Abstract
Efforts in genome sequencing in the Aspergillus genus have led to the development of quality reference genomes for several important species including A. nidulans, A. fumigatus, and A. oryzae. However, less progress has been made for A. flavus. As part of the effort of the USDA-ARS Annual Aflatoxin Workshop Fungal Genome Project, the isolate NRRL3357 was sequenced and resulted in a scaffold-level genome released in 2005. Our goal has been biologically driven, focusing on two areas: isolate variation in aflatoxin production and drought stress exacerbating aflatoxin production by A. flavus. Therefore, we developed two reference pseudomolecule genome assemblies derived from chromosome arms for two isolates: AF13, a MAT1-2, highly stress tolerant, and highly aflatoxigenic isolate; and NRRL3357, a MAT1-1, less stress tolerant, and moderate aflatoxin producer in comparison to AF13. Here, we report these two reference-grade assemblies for these isolates through a combination of PacBio long-read sequencing and optical mapping, and coupled them with comparative, functional, and phylogenetic analyses. This analysis resulted in the identification of 153 and 45 unique genes in AF13 and NRRL3357, respectively. We also confirmed the presence of a unique 310 Kb insertion in AF13 containing 60 genes. Analysis of this insertion revealed the presence of a bZIP transcription factor, named atfC, which may contribute to isolate pathogenicity and stress tolerance. Phylogenomic analyses comparing these and other available assemblies also suggest that the species complex of A. flavus is polyphyletic.
Keywords: Aspergillus flavus, aflatoxin, reference genomes, phylogenomics, polyphyletic
Of the secondary metabolite biosynthetic clusters identified in fungi, there are few as well characterized as aflatoxin biosynthesis in Aspergillus flavus and related Aspergillus species. From the time of its discovery in the 1960s (Amaike and Keller 2011; Forgacs and Carll 1962), the process of aflatoxin production has been under constant investigation. Identification of the bulk of the biosynthetic pathway occurred throughout the 1990s in other species, specifically A. nidulans (Brown et al. 1996). In the early 2000s, the individual genes in the biosynthetic cluster were fully described in A. parasiticus and later in A. flavus (Yu et al. 2004a, 2004b). The characterization of the aflatoxin cluster, however, was only the beginning of a large scale effort to sequence the entire genome of this important pathogen to learn more about its biology, plant and human pathogenicity, and the functional regulation of the production of aflatoxin and other toxic secondary metabolites produced by A. flavus and related fungi.
In 2003, efforts in sequencing the A. flavus genome were initiated with the goal of producing a draft genome for the aflatoxigenic isolate NRRL3357, a MAT1-1, L-strain isolated from peanut in Georgia (Payne et al. 2006; 2007; Yu et al. 2008). This genome, developed through Sanger sequencing at 5x coverage, was released to the National Center for Biotechnology Information (NCBI) with 2,761 scaffolds with an N50 of 2.388 Mb and a total length of 36.892 Mb (Nierman et al. 2015). In 2010, the genome was further revised due to contaminant sequences identified in the dataset to a final total of 331 scaffolds in the present assembly (GCA_000006275.2). This isolate has since been adopted as “type” strain for A. flavus and has seen near ubiquitous usage by the aflatoxin research community as a standard isolate for biological investigation of aflatoxin production. This isolate along with others such as AF13, a highly aflatoxigenic, MAT1-2, L-strain fungus from cotton field soils in Arizona (Cotty 1989), have been used in laboratory and field evaluations of breeding germplasm for resistance to A. flavus colonization and reduced aflatoxin contamination (Fountain et al. 2019a; Guo et al. 1995).
In addition to NRRL3357, several other isolates of A. flavus have also been sequenced and used for draft de novo genome assemblies. In 2015, AF70, a MAT1-2, S-strain from cotton field soils in Arizona, was sequenced using an Illumina platform (GCA_000952835.1). This genome was described in Gilbert et al. (2018) and compared to NRRL3357, where significant polymorphisms were identified potentially affecting both secondary metabolism and morphological development between the two morphotypes (S v. L strains; S – Small Sclerotia < 400µm; L – Large Sclerotia > 400µm) and mating type loci (MAT; MAT1-1 v. MAT1-2). Gene content was similar between these two isolates with 13,487 predicted in NRRL3357 and 13,118 in AF70 as were the overall lengths of the two draft assemblies. Similar levels of distinction between S and L strains were also observed by Ohkura et al. (2018) who sequenced three S strains (AF12, AF70, and AZS) and three L strains (BS01, DV901, and MC04). With the advent of less expensive, more rapid, and more powerful sequencing technologies, there has been an increase in the number of A. flavus isolate draft genome assemblies in public databases. At the time of this publication (February 2020), there are 60 released isolate draft assemblies in the NCBI Genbank (Table S1). These draft assemblies have primarily been sequenced with Illumina platforms and have an average of 997 contigs ranging in total length from 35.094 Mb to 40.273 Mb in length (Table S1). Very recently, a chromosome-level assembly of NRRL3357 with 8 chromosomes and a length of 37.749Mb was released by the University of California, Berkeley (GCA_009017415.1; Table S1).
In addition to A. flavus, the genomes of other Aspergillus species have also been sequenced. A. nidulans FGSC A4, A. oryzae RIB40, and A. fumigatus Af293 were all sequenced and assembled in 2005 using Sanger technology (Galagan et al. 2005; Machida et al. 2005; Nierman et al. 2005; Payne et al. 2006). Later in 2007 the genome for A. niger CBS 513.88, also produced using Sanger sequencing, was released (Pel et al. 2007). These genomes are all comprised of 8 chromosomes. Interestingly, A. fumigatus and A. nidulans have shorter overall lengths, 29.385 Mb and 29.828 Mb, respectively, compared to the other species which have genome sizes >34 Mb. This has led to the hypothesis that these species represent either earlier evolutionary development of the species complex with additional genome content being acquired through partial genome duplications, introgressions, or horizontal gene transfer (HGT); or that these species represent a distinct evolutionary event separate from that of the A. oryzae lineage which contains A. flavus (Galagan et al. 2005).There are 71 other species of Aspergillus fungi with draft or complete genome assemblies in NCBI’s Genbank (February 2020). This abundance of information provides extensive opportunities for investigating the biology and evolutionary history of this genus of fungi. However, despite this surge in information and the importance of A. flavus as a threat to food safety and security (Amaike and Keller 2011), there remains no complete reference genome, defined as a genome with coverage of the entire chromosomes (with expected error and gaps present) coupled with accurate annotation of associated genes, for this and other diverse isolates of this fungus.
The currently available genomes have been invaluable for and have enabled genomics-assisted experiments including transcriptome sequencing and the characterization of genes involved in a number of primary and secondary metabolic pathways. Still, an understanding of A. flavus phenotypic diversity has been hindered by the lack of suitable and diverse references. In addition, since reference-guided sequencing analyses rely on their reference for the identification and annotation of putative genes for analyses, the limitation of having only a single reference assembly for A. flavus becomes an issue given the potential for having several hundred unique genes in different isolates as seen in the comparison of NRRL3357 and AF70 (Gilbert et al. 2018) or among isolates with distinct morphologies (Ohkura et al. 2018). Therefore, to address these concerns, and to investigate the structure and evolutionary history of this pathogen, here we present two novel chromosome arm reference genome assemblies for the A. flavus isolates AF13 and NRRL3357. These isolates were chosen based on two biologically-driven questions: (1) what are the causes of variation in A. flavus isolates’ aflatoxin production; and (2) why do these isolates exhibit contrasting responses to reactive oxygen species (ROS), reactive compounds associated with drought stress which exacerbate aflatoxin production by A. flavus (Fountain et al. 2019b; Yang et al.2018)?
These genomes were sequenced using PacBio sequencing and scaffolds were bridged using optical mapping to produce full chromosome arms. Comparative genomics resulted in the identification of structural variation between these isolates representing the recent evolutionary acquisition of novel genes in AF13 compared to NRRL3357. The utility of these novel reference genomes in gene annotation is also demonstrated through the refinement of splice-site identification and annotation of transcriptome datasets. Comparative analysis of these references also resulted in the identification of a novel bZIP transcription factor gene, annotated atfC, which may contribute to stress tolerance in A. flavus under drought stress conditions. Phylogenomics analyses also show that the A. flavus section Flavi is polyphyletic, and that AF13 represents a distinct but closely related sister clade of NRRL3357. These reference genomes represent a valuable asset for use by the Aspergillus research community, and will serve as a starting point for continuing research into the biology of these organisms, particularly for stress biology related to oxidative stress and aflatoxin production.
Materials and Methods
Isolate collection and culturing
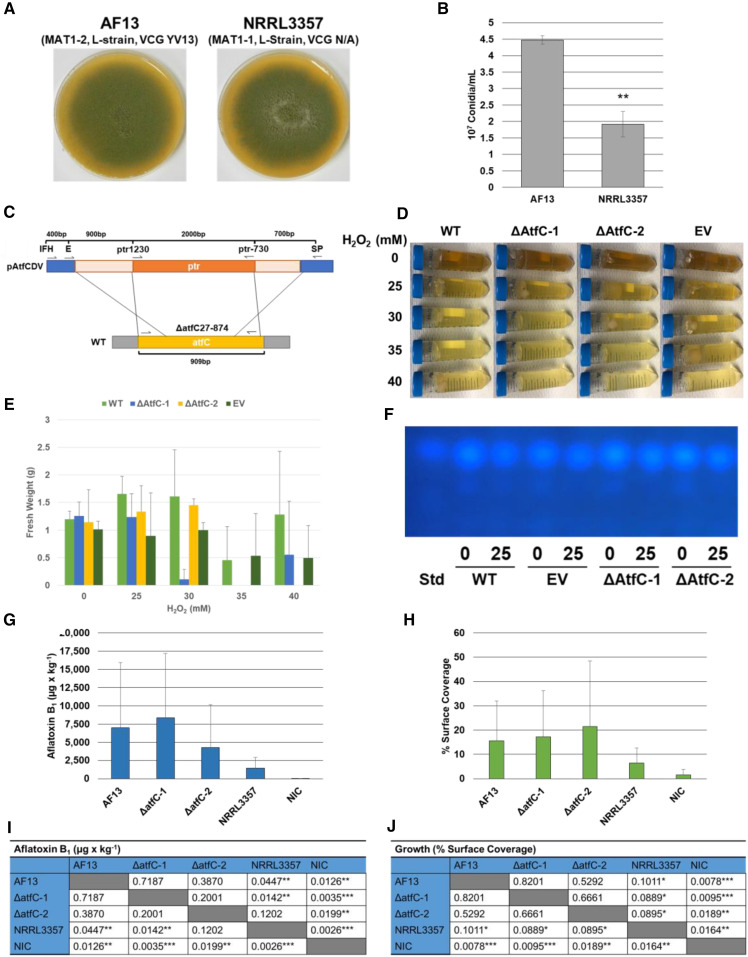
For isolates used for genome sequencing and assembly, NRRL3357 was obtained from the USDA-ARS Northern Regional Research Center, Peoria, IL, USA; and AF13 was obtained from Kenneth Damann, Department of Plant Pathology and Crop Physiology, Louisiana State University Agricultural Center, Baton Rouge, LA, USA. Additional isolates collected for re-sequencing and comparisons are as follows. A1, A9, AF36 (NRRL18543), Afla-Guard (NRRL21882), Tox4, VCG1, and VCG4 were obtained from K. Damann. K49 (NRRL30797) and K54A were obtained from Hamed Abbas, USDA-ARS Biological Control of Pests Research Unit, Stoneville, MS, USA. All isolates were received on potato dextrose agar (PDA) plates, and were transferred to V8 agar (20% V8, 1% CaCO3, 3% agar) to stimulate conidiation. For long term storage, 5 – 6 agar plugs were taken from the growing edge of the plates, and placed into amber vials containing 5 mL of either sterile water or 20% glycerol and stored at 4° and -20°, respectively. These conidial suspensions (∼107 conidia/mL) were used as inoculum for subsequent experiments. Phenotypic differences between AF13 and NRRL3357 were evaluated on V8 agar. Differences in conidia production between these isolates were evaluated by washing V8 agar plates of each isolate with 25mL of 0.1% (v/v) Tween 20, and the concentration obtained for each conidial suspension was measured using a hemocytometer. This evaluation was performed three times.
Standard and high molecular weight DNA isolation
For short read sequencing of the isolate collection, a normal CTAB DNA isolation was done as follows. Each isolate was cultured in yeast extract – sucrose (YES, 2% yeast extract, 1% sucrose) for five days at 30° in the dark. Mycelial mats from each culture were collected and ground in a chilled mortar and pestle with liquid nitrogen. The ground mycelia (1-2 g) was then combined with 15 mL of CTAB extraction buffer (0.1M Tris pH 8.0, 1.4M NaCl, 20mM EDTA, 2% (w/v) CTAB, 4% (w/v) polyvinylpyrrolidone (PVP-40), and 0.5% (v/v) β-mercaptoethanol), mixed by inversion, and incubated in a water bath at 65° for 45 min with occasional inversion. The lysate was then combined with 15 mL of chloroform:isoamyl alcohol (24:1), mixed by inversion, and centrifuged at 8,000 × g for 15 min at 4°. The upper phase was then transferred to a new 50 mL centrifuge tube. The chloroform separation was then performed a second time, and the upper phase was then combined with one volume of cold isopropanol for DNA precipitation. The DNA was then pelleted by centrifuging at 8,000 × g for 15 min at 4°, and washed with 70% ethanol. The pellets were then dried and suspended in 100 µL TE buffer (10mM Tris pH 8.0, 1 mM EDTA pH 8.0). RNaseA was then added to a final concentration of 5 µg/mL and the samples were incubated at 37° for 1 hr. The obtained DNA was then stored at -20° until used.
For long read sequencing of AF13 and NRRL3357, high molecular weight (HMW) DNA was isolated using a modified version of the CTAB protocol. Ground mycelium (1-2 g) was combined with 15 mL CTAB buffer as previously described, but with the addition of 75 µL proteinase K (20 mg/mL) to each sample to improve cell lysis along with the addition of 20 µL RNaseA (10 mg/mL). The samples were then incubated at 60° for 45 min with occasional gentle agitation. The temperature was increased to 70° for 15 min to begin inactivating proteinase K. The lysate was combined with 15 mL of phenol:chloroform:isoamyl alcohol (25:24:1), mixed by gentle inversion, and centrifuged at 8,000 × g for 15 min at 4°. The upper phase was then transferred to a new 50 mL centrifuge tube using a large bore pipet, and was combined with 15 mL of chloroform:isoamyl alcohol (24:1), mixed by gentle inversion, and again centrifuged. The resultant upper aqueous phase was transferred to a new tube and DNA was precipitated with one volume of cold isopropanol and 2 mL 7.5 M ammonium acetate. The DNA was pelleted by centrifugation and washed with 70% ethanol. After drying, the pelleted DNA was then dissolved in 500 µL TE buffer and stored at -20° until use. DNA isolated using either method was quantified with both a Nanodrop ND-1000 spectrophotometer (ThermoFisher, Waltham, MA, USA) and a Qubit 3.0 fluorometer (ThermoFisher), and checked using gel electrophoresis.
DNA sequencing
Isolated DNA for short read sequencing was frozen and shipped to the Novogene Corporation (Sacramento, CA, USA). Sequencing was carried out as described in Fountain et al. (2020) using a HiSeq 4000 platform (Illumina, San Diego, CA, USA). For long read sequencing, HMW DNA from AF13 and NRRL3357 were frozen and shipped to the USDA-ARS Genomics and Bioinformatics Research Unit, Stoneville, MS, USA for sequencing. Sequencing was carried out on a PacBio RSII platform (Pacific Biosciences, Menlo Park, CA, USA). These PacBio reads were then used in conjunction with optical mapping for reference assembly construction.
Optical mapping
In order to bridge contigs in the assembled PacBio genomes for AF13 and NRRL3357 to assemble full chromosomes, and given the lack of a published genetic map for A. flavus, optical mapping was performed at the Emory Integrated Genomics Core at Emory University, Atlanta, GA, USA. A modified protocol was developed for HMW DNA isolation from A. flavus protoplasts. The protocol used for protoplast generation and preparation was based on those used by Cary et al. (2006), Liu and Friesen (2012), and Yang et al. (2016). Briefly, conidia from each isolate were grown on V8 agar for five days. Plugs were taken from the growing edge of the generated colonies and were placed into amber vials containing 5 mL of sterile water. With this, 1 mL of each inoculum (∼106 conidia/mL) was added into 250 mL of potato dextrose broth (PDB) in a 1 L media bottle which was capped and sealed with parafilm. After culturing for 12 hr at 30° in the dark, mycelia were isolated by vacuum filtration through two layers of Miracloth (Millipore-Sigma, Burlington, MA, USA). The isolated mycelia were then washed three times with sterile water and transferred to a sterile 50 mL centrifuge tube. Enzymatic digestion of the fungal cell walls was then carried out by adding 40 mL of enzyme solution to mycelia from each isolate. This digestion solution was prepared by combining 4 mL 0.2 M NaPO4 pH 5.8, 0.8 mL 1.0 M CaCl2, 2.8g NaCl, 139.48 µL β-glucuronidase (24,377 U/mL; Sigma G8420), 400 mg lysing enzyme (Sigma L1412), 100 mg driselase (Sigma D9515), and 34 mL sterile water. The solution was gently stirred for 5 – 10 min to allow the materials to completely dissolve, followed by centrifugation at 2,000 × g for 10 min at 4°, and filter sterilization of the resultant supernatant. Digestion of the mycelia was carried out over 3 hr at 30° with gentle shaking (80 rpm).
The resultant digestions were then filtered through four layers of Miracloth to separate the protoplasts from undigested mycelial fragments, and stored on ice for the remainder of the procedure. The protoplasts were then pelleted by centrifugation at 300 × g for 10 min at 4°, washed with 20 mL of mycelia wash solution (MWS; 0.7M KCl, 10mM CaCl2), pelleted and washed in 500 µL of cell buffer from the Bionano Prep Blood and Cell Culture DNA Isolation Kit (Bionano Genomics, San Diego, CA, USA). The protoplasts were then pelleted again and resuspended in 66 µL of cell buffer to a final concentration of at least 109 protoplasts/sample (>6 µg DNA content) for use in agarose plug generation. Throughout the procedure following digestion filtration, the protoplasts were quantified and evaluated for viability using a hemocytometer and a Countess automated cell counter (ThermoFisher). For cell lysis and HMW DNA isolation, the protoplasts were then cast into agarose plugs. For each plug, 66 µL of cell suspension was combined with 40µL of molten 2% low melting point agarose, mixed with a wide bore pipette, and placed into a plug mold (Cat# 1703713, Bio-Rad, Hercules, CA, USA) at 4° for plug solidification. Proteinase K digestion, RNaseA digestion, washing, and HMW DNA isolation were then performed using the Bionano DNA isolation kit according to the manufacturer’s instructions. The integrity of the isolated HWM DNA was evaluated using pulse field gel electrophoresis (PFGE). Labeling of the HWM DNA for use in sequencing was done using the Bionano Prep DLS (Direct Label and Stain) Labeling Kit (Bionano Genomics) according to the manufacturer’s instructions. Sequencing was then carried out on a Saphyr platform (Bionano Genomics).
Genome assembly
PacBio reads were assembled using Mecat. This assembly resulted in 16 fully contiguous sequences representing all chromosome arms (broken only by centromeric sequence). Chromosome arms were paired, and chromosome numbers assigned using collinearity with the A. oryzae RIB40 sequence (GCA_000184455.3), which was produced previously based on optical maps. Chromosome 6 (Chr6) and Chr2 are involved in a reciprocal translocation. We assigned the portion of the PacBio contig closest to the centromere to its respective A. oryzae chromosome based on the most parsimonious explanation of two chromosome breaks and translocation. Fifty “N” characters were placed between chromosome arms as a stand-in for actual centromere sequence.
Gene annotation and presence/absence variation
The evidence-based gene prediction pipeline, MAKER, was used for genome annotation. MAKER aligns expressed sequence tags (ESTs) and protein evidence to a genome, produces ab-intio gene predictions, and identifies repeats (Cantarel et al. 2008). Expressed sequence tag and protein evidence were obtained from the Aspergillus Genome Database (AspGD) (Arnaud et al. 2012). The AspGD is a central repository for gene annotation and protein information for Aspergillus species. Specifically, sequences from A. flavus NRRL 3357 and A. oryzae RIB40 with no introns for all open reading frames (ORFs) were used as EST evidence and protein evidence was provided by translations of all ORFs of A. fumigatus Af293, A. niger CBS 513 88, and A. nidulans FGSC A4. The Augustus (Stanke et al. 2004) trained dataset of A. oryzae was used for ab-initio gene prediction and repeat soft-masking was performed using the Aspergillus repeat library from RepBase (Bao et al. 2015). The Galaxy tool (Afgan et al. 2018) version 2.31.9.1 of MAKER was run on a Galaxy SlipStream server (BioTeam Inc. Middleton, MA) to perform above mentioned MAKER pipeline. Annotation of secondary metabolite gene clusters was performed using the web-based application antiSMASH (v5.0; Blin et al. 2019). Annotation was performed for tRNAs using tRNAscan-SE (v2.0.5; Chan and Lowe 2019), and for rRNAs using Barrnap (v0.8; https://github.com/tseemann/barrnap, last accessed August 5, 2020).
In order to connect pre-existing annotations with new annotations and examine variation in gene content, coding sequences were extracted from GFF files. The AF13 and NRRL3357 coding sequences (CDS) was combined with the AFL1 reference transcriptome derived from NCBI_Assembly GCF_000006275.2 (JCVI-afl1-v2.0). This combined set was searched against itself using nucmer (version 3.1) with maxmatch flag. Results were filtered based on overall alignment length across all sub-matches in the same orientation between the pairs of sequences. If the overall alignment length was >80% of the longest sequence in the pair, then the pair was retained. This length criterion was based on manual curation and designed to cluster alternative transcripts and homologs that are likely to have very similar function. A pairwise matrix of all sequences was built using this overall alignment length as a distance criterion. mcl (version 14) was then used to cluster sequences based on this matrix.
Insertion/deletions inference
Indels were identified from whole genome alignments and were polarized relative to the outgroup, A. oryzae RIB40, using a custom program. Columns in the whole chromosome alignments that involved >50 consecutive gaps (in any sequence) were extracted along with +/− 50 bp of flanking sequence. Gaps were analyzed further if the left and right flanking regions aligned with >90% columns being identical. If AF13 and NRRL3357 shared 95% identity in the gapped region, the structural variant (SV) was not considered further. Alternatively, if there was variation between AF13 and NRRL3357 and one matched the outgroup with >95% identity, then the event was inferred to have occurred in the non-matching sequence. This approach captured the biological reality that mutations creating long (>50 bp) SVs rarely involve only insertion or deletion of DNA but a combination of both. To that end, we also characterized the degree to which mutations represents a net gain or loss of DNA. The length of the entire gapped region was divided by the length of novel sequence introduced in the gap such that values approaching 0 are, in effect, deletions and values approaching 1 are insertions (a small number of SVs with gap values between 0.49 and 0.51 were removed after manual curation indicated these “perfectly balanced” indels represent unwarranted gap openings).
Phylogenetic analyses
Illumina short read data were obtained from the results of the “DNA Sequencing” section above. Assembled contigs for A. flavus isolates 206-4, 26-3, 3-2, 40-5. 54-2, 61-4, 72-5, 78-6, 79-2, CA14, CS0504, CS1137, JAU2, NRRL21882, NRRL18543, NRRL30797, and WRRL1519 were obtained from NCBI. All lines, short reads and contigs, were aligned to the AF13 reference using BWA v 0.7.1 with standard parameters. These alignments were sorted and indexed, and read depth per position was calculated and visualized via IGVtools 2.7.2. These alignments were then used to call short indels and SNPs using the BCFtools ‘mpileup’ and ‘call’ commands (version 1.9-274-g7db9558+). The samples were treated as haploid, and a multiallelic model was used, allowing for more than 2 alleles per position to be called. These variants were filtered to exclude sites that were present in fewer than 29 lines. A phylogenetic tree was created to visualize relationships from this filtered variant data using the UPGMA method in TASSEL 5.
RNA sequencing
To facilitate annotation of the newly developed genomes and to explore the signaling responses of A. flavus to drought-related oxidative stress, an RNA sequencing experiment was conducted. The AF13 isolate was cultured on V8 agar for five days, and conidia were harvested as inoculum (107 conidia/mL). The isolate was cultured in 50 mL of YES liquid medium in a 125 mL Erlenmeyer flask capped with sterile cotton for 48 hr at 30° with shaking at 150 rpm. After 48 hr, hydrogen peroxide (H2O2) was added to a final concentration of 30 mM in each treated culture. Control cultures received no H2O2. Mycelia were then collected at 0, 3, 6, and 9 hr after H2O2 amendment and flash frozen in liquid nitrogen and stored at -80°. Four replicate cultures were collected at 0, 3, and 6 hr for both treated and control samples, and two replicated cultures were collected at 9 hr for both. This yielded a total of 24 samples for RNA sequencing.
The collected mycelia were ground to a fine powder using a Bullet Blender 24 (Next Advance, Troy, NY, USA), and total RNA was isolated using a RNeasy Plant Mini Kit with on-column DNase digestion (Qiagen, Hilden, Germany). Sample quantity and quality were estimated using a Nanodrop ND-1000 spectrophotometer (ThermoFisher) and gel electrophoresis. Isolated total RNA was then frozen and shipped to the Novogene Corporation for quality checks, library preparation, and sequencing. RNA integrity numbers (RINs) were measured using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) and samples used for sequencing had RINs > 7.0. Library preparation was done using a TruSeq Library Prep Kit (Illumina) according to the manufacturer’s instructions. Prepared libraries were quantified using a Qubit 2.0 fluorometer (ThermoFisher), and sequenced on a HiSeq 4000 platform (Illumina).
Transcriptome analysis
Differential expression analysis was done using kallisto pseudoaligner with 10 boostrap iterations (Bray et al. 2016). Raw counts were then analyzed using DESeq2 (Love et al. 2014). Two different models were tested. First, the effect of oxidative stress was tested using the full model, gene ∼ trt + time, and the reduced model, gene ∼ trt. Second, the effect of time under stress was tested using only the 0 time point and inoculated samples using the full model, gene ∼ time, and the reduced model gene ∼1. Genes were determined to be differentially expressed with the adjusted P < 0.05 using a Bonferroni multiple testing correction.
Functional characterization of atfC and isolate phenotyping
atfC disruption mutant generation:
Annotation of the 310 Kb insertion in AF13 identified a putative bZIP transcription factor gene homologous with atfA and atfB. This transcription factor, dubbed atfC, was functionally evaluated for its influence on stress tolerance and aflatoxin production. Disruption of atfC was carried out using a double-crossover recombination approach as previously described by Chang et al. (2010). Briefly, the disruption vector was constructed through the introduction of the ptrA (pyrithiamine (PT) resistance) marker amplified from the pPTR1 vector (TaKaRa Bio, Japan), combined with 0.9 and 0.7 kb fragments of the 5′ and 3′ ends of atfC, respectively, including some flanking sequences using PCR to generate the pAtfCDV vector. Protoplasts of AF13, generated as previously described (Chang et al. 2010), were then transformed using polyethylene glycol (PEG) as described by Horng et al. (1990) with minor modifications, and selected on CZ regeneration medium containing 0.6 M KCl, 5 mM (NH4)2SO4, and 0.1 µg PT/mL for up to 10 days at 30°. The insertion and orientation of ptrA into atfC in AF13 were then evaluated using diagnostic PCR and gel electrophoresis. Empty transformation vectors lacking the disruption construct were used as controls in the experiment. In addition to disruption, an additional copy of atfC along with its native promoter and terminator sequences (1.0 kb up and down-stream of the coding region) was introduced into NRRL3357 and AF13 wild type (WT) isolates to examine introduction and dosage effects, respectively. In addition to PCR amplicon size, insertions and deletions of atfC were also confirmed using Sanger sequencing. Overall, two isolates were identified for each event and used for downstream phenotypic characterization.
Phenotypic characterization and H2O2-stress tolerance:
Once obtained, the isolates along with the WTs were screened for gross morphological effects of their respective mutations on different media including Czapek-Dox agar, PDA, and V8 agar. The isolates were then examined for effects on oxidative stress tolerance by culturing them on a gradient of H2O2-amended YES liquid medium ranging from 0 – 50 mM H2O2 for seven days at 30° in the dark as previously described (Fountain et al. 2015a) in either stationary in 125mL Erlenmeyer flasks or with shaking at 150 rpm in 50 mL conical bottom tubes. Culture medium was also sampled from each isolate and developed using thin layer chromatography as previously described (Fountain et al. 2015a; 2019b) to examine for effects on aflatoxin production under increasing oxidative stress.
Pathogenicity and aflatoxin assays on peanut kernels:
Effects on pathogenicity and aflatoxin production in vitro were evaluated for the WT and disrupted isolates of AF13 using a kernel screening assay as described by Guo et al. (1995) with modifications. Seeds of the peanut cultivar Tifrunner with intact testa and free of visible damage were surface sterilized using UV exposure for 60 min. To examine each isolate, sterilized seeds were immersed in an inoculum containing 105 conidia/mL in 0.1% (v/v) Tween 20. The seeds (four seeds per well) were then transferred to sterile 6-well cell culture plates which were then placed into moist chambers and incubated for five days at 28° in the dark. The seeds were then evaluated for visible fungal growth and conidiation as an indicator of isolate pathogenicity. The seeds were then collected, ground into powder, and placed into 2mL tubes. The tubes were weighed and a 1.0 mL solution of 5% (w/v) NaCl and 80% (v/v) methanol was added to each tube. The tubes were then vortexed, kept at room temperature for 30 min, and centrifuged at 10,000 rpm for 10 min for aflatoxin extraction. For quantification, 100 µL of extraction supernatant was added to 400 µL of HPLC-grade water in 2 mL tubes and vortexed. The resultant solution was tested for aflatoxin concentration using a VICAM Series-4EX Fluorometer (Vicam, Milford, MA, USA) with Afla B columns according to the manufacturer’s instructions. Obtained data were then normalized based on seed weight and dilution, and analyzed by ANOVA with post-hoc grouping and non-parametric transformation using R (v3.5.2).
Data availability
Analyzed data are provided in the attached supplementary files. The assemblies and associated metadata are available through NCBI Bioproject IDs PRJNA606291 for NRRL3357 and PRJNA606266 for AF13. Genome assembly accession numbers at NCBI are GCA_014117465.1 for NRRL3357 and GCA_014117485.1 for AF13. Fungal isolates are available upon request by contacting the corresponding author. Supplemental material available at figshare: https://doi.org/10.25387/g3.12816593.
Results
Chromosome-level assemblies for two isolates of A. flavus
Two reference genome assemblies were generated for AF13 and NRRL3357 (Figure S1). Using PacBio sequencing, for AF13, a total of 7.73 Gb of sequencing data were generated with an average read length of 12,822 bp and read N50 of 21,750 bp. For NRRL3357, 7.97 Gb of sequencing data were generated with an average and N50 read length of 10,437 and 18,750 bp, respectively. These data were sufficient for 210 and 216X coverage for AF13 and NRRL3357. For assembly, reads >15kb in length were used yielding ∼70X coverage for each isolate assembly. Overall, 19 and 69 contigs were generated for AF13 and NRRL3357, respectively, and these contigs were then further assembled into 19 and 17 scaffolds (Table 1). When further assembled, these scaffolds approached chromosome-length assemblies generating eight pseudomolecules for each isolate (Table 2). Large variants detected between assemblies and linkage between scaffolds to generate chromosome arms were validated using Bionano optical mapping. Chromosomal assignments were based on homology and alignment with the related A. oryzae RIB40 genome (GCA_000184455.3). RIB40 alignments were also used to confirm scaffold ordering. Lengths of the assembled chromosomes ranged from 6.783 to 3.015 Mb for AF13 and 6.387 to 3.033 Mb for NRRL3357 (Table 2). Final lengths of the assembled genomes were 37.439 Mb for AF13 and 36.996 Mb for NRRL3357, which are comparable to those obtained for other A. flavus assemblies in public databases (Table S1).
Table 1. Assembled contig and scaffold descriptor statistics for AF13 and NRRL3357.
| Descriptor | AF13 | NRRL3357 | ||
|---|---|---|---|---|
| Length (Mb) | n | Length (Mb) | n | |
| Contigs | ||||
| N50 | 2.579 | 6 | 1.998 | 7 |
| N60 | 2.145 | 8 | 1.827 | 9 |
| N80 | 1.929 | 11 | 0.659 | 17 |
| N90 | 1.876 | 13 | 0.357 | 25 |
| Total | 37.599 | 19 | 38.645 | 69 |
| Average/Contig | 1.979 | 0.560 | ||
| Scaffolds | ||||
| N50 | 2.388 | 6 | 2.398 | 6 |
| N60 | 2.169 | 8 | 2.114 | 8 |
| N80 | 1.929 | 12 | 1.927 | 11 |
| N90 | 1.876 | 13 | 1.823 | 13 |
| Total | 37.439 | 19 | 36.996 | 17 |
| Average/Scaffold | 1.979 | 2.179 | ||
| Largest Scaffold | 4.615 | 4.517 | ||
| Gaps | 0 | 0 | ||
Table 2. Assembled chromosomes for AF13 and NRRL3357.
| AF13 | NRRL3357 | |||||
|---|---|---|---|---|---|---|
| Chromosome | Length (bp) | GC (%) | Predicted Genes | Length (bp) | GC (%) | Predicted Genes |
| Chr1 | 6,783,352 | 47.90 | 2,146 | 6,386,556 | 48.07 | 2,075 |
| Chr2 | 6,263,604 | 48.12 | 2,026 | 6,246,150 | 48.09 | 2,031 |
| Chr3 | 5,029,825 | 48.16 | 1,619 | 5,100,955 | 48.02 | 1,636 |
| Chr4 | 4,650,921 | 47.85 | 1,489 | 4,658,713 | 48.08 | 1,518 |
| Chr5 | 4,535,909 | 47.61 | 1,483 | 4,453,722 | 48.23 | 1,472 |
| Chr6 | 4,021,220 | 47.87 | 1,321 | 3,936,580 | 48.24 | 1,290 |
| Chr7 | 3,015,401 | 48.15 | 933 | 3,033,036 | 47.90 | 941 |
| Chr8 | 3,138,692 | 47.63 | 1,037 | 3,179,870 | 47.39 | 1,046 |
| Average/Chr | 4,679,866 | 47.91 | 1,507 | 4,624,448 | 48.00 | 1,501 |
| Unmapped (bp) | 159,798 | — | — | 53,376 | — | — |
| Total | 37,438,924 | — | 12,054 | 36,995,582 | — | 12,009 |
Indel and structural analyses reveal a novel 310kb insertion between the assemblies
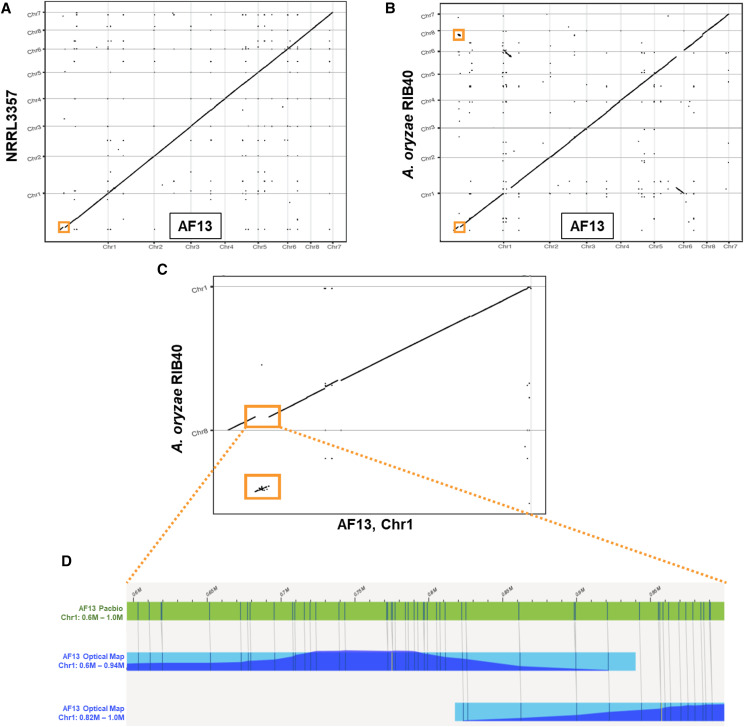
Structural and indel variation between the assemblies (Table S2) was evaluated leading to the discovery of a large, 310 Kb insertion present on Chromosome 1 of AF13 ranging from 655,567 – 967,172 bp that was completely absent from NRRL3357 (Figure 1A; Figure S2). This insertion shared homology with a similarly sized region on Chromosome 8 of the A. oryzae RIB40 genome, but limited homology with other Aspergilli and Eurotiomycete fungi suggesting that this region may be either derived from A. oryzae by horizontal transfer, represent a degenerate version of the A. oryzae Chromosome 8 region, or may represent a distinct lineage of A. flavus following speciation from A. oryzae (Figure 1B,C). The presence of this insertion, however, could be the product of sequence assembly artifacts. Therefore, Bionano optical mapping was used to confirm the presence of the insertion in the AF13 genome which clearly demonstrated that the insertion was genuine (Figure 1D).
Figure 1.
Whole genome alignment and structural confirmation using optical mapping. A. Dotplot showing a comparison between AF13 and NRRL3357. A large insertion (310 Kb) can be observed on Chromosome 1. B. Comparison between AF13 and A. oryzae RIB40 at the insert position (enlarged in C) clearly showed alignment to a region on A. oryzae Chromosome 8 for the insertion. Otherwise, the genomes shared a similar structure with the exception of a translocation on Chromosomes 6 and 2. D. Bionano optical mapping reads (blue) aligned to assembled PacBio contigs (green) show sufficient read depth in the region to confirm the presence of the insertion and validate the AF13 assembly.
Phylogenomics and prevalence of the 310 Kb insertion among other Aspergillus genomes
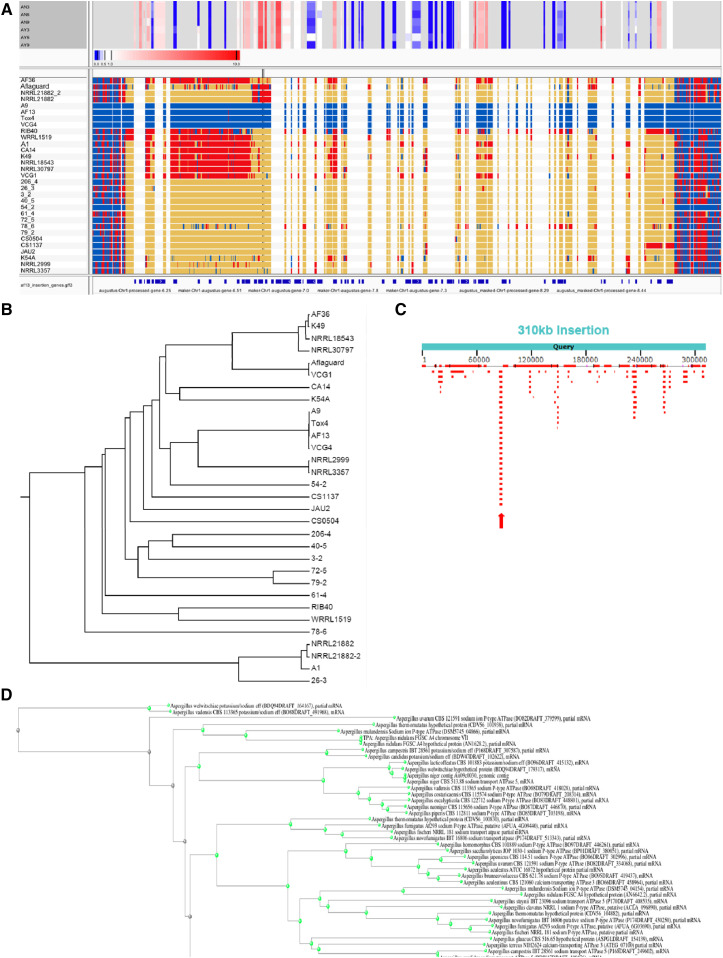
To examine the prevalence of the 310 Kb insertion within the species, available A. flavus genomes were collected from NCBI (Table S1) for comparative analyses with the AF13 assembly. In addition to these, the genomes of 10 additional isolates, including one A. parasiticus isolate, were sequenced using Illumina sequencing and used for comparative analyses (Fountain et al. 2020; Table S1). Based on blastn searches and structural comparisons (Figure 1) the insert was found to bear a relatively high level of similarity of a similar sized region of Chromosome 8 in the A. oryzae RIB40 genome. Therefore, this genome was also included in the comparative analysis.
Single nucleotide polymorphism (SNP) calling was performed relative to the AF13 genome for each isolate with a focus on the insertion and the sequences immediately surrounding it (Figure 2A; Figure S3). The isolates A9, Tox4, and VCG4 (likely clonal to Tox4), were identical to AF13 for all loci and SNP calls throughout the insert region. Most of the isolates, including NRRL3357, showed little to no alignment of their contigs with the insert region yielding no detectable SNP calls. However, several isolates showed alignment and SNP detection, but with calls being predominantly non-AF13. These calls were concentrated in the first 100 Kb of the insert region and could be observed in the isolates A1, AF36, CA14, K49, NRRL18543 (AF36), NRRL30797 (K49), VCG1, and WRRL1519. This region contains 23 genes including NADH oxidase, glycoside hydrolase pyruvate decarboxylase, and extracellular endo-1,5-alpha-L-arabinase. The three available sequences for Afla-Guard (Aflaguard-2, NRRL21882, and NRRL21882_2) also showed partial alignment within the first 100 Kb, but to a lesser extent than those previously mentioned. RIB40 showed alignment for a majority of the insert and exhibited both AF13 and non-AF13 SNP calls. Regions surrounding the insert were also identical to AF13 in A9, Tox4, and VCG4, however generally exhibiting an even mixture of AF13 and non-AF13 SNP calls in the remaining isolates.
Figure 2.
Variation in the 310Kb insertion gives insights into its origins and distribution within the species Aspergillus flavus. A. SNP calls within the insertion were evaluated. In the SNP plot, blue – AF13 calls; red – Non-AF13 calls; and yellow – no calls. The bounds of the insertion are visually apparent as an extended row of yellow (‘no call’) in strains lacking the insertion. Above the SNP calls, gene expression levels are displayed in the heatmap with box size corresponding to the position of each annotated gene in the insertion. Transcript expression levels for the annotated genes within the insertion in AF13 in response to oxidative stress over time (0 – 9 hr) are indicated above the SNP plots according to the inset scale. The positions of annotated genes within the insertion can be seen in the lowermost track below the SNP plots. Partial insertions can be observed in several biological control isolates. B. Neighbor-joining tree based on genome-wide SNP calls. AF13 and related isolates appear polyphyletic to the other A. flavus isolates. C. A single conserved gene, a Na ATPase, was identified in the insertion shown by BLAST hit alignments relative to the insertion (note the stacked hits for this gene indicated by the red arrow). D. Hits from related Aspergillus species were used to build a neighbor-joining tree. Maximum homology was only 84.35%, and the tree suggests that the insertion may be ancestral to the speciation of A. flavus and A. oryzae.
Using these genome-wide variant calls, a tree was constructed using an unweighted pair group method with arithmetic means to visualize the genetic relationship among the isolates (Figure 2B). Rooting was done based on the NRRL21882 lineage based on results from unrooted trees. As expected, the related isolates segregated into their own respective clades such as AF36, K49, NRRL18543, and NRRL30797. AF13 and its related isolates A9, Tox4, and VCG4 shared a sister lineage to NRRL3357 in the tree. However, A. flavus NRRL3357 and A. parasiticus NRRL2999 paired into their own clade, as did A. flavus WRRL1519 and A. oryzae RIB40.
In addition to variant calls, BLASTN analysis yielded a number of hits with high coverage. Alignment of the hits resulted in the identification of a Na P-type ATPase, maker-Chr1-augustus-gene-7.0, that was seemingly conserved across several Aspergillus spp. (Figure 2C). A portion of the insert containing this gene, 2,874 bp in length, was then searched in the nr database with blastn. The results showed a hit for a region on Chromosome 3 in A. flavus NRRL3357 (CP044620) with 100% coverage and 84.35% identity. The same could be found with the NRRL3357 assembly presented here. The same was observed for A. oryzae RIB40 SC023 with 100% coverage and 84.18% identity. A similar hit could also be found for the original Sanger sequenced assembly for NRRL3357 AFLA_110050. Interestingly, a second hit for this gene could be found in a similar location on Chromosome 3 in AF13, augustus-Chr3-processed-gene-16.18. This gene shared a 99.38% identity with the AFLA_110050 gene in the NRRL3357 Sanger assembly and similar levels in the current NRRL3357 assembly. Using the distance tree tool associated with NCBI blastn, a neighbor-joining tree was generated based on the top 100 alignments to the Na P-type ATPase from the insert (Figure 2D). This tree showed the genes found on Chromosome 3 in NRRL3357 to share a clade with A. oryzae RIB40, A. sojae SMF134, A. bombycis, and A. nomius NRRL13137 while the query AF13 sequence from the insertion was more ancestral sharing a common ancestor with this clade of Chromosome 3 hits from these species.
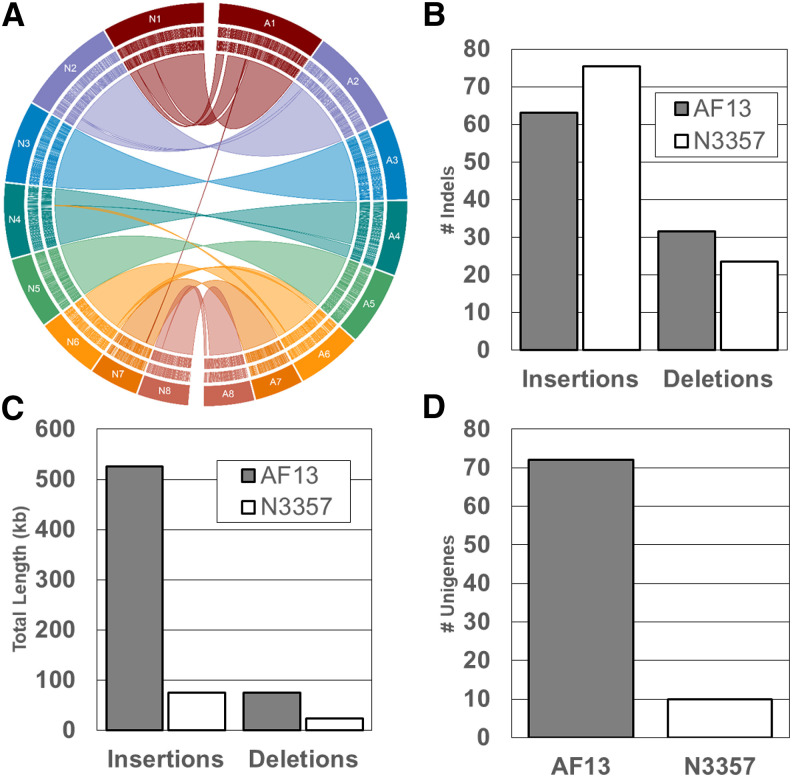
Diverse, unique genes identified between assemblies
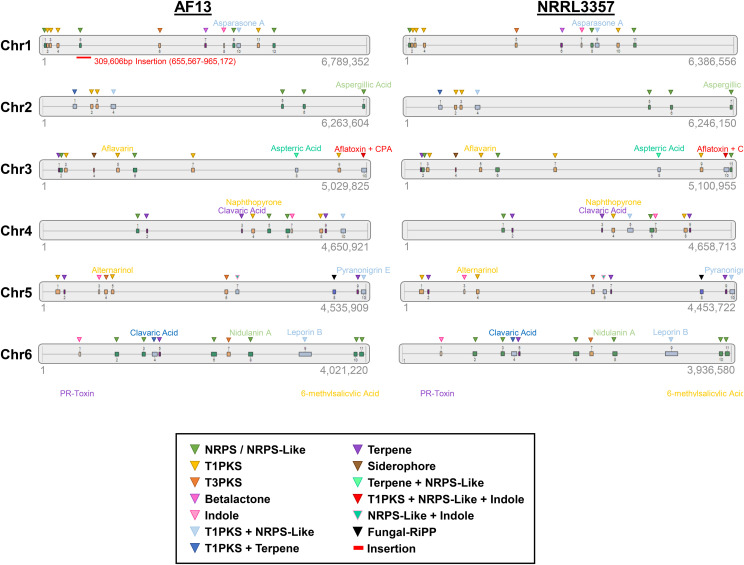
Given the distinct genetic relationship and observed phenotypes between AF13 and NRRL3357, the specific genes underlying these differences were investigated. Comparison of the two reference assemblies resulted in the identification of a number of unique genes (Figure 3; Figure S4). Based on indel analyses, AF13 was found to contain 153 unique genes interspersed throughout the genome. These genes could be subdivided into two groups, presence/absence and indel-associated genes. Among the 81 presence/absence genes (Table S3), most encoded for products involved in transmembrane transport, oxidation-reduction processes, and protein phosphorylation as indicated by GO biological process annotations. Of these genes, one Zn(II)2Cys6 transcription factor was identified along with the MAT1-2 mating type locus gene. In addition, benzoate 4-monooxygenase, S-adenosyl-L-methionine (SAM)-dependent methyltransferase, alkaline serine protease (PR1), and indoleamine 2,3-dioxygenase genes were also found among this group.
Figure 3.
Indel analysis of the AF13 and NRRL3357 genome assemblies. A. Chromosome alignments between the assemblies showing indel locations. B. Insertion and deletion counts. C. Total length of the identified insertions and deletions in each assembly. D. Total number of indel-associated unique genes.
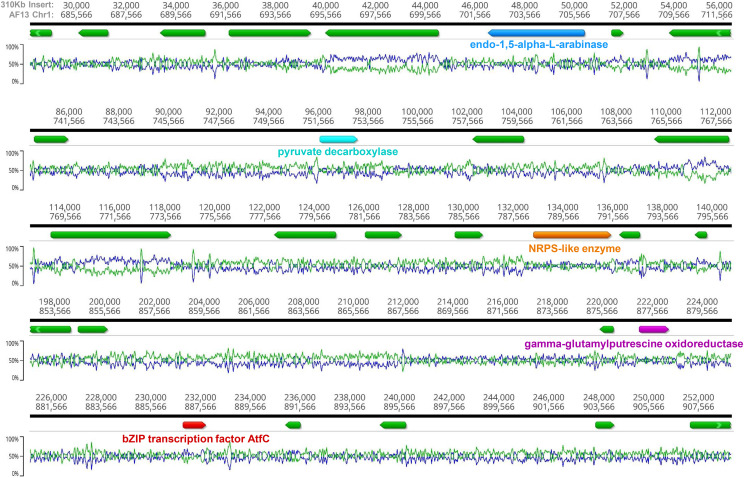
Of the 72 indel-associated genes unique to AF13 (Table S4), a majority were associated with a large 310 Kb insertion on Chromosome 1 (Figure 4). This insertion contains 60 genes of which 26 were expressed with an FPKM ≥ 2 in AF13 under oxidative stress over time in at least one replicate (Table S5). Differential expression analyses (Figure 2A, Table S5) identified 11 differentially expressed genes which were mostly up-regulated early in response to stress, but then leveled off over time. Among these genes, gamma-glutamylputresine oxidoreductase was found to be slightly down-regulated by oxidative stress while a cyclin-dependent kinase regulator Pho80, and a hypothetical protein AFLA70_740g000270 were significantly up-regulated by stress. The insert also included a novel non-ribosomal polyketide synthetase (NRPS)-like gene, however this gene was not expressed in the examined conditions. Also of interest were several constitutively expressed genes including a pyruvate decarboxylase, an extracellular endo-1,5-alpha-L-arabinase, and a novel bZIP transcription factor (augustus-Chr1-processed-gene-8.26-mRNA-1) putatively annotated here as atfC. The remaining indel-associated genes outside the insertion were dispersed among loci on Chromosomes 3 (1), 4 (9), 5 (2), 6 (1), and 8 (1). Genes of interest included a novel polyketide synthase, alanine racemase TOXG, and an acetyl-CoA synthetase-like protein gene.
Figure 4.
Composition and unique genes contained within the 310Kb insertion identified on Chromosome 1 of AF13. This plot of some select regions of the insertion contains colored arrows indicating genes of interest within the insertion. Relative position within the insertion and AF13 Chromosome1 are listed on the top of the plot. The line graphs show G/C (blue) and A/T (green) content along the sequence. A novel bZIP transcription factor, annotated here atfC, can be seen in red.
In comparison to AF13, NRRL3357 was found to contain fewer (45) unique genes. Among the 35 presence/absence genes (Table S6), GO analysis showed enrichment for oxidation-reduction and transcriptional regulation among the genes. These genes included three that encoded transcription factors: a Zn(II)2Cys6, a C6 (Fcr1), and a C2H2 transcription factor. In addition to these, other genes of interest included a synaptic vesicle transporter and a dihydrofolate reductase. Among the 10 indel-associated genes (Table S7), genes of interest included those encoding for a C6 zinc finger protein, formiminoglutamate hydrolase, copper amine oxidase, and 1-aminocyclopropane-1-carboxylate oxidase (ACC). All of these genes are located on Chromosome 5 inside a 19Kb insertion.
Secondary metabolite gene clusters
To identify secondary metabolite gene clusters present in the assemblies, antiSMASH was used to identify core biosynthetic genes within each assembly (Figure 5, Tables S8 and S9). The AF13 assembly contained 80 secondary metabolite gene regions consisting of 36 non-ribosomal polyketide synthetase (NRPS), 29 type 1 polyketide synthase, 13 terpene, 6 indole, 4 type 3 polyketide synthase, and one each of betalactone, fungal ribosomally-synthesized and posttranslationally-modified peptides (RiPP), and siderophore genes. Likewise, the NRRL3357 assembly contained 78 secondary metabolite gene regions consisting of 36 non-ribosomal polyketide synthetase (NRPS), 28 type 1 polyketide synthase, 15 terpene, 7 indole, 3 type 3 polyketide synthase, and one each of betalactone, fungal RiPP, and siderophore genes. Of the detected secondary metabolite core biosynthetic genes, five were unique to AF13 occurring on Chromosomes 1, 4, 5, and 7; and three were unique to NRRL3357 occurring on Chromosomes 1, 3, 4, 5, and 7. While most encoded for unknown gene products, the unique NRPS and type 1 polyketide synthase genes on Chromosome 4 of AF13 are homologous to citrinin biosynthetic genes. In NRRL3357, the unique terpene metabolite gene located on Chromosome 5 encodes for a geranyl-geranyl pyrophosphate synthase which is involved in the synthesis of several key precursor compounds for the production of terpenoid secondary metabolites, though the specific metabolite this gene is associated with is unknown.
Figure 5.
Secondary metabolite gene cluster prediction in the AF13 and NRRL3357 assemblies. Physical positions of secondary metabolite biosynthetic gene clusters identified by antiSMASH are plotted on each chromosome of the assemblies (gray bars, not to scale). The location and type of the core biosynthetic gene identified in each cluster are indicated by the colored triangles according the legend. The location of the 310 Kb insertion on AF13 Chromosome 1 is indicated by a red bar and associated text. Annotations of several secondary metabolites of interest identified by the analysis are listed above the triangles denoting their positions. Numbers below each chromosome plot indicate the lengths of each chromosome.
atfC, a novel bZIP transcription factor gene in A. flavus:
The putative bZIP transcription factor gene atfC identified within the 310Kb insertion in AF13 shares 74.84% similarity with the NRRL3357 atf21 gene (atfB; AFLA_094010). Given the similarity between this novel transcription factor and atf21, which has been shown to coordinate secondary metabolism and aflatoxin production along with stress responses and developmental processes in Aspergillus spp. (Roze et al. 2011, 2013), functional analyses were performed to determine the potential function of atfC. Two independent deletion mutant isolates were generated, ΔatfC-1 and ΔatfC-2 (Figure S5). No gross morphological differences were observed when culturing the mutant isolates on V8 agar (Figure S6). However, obvious phenotypic differences including aerial mycelial growth and differences in conidia production can be observed between WT AF13 and NRRL3357 (Figure 6A,B). These mutants were evaluated for aflatoxin production and oxidative stress tolerance by culturing them on YES medium amended with different concentrations of H2O2 ranging from 25 to 45 mM for five days in the dark. This range was selected based on previously observed oxidative stress tolerance ranges for AF13 (Fountain et al. 2015a). There were no observable effects on aflatoxin production in ΔAtfC-1 and ΔAtfC-2 compared to wildtype AF13 (WT) and the empty vector (EV) control (Figure 6C). The H2O2 gradient study showed a significant reduction in fungal biomass under increasing levels of oxidative stress in ΔAtfC-1 and ΔAtfC-2 in comparison to the WT and EV controls when cultured in 50mL conical tubes with shaking (Figure 6D, E). However, this reduction was not observed consistently when culturing in 125 mL Erlenmeyer flasks which showed little differences between the mutant and control isolates representing possible artifacts in the system (Figure S7). In addition, observed growth of ΔAtfC-1 at 40mM was unexpected given it was completely inhibited at 35mM (Figure 6E). These observations may indicate possible escape of inoculum from H2O2 stress, and represent a possible artifact of the system.
Figure 6.
Isolate phenotypic evaluations and effects of the deletion of atfC in AF13 on oxidative stress tolerance and pathogenicity. A. Wild type AF13 (WT) and NRRL3357 cultures on V8 agar. B. Conidia counts AF13 and NRRL3357 conidial suspensions. NRRL3357 produced significantly fewer conidia than AF13. C. A double recombination strategy was employed for the deletion of the wild type atfC gene in AF13. This is elaborated on in Figure S5. D & E. Deletion mutants of atfC were grown in the dark with shaking at 150 rpm for five days in yeast-extract sucrose (YES) medium supplemented with increasing levels of H2O2 and compared with growth of AF13 (WT) and empty vector (EV) controls. Mycelia fresh weights indicated compromised oxidative stress tolerance in the mutant isolates, particularly for ΔatfC-2. F. Aflatoxin production was examined using thin layer chromatography (TLC) and no significant effects on aflatoxin were observed in the mutant isolates. G. Kernel screening assay (KSA) on the peanut cultivar Tifrunner. Comparison of the isolates (I) showed that AF13 had significantly greater aflatoxin production compared to NRRL3357. Mutant ΔatfC-2 showed aflatoxin levels comparable to NRRL3357 suggesting compromised aflatoxin production. H. Fungal growth in terms of percentage of kernel surface area covered by visible conidia. AF13 and the mutants showed marginally significantly more growth than NRRL3357 (J). In I and J, p-values are the results of two-tailed T-tests assuming equal variance. *P ≤ 0.10; **P ≤ 0.05; ***P ≤ 0.01.
In addition to stress responsiveness, the mutant isolates were also evaluated for plant pathogenicity and aflatoxin production during peanut kernel colonization. The mutant isolates ΔatfC-1 and ΔatfC-2, the wild type (WT) AF13, and NRRL3357 were inoculated onto seeds of the peanut cultivar Tifrunner which is moderately susceptible to A. flavus infection with increased aflatoxin contamination (Figure 6G, I). A non-inoculated control was also included as a reference for possible latent A. flavus seed infections. As a baseline comparison, AF13 and NRRL3357 were evaluated and AF13 was found to exhibit greater levels of kernel colonization, moderately significant with P = 0.1011, and aflatoxin contamination, significant with P = 0.0447, in comparison to NRRL3357. The ΔatfC-1 and ΔatfC-2 mutant isolates showed somewhat contrasting phenotypes with ΔatfC-1 exhibiting near WT levels of aflatoxin production while ΔatfC-2 showed reduced aflatoxin contamination similar to that observed for NRRL3357. Neither mutant event showed a significant effect on fungal colonization, but a marginally significant difference could be observed between AF13 and the mutants, and NRRL3357 (Figure 6H, J).
Discussion
These assemblies represent a significant improvement in quality in comparison to the original scaffold-level reference genome for NRRL3357 (GCA_000006275.2) (Nierman et al. 2015; Payne et al. 2006; 2007; Yu et al. 2008). While some individual scaffolds of the previously assembled genome represented arms of chromosomes based on comparisons with A. oryzae (Machida et al. 2005; Payne et al. 2006), the current assembly has allowed for a complete picture of the full-length chromosome of NRRL3357. In comparison to the recently released NRRL3357 assembly by UC Berkeley (Skerker et al., GCA_009017425.1), those presented here share comparable lengths for both individual chromosomes, 6.387 – 3.033 Mb in the present assembly and 6.510 – 3.252 Mb in the UC Berkeley assembly, and overall, 36.996 Mb in the present assembly and 37.749 Mb in the UC Berkeley assembly.
In addition to NRRL3357, AF13 was also used to generate a chromosome-arm genome assembly. This isolate is distinct from NRRL3357 in several areas. First, this isolate is from a distinct geographical and cropping system origin. AF13 was originally isolated from cotton field soils in Yuma Valley, Arizona, USA (Cotty 1989) while NRRL3357 was isolated from peanuts with visible mold in Georgia, USA (Wicklow and Shotwell 1983). Second, these isolates represent distinct mating types and vegetative compatibility groups (VCGs) with AF13 being MAT1-2 and a member of VCG YV13 while NRRL3357 is a MAT1-1 isolate with an as yet unreported VCG classification (Chang et al. 2012; Cotty 1989; Ehrlich et al. 2007; Olarte et al. 2013). Finally, these isolates display contrasting growth behaviors and aflatoxin production capabilities in in vitro assays. Here, AF13 was shown to exhibit significantly greater levels of aflatoxin production during in vitro seed colonization assays than NRRL3357 (Figure 6). The AF13 genome assembly was comparable to these other assemblies with a total size of 37.439 Mb. Lengths of individual chromosomes were also similar with the other assemblies ranging from 6.387 to 3.033 Mb (Table 1). However, the primary differences between these genomes came in terms of unique gene content. The AF13 assembly contained 153 unique genes compared to only 45 unique genes in NRRL3357 (Figure 3). These contrasting phenotypes and novel gene content make AF13 a useful and novel reference genome candidate and will prove useful for future studies.
Unique gene content in AF13 was also of particular interest given the observed higher levels of seed colonization and aflatoxin production in comparison to NRRL3357 (Figure 6). AF13 has been previously shown to exhibit high levels of maize pathogenicity (Guo et al. 1995; Fountain et al. 2015b; Mellon et al. 2005), and a high degree of oxidative stress tolerance (Fountain et al. 2015a). Of the presence-absence and indel-associated unique genes in AF13, a benzoate 4-monooxygenase gene (maker-Chr5-augustus-gene-38.22), an indoleamine 2,3-dioxygenase (augustus-Chr6-processed-gene-39.60), an acetyl-CoA synthetase-like protein (augustus_masked-Chr4-processed-gene-43.76), and an alanine racemase TOXG (augustus_masked-Chr4-processed-gene-43.29) gene were of interest for their potential roles in stress responses and mycotoxin production.
Benzoate-4-monooxygenase, a cytochrome p450 monooxygenase, was previously found to be up-regulated in AF13 in response to H2O2-induced oxidative stress (Fountain et al. 2016a, 2016b). Aminobenzoate derivatives including methyl benzoate, ethyl benzoate, salicylic acid, and trans-cinnamic acid have been demonstrated to inhibit both growth and aflatoxin production in A. flavus cultures (Chipley and Uraih 1980). Therefore, degradation of benzoic acid by this monooxygenase in AF13 in addition to mechanisms present in other loci in the genome may partially account for the increased level of aflatoxin production observed in AF13 compared to NRRL3357. Indoleamine 2,3-dioxygenase functions as an initial reaction in the catabolism of tryptophan to kynurenine. Previously we showed that NRRL3357 had significant increases in kynurenine accumulation in response to oxidative stress over time (Fountain et al. 2019b). Inhibition of kynurenine catabolism by kynurenine 3-monooxygenase has been shown to result in increased oxidative stress tolerance in fungi (Zhang et al. 2018). Presence of an additional copy of this gene may contribute to increased stress tolerance in AF13 under certain conditions. For the acetyl-CoA synthetase, acetyl CoA serves as the primary substrate used for the production of polyketide mycotoxins like aflatoxin (Abdollahi and Buchanan 1981; Buchanan and Ayres 1977). The presence of an additional copy of this gene in AF13 may also contribute to increased levels of aflatoxin production (Figure 6). Finally, the alanine racemase TOXG gene is a component of HC toxin production, a mycotoxin that has been shown to be involved in maize pathogenicity in Cochliobolus carbonum (Walton 2006). Comparison of the sequence of this gene by blastn showed significant homology only to Uncinocarpus reesii (Coverage: 93%, ID: 74.62%), Coccidioides posadasii (Coverage: 78%, ID: 69.71%), and Coccidioides immitis (Coverage: 78%, ID: 69.61%). No significant homologs could be found among the Aspergilli. This is interesting given that C. posadasii and C. immitis are both the causal agents of San Joaquin valley fever (coccidiodomycosis), and are endemic to the Southwestern US (Cole and Hung 2001). The model U. reesii is a non-pathogenic species used for studying C. posadasii, C. immitis, and related pathogens (Pan et al. 1994). This may provide for enhanced pathogenicity in AF13 for maize colonization. It also suggests that the TOXG-containing insertion on Chromosome 4 has been acquired by horizontal gene transfer (HGT) from a Coccidioides sp. given their co-localization both to soil environments, and to the Southwestern US in origin (Cotty 1989). Novel secondary metabolite clusters identified in AF13 may provide similar advantages, however, none of the detected novel clusters had a defined function based on homology to those in public databases (Figure 5).
The starkest finding of the indel and structural comparative analyses between the assemblies was the identification of a large 310 Kb insertion unique to Chromosome 1 of AF13. This insertion contained diverse assortment of genes including those encoding a gamma-glutamylputrescine oxidoreductase (puuB, augustus-Chr1-processed-gene-8.25), and a novel bZIP transcription factor (atfC, augustus-Chr1-processed-gene-8.26). The puuB gene functions in the degradation of putrescine, a polyamine compound that serves as a precursor for the biosynthesis of spermidine and spermine. Recycling of putrescine to succinate would allow for its use in energy metabolism, however this gene was found to be significantly downregulated in AF13 in response to oxidative stress (Figure 2A). Preventing putrescine degradation may promote additional spermidine and spermine production, both of which have been shown to accumulate in response to oxidative stress in A. flavus, and to be required for normal growth, development, and aflatoxin production in in vitro assays (Fountain et al. 2019b; Majumdar et al. 2018). Polyamine metabolism here may also be connected to the previously described benzoate-4-monooxygenase system. The product of benzoate-4-monooxygenase, 4(p)-hydroxybenzoate, is also a precursor of folate biosynthesis which feeds the biosynthesis of SAM, a regulator of polyamine metabolism (Bistulfi et al. 2009; Lozoya et al. 2018). Therefore, polyamine metabolism may form the basis of a significant antioxidant mechanism employed to a greater extent in AF13 and warrants further investigation.
The novel bZIP transcription factor, annotated here as AtfC, shares homology with the previously characterized A. flavus bZIP transcription factors AtfA (51.19%) and Atf21/AtfB (74.84%). These transcription factors have been shown to regulate the production of aflatoxin and its precursors in response to oxidative stress in Aspergillus spp., and to coordinate oxidative stress responsive genes including catalase (Baidya et al. 2014; Roze et al. 2011, 2013). Silencing of atfA expression in A. nidulans has been shown to compromise tolerance to oxidative stress induced by several compounds including H2O2, menadione sodium bisulphite, and tert-butylhydroperoxide (Balázs et al. 2010; Emri et al. 2015). Silencing of atfB expression in A. parasiticus was also shown to compromise aflatoxin cluster and virulence-related gene expression and inhibit conidia production (Wee et al. 2017). Previously, the expression of these genes was observed to increase in NRRL3357 and AF13 in response to increasing oxidative stress at later timepoints in culture (Fountain et al. 2016b). Taking these facts together, therefore, the possibility of expression of a third as yet undescribed activating transcription factor (ATF) warrants investigation in AF13. Silencing of atfC in AF13 resulted in compromised oxidative stress tolerance to a varying degree between assays and the generated mutants (Figure 6) and had no obvious morphological effect in comparison to the WT or the empty vector control isolates (Figure S6). The ΔatfC-2 mutant did show a significant reduction in aflatoxin production in the kernel assay in comparison to the WT AF13 isolate to a level comparable to NRRL3357 (Figure 6). This suggests that AtfC may act as a supplement to the transcriptional regulation provided by AtfA and Atf21/AtfB, though the specific mechanism as to how this is accomplished is unknown and requires further investigation.
The prevalence of this potentially advantageous insertion was investigated among the available genome assemblies for A. flavus and closely related species including A. oryzae and A. parasiticus. The genomes of 10 additional isolates (nine A. flavus and one A. parasiticus) were sequenced and used for the evaluation of diversity within the insert and genome-wide (Fountain et al. 2020) along with several obtained from NCBI (Table S1). Plotting SNPs along the insertion, clear patterns can be observed regarding AF13 and non-AF13 calls which point to the distribution of some portions of the insertion among the examined isolate genomes (Figure 2A). However, careful examination showed that only the first ∼100 Kb of the insertion were present mainly in atoxigenic biological control isolates such as AF36 (NRRL18543), K49 (NRRL30797), and WRRL1519 (Chang et al. 2012; Pennerman et al. 2018; Yin et al. 2018), and even then exhibited significant levels of polymorphism compared to AF13 and its related isolates. This region contained several genes involved both in energy production, defense responses, and in the catabolism of pectin, all of which are potentially beneficial to saprophytic and plant pathogenic fungi. Therefore, this region may contribute to the efficacy of these isolates as biological controls in competition with native aflatoxigenic A. flavus populations in field environments.
In examining the insertion for orthologs in other Aspergillus spp. by blastn analysis, it was found that a single Na ATPase gene (maker-Chr1-augustus-gene-7.0) was conserved across multiple Aspergilli and was used for construction of neighbor-joining tree (Figure 2B). This gene, which has a homolog on Chromosome 3 in both AF13 and NRRL3357, is distinct from its orthologs within the genus. The relatively low degree of homology with A. flavus and A. oryzae, which shared the most homology overall for the insertion on Chromosome 8 of the A. oryzae RIB40 genome (Figure 1), does suggest that this gene, and therefore the insertion, may be ancestral to speciation between A. flavus and A. oryzae, and preserved at least in part in lineages of both species. This assertion is further supported by the examination of genome-wide variants and the construction of a rooted neighbor-joining phylogenetic tree in this analysis (Figure 2B). Here, A. oryzae RIB40 and most A. flavus clades diverged after the separation of the NRRL21882 lineage. Given that NRRL21882 contains only a small portion of the insertion, it seems likely that in the other clade containing AF13 and RIB40, the insertion was preserved being passed along in part to the AF36 clade and to the AF13 clade, and not to the remainder including NRRL3357. This may also be true for the aflatoxin gene cluster, and not only for the insertion given that all the members of the NRRL21882 clade are atoxigenic isolates while the remaining isolates and species within the tree contain at least partial aflatoxin clusters (Chang et al. 2005; Faustinelli et al. 2016; 2017).
Surprising here is the level of similarity observed between A. flavus NRRL3357 and A. parasiticus NRRL2999, and between A. flavus WRRL1519 and A. oryzae RIB40 (Figure 2B). This close relationship is supported in the literature with WRRL1519 having been previously reported to be more genetically related to A. oryzae than other A. flavus isolates (Chang 2019). This same report by Chang (2019) also supports the hypothesis that NRRL21882 is more genetically related to A. oryzae compared to other toxigenic L-strains of A. flavus which concurs with the phylogenetic analysis here. At the genus level, Aspergillus has been clearly demonstrated to be monophyletic in relation to other related members of the Eurotiales and Trichocomaceae such as Penicillium (Frisvad et al. 2019; Kocsubé et al. 2016; Samson et al. 2014). However, within the species there has been more variation in classification over time with A. oryzae and A. parasiticus being previously referred to as subspecies within A. flavus (Kurtzman et al. 1986). Distinctions based on morphological characteristics in addition to sequencing of conserved genes such as internal transcribed spacer (ITS) rRNA sequences have since been used to classify these as distinct species from A. flavus (Kumeda and Asao 1996; Machida et al. 2008; Peterson 2008; Varga et al. 2011). Making this distinction, the tree presented here concurs and supports the proposal that A. flavus is comprised of a polyphyletic collection of related isolates, subspecies, and species as presented in the literature (Chang et al. 2006; Chang 2019; Geiser et al. 1998, 2000; Gonçalves et al. 2012; Moore et al. 2009; Okoth et al. 2018). However, the close relationship of these distinct species with A. flavus isolates described in the present study from the genomics perspective does cast doubt on the classification of these as distinct species rather than as subspecies of A. flavus. In comparison to ITS, whole genome sequencing allows for the evaluation of evolutionary changes throughout the entire genome, and should result in increased statistical power to delineate species and subdivisions within them (Baumsteiger et al. 2017). Addressing these classification issues will require the increasing prevalence of genomics information for isolates within this species, and studies comparing the results of genomics analyses and traditional ITS barcoding along with evaluating the reliability of common morphological characteristics for use in delineating species.
In conclusion, these newly generated, high quality, reference genomes for AF13 and NRRL3357 will provide new tools in the toolbox for genomics-assisted research into these important fungi. Comparative genomics analyses here have also identified genes and components of these isolate genomes which may contribute to plant pathogenicity, aflatoxin production, and biocontrol efficacy. They also provide a foundation for the beginnings of a pangenomic understanding of A. flavus by providing insights into novel gene content and structural variants which do not present in the previous reference isolate, NRRL3357. This novel gene content may prove useful in the elucidation and development of host resistance mechanisms against A. flavus colonization, biological control selection and screening, and field and storage-focused control measures to mitigate aflatoxin contamination.
Acknowledgments
We would like to thank Billy Wilson, Sheron Simpson, and Leslie Scharfenstein for technical assistance in the laboratory. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the USDA. The USDA is an equal opportunity employer and provider.
Footnotes
Supplemental material available at figshare: https://doi.org/10.25387/g3.12816593.
Communicating editor: B. Andrews
Literature Cited
- Abdollahi A., and Buchanan R. L., 1981. Regulation of aflatoxin biosynthesis: induction of aflatoxin production by various carbohydrates. J. Food Sci. 46: 633–635. 10.1111/j.1365-2621.1981.tb04928.x [DOI] [Google Scholar]
- Afgan E., Baker D., Batut B., van den Beek M., Bouvier D. et al. , 2018. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 46: W537–W544. 10.1093/nar/gky379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaike S., and Keller N. P., 2011. Aspergillus flavus. Annu. Rev. Phytopathol. 49: 107–133. 10.1146/annurev-phyto-072910-095221 [DOI] [PubMed] [Google Scholar]
- Arnaud, M. B., Cerqueira G. C., Inglis D. O., Skrzypek M. S., J. Binkley et al., 2012 The Aspergillus Genome Database (AspGD): recent developments in comprehensive multispecies curation, comparative genomics and community resources. Nucleic Acids Res. 40: D653–D659. 10.1093/nar/gkr875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baidya S., Duran R. M., Lohmar J. M., Harris-Coward P. Y., Cary J. W. et al. , 2014. VeA is associated with the response to oxidative stress in the aflatoxin producer Aspergillus flavus. Eukaryot. Cell 13: 1095–1103. 10.1128/EC.00099-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balázs A., Pócsi I., Hamari Z., Leiter É., Emri T. et al. , 2010. AtfA bZIP-type transcription factor regulates oxidative and osmotic stress responses in Aspergillus nidulans. Mol. Genet. Genomics 283: 289–303. 10.1007/s00438-010-0513-z [DOI] [PubMed] [Google Scholar]
- Bao W., Kojima K. K., and Kohany O., 2015. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob. DNA 6: 11 10.1186/s13100-015-0041-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumsteiger J., Moyle P. B., Aguilar A., O’Rourke S. M., and Miller M. R., 2017. Genomics clarifies taxonomic boundaries in a difficult species complex. PLoS One 12: e0189417 10.1371/journal.pone.0189417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bistulfi G., Diegelman P., Foster B. A., Kramer D. L., Porter C. W. et al. , 2009. Polyamine biosynthesis impacts cellular folate requirements necessary to maintain S-adenosylmethionine and nucleotide pools. FASEB J. 23: 2888–2897. 10.1096/fj.09-130708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blin K., Shaw S., Steinke K., Villebro R., Ziemert N. et al. , 2019. antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 47: W81–W87. 10.1093/nar/gkz310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray N. L., Pimentel H., Melsted P., and Pachter L., 2016. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34: 525–527. 10.1038/nbt.3519 [DOI] [PubMed] [Google Scholar]
- Brown D. W., Yu J. H., Kelkar H. S., Fernandes M., Nesbitt T. C. et al. , 1996. Twenty-five coregulated transcripts define a sterigmatocystin gene cluster in Aspergillus nidulans. Proc. Natl. Acad. Sci. USA 93: 1418–1422. 10.1073/pnas.93.4.1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan R. L., and Ayres J. C., 1977. Effect of various glycolytic and TCA intermediates on aflatoxin production. J. Food Saf. 1: 19–28. 10.1111/j.1745-4565.1977.tb00256.x [DOI] [Google Scholar]
- Cantarel B. L., Korf I., Robb S. M., Parra G., Ross E. et al. , 2008. MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 18: 188–196. 10.1101/gr.6743907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cary J. W., Ehrlich K. C., Bland J. M., and Montalbano B. G., 2006. The aflatoxin biosynthesis cluster gene, aflX, encodes an oxidoreductase involved in conversion of versicolorin A to demethylsterigmatocystin. Appl. Environ. Microbiol. 72: 1096–1101. 10.1128/AEM.72.2.1096-1101.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan P. P., and Lowe T. W., 2019. tRNAscan-SE: Searching for tRNA genes in genomic sequences. Methods Mol. Biol. 1962: 1–14. 10.1007/978-1-4939-9173-0_1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang P. K., 2019. Genome‐wide nucleotide variation distinguishes Aspergillus flavus from Aspergillus oryzae and helps to reveal origins of atoxigenic A. flavus biocontrol strains. J. Appl. Microbiol. 127: 1511–1520. 10.1111/jam.14419 [DOI] [PubMed] [Google Scholar]
- Chang P. K., Horn B. W., and Dorner J. W., 2005. Sequence breakpoints in the aflatoxin biosynthesis gene cluster and flanking regions in nonaflatoxigenic Aspergillus flavus isolates. Fungal Genet. Biol. 42: 914–923. 10.1016/j.fgb.2005.07.004 [DOI] [PubMed] [Google Scholar]
- Chang P. K., Abbas H. K., Weaver M. A., Ehrlich K. C., Scharfenstein L. L. et al. , 2012. Identification of genetic defects in the atoxigenic biocontrol strain Aspergillus flavus K49 reveals the presence of a competitive recombinant group in field populations. Int. J. Food Microbiol. 154: 192–196. 10.1016/j.ijfoodmicro.2012.01.005 [DOI] [PubMed] [Google Scholar]
- Chang P. K., Ehrlich K. C., and Hua S. S. T., 2006. Cladal relatedness among Aspergillus oryzae isolates and Aspergillus flavus S and L morphotype isolates. Int. J. Food Microbiol. 108: 172–177. 10.1016/j.ijfoodmicro.2005.11.008 [DOI] [PubMed] [Google Scholar]
- Chang P. K., Scharfenstein L. L., Wei Q., and Bhatnagar D., 2010. Development and refinement of a high-efficiency gene-targeting system for Aspergillus flavus. J. Microbiol. Methods 81: 240–246. 10.1016/j.mimet.2010.03.010 [DOI] [PubMed] [Google Scholar]
- Chipley J. R., and Uraih N., 1980. Inhibition of Aspergillus growth and aflatoxin release by derivatives of benzoic acid. Appl. Environ. Microbiol. 40: 352–357. 10.1128/AEM.40.2.352-357.1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole G. T., and Hung C. Y., 2001. The parasitic cell wall of Coccidioides immitis. Sabouraudia 39: 31–40. 10.1080/mmy.39.1.31.40 [DOI] [PubMed] [Google Scholar]
- Cotty P. J., 1989. Virulence and cultural characteristics of two Aspergillus flavus strains pathogenic on cotton. Phytopathology 79: 808–814. 10.1094/Phyto-79-808 [DOI] [Google Scholar]
- Ehrlich K. C., Montalbano B. G., and Cotty P. J., 2007. Analysis of single nucleotide polymorphisms in three genes shows evidence for genetic isolation of certain Aspergillus flavus vegetative compatibility groups. FEMS Microbiol. Lett. 268: 231–236. 10.1111/j.1574-6968.2006.00588.x [DOI] [PubMed] [Google Scholar]
- Emri T., Szarvas V., Orosz E., Antal K., Park H. et al. , 2015. Core oxidative stress response in Aspergillus nidulans. BMC Genomics 16: 478 10.1186/s12864-015-1705-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faustinelli P. C., Palencia E. R., Sobolev V. S., Horn B. W., Sheppard H. T. et al. , 2017. Study of the genetic diversity of the aflatoxin biosynthesis cluster in Aspergillus section Flavi using insertion/deletion markers in peanut seeds from Georgia, USA. Mycologia 109: 200–209. 10.1080/00275514.2017.1307095 [DOI] [PubMed] [Google Scholar]
- Faustinelli P. C., Wang X. M., Palencia E. R., and Arias R. S., 2016. Genome sequences of eight Aspergillus flavus spp. and one A. parasiticus sp., isolated from peanut seeds in Georgia. Genome Announc. 4: e00278–16 10.1128/genomeA.00278-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forgacs J., and Carll W. T., 1962. Mycotoxicoses, pp. 273–382 in Advances in Veterinary Science, edited by Bradly C. A., and Jungherr E. L.. Academic, New York. [Google Scholar]
- Fountain J. C., Scully B. T., Chen Z. Y., Gold S. E., Glenn A. E. et al. , 2015a Effects of hydrogen peroxide on different toxigenic and atoxigenic isolates of Aspergillus flavus. Toxins (Basel) 7: 2985–2999. 10.3390/toxins7082985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fountain J. C., Abbas H. K., Scully B. T., Li H., Lee R. D. et al. , 2019a Evaluation of maize inbred lines and topcross progeny for resistance to pre-harvest aflatoxin contamination. Crop J. 7: 118–125. 10.1016/j.cj.2018.10.001 [DOI] [Google Scholar]
- Fountain J. C., Yang L., Pandey M. K., Bajaj P., Alexander D. et al. , 2019b Carbohydrate, glutathione, and polyamine metabolism are central to Aspergillus flavus oxidative stress responses over time. BMC Microbiol. 19: 209 10.1186/s12866-019-1580-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fountain J. C., Bajaj P., Pandey M. K., Nayak S. N., Yang L. et al. , 2016a Oxidative stress and carbon metabolism influence Aspergillus flavus transcriptome composition and secondary metabolite production. Sci. Rep. 6: 38747 10.1038/srep38747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fountain J. C., Bajaj P., Nayak S. N., Yang L., Pandey M. K. et al. , 2016b Responses of Aspergillus flavus to oxidative stress are related to fungal development regulator, antioxidant enzyme, and secondary metabolite biosynthetic gene expression. Front. Microbiol. 7: 2048 10.3389/fmicb.2016.02048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fountain J. C., Raruang Y., Luo M., Brown R. L., Guo B. et al. , 2015b Potential roles of WRKY transcription factors in regulating host defense responses during Aspergillus flavus infection of immature maize kernels. Physiol. Mol. Plant Pathol. 89: 31–40. 10.1016/j.pmpp.2014.11.005 [DOI] [Google Scholar]
- Fountain J. C., Clevenger J. P., Nadon B., Wang H., Abbas H. K. et al. , 2020. Draft genome sequences of one Aspergillus parasiticus and nine A. flavus isolates with varying stress tolerance and aflatoxin production. Microbiol. Resour. Announc. (Submitted). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisvad J. C., Hubka V., Ezekiel C. N., Hong S. B., Nováková A. et al. , 2019. Taxonomy of Aspergillus section Flavi and their production of aflatoxins, ochratoxins and other mycotoxins. Stud. Mycol. 93: 1–63. 10.1016/j.simyco.2018.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galagan J. E., Calvo S. E., Cuomo C., Ma L. J., Wortman J. R. et al. , 2005. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 438: 1105–1115. 10.1038/nature04341 [DOI] [PubMed] [Google Scholar]
- Geiser D. M., Pitt J. I., and Taylor J. W., 1998. Cryptic speciation and recombination in the aflatoxin-producing fungus Aspergillus flavus. Proc. Natl. Acad. Sci. USA 95: 388–393. 10.1073/pnas.95.1.388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiser D. M., Dorner J. W., Horn B. W., and Taylor J. W., 2000. The phylogenetics of mycotoxin and sclerotium production in Aspergillus flavus and Aspergillus oryzae. Fungal Genet. Biol. 31: 169–179. 10.1006/fgbi.2000.1215 [DOI] [PubMed] [Google Scholar]
- Gilbert M. K., Mack B. M., Moore G. G., Downey D. L., Lebar M. D. et al. , 2018. Whole genome comparison of Aspergillus flavus L-morphotype strain NRRL 3357 (type) and S-morphotype strain AF70. PLoS One 13: e0199169 10.1371/journal.pone.0199169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves S. S., Cano J. F., Stchigel A. M., Melo A. S., Godoy-Martinez P. C. et al. , 2012. Molecular phylogeny and phenotypic variability of clinical and environmental strains of Aspergillus flavus. Fungal Biol. 116: 1146–1155. 10.1016/j.funbio.2012.08.006 [DOI] [PubMed] [Google Scholar]
- Guo B. Z., Russin J. S., Cleveland T. E., Brown R. L., and Widstrom N. W., 1995. Wax and cutin layers in maize kernels associated with resistance to aflatoxin production by Aspergillus flavus. J. Food Prot. 58: 296–300. 10.4315/0362-028X-58.3.296 [DOI] [PubMed] [Google Scholar]
- Horng J. S., Chang P. K., Pestka J. J., and Linz J. E., 1990. Development of a homologous transformation system for Aspergillus parasiticus with the gene encoding nitrate reductase. Mol. Gen. Genet. MGG. 224: 294–296. 10.1007/BF00271564 [DOI] [PubMed] [Google Scholar]
- Kocsubé S., Perrone G., Magistà D., Houbraken J., Varga J. et al. , 2016. Aspergillus is monophyletic: evidence from multiple gene phylogenies and extrolites profiles. Stud. Mycol. 85: 199–213. 10.1016/j.simyco.2016.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumeda Y., and Asao T., 1996. Single-strand conformation polymorphism analysis of PCR-amplified ribosomal DNA internal transcribed spacers to differentiate species of Aspergillus section Flavi. Appl. Environ. Microbiol. 62: 2947–2952. 10.1128/AEM.62.8.2947-2952.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtzman C. P., Smiley M. J., Robnett C. J., and Wicklow D. T., 1986. DNA relatedness among wild and domesticated species in the Aspergillus flavus group. Mycologia 78: 955–959. 10.1080/00275514.1986.12025355 [DOI] [Google Scholar]
- Liu Z., and Friesen T. L., 2012. Polyethylene glycol (PEG)-mediated transformation in filamentous fungal pathogens. Methods Mol Biol 835: 365–375. . 10.1007/978-1-61779-501-5_21 [DOI] [PubMed] [Google Scholar]
- Love M. I., Huber W., and Anders S., 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15: 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozoya O. A., Martinez-Reyes I., Wang T., Grenet D., Bushel P. et al. , 2018. Mitochondrial nicotinamide adenine dinucleotide reduced (NADH) oxidation links the tricarboxylic acid (TCA) cycle with methionine metabolism and nuclear DNA methylation. PLoS Biol. 16: e2005707 10.1371/journal.pbio.2005707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida M., Asai K., Sano M., Tanaka T., Kumagai T. et al. , 2005. Genome sequencing and analysis of Aspergillus oryzae. Nature 438: 1157–1161. 10.1038/nature04300 [DOI] [PubMed] [Google Scholar]
- Machida M., Yamada O., and Gomi K., 2008. Genomics of Aspergillus oryzae: learning from the history of Koji mold and exploration of its future. DNA Res. 15: 173–183. 10.1093/dnares/dsn020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar R., Lebar M., Mack B., Minocha R., Minocha S. et al. , 2018. The Aspergillus flavus spermidine synthase (spds) gene, is required for normal development, aflatoxin production, and pathogenesis during infection of maize kernels. Front. Plant Sci. 9: 317 10.3389/fpls.2018.00317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellon J. E., Dowd M. K., and Cotty P. J., 2005. Substrate utilization by Aspergillus flavus in inoculated whole corn kernels and isolated tissues. J. Agric. Food Chem. 53: 2351–2357. 10.1021/jf040276g [DOI] [PubMed] [Google Scholar]
- Moore G. G., Singh R., Horn B. W., and Carbone I., 2009. Recombination and lineage‐specific gene loss in the aflatoxin gene cluster of Aspergillus flavus. Mol. Ecol. 18: 4870–4887. 10.1111/j.1365-294X.2009.04414.x [DOI] [PubMed] [Google Scholar]
- Nierman W. C., Pain A., Anderson M. J., Wortman J. R., Kim H. S. et al. , 2005. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 438: 1151–1156. 10.1038/nature04332 [DOI] [PubMed] [Google Scholar]
- Nierman W. C., Yu J., Fedorova-Abrams N. D., Losada L., Cleveland T. E. et al. , 2015. Genome sequence of Aspergillus flavus NRRL 3357, a strain that causes aflatoxin contamination of food and feed. Genome Announc. 3: e00168–15 10.1128/genomeA.00168-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkura M., Cotty P. J., and Orbach M. J., 2018. Comparative genomics of Aspergillus flavus S and L morphotypes yield insights into niche adaptation. G3 (Bethesda) 8: 3915–3930. 10.1534/g3.118.200553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okoth S., De Boevre M., Vidal A., Diana Di Mavungu J., Landschoot S. et al. , 2018. Genetic and toxigenic variability within Aspergillus flavus population isolated from maize in two diverse environments in Kenya. Front. Microbiol. 9: 57 10.3389/fmicb.2018.00057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olarte, R. A., B. W. Horn, C. J. Worthington, R. Singh, and I. Carbone, 2013 Population shifts and mating-type heterokaryosis in Aspergillus flavus. 9th International Aspergillus Meeting. March 11–12, 2013. Pacific Grove, CA. [Google Scholar]
- Pan S., Sigler L., and Cole G. T., 1994. Evidence for a phylogenetic connection between Coccidioides immitis and Uncinocarpus reesii (Onygenaceae). Microbiol. 140: 1481–1494. 10.1099/00221287-140-6-1481 [DOI] [PubMed] [Google Scholar]
- Payne G. A., Yu J., Nierman W. C., Machida M., Bhatnagar D. et al. , 2007. A First Glance into the Genome Sequence of Aspergillus flavus, pp. 35–44 in The Aspergilli, CRC Press, Boca Raton, Florida. [Google Scholar]
- Payne G. A., Nierman W. C., Wortman J. R., Pritchard B. L., Brown D. et al. , 2006. Whole genome comparison of Aspergillus flavus and A. oryzae. Med. Mycol. 44: S9–S11. 10.1080/13693780600835716 [DOI] [PubMed] [Google Scholar]
- Pel H. J., De Winde J. H., Archer D. B., Dyer P. S., Hofmann G. et al. , 2007. Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat. Biotechnol. 25: 221–231. 10.1038/nbt1282 [DOI] [PubMed] [Google Scholar]
- Pennerman K. K., Gonzalez J., Chenoweth L. R., Bennett J. W., Yin G. et al. , 2018. Biocontrol strain Aspergillus flavus WRRL 1519 has differences in chromosomal organization and an increased number of transposon-like elements compared to other strains. Mol. Genet. Genomics 293: 1507–1522. 10.1007/s00438-018-1474-x [DOI] [PubMed] [Google Scholar]
- Peterson S. W., 2008. Phylogenetic analysis of Aspergillus species using DNA sequences from four loci. Mycologia 100: 205–226. 10.1080/15572536.2008.11832477 [DOI] [PubMed] [Google Scholar]
- Roze L. V., Chanda A., Wee J., Awad D., and Linz J. E., 2011. Stress-related transcription factor AtfB integrates secondary metabolism with oxidative stress response in Aspergilli. J. Biol. Chem. 286: 35137–35148. 10.1074/jbc.M111.253468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roze L. V., Hong S. Y., and Linz J. E., 2013. Aflatoxin biosynthesis: current frontiers. Annu. Rev. Food Sci. Technol. 4: 293–311. 10.1146/annurev-food-083012-123702 [DOI] [PubMed] [Google Scholar]
- Samson R. A., Visagie C. M., Houbraken J., Hong S. B., Hubka V. et al. , 2014. Phylogeny, identification and nomenclature of the genus Aspergillus. Stud. Mycol. 78: 141–173. 10.1016/j.simyco.2014.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M., Steinkamp R., Waack S., and Morgenstern B., 2004. AUGUSTUS: a web server for gene finding in eukaryotes. Nucleic Acids Res. 32: W309–W312. 10.1093/nar/gkh379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga J., Frisvad J. C., and Samson R. A., 2011. Two new aflatoxin producing species, and an overview of Aspergillus section Flavi. Stud. Mycol. 69: 57–80. 10.3114/sim.2011.69.05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton J. D., 2006. HC-toxin. Phytochemistry 67: 1406–1413. 10.1016/j.phytochem.2006.05.033 [DOI] [PubMed] [Google Scholar]
- Wee J., Hong S. Y., Roze L., Day D., Chanda A. et al. , 2017. The fungal bZIP transcription factor AtfB controls virulence-associated processes in Aspergillus parasiticus. Toxins (Basel) 9: 287 10.3390/toxins9090287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicklow D. T., and Shotwell O. L., 1983. Intrafungal distribution of aflatoxins among conidia and sclerotia of Aspergillus flavus and Aspergillus parasiticus. Can. J. Microbiol. 29: 1–5. 10.1139/m83-001 [DOI] [PubMed] [Google Scholar]
- Yang K., Liang L., Ran F., Liu Y., Li Z. et al. , 2016. The DmtA methyltransferase contributes to Aspergillus flavus conidiation, sclerotial production, aflatoxin biosynthesis and virulence. Sci. Rep. 6: 23259 10.1038/srep23259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Fountain J. C., Ji P., Ni X., Chen S. et al. , 2018. Deciphering drought‐induced metabolic responses and regulation in developing maize kernels. Plant Biotechnol. J. 16: 1616–1628. 10.1111/pbi.12899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin G., Hua S. S. T., Pennerman K. K., Yu J., Bu L. et al. , 2018. Genome sequence and comparative analyses of atoxigenic Aspergillus flavus WRRL 1519. Mycologia 110: 482–493. 10.1080/00275514.2018.1468201 [DOI] [PubMed] [Google Scholar]
- Yu J., Whitelaw C. A., Nierman W. C., Bhatnagar D., and Cleveland T. E., 2004b Aspergillus flavus expressed sequence tags for identification of genes with putative roles in aflatoxin contamination of crops. FEMS Microbiol. Lett. 237: 333–340. [DOI] [PubMed] [Google Scholar]
- Yu J., Payne G. A., Nierman W. C., Machida M., Bennett J. W. et al. , 2008. Aspergillus flavus genomics as a tool for studying the mechanism of aflatoxin formation. Food Addit. Contam. 25: 1152–1157. 10.1080/02652030802213375 [DOI] [PubMed] [Google Scholar]
- Yu J., Chang P. K., Ehrlich K. C., Cary J. W., Bhatnagar D. et al. , 2004a Clustered pathway genes in aflatoxin biosynthesis. Appl. Environ. Microbiol. 70: 1253–1262. 10.1128/AEM.70.3.1253-1262.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K., Yuan X., Zang J., Wang M., Zhao F. et al. , 2018. The kynurenine 3-monooxygenase encoding gene, BcKMO, is involved in the growth, development, and pathogenicity of Botrytis cinerea. Front. Microbiol. 9: 1039 10.3389/fmicb.2018.01039 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Analyzed data are provided in the attached supplementary files. The assemblies and associated metadata are available through NCBI Bioproject IDs PRJNA606291 for NRRL3357 and PRJNA606266 for AF13. Genome assembly accession numbers at NCBI are GCA_014117465.1 for NRRL3357 and GCA_014117485.1 for AF13. Fungal isolates are available upon request by contacting the corresponding author. Supplemental material available at figshare: https://doi.org/10.25387/g3.12816593.