiao et al.27 identified the STAT4 rs7574865 polymorphism as a risk factor for HCC, but failed to find any significant association between HCC risk and other polymorphisms in genes related to the IL-12 signaling pathway, including IL12A rs568408 and IL12B rs3212227. In contrast, the current meta-analysis indicated that rs568408 polymorphisms in the IL-12 gene were significantly correlated with HCC risk, suggesting that the AA wild-type IL-12 genotype may protect against HCC tumorigenesis, especially among individuals of Chinese ethnicity. All 1,342 cases and 2,026 controls included in this meta-analysis were infected with HBV or hepatitis C virus (HCV), further suggesting that the IL-12 rs568408 polymorphism might increase the risk of HBV or HCV infection. Regarding the IL-12 rs3212227 polymorphism, several studies21 found no significant association between this polymorphism and HCC susceptibility. However, Saxena et al.25 concluded that mutation of the rs3212227 C allele increased the risk of HCC. Similarly, a previous meta-analysis28 reported a significant association between the rs3212227 polymorphism and HCC tumorigenesis.
Notably, the current meta-analysis included seven studies with 2375 HCC patients and 3445 controls. Most of the included studies reported a significant association between the rs3212227 polymorphism and an increased risk of HCC, especially in Asian patients. Overall, the findings indicated that the IL-12 rs3212227 polymorphism contributes to the development of HCC.
This study demonstrated a clear increase in HCC risk associated with the IL-12 rs568408 polymorphism under all the genetic models investigated, further suggesting that IL-12 may contribute to the etiology of HCC. IL-12 has been shown to be involved in Th1 cell development and interferon-gamma (IFN-γ) production, as well as cell-mediated cytotoxicity,29 and has demonstrated antitumor effects in animal models of melanoma,30 renal carcinoma,31 and colon carcinoma.32 A previous study33 demonstrated that IL-12 enhanced tumor control via the inhibition of tumor angiogenesis and the expansion of intertumoral T regulatory cell populations. As a tumor suppressor, IL-12 increases production of IFN-γ, which increases p53 expression, contributing to tumor cell apoptosis.34 Wang et al.35 showed that IL-12 downregulated the expression of STAT3 and promoted macrophage polarization to the M1-like phenotype, resulting in HCC tumor growth.
Inherited genetic variations in IL12A and IL12B may dramatically alter the cytokine expression and protein structure. IL-12 has been shown to positively regulate the expression of the interferon regulatory factors IRF1 and IRF4, which have demonstrated tumor suppressor activity in cancer.36,37 Moreover, the results of several animal studies indicated that IL-12 regulates the release of cytokines via non-specific immune mechanisms, inhibiting carcinogenesis in vivo.38,39 IL-12 is associated with the clearance of HBV and is highly expressed in hepatic cells in individuals infected with HBV.40,41 Many IL-12 SNPs were recently found to be associated with HCC susceptibility, including rs321222723,42 and rs568408.22 SNPs in the IL-12 gene regulate gene expression levels, leading to dysregulation of the immune response to infection and tumorigenesis.9 These SNPs may also disrupt signaling pathways, dysregulate exonic mRNA, and alter protein expression, contributing to the occurrence of HCC.43
This study had several limitations. First, the control populations in the included studies did not necessarily comprise healthy individuals. Second, the P-values for the HWE test for the rs568408 polymorphism carried out by Liu et al.22 and Tan et al.9 were <0.05, similar to the results of Elsayed et al.,21 which may have led to increased heterogeneity among the studies. Third, we did not adjust for factors that may have influenced the comparison, such as cancer status, sex, age, and of alcohol use. Fourth, the assays used to detect genetic polymorphisms may have varied in terms of sensitivity and specificity, which may have affected the reliability of the results. Fifth, the major studies investigating rs3212227 polymorphism focused entirely on Asian populations, suggesting that larger sample sizes and more sophisticated designs are needed to validate our findings.
This meta-analysis showed that IL-12 polymorphisms were strongly associated with HCC risk. The rs3212227 polymorphism appeared to be related to a higher risk of HCC, while the rs568408 polymorphism also showed a significant association with HCC, suggesting that rs568408 may also lead to increased susceptibility to HCC.
Declaration of conflicting interest
The authors declare that there is no conflict of interest.
Funding
The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by grants from the National Natural Science Foundation of China [81960511]; the Guangxi Natural Science Foundation [2018GXNSFDA281043]; and Open Foundation of Guangxi Key Laboratory of Biological Target Diagnosis and Treatment in 2018.
ORCID iD
Xingcai Chen https://orcid.org/0000-0002-9489-5014
References
- 1.Yeh FS, Yu MC, Mo CC, et al. Hepatitis B virus, aflatoxins, and hepatocellular carcinoma in southern Guangxi, China. Cancer Res 1989; 49: 2506–2509. [PubMed] [Google Scholar]
- 2.Yang HI, Lu SN, Liaw YF, et al. Hepatitis B e antigen and the risk of hepatocellular carcinoma. N Engl J Med 2002; 347: 168–174. [DOI] [PubMed] [Google Scholar]
- 3.Pan X, Wang G. Correlations of IL-23R gene polymorphism with clinicopathological characteristics and prognosis of hepatocellular carcinoma patients after interventional therapy. Genomics 2019; 111: 930–935. [DOI] [PubMed] [Google Scholar]
- 4.Yang Y, Luo C, Feng R, et al. The TNF-α, IL-1B and IL-10 polymorphisms and risk for hepatocellular carcinoma: a meta-analysis. J Cancer Res Clin Oncol 2011; 137: 947–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kalvakolanu DV. Cytokine signaling in cancer: Novel players and pathways. Cytokine 2017; 89: 1–3. [DOI] [PubMed] [Google Scholar]
- 6.Zekri ARN, El Deeb S, Bahnassy AA, et al. Role of relevant immune-modulators and cytokines in hepatocellular carcinoma and premalignant hepatic lesions. World J Gastroenterol 2018; 24: 1228–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O’Shea JJ, Gadina M, Siegel RM. Cytokines and cytokine receptors In: Rich RR, Fleisher TA, Shearer WT, et al. (eds) Clinical Immunology: Principles and Practice. 5th ed Elsevier, 2019, pp.127–155. [Google Scholar]
- 8.Yan J, Smyth MJ, Teng MW. Interleukin (IL)-12 and IL-23 and their conflicting roles in cancer. Cold Spring Harb Perspect Biol 2018: 10: a028530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tan A, Gao Y, Yao Z, et al. Genetic variants in IL12 influence both hepatitis B virus clearance and HBV-related hepatocellular carcinoma development in a Chinese male population. Tumour Biol 2016; 37: 6343–6348. [DOI] [PubMed] [Google Scholar]
- 10.Jiang J, Zhang Y, Peng K, et al. Combined delivery of a TGF-β inhibitor and an adenoviral vector expressing interleukin-12 potentiates cancer immunotherapy. Acta Biomater 2017; 61: 114–123. [DOI] [PubMed] [Google Scholar]
- 11.Veinalde R, Grossardt C, Hartmann L, et al. Oncolytic measles virus encoding interleukin-12 mediates potent antitumor effects through T cell activation. Oncoimmunology 2017; 6: e1285992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Naif HM. Association of gene polymorphism and serum levels of IL-12p40 with the susceptibility to non-Hodgkin’s lymphoma in Iraq. Journal of Contemporary Medical Sciences 2017; 3. [Google Scholar]
- 13.Taheri M, Naderi M, Hashemi M, et al. Association between IL12A rs568408, IL12B rs3212227 and IL-12 receptor rs383483 polymorphisms and risk of pulmonary tuberculosis. Archives of Clinical Infectious Diseases 2017; 12. DOI: 10.5812/archcid.39318. [Google Scholar]
- 14.Paradowska‐Gorycka A, Sowinska A, Stypińska B, et al. IL‐12B gene polymorphisms and IL‐12 p70 serum levels among patients with rheumatoid arthritis. Scand J Immunol 2017; 85: 147–154. [DOI] [PubMed] [Google Scholar]
- 15.Zheng Y, Wang M, Tian T, et al. Role of interleukin-12 gene polymorphisms in the onset risk of cancer: a meta-analysis. Oncotarget 2017; 8: 29795–29807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sepesi B, Ye Y, Mitchell KG, et al. Genetic variants in cytokine signaling pathways and clinical outcomes in early-stage lung cancer patients. J Thorac Cardiovasc Surg 2018; 155: 2635–2645.e15. [DOI] [PubMed] [Google Scholar]
- 17.Shi X, Jia Y, Xie X, et al. Single-nucleotide polymorphisms of the IL-12 gene lead to a higher cancer risk: a meta-analysis based on 22,670 subjects. Genes Genet Syst 2018: 92: 173–187. [DOI] [PubMed] [Google Scholar]
- 18.Ben-Selma BW, Laribi AB, Alibi S, et al. Interaction analysis of IL-12A and IL-12B gene variants with chronic hepatitis B infection in Tunisian patients. Immunol Lett 2020; 225: 50–56. [DOI] [PubMed] [Google Scholar]
- 19.Wang HW, Gao HL, Wei XX, et al. Up-regulation of IL-12 expression in patients with chronic hepatitis B is mediated by the PI3K/Akt pathway. Mol Cell Biochem 2015; 407: 135–142. [DOI] [PubMed] [Google Scholar]
- 20.Yin D, Wang Y, Sai W, et al. HBx-induced miR-21 suppresses cell apoptosis in hepatocellular carcinoma by targeting interleukin-12. Oncol Rep 2016; 36: 2305–2312. [DOI] [PubMed] [Google Scholar]
- 21.Elsayed HM, Nabiel Y, Sheta T. IL12 gene polymorphism in association with hepatocellular carcinoma in HCV-infected Egyptian patients. Immunol Invest 2017; 46: 123–133. [DOI] [PubMed] [Google Scholar]
- 22.Liu L, Xu Y, Liu Z, et al. IL12 polymorphisms, HBV infection and risk of hepatocellular carcinoma in a high‐risk Chinese population. Int J Cancer 2011; 128: 1692–1696. [DOI] [PubMed] [Google Scholar]
- 23.Nieters A, Yuan JM, Sun CL, et al. Effect of cytokine genotypes on the hepatitis B virus‐hepatocellular carcinoma association. Cancer 2005; 103: 740–748. [DOI] [PubMed] [Google Scholar]
- 24.Ognjanovic S, Yuan JM, Chaptman AK, et al. Genetic polymorphisms in the cytokine genes and risk of hepatocellular carcinoma in low-risk non-Asians of USA. Carcinogenesis 2009; 30: 758–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saxena R, Chawla YK, Verma I, et al. Effect of IL-12B, IL-2, TGF-β1, and IL-4 polymorphism and expression on hepatitis B progression. J Interferon Cytokine Res 2014; 34: 117–128. [DOI] [PubMed] [Google Scholar]
- 26.Yan Y, Xiaoqiang Q, Hongping Y, et al. Polymorphism of TGF-β1 and IL-12B gene and risk of hepatocellular cancer in Guangxi—a case-control study. Chin J Public Health 2011: 1383–1385. [Google Scholar]
- 27.Xiao Y, Liu G, Gong L. Systematic review and meta-analysis on the association between polymorphisms in genes of IL-12 signaling pathway and hepatocellular carcinoma risk. J Cancer 2018; 9: 3583–3592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen H, Cheng S, Wang J, et al. Interleukin-12B rs3212227 polymorphism and cancer risk: a meta-analysis. Mol Biol Rep 2012; 39:10235–10242. [DOI] [PubMed] [Google Scholar]
- 29.Zundler S, Neurath MF. Interleukin-12: Functional activities and implications for disease. Cytokine Growth Factor Rev 2015; 26:559–568. [DOI] [PubMed] [Google Scholar]
- 30.Lampreht Tratar U, Kos S, Kamensek U. Antitumor effect of antibiotic resistance gene-free plasmids encoding interleukin-12 in canine melanoma model. Cancer Gene Ther 2018; 25: 260–273. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Y, Luo CL, He BC, et al. Exosomes derived from IL-12-anchored renal cancer cells increase induction of specific antitumor response in vitro: a novel vaccine for renal cell carcinoma. Int J Oncol 2010; 36: 133–140. [PubMed] [Google Scholar]
- 32.Tasaki K, Yoshida Y, Maeda T, et al. Protective immunity is induced in murine colon carcinoma cells by the expression of interleukin-12 or interleukin-18, which activate type 1 helper T cells. Cancer Gene Ther 2000; 7: 247–254. [DOI] [PubMed] [Google Scholar]
- 33.Cao X, Leonard K, Collins LI, et al. Interleukin 12 Stimulates IFN-γ–Mediated Inhibition of Tumor-Induced Regulatory T-Cell Proliferation and Enhances Tumor Clearance. Cancer Res 2009; 69: 8700–8709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takaoka A, Hayakawa S, Yanai H, et al. Integration of interferon-α/β signalling to p53 responses in tumour suppression and antiviral defence. Nature 2003; 424: 516–523. [DOI] [PubMed] [Google Scholar]
- 35.Wang Q, Cheng F, Ma TT, et al. Interleukin-12 inhibits the hepatocellular carcinoma growth by inducing macrophage polarization to the M1-like phenotype through downregulation of Stat-3. Mol Cell Biochem 2016; 415: 157–168. [DOI] [PubMed] [Google Scholar]
- 36.Lehtonen A, Lund R, Lahesmaa R, et al. IFN-α and IL-12 activate IFN regulatory factor 1 (IRF-1), IRF-4, and IRF-8 gene expression in human NK and T cells. Cytokine 2003; 24: 81–90. [DOI] [PubMed] [Google Scholar]
- 37.Pathak S, Ma S, Trinh L, et al. IRF4 is a suppressor of c-Myc induced B cell leukemia. PLoS One 2011; 6: e22628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Noguchi Y, Jungbluth A, Richards EC, et al. Effect of interleukin 12 on tumor induction by 3-methylcholanthrene. Proc Natl Acad Sci U S A 1996; 93: 11798–11801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nanni P, Nicoletti G, De Giovanni C, et al. Combined allogeneic tumor cell vaccination and systemic interleukin 12 prevents mammary carcinogenesis in HER-2/neu transgenic mice. J Exp Med 2001; 194: 1195–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Becskei A, Grusby MJ. Contribution of IL‐12R mediated feedback loop to Th1 cell differentiation. FEBS Lett 2007; 581: 5199–5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoshimoto T, Takeda K, Tanaka T, et al. IL-12 up-regulates IL-18 receptor expression on T cells, Th1 cells, and B cells: synergism with IL-18 for IFN-γ production. J Immunol 1998; 161: 3400–3407. [PubMed] [Google Scholar]
- 42.Han SS, Cho EY, Lee TS, et al. Interleukin-12 p40 gene (IL12B) polymorphisms and the risk of cervical cancer in Korean women. Eur J Obstet Gynecol Reprod Biol 2008; 140: 71–75. [DOI] [PubMed] [Google Scholar]
- 43.Howell WM, Rose-Zerilli MJ. Cytokine gene polymorphisms, cancer susceptibility, and prognosis. J Nutr 2007; 137: 194S–199S. [DOI] [PubMed] [Google Scholar]