Abstract
A Rosaceae family-level candidate gene approach was used to identify genes associated with sugar content in blackberry (Rubus subgenus Rubus). Three regions conserved among apple (Malus × domestica), peach (Prunus persica), and alpine strawberry (Fragaria vesca) were identified that contained previously detected sweetness-related quantitative trait loci (QTL) in at least two of the crops. Sugar related genes from these conserved regions and 789 sugar-associated apple genes were used to identify 279 Rubus candidate transcripts. A Hyb-Seq approach was used in conjunction with PacBio sequencing to generate haplotype level sequence information of sugar-related genes for 40 cultivars with high and low soluble solids content from the University of Arkansas and USDA blackberry breeding programs. Polymorphisms were identified relative to the ‘Hillquist’ blackberry (R. argutus) and ORUS 4115-3 black raspberry (R. occidentalis) genomes and tested for their association with soluble solids content (SSC). A total of 173 alleles were identified that were significantly (α = 0.05) associated with SSC. KASP genotyping was conducted for 92 of these alleles on a validation set of blackberries from each breeding program and 48 markers were identified that were significantly associated with SSC. One QTL, qSSC-Ruh-ch1.1, identified in both breeding programs accounted for an increase of 1.5 °Brix and the polymorphisms were detected in the intron space of a sucrose synthase gene. This discovery represents the first environmentally stable sweetness QTL identified in blackberry. The approach demonstrated in this study can be used to develop breeding tools for other crops that have not yet benefited directly from the genomics revolution.
Keywords: Fruit sweetness, Rubus spp., RosBREED, Marker-assisted breeding, Marker-assisted selection
Blackberries (Rubus subgenus Rubus) are the fourth most economically important U.S. berry crop, accounting for over $650 million in sales during 2019 (CA Strawberry Commission 2019). Cultivated blackberries are primarily hybrids of two or more Rubus species and belong to the family Rosaceae (Clark and Finn 2011). This family is diverse and contains many important crops such as apple (Malus ×domestica), peach (Prunus persica), pear (Pyrus communis), and strawberry (Fragaria ×ananassa). Similar to other members of the Rosaceae, blackberry is highly prized worldwide for its sweet fruit (Clark and Finn 2011; Zurn et al. 2018).
One of the major challenges in bringing cultivated blackberry to market is a lack of cultivars that are high in sugar content and retain firmness (Clark and Finn 2011). Marker-assisted selection (MAS) is particularly effective for evaluation of genetic potential for traits with low heritability but for which most of that genetic influence is determined by one or a few loci (Ru et al. 2016), such as apple fruit fructose content (Guan et al. 2015). Currently, blackberry lacks genomic resources such as dense genetic maps, large mapping populations, and high-throughput marker genotyping assays that are needed to fuel quantitative trait locus (QTL) discovery and DNA test development underlying MAS (Garcia-Seco et al. 2015; Foster et al. 2019). High ploidy in cultivated blackberry (2n = 2x-12x = 14-84) further complicates the development of genomic resources and genetic analyses (Foster et al. 2019). Recently, QTL related to sugar content and soluble solids content (SSC) in fruit were identified in strawberry, apple, and peach (Etienne et al. 2002; Zorrilla-Fontanesi et al. 2011; Lerceteau-Köhler et al. 2012; Verma et al. 2017). SSC is often used as a proxy for sugar content and sweetness in berry crops during breeding as the majority of soluble solids in fruit are sugars (Zorrilla-Fontanesi et al. 2011).
Within the Rosaceae family, a high degree of synteny is observed among species due to shared evolutionary ancestry (Vilanova et al. 2008; Sargent et al. 2009; Illa et al. 2011; Bushakra et al. 2012; Jung et al. 2012; Edger et al. 2019; Hardigan et al. 2020). When comparing the Fragaria and Prunus genomes, Vilanova et al. (2008) noted a clear pattern of synteny between the two genera. Yamamoto et al. (2004) showed that linkage maps of Japanese pear (Pyrus pyrifolia) and European pear (P. communis) had conserved marker order for intergeneric markers on the apple consensus map. Moreover, markers have been identified that amplify syntenic regions in Malus, Fragaria, and Prunus (Sargent et al. 2009). These early findings using linkage mapping approaches are supported by the recent sequencing and assembly of many rosaceous crop genomes (VanBuren et al. 2016, 2018; Hibrand Saint-Oyant et al. 2018; Jibran et al. 2018; Edger et al. 2019; Raymond et al. 2018; Linsmith et al. 2019; Hardigan et al. 2020).
Genes and pathways for sweetness and fruit ripening have been shown to be conserved within Rosaceae and other plant families (Le Dantec et al. 2010; Wei et al. 2014). Sugar transport genes play a vital role in the long-distance transport of sugar and in the allocation of sugar into source and sink cells in developing fruit (Le Dantec et al. 2010; Wei et al. 2014). Wei et al. (2014) identified sugar transport genes in Malus and found them to be conserved in Arabidopsis and Vitis. A study of peach genes identified 59 candidate genes (CGs) associated with fruit quality, including sweetness (Le Dantec et al. 2010). Primers were designed for 55 of these CGs and were tested in strawberry and two-thirds of them produced amplicons, demonstrating that many of the genes involved in sugar production, degradation, conversion, and transport are conserved among Rosaceae species.
The development of DNA-based genetic markers for assisting plant breeding that began in the 1980s (Xu and Crouch 2008) has changed substantially with the invention of next-generation sequencing (NGS). NGS has allowed a multitude of tools and approaches to be developed for identifying polymorphisms in DNA sequence for use in MAS. One such method commonly used to detect polymorphisms is amplicon sequencing or targeted amplicon sequencing (Fritsch et al. 2016; Shirasawa et al. 2016; Onda et al. 2018). Amplicon sequencing produces PCR products that flank or span a polymorphism of interest and can be used to identify polymorphisms reliably and rapidly for known regions of interest with few limitations (Fritsch et al. 2016). With amplicon sequencing, the user only gains insight into a single region between the forward and reverse primers (Ranjan et al. 2016). This limited window could exclude other polymorphisms, that could be contained on adjacent exons, introns, and neighboring genes. Moreover, amplicon sequencing can be ineffective for regions or genes with high levels of sequence divergence. Another approach used for detecting polymorphisms is RNA sequencing (RNA-Seq; Wang et al. 2013; Garcia-Seco et al. 2015; Salazar et al. 2015). RNA-Seq is good for capturing whole mRNA transcripts, but it can be cost-prohibitive as low-level transcripts require very deep sequencing for reliable capture (Ozsolak and Milos 2011). An alternative to the aforementioned sequencing methods is Hyb-Seq (Weitemier et al. 2014). Hyb-Seq can target and capture long genomic sequences that contain sequence variant information in the targeted and flanking regions. This target capture approach can and has been used to cost-effectively capture low-copy nuclear genes (Kamneva et al. 2017). Hyb-Seq targets and captures sequences using biotinylated RNA baits. The baits can be designed from closely related species to capture syntenic genes and regions (Weitemier et al. 2014; Carter et al. 2019). Because baits can be designed from related species and polymorphisms and corresponding flanking information can be captured, Hyb-Seq is a promising approach for blackberry given the genomic complexity associated with its interspecific hybrid nature and the lack of available genomic resources.
Many of the genes and pathways mediating sugar content are likely conserved across Rosaceae and might be useful to identify associated regions in blackberry. As such, polymorphisms associated with SSC in blackberry were identified using a homologous gene-based approach and markers were developed and validated for use in DNA-informed breeding.
Materials and Methods
Germplasm and Phenotyping
Blackberry crosses were made in 2011, 2012, and 2013 at the University of Arkansas System Division of Agriculture (UA) and the USDA-ARS Horticultural Crops Research Unit (HCRU) breeding programs (Supplementary Table S1; Zurn et al. 2018). Populations and parents developed by UA were planted at the UA Fruit Research Station (Clarksville, AR) and those developed by the USDA-ARS HCRU program were planted at Oregon State University’s Lewis-Brown Farm (Corvallis, OR). Parentage for all individuals was previously verified using a microsatellite fingerprinting set (Zurn et al. 2018).
Parents and offspring were evaluated for two years (2015 and 2016) for SSC. In the morning before temperatures exceeded 27°, 15 berries were harvested from each plant at the shiny-black stage. Berries were frozen following harvest until ripe berries were obtained from all plants. After all berries were collected, the 15 berries from each genotype were divided into three replicates and juiced. Frozen juice from the USDA-ARS HCRU program was sent via overnight shipping to UA where it was thawed overnight before measurement. The berry juice from each sample was measured using an Abbe Mark II refractometer (Bausch and Lomb Inc., Rochester, NY, U.S.A.). Historical SSC data for the parental germplasm and important cultivars released from each breeding program were also collected from annual breeding records. Mean SSC was calculated for each individual and 20 individuals from each of the two breeding programs with high and low SSC were chosen (Table 1), to maximize the likelihood of discovering polymorphisms associated with SSC. High SSC was defined as a mean SSC greater than 11.5 °Brix, and low SSC was a mean SSC less than or equal to 11.5 °Brix.
Table 1. Summary of sequenced blackberries from the University of Arkansas System Division of Agriculture (UA) and the USDA-ARS Horticultural Crops Research Unit (HCRU) breeding programs. Mean historical soluble solids content (SSC), circular consensus sequences (CCS) generated during sequencing are presented, and groupings determined via K-means clustering using markers identified from the Hillquist V1 (HV1) and R. occidentalis V3 (RoV3) genome assemblies.
| Program | Name | Mean SSC (°Brix) | No. CCS Reads | Mean CCS Read Length | Median CCS Read Length | HV1 Group | RoV3 Group |
|---|---|---|---|---|---|---|---|
| HCRU | ORUS 4647M | 7.6 | 10,745 | 2,678.00 | 2,767 | 1 | 3 |
| HCRU | ORUS 4540N | 8.0 | 17,995 | 2,723.20 | 2,720 | 1 | 3 |
| HCRU | ORUS 4647L | 8.2 | 62 | 1,628.70 | 1,493.5 | 2 | 2 |
| HCRU | ORUS 4647R | 8.6 | 32,015 | 2,880.20 | 2,936 | 1 | 3 |
| HCRU | ORUS 4647U | 8.9 | 6,914 | 1,802.90 | 1,561 | 1 | 3 |
| HCRU | Kotata | 9.3 | 4,310 | 1,109.10 | 1,029 | 1 | 4 |
| HCRU | Bassettberry | 9.7 | 5,185 | 1,924.70 | 1,646 | 1 | 3 |
| HCRU | Ollalie | 9.7 | 1,640 | 1,118.00 | 1,067 | 2 | 4 |
| HCRU | Silvan | 9.7 | 9,542 | 3,116.70 | 3,603 | 1 | 3 |
| HCRU | Black Diamond | 10.5 | 4,509 | 2,492.60 | 2,369 | 1 | 3 |
| HCRU | ORUS 1932-1 | 11.5 | 9,484 | 2,963.70 | 3,258 | 1 | 3 |
| HCRU | Marion | 12.2 | 5,707 | 1,173.30 | 1,095 | 1 | 4 |
| HCRU | Nightfall | 12.2 | 9,522 | 2,676.80 | 2,439 | 3 | 3 |
| HCRU | Columbia Star | 12.8 | 3,988 | 2,703.30 | 2,689.5 | 1 | 3 |
| HCRU | Waldo | 13.7 | 6,397 | 2,478.10 | 2,143 | 1 | 3 |
| HCRU | ORUS 4540A | 15.1 | 4,761 | 2,038.20 | 1,702 | 3 | 1 |
| HCRU | ORUS 4540I | 15.6 | 16,413 | 3,108.20 | 3,386 | 1 | 3 |
| HCRU | ORUS 4674C | 16.3 | 2,840 | 1,085.90 | 973.5 | 1 | 4 |
| HCRU | ORUS 4674J | 17.1 | 2,289 | 1,119.00 | 1,017 | 1 | 4 |
| HCRU | ORUS 4660T | 18.4 | 7,681 | 2,320.20 | 2,208 | 3 | 3 |
| UA | Choctaw | 7.4 | 130 | 1,662.60 | 1,464 | 2 | 2 |
| UA | Comanche | 7.5 | 22,542 | 3,465.50 | 3,758 | 3 | 1 |
| UA | A-2562T | 8.3 | 28,239 | 2,112.40 | 1,888 | 1 | 1 |
| UA | APF-329 | 8.4 | 9,420 | 2,947.00 | 3,009.5 | 3 | 1 |
| UA | A-2418T | 8.6 | 22,640 | 2,878.10 | 2,942 | 1 | 1 |
| UA | APF-326TN | 8.6 | 122 | 1,218.30 | 1,152.5 | 2 | 2 |
| UA | Cheyenne | 8.6 | 4,332 | 1,061.60 | 977.5 | 2 | 4 |
| UA | APF-236T | 8.7 | 37,369 | 3,062.30 | 3,125 | 3 | 1 |
| UA | APF-306T | 8.7 | 6,833 | 2,741.90 | 2,679 | 3 | 1 |
| UA | Kiowa | 9.0 | 13,368 | 2,425.60 | 2,200 | 3 | 1 |
| UA | A-2421 | 11.6 | 3,260 | 1,140.70 | 1,050 | 1 | 4 |
| UA | A-2548T | 11.7 | 2,984 | 1,148.80 | 1,054 | 1 | 4 |
| UA | Osage | 11.7 | 32,607 | 3,432.50 | 3,681 | 3 | 1 |
| UA | A-2444T | 11.9 | 27,512 | 2,359.00 | 2,081 | 3 | 1 |
| UA | A-2552T | 12.0 | 14,495 | 2,918.80 | 2,960 | 1 | 1 |
| UA | Ponca | 12.2 | 2,904 | 877.5 | 789 | 2 | 4 |
| UA | A-2542T | 12.2 | 23,180 | 2,967.50 | 3,005 | 1 | 1 |
| UA | A-2496 | 12.3 | 7,112 | 1,815.00 | 1,555 | 1 | 1 |
| UA | A-2487T | 12.4 | 5,154 | 2,831.80 | 2,876 | 3 | 1 |
| UA | APF-318 | 14.3 | 3,965 | 1,789.20 | 1,602 | 1 | 1 |
Hyb-Seq Bait Design and Sequencing
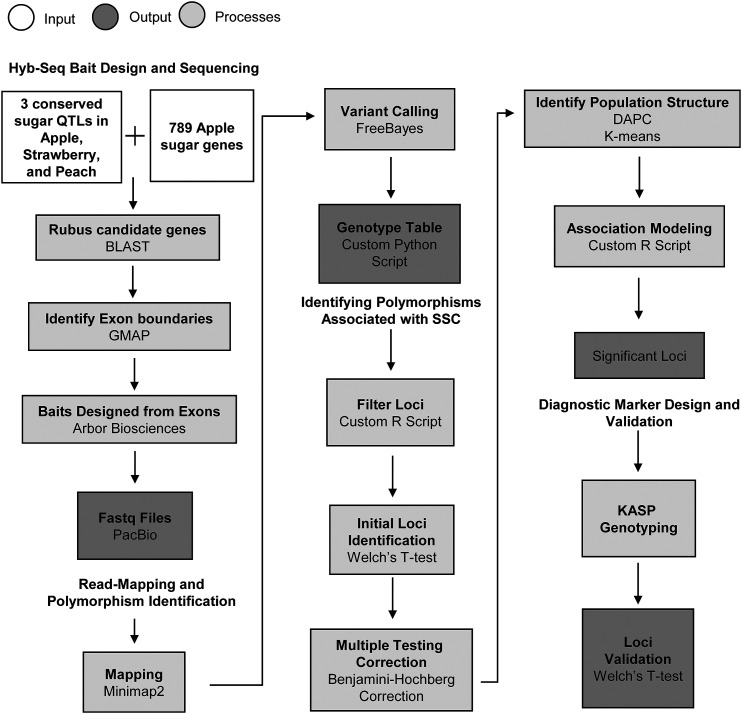
A set of 789 unique genes from the Malus domestica v3.0.a1 assembly that were associated with sugar content (Li et al. 2012, 2016) were BLAST-searched (Altschul et al. 1990) against the Rubus RefTrans v2 transcripts from the Genome Database for Rosaceae (GDR; Jung et al. 2019) and filtered with an e-value cutoff of 0.01 (Figure 1). Data mining was also performed using the GDR’s tools and collated information (Jung et al. 2019) to identify QTL associated with sweetness-related traits for Fragaria, Malus, and Prunus (Quilot et al. 2004; Lerceteau-Köhler et al. 2012; Guan et al. 2015; Jung et al. 2019). The physical regions of the QTL were identified using the genomic location of SNP markers that are associated with QTL. Syntenic regions that are conserved across the Prunus persica v1 (Ppv1; The International Peach Genome Initiative 2013), Fragaria vesca v1 (Fvv1; Shulaev et al. 2011), and Malus ×domestica v1 primary (Mdv1; Velasco et al. 2010) genome assemblies, identified by the Mercator program (Dewey et al. 2007) and made available on GDR, were further mined to identify regions that contain sugar-related QTL from at least two of the species (Table 2). Genes were extracted from the Fragaria vesca v2.0.a1 genome assembly for three syntenic regions that had sugar-related QTL reported in two crops (Table 2) and BLAST2GO was used to re-annotate the extracted genes to identify those related to sugar content (Conesa and Götz 2008). Fragaria candidate gene sequences were BLAST-searched against the Rubus RefTrans v2 transcripts to identify orthologous genes in Rubus. Identified Rubus genes were mapped to the R. occidentalis v1.1 genome (Jibran et al. 2018) with GMAP version 2018-05-30 to identify intron and exon position boundaries (Wu et al. 2016). Exon sequences less than 50 nucleotides in length were removed and the remaining exon sequences were sent to Arbor Biosciences (Ann Arbor, MI, U.S.A.) for bait design. Arbor Biosciences designed baits to fit a 2X tiling density for the submitted exon sequences.
Figure 1.
Schematic of experimental workflow.
Table 2. Syntenic regions identified that contain sugar-related QTL in at least two of the investigated crops (apple, strawberry, and peach). The physical positions described are for the Prunus persica v1 (Ppv1; The International Peach Genome Initiative 2013), Fragaria vesca v1 (Fvv1; Shulaev et al. 2011), and Malus ×domestica v1 (Mdv1; Velasco et al. 2010) genome assemblies. References and associated markers are presented for each syntenic QTL identified.
| Region | Physical Positions | QTL | Crop | Associated/Delimiting Marker(s) | QTL References |
|---|---|---|---|---|---|
| 1 | Ppv1 scaffold_1:10507957-10972280 | qFRUC.SP-G1 | Peach | PC102 | Quilot et al. (2004) |
| Fvv1 LG4:18318384-19134134 | SSC | Apple | ss475880868-ss475882452 | Guan et al. (2015) | |
| Mdv1 Chr13:17977974-18806402 | |||||
| 2 | Ppv1 scaffold_6:24867717-25235545 | SSC | Strawberry | AX-89805813-AX-89906546 | V. Whitaker personal communication |
| Fvv1 LG6:7484789-7911306 | Sucrose | Apple | ss475880518-ss475880556 | Guan et al. (2015) | |
| Mdv1 Chr12:30215039-30750693 | |||||
| 3 | Ppv1 scaffold_2:25475692-25774711 | qSUCR.CCF-LGVIIa-f | Strawberry | BFACT044 | Lerceteau-Köhler et al. (2012) |
| Fvv1 LG7:20618518-21128712 | Sorbitol | Apple | ss475876853-ss475876937 | Guan et al. (2015) | |
| Mdv1 Chr1:34158601-34656509 | Fructose | Apple | ss475876857-ss475876937 | ||
| Fructose | Apple | ss475883868-ss475876937 | |||
| Glucose-2012(20wk) | Apple | ss475876868-ss475876937 | |||
| Glucose-2012 (10wk) | Apple | ss475876871-ss475876937 | |||
| Sorbitol | Apple | ss475882286-ss475876937 |
Young actively growing leaf tissue or the youngest possible leaf material was collected from the 40 chosen breeding selections and cultivars with low and high SSC. For individuals grown by the UA program, tissue was shipped overnight on ice to the USDA-ARS National Clonal Germplasm Repository (NCGR) in Corvallis, OR. Approximately 30-50 mg of tissue from each individual was sampled into a 96-well plate and flash-frozen in liquid nitrogen. Samples were stored at -80° until DNA extraction was conducted. Prior to extraction, samples were ground using a mixer mill (MM 301; Retsch International, Hann, Germany). DNA was extracted using the E-Z 96 Plant DNA Kit (Omega BioTek Inc., Norcross, GA, U.S.A.) following the modifications proposed by Gilmore et al. (2011). DNA was quantified with a Quant-iT PicoGreenTM dsDNA Assay Kit (Thermo Fisher Scientific, Waltham, MA, U.S.A.) and a Tecan Infinite M Plex multimode plate reader (Tecan Group Ltd, Zürich, Switzerland). For each sequenced sample, 1 µg of total DNA was sent to Arbor Biosciences for Hyb-Seq. Captured genomic DNA from the 40 samples were sequenced at Arbor Bioscientific using a PacBio instrument. The raw reads were processed into high-quality circular consensus reads (CCS) that were polished with the Arrow algorithm, available through PacBio tools.
Read-Mapping and Polymorphism Identification
The CCS reads for the 40 samples were individually mapped to both the ORUS 4115-3 black raspberry R. occidentalis v3.0 (R. occidentalis v3.0, Van Buren et al. 2018) and the ‘Hillquist’ blackberry v1 genomes (‘Hillquist’ V1, Worthington et al. 2020) with Minimap2 2.15-r915-dirty, using the settings for PacBio genomic reads (Li 2018). Files generated by Minimap2 were converted to bam files, sorted, and indexed with SAMtools 1.9 (Li et al. 2009). The bam files for each assembly were used with Freebayes v1.2.0-4-gd15209e to identify structural variants (Garrison and Marth 2012). Freebayes was set to the recommended settings for PacBio reads with the correct ploidy reflecting each sample. A custom Python script was created to take the output VCF files from Freebayes and to create a genotype table for each reference. The tables contained loci named by the chromosome or contig, the position for each polymorphism, and the genotypic information for all 40 samples. Read depth was calculated for all positions and samples that had missing data using SAMtools 1.9 depth command. If no reads for a region were present, it was recorded as missing.
Identifying Polymorphisms Associated With SSC
Loci identified for each assembly were filtered to have less than 20% missing data and to have between two and four alleles present across all samples. Significant loci were identified using a similar process as Wei et al. (2006). In Wei et al. (2006), markers associated with disease resistance were identified for sugarcane, which is also a complex autopolyploid like blackberry. A custom R script was used to determine the presence and absence of each locus-allele in each of the 40 samples. Each locus was initially examined individually using Welch’s T-test. A Benjamini-Hochberg correction was applied to correct for error resulting from multiple testing and to identify significant loci (α = 0.05). Each of the significant loci were fitted to two general linear models to correct for false associations due to population structure associated with each breeding program:
| (1) |
| (2) |
Groups describing population structure were established using a discriminant analysis of principal components (DAPC) approach based on k-means clustering using the ‘find.clusters’ function in the R package ‘adegenet’ (Jombart 2008; Jombart and Ahmed 2011). Alleles with less than 20% missing data were used for DAPC. A significant (α = 0.05) group × allele interaction would indicate the effects of the locus differed among population groups. Significant (α = 0.05) within-group variance would suggest the allele-trait association was independent of population structure. Alleles that did not have significant group × allele interactions and had significant within-group variance (i.e., allele within group) were chosen for diagnostic marker design and validation.
Diagnostic Marker Design and Validation
Two sets of 96 individuals, one from each breeding program, representing high and low SSC within each family were chosen for allele validation (Supplementary Table S1). Leaf tissue was obtained and processed as described for the PacBio sequencing and lyophilized. Lyophilized tissue and DNA consensus sequences consisting of the potential diagnostic alleles and their flanking sequences were submitted to LGC Ltd (Teddington, United Kingdom) for KASP marker design, DNA extraction, and assay execution. Diagnostic alleles were composed of the significant target allele and a second allele that could be the reference and/or an alternative allele. Due to ploidy variation, high diversity, and the complexity of genetic sequences represented by the 40 sequenced samples in a given region, some consensus sequences were designed with a preference toward the target diagnostic allele. Genotypic data were received from LGC and curated using the LGC KlusterCaller software. Alleles were validated for diagnostic ability in each environment (location-year) using Welch’s T-test and a Benjamini-Hochberg correction (α = 0.05).
Characterization of Chromosome 1 QTL
Positions of alleles of the chromosome 1 QTL were determined using the JBrowse tool on the GDR (Jung et al. 2019). If an allele was determined to be in the exon or intron of a gene, the gene sequence was extracted and conserved protein domains were identified to predict gene function using the conserved domain database (Lu et al. 2020). Haplotype sequence information was also extracted from the Integrated Genome Viewer (IGV, Robinson et al. 2017) for the significant alleles. Haplotype sequences were also used in conjunction with the conserved domain database to compare and validate the results. The gene and haplotype sequences were subjected to a BLAST search to determine if similar gene functions were found in other species beyond Rosaceae
Identification of regions in Rubus occidentalis v3.0 that are syntenic to the sugar-related QTL containing regions in peach, apple and strawberry
Synteny among the R. occidentalis v3.0 and the newest apple (M. × domestica GDDH13 v1.1; Daccord et al. 2017), peach (P. persica v2.0; Verde et al. 2013), and strawberry (F. vesca v4.0; Edger et al. 2017) genome assemblies was identified for the three conserved sugar-related QTL regions (Table 2). The synteny analysis in GDR was conducted using MCScanX (Wang et al. 2012) with default settings. The genomic sequences from old genome assemblies of apple, peach and strawberry (Table 2) were first BLAST-searched to the corresponding new genomes, then the syntenic regions in R. occidentalis v3.0 was identified using the synteny browser in GDR (Jung et al. 2019).
Data Availability
Raw CCS reads have been deposited to the National Center for Biotechnology Information under BioProject number PRJNA633906. Genotypic Data from the resulting KASP assay and phenotypic data are provided in Supplementary Table S2. VCF files, Python code, and R scripts are available at the following github repository: github.com/Bassil-Lab/Zurn-et-al-2020-G3-Blackberry-SSC. KASP assays (Supplementary Table S3) are available through LGC. Supplemental material available at figshare: https://doi.org/10.25387/g3.12767864.
Results
Phenotypic Data
A high degree of variability was observed among progeny from the UA and USDA-ARS HCRU breeding programs (Supplementary Table S1). For the six UA populations evaluated in this study, SSC ranged from 5.3 – 14.8 °Brix in 2015 and from 4.6 – 16.2 °Brix in 2016. The mean SSC for the UA progeny was 9.9 and 9.8 °Brix in 2015 and 2016, respectively. Soluble solids content in the eight USDA-ARS HCRU populations ranged from 6.0 – 20.2 °Brix in 2015 and 5.8 – 18.9 °Brix in 2016. The mean SSC for the USDA-ARS HCRU populations was 12.4 and 11.7 °Brix in 2015 and 2016, respectively.
Rubus SSC Candidate Gene Identification and Bait Design
Among the Fragaria genes in the three conserved syntenic regions (table 2), seven genes were identified with functions associated with sweetness, including beta-amylase 3 and sugar transport genes. A BLAST-search of the seven Fragaria genes and the 789 Malus sweetness-associated genes against the Rubus RefTrans v2 transcripts identified 279 unique genes putatively associated with sweetness in Rubus. Mapping these genes to the R. occidentalis v1.1 reference genome identified 2,122 exon sequences with start and stop boundaries. Arbor Biosciences designed 9,355 baits with 2X tiling density for 2,114 of the 2,122 exon sequences (99.6% of the exons). Despite having eight exons with no baits designed, total target region coverage was still high at 98.8% of the total length of the submitted exons.
Sequencing and Polymorphism Identification
Sequencing and filtering of the captured genomic reads for the 40 samples produced 430,167 high-quality CCS reads (Table 1). The number of CCS reads were variable and ranged from 62 to 37,369 reads per individual. The mean and median read lengths were 2,661 and 2,610 nucleotides, respectively. The quality of the CCS reads was high and a mean phred score of 40 was observed. A total of 929,430 and 1,324,854 loci were found that had alleles different from the reference when mapping CCS reads to the ‘Hillquist’ v1 and R. occidentalis v3.0 assemblies, respectively.
Identifying Polymorphisms Associated With SSC
After filtering on missing data, 12,945 and 15,194 loci were available for investigation that were identified in the ‘Hillquist’ V1 and R. occidentalis v3.0 genome assemblies, respectively. After the first round of statistical testing, 467 loci from the ‘Hillquist’ V1 assembly and 312 loci from the R. occidentalis v3.0 assembly were identified to be significant (α = 0.05).
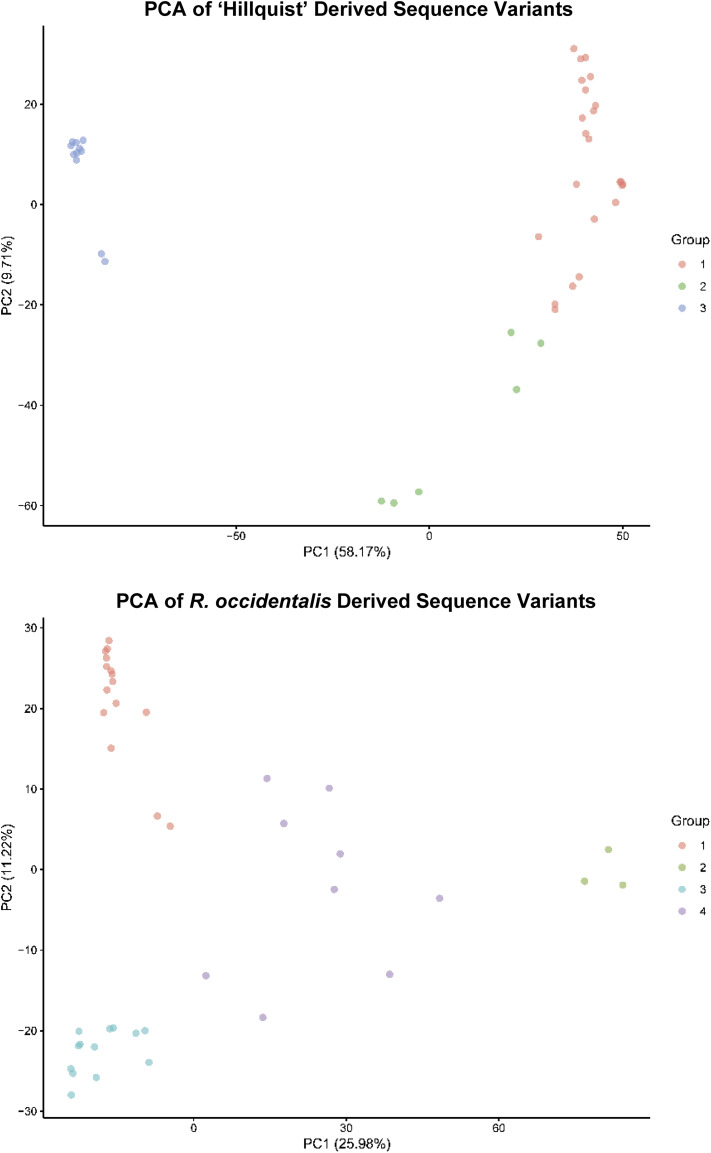
The sequenced individuals were clustered into three and four sub-groups during a discriminant analysis of principal components using loci identified in the ‘Hillquist’ V1 and R. occidentalis v3.0 assemblies, respectively (Figure 2). Population structure was used to model the previously identified significant loci to determine independence of population structure and if allele effects differed between groups. No allele effect differences (α = 0.05) were identified between groups when using model 1, indicating that the effects of the identified loci were independent of population structure. Model 2 identified 64 alleles identified from the ‘Hillquist’ V1 assembly that were significantly (α = 0.05) associated with SSC regardless of population structure. For loci identified using the R. occidentalis v3.0 assembly, 109 alleles were found to be significant (α = 0.05) regardless of population structure.
Figure 2.
Principal Component Analysis (PCA) results of sequence variants with less than 20% missing data identified in 40 sequenced blackberry cultivars and advanced selections using the ‘Hillquist’ blackberry v1 and ORUS 4115-3 black raspberry R. occidentalis v3.0 assemblies. Discriminant analysis of principal components identified three and four groups for the ‘Hillquist’ and R. occidentalis derived variants, respectively.
Diagnostic Marker Validation
A total of 111 KASP assays (Supplementary Table S3) representing 92 loci could be designed for the 173 significant loci identified from the two assemblies. Low GC content, dimer formation, low/high annealing temperature, or large amounts of sequence variation near the target polymorphism prevented primer design for the unrepresented targeted loci. Twenty-seven of the markers (24.3%) performed poorly or were monomorphic and were subsequently removed. Evaluation of the remaining 84 markers (Supplementary Table S2) for their association with SSC in the UA and USDA-ARS HCRU offspring populations during the 2015 and 2016 growing seasons identified a total of 48 alleles that remained significant after validation (Supplementary Table S4). Overall, most of the alleles identified had a negative influence on SSC, with only 16 being associated with an increase in SSC. Fewer alleles were found to be associated with SSC in the UA populations compared to the USDA-ARS HCRU populations.
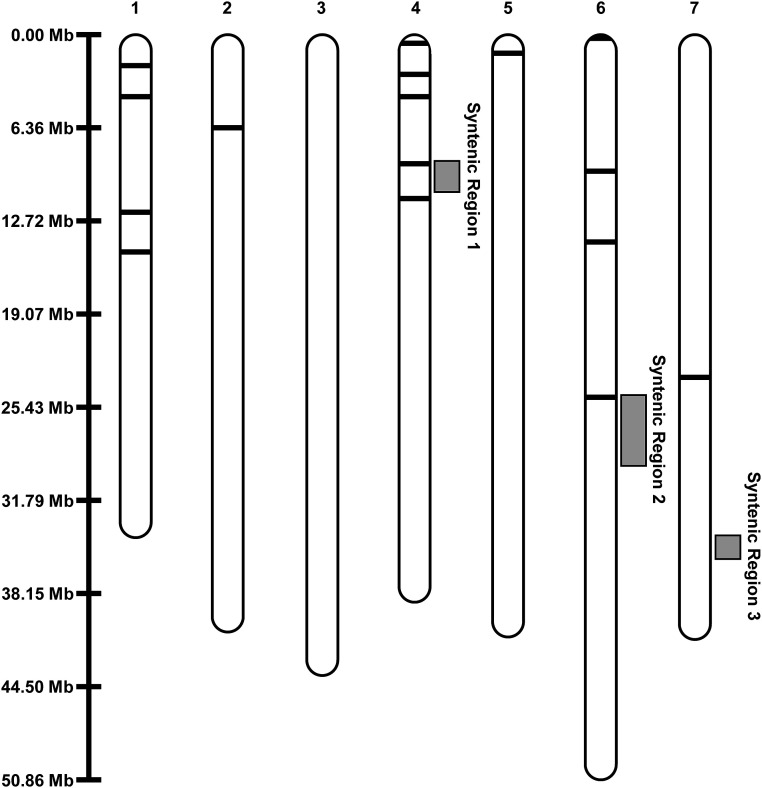
The 48 significant alleles identified in each assembly mapped to 16 regions across six of the seven chromosomes (all but Ro03) in the R. occidentalis v3.0 assembly (Figure 3; Supplementary Table S4). Markers associated with SSC were found in syntenic region 1 on chromosome 4 and syntenic region 2 on chromosome 6. The markers found in these regions were only significant in one environment (Oregon 2016) with the exception of marker BBS_SNP29, which was significant in two environments (Oregon 2015 and Oregon 2016; Supplementary Table S4). Three alleles in markers BBS_SNP45, BBS_INDL31, and BBS_SNP46 were found in a 736 bp region on chromosome 1 that were significant in three environments (Supplementary Table S4). The QTL associated with these markers accounted for a 1.46 °Brix difference in SSC and was named qSSC-Ruh-ch1.1. Additionally, 15 QTL regions were significant in both of the Oregon environments (Supplementary Table S4).
Figure 3.
Significant SSC-associated marker positions relative to the three syntenic QTL regions (Table 2) in the black raspberry ORUS 4115-3 genome (R. occidentalis v3.0 assembly). The synteny viewer tool on the Genomic Database for the Rosaceae (GDR; https://www.rosaceae.org/) is suggested for an in-depth view of the synteny for these regions.
The genotypic data for the three significant alleles on chromosome 1 were consistent for 11 of the 13 samples that were genotyped using sequencing and KASP assays (Table 3). In two samples, ORUS 4674C and A-2487T, two KASP markers were not able to capture both diagnostic alleles that were identified during sequencing (Table 3).
Table 3. Genotype comparison of 13 samples that were genotyped with the KASP assay and sequencing at positions 14,978,562, 14,978,613, and 14,979,298 on Chromosome 1 from the Rubus occidentalis v3.0 genome assembly. Eleven of the 13 samples had the same genotypes with both methods while two samples, ORUS 4674C and A-2487T, had homozygous genotypes of the targeted allele at two and three of the positions investigated, respectively.
| Sample Name | Ro01 14,978,562 | Ro01 14,978,613 | Ro01 14,979,298 | |||
|---|---|---|---|---|---|---|
| Sequencing | KASP | Sequencing | KASP | Sequencing | KASP | |
| ORUS 4540A | A:T | A:T | AC:GT | AC:GT | T:A | T:A |
| ORUS 4647U | A:A | A:A | AC:AC | AC:AC | T:T | T:T |
| ORUS 4674C | A:T | T:T | AC:GT | GT:GT | T:A | T:A |
| Marion | A:T | A:T | AC:GT | AC:GT | T:A | T:A |
| Nightfall | A:A | A:A | AC:AC | AC:AC | T:T | T:T |
| Waldo | A:G | A:A | AC:AC | AC:AC | T:T | T:T |
| Bassettberry | A:G | A:A | AC:AA | AC:AC | T:T | T:T |
| ORUS 4647L | A:A | A:A | AC:AC | AC:AC | T:T | T:T |
| ORUS 4647M | A:A | A:A | AC:AC | AC:AC | T:T | T:T |
| ORUS 4647R | A:A | A:A | AC:AC | AC:AC | T:T | T:T |
| Silvan | A:A | A:A | AC:AA | AC:AC | T:T | T:T |
| Osage | A:T | A:T | AC:GT | AC:GT | T:A | T:A |
| A-2487T | A:T | T:T | AC:GT | GT:GT | T:A | A:A |
Characterization of Chromosome 1 QTL
All three significant SSC-associated alleles in qSSC-Ruh-ch1.1 were associated with a single ohnolog. The physical gene space for the three alleles in the 736 bp region on chromosome 1 contained two overlapping genes: maker-Ro01-snap-gene-149.62 and maker_Ro01_snap_gene-149.66. Maker-Ro01-snap-gene-149.62 was found to have protein domains associated with aldo-keto reductase, glycosyltransferase, or sucrose synthase genes. Maker_Ro01_snap_gene-149.66 and the haplotype sequences had domains associated with glycosyltransferase and sucrose synthase genes. A BLAST-search using both genes and the associated haplotype identified sucrose synthase genes in rosaceous species as the candidate with the highest proportion of identity (94%) and lowest e-values (0). The gene maker-Ro01-snap-gene-149.62 is possibly a chimeric sequence generated during assembly or a pseudogene because it had domain hits to two different genes, contained a small number of short exons (6), and was mostly composed of large introns. The three alleles were likely associated with maker_Ro01_snap_gene-149.66. This gene contains 14 introns and 15 exons in the R. occidentalis v3.0 assembly. The first two alleles were located at positions 14,978,562 and 14,978,613 in the fourth intron of maker_Ro01_snap_gene-149.66 while the third allele was located in the fifth intron at position 14,979,298.
Discussion
The Hyb-Seq approach together with exploiting synteny among Rosaceae species effectively identified candidate genes for sweetness in blackberry. This approach captured targeted genomic sequences used to detect polymorphisms among breeding individuals and identified 173 alleles for investigation. This experiment can serve as a model approach to rapidly create tools useful for MAS in systems with scarce genomic resources, which was the intent of the RosBREED project (Iezzoni et al. 2017). One advantage of the approach was using PacBio sequencing in conjunction with Hyb-Seq. At low coverage, PacBio sequencing is somewhat error-prone (Jiao et al. 2013; Westbrook et al. 2015; Frank et al. 2016). This flaw can be overcome by sequencing the same circularized DNA molecule multiple times to form accurate consensus sequence (Jiao et al. 2013; Westbrook et al. 2015; Frank et al. 2016). This approach substantially reduces base call errors and significantly improves read accuracy (Jiao et al. 2013). Reducing sequence errors is especially important when identifying polymorphisms associated with traits because base errors can be incorrectly classified as polymorphisms. The average CCS Phred quality score in the present study was high at 40, so the likelihood that a base call error affected downstream analysis is therefore low. Moreover, the long reads generated by PacBio sequencing enabled identification of individual haplotypes within each of the sequenced samples. Considering the variable ploidy and the complex genetics found in cultivated blackberry (Clark and Finn 2011), the haplotypes present at a locus can be difficult to identify and define with short-read sequencing. To reduce consensus sequence complexity and increase primer design success, identification and preference of individual haplotypes that contained the target allele was important for diagnostic marker development.
Despite identifying 173 target alleles, KASP assays could only be developed for 92 alleles. This was mainly caused by additional polymorphisms being situated near the target allele or low GC contents. Polymorphisms near the allele of interest might explain why KASP markers BBS_SNP45, BBS_INDL31, and BBS_SNP46 designed for positions Ro01 14,978,562, Ro01 14,978,613, and Ro01 14,979,298, respectively, were only able to capture the target diagnostic allele in some of the 13 samples investigated (Table 3). Viewing the physical positions through IGV confirmed that reads from some of the 40 samples possibly representing the non-target allele contained the start of a major insertion or deletion within the flanking 25 base pairs of the target. These polymorphisms could interfere with annealing of the forward KASP primer of the non-target alleles, producing only homozygous calls for the target alleles in a sample. It is also possible that off-target amplification could have influenced the scoring of these markers, given how large and conserved some of the targeted gene families are. Markers may have amplified paralogous sequences in addition to the target locations. Given the complexity of the blackberry genome, it would be difficult to separate these off-target amplification events from on-target amplification of ohnologous loci. Therefore, the KASP assays in these cases can only be used to assess the presence or absence of the target allele and should not be used to assign dosage. For the 81 alleles associated with SSC for which a KASP marker assay could not be designed, sequencing-based genotyping methodologies may be the only effective way to validate these alleles.
The 48 significant markers identified in this study were located in 16 regions in the R. occidentalis v3.0 genome assembly (Figure 3); many of these regions were only significant in a single testing environment (Supplementary Table S4). This outcome is not unexpected and has been observed during SSC studies in other rosaceous crops (Etienne et al. 2002; Zorrilla-Fontanesi et al. 2011; Lerceteau-Köhler et al. 2012; Verma et al. 2017). Fruit SSC is known to have low heritability and individual SSC QTL often explain less than 10% of the phenotypic variation (Etienne et al. 2002; Zorrilla-Fontanesi et al. 2011; Lerceteau-Köhler et al. 2012; Verma et al. 2017). More QTL were identified in the USDA-ARS HCRU than in the UA breeding program. The USDA-ARS HCRU breeding program releases cultivars of varying ploidies for the processing market and features trailing germplasm from Western North America and erect and semi-erect germplasm from Eastern North America. In contrast, the UA breeding program has historically focused on erect germplasm from Eastern North America for the fresh market (Clark and Finn 2011). The differences in market focuses has caused the USDA-ARS HCRU breeding program to focus on higher acid and sugar content while the UA program has placed emphasis on postharvest storage and transportation capacity (Clark and Finn 2011). As such, the increased number of QTL may be associated with the increased diversity of the germplasm found in the USDA-ARS HCRU breeding program. The markers BBS_SNP45, BBS_INDL31, and BBS_SNP46 identified on chromosome 1 compose a QTL that was detected in three environments and accounted for a 1.5 °Brix increase in SSC. This QTL, qSSC-Ruh-ch1.1, was detected in germplasm from both the UA and the HCRU breeding programs. qSSC-Ruh-ch1.1 is expected to be quite stable as these two breeding programs were reported to have very genetically distinct germplasm driven by geographical differences (Zurn et al. 2018).
The gene space for qSSC-Ruh-ch1.1 was identified, and all three alleles were located in introns flanking the fifth exon in the gene marker-Ro01-snap-gene-149.66 from the R. occidentalis v3.0 assembly. Conserved domain and BLAST searches for the genes associated with the QTL revealed highest homology to sucrose synthase (SUS) genes from the glycosyltransferase-4 subfamily of the glycosyltransferase super family (reviewed by Stein and Granot 2019). In plants, many genes including SUS genes have been reported to control the accumulation of sugars and starch in plants such as Arabidopsis, rice, maize, and apple (Tsai et al. 1970; Perez et al. 1975; Reidel et al. 2008; Rennie et al. 2012; Raynaud et al. 2016; Wang et al. 2018). Sucrose synthase genes catalyze the reversible cleavage of sucrose into fructose and either uridine diphosphate glucose (UDP-G) or adenosine diphosphate glucose (ADP-G). Plant SUS genes are divided into three separate clades (SUS I, SUS II, and SUS III), are ubiquitous in monocots and dicots, and range widely in number among species. In rosaceous crops, the number of SUS genes vary from six in peach (Prunus persica; Zhang et al. 2015) to 11 in apple (Malus ×domestica; Tong et al. 2018) and black raspberry (R. occidentalis; Van Buren et al. 2018) and 30 in Chinese pear (Pyrus ×bretschneideri; Abdullah et al. 2018). Isoforms of SUS genes in the Rosaceae are differentially expressed in different tissues (Zhang et al. 2015; Zhao et al. 2017; Abdullah et al. 2018). Some isoforms are expressed during fruit development in pear (PbSS5, PbSS3, and PbSS24; Abdullah et al. 2018) and strawberry (FaSS1; Zhao et al. 2017). In strawberry, downregulation of FaSS1 significantly delayed fruit ripening and resulted in decreased sucrose content (Zhao et al. 2017), indicating a possible role in controlling fruit sweetness. Sucrose synthase, sucrose-phosphate synthase (SPS), and acid invertase activity have also been analyzed for their effects on SSC and sugars in Asian pear fruit (Moriguchi et al. 1992). Sucrose synthase transcript levels and sucrose content were highly correlated in 23 Asian pear cultivars. The true function of this haplotype in blackberry is unknown due to scarce genomic resources in Rubus and a lack of functional understanding of glycosyltransferase genes and their role within different plant species (Tong et al. 2018; Thirugnanasambandam et al. 2019). Still, synteny and gene conservation within Rosaceae suggest that this Rubus SUS gene imparts similar sugar metabolic function as pear and strawberry SUS genes.
When examining where the syntenic regions mapped to on the R. occidentalis v3.0 assembly, very few significant markers were identified that were associated with these regions (Table 2; Figure 2). Thirteen markers were associated with syntenic region 1 (Figure 2; Supplementary Table S4). Diagnostic alleles for three of these markers (BBS_SNP23, BBS_SNP24, and BBS_SNP88) were associated with an increase in SSC while the remaining 10 were associated with a decrease in SSC. The marker BBS_INDL12 on chromosome 6 also appeared to be associated with syntenic region 2, with the presence of the diagnostic allele being associated with a 1.4 °Brix increase in SSC. In each of these cases, the significant effect was only detected in a single environment. The presence of associated markers in the syntenic regions suggests that QTL for conserved pathways may be transferable across genera within a family. Given these markers were significant in only a single environment, additional testing and possibly gene cloning is needed to confirm the transferability across genera of the associated QTL. Traits associated with flower petal formation and fruit aromatics and flavor components could be investigated in the future. While tempting, disease resistance QTL, with the exception of MLO-mediated powdery mildew resistance (reviewed in Kusch and Panstruga 2017), should be avoided for this approach as unique host-pathogen interactions are expected to govern the evolution of resistance genes within individual species.
Genomic tools are rapidly being developed for many agriculturally important crops. Despite this, a number of regionally important crops remain that lacks the resources available to agronomic crops. The genomics resource deficiency of cultivated blackberry is fortunately beginning to be overcome with exploitation of large genomics resources available for related crops such as apple, peach, and strawberry. Such resources can be leveraged to target genes associated with pathways conserved at a family level. Using family-level information, genomic and bioinformatics scientists can develop new tools to assist breeders for crops that have not yet benefited from the genomics revolution.
Acknowledgments
The authors would like to thank April Nyberg, Jaimie Green, Mary Peterson, Jaime Willard, David Gilmore, Tralena Kay Buck, Dan Chapman, and staff at the UA System Division of Agriculture Fruit Research Station for their technical support. The researchers would also like to thank Drs. David Chagné and Susan Thomson for useful discussion on analyzing the CCS data. This research was funded through the USDA’s National Institute of Food and Agriculture – Specialty Crop Research Initiative project ‘RosBREED: Combining Disease Resistance and Horticultural Quality in New Rosaceous Cultivars’ (2014-51181-22378) and by the USDA-ARS CRIS projects 2072-21000-049-00D and 2072-21220-003-00D.
Footnotes
Supplemental material available at figshare: https://doi.org/10.25387/g3.12767864.
Communicating editor: J.-L. Jannink
Literature Cited
- Abdullah M., Cao Y., Cheng X., Meng D., Chen Y. et al. , 2018. The sucrose synthase gene family in Chinese pear (Pyrus bretschneideri Rehd.): structure, expression, and evolution. Molecules 23: 1144 10.3390/molecules23051144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., and Lipman D. J., 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Bushakra J. M., Sargent D. J., Cabrera A., Crowhurst R., Lopez Girona E. et al. , 2012. Rosaceae conserved orthologous set (RosCos) markers as a tool to assess genome synteny between Malus and Fragaria. Tree Genet. Genomes 8: 643–658. 10.1007/s11295-011-0450-y [DOI] [Google Scholar]
- CA Strawberry Commission, 2019 Retail category trends report – 12/29/2019 data. https://www.calstrawberry.com/en-us/market-data/retail-category-trends
- Carter K. A., Liston A., Bassil N. V., Alice L. A., Bushakra J. M. et al. , 2019. Target capture sequencing unravels Rubus evolution. Front Plant Sci 10: 1615 10.3389/fpls.2019.01615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark J. R., and Finn C. E., 2011. Blackberry breeding and genetics. Fruit Veg Cereal Sci Biotechnol 5: 27–43. [Google Scholar]
- Conesa A., and Götz S., 2008. Blast2GO: A comprehensive suite for functional analysis in plant genomics. Int. J. Plant Genomics 2008: 619832 10.1155/2008/619832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daccord N., Celton J. M., Linsmith G., Becker C., Choisne N. et al. , 2017. High-quality de novo assembly of the apple genome and methylome dynamics of early fruit development. Nat. Genet. 49: 1099–1106. 10.1038/ng.3886 [DOI] [PubMed] [Google Scholar]
- Dewey C. N., 2007. Aligning multiple whole genomes with Mercator and MAVID. Methods Mol. Biol. 395: 221-236. 10.1007/978-1-59745-514-5_14 [DOI] [PubMed] [Google Scholar]
- Edger P. P., Poorten T. J., VanBuren R., Hardigan M. A., Colle M. et al. , 2019. Origin and evolution of the octoploid strawberry genome. Nat. Genet. 51: 541–547. 10.1038/s41588-019-0356-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edger P. P., Van R., Buren M., Colle T. J., Poorten T. J. et al. , 2017. Single-molecule sequencing and optical mapping yields an improved genome of woodland strawberry (Fragaria vesca) with chromosome-scale contiguity. Gigascience 7: gix124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne C., Rothan C., Moing A., Plomion C., Bodénès C. et al. , 2002. Candidate genes and QTLs for sugar and organic acid content in peach. [Prunus persica (L.) Batsch] Theor. Appl. Genet. 105: 145–159. 10.1007/s00122-001-0841-9 [DOI] [PubMed] [Google Scholar]
- Foster T. M., Bassil N. V., Dossett M., Worthington M. L., and Graham J., 2019. Genetic and genomic resources for Rubus breeding: a roadmap for the future. Hortic. Res. 6: 116 10.1038/s41438-019-0199-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J. A., Pan Y., Tooming-Klunderud A., Eijsink V. G. H., McHardy A. C. et al. , 2016. Improved metagenome assemblies and taxonomic binning using long-read circular consensus sequence data. Sci. Rep. 6: 25373 10.1038/srep25373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsch L., Soeur R., Hansen C., Fischer R., Schillberg S. et al. , 2016. Next-generation sequencing of amplicons is a rapid and reliable method for the detection of polymorphisms relevant for barley breeding. Mol. Breed. 36: 83 10.1007/s11032-016-0507-6 [DOI] [Google Scholar]
- Garcia-Seco D., Zhang Y., Gutierrez-Mañero F. J., Martin C., and Ramos-Solano B., 2015. RNA-Seq analysis and transcriptome assembly for blackberry (Rubus sp. Var. Lochness) fruit. BMC Genomics 16: 5 10.1186/s12864-014-1198-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison E., and Marth G., 2012. Haplotype-based variant detection from short-read sequencing. ArXiv:1207.3907
- Gilmore B. S., Bassil N. V., and Hummer K. E., 2011. DNA extraction protocols from dormant buds of twelve woody plant genera. J. Am. Pomolog. Soc. 65: 201–206. www.ars.usda.gov/research/publications/publication/?seqNo115=270236 [Google Scholar]
- Guan Y., Peace C., Rudell D., Verma S., and Evans K., 2015. QTLs detected for individual sugars and soluble solids content in apple. Mol. Breed. 35: 135 10.1007/s11032-015-0334-1 [DOI] [Google Scholar]
- Hardigan M. A., Feldmann M. J., Lorant A., Bird K. A., Famula R. et al. , 2020. Genome synteny has been conserved among the octoploid progenitors of cultivated strawberry over millions of years of evolution. Front Plant Sci 10: 1789 10.3389/fpls.2019.01789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibrand Saint-Oyant L., Ruttink T., Hamama L., Kirov I., Lakhwani D. et al. , 2018. A high-quality genome sequence of Rosa chinensis to elucidate ornamental traits. Nat. Plants 4: 473–484. 10.1038/s41477-018-0166-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iezzoni A., Peace C., Main D., Bassil N., Coe M. et al. , 2017. RosBREED2: progress and future plans to enable DNA-informed breeding in the Rosaceae. Acta Hortic. (1172): 115–118. 10.17660/ActaHortic.2017.1172.20 [DOI] [Google Scholar]
- Illa E., Sargent D. J., Lopez Girona E., Bushakra J., Cestaro A. et al. , 2011. Comparative analysis of rosaceous genomes and the reconstruction of a putative ancestral genome for the family. BMC Evol. Biol. 11: 9 10.1186/1471-2148-11-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao X., Zheng X., Ma L., Kutty G., Gogineni E. et al. , 2013. A benchmark study on error assessment and quality control of CCS reads derived from the PacBio RS. J. Data Mining Genomics Proteomics 4: 16008 10.4172/2153-0602.1000136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jibran R., Dzierzon H., Bassil N., Bushakra J. M., Edger P. P. et al. , 2018. Chromosome-scale scaffolding of the black raspberry (Rubus occidentalis L.) genome based on chromatin interaction data. Hortic. Res. 5: 8 10.1038/s41438-017-0013-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jombart T., 2008. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24: 1403–1405. 10.1093/bioinformatics/btn129 [DOI] [PubMed] [Google Scholar]
- Jombart T., and Ahmed I., 2011. adegenet 1.3–1: new tools for the analysis of genome-wide SNP data. Bioinformatics 27: 3070–3071. 10.1093/bioinformatics/btr521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung S., Cestaro A., Troggio M., Main D., Zheng P. et al. , 2012. Whole genome comparisons of Fragaria, Prunus and Malus reveal different modes of evolution between Rosaceous subfamilies. BMC Genomics 13: 129 10.1186/1471-2164-13-129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung S., Lee T., Cheng C., Buble K., Zheng P. et al. , 2019. 15 years of GDR: New data and functionality in the Genome Database for Rosaceae. Nucleic Acids Res. 47: D1137–D1145. 10.1093/nar/gky1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamneva O. K., Syring J., Liston A., and Rosenberg N. A., 2017. Evaluating allopolyploid origins in strawberries (Fragaria) using haplotypes generated from target capture sequencing. BMC Evol. Biol. 17: 180 10.1186/s12862-017-1019-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusch S., and Panstruga R., 2017. mlo-based resistance: an apparently universal “weapon” to defeat powdery mildew disease. Mol. Plant Microbe Interact. 30: 179–189. 10.1094/MPMI-12-16-0255-CR [DOI] [PubMed] [Google Scholar]
- Le Dantec L., Cardinet G., Bonet J., Fouché M., Boudehri K. et al. , 2010. Development and mapping of peach candidate genes involved in fruit quality and their transferability and potential use in other Rosaceae species. Tree Genet. Genomes 6: 995–1012. 10.1007/s11295-010-0308-8 [DOI] [Google Scholar]
- Lerceteau-Köhler E., Moing A., Guérin G., Renaud C., Petit A. et al. , 2012. Genetic dissection of fruit quality traits in the octoploid cultivated strawberry highlights the role of homoeo-QTL in their control. Theor. Appl. Genet. 124: 1059–1077. 10.1007/s00122-011-1769-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34: 3094–3100. 10.1093/bioinformatics/bty191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Wysoker A., Fennell T., Ruan J., Homer N. et al. , 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Feng F., and Cheng L., 2012. Expression patterns of genes involved in sugar metabolism and accumulation during apple fruit development. PLoS One 7: e33055 10.1371/journal.pone.0033055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Li D., Feng F., Zhang S., Ma F. et al. , 2016. Proteomic analysis reveals dynamic regulation of fruit development and sugar and acid accumulation in apple. J. Exp. Bot. 67: 5145–5157. 10.1093/jxb/erw277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linsmith G., Rombauts S., Montanari S., Deng C. H., Celton J. et al. , 2019. Pseudo-chromosome length genome assembly of a double haploid “Bartlett” pear (Pyrus communis L.). Gigascience 8: giz138 10.1093/gigascience/giz138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S., Wang J., Chitsaz F., Derbyshire M. K., Geer R. C. et al. , 2020. CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res. 48: D265–D268. 10.1093/nar/gkz991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriguchi T., Abe K., Sanada T., and Yamaki S., 1992. Levels and role of sucrose synthase, sucrose-phosphate synthase, and acid invertase in sucrose accumulation in fruit of Asian pear. J. Am. Soc. Hortic. Sci. 117: 274–278. 10.21273/JASHS.117.2.274 [DOI] [Google Scholar]
- Onda Y., Takahagi K., Shimizu M., Inoue K., and Mochida K., 2018. Multiplex PCR targeted amplicon sequencing (MTA-Seq): Simple, flexible, and versatile SNP genotyping by highly multiplexed PCR Amplicon Sequencing. Front Plant Sci 9: 201 10.3389/fpls.2018.00201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozsolak F., and Milos P. M., 2011. RNA sequencing: advances, challenges and opportunities. Nat. Rev. Genet. 12: 87–98. 10.1038/nrg2934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quilot B., Wu B. H., Kervella J., Génard M., Foulongne M. et al. , 2004. QTL analysis of quality traits in an advanced backcross between Prunus persica cultivars and the wild relative species P. davidiana. Theor. Appl. Genet. 109: 884–897. 10.1007/s00122-004-1703-z [DOI] [PubMed] [Google Scholar]
- Perez C. M., Perdon A. A., Resurreccion A. P., Villareal R. M., and Juliano B. O., 1975. Enzymes of carbohydrate metabolism in the developing rice grain. Plant Physiol. 56: 579–583. 10.1104/pp.56.5.579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjan R., Rani A., Metwally A., McGee H. S., and Perkins D. L., 2016. Analysis of the microbiome: advantages of whole genome shotgun vs. 16S amplicon sequencing. Biochem. Biophys. Res. Commun. 469: 967–977. 10.1016/j.bbrc.2015.12.083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond O., Gouzy J., Just J., Badouin H., Verdenaud M. et al. , 2018. The Rosa genome provides new insights into the domestication of modern roses. Nat. Genet. 50: 772–777. 10.1038/s41588-018-0110-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynaud S., Ragel P., Rojas T., and Mérida Á., 2016. The N-terminal part of Arabidopsis thaliana starch synthase 4 determines the localization and activity of the enzyme. J. Biol. Chem. 291: 10759–10771. 10.1074/jbc.M115.698332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reidel E. J., Turgeon R., and Cheng L., 2008. A maltose transporter from apple is expressed in source and sink tissues and complements the arabidopsis maltose export-defective mutant. Plant Cell Physiol. 49: 1607–1613. 10.1093/pcp/pcn134 [DOI] [PubMed] [Google Scholar]
- Rennie E. A., Hansen S. F., Baidoo E. E. K., Hadi M. Z., Keasling J. D. et al. , 2012. Three members of the Arabidopsis glycosyltransferase family 8 are xylan glucuronosyltransferases. Plant Physiol. 159: 1408–1417. 10.1104/pp.112.200964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J. T., Thorvaldsdóttir H., Wenger A. M., Zehir A., and Mesirov J. P., 2017. Variant review with integrative genomics viewer. Cancer Res. 77: e31–e34. 10.1158/0008-5472.CAN-17-0337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ru S., Hardner C., Carter P. A., Evans K., Main D. et al. , 2016. Modeling of genetic gain for single traits from marker-assisted seedling selection in clonally propagated crops. Hortic. Res. 3: 16015 10.1038/hortres.2016.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar J. A., Rubio M., Ruiz D., Tartarini S., Martínez-Gómez P. et al. , 2015. SNP development for genetic diversity analysis in apricot. Tree Genet. Genomes 11: 15 10.1007/s11295-015-0845-2 [DOI] [Google Scholar]
- Sargent D. J., Marchese A., Simpson D. W., Howad W., Fernández-fernández F. et al. , 2009. Development of “universal” gene-specific markers from Malus spp. cDNA sequences, their mapping and use in synteny studies within Rosaceae. Tree Genet. Genomes 5: 133–145. 10.1007/s11295-008-0178-5 [DOI] [Google Scholar]
- Shirasawa K., Kuwata C., Watanabe M., Fukami M., Hirakawa H. et al. , 2016. Target amplicon sequencing for genotyping genome-wide single nucleotide polymorphisms identified by whole-genome resequencing in peanut. Plant Genome 9: 1–8. 10.3835/plantgenome2016.06.0052 [DOI] [PubMed] [Google Scholar]
- Shulaev V., Sargent D. J., Crowhurst R. N., Mockler T. C., Folkerts O. et al. , 2011. The genome of woodland strawberry (Fragaria vesca). Nat. Genet. 43: 109–116. 10.1038/ng.740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein O., and Granot D., 2019. An overview of sucrose synthases in plants. Front Plant Sci 10: 95 10.3389/fpls.2019.00095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International Peach Genome Initiative , 2013. The high-quality draft genome of peach (Prunus persica) identifies unique patterns of genetic diversity, domestication and genome evolution. Nat. Genet. 45: 487–494. 10.1038/ng.2586 [DOI] [PubMed] [Google Scholar]
- Thirugnanasambandam P. P., Mason P. J., Hoang N. V., Furtado A., Botha F. C. et al. , 2019. Analysis of the diversity and tissue specificity of sucrose synthase genes in the long read transcriptome of sugarcane. BMC Plant Biol. 19: 160 10.1186/s12870-019-1733-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong X., Wang Z., Ma B., Zhang C., Zhu L. et al. , 2018. Structure and expression analysis of the sucrose synthase gene family in apple. J. Integr. Agric. 17: 847–856. 10.1016/S2095-3119(17)61755-6 [DOI] [Google Scholar]
- Tsai C.Y., Salamini F., and Nelson O.E., 1970. Enzymes of carbohydrate metabolism in the developing endosperm of maize. Plant Physiol. 46: 299–306. 10.1104/pp.46.2.299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanBuren R., Bryant D., Bushakra J. M., Vining K. J., Edger P. P. et al. , 2016. The genome of black raspberry (Rubus occidentalis). Plant J. 87: 535–547. 10.1111/tpj.13215 [DOI] [PubMed] [Google Scholar]
- VanBuren R., Wai C. M., Colle M., Wang J., Sullivan S. et al. , 2018. A near complete, chromosome-scale assembly of the black raspberry (Rubus Occidentalis) genome. Gigascience 7: giy094 10.1093/gigascience/giy094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco R., Zharkikh A., Affourtit J., Dhingra A., Cestaro A. et al. , 2010. The genome of the domesticated apple (Malus ×domestica Borkh.). Nat. Genet. 42: 833–839. 10.1038/ng.654 [DOI] [PubMed] [Google Scholar]
- Verde I., Jenkins J., Dondini L., Micali S., Pagliarani G. et al. , 2017. The Peach v2.0 release: high-resolution linkage mapping and deep resequencing improve chromosome-scale assembly and contiguity. BMC Genomics 18: 225 10.1186/s12864-017-3606-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma S., Zurn J. D., Salinas N., Mathey M., Denoyes B. et al. , 2017. Clarifying sub-genomic positions of QTLs for flowering habit and fruit quality in U.S. strawberry (Fragaria ×ananassa) breeding populations using pedigree-based QTL analysis. Hortic. Res. 4: 17062 10.1038/hortres.2017.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilanova S., Sargent D. J., Arús P., and Monfort A., 2008. Synteny conservation between two distantly-related Rosaceae genomes: Prunus (the stone fruits) and Fragaria (the strawberry). BMC Plant Biol. 8: 67 10.1186/1471-2229-8-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Tang H., Debarry J. D., Tan X., Li J. et al. , 2012. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40:e49 10.1093/nar/gkr1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Zhao S., Gu C., Zhou Y., Zhou H. et al. , 2013. Deep RNA-Seq uncovers the peach transcriptome landscape. Plant Mol. Biol. 83: 365–377. 10.1007/s11103-013-0093-5 [DOI] [PubMed] [Google Scholar]
- Wang M., Zhang T., Peng H., Luo S., Tan J. et al. , 2018. Rice premature leaf senescence 2, encoding a glycosyltransferase (GT), is involved in leaf senescence. Front Plant Sci 9: 560 10.3389/fpls.2018.00560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X., Jackson P. A., McIntyre C. L., Aitken K. S., and Croft B., 2006. Associations between DNA markers and resistance to diseases in sugarcane and effects of population substructure. Theor. Appl. Genet. 114: 155–164. 10.1007/s00122-006-0418-8 [DOI] [PubMed] [Google Scholar]
- Wei X., Liu F., Chen C., Ma F., and Li M., 2014. The Malus domestica sugar transporter gene family: identifications based on genome and expression profiling related to the accumulation of fruit sugars. Front Plant Sci 5: 569 10.3389/fpls.2014.00569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitemier K., Straub S. C. K., Cronn R. C., Fishbein M., Schmickl R. et al. , 2014. Hyb-Seq: Combining target enrichment and genome skimming for plant phylogenomics. Appl. Plant Sci. 2: 1400042 10.3732/apps.1400042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- HC class I allele discovery by PacBio circular consensus sequencing. Hum. Immunol. 76: 891–896. 10.1016/j.humimm.2015.03.022 [DOI] [PubMed] [Google Scholar]
- Worthington M. L., Aryal R., Bassil N. V., Mead D., Fernandez G. E. et al. , 2020. Development of new genomic resources and tools for molecular breeding in blackberry. Acta Hortic. (1277): 39–46. 10.17660/ActaHortic.2020.1277.6 [DOI] [Google Scholar]
- Wu T. D., Reeder J., Lawrence M., Becker G., and Brauer M. J., 2016. GMAP and GSNAP for genomic sequence alignment: Enhancements to speed, accuracy, and functionality. Methods Mol. Biol. 1418: 283–334. 10.1007/978-1-4939-3578-9_15 [DOI] [PubMed] [Google Scholar]
- Xu Y., and Crouch J. H., 2008. Marker-assisted selection in plant breeding: from publications to practice. Crop Science 48: 391–407. [Google Scholar]
- Yamamoto T., Kimura T., Saito T., Kotobuki K., Matsuta N. et al. , 2004. Genetic linkage maps of Japanese and European pears aligned to the apple consensus map. Acta Hortic. 663: 51–56. 10.1007/s00122-002-0966-5 [DOI] [Google Scholar]
- Zhang C., Yu M., Ma R., Shen Z., Zhang B. et al. , 2015. Structure, expression profile, and evolution of the sucrose synthase gene family in peach (Prunus persica). Acta Physiol. Plant. 37: 81 10.1111/febs.12595 [DOI] [Google Scholar]
- Zhao C., Hua L., Liu X., Li Y., Shen Y. et al. , 2017. Sucrose synthase FaSS1 plays an important role in the regulation of strawberry fruit ripening. Plant Growth Regul. 81: 175–181. https://link.springer.com/article/10.1007%2Fs10725-016-0189-4 [Google Scholar]
- Zorrilla-Fontanesi Y., Cabeza A., Domínguez P., Medina J. J., Valpuesta V. et al. , 2011. Quantitative trait loci and underlying candidate genes controlling agronomical and fruit quality traits in octoploid strawberry (Fragaria ×ananassa). Theor. Appl. Genet. 123: 755–778. 10.1007/s00122-011-1624-6 [DOI] [PubMed] [Google Scholar]
- Zurn J. D., Carter K. A., Yin M. H., Worthington M., Clark J. R. et al. , 2018. Validating blackberry seedling pedigrees and developing an improved multiplexed microsatellite fingerprinting set. J. Am. Soc. Hortic. Sci. 143: 381–390. 10.21273/JASHS04474-18 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Raw CCS reads have been deposited to the National Center for Biotechnology Information under BioProject number PRJNA633906. Genotypic Data from the resulting KASP assay and phenotypic data are provided in Supplementary Table S2. VCF files, Python code, and R scripts are available at the following github repository: github.com/Bassil-Lab/Zurn-et-al-2020-G3-Blackberry-SSC. KASP assays (Supplementary Table S3) are available through LGC. Supplemental material available at figshare: https://doi.org/10.25387/g3.12767864.