Abstract
Social wasps of the genus Vespula have spread to nearly all landmasses worldwide and have become significant pests in their introduced ranges, affecting economies and biodiversity. Comprehensive genome assemblies and annotations for these species are required to develop the next generation of control strategies and monitor existing chemical control. We sequenced and annotated the genomes of the common wasp (Vespula vulgaris), German wasp (Vespula germanica), and the western yellowjacket (Vespula pensylvanica). Our chromosome-level Vespula assemblies each contain 176–179 Mb of total sequence assembled into 25 scaffolds, with 10–200 unanchored scaffolds, and 16,566–18,948 genes. We annotated gene sets relevant to the applied management of invasive wasp populations, including genes associated with spermatogenesis and development, pesticide resistance, olfactory receptors, immunity and venom. These genomes provide evidence for active DNA methylation in Vespidae and tandem duplications of venom genes. Our genomic resources will contribute to the development of next-generation control strategies, and monitoring potential resistance to chemical control.
Keywords: Vespula germanica, Vespula pensylvanica, Vespula vulgaris, Hymenoptera, social insects, genomes
Social wasps (Hymenoptera: Vespidae) are remarkable because their highly eusocial lifestyle appears to have evolved independently of other eusocial Hymenoptera (Piekarski et al. 2018; Hines et al. 2007; Peters et al. 2017). The eusocial lifestyle is characterized by overlapping generations of adults living together, cooperative care of offspring and reproductive division of labor (Michener and Brothers 1974). Along with their foraging flexibility and predatory ability, eusociality appears to play a major role in the ecological success of social waps (Grimaldi and Engel 2005; Lester and Beggs 2019).
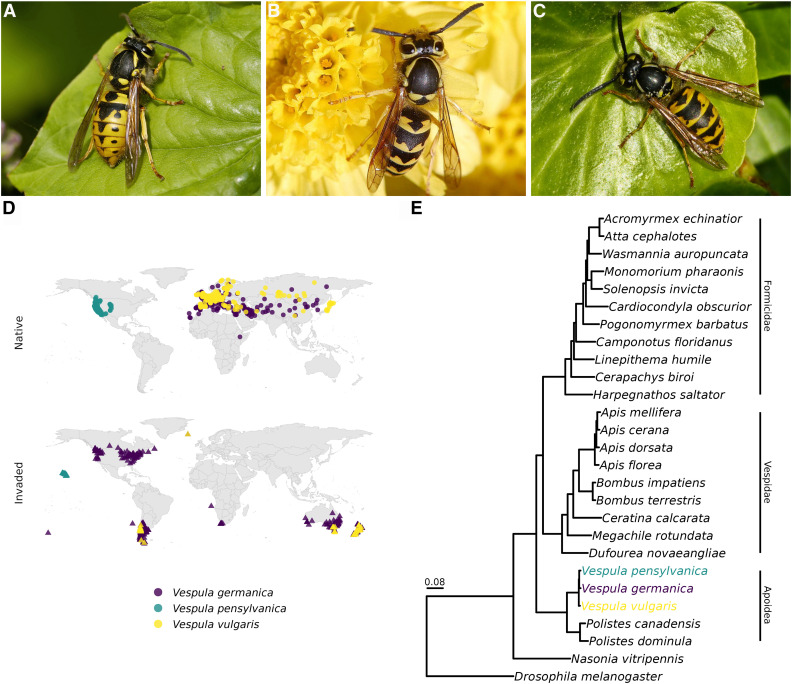
Vespid wasps can be effective pollinators of plants including ivy or orchid species (Cheng et al. 2009; Jacobs et al. 2010). They are generalist predators and effect biological control of some pest species (Donovan 2003). Their ecological success can also be problematic. Invasive colonies of common wasps (Vespula vulgaris) can contain up to 230,000 workers, while nests of the western yellowjacket (Vespula pensylvanica) containing up to half a million individuals have been observed (Lester and Beggs 2019). Colonies are smaller in the native ranges (Archer and Turner 2014 p. 61). In New Zealand’s native beech forests, Vespid wasp populations can reach up to 40 nests per hectare and have a biomass similar to, or greater than, the combined biomasses of birds and mammals (Thomas et al. 1990; Lester et al. 2017). These invasive Vespid populations (Figure 1; Lester and Beggs 2019) have major impacts on ecosystems because of their large colony sizes, reproductive capacity and flexible predation.
Figure 1.
Vespula species are major invasive pests. A–C. Examples of (A) Vespula germanica, (B) Vespula pensylvanica (Jon Sullivan, public domain), and (C) Vespula vulgaris (Sid Mosdell, CC BY 2.0). D. Native and invaded ranges of the sequenced Vespula species. E. Rooted phylogenetic species tree of Vespula species and other sequenced Hymenoptera, with Drosophila melanogaster as an outspecies, derived from 415 orthogroups using OrthoFinder.
Current control methods for vespid wasps are limited, with pesticides containing Fipronil being the most common and widespread chemical control method (Edwards et al. 2017). The use of this neurotoxic pesticide over large areas, and over consecutive years, may select for resistance. Fipronil resistance has been observed in other economically important insect pests (Matsumura et al. 2008; reviewed in Feyereisen et al. 2015). Next-generation pest control technologies, including gene drives, have been proposed as part of an alternative solution for controlling or eradicating invasive social wasps (Dearden et al. 2018). Targets for genetic modification include developmental genes associated with wasp fitness or fecundity (Lester and Beggs 2019; Dearden et al. 2018). Gene drives that have immune system targets have been proposed and developed in the laboratory for other pests such as mosquitoes (e.g., Gantz et al. 2015; Dong et al. 2018). Detailed knowledge of Vespula genomes is required to ensure that targeted control of Vespids does not affect other beneficial Hymenoptera, such as honeybees or biocontrol agents.
Five genomes from the Polistes genus of the Vespidae subclade are available in the NCBI Genome database. Genomes for Polistes canadensis and Polistes dominula were assembled from short reads (Standage et al. 2016; Patalano et al. 2015). Genomes for Polistes dorsalis, Polistes fuscatus and Polistes metricus, based on Pacific Biosciences single-molecule sequencing, were recently deposited in the NCBI database (Miller et al. 2020). The three long-read genomes have improved contiguity compared to the short-read genomes produced for P. dominula and P. canadensis (Standage et al. 2016; Patalano et al. 2015; Miller et al. 2020). Here, we report chromosome-level assemblies for three Vespula species that are invasive pests in their introduced ranges (Figure 1; Lester and Beggs 2019). We undertake manual and computational annotation and phylogenomic analyses, with emphasis on sets of genes relevant to chemical control, olfaction, venom and DNA methylation. These resources will be useful for understanding the biology of vespid wasps, developing next-generation control strategies, and monitoring resistance to current chemical control.
Materials and Methods
Genome assembly and scaffolding
Vespula pensylvanica samples were collected in Volcano, Hawaii in August 2017 (lat 19.43, lon -155.21). The V. pensylvanica assembly was produced by Dovetail Genomics (https://dovetailgenomics.com/), starting with an Illumina library generated from a single haploid male to produce 115.5 Gb of 150bp paired-end reads. After trimming adaptors and removing bases or truncating reads with low quality scores, the draft genome was assembled with Meraculous 2.2.6 using a kmer size of 55, which produced the best fit with a constrained heterozygous model (Chapman et al. 2011, 2016). Chromosome-scale scaffolds were generated using Chicago and Dovetail Hi-C data from diploid females, and implemented using Dovetail’s proprietary HiRise software.
We collected V. vulgaris samples in May 2015, from Richmond Hill Forest Park, New Zealand (lat -41.3292, lon 173.4637). V. germanica samples were collected in March 2018, from Lincoln, New Zealand (lat -43.6384, lon 172.4784). We used libraries generated from a single, haploid male to produce 17.1 Gb of 150b paired-end reads and 27.3 Gb of 125b paired-end reads, respectively. After trimming adaptor sequences, removing contaminants and verifying pairing with BBMap 38.00 (Bushnell 2014), we assembled draft genomes with Meraculous 2.2.6 (Chapman et al. 2011, 2016). We repeated assembly with a range of parameters, and used BUSCO analysis (Simão et al. 2015), assembly size and contiguity statistics to choose the best set of parameters for each dataset. Code for running and assessing the assemblies is hosted at https://github.com/tomharrop/vger-illumina and https://github.com/tomharrop/vvul-illumina. Draft genomes were scaffolded by Phase Genomics using Hi-C data generated from pools of 20 larvae. Chromatin conformation capture data were generated using Phase Genomics Proximo Hi-C 2.0 Kit, which is a commercially available version of the Hi-C protocol (Lieberman-Aiden et al. 2009). The Phase Genomics Proximo Hi-C genome scaffolding platform was used to create chromosome-scale scaffolds from the corrected assembly as described Bickhart et al. (2017).
Genome curation
Command-line arguments and scripts can be found at https://github.com/jguhlin/vespula_paper. Assembled genomes were cleared of contamination by removing contigs which had BLAST taxonomy results which did not include Polistes, Vespula, or the word “wasp”. Contigs without BLAST results were kept if they contained predicted genes found in a Hymenoptera orthogroup from an initial gene prediction. Remaining contigs were only kept if they fell within 2 standard deviations of mean GC% of our kept contigs. The largest 25 chromosomes of V. pensylvanica were renamed to chromosomes and ordered according to size. Our V. vulgaris and V. germanica assemblies were aligned to the V. pensylvanica genome using D-GENIES, which inserts contigs into syntenic locations, flanked by 100 Ns, to assign chromosome names to those most related to those in V. pensylvanica (Cabanettes and Klopp 2018). Scaffolds were numbered with four digits in order of size.
Repeat masking
Repeats were identified using RepeatModeler 2.0.1 (github.com/Dfam-consortium/RepeatModeler) and RepeatMasker 4.0.9 (repeatmasker.org/RMDownload.html) via the funannotate pipeline (Palmer and Stajich 2019).
RNA sequencing
Vespula vulgaris queens, workers, and larvae were sampled from mature nests in the native range of Belgium and the introduced range in New Zealand and total RNA transcriptome data were generated as described by Gruber et al. (2019).
Gene prediction
We performed iterative gene prediction using the Funannotate pipeline v1.6.0, manual annotation, and extrinsic protein evidence (Palmer and Stajich 2019). For V. vulgaris we used RNA-seq reads (described in the previous section) from V. vulgaris queens, workers, and larvae as additional evidence. The reads were trimmed with sickle (github.com/najoshi/sickle) and aligned to our assembly using STAR in two-pass mode (Dobin et al. 2012). Gene predictions were performed on the assembly using funannotate predict with the RNA alignments and extrinsic protein of all Vespula proteins from NCBI, Apis mellifera, Nasonia vitripennis, and the UniProt SWISS-PROT database (Boutet et al. 2007; Pruitt et al. 2005; The Honeybee Genome Sequencing Consortium 2006; Werren et al. 2010).
Initial predictions were evaluated with GeneValidator with UniProt SWISS-PROT database as the high-quality targets. Genes whose protein predictions scored > 90 were used to train Augustus via the optimize_augustus.pl script (Drăgan et al. 2016). The prediction step of funannotate was then re-run as before using the trained vvulg AUGUSTUS species definition. This allowed retention of high-quality gene predictions from the target species to be used as a training set for AUGUSTUS gene prediction, a component of the funannotate pipeline. This process was repeated for V. pensylvanica and V. germanica using V. vulgaris species definition as the initial AUGUSTUS species in the first iteration of gene prediction, generating a species-specific configuration in the following round. This two-step gene prediction with validation and training using high-confidence gene calls between the first and second round allowed for the creation of species-specific AUGUSTUS models.
Manual curation
Genes were manually curated in WebApollo (Lee et al. 2013). These manual annotations took precedence over intersecting computational gene predictions. Manual annotation was performed on V. vulgaris and lifted over to V. germanica and V. pensylvanica where possible.
Gene family specific predictions
Gene-family specific predictions were enhanced using AUGUSTUS-PPX for the LGIC and Olfactory families (Keller et al. 2011). Protein sequences of interest from external sources were clustered based on bitscore using BLAST+ and MCL (Dongen 2000). Clusters of protein sequences were converted to protein profiles via AUGUSTUS tool msa2prfl.pl Assemblies were searched with fastBlockSearch and gene prediction was performed on matched regions with an additional flanking sequence of 1kbp. These predictions took precedence over intersecting computationally predicted genes.
Annotation
Further annotation was performed with funannotate using InterProScan 5.32-71.0 (Mitchell et al. 2019). Genes were renamed using custom scripts. Protein predictions were compared with GeneValidator to both our Hymenoptera + Drosophila Protein Set and UniProt-SwissProt to generate GV scores and statistics. Proteins were compared with the publicly available genomes from Hymenoptera base using OrthoFinder (Elsik et al. 2016; Emms and Kelly 2019).
Methylation analysis
Nucleotide and dinucleotide content of gene body sequences were calculated using a custom perl script. CpG[o/e] was calculated as the number of CpG dinucleotides divided by the number of C nucleotides times the number of G nucleotides. The number of components in CpG[o/e] distributions was estimated in R (R Core Team 2015) using mclust model-based clustering (Scrucca et al. 2016). The best fitting model was identified among several non-nested models using Bayesian information criteria (BIC).
Data availability
Raw sequence data are hosted in the NCBI Sequence Read Archive under accession PRJNA643352. Assembled genomes are available on GenBank under accessions JACSDY000000000 (Vespula pensylvanica), JACSDZ000000000 (Vespula germanica) and JACSEA000000000 (Vespula vulgaris). Supplemental material available at figshare: https://doi.org/10.25387/g3.12885599.
Results and discussion
Genome assemblies and annotation
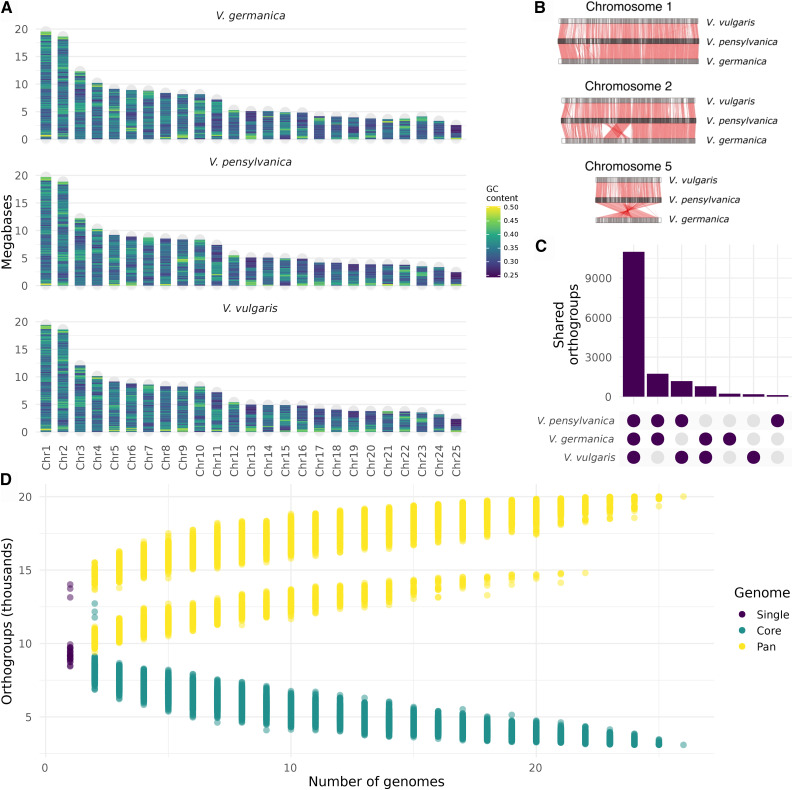
We used a combination of short-read Illumina sequencing and Hi-C scaffolding to assemble draft genomes for V. germanica, V. pensylvanica, and V. vulgaris. The genomes each contain 176–179 Mb of total sequence assembled into 25 superscaffolds (Figure 2A; Supplementary table 1; N50 length 8.30–8.53 Mb), which likely represent the 25 chromosomes observed in Vespula karyotypes (Hoshiba et al. 1989). Each draft genome also contains 10–200 unanchored scaffolds (N50 lengths 1.77–2.28 kb; Supplementary table 2). These genomes are similar in size to the genomes of the closely related European paper wasp, Polistes dominula (Standage et al. 2016), and the red paper wasp, Polistes canadensis (Patalano et al. 2015). However, the contiguity in our Vespula assemblies is higher than Illumina-based assemblies of other Vespidae, and comparable to the latest-generation Apis mellifera assembly (Table 1). We ordered and named scaffolds in the Vespula assemblies based on scaffold length in V. pensylvanica. The three genomes are highly syntenic, with evidence of some structural rearrangements (Figure 2B; Supplementary Figure 1). Repeat masking masked 17.86%, 18.75% and 18.71% of the V. vulgaris, V. pensylvanica, and V. germanica genomes, respectively. We predicted 16,751, 17,854, and 19,142 genes for V. vulgaris, V. germanica, and V. pensylvanica, respectively. We found between 92.2% and 96.0% of expected single-copy orthologs using BUSCO with the Hymenoptera lineage dataset (Simão et al. 2015). The contiguity of our Vespula assemblies and completeness of our annotations indicates that the combination of short-read sequencing and Hi-C scaffolding on haploid material is an effective strategy for assembling high-quality hymenopteran genomes.
Figure 2.
A. Assembled chromosome lengths of the three Vespula species. Chromosomes were named in order of scaffold length in V. pensylvanica. B. Synteny between selected chromosomes of Vespula species. Chromosome 1 has minor translocations and inversions but is syntenic overall. Chromosome 2 has a larger inversion between V. pensylvanica and V. germanica that is not present in V. vulgaris. Chromosome 5 has a large translocation and inversion between V. pensylvanica and V. germanica. Chromosome lengths are scaled to chromosome 1. C. Shared orthogroups between the three Vespula species. D. The pan and core genomes of Hymenoptera. The core genome (the set of orthogroups present in all genomes sampled) decreases as more genomes are sampled, to a final core genome size of 3,092 orthogroups when all 26 hymenopteran genomes are compared. The pan genome (the total set of orthogroups present in one or more of the sampled genomes) does not continue to grow as more genomes are sampled, indicating a closed hymenopteran pan genome. We predicted more orthogroups (13,141–14,022 orthogroups) in Vespula species than in non-Vespula Hymenoptera (median 9,193 orthogroups), which resulted in a larger pan genome in comparisons that included Vespula species. We only analyzed genes that were assigned to an orthogroup, and we plotted a random subset of 1,000 comparisons for genome numbers that resulted in more than 1000 comparisons.
Table 1. Comparison of Vespidae and honeybee genome assembles. 1. Apart from our Vespula assemblies, repeat content was counted from lowercase nucleotides in published assemblies.
| Species | Sequencing Strategy | Total Sequence | Largest | Scaffolds/ contigs | N50 length | Ns | Gaps | Repeats1 (%) |
|---|---|---|---|---|---|---|---|---|
| Vespula vulgaris | Illumina + Hi-C | 176,275,134 | 19,426,332 | 35 | 8,304,510 | 4,147,610 | 49,679 | 17.12 |
| Vespula germanica | Illumina + Hi-C | 178,312,246 | 19,524,135 | 133 | 8,396,154 | 1,783,864 | 18,963 | 18.89 |
| Vespula pensylvanica | Illumina + Hi-C | 179,379,562 | 19,704,315 | 225 | 8,532,720 | 444,951 | 4,987 | 19.15 |
| Polistes canadensis | Illumina | 211,202,212 | 3,185,661 | 3,836 | 521,566 | 14,106,256 | 15,755 | 41.60 |
| Polistes dominula | Illumina | 208,026,220 | 7,126,315 | 1,483 | 1,625,592 | 7,426,626 | 14,286 | 44.50 |
| Polistes fuscatus | PacBio Sequel + Illumina + Dovetail | 219,116,742 | 19,629,704 | 187 | 9,116,088 | 4,436,170 | 1,204 | 41.79 |
| Polistes metricus | PacBio Sequel + Illumina | 219,838,961 | 15,979,625 | 216 | 4,634,047 | 1,082,619 | 459 | 44.48 |
| Polistes dorsalis | 10x Genomics | 209,288,276 | 20,305,868 | 5,129 | 5,372,633 | 4,771,923 | 8,816 | 42.35 |
| Apis mellifera 4.5 | Sanger + SOLiD + 454 | 250,287,000 | 29,893,408 | 5,321 | 13,219,345 | 21,165,099 | 12,690 | 5.28 |
| Apis mellifera HAv3.1 | PacBio + 10x Chromium + BioNano + Hi-C | 225,250,884 | 27,754,200 | 177 | 13,619,445 | 1,313,614 | 51 | 44.63 |
To predict divergence time of Vespula species from Polistes, we reconstructed a phylogeny using other hymenopteran genomes. Based on a published estimate for Hymenoptera (Peters et al. 2017), the divergence time of Vespid wasps from their last common ancestor with P. dominula is estimated to be 51 million years (95% CI: 34–71 million years). Mitochondrial genomes suggest a divergence time of 75 mya (Huang et al. 2019). Scaling our ultrametric phylogenetic tree to the former estimate places separation of V. vulgaris from V. pensylvanica and V. germanica at ∼6 mya, and separation of V. pensylvanica from V. germanica at 4.5 mya (Supplementary Figure 2).
We manually curated 361 gene models in V. vulgaris and used these curations to improve automated prediction steps for the other two species. During manual curation, we focused on a range of gene sets relevant to the evolution and applied management of invasive Vespula spp., including olfactory receptors, pesticide resistance, immunity and viruses, venom, and spermatogenesis and development.
To investigate relationships between Vespula genes, we clustered predicted proteins into orthogroups with predicted proteins from the Hymenoptera Genome Database, using Drosophila melanogaster as the outgroup (Hoskins et al. 2015; Elsik et al. 2016; Emms and Kelly 2019). Each orthogroup contains a set of genes putatively descended from a single gene in the last common ancestor of the species represented in the orthogroup. Between 82.6% and 88.4% of our predicted Vespula proteins belonged to orthogroups. V. vulgaris shares 12,560 and 12,084 orthogroups with V. pensylvanica and V. germanica, respectively, and V. pensylvanica shares 13,209 orthogroups with V. germanica (Figure 2C). Orthogroups including other hymenopteran species allowed us to predict the core- and pan-genomes for Hymenoptera (Figure 2D). This analysis suggests that Hymenoptera have a closed pan-genome, because as we include more genomes the rate of discovery of new orthogroups decreases. We also observed more orthogroups in our Vespid genomes than in other Hymenoptera, which could indicate over-prediction resulting from our annotation.
93.7–94.5% of predicted orthogroups in P. canadensis and 96.0–96.9% in P. dominula clustered into orthogroups that contained a Vespula gene. Vespula genes from our annotations were present in 58.9–62.8% of all orthogroups across Hymenoptera and D. melanogaster, compared to 40.7% and 41.2% for P. canadensis and P. dominula, respectively (Table 2). Other hymenopteran species had a member in 37.8–44.5% of othogroups, indicating gene families may be missing in predictions among other species. Most of the genes we annotated were in shared orthogroups, with 100 genes (0.1–0.4% per species) in species-specific orthogroups. Other hymenopteran genomes had 0–3.3% (mean 1.7%) of genes in species-specific orthogroups. Our annotation results suggest that Vespid wasps have more genes than other Hymenoptera and/or gene annotation in other Hymenoptera is incomplete. This could be resolved by re-annotation of other hymenopteran genes using a comparative approach.
Table 2. Gene content and orthogroup representation for selected hymenopteran genomes.
| Species | Genes | Orthogroups represented | Orthogroups represented (%) | Species-specific orthogroups | Genes in species-specific orthogroups (%) |
|---|---|---|---|---|---|
| Vespula vulgaris | 16,751 | 13,141 | 58.9% | 5 | 0.1% |
| Vespula germanica | 17,854 | 13,739 | 61.6% | 23 | 0.4% |
| Vespula pensylvanica | 19,142 | 14,022 | 62.8% | 11 | 0.1% |
| Polistes canadensis | 10,518 | 9,086 | 40.7% | 12 | 0.2% |
| Polistes dominula | 11,069 | 9,193 | 41.2% | 27 | 0.5% |
| Apis mellifera | 14,064 | 8,949 | 40.1% | 35 | 0.6% |
| Nasonia vitripennis | 14,647 | 8,901 | 39.9% | 616 | 11.1% |
Evidence for active DNA methylation
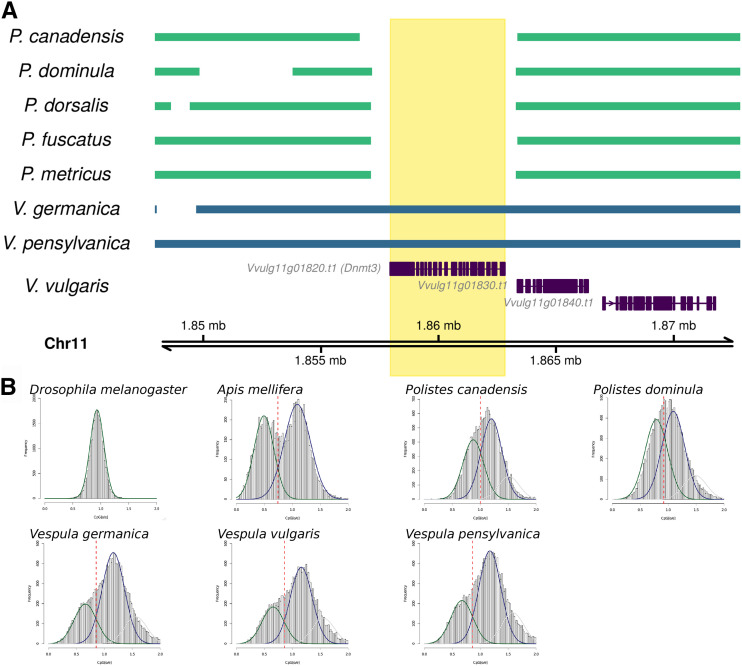
DNA methylation has been functionally linked to caste specification in honeybees and ants and division of labor in honeybees (Herb et al. 2012; Kucharski et al. 2008; Bonasio et al. 2012; Guan et al. 2013). DNA methylation may be integral for aspects of eusociality (reviewed by Li-Byarlay et al. 2013), although recent studies have found no consistent link (Bewick et al. 2017; Glastad et al. 2017). In mice, Dnmt3 enzymes catalyze de novo DNA methylation (reviewed by Goll and Bestor 2005). The genomes of P. canadensis and P. dominula do not encode a Dnmt3 homolog (Standage et al. 2016; Patalano et al. 2015; Bewick et al. 2017). While this manuscript was under revision, three new Polistes genomes based on Pacific Biosciences single-molecule sequencing were deposited in the NCBI database (Miller et al. 2020). Genome annotations have not been reported for the three long-read Polistes genomes. We aligned all five Polistes genomes and our V. germanica and V. pensylvanica genomes against Chr11 of the V. vulgaris genome, which encodes Dnmt3. None of the five Polistes genomes have a region that aligns to the V. vulgaris Dnmt3 locus (Figure 3A). All three Vespula genomes encode an ortholog of Dnmt3, indicating the lack of this gene in Polistes is due to gene loss following the divergence from the Vespula lineage approximately 50 million years ago (Peters et al. 2017). Vespula genomes all contain a single ortholog of Dnmt1 and have evidence of active DNA methylation (Figure 3B; Supplementary Figure 3).
Figure 3.
A. Whole-genome alignments of five Polistes genomes and genomes of V. germanica and V. pensylvanica against Chr11 of the V. vulgaris genome, showing the Dnmt3 locus (Vvulg11g01820) and the surrounding 20 kb. None of the Polistes assemblies have regions that are homologous to the Dnmt3 locus, and annotations for P. canadensis and P. dominula do not include a Dnmt3 gene (Standage et al. 2016; Patalano et al. 2015; Bewick et al. 2017). All three Vespula genomes encode an ortholog of Dnmt3. B. Frequency histograms of CpG[o/e] observed in coding sequences of Drosophila melanogaster, Apis mellifera, Polistes canadensis, Polistes dominula, Vespula germanica, Vespula vulgaris and Vespula pensylvanica. The y-axis depicts the number of genes (Frequency) with CpG[o/e] values given on the x-axis. The distribution of CpG[o/e] in Vespid wasps is a trimodal distribution, with a low-CpG[o/e] peak consistent with the presence of historical DNA methylation in all five Vespid species.
Gene content and duplications
We found more olfactory receptor (OR) genes in Vespula genomes than in the genomes of P. dominula and P. canadensis. We predicted 120 OR genes in V. vulgaris, 133 in V. germanica, and 102 in V. pensylvanica. Annotations for P. dominula and P. canadensis contain 94 and 72 OR genes, respectively (Supplementary table 3). In contrast, honeybee and Nasonia genomes encode more OR genes (170 and 301, respectively; Robertson and Wanner 2006; Robertson et al. 2010). Vespula OR genes cluster into 28 orthogroups. The co-receptor Orco is present in all genomes and forms a stable orthogroup (orthogroup 7148; Larsson et al. 2004; Jones et al. 2005). In contrast, there are significant expansions of particular OR orthogroups in the Vespid wasps, and these differ from the groups expanded in Nasonia and honeybee (Robertson et al. 2010; Robertson and Wanner 2006). Orthogroup members are arranged in expanded tandem arrays on chromosome 3 (orthogroup 51), chromosome 13 (orthogroup 2434), and chromosome 25 (orthogroup 232) of Vespula genomes. In these clusters, numbers of genes vary between species, implying that duplications and deletions are recent and ongoing. Variation in olfactory receptors between wasp species, and between these wasps and other Hymenoptera may indicate species-specific olfactory biology. These may be key to understanding the social behavior and pheromone signaling systems present within these species.
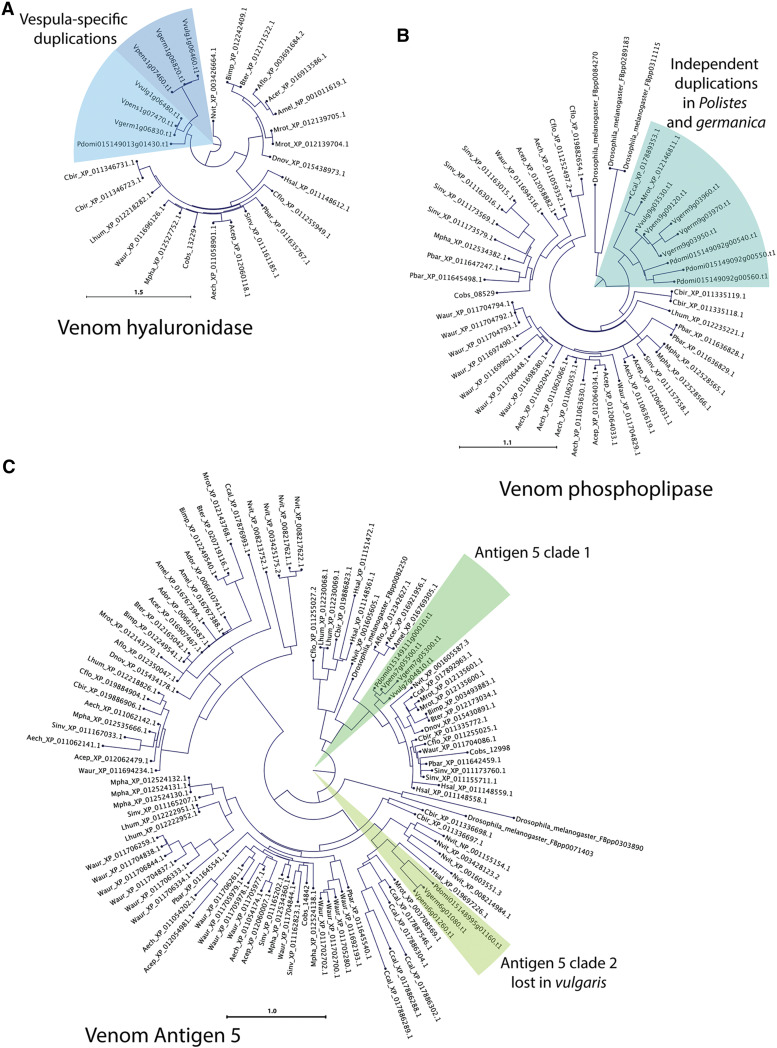
Some of the genes encoding venom components also have variable copy numbers in Vespula genomes. The major allergens in Vespula venoms are phospholipase A1, hyaluronidase, and antigen 5 (Biló et al. 2005; Kolarich et al. 2007). Phospholipase A1 is found in three tandem copies in the P. dominula and V. germanica genomes (chromosome 9 in Vespula), and one copy in each of the V. pensylvanica and V. vulgaris genomes. The phylogenetic placement of these duplicates in Polistes and V. germanica (Figure 4) implies that these are independent amplifications. The hyluronidase gene is duplicated in our three Vespid genomes, but not P. dominula. These are tandem duplications that appear to have been present in the last common ancestor of the three Vespula species. P. dominula, V. germanica and V. pensylvanica also have two Antigen 5 genes, but these duplications appear ancient before the common ancestor of Hymenoptera (Figure 4). In Vespula species, one copy is on chromosome 6 and one is on chromosome 7. In V. vulgaris, the chromosome 6 copy is absent. Duplication of venom genes in Vespids is no surprise, given the importance of venom to their life cycle.
Figure 4.
Ultrametric trees of orthogroups encoding Vespid wasp venom components in insect genomes. A. Venom hyaluronidase phylogram showing duplication of this gene in Vespid genomes but not Polistes. B. Venom phospholipase phylogram indicating independent duplications in Polistes and V. germanica from a single gene ancestor in related Hymenoptera. C. Antigen 5 phylogram, indicating two genes encode this venom component in Vespid wasps and Polisties, one of which is missing from the V. vulgaris genome.
Primary chemical control of Vespula populations is through the use of baits containing a low concentration of the phenylpyrazole insecticide, Fipronil (Lester and Beggs 2019; Edwards et al. 2017). We used targeted prediction to identify ligand-gated ion channel (LGIC), olfactory receptor, and spermatogenesis genes using Augustus protein profiles (Keller et al. 2011). Our annotation of the Fipronil target site, the GABA receptor Resistant to dieldrin (Rdl), did not suggest the presence of the classical Ala301 mutation that confers high resistance. Vespula LGIC receptors are highly conserved, with one-to-one orthology in Apis and Bombus (Supplementary Figure 4). This suggests that any chemicals targeting Vespula LGICs will also affect bees, as is the case with Fipronil.
Conclusions
We have produced chromosome-level genome assemblies for three invasive social wasps in the Vespula genus. Our approach of short-read sequencing and Hi-C scaffolding using haploid material allowed us to produce assemblies that exceed the genome quality targets suggested by the i5k insect genome sequencing initiative (scaffold N50 length > 300 kb; Richards and Murali 2015). Using manual curation and computational prediction, we have identified genes that may encode specific biology suitable for targeting with next-generation control technologies, and genes that may be affected by selection by current chemical controls.
These are the first three genomes from this branch of the Aculeata subclade, which will be useful in phylogenetic comparisons of the remarkable life history characteristics of Hymenoptera. In particular, these genomes will be valuable for understanding the evolution of eusociality, which has appeared twice in Vespid wasps independently of other Hymenoptera (Piekarski et al. 2018; Hines et al. 2007; Peters et al. 2017). Comparing the genomes of Vespid wasps, which are highly eusocial, with the closely related paper wasps, which are primitively eusocial, may also help our understanding of how evolution elaborates mechanisms of colonial living.
Vespid wasps are significant invasive pests in many parts of the world. These genomes will be of major importance for applied management of Vespula, in programs using both chemical control methods and for next-generation applications. Our assemblies will provide species-specific targets for novel control methods, such as RNA interference, gene drives and the deployment of damaging viruses. The genomic resources we have developed will also be essential for monitoring the effects of next-generation control methods and measuring genomic variation across natural populations.
Acknowledgments
The authors would like to thank P.M. Dearden for editing the manuscript. New Zealand Genomics Ltd for generation of the Common wasp genome data. The New Zealand National eScience Infrastructure (NeSI) for computational support. D. Hart and D.J. Champion for I.T. support and B.P. Dearden for critical discussions in the formulation of this work.
Footnotes
Supplemental material available at figshare: https://doi.org/10.25387/g3.12885599.
Communicating editor: Susan Lott
Literature Cited
- Archer, M. E., and J. Turner, 2014 Handbooks for the identification of British insects. Vol. 6, Pt. 6: The vespoid wasps (Tiphiidae, Mutillidae, Sapygidae, Scoliidae and Vespidae) of the British Isles. Field Studies Council, Telford. ISBN:9780901546982 [Google Scholar]
- Bewick A. J., Vogel K. J., Moore A. J., and Schmitz R. J., 2017. Evolution of DNA Methylation across Insects. Mol. Biol. Evol. 34: 654–665. 10.1093/molbev/msw264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickhart D. M., Rosen B. D., Koren S., Sayre B. L., Hastie A. R. et al. , 2017. Single-molecule sequencing and chromatin conformation capture enable de novo reference assembly of the domestic goat genome. Nat. Genet. 49: 643–650. 10.1038/ng.3802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biló B. M., Rueff F., Mosbech H., Bonifazi F., and Oude‐Elberink J. N. G., 2005. Diagnosis of Hymenoptera venom allergy. Allergy 60: 1339–1349. 10.1111/j.1398-9995.2005.00963.x [DOI] [PubMed] [Google Scholar]
- Bonasio R., Li Q., Lian J., Mutti N., Jin L. et al. , 2012. Genome-wide and Caste-Specific DNA Methylomes of the Ants Camponotus floridanus and Harpegnathos saltator. Curr. Biol. 22: 1755–1764. 10.1016/j.cub.2012.07.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutet, E., D. Lieberherr, M. Tognolli, M. Schneider, and A. Bairoch, 2007 UniProtKB/Swiss-Prot, pp. 89–112 in Plant Bioinformatics: Methods and Protocols, edited by D. Edwards. Methods in Molecular Biology, Humana Press, Totowa, NJ. doi: 10.1007/978–1-59745–535–0_4. 10.1007/978-1-59745-535-0_4 [DOI] [PubMed] [Google Scholar]
- Bushnell, B., 2014 BBMap: A Fast, Accurate, Splice-Aware Aligner: Lawrence Berkeley National Lab. (LBNL), Berkeley, CA LBNL-7065E.
- Cabanettes F., and Klopp C., 2018. D-GENIES: dot plot large genomes in an interactive, efficient and simple way. PeerJ 6: e4958 10.7717/peerj.4958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman J. A., Ho I. Y., Goltsman E., and Rokhsar D. S., 2016. Meraculous2: fast accurate short-read assembly of large polymorphic genomes. arXiv:1608.01031v2.
- Chapman J. A., Ho I., Sunkara S., Luo S., Schroth G. P. et al. , 2011. Meraculous: De Novo Genome Assembly with Short Paired-End Reads. PLoS One 6: e23501 10.1371/journal.pone.0023501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J., Shi J., Shangguan F.-Z., Dafni A., Deng Z.-H. et al. , 2009. The pollination of a self-incompatible, food-mimic orchid, Coelogyne fimbriata (Orchidaceae), by female Vespula wasps. Ann. Bot. 104: 565–571. 10.1093/aob/mcp029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dearden P. K., Gemmell N. J., Mercier O. R., Lester P. J., Scott M. J. et al. , 2018. The potential for the use of gene drives for pest control in New Zealand: a perspective. J. R. Soc. N. Z. 48: 225–244. 10.1080/03036758.2017.1385030 [DOI] [Google Scholar]
- Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C. et al. , 2012. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y., Simoes M. L., Marois E., and Dimopoulos G., 2018. CRISPR/Cas9 -mediated gene knockout of Anopheles gambiae FREP1 suppresses malaria parasite infection. PLoS Pathog. 14: e1006898 10.1371/journal.ppat.1006898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan B. J., 2003. Potential manageable exploitation of social wasps, Vespula spp. (Hymenoptera: Vespidae), as generalist predators of insect pests. Int. J. Pest Manage. 49: 281–285. 10.1080/0967087031000123698 [DOI] [Google Scholar]
- Drăgan M.-A., Moghul I., Priyam A., Bustos C., and Wurm Y., 2016. GeneValidator: identify problems with protein-coding gene predictions. Bioinformatics 32: 1559–1561. 10.1093/bioinformatics/btw015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards E., Toft R., Joice N., and Westbrooke I., 2017. The efficacy of Vespex wasp bait to control Vespula species (Hymenoptera: Vespidae) in New Zealand. Int. J. Pest Manage. 63: 266–272. 10.1080/09670874.2017.1308581 [DOI] [Google Scholar]
- Elsik C. G., Tayal A., Diesh C. M., Unni D. R., Emery M. L. et al. , 2016. Hymenoptera Genome Database: integrating genome annotations in HymenopteraMine. Nucleic Acids Res. 44: D793–D800. 10.1093/nar/gkv1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms D. M., and Kelly S., 2019. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20: 238 10.1186/s13059-019-1832-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feyereisen R., Dermauw W., and Van Leeuwen T., 2015. Genotype to phenotype, the molecular and physiological dimensions of resistance in arthropods. Pestic. Biochem. Physiol. 121: 61–77. 10.1016/j.pestbp.2015.01.004 [DOI] [PubMed] [Google Scholar]
- Gantz V. M., Jasinskiene N., Tatarenkova O., Fazekas A., Macias V. M. et al. , 2015. Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi. Proc. Natl. Acad. Sci. USA 112: E6736–E6743. 10.1073/pnas.1521077112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glastad K. M., Arsenault S. V., Vertacnik K. L., Geib S. M., Kay S. et al. , 2017. Variation in DNA Methylation Is Not Consistently Reflected by Sociality in Hymenoptera. Genome Biol. Evol. 9: 1687–1698. 10.1093/gbe/evx128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goll M. G., and Bestor T. H., 2005. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 74: 481–514. 10.1146/annurev.biochem.74.010904.153721 [DOI] [PubMed] [Google Scholar]
- Grimaldi D. A., and Engel M. S., 2005. Evolution of the insects. Cambridge University Press, Cambridge [U.K.]; New York. [Google Scholar]
- Gruber M. A. M., Quinn O., Baty J. W., Dobelmann J., Haywood J. et al. , 2019. Fitness and microbial networks of the common wasp, Vespula vulgaris (Hymenoptera: Vespidae), in its native and introduced ranges. Ecol. Entomol. 44: 512–523. 10.1111/een.12732 [DOI] [Google Scholar]
- Guan C., Barron A. B., He X. J., Wang Z. L., Yan W. Y. et al. , 2013. A Comparison of Digital Gene Expression Profiling and Methyl DNA Immunoprecipitation as Methods for Gene Discovery in Honeybee (Apis mellifera) Behavioural Genomic Analyses. PLoS One 8: e73628 10.1371/journal.pone.0073628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herb B. R., Wolschin F., Hansen K. D., Aryee M. J., Langmead B. et al. , 2012. Reversible switching between epigenetic states in honeybee behavioral subcastes. Nat. Neurosci. 15: 1371–1373. 10.1038/nn.3218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines H. M., Hunt J. H., O’Connor T. K., Gillespie J. J., and Cameron S. A., 2007. Multigene phylogeny reveals eusociality evolved twice in vespid wasps. Proc. Natl. Acad. Sci. USA 104: 3295–3299. 10.1073/pnas.0610140104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshiba, H., M. Matsuura, and H. T. Imai, 1989 Karyotype evolution in the social wasps. Jpn J Genet., 遺伝学雑誌 64: 209–222. 10.1266/jjg.64.209. 10.1266/jjg.64.209 [DOI]
- Hoskins R. A., Carlson J. W., Wan K. H., Park S., Mendez I. et al. , 2015. The Release 6 reference sequence of the Drosophila melanogaster genome. Genome Res. 25: 445–458. 10.1101/gr.185579.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P., Carpenter J. M., Chen B., and Li T.-J., 2019. The first divergence time estimation of the subfamily Stenogastrinae (Hymenoptera: Vespidae) based on mitochondrial phylogenomics. Int. J. Biol. Macromol. 137: 767–773. 10.1016/j.ijbiomac.2019.06.239 [DOI] [PubMed] [Google Scholar]
- Jacobs J. H., Clark S. J., Denholm I., Goulson D., Stoate C. et al. , 2010. Pollinator effectiveness and fruit set in common ivy, Hedera helix (Araliaceae). Arthropod-Plant Interact. 4: 19–28. 10.1007/s11829-009-9080-9 [DOI] [Google Scholar]
- Jones W. D., Nguyen T.-A. T., Kloss B., Lee K. J., and Vosshall L. B., 2005. Functional conservation of an insect odorant receptor gene across 250 million years of evolution. Curr. Biol. 15: R119–R121. 10.1016/j.cub.2005.02.007 [DOI] [PubMed] [Google Scholar]
- Keller O., Kollmar M., Stanke M., and Waack S., 2011. A novel hybrid gene prediction method employing protein multiple sequence alignments. Bioinformatics 27: 757–763. 10.1093/bioinformatics/btr010 [DOI] [PubMed] [Google Scholar]
- Kolarich D., Loos A., Léonard R., Mach L., Marzban G. et al. , 2007. A proteomic study of the major allergens from yellow jacket venoms. Proteomics 7: 1615–1623. 10.1002/pmic.200600800 [DOI] [PubMed] [Google Scholar]
- Kucharski R., Maleszka J., Foret S., and Maleszka R., 2008. Nutritional Control of Reproductive Status in Honeybees via DNA Methylation. Science 319: 1827–1830. 10.1126/science.1153069 [DOI] [PubMed] [Google Scholar]
- Larsson M. C., Domingos A. I., Jones W. D., Chiappe M. E., Amrein H. et al. , 2004. Or83b Encodes a Broadly Expressed Odorant Receptor Essential for Drosophila Olfaction. Neuron 43: 703–714. 10.1016/j.neuron.2004.08.019 [DOI] [PubMed] [Google Scholar]
- Lee E., Helt G. A., Reese J. T., Munoz-Torres M. C., Childers C. P. et al. , 2013. Web Apollo: a web-based genomic annotation editing platform. Genome Biol. 14: R93 10.1186/gb-2013-14-8-r93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lester P. J., and Beggs J. R., 2019. Invasion Success and Management Strategies for Social Vespula Wasps. Annu. Rev. Entomol. 64: 51–71. 10.1146/annurev-ento-011118-111812 [DOI] [PubMed] [Google Scholar]
- Lester P. J., Haywood J., Archer M. E., and Shortall C. R., 2017. The long-term population dynamics of common wasps in their native and invaded range. J. Anim. Ecol. 86: 337–347. 10.1111/1365-2656.12622 [DOI] [PubMed] [Google Scholar]
- Li-Byarlay H., Li Y., Stroud H., Feng S., Newman T. C. et al. , 2013. RNA interference knockdown of DNA methyl-transferase 3 affects gene alternative splicing in the honey bee. Proc. Natl. Acad. Sci. USA 110: 12750–12755. 10.1073/pnas.1310735110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E., van Berkum N. L., Williams L., Imakaev M., Ragoczy T. et al. , 2009. Comprehensive mapping of long range interactions reveals folding principles of the human genome. Science 326: 289–293. 10.1126/science.1181369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura M., Takeuchi H., Satoh M., Sanada-Morimura S., Otuka A. et al. , 2008. Species-specific insecticide resistance to imidacloprid and fipronil in the rice planthoppers Nilaparvata lugens and Sogatella furcifera in East and South-east Asia. Pest Manag. Sci. 64: 1115–1121. 10.1002/ps.1641 [DOI] [PubMed] [Google Scholar]
- Michener C. D., and Brothers D. J., 1974. Were workers of eusocial hymenoptera initially altruistic or oppressed? Proc. Natl. Acad. Sci. USA 71: 671–674. 10.1073/pnas.71.3.671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S. E., Legan A. W., Henshaw M. T., Ostevik K. L., Samuk K. et al. , 2020. Evolutionary dynamics of recent selection on cognitive abilities. Proc. Natl. Acad. Sci. USA 117: 3045–3052. 10.1073/pnas.1918592117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell A. L., Attwood T. K., Babbitt P. C., Blum M., Bork P. et al. , 2019. InterPro in 2019: improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res. 47: D351–D360. 10.1093/nar/gky1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer, J., and J. Stajich, 2019 nextgenusfs/funannotate: funannotate v1.6.0 Zenodo. 10.5281/zenodo.3354704. [DOI]
- Patalano S., Vlasova A., Wyatt C., Ewels P., Camara F. et al. , 2015. Molecular signatures of plastic phenotypes in two eusocial insect species with simple societies. Proc. Natl. Acad. Sci. USA 112: 13970–13975. 10.1073/pnas.1515937112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters R. S., Krogmann L., Mayer C., Donath A., Gunkel S. et al. , 2017. Evolutionary History of the Hymenoptera. Curr. Biol. 27: 1013–1018. 10.1016/j.cub.2017.01.027 [DOI] [PubMed] [Google Scholar]
- Piekarski P. K., Carpenter J. M., Lemmon A. R., Moriarty Lemmon E., and Sharanowski B. J., 2018. Phylogenomic evidence overturns current conceptions of social evolution in wasps (Vespidae). Mol. Biol. Evol. 35: 2097–2109. 10.1093/molbev/msy124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt K. D., Tatusova T., and Maglott D. R., 2005. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 33: D501–D504. 10.1093/nar/gki025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team , 2015. R: A Language and Environment for Statistical Computing.
- Richards S., and Murali S. C., 2015. Best practices in insect genome sequencing: what works and what doesn’t. Curr. Opin. Insect Sci. 7: 1–7. 10.1016/j.cois.2015.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson H. M., Gadau J., and Wanner K. W., 2010. The insect chemoreceptor superfamily of the parasitoid jewel wasp Nasonia vitripennis. Insect Mol. Biol. 19: 121–136. 10.1111/j.1365-2583.2009.00979.x [DOI] [PubMed] [Google Scholar]
- Robertson H. M., and Wanner K. W., 2006. The chemoreceptor superfamily in the honey bee, Apis mellifera: Expansion of the odorant, but not gustatory, receptor family. Genome Res. 16: 1395–1403. 10.1101/gr.5057506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scrucca L., Fop M., Murphy T. B., and Raftery A. E., 2016. mclust 5: Clustering, Classification and Density Estimation Using Gaussian Finite Mixture Models. R J. 8: 289–317. 10.32614/RJ-2016-021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão F. A., Waterhouse R. M., Ioannidis P., Kriventseva E. V., and Zdobnov E. M., 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31: 3210–3212. 10.1093/bioinformatics/btv351 [DOI] [PubMed] [Google Scholar]
- Standage D. S., Berens A. J., Glastad K. M., Severin A. J., Brendel V. P. et al. , 2016. Genome, transcriptome and methylome sequencing of a primitively eusocial wasp reveal a greatly reduced DNA methylation system in a social insect. Mol. Ecol. 25: 1769–1784. 10.1111/mec.13578 [DOI] [PubMed] [Google Scholar]
- The Honeybee Genome Sequencing Consortium , 2006. Insights into social insects from the genome of the honeybee Apis mellifera. Nature 443: 931–949. 10.1038/nature05260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C. D., Moller H., Plunkett G. M., and Harris R. J., 1990. The prevalence of introduced Vespula vulgaris wasps in a New Zealand beech forest community. N. Z. J. Ecol. 13: 63–72. [Google Scholar]
- van Dongen S., 2000. Graph Clustering by Flow Simulation. PhD thesis, University of Utrecht. (http://www.library.uu.nl/digiarchief/dip/diss/1895620/inhoud.htm). [Google Scholar]
- Werren J. H., Richards S., Desjardins C. A., Niehuis O., Gadau J. et al. , 2010. Functional and Evolutionary Insights from the Genomes of Three Parasitoid Nasonia Species. Science 327: 343–348. 10.1126/science.1178028 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Raw sequence data are hosted in the NCBI Sequence Read Archive under accession PRJNA643352. Assembled genomes are available on GenBank under accessions JACSDY000000000 (Vespula pensylvanica), JACSDZ000000000 (Vespula germanica) and JACSEA000000000 (Vespula vulgaris). Supplemental material available at figshare: https://doi.org/10.25387/g3.12885599.