Short syntheses to high Fsp 3 index natural-product analogues such as iminosugars are of paramount importance in the investigation of their biological activities and reducing the use of protecting groups is an advantageous synthetic strategy. In this case only an isopropylidene group was employed towards the synthesis of seven-membered ring iminosugars.
Keywords: crystal structure, iminosugar, d-glucose, tosylation, azide, regioselectivity, glycosidase inhibition
Abstract
Short syntheses to high Fsp 3 index natural-product analogues such as iminosugars are of paramount importance in the investigation of their biological activities and reducing the use of protecting groups is an advantageous synthetic strategy. An isopropylidene group was employed towards the synthesis of seven-membered ring iminosugars and the title compound, C9H15N3O5, was crystallized as an intermediate, in which the THF ring is twisted and the dioxolane ring adopts an envelope conformation: the dihedral angle between the rings is 67.50 (13)°. In the crystal, the hydroxyl groups participate in O—H⋯(O,O) and O—H⋯N hydrogen-bonding interactions, which generate chains of molecules propagating parallel to the a-axis direction. There is a notable non-classical C—H⋯O hydrogen bond, which cross-links the [100] chains into (001) sheets.
Chemical context
The installation of various functionalities via N- and/or O-alkylation has been shown to impart improved biological profiles and potencies to iminosugars (Šesták et al., 2018 ▸; Prichard et al., 2018 ▸; Simone et al., 2012 ▸; Sayce et al., 2016 ▸, Woodhouse et al., 2008 ▸; Johnson & Houston, 2002 ▸). Diminishing the number of synthetic steps to the iminosugar building blocks that are precursors to their alkylated congeners is advantageous. Many iminosugar syntheses start from monosaccharide starting materials (Wood et al., 2018 ▸; Lee et al., 2012 ▸; Rasmussen & Jensen, 2011 ▸). Reducing the number of protecting groups and removing the need for purification by chromatography are useful strategies to a more expedited synthesis of analogues (Katritzky et al., 1991 ▸; Steiner et al., 2009 ▸; Liu et al., 2014 ▸).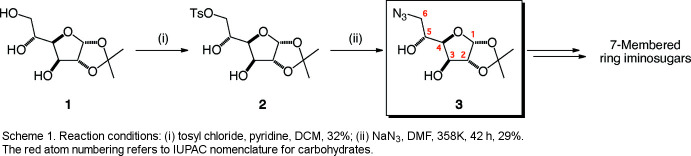
In the present study, the only protecting group that was used to synthesize seven-membered ring iminosugars was an isopropylidene group (acetonide) to make intermediate 1 from d-glucose. Selective tosylation of the primary hydroxyl group, followed by nucleophilic displacement with sodium azide afforded the title compound 3, C9H15N3O5 (Tsuchiya et al., 1981 ▸; Fleet et al., 1989 ▸), see Scheme 1 ▸.
Primary alcohols can be tosylated regioselectively over secondary alcohols (Johnson et al., 1963 ▸). There are examples of monotosylation of monosaccharides and analogues using di-n-butyltin oxide and dimethylaminopyridine as catalyst (Tsuda et al., 1991 ▸) and of cyclodextrins (Yamamura & Fujita, 1991 ▸; Ashton et al., 1991 ▸; Fujita et al., 1992 ▸). Any mechanistic ambiguities that may have arisen from the SN2 reaction with azide ions was clarified by X-ray crystallographic analysis, which confirmed the structure of the title compound as described below.
Structural commentary
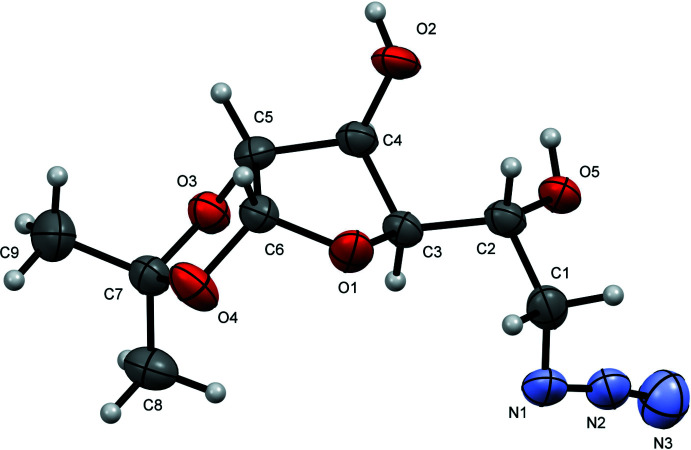
In compound 3 (Fig. 1 ▸), the tetrahydrofuran (THF) ring is best described as twisted with atoms C3 and C4 displaced by 0.169 (3) and −0.384 (2) Å, respectively, from the plane through C5/C6/O1. The fused dioxolane ring adopts an envelope conformation with O3 displaced by 0.402 (2) Å from the mean plane of the other ring atoms (C5/C6/O4/C7; r.m.s. deviation = 0.005 Å). The dihedral angle between the five-membered rings (all atoms) is 67.50 (13)°. The hydroxyl group O2—H2A and the acetonide oxygen atom O3 project axially from the THF ring, lying respectively above and below in a trans arrangement from one another [O2—C4—C5—O3 = 164.46 (18)°]. The other two groups projecting from the THF ring are O4 of the acetonide and the side chain attached to C3, which sit equatorially. The absolute structure of 3 was not definitively established in the refinement but the configurations of the stereogenic atoms (C2 R, C3 R, C4 S, C5 R and C6 R) were set to match those of the starting material.
Figure 1.
The molecular structure of 3 showing 50% displacement ellipsoids.
Supramolecular features
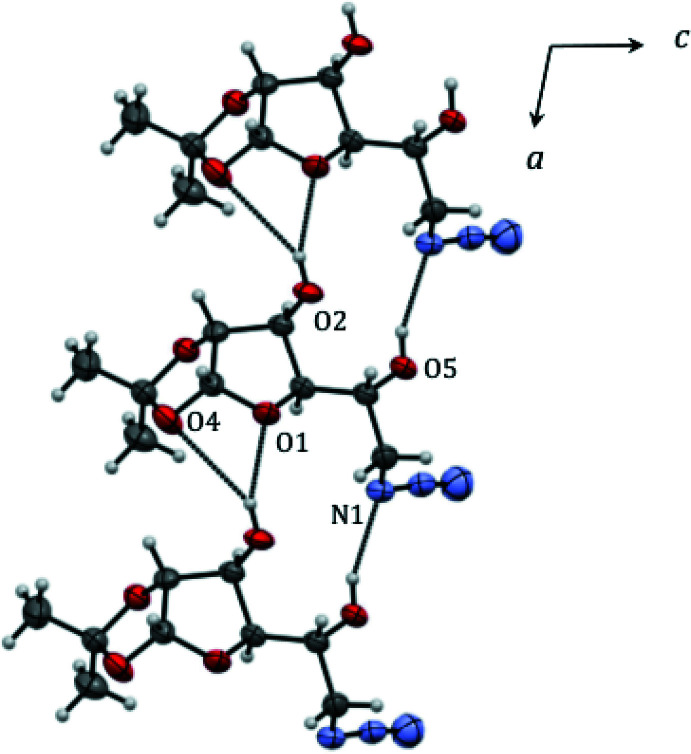
There are no intramolecular hydrogen-bonding interactions in 3 but both hydroxyl groups participate in intermolecular hydrogen-bonding interactions (Table 1 ▸, Fig. 2 ▸), which generate chains propagating parallel to the a-axis direction. The O5 hydroxyl group donates a hydrogen bond to the proximal azide N atom [O5—H5A⋯N1i; H⋯N = 2.12 Å; O—H⋯N = 157°; symmetry code: (i) x − 1, y, z]. The other group (O2) is involved in an asymmetric, bifurcated hydrogen-bond to the THF ring O atom (O2—H2A⋯O1i; 2.09 Å; 154°) and a weaker contact with one of the dioxolane O-atoms (O2—H2A⋯O4i; 2.71 Å; 150°). There is a notable non-classical hydrogen-bond [C6—H6⋯O5ii; 2.38 Å; 159°; symmetry code: (ii) −x, y − 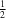 , −z + 2], which cross-links the [100] chains into (001) sheets.
, −z + 2], which cross-links the [100] chains into (001) sheets.
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O2—H2A⋯O1i | 0.84 | 2.09 | 2.871 (2) | 154 |
| O2—H2A⋯O4i | 0.84 | 2.71 | 3.462 (2) | 150 |
| O5—H5A⋯N1i | 0.84 | 2.12 | 2.910 (3) | 157 |
| C6—H6⋯O5ii | 1.00 | 2.38 | 3.332 (3) | 159 |
Symmetry codes: (i) 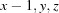 ; (ii)
; (ii) 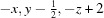 .
.
Figure 2.
Partial packing diagram for 3 showing hydrogen bonds as dashed lines.
Database survey
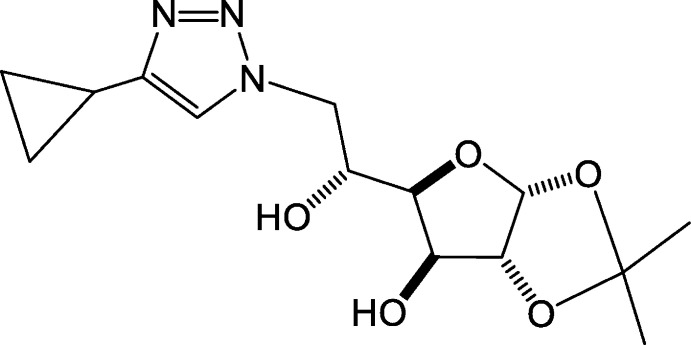
The most closely related crystal structure in the literature is 4 (Fig. 3 ▸), the 4-cyclopropyl-1,2,3-triazole derivative of compound 3 [Zhang et al., 2013 ▸, Cambridge Structural Database (Groom et al., 2016 ▸) refcode NINQOS] synthesized from a copper-catalysed azide–alkyne cycloaddition of the tribenzyl ether analogue of 3 followed by deprotection with NH3/NaOH (Pradere et al., 2008 ▸). Conversion of the azide to a triazole removes the hydrogen-bonding capability of the proximal N atom and the packing in this structure is distinctly different with a hydrogen-bonded network being present. Other points of difference in structure 4 relative to 3 include the free hydroxyl group on the THF ring, which adopts an axial conformation, and the dioxolane ring methyl groups tilted closer to the THF ring.
Figure 3.
Structure of 4 (see text).
Other examples of crystal structures of α-d-glucofuranose derivatives constrained by a 1,2-O-isopropylidene or analogous protecting group include: 3-O-ethyl-3-C-nitromethyl-1,2;5,6-di-O-isopropylidene-α-d-glucofuranose (Ivanovs et al., 2016 ▸; QENNEF) and 3-O-benzyl-1,2-O-isopropylidene-5-O-methanesulfonyl-6-O-triphenyl-methyl-α-d-glucofuranose and its azide displacement product (Clarke et al., 2018 ▸; QIBFUF). A general observation is that groups departing from O-3 take up axial or quasi-axial orientations relative to the THF ring in all cases examined and as is the case for the 3-O-ethyl group in QENNEF and the benzyl groups in QIBFUF and (4R)-4-carbamoyl-4-[(4R)-3-O-benzyl-1,2-O-isopropylidene-β-l-threofuranos-4-C-yl]-oxazolidin-2-one (Steiner et al., 2009 ▸) and the tosylate group in 1,2:5,6-di-O-isopropylidene-3-O-toluenesulfonyl-α-d-glucofuranose (Mamat et al., 2012 ▸). The impact of perfluorination on the conformation of monosaccharide derivatives was probed on (R/S)-N-benzyl-N-(5-deoxy-1,2-O-isopropylidene-3-O-methyl-α-d-xylofuranos-5-yl)-2,3,3,3-tetrafluoropropanamide and analogous compounds (Bilska-Markowska et al., 2017 ▸). The crystal structures of α-d-glucofuranose-1,2:3,5-bis(phenyl)boronate and α-d-glucofuranose-1,2:3,5-bis(p-tolyl)boronate highlight modulation in structures according to a temperature gradient (Chandran & Nangia, 2006 ▸). The structure of chloro(cyclopentadienyl)bis(1,2:5,6-di-O-isopropylidene-α-d-glucofuranos-3-O-yl)titanate provides insight into the use of monosaccharides as ligands in complexes. The titanium atom is bonded to two monosaccharide OH-3, in axial positions, a cyclopentadienyl and a chloride ligand, to take up a three-legged piano stool arrangement (Riediker et al., 1989 ▸). The unit cell of (R)-3-deoxy-1,2:5,6-di-O-isopropylidene-α-d-glucofuranos-3-yl-tert-butanesulfinate contains four symmetry-independent molecules with the tert-butyl and glucose moieties turned away from each other in order to minimize steric repulsion (Chelouan et al., 2018 ▸).
Synthesis and crystallization
1,2- O -Isopropylidene-6- O - p -toluenesulfonyl-α-d-glucofuranose, 2:
A solution of freshly recrystallized tosyl chloride (0.479 g, 2.55 mmol) in DCM (1.6 ml) was added dropwise over 20 min to a stirring solution of 1,2-O-isopropylidene-α-d-glucofuranose 1 (0.513 g, 2.32 mmol) in pyridine (3.8 ml) and DCM (4.2 ml), under an atmosphere of nitrogen. The reaction was stirred at room temperature for 48 h. TLC analysis (EtOAc/cyclohexane 2:3) revealed the formation of one product (R f = 0.45). After adding DCM (10 ml), the reaction mixture was washed with 1 M HCl (1 ml). The DCM layer was dried to give 1,2-O-isopropylidene-6-O-p-toluenesulfonyl-α-d-glucofuranose 2 (0.282 g, 32%) as an off-white crystalline solid. δH (CDCl3, 400 MHz) 7.79 (2H, d, J = 8.3 Hz, 2 Ar-H), 7.35 (2H, d, J = 8.1 Hz, 2 Ar-H), 5.88 (1H, d, J = 3.6 Hz, H-1), 4.50 (1H, d, J = 3.6 Hz, H-2), 4.36 (1H, d, J = 2.7 Hz, H-3), 4.29 (1H, dd, J = 10.2, 2.5 Hz, H-6), 4.19 (1H, td, J = 7.7, 2.5 Hz, H-5), 4.11 (1H, dd, J = 10.2, 6.8 Hz, H-6′), 4.01 (1H, dd, J = 7.7, 2.7 Hz, H-4), 2.45 (3H, s, Ar—CH3), 1.45, 1.29 (6H, 2 s, 2 acetonide CH3). δC (acetone-d 6, 100 MHz): 145.2 (ArCq—S), 132.3 (ArCq—CH3), 130.0 (2 ArC), 128.0 (2 ArC), 111.9 (Cq acetonide), 105.1 (C-1), 85.0 (C-2), 79.4 (C-4), 75.0 (C-3), 72.1 (C-6), 68.0 (C-5), 26.8 (acetonide CH3), 26.2 (acetonide CH3), 21.7 (Ar—CH3); νmax (cm−1): 3426, 3322, 2979, 2928, 1378, 1215, 1162, 1058, 1037, 1007, 962, 883, 850, 673, 657, 626.
6-Azido-6-deoxy-1,2- O -isopropylidene-α-d-glucofuranose, 3:
Sodium azide (0.290 g, 4.46 mmol) was added to a stirring solution of 2 (1.668 g, 4.45 mmol) in DMF (18 ml) at room temperature. The reaction mixture was then heated to 358 K for 42 h. TLC analysis (EtOAc/cyclohexane 2:3) revealed complete consumption of the starting material (R f = 0.45) and the formation of one product (R f = 0.30). The crude product was dried and successively dissolved in 1,4-dioxane with addition of hexane to yield an off-white precipitate, which was filtered off. The remaining filtrate contained 6-azido-6-deoxy-1,2-O-isopropylidene-α-d-glucofuranose, 3. Crystallization was achieved overnight at 248 K, after dissolution in diethyl ether with addition of hexane. The ether–hexane solution was recrystallized to obtain 2nd and 3rd crops of product to yield a combined 0.319 g (29%) of product 3 as a white crystalline solid. δH (CDCl3, 400 MHz): 5.95 (1H, d, J = 3.7 Hz, H-1), 4.53 (1H, d, J = 3.6 Hz, H-2), 4.37 (1H, d, J = 2.8 Hz, H-3), 4.16 (1H, td, J = 6.6, 3.6 Hz, H-5), 4.05 (1H, dd, J = 6.6, 2.8 Hz, H-4), 3.61 (1H, dd, J = 12.7, 3.5 Hz, H-6), 3.55 (1H, dd, J = 12.7, 6.5 Hz, H-6′), 1.49, 1.32 (6H, 2 s, 2 acetonide CH3). νmax (cm−1): 3442, 2992, 2938, 2109, 1385, 1376, 1215, 1164, 1066, 1048, 1008, 955, 881, 854, 788, 674.
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸. All H atoms were positioned geometrically (O—H = 0.84, C—H = 0.98–1.00 Å) and refined as riding with U iso(H) = 1.2U eq(O,C) or 1.5U eq(C-methyl).
Table 2. Experimental details.
| Crystal data | |
| Chemical formula | C9H15N3O5 |
| M r | 245.24 |
| Crystal system, space group | Monoclinic, P21 |
| Temperature (K) | 190 |
| a, b, c (Å) | 5.7615 (4), 9.7752 (8), 10.6833 (9) |
| β (°) | 101.255 (8) |
| V (Å3) | 590.11 (8) |
| Z | 2 |
| Radiation type | Cu Kα |
| μ (mm−1) | 0.97 |
| Crystal size (mm) | 0.40 × 0.30 × 0.02 |
| Data collection | |
| Diffractometer | Rigaku Xcalibur, EosS2, Gemini ultra |
| Absorption correction | Multi-scan (CrysAlis PRO; Rigaku, 2015 ▸) |
| T min, T max | 0.741, 1 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 3705, 1788, 1674 |
| R int | 0.044 |
| θmax (°) | 61.5 |
| (sin θ/λ)max (Å−1) | 0.570 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.036, 0.086, 1.08 |
| No. of reflections | 1788 |
| No. of parameters | 156 |
| No. of restraints | 1 |
| H-atom treatment | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.13, −0.15 |
| Absolute structure | Flack (1983 ▸) |
| Absolute structure parameter | −0.4 (3) |
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S2056989020012438/hb7940sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989020012438/hb7940Isup2.hkl
CCDC reference: 1968033
Additional supporting information: crystallographic information; 3D view; checkCIF report
supplementary crystallographic information
Crystal data
| C9H15N3O5 | F(000) = 260 |
| Mr = 245.24 | Dx = 1.38 Mg m−3 |
| Monoclinic, P21 | Cu Kα radiation, λ = 1.54184 Å |
| Hall symbol: P 2yb | Cell parameters from 1783 reflections |
| a = 5.7615 (4) Å | θ = 6.2–60.6° |
| b = 9.7752 (8) Å | µ = 0.97 mm−1 |
| c = 10.6833 (9) Å | T = 190 K |
| β = 101.255 (8)° | Plate, colourless |
| V = 590.11 (8) Å3 | 0.40 × 0.30 × 0.02 mm |
| Z = 2 |
Data collection
| Rigaku Xcalibur, EosS2, Gemini ultra diffractometer | 1788 independent reflections |
| Radiation source: fine-focus sealed X-ray tube | 1674 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.044 |
| Detector resolution: 8.0217 pixels mm-1 | θmax = 61.5°, θmin = 4.2° |
| ω scans | h = −6→6 |
| Absorption correction: multi-scan (CrysAlisPro; Rigaku, 2015) | k = −11→10 |
| Tmin = 0.741, Tmax = 1 | l = −12→12 |
| 3705 measured reflections |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.036 | H-atom parameters constrained |
| wR(F2) = 0.086 | w = 1/[σ2(Fo2) + (0.0346P)2 + 0.0212P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.08 | (Δ/σ)max < 0.001 |
| 1788 reflections | Δρmax = 0.13 e Å−3 |
| 156 parameters | Δρmin = −0.14 e Å−3 |
| 1 restraint | Absolute structure: Flack (1983) |
| Primary atom site location: structure-invariant direct methods | Absolute structure parameter: −0.4 (3) |
Special details
| Geometry. All s.u.'s (except the s.u. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell s.u.'s are taken into account individually in the estimation of s.u.'s in distances, angles and torsion angles; correlations between s.u.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell s.u.'s is used for estimating s.u.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > 2σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.3329 (4) | 0.6279 (3) | 1.2308 (2) | 0.0428 (6) | |
| H1A | 0.3365 | 0.5953 | 1.3189 | 0.051* | |
| H1B | 0.4098 | 0.5574 | 1.1863 | 0.051* | |
| C2 | 0.0786 (4) | 0.6435 (2) | 1.1633 (2) | 0.0345 (5) | |
| H2 | −0.0043 | 0.5539 | 1.1654 | 0.041* | |
| C3 | 0.0532 (4) | 0.6893 (3) | 1.0256 (2) | 0.0329 (5) | |
| H3 | 0.1366 | 0.7784 | 1.0212 | 0.04* | |
| C4 | −0.2022 (4) | 0.6991 (2) | 0.9528 (2) | 0.0340 (5) | |
| H4 | −0.2721 | 0.7914 | 0.9614 | 0.041* | |
| C5 | −0.1750 (4) | 0.6702 (2) | 0.8164 (2) | 0.0342 (5) | |
| H5 | −0.3207 | 0.6292 | 0.7634 | 0.041* | |
| C6 | 0.0389 (4) | 0.5759 (3) | 0.8297 (2) | 0.0357 (5) | |
| H6 | −0.0087 | 0.4799 | 0.8045 | 0.043* | |
| C7 | 0.0728 (4) | 0.7525 (3) | 0.6883 (2) | 0.0393 (6) | |
| C8 | 0.2544 (5) | 0.8648 (3) | 0.7005 (3) | 0.0524 (7) | |
| H8A | 0.3755 | 0.8406 | 0.6514 | 0.079* | |
| H8B | 0.3287 | 0.8765 | 0.7905 | 0.079* | |
| H8C | 0.1774 | 0.9504 | 0.6675 | 0.079* | |
| C9 | −0.0399 (6) | 0.7197 (4) | 0.5530 (3) | 0.0654 (9) | |
| H9A | 0.0828 | 0.6935 | 0.5056 | 0.098* | |
| H9B | −0.1246 | 0.8003 | 0.513 | 0.098* | |
| H9C | −0.1514 | 0.6438 | 0.5518 | 0.098* | |
| N1 | 0.4698 (3) | 0.7562 (3) | 1.2362 (2) | 0.0498 (6) | |
| N2 | 0.4455 (3) | 0.8362 (3) | 1.3223 (2) | 0.0464 (5) | |
| N3 | 0.4413 (5) | 0.9168 (3) | 1.3977 (3) | 0.0654 (7) | |
| O1 | 0.1584 (3) | 0.58380 (19) | 0.95897 (16) | 0.0426 (4) | |
| O2 | −0.3347 (3) | 0.5933 (2) | 0.99674 (16) | 0.0460 (4) | |
| H2A | −0.4782 | 0.6016 | 0.9625 | 0.069* | |
| O3 | −0.0999 (3) | 0.79118 (16) | 0.76079 (15) | 0.0386 (4) | |
| O4 | 0.1823 (3) | 0.63255 (19) | 0.74985 (18) | 0.0497 (5) | |
| O5 | −0.0267 (3) | 0.74186 (18) | 1.23341 (15) | 0.0390 (4) | |
| H5A | −0.1747 | 0.7349 | 1.2142 | 0.059* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.0349 (12) | 0.0567 (16) | 0.0369 (12) | 0.0109 (11) | 0.0075 (9) | 0.0070 (11) |
| C2 | 0.0268 (11) | 0.0403 (13) | 0.0379 (12) | 0.0022 (10) | 0.0099 (9) | 0.0035 (10) |
| C3 | 0.0271 (12) | 0.0395 (12) | 0.0340 (12) | 0.0030 (9) | 0.0105 (9) | −0.0009 (10) |
| C4 | 0.0246 (12) | 0.0409 (13) | 0.0376 (12) | 0.0017 (9) | 0.0086 (10) | 0.0029 (10) |
| C5 | 0.0257 (12) | 0.0402 (14) | 0.0352 (12) | −0.0063 (9) | 0.0027 (9) | −0.0005 (10) |
| C6 | 0.0323 (12) | 0.0412 (13) | 0.0349 (12) | −0.0046 (10) | 0.0094 (9) | −0.0029 (10) |
| C7 | 0.0339 (13) | 0.0491 (14) | 0.0368 (13) | 0.0019 (11) | 0.0112 (10) | 0.0015 (11) |
| C8 | 0.0435 (14) | 0.0534 (18) | 0.0639 (18) | −0.0069 (12) | 0.0193 (13) | 0.0006 (14) |
| C9 | 0.0598 (18) | 0.094 (3) | 0.0432 (16) | −0.0117 (17) | 0.0118 (13) | −0.0108 (15) |
| N1 | 0.0259 (11) | 0.0806 (18) | 0.0433 (12) | −0.0012 (10) | 0.0075 (9) | −0.0069 (12) |
| N2 | 0.0298 (10) | 0.0663 (16) | 0.0419 (13) | 0.0060 (10) | 0.0042 (9) | 0.0052 (12) |
| N3 | 0.0656 (16) | 0.0718 (17) | 0.0578 (16) | 0.0104 (13) | 0.0099 (12) | −0.0061 (15) |
| O1 | 0.0278 (8) | 0.0598 (11) | 0.0404 (9) | 0.0128 (8) | 0.0071 (6) | −0.0047 (8) |
| O2 | 0.0220 (7) | 0.0654 (11) | 0.0521 (10) | −0.0030 (8) | 0.0110 (7) | 0.0101 (9) |
| O3 | 0.0346 (8) | 0.0432 (10) | 0.0396 (9) | 0.0038 (7) | 0.0110 (7) | 0.0045 (7) |
| O4 | 0.0467 (10) | 0.0532 (11) | 0.0569 (11) | 0.0106 (8) | 0.0292 (8) | 0.0106 (8) |
| O5 | 0.0269 (8) | 0.0532 (11) | 0.0389 (9) | 0.0031 (7) | 0.0113 (7) | −0.0032 (8) |
Geometric parameters (Å, º)
| C1—N1 | 1.477 (4) | C6—O1 | 1.420 (3) |
| C1—C2 | 1.509 (3) | C6—H6 | 1 |
| C1—H1A | 0.99 | C7—O3 | 1.427 (3) |
| C1—H1B | 0.99 | C7—O4 | 1.429 (3) |
| C2—O5 | 1.425 (3) | C7—C9 | 1.499 (4) |
| C2—C3 | 1.517 (3) | C7—C8 | 1.505 (4) |
| C2—H2 | 1 | C8—H8A | 0.98 |
| C3—O1 | 1.451 (3) | C8—H8B | 0.98 |
| C3—C4 | 1.527 (3) | C8—H8C | 0.98 |
| C3—H3 | 1 | C9—H9A | 0.98 |
| C4—O2 | 1.419 (3) | C9—H9B | 0.98 |
| C4—C5 | 1.522 (3) | C9—H9C | 0.98 |
| C4—H4 | 1 | N1—N2 | 1.236 (3) |
| C5—O3 | 1.428 (3) | N2—N3 | 1.131 (4) |
| C5—C6 | 1.523 (3) | O2—H2A | 0.84 |
| C5—H5 | 1 | O5—H5A | 0.84 |
| C6—O4 | 1.411 (3) | ||
| N1—C1—C2 | 113.2 (2) | O4—C6—C5 | 105.36 (19) |
| N1—C1—H1A | 108.9 | O1—C6—C5 | 106.73 (17) |
| C2—C1—H1A | 108.9 | O4—C6—H6 | 111.6 |
| N1—C1—H1B | 108.9 | O1—C6—H6 | 111.6 |
| C2—C1—H1B | 108.9 | C5—C6—H6 | 111.6 |
| H1A—C1—H1B | 107.8 | O3—C7—O4 | 105.03 (18) |
| O5—C2—C1 | 106.9 (2) | O3—C7—C9 | 111.3 (2) |
| O5—C2—C3 | 109.89 (18) | O4—C7—C9 | 109.7 (2) |
| C1—C2—C3 | 113.22 (17) | O3—C7—C8 | 107.8 (2) |
| O5—C2—H2 | 108.9 | O4—C7—C8 | 108.8 (2) |
| C1—C2—H2 | 108.9 | C9—C7—C8 | 113.7 (2) |
| C3—C2—H2 | 108.9 | C7—C8—H8A | 109.5 |
| O1—C3—C2 | 107.18 (18) | C7—C8—H8B | 109.5 |
| O1—C3—C4 | 104.35 (18) | H8A—C8—H8B | 109.5 |
| C2—C3—C4 | 114.43 (17) | C7—C8—H8C | 109.5 |
| O1—C3—H3 | 110.2 | H8A—C8—H8C | 109.5 |
| C2—C3—H3 | 110.2 | H8B—C8—H8C | 109.5 |
| C4—C3—H3 | 110.2 | C7—C9—H9A | 109.5 |
| O2—C4—C5 | 110.12 (19) | C7—C9—H9B | 109.5 |
| O2—C4—C3 | 108.23 (18) | H9A—C9—H9B | 109.5 |
| C5—C4—C3 | 101.95 (16) | C7—C9—H9C | 109.5 |
| O2—C4—H4 | 112 | H9A—C9—H9C | 109.5 |
| C5—C4—H4 | 112 | H9B—C9—H9C | 109.5 |
| C3—C4—H4 | 112 | N2—N1—C1 | 115.41 (19) |
| O3—C5—C4 | 109.89 (18) | N3—N2—N1 | 173.0 (3) |
| O3—C5—C6 | 103.56 (16) | C6—O1—C3 | 110.24 (17) |
| C4—C5—C6 | 104.82 (18) | C4—O2—H2A | 109.5 |
| O3—C5—H5 | 112.6 | C7—O3—C5 | 107.83 (18) |
| C4—C5—H5 | 112.6 | C6—O4—C7 | 110.02 (17) |
| C6—C5—H5 | 112.6 | C2—O5—H5A | 109.5 |
| O4—C6—O1 | 109.68 (18) | ||
| N1—C1—C2—O5 | −60.8 (2) | C4—C5—C6—O1 | −15.1 (2) |
| N1—C1—C2—C3 | 60.3 (3) | C2—C1—N1—N2 | 81.8 (3) |
| O5—C2—C3—O1 | −178.66 (18) | C1—N1—N2—N3 | 172 (2) |
| C1—C2—C3—O1 | 61.9 (3) | O4—C6—O1—C3 | 106.5 (2) |
| O5—C2—C3—C4 | −63.5 (3) | C5—C6—O1—C3 | −7.2 (2) |
| C1—C2—C3—C4 | 177.1 (2) | C2—C3—O1—C6 | 148.18 (18) |
| O1—C3—C4—O2 | 81.9 (2) | C4—C3—O1—C6 | 26.5 (2) |
| C2—C3—C4—O2 | −34.9 (3) | O4—C7—O3—C5 | −28.5 (2) |
| O1—C3—C4—C5 | −34.2 (2) | C9—C7—O3—C5 | 90.2 (3) |
| C2—C3—C4—C5 | −151.02 (19) | C8—C7—O3—C5 | −144.4 (2) |
| O2—C4—C5—O3 | 164.46 (17) | C4—C5—O3—C7 | 139.25 (18) |
| C3—C4—C5—O3 | −80.8 (2) | C6—C5—O3—C7 | 27.7 (2) |
| O2—C4—C5—C6 | −84.8 (2) | O1—C6—O4—C7 | −115.1 (2) |
| C3—C4—C5—C6 | 29.9 (2) | C5—C6—O4—C7 | −0.5 (2) |
| O3—C5—C6—O4 | −16.5 (2) | O3—C7—O4—C6 | 17.4 (2) |
| C4—C5—C6—O4 | −131.67 (19) | C9—C7—O4—C6 | −102.3 (2) |
| O3—C5—C6—O1 | 100.11 (19) | C8—C7—O4—C6 | 132.7 (2) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O2—H2A···O1i | 0.84 | 2.09 | 2.871 (2) | 154 |
| O2—H2A···O4i | 0.84 | 2.71 | 3.462 (2) | 150 |
| O5—H5A···N1i | 0.84 | 2.12 | 2.910 (3) | 157 |
| C6—H6···O5ii | 1.00 | 2.38 | 3.332 (3) | 159 |
Symmetry codes: (i) x−1, y, z; (ii) −x, y−1/2, −z+2.
Funding Statement
This work was funded by B18 Project grant . Faculty of Science University of Newcastle grant . Priority Research Centre for Drug Development, University of Newcastle grant .
References
- Altomare, A., Cascarano, G., Giacovazzo, C., Guagliardi, A., Burla, M. C., Polidori, G. & Camalli, M. (1994). J. Appl. Cryst. 27, 435.
- Ashton, P. R., Ellwood, P., Staton, I. & Stoddart, J. F. (1991). J. Org. Chem. 56, 7274–7280.
- Bilska-Markowska, M., Siodla, T., Patyk-Kaźmierczak, S., Katrusiak, A. & Koroniak, H. (2017). New J. Chem. 41, 12631–12644.
- Chandran, S. K. & Nangia, A. (2006). CrystEngComm, 8, 581–585.
- Chelouan, A., Bao, S., Friess, S., Herrera, A., Heinemann, F. W., Escalona, A., Grasruck, A. & Dorta, R. (2018). Organometallics, 37, 3983–3992.
- Clarke, Z., Barnes, E., Prichard, K. L., Mares, L. J., Clegg, J. K., McCluskey, A., Houston, T. A. & Simone, M. I. (2018). Acta Cryst. E74, 862–867. [DOI] [PMC free article] [PubMed]
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Flack, H. D. (1983). Acta Cryst. A39, 876–881.
- Fleet, G. W. J., Ramsden, N. G. & Witty, D. R. (1989). Tetrahedron, 45, 327–336.
- Fujita, K. E., Ohta, K., Masunari, K., Obe, K. & Yamamura, H. (1992). Tetrahedron Lett. 33, 5519–5520.
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- IUPAC (1996). Pure Appl. Chem. 68, 1919–2008.
- Ivanovs, I., Bērziņa, S., Lugiņina, J., Belyakov, S. & Rjabovs, V. (2016). Heterocycl. Commun. 22, 95–98.
- Johnson, L. L. & Houston, T. A. (2002). Tetrahedron Lett. 43, 8905–8908.
- Johnson, W. S., Collins, J. C., Pappo, R., Rubin, M. B., Kropp, P. J., Johns, W. F., Pike, J. E. & Bartmann, W. (1963). J. Am. Chem. Soc. 85, 1409–1430.
- Katritzky, A. R., Rachwal, S. & Hitchings, G. J. (1991). Tetrahedron, 47, 2683–2732.
- Lee, J. C., Francis, S., Dutta, D., Gupta, V., Yang, Y., Zhu, J., Tash, J. S., Schönbrunn, E. & Georg, G. I. (2012). J. Org. Chem. 77, 3082–3098. [DOI] [PMC free article] [PubMed]
- Liu, Z., Yoshihara, A., Wormald, M. R., Jenkinson, S. F., Gibson, V., Izumori, K. & Fleet, G. W. J. (2014). Org. Lett. 16, 5663–5665. [DOI] [PubMed]
- Mamat, C., Peppel, T. & Köckerling, M. (2012). Crystals, 2, 105–109.
- Pradere, U., Roy, V., McBrayer, T. R., Schinazi, R. F. & Agrofoglio, L. A. (2008). Tetrahedron, 64, 9044–9051. [DOI] [PMC free article] [PubMed]
- Prichard, K., Campkin, D., O’Brien, N., Kato, A., Fleet, G. W. J. & Simone, M. I. (2018). Chem. Biol. Drug Des. 92, 1171–1197. [DOI] [PubMed]
- Rasmussen, T. S. & Jensen, H. H. (2011). Carbohydr. Res. 346, 2855–2861. [DOI] [PubMed]
- Riediker, M., Hafner, A., Piantini, U., Rihs, G. & Togni, A. (1989). Angew. Chem. Int. Ed. Engl. 28, 499–500.
- Rigaku (2015). CrysAlis PRO. Rigaku Corporation, Tokyo, Japan.
- Sayce, A. C., Alonzi, D. S., Killingbeck, S. S., Tyrrell, B. E., Hill, M. L., Caputo, A. T., Iwaki, R., Kinami, K., Ide, D., Kiappes, J. L., Beatty, P. R., Kato, A., Harris, E., Dwek, R. A., Miller, J. L. & Zitzmann, N. (2016). PLoS Negl. Trop. Dis. 10, e0004524. [DOI] [PMC free article] [PubMed]
- Šesták, S., Bella, M., Klunda, T., Gurská, S., Džubák, P., Wöls, F., Wilson, I. B. H., Sladek, V., Hajdúch, M., Poláková, M. & Kóňa, J. (2018). ChemMedChem, 13, 373–383. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Simone, M. I., Soengas, R. G., Jenkinson, S. F., Evinson, E. L., Nash, R. J. & Fleet, G. W. J. (2012). Tetrahedron Asymmetry, 23, 401–408.
- Steiner, A. J., Stütz, A. E., Tarling, C. A., Withers, S. G. & Wrodnigg, T. M. (2009). Aust. J. Chem. 62, 553–557.
- Steiner, B., Langer, V. & Koóš, M. (2009). Carbohydr. Res. 344, 2079–2082. [DOI] [PubMed]
- Tsuchiya, T., Miyake, T., Kageyama, S., Umezawa, S., Umezawa, H. & Takita, T. (1981). Tetrahedron Lett. 22, 1413–1416.
- Tsuda, Y., Nishimura, M., Kobayashi, T., Sato, Y. & Kanemitsu, K. (1991). Chem. Pharm. Bull. 39, 2883–2887.
- Wood, A., Prichard, K. L., Clarke, Z., Houston, T. A., Fleet, G. W. J. & Simone, M. I. (2018). Eur. J. Org. Chem. pp. 6812–6829.
- Woodhouse, S. D., Smith, C., Michelet, M., Branza-Nichita, N., Hussey, M., Dwek, R. A. & Zitzmann, N. (2008). Antimicrob. Agents Chemother. 52, 1820–1828. [DOI] [PMC free article] [PubMed]
- Yamamura, H. & Fujita, K. (1991). Chem. Pharm. Bull. 39, 2505–2508.
- Zhang, Q., He, P., Zhou, G., Yu, K. & Liu, H. (2013). Acta Cryst. E69, o1386. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S2056989020012438/hb7940sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989020012438/hb7940Isup2.hkl
CCDC reference: 1968033
Additional supporting information: crystallographic information; 3D view; checkCIF report