The synthesis and crystal structures of three new disubstituted [13]-macrodilactones, trans-4,8-dimethyl-1,10-dioxacyclotridec-5-ene-2,9-dione, cis-4-(4-bromophenyl)-13-methyl-1,10-dioxacyclotridec-5-ene-2,9-dione, and trans-11-methyl-4-phenyl-1,10-dioxacyclotridec-5-ene-2,9-dione are reported and their conformations are put in the context of other [13]-macrodilactone structures reported previously, showing that the number, location, and relative disposition of groups attached at the termini of planar units of the [13]-macrodilactones subtly influence their aspect ratios.
Keywords: macrocycle, conformation, planar chirality, crystal structure
Abstract
The synthesis and crystal structures of three new disubstituted [13]-macrodilactones, namely, trans-4,8-dimethyl-1,10-dioxacyclotridec-5-ene-2,9-dione, C13H20O4, I, cis-4-(4-bromophenyl)-13-methyl-1,10-dioxacyclotridec-5-ene-2,9-dione C18H21BrO4, II, and trans-11-methyl-4-phenyl-1,10-dioxacyclotridec-5-ene-2,9-dione, C18H22O4, III, are reported and their conformations are put in the context of other [13]-macrodilactone structures reported previously. Together, they show that the number, location, and relative disposition of groups attached at the termini of planar units of the [13]-macrodilactones subtly influence their aspect ratios.
Chemical context
Macrocyclic rings adopt particular conformations by balancing the contributions of multiple, local domain features. We have studied the synthesis, structure, and function of a specific family of macrocycles, the [13]-macrodilactones. These macrocycles, which are made more rigid by ester and alkene planar units, minimize transannular interactions of substituents at stereogenic centers along their backbone. The overall effect of the number of atoms in the ring, the planar units, and the stereogenic centers promotes the adoption of a conformation that contains an element of planar chirality.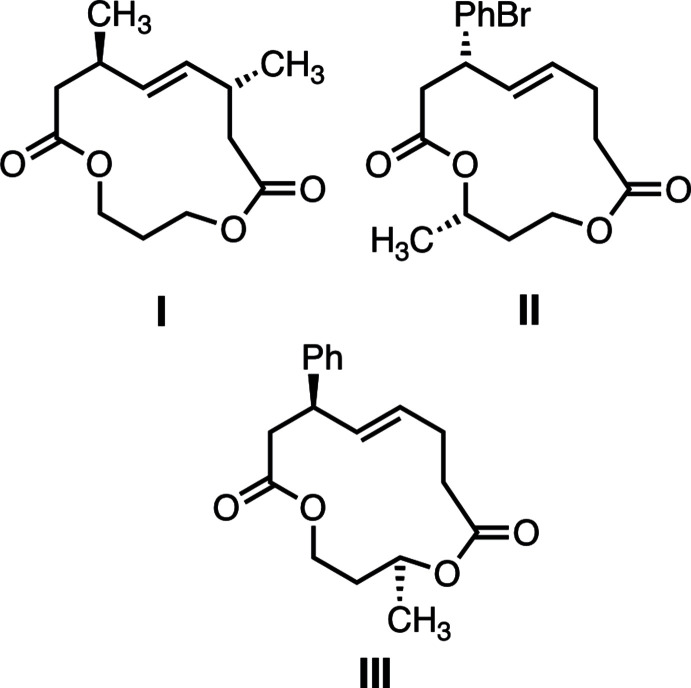
The modularity of macrocycles lends to their attractiveness as scaffolds for the development of new bioactive compounds (Whitty et al., 2017 ▸; Yudin, 2015 ▸; Driggers et al., 2008 ▸). Macrocycles have mini-domains of a few atoms that can influence the conformation of the ring, modulate rigidity/flexibility, and tune their physicochemical and biochemical properties (Whitty et al., 2016 ▸; Larsen et al., 2015 ▸). For the [13]-macrodilactone motif exemplified by trans-4,8-dimethyl-1,10-dioxacyclotridec-5-ene-2,9-dione I in Fig. 1 ▸, two four-atom ester units (C13/O1/C2/C3 and C8/C9/O10/C11) are linked via a central carbon (C12) and one trans-2-butenyl moiety (atoms C4–C7). By virtue of the planarity of the multi-atom units, the ring is significantly stiffened compared to a saturated thirteen-membered ring. The conformation, which is informally referred to as the ‘ribbon’ conformation, arises from a balance between the number of atoms that make up the ring and the nature of the planar units that reduce its flexibility. There is a planar chirality associated with the macrocycle (only the R-configured plane, pR, form is shown for I) that arises from the asymmetry around the alkene unit as it orients itself perpendicular to the mean plane of the macrocycle. The planar chirality of the [13]-macrodilactones is directly analogous to that of E-cyclooctene (Fyvie & Peczuh, 2008a ▸,b ▸; Eliel & Wilen, 1994 ▸). Viewed from above (parallel to the alkene), the ribbon appears roughly triangular (Fig. 1 ▸ c, top), with a long axis and a short axis.
Figure 1.
Ribbon motif of [13]-macrodilactones. (a) Structure and number of [13]-macrodilactones using compound I as an illustration. (b) Schematic of the ring showing the three planar units: two esters (brown) and alkene (blue). Key atoms at the termini of the planar units are marked with asterisks. (c) Molecular structure of I from X-ray data.
Here we report on the synthesis and solid-state structural characterization of [13]-macrodilactones I, II, and III. These new structures, along with eight more previously reported [13]-macrodilactone structures, are analyzed to assess how substitution at specific atoms of the backbone influences the conformation.
Structural commentary
Each of the new structures has two stereogenic centers. The synthetic routes were not stereoselective, and the products were isolated as racemates. Consequently, each compound crystallized as a racemate. The stereogenic centers of compounds I, II, and III seen in Fig. 2 ▸ establish only the relative stereochemistry observed in the asymmetric unit for each one. The only common feature of I, II, and III is the [13]-macrodilactone core. All bond distances and angles are in the expected ranges and unexceptional. A more in depth analysis of molecular aspect ratios can be found in the Database survey.
Figure 2.

The molecular structures of I, II, and III with 50% displacement ellipsoid probability levels. Note that the structures here are of the pS-configured planar chirality.
Database survey
A survey of the Cambridge Structural Database (CSD) yielded a total of 17 structures of [13]-macrodilactones, counting the three new structures reported here (Table 1 ▸). Of these structures, 11 share the same fundamental ribbon conformation described earlier. Comparison of their structures in light of their substitution patterns along the macrocyclic backbone revealed subtle differences in their conformations. Aspect ratio, defined as the ratio of the C12-to-centroid of C5 and C6 (length, or long-axis) and the C2-to-C9 carbonyl carbon distance (width, or short-axis) of the macrocyclic ring, was our metric to express the changes in conformation. By virtue of the cyclic structure, compression along one axis leads to expansion along the complementary one, and vice versa, affecting the aspect. Note that structure e (Fig. 3 ▸) is of the unsubstituted [13]-macrodilactone, containing no pendant groups along its backbone; it represents a reference point for comparisons amongst the other substituted macrocycles.
Table 1. Substitution patterns and refcodes for [13]-macrodilactones.
Cpd. = compound identifier in Fig. 3 ▸, Conf. = conformer adopted in the crystal structure and Subs. = substituted positions on [13]-macrodilactone.
| Entry | Cpd. | Conf. | Subs. | cis/trans | Refcode | Citation |
|---|---|---|---|---|---|---|
| 1 | a | ribbon | 11,13 | trans | URILEO | Ma & Peczuh (2013 ▸) |
| 2 | b | ribbon | 11 (mono) | – | KOHLAV | Fyvie & Peczuh (2008a ▸) |
| 3 | c | ribbon | 3 (mono) | – | XUFKOA | Magpusao, Rutledgeet al. (2015 ▸) |
| 4 | – | ribbon | D-gluco | trans | XOCWIW | Fyvie & Peczuh (2008b ▸) |
| 5 | d | ribbon | 3,8 | trans | XUFLAN | Magpusao, Rutledge et al. (2016 ▸) |
| 6 | e | ribbon | – | – | IJEHAI | Magpusao, Rutledge et al. (2016 ▸) |
| 7 | f | ribbon | 3,11 | cis | IJEHOW | Magpusao, Rutledge et al. (2016 ▸) |
| 8 | g | ribbon | 4,13 | cis | II | This work (CCDC 1944827) |
| 9 | h | ribbon | 4,8 | trans | I | This work (CCDC 1944826) |
| 10 | i | ribbon | 4 (mono) | – | ECOYED | Rutledge, Hamlin et al. (2017 ▸) |
| 11 | j | ribbon | 3,13 | trans | IJEHEM | Magpusao, Rutledge et al. (2016 ▸) |
| 10 | – | other | 4 (mono) | – | ECOYED | |
| 12 | – | other | 11,13 | cis | URILAK | Ma & Peczuh (2013 ▸) |
| 3 | c | other | 3 (mono) | – | XUFKOA | |
| 13 | – | other | 11,13 | cis | URILAK | Ma & Peczuh (2013 ▸) |
| 14 | – | other | 3,11 | trans | IJEHUC | Magpusao, Rutledge et al. (2016 ▸) |
| 15 | k | ribbon | 4,11 | trans | III | This work (CCDC 1944828) |
| 16 | – | other | 3,8 | cis | XUFKUG | Magpusao, Rutledge et al. (2016 ▸) |
| 17 | – | other | 8,11 | cis | IJEHIQ | Magpusao, Rutledge et al. (2016 ▸) |
Figure 3.
Length, width, and aspect ratios of pS-configured [13]-macrodilactones in the ribbon conformation. The inset shows the distances measured in the macrocycles. All error bars are shown to 3σ, but some are smaller than the marker chosen to represent the point. The boxes highlight data from compounds I (h), II (g), and III (k) in this report. All distances are shown in Å.
Subtle differences in the aspect ratios of the [13]-macrodilactones depicted in Fig. 3 ▸ were attributed to the location and number but not size of groups attached to the ring, which we found remarkable. For example, the positioning of a single substituted atom affected the aspect ratio as exemplified by b, c, and i. Trends for di-substituted [13]-macrodilactones separated into two groups. In the first group are the ‘symmetrical’ di-substituted compounds: trans-11,13- (a), trans −3,8- (d), and trans-4,8- (h, compound I). The trend for this group largely follows that in the monosubstituted series. That is, the aspect ratio increased slightly when substitutions were made on either end of the ester units but decreased upon substitution at the allylic carbons. The second group of di-substituted macrocycles is a catch-all that collects compounds where the substituted carbons are either on the same side of the ring relative to its long-axis [cis-4,13- (g, compound II) and trans-3,13 (j)] or opposite sides [cis-3,11 (f) and trans-4,11 (k, compound III)]. These compounds pit substitutions at a site (C11/13) that stretches the long-axis with sites that either also extend (C3/C8) or compress (C4/C7) it. A clear rationale to explain relationships between these substitution patterns and their aspect ratios was not apparent. One observation was that any substitution at the allylic positions tended to compress the aspect ratios of all the [13]-macrodilactones. Aspect ratios ranged from 1.16 on the low end (q, compound III) to 1.32 (a) on the high end. That represents a 14% change in aspect ratio linked only to the number and location of substituted carbons along the backbone of the [13]-macrodilactone structure.
Computational analysis of conformations
To ascertain whether the solid-state structures were representative of their local minimum-energy conformations, gas-phase computational optimizations were performed on the new [13]-macrodilactones I–III and also compounds a and e in Fig. 3 ▸. The pR conformers from the X-ray data were optimized via DFT using the Schrödinger Maestro application Jaguar (Bochevarov et al., 2013 ▸). Four different levels of theory, chosen because of their large number of basis functions and their inclusion of bromine orbitals, were used with the calculation, B3LYD-D3/CC-PVDC, M06-2X-D3/CC-PVDZ, B3LYD-D3/LACVP**, and M06-2X-D3/LACVP**. B3LYD-D3 was chosen because of its widespread use and M06-2X-D3 was chosen for its ability to accurately describe non-covalent interactions within the macrocycles (Grimme, 2011 ▸). Single point energy calculations were run on both the conformer from the crystallographic data and the conformer optimized at the B3LYD-D3/CC-PVDC level of theory. RMSD values comparing the 13 non-hydrogen atoms of the macrocycle ring were calculated comparing crystallographic structure and DFT-optimized conformers (Fig. 4 ▸). Values that express the average RMSD across the different DFT calculations ranged from 0.055 to 0.251 Å (Table S3 in the supporting information). The low values suggest that [13]-macrodilactones do not significantly change their conformations upon optimization.
Figure 4.
Overlays of crystallographic (green) and the DFT-optimized (black) structures for compounds a, f, g (II), h (I), and k (III).
Supramolecular features
The crystal structures of I, II and III were searched for non-bonded interactions that may influence the measurements reported in Fig. 2 ▸. The program Mercury (Macrae, et al., 2020 ▸) highlighted close contacts, which are defined as less than or equal to the sum of the van der Waals radii of atom pairs (Rowland et al., 1996 ▸). In I, a contact of 3.188 (4) Å between the carbonyl oxygen O17 and methylene carbon C11 in the molecule generated by the c-glide operation was identified. Molecules associated with this contact line up along [001]. In II, a Br⋯Br contact of 3.6466 (2) Å was identified where Br1 is near the crystallographic 21 screw axis. Sequential applications of this symmetry operation generate a zigzag pattern of Br⋯Br contacts along [010]. In III, a contact of 3.386 (4) Å between C3 and the para carbon, C18, of the pendant phenyl ring is generated by the the 21 screw axis and repeats along [001]. These distances fall well within what is observed in other solid state structures and no attractive interactions were found in I, II, or III (see Figures S1 to S7 in the supporting information).
Synthesis and crystallization
As shown in Fig. 5 ▸, [13]-macrodilactones I, II, and III were prepared by an established synthetic route that entailed sequential acylation reactions, followed by macrocyclization via ring closing metathesis (RCM) (Magpusao et al., 2015 ▸, 2016 ▸). Because the syntheses were not stereo-controlled and each of the new compounds contained two stereogenic centers, two diastereomeric products (each racemic) arose for each macrocycle. The diastereomers of I, II, and III in Fig. 4 ▸ are the ones that gave rise to the ‘ribbon’ conformers presented herein. See the supporting information for additional details on the synthesis of compounds I–III. Single crystals of the compounds were prepared by slow diffusion of hexane vapor into ethyl acetate:hexanes solutions of the compounds.
Figure 5.
Synthesis of macrocycles I–III.
General
3-Bromo-phenyl-4-pentenoic acid, 3-phenyl-4-pentenoic acid and monoacylated 3-methyl-3-hydroxypropyl-4-pentenoate were prepared as previously described (Magpusao et al., 2015 ▸). Unless stated otherwise, all acylations were conducted at 273 K and allowed to warm to room temperature over 12 h. Reactions were monitored using TLC. Spots on TLC plates were visualized with UV light and p-anisaldehyde or ceric ammonium molybdate (CAM) stains. Chromatography was performed on silica gel and solvent systems were based on the R f values. 1H NMR spectra were referenced to CDCl3 proton (δ H 7.27 ppm) and 13C NMR to the CDCl3 carbon (δ C 77.2 ppm).
1,3-Di-(3-methyl-4-pentenoyloxy)-propane
Into a 25 mL round-bottom flask were added dicyclohexylcarbodiimide (DCC) (1.08 eq.), N,N-dimethylaminopyridine (DMAP) (0.3 eq.) and dichloromethane (DCM) (7mL) and the solution was cooled to 273 K under nitrogen. Then 3-methyl-4-pentenoic acid (1.0 eq.) was added and the mixture stirred at the same temperature for 30 minutes until a white suspension formed. A solution of 1,3-butanediol (0.5 eq.) in DCM (3 mL) was then added and the mixture was stirred overnight at room temperature. After completion of the reaction, the mixture was filtered through a short pad of celite, rinsed with DCM, and solvent from the filtrates was removed under reduced pressure. The residue was then dissolved in cold ether to precipitate excess dicyclohexyl urea (DCU), filtered through a pad of celite, and rinsed with additional ether. Ether from the combined filtrates was removed under reduced pressure and the residue was purified by column chromatography (10:90 EtOAc:Hex) to give a clear colorless oil (43%). R f 0.47 (10:90 EtOAc:Hex); 1H NMR (CDCl3) 400 MHz δ 5.73 (ddd, J = 17.2, 10.2, 7.3 Hz, 2H), 4.96 (dd, J = 22.3, 17.2 Hz, 4H), 4.13 (dd, J = 6.33, 6.33 Hz, 4H), 2.65 (s, J = 7.0, 7.0, 7.0, 7.0, 7.0, 7.0, 7.0 Hz, 2H), 2.29 (dddd or dq, J = 22.1, 14.8, 14.8, 14.8 Hz, 4H), 1.94 (dddd or dq, J = 12.5, 6.2, 6.2, 6.2 Hz, 2H), 1.03 (d, J = 6.6 Hz, 6H); 13C NMR (CDCl3) 100 MHz δ 172.4, 142.5, 113.5, 61.0, 41.4, 34.6, 28.2, 19.9; TOF HRMS (DART) m/z calculated for C15H24O4 [MH]+ calculated 269.1753, found 269.1749.
Sequential acylation method
To a 25 mL round-bottom flask was added DCM (7 mL), dicyclohexylcarbodiimide (DCC) (1.08 eq.) and N,N-dimethylaminopyridine (DMAP) (0.3 eq.) and the mixture was cooled to 273 K. 4-Pentenoic acid (1.0 eq.) was added and the mixture was stirred at the same temperature for 30 min until a white suspension was observed. Then, 1,3-propanediol or 1,3-butanediol (1.0 eq.) in DCM (3 mL) was added and the mixture was stirred overnight at room temperature. After completion of the reaction, the mixture was filtered through a short pad of celite, rinsed with DCM and solvent from the filtrates was removed under reduced pressure. The residue was then dissolved in cold ether to precipitate excess DCU, filtered through a pad of celite, and rinsed with additional ether. The crude residue was purified by silica gel column chromatography (15:85 EtOAc:Hex) to give the monoacylated product. The same procedure, where the mono-acylated alcohol was used in place of the diol, was then followed for the second acylation.
1-[3-(p-Bromophenyl)-4-pentenoyloxy]-1-methylpropyl-4-pentenoate
Synthesis followed the sequential acylation method above to give the compound in 70% yield as a colorless oil. R f 0.56 (hexanes: EtOAc 80:20); 1H NMR (CDCl3) 400 MHz δ 7.42 (d, J = 8.31 Hz, 2H), 7.09 (d, J = 8.3 Hz, 2H), 5.93 (dddd, J = 16.8, 10.3, 6.5, 1.7 Hz, 1H), 5.8 (m, 1H), 5.01 (m, 5H), 4.02 (m, 2H), 3.81 (ddd, J = 7.5, 7.5, 7.5 Hz, 1H), 2.73 (m, 1H), 2.64 (ddd, J = 15.3, 7.9, 0 Hz, 1H) 2.38 (m, 4H), 1.81 (m, 2H), 1.17 (d, J = 6.3 Hz, 2H), 1.12 (d, J = 6.3 Hz, 1H); 13C NMR (CDCl3) 100 MHz δ 173.1, 171.2, 141.5, 139.9, 136.8, 131.8, 129.6, 120.8, 115.7, 115.4, 68.3, 60.7, 45.3, 40.4, 34.9, 33.7, 29.0, 20.1.
3-(3-Phenyl-4-pentenoyloxy)-1-methylpropyl-4-pentenoate
The synthesis followed the sequential acylation method above to give the compound in 74% yield as a yellow oil. R f 0.22 (hexanes: EtOAc 95:5); 1H NMR (CDCl3) 400 MHz δ 7.32 (m, 2H), 7.21 (m, 3H), 5.98 (ddd, J = 18.2, 10.14, 7.8 Hz, 1H), 5.81 (m, 1H), 5.0 (m, 5H), 4.21 (m, 2H), 3.86 [ddd (or dt), J = 7.4, 7.4, 7.4 Hz, 1H], 2.80 (dd, J = 15.1, 8.12 Hz, 1H), 2.70 (dd, J = 15.1, 7.4 Hz, 1H), 2.40 (m, 4H), 1.78 (m, 2H) 1.21 (d, 3H, J = 5.6 Hz); 13C NMR (CDCl3) 100 MHz δ 172.6, 171.8, 142.5, 140.4, 136.8, 128.7, 127.7, 126.8, 115.7, 115.0, 67.9, 60.9, 45.7, 40.3, 34.9, 33.9, 29.0, 20.2.
General RCM Method
Under an atmosphere of nitrogen, Grubbs’ second-generation catalyst (0.10 eq.) was added to a solution of the diene in sufficient toluene so that the [diene] ≤ 10 mM. The mixture was heated to 383 K for 18 h. When the reaction was complete, the toluene was removed under reduced pressure to give a residue that was purified by column chromatography.
trans-4,8-Dimethyl-1,10-dioxacyclotridec-5-ene-2,9-dione (I)
Followed the general method of RCM in 21% overall yield (3:2 cis:trans) as a white solid. Compound I is the trans isomer; R f 0.45 (hexanes: EtOAc 80:20) (higher R f, trans); 1HNMR (CDCl3) 400 MHz δ 5.80 (dd, J = 5.9, 2.6 Hz, 2H), 4.43 (ddd, J = 11.1, 8.8, 6.8 Hz, 2H), 3.91 (dd, J = 4.0, 4.0 Hz, 1H), 3.88 (dd, J = 4.0, 4.0 Hz, 1H), 2.61 (m, 2H), 2.30 (dd, J = 13.4, 3.2 Hz, 2H), 2.11 (dd, J = 12.7, 12.7 Hz, 2H), 2.00 (dddd, J = 9.5, 9.5, 4.0, 4.0 Hz, 2H), 1.03 (d, J = 6.9 Hz, 6H); 13C NMR (CDCl3) 100 MHz δ 173.2, 134.3, 59.8, 42.5, 35.9, 25.6, 21.6; TOF HRMS (DART) m/z [M+H]+ calculated for C13H21O4, calculated 241.1440, found 241.1440.
cis-4-Phenyl-13-methyl-1,10-dioxacyclotridec-5-ene-2,9-dione (II)
Followed the general method of RCM in overall 44% yield (2:3 cis:trans). Compound II is the cis isomer and was isolated as a white solid. m.p. 370–374 K; R f 0.46 (hexanes: EtOAc 80:20) (higher R f, cis); 1H NMR (CDCl3) 400 MHz δ 7.43 (d, J = 8.5 Hz, 2H), 7.10 (d, J = 8.5 Hz, 2H), 5.59 (ddd, J = 15.0, 8.7, 5.4 Hz, 1H), 5.44 (dd, J = 15.2, 9.0 Hz, 1H), 5.11 (m, 1H), 4.35 (ddd, J = 11.9, 11.9, 3.7 Hz, 1H), 3.95 (ddd, J = 11.3, 5.2, 2.4 Hz, 1H), 3.81 (ddd, J = 8.3, 8.3, 8.3 Hz, 1H), 2.55 (m, 2H), 2.32 (m, 4H), 2.04 (m, 1H), 1.81 (dddd or ddt, J = 14.6, 6.0, 3.3, 3.3 Hz, 1H), 1.30 (d, J = 6.2 Hz, 3H); 13C NMR (CDCl3) 100 MHz δ 174.0, 171.7, 142.2, 133.2, 132.0, 130.3, 129.0, 67.5, 60.5, 45.5, 41.5, 34.5, 33.5, 29.0, 20.7.
trans-4-Phenyl-11-methyl-1,10-dioxacyclotridec-5-ene-2,9-dione (III)
Followed the general method of RCM in 89% overall yield 2:3 cis:trans). Compound III is the trans isomer, a yellow solid. m.p 359–362 K; R f 0.45 (hexanes: EtOAc 80:20) (higher R f, trans); 1H NMR (CDCl3) 400 MHz δ 7.33 (m, 2H), 7.24 (m, 3H), 5.59 (dd, J = 15.5, 8.5 Hz, 1H), 5.53 (ddd, J = 15.9, 8.7, 5.1 Hz, 1H), 5.14 (m, 1H), 4.35 (ddd, J = 14.9, 11.4, 3.5 Hz, 1H), 3.94 (ddd, J = 12.4, 7.3, 1.3 Hz, 1H), 3.74 (ddd, J = 11.9, 8.8, 3.8, 1H), 2.61 (dd, J = 12.9, 12.9 Hz, 1H), 2.56 (dd, J = 12.6, 4.0 Hz, 1H), 2.42 (m, 2H), 2.29 (m, 1H), 2.20 (m, 1H), 2.1 (dddd, J = 18.6, 9.5, 5.7, 4.1 Hz, 1H), 1.8 (m, 1H), 1.31 (d, J = 6.19 Hz, 3H); 13C NMR (CDCl3) 100 MHz δ 173.0, 172.9, 143.3, 134.5, 129.0, 128.9, 127.1, 126.9, 66.8, 60.6, 47.1, 41.9, 34.3, 33.4, 28.1, 20.7.
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸. For all three structures, no evidence of disorder was found and no special restraints or constraints were required to achieve a stable refinement model. The hydrogen atoms were first found in the difference map, then generated geometrically and refined as riding atoms with C—H distances = 0.95–0.99 Å and U iso(H) = 1.2U eq(C) for CH and CH2 groups and U iso(H) = 1.5U eq(C) for CH3 groups.
Table 2. Experimental details.
| (I) | (II) | (III) | |
|---|---|---|---|
| Crystal data | |||
| Chemical formula | C13H20O4 | C18H21BrO4 | C18H22O4 |
| M r | 240.29 | 381.26 | 302.35 |
| Crystal system, space group | Monoclinic, P21/c | Monoclinic, P21/n | Orthorhombic, P n a21 |
| Temperature (K) | 93 | 93 | 93 |
| a, b, c (Å) | 8.0547 (6), 18.7875 (14), 8.9660 (7) | 15.3128 (3), 5.55594 (11), 20.5689 (4) | 11.2952 (8), 20.9595 (15), 6.6840 (5) |
| α, β, γ (°) | 90, 102.530 (6), 90 | 90, 95.7658 (18), 90 | 90, 90, 90 |
| V (Å3) | 1324.49 (18) | 1741.08 (6) | 1582.4 (2) |
| Z | 4 | 4 | 4 |
| Radiation type | Mo Kα | Cu Kα | Mo Kα |
| μ (mm−1) | 0.09 | 3.37 | 0.09 |
| Crystal size (mm) | 0.18 × 0.17 × 0.16 | 0.20 × 0.19 × 0.10 | 0.32 × 0.20 × 0.20 |
| Data collection | |||
| Diffractometer | Rigaku Mercury275R CCD | Rigaku Saturn 944+ CCD | Rigaku Mercury275R CCD |
| Absorption correction | Multi-scan (REQAB; Jacobson, 1998 ▸) | Multi-scan (CrysAlis PRO; Rigaku OD, 2015 ▸) | Multi-scan (REQAB; Jacobson, 1998 ▸) |
| T min, T max | 0.705, 1.000 | 0.823, 1.000 | 0.815, 1.000 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 17742, 2341, 1835 | 59208, 3075, 3000 | 26669, 3664, 3410 |
| R int | 0.131 | 0.040 | 0.045 |
| (sin θ/λ)max (Å−1) | 0.595 | 0.596 | 0.653 |
| Refinement | |||
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.085, 0.230, 1.09 | 0.027, 0.062, 1.10 | 0.050, 0.128, 1.15 |
| No. of reflections | 2341 | 3075 | 3664 |
| No. of parameters | 156 | 209 | 201 |
| No. of restraints | 0 | 0 | 1 |
| H-atom treatment | H-atom parameters constrained | H-atom parameters constrained | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.53, −0.27 | 0.51, −0.52 | 0.26, −0.20 |
| Absolute structure | – | – | Flack x determined using 1473 quotients [(I +)−(I −)]/[(I +)+(I −)] (Parsons et al., 2013 ▸) |
| Absolute structure parameter | – | – | 0.4 (5) |
Supplementary Material
Crystal structure: contains datablock(s) global, I, II, III. DOI: 10.1107/S2056989020012037/mw2168sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989020012037/mw2168Isup2.hkl
Structure factors: contains datablock(s) II. DOI: 10.1107/S2056989020012037/mw2168IIsup3.hkl
Structure factors: contains datablock(s) III. DOI: 10.1107/S2056989020012037/mw2168IIIsup4.hkl
PDF document (Supplementary data associate with the manuscript: tables, computed structure coordinates, NMR spectra). DOI: 10.1107/S2056989020012037/mw2168sup5.pdf
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
The authors thank Trevor Hamlin for helpful conversations.
supplementary crystallographic information
trans-4,8-Dimethyl-1,10-dioxacyclotridec-5-ene-2,9-dione (I) . Crystal data
| C13H20O4 | F(000) = 520 |
| Mr = 240.29 | Dx = 1.205 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| a = 8.0547 (6) Å | Cell parameters from 687 reflections |
| b = 18.7875 (14) Å | θ = 2.2–27.9° |
| c = 8.9660 (7) Å | µ = 0.09 mm−1 |
| β = 102.530 (6)° | T = 93 K |
| V = 1324.49 (18) Å3 | Prism, colorless |
| Z = 4 | 0.18 × 0.17 × 0.16 mm |
trans-4,8-Dimethyl-1,10-dioxacyclotridec-5-ene-2,9-dione (I) . Data collection
| Rigaku Mercury275R CCD diffractometer | 2341 independent reflections |
| Radiation source: Sealed Tube | 1835 reflections with I > 2σ(I) |
| Detector resolution: 6.8 pixels mm-1 | Rint = 0.131 |
| ω scans | θmax = 25.0°, θmin = 2.2° |
| Absorption correction: multi-scan (REQAB; Jacobson, 1998) | h = −9→9 |
| Tmin = 0.705, Tmax = 1.000 | k = −22→22 |
| 17742 measured reflections | l = −10→10 |
trans-4,8-Dimethyl-1,10-dioxacyclotridec-5-ene-2,9-dione (I) . Refinement
| Refinement on F2 | Primary atom site location: dual |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.085 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.230 | H-atom parameters constrained |
| S = 1.09 | w = 1/[σ2(Fo2) + (0.1386P)2 + 0.4739P] where P = (Fo2 + 2Fc2)/3 |
| 2341 reflections | (Δ/σ)max < 0.001 |
| 156 parameters | Δρmax = 0.53 e Å−3 |
| 0 restraints | Δρmin = −0.27 e Å−3 |
trans-4,8-Dimethyl-1,10-dioxacyclotridec-5-ene-2,9-dione (I) . Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > 2sigma(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. The hydrogen atoms were first found in the difference map, then generated geometrically and refined as riding atoms with C-H distances = 0.95 - 0.99 angstroms and Uiso(H) = 1.2 times Ueq(C) for CH and CH2 groups and Uiso(H) = 1.5 times Ueq(C) for CH3 groups. |
trans-4,8-Dimethyl-1,10-dioxacyclotridec-5-ene-2,9-dione (I) . Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.5209 (3) | 0.33574 (11) | 0.6049 (2) | 0.0443 (6) | |
| O10 | 0.8844 (3) | 0.38480 (10) | 0.5944 (2) | 0.0426 (6) | |
| O14 | 0.3382 (3) | 0.42433 (13) | 0.5285 (3) | 0.0583 (7) | |
| O17 | 0.9157 (3) | 0.31352 (11) | 0.4047 (2) | 0.0441 (6) | |
| C2 | 0.3918 (4) | 0.36655 (17) | 0.5079 (3) | 0.0419 (7) | |
| C3 | 0.3224 (4) | 0.32154 (16) | 0.3709 (3) | 0.0409 (7) | |
| H3A | 0.2054 | 0.3064 | 0.3736 | 0.049* | |
| H3B | 0.3932 | 0.2782 | 0.3740 | 0.049* | |
| C4 | 0.3199 (4) | 0.36195 (15) | 0.2221 (3) | 0.0387 (7) | |
| H4 | 0.2743 | 0.4108 | 0.2317 | 0.046* | |
| C5 | 0.4992 (3) | 0.36868 (15) | 0.1969 (3) | 0.0378 (7) | |
| H5 | 0.5469 | 0.3282 | 0.1584 | 0.045* | |
| C6 | 0.5933 (4) | 0.42558 (15) | 0.2237 (3) | 0.0372 (7) | |
| H6 | 0.5432 | 0.4665 | 0.2580 | 0.045* | |
| C7 | 0.7736 (4) | 0.43256 (15) | 0.2055 (3) | 0.0384 (7) | |
| H7 | 0.8039 | 0.3891 | 0.1526 | 0.046* | |
| C8 | 0.8940 (4) | 0.43846 (15) | 0.3605 (3) | 0.0401 (7) | |
| H8A | 1.0097 | 0.4494 | 0.3460 | 0.048* | |
| H8B | 0.8573 | 0.4784 | 0.4179 | 0.048* | |
| C9 | 0.8998 (3) | 0.37194 (15) | 0.4516 (3) | 0.0369 (7) | |
| C11 | 0.8732 (4) | 0.32312 (17) | 0.6890 (3) | 0.0450 (8) | |
| H11A | 0.9884 | 0.3077 | 0.7422 | 0.054* | |
| H11B | 0.8174 | 0.2832 | 0.6252 | 0.054* | |
| C12 | 0.7703 (4) | 0.34428 (18) | 0.8036 (3) | 0.0464 (8) | |
| H12A | 0.7449 | 0.3013 | 0.8584 | 0.056* | |
| H12B | 0.8387 | 0.3768 | 0.8797 | 0.056* | |
| C13 | 0.6064 (4) | 0.38033 (17) | 0.7309 (3) | 0.0460 (8) | |
| H13A | 0.5341 | 0.3861 | 0.8064 | 0.055* | |
| H13B | 0.6293 | 0.4280 | 0.6928 | 0.055* | |
| C15 | 0.2026 (4) | 0.32451 (17) | 0.0917 (4) | 0.0468 (8) | |
| H15A | 0.2431 | 0.2758 | 0.0830 | 0.070* | |
| H15B | 0.2010 | 0.3503 | −0.0037 | 0.070* | |
| H15C | 0.0874 | 0.3231 | 0.1109 | 0.070* | |
| C16 | 0.7922 (4) | 0.49735 (17) | 0.1094 (4) | 0.0498 (8) | |
| H16A | 0.7628 | 0.5403 | 0.1602 | 0.075* | |
| H16B | 0.7159 | 0.4928 | 0.0087 | 0.075* | |
| H16C | 0.9100 | 0.5008 | 0.0974 | 0.075* |
trans-4,8-Dimethyl-1,10-dioxacyclotridec-5-ene-2,9-dione (I) . Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0427 (12) | 0.0569 (13) | 0.0360 (11) | 0.0046 (9) | 0.0147 (10) | −0.0001 (9) |
| O10 | 0.0475 (12) | 0.0532 (12) | 0.0304 (11) | 0.0040 (9) | 0.0158 (9) | −0.0037 (8) |
| O14 | 0.0538 (14) | 0.0732 (16) | 0.0481 (14) | 0.0204 (12) | 0.0117 (11) | −0.0121 (11) |
| O17 | 0.0451 (12) | 0.0523 (13) | 0.0410 (12) | 0.0014 (9) | 0.0231 (10) | −0.0055 (9) |
| C2 | 0.0361 (15) | 0.0590 (19) | 0.0358 (16) | 0.0042 (13) | 0.0194 (13) | 0.0013 (13) |
| C3 | 0.0316 (15) | 0.0555 (18) | 0.0400 (17) | 0.0003 (12) | 0.0176 (13) | 0.0009 (12) |
| C4 | 0.0322 (15) | 0.0514 (17) | 0.0364 (15) | 0.0003 (12) | 0.0161 (12) | −0.0003 (12) |
| C5 | 0.0335 (15) | 0.0539 (17) | 0.0308 (14) | 0.0020 (12) | 0.0175 (12) | −0.0032 (12) |
| C6 | 0.0385 (15) | 0.0481 (16) | 0.0297 (14) | 0.0040 (12) | 0.0179 (12) | 0.0002 (11) |
| C7 | 0.0354 (15) | 0.0519 (17) | 0.0337 (15) | −0.0015 (12) | 0.0202 (12) | −0.0026 (11) |
| C8 | 0.0352 (15) | 0.0516 (17) | 0.0391 (16) | −0.0020 (12) | 0.0205 (13) | −0.0053 (12) |
| C9 | 0.0248 (13) | 0.0525 (18) | 0.0360 (15) | 0.0018 (11) | 0.0120 (12) | −0.0047 (12) |
| C11 | 0.0493 (18) | 0.0552 (18) | 0.0341 (16) | 0.0086 (13) | 0.0170 (14) | 0.0017 (12) |
| C12 | 0.0477 (18) | 0.063 (2) | 0.0306 (15) | 0.0076 (14) | 0.0136 (14) | −0.0013 (13) |
| C13 | 0.0502 (18) | 0.0589 (19) | 0.0320 (15) | 0.0054 (14) | 0.0161 (14) | −0.0041 (12) |
| C15 | 0.0391 (17) | 0.0627 (19) | 0.0418 (17) | −0.0038 (14) | 0.0161 (14) | −0.0009 (14) |
| C16 | 0.0486 (19) | 0.065 (2) | 0.0422 (17) | −0.0030 (15) | 0.0247 (16) | 0.0033 (14) |
trans-4,8-Dimethyl-1,10-dioxacyclotridec-5-ene-2,9-dione (I) . Geometric parameters (Å, º)
| O1—C2 | 1.334 (4) | C7—H7 | 1.0000 |
| O1—C13 | 1.454 (4) | C8—C9 | 1.488 (4) |
| O10—C9 | 1.335 (3) | C8—H8A | 0.9900 |
| O10—C11 | 1.450 (4) | C8—H8B | 0.9900 |
| O14—C2 | 1.197 (4) | C11—C12 | 1.506 (4) |
| O17—C9 | 1.192 (3) | C11—H11A | 0.9900 |
| C2—C3 | 1.496 (4) | C11—H11B | 0.9900 |
| C3—C4 | 1.532 (4) | C12—C13 | 1.501 (4) |
| C3—H3A | 0.9900 | C12—H12A | 0.9900 |
| C3—H3B | 0.9900 | C12—H12B | 0.9900 |
| C4—C15 | 1.508 (4) | C13—H13A | 0.9900 |
| C4—C5 | 1.515 (4) | C13—H13B | 0.9900 |
| C4—H4 | 1.0000 | C15—H15A | 0.9800 |
| C5—C6 | 1.302 (4) | C15—H15B | 0.9800 |
| C5—H5 | 0.9500 | C15—H15C | 0.9800 |
| C6—C7 | 1.502 (4) | C16—H16A | 0.9800 |
| C6—H6 | 0.9500 | C16—H16B | 0.9800 |
| C7—C8 | 1.516 (4) | C16—H16C | 0.9800 |
| C7—C16 | 1.517 (4) | ||
| C2—O1—C13 | 115.3 (2) | H8A—C8—H8B | 107.9 |
| C9—O10—C11 | 116.5 (2) | O17—C9—O10 | 123.0 (3) |
| O14—C2—O1 | 123.3 (3) | O17—C9—C8 | 124.9 (3) |
| O14—C2—C3 | 123.8 (3) | O10—C9—C8 | 112.1 (2) |
| O1—C2—C3 | 112.9 (3) | O10—C11—C12 | 107.5 (2) |
| C2—C3—C4 | 111.5 (2) | O10—C11—H11A | 110.2 |
| C2—C3—H3A | 109.3 | C12—C11—H11A | 110.2 |
| C4—C3—H3A | 109.3 | O10—C11—H11B | 110.2 |
| C2—C3—H3B | 109.3 | C12—C11—H11B | 110.2 |
| C4—C3—H3B | 109.3 | H11A—C11—H11B | 108.5 |
| H3A—C3—H3B | 108.0 | C13—C12—C11 | 112.7 (2) |
| C15—C4—C5 | 112.4 (2) | C13—C12—H12A | 109.1 |
| C15—C4—C3 | 109.4 (2) | C11—C12—H12A | 109.1 |
| C5—C4—C3 | 109.8 (2) | C13—C12—H12B | 109.1 |
| C15—C4—H4 | 108.4 | C11—C12—H12B | 109.1 |
| C5—C4—H4 | 108.4 | H12A—C12—H12B | 107.8 |
| C3—C4—H4 | 108.4 | O1—C13—C12 | 107.5 (2) |
| C6—C5—C4 | 125.2 (2) | O1—C13—H13A | 110.2 |
| C6—C5—H5 | 117.4 | C12—C13—H13A | 110.2 |
| C4—C5—H5 | 117.4 | O1—C13—H13B | 110.2 |
| C5—C6—C7 | 126.1 (3) | C12—C13—H13B | 110.2 |
| C5—C6—H6 | 117.0 | H13A—C13—H13B | 108.5 |
| C7—C6—H6 | 117.0 | C4—C15—H15A | 109.5 |
| C6—C7—C8 | 110.4 (2) | C4—C15—H15B | 109.5 |
| C6—C7—C16 | 110.4 (2) | H15A—C15—H15B | 109.5 |
| C8—C7—C16 | 109.8 (2) | C4—C15—H15C | 109.5 |
| C6—C7—H7 | 108.7 | H15A—C15—H15C | 109.5 |
| C8—C7—H7 | 108.7 | H15B—C15—H15C | 109.5 |
| C16—C7—H7 | 108.7 | C7—C16—H16A | 109.5 |
| C9—C8—C7 | 112.3 (2) | C7—C16—H16B | 109.5 |
| C9—C8—H8A | 109.1 | H16A—C16—H16B | 109.5 |
| C7—C8—H8A | 109.1 | C7—C16—H16C | 109.5 |
| C9—C8—H8B | 109.1 | H16A—C16—H16C | 109.5 |
| C7—C8—H8B | 109.1 | H16B—C16—H16C | 109.5 |
| C13—O1—C2—O14 | −6.6 (4) | C6—C7—C8—C9 | 66.3 (3) |
| C13—O1—C2—C3 | 173.1 (2) | C16—C7—C8—C9 | −171.8 (2) |
| O14—C2—C3—C4 | 53.8 (4) | C11—O10—C9—O17 | −5.1 (4) |
| O1—C2—C3—C4 | −125.8 (2) | C11—O10—C9—C8 | 174.7 (2) |
| C2—C3—C4—C15 | −162.7 (2) | C7—C8—C9—O17 | 47.6 (4) |
| C2—C3—C4—C5 | 73.6 (3) | C7—C8—C9—O10 | −132.2 (2) |
| C15—C4—C5—C6 | 136.5 (3) | C9—O10—C11—C12 | −151.1 (3) |
| C3—C4—C5—C6 | −101.5 (3) | O10—C11—C12—C13 | 49.6 (4) |
| C4—C5—C6—C7 | 177.4 (3) | C2—O1—C13—C12 | −166.3 (2) |
| C5—C6—C7—C8 | −109.7 (3) | C11—C12—C13—O1 | 50.5 (4) |
| C5—C6—C7—C16 | 128.7 (3) |
cis-4-(4-Bromophenyl)-13-methyl-1,10-dioxacyclotridec-5-ene-2,9-dione (II) . Crystal data
| C18H21BrO4 | F(000) = 784 |
| Mr = 381.26 | Dx = 1.454 Mg m−3 |
| Monoclinic, P21/n | Cu Kα radiation, λ = 1.54184 Å |
| a = 15.3128 (3) Å | Cell parameters from 41470 reflections |
| b = 5.55594 (11) Å | θ = 2.1–66.6° |
| c = 20.5689 (4) Å | µ = 3.37 mm−1 |
| β = 95.7658 (18)° | T = 93 K |
| V = 1741.08 (6) Å3 | Block, colorless |
| Z = 4 | 0.20 × 0.19 × 0.10 mm |
cis-4-(4-Bromophenyl)-13-methyl-1,10-dioxacyclotridec-5-ene-2,9-dione (II) . Data collection
| Rigaku Saturn 944+ CCD diffractometer | 3075 independent reflections |
| Radiation source: microfocus rotating anode | 3000 reflections with I > 2σ(I) |
| Detector resolution: 22.2 pixels mm-1 | Rint = 0.040 |
| ω scans | θmax = 66.8°, θmin = 3.4° |
| Absorption correction: multi-scan (CrysAlisPro; Rigaku OD, 2015) | h = −18→18 |
| Tmin = 0.823, Tmax = 1.000 | k = −6→6 |
| 59208 measured reflections | l = −24→24 |
cis-4-(4-Bromophenyl)-13-methyl-1,10-dioxacyclotridec-5-ene-2,9-dione (II) . Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.027 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.062 | H-atom parameters constrained |
| S = 1.10 | w = 1/[σ2(Fo2) + (0.0186P)2 + 2.082P] where P = (Fo2 + 2Fc2)/3 |
| 3075 reflections | (Δ/σ)max = 0.001 |
| 209 parameters | Δρmax = 0.51 e Å−3 |
| 0 restraints | Δρmin = −0.52 e Å−3 |
cis-4-(4-Bromophenyl)-13-methyl-1,10-dioxacyclotridec-5-ene-2,9-dione (II) . Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > 2sigma(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. The hydrogen atoms were first found in the difference map, then generated geometrically and refined as riding atoms with C-H distances = 0.95 - 0.99 angstroms and Uiso(H) = 1.2 times Ueq(C) for CH and CH2 groups and Uiso(H) = 1.5 times Ueq(C) for CH3 groups. |
cis-4-(4-Bromophenyl)-13-methyl-1,10-dioxacyclotridec-5-ene-2,9-dione (II) . Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Br1 | 0.67994 (2) | 1.25106 (4) | 0.26935 (2) | 0.02712 (8) | |
| O1 | 0.27479 (8) | 0.5670 (2) | 0.52299 (6) | 0.0222 (3) | |
| O10 | 0.08625 (9) | 0.6437 (3) | 0.45484 (7) | 0.0297 (3) | |
| O15 | 0.32471 (11) | 0.2793 (3) | 0.45930 (8) | 0.0361 (4) | |
| O22 | 0.08714 (12) | 1.0133 (3) | 0.40964 (8) | 0.0466 (4) | |
| C2 | 0.32629 (13) | 0.4847 (4) | 0.47904 (9) | 0.0246 (4) | |
| C3 | 0.38595 (13) | 0.6777 (4) | 0.45684 (9) | 0.0253 (4) | |
| H3A | 0.4470 | 0.6480 | 0.4759 | 0.030* | |
| H3B | 0.3674 | 0.8369 | 0.4721 | 0.030* | |
| C4 | 0.38196 (12) | 0.6768 (4) | 0.38145 (9) | 0.0245 (4) | |
| H4 | 0.3863 | 0.5058 | 0.3669 | 0.029* | |
| C5 | 0.29466 (13) | 0.7741 (4) | 0.35257 (9) | 0.0271 (4) | |
| H5 | 0.2847 | 0.9422 | 0.3559 | 0.032* | |
| C6 | 0.23075 (14) | 0.6403 (4) | 0.32272 (10) | 0.0311 (5) | |
| H6 | 0.2416 | 0.4733 | 0.3177 | 0.037* | |
| C7 | 0.14240 (14) | 0.7363 (4) | 0.29646 (10) | 0.0334 (5) | |
| H7A | 0.1272 | 0.6740 | 0.2517 | 0.040* | |
| H7B | 0.1452 | 0.9140 | 0.2940 | 0.040* | |
| C8 | 0.07030 (13) | 0.6635 (5) | 0.33969 (10) | 0.0339 (5) | |
| H8A | 0.0120 | 0.7011 | 0.3167 | 0.041* | |
| H8B | 0.0731 | 0.4879 | 0.3478 | 0.041* | |
| C9 | 0.08167 (14) | 0.7959 (4) | 0.40389 (11) | 0.0316 (5) | |
| C11 | 0.10483 (14) | 0.7529 (4) | 0.51898 (10) | 0.0313 (5) | |
| H11A | 0.0512 | 0.8289 | 0.5328 | 0.038* | |
| H11B | 0.1506 | 0.8782 | 0.5178 | 0.038* | |
| C12 | 0.13630 (13) | 0.5550 (4) | 0.56615 (9) | 0.0275 (4) | |
| H12A | 0.0858 | 0.4510 | 0.5737 | 0.033* | |
| H12B | 0.1582 | 0.6286 | 0.6085 | 0.033* | |
| C13 | 0.20832 (12) | 0.3996 (4) | 0.54275 (9) | 0.0231 (4) | |
| H13 | 0.1840 | 0.3031 | 0.5041 | 0.028* | |
| C14 | 0.24961 (14) | 0.2320 (4) | 0.59525 (10) | 0.0290 (5) | |
| H14A | 0.2794 | 0.3270 | 0.6310 | 0.043* | |
| H14B | 0.2923 | 0.1275 | 0.5767 | 0.043* | |
| H14C | 0.2038 | 0.1331 | 0.6120 | 0.043* | |
| C16 | 0.45741 (12) | 0.8154 (4) | 0.35657 (9) | 0.0213 (4) | |
| C17 | 0.48886 (12) | 1.0298 (4) | 0.38536 (9) | 0.0239 (4) | |
| H17 | 0.4642 | 1.0888 | 0.4228 | 0.029* | |
| C18 | 0.55561 (13) | 1.1589 (4) | 0.36022 (9) | 0.0240 (4) | |
| H18 | 0.5769 | 1.3044 | 0.3803 | 0.029* | |
| C19 | 0.59046 (12) | 1.0712 (4) | 0.30541 (9) | 0.0217 (4) | |
| C20 | 0.56177 (13) | 0.8585 (4) | 0.27595 (9) | 0.0241 (4) | |
| H20 | 0.5871 | 0.7998 | 0.2387 | 0.029* | |
| C21 | 0.49528 (13) | 0.7323 (4) | 0.30178 (9) | 0.0239 (4) | |
| H21 | 0.4750 | 0.5859 | 0.2818 | 0.029* |
cis-4-(4-Bromophenyl)-13-methyl-1,10-dioxacyclotridec-5-ene-2,9-dione (II) . Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Br1 | 0.02422 (12) | 0.02824 (13) | 0.03034 (13) | −0.00158 (8) | 0.00974 (8) | 0.00148 (8) |
| O1 | 0.0221 (6) | 0.0222 (7) | 0.0230 (6) | −0.0016 (6) | 0.0054 (5) | −0.0003 (5) |
| O10 | 0.0267 (7) | 0.0356 (8) | 0.0258 (7) | −0.0019 (6) | −0.0033 (6) | −0.0008 (6) |
| O15 | 0.0445 (9) | 0.0246 (8) | 0.0427 (9) | 0.0027 (7) | 0.0216 (7) | 0.0000 (7) |
| O22 | 0.0656 (12) | 0.0349 (10) | 0.0395 (9) | 0.0091 (9) | 0.0059 (8) | −0.0002 (8) |
| C2 | 0.0246 (10) | 0.0255 (11) | 0.0242 (9) | 0.0040 (8) | 0.0048 (8) | 0.0025 (8) |
| C3 | 0.0221 (9) | 0.0291 (11) | 0.0250 (10) | −0.0001 (8) | 0.0035 (8) | 0.0038 (8) |
| C4 | 0.0230 (10) | 0.0272 (11) | 0.0237 (10) | −0.0003 (8) | 0.0041 (8) | 0.0008 (8) |
| C5 | 0.0228 (10) | 0.0340 (12) | 0.0245 (10) | 0.0015 (9) | 0.0030 (8) | 0.0028 (9) |
| C6 | 0.0331 (11) | 0.0340 (12) | 0.0264 (10) | 0.0019 (10) | 0.0037 (8) | −0.0019 (9) |
| C7 | 0.0300 (11) | 0.0421 (13) | 0.0268 (10) | −0.0016 (10) | −0.0027 (9) | −0.0007 (10) |
| C8 | 0.0237 (10) | 0.0476 (14) | 0.0292 (11) | −0.0040 (10) | −0.0039 (8) | −0.0011 (10) |
| C9 | 0.0222 (10) | 0.0410 (14) | 0.0315 (11) | 0.0060 (9) | 0.0026 (8) | 0.0016 (10) |
| C11 | 0.0299 (11) | 0.0360 (12) | 0.0277 (11) | 0.0070 (10) | 0.0017 (9) | −0.0070 (9) |
| C12 | 0.0235 (10) | 0.0353 (12) | 0.0241 (10) | −0.0010 (9) | 0.0047 (8) | −0.0027 (9) |
| C13 | 0.0241 (9) | 0.0239 (10) | 0.0214 (9) | −0.0055 (8) | 0.0034 (7) | 0.0004 (8) |
| C14 | 0.0356 (11) | 0.0287 (11) | 0.0225 (10) | −0.0013 (9) | 0.0023 (8) | 0.0034 (9) |
| C16 | 0.0194 (9) | 0.0227 (10) | 0.0217 (9) | 0.0025 (8) | 0.0014 (7) | 0.0035 (8) |
| C17 | 0.0234 (9) | 0.0260 (10) | 0.0230 (9) | 0.0023 (8) | 0.0065 (7) | −0.0018 (8) |
| C18 | 0.0246 (10) | 0.0220 (10) | 0.0254 (9) | 0.0000 (8) | 0.0025 (8) | −0.0018 (8) |
| C19 | 0.0181 (9) | 0.0245 (10) | 0.0226 (9) | 0.0021 (8) | 0.0024 (7) | 0.0043 (8) |
| C20 | 0.0264 (10) | 0.0257 (10) | 0.0207 (9) | 0.0044 (8) | 0.0052 (8) | −0.0004 (8) |
| C21 | 0.0270 (10) | 0.0221 (10) | 0.0223 (9) | 0.0002 (8) | 0.0013 (8) | −0.0006 (8) |
cis-4-(4-Bromophenyl)-13-methyl-1,10-dioxacyclotridec-5-ene-2,9-dione (II) . Geometric parameters (Å, º)
| Br1—C19 | 1.9053 (19) | C8—H8B | 0.9900 |
| O1—C2 | 1.338 (2) | C11—C12 | 1.513 (3) |
| O1—C13 | 1.466 (2) | C11—H11A | 0.9900 |
| O10—C9 | 1.343 (3) | C11—H11B | 0.9900 |
| O10—C11 | 1.454 (2) | C12—C13 | 1.516 (3) |
| O15—C2 | 1.210 (3) | C12—H12A | 0.9900 |
| O22—C9 | 1.216 (3) | C12—H12B | 0.9900 |
| C2—C3 | 1.509 (3) | C13—C14 | 1.515 (3) |
| C3—C4 | 1.546 (3) | C13—H13 | 1.0000 |
| C3—H3A | 0.9900 | C14—H14A | 0.9800 |
| C3—H3B | 0.9900 | C14—H14B | 0.9800 |
| C4—C5 | 1.508 (3) | C14—H14C | 0.9800 |
| C4—C16 | 1.519 (3) | C16—C17 | 1.395 (3) |
| C4—H4 | 1.0000 | C16—C21 | 1.397 (3) |
| C5—C6 | 1.329 (3) | C17—C18 | 1.390 (3) |
| C5—H5 | 0.9500 | C17—H17 | 0.9500 |
| C6—C7 | 1.503 (3) | C18—C19 | 1.384 (3) |
| C6—H6 | 0.9500 | C18—H18 | 0.9500 |
| C7—C8 | 1.540 (3) | C19—C20 | 1.379 (3) |
| C7—H7A | 0.9900 | C20—C21 | 1.385 (3) |
| C7—H7B | 0.9900 | C20—H20 | 0.9500 |
| C8—C9 | 1.506 (3) | C21—H21 | 0.9500 |
| C8—H8A | 0.9900 | ||
| C2—O1—C13 | 116.31 (15) | C12—C11—H11A | 110.2 |
| C9—O10—C11 | 115.81 (18) | O10—C11—H11B | 110.2 |
| O15—C2—O1 | 123.73 (19) | C12—C11—H11B | 110.2 |
| O15—C2—C3 | 124.10 (18) | H11A—C11—H11B | 108.5 |
| O1—C2—C3 | 112.16 (17) | C11—C12—C13 | 113.90 (16) |
| C2—C3—C4 | 109.70 (17) | C11—C12—H12A | 108.8 |
| C2—C3—H3A | 109.7 | C13—C12—H12A | 108.8 |
| C4—C3—H3A | 109.7 | C11—C12—H12B | 108.8 |
| C2—C3—H3B | 109.7 | C13—C12—H12B | 108.8 |
| C4—C3—H3B | 109.7 | H12A—C12—H12B | 107.7 |
| H3A—C3—H3B | 108.2 | O1—C13—C14 | 109.62 (15) |
| C5—C4—C16 | 111.08 (17) | O1—C13—C12 | 105.93 (16) |
| C5—C4—C3 | 109.77 (16) | C14—C13—C12 | 112.85 (16) |
| C16—C4—C3 | 112.43 (16) | O1—C13—H13 | 109.5 |
| C5—C4—H4 | 107.8 | C14—C13—H13 | 109.5 |
| C16—C4—H4 | 107.8 | C12—C13—H13 | 109.5 |
| C3—C4—H4 | 107.8 | C13—C14—H14A | 109.5 |
| C6—C5—C4 | 124.3 (2) | C13—C14—H14B | 109.5 |
| C6—C5—H5 | 117.8 | H14A—C14—H14B | 109.5 |
| C4—C5—H5 | 117.8 | C13—C14—H14C | 109.5 |
| C5—C6—C7 | 124.2 (2) | H14A—C14—H14C | 109.5 |
| C5—C6—H6 | 117.9 | H14B—C14—H14C | 109.5 |
| C7—C6—H6 | 117.9 | C17—C16—C21 | 118.13 (18) |
| C6—C7—C8 | 111.78 (18) | C17—C16—C4 | 122.13 (17) |
| C6—C7—H7A | 109.3 | C21—C16—C4 | 119.71 (18) |
| C8—C7—H7A | 109.3 | C18—C17—C16 | 121.21 (18) |
| C6—C7—H7B | 109.3 | C18—C17—H17 | 119.4 |
| C8—C7—H7B | 109.3 | C16—C17—H17 | 119.4 |
| H7A—C7—H7B | 107.9 | C19—C18—C17 | 118.67 (19) |
| C9—C8—C7 | 110.59 (18) | C19—C18—H18 | 120.7 |
| C9—C8—H8A | 109.5 | C17—C18—H18 | 120.7 |
| C7—C8—H8A | 109.5 | C20—C19—C18 | 121.85 (18) |
| C9—C8—H8B | 109.5 | C20—C19—Br1 | 119.21 (14) |
| C7—C8—H8B | 109.5 | C18—C19—Br1 | 118.94 (15) |
| H8A—C8—H8B | 108.1 | C19—C20—C21 | 118.62 (18) |
| O22—C9—O10 | 123.5 (2) | C19—C20—H20 | 120.7 |
| O22—C9—C8 | 124.8 (2) | C21—C20—H20 | 120.7 |
| O10—C9—C8 | 111.6 (2) | C20—C21—C16 | 121.52 (19) |
| O10—C11—C12 | 107.42 (17) | C20—C21—H21 | 119.2 |
| O10—C11—H11A | 110.2 | C16—C21—H21 | 119.2 |
| C13—O1—C2—O15 | −5.6 (3) | C2—O1—C13—C12 | −155.41 (15) |
| C13—O1—C2—C3 | 174.13 (15) | C11—C12—C13—O1 | 50.0 (2) |
| O15—C2—C3—C4 | 47.6 (3) | C11—C12—C13—C14 | 169.98 (18) |
| O1—C2—C3—C4 | −132.13 (17) | C5—C4—C16—C17 | 84.5 (2) |
| C2—C3—C4—C5 | 71.6 (2) | C3—C4—C16—C17 | −38.9 (3) |
| C2—C3—C4—C16 | −164.23 (16) | C5—C4—C16—C21 | −93.3 (2) |
| C16—C4—C5—C6 | 128.0 (2) | C3—C4—C16—C21 | 143.29 (18) |
| C3—C4—C5—C6 | −107.0 (2) | C21—C16—C17—C18 | 0.4 (3) |
| C4—C5—C6—C7 | 177.34 (19) | C4—C16—C17—C18 | −177.39 (18) |
| C5—C6—C7—C8 | −105.8 (3) | C16—C17—C18—C19 | 0.3 (3) |
| C6—C7—C8—C9 | 70.3 (3) | C17—C18—C19—C20 | −1.1 (3) |
| C11—O10—C9—O22 | −4.3 (3) | C17—C18—C19—Br1 | 178.45 (14) |
| C11—O10—C9—C8 | 174.89 (16) | C18—C19—C20—C21 | 1.0 (3) |
| C7—C8—C9—O22 | 54.0 (3) | Br1—C19—C20—C21 | −178.54 (14) |
| C7—C8—C9—O10 | −125.2 (2) | C19—C20—C21—C16 | −0.2 (3) |
| C9—O10—C11—C12 | −161.83 (17) | C17—C16—C21—C20 | −0.5 (3) |
| O10—C11—C12—C13 | 49.7 (2) | C4—C16—C21—C20 | 177.35 (18) |
| C2—O1—C13—C14 | 82.6 (2) |
trans-11-Methyl-4-phenyl-1,10-dioxacyclotridec-5-ene-2,9-dione (III) . Crystal data
| C18H22O4 | Dx = 1.269 Mg m−3 |
| Mr = 302.35 | Mo Kα radiation, λ = 0.71073 Å |
| Orthorhombic, Pna21 | Cell parameters from 576 reflections |
| a = 11.2952 (8) Å | θ = 2.6–27.5° |
| b = 20.9595 (15) Å | µ = 0.09 mm−1 |
| c = 6.6840 (5) Å | T = 93 K |
| V = 1582.4 (2) Å3 | Prism, colorless |
| Z = 4 | 0.32 × 0.20 × 0.20 mm |
| F(000) = 648 |
trans-11-Methyl-4-phenyl-1,10-dioxacyclotridec-5-ene-2,9-dione (III) . Data collection
| Rigaku Mercury275R CCD diffractometer | 3664 independent reflections |
| Radiation source: Sealed Tube | 3410 reflections with I > 2σ(I) |
| Detector resolution: 6.8 pixels mm-1 | Rint = 0.045 |
| ω scans | θmax = 27.7°, θmin = 2.7° |
| Absorption correction: multi-scan (REQAB; Jacobson, 1998) | h = −14→14 |
| Tmin = 0.815, Tmax = 1.000 | k = −27→27 |
| 26669 measured reflections | l = −8→8 |
trans-11-Methyl-4-phenyl-1,10-dioxacyclotridec-5-ene-2,9-dione (III) . Refinement
| Refinement on F2 | Hydrogen site location: inferred from neighbouring sites |
| Least-squares matrix: full | H-atom parameters constrained |
| R[F2 > 2σ(F2)] = 0.050 | w = 1/[σ2(Fo2) + (0.0578P)2 + 0.8966P] where P = (Fo2 + 2Fc2)/3 |
| wR(F2) = 0.128 | (Δ/σ)max < 0.001 |
| S = 1.15 | Δρmax = 0.26 e Å−3 |
| 3664 reflections | Δρmin = −0.20 e Å−3 |
| 201 parameters | Extinction correction: SHELXL2014/7 (Sheldrick 2015b), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| 1 restraint | Extinction coefficient: 0.012 (3) |
| Primary atom site location: dual | Absolute structure: Flack x determined using 1473 quotients [(I+)-(I-)]/[(I+)+(I-)] (Parsons et al., 2013) |
| Secondary atom site location: difference Fourier map | Absolute structure parameter: 0.4 (5) |
trans-11-Methyl-4-phenyl-1,10-dioxacyclotridec-5-ene-2,9-dione (III) . Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > 2sigma(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. The hydrogen atoms were first found in the difference map, then generated geometrically and refined as riding atoms with C-H distances = 0.95 - 0.99 angstroms and Uiso(H) = 1.2 times Ueq(C) for CH and CH2 groups and Uiso(H) = 1.5 times Ueq(C) for CH3 groups. |
trans-11-Methyl-4-phenyl-1,10-dioxacyclotridec-5-ene-2,9-dione (III) . Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.15097 (18) | 0.56820 (10) | 0.5040 (3) | 0.0233 (4) | |
| O10 | 0.08596 (17) | 0.70330 (9) | 0.6331 (3) | 0.0221 (4) | |
| O14 | 0.13825 (19) | 0.55047 (11) | 0.1764 (4) | 0.0299 (5) | |
| O15 | 0.2504 (2) | 0.73924 (11) | 0.7780 (4) | 0.0338 (5) | |
| C2 | 0.1964 (2) | 0.55372 (12) | 0.3251 (5) | 0.0227 (6) | |
| C3 | 0.3271 (2) | 0.54399 (13) | 0.3346 (5) | 0.0238 (6) | |
| H3A | 0.3477 | 0.5027 | 0.2718 | 0.029* | |
| H3B | 0.3528 | 0.5427 | 0.4761 | 0.029* | |
| C4 | 0.3912 (2) | 0.59802 (13) | 0.2261 (4) | 0.0221 (6) | |
| H4 | 0.3547 | 0.6031 | 0.0907 | 0.026* | |
| C5 | 0.3778 (2) | 0.65980 (13) | 0.3371 (5) | 0.0217 (5) | |
| H5 | 0.4104 | 0.6627 | 0.4679 | 0.026* | |
| C6 | 0.3236 (2) | 0.71018 (13) | 0.2639 (5) | 0.0228 (5) | |
| H6 | 0.2880 | 0.7067 | 0.1356 | 0.027* | |
| C7 | 0.3150 (3) | 0.77242 (14) | 0.3707 (5) | 0.0249 (6) | |
| H7A | 0.3313 | 0.8074 | 0.2751 | 0.030* | |
| H7B | 0.3761 | 0.7740 | 0.4767 | 0.030* | |
| C8 | 0.1937 (3) | 0.78283 (13) | 0.4646 (5) | 0.0236 (6) | |
| H8A | 0.1850 | 0.8280 | 0.5058 | 0.028* | |
| H8B | 0.1309 | 0.7730 | 0.3660 | 0.028* | |
| C9 | 0.1814 (3) | 0.74016 (13) | 0.6434 (5) | 0.0231 (6) | |
| C11 | 0.0719 (2) | 0.65729 (13) | 0.7937 (4) | 0.0227 (6) | |
| H11 | 0.1511 | 0.6395 | 0.8309 | 0.027* | |
| C12 | −0.0051 (3) | 0.60478 (14) | 0.7118 (5) | 0.0249 (6) | |
| H12A | −0.0009 | 0.5679 | 0.8039 | 0.030* | |
| H12B | −0.0882 | 0.6199 | 0.7106 | 0.030* | |
| C13 | 0.0263 (2) | 0.58247 (14) | 0.5055 (5) | 0.0252 (6) | |
| H13A | −0.0200 | 0.5439 | 0.4709 | 0.030* | |
| H13B | 0.0082 | 0.6161 | 0.4063 | 0.030* | |
| C15 | 0.5218 (2) | 0.58492 (13) | 0.1992 (4) | 0.0215 (5) | |
| C16 | 0.5869 (2) | 0.54591 (13) | 0.3230 (5) | 0.0238 (6) | |
| H16 | 0.5477 | 0.5230 | 0.4262 | 0.029* | |
| C17 | 0.7081 (3) | 0.53929 (14) | 0.3008 (5) | 0.0261 (6) | |
| H17 | 0.7515 | 0.5124 | 0.3885 | 0.031* | |
| C18 | 0.7656 (2) | 0.57231 (13) | 0.1494 (5) | 0.0258 (6) | |
| H18 | 0.8487 | 0.5678 | 0.1318 | 0.031* | |
| C19 | 0.7017 (3) | 0.61141 (14) | 0.0255 (5) | 0.0260 (6) | |
| H19 | 0.7411 | 0.6342 | −0.0778 | 0.031* | |
| C20 | 0.5811 (3) | 0.61804 (14) | 0.0487 (5) | 0.0250 (6) | |
| H20 | 0.5382 | 0.6454 | −0.0384 | 0.030* | |
| C21 | 0.0176 (3) | 0.68939 (15) | 0.9724 (5) | 0.0283 (6) | |
| H21A | 0.0712 | 0.7226 | 1.0218 | 0.043* | |
| H21B | 0.0041 | 0.6578 | 1.0781 | 0.043* | |
| H21C | −0.0581 | 0.7087 | 0.9340 | 0.043* |
trans-11-Methyl-4-phenyl-1,10-dioxacyclotridec-5-ene-2,9-dione (III) . Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0202 (9) | 0.0267 (10) | 0.0231 (10) | 0.0027 (8) | 0.0020 (8) | 0.0009 (8) |
| O10 | 0.0216 (9) | 0.0228 (9) | 0.0219 (10) | −0.0017 (7) | 0.0008 (8) | 0.0030 (8) |
| O14 | 0.0291 (11) | 0.0330 (11) | 0.0276 (11) | 0.0007 (9) | −0.0002 (10) | −0.0049 (9) |
| O15 | 0.0318 (11) | 0.0428 (12) | 0.0268 (11) | −0.0113 (10) | −0.0055 (9) | 0.0017 (10) |
| C2 | 0.0238 (13) | 0.0185 (12) | 0.0258 (14) | −0.0023 (10) | 0.0036 (12) | −0.0009 (11) |
| C3 | 0.0218 (13) | 0.0204 (12) | 0.0291 (14) | 0.0008 (10) | 0.0050 (12) | 0.0017 (11) |
| C4 | 0.0212 (12) | 0.0207 (12) | 0.0244 (14) | −0.0001 (10) | 0.0024 (11) | 0.0007 (10) |
| C5 | 0.0213 (12) | 0.0223 (12) | 0.0215 (12) | −0.0002 (10) | 0.0023 (11) | 0.0001 (10) |
| C6 | 0.0230 (12) | 0.0234 (12) | 0.0221 (13) | −0.0003 (10) | 0.0019 (11) | −0.0012 (11) |
| C7 | 0.0247 (13) | 0.0198 (12) | 0.0302 (16) | 0.0003 (11) | 0.0055 (12) | 0.0009 (11) |
| C8 | 0.0229 (13) | 0.0206 (12) | 0.0273 (14) | 0.0020 (10) | 0.0011 (11) | 0.0026 (11) |
| C9 | 0.0235 (13) | 0.0207 (13) | 0.0249 (13) | −0.0006 (10) | 0.0016 (11) | 0.0002 (11) |
| C11 | 0.0219 (12) | 0.0245 (13) | 0.0218 (14) | 0.0006 (10) | 0.0012 (11) | 0.0056 (11) |
| C12 | 0.0243 (13) | 0.0242 (13) | 0.0262 (14) | −0.0011 (11) | 0.0046 (11) | 0.0028 (11) |
| C13 | 0.0212 (13) | 0.0274 (13) | 0.0272 (15) | −0.0002 (10) | 0.0034 (12) | −0.0020 (12) |
| C15 | 0.0218 (12) | 0.0207 (12) | 0.0221 (13) | −0.0006 (10) | 0.0017 (11) | −0.0015 (10) |
| C16 | 0.0234 (13) | 0.0250 (13) | 0.0230 (13) | −0.0007 (10) | 0.0005 (12) | 0.0018 (11) |
| C17 | 0.0243 (13) | 0.0255 (13) | 0.0284 (15) | 0.0016 (10) | 0.0007 (11) | −0.0017 (11) |
| C18 | 0.0204 (12) | 0.0249 (13) | 0.0321 (15) | −0.0013 (10) | 0.0014 (12) | −0.0060 (12) |
| C19 | 0.0259 (13) | 0.0244 (13) | 0.0278 (14) | −0.0030 (11) | 0.0050 (12) | −0.0005 (11) |
| C20 | 0.0260 (13) | 0.0226 (12) | 0.0264 (14) | 0.0003 (11) | 0.0042 (11) | 0.0009 (11) |
| C21 | 0.0313 (14) | 0.0307 (14) | 0.0230 (14) | 0.0012 (12) | 0.0039 (12) | 0.0004 (12) |
trans-11-Methyl-4-phenyl-1,10-dioxacyclotridec-5-ene-2,9-dione (III) . Geometric parameters (Å, º)
| O1—C2 | 1.336 (4) | C11—C21 | 1.502 (4) |
| O1—C13 | 1.440 (3) | C11—C12 | 1.506 (4) |
| O10—C9 | 1.328 (3) | C11—H11 | 1.0000 |
| O10—C11 | 1.452 (3) | C12—C13 | 1.498 (4) |
| O14—C2 | 1.194 (4) | C12—H12A | 0.9900 |
| O15—C9 | 1.191 (4) | C12—H12B | 0.9900 |
| C2—C3 | 1.491 (4) | C13—H13A | 0.9900 |
| C3—C4 | 1.527 (4) | C13—H13B | 0.9900 |
| C3—H3A | 0.9900 | C15—C16 | 1.376 (4) |
| C3—H3B | 0.9900 | C15—C20 | 1.394 (4) |
| C4—C5 | 1.500 (4) | C16—C17 | 1.383 (4) |
| C4—C15 | 1.512 (4) | C16—H16 | 0.9500 |
| C4—H4 | 1.0000 | C17—C18 | 1.388 (4) |
| C5—C6 | 1.315 (4) | C17—H17 | 0.9500 |
| C5—H5 | 0.9500 | C18—C19 | 1.370 (4) |
| C6—C7 | 1.490 (4) | C18—H18 | 0.9500 |
| C6—H6 | 0.9500 | C19—C20 | 1.378 (4) |
| C7—C8 | 1.523 (4) | C19—H19 | 0.9500 |
| C7—H7A | 0.9900 | C20—H20 | 0.9500 |
| C7—H7B | 0.9900 | C21—H21A | 0.9800 |
| C8—C9 | 1.499 (4) | C21—H21B | 0.9800 |
| C8—H8A | 0.9900 | C21—H21C | 0.9800 |
| C8—H8B | 0.9900 | ||
| C2—O1—C13 | 115.4 (2) | C21—C11—C12 | 112.3 (2) |
| C9—O10—C11 | 115.9 (2) | O10—C11—H11 | 109.5 |
| O14—C2—O1 | 123.2 (3) | C21—C11—H11 | 109.5 |
| O14—C2—C3 | 124.9 (3) | C12—C11—H11 | 109.5 |
| O1—C2—C3 | 111.9 (3) | C13—C12—C11 | 115.2 (2) |
| C2—C3—C4 | 110.4 (2) | C13—C12—H12A | 108.5 |
| C2—C3—H3A | 109.6 | C11—C12—H12A | 108.5 |
| C4—C3—H3A | 109.6 | C13—C12—H12B | 108.5 |
| C2—C3—H3B | 109.6 | C11—C12—H12B | 108.5 |
| C4—C3—H3B | 109.6 | H12A—C12—H12B | 107.5 |
| H3A—C3—H3B | 108.1 | O1—C13—C12 | 107.6 (2) |
| C5—C4—C15 | 108.3 (2) | O1—C13—H13A | 110.2 |
| C5—C4—C3 | 110.9 (2) | C12—C13—H13A | 110.2 |
| C15—C4—C3 | 112.6 (2) | O1—C13—H13B | 110.2 |
| C5—C4—H4 | 108.3 | C12—C13—H13B | 110.2 |
| C15—C4—H4 | 108.3 | H13A—C13—H13B | 108.5 |
| C3—C4—H4 | 108.3 | C16—C15—C20 | 118.2 (3) |
| C6—C5—C4 | 123.8 (3) | C16—C15—C4 | 123.9 (3) |
| C6—C5—H5 | 118.1 | C20—C15—C4 | 117.7 (2) |
| C4—C5—H5 | 118.1 | C15—C16—C17 | 121.6 (3) |
| C5—C6—C7 | 123.7 (3) | C15—C16—H16 | 119.2 |
| C5—C6—H6 | 118.1 | C17—C16—H16 | 119.2 |
| C7—C6—H6 | 118.1 | C16—C17—C18 | 119.4 (3) |
| C6—C7—C8 | 112.5 (2) | C16—C17—H17 | 120.3 |
| C6—C7—H7A | 109.1 | C18—C17—H17 | 120.3 |
| C8—C7—H7A | 109.1 | C19—C18—C17 | 119.5 (3) |
| C6—C7—H7B | 109.1 | C19—C18—H18 | 120.2 |
| C8—C7—H7B | 109.1 | C17—C18—H18 | 120.2 |
| H7A—C7—H7B | 107.8 | C18—C19—C20 | 120.8 (3) |
| C9—C8—C7 | 109.1 (2) | C18—C19—H19 | 119.6 |
| C9—C8—H8A | 109.9 | C20—C19—H19 | 119.6 |
| C7—C8—H8A | 109.9 | C19—C20—C15 | 120.4 (3) |
| C9—C8—H8B | 109.9 | C19—C20—H20 | 119.8 |
| C7—C8—H8B | 109.9 | C15—C20—H20 | 119.8 |
| H8A—C8—H8B | 108.3 | C11—C21—H21A | 109.5 |
| O15—C9—O10 | 124.1 (3) | C11—C21—H21B | 109.5 |
| O15—C9—C8 | 123.5 (3) | H21A—C21—H21B | 109.5 |
| O10—C9—C8 | 112.4 (3) | C11—C21—H21C | 109.5 |
| O10—C11—C21 | 109.6 (2) | H21A—C21—H21C | 109.5 |
| O10—C11—C12 | 106.2 (2) | H21B—C21—H21C | 109.5 |
| C13—O1—C2—O14 | −2.5 (4) | O10—C11—C12—C13 | 44.7 (3) |
| C13—O1—C2—C3 | 176.1 (2) | C21—C11—C12—C13 | 164.5 (2) |
| O14—C2—C3—C4 | 67.8 (4) | C2—O1—C13—C12 | −173.5 (2) |
| O1—C2—C3—C4 | −110.8 (3) | C11—C12—C13—O1 | 51.2 (3) |
| C2—C3—C4—C5 | 68.5 (3) | C5—C4—C15—C16 | 96.9 (3) |
| C2—C3—C4—C15 | −169.9 (2) | C3—C4—C15—C16 | −26.2 (4) |
| C15—C4—C5—C6 | 118.9 (3) | C5—C4—C15—C20 | −78.3 (3) |
| C3—C4—C5—C6 | −117.0 (3) | C3—C4—C15—C20 | 158.6 (3) |
| C4—C5—C6—C7 | −177.2 (3) | C20—C15—C16—C17 | 0.1 (4) |
| C5—C6—C7—C8 | −102.7 (3) | C4—C15—C16—C17 | −175.1 (3) |
| C6—C7—C8—C9 | 72.7 (3) | C15—C16—C17—C18 | −0.5 (4) |
| C11—O10—C9—O15 | −3.5 (4) | C16—C17—C18—C19 | 0.6 (4) |
| C11—O10—C9—C8 | 175.8 (2) | C17—C18—C19—C20 | −0.4 (5) |
| C7—C8—C9—O15 | 54.4 (4) | C18—C19—C20—C15 | −0.1 (5) |
| C7—C8—C9—O10 | −125.0 (3) | C16—C15—C20—C19 | 0.2 (4) |
| C9—O10—C11—C21 | 82.1 (3) | C4—C15—C20—C19 | 175.6 (3) |
| C9—O10—C11—C12 | −156.3 (2) |
Funding Statement
This work was funded by National Science Foundation grant 0957626.
References
- Bochevarov, A. D., Harder, E., Hughes, T. F., Greenwood, J. R., Braden, D. A., Philipp, D. M., Rinaldo, D., Halls, M. D., Zhang, J. & Friesner, R. A. (2013). Int. J. Quantum Chem. 113, 2110–2142.
- Dolomanov, O. V., Bourhis, L. J., Gildea, R. J., Howard, J. A. K. & Puschmann, H. (2009). J. Appl. Cryst. 42, 339–341.
- Driggers, E. M., Hale, S. P., Lee, J. & Terrett, N. K. (2008). Nat. Rev. Drug Discov. 7, 608–624. [DOI] [PubMed]
- Eliel, E. L. & Wilen, S. H. (1994). Chirality in Molecules Devoid of Chiral Centers, in Stereochemistry of Organic Compounds, pp 1172–1175. New York: John Wiley & Sons.
- Fyvie, W. S. & Peczuh, M. W. (2008a). Chem. Commun. pp. 4028–4030. [DOI] [PubMed]
- Fyvie, W. S. & Peczuh, M. W. (2008b). J. Org. Chem. 73, 3626–3629. [DOI] [PubMed]
- Grimme, S. (2011). WIREs Comput. Mol. Sci. 1, 211–228.
- Jacobson, R. (1998). REQAB. Rigaku Corporation, Tokyo, Japan.
- Larsen, E. M., Wilson, M. R. & Taylor, R. E. (2015). Nat. Prod. Rep. 32, 1183–1206. [DOI] [PMC free article] [PubMed]
- Ma, J. & Peczuh, M. W. (2013). J. Org. Chem. 78, 7414–7422. [DOI] [PubMed]
- Macrae, C. F., Sovago, I., Cottrell, S. J., Galek, P. T. A., McCabe, P., Pidcock, E., Platings, M., Shields, G. P., Stevens, J. S., Towler, M. & Wood, P. A. (2020). J. Appl. Cryst. 53, 226–235. [DOI] [PMC free article] [PubMed]
- Magpusao, A. N., Rutledge, K., Mercado, B. & Peczuh, M. W. (2015). Org. Biomol. Chem. 13, 5086–5089. [DOI] [PubMed]
- Magpusao, A. N., Rutledge, K. M., Hamlin, T. A., Lawrence, J.-M., Mercado, B. Q., Leadbeater, N. E. & Peczuh, M. W. (2016). Chem. Eur. J. 22, 6001–6011. [DOI] [PubMed]
- Parsons, S., Flack, H. D. & Wagner, T. (2013). Acta Cryst. B69, 249–259. [DOI] [PMC free article] [PubMed]
- Rigaku (2011). CrystalClear-SM Expert. Rigaku Corporation, Tokyo, Japan.
- Rigaku OD (2015). CrysAlis PRO. Rigaku Oxford Diffraction, Yarnton, England.
- Rowland, R. S. & Taylor, R. (1996). J. Phys. Chem. 100, 7384–7391.
- Rutledge, K. M., Hamlin, T. A., Baldisseri, D., Bickelhaupt, F. M. & Peczuh, M. W. (2017). Chem. Asian J. 12, 2623–2633. [DOI] [PubMed]
- Sheldrick, G. M. (2014). CIFTAB2014/2. University of Göttingen, Germany.
- Sheldrick, G. M. (2015a). Acta Cryst. A71, 3–8.
- Sheldrick, G. M. (2015b). Acta Cryst. C71, 3–8.
- Whitty, A., Viarengo, L. A. & Zhong, M. (2017). Org. Biomol. Chem. 15, 7729–7735. [DOI] [PubMed]
- Whitty, A., Zhong, M., Viarengo, L. A., Beglov, D., Hall, D. R. & Vajda, S. (2016). Drug Discovery Today, 21, 712–717. [DOI] [PMC free article] [PubMed]
- Yudin, A. K. (2015). Chem. Sci. 6, 30–49. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I, II, III. DOI: 10.1107/S2056989020012037/mw2168sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989020012037/mw2168Isup2.hkl
Structure factors: contains datablock(s) II. DOI: 10.1107/S2056989020012037/mw2168IIsup3.hkl
Structure factors: contains datablock(s) III. DOI: 10.1107/S2056989020012037/mw2168IIIsup4.hkl
PDF document (Supplementary data associate with the manuscript: tables, computed structure coordinates, NMR spectra). DOI: 10.1107/S2056989020012037/mw2168sup5.pdf
Additional supporting information: crystallographic information; 3D view; checkCIF report






