The crystal structure of the title compound shows a disorder of the methyl and methoxycarbonyl groups of one alanine residue. Compared to previously reported peptide biphenyl hybrids, the backbone torsion angles are different.
Keywords: crystal structure, hydrogen bonding, peptide dimethyl biphenyl hybrids, Pro-Phe-Ala
Abstract
The synthesis and crystal structure of peptide 6,6′-dimethyl biphenyl hybrid are described. The title compound was synthesized by reaction between 6,6′-dimethyl-[1,1′-biphenyl]-2,2′-dicarbonyl dichloride in CH2Cl2, amine HN–proline–phenylalanine–alanine–COOMe and Et3N at 273 K under N2 atmosphere and characterized by single-crystal X-ray diffraction. The asymmetric unit contains one peptide molecule and a quarter of a water molecule. A disorder of a methyl and methoxycarbonyl group of one alanine residue is observed with occupancy ratio 0.502 (6):0.498 (6). The structure is consolidated by intra- and intermolecular hydrogen bonds.
Chemical context
Since the first application in 1922 of peptides in the treatment of diabetes with insulin (Banting et al., 1922 ▸), the chemistry of peptides has become a very important domain in the search of new therapeutic drugs. From 2011 to 2018, the global market of drugs has increased from US $ 14.1 to 24.4 billion. With more than 140 peptides in clinical trials, the number of peptide-based drugs is expected to grow significantly (Fosgerau et al., 2015 ▸). Despite their tremendous potential, applications of peptides for pharmaceutical purposes are limited by their instability toward enzymatic systems, short half-life, rapid renal clearance, and formulation challenges (Otvos et al., 2014 ▸). These problems can be overcome by modifying the linear peptide to enhance the stability and therefore the selectivity and affinity. The biphenyl structure is present in numerous pharmaceuticals and bioactive compounds, as illustrated by the glycopeptide antibiotic vancomycin, the proteasome inhibitor TMC-95A (Kaiser et al., 2004 ▸) and arylomycins (Schimana et al., 2002 ▸). A statistical analysis of NMR data indicates that compounds containing the biphenyl structure can bind a wide range of proteins with high levels of specificity (Hajduk et al., 2000 ▸). Coupling of a small protein chain to the biphenyl structure is a strategy to create a new family of peptidomimetic compounds, which can be used in medicinal chemistry because of its specific conformation and its particular hydrogen-bonding interactions.
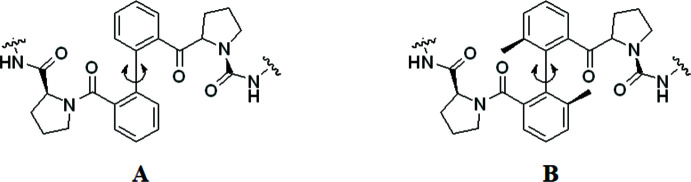
The synthesis and biological activity as calpain inhibitor of peptide–biphenyl hybrids type I have been reported by Montero and Mann (Montero et al., 2004a
▸,b
▸; Mann et al., 2002 ▸). Amine et al. (2002 ▸) synthesized a bis amido–copper(II) complex from N-containing tetradentate ligands having two amido groups with a biphenyl skeleton, which is used as a DNA cleaving agent. Recently, we have reported crystallographic studies of a peptide-biphenyl hybrid A (Fig. 1 ▸) with tripeptide Pro–Phe–Ala (Le et al., 2020 ▸).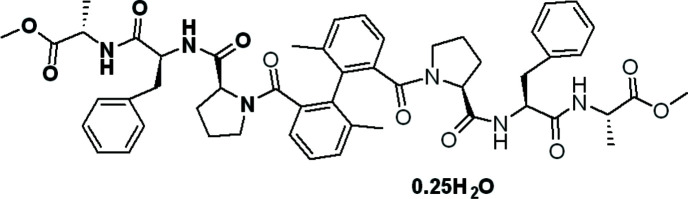
Figure 1.
Peptide–biphenyl hybrids A and B.
We report herein the synthesis and crystallographic study of a peptide-2,2′-biphenyl B (Fig. 1 ▸) with the introduction of two methyl groups at the 6-6′ positions to prevent free rotation around the central aryl–aryl bond.
Structural commentary
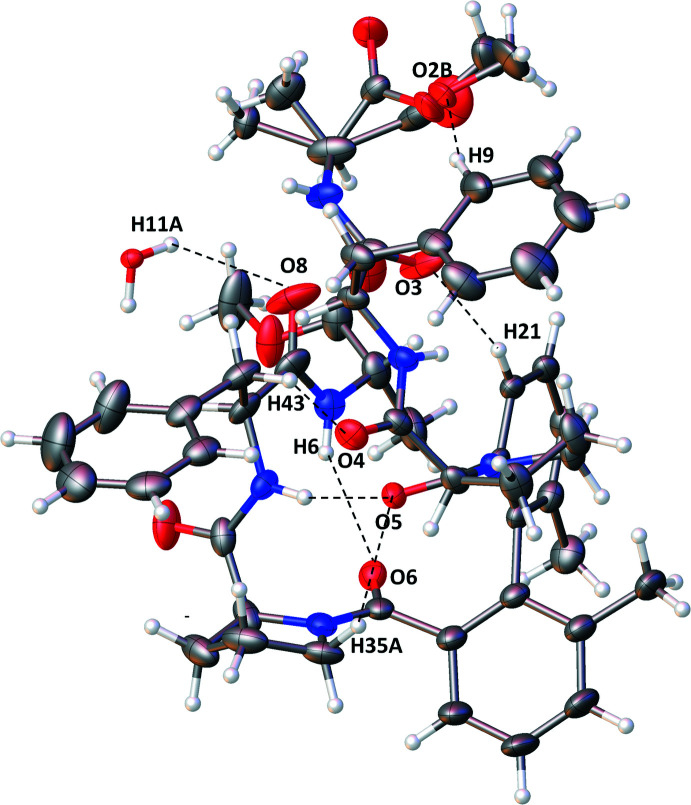
The compound dimethyl 2,2′-[((2S,2′S)-2,2′-{[(2S,2′S)-1,1′-(6,6′-dimethyl-[1,1′-biphenyl]-2,2′-dicarbonyl)bis(pyrrolidine-1,2-diyl-2-carbonyl)]bis(azanediyl)}bis(3-phenylpropanoyl))bis(azanediyl)](2S,2′S)-dipropionate (Fig. 2 ▸) crystallizes in the monoclinic space group C2 with one molecule of peptide biphenyl hybrid accompanied by a quarter of a water molecule in the asymmetric unit. Two methyl groups have been introduced to the biphenyl rings at the 6,6′ position in order to limit the rotation of the two central phenyl rings in solution. In the solid state, the dihedral angle between biphenyl rings C20–C25 and C27-C32 is 73.8 (3)°. However, this value is similar to that of a previous compound not bearing the methyl groups (C50H56N6O10·0.5H2O; Le et al., 2020 ▸). A disorder of a methyl and methoxycarbonyl group of alanine is observed in the crystal structure and was refined with an occupancy ratio of 0.502 (6):0.498 (6).
Figure 2.
A view of the molecular structure of the title compound showing displacement ellipsoids drawn at the 50% probability level and hydrogen bonds (dashed lines) within the asymmetric unit. H atoms are shown as small circles of arbitrary radii.
The backbone conformation of the two tripeptide fragments is characterized by the torsion angles ω, φ, ψ (see Table 1 ▸). The torsion angles φ and ψ of amino acids Ala1, Ala2, Phe2 correspond with the usual α-helix (right-handed) region of the Ramachandran plot, and only the torsion angles of amino acid Phe1 fall into the corresponding type β-sheet Ramachandran plot region. For both prolines, the related torsion angles lie in the α region of the Ramachandran plot for proline.
Table 1. Backbone torsion angles ω, φ, ψ (°) for the two tripeptide fragments.
| C20—C19—N3—C15 | 178.3 (2) | C32—C34—N4—C38 | −164.6 (2) |
| C19—N3—C15—C14 | −73.4 (3) | C34—N4—C38—C39 | −69.1 (3) |
| N3—C15—C14—N2 | −17.5 (3) | N4—C38—C39—N5 | −14.4 (4) |
| C15—C14—N2—C6 | 176.5 (2) | C38—C39—N5—C40 | −177.2 (2) |
| C14—N2—C6—C5 | −163.0 (2) | C39—N5—C40—C48 | −106.8 (3) |
| N2—C6—C5—N1 | 171.4 (2) | N5—C40—C48—N6 | 18.6 (3) |
| C6—C5—N1—C3 | −174.8 (3) | C40—C48—N6—C49 | 179.1 (2) |
| C5—N1—C3—C2B | −58.0 (5) | C48—N6—C49—C51 | −60.9 (3) |
| N1—C3—C2B—O2B | −39.6 (13) | N6—C49—C51—O9 | −35.0 (4) |
There are six intramolecular hydrogen bonds formed in the structure of the title compound (Table 2 ▸). Two hydrogen bonds are formed between the NH and CO groups with H⋯O distances of 2.07 Å for N5—H5 ⋯O5 and 2.42 Å for N6—H6⋯O6. The latter value is noticeably longer than the values observed (from 2.04 to 2.29 Å) in other reported peptides (Ranganathan et al., 1997 ▸; Le et al., 2020 ▸). Four other intramolecular bonds are formed between CH and CO groups with distances from 2.35 to 2.59 Å.
Table 2. Hydrogen-bond geometry (Å, °).
Cg3 and Cg5 are the centroids of the C8–C13 and C27–C32 rings, respectively.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N5—H5⋯O5 | 0.88 | 2.07 | 2.923 (3) | 162 |
| N6—H6⋯O6 | 0.88 | 2.42 | 3.233 (3) | 154 |
| C9—H9⋯O2B | 0.95 | 2.35 | 3.270 (18) | 164 |
| C21—H21⋯O3 | 0.95 | 2.44 | 3.352 (4) | 161 |
| C35—H35A⋯O5 | 0.99 | 2.51 | 3.171 (4) | 124 |
| C43—H43⋯O4 | 0.95 | 2.59 | 3.443 (4) | 149 |
| N1—H1⋯O4i | 0.88 | 2.01 | 2.865 (3) | 163 |
| C1B—H1BB⋯O10ii | 0.98 | 2.46 | 2.913 (16) | 108 |
| C30—H30⋯O8iii | 0.95 | 2.46 | 3.222 (4) | 137 |
| C35—H35⋯O7iv | 0.99 | 2.39 | 3.228 (4) | 142 |
| C52—H52B⋯O10v | 0.98 | 2.60 | 3.559 (5) | 166 |
| O11—H11A⋯O8 | 0.87 | 2.48 | 3.136 (6) | 133 |
| C13—H13⋯O11vi | 0.95 | 2.52 | 3.155 (7) | 124 |
| C36—H36B⋯Cg3vi | 0.99 | 2.94 | 3.845 (4) | 152 |
| C4A—H4AC⋯Cg5vii | 1.05 (8) | 2.93 (7) | 3.770 (8) | 135 (5) |
Symmetry codes: (i) 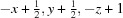 ; (ii)
; (ii) 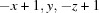 ; (iii)
; (iii)  ; (iv)
; (iv)  ; (v)
; (v) 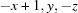 ; (vi)
; (vi)  ; (vii)
; (vii)  .
.
Supramolecular features
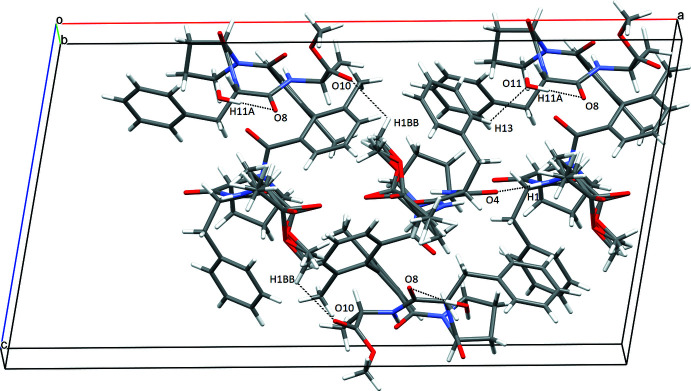
In the crystal, the packing is characterized by N—H⋯O, O—H⋯O and C—H⋯O hydrogen bonding (see Table 2, Fig. 3 ▸
▸). The strongest intermolecular interaction is formed between NH and CO groups of two neighboring peptide residues [N1—H1⋯O4i, with d = 2.01 Å; symmetry code: (i) 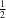 − x,
− x, 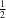 + y, 1 − z]. Furthermore, there are six additional hydrogen bonds linking the molecules. Two contacts are established between the water molecule and two tripeptides (O11—H11A⋯O8; C13— H13⋯O11). Four C—H⋯O=C contacts with H⋯O distances ranging from 2.39 to 2.60 Å further consolidate the crystal packing. In addition, the molecules are linked by two intermolecular C—H ⋯π interactions, one between a proline H atom and the phenyl ring of a phenylalanine residue, the other between a H atom of the disordered methyl group and a phenyl ring of the central biphenyl fragment.
+ y, 1 − z]. Furthermore, there are six additional hydrogen bonds linking the molecules. Two contacts are established between the water molecule and two tripeptides (O11—H11A⋯O8; C13— H13⋯O11). Four C—H⋯O=C contacts with H⋯O distances ranging from 2.39 to 2.60 Å further consolidate the crystal packing. In addition, the molecules are linked by two intermolecular C—H ⋯π interactions, one between a proline H atom and the phenyl ring of a phenylalanine residue, the other between a H atom of the disordered methyl group and a phenyl ring of the central biphenyl fragment.
Figure 3.
Crystal packing of the title compound, indicating some intermolecular hydrogen bonds (dashed lines).
Database survey
A search of the Cambridge Structural Database (version 5.41 with update of March 2020; Groom et al., 2016 ▸) for peptide–dimethyl biphenyl hybrids was conducted. There are seven dimethyl biphenyl hybrid structures with only one amino acid, including JITYET (Linden & Rippert, 2018a ▸), JITZEU (Linden & Rippert, 2018b ▸), JITYOD (Linden & Rippert, 2018c ▸), NOSPUG & NOSQAN (Weigand & Feigel, 1998 ▸), PITSUJ (Linden et al., 2018d ▸) and NIKJOI (Samadi et al., 2013 ▸). For these structures the dihedral angles between the dimethyl biphenyl rings varies from 82.0 to 95.8o, larger than the corresponding angle of the title compound.
Synthesis and crystallization
To a round-bottom flask was added 6,6′-dimethyl-[1,1′-biphenyl]-2,2′-dicarboxylic acid (1 eq.) and SOCl2 (3 eq.) respectively under a nitrogen atmosphere. The mixture was heated under reflux for 4 h and was then evaporated under vacuum. The acid chloride was used in the next step without further purification.
To a round-bottom flask was added amine HN–proline–phenylalanine–alanine–COOMe (1 eq.), Et3N (2 eq.) and anhydrous CH2Cl2 (50mL). To this solution was added a solution of (6,6′-dimethyl-[1,1′-biphenyl]-2,2′-dicarbonyl dichloride in CH2Cl2 at 273 K under an N2 atmosphere. After completion of the reaction, the mixture was washed with 1 N HCl solution, water and a solution of brine, respectively. The organic phase was dried over Na2SO4, filtered and evaporated under reduced pressure. The crude product was then purified by flash chromatography (AcOEt/hexane 3:2) to give a white solid (60% yield). The compound was recrystallized by slow evaporation in methanol to give crystals suitable for X-ray diffraction.
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 3 ▸. The methyl and methoxycarbonyl groups of alanine show two conformations with refined occupancy factors converging to 0.502 (6) and 0.498 (6). Geometrical restraints were applied to the disordered atoms. H atoms were placed at calculated positions (C—H = 0.95–1.08 Å and N—H = 0.88 Å), with isotropic displacement parameters U iso(H) = 1.5U eq(C) for methyl H atoms and 1.2U eq(C,N) for all other H atoms. The solvent water molecule is disordered and was refined with a site occupation factor fixed to 0.25. The H atoms of the water molecule were located in difference-Fourier maps and refined in riding-model approximation with U iso(H) = 1.5U eq(O).
Table 3. Experimental details.
| Crystal data | |
| Chemical formula | C52H60N6O10·0.25H2O |
| M r | 933.56 |
| Crystal system, space group | Monoclinic, C2 |
| Temperature (K) | 100 |
| a, b, c (Å) | 27.505 (3), 12.3814 (12), 14.6346 (14) |
| β (°) | 99.999 (3) |
| V (Å3) | 4908.2 (8) |
| Z | 4 |
| Radiation type | Mo Kα |
| μ (mm−1) | 0.09 |
| Crystal size (mm) | 0.3 × 0.2 × 0.1 |
| Data collection | |
| Diffractometer | Bruker D8 Quest CMOS |
| Absorption correction | Multi-scan (SADABS-; Bruker, 2013 ▸) |
| T min, T max | 0.713, 0.745 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 84318, 9371, 7959 |
| R int | 0.062 |
| (sin θ/λ)max (Å−1) | 0.611 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.038, 0.086, 1.06 |
| No. of reflections | 9371 |
| No. of parameters | 693 |
| No. of restraints | 4 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.28, −0.16 |
| Absolute structure | Flack x determined using 3323 quotients [(I +)−(I −)]/[(I +)+(I −)] (Parsons et al., 2013 ▸) |
| Absolute structure parameter | −0.1 (3) |
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989020012931/vm2240sup1.cif
Supporting information file. DOI: 10.1107/S2056989020012931/vm2240Isup3.cdx
Supporting information file. DOI: 10.1107/S2056989020012931/vm2240Isup4.cdx
Supporting information file. DOI: 10.1107/S2056989020012931/vm2240sup5.docx
Supporting information file. DOI: 10.1107/S2056989020012931/vm2240sup6.docx
Supporting information file. DOI: 10.1107/S2056989020012931/vm2240sup7.tif
Supporting information file. DOI: 10.1107/S2056989020012931/vm2240sup8.tif
CCDC reference: 2026794
Additional supporting information: crystallographic information; 3D view; checkCIF report
supplementary crystallographic information
Crystal data
| C52H60N6O10·0.25H2O | F(000) = 1986 |
| Mr = 933.56 | Dx = 1.263 Mg m−3 |
| Monoclinic, C2 | Mo Kα radiation, λ = 0.71073 Å |
| a = 27.505 (3) Å | Cell parameters from 9371 reflections |
| b = 12.3814 (12) Å | θ = 2.8–25.8° |
| c = 14.6346 (14) Å | µ = 0.09 mm−1 |
| β = 99.999 (3)° | T = 100 K |
| V = 4908.2 (8) Å3 | Needle, clear light colourless |
| Z = 4 | 0.3 × 0.2 × 0.1 mm |
Data collection
| Bruker D8 Quest CMOS diffractometer | 7959 reflections with I > 2σ(I) |
| φ and ω scans | Rint = 0.062 |
| Absorption correction: multi-scan (SADABS-; Bruker, 2013) | θmax = 25.8°, θmin = 2.8° |
| Tmin = 0.713, Tmax = 0.745 | h = −33→33 |
| 84318 measured reflections | k = −15→15 |
| 9371 independent reflections | l = −17→17 |
Refinement
| Refinement on F2 | Hydrogen site location: mixed |
| Least-squares matrix: full | H atoms treated by a mixture of independent and constrained refinement |
| R[F2 > 2σ(F2)] = 0.038 | w = 1/[σ2(Fo2) + (0.0359P)2 + 2.1291P] where P = (Fo2 + 2Fc2)/3 |
| wR(F2) = 0.086 | (Δ/σ)max < 0.001 |
| S = 1.06 | Δρmax = 0.28 e Å−3 |
| 9371 reflections | Δρmin = −0.16 e Å−3 |
| 693 parameters | Absolute structure: Flack x determined using 3323 quotients [(I+)-(I-)]/[(I+)+(I-)] (Parsons et al., 2013) |
| 4 restraints | Absolute structure parameter: −0.1 (3) |
| Primary atom site location: dual |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| O5 | 0.33318 (6) | 0.37671 (15) | 0.30911 (12) | 0.0262 (4) | |
| O4 | 0.25033 (7) | 0.41245 (17) | 0.49754 (15) | 0.0367 (5) | |
| O6 | 0.37283 (7) | 0.35432 (16) | 0.07943 (13) | 0.0314 (4) | |
| N3 | 0.36969 (8) | 0.32535 (18) | 0.45153 (14) | 0.0252 (5) | |
| N2 | 0.32176 (8) | 0.50351 (19) | 0.50273 (15) | 0.0291 (5) | |
| H2 | 0.3537 | 0.4960 | 0.5038 | 0.035* | |
| O7 | 0.25946 (8) | 0.5451 (2) | −0.00116 (15) | 0.0531 (6) | |
| O3 | 0.38550 (8) | 0.64671 (17) | 0.4803 (2) | 0.0552 (7) | |
| O9 | 0.40799 (7) | 0.7684 (2) | 0.02936 (16) | 0.0536 (7) | |
| N1 | 0.33544 (8) | 0.7906 (2) | 0.48045 (17) | 0.0351 (6) | |
| H1 | 0.3070 | 0.8150 | 0.4917 | 0.042* | |
| N5 | 0.29721 (8) | 0.50901 (18) | 0.14594 (15) | 0.0277 (5) | |
| H5 | 0.3085 | 0.4583 | 0.1862 | 0.033* | |
| N4 | 0.29975 (8) | 0.2897 (2) | 0.10755 (15) | 0.0302 (5) | |
| O2A | 0.4152 (8) | 0.8404 (16) | 0.5865 (9) | 0.052 (4) | 0.498 (6) |
| O10 | 0.47865 (9) | 0.8022 (2) | 0.12726 (19) | 0.0665 (8) | |
| C19 | 0.37162 (9) | 0.35712 (19) | 0.36445 (17) | 0.0221 (5) | |
| O8 | 0.36763 (10) | 0.74704 (19) | 0.2248 (2) | 0.0667 (8) | |
| N6 | 0.38529 (8) | 0.6049 (2) | 0.14123 (16) | 0.0335 (5) | |
| H6 | 0.3742 | 0.5463 | 0.1106 | 0.040* | |
| C20 | 0.42128 (9) | 0.3761 (2) | 0.33809 (17) | 0.0224 (5) | |
| C14 | 0.29481 (10) | 0.4139 (2) | 0.49355 (18) | 0.0277 (6) | |
| C22 | 0.49273 (10) | 0.4901 (2) | 0.34994 (19) | 0.0300 (6) | |
| H22 | 0.5116 | 0.5499 | 0.3769 | 0.036* | |
| C21 | 0.44813 (10) | 0.4663 (2) | 0.37539 (19) | 0.0264 (6) | |
| H21 | 0.4355 | 0.5113 | 0.4184 | 0.032* | |
| C39 | 0.27589 (10) | 0.4803 (3) | 0.05996 (19) | 0.0336 (7) | |
| C34 | 0.34964 (10) | 0.2867 (2) | 0.11713 (18) | 0.0292 (6) | |
| C25 | 0.43896 (10) | 0.3099 (2) | 0.27349 (17) | 0.0264 (6) | |
| C15 | 0.32173 (10) | 0.3090 (2) | 0.48016 (18) | 0.0290 (6) | |
| H15 | 0.3004 | 0.2638 | 0.4327 | 0.035* | |
| C23 | 0.50992 (10) | 0.4261 (2) | 0.2846 (2) | 0.0339 (7) | |
| H23 | 0.5403 | 0.4441 | 0.2659 | 0.041* | |
| C32 | 0.37479 (11) | 0.1926 (2) | 0.1698 (2) | 0.0342 (7) | |
| C5 | 0.34545 (10) | 0.6858 (2) | 0.4894 (2) | 0.0362 (7) | |
| C42 | 0.22491 (12) | 0.6148 (3) | 0.2431 (2) | 0.0393 (7) | |
| C8 | 0.32288 (11) | 0.5941 (3) | 0.6886 (2) | 0.0368 (7) | |
| C18 | 0.41107 (10) | 0.2861 (3) | 0.52286 (19) | 0.0355 (7) | |
| H18A | 0.4391 | 0.3375 | 0.5316 | 0.043* | |
| H18B | 0.4228 | 0.2143 | 0.5062 | 0.043* | |
| C24 | 0.48386 (10) | 0.3365 (2) | 0.24582 (19) | 0.0326 (7) | |
| C6 | 0.30370 (9) | 0.6123 (2) | 0.5111 (2) | 0.0303 (6) | |
| H6A | 0.2743 | 0.6231 | 0.4612 | 0.036* | |
| C28 | 0.43711 (12) | 0.1101 (2) | 0.2858 (2) | 0.0382 (7) | |
| C38 | 0.27062 (10) | 0.3606 (3) | 0.03950 (19) | 0.0348 (7) | |
| H38 | 0.2789 | 0.3459 | −0.0232 | 0.042* | |
| C27 | 0.41520 (10) | 0.2032 (2) | 0.2421 (2) | 0.0306 (6) | |
| C7 | 0.28776 (10) | 0.6356 (3) | 0.6048 (2) | 0.0360 (7) | |
| H7A | 0.2831 (11) | 0.715 (3) | 0.610 (2) | 0.043* | |
| H7B | 0.2561 (12) | 0.604 (3) | 0.606 (2) | 0.043* | |
| C40 | 0.30215 (11) | 0.6212 (2) | 0.1744 (2) | 0.0344 (7) | |
| H40 | 0.2820 | 0.6644 | 0.1237 | 0.041* | |
| C48 | 0.35480 (12) | 0.6626 (2) | 0.1841 (2) | 0.0395 (7) | |
| C51 | 0.44291 (12) | 0.7469 (3) | 0.1020 (2) | 0.0465 (8) | |
| C41 | 0.27949 (12) | 0.6391 (2) | 0.2612 (2) | 0.0403 (7) | |
| H41A | 0.2962 | 0.5918 | 0.3116 | 0.048* | |
| H41B | 0.2848 | 0.7150 | 0.2818 | 0.048* | |
| C49 | 0.43632 (11) | 0.6369 (3) | 0.1441 (2) | 0.0381 (7) | |
| H49 | 0.4534 | 0.6375 | 0.2103 | 0.046* | |
| C31 | 0.35836 (13) | 0.0900 (2) | 0.1390 (2) | 0.0456 (8) | |
| H31 | 0.3315 | 0.0830 | 0.0888 | 0.055* | |
| C17 | 0.38721 (11) | 0.2796 (3) | 0.6093 (2) | 0.0461 (8) | |
| H17A | 0.3879 | 0.3506 | 0.6406 | 0.055* | |
| H17B | 0.4040 | 0.2256 | 0.6538 | 0.055* | |
| C43 | 0.20546 (13) | 0.5304 (3) | 0.2867 (2) | 0.0406 (8) | |
| H43 | 0.2267 | 0.4876 | 0.3304 | 0.049* | |
| C26 | 0.50273 (12) | 0.2713 (3) | 0.1714 (2) | 0.0459 (8) | |
| H26A | 0.5078 | 0.1962 | 0.1920 | 0.069* | |
| H26B | 0.5341 | 0.3018 | 0.1604 | 0.069* | |
| H26C | 0.4785 | 0.2737 | 0.1139 | 0.069* | |
| C35 | 0.26820 (12) | 0.2261 (3) | 0.1605 (2) | 0.0403 (8) | |
| H35A | 0.2818 | 0.2267 | 0.2278 | 0.048* | |
| H35B | 0.2648 | 0.1504 | 0.1386 | 0.048* | |
| C9 | 0.36689 (11) | 0.6476 (3) | 0.7211 (3) | 0.0515 (9) | |
| H9 | 0.3753 | 0.7096 | 0.6890 | 0.062* | |
| C30 | 0.38083 (14) | −0.0007 (3) | 0.1809 (3) | 0.0563 (10) | |
| H30 | 0.3697 | −0.0703 | 0.1592 | 0.068* | |
| C36 | 0.21906 (12) | 0.2852 (3) | 0.1399 (2) | 0.0492 (9) | |
| H36A | 0.2182 | 0.3463 | 0.1833 | 0.059* | |
| H36B | 0.1912 | 0.2358 | 0.1441 | 0.059* | |
| C44 | 0.15538 (13) | 0.5070 (3) | 0.2676 (2) | 0.0488 (9) | |
| H44 | 0.1427 | 0.4490 | 0.2990 | 0.059* | |
| C16 | 0.33427 (11) | 0.2449 (3) | 0.5705 (2) | 0.0434 (8) | |
| H16A | 0.3324 | 0.1662 | 0.5584 | 0.052* | |
| H16B | 0.3117 | 0.2637 | 0.6138 | 0.052* | |
| C29 | 0.41922 (14) | 0.0090 (3) | 0.2539 (2) | 0.0520 (9) | |
| H29 | 0.4339 | −0.0543 | 0.2834 | 0.062* | |
| C33 | 0.48036 (13) | 0.1160 (3) | 0.3656 (2) | 0.0466 (8) | |
| H33A | 0.5112 | 0.1201 | 0.3410 | 0.070* | |
| H33B | 0.4807 | 0.0514 | 0.4044 | 0.070* | |
| H33C | 0.4771 | 0.1804 | 0.4030 | 0.070* | |
| C37 | 0.21720 (11) | 0.3253 (3) | 0.0408 (2) | 0.0492 (9) | |
| H37A | 0.1941 | 0.3867 | 0.0268 | 0.059* | |
| H37B | 0.2070 | 0.2668 | −0.0047 | 0.059* | |
| C50 | 0.46145 (12) | 0.5544 (3) | 0.0915 (2) | 0.0498 (9) | |
| H50A | 0.4560 | 0.4818 | 0.1145 | 0.075* | |
| H50B | 0.4970 | 0.5693 | 0.1008 | 0.075* | |
| H50C | 0.4476 | 0.5587 | 0.0252 | 0.075* | |
| C13 | 0.31123 (13) | 0.5060 (3) | 0.7362 (2) | 0.0546 (9) | |
| H13 | 0.2811 | 0.4689 | 0.7158 | 0.065* | |
| C3 | 0.37093 (12) | 0.8660 (3) | 0.4522 (3) | 0.0506 (9) | |
| H3A | 0.3893 | 0.8282 | 0.4082 | 0.061* | 0.498 (6) |
| H3B | 0.3776 | 0.8469 | 0.3891 | 0.061* | 0.502 (6) |
| C45 | 0.12403 (14) | 0.5668 (3) | 0.2040 (3) | 0.0587 (10) | |
| H45 | 0.0898 | 0.5502 | 0.1903 | 0.070* | |
| C47 | 0.19287 (15) | 0.6750 (4) | 0.1803 (3) | 0.0668 (12) | |
| H47 | 0.2053 | 0.7342 | 0.1500 | 0.080* | |
| C52 | 0.41693 (14) | 0.8637 (4) | −0.0232 (3) | 0.0722 (13) | |
| H52A | 0.3905 | 0.8713 | −0.0770 | 0.108* | |
| H52B | 0.4487 | 0.8563 | −0.0443 | 0.108* | |
| H52C | 0.4177 | 0.9278 | 0.0163 | 0.108* | |
| C11 | 0.38662 (15) | 0.5233 (4) | 0.8454 (3) | 0.0675 (12) | |
| H11 | 0.4085 | 0.4988 | 0.8989 | 0.081* | |
| C10 | 0.39836 (14) | 0.6128 (4) | 0.7984 (3) | 0.0665 (12) | |
| H10 | 0.4283 | 0.6505 | 0.8195 | 0.080* | |
| C12 | 0.34359 (16) | 0.4694 (4) | 0.8155 (3) | 0.0695 (12) | |
| H12 | 0.3354 | 0.4074 | 0.8480 | 0.083* | |
| C46 | 0.14300 (16) | 0.6512 (4) | 0.1605 (3) | 0.0794 (14) | |
| H46 | 0.1217 | 0.6935 | 0.1164 | 0.095* | |
| O1B | 0.46376 (18) | 0.8694 (5) | 0.5125 (5) | 0.082 (2) | 0.502 (6) |
| C1A | 0.4577 (7) | 0.8657 (13) | 0.6565 (10) | 0.065 (4) | 0.498 (6) |
| H1AA | 0.4865 | 0.8781 | 0.6266 | 0.098* | 0.498 (6) |
| H1AB | 0.4645 | 0.8052 | 0.7000 | 0.098* | 0.498 (6) |
| H1AC | 0.4510 | 0.9308 | 0.6902 | 0.098* | 0.498 (6) |
| C4A | 0.3372 (3) | 0.9666 (6) | 0.3947 (6) | 0.0338 (16) | 0.498 (6) |
| H4AA | 0.314 (3) | 1.001 (6) | 0.437 (5) | 0.051* | 0.498 (6) |
| H4AB | 0.318 (3) | 0.951 (6) | 0.348 (5) | 0.051* | 0.498 (6) |
| H4AC | 0.365 (3) | 1.022 (6) | 0.380 (5) | 0.051* | 0.498 (6) |
| C2B | 0.4233 (4) | 0.8525 (9) | 0.5327 (8) | 0.051 (3) | 0.502 (6) |
| O11 | 0.2727 (2) | 0.8908 (5) | 0.1778 (4) | 0.0166 (14) | 0.25 |
| H11A | 0.3048 | 0.8916 | 0.1906 | 0.025* | 0.25 |
| H11B | 0.2657 | 0.8381 | 0.1383 | 0.025* | 0.25 |
| C4B | 0.3574 (3) | 0.9738 (5) | 0.4551 (7) | 0.049 (2) | 0.502 (6) |
| H4BA | 0.3299 | 0.9887 | 0.4044 | 0.074* | 0.502 (6) |
| H4BB | 0.3856 | 1.0197 | 0.4483 | 0.074* | 0.502 (6) |
| H4BC | 0.3471 | 0.9891 | 0.5146 | 0.074* | 0.502 (6) |
| C2A | 0.4049 (2) | 0.9141 (6) | 0.5214 (4) | 0.0317 (15) | 0.498 (6) |
| O2B | 0.4184 (6) | 0.8407 (16) | 0.6190 (9) | 0.044 (3) | 0.502 (6) |
| O1A | 0.42431 (16) | 1.0015 (3) | 0.5223 (3) | 0.0492 (16) | 0.498 (6) |
| C1B | 0.4595 (6) | 0.8377 (14) | 0.6925 (11) | 0.081 (5) | 0.502 (6) |
| H1BA | 0.4832 | 0.7835 | 0.6788 | 0.121* | 0.502 (6) |
| H1BB | 0.4483 | 0.8188 | 0.7504 | 0.121* | 0.502 (6) |
| H1BC | 0.4754 | 0.9088 | 0.6989 | 0.121* | 0.502 (6) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O5 | 0.0269 (9) | 0.0271 (10) | 0.0227 (9) | 0.0019 (8) | −0.0009 (8) | −0.0012 (8) |
| O4 | 0.0254 (10) | 0.0376 (12) | 0.0472 (12) | −0.0104 (9) | 0.0069 (8) | −0.0008 (10) |
| O6 | 0.0302 (10) | 0.0322 (11) | 0.0318 (10) | 0.0050 (9) | 0.0054 (8) | −0.0008 (9) |
| N3 | 0.0254 (12) | 0.0256 (12) | 0.0232 (11) | −0.0044 (9) | 0.0002 (9) | 0.0021 (9) |
| N2 | 0.0221 (11) | 0.0308 (13) | 0.0348 (13) | −0.0061 (10) | 0.0056 (9) | −0.0074 (11) |
| O7 | 0.0503 (14) | 0.0743 (17) | 0.0343 (11) | 0.0200 (12) | 0.0064 (10) | 0.0256 (12) |
| O3 | 0.0292 (12) | 0.0309 (12) | 0.114 (2) | −0.0076 (9) | 0.0349 (13) | −0.0243 (13) |
| O9 | 0.0327 (12) | 0.0701 (18) | 0.0595 (15) | 0.0071 (11) | 0.0126 (11) | 0.0410 (13) |
| N1 | 0.0259 (12) | 0.0284 (13) | 0.0554 (16) | −0.0076 (10) | 0.0196 (11) | −0.0116 (12) |
| N5 | 0.0354 (13) | 0.0248 (12) | 0.0246 (12) | 0.0065 (10) | 0.0099 (10) | 0.0053 (10) |
| N4 | 0.0309 (12) | 0.0303 (13) | 0.0290 (12) | −0.0028 (10) | 0.0040 (10) | −0.0103 (10) |
| O2A | 0.041 (5) | 0.038 (5) | 0.065 (10) | −0.016 (3) | −0.026 (7) | 0.003 (8) |
| O10 | 0.0492 (15) | 0.0653 (18) | 0.0834 (19) | −0.0116 (14) | 0.0069 (13) | 0.0320 (15) |
| C19 | 0.0295 (14) | 0.0124 (12) | 0.0231 (13) | −0.0013 (10) | 0.0014 (11) | −0.0028 (10) |
| O8 | 0.0794 (18) | 0.0229 (12) | 0.110 (2) | −0.0139 (12) | 0.0500 (16) | −0.0127 (13) |
| N6 | 0.0345 (13) | 0.0342 (13) | 0.0325 (13) | −0.0013 (11) | 0.0081 (10) | 0.0046 (11) |
| C20 | 0.0277 (13) | 0.0161 (12) | 0.0222 (13) | 0.0028 (10) | 0.0005 (10) | 0.0058 (11) |
| C14 | 0.0282 (15) | 0.0322 (15) | 0.0219 (14) | −0.0075 (12) | 0.0020 (11) | 0.0003 (12) |
| C22 | 0.0305 (15) | 0.0233 (15) | 0.0342 (15) | −0.0039 (12) | 0.0000 (12) | 0.0053 (12) |
| C21 | 0.0305 (15) | 0.0200 (13) | 0.0269 (14) | 0.0013 (11) | 0.0000 (11) | 0.0012 (11) |
| C39 | 0.0246 (14) | 0.052 (2) | 0.0257 (15) | 0.0092 (13) | 0.0095 (12) | 0.0095 (14) |
| C34 | 0.0337 (15) | 0.0254 (14) | 0.0272 (14) | 0.0013 (12) | 0.0019 (12) | −0.0103 (12) |
| C25 | 0.0307 (14) | 0.0229 (14) | 0.0241 (13) | 0.0047 (11) | 0.0006 (11) | 0.0036 (11) |
| C15 | 0.0274 (14) | 0.0286 (15) | 0.0300 (15) | −0.0091 (11) | 0.0020 (11) | 0.0045 (12) |
| C23 | 0.0269 (14) | 0.0369 (17) | 0.0379 (17) | −0.0005 (13) | 0.0057 (12) | 0.0097 (14) |
| C32 | 0.0427 (17) | 0.0248 (15) | 0.0337 (16) | 0.0039 (13) | 0.0028 (13) | −0.0065 (12) |
| C5 | 0.0258 (15) | 0.0318 (17) | 0.054 (2) | −0.0077 (12) | 0.0160 (14) | −0.0184 (14) |
| C42 | 0.0492 (18) | 0.0316 (16) | 0.0422 (17) | 0.0097 (14) | 0.0227 (14) | −0.0042 (14) |
| C8 | 0.0328 (15) | 0.0426 (18) | 0.0371 (16) | 0.0007 (13) | 0.0116 (13) | −0.0186 (15) |
| C18 | 0.0313 (15) | 0.0407 (17) | 0.0315 (15) | −0.0012 (13) | −0.0032 (12) | 0.0135 (14) |
| C24 | 0.0320 (15) | 0.0343 (17) | 0.0319 (15) | 0.0077 (13) | 0.0070 (12) | 0.0033 (13) |
| C6 | 0.0210 (13) | 0.0302 (15) | 0.0406 (16) | −0.0080 (12) | 0.0084 (11) | −0.0110 (13) |
| C28 | 0.0498 (18) | 0.0244 (15) | 0.0389 (17) | 0.0049 (14) | 0.0033 (14) | −0.0024 (14) |
| C38 | 0.0255 (14) | 0.057 (2) | 0.0208 (13) | 0.0045 (14) | 0.0019 (11) | −0.0098 (14) |
| C27 | 0.0378 (16) | 0.0226 (14) | 0.0316 (15) | 0.0033 (12) | 0.0069 (12) | −0.0036 (12) |
| C7 | 0.0213 (14) | 0.043 (2) | 0.0457 (18) | −0.0077 (13) | 0.0109 (13) | −0.0152 (15) |
| C40 | 0.0463 (17) | 0.0194 (15) | 0.0407 (16) | 0.0077 (12) | 0.0168 (13) | 0.0088 (12) |
| C48 | 0.055 (2) | 0.0228 (16) | 0.0453 (18) | 0.0042 (14) | 0.0211 (15) | 0.0123 (14) |
| C51 | 0.0326 (17) | 0.057 (2) | 0.051 (2) | 0.0011 (16) | 0.0109 (15) | 0.0229 (17) |
| C41 | 0.058 (2) | 0.0239 (16) | 0.0442 (18) | 0.0049 (14) | 0.0228 (15) | 0.0003 (13) |
| C49 | 0.0352 (16) | 0.0478 (19) | 0.0303 (15) | −0.0007 (14) | 0.0030 (12) | 0.0116 (14) |
| C31 | 0.059 (2) | 0.0280 (18) | 0.0443 (19) | 0.0017 (15) | −0.0071 (15) | −0.0127 (14) |
| C17 | 0.0407 (17) | 0.065 (2) | 0.0304 (16) | −0.0062 (16) | −0.0007 (13) | 0.0203 (16) |
| C43 | 0.056 (2) | 0.0351 (18) | 0.0337 (16) | 0.0022 (15) | 0.0160 (14) | −0.0066 (14) |
| C26 | 0.0419 (18) | 0.052 (2) | 0.0469 (19) | 0.0102 (16) | 0.0174 (15) | −0.0010 (16) |
| C35 | 0.0485 (19) | 0.0351 (17) | 0.0394 (17) | −0.0185 (15) | 0.0138 (14) | −0.0148 (14) |
| C9 | 0.0289 (17) | 0.046 (2) | 0.075 (2) | 0.0047 (14) | −0.0034 (16) | −0.0197 (18) |
| C30 | 0.079 (3) | 0.0222 (17) | 0.060 (2) | 0.0023 (17) | −0.0101 (19) | −0.0131 (16) |
| C36 | 0.0418 (18) | 0.049 (2) | 0.062 (2) | −0.0186 (16) | 0.0242 (16) | −0.0270 (18) |
| C44 | 0.060 (2) | 0.042 (2) | 0.051 (2) | −0.0074 (17) | 0.0283 (17) | −0.0189 (17) |
| C16 | 0.0408 (18) | 0.051 (2) | 0.0391 (18) | −0.0072 (15) | 0.0079 (14) | 0.0166 (15) |
| C29 | 0.071 (2) | 0.0218 (16) | 0.058 (2) | 0.0099 (16) | −0.0032 (18) | −0.0010 (16) |
| C33 | 0.057 (2) | 0.0244 (16) | 0.052 (2) | 0.0060 (15) | −0.0065 (16) | 0.0037 (15) |
| C37 | 0.0285 (16) | 0.066 (2) | 0.052 (2) | −0.0036 (15) | 0.0053 (14) | −0.0270 (18) |
| C50 | 0.0316 (17) | 0.073 (2) | 0.0423 (19) | 0.0083 (17) | 0.0006 (14) | 0.0043 (18) |
| C13 | 0.055 (2) | 0.075 (3) | 0.0339 (18) | −0.022 (2) | 0.0101 (15) | −0.0034 (18) |
| C3 | 0.048 (2) | 0.0403 (19) | 0.073 (2) | −0.0194 (16) | 0.0389 (18) | −0.0190 (18) |
| C45 | 0.047 (2) | 0.069 (3) | 0.063 (2) | 0.009 (2) | 0.0205 (19) | −0.013 (2) |
| C47 | 0.060 (2) | 0.064 (3) | 0.085 (3) | 0.023 (2) | 0.035 (2) | 0.034 (2) |
| C52 | 0.047 (2) | 0.088 (3) | 0.089 (3) | 0.017 (2) | 0.032 (2) | 0.066 (3) |
| C11 | 0.059 (3) | 0.093 (4) | 0.046 (2) | 0.008 (2) | −0.0040 (18) | −0.026 (2) |
| C10 | 0.042 (2) | 0.062 (3) | 0.087 (3) | 0.0087 (19) | −0.013 (2) | −0.031 (2) |
| C12 | 0.079 (3) | 0.091 (3) | 0.039 (2) | −0.013 (2) | 0.013 (2) | 0.005 (2) |
| C46 | 0.053 (3) | 0.095 (4) | 0.094 (3) | 0.031 (2) | 0.022 (2) | 0.029 (3) |
| O1B | 0.034 (3) | 0.090 (5) | 0.129 (6) | −0.012 (3) | 0.029 (3) | 0.011 (4) |
| C1A | 0.067 (8) | 0.054 (7) | 0.063 (8) | −0.011 (6) | −0.020 (6) | 0.018 (5) |
| C4A | 0.029 (4) | 0.035 (4) | 0.038 (4) | 0.007 (3) | 0.007 (3) | 0.009 (3) |
| C2B | 0.040 (5) | 0.023 (5) | 0.095 (9) | −0.004 (4) | 0.027 (6) | −0.008 (5) |
| O11 | 0.016 (3) | 0.015 (3) | 0.019 (3) | −0.002 (3) | 0.005 (3) | −0.001 (3) |
| C4B | 0.051 (5) | 0.035 (4) | 0.069 (6) | −0.002 (3) | 0.028 (5) | 0.012 (4) |
| C2A | 0.024 (3) | 0.025 (4) | 0.048 (4) | 0.000 (3) | 0.012 (3) | 0.001 (3) |
| O2B | 0.023 (4) | 0.034 (4) | 0.065 (8) | −0.008 (3) | −0.017 (6) | −0.012 (6) |
| O1A | 0.041 (3) | 0.027 (3) | 0.074 (3) | −0.017 (2) | −0.006 (2) | 0.009 (2) |
| C1B | 0.039 (5) | 0.087 (11) | 0.101 (12) | −0.018 (6) | −0.031 (7) | −0.012 (9) |
Geometric parameters (Å, º)
| O5—C19 | 1.239 (3) | C49—C50 | 1.517 (5) |
| O4—C14 | 1.235 (3) | C31—H31 | 0.9500 |
| O6—C34 | 1.239 (3) | C31—C30 | 1.374 (5) |
| N3—C19 | 1.343 (3) | C17—H17A | 0.9900 |
| N3—C15 | 1.466 (3) | C17—H17B | 0.9900 |
| N3—C18 | 1.486 (3) | C17—C16 | 1.530 (4) |
| N2—H2 | 0.8800 | C43—H43 | 0.9500 |
| N2—C14 | 1.328 (3) | C43—C44 | 1.388 (5) |
| N2—C6 | 1.449 (4) | C26—H26A | 0.9800 |
| O7—C39 | 1.228 (4) | C26—H26B | 0.9800 |
| O3—C5 | 1.232 (3) | C26—H26C | 0.9800 |
| O9—C51 | 1.330 (4) | C35—H35A | 0.9900 |
| O9—C52 | 1.452 (4) | C35—H35B | 0.9900 |
| N1—H1 | 0.8800 | C35—C36 | 1.521 (5) |
| N1—C5 | 1.329 (4) | C9—H9 | 0.9500 |
| N1—C3 | 1.461 (4) | C9—C10 | 1.369 (5) |
| N5—H5 | 0.8800 | C30—H30 | 0.9500 |
| N5—C39 | 1.341 (4) | C30—C29 | 1.371 (5) |
| N5—C40 | 1.450 (4) | C36—H36A | 0.9900 |
| N4—C34 | 1.355 (3) | C36—H36B | 0.9900 |
| N4—C38 | 1.459 (4) | C36—C37 | 1.526 (5) |
| N4—C35 | 1.486 (4) | C44—H44 | 0.9500 |
| O2A—C1A | 1.449 (14) | C44—C45 | 1.371 (5) |
| O2A—C2A | 1.314 (15) | C16—H16A | 0.9900 |
| O10—C51 | 1.203 (4) | C16—H16B | 0.9900 |
| C19—C20 | 1.501 (4) | C29—H29 | 0.9500 |
| O8—C48 | 1.225 (4) | C33—H33A | 0.9800 |
| N6—H6 | 0.8800 | C33—H33B | 0.9800 |
| N6—C48 | 1.338 (4) | C33—H33C | 0.9800 |
| N6—C49 | 1.452 (4) | C37—H37A | 0.9900 |
| C20—C21 | 1.397 (4) | C37—H37B | 0.9900 |
| C20—C25 | 1.401 (4) | C50—H50A | 0.9800 |
| C14—C15 | 1.524 (4) | C50—H50B | 0.9800 |
| C22—H22 | 0.9500 | C50—H50C | 0.9800 |
| C22—C21 | 1.375 (4) | C13—H13 | 0.9500 |
| C22—C23 | 1.387 (4) | C13—C12 | 1.409 (5) |
| C21—H21 | 0.9500 | C3—H3A | 1.0000 |
| C39—C38 | 1.513 (5) | C3—H3B | 1.0000 |
| C34—C32 | 1.498 (4) | C3—C4A | 1.688 (8) |
| C25—C24 | 1.404 (4) | C3—C2B | 1.704 (13) |
| C25—C27 | 1.510 (4) | C3—C4B | 1.389 (7) |
| C15—H15 | 1.0000 | C3—C2A | 1.388 (7) |
| C15—C16 | 1.529 (4) | C45—H45 | 0.9500 |
| C23—H23 | 0.9500 | C45—C46 | 1.373 (6) |
| C23—C24 | 1.388 (4) | C47—H47 | 0.9500 |
| C32—C27 | 1.402 (4) | C47—C46 | 1.384 (6) |
| C32—C31 | 1.396 (4) | C52—H52A | 0.9800 |
| C5—C6 | 1.540 (4) | C52—H52B | 0.9800 |
| C42—C41 | 1.509 (5) | C52—H52C | 0.9800 |
| C42—C43 | 1.379 (4) | C11—H11 | 0.9500 |
| C42—C47 | 1.377 (5) | C11—C10 | 1.372 (6) |
| C8—C7 | 1.513 (4) | C11—C12 | 1.363 (6) |
| C8—C9 | 1.389 (4) | C10—H10 | 0.9500 |
| C8—C13 | 1.363 (5) | C12—H12 | 0.9500 |
| C18—H18A | 0.9900 | C46—H46 | 0.9500 |
| C18—H18B | 0.9900 | O1B—C2B | 1.218 (10) |
| C18—C17 | 1.526 (4) | C1A—H1AA | 0.9800 |
| C24—C26 | 1.517 (4) | C1A—H1AB | 0.9800 |
| C6—H6A | 1.0000 | C1A—H1AC | 0.9800 |
| C6—C7 | 1.537 (4) | C4A—H4AA | 1.05 (7) |
| C28—C27 | 1.403 (4) | C4A—H4AB | 0.81 (8) |
| C28—C29 | 1.395 (5) | C4A—H4AC | 1.08 (8) |
| C28—C33 | 1.517 (4) | C2B—O2B | 1.301 (17) |
| C38—H38 | 1.0000 | O11—H11A | 0.8705 |
| C38—C37 | 1.537 (4) | O11—H11B | 0.8699 |
| C7—H7A | 0.99 (4) | C4B—H4BA | 0.9800 |
| C7—H7B | 0.96 (3) | C4B—H4BB | 0.9800 |
| C40—H40 | 1.0000 | C4B—H4BC | 0.9800 |
| C40—C48 | 1.519 (4) | C2A—O1A | 1.205 (8) |
| C40—C41 | 1.525 (4) | O2B—C1B | 1.419 (13) |
| C51—C49 | 1.518 (5) | C1B—H1BA | 0.9800 |
| C41—H41A | 0.9900 | C1B—H1BB | 0.9800 |
| C41—H41B | 0.9900 | C1B—H1BC | 0.9800 |
| C49—H49 | 1.0000 | ||
| C19—N3—C15 | 119.8 (2) | C16—C17—H17B | 111.2 |
| C19—N3—C18 | 127.6 (2) | C42—C43—H43 | 119.4 |
| C15—N3—C18 | 111.8 (2) | C42—C43—C44 | 121.1 (3) |
| C14—N2—H2 | 116.9 | C44—C43—H43 | 119.4 |
| C14—N2—C6 | 126.3 (2) | C24—C26—H26A | 109.5 |
| C6—N2—H2 | 116.9 | C24—C26—H26B | 109.5 |
| C51—O9—C52 | 114.9 (3) | C24—C26—H26C | 109.5 |
| C5—N1—H1 | 119.5 | H26A—C26—H26B | 109.5 |
| C5—N1—C3 | 121.0 (2) | H26A—C26—H26C | 109.5 |
| C3—N1—H1 | 119.5 | H26B—C26—H26C | 109.5 |
| C39—N5—H5 | 119.1 | N4—C35—H35A | 111.2 |
| C39—N5—C40 | 121.9 (2) | N4—C35—H35B | 111.2 |
| C40—N5—H5 | 119.1 | N4—C35—C36 | 102.8 (3) |
| C34—N4—C38 | 120.9 (2) | H35A—C35—H35B | 109.1 |
| C34—N4—C35 | 127.2 (3) | C36—C35—H35A | 111.2 |
| C38—N4—C35 | 112.0 (2) | C36—C35—H35B | 111.2 |
| C2A—O2A—C1A | 114.1 (14) | C8—C9—H9 | 119.3 |
| O5—C19—N3 | 120.5 (2) | C10—C9—C8 | 121.4 (4) |
| O5—C19—C20 | 120.8 (2) | C10—C9—H9 | 119.3 |
| N3—C19—C20 | 118.5 (2) | C31—C30—H30 | 119.9 |
| C48—N6—H6 | 119.3 | C29—C30—C31 | 120.2 (3) |
| C48—N6—C49 | 121.5 (3) | C29—C30—H30 | 119.9 |
| C49—N6—H6 | 119.3 | C35—C36—H36A | 111.1 |
| C21—C20—C19 | 117.9 (2) | C35—C36—H36B | 111.1 |
| C21—C20—C25 | 120.6 (2) | C35—C36—C37 | 103.1 (2) |
| C25—C20—C19 | 121.3 (2) | H36A—C36—H36B | 109.1 |
| O4—C14—N2 | 123.2 (3) | C37—C36—H36A | 111.1 |
| O4—C14—C15 | 120.1 (2) | C37—C36—H36B | 111.1 |
| N2—C14—C15 | 116.7 (2) | C43—C44—H44 | 119.7 |
| C21—C22—H22 | 120.3 | C45—C44—C43 | 120.6 (3) |
| C21—C22—C23 | 119.4 (3) | C45—C44—H44 | 119.7 |
| C23—C22—H22 | 120.3 | C15—C16—C17 | 103.5 (2) |
| C20—C21—H21 | 119.9 | C15—C16—H16A | 111.1 |
| C22—C21—C20 | 120.1 (3) | C15—C16—H16B | 111.1 |
| C22—C21—H21 | 119.9 | C17—C16—H16A | 111.1 |
| O7—C39—N5 | 123.8 (3) | C17—C16—H16B | 111.1 |
| O7—C39—C38 | 119.0 (3) | H16A—C16—H16B | 109.0 |
| N5—C39—C38 | 117.2 (2) | C28—C29—H29 | 119.4 |
| O6—C34—N4 | 121.7 (3) | C30—C29—C28 | 121.2 (3) |
| O6—C34—C32 | 121.8 (2) | C30—C29—H29 | 119.4 |
| N4—C34—C32 | 116.3 (3) | C28—C33—H33A | 109.5 |
| C20—C25—C24 | 118.9 (2) | C28—C33—H33B | 109.5 |
| C20—C25—C27 | 122.3 (2) | C28—C33—H33C | 109.5 |
| C24—C25—C27 | 118.3 (2) | H33A—C33—H33B | 109.5 |
| N3—C15—C14 | 113.6 (2) | H33A—C33—H33C | 109.5 |
| N3—C15—H15 | 109.1 | H33B—C33—H33C | 109.5 |
| N3—C15—C16 | 103.9 (2) | C38—C37—H37A | 111.1 |
| C14—C15—H15 | 109.1 | C38—C37—H37B | 111.1 |
| C14—C15—C16 | 111.9 (2) | C36—C37—C38 | 103.3 (2) |
| C16—C15—H15 | 109.1 | C36—C37—H37A | 111.1 |
| C22—C23—H23 | 119.2 | C36—C37—H37B | 111.1 |
| C22—C23—C24 | 121.7 (3) | H37A—C37—H37B | 109.1 |
| C24—C23—H23 | 119.2 | C49—C50—H50A | 109.5 |
| C27—C32—C34 | 123.4 (2) | C49—C50—H50B | 109.5 |
| C31—C32—C34 | 116.5 (3) | C49—C50—H50C | 109.5 |
| C31—C32—C27 | 120.0 (3) | H50A—C50—H50B | 109.5 |
| O3—C5—N1 | 123.2 (3) | H50A—C50—H50C | 109.5 |
| O3—C5—C6 | 120.2 (3) | H50B—C50—H50C | 109.5 |
| N1—C5—C6 | 116.6 (2) | C8—C13—H13 | 119.7 |
| C43—C42—C41 | 121.6 (3) | C8—C13—C12 | 120.5 (3) |
| C47—C42—C41 | 120.7 (3) | C12—C13—H13 | 119.7 |
| C47—C42—C43 | 117.7 (3) | N1—C3—H3A | 108.7 |
| C9—C8—C7 | 120.8 (3) | N1—C3—H3B | 110.4 |
| C13—C8—C7 | 120.8 (3) | N1—C3—C4A | 106.0 (3) |
| C13—C8—C9 | 118.3 (3) | N1—C3—C2B | 105.5 (4) |
| N3—C18—H18A | 111.3 | C4A—C3—H3A | 108.7 |
| N3—C18—H18B | 111.3 | C2B—C3—H3B | 110.4 |
| N3—C18—C17 | 102.3 (2) | C4B—C3—N1 | 114.2 (4) |
| H18A—C18—H18B | 109.2 | C4B—C3—H3B | 110.4 |
| C17—C18—H18A | 111.3 | C4B—C3—C2B | 105.8 (6) |
| C17—C18—H18B | 111.3 | C2A—C3—N1 | 117.8 (4) |
| C25—C24—C26 | 120.6 (3) | C2A—C3—H3A | 108.7 |
| C23—C24—C25 | 119.2 (3) | C2A—C3—C4A | 106.8 (5) |
| C23—C24—C26 | 120.2 (3) | C44—C45—H45 | 120.6 |
| N2—C6—C5 | 104.6 (2) | C44—C45—C46 | 118.8 (4) |
| N2—C6—H6A | 107.9 | C46—C45—H45 | 120.6 |
| N2—C6—C7 | 113.9 (3) | C42—C47—H47 | 119.3 |
| C5—C6—H6A | 107.9 | C42—C47—C46 | 121.3 (4) |
| C7—C6—C5 | 114.3 (2) | C46—C47—H47 | 119.3 |
| C7—C6—H6A | 107.9 | O9—C52—H52A | 109.5 |
| C27—C28—C33 | 121.9 (3) | O9—C52—H52B | 109.5 |
| C29—C28—C27 | 119.1 (3) | O9—C52—H52C | 109.5 |
| C29—C28—C33 | 119.0 (3) | H52A—C52—H52B | 109.5 |
| N4—C38—C39 | 115.6 (2) | H52A—C52—H52C | 109.5 |
| N4—C38—H38 | 109.3 | H52B—C52—H52C | 109.5 |
| N4—C38—C37 | 103.5 (3) | C10—C11—H11 | 120.0 |
| C39—C38—H38 | 109.3 | C12—C11—H11 | 120.0 |
| C39—C38—C37 | 109.6 (2) | C12—C11—C10 | 120.1 (4) |
| C37—C38—H38 | 109.3 | C9—C10—C11 | 119.9 (4) |
| C32—C27—C25 | 123.8 (2) | C9—C10—H10 | 120.1 |
| C32—C27—C28 | 119.2 (3) | C11—C10—H10 | 120.1 |
| C28—C27—C25 | 116.9 (2) | C13—C12—H12 | 120.1 |
| C8—C7—C6 | 114.8 (2) | C11—C12—C13 | 119.8 (4) |
| C8—C7—H7A | 110.1 (19) | C11—C12—H12 | 120.1 |
| C8—C7—H7B | 107.6 (19) | C45—C46—C47 | 120.5 (4) |
| C6—C7—H7A | 108.4 (19) | C45—C46—H46 | 119.7 |
| C6—C7—H7B | 109.5 (19) | C47—C46—H46 | 119.7 |
| H7A—C7—H7B | 106 (3) | O2A—C1A—H1AA | 109.5 |
| N5—C40—H40 | 106.6 | O2A—C1A—H1AB | 109.5 |
| N5—C40—C48 | 113.0 (2) | O2A—C1A—H1AC | 109.5 |
| N5—C40—C41 | 110.3 (2) | H1AA—C1A—H1AB | 109.5 |
| C48—C40—H40 | 106.6 | H1AA—C1A—H1AC | 109.5 |
| C48—C40—C41 | 113.2 (3) | H1AB—C1A—H1AC | 109.5 |
| C41—C40—H40 | 106.6 | C3—C4A—H4AA | 110 (4) |
| O8—C48—N6 | 122.2 (3) | C3—C4A—H4AB | 117 (5) |
| O8—C48—C40 | 121.5 (3) | C3—C4A—H4AC | 102 (4) |
| N6—C48—C40 | 116.2 (3) | H4AA—C4A—H4AB | 102 (6) |
| O9—C51—C49 | 112.6 (3) | H4AA—C4A—H4AC | 113 (5) |
| O10—C51—O9 | 124.8 (3) | H4AB—C4A—H4AC | 112 (7) |
| O10—C51—C49 | 122.3 (3) | O1B—C2B—C3 | 120.9 (9) |
| C42—C41—C40 | 111.3 (3) | O1B—C2B—O2B | 120.6 (13) |
| C42—C41—H41A | 109.4 | O2B—C2B—C3 | 117.7 (9) |
| C42—C41—H41B | 109.4 | H11A—O11—H11B | 104.4 |
| C40—C41—H41A | 109.4 | C3—C4B—H4BA | 109.5 |
| C40—C41—H41B | 109.4 | C3—C4B—H4BB | 109.5 |
| H41A—C41—H41B | 108.0 | C3—C4B—H4BC | 109.5 |
| N6—C49—C51 | 114.6 (2) | H4BA—C4B—H4BB | 109.5 |
| N6—C49—H49 | 108.6 | H4BA—C4B—H4BC | 109.5 |
| N6—C49—C50 | 108.9 (3) | H4BB—C4B—H4BC | 109.5 |
| C51—C49—H49 | 108.6 | O2A—C2A—C3 | 105.3 (9) |
| C50—C49—C51 | 107.5 (3) | O1A—C2A—O2A | 125.2 (10) |
| C50—C49—H49 | 108.6 | O1A—C2A—C3 | 129.3 (6) |
| C32—C31—H31 | 119.9 | C2B—O2B—C1B | 122.3 (15) |
| C30—C31—C32 | 120.2 (3) | O2B—C1B—H1BA | 109.5 |
| C30—C31—H31 | 119.9 | O2B—C1B—H1BB | 109.5 |
| C18—C17—H17A | 111.2 | O2B—C1B—H1BC | 109.5 |
| C18—C17—H17B | 111.2 | H1BA—C1B—H1BB | 109.5 |
| C18—C17—C16 | 103.0 (2) | H1BA—C1B—H1BC | 109.5 |
| H17A—C17—H17B | 109.1 | H1BB—C1B—H1BC | 109.5 |
| C16—C17—H17A | 111.2 | ||
| C20—C19—N3—C15 | 178.3 (2) | C32—C34—N4—C38 | −164.6 (2) |
| C19—N3—C15—C14 | −73.4 (3) | C34—N4—C38—C39 | −69.1 (3) |
| N3—C15—C14—N2 | −17.5 (3) | N4—C38—C39—N5 | −14.4 (4) |
| C15—C14—N2—C6 | 176.5 (2) | C38—C39—N5—C40 | −177.2 (2) |
| C14—N2—C6—C5 | −163.0 (2) | C39—N5—C40—C48 | −106.8 (3) |
| N2—C6—C5—N1 | 171.4 (2) | N5—C40—C48—N6 | 18.6 (3) |
| C6—C5—N1—C3 | −174.8 (3) | C40—C48—N6—C49 | 179.1 (2) |
| C5—N1—C3—C2B | −58.0 (5) | C48—N6—C49—C51 | −60.9 (3) |
| N1—C3—C2B—O2B | −39.6 (13) | N6—C49—C51—O9 | −35.0 (4) |
Hydrogen-bond geometry (Å, º)
Cg3 and Cg5 are the centroids of the C8–C13 and C27–C32 rings, respectively.
| D—H···A | D—H | H···A | D···A | D—H···A |
| N5—H5···O5 | 0.88 | 2.07 | 2.923 (3) | 162 |
| N6—H6···O6 | 0.88 | 2.42 | 3.233 (3) | 154 |
| C9—H9···O2B | 0.95 | 2.35 | 3.270 (18) | 164 |
| C21—H21···O3 | 0.95 | 2.44 | 3.352 (4) | 161 |
| C35—H35A···O5 | 0.99 | 2.51 | 3.171 (4) | 124 |
| C43—H43···O4 | 0.95 | 2.59 | 3.443 (4) | 149 |
| N1—H1···O4i | 0.88 | 2.01 | 2.865 (3) | 163 |
| C1B—H1BB···O10ii | 0.98 | 2.46 | 2.913 (16) | 108 |
| C30—H30···O8iii | 0.95 | 2.46 | 3.222 (4) | 137 |
| C35—H35···O7iv | 0.99 | 2.39 | 3.228 (4) | 142 |
| C52—H52B···O10v | 0.98 | 2.60 | 3.559 (5) | 166 |
| O11—H11A···O8 | 0.87 | 2.48 | 3.136 (6) | 133 |
| C13—H13···O11vi | 0.95 | 2.52 | 3.155 (7) | 124 |
| C36—H36B···Cg3vi | 0.99 | 2.94 | 3.845 (4) | 152 |
| C4A—H4AC···Cg5vii | 1.05 (8) | 2.93 (7) | 3.770 (8) | 135 (5) |
Symmetry codes: (i) −x+1/2, y+1/2, −z+1; (ii) −x+1, y, −z+1; (iii) x, y−1, z; (iv) −x+1/2, y−1/2, −z; (v) −x+1, y, −z; (vi) −x+1/2, y−1/2, −z+1; (vii) x, y+1, z.
Funding Statement
This work was funded by Asia Research Center-Vietnam National University grant CA.20.7A. Korea Foundation for Advanced Studies grant CA.20.7A.
References
- Amine, A., Atmani, Z., El Hallaoui, A., Giorgi, M., Pierrot, M. & Réglier, M. (2002). Bioorg. Med. Chem. Lett. 12, 57–60. [DOI] [PubMed]
- Banting, F. G., Best, C. H., Collip, J. B., Campbell, W. R. & Fletcher, A. A. (1922). Can. Med. Assoc. J. 12, 141–146. [PMC free article] [PubMed]
- Bruker (2013). APEX2, SAINT and SADABS. Bruker AXS Inc., Madison, Wisconsin, USA.
- Dolomanov, O. V., Bourhis, L. J., Gildea, R. J., Howard, J. A. K. & Puschmann, H. (2009). J. Appl. Cryst. 42, 339–341.
- Fosgerau, K. & Hoffmann, T. (2015). Drug Discovery Today, 20, 122–128. [DOI] [PubMed]
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Hajduk, P. J., Bures, M., Praestgaard, J. & Fesik, S. W. (2000). J. Med. Chem. 43, 3443–3447. [DOI] [PubMed]
- Kaiser, M., Groll, M., Siciliano, C., Assfalg-Machleidt, I., Weyher, E., Kohno, J., Milbradt, A. G., Renner, C., Huber, R. & Moroder, L. (2004). ChemBioChem, 5, 1256–1266. [DOI] [PubMed]
- Le, T. Q., Nguyen, X. T., Nguyen, H. H., Mac, D. H. & Bui, T. T. T. (2020). Acta Cryst. E76, 257–260. [DOI] [PMC free article] [PubMed]
- Linden, A., Furegati, M. & Rippert, A. J. (2018d). Private communication (CCDC refcode 1885480). CCDC, Cambridge, England.
- Linden, A. & Rippert, A. J. (2018a). Private communication (CCDC refcode 1884542). CCDC, Cambridge, England.
- Linden, A. & Rippert, A. J. (2018b). Private communication (CCDC refcode 1884572). CCDC, Cambridge, England.
- Linden, A. & Rippert, A. J. (2018c). Private communication (CCDC refcode 1884549). CCDC, Cambridge, England.
- Mann, E., Montero, A., Maestro, M. & Herradón, B. (2002). Helv. Chim. Acta, 85, 3624–3638.
- Montero, A., Albericio, F., Royo, M. & Herradón, B. (2004b). Org. Lett. 6, 4089–4092. [DOI] [PubMed]
- Montero, A., Mann, E., Chana, A. & Herradón, B. (2004a). Chem. Biodiv. 1, 442–457. [DOI] [PubMed]
- Otvos, L. Jr & Wade, J. D. (2014). Front. Chem, 2, 62. [DOI] [PMC free article] [PubMed]
- Parsons, S., Flack, H. D. & Wagner, T. (2013). Acta Cryst. B69, 249–259. [DOI] [PMC free article] [PubMed]
- Ranganathan, D., Kurur, S., Madhusudanan, K. P. & Karle, I. L. (1997). Tetrahedron Lett. 38, 4659–4662.
- Samadi, S., Nazari, S., Arvinnezhad, H., Jadidi, K. & Notash, B. (2013). Tetrahedron, 69, 6679–6686.
- Schimana, J., Gebhardt, K., Holtzel, A., Schmid, D. G., Sussmuth, R., Muller, J., Pukall, R. & Fiedler, H.-P. (2002). J. Antibiot. 55, 565–570. [DOI] [PubMed]
- Sheldrick, G. M. (2015a). Acta Cryst. A71, 3–8.
- Sheldrick, G. M. (2015b). Acta Cryst. C71, 3–8.
- Weigand, C. & Feigel, M. (1998). Chem. Commun. pp. 679–680.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989020012931/vm2240sup1.cif
Supporting information file. DOI: 10.1107/S2056989020012931/vm2240Isup3.cdx
Supporting information file. DOI: 10.1107/S2056989020012931/vm2240Isup4.cdx
Supporting information file. DOI: 10.1107/S2056989020012931/vm2240sup5.docx
Supporting information file. DOI: 10.1107/S2056989020012931/vm2240sup6.docx
Supporting information file. DOI: 10.1107/S2056989020012931/vm2240sup7.tif
Supporting information file. DOI: 10.1107/S2056989020012931/vm2240sup8.tif
CCDC reference: 2026794
Additional supporting information: crystallographic information; 3D view; checkCIF report