Abstract
Regulation at the post‐transcriptional level is an important mode of gene expression control in bacteria. Small RNA regulators (sRNAs) that act via intramolecular base‐pairing with target mRNAs are key players in this process and most often sequester the target's ribosome binding site (RBS) to down‐regulate translation initiation. Over the past few years, several exceptions from this mechanism have been reported, revealing that sRNAs are able to influence translation initiation from a distance. In this issue of Molecular Microbiology, Azam and Vanderpool show that repression of the manY mRNA by the sRNA SgrS relies on an unconventional mechanism involving a translational enhancer element and ribosomal protein S1. Binding of S1 to an AU‐rich sequence within the 5ʹ untranslated region of the manY transcript promotes translation of the mRNA, and base‐pairing of SgrS to the same site can interfere with this process. Therefore, instead of blocking translation initiation by occluding the manY RBS, SgrS reduces ManY synthesis by inhibiting S1‐dependent translation activation. These findings increase the base‐pairing window for sRNA‐mediated gene expression control in bacteria and highlight the role of ribosomal protein S1 for translation initiation.
Keywords: enhancer, Hfq, post‐transcriptional regulation, small RNA, translation initiation
Small RNAs (sRNAs) are key players in the post‐transcriptional control of bacterial gene expression. Typically, sRNAs base‐pair to the translation initiation site of their target mRNAs, which blocks access of the ribosome and results in repression of target translation. The SgrS sRNA from Escherichia coli is able to influence translation initiation from a distance: Upstream of the ribosome binding site, SgrS competes with ribosomal protein S1 for binding to an AU‐rich site on the manY transcript. Binding of S1 stimulates manY translation, and binding of SgrS interferes with this process.
![]()
1. INTRODUCTION
Control of de novo transcript synthesis in bacteria usually occurs through the interaction of transcription factors with DNA sequences close to or overlapping with promoter elements. However, transcription control can also occur at a distance employing auxiliary regulatory mechanisms (Xu and Hoover, 2001). In analogy, post‐transcriptional gene control through small regulatory RNAs (sRNAs) has long been limited to interactions occurring around the Shine‐Dalgarno sequence and start codon of the target transcript; a view that has now been challenged by several reports.
sRNAs are found everywhere on the bacterial chromosome and on plasmids. Despite considerable diversity in size and structure, the majority of studied sRNAs control gene expression by a common mechanism involving the interaction with an RNA chaperone, e.g., Hfq and/or ProQ, and base‐pairing with trans‐encoded target mRNAs (Hör et al., 2018). sRNAs can activate and repress target mRNAs, and a single sRNA may perform both of these functions (Papenfort and Vanderpool, 2015; Kavita et al., 2018). The base‐pairing sequences of activated targets are typically located upstream of translation control elements, whereas target repression most often occurs at the ribosome binding site. Studies focusing on the regulation of ompN by the RybB sRNA revealed a five codon window downstream of the start codon suitable for translation repression, which overlaps with the sequence space occluded by the 30S ribosomal subunit during translation initiation (Bouvier et al., 2008). While this mode of action may well account for a large fraction of the reported sRNA‐target mRNA interactions, translation initiation control by sRNAs can also occur through alternative mechanisms that are independent of RNA duplex formation at the RBS. While this commentary will only focus on non‐canonical mechanisms of gene repression by sRNAs, it should be pointed out that activation of target genes by sRNAs normally occurs through base‐pairing events upstream of the RBS (Papenfort and Vanderpool, 2015).
2. REPRESSION UPSTREAM OF THE RBS
The 5ʹ UTR (untranslated region) of an mRNA is key for translation initiation control and frequently the site for post‐transcriptional regulation. For example, riboswitches and RNA thermometers are typically contained in 5ʹ UTRs (Serganov and Nudler, 2013), and so are translation enhancer elements and ribosome standby sites. Translation enhancer elements and ribosome standby sites both promote the translation of their respective mRNAs, however, the underlying molecular mechanisms vary. Standby sites are single‐stranded sequences that allow unspecific binding of 30S ribosomes to mRNAs and thereby antagonize the inhibitory effect of otherwise stable RNA structures sequestering the RBS (Unoson and Wagner, 2007). In contrast, translational enhancer sequences seem to promote ribosome liberation during translation initiation (Takahashi et al., 2013). Despite these differences, accumulating evidence suggests that these auxiliary mechanisms of translation initiation require the aid of additional protein factors such as ribosomal protein S1. S1 is an RNA chaperone that unfolds structured mRNAs and, for example, facilitates translation initiation at the ribosome standby sites of the tisAB and lpp mRNAs (Andreeva et al., 2018; Romilly et al., 2019). However, it also interacts with unstructured, AU‐rich enhancer sequences upstream of RBSs and activates translation of the associated mRNAs (Cifuentes‐Goches et al., 2019). Work by Azam and Vanderpool (2020) presented in this issue of Molecular Microbiology now shows that the SgrS sRNA can meddle with this regulatory mechanism and inhibit translation initiation by interfering with binding of ribosomal protein S1 to an AU‐rich enhancer sequence located in the 5ʹ UTR of the manY mRNA (Figure 1). Similarly, masking of the tisAB ribosome standby site by the IstR sRNA inhibits translation of the transcript (Darfeuille et al., 2007), and so does base‐pairing of the GcvB sRNA with CA‐rich translational enhancers of the gltI and yifK mRNAs (Sharma et al., 2007; Yang et al., 2014). A common theme for these cases is therefore the sequestration of translation activator sequences upstream of the RBS by the sRNAs, however, for the gltI and yifK mRNAs it is currently not clear if RNA chaperones such as ribosomal protein S1 are also required in these processes.
FIGURE 1.
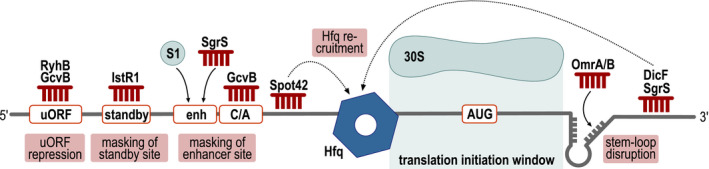
Repression of translation initiation through sRNA base‐pairing outside the RBS. Bacterial sRNAs employ various regulatory mechanisms to regulate the expression of target outside the window of translation initiation. See main text for details
Another RNA chaperone associated with the activity of sRNAs is Hfq. The principle role of Hfq is to act as a molecular matchmaker facilitating RNA duplex formation of sRNAs with their target mRNAs (Woodson et al., 2018). Cellular concentrations of Hfq are limiting and successful use of this resource is achieved by active cycling of RNAs on Hfq (Wagner, 2013). As pointed out above, in the majority of cases the base‐pairing site of the sRNA overlaps with the RBS, which inhibits 30S ribosome recruitment. However, rather than blocking translation initiation themselves, sRNAs can also exert their regulatory function by depositing Hfq at the RBS (Figure 1). Three examples for this mechanisms have been reported so far: whereas Spot42 interacts with the sdhC mRNA upstream of the start codon (Desnoyers and Masse, 2012), the DicF and SgrS sRNAs form RNA‐duplexes with the coding sequences of their targets (Azam and Vanderpool, 2018).
Regulation of gene expression through cis‐acting elements can also involve the activity of additional coding sequences, such as uORFs (upstream open reading frames). uORFs are typically short and therefore frequently overlooked in standard genome annotations (Orr et al., 2019). In Escherichia coli, efficient translation of the fur mRNA, encoding a master regulator of iron homeostasis, is linked to a 28 aa uORF called uof (upstream of Fur). Fur is a transcriptional repressor of the RyhB sRNA, and, vice versa, RyhB prevents fur translation (Vecerek et al., 2007). Detailed analyses of the molecular mechanism underlying RyhB‐mediated repression of fur revealed that RyhB blocks translation initiation of uof and thereby also down‐regulates Fur synthesis (Figure 1). In analogy, GcvB sRNA inhibits the production of the threonine biosynthesis operon by interfering with translation initiation of the thrL leader peptide mRNA and similarly repression of ilvB by GcvB might be due to targeting of the cognate ivbL leader peptide (Sharma et al., 2011). Various ribosome profiling approaches, e.g., Ribo‐RET (retapamulin‐enhanced Ribo‐seq analysis; [Meydan et al., 2019]), have now exposed several previously unknown uORFs, and it will be interesting to test if and how these are involved in post‐transcriptional control mechanisms by sRNAs.
3. REGULATION DOWNSTREAM OF THE RBS
Given that elongating ribosomes function as highly processive helicases which should easily be able to unwind sRNA‐mRNA hybrids (Takyar et al., 2005), the deeper coding sequence of bacterial transcripts has originally not been considered as a promising base‐pairing site. Nevertheless, a number of sRNAs have been identified to regulate expression of their targets through binding downstream of the fifth codon, and thus outside of the region where mRNAs are sensitive to inhibition of translation initiation (Bouvier et al., 2008). Both MicC (Pfeiffer et al., 2009) and SdsR (Fröhlich et al., 2012) do not directly interfere with translation initiation or elongation, and instead act through induction of an RNaseE‐dependent cleavage of the ompD mRNA immediately downstream of the sRNA binding site, resulting in rapid decay of the transcript. Likewise, down‐regulation of fadL and ompA mRNAs by RybB, and lpxR mRNA by MicF (Papenfort et al., 2010; Corcoran et al., 2012), respectively, is based on a similar mechanism.
In contrast to the latter examples, some sRNAs are able to not only destabilize their target mRNAs, but to influence translation initiation from binding sites within the CDS. OmrA and B, two homologous OmpR‐controlled sRNAs, act as repressors of numerous mRNAs through occlusion of the RBS (Jagodnik et al., 2017). Instead, base‐pairing of the conserved 5ʹ end of OmrA/B within the CDS of the fepA transcript interferes with the formation of a short stem‐loop within the mRNA. This cis‐regulatory element increases efficiency of translation initiation, probably by acting as a roadblock to optimize positioning of the 30S subunit. Similar stem‐loop structures have been predicted downstream of the AUG in additional mRNAs, suggesting that this mechanism could be more prevalent than previously anticipated. Association of OmrA/B with Hfq increases the stability of the sRNAs, but the RNA chaperone is, at least in vitro, dispensable for repression of fepA. Other sRNAs binding within the CDS leverage Hfq to influence translation initiation of their mRNA targets, similar to what is observed for Spot42 base‐pairing to the 5’ UTR of sdhC mRNA (see above and [Desnoyers and Masse, 2012]). Both DicF (encoded by the E. coli prophage Qin) and SgrS bind more than 20 nt downstream of the start codon within the manX mRNA, the first cistron of the manXYZ operon. Association of the Hfq‐dependent sRNAs results in deposition of the RNA chaperone more upstream at a site close to the RBS, where it interferes with ribosome binding of manX (Azam and Vanderpool, 2018). Based on the idea that the majority of sRNAs recognize their targets within the 5ʹ UTR or the very beginning of the CDS, binding sites further downstream of the RBS have potentially been overlooked in the past. The advent of global approaches dedicated to the unbiased mapping of base‐pairing sites, including for example CLASH or RIL‐seq (Tree et al., 2014; Melamed et al., 2020), will likely lead to the discovery of more examples of long‐distance regulation, and potentially uncover yet unknown mechanisms.
4. CONCLUDING REMARKS
Interference with translation initiation is the most prevalent mechanism employed by bacterial sRNAs to repress their target transcripts. However, research of the past decade has revealed a repertoire of mechanisms different from the canonical model. sRNAs interacting with sequences up‐ and downstream of the RBS can influence ribosome assembly from a distance by interfering with the activity of standby or enhancer sites, the regulation of uORF translation, the recruitment of protein factors, or structural rearrangements. Yet, more examples for sRNA‐mediated repression through distant base‐pairing sites exist, which include the OmrA/B‐csgD, RybB‐ompC, RybB‐ompF, CpxQ‐cfa, MicV‐ushA, and ChvR‐chvT mRNA interactions (Holmqvist et al., 2010; Papenfort et al., 2010; Fröhlich et al., 2018; Bianco et al., 2019; Peschek et al., 2019). The exact regulatory mechanisms for all these latter sRNA‐target pairs still remains to be determined and it will be exciting to uncover the ever increasing versatility of bacterial non‐coding RNAs within the next years.
CONFLICT OF INTEREST
The authors have no conflicts of interest to declare.
ACKNOWLEDGMENTS
Work in the Papenfort laboratory is supported by the DFG (SPP2002, TRR174, PA2820/1‐2, and EXC2051‐ID390713860), the Human Frontier Science Program (CDA00024/2016‐C), the European Research Council (StG‐758212), and the Vallee Foundation.
Fröhlich KS, Papenfort K. Regulation outside the box: New mechanisms for small RNAs. Mol Microbiol. 2020;114:363–366. 10.1111/mmi.14523
Contributor Information
Kathrin S. Fröhlich, Email: kathrin.froehlich@uni-jena.de.
Kai Papenfort, Email: kai.papenfort@uni-jena.de.
REFERENCES
- Andreeva, I. , Belardinelli, R. and Rodnina, M.V. (2018) Translation initiation in bacterial polysomes through ribosome loading on a standby site on a highly translated mRNA. Proceedings of the National Academy of Sciences of the United States of America, 115, 4411–4416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azam, M.S. and Vanderpool, C.K. (2018) Translational regulation by bacterial small RNAs via an unusual Hfq‐dependent mechanism. Nucleic Acids Research, 46, 2585–2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azam, M.S. and Vanderpool, C.K. (2020) Translation inhibition from a distance: the small RNA SgrS silences a ribosomal protein S1‐dependent enhancer. Molecular Microbiology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco, C.M. , Fröhlich, K.S. and Vanderpool, C.K. (2019) Bacterial cyclopropane fatty acid synthase mRNA is targeted by activating and repressing small RNAs. Journal of Bacteriology, 201(19), e00461‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier, M. , Sharma, C.M. , Mika, F. , Nierhaus, K.H. and Vogel, J. (2008) Small RNA binding to 5' mRNA coding region inhibits translational initiation. Molecular Cell, 32, 827–837. [DOI] [PubMed] [Google Scholar]
- Cifuentes‐Goches, J.C. , Hernandez‐Ancheyta, L. , Guarneros, G. , Oviedo, N. and Hernandez‐Sanchez, J. (2019) Domains two and three of Escherichia coli ribosomal S1 protein confers 30S subunits a high affinity for downstream A/U‐rich mRNAs. Journal of Biochemistry, 166, 29–40. [DOI] [PubMed] [Google Scholar]
- Corcoran, C.P. , Podkaminski, D. , Papenfort, K. , Urban, J.H. , Hinton, J.C. and Vogel, J. (2012) Superfolder GFP reporters validate diverse new mRNA targets of the classic porin regulator, MicF RNA. Molecular Microbiology, 84, 428–445. [DOI] [PubMed] [Google Scholar]
- Darfeuille, F. , Unoson, C. , Vogel, J. and Wagner, E.G. (2007) An antisense RNA inhibits translation by competing with standby ribosomes. Molecular Cell, 26, 381–392. [DOI] [PubMed] [Google Scholar]
- Desnoyers, G. and Masse, E. (2012) Noncanonical repression of translation initiation through small RNA recruitment of the RNA chaperone Hfq. Genes & Development, 26, 726–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fröhlich, K.S. , Forstner, K.U. and Gitai, Z. (2018) Post‐transcriptional gene regulation by an Hfq‐independent small RNA in Caulobacter crescentus . Nucleic Acids Research, 46, 10969–10982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fröhlich, K.S. , Papenfort, K. , Berger, A.A. and Vogel, J. (2012) A conserved RpoS‐dependent small RNA controls the synthesis of major porin OmpD. Nucleic Acids Research, 40, 3623–3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmqvist, E. , Reimegard, J. , Sterk, M. , Grantcharova, N. , Romling, U. and Wagner, E.G. (2010) Two antisense RNAs target the transcriptional regulator CsgD to inhibit curli synthesis. EMBO Journal, 29, 1840–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hör, J. , Gorski, S.A. and Vogel, J. (2018) Bacterial RNA biology on a genome scale. Molecular Cell, 70, 785–799. [DOI] [PubMed] [Google Scholar]
- Jagodnik, J. , Chiaruttini, C. and Guillier, M. (2017) Stem‐loop structures within mRNA coding sequences activate translation initiation and mediate control by small regulatory RNAs. Molecular Cell, 68(158–170), e153. [DOI] [PubMed] [Google Scholar]
- Kavita, K. , de Mets, F. and Gottesman, S. (2018) New aspects of RNA‐based regulation by Hfq and its partner sRNAs. Current Opinion in Microbiology, 42, 53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melamed, S. , Adams, P.P. , Zhang, A. , Zhang, H. and Storz, G. (2020) RNA‐RNA Interactomes of ProQ and Hfq reveal overlapping and competing roles. Molecular Cell, 77, 411–425.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meydan, S. , Marks, J. , Klepacki, D. , Sharma, V. , Baranov, P.V. , Firth, A.E. , et al (2019) Retapamulin‐assisted ribosome profiling reveals the alternative bacterial proteome. Molecular Cell, 74, 481–493.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr, M.W. , Mao, Y. , Storz, G. and Qian, S.B. (2019) Alternative ORFs and small ORFs: shedding light on the dark proteome. Nucleic Acids Research, 48(3), 1029–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenfort, K. , Bouvier, M. , Mika, F. , Sharma, C.M. and Vogel, J. (2010) Evidence for an autonomous 5ʹ target recognition domain in an Hfq‐associated small RNA. Proceedings of the National Academy of Sciences of the United States of America, 107, 20435–20440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenfort, K. and Vanderpool, C.K. (2015) Target activation by regulatory RNAs in bacteria. FEMS Microbiology Reviews, 39, 362–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschek, N. , Hoyos, M. , Herzog, R. , Forstner, K.U. and Papenfort, K. (2019) A conserved RNA seed‐pairing domain directs small RNA‐mediated stress resistance in enterobacteria. EMBO Journal, 38, e101650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer, V. , Papenfort, K. , Lucchini, S. , Hinton, J.C. and Vogel, J. (2009) Coding sequence targeting by MicC RNA reveals bacterial mRNA silencing downstream of translational initiation. Nature Structural & Molecular Biology, 16, 840–846. [DOI] [PubMed] [Google Scholar]
- Romilly, C. , Deindl, S. and Wagner, E.G.H. (2019) The ribosomal protein S1‐dependent standby site in tisB mRNA consists of a single‐stranded region and a 5' structure element. Proceedings of the National Academy of Sciences of the United States of America, 116, 15901–15906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov, A. and Nudler, E. (2013) A decade of riboswitches. Cell, 152, 17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, C.M. , Darfeuille, F. , Plantinga, T.H. and Vogel, J. (2007) A small RNA regulates multiple ABC transporter mRNAs by targeting C/A‐rich elements inside and upstream of ribosome‐binding sites. Genes & Development, 21, 2804–2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, C.M. , Papenfort, K. , Pernitzsch, S.R. , Mollenkopf, H.J. , Hinton, J.C. and Vogel, J. (2011) Pervasive post‐transcriptional control of genes involved in amino acid metabolism by the Hfq‐dependent GcvB small RNA. Molecular Microbiology, 81, 1144–1165. [DOI] [PubMed] [Google Scholar]
- Takahashi, S. , Furusawa, H. , Ueda, T. and Okahata, Y. (2013) Translation enhancer improves the ribosome liberation from translation initiation. Journal of the American Chemical Society, 135, 13096–13106. [DOI] [PubMed] [Google Scholar]
- Takyar, S. , Hickerson, R.P. and Noller, H.F. (2005) mRNA helicase activity of the ribosome. Cell, 120, 49–58. [DOI] [PubMed] [Google Scholar]
- Tree, J.J. , Granneman, S. , McAteer, S.P. , Tollervey, D. and Gally, D.L. (2014) Identification of bacteriophage‐encoded anti‐sRNAs in pathogenic Escherichia coli . Molecular Cell, 55, 199–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unoson, C. and Wagner, E.G. (2007) Dealing with stable structures at ribosome binding sites: bacterial translation and ribosome standby. RNA Biology, 4, 113–117. [DOI] [PubMed] [Google Scholar]
- Vecerek, B. , Moll, I. and Blasi, U. (2007) Control of Fur synthesis by the non‐coding RNA RyhB and iron‐responsive decoding. EMBO Journal, 26, 965–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, E.G. (2013) Cycling of RNAs on Hfq. RNA Biology, 10, 619–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson, S.A. , Panja, S. and Santiago‐Frangos, A. (2018) Proteins that chaperone RNA regulation. Microbiology Spectrum, 6(4), RWR‐0026‐2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, H. and Hoover, T.R. (2001) Transcriptional regulation at a distance in bacteria. Current Opinion in Microbiology, 4, 138–144. [DOI] [PubMed] [Google Scholar]
- Yang, Q. , Figueroa‐Bossi, N. and Bossi, L. (2014) Translation enhancing ACA motifs and their silencing by a bacterial small regulatory RNA. PLoS Genetics, 10, e1004026. [DOI] [PMC free article] [PubMed] [Google Scholar]


