Abstract
Objective:
We aim to identify several microRNAs (miRNAs/miRs)-messenger RNAs (mRNAs) biomarkers correlated to nasopharyngeal carcinoma (NPC) based on an integrated analysis of miRNA and mRNAs microarray expression profiles.
Methods:
The available mRNA and miRNA microarray datasets were retrieved from Gene Expression Omnibus (GEO) database according to pre-determined screening criteria. Differentially expressed miRNA and mRNAs (DEmiRNAs and DEmRNAs) were extracted between NPC and noncancerous nasopharyngeal tissues. The target genes of DEmiRNAs were predicted with miRTarBase followed by the construction of DEmiRNAs-target DEmRNAs network, and functional analyses were performed. The DEmiRNAs expressions were validated and the performance of these DEmiRNAs was assessed by the area under the curve (AUC) values. Finally, the correlations between DEmiRNAs and specific clinical factors were analyzed.
Results:
There were 1140 interaction pairs (including let-7d/f-MYC/HMGA2 and miR-452-ITGA9) in DEmiRNAs-target DEmRNAs network. The GO annotation analysis showed that several genes such as MYC, HMGA2 and ITGA9 primarily participated in cellular process. KEGG analysis showed that these targets were associated with cell cycle and cancer-related pathways. Down-regulated let-7(-d and –f) and up-regulated miR-452 were verified in datasets. The AUC values of these 3 DEmiRNAs (let-7d, let-7-f and miR-452) was 0.803, 0.835 and 0.735, respectively. Besides, miR-452 was significantly related to survival rate of NPC patients.
Conclusion:
The findings implied let-7d/f-MYC/HMGA2 and miR-452-ITGA9 might be promising targets for the detection and treatment of NPC.
Keywords: nasopharyngeal carcinoma, mRNA, microRNA, biomarker
Introduction
Nasopharyngeal carcinoma (NPC) is a metastatic malignant tumor from the nasopharynx epithelium.1 The epidemiological evidence suggested that there was a considerable risk for NPC occurrence in Southeast Asia with the incidence rate of approximately 50/100,000 deaths each year.2 The latest statistics indicated that around 130,000 NPC new cases and 73,000 deaths were estimated across the world during 2018.3 Although advanced diagnostic tools, effective surgical operation and pharmacological therapy dramatically reduced the occurrence of NPC over past few decades,4-7 the corresponding clinical outcomes has been unsatisfied due to delayed diagnosis, distant metastasis, high recurrence and lack of targeted agents. Encouragingly, high-throughput sequencing technology has considerably facilitated molecular genetic analysis of various diseases including cancers. Consequently, increasing investigators have concentrated on uncovering the molecular mechanisms of NPC initiation and development to develop promising therapeutic strategies for NPC treatment.8,9
MicroRNAs (miRNAs) belong to a subclass of short non-coding RNAs with approximately 22 nucleotides, which can regulate gene expression by binding to the 3’-untranslated region (UTR) of target messenger RNAs (mRNAs).10 They function as tumor repressors or oncogenes and play imperative roles in tumor biological behaviors including cell proliferation, migration, apoptosis and metastasis.11-14 Notably, an increasing body of research has demonstrated that numerous miRNAs were closely associated with NPC occurrence and development according to transcriptome analyses based on microarray expression profiles.15,16 For example, Chen et al found that miR-17-5p was up-regulated in NPC tissues compared with control tissues and could promote NPC cell proliferation by inhibiting p21 expression.17 Wu et al argued that let-7a down-regulated HMGA2(high mobility group AT-hook 2) expression level, retarding cell invasion and epithelial-mesenchymal transition process of NPC clinical samples.18 Zhou et al analyzed the microarray data from plasma of NPC patients and highlighted that miR-548 and miR940 were potential diagnostic biomarkers for NPC with high sensitivity and specificity.19 Furthermore, several investigators conducted an integrated analysis of miRNA and mRNA microarray data and unraveled that hsa-miR-423-5p/MYC(a proto-oncogene) signature was pivotal for NPC diagnosis,20 which offered novel perspectives to extract high-efficiency therapeutic targets for the treatment of NPC. Unfortunately, the detailed molecular mechanisms of NPC have not completely clarified.
Therefore, we searched available miRNA and mRNA microarray datasets from NPC tissues from the National Center for Biotechnology Information Gene Expression Omnibus (GEO) database to perform an integrated bioinformatics analysis. Specifically, the miRNA-mRNA regulatory analysis and functional analysis were performed to identify potential biomarkers related to NPC. Then, the candidate signatures were validated and evaluated. Finally, the correlations between key miRNAs and clinical parameters were predicted. This study will provide a deeper understanding for pathological mechanisms of NPC progression.
Materials and Methods
Data Acquisition
The miRNAs and mRNAs sequencing datasets were retrieved and downloaded from GEO21 repository using the searching terms of “nasopharyngeal neoplasms” [MeSH Terms] OR “Nasopharyngeal carcinoma” [All Fields] AND “Homo sapiens” [porgn] AND “gse” [Filter]. The available datasets were selected according to the following criteria: 1) the type of studies with eligible datasets was restricted to expression profiling by array or non-coding RNA profiling by array; 2) all data were whole-genome mRNA expression profiles; and 3) these datasets were generated from NPC tissues and noncancerous nasopharyngeal tissues (controls), and all tissues were not treated with stimulant medications. Finally, 5 mRNAs datasets were obtained, including GSE64634 (12 NPC and 4 normal healthy nasopharyngeal tissue specimens), GSE53819 (18 NPC primary tumor tissues and 18 non-cancerous nasopharyngeal tissues), GSE34573 (15 NPC biopsy tissues and 3 normal nasopharyngeal biopsy tissues), GSE13597 (25 undifferentiated NPC biopsy tissues and 3 non-malignant nasopharyngeal biopsy tissues) and GSE12452 (31 NPC tissues and 10 normal nasopharyngeal tissues). Besides, 5 miRNAs microarray datasets were also extracted, including GSE43039 (20 chronic nasopharyngitis tissues and 20 NPC tissues), GSE70970 (246 NPC biopsy tissues and 17 nasopharyngeal epithelium tissues), GSE32906 (16 NPC tissues and 6 normal healthy nasopharyngeal epithelial tissues), GSE32960 (312 NPC tissues and 18 normal nasopharyngeal tissues), and GSE36682 (62 NPC tissues and 18 nasopharyngitis tissues). Table 1 showed the baseline information about mRNA and miRNA datasets.
Table 1.
A List of mRNA and miRNA Datasets Information for NPC.
| GEO ID | Microarray type | Platform | Samples(N: P) | Year |
|---|---|---|---|---|
| GSE64634 | mRNA | GPL570 [HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array | 4 normal healthy nasopharyngeal tissues:12 NPC tissues | 2017 |
| GSE53819 | mRNA | GPL6480Agilent-014850 Whole Human Genome Microarray 4x44 K G4112F (Probe Name version) | 18 non-cancerous nasopharyngeal tissues:18 NPC primary tumor tissues | 2014 |
|
GSE34573 |
mRNA | GPL570 [HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array | 3 normal nasopharyngeal biopsy tissues:15 nasopharyngeal carcinoma biopsy tissues | 2012 |
| GSE13597 | mRNA | GPL96[HG-U133A] Affymetrix Human Genome U133A Array | 3 nasopharyngeal carcinoma biopsy tissues:25 undifferentiated NPC biopsy tissues | 2009 |
| GSE12452 | mRNA | GPL570[HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array | 10 normal nasopharyngeal tissues:31 NPC tissues | 2008 |
| GSE43039 | miRNA | GPL16414CCDTM-miRNA850-version 4p1.4 | 20 chronic nasopharyngitis tissues:20 NPC tissues | 2015 |
| GSE70970 | miRNA | GPL20699 nCounter® Human miRNA Assay (v1.0, Nanostring) | 17 nasopharyngeal epithelium tissues:246 nasopharyngeal carcinoma biopsy tissues | 2015 |
| GSE32906 | miRNA | GPL11350 Illumina Custom Prostate Cancer DASL Panel miRNA | 6 normal healthy nasopharyngeal epithelial tissues:16 NPC tissues | 2014 |
| GSE32960 | miRNA | GPL14722microRNA array | 18 normal nasopharyngeal tissues:312 NPC tissues | 2012 |
| GSE36682 | miRNA | GPL15311 Human miRNA 1K | 6 nasopharyngitis tissues:62 NPC tissues | 2012 |
mRNA, messenger mRNA; miRNA, microRNA; NPC, nasopharyngeal carcinoma; N, the number of normal nasopharyngeal tissue specimens; P, the number of nasopharyngeal carcinomas tissue specimens
Data Pre-Processing and Identification of Differentially Expressed RNAs (DERs)
For data pre-processing, the annotated mRNAs and miRNAs in each sequencing platform were obtained and over-lapped. Herein, a total of 12180 over-lapping genes among mRNA microarray datasets were identified while there were 196 over-lapping miRNAs across miRNA expression profiles. Then, gene or miRNA distribution matrix were obtained. If the data does not conform to a normal distribution, log2 transformation will be conducted to standardize data. Subsequently, we used the metaDE package in R to extract differentially expressed miRNA and mRNAs (DEmiRNAs and DEmRNAs) between NPC and control samples using false discovery rate (FDR) < 0.05 as the screening cutoff.22 Finally, the hierarchical clustering analysis was conducted with the pheatmap package (https://cran.r-project.org/package=pheatmap) in R language.
Prediction of Target Genes for DEmiRNAs and Functional Analyses
Admittedly, miRNAs exert significant roles at post-transcription processes via regulating mRNA levels. The updated miRTarBase (version 7.0; http://miRTarBase.mbc.nctu.edu.tw/) database contains thousands of experimentally verified miRNA-target interactions (MTIs) involving 29 species. More specifically, it collects 2599 miRNAs and 15064 corresponding targets for human beings. Herein, this database was employed to predict the target genes for screened DEmiRNAs and the Cytoscape software23 was utilized to construct the DEmiRNA-target DEmRNA regulatory network. We would regard those target DEmRNAs as candidate genes for the following analyses.
Afterward, we further explored the underlying biological roles of these genes, the Gene ontology (GO) analysis was firstly performed on the basis of 3 categories molecular function (MF), cellular component (CC) and biological process (BP) using BiNGO plugin in cytoscape software. Notably, the P value of each gene was calculated with hypergeometric test and the Benjamini & Hochberg method was applied to adjust the FDR value. Finally, adjusted FDR < 0.05 was considered as the cutoff for significant enrichment. In addition, KOBAS (version 3.0; http://kobas.cbi.pku.edu.cn/index.php) as a web server has been successfully used to carry out functional gene set enrichment analysis. Here, the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of candidate genes was also conducted by KOBAS3.0 according to the screening criterion of adjusted FDR < 0.05.
Validation of DEmiRNAs in Plasma Samples of NPC Patients
To examine whether DEmiRNAs in NPC tissues can be detected in datasets from plasma samples of patients undergoing NPC, the relevant microarray datasets were searched from GEO database. Finally, 2 datasets (GSE43329 and GSE48442) from plasma samples from NPC patients were obtained. Specifically, there were 31 NPC cases and 19 healthy control plasma samples in GSE43329 dataset while GSE48442 comprised of 20 NPC cases and 10 control samples. Following this, the miRNA expression levels were computed by t-test and the receiver operating characteristic (ROC) analysis was implemented to evaluate the performance of DEmiRNAs using the pROC package (https://cran.r-project.org/web/packages/pROC/index.html) in R language. P value < 0.05 was considered as the cutoff of the significant difference.
Association Analysis Between DEmiRNAs and Clinical Parameters
To further explore whether there are correlations between DEmiRNAs and some clinical factors, the association analysis was carried out. The clinical information was respectively collected from mRNA and miRNA datasets. As indicated in Table S1, the clinical data in GSE64634 and GSE13597 were unavailable. GSE53819 contained the information about age and gender. Two datasets (GSE13597 and GSE12452) included NPC stage data. Therefore, the clinical data was incomplete in mRNA datasets. For miRNA datasets, GSE32906 and GSE32960 contained NPC stage information (GSE32906: 4 patients in stage I-II, III and IV, respectively; GSE32960: 12, 86, 91 123 patients in stage I, II, III and IV, respectively; Table S1). Additionally, GSE36682 included the survival data and metastasis (45 patients without NPC metastasis and 17 patients with metastasis; Table S1). Herein, we only focused on the association analysis between DEmiRNAs and 2 clinical parameters (stage and overall survival rate).
Results
Screening of DEmiRNA and DEmRNAs
A total of 28 DEmiRNAs were identified between NPC tissues and controls based on the pre-determined criteria, which consisted of 13 up-regulated miRNAs and 15 down-regulated miRNAs (Table 2). Additionally, 2593 DEmRNAs, including 1399 up-regulated genes and 1194 down-regulated genes, were also obtained after microarray data mining and standardization (Table S2). The Figure S1 displayed the differential expression levels of all genes in NPC tissues among all datasets and the top 20 DEmRNAs were exhibited in Table 3.
Table 2.
The Differentially Expressed miRNAs in NPC.
| miRNA | FDR | Regulation | miRNA | FDR | Regulation |
|---|---|---|---|---|---|
| hsa-miR-93 | 0 | up | hsa-miR-26b | 0 | down |
| hsa-miR-107 | 8.12E-10 | up | hsa-miR-203 | 0 | down |
| hsa-miR-196b | 5.03E-07 | up | hsa-miR-150 | 1.67E-14 | down |
| hsa-miR-184 | 8.57E-07 | up | hsa-miR-100 | 4.66E-14 | down |
| hsa-miR-210 | 2.14E-06 | up | hsa-let-7f | 5.80E-14 | down |
| hsa-miR-29a | 2.73E-06 | up | hsa-miR-34b | 9.52E-14 | down |
| hsa-let-7a | 1.30E-04 | up | hsa-let-7d | 2.51E-13 | down |
| hsa-miR-452 | 1.51E-04 | up | hsa-miR-145 | 3.34E-13 | down |
| hsa-miR-18b | 3.43E-04 | up | hsa-miR-30c | 1.65E-11 | down |
| hsa-miR-542-3p | 5.98E-04 | up | hsa-miR-29c | 2.70E-11 | down |
| hsa-miR-505 | 9.23E-04 | up | hsa-let-7g | 3.70E-11 | down |
| hsa-miR-25 | 5.14E-03 | up | hsa-miR-26a | 7.48E-11 | down |
| hsa-let-7b | 1.21E-02 | up | hsa-miR-30b | 5.99E-07 | down |
| hsa-miR-31 | 1.51E-04 | down | hsa-miR-146a | 1.81E-05 | down |
miRNA, microRNA; NPC, nasopharyngeal carcinoma; FDR, false discovery rate
Table 3.
The Top 40 Differentially Expressed mRNAs in NPC.
| Gene ID | Gene symbol | Regulation | Gene ID | Gene symbol | Regulation |
|---|---|---|---|---|---|
| 9837 | GINS1 | up | 9940 | DLEC1 | down |
| 9055 | PRC1 | up | 9576 | SPAG6 | down |
| 84823 | LMNB2 | up | 94025 | MUC16 | down |
| 83990 | BRIP1 | up | 9071 | CLDN10 | down |
| 79412 | KREMEN2 | up | 8991 | SELENBP1 | down |
| 7444 | VRK2 | up | 8537 | BCAS1 | down |
| 7130 | TNFAIP6 | up | 8382 | NME5 | down |
| 6373 | CXCL11 | up | 80736 | SLC44A4 | down |
| 5954 | RCN1 | up | 79838 | TMC5 | down |
| 5932 | RBBP8 | up | 79747 | ADGB | down |
| 54517 | PUS7 | up | 79740 | ZBBX | down |
| 51514 | DTL | up | 79645 | EFCAB1 | down |
| 5111 | PCNA | up | 79083 | MLPH | down |
| 3685 | ITGAV | up | 7802 | DNALI1 | down |
| 3627 | CXCL10 | up | 7356 | SCGB1A1 | down |
| 3206 | HOXA10 | up | 7348 | UPK1B | down |
| 2335 | FN1 | up | 6590 | SLPI | down |
| 1503 | CTPS1 | up | 6286 | S100P | down |
| 10635 | RAD51AP1 | up | 60494 | CCDC81 | down |
| 1063 | CENPF | up | 5858 | PZP | down |
mRNA, messenger mRNA; NPC, nasopharyngeal carcinoma
Prediction of DEmiRNA Targets and Functional Analyses
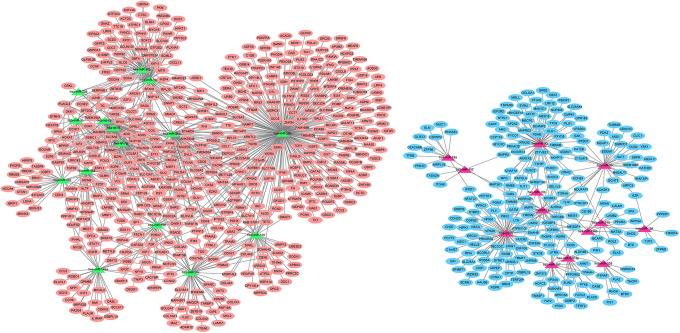
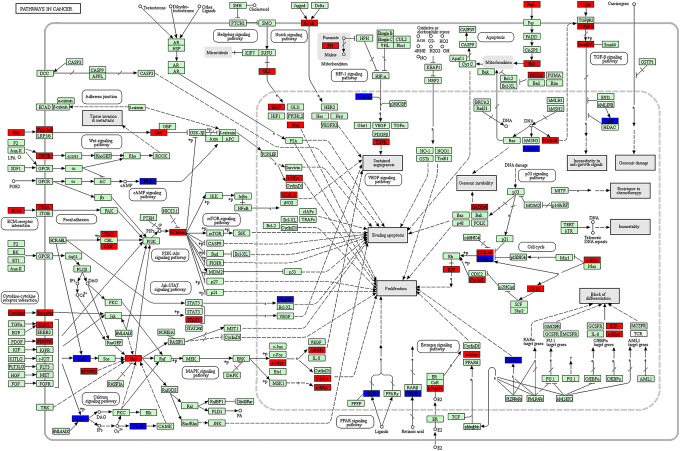
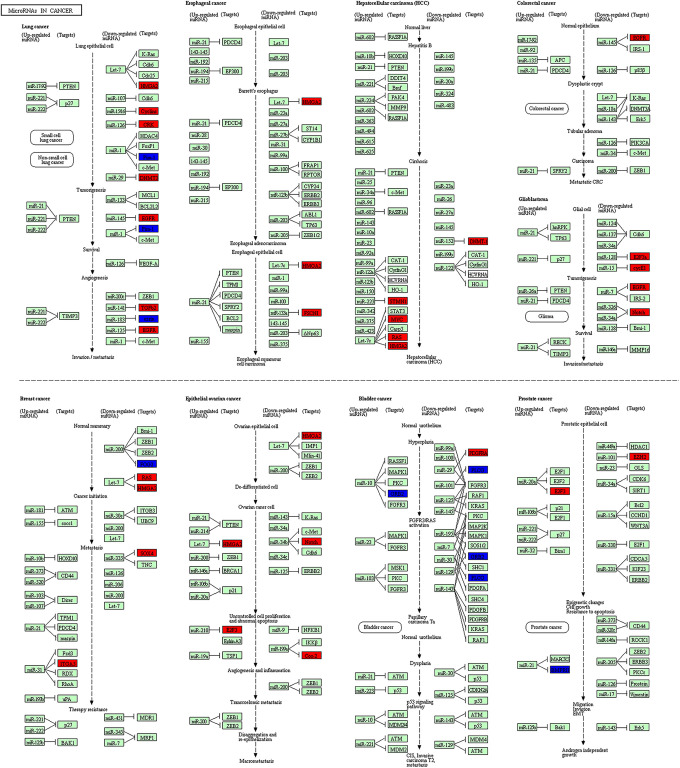
Firstly, we determined 751 target genes for DEmiRNAs based on the miRTarBase 7.0. Then, the DEmiRNAs-target DEmRNAs network involving 1140 interaction pairs was established, of which, there were 280 up-regulated DEmiRNAs-down-regulated DEmRNAs interactions such as miR-452-ITGA9 (integrin subunit alpha 9) and 860 down-regulated DEmiRNAs-up-regulated DEmRNAs relationships including let-7d- MYC/HMGA2 and let-7f-MYC/HMGA2 (Figure 1). The GO analyses of target genes suggested that they were predominately correlated with GO-BP terms of DNA replication, cellular process and nucleic acid metabolic process (Table 4). Meanwhile, these genes were responsible for several GO-CC terms such as membrane-enclosed lumen and intracellular part (Table 4). Besides, GO-MF analysis indicated that these genes primarily participated in protein binding, adenyl ribonucleotide binding and protein kinase activity (Table 4). We also found that several genes such as MYC, HMGA2 and ITGA9 were predominately associated with cell cycle process. In addition, our KEGG analysis implied that DEmiRNA targets were significantly enriched in multiple pathways including cell cycle, pathways in cancer, p53 signaling pathway, PI3K-Akt signaling pathway and microRNAs in cancer. Notably, MYC and ITGA9 were crucial gene signatures in pathway-related to cancers while let-7-HMGA2/axis was involved in various cancers such as lung cancer, esophageal cancer and breast cancer (Figure 2 and 3)
Figure 1.
The DEmiRNAs-target DEmRNAs network. The orange ovals show the up-regulated target genes for DEmiRNAs and blue ovals indicates the down-regulated target genes for DEmiRNAs in nasopharyngeal carcinoma tissues. The red triangles indicate up-regulated miRNAs while green triangles denote down-regulated miRNAs in nasopharyngeal carcinoma tissues. DEmiRNAs: differentially expressed microRNAs.
Table 4.
The Functional Analyses of Target Genes for Differentially Expressed miRNAs in NPC.
| Category | Term | Count | P-value | Genes |
|---|---|---|---|---|
| GOTERM_BP (top 5) | DNA replication | 36 | 1.46E-10 | SLBP, PRIM2, PCNA, RRM1, RRM2, CDC7, CDC6, DNA2, DTL, MCM2,,,,,, |
| cellular process | 498 | 6.77E-10 | MAPKBP1, MYC, SOX11, HMGA2, ITGA9, CXCL9, CAD, IFNG, TERF2IP, NDUFS2,,,,,, | |
| nucleic acid metabolic process | 121 | 4.85E-09 | CCNT1, MPG, DSCC1, YARS2, NARS, ZFP36, PTTG1, SNRPD1, CHEK1, NARS2,,,,,, | |
| DNA metabolic process | 60 | 4.85E-09 | MPG, DSCC1, CHAF1A, SUMO1, PTTG1, CHEK1, PIM1, DNMT3B, TK1, KPNA2,,,,,, | |
| macromolecule biosynthetic process | 96 | 4.85E-09 | BMPR2, CCNT1, TUSC3, DSCC1, YBX1, MED16, FOXM1, TRAM2, YARS2, NARS,,,,,, | |
| GOTERM_CC (top 5) | membrane-enclosed lumen | 151 | 3.02E-17 | SPARC, MYC, CHEK2, CHEK1, NARS2, PIM1, OIP5, KPNA2, PHLDA1, PDK2,,,,,, |
| intracellular part | 556 | 1.06E-16 | ATP5C1, GABPB1, ELK3, MPRIP, MYC, DPYSL2, AKT3, NAMPT, STMN1, RPL37|HOXA7, RKACB,,,,,, | |
| organelle lumen | 146 | 1.57E-16 | SPARC, CCNT1, MPG, DSCC1, MYC, CHEK2, CHEK1, NARS2, PIM1, OIP5,,,,,, | |
| intracellular organelle lumen | 143 | 2.88E-16 | CCNT1, MPG, DSCC1, GMNN, GOLIM4, ATP5C1, YARS2, SNRPD1, MYC, CHEK2,,,,,, | |
| intracellular | 566 | 2.97E-16 | ATP5C1, GABPB1, ELK3, MPRIP, MYC, DPYSL2, AKT3, NAMPT, STMN1, RPL37,,,,,, | |
| GOTERM_MF (top 5) | protein binding | 473 | 8.16E-20 | MAPKBP1, SERPINE1, GABPB1, ELK3, MPRIP, MYC, HMGA2, ISG15, NUP153, POU2F3,,,,,, |
| binding | 596 | 3.60E-08 | MAPKBP1, SERPINE1, MYC, HMGA2, ISG15, NUP153, POU2F3, IGF2 R, SMAD7, BMP2,,,,,, | |
| platelet-derived growth factor binding | 5 | 1.26E-02 | PDGFRA, COL3A1, COL1A2, COL5A1, COL4A1 | |
| ATP binding | 94 | 1.26E-02 | ERCC6 L, BMPR2, CHD7, OLA1, KIF11, PIK3C2A, PKD2, YARS2, OASL, IFIH1,,,,,, | |
| adenyl ribonucleotide binding | 95 | 1.26E-02 | ERCC6 L, BMPR2, CHD7, OLA1, KIF11, PIK3C2A, PKD2, YARS2, IFIH1, NARS,,,,,, |
GO, Gene ontology; BP, biological process; MF, molecular function; CC, cellular component; NPC, nasopharyngeal carcinoma
Figure 2.
Pathway in cancer from the Kyoto Encyclopedia of Genes and Genomes enrichment analysis. The red color shows the up-regulated genes while blue color represents the down-regulated genes in nasopharyngeal carcinoma tissues. The green color indicates genes which had no significant difference between nasopharyngeal carcinoma tissues and control tissues.
Figure 3.
MicroRNAs in cancer from the Kyoto Encyclopedia of Genes and Genomes enrichment analysis. The red color shows the up-regulated genes while blue color represents the down-regulated genes in nasopharyngeal carcinoma tissues. The green color indicates genes which had no significant difference between nasopharyngeal carcinoma tissues and control tissues.
Confirmation of DEmiRNAs in NPC Plasma Samples
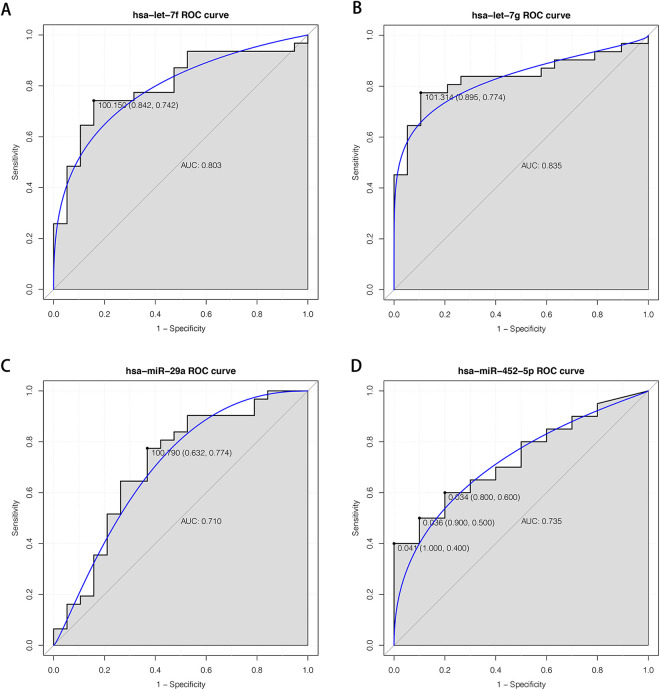
For GSE43329 dataset, we found that let-7b, let-7f, let-7 g, miR-107 and miR-29a were dramatically under-expressed in NPC plasma compared to normal controls (P < 0.05; (Table S3). Moreover, let-7f and let-7 g were down-regulated in both validation dataset from NPC plasma samples and 5 training datasets from NPC tissue samples (Table S3). Besides, the area under curve (AUC) values for let-7f, let-7 g and miR-29a were 0.803, 0.835 and 0.710, respectively (Figure 4). For GSE48442 dataset, miR-452 was significantly differentially expressed in NPC plasma samples compared to normal controls (Table S4). We also noted that this miRNA was up-regulated in validation dataset from NPC plasma samples and 5 training datasets from NPC tissue samples (Table S4). The AUC value for miR-452 was 0.735 (Figure S4).
Figure 4.
The ROC curves of key miRNAs in GSE43329 and GSE48442 datasets. The X-axis indicated 1-specificity and Y-axis represented sensitivity. (A) ROC curve of let-7f in GSE43329 dataset; (B) ROC curve of let-7 g in GSE43329 dataset; (C)ROC curve of miR-29a in GSE43329 dataset; (D) ROC curve of miR-452 in GSE48442 dataset. ROC: receiver operating characteristic.
Correlation Analysis between DEmiRNAs and Clinical Parameters
The relationships between DEmiRNAs and 2 clinical parameters (stage and overall survival) were analyzed (Figure S2-4). The results showed that miR-150 and miR-29a were associated with NPC stage (miR-150: P = 0.012; cor = -0.142; miR-29a: P = 0.046; cor = -0.113; Figure S3). Additionally, miR-93 (P = 0.008), miR-25 (P = 0.048), and miR-452 (P = 0.029) were markedly correlated with survival rate of NPC patients (Figure S4). Moreover, lower expressions of these 3 miRNAs showed higher survival rates.
Discussion
NPC has been recognized as a serious public health issue around the globe due to the high prevalence and mortality.24 A growing number of researchers have concentrated on identifying potential miRNA signatures related to NPC in the past decade. For example, Li et al carried out a propensity-score-matched miRNA microarray analysis and found that miR-142-3p was strongly related to the metastasis of NPC metastasis.25 Recently, Zhao et al analyzed miRNA expression profiles in NPC and suggested that miR-1278 played essential roles in clinical survival of NPC patients and chemotherapy response.26 However, these studies generally involve a relatively small sample size. Moreover, few researchers systematically performed miRNA and mRNA expression profiling analysis in NPC. Therefore, we will conduct an integrated analysis of miRNA-mRNA expression to decipher the underlying molecular mechanisms of NPC. Our results showed that 28 DEmiRNAs (13 up-regulated and 15 down-regulated miRNAs) and 3593 DEmRNAs (1399 up-regulated and 1194 down-regulated mRNAs) were identified between NPC and control tissues from miRNA and mRNA microarray datasets, respectively. Moreover, there were 1140 interaction pairs such as let-7d/f- MYC/HMGA2 and miR-452-ITGA9 among the DEmiRNAs-target DEmRNAs regulatory network. The GO annotation analysis of target genes for DEmiRNAs revealed that several genes including MYC, HMGA2 and ITGA9 played prominent roles in cell cycle process. Meanwhile, KEGG enrichment analysis showed that these targets were mainly focused on cell cycle and cancer-related pathways. Notably, we also found that MYC and ITGA9 were involved in pathway in cancers while let-HMGA2 were implicated with various cancers such as lung cancer, esophageal cancer and breast cancer. More importantly, let-7d and let-7f were also observed to be markedly under-expressed while miR-452 was significantly up-regulated in plasma samples from NPC patients. Furthermore, the AUC values for these 3 miRNAs were all more than 0.7, suggesting that they were possibly critical biomarkers for NPC detection.
Extensive studies have demonstrated that miRNAs might serve as oncogenes and contribute to the initiation and progression of NPC through regulating their target genes that participate in numerous cellular processes.27,28 We carried out an integrated analysis of miRNAs and mRNAs microarray datasets and our findings indicated that down-regulated let-7d, let-7f and their targets genes (up-regulated MYC and HMGA2) played essential roles in carcinogenesis. Besides, let-7d and let-7f were also validated to be under-expressed in NPC plasma samples in comparison with healthy controls from other datasets, which suggested that plasma let-7d and let-7f level may be potential biomarkers for NPC detection. Overwhelming evidence has demonstrated that miRNA let-7 family (-a, -b, -c, -d, -e, -f, -g and –i) exerted regulatory roles at the transcriptional and post-transcriptional levels among species and their aberrant expression might be closely linked with the pathogenesis of cancers such as NPC.29,30 Luan et al conducted a meta-analysis of 8 miRNA studies for NPC and pointed out that let-7d was remarkably decreased in NPC tumor tissues, which was consistent with our results from bioinformatics analysis.31 Similarly, Wong et al previously also found that there were lower levels of let-7d and let-7f in NPC cells than normal epithelial cells according to the real-time quantitative PCR, More interestingly, they suggested that increased let-7 levels could down-regulate the MYC protein expression in NPC cell lines.32 This also provided experimental evidence for our result that a strong relationship between let-7(-d and -f) and MYC were observed based on miRNAs-mRNAs network. Earlier research has showed that MYC was a key cell progression-related gene and its knockdown could suppress cell proliferation and growth in NPC cell lines.33 Accordingly, we also noted that let-7d and let-7f strongly interacted with up-regulated HMGA2 and this gene was implicated with cellular processes. Xia et al argued that the HMGA2 level was obviously enhanced in NPC tissues and the association analysis between HMGA2 and clinic-pathological factors indicated that HMGA2 contributed to NPC development and metastasis.34 Additionally, our evaluation analysis for DEmiRNAs also revealed that let-7d and let-7f had higher AUC values (AUC > 0.7), implying that these 2 miRNAs were probably promising signatures for NPC diagnosis and treatment. Taken together, we inferred that let-7(-d and –f)/MYC and let-7(-d and –f)/HMGA2 axis might serve as novel therapeutic targets for NPC.
MiR-452, another vital regulator, was over-expressed in NPC tissues compared with normal controls, which was further confirmed in NPC plasma samples. Moreover, a lower expression of miR-452 represented a higher survival rate. Existing studies suggested that miR-452 played key roles in pathogenic mechanisms of a wide variety of cancers, including non-small cell lung cancer, prostate cancer and hepatocellular carcinoma.35-37 However, the potential effects of miR-452 on NPC initiation have not been elaborated. Herein, a close correlation between miR-452 and down-regulated ITGA9 was detected and functional analyses implied that this gene was primarily responsible for cellular processes. ITGA9 was located at 3p21A region and a linkage analysis showed that the mutations in this segment might induce NPC formation.38 Later on, Ng et al genotyped 111 patients suffering from NPC and 260 healthy individuals on the basis of a genome-wide association analysis and they emphasized that a variant (SNP rs189897 in ITGA9) conferred a risk for NPC development among a Malaysian Chinese population.39 Nawaz et al discovered ITGA9 as critical tumor suppressor gene could control cell growth and differentiation and highly methylated in NPC cells,40 which was further verified in following investigation that unraveled ITGA9 was under-expressed in NPC cell lines and its methylation was also observed by the methylation specific PCR.41 Therefore, we speculated that MiR-452/ITGA9 axis could be regarded as an early predictor in the diagnosis of NPC.
Although several prominent gene and miRNAs biomarkers have been identified using bioinformatics methods, there are still limitations in our current analysis. Firstly, a comprehensive bioinformatics analysis based on a larger sample size needs to be carried out to confirm our results. Secondly, the corresponding experiments such as qPCR array and NPC cell experiments require to verify several potential RNA transcripts and corresponding signaling pathway. Finally, the extensive clinical information will also be requisite to integrate into following analysis to evaluate prognosis.
Conclusion
In summary, we performed an integrated analysis of miRNAs and mRNAs to identify potential biomarkers for NPC treatment. The results showed that let-7d-MYC/HMGA, let-7f-MYC/HMGA2 and miR-452/ITGA9 axis were possibly associated with NPC progression, which will provide novel insights into developing novel therapeutic strategies for diagnosis and treatment of NPC. However, a detailed bioinformatics analysis and experimental validation also need to be undertaken in following investigation.
Supplemental Material
Supplemental Material, Figure_S1 for An Integrated Analysis of mRNAs and miRNAs Microarray Profiles to Screen miRNA Signatures Involved in Nasopharyngeal Carcinoma by Lei Liu, Hailing Wang, Chaohui Yan and Shudong Tao in Technology in Cancer Research & Treatment
Supplemental Material, Figure_S2 for An Integrated Analysis of mRNAs and miRNAs Microarray Profiles to Screen miRNA Signatures Involved in Nasopharyngeal Carcinoma by Lei Liu, Hailing Wang, Chaohui Yan and Shudong Tao in Technology in Cancer Research & Treatment
Supplemental Material, Figure_S3 for An Integrated Analysis of mRNAs and miRNAs Microarray Profiles to Screen miRNA Signatures Involved in Nasopharyngeal Carcinoma by Lei Liu, Hailing Wang, Chaohui Yan and Shudong Tao in Technology in Cancer Research & Treatment
Supplemental Material, Figure_S4 for An Integrated Analysis of mRNAs and miRNAs Microarray Profiles to Screen miRNA Signatures Involved in Nasopharyngeal Carcinoma by Lei Liu, Hailing Wang, Chaohui Yan and Shudong Tao in Technology in Cancer Research & Treatment
Supplementary_tables for An Integrated Analysis of mRNAs and miRNAs Microarray Profiles to Screen miRNA Signatures Involved in Nasopharyngeal Carcinoma by Lei Liu, Hailing Wang, Chaohui Yan and Shudong Tao in Technology in Cancer Research & Treatment
Abbreviations
- NPC
Nasopharyngeal carcinoma
- miRNAs/miRs
MicroRNAs
- mRNAs
Messenger RNAs
- DEmiRNAs:
Differentially expressed miRNAs
- DEmRNAs
Differentially expressed mRNAs
- GEO
Gene Expression Omnibus
- GO
Gene ontology
- ROC
Receiver operating characteristic
- AUC
Area under the curve
- DERs
Differentially expressed RNAs
- MTIs
miRNA-target interactions
- MF
Molecular function
- CC
Cellular component
- BP
Biological process
- FDR
False discovery rate
Footnotes
Authors’ Note: Lei Liu and Hailing Wang contributed equally to this work.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
ORCID iD: Chaohui Yan  https://orcid.org/0000-0002-4958-6626
https://orcid.org/0000-0002-4958-6626
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Chua ML, Wee JT, Hui EP, Chan AT. Nasopharyngeal carcinoma. Lancet. 2016;387(10022):1012–1024. [DOI] [PubMed] [Google Scholar]
- 2. Razak AR, Siu LL, Liu FF, Ito E, O’Sullivan B, Chan K. Nasopharyngeal carcinoma: the next challenges. Eur J Cancer. 2010;46(11):1967–1978. [DOI] [PubMed] [Google Scholar]
- 3. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68(6):394–424. [DOI] [PubMed] [Google Scholar]
- 4. Gao Y, Zhu SY, Dai Y, Lu BF, Lu L. Diagnostic accuracy of sonography versus magnetic resonance imaging for primary nasopharyngeal carcinoma. J Ultrasound Med. 2014;33(5):827–834. [DOI] [PubMed] [Google Scholar]
- 5. Shayah A, Wickstone L, Kershaw E, Agada F. The role of cross-sectional imaging in suspected nasopharyngeal carcinoma. Ann R Coll Surg Engl. 2019;101(5):325–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ma BB, Hui EP, Chan AT. Investigational drugs for nasopharyngeal carcinoma. Expert Opin Investig Drugs. 2017;26(6):677–685. [DOI] [PubMed] [Google Scholar]
- 7. Kaidar-Person O, Gil Z, Billan S. Precision medicine in head and neck cancer. Drug Resist Updat. 2018;40:13–16. [DOI] [PubMed] [Google Scholar]
- 8. Zeng YX, Jia WH. Familial nasopharyngeal carcinoma. Semin Cancer Biol. 2002. 12(6):443–450. [DOI] [PubMed] [Google Scholar]
- 9. Zheng ZQ, Li ZX, Zhou GQ, et al. Long non-coding RNA FAM225A promotes nasopharyngeal carcinoma tumorigenesis and metastasis by acting as ceRNA to sponge miR-590-3p/miR-1275 and upregulate ITGB3. Cancer Res. 2019;79(18):4612–4626. doi:10.1158/0008-5472 [DOI] [PubMed] [Google Scholar]
- 10. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell Physiol Biochem. 2004;116(2);281–297. [DOI] [PubMed] [Google Scholar]
- 11. Chan SH, Wang LH. Regulation of cancer metastasis by microRNAs. J Biomed Sci. 2015;22(1):1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nogales-Cadenas R, Cai Y, Lin JR, et al. MicroRNA expression and gene regulation drive breast cancer progression and metastasis in PyMT mice. Breast Cancer Res. 2016;18(1):75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mansoori B, Mohammadi A, Naghizadeh S, et al. miR-330 suppresses EMT and induces apoptosis by downregulating HMGA2 in human colorectal cancer. J Cell Physiol. 2019;235(2):920–931. doi:10.1002/jcp.29007 [DOI] [PubMed] [Google Scholar]
- 14. Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122(1):6–7. [DOI] [PubMed] [Google Scholar]
- 15. Liang TS, Zheng YJ, Wang J, Zhao JY, Yang DK, Liu ZS. MicroRNA-506 inhibits tumor growth and metastasis in nasopharyngeal carcinoma through the inactivation of the Wnt/β-catenin signaling pathway by down-regulating LHX2. J Exp Clin Cancer Res. 2019;38(1):97. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16. Liu FF. Nasopharyngeal cancer: new frontiers from the laboratory to the clinic. Int J Radiat Oncol Biol Phys. 2007;69(2):S122–124. [DOI] [PubMed] [Google Scholar]
- 17. Chen C, Lu Z, Yang J, et al. MiR-17-5p promotes cancer cell proliferation and tumorigenesis in nasopharyngeal carcinoma by targeting p21. Cancer Med. 2016;5(12):3489–3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wu A, Wu K, Li J, et al. Let-7a inhibits migration, invasion and epithelial-mesenchymal transition by targeting HMGA2 in nasopharyngeal carcinoma. J Transl Med. 2015;13(1):105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhuo X, Zhou W, Li D, et al. Plasma microRNA expression signature involving miR-548q, miR-630 and miR-940 as biomarkers for nasopharyngeal carcinoma detection. Cancer Biomark. 2018;23(4):579–587. [DOI] [PubMed] [Google Scholar]
- 20. ian H, Chen S, Zhang C, Li M, Zheng H. MYC and hsa-miRNA-423-5p as biomarkers in nasopharyngeal carcinoma revealed by miRNA-mRNA-pathway network integrated analysis. Mol Med Rep. 2017;16(2):1039–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Barrett T, Troup DB, Wilhite SE. NCBI GEO: mining tens of millions of expression profiles—database and tools update. Nucleic Acids Res. 2007;35(Database issue):D760–D765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Marot G, Foulley JL, Mayer CD, Jaffrézic F. Moderated effect size and P-value combinations for microarray meta-analyses. Bioinformatics. 2009;25(20):2692–2699. [DOI] [PubMed] [Google Scholar]
- 23. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bruce JP, Yip K, Bratman SV, Ito E, Liu FF. Nasopharyngeal cancer: molecular landscape. J Clin Oncol. 2015;33(29):3346–3355. [DOI] [PubMed] [Google Scholar]
- 25. Li Y, He Q, Wen X, et al. EZH2-DNMT1-mediated epigenetic silencing of miR-142-3p promotes metastasis through targeting ZEB2 in nasopharyngeal carcinoma. Cell Death Differ. 2019;26(6):1089–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhao Y, Wang P, Wu Q. MiR-1278 sensitizes nasopharyngeal carcinoma cells to cisplatin and suppresses autophagy via targeting ATG2B. Mol Cell Probes. 2020;101597. [DOI] [PubMed] [Google Scholar]
- 27. Lee KT, Tan JK, Lam AK, Gan SY. MicroRNAs serving as potential biomarkers and therapeutic targets in nasopharyngeal carcinoma: a critical review. Crit Rev Oncol Hematol. 2016;103:1–9. [DOI] [PubMed] [Google Scholar]
- 28. Zhao CX, Zhu W, Ba ZQ, et al. The regulatory network of nasopharyngeal carcinoma metastasis with a focus on EBV, lncRNAs and miRNAs. Am J Cancer Res. 2018;8(11):2185–2209. [PMC free article] [PubMed] [Google Scholar]
- 29. Roush S, Slack FJ. The let-7 family of microRNAs. Trends Cell Biol. 2008;18(10):505–516. [DOI] [PubMed] [Google Scholar]
- 30. Li T, Chen JX, Fu XP, et al. Microrna expression profiling of nasopharyngeal carcinoma. Oncol Rep. 2011;25(5):1353–1363. [DOI] [PubMed] [Google Scholar]
- 31. Luan J, Wang J, Su Q, Chen X, Jiang G, Xu X. Meta-analysis of the differentially expressed microRNA profiles in nasopharyngeal carcinoma. Oncotarget. 2016;7(9):10513–10521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wong TS, Man OY, Tsang CM. MicroRNA let-7 suppresses nasopharyngeal carcinoma cells proliferation through downregulating c-Myc expression. J Cancer Res Clin Oncol. 2011;137(3):415–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Niu Z, Liu H, Zhou M, et al. Knockdown of c-Myc inhibits cell proliferation by negatively regulating the Cdk/Rb/E2F pathway in nasopharyngeal carcinoma cells. Acta Biochim Biophys Sin (Shanghai). 2015;47(3):83–91. [DOI] [PubMed] [Google Scholar]
- 34. Xia YY, Yin L, Tian H, et al. HMGA2 is associated with epithelial-mesenchymal transition and can predict poor prognosis in nasopharyngeal carcinoma. Onco Targets Ther. 2015;8:169–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kristensen H, Haldrup C, Strand S, et al. Hypermethylation of the GABRE∼miR-452∼miR-224 promoter in prostate cancer predicts biochemical recurrence after radical prostatectomy. Clin Cancer Res. 2014;20(8):2169–2181. [DOI] [PubMed] [Google Scholar]
- 36. Zheng Q, Sheng Q, Jiang C, et al. MicroRNA-452 promotes tumorigenesis in hepatocellular carcinoma by targeting cyclin-dependent kinase inhibitor 1B. Mol Cell Biochem. 2014;389(1-2):187–195. [DOI] [PubMed] [Google Scholar]
- 37. He Z, Xia Y, Pan C, et al. Up-regulation of MiR-452 inhibits metastasis of non-small cell lung cancer by regulating BMI1. Cell Physiol Biochem. 2015;37(1):387–398. [DOI] [PubMed] [Google Scholar]
- 38. Xiong W, Zeng ZY, Xia JH, et al. A susceptibility locus at chromosome 3p21 linked to familial nasopharyngeal carcinoma. Cancer Res. 2004;64(6):1972–1974. [DOI] [PubMed] [Google Scholar]
- 39. Ng CC, Yew PY, Puah SM. A genome-wide association study identifies ITGA9 conferring risk of nasopharyngeal carcinoma. J Hum Genet. 2009;54(7):392–397. [DOI] [PubMed] [Google Scholar]
- 40. Nawaz I, Moumad K, Martorelli D. Detection of nasopharyngeal carcinoma in Morocco (North Africa) using a multiplex methylation-specific PCR biomarker assay. Clin Epigenetics. 2015;7(1):89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nawaz I, Hu LF, Du ZM. Integrin α9 gene promoter is hypermethylated and downregulated in nasopharyngeal carcinoma. Oncotarget. 2015;6(31):31493–314507. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material, Figure_S1 for An Integrated Analysis of mRNAs and miRNAs Microarray Profiles to Screen miRNA Signatures Involved in Nasopharyngeal Carcinoma by Lei Liu, Hailing Wang, Chaohui Yan and Shudong Tao in Technology in Cancer Research & Treatment
Supplemental Material, Figure_S2 for An Integrated Analysis of mRNAs and miRNAs Microarray Profiles to Screen miRNA Signatures Involved in Nasopharyngeal Carcinoma by Lei Liu, Hailing Wang, Chaohui Yan and Shudong Tao in Technology in Cancer Research & Treatment
Supplemental Material, Figure_S3 for An Integrated Analysis of mRNAs and miRNAs Microarray Profiles to Screen miRNA Signatures Involved in Nasopharyngeal Carcinoma by Lei Liu, Hailing Wang, Chaohui Yan and Shudong Tao in Technology in Cancer Research & Treatment
Supplemental Material, Figure_S4 for An Integrated Analysis of mRNAs and miRNAs Microarray Profiles to Screen miRNA Signatures Involved in Nasopharyngeal Carcinoma by Lei Liu, Hailing Wang, Chaohui Yan and Shudong Tao in Technology in Cancer Research & Treatment
Supplementary_tables for An Integrated Analysis of mRNAs and miRNAs Microarray Profiles to Screen miRNA Signatures Involved in Nasopharyngeal Carcinoma by Lei Liu, Hailing Wang, Chaohui Yan and Shudong Tao in Technology in Cancer Research & Treatment