Abstract
Introgression from wild relatives is a valuable source of novel allelic diversity for breeding. We investigated the genomic patterns of introgression from Aegilops tauschii, the diploid ancestor of the wheat D genome, into winter wheat (Triticum aestivum) cultivars. The population of 351 BC1F3:5 lines was selected based on phenology from crosses between six hexaploid wheat lines and 21 wheat-Ae. tauschii octoploids. SNP markers developed for this population and a diverse panel of 116 Ae. tauschii accessions by complexity-reduced genome sequencing were used to detect introgression based on the identity-by-descent analysis. Overall, introgression frequency positively correlated with recombination rate, with a high incidence of introgression at the ends of chromosomes and low in the pericentromeric regions, and was negatively related to sequence divergence between the parental genomes. Reduced introgression in the pericentromeric low-recombining regions spans nearly 2/3 of each chromosome arm, suggestive of the polygenic nature of introgression barriers that could be associated with multilocus negative epistasis between the alleles of wild and cultivated wheat. On the contrary, negative selection against the wild allele of Tg, controlling free-threshing trait and located in the high-recombining chromosomal region, led to reduced introgression only within ∼10 Mbp region around Tg. These results are consistent with the effect of selection on linked variation described by the Hill-Robertson effect, and offer insights into the introgression population development for crop improvement to maximize retention of introgressed diversity across entire genome.
Keywords: Aegilops tauschii, wild relative, introgression, hexaploid wheat, recombination, tenacious glume
Wild relatives of wheat show a broad range of adaptation to diverse climatic and biotic stress factors, and for decades served as a source of beneficial allelic diversity in breeding programs for improving drought tolerance (Peleg et al. 2005; Reynolds et al. 2007), disease resistance (Huang et al. 2003; Periyannan et al. 2013; Saintenac, Zhang, et al. 2013), grain quality (Uauy et al. 2006) and yield potential (Cooper et al. 2012; Reynolds et al. 2017). The availability of high-quality genome assemblies for a number of wild relatives (Ling et al. 2013; Avni et al. 2017; Luo et al. 2017) combined with inexpensive next-generation sequencing technologies allowing for quick genome-wide genotyping of large populations stimulated substantial interest in introgression of the wild relatives’ diversity as a strategy for crop improvement. Allohexaploid bread wheat, Triticum aestivum (2n = 6x = 42, genome formula AABBDD), resulted from hybridization between tetraploid Triticum turgidum (2n = 4x = 28, genome formula AABB) and Aegilops tauschii (2n = 2x = 14, genome formula DD) (Kihara 1944; Luo et al. 2007; Wang et al. 2013). These two wild relatives serve as the primary sources of allelic diversity for introgression into common wheat (Gill and Raupp 1987; Qi et al. 2007; Reynolds et al. 2007; Dreisigacker et al. 2008). Introgression from these species, which share homologous genomes with wheat, could be easily achieved by either direct crosses between wheat and wild relatives, or crosses between wheat and synthetic wheat lines produced by hybridizing tetraploid wheat and Ae. tauschii (Qi et al. 2007; Ogbonnaya et al. 2013). The wild diploid and tetraploid relatives of wheat that carry homeologous genomes, such as T. monococcum (Am genome), T. urartu (Au genome), T. timopheevii (2n = 4x = 28, AAGG) or Ae. speltoides (2n = 2x = 14, SS) are considered as secondary sources of genetic diversity for introgression. Effective introgression from these species requires recombining the homeologous chromosomes of common wheat and wild relatives in the absence of Ph1 gene that controls pairing between homeologs (Sears 1977).
The retention of wild relative introgression in the populations of cultivated crops could be affected by selection for fitness or traits related to agronomic performance. There was evidence for reduced historic introgression from wild relatives into wheat and maize cultivars around genes controlling major domestication traits (Wang et al. 2017; He et al. 2019). For example, in wheat, the region of the genome harboring domestication gene Q (Simons et al. 2006) showed reduced genetic diversity and no evidence of introgression from sympatric populations of wild emmer wheat (He et al. 2019), consistent with selection against alleles affecting the domestication phenotype. Interaction between introgressed alleles and the genetic backgrounds of recipient lines could result in hybrid sterility, or influence the general phenology or agronomic performance resulting in the loss or retention of introgressed genome segments (Rieseberg et al. 1999; Jiang et al. 2000). In sunflower, variation in the frequency and distribution of introgression across genome was attributed to reduced introgression around the structurally re-arranged regions and alleles linked with pollen sterility (Rieseberg et al. 1999). A multilocus epistatic interaction was proposed as a possible explanation for restricted introgression in the interspecific populations of polyploid cotton (Jiang et al. 2000). Selection on the introgressed variants is also expected to affect linked variation, with the size of the affected region depending on the distribution of recombination rate across genome (Hill and Robertson 1966; Martin et al. 2019). Therefore, from the practical perspective, understanding the genomic patterns of wild relative introgression into cultivated crops will be crucial for the effective deployment of novel diversity for crop improvement.
Genotyping approaches based on next-generation sequencing of complexity-reduced genomic libraries substantially accelerated analysis of genetic diversity in large crop genomes (Elshire et al. 2011; Saintenac, Jiang, et al. 2011, 2013; Poland et al. 2012; Jordan et al. 2015, 2018). The high proportion of missing data in the low-coverage sequencing datasets can be compensated by genotype imputation using the recently developed reference genome (The International Wheat Genome Sequencing Consortium (IWGSC) 2018). Imputation of ungenotyped SNP markers from a reference panel into a target population takes advantage of regions of identity-by-descent (IBD), thus allowing for the interpolation of SNPs into the target population (Browning and Browning 2013). The power and resolution of association studies have been shown to improve after imputation (Browning and Browning 2012; Jordan et al. 2015; Nyine et al. 2019).
In this study, we developed a population of winter wheat lines carrying introgression from a diverse set of Ae. tauschii accessions selected to represent both the broad genetic and geographic diversity of the species. The boundaries of the introgressed segments in the wheat genome were inferred using the IBD analyses based on the SNP datasets generated by complexity-reduced sequencing of 378 samples including 21 Ae. tauschii accessions, 6 hexaploid wheat lines and 351 introgression lines. The ungenotyped D-genome SNPs in the introgression population were imputed from a reference panel of 116 Ae. tauschii accessions to improve IBD detection. The distribution of introgression across the genome was investigated to assess its overall effect on genetic diversity, and to evaluate the impact of recombination rate variation and early selection for uniform phenological and developmental characteristics on the introgression frequency in different parts of the wheat genome. The effect of phenotypic selection against a non-adaptive allele contributed by Ae. tauschii was investigated around the Tg gene on chromosome arm 2DS, which controls tenacious glume trait affecting grain threshability (Sood et al. 2009).
Materials and Methods
Plant material
The study population consisted of 351 BC1F3:5 Ae. tauschii introgression lines developed by crossing synthetic Ae. tauschii-wheat octoploid lines with hexaploid wheat recurrent parents. The octoploid lines were developed by crossing five hexaploid wheat parents with 21 Ae. tauschii accessions (Supporting Information Table S1, File S1). The resulting F1 hybrid plants regenerated from rescued embryos were treated with colchicine to produce the synthetic octoploids (Dale et al. 2017). The synthetic octoploids were backcrossed once to the respective hexaploid wheat parents or to another wheat line. The BC1F1 plants were self-pollinated and advanced by single seed descent to the BC1F3 generation. Seeds from individual BC1F3 plants were bulked and grown in single rows in the field at the Kansas State University Ashland Research Farm near Manhattan, KS in the 2016-17 growing season. A total of thirty-one families were represented in this experiment. The number of lines per family ranged from 42 to 137, and resulted in a total of 2,861 lines that were planted. The 351 lines used in our study were selected in 2017 from this set of lines. Selection criteria included production of sufficient seed to allow yield testing, general fitness, threshability to allow mechanical harvest and phenology similar to the elite hexaploid parent(s). In addition, 116 diverse Ae. tauschii accessions representing Ae. tauschii ssp. tauschii and Ae. tauschii ssp. strangulata from different geographical locations were used as the reference panel to impute ungenotyped SNPs in the study population to improve detection of region identical-by-descent (IBD) (Supporting Information Table S2).
Sequencing complexity-reduced genomic libraries
DNA from the Ae. tauschii introgression population and the reference panel samples was extracted using DNeasy 96 Plant DNA extraction kit (Qiagen) following the manufacturer’s protocol. The quality and concentration of the DNA was assessed using PicoGreen dsDNA assay kit (Life Technologies). Input DNA was normalized to 400 ng (20ul of 20ng/ul) using Qiagility robot (Qiagen). Genotyping by sequencing (GBS) libraries were constructed using the protocol described by Saintenac et al. (2013), and then subjected to size selection using the Pippin Prep system (Sage Scientific) to enrich for 270-330 bp fragments. In total, five libraries were produced, representing 80 barcoded accessions each. Each library was sequenced on Illumina NextSeq 500 using a 1 × 75 bp kit for the introgression lines and 1 × 100 bp kit for the reference panel following the Illumina protocol. TASSEL 5.0 GBS v2 pipeline (Glaubitz et al. 2014) was used to generate SNPs from the fastq files of the introgression lines and the reference panel. In brief, the raw GBS sequence reads were aligned to the Chinese Spring reference sequence v1.0 (The International Wheat Genome Sequencing Consortium (IWGSC) 2018) using Burrow’s Wheeler Alignment (BWA) software v0.7.17. TASSEL 5.0 GBS v2 default parameters were used in all steps (Glaubitz et al. 2014), except the KmerLength for the introgression population was set at 35 to account for shorter read length.
SNP genotyping and imputation
SNPs for the reference Ae. tauschii panel with minor allele frequency (MAF) less than 0.02 and maximum missingness greater than 50% were filtered out using vcf-filter tools v0.1.13. The missing SNPs were imputed using the program Beagle v.5.0 (Browning and Browning 2013) with default parameters (File S2). SNPs from Ae. tauschii derived introgression lines were filtered in two steps. First, SNPs from all sub-genomes (A, B and D) with minor allele frequency (MAF) less than 0.05 and maximum missingness greater than 30% were filtered out using vcf-filter tools v0.1.13. The missing SNPs were imputed using the program Beagle v.5.0 with default parameters. In the next step, all A and B genome SNPs, and D genome SNPs with MAF less than 0.01 were excluded from the raw vcf file using vcf-filter tools v0.1.13. The program conform-gt (https://faculty.washington.edu/browning/conform-gt.html) was used to check the concordance of D genome SNP positions between the introgression population and the reference panel based on the Chinese Spring genome v1.0 coordinates (IWGSC, 2018). Missing and ungenotyped SNPs in the D genome of the introgression population were imputed from the reference panel using Beagle v.5.0 (File S3).
Principal component analysis (PCA)
The population structure of the diverse Ae. tauschii accessions and the introgression population was analyzed using the 27,880 D genome SNPs segregating in both populations. The SNP dataset was converted to the hapmap format and imported into TASSEL v.5.0, which was used to calculate the principal components. The first two components were plotted to show the distribution and clustering of the reference panel accessions in relation to the 21 parental Ae. tauschii accessions and the entire introgression population. In addition, a total of 13,970 SNPs (File S4), including 4,039, 4,255, 5,222 and 454 from A, B, D genomes and unanchored scaffolds, respectively, were used to evaluate the distribution of Ae. tauschii-derived introgression lines on the first two principal components using the wheat parents as grouping factors.
Genetic diversity
To evaluate the effect of introgression on SNP diversity, the mean number of base differences for each D genome SNP site in all pairwise comparisons (π) among Ae. tauschii accessions, introgression lines, and hexaploid wheat lines were calculated using vcftools v0.1.13 and summarized using basic R functions (R Development Core Team 2011). The π values for each chromosome were interpolated by calculating the average of ten SNPs sliding forward by two SNPs using a Perl script and plotted using R package ‘ggplot2’ v3.2.1. The set of D genome SNPs used in this analysis was 5,222 that were retained after filtering out SNPs with minor allele frequency less than 0.05 and maximum missingness greater than 30% (File S4).
Recombination hotspots
The distribution of recombination hotspots was analyzed using the imputed D-genome SNPs split into subsets based on families. A combination of custom Perl and R scripts (Nyine et al. 2018), were used to convert the SNP alleles to 0, 1, and 2, of which, 0 is homozygous major allele, 1 is heterozygous and 2 is homozygous minor allele. Regions containing monomorphic SNPs were eliminated by the R script. A total of 16 families each having at least 10 progenies plus the respective parents were used in this analysis. A separate custom Perl script was used to count the number of allele phase transitions in each chromosome per individual and recode the flanking SNP positions as break points (Jordan et al. 2018). The number of recombination breakpoints (RBP) per 2 Mb sliding window with 1 Mb step size in each chromosome per family was obtained using bedmap option from BEDOPS v2.4.35 (Neph et al. 2012). The total RBP per 2 Mbp window across the 16 families was determined and the windows within the 95th percentile were considered as recombination hotspots. The centromere position on each chromosome was based on the Chinese Spring reference genome v1.0 (The International Wheat Genome Sequencing Consortium (IWGSC) 2018; Su et al. 2019). Kruskal Wallis test was used to test for significant differences in the distribution of recombination breakpoints in each family.
In order to investigate the effect of sequence divergence (SD) and structural re-arrangements on recombination, we compared hexaploid wheat (Chinese Spring) and the diploid relative, Ae. tauschii ssp. strangulata (AL8/79) D genomes at the protein level. High confidence D genome gene protein sequences from Chinese Spring v.1.0 and Ae. tauschii v.4.0 (Luo et al. 2017) were used. The annotation of the Ae. tauschii genome was downloaded from http://aegilops.wheat.ucdavis.edu/ATGSP/annotation/. Local protein BLAST databases were created for each dataset using BLAST+ v2.7.1. Reciprocal blastp was performed between the two species’ genome proteins using default parameters. A Perl script was used to filter out blast hits with percent identity less than 95 and gap opens greater than 0. A file consisting of species chromosome identity, gene name, gene start and end positions was generated from the respective gff3 file. MCScanX software (Wang et al. 2012) was used to generate the dot plot and dual synteny plot that were used to compare the structural differences between the genome of T. aesitvum and Ae. tauschii.
The difference in recombination rate between lines carrying introgression from Ae. tauschii ssp. strangulata or Ae. tauschii ssp. tauschii was assessed by comparing the sequence divergence (SD) of the parents with introgression frequency (IF) and the total RBPs estimated from the progenies in six introgression families. A total of 5,222 high stringency filtered D genome SNPs were used to determine the RBP and SD while IBD was used to infer introgression frequency in 5 Mbp chromosome windows. SD in our study corresponds to the average number of polymorphic sites between Ae. tauschii and hexaploid wheat parents within 5 Mbp windows. Chromosomes were divided into 2/3 pericentromeric and distal regions. Mann-Whitney U-test was used to determine if significant difference in SD, IF and RBP existed between ssp. strangulata and ssp. tauschii-derived families.
Identity by descent detection (IBD)
Introgression of Ae. tauschii genome in hexaploid wheat was inferred using IBD. The D genome SNPs from each chromosome were separated and used as input genotype (gt) data for IBD detection. The program Beagle v.4.1 was used to detect IBD segments between introgression lines, hexaploid wheat and Ae. tauschii parents using default parameters. The R-package ggplot2 v3.2.1 was used to generate a density plot of IBD segment start per chromosome to show the distribution pattern. All chromosomes were scaled by dividing the IBD values by the individual chromosome length and then multiplied by 100.
The efficiency of introgression was estimated as the percentage of observed proportion of Ae. tauschii genome (introgression frequency) in the introgression lines as inferred by IBD to the expected proportion of Ae. tauschii in BC1F3:5. Assuming that recombination events between Ae. tauschii and hexaploid wheat D genomes occurred normally in each chromosome, the expected proportion of Ae. tauschii genome in the BC1F3:5 introgression lines was approximated at 25%. The observed proportion of introgression was obtained by dividing the total length of IBD segments from Ae. tauschii shared with each line by the genome size of Ae. tauschii (4.3 Gb) and multiplied by 100. The result was divided by 25 and multiplied by 100 to get the percentage introgression efficiency. The average, standard deviation, minimum and maximum IBD length shared between introgression lines, introgression lines and hexaploid wheat, introgression lines and Ae. tauschii parents were determined, and divided by the chromosome size.
The relationship between IBD and the Tg gene on chromosome arm 2DS was explored. The IBD count per 1 kb sliding window was used to compare the frequency of introgression in the Tg region. Genes within the Tg region (21.8 Mbp to 23.3 Mbp) and their functional annotation were extracted from the Chinese Spring reference gene annotation file. Introgression lines were phenotyped for the tenacious glume trait. The results were used to confirm the presence or absence of wild type alleles depending on whether the introgression segment spanned the Tg gene region or not. Genome-wide association analysis of tenacious glume trait with the 27,880 SNP markers was done using GAPIT function in R. A mixed linear model was used and the population structure was controlled using the first three principal components calculated from the markers. A Manhattan plot of negative log10 of false discovery rate (FDR) transformed P-values from the D chromosomes was generated in R using ‘qqman’ package.
Data availability
All supplemental material and relevant data are available at FigShare. Raw sequence reads were deposited at NCBI’s Sequence Read Archive (SRA) under BioProjectID PRJNA637316 and PRJNA637416 and will be accessible upon release through the links below. https://www.ncbi.nlm.nih.gov/sra/PRJNA637316, https://www.ncbi.nlm.nih.gov/sra/PRJNA637416. Supplemental material available at figshare:https://doi.org/10.25387/g3.12724943.
Results
Genotyping and SNP imputation
A total of ∼315 million high-quality NGS reads with barcodes were generated with an average of ∼2.8 million reads per sample from the diverse Ae. tauschii accessions (Supporting Information Table S2). Eighty-eight percent (88%) of the reads were aligned to the Chinese Spring reference sequence v.1.0 (The International Wheat Genome Sequencing Consortium (IWGSC) 2018) with an average of ∼2.4 million reads per sample. The number of SNP sites generated from the TASSEL v. 5.0 GBS v.2 pipeline was 148,430. After filtering out SNPs with MAF less than 0.02, and maximum missingness greater than 50%, the number of retained SNPs was 96,056.
Similarly, ∼1.1 billion high quality reads with barcodes were generated from the introgression population, with an average of ∼3.0 million reads per sample (Supporting Information Table S1). Ninety-six percent (96%) of the reads were aligned to the Chinese Spring reference v1.0 with an average of ∼2.9 million reads per sample. Samples with less than 91,000 reads were excluded from downstream analysis. The number of unfiltered SNPs generated by the TASSEL v.5.0 GBS v.2 pipeline was 281,846. A total of 13,970 SNPs from the A, B, D genomes and unanchored scaffolds were retained after filtering out SNPs with MAF less than 0.05 and maximum missingness greater than 30%. The number of SNPs from the D genome was 37.4% of the filtered SNP dataset. A second round of filtering was performed on the D genome SNPs to remove sites with MAF less than 0.01 resulting in 142,740 SNPs, out of which, 27,880 also segregated in the diverse set of Ae. tauschii accessions (henceforth, reference panel). Using the program Beagle v.5.0 (Browning and Browning 2013), 85,752 SNPs were imputed from the reference panel into the Ae. tauschii-derived introgression population.
Principle component analysis
We used the analysis of principal components to assign the Ae. tauschii accessions used as parents for introgression population development to the previously identified lineages (Wang et al. 2013). The 137 Ae. tauschii accessions (116 reference panel plus 21 parental accessions) formed three distinct clusters when the first two PCs calculated from 27,880 SNPs were plotted (Fig. S1), with 79.2% of variance explained by the first two PCs. One of the three clusters included accessions known to belong to Ae. tauschii ssp. strangulata or lineage 2 (L2) (Wang et al. 2013). The remaining two clusters included accessions that belong to Ae. tauschii ssp. tauschii or lineage 1 (labeled L1a and L1b in Fig. S1). Cluster L1a corresponds to lineage 1E from Wang et al. (2013) study, and includes accessions coming from Afghanistan (AFG), Turkmenistan (TKM), Iran (IRN), Pakistan (PAK) and Tajikistan (TJK) (Table S3). Fifteen of the Ae. tauschii parents used to generate the introgression population were assigned to this cluster. More than two thirds of the accessions in cluster L1b, corresponding to previously identified lineage 1W (Wang et al. 2013), were from Turkey (TUR) with only a few accessions from Armenia (ARM), IRN, TJK and PAK. Consistent with previous findings showing that lineages 1E and 1W are less genetically diverse than linage 2 (Wang et al. 2013), L1a and L1b groups were separated along PC2-axis, which explains only 2.7% of variance (Fig. S1). Three parents of the introgression population were present in this cluster. Cluster L2 consisted of Ae. tauschii accessions mostly collected from Iran (IRN), although a few accessions from Azerbaijan (AZE), Turkmenistan (TKM), Syria (SYR) and TUR were present. Three parents of the introgression population parents clustered in this group and two of them (TA1642, TA2378) are known to belong to Ae. tauschii ssp. strangulata or lineage 2 (Wang et al. 2013; Singh et al. 2019).
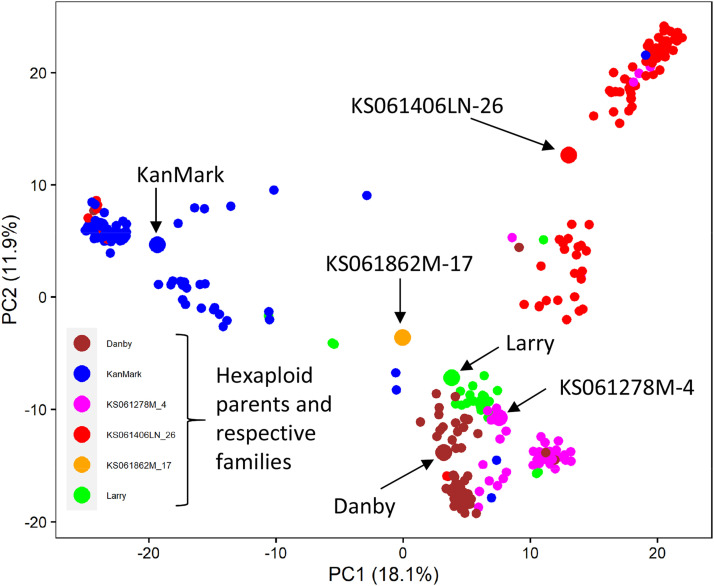
When the introgression lines were plotted on the first two PCs together with Ae. tauschii accessions and hexaploid wheat parents, clusters L1a and L1b were less distinguishable from each other (Fig. S2). The total variance explained by the first two PCs was 56.7%. The cluster including Ae. tauschii accessions from lineage L2 was located in close proximity to cluster including hexaploid wheat parents (HW) consistent with earlier studies demonstrating the origin of the wheat D genome from the Ae. tauschii ssp. strangulata lineage (Dvorak et al. 1998; Wang et al. 2013). The introgression lines did not form a clearly distinguishable cluster, and were scattered broadly, indicating a wide range of variation in the proportion of the genome introgressed from the different lineages of Ae. tauschii (Fig. S2). Many introgression lines clustered closer to the hexaploid wheat parents indicating that the greater proportion of genome in the BC1F3:5 lines comes from hexaploid wheat. This trend is likely associated with the loss of the introgressed segments as a result of backcrossing to the hexaploid parents and selection during population development. When the introgression lines were compared with the hexaploid wheat parents using 13,970 SNPs from all three sub-genomes, clustering was mostly consistent with the pedigree based on hexaploid parents (Figure 1). The rare cases of co-clustering of introgression lines from different families were likely due to the misclassification of introgression lines’ pedigree or the shared Ae. tauschii parents.
Figure 1.
Distribution of the introgression lines and the hexaploid wheat parents on the first two principal components based on SNP markers from A, B, D genomes and unanchored scaffolds.
Distribution of SNP diversity in the introgression population
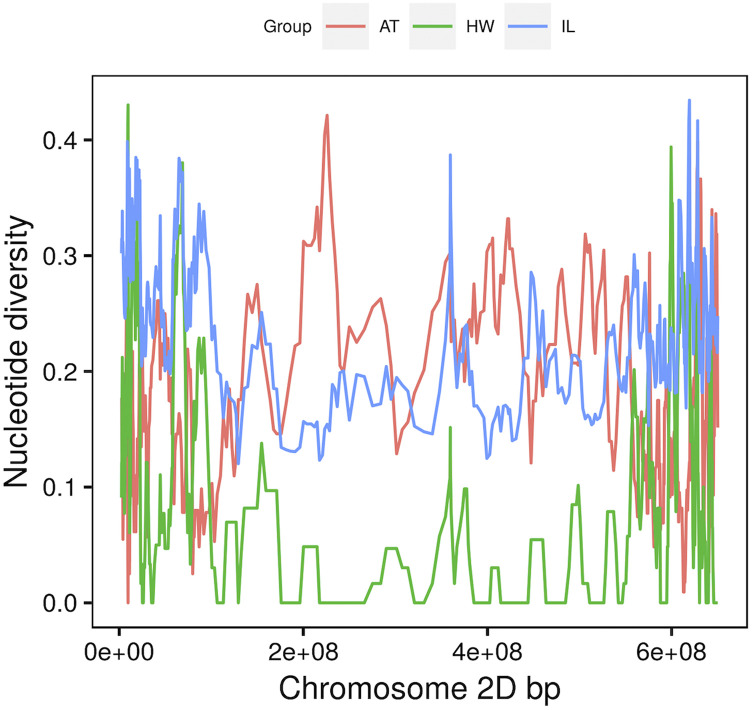
To assess the effect of wild relative introgression on genetic diversity in wheat, we estimated SNP diversity (π) in the populations of Ae. tauschii, hexaploid wheat parents, and the introgression lines. The average per-site estimate of π for the diverse Ae. tauschii populations was 0.304. Consistent with previous studies (Akhunov et al. 2010; Jordan et al. 2015; Balfourier et al. 2019; Pont et al. 2019), a cross-population diversity comparison showed a low average SNP diversity in the wheat D genome across all chromosomes (Table 1). The lowest diversity was found in the hexaploid wheat parents with the chromosome mean ranging from 0.003 to 0.012 as compared to Ae. tauschii parents that ranged from 0.118 to 0.131. For most chromosome regions, the levels of SNP diversity in the introgression population were intermediate between the levels of diversity in the parental populations of wheat and Ae. tauschii, but tended toward the Ae. tauschii with maximum mean π of 0.122 on chromosome 4D (Figure 2 and Fig. S3). Analysis of variance showed significant differences in π values between Ae. tauschii, hexaploid wheat, and introgression lines, and between chromosomes (P < 0.001). The SNP diversity of the introgression lines for most regions of chromosome 4D and 5D were higher than those of Ae. tauschii parents (Fig. S3). Taken together, these results indicate that Ae. tauschii introgression substantially increased the SNP diversity of the recurrent hexaploid wheat parents.
Table 1. A comparison of nucleotide diversity of Ae. tauschii derived introgression lines and their parents.
| Group | Min. | Mean | Max. |
|---|---|---|---|
| chr1D_AT | 0.000 | 0.118 | 0.626 |
| chr1D_HW | 0.000 | 0.012 | 0.545 |
| chr1D_IL | 0.000 | 0.085 | 0.501 |
| chr2D_AT | 0.000 | 0.122 | 0.654 |
| chr2D_HW | 0.000 | 0.011 | 0.546 |
| chr2D_IL | 0.000 | 0.066 | 0.504 |
| chr3D_AT | 0.000 | 0.128 | 0.664 |
| chr3D_HW | 0.000 | 0.0101 | 0.545 |
| chr3D_IL | 0.000 | 0.048 | 0.500 |
| chr4D_AT | 0.000 | 0.129 | 0.595 |
| chr4D_HW | 0.000 | 0.003 | 0.545 |
| chr4D_IL | 0.000 | 0.122 | 0.606 |
| chr5D_AT | 0.000 | 0.119 | 0.659 |
| chr5D_HW | 0.000 | 0.007 | 0.545 |
| chr5D_IL | 0.000 | 0.118 | 0.614 |
| chr6D_AT | 0.000 | 0.131 | 0.627 |
| chr6D_HW | 0.000 | 0.011 | 0.545 |
| chr6D_IL | 0.000 | 0.095 | 0.501 |
| chr7D_AT | 0.000 | 0.127 | 0.652 |
| chr7D_HW | 0.000 | 0.011 | 0.545 |
| chr7D_IL | 0.000 | 0.081 | 0.501 |
AT are Aegilops tauschii parents, HW are hexaploid wheat parents and IL are the introgression lines.
Figure 2.
Variation in nucleotide diversity for chromosome 2D based on per estimates of π for Ae. tauschii accessions (AT), hexaploid wheat (HW) lines and introgression lines (IL).
Genomic patterns of introgression and recombination rate
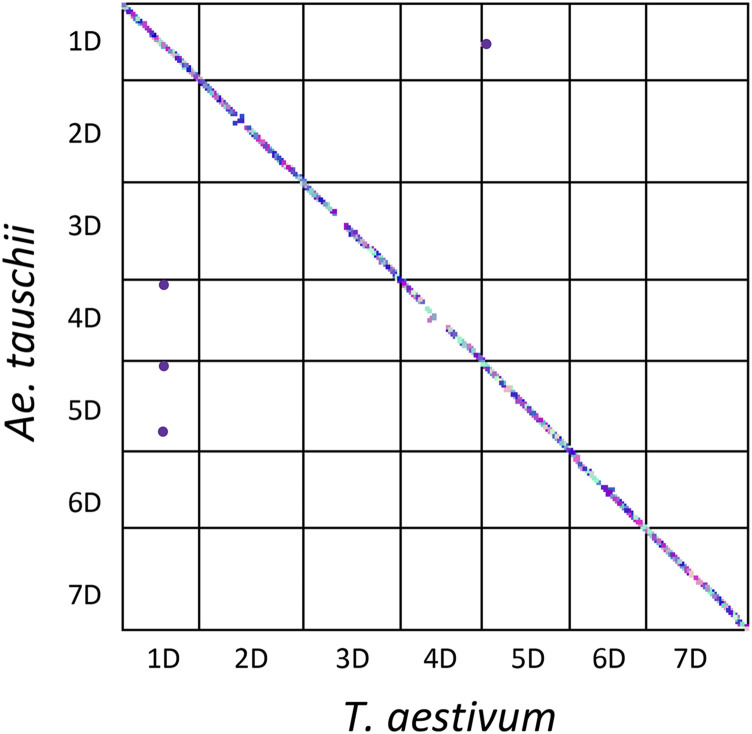
One of the factors affecting the frequency of introgression across the genome are structural re-arrangements (Rieseberg et al. 1995, 1999). Comparative analysis of gene order along the chromosomes showed that more than 99% of the genes from T. aestivum are perfectly collinear to those of Ae. tauschii ssp. strangulata suggesting a lack of major structural re-arrangements (Figure 3). Only small-scale inversions were observed on chromosomes 2D, 4D and 6D in the regions near the centromeres, and four genes were found in non-syntenic positions between the wheat (1D and 5D) and Ae. tauschii (1D, 4D and 5D) chromosomes. Therefore, it is unlikely that structural rearrangements could have a significant impact on the genomic patterns of introgression in the wheat-Ae. tauschii crosses.
Figure 3.
Gene collinearity and structural variation between the T. aestivum and Ae. tauschii chromosomes. Each dot on the dot-plot corresponds to a single gene and the dots off diagonal represent the non-syntenic genes between genomes of the two species.
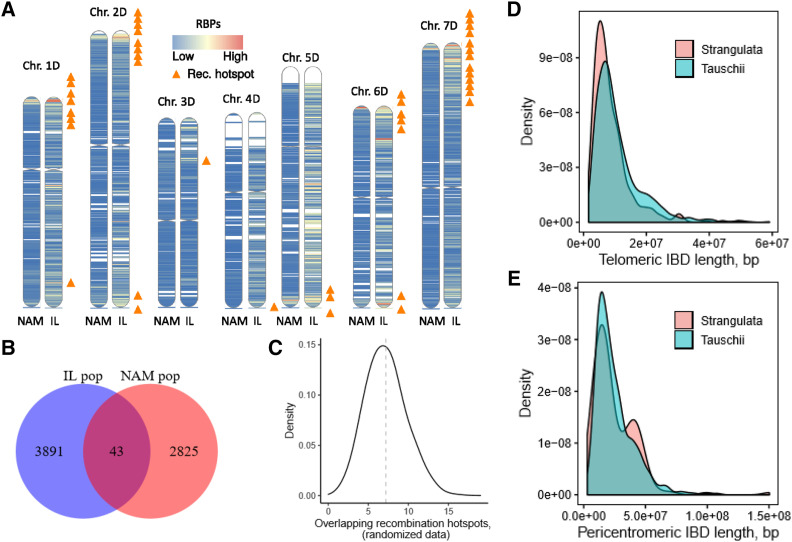
Interspecific introgression may also be influenced by the distribution of recombination rate along the chromosomes (Martin et al. 2019). Since strong depression of recombination in the pericentromeric regions creates steep recombination rate gradient along the centromere-telomere axis of large wheat chromosomes (Saintenac, Faure, et al. 2011; Jordan et al. 2018; Gardiner et al. 2019), we should expect distinct patterns of introgression in the pericentromeric and distal regions. To investigate the relationship between recombination and introgression, first, we calculated the number of recombination breakpoints (RBP) in 2 Mbp windows across the wheat genome across all families. Using the 95th percentile of window-based RBP estimates as a threshold, we identified 200 genomic regions with a high recombination rate (Table 2, Table S4). To test whether the distribution of these regions with elevated recombination is random or associated with specific genomic regions, we compared their locations with the positions of high-recombining regions previously detected in the nested association mapping (NAM) population (Jordan et al. 2018) (Figure 4a). We detected 43 overlapping regions with elevated recombination between these two populations (Table S4), mostly located in the distal chromosomal regions. This level of overlap was 6 times higher than the average overlap (7.2 RBPs) observed between the randomized datasets generated by permuting 2 Mbp windows in both introgression and NAM populations (Figure 4b and 4c), suggestive of the presence of region-specific genomic features promoting recombination.
Table 2. Summary of 2 Mb windows recombination hotspots at 95th percentile of total recombination breakpoints from 16 families of the introgression population.
| Chromosome | No. of windows | Min recombination | Max recombination |
|---|---|---|---|
| chr1D | 25 | 755 | 1968 |
| chr2D | 33 | 705 | 1441 |
| chr3D | 31 | 507 | 1136 |
| chr4D | 26 | 736 | 1499 |
| chr5D | 29 | 1052 | 1552 |
| chr6D | 24 | 899 | 1940 |
| chr7D | 32 | 869 | 1783 |
Figure 4.
Effect of chromosome physical location on recombination rate and introgression. (a) A comparison of normalized recombination rate in 2 Mbp windows between nested association mapping (NAM) (Jordan et al. 2018) and Ae. tauschii introgression (IL) populations. The orange triangles show the overlapping windows with high recombination rate in the two populations at 95th percentile. (b) A venn diagram showing the total number of 2 Mbp windows with at least one recombination event across the NAM and IL population while the intersection represents the overlaps at 95th percentile. (c) Distribution of 2 Mbp window recombination hotspots permutation overlaps between the two populations. (d) Distribution of the introgressed segment’s length from Ae. tauschii ssp. strangulata and Ae. tauschii ssp. tauschii in the distal regions of the chromosomes. (e) Distribution of the introgressed segment’s length from Ae. tauschii ssp. strangulata and Ae. tauschii ssp. tauschii in the 2/3 pericentromeric regions of the chromosomes.
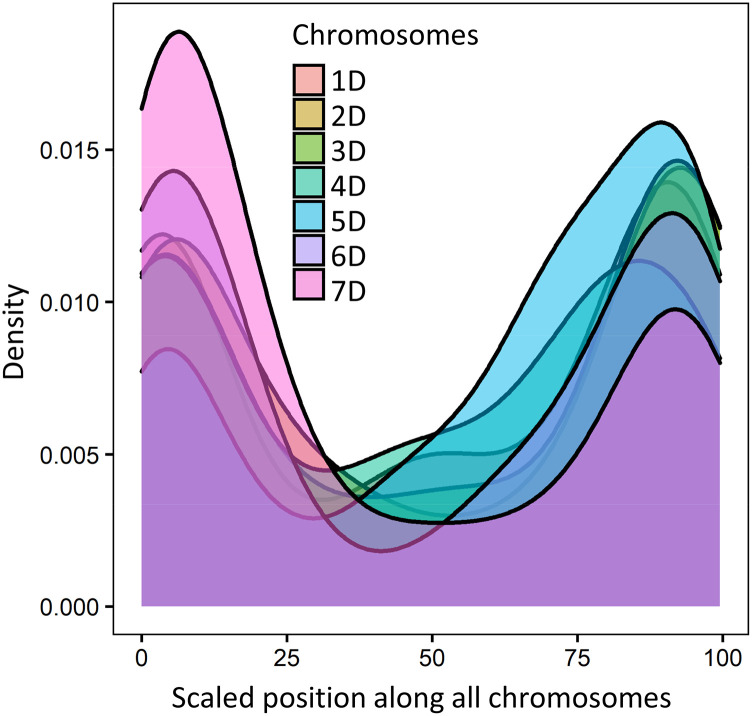
Second, we assessed the frequency of introgression along the wheat chromosomes. The IBD analysis was used to infer the boundaries of segments introgressed from Ae. tauschii into each line. A density plot of IBD segments along the chromosomes of the introgression population showed a U-shaped distribution (Figure 5). The frequency of IBD segments positively correlated with the distribution of recombination rate (Jordan et al. 2018) and increased from the centromeres toward the telomeric regions of the chromosomes except on chromosome 1DL and 5DL in some families (Table S5). There was no chromosome preference during introgression. Variation in the number of introgressed segments per line was observed across chromosomes with the proportion of introgressed Ae. tauschii genome per line ranging from 0.05 to 25.5% (Table S6), deviating from the expected 25% of Ae. tauschii genome in the BC1F3:5 lines. Therefore, the efficiency of introgression in the population ranged from 0.2 to 100%. Some lines had single introgression while others had multiple introgressions per chromosome (Table S7). The IBD segments shared between the introgression lines and wheat parents were on average 5.ninefold longer than those shared with the Ae. tauschii parents (Table 3), and significantly different at the 95% confidence level based on the t-test statistics (P < 2.2e-16). The average length of IBD segments, expressed as a percent of the D genome size, shared between introgression lines and Ae. tauschii varied from 2.08 to 3.42% with a minimum of 0.25% and a maximum of 57.19%. Similarly, the average percent length of IBD segments shared between the chromosomes of introgression lines and hexaploid wheat parents ranged between 8.59% and 25.74% with a minimum of 0.36% and a maximum of 97.06%.
Figure 5.
Density plots of identity by descent segments positions along the seven D genome chromosomes of the introgression lines derived from hexploid wheat and Ae. tauschii crosses. Chromosomes were scaled by dividing the IBD values by the individual chromosome length and then multiplied by 100.
Table 3. Percentage length of identity by descent segments shared between introgression lines, hexaploid wheat and Ae. tauschii accessions.
| Chromosome | Mean (%) | SD (%) | Min (%) | Max (%) |
|---|---|---|---|---|
| chr1Da | 3.03 | 5.15 | 0.30 | 57.19 |
| chr1Db | 12.77 | 12.70 | 0.80 | 57.19 |
| chr1Dc | 9.31 | 12.02 | 0.31 | 85.88 |
| chr2Da | 2.08 | 1.82 | 0.28 | 23.09 |
| chr2Db | 8.59 | 12.41 | 0.53 | 55.74 |
| chr2Dc | 9.17 | 13.88 | 0.25 | 90.78 |
| chr3Da | 2.10 | 1.80 | 0.31 | 19.71 |
| chr3Db | 23.84 | 28.01 | 0.77 | 97.06 |
| chr3Dc | 14.66 | 21.85 | 0.25 | 98.36 |
| chr4Da | 3.42 | 2.34 | 0.35 | 17.85 |
| chr4Db | 25.74 | 23.17 | 3.86 | 84.17 |
| chr4Dc | 11.65 | 13.67 | 0.30 | 100.00 |
| chr5Da | 2.27 | 1.63 | 0.33 | 17.47 |
| chr5Db | 11.08 | 7.16 | 1.86 | 49.01 |
| chr5Dc | 7.38 | 9.66 | 0.31 | 85.24 |
| chr6Da | 3.01 | 2.59 | 0.37 | 17.89 |
| chr6Db | 14.43 | 15.86 | 0.47 | 88.21 |
| chr6Dc | 9.94 | 13.66 | 0.00 | 95.94 |
| chr7Da | 2.32 | 2.21 | 0.25 | 15.71 |
| chr7Db | 11.72 | 18.33 | 0.36 | 73.30 |
| chr7Dc | 8.19 | 12.92 | 0.24 | 92.46 |
Relationship between sequence divergence, recombination and introgression length and frequency
The low rate of introgression in the pericentromeric regions may be the consequence of negative multilocus epistasis (Jiang et al. 2000; Martin et al. 2019) associated with introgression of large chromosomal segments. We compared the distribution of introgression segment’s length in the distal and pericentromeric regions of chromosomes between families derived from crosses with Ae. tauschii ssp. strangulata (direct donor of the wheat D genome), which shows a low level of divergence from the wheat D genome, and with Ae. tauschii ssp. tauschii, which shows higher levels of divergence from both the wheat D genome and Ae. tauschii ssp. strangulata (Figures 4d and 4e) (Wang et al. 2013). No substantial differences in the length of introgressed segments were observed between the two subspecies in the distal regions. The mean length of introgressed segments was 8.8 Mbp (2.0 – 49.1 Mbp) for Ae. tauschii ssp. strangulata and 10.9 Mbp (1.6 – 59.1 Mbp) for Ae. tauschii ssp. tauschii. However, in the pericentromeric regions, we found an increase in the length of introgressed segments from Ae. tauschii ssp. strangulata (2.8 – 150 Mbp) compared to that from Ae. tauschii ssp. tauschii (3.9 – 107 Mbp) (Figure 4e). Even though the mean length of introgression was not significantly different (24.5 Mbp and 24.7 Mbp, respectively) between the subspecies, Ae. tauschii ssp. strangulata showed bi-modal introgression size distribution, with a second density peak corresponding to introgressed segments with the mean of ∼40 Mbp (Figure 4e).
We also compared the parental sequence divergence (SD) in the 2/3 pericentromeric and telomeric regions with IF between three Ae. tauschii ssp. strangulata derived families (FAM92, FAM93 and FAM96) and three Ae. tauschii ssp. tauschii derived families (FAM97, FAM98 and FAAM99) (Fig. S4). The mean SD was significantly different between the two subspecies with ssp. strangulata exhibiting a fourfold lower SD (1.47 SNPs / 5 Mbp window) than ssp. tauschii (5.78 SNPs / 5 Mbp window) in the pericentromeric region with Mann-Whitney U-test p-value < 2.2e-16. The low SD in the pericentromeric region of ssp. strangulata and hexaploid wheat parents was associated with a twofold increase in IF in the progenies. Similarly, SD and IF were significantly different in the telomeric regions of the two subspecies with Mann-Whitney U-test p-value = 1.105e-15. The mean telomeric SD between hexaploid wheat and Ae. tauschii ssp. strangulata parents (10.6 SNPs / 5 Mbp window) was ∼2-folds lower than that between hexaploid wheat and Ae. tauschii ssp. tauschii parents (19.45 SNPs / 5 Mbp window). This resulted into a 1.threefold increase in introgression frequency from ssp. strangulata (4.07) compared to that from ssp. tauschii (3.06). Taken together, these results suggest that the lower levels of divergence between parental genomes is linked with improved introgression efficiency, likely due to reduced negative multilocus epistasis.
The sequence divergence between the parental genomes might influence the frequency of homologous recombination (Opperman et al. 2004). In the pericentromeric regions, the mean number of RBPs estimated from ssp. strangulata (0.89) was 4-times lower than that estimated from ssp. tauschii derived families (3.44) (Fig. S4). Similarly, in the distal chromosomal regions, the estimated mean number of RBPs was higher in ssp. tauschii (5.94) than in ssp. strangulata (2.62) derived families. The high number of RBPs observed in ssp. tauschii compared to ssp. strangulata derived families may be explained by biases resulting from selection in the progenies of each family, and the low level of SNP diversity between the wheat D genome and Ae. tauschii ssp. strangulata, which is considered to be the donor of the wheat D genome (Dvorak et al. 1998), resulting in an underestimation of the total number of crossovers in the ssp. strangulata derived families. It is also possible that the increase in the levels of interhomolog polymorphism may stimulate recombination. In Arabidopsis, an increase in crossovers was observed when heterozygous regions were juxtaposed with homozygous regions (Ziolkowski et al. 2015), suggesting that the genomic distribution of interhomolog divergence have substantial effect on distribution of recombination rate.
Patterns of introgression near the Tg locus
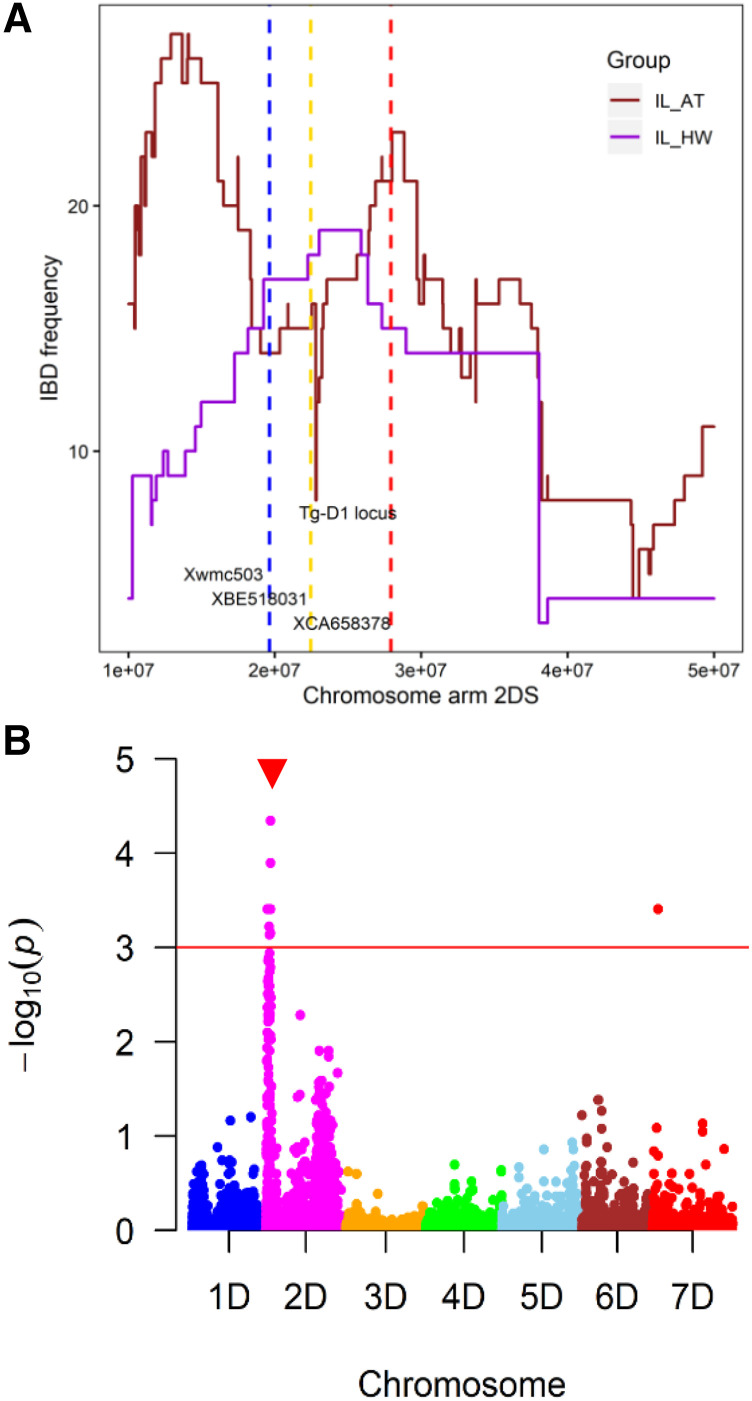
The free-threshing trait in wheat is controlled by the recessive variant of the Tg gene and the dominant variant of the Q gene (Simons et al. 2006; Sood et al. 2009; Dvorak et al. 2012; Faris et al. 2014). To understand the effect of selection against the hulled trait controlled by the dominant variant of Tg from Ae. tauschii, we investigated the patterns of IBD around the Tg locus shared by the introgression population with the Ae. tauschii parental genomes. We expected that the selection for mechanical seed threshing imposed during the introgression population development should result in a deficiency of the dominant Tg allele from Ae. tauschii, and increased frequency of the recessive tg allele contributed by the hexaploid wheat parents. To test this hypothesis, we identified the boundaries of the Tg-D1 locus on chromosome 2D using two markers based on the expressed sequence tags (ESTs), CA658378 and BE518031 (Dvorak et al. 2012). These markers mapped to the reference wheat genome at positions 22,447,585 bp (BE518031) and 27,924,545 bp (CA658378). Additionally, the sequences of microsatellite markers Xgwm455, Xgwm296, Xgwm261 and Xwmc503 linked to Tg were aligned to the Chinese Spring reference v.1.0. Marker Xwmc503 closest to Tg gene mapped at 19.6 Mbp on 2DS (Table S8). Based on Sood et al. (2009) genetic map, the Tg gene is located 2.2 cM away from marker Xwmc503, implying that the Tg gene is located approximately at position 21.8 Mbp, which is consistent with the genomic interval flanked by markers based on CA658378 and BE518031 sequences. A count of IBD segments within 1-kbp sliding windows showed a steep decline in IBD between introgression lines and Ae. tauschii parents within the Tg gene region (Figure 6a). This trend was accompanied by an increase in the proportion of IBD segments shared between the introgression lines and the hexaploid wheat parents, indicating a selection pressure for free-threshing trait during population development. The lowest decline in IBD segments count was observed around 22.8 Mbp. There are 40 high confidence genes within the 21.8 Mbp to 23.3 Mbp interval (Table S9), including two transcription factors from the bZIP and GRAS families.
Figure 6.
Location of Tg locus on chromosome arm 2DS as inferred by identity by descent (IBD) analysis and genome-wide association mapping. a). Shows the frequency of introgression from Ae. tauschii into hexploid wheat as inferred by IBD for chromosome arm 2DS region containing tenacious glume (Tg) gene. The IBD segments were counted per 1-kb sliding window. Where IL_AT and IL_HW are the trends of IBD frequency between introgression lines and Ae. tauschii, and hexaploid wheat, respectively. The blue line shows the position of marker Xmwc503 (Sood et al. 2009), gold and red lines indicate the positions of EST markers XBE518031 and XCA658378, respectively, flanking the Tg-D1 locus. b). Manhattan plot showing the position of significant SNPs on 2DS (red arrow) and the red horizontal line shows the SNPs that are significant at an FDR q-value of 0.001.
To verify the impact of introgression on free-threshing, we phenotyped a subset of introgression lines for tenacious glume trait and compared the results with the IBD map (Figure 6a). Most lines that had introgression spanning the Tg gene region were positive for tenacious glume trait (Table S10). Some lines that were shown to carry introgression at the Tg locus were not available for phenotyping because they were removed during harvesting by combine. As expected, lines showing no or small overlap of the introgressed segments with the Tg region were negative for tenacious glume trait.
Genome-wide association mapping was used to identify markers associated with the threshability trait. Using a mixed linear model while controlling for the population structure, we observed that the majority of the significant SNPs associated with tenacious glume trait in the introgression population were located on chromosome arm 2DS (Figure 6b), which was consistent with the results of the IBD analysis. At a threshold FDR q-value of 0.05, 84 SNPs distributed along the Tg locus on 2DS showed a significant association with the trait, consistent with the previously reported maps (Table S11).
Discussion
Loss of genetic diversity associated with domestication and improvement (Haudry et al. 2007; Akhunov et al. 2010; Özkan et al. 2011; Xu et al. 2012; Hufford et al. 2012) can potentially reduce the adaptive potential of cultivated crops. Wild relatives found in regions with diverse agroecological environments are a valuable source of adaptive diversity that can improve resilience to biotic and abiotic stressors, or positively impact yield and quality traits (Uauy et al. 2006; Reynolds et al. 2007; Sohail et al. 2011; Cooper et al. 2012; Saintenac, Jiang, et al. 2013b; Periyannan et al. 2013; Wulff and Moscou 2014; Chen et al. 2015). However, effective deployment of wild relative diversity in breeding requires a better understanding of the factors that represent a barrier for introgression into adapted germplasm. In this study, we used the progenies of crosses between bread wheat and lines carrying the genome of the diploid wild relative Ae. tauschii to investigate factors that have potential to impact the genomic patterns of introgression in the D genome of wheat.
As expected, we observed an increase in the number of polymorphic sites after introgression, with the increase being higher in the families derived from crosses involving Ae. tauschii ssp. tauschii rather than in the families derived from crosses with Ae. tauschii ssp. strangulata. The latter is consistent with the origin of the wheat D genome from Ae. tauschii ssp. strangulata lineage (Dvorak et al. 2012; Wang et al. 2013). In the families derived from both lineages of Ae. tauschii, we observed an uneven distribution of introgressed diversity along the chromosomes. The frequency of IBD regions shared with Ae. tauschii along all chromosomes showed a U-shaped distribution with lower incidence of introgression in the pericentromeric regions, indicating that the pericentromeric regions of chromosomes represent a more significant barrier to introgression than the distal chromosomal regions.
The introgression frequency correlated negatively with the length of IBD regions, sequence divergence between the parental genomes and positively with recombination rate, suggesting that longer introgressed segments in the low-recombining pericentromeric regions had a lower chance of being inherited in the progeny of crosses between Ae. tauschii-derived octoploids and wheat. These chromosomal patterns of introgression efficiency and length suggest that introgression was strongly affected by the distribution of recombination rate along chromosomes. This conclusion is consistent with recent research on butterflies that showed that the rate of inter-species introgression could be predicted by variation in recombination rate (Martin et al. 2019). It is likely that selection applied at BC1F3:4 generation to maintain uniform phenology, threshability, flowering time, and developmental characteristics inadvertently eliminated many lines carrying large introgressed regions in the pericentromeric regions. Our results also suggest that introgression from less divergent wild relative have lower probability of being eliminated from population than introgression from more divergent wild relative. Negative selection against introgression was previously observed in the interspecific crosses of polyploid cotton (Jiang et al. 2000), and attributed to multilocus negative epistasis between introgressed alleles and the genetic backgrounds of recipient lines. According to theory, introgressions that carry alleles having a negative impact on the selected traits will be removed from the population, with the size of the affected region defined by the recombination rate (Hill and Robertson 1966). It appears that a negative interaction between alleles located within the large introgressions in the low-recombining pericentromeric region and alleles of the adapted recurrent parent affected phenotypes targeted by selection, resulting in a loss of certain genotypes during population development. Thus, the limited number of recombination events at the BC1F2 generation, especially in the large pericentromeric regions of wheat chromosomes, resulted in linkage drag that affected a substantial proportion of the genome.
On the contrary, the terminal regions of the wheat chromosomes showed a high rate of introgression consistent with the theoretical predictions of the effect of selection on linked variation (Hill and Robertson 1966). In these regions, negatively selected alleles could be separated from the introgressed segments, as was clearly demonstrated for the Tg locus controlling free-threshing trait in wheat (Jantasuriyarat et al. 2004; Sood et al. 2009; Dvorak et al. 2012; Faris et al. 2014). Since this gene is located in the high-recombining terminal region of chromosome, we did not observe a substantial effect of selection against the wild-type allele on the frequency of introgression from Ae. tauschii outside of the Tg locus. The high recombination rate allowed for separating the negatively selected dominant Tg allele from the introgression and mapping the Tg gene locus to a 1.5 Mbp genomic interval, which was further confirmed by genome-wide association mapping.
Consistent with earlier studies in the field of speciation genomics (Martin and Jiggins 2017; Martin et al. 2019), barriers to introgression in wheat likely have polygenic nature with multiple negatively selected alleles affecting agronomic performance traits distributed across entire genome. As a result, the unintended consequence of selection applied at the early generation of interspecific crosses is the low rate of introgression in the large low-recombining regions of wheat chromosomes. This outcome suggests that multiple genes with a strong combined effect on adaptive traits are present in these regions, and identification of beneficial alleles in these regions will be complicated by linkage drag.
It is common practice for germplasm development programs to subject lines to selection pressure from early stages in population development. This approach is consistent with the goal of quickly identifying high performing lines to support commercial breeding, and allows for rapid exploitation of beneficial alleles in the chromosomal regions with high recombination. While this is a worthy objective, our results are a clear justification for a two-tiered approach to germplasm development aimed to maximize the exploitation of genetic diversity present in wild relatives. The first step is to ensure that maximum diversity is maintained in the introgression population. This could be achieved by applying low selection pressure and genotyping early generation populations to select subsets of lines carrying introgressions covering the genome. The second step is to maintain introgressed segments in the heterozygous state through random mating using approaches based on genetic male sterility or chemical hybridizing agents (Qi and Gill 2001; Singh et al. 2015; Guttieri 2019), that are warranted in self-pollinated species. This step would enhance effective recombination and increase the probability of separating beneficial alleles from the influence of linked negatively selected alleles in the regions of low recombination.
Recently, genetic factors controlling crossover rate across the genome and in the pericentromeric regions of wheat chromosomes have been identified (Jordan et al. 2018; Gardiner et al. 2019). The discovery of these genetic factors could also facilitate strategies to further increase the efficiency of introgression, and selection for favorable introgressed alleles in the low recombining regions. Failure to engage such strategies will result in the substantial loss of introgressed diversity across large portions of the genome, reducing the potential long-term impact of germplasm development programs.
Acknowledgments
This project was supported by the Agriculture and Food Research Initiative Competitive Grant 2016-67013-24473 from the USDA National Institute of Food and Agriculture. We would like to thank Bliss Betzen for preparing genomic libraries, Alina Akhunova and KSU Integrated Genomics Facility for sequencing genomic libraries, and Jon Raupp from Wheat Genetics Resources Center for providing seeds of 21 accessions of Aegilops tauschii used for developing introgression population.
Footnotes
Supplemental material available at figshare: https://doi.org/10.25387/g3.12724943.
Communicating editor: A. Paterson
Literature Cited
- Akhunov E. D., Akhunova A. R., Anderson O. D., Anderson J. A., Blake N. et al. , 2010. Nucleotide diversity maps reveal variation in diversity among wheat genomes and chromosomes. BMC Genomics 11: 702 10.1186/1471-2164-11-702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avni R., Nave M., Barad O., Baruch K., Twardziok S. O. et al. , 2017. Wild emmer genome architecture and diversity elucidate wheat evolution and domestication. Science 357: 93–97. 10.1126/science.aan0032 [DOI] [PubMed] [Google Scholar]
- Balfourier F., Bouchet S., Robert S., DeOliveira R., Rimbert H. et al. , 2019. Worldwide phylogeography and history of wheat genetic diversity. Sci. Adv. 5: eaav0536 10.1126/sciadv.aav0536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning S. R., and Browning B. L., 2012. Identity by descent between distant relatives: detection and applications. Annu. Rev. Genet. 46: 617–633. 10.1146/annurev-genet-110711-155534 [DOI] [PubMed] [Google Scholar]
- Browning B. L., and Browning S. R., 2013. Improving the accuracy and efficiency of identity-by-descent detection in population data. Genetics 194: 459–471. 10.1534/genetics.113.150029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Rouse M. N., Zhang W., Jin Y., Akhunov E. et al. , 2015. Fine mapping and characterization of Sr21, a temperature-sensitive diploid wheat resistance gene effective against the Puccinia graminis f. sp. tritici Ug99 race group. Theor. Appl. Genet. 128: 645–656. 10.1007/s00122-015-2460-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper J. K., Ibrahim M. H., Rudd J., Malla S., Hays D. B. et al. , 2012. Increasing hard winter wheat yield potential via synthetic wheat: I. path-coefficient analysis of yield and its components. Crop Sci. 52: 2014–2022. 10.2135/cropsci2011.07.0382 [DOI] [Google Scholar]
- Dale Z., Jie H., Luyu H., Cancan Z., Yun Z. et al. , 2017. An advanced backcross population through synthetic octaploid wheat as a “Bridge”: Development and QTL detection for seed dormancy. Front. Plant Sci. 8: 2123 10.3389/fpls.2017.02123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreisigacker S., Kishii M., Lage J., and Warburton M., 2008. Use of synthetic hexaploid wheat to increase diversity for CIMMYT bread wheat improvement. Aust. J. Agric. Res. 59: 413–420. 10.1071/AR07225 [DOI] [Google Scholar]
- Dvorak J., Deal K. R., Luo M.-C., You F. M., von Borstel K. et al. , 2012. The origin of spelt and free-threshing hexaploid wheat. J. Hered. 103: 426–441. 10.1093/jhered/esr152 [DOI] [PubMed] [Google Scholar]
- Dvorak J., Luo M. C., Yang Z. L., and Zhang H. B., 1998. The structure of the Aegilops tauschii genepool and the evolution of hexaploid wheat. Theor. Appl. Genet. 97: 657–670. 10.1007/s001220050942 [DOI] [Google Scholar]
- Elshire R. J., Glaubitz J. C., Sun Q., Poland J. A., Kawamoto K. et al. , 2011. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One 6: e19379 10.1371/journal.pone.0019379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faris J. D., Zhang Q., Chao S., Zhang Z., and Xu S. S., 2014. Analysis of agronomic and domestication traits in a durum × cultivated emmer wheat population using a high-density single nucleotide polymorphism-based linkage map. Theor. Appl. Genet. 127: 2333–2348. 10.1007/s00122-014-2380-1 [DOI] [PubMed] [Google Scholar]
- Gardiner L., Wingen L. U., Bailey P., Joynson R., Brabbs T. et al. , 2019. Analysis of the recombination landscape of hexaploid bread wheat reveals genes controlling recombination and gene conversion frequency. Genome Biol. 20: 69 10.1186/s13059-019-1675-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill B. S., and Raupp W. J., 1987. Direct Genetic Transfers from Aegilops squarrosa L. to Hexaploid Wheat. Crop Sci. 27: 445–450. 10.2135/cropsci1987.0011183X002700030004x [DOI] [Google Scholar]
- Glaubitz J. C., Casstevens T. M., Lu F., Harriman J., Elshire R. J. et al. , 2014. TASSEL-GBS: A High Capacity Genotyping by Sequencing Analysis Pipeline. PLoS One 9: e90346 10.1371/journal.pone.0090346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttieri M. J., 2019. Ms3 dominant genetic male sterility for wheat improvement with molecular breeding. Crop Sci. 10.2135/cropsci2019.06.0378 [DOI] [Google Scholar]
- Haudry, a, a Cenci, C. Ravel, T. Bataillon, D. Brunel et al, 2007. Grinding up wheat: a massive loss of nucleotide diversity since domestication. Mol. Biol. Evol. 24: 1506–17. [DOI] [PubMed] [Google Scholar]
- He F., Pasam R., Shi F., Kant S., Keeble-Gagnere G. et al. , 2019. Exome sequencing highlights the role of wild relative introgression in shaping the adaptive landscape of the wheat genome. Nat. Genet. 51: 896–904. 10.1038/s41588-019-0382-2 [DOI] [PubMed] [Google Scholar]
- Hill W., and Robertson A., 1966. The effect of linkage on limits to artificial selection. Genet. Res. 8: 269–294. 10.1017/S0016672300010156 [DOI] [PubMed] [Google Scholar]
- Huang L., Brooks S. A., Li W., Fellers J. P., Trick H. N. et al. , 2003. Map-based cloning of leaf rust resistance gene Lr21 from the large and polyploid genome of bread wheat. Genetics 164: 655–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hufford M. B., Xu X., van Heerwaarden J., Pyhäjärvi T., Chia J.-M. et al. , 2012. Comparative population genomics of maize domestication and improvement. Nat. Genet. 44: 808–811. 10.1038/ng.2309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jantasuriyarat C., Vales M. I., Watson C. J. W., and Riera-Lizarazu O., 2004. Identification and mapping of genetic loci affecting the free-threshing habit and spike compactness in wheat (Triticum aestivum L.). Theor. Appl. Genet. 108: 261–273. 10.1007/s00122-003-1432-8 [DOI] [PubMed] [Google Scholar]
- Jiang C. X., Chee P. W., Draye X., Morrell P. L., Smith C. W. et al. , 2000. Multilocus interactions restrict gene introgression in interspecific populations of polyploid Gossypium (cotton). Evolution (N. Y.) 54: 798–814. [DOI] [PubMed] [Google Scholar]
- Jordan K. W., Wang S., He F., Chao S., Lun Y. et al. , 2018. The genetic architecture of genome‐wide recombination rate variation in allopolyploid wheat revealed by nested association mapping. Plant J. 95: 1039–1054. 10.1111/tpj.14009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan K., Wang S., Lun Y., Gardiner L., MacLachlan R. et al. , 2015. A haplotype map of allohexaploid wheat reveals distinct patterns of selection on homoeologous genomes. Genome Biol. 16: 48 10.1186/s13059-015-0606-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara H., 1944. Discovery of the DD-analyser, one of the ancestors of Triticum vulgare. Agricuture Hortic. 19: 889–890. [Google Scholar]
- Ling H.-Q., Zhao S., Liu D., Wang J., Sun H. et al. , 2013. Draft genome of the wheat A-genome progenitor Triticum urartu. Nature 496: 87–90. 10.1038/nature11997 [DOI] [PubMed] [Google Scholar]
- Luo M.-C., Gu Y. Q., Puiu D., Wang H., Twardziok S. O. et al. , 2017. Genome sequence of the progenitor of the wheat D genome Aegilops tauschii. Nature 551: 498–502. 10.1038/nature24486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M.-C., Yang Z.-L., You F. M., Kawahara T., Waines J. G. et al. , 2007. The structure of wild and domesticated emmer wheat populations, gene flow between them, and the site of emmer domestication. Theor. Appl. Genet. 114: 947–959. 10.1007/s00122-006-0474-0 [DOI] [PubMed] [Google Scholar]
- Martin S. H., Davey J. W., Salazar C., and Jiggins C. D., 2019. Recombination rate variation shapes barriers to introgression across butterfly genomes. PLoS Biol. 17: e2006288 10.1371/journal.pbio.2006288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S. H., and Jiggins C. D., 2017. Interpreting the genomic landscape of introgression. Curr. Opin. Genet. Dev. 47: 69–74. 10.1016/j.gde.2017.08.007 [DOI] [PubMed] [Google Scholar]
- Neph S., Kuehn M. S., Reynolds A. P., Haugen E., Thurman R. E. et al. , 2012. BEDOPS: High-performance genomic feature operations. Bioinformatics 28: 1919–1920. 10.1093/bioinformatics/bts277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyine M., Uwimana B., Blavet N., Hřibová E., Vanrespaille H. et al. , 2018. Genomic Prediction in a Multiploid Crop: Genotype by Environment Interaction and Allele Dosage Effects on Predictive Ability in Banana. Plant Genome 11: 170090 10.3835/plantgenome2017.10.0090 [DOI] [PubMed] [Google Scholar]
- Nyine M., Wang S., Kiani K., Jordan K., Liu S. et al. , 2019. Genotype Imputation in Winter Wheat Using First-Generation Haplotype Map SNPs Improves Genome-Wide Association Mapping and Genomic Prediction of Traits. G3 (Bethesda) 9: 125–133. 10.1534/g3.118.200664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogbonnaya F. C., Abdalla O., Mujeeb-Kazi A., Kazi A. G., Xu S. S. et al. , 2013. Synthetic Hexaploids: Harnessing Species of the Primary Gene Pool for Wheat Improvement. Plant Breed. Rev. 37: 35–122. 10.1002/9781118497869.ch2 [DOI] [Google Scholar]
- Opperman R., Emmanuel E., and Levy A. A., 2004. The effect of sequence divergence on recombination between direct repeats in arabidopsis. Genetics 168: 2207–2215. 10.1534/genetics.104.032896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özkan H., Willcox G., Graner A., Salamini F., and Kilian B., 2011. Geographic distribution and domestication of wild emmer wheat (Triticum dicoccoides). Genet. Resour. Crop Evol. 58: 11–53. 10.1007/s10722-010-9581-5 [DOI] [Google Scholar]
- Peleg Z., Fahima T., Abbo S., Krugman T., Nevo E. et al. , 2005. Genetic diversity for drought resistance in wild emmer wheat and its ecogeographical associations. Plant Cell Environ. 28: 176–191. 10.1111/j.1365-3040.2005.01259.x [DOI] [Google Scholar]
- Periyannan S., Moore J., Ayliffe M., Bansal U., Wang X. et al. , 2013. The Gene Sr33, an Ortholog of Barley Mla Genes, Encodes Resistance to Wheat Stem Rust Race Ug99. Science 341: 786–788. 10.1126/science.1239028 [DOI] [PubMed] [Google Scholar]
- Poland J. A., Brown P. J., Sorrells M. E., and Jannink J.-L., 2012. Development of high-density genetic maps for barley and wheat using a novel two-enzyme genotyping-by-sequencing approach. PLoS One 7: e32253 10.1371/journal.pone.0032253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pont C., Leroy T., Seidel M., Tondelli A., Duchemin W. et al. , 2019. Tracing the ancestry of modern bread wheats. Nat. Genet. 51: 905–911. 10.1038/s41588-019-0393-z [DOI] [PubMed] [Google Scholar]
- Qi L., Friebe B., Zhang P., and Gill B. S., 2007. Homoeologous recombination, chromosome engineering and crop improvement. Chromosome Res. 15: 3–19. 10.1007/s10577-006-1108-8 [DOI] [PubMed] [Google Scholar]
- Qi L. L., and Gill B. S., 2001. High-density physical maps reveal that the dominant male-sterile gene Ms3 is located in a genomic region of low recombination in wheat and is not amenable to map-based cloning. Theor. Appl. Genet. 103: 998–1006. 10.1007/s001220100699 [DOI] [Google Scholar]
- R Development Core Team , 2011. R: A Language and Environment for Statistical Computing (R. D. C. Team, Ed.). R Foundation for Statistical Computing, Vienna, Austria: 409. [Google Scholar]
- Reynolds M., Dreccer F., and Trethowan R., 2007. Drought-adaptive traits derived from wheat wild relatives and landraces. J. Exp. Bot. 58: 177–186. 10.1093/jxb/erl250 [DOI] [PubMed] [Google Scholar]
- Reynolds M. P., Pask A. J. D., Hoppitt W. J. E., Sonder K., Sukumaran S. et al. , 2017. Strategic crossing of biomass and harvest index—source and sink—achieves genetic gains in wheat. Euphytica 213: 213–257. 10.1007/s10681-017-2040-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg L. H., Linder C. R., and Seiler G. J., 1995. Chromosomal and genic barriers to introgression in helianthus. Genetics 141: 1163–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg L. H., Whitton J., and Gardner K., 1999. Hybrid zones and the genetic architecture of a barrier to gene flow between two sunflower species. Genetics 152: 713–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saintenac C., Faure S., Remay A., Choulet F., Ravel C. et al. , 2011a Variation in crossover rates across a 3-Mb contig of bread wheat (Triticum aestivum) reveals the presence of a meiotic recombination hotspot. Chromosoma 120: 185–198. 10.1007/s00412-010-0302-9 [DOI] [PubMed] [Google Scholar]
- Saintenac C., Jiang D., and Akhunov E. D., 2011b Targeted analysis of nucleotide and copy number variation by exon capture in allotetraploid wheat genome. Genome Biol. 12: R88 10.1186/gb-2011-12-9-r88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saintenac C., Jiang D., Wang S., and Akhunov E., 2013a Sequence-based mapping of the polyploid wheat genome. G3 (Bethesda) 3: 1105–1114 (Bethesda) 10.1534/g3.113.005819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saintenac C., Zhang W., Salcedo A., Rouse M. N., Trick H. N. et al. , 2013b Identification of wheat gene Sr35 that confers resistance to Ug99 stem rust race group. Science 341: 783–786. 10.1126/science.1239022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears E. R., 1977. An induced mutant with homoeologous pairing in common wheat. Can. J. Genet. Cytol. 19: 585–593. 10.1139/g77-063 [DOI] [Google Scholar]
- Simons K. J., Fellers J. P., Trick H. N., Zhang Z., Tai Y.-S. et al. , 2006. Molecular characterization of the major wheat domestication gene Q. Genetics 172: 547–555. 10.1534/genetics.105.044727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S., Srivastava R., and Kumar J., 2015. Male sterility systems in wheat and opportunities for hybrid wheat development. Acta Physiol. Plant. 37: 1713 10.1007/s11738-014-1713-7 [DOI] [Google Scholar]
- Singh N., Wu S., Tiwari V., Sehgal S., Raupp J. et al. , 2019. Genomic analysis confirms population structure and identifies inter-lineage hybrids in Aegilops tauschii. Front. Plant Sci. 10: 9 10.3389/fpls.2019.00009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohail Q., Inoue T., Tanaka H., Eltayeb A. E., Matsuoka Y. et al. , 2011. Applicability of Aegilops tauschii drought tolerance traits to breeding of hexaploid wheat. Breed. Sci. 61: 347–357. 10.1270/jsbbs.61.347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sood S., Kuraparthy V., Bai G., and Gill B. S., 2009. The major threshability genes soft glume (sog) and tenacious glume (Tg), of diploid and polyploid wheat, trace their origin to independent mutations at non-orthologous loci. Theor. Appl. Genet. 119: 341–351. 10.1007/s00122-009-1043-0 [DOI] [PubMed] [Google Scholar]
- Su H., Liu Y., Liu C., Shi Q., Huang Y. et al. , 2019 Centromere Satellite Repeats Have Undergone Rapid Changes in Polyploid Wheat Subgenomes. Plant Cell 31: tpc.00133.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International Wheat Genome Sequencing Consortium (IWGSC), 2018 Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361: eaar7191. [DOI] [PubMed]
- Uauy C., Distelfeld A., Fahima T., Blechl A., and Dubcovsky J., 2006. A NAC Gene Regulating Senescence Improves Grain Protein, Zinc, and Iron Content in Wheat. Science 314: 1298–1301. 10.1126/science.1133649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Beissinger T. M., Lorant A., Ross-Ibarra C., Ross-Ibarra J. et al. , 2017. The interplay of demography and selection during maize domestication and expansion. Genome Biol. 18: 215 10.1186/s13059-017-1346-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Luo M.-C., Chen Z., You F. M., Wei Y. et al. , 2013. Aegilops tauschii single nucleotide polymorphisms shed light on the origins of wheat D-genome genetic diversity and pinpoint the geographic origin of hexaploid wheat. New Phytol. 198: 925–937. 10.1111/nph.12164 [DOI] [PubMed] [Google Scholar]
- Wang Y., Tang H., Debarry J. D., Tan X., Li J. et al. , 2012. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40: e49 10.1093/nar/gkr1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulff B. B. H., and Moscou M. J., 2014. Strategies for transferring resistance into wheat: from wide crosses to GM cassettes. Front. Plant Sci. 5: 692 10.3389/fpls.2014.00692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Liu X., Ge S., Jensen J. D., Hu F. et al. , 2012. Resequencing 50 accessions of cultivated and wild rice yields markers for identifying agronomically important genes. Nat. Biotechnol. 30: 105–111. 10.1038/nbt.2050 [DOI] [PubMed] [Google Scholar]
- Ziolkowski P. A., Berchowitz L. E., Lambing C., Yelina N. E., Zhao X. et al. , 2015. Juxtaposition of heterozygous and homozygous regions causes reciprocal crossover remodelling via interference during Arabidopsis meiosis. eLife 4: e03708 10.7554/eLife.03708 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All supplemental material and relevant data are available at FigShare. Raw sequence reads were deposited at NCBI’s Sequence Read Archive (SRA) under BioProjectID PRJNA637316 and PRJNA637416 and will be accessible upon release through the links below. https://www.ncbi.nlm.nih.gov/sra/PRJNA637316, https://www.ncbi.nlm.nih.gov/sra/PRJNA637416. Supplemental material available at figshare:https://doi.org/10.25387/g3.12724943.