Abstract
Objective
To explore the related influencing mechanism of miR-384 and miR-134-5p acting on Yin Yang 1 (YY1) signaling transduction on the biological function of gastric cancer (GC) cells.
Methods
miR-384, miR-134-5p and YY1 levels in human GC cell lines KATO III, MKN-45, SNU-1 and normal gastric cell line GES-1 were measured by polymerase chain reaction (PCR). Dual luciferase reporter (DLR) gene assay and Western blot (WB) were employed for correlation analysis between miR-384, miR-134-5p and YY1. miR-384-inhibitor, miR-384-mimics, empty plasmid (miRNA-NC) and sh-YY1 were transfected into KATO III cells. Cell proliferation was determined by 3-(4,5-Dimethylthiazolyl-2)-2,5-Diphenyl Tetrazolium Bromide (MTT), cell invasion was measured by Transwell, and apoptosis was analyzed by flow cytometry (FC).
Results
In KATO III, MKN-45 and SNU-1 cell lines, YY1 was upregulated while miR-384 and miR-134-5p were downregulated (P<0.001). The expression of miR-134-5p in the miR-134-5p-inhibitor group was significantly lower (P<0.001), while that in the miR-134-5p-mimics group was significantly higher (P<0.001). The expression of miR-384 in the miR-384-inhibitor group was significantly lower (P<0.001), and that in the miR-384-mimics group was significantly higher as compared to the NC group (P<0.001). Both miR-384 and miR-134-5p overexpression could inhibit cell proliferation and invasion, and promote apoptosis. As detected by WB, overexpressed miR-384 and miR-134-5p inhibited the expression of EMT-related molecular markers. Compared with sh-YY1, the number of cells in S phase decreased, the pro-apoptotic proteins boosted statistically, and the anti-apoptotic proteins declined notably after transfecting miR-134-5p-mimics/sh-YY1 or miR-384-mimics/sh-YY1 (P<0.05). The tumor growth rate of nude mice in miR-134-5p/sh-YY1 and miR-384/sh-YY1 groups were significantly lower than those in sh-YY1 group (all P<0.001).
Conclusion
By targeting YY1 signaling transduction, miR-134-5p and miR-384 can alter the growth and apoptosis of GC cells, which are promising targets for new therapeutics of GC.
Keywords: miR-384, miR-134-5p, YY1, gastric cancer cells, proliferation
Introduction
As one of the most pervasive malignancies, gastric cancer (GC) is the second leading cause of cancer-related death worldwide.1,2 Although recent years have witnessed the significant decline in mortality of GC, the number of patients diagnosed each year increases constantly due to population growth.3 What is notable is that the lack of suitable screening and treatment biomarkers has resulted in dreadful survival rate of GC patients. Therefore, the call for investigation in the mechanisms underlying the onset and progression of GC is pressing.4
Endogenous RNAs as they are, microRNAs (miRNAs) can reversely regulate approximately 30% of human genes through binding to the complementary 3 ‘untranslated region (3ʹ-UTR).5 According to reports, the role of miRNAs varies in different cancers.6,7 Of these, miR-384 and miR-134-5p are recently discovered miRNAs associated with GC. miR-384 plays an inhibitory role in gastric cancer, suggesting that miR-384/MTDH axis may be a potential therapeutic target for gastric cancer.8 LncRNA LUCAT1 can promote proliferation and invasion of gastric cancer through the regulation of miR-134-5p/YWHAZ axis.9 MiR-384, which possesses tumor inhibitory effect on breast cancer (BC) and non-small cell lung cancer (NSCLC), is found to be down-regulated in multiple human cancers.10 While miR-134-5p overexpression has been confirmed to be capable of reducing SGC7901 cells’ proliferation, colony formation and invasion.11 All these studies verify the value of miRNAs in regulating GC progression. As an ubiquitous transcription factor, Yin Yang 1 (YY1) can drive tumorigenesis, metastasis and drug resistance, and targeting YY1 is deemed to be a potential therapeutic option for various malignant tumors.12,13 Apart from that, YY1 has been shown to be a new transcription factor regulating miRNAs.14,15 However, it is still unknown whether miR-384 and miR-134-5p act on the role and potential mechanism of YY1 in the occurrence and invasion of GC. In order to continuously elucidate the basic mechanism of the initiation and progression of GC, this study is therefore carried out to explore the mechanism of miR-384 and miR-134-5p acting on YY1 to regulate the biological behaviors of KATO III.
Materials and Methods
Main Instruments and Reagents
Human GC cell lines KATO III, MKN-45, SNU-1 and human normal gastric GES 1 cell line (BeNa Culture Collection); Real-time fluorescence quantitative PCR instrument (ABI 7500, Applied Biosystems, USA); Lip 2000 (TaKaRa, Dalian, China), Trizol total RNA isolation kit (TaKaRa, Dalian, China); Annexin V/PI apoptosis detection kit (TaKaRa, Dalian, China); MTT kit (Zeye Biological Technology Co., Ltd., Shanghai, China); SYBR Green qPCR Mix (TOYOBO, Japan); MTT kit (Zeye Biotechnology Co., Ltd., Shanghai, China). Electrochemiluminescence (ECL) reagent (ECL-P-100, Yanxi Biotechnology Co., Ltd., Shanghai, China), bicinchoninic acid (BCA) kit (Fermentas, Canada); Multiskan GO Full Wavelength Microplate Reader (Thermo Fisher Scientific Co., Ltd., China). The design and synthesis of all primer sequences were conducted by Sangong Bioengineering Co., Ltd., Shanghai, China (Table 1).
Table 1.
Primer Sequences
| Gene | Forward | Reverse |
|---|---|---|
| miR-384 | 5ʹ-TGAAAATGTGGACTAGAGCCAGA’ | 5ʹ-CAGACACTCCAGAACAGGGC-3’ |
| miR-134-5p | 5′-CCGCTCGAGCCGGCCTTCCAACCTTTGTC-3′ | 5′-GAATGCGGCCGCTCCCATCATCAATATTTATTGAGCATTTAC-3′ |
| YY1 | 5ʹ-GGCCGGCGTACAGTATAGATGA-3’ | 5ʹ-GTGCAGGGTCCGAGGT-3’ |
| U6 | 5ʹ-CTCGCTTCGGCAGCACA-3’ | 5ʹ-AACGCTTCACGAATTTGCGT-3’ |
| GAPDH | 5ʹ-GGTCTCCTCTGACTTCAACA-3’ | 5ʹ-GTGAGGGTCTCTCTCTTCCT-3’ |
Detection Methods
Cell Culture and Transfection Experiments
Placed in Dulbecco’s Modified Eagle’s Medium (DMEM) containing 10% fetal bovine tissue and penicillin-streptomycin mixed solution, the GC cell lines were cultured in a cell incubator with 5% CO2 under 37°C constant temperature and saturated humidity. Following the instructions of LipofectamineTM 2000 transfection kit, the cells were then transfected with miR-134-5p-inhibitor, miR-134-5p-mimics, empty plasmid (NC), miR-384-mimics and miR-384-inhibitor, respectively. The cells with the greatest difference in mRNA expression were then selected and transfected with the primers, and cultured in a renewed medium containing 10% fetal bovine tissue 6 hours after transfection. After verifying the transfection efficiency with quantitative real-time polymerase chain reaction (qRT-PCR), the cells were collected for subsequent experiments.
QRT-PCR Detection
mRNA expression in tissues and cells was examined by qRT-PCR. The total RNA was isolated from tissues according to Trizol reagent instructions and dissolved in 20 μL DEPC water. Then, reverse transcription was performed on the total RNA using a reverse transcription kit, with the synthesized cDNA taking as the template for qRT-PCR amplification. PCR reaction conditions: 95°C/5 s (40 cycles), denaturation: 95°C/10 s, annealing: 60°C/20 s, extension: 72°C/15 s. Each sample was equipped with three replicate wells. The target genes’ relative amount was calculated according to the result parameters, and their relative quantification was worked out by 2-ΔCt.
Western Blot (WB) Detection
The lysed cells were collected and placed in a centrifuge tube, where they were centrifuged for 10 min at 12,000×g at 4°C, and the obtained supernatant was collected as a protein sample. The protein concentration was determined by BCA method. Then, lysis buffer was added to dilute the protein sample to a concentration of 20 mg/mL. After that, separation gel (8.00%) and laminated gel (5.00%) were prepared for SDS-PAGE electrophoresis, and then the protein was transferred to a Polyvinylidene Fluoride (PVDF) membrane. Followed by the addition of cyclin D1, β-catenin, c-myc (1:1000) primary antibody, and internal reference β-actin (1:3000) before blocking it at 4°C all night long. After the addition of horseradish peroxidase (HRP)-labeled goat anti-mouse secondary antibody (1:5000), the membrane was incubated at 37°C for 1 h, and then rinsed 3 times with TBST for 5 min each. After that, it was developed with the ECL kit in the dark after the excess liquid on the membrane was blotted. After scanning the protein bands, the grayscale values were analyzed with the aid of Quantity One software obtained from Molecular Devices Corp, The Bay Area, CA, USA.
MTT Assay for Cell Proliferation
Twenty-four hours after transfection, the cells were harvested and adjusted to 5 × 103 cells/well. Then the cells were inoculated at 37°C on 96-well plates. After that, 20 μL MTT solution with a concentration of 5 μmg/mL was added at each time point, followed by the removal of MTT and the addition of 150 μL of dimethyl sulfoxide to the wells. Subsequently, the OD value of each group of cells was measured using a spectrophotometer.
Transwell Invasion Experiment
Transwell chamber was coated with Matrigel and left to stand at 37°C for 30 min. After adjusting the cells to 4 × 105 cells/mL in tissue-free DMEM medium, 200 μL of it was put into the upper chamber, while 800 μL of DMEM (10% fetal bovine tissue) was added into the lower one. After continuous culture for 24–48 h, the chamber was taken out, fixed in 4% paraformaldehyde and stained with hematoxylin before wiping the cells remained in the upper chamber. The cells passing through the chamber basilar membrane, which reflected the cell invasion capacity, were finally counted under an optical microscope with 10 high-power fields randomly selected.
Cell Cycle Analysis
As mentioned earlier, the cell’s DNA content was analyzed using FC. After collecting the NC and transfected cells, they were rinsed with PBS (phosphate buffered saline) twice, immobilized with 70% ethanol, and left at −20°C until analyzed. Then the cells were dyed with 20 μg/mL PI containing 20 μg/mL RNase (without DNase) for 2 h, and analyzed by FC. The percentage of cells in G1, S, and G2 phases was determined by Multicycle Cell Cycle Software.
Apoptosis Experiment
The cells were processed for 1-minute digestion with 0.25% trypsin and PBS rinsing for 3 times (5 min each time) 48 h after transfection. Then they were resuspended in 100 μL AnnexinV binding buffer to configure to 1×106 cells/mL, added with 5 μL of 20 μg/mL Annexin-V-Fluorescein isothiocyanate (FITC), and cultured at 4°C for 15 min. Followed by the addition of 5 μL propidium iodide (PI) staining solution, and then the culture was resumed for 2 min at 4°C. Cell apoptosis was measured by FC within 30 min.
Xenograft Tumor Model
Female BALB/C nude mice aged 4 weeks were raised in a sterile environment, and then stable miR-134-5p/sh-YY1, miR-384/sh-YY1 and sh-YY1 and their control plasmids 3 x 106 KATO III cells were injected subcutaneously into the left abdomen of nude mice. Tumor growth was detected every 7 days in each group of 5 nude mice. After injection for 28 days, the mice were killed by cervical dislocation and the size and mass of tumors in vivo were accurately measured.
Statistical Methods
The data were statistically analyzed using SPSS 19.0 (SPSS, Inc, Chicago, IL, USA). Data in line with normal distribution were expressed as mean ± standard deviation (mean ± SD). The measurement data were compared by independent sample t-test between groups. Multi-time points data were compared by repeated measures analysis of variance (ANOVA), and Bonferroni method was used for post hoc testing. Mean comparison among multiple groups was carried out by one-way ANOVA, and post hoc tests were done by LSD-t test. The receiver operating characteristic (ROC) curve was utilized for diagnostic value assessment. Pearson test was employed for correlation analysis. P<0.001 meant that the difference was statistically significant.
Results
miR-134-5p and miR-384 Expression in Cells and Their Effects on Cell EMT Function
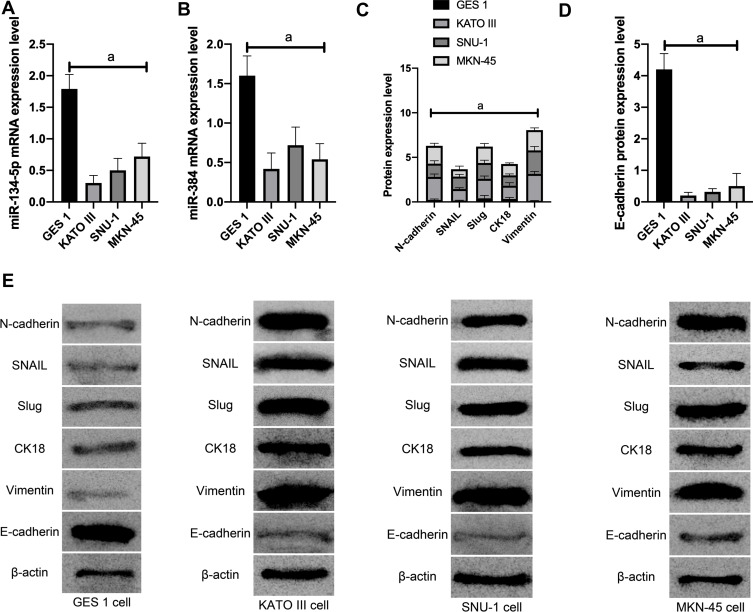
(1) MiR-134-5p and miR-384 expression in each cell line was detected by qRT-PCR. Compared with normal gastric cells GES-1, miR-134-5p and miR-384 mRNA expression levels were notably down-regulated in KATO III, SNU-1 and MKN-45 cells (P<0.001).
(2) WB was utilized to detect the expression of EMT-related marker molecules in the cell lines of each group. Compared with GES 1 of normal human gastric cells, N-cadherin, SNAIL, Slug, CK18 and Vimentin were dramatically up-regulated (P<0.001) and E-cadherin was noticeably down-regulated (P <0.001) in KATO III, SNU-1 and MKN-45 cells (Figure 1).
Figure 1.
The effect of miR-134-5p and miR-384 on cell EMT function. A: mRNA expression of miR-134-5p in cells; B: mRNA expression of miR-384 in cells; C: expression of EMT related marker molecules; D: expression of EMT related marker molecules; E: EMT related marker molecules WB picture; a means P<0.001
Transfection
KATO III, which showed the greatest expression difference, was selected for transfection. As demonstrated by QRT-PCR, miR-134-5p was significantly downregulated in the miR-134-5p-inhibitor group (P<0.001) while was remarkably upregulated in the miR-134-5p-mimics group after transfection (P<0.001) compared with the NC group. MTT results revealed that the proliferation capacity of the miR-134-5p-inhibitor group was obviously enhanced, and that of the miR-134-5p-mimics group was weakened compared with the NC group (both P<0.001). FC results demonstrated that the apoptosis rate of the miR-134-5p-inhibitor group was notably reduced, and that of the miR-134-5p-mimics group was remarkably increased when compared with the NC group (P<0.001). Transwell experiment results identified that the cell invasion capacity was markedly enhanced in the miR-134-5p-inhibitor group than in the NC group, and that in the miR-134-5p-mimics group was evidently reduced (P<0.001) than the NC group. WB results exhibited that in KATO III, N-cadherin, SNAIL, Slug, CK18 and Vimentin were notably increased (P<0.001), while E-cadherin was statistically reduced in the miR-134-5p-inhibitor group compared with the NC group (P<0.001) (Figure 2).
We selected the most differentially expressed cell line KATO III for transfection. As revealed by qRT-PCR, among miR-384-inhibitor, miR-384-mimics, and NC groups, miR-384 showed the highest level in the miR-384-mimics group, followed by the NC groups, and the highest in the miR-384-inhibitor group after transfection (P<0.001) (Figure 3).
Figure 2.
Effects of miR-134-5p on the biological functions of KATO III cells. A: The expression of miR-134-5p after transfection of KATO III cells; B: The proliferation of KATO III cells after transfection. C: KATO III cell cycle after transfection. D: KATO III cell apoptosis after transfection. E: KATO III cell invasion after transfection; F: EMT related marker molecule expression after transfection. G: WB picture.
Note: a means P<0.001.
Figure 3.
Effects of miR-384 on the biological function of KATO III cells. A: miR-384 expression after transfection of KATO III cells; B: proliferation of KATO III cells after transfection. C: KATO III cell cycle after transfection. D: KATO III cell apoptosis after transfection. E: KATO III cell invasion after transfection; F: EMT related marker molecule expression after transfection. G: WB picture.
Note: a means P<0.001.
MTT results revealed that the proliferation capacity of the miR-384-inhibitor group was dramatically enhanced, while that of the miR-384-mimics group was reduced (both P<0.001) compared with the NC group. FC results displayed that the apoptosis rate of the miR-384-inhibitor group was noticeably declined, while that of the miR-384-mimics group was notably elevated compared with the NC group (P<0.001). According to Transwell experiment results, the cell invasion capacity of the miR-384-inhibitor group was significantly enhanced while that of the miR-384-mimics group was markedly reduced compared with the NC group (P<0.001). The results of WB showed that in KATO III, SNAIL, Slug, CK18 and Vimentin were evidently up-regulated (P<0.001) and E-cadherin was obviously down-regulated in the miR-384-inhibitor group compared with the NC group (P<0.001) (Figure 3).
YY1 Was a Functional Target for miR-134-5p and miR-384 to Inhibit GC Cells
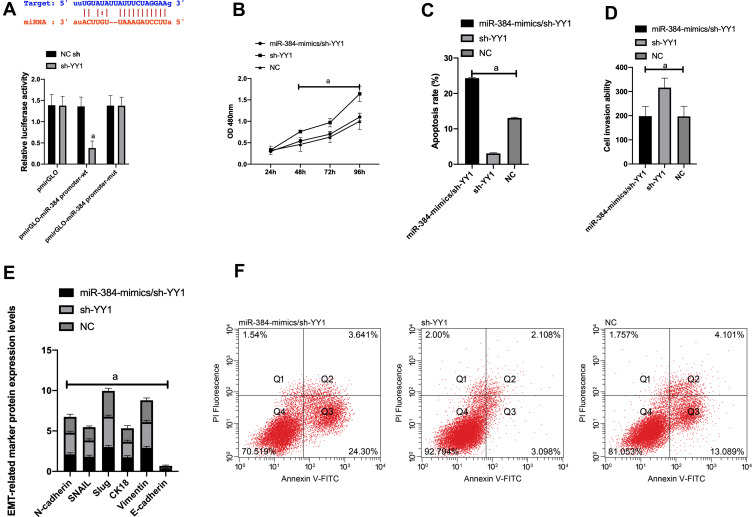
First of all, it was predicted by Targetscan6.2 that YY1 had complementary binding sequences with both miR-134-5p and miR-384 in its 3ʹ-UTR. Further, we conducted a dual luciferase activity assay to find that miR-384 or miR-134-5p overexpression could statistically reduce the luciferase activity of YY1-Wt transfected cells (P<0.001), but had nothing to do with YY1-Mut luciferase activity (P>0.05). After WB detection, it was observed that the YY1 in KATO III was noticeably down-regulated after transfecting miR-134-5p-mimics and miR-384-mimics, while was greatly up-regulated in KATO III after transfection of miR-134-5p-inhibitor and miR-384-inhibitor (P<0.001) (Figures 4 and 5).
Figure 4.
Dual luciferin activity detection. A: There is a binding site between miR-134-5p and YY1, relative luciferase activity-dual luciferase report test. B: KATO III cell viability after transfection. C: Apoptosis. D: Cell invasion. E: EMT situation after transfection. F: Cell flow cytometry.
Note: a means P<0.001.
Figure 5.
Dual luciferin activity detection. A: There is a binding site between miR-384 and YY1, relative luciferase activity-dual luciferase report test. B: KATO III cell viability after transfection. C: Apoptosis. D: Cell invasion. E: EMT situation after transfection. F: Cell flow cytometry.
Note: a means P<0.001.
(1) In order to verify the correlation between YY1 and miR-134-5p, we cotransfected miR-134-5p-mimics and sh-YY1 into KATO III to explore the role of YY1 in GC. WB identified an evident increase in YY1 protein level in the sh-YY1 group compared with the NC group (P<0.001). The inhibitory effect of miR-134-5p-mimics on YY1 level could be reversed by sh-YY1 transfection. Functional analysis identified that the proliferation and invasion capacity of KATO III enhanced observably and the apoptosis capacity decreased dramatically after transfection of sh-YY1 (both P<0.01). Transfection of miR-134-5p-mimics/sh-YY1 partially reversed the inhibitory effect of miR-134-5p-mimics on apoptosis and EMT of KATO III. All these demonstrated that YY1 was a functional target of miR-134-5p in KATO III.
(2) We further probed into the effect of YY1 on GC by co-transfecting miR-384-mimics and sh-YY1 into KATO III to verify the correlation between YY1 and miR-384. WB results revealed a marked increase in YY1 protein level in the sh-YY1 group compared with the NC group (P<0.001). The inhibition of miR-384-mimics on YY1 level could be reversed by transfection with sh-YY1. Functional analysis identified that after sh-YY1 transfection, the proliferation and invasion capacity of KATO III enhanced evidently and the apoptosis capacity decreased dramatically (both P<0.01). Transfection of miR-384-mimics/sh-YY1 could partially reverse the inhibitory effect of miR-384-mimics on KATO III cell apoptosis and EMT. All these findings indicated the role of YY1 as a functional target of miR-384 in KATO III.
Effects of miR-134-5p/YY1, miR-384/YY1 on KATO III Cell Cycle and Apoptosis-Related Proteins
Further, miR-134-5p-mimics/sh-YY1 and miR-384-mimics/sh-YY1 were transfected into KATO III to detect the biological function of the cells.
Compared with NC, the cells in the S phase increased, pro-apoptotic proteins Bax and Bak decreased remarkably, and Bcl-2, APR3 and Bcl-xl increased dramatically. Whereas, the number of S phase cells decreased, Bax and Bak increased evidently compared with sh-YY1, while Bcl-2, APR3 and Bcl-xl reduced notably (P<0.05) after transfecting miR-134-5p-mimics/sh-YY1 (Figure 6).
Figure 6.
KATO III cell cycle and apoptosis-related proteins. A: Cell cycle changes after miR-134-5p-mimics/sh-YY1 transfection; B: Apoptotic protein; C: WB diagram; D: Cell cycle changes after miR-384-mimics/sh-YY1 transfection ; E: apoptotic protein; F: WB picture; a means P<0.001.
Compared with NC, the S-phase cells increased, pro-apoptotic proteins Bax and Bak decreased remarkably, and Bcl-2, APR3 and Bcl-xl increased dramatically. After transfection of miR-384-mimics/sh-YY1, the S-phase cells increased, Bax and Bak increased evidently compared with sh-YY1, while Bcl-2, APR3 and Bcl-xl reduced notably (P<0.05).
Effects of YY1 Inhibition on Tumor Growth in Nude Mice
We established a tumor formation model by injecting KATO III cells transfected with miR-134-5p/sh-YY1, miR-384/sh-YY1 and sh-YY1 into the abdomen of nude mice. It was found that miR-134-5p/sh-YY1 and miR-384/sh-YY1 groups presented significantly lower tumor growth rate than the sh-YY1 group, and the tumor size and weight obtained after the nude mice were killed were notably lower than those of sh-YY1 group (all P<0.001), with markedly reduced YY1 expression, suggesting that YY1 inhibition can inhibit tumor growth in vivo (Figure 7).
Figure 7.
Effect of tumor growth in nude mice. A: The expression of YY1 in tumor tissue is low. B: Tumor growth rate. C: Tumor weight. D: Tumor volume.
Note: a means P<0.001.
Discussion
The role of specific miRNAs in regulating signaling pathways has attracted extensive attention in the clinical development of new molecular target therapy.16 miRNA presents a unique expression state in different cancer types, which provides a new theoretical basis for GC diagnosis and treatment in molecular biology.17,18 Therefore, we explored the effects of miR-384 and miR-134-5p acting on YY1 signaling transduction on the biological function of GC cells.
In this study, we adopted qRT-PCR technology in cell experiments to find that, compared with normal human gastric cells, miR-134-5p and miR-384 were both downregulated in GC cell lines. Studies have revealed that miR-384 decreased significantly in laryngeal carcinoma, breast cancer and GC cells,19–21 and Lai et al confirmed that overexpression of miR-384 inhibited hepatocellular carcinoma progression.22 Subsequently, we silenced and overexpressed miR-134-5p and miR-384 in KATO III to find that miR-384 or miR-134-5p overexpression could inhibit cell proliferation and invasion, promote apoptosis, and inhibit cell EMT-related molecular markers. Further, we transfected miR-134-5p-mimics, miR-134-5p-inhibitor, miR-384-mimics and miR-384-inhibitor sequences into KATO III to observe the change in cell biological function. It was found that the cell proliferation and invasion capacity was observably inhibited after miR-134-5p or miR-384 overexpression, which suggested that the two may be potential targets for the treatment of GC that can be utilized to affect the proliferation, invasion and EMT of GC cells. In addition, a remarkably down-regulated miR-384 in GC cell lines was also reported.23 By directly targeting MTDH, it was shown to be able to inhibit the proliferation, migration and invasion of GC cells.24 There is also evidence showing that the regulation of miR-134-5p/YWHAZ axis promoted the proliferation and invasion of GC. The preceding research further suggest that miR-134-5p/miR-384 can influence GC through the exploration of miRNA target gene changes.25
We noticed that miR-134-5p and miR-384 both had adjacent binding sites to YY1. As a type of small non-coding RNAs, miRNAs mainly target binding sites in the 3ʹ-UTR. Recently, endogenous miRNAs are reported to be able to identify complementary genomic sites in human gene promoter and interfere with the formation and regulation of heterochromatin in gene transcription. In the present study, DLR was utilized to evaluate the correlation between miR-384 and miR-134-5p. The results showed that miR-384 or miR-134-5p overexpression led to evidently boosted YY1-Wt luciferase activity, but it had little effect on YY1-Mut luciferase activity, suggesting the presence of targeted regulatory network between miR-134-5p, miR-384 and YY1. In HCC827 cells, miR-186 was also found to be down-regulated, while YY1 was up-regulated. Moreover, miR-186 can act on the target gene YY1 in the treatment of human NSCLC.26 Hence, we further transfected miR-134-5p-mimics/sh-YY1 and miR-384-mimics/sh-YY1 into KATO III to detect the biological function of the cells. It was found that after transfection of sh-YY1, the S-phase cells increased, the apoptosis capacity decreased, pro-apoptotic proteins notably decreased, and the inhibitory apoptotic proteins markedly increased. While after miR-134-5p-mimics/sh-YY1 or miR-384-mimics/sh-YY1 transfection, the S-phase cells decreased, pro-apoptotic proteins increased notably, and anti-apoptotic proteins reduced noticeably compared with sh-YY1. This suggested that to some extent, overexpression of miR-134-5p or miR-384 could restore the inhibitory effect of sh-YY1-induced apoptosis on cancer cells.
In this study, we first determined the expression of miR-384 and miR-134-5p in KATO III, and further transfected KATO III with synthesized miRNAs to inquire into the biological roles of miR-384 and miR-134-5p in the biological behaviors. Moreover, functional experiments were carried out to investigate the mechanism of miR-384 and miR-134-5p targeting YY1 in regulating the behavior of KATO III. However, there are still some limitations in this study. To begin with, whether other related miRNAs can affect the occurrence and development of tumors through other signaling pathways needs further study. Moreover, there is a lack of appropriate bioinformatics research on the regulatory network of YY1. Therefore, we hope to explore the regulatory network of YY1 through bioinformatics analysis in future studies, so as to provide more basis for our experiments.
Conclusion
To sum up, miR-134-5p and miR-384 can change the growth and apoptosis of GC cells by targeting YY1 signaling transduction, which are promising potential clinical therapeutic targets for GC.
Disclosure
The authors report no conflicts of interest for this work.
References
- 1.Mei B, Chen J, Yang N, Peng Y. The regulatory mechanism and biological significance of the Snail-miR590-VEGFR-NRP1 axis in the angiogenesis, growth and metastasis of gastric cancer. Cell Death Dis. 2020;11(4):241. doi: 10.1038/s41419-020-2428-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu Y, Shi F, Wang M, Ding J. Knockdown of Rab9 suppresses the progression of gastric cancer through regulation of Akt signaling pathway. Technol Cancer Res Treat. 2020;19:1533033820915958. doi: 10.1177/1533033820915958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ye SP, Shi J, Liu DN, et al. Robotic- versus laparoscopic-assisted distal gastrectomy with D2 lymphadenectomy for advanced gastric cancer based on propensity score matching: short-term outcomes at a high-capacity center. Sci Rep. 2020;10(1):6502. doi: 10.1038/s41598-020-63616-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu H, Liu B, Chen Z, Li G, Zhang Z. MSC-induced lncRNA HCP5 drove fatty acid oxidation through miR-3619-5p/AMPK/PGC1alpha/CEBPB axis to promote stemness and chemo-resistance of gastric cancer. Cell Death Dis. 2020;11(4):233. doi: 10.1038/s41419-020-2426-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang S, Guo Y, Li Z, et al. A systematic review of metabolomic profiling of gastric cancer and esophageal cancer. Cancer Biol Med. 2020;17(1):181–198. doi: 10.20892/j.issn.2095-3941.2019.0348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tian YZ, Liu YP, Tian SC, Ge SY, Wu YJ, Zhang BL. Antitumor activity of ginsenoside Rd in gastric cancer via up-regulation of Caspase-3 and Caspase-9. Pharmazie. 2020;75(4):147–150. doi: 10.1691/ph.2020.9931 [DOI] [PubMed] [Google Scholar]
- 7.Chen JQ, Huang ZP, Li HF, Ou YL, Huo F, Hu LK. MicroRNA-520f-3p inhibits proliferation of gastric cancer cells via targeting SOX9 and thereby inactivating Wnt signaling. Sci Rep. 2020;10(1):6197. doi: 10.1038/s41598-020-63279-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang F. miR-384 targets metadherin gene to suppress growth, migration, and invasion of gastric cancer cells. J Int Med Res. 2019;47(2):926–935. doi: 10.1177/0300060518817171 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Chi J, Liu T, Shi C, et al. Long non-coding RNA LUCAT1 promotes proliferation and invasion in gastric cancer by regulating miR-134-5p/YWHAZ axis. Biomed Pharmacother. 2019;118:109201. doi: 10.1016/j.biopha.2019.109201 [DOI] [PubMed] [Google Scholar]
- 10.Yan Z, Yang Q, Xue M, Wang S, Hong W, Gao X. YY1-induced lncRNA ZFPM2-AS1 facilitates cell proliferation and invasion in small cell lung cancer via upregulating of TRAF4. Cancer Cell Int. 2020;20(1):108. doi: 10.1186/s12935-020-1157-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rong Z, Wang Z, Wang X, Qin C, Geng W. Molecular interplay between linc01134 and YY1 dictates hepatocellular carcinoma progression. J Exp Clin Cancer Res. 2020;39(1):61. doi: 10.1186/s13046-020-01551-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sarvagalla S, Kolapalli SP, Vallabhapurapu S. The two sides of YY1 in cancer: a friend and a foe. Front Oncol. 2019;9:1230. doi: 10.3389/fonc.2019.01230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qiao K, Ning S, Wan L, et al. LINC00673 is activated by YY1 and promotes the proliferation of breast cancer cells via the miR-515-5p/MARK4/Hippo signaling pathway. J Exp Clin Cancer Res. 2019;38(1):418. doi: 10.1186/s13046-019-1421-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang ZC, Wang TR, Li RT, et al. Construction of hybrid DNAs@CP for the rapid synchronous sensing of multiplex microRNAs based on experimental studies and molecular simulation. J Inorg Biochem. 2020;208:111076. doi: 10.1016/j.jinorgbio.2020.111076 [DOI] [PubMed] [Google Scholar]
- 15.Lu S, Wang MS, Chen PJ, Ren Q, Bai P. miRNA-186 inhibits prostate cancer cell proliferation and tumor growth by targeting YY1 and CDK6. Exp Ther Med. 2017;13(6):3309–3314. doi: 10.3892/etm.2017.4387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu XM, Zhang HJ. miRNAs as new molecular insights into inflammatory bowel disease: crucial regulators in autoimmunity and inflammation. World J Gastroenterol. 2016;22(7):2206–2218. doi: 10.3748/wjg.v22.i7.2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang L, Sun J, Cao H. MicroRNA-384 regulates cell proliferation and apoptosis through directly targeting WISP1 in laryngeal cancer. J Cell Biochem. 2019;120(3):3018–3026. doi: 10.1002/jcb.27323 [DOI] [PubMed] [Google Scholar]
- 18.Ma Q, Qi X, Lin X, Li L, Chen L, LncRNA HW. SNHG3 promotes cell proliferation and invasion through the miR-384/hepatoma-derived growth factor axis in breast cancer. Hum Cell. 2020;33(1):232–242. doi: 10.1007/s13577-019-00287-9 [DOI] [PubMed] [Google Scholar]
- 19.Wang L, Su K, Wu H, Li J, Song D. LncRNA SNHG3 regulates laryngeal carcinoma proliferation and migration by modulating the miR-384/WEE1 axis. Life Sci. 2019;232:116597. doi: 10.1016/j.lfs.2019.116597 [DOI] [PubMed] [Google Scholar]
- 20.Lai YY, Shen F, Cai WS, et al. MiR-384 regulated IRS1 expression and suppressed cell proliferation of human hepatocellular carcinoma. Tumour Biol. 2016;37(10):14165–14171. doi: 10.1007/s13277-016-5233-5 [DOI] [PubMed] [Google Scholar]
- 21.Hong Z, Fu W, Wang Q, Zeng Y, Qi L. MicroRNA-384 is lowly expressed in human prostate cancer cells and has anti-tumor functions by acting on HOXB7. Biomed Pharmacother. 2019;114:108822. doi: 10.1016/j.biopha.2019.108822 [DOI] [PubMed] [Google Scholar]
- 22.Song H, Rao Y, Zhang G, Kong X. MicroRNA-384 inhibits the growth and invasion of renal cell carcinoma cells by targeting astrocyte elevated gene 1. Oncol Res. 2018;26(3):457–466. doi: 10.3727/096504017X15035025554553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang Z, Dong X, Pu M, et al. LBX2-AS1/miR-219a-2-3p/FUS/LBX2 positive feedback loop contributes to the proliferation of gastric cancer. Gastric Cancer. 2020;23(3):449–463. doi: 10.1007/s10120-019-01019-6 [DOI] [PubMed] [Google Scholar]
- 24.Huang T, Wang G, Yang L, et al. MiR-186 inhibits proliferation, migration, and invasion of non-small cell lung cancer cells by downregulating Yin Yang 1. Cancer Biomark. 2017;21(1):221–228. doi: 10.3233/CBM-170670 [DOI] [PubMed] [Google Scholar]
- 25.Han S, Lu Q, Apr WN. 3 accelerates the senescence of human retinal pigment epithelial cells. Mol Med Rep. 2016;13(4):3121–3126. doi: 10.3892/mmr.2016.4926 [DOI] [PubMed] [Google Scholar]
- 26.Alonso RS, Solari HP, de Franca Damasceno E, Burnier MNN, Ventura MP. The chemotactic properties of various topical brimonidine tartrate ophthalmic preparations. BMC Pharmacol Toxicol. 2020;21(1):24. doi: 10.1186/s40360-020-0401-z [DOI] [PMC free article] [PubMed] [Google Scholar]