The SINAT E3 ubiquitin ligases regulate ABA signaling by modulating the protein stability of ESCRT components FREE1 and VPS23A.
Abstract
In plants, the ubiquitin-proteasome system, endosomal sorting, and autophagy are essential for protein degradation; however, their interplay remains poorly understood. Here, we show that four Arabidopsis (Arabidopsis thaliana) E3 ubiquitin ligases, SEVEN IN ABSENTIA OF ARABIDOPSIS THALIANA1 (SINAT1), SINAT2, SINAT3, and SINAT4, regulate the stabilities of FYVE DOMAIN PROTEIN REQUIRED FOR ENDOSOMAL SORTING1 (FREE1) and VACUOLAR PROTEIN SORTING23A (VPS23A), key components of the endosomal sorting complex required for transport-I, to modulate abscisic acid (ABA) signaling. GFP-SINAT1, GFP-SINAT2, and GFP-SINAT4 primarily localized to the endosomal and autophagic vesicles. SINATs controlled FREE1 and VPS23A ubiquitination and proteasomal degradation. SINAT overexpressors showed increased ABA sensitivity, ABA-responsive gene expression, and PYRABACTIN RESISTANCE1-LIKE4 protein levels. Furthermore, the SINAT-FREE1/VPS23A proteins were codegraded by the vacuolar pathway. In particular, during recovery post-ABA exposure, SINATs formed homo- and hetero-oligomers in vivo, which were disrupted by the autophagy machinery. Taken together, our findings reveal a novel mechanism by which the proteasomal and vacuolar turnover systems regulate ABA signaling in plants.
INTRODUCTION
The ubiquitin-proteasome system (UPS), multi-vesicle body (MVB)–mediated vacuolar sorting, and the autophagy–vacuole pathway are predominant routes for protein quality control (Vierstra, 2009; Dikic, 2017; Gao et al., 2017; Marshall and Vierstra, 2018). Ubiquitination is a common signal used by all three pathways to mark abnormal proteins for proteasomal or vacuolar degradation (Tian and Xie, 2013; Dikic, 2017). In particular, degradation by the UPS involves covalent attachment of ubiquitin to soluble proteins that are then recognized and degraded via the 26S proteasome (Vierstra, 2009). The UPS is the primary proteolytic mechanism for short-lived, misfolded, and damaged proteins and is crucial for the maintenance of cellular functions (Vierstra, 2009). The MVB-mediated vacuolar sorting pathway depends on the endosomal sorting complex required for transport (ESCRT) machinery, which transports ubiquitinated membrane proteins to the vacuole for degradation (Raiborg and Stenmark, 2009; Gao et al., 2017). By contrast, autophagy recognizes and removes large cytoplasmic components, including aggregated proteins, dysfunctional organelles, and invading pathogens in double-membrane vesicles, termed autophagosomes, and eventually delivers the cargoes into the vacuole for recycling (Floyd et al., 2012; Marshall and Vierstra, 2018; Qi et al., 2020b).
ESCRT is an evolutionarily conserved system that is essential for MVB biogenesis and the vacuolar sorting of membrane proteins (Raiborg and Stenmark, 2009; Paez Valencia et al., 2016; Gao et al., 2017). During this process, four protein complexes, ESCRT-0, ESCRT-I, ESCRT-II, and ESCRT-III, are sequentially assembled into the early endosome membrane. There, they promote enrichment and invagination of the ubiquitinated proteins by forming intraluminal vesicles and, subsequently, mature MVBs. The outer membranes of mature MVBs fuse with the vacuole or lysosome to release ubiquitinated membrane proteins for degradation, while the ESCRT complexes are released from the MVB membrane to the cytosol for reuse (Raiborg and Stenmark, 2009).
In mammalian cells, the activities of several ESCRT components are controlled by posttranslational modifications, including phosphorylation and ubiquitination. In particular, phosphorylation of intramolecular interaction sites within the Pro-rich domain of the ESCRT protein ALG2 INTERACTING PROTEIN X (ALIX) induces an active conformation change that permits its function in cytokinetic abscission and retroviral budding (Sun et al., 2016). Moreover, the Pkh1/Pkh2 (Pkb-activating kinase homolog) kinase-dependent phosphorylation of mammalian Vps27 facilities ESRCT-I assembly and endosome recruitment (Morvan et al., 2012). Evidence also suggests that monoubiquitination of ESCRT proteins impairs their ability to bind ubiquitinated targets. For instance, the E3 ligase Mahogunin catalyzes the monoubiquitination of a key component of ESCRT-I, tumor suppressor gene101 (Tsg101), switching ESCRT-I between its active and inactive forms in a manner that is independent of proteasome degradation (Kim et al., 2007).
To date, most isoforms of ESCRT components have been identified in plants (Gao et al., 2017). Among these, VACUOLE PROTEIN SORTING23A (VPS23A) and FYVE DOMAIN PROTEIN REQUIRED FOR ENDOSOMAL SORTING1 (FREE1) are key components of ESCRT-I in Arabidopsis (Arabidopsis thaliana) and can bind ubiquitin (Spitzer et al., 2006; Gao et al., 2014). The plant-unique FREE1 interacts with VPS23A and plays dual roles in MVB biogenesis and autophagy dynamics (Gao et al., 2014, 2015). The VPS23A knockout mutants and FREE1-inducible RNA interference knockdown lines show impaired vacuole biogenesis and accumulate ubiquitinated proteins (Spitzer et al., 2006; Gao et al., 2014; Yu et al., 2016), indicative of the essential roles that VPS23A and FREE1 play in vacuolar protein sorting. Moreover, deletion of FREE1 leads to an embryo-lethality phenotype (Gao et al., 2014). By contrast, the vps23a mutant displays only minor defects in trichome morphogenesis and cytokinesis, suggesting that VPS23A might share functional redundancy with its homolog VPS23B in the regulation of plant growth and development (Spitzer et al., 2006).
Increasing evidence reveals that the ESCRT machinery plays crucial roles in the modulation of cellular signaling by recycling membrane proteins such as the auxin carriers PIN-FORMED1 (PIN1), PIN2, and AUXIN RESISTANT1 (AUX1; Spitzer et al., 2009); the pattern recognition receptor FLAGELLIN-SENSITIVE2 (FLS2; Spallek et al., 2013); and the metal transporter IRON-REGULATED TRANSPORTER1 (IRT1; Barberon et al., 2014). Moreover, YUC1 flavin monooxygenase, a soluble protein in the auxin biosynthesis pathway, is degraded through an ESCRT-dependent mechanism (Ge et al., 2019), suggesting that ESCRT targets both membrane and soluble proteins as cargoes. More recently, two independent studies have demonstrated that upon recognition by FREE1 and VPS23A, the ubiquitinated abscisic acid (ABA) receptors PYRABACTIN RESISTANCE1 (PYR1)/PYR1-LIKE4 (PYL4) are targeted for vacuolar degradation (Belda-Palazon et al., 2016; Yu et al., 2016). Consistent with this, mutants of FREE1 or VPS23A accumulate PYR1/PYL4 proteins and are hypersensitive to ABA treatment (Belda-Palazon et al., 2016; Yu et al., 2016). Furthermore, upon ABA exposure, the kinase SNF-RELATED PROTEIN KINASE2 (SnRK2) phosphorylates FREE1, which translocates to the nucleus and interacts with transcription factors ABA-RESPONSIVE ELEMENT BINDING FACTOR4 (ABF4) and ABA-INSENSITIVE5 (ABI5) to attenuate their transcriptional activity (Li et al., 2019). However, it is unclear how the protein stabilities of FREE1 and VPS23A are maintained in plant responses to ABA.
SEVEN IN ABSENTIA (SINA) is a RING–type E3 ubiquitin ligase originally identified in Drosophila melanogaster, where it functions in neuronal specification of the R7 photoreceptor cell in the eye (Carthew and Rubin, 1990). Later, SINA homologs were characterized from mouse, human, and plants (Della et al., 1993; Hu et al., 1997; Xie et al., 2002). Typically, SINAT proteins contain an N-terminal RING domain for proteolysis and a C-terminal TUMOR NECROSIS FACTOR RECEPTOR-ASSOCIATED FACTOR (TRAF) domain for protein–protein interactions and oligomerization (Hu and Fearon, 1999; Park et al., 1999; Polekhina et al., 2002; Xie et al., 2002). Animal and plant TRAF proteins share a conserved meprin and TRAF homology (MATH) domain with the extracellular meprin proteins, and this domain has diverse physiological functions (Sunnerhagen et al., 2002; Oelmüller et al., 2005). Arabidopsis contains six SEVEN IN ABSENTIA OF ARABIDOPSIS THALIANA (SINAT) members (SINAT1 to SINAT6), which function in biological activities such as hormone signaling, inflorescence development, abiotic stress responses, and autophagy (Zhang et al., 2019). In the Arabidopsis Columbia-0 ecotype, four SINATs (SINAT1 to SINAT4) harbor RING finger and Zinc finger domains at the amino termini, which give them E3 ubiquitin ligase activity. The other two SINATs (SINAT5 and SINAT6) are truncated within these domains due to alternative splicing (Qi et al., 2017). By contrast, in the Landsberg ecotype, the SINAT5 protein possesses a complete catalytic RING domain and active E3 activity (Xie et al., 2002), indicating evolutionary variation in the SINAT protein family in Arabidopsis ecotypes. Additional evidence suggests that SINATs target substrates for proteasomal and autophagic degradation (Nolan et al., 2017; Qi et al., 2017, 2020a; Yang et al., 2017) and are thus potentially important communication nodes in mediating crosstalk between the proteasomal and autophagic degradation pathways.
In this study, we performed an immunoprecipitation–mass spectrometry (IP-MS) screen to identify the SINAT1 interaction network and identified FREE1 and VPS23A as potential targets of SINAT proteins. Moreover, we observed that SINATs mediated the ubiquitination and degradation of FREE1 and VPS23A and contributed to the modulation of ABA signaling. In addition, SINATs formed homo- and hetero-oligomers in vivo, which were predominantly disrupted by autophagy during recovery following ABA treatment, revealing a mechanism for clearance of SINAT proteins post-ABA signaling.
RESULTS
Subcellular Localization of SINAT Family Proteins
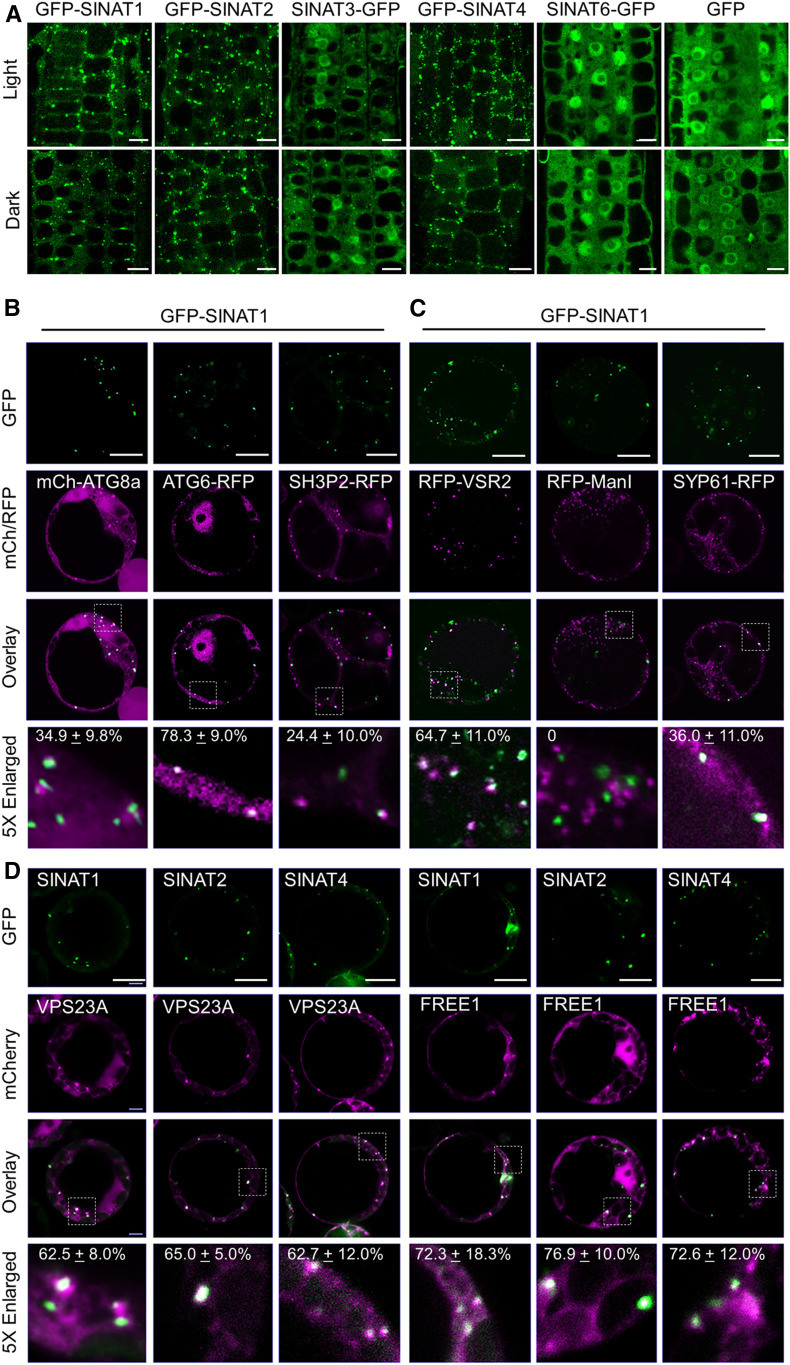
To improve our understanding of the cellular roles of SINAT family proteins, we first analyzed their subcellular localization. To this end, we generated constructs encoding Arabidopsis SINAT1 to SINAT4 and SINAT6 fused to the N-terminal or C-terminal of GFP (SINATs-GFP or GFP-SINATs) for transient expression in the wild-type Arabidopsis protoplasts. Following overnight culture, cells were subjected to confocal laser scanning microscopy to detect GFP fluorescence. We observed that the GFP-SINAT fusions showed distinct subcellular distributions. In particular, GFP-SINAT1, GFP-SINAT2, and GFP-SINAT4 predominantly localized to punctate structures in the cytoplasm, whereas GFP-SINAT3 signals were detected in the cytoplasm and nucleus in addition to the punctate signals (Supplemental Figure 1A). By contrast, GFP-SINAT6 was distributed in the cytoplasm and nucleus (Supplemental Figure 1A). The GFP-SINATs and SINATs-GFP fusions showed similar patterns of localization. As a control, GFP fluorescent signals from the empty vector (GFP) were detected in the cytoplasm and nucleus (Supplemental Figure 1A).
We next confirmed the subcellular localizations of SINATs by generating stable transgenic Arabidopsis expressing GFP-SINAT1, GFP-SINAT2, SINAT3-GFP, GFP-SINAT4, and SINAT6-GFP fusions under the control of the UBQ10 promoter. Compared to the GFP vector control and SINAT6-GFP lines, we found that GFP-SINAT1, GFP-SINAT2, and GFP-SINAT4 primarily localized to punctate structures, while SINAT3-GFP showed both punctate and nuclear signals in the root cells of 16-h-light-/8-h-dark-grown seedlings (Figure 1A). Moreover, 12 h of constant darkness treatment did not substantially change the GFP fluorescence of GFP-SINAT fusion proteins (Figure 1A). The different subcellular distributions of SINAT1 and SINAT6 were further validated by analyses of transgenic lines expressing ProSINAT1:SINAT1-GFP and ProSINAT6:SINAT6-GFP in which the GFP-fused SINAT proteins were driven by their native promoters (Supplemental Figure 1B).
Figure 1.
Subcellular Localization of SINAT Proteins.
(A) Examination of GFP-SINAT1, GFP-SINAT2, SINAT3-GFP, and GFP-SINAT4 subcellular distribution patterns. Transgenic seedlings expressing GFP-fused SINAT proteins (GFP-SINAT1-HA, GFP-SINAT2-HA, SINAT3-GFP-HA, GFP-SINAT4-HA, GFP-SINAT5-HA, and SINAT6-GFP-HA) were grown under normal growth conditions (16-h-light/8-h-dark cycle) for 5 d. Before being imaged by confocal microscopy, seedlings were cultured under light or dark for an additional 12 h.
(B) Examination of GFP-SINAT1 puncta colocalization with autophagosomes. GFP-SINAT1-HA was cotransfected with the autophagosome markers mCh-ATG8a, ATG6-RFP, or SH3P2-RFP, in cultured Arabidopsis PSB-D cells under constant darkness.
(C) Examination of GFP-SINAT1 colocalization with MVB, TGN, and the cis-Golgi apparatus. GFP-SINAT1-HA was cotransfected with RFP-fused markers VSR2 (MVB), mannosidase I (ManI; cis-Golgi), or SYP61 (TGN) in cultured Arabidopsis PSB-D cells under constant darkness.
(D) Examination of colocalization of the SINAT puncta with VPS23A and FREE1. GFP-fused SINAT proteins (GFP-SINAT1-HA, GFP-SINAT2-HA, and GFP-SINAT4-HA) were coexpressed with FLAG-mCh-VPS23a or FLAG-mCh-FREE1 in Arabidopsis mesophyll protoplasts overnight under constant darkness. GFP, RFP, and mCh signals were detected using confocal microscopy.
The numbers in the 5× enlarged images of (B), (C), and (D) indicate the colocalization ratio (the number of colocalization puncta to total GFP-SINAT puncta). The percentages are means ± sd (n = 3) of three independent experiments. For each experiment, four to six cells were collected for analysis. Bars in (A) to (D) = 10 μm.
The punctate structures of GFP-SINAT1, GFP-SINAT2, and GFP-SINAT4 resemble autophagosomes (Yoshimoto et al., 2004; Qi et al., 2017). To determine the potential colocalization of SINAT proteins to autophagosomes, we coexpressed GFP-SINAT1 and GFP-SINAT2 with mCherry (mCh)- or red fluorescent protein (RFP)–labeled autophagy-related gene (ATG) proteins mCh-ATG8a, ATG6-RFP, and SH3 DOMAIN-CONTAINING PROTEIN 2 (SH3P2)-RFP in protoplasts (Zhuang et al., 2013; Qi et al., 2017). We observed that the GFP-SINAT1 and GFP-SINAT2 puncta predominantly colocalized with ATG6-RFP signals and partially overlapped with mCh-ATG8a and SH3P2-RFP (Figure 1B; Supplemental Figure 1C).
We next examined whether the SINAT puncta were associated with the endomembrane trafficking system, which frequently forms puncta in the cytosol. The GFP-SINAT1 and GFP-SINAT2 signals partially associated with the MVB marker RFP-VACUOLAR SORTING RECEPTOR 2 (VSR2) and the trans-Golgi network (TGN) resident protein SYNTAXIN OF PLANTS 61 (SYP61)-RFP (Figure 1C; Supplemental Figure 1D) but were distinct from the cis-Golgi apparatus marker RFP-α-mannosidase I or the peroxisomal targeted RFP SRL-RFP (Figure 1C; Supplemental Figure 1D; Reumann, 2004). The partial colocalization with various organelles suggests that SINATs might play multiple roles in the endomembrane system.
To test the potential colocalization of SINATs with ESCRT-I components, we generated mCh-fused FREE1 and VPS23A and cotransfected them with GFP-SINAT1, GFP-SINAT2, and GFP-SINAT4, respectively, into the wild-type protoplasts. Consistent with previous findings (Spitzer et al., 2006; Gao et al., 2014), we observed that mCh-FREE1 and mCherry-VPS23A were both distributed in puncta and in the cytoplasm (Figure 1D). Particularly, GFP-SINAT1, GFP-SINAT2, and GFP-SINAT4 expression predominantly overlapped with the mCh-labeled punctate structures (Figure 1D). Taken together, our data demonstrate that SINAT1, SINAT2, and SINAT4 are primarily distributed in the endomembrane system, especially in the autophagosomes and ESCRT-I–associated vesicles.
Screening of SINAT1-Interacting Proteins by IP-MS Analysis
To evaluate the protein network downstream of SINATs, we screened for interactors of SINAT1 by IP-MS. Total proteins were extracted from 7-d-old transgenic seedlings expressing GFP-SINAT1-hemagglutinin (HA) grown on Murashige and Skoog (MS) medium and were immunoprecipitated by anti-GFP agarose beads and then subjected to in-gel digestion and MS analysis. The transgenic seedlings expressing free GFP-HA were used as a control. In total, we identified 34 candidate interacting proteins that were predominately associated with either the ubiquitin-proteasome or the endomembrane system (Table 1). As expected, several typical ubiquitin-proteasome components, such as ubiquitin and 26S proteasome subunits, were putative SINAT1 interactors (Table 1). In particular, the specific peptides of SINAT1 and three other SINAT proteins, SINAT2, SINAT3, and SINAT4, emerged as highly probable interactors (Table 1), suggesting that SINAT proteins might form homo- or hetero-oligomers in planta. Moreover, IP-MS identified a large number of endomembrane trafficking–related proteins, including ESCRT-I components (FREE1 and VPS23A), retromer components, coatomer subunits for endoplasmic reticulum–Golgi transport, exocyst elements, clathrin heavy chains, and adaptor protein (AP)-2 members (Table 1). Given recent evidence showing the inter-connections among the ubiquitin-proteasome, autophagy, and ESCRT pathways (Gao et al., 2015; Qi et al., 2017), our findings imply that SINAT1 might function as a key regulator of endomembrane systems.
Table 1. Screening of SINAT1-Interacting Proteins by IP-MS Analysis.
| Protein Type | Gene Locus | Description | Score (–10lgP) | Coverage (%) | Unique Peptides |
|---|---|---|---|---|---|
| Bait | AT2G41980 | SINAT1 | 335 | 61 | 17 |
| SINAT homologs | AT3G58040 | SINAT2 | 259 | 34 | 9 |
| AT3G61790 | SINAT3 | 91 | 10 | 3 | |
| AT4G27880 | SINAT4 | 134 | 15 | 3 | |
| UPS | AT3G09790 | Ubiquitin 8 | 167 | 14 | 2 |
| AT1G65350 | Polyubiquitin 13 | 158 | 14 | 5 | |
| AT2G20580 | 26S proteasome non-ATPase regulatory subunit 2 homolog A | 115 | 6 | 4 | |
| AT1G20200 | 26S proteasome non-ATPase regulatory subunit 3 homolog A | 80 | 8 | 4 | |
| AT4G24820 | 26S proteasome non-ATPase regulatory subunit 6 homolog | 71 | 6 | 2 | |
| AT4G29040 | 26S proteasome regulatory subunit 4 homolog A | 82 | 6 | 2 | |
| AT5G58290 | 26S protease regulatory subunit 6B homolog | 116 | 14 | 4 | |
| AT3G05530 | 26S protease regulatory subunit 6A homolog A | 75 | 10 | 3 | |
| AT5G02310 | E3 ubiquitin-protein ligase PRT6 | 48 | 1 | 2 | |
| AT4G25160 | U-box domain-containing protein 35 | 22 | 1 | 1 | |
| Endomembrane trafficking system | AT1G20110 | ESCRT-I component FREE1 | 254 | 20 | 10 |
| AT3G12400 | ESCRT-I component VPS23A | 26 | 4 | 1 | |
| AT3G52100 | RING/FYVE/PHD zinc finger-containing protein | 23 | 2 | 1 | |
| AT4G27690 | Vacuolar protein sorting-associated protein 26B | 106 | 10 | 3 | |
| AT2G17790 | Vacuolar protein sorting-associated protein 35A | 32 | 2 | 1 | |
| AT3G51310 | Vacuolar protein sorting-associated protein 35C | 21 | 1 | 1 | |
| AT1G78900 | V-type proton ATPase catalytic subunit A | 133 | 14 | 8 | |
| AT4G39080 | V-type proton ATPase subunit A3 | 48 | 3 | 2 | |
| AT1G76030 | V-type proton ATPase subunit B1 | 64 | 6 | 2 | |
| AT5G22770 | AP-2 complex subunit alpha-1 | 117 | 7 | 6 | |
| AT5G46630 | AP-2 complex subunit mu | 81 | 4 | 2 | |
| AT3G11130 | Clathrin heavy chain 1 | 189 | 11 | 3 | |
| AT3G08530 | Clathrin heavy chain 2 | 180 | 10 | 1 | |
| AT1G62020 | Coatomer subunit alpha-1 | 211 | 16 | 4 | |
| AT2G21390 | Coatomer subunit alpha-2 | 208 | 19 | 5 | |
| AT4G31480 | Coatomer subunit beta-1 | 108 | 11 | 8 | |
| AT1G52360 | Coatomer subunit beta-2 | 43 | 3 | 3 | |
| AT1G47550 | Exocyst complex component SEC3A | 55 | 2 | 2 | |
| AT3G10380 | Exocyst complex component SEC8 | 37 | 1 | 1 | |
| AT5G47480 | RGPR-related protein; SEC16A homolog | 69 | 17 | 13 | |
| AT1G24460 | TGN-localized SYP41-interacting protein | 22 | 1 | 1 |
SINATs Interact with the ESCRT-I Components FREE1 and VPS23A in Vivo
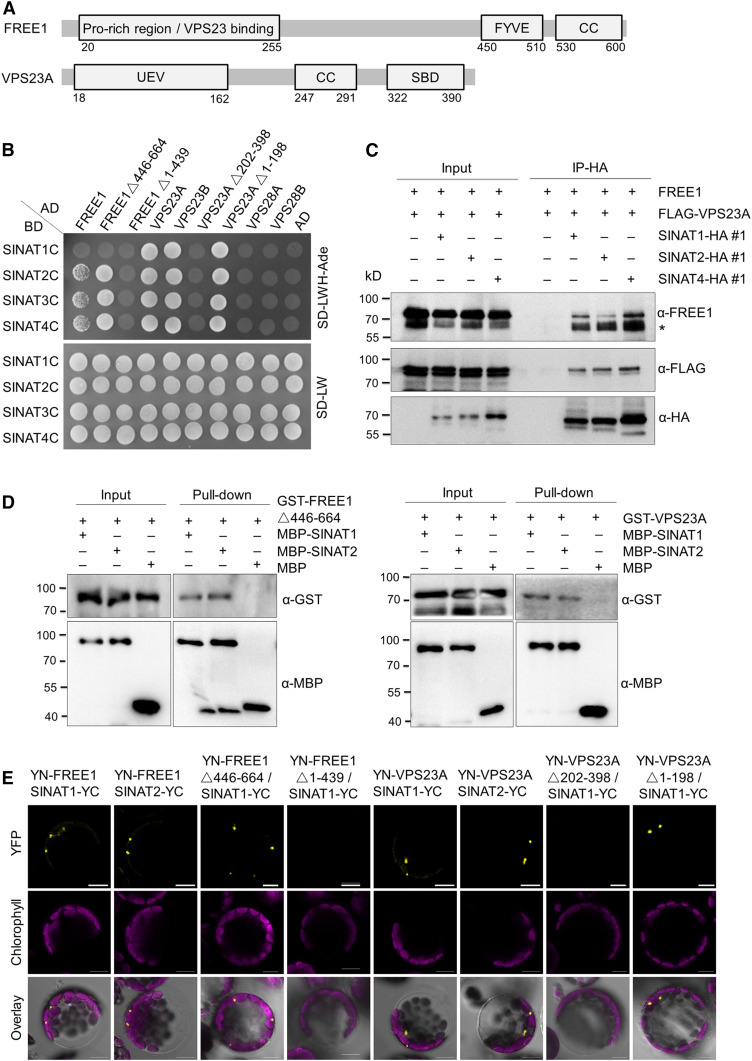
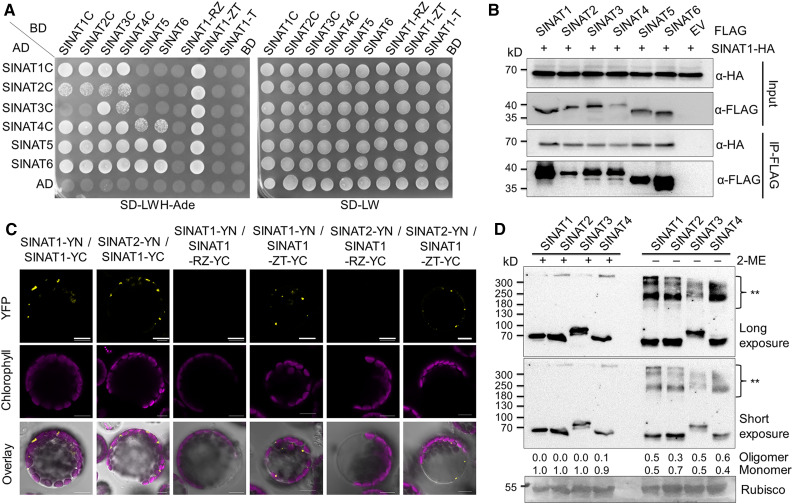
To validate the potential protein–protein interaction between SINATs and ESCRT-I components, particularly FREE1 and VPS23A, we first performed yeast two-hybrid (Y2H) analysis to confirm their association. To avoid the influence of SINAT E3 ligase activity in the protein interaction assays, we made constructs to generate mutated SINATs by changing sequences encoding a conserved Cys (C) to a Ser (S) in the RING finger domain (Supplemental Figure 2A; Xie et al., 2002) to impair their E3 ligase activity and used these in Y2H assays. These mutant SINATs (SINAT1C, SINAT2C, SINAT3C, and SINAT4C), which showed increased stability during Y2H according to preliminary assays, were therefore fused with the binding domain (BD) domain and cotransformed with the activation domain (AD) fused to FREE1 and VPS23A into yeast (Saccharomyces cerevisiae) strain AH109. The cotransformation of SINAT-BDs with other vacuolar protein sorting-associated proteins VPS23B-AD, VPS28A-AD, and VPS28B-AD was performed as controls. In yeast cells, SINAT1, SINAT2, SINAT3, and SINAT4 interacted with FREE1, VPS23A, and VPS23B, except the combination SINAT1/FREE1 (Figure 2B). By contrast, SINATs did not interact with VPS28A and VPS28B in Y2H assays (Figure 2B), suggesting that SINATs specifically target components within the ESCRT.
Figure 2.
SINATs Interact with ESCRT-I Components FREE1 and VPS23A in Vivo.
(A) Domain structure of FREE1 and VPS23A proteins. CC, coiled coil; UEV, ubiquitin E2 variant domain; SBD, steadiness box domain.
(B) Y2H assay for interactions of SINATs with FREE1 and VPS23A. Full-length or truncated FREE1, VPS23A, VPS23B, and two VPS28 proteins (VPS28A and VPS28A) were fused with GAL4-AD and cotransformed with mutant BD-SINATs (SINAT1-C60S, SINAT2-C63S, SINAT3-C66S, and SINAT4-C67S) into the YH109 yeast strain. The transformants were selected on SD-Trp-Leu-His-Ade medium (SD-LWH-Ade). AD and BD indicate the empty AD and BD plasmids, respectively.
(C) In vivo Co-IP assay for the association of SINATs (SINAT1, SINAT2, and SINAT4) with FREE1 and VPS23A. Total protein was extracted from 1-week-old transgenic seedlings expressing FLAG-mCh-VPS23A with HA-fused SINAT proteins (GFP-SINAT1-HA, GFP-SINAT2-HA, and GFP-SINAT4-HA) and incubated with anti-HA magnetic beads. The anti-FREE1, anti-FLAG, and anti-HA antibodies were used to detect FREE1, FLAG-mCh-VPS23A, and GFP-SINAT-HA proteins, respectively. Numbers on the left indicate the molecular weight (kD) of each band. Asterisk (*) indicates unspecific bands.
(D) In vitro pull-down assay for physical interaction between SINATs (SINAT1 and SINAT2) and FREE1 or VPS23A. Recombinant MBP-SINAT1, MBP-SINAT2, GST-FREE1△446-664, and GST-VPS23A proteins purified from E. coli were incubated in tubes as indicated, followed by pull-down with amylose resin. The proteins in the eluate were detected using anti-GST and anti-MBP antibodies.
(E) BiFC assay for interaction between SINAT proteins and FREE1 or VPS23A. Full-length or truncated FREE1 and VPS23A were fused with nYFP and coexpressed with cYFP-fused SINAT1 or SINAT2 in protoplasts isolated from Arabidopsis rosette leaves. Signals were detected by confocal microscopy. Bars = 10 μm.
When FREE1 was truncated within the N-terminal region (amino acids 1 to 439) and therefore lacked the VPS23A-interacting domain or C-terminal region (amino acids 446 to 664) containing a FYVE domain and coiled-coil motif (Figure 2A), we observed that the N-terminal region of FREE1 specifically interacted with SINATs (Figure 2B). Similarly, when the N-terminal region (amino acids 1 to 198) of VPS23A was truncated, containing the ubiquitin E2 variant domain, or the C-terminal region (amino acids 202 to 398) containing a coiled-coil region as well as a steadiness box domain (Figure 2A), only the C-terminal fragment positively interacted with SINATs in Y2H assays (Figure 2B).
We then confirmed the interaction between SINATs and FREE1/VPS23A by an in planta coimmunoprecipitation (Co-IP) assay. When FLAG-tagged SINAT proteins were transiently expressed in protoplasts isolated from HA-GFP-VPS23A transgenic plants, we observed that the VPS23A fusion protein could be immunoprecipitated by all six SINAT-FLAG proteins, but not by anti-FLAG magnetic beads (Supplemental Figure 3A). Similarly, endogenous FREE1 could be pulled down in vivo by all six SINAT proteins in the SINATs-HA transgenic seedlings, but not in the wild-type plants (Supplemental Figure 3B). In the positive controls, FREE1 showed associations with VPS23A and PYL4 (Supplemental Figure 3B). To investigate the interactions between SINATs and FREE1/VPS23A in planta, we performed a Co-IP assay using the stable transgenic lines that coexpressed FLAG-mCh-VPS23A with GFP-SINAT1-HA, GFP-SINAT2-HA, or GFP-SINAT4-HA. When SINAT fusion proteins were enriched using anti-HA affinity beads, FREE1 and VPS23A were both immunoprecipitated (Figure 2C).
To test whether FREE1 and VPS23A bind SINATs directly, we performed in vitro pull-down assays. To this end, the glutathione S-transferase (GST)–fused FREE1 and VPS23A proteins were purified from Escherichia coli and then incubated with maltose binding protein (MBP)–fused SINAT1, SINAT2, or free MBP. As shown in Figure 2D, both GST-FREE1 and GST-VPS23A were pulled down by MBP-SINAT1 and MBP-SINAT2, but not free MBP, supporting the physical associations of FREE1 and VPS23A with SINATs in vitro.
Next, we used bimolecular fluorescence complementation (BiFC) analyses to confirm the protein–protein interactions between SINATs and FREE1/VPS23A. For this, we constructed fusions of FREE1 or VPS23A with split yellow fluorescent protein (YFP) N terminus (YN) or C terminus (YC), YN-FREE1, or YN-VPS23A and transiently coexpressed these fusions with SINAT1-YC, SINAT2-YC, SINAT3-YC, and SINAT4-YC in Arabidopsis protoplasts. For all protein combinations, reconstituted YFP signals were detected as puncta (Figure 2E; Supplemental Figure 3C). When we coexpressed the domain-truncated YN-FREE1 or YN-VPS23A with SINAT1-YC, SINAT1 specifically interacted with the N-terminal region of FREE1 and the C-terminal region of VPS23A (Figure 2E), consistent with the Y2H results (Figure 2B). As controls, neither YN-VPS28 nor YN-VPS37 could reconstitute YFP signals with SINAT-YC constructs (Supplemental Figure 3C).
Given the physical interactions between SINATs and both FREE1 and VPS23A, we also examined whether these three proteins gathered in the same subcellular locations. As shown in Supplemental Figure 3D, the BiFC signals of YN-FREE1 with YC-VPS23A or SINATs-YC colocalized with the mCh-fused SINAT1, SINAT2, and VPS23A. Taken together, these findings support the idea that SINATs physically interact with FREE1 and VPS23A in vitro and in vivo.
SINATs Mediate the Ubiquitination and Degradation of FREE1 and VPS23A
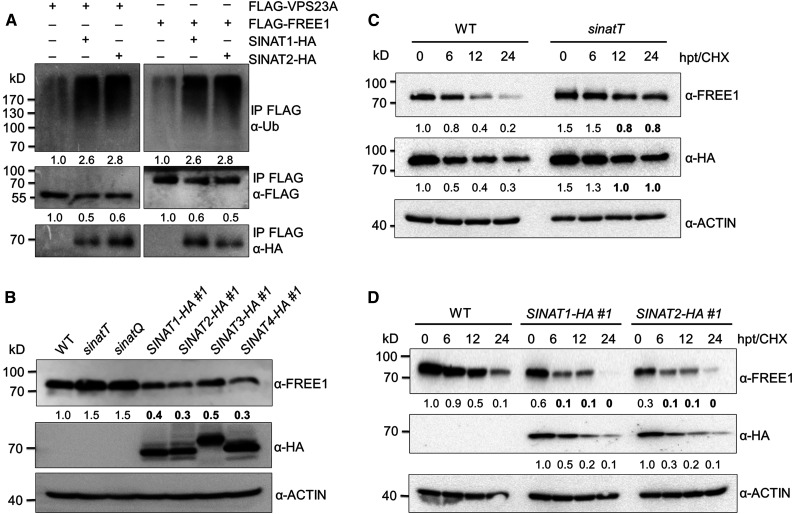
To investigate the significance of the protein association between SINATs and FREE1/VPS23A, we analyzed the ubiquitination of FREE1 and VPS23A in the presence or absence of SINAT1 and SINAT2. The constructs encoding FLAG-tagged FREE1 or VPS23A were transfected into the wild-type Arabidopsis protoplasts in the presence or absence of HA-tagged SINAT1 or SINAT2. After overnight culture, total protein was extracted from collected cells and FLAG-FREE1 or FLAG-VPS23A were enriched using anti-FLAG magnetic beads. The eluate was then immunoblotted using anti-ubiquitin antibodies to detect FREE1- and VPS23A-specific ubiquitin conjugates. Compared to the controls in the absence of SINAT1 or SINAT2, the ubiquitination levels of both FLAG-FREE1 and FLAG-VPS23A were enhanced in the presence of SINAT1 or SINAT2 (Figure 3A; Supplemental Figure 4).
Figure 3.
SINATs Mediate Ubiquitination and Degradation of FREE1 and VPS23A.
(A) In vivo ubiquitination assays for VPS23A and FREE1. FLAG-tagged VPS23A and FREE1 were coexpressed with GFP-SINAT1-HA or GFP-SINAT2-HA, respectively, in the wild-type (WT) Arabidopsis protoplasts. Total protein from each sample was extracted in IP buffer and incubated with anti-FLAG magnetic beads. Proteins in the eluate were detected by anti-ubiquitin (Ub), anti-FLAG, and anti-HA antibodies. The relative intensities of Ub-FLAG-VPS23A, Ub-FLAG-FREE1, FLAG-VPS23A, and FLAG-FREE1 are shown below.
(B) FREE1 protein level in the wild-type (WT), sinat mutants, and SINAT overexpression lines. Total proteins were extracted from 7-d-old seedlings of WT, sinatT (sinat1 sinat2 sinat4), sinatQ (sinat1 sinat2 sinat3 sinat4), and SINAT-overexpression lines (SINAT1-HA #1, SINAT2-HA #1, SINAT3-HA #1, and SINAT4-HA #1). The anti-FREE1 and anti-HA antibodies were used in immunoblots to detect FREE1 and SINAT proteins.
(C) Protein stabilities of FREE1 and VPS23A in the sinatT mutant. Transgenic seedlings (7 d old) expressing HA-GFP-VPS23A in the wild-type (WT) or sinatT mutant backgrounds were treated with 500 μM CHX for the indicated times (0, 6, 12, and 24 h). Total proteins were extracted with IP buffer, and anti-FREE1 and anti-HA antibodies were used to detect FREE1 and HA-GFP-VPS23A, respectively.
(D) Degradation of FREE1 in SINAT-overexpression plants. The wild-type (WT), SINAT1-HA #1, and SINAT2-HA #1 seedlings (7 d old) were treated with 500 μM CHX for the indicated times (0, 6, 12, and 24 h).
Total proteins were extracted with IP buffer, and anti-FREE1 and anti-HA antibodies were used to detect FREE1 and GFP-SINATs-HA proteins, respectively. The level of ACTIN was used as a control. The relative intensities of FREE1, HA-GFP-VPS23A, and GFP-SINATs-HA proteins are shown below. The bold numbers show remarkably decreased protein levels compared with that of controls. Numbers on the left indicate the molecular weight (kD) of each band. hpt, hours post treatment.
To determine the role of SINATs in regulating FREE1 and VPS23A stability, we first examined the endogenous FREE1 protein level in sinat mutants, including the sinat1 sinat2 sinat4 (termed sinatT) triple mutant and sinat1 sinat2 sina3 sinat4 (sinatQ) quadruple mutant (Qi et al., 2017, 2020a), and in the SINAT-overexpression lines (SINAT1-HA, SINAT2-HA, SINAT3-HA, and SINAT4-HA; Supplemental Figure 5). Using anti-FREE1–specific antibodies, we observed that FREE1 protein accumulated in the sinatT and sinatQ mutants but was reduced in all SINAT-HA lines (Figure 3B), suggesting that SINAT proteins negatively regulate FREE1 protein stability. We further examined and compared the half-lives of FREE1 and HA-VPS23A proteins in the wild-type and sinatT mutant backgrounds by crossing the HA-GFP-VPS23A transgenic line into the sinatT mutant background. Following exposure to cycloheximide (CHX) for various times (0, 6, 12, and 24 h), the degradation of FREE1 and VPS23A proteins was reduced in the sinatT mutant compared to that in the wild type (Figure 3C). By contrast, we observed that the degradation of FREE1 was clearly enhanced in SINAT1-HA and SINAT2-HA lines at all time points (Figure 3C). These findings imply that SINAT proteins control the ubiquitination and degradation of FREE1 and VPS23A. Notably, from 6 to 24 h post-CHX treatment, the protein levels of SINAT1 and SINAT2 were also considerably reduced (Figure 3D), suggesting that an alternative degradation pathway might contribute to the regulation of SINAT protein stability.
Overexpression of SINATs Confers Increased ABA Sensitivity
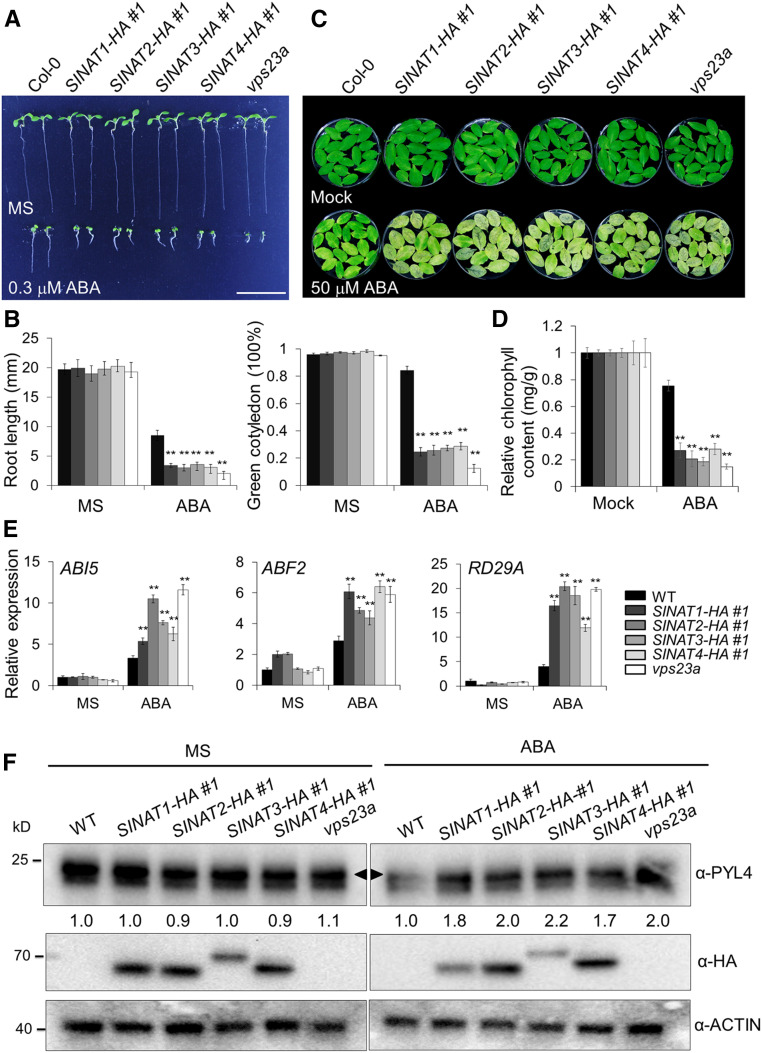
FREE1 and VPS23A play crucial roles in ABA signaling by modulating the stability of PYR/PYL/RCAR ABA receptors (Belda-Palazon et al., 2016; Yu et al., 2016). Because the vps23a mutants and free1 knockdown alleles are hypersensitive to ABA treatment (Belda-Palazon et al., 2016; Yu et al., 2016), we examined the phenotypes of SINAT-HA overexpression lines upon ABA exposure. To this end, seeds of the wild-type and SINAT-HA lines (SINAT1-HA, SINAT2-HA, SINAT3-HA, and SINAT4-HA) were germinated on MS medium supplemented with or without 0.3 μM ABA and the vps23a mutant (Yu et al., 2016) was used as a control. All genotypes showed few morphological differences compared with the wild type on MS medium. By contrast, similar to the vps23a mutant, all four SINAT-HA lines showed enhanced sensitivity to ABA treatment compared with the wild-type seedlings (Figure 4A; Supplemental Figure 6A). After germination under normal conditions for 7 d, all seedlings of SINAT-HAs and vps23a showed inhibited growth, as indicated by a significantly reduced root length and proportion of seedlings with green cotyledons (Figure 4B). Consistent with previous findings (Yu et al., 2016), we observed that the germination rates of SINAT-HA lines as well as the vps23a mutant showed no significant difference from the wild type on 0 and 0.3 μM ABA for up to 7 d (Supplemental Figure 6B), suggesting that SINATs are dispensable for ABA signaling during seed germination. Furthermore, we examined the ABA responses in mature SINAT-HA plants by observing ABA-induced leaf senescence (Gao et al., 2016; Li et al., 2019). As shown in Figures 4C and 4D, compared to wild-type leaves, all of the SINAT-HA lines and vps23a mutant showed accelerated leaf senescence and significantly reduced chlorophyll contents upon 50 μM ABA treatment for 3 d.
Figure 4.
SINAT Overexpression Confers Increased ABA Sensitivity.
(A) Sensitivities of SINAT-overexpression lines to ABA. Seeds of the wild-type (WT) and SINAT-overexpression lines (SINAT1-HA #1, SINAT2-HA #1, SINAT3-HA #1, and SINAT4-HA #1) and vps23a mutant were sown on MS medium with or without 0.3 μM ABA for 7 d. Bar = 10 mm.
(B) Measurements of root length and proportion of green cotyledons in (A). Data are means ± sd (n = 3) of three independent experiments (biological replicates). For each experiment, more than 20 seedlings were measured for root length and more than 100 seedlings were assayed for the green cotyledon frequency. Asterisks indicate significant differences from WT (*P < 0.05; **P < 0.01 by Student’s t test).
(C) Leaf senescence in SINAT-overexpression lines. The detached rosette leaves of the 4-week-old wild type (WT), SINAT-overexpression lines (SINAT1-HA #1, SINAT2-HA #1, SINAT3-HA #1, and SINAT4-HA #1), and vps23a mutant were incubated in distilled water (mock) or 50 µM ABA for 3 d under continuous darkness.
(D) Measurement of chlorophyll contents in (C). The experiments were repeated three times (biological replicates) with similar results, and the representative data from one replicate are shown. Data are means ± sd (n = 3) of three technical replicates. Asterisks indicate significant differences from the wild type (WT; **P < 0.01 by Student’s t test).
(E) RT-qPCR analysis showing ABA-response gene expression in SINAT-overexpression lines. The wild type (WT), SINAT-OE lines (SINAT1-HA #1, SINAT2-HA #1, SINAT3-HA #1, and SINAT4-HA #1), and vps23a (7 d old) grown on MS medium with or without 0.3 μM ABA were harvested for total RNA extraction. Transcript levels of ABI5, ABF2, and RD19A genes in SINAT-overexpression lines relative to the WT were normalized to that of ACTIN2. The experiments were repeated three times (biological replicates) with similar results, and representative data from one replicate are shown. Data are means ± sd (n = 3) of three technical replicates. Asterisks indicate significant differences from the WT (**P < 0.01 by the Student’s t test).
(F) Accumulation of PYL4 protein in SINAT-overexpression seedlings compared with the wild type (WT) under ABA treatment. The 7-d-old seedlings of WT and SINAT-overxpression lines (SINAT1-HA #1, SINAT2-HA #1, SINAT3-HA #1, and SINAT4-HA #1), grown on MS medium with or without 0.3 μM of ABA, were collected for total protein extraction and immunoblot analysis by anti-PYL4, anti-HA, and anti-ACTIN antibodies, respectively. The arrowhead indicates a specific band of PYL4. The relative intensities of PYL4 proteins in SINAT-overexpression lines and vps23a mutant compared with the WT were normalized to that of ACTIN and shown below the anti-PYL4 row. Numbers on the left indicate the molecular weight (kD) of each band.
Consistent with the increased ABA sensitivity in SINAT-HA and vps23a mutant plants, RT-qPCR analyses showed that the transcripts of ABA downstream transcriptional factors, including ABI5, ABF2, and RD29A, were significantly upregulated in the SINAT-HA lines compared to the wild type (Figure 4E). Immunoblotting data suggested that the ABA receptor PYL4 proteins increased considerably in the SINAT-HA lines and vps23a mutant compared to the wild type when grown on MS medium supplemented with ABA (Figure 4F), suggesting that overexpression of SINAT1, SINAT2, SINAT3, and SINAT4 enhances the ABA response by increasing the stabilities of ABA receptors.
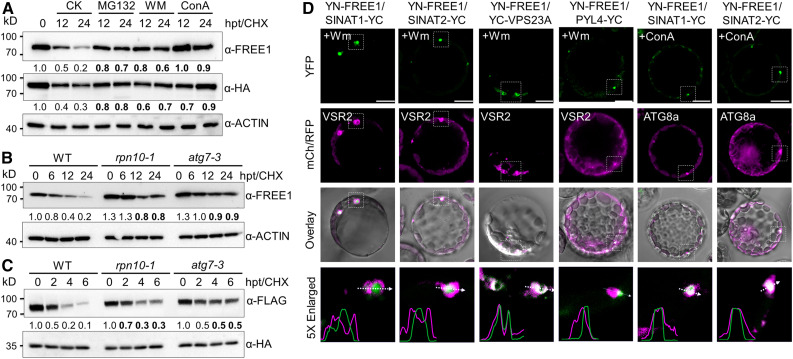
FREE1 and VPS23A Are Codegraded with SINATs via Vacuolar Pathways
To explore further the nature of FREE1 and VPS23A degradation, we applied the proteasome inhibitor MG132, the vacuole-type ATPase inhibitor concanamycin A (ConA), or the phosphatidylionsitol-3-kinase/phosphatidylionsitol-4-kinase inhibitor wortmannin (WM; blocking both MVB pathway and autophagy pathway) to transgenic seedlings expressing HA-GFP-VPS23A. The FREE1 and HA-VPS23A proteins were degraded in the presence of CHX for 12 and 24 h under normal conditions (Figure 5A). By contrast, MG132, WM, and ConA application strongly inhibited the degradation of both proteins (Figure 5A), suggesting that FREE1 and HA-VPS23A are degraded via both proteasomal and vacuolar pathways. To confirm this, we compared the half-lives of FREE1 and HA-VPS23A proteins in the proteasomal mutant rpn10-1 (Smalle et al., 2003) and autophagy mutant atg7-3 (Chen et al., 2015) with that of the wild type. In response to CHX treatment at 12 and 24 h, both FREE1 and HA-VPS23A proteins were more stable in the rpn10-1 and atg7-3 mutants than in the wild-type plants (Figures 5B and 5C).
Figure 5.
Codegradation of FREE1 with SINATs via MVB- and Autophagy-Mediated Vacuole Sorting.
(A) Degradation patterns of FREE1 and VPS23A in vivo. Transgenic seedlings (7 d old) expressing HA-GFP-VPS23A were treated with 500 μM CHX supplemented with 50 μM MG132, 10 μM WM, or 1 μM ConA for the indicated times (0, 12, and 24 h). Anti-FREE1 and anti-HA antibodies were used to detect FREE1 and VPS23A proteins. CK, mock treatment control; hpt, hours posttreatment.
(B) FREE1 protein stability in the wild type (WT) and rpn10-1 and atg7-3 mutants. Seedlings (7 d old) of the WT, rpn10-1, and atg7-3 were treated with 500 μM CHX for the indicated times (0, 6, 12, and 24 h). Anti-FREE1 antibodies were used for FREE1 detection. hpt, hours posttreatment.
(C) VPS23A protein stability in the wild-type (WT) and rpn10-1 and atg7-3 mutants. Plasmids of FLAG-VPS23A and GFP-HA were transfected into protoplasts made from the WT and rpn10-1 and atg7-3 mutants. CHX (10 μM) was added into medium for the indicated times (0, 2, 4, and 6 h) after overnight culture. Total protein was extracted with IP buffer and blotted with anti-FLAG and anti-HA antibodies for FLAG-VPS23A and GFP-HA detection, respectively. The levels of ACTIN or GFP were detected as controls. The relative intensities of FREE1 and VPS23A proteins normalized to ACTIN or GFP are shown below. Numbers on the left indicate the molecular weight (kD) of each band. hpt, hours posttreatment.
(D) BiFC assay showing the subcellular interactions of FREE1 and SINATs. nYFP-FREE1 was coexpressed with cYFP-fused SINAT1, SINAT2, VPS23A, and PYL4 proteins in Arabidopsis protoplasts. RFP-VSR2 or mCh-ATG8a was cotransfected as MVB or autophagosome markers, respectively. Cells were incubated with 5 μM WM or 1 μM ConA overnight and then imaged using confocal microscopy. Profile analysis was conducted using ImageJ. Bars = 10 μm.
The application of proteasome and vacuole inhibitors may cause indirect effects on the protein stabilities of FREE1 and VPS23A. To further confirm the codegradation of SINATs and FREE1 proteins, we applied BiFC assays to determine the subcellular interactions of FREE1 and SINATs. Using transgenic seedlings, we observed that GFP-SINAT2 and GFP-VPS23A colocalized with the autophagosome marker mCh-ATG8f (Supplemental Figure 7B). Moreover, the GFP-SINAT2 and GFP-FREE1 puncta were located on the membrane and in the lumen of MVBs (Supplemental Figure 7A). To investigate whether SINAT proteins are involved in the vacuolar degradation of FREE1, we coexpressed the YN-FREE1 and SINAT1-YC or SINAT2-YC fusions with the MVB marker RFP-VSR2 and the autophagy marker mCh-ATG8a in Arabidopsis protoplasts. We used WM and ConA treatments to inhibit MVB fusion with the vacuole and vacuolar degradation, respectively, aiming to improve the visualization of MVB and autophagosomes (Spallek et al., 2013; Cai et al., 2014; Cui et al., 2016). The reconstituted YFP signals of YN-FREE1/SINAT1-YC and YN-FREE1/SINAT2-YC were encircled by the RFP-VSR2–labeled MVBs after application of WM for 12 h (Figure 5D). As controls, the interacting signals of YN-FREE1 with YC-VPS23A, as well as YN-FREE1 with PYL4-YC, were examined and were located primarily at the outer membrane of MVBs (Figure 5D). By contrast, the fluorescent signals of YN-FREE1/SINAT1-YC and YN-FREE1/SINAT2-YC colocalized with mCh-ATG8a upon ConA exposure for 12 h (Figure 5D). These findings suggest that SINATs are codegraded with FREE1/VPS23A proteins via the MVB- and autophagy-mediated vacuolar degradation pathways.
SINATs Form Homo- and Hetero-Oligomers in Vivo
SINATs contain a TRAF domain and in mammalian TRAF proteins, the TRAF domain is required for trimeric self-association and interaction with receptors (Park et al., 1999). Given that SINAT1, SINAT2, SINAT3, and SINAT4 were detected as potential SINAT1 interactors by IP-MS (Table 1), we speculated that SINAT proteins might form homo- or heterodimers or homo- and hetero-oligomers under specific physiological conditions. To test this, we first conducted Y2H assays to explore the potential interaction between different SINAT proteins. By fusing four mutated SINAT proteins (SINAT1C, SINAT2C, SINAT3C, SINAT4C) and SINAT5 and SINAT6 with the GAL4-AD or DNA-BD, we generated a 6 × 6 interaction assay and found that each pair of SINAT proteins showed interactions in at least one AD–BD combination (Figure 6A). Domain truncation analyses showed that the substrate binding domains, including the ZINC finger and TRAF domain, were necessary for the interactions of SINAT1 and SINAT2 with other SINATs (Figure 6A; Supplemental Figure 8A).
Figure 6.
SINAT Proteins Form Homo- and Hetero-Oligomers in Vivo.
(A) Y2H assay showing mutual interactions between SINAT proteins. Mutated SINAT proteins SINAT1-C60S, SINAT2-C63S, SINAT3-C66S, SINAT4-C67S, SINAT5, and SINAT6 were fused with AD or BD and then paired into a 6 × 6 array for the interaction assay. Three truncated forms of SINAT1 (SINAT1-RZ, SINAT1-ZT, and SINAT1-T) were also fused with BD and cotransformed with six AD-SINATs to identify the binding domain. The plasmids were coexpressed in the YH109 yeast strain and selected on SD-Trp-Leu-His-Ade medium (SD-LWH-Ade). AD and BD indicate the empty AD and BD plasmids, respectively. R, RING finger; T, TRAF domain; Z, ZINC finger.
(B) Co-IP assay for physical interaction of SINAT1 with six SINAT proteins (SINAT1, SINAT2, SINAT3, SINAT4, SINAT5, and SINAT6). Plasmids of FLAG-tagged SINAT1, SINAT2, SINAT3, SINAT4, SINAT5, SINAT6, or empty vector (EV) were transfected into Arabidopsis protoplasts isolated from a GFP-SINAT1-HA transgenic line. Protoplasts were cultured overnight and were then lysed with IP buffer and incubated with anti-FLAG magnetic beads.
(C) BiFC assay for the association between SINAT proteins. nYFP-fused SINAT1 or SINAT2 was coexpressed with cYFP-fused SINAT1, SINAT1-RZ, or SINAT1-ZT in the wild-type (WT) Arabidopsis protoplasts. Signals were detected after overnight culture by confocal microscopy. The reconstituted YFP signal suggests an interaction between two proteins. Bars = 10 μm.
(D) Oligomer formation of SINAT proteins in vivo. Total proteins were extracted from 1-week-old GFP-SINAT1-HA, GFP-SINAT2-HA, SINAT3-GFP-HA, and GFP-SINAT4-HA transgenic seedlings with IP buffer and were denatured with SDS sample buffer with or without 1% 2-ME. SINAT proteins were detected with anti-HA antibodies. A Ponceau-stained membrane shown below the blot indicates the loading protein amounts. The relative intensities of oligomers and monomers normalized to ribulose-1,5-bis-phosphate carboxylase/oxygenase (Rubisco) are shown below, with each 2-ME–containing samples set as 1. Double asterisk (**) indicates oligomers of SINAT proteins. Numbers on the left indicate the molecular weight (kD) of each band.
We also performed Co-IP and BiFC assays to confirm the interaction between SINAT1 and other SINAT proteins. When FLAG-tagged SINAT proteins were transiently expressed in protoplasts from transgenic Arabidopsis lines expressing GFP-SINAT1-HA, we observed that GFP-SINAT1-HA coimmunoprecipitated with all six SINAT-FLAG proteins, but not with pure anti-FLAG magnetic beads (empty vector; Figure 6B). Consistent with these Co-IP results, both SINAT1-YN and SINAT2-YN bound SINAT1, SINAT2, SINAT3, and SINAT4 fused to YC, to reconstitute YFP signals (Figure 6C; Supplemental Figure 8D). Domain-truncation analyses further suggested that the Zinc finger and TRAF domains together, rather than the RING finger and Zinc finger domains, of SINAT1-YC could reconstitute YFP signals with SINAT1-YN and SINAT2-YN in BiFC assays (Figure 6C). As negative controls, neither SINATs-YN nor SINATs-YC reconstituted YFP signals with free YC or YN (Supplemental Figure 8D). Notably, similar to the subcellular localizations of SINAT1 (Figure 1A; Supplemental Figure 1A), all of the BiFC signals were observed as punctate dots within the cytoplasm (Figure 6C; Supplemental Figure 8D).
To investigate the functional significance of the interactions among different SINAT proteins, we performed immunoblot analyses to examine whether they formed dimers or oligomers in vivo. For this experiment, total proteins were extracted from 1-week-old transgenic seedlings expressing SINAT1-HA, SINAT2-HA, SINAT3-HA, or SINAT4-HA and were denatured with SDS buffer with or without 1% 2-mercaptoethanol (2-ME), which reduces disulfide linkages. Using anti-HA antibodies, we observed that in the presence of 2-ME, all SINAT-HA protein bands were detected at ∼70 kD, as expected (Figure 6D). In the absence of 2-ME, however, some slower migrating bands larger than 180 kD were observed (Figure 6D). This phenomenon was also observed in the ProSINAT1:SINAT1-HA transgenic lines driven by the native SINAT1 promoter (Supplemental Figure 8B), suggesting that SINAT proteins might function as oligomers in endogenous conditions, organized mainly by disulfide linkages.
Considering the E3 protein ligase nature of SINATs, those slower migrating bands might also be ubiquitin conjugates. To distinguish disulfide-linked oligomerization from other possible modifications such as ubiquitination, we performed two-dimensional SDS-PAGE with no 2-ME in the first dimension and 2-ME in the second dimension. The high molecular weight bands of SINAT1-HA in one-dimensional (1D) SDS-PAGE detached into low molecular weight bands identical to the size of SINAT1 monomers when electrophoresed in the second dimension (Supplemental Figure 8C). By contrast, the ubiquitin conjugates of SINAT1-HA formed a diagonal ladder after two-dimensional SDS-PAGE (Supplemental Figure 8C). Together, our data suggest that SINAT proteins form oligomers under endogenous conditions.
Autophagy Mediates Turnover of SINAT Oligomers during Recovery following ABA Treatment
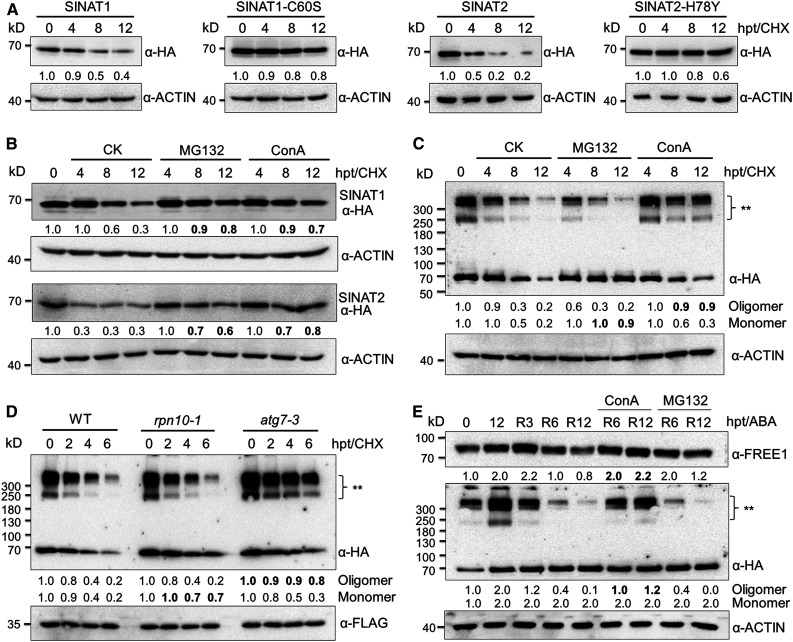
We previously demonstrated that recombinant SINAT1 and SINAT2 undergo self-ubiquitination in vitro and that SINAT1 and SINAT2 are rapidly degraded upon nutrient starvation (Qi et al., 2017). To further examine the effects of auto-ubiquitination on the regulation of SINAT protein stability, we examined their half-lives following treatment with the translational inhibitor CHX. Using the SINAT-HA lines, we observed that the protein levels of SINAT1-HA, SINAT2-HA, SINAT3-HA, and SINAT4-HA decreased after 4 h of CHX treatment (Figure 7A; Supplemental Figure 9A). By contrast, the SINAT5-HA and SINAT6-HA proteins were stable in similar conditions (Supplemental Figure 9A). Next, we constructed two mutant forms of SINAT1 and SINAT2, SINAT1-C60S and SINAT2-H78Y, that were altered by site-directed mutagenesis at the catalytic sites of the RING finger domain (Supplemental Figure 2A; Xie et al., 2002). SINAT1-C60S and SINAT2-H78Y possessed much longer half-lives than the wild-type SINAT1 and SINAT2 (Figure 7A), indicating that their E3 ligase activities are important to maintain SINAT protein levels.
Figure 7.
Differential Degradation of SINAT Proteins by Proteasomal or Vacuolar Pathway.
(A) Half-life measurement of SINAT1 and SINAT2. Transgenic seedlings (1-week-old) expressing GFP-SINAT1-HA, GFP-SINAT1(C60S)-HA, GFP-SINAT2-HA, and GFP-SINAT2(H78Y)-HA were treated with 500 μM CHX for 0, 4, 8, or 12 h.
(B) Chemical treatment showing the degradation patterns of SINAT proteins. GFP-SINAT1-HA and GFP-SINAT2-HA transgenic seedlings (1-week-old) treated with 500 μM CHX were incubated with or without 50 μM MG132 or 1 μM ConA for the indicated times (0, 4, 8, and 12 h).
(C) Chemical treatment showing the differential degradation preference of GFP-SINAT1-HA monomers and oligomers. GFP-SINAT1-HA transgenic seedlings (1 week old) treated with 500 μM CHX were incubated with or without 50 MG132 or 1 μM ConA for the indicated times (0, 4, 8, and 12 h).
(D) Degradation pattern of GFP-SINAT1-HA monomers and oligomers in the wild type (WT), rpn10-1, and atg7-3. Plasmids of GFP-SINAT1-HA and RFP-FLAG were transfected into protoplasts isolated from WT and rpn10-1 and atg7-3 mutants. After overnight culture, protoplast cells were treated with 10 μM CHX for the indicated times (0, 2, 4, and 6 h).
(E) Codegradation of SINAT oligomers with FREE1 during prolonged ABA treatment and a recovery period. GFP-SINAT2-HA transgenic seedlings (1 week old) grown on MS medium were treated with 50 μM ABA for 12 h and allowed to recover for 3 h (R3), 6 h (R6), or 12 h (R12). ConA (1 μM) or 50 μM MG132 was supplied during the recovery period to identify the degradation pathway.
Total proteins were extracted with IP buffer. GFP-SINATs-HA, RFP-FLAG, FREE1, and ACTIN were detected by anti-HA, anti-FLAG, anti-FREE1, and anti-ACTIN antibodies, respectively. For SINAT oligomer detection in (C) to (E), 2-ME was omitted from the SDS sample buffer. The expression levels of ACTIN or RFP were used as controls. The relative intensities of GFP-SINATs-HA and FREE1 proteins normalized to ACTIN or RFP are shown below. Double asterisk (**) indicates oligomers of GFP-SINATs-HA. Numbers on the left indicate the molecular weight (kD) of each band. hpt, hours posttreatment.
To explore whether the different monomer and oligomer forms of SINAT proteins are degraded via distinct proteolytic machineries, we first used the proteasomal-specific inhibitor MG132 and the vacuolar-specific inhibitor ConA to treat 1-week-old SINAT1-HA, SINAT2-HA, SINAT3-HA, and SINAT4-HA transgenic seedlings. Immunoblotting showed that MG132 and ConA suppressed the degradation of SINAT1, SINAT2, SINAT3, and SINAT4 in the presence of CHX (Figure 7B; Supplemental Figure 9B), suggesting that both proteasomal and vacuolar pathways contribute to the turnover of SINAT-HA proteins. We also expressed the SINAT1-HA and SINAT2-HA fusions in the ESCRT mutant vps23a, the autophagy mutant atg7-3, and the proteasome mutant rpn10-1 backgrounds. Upon treatment with CHX, SINAT1-HA and SINAT2-HA proteins showed delayed degradation in the vps23a, atg7-3, and rpn10-1 mutants compared to that in the wild-type plants (Supplemental Figures 9D and 9E). This further supports the idea that the turnover of SINAT proteins is controlled via both proteasome and vacuole degradation systems.
In yeast, the oligomerization of protein aggregates as well as ubiquitin receptors guide cargoes for autophagic degradation (Lu et al., 2017). Because SINAT proteins form homo- and hetero-oligomers in vivo, we tested how SINAT aggregates were selected for autophagic degradation. Following exposure to CHX for 8 h, the levels of both monomers and oligomers of SINAT1-HA and SINAT2-HA, which were detected by removing 2-ME in the SDS sample buffer, declined remarkably (Figure 7C; Supplemental Figure 9C). Notably, application of MG132 inhibited the decrease in monomeric SINAT1-HA and SINAT2-HA, whereas ConA treatment protected SINAT1-HA and SINAT2-HA oligomers from degradation (Figure 7C; Supplemental Figure 9C). Consistent with the selective response to inhibitor treatment (Figure 7C; Supplemental Figure 9C), the degradation of SINAT monomers was delayed in the rpn10-1 mutant, whereas the decrease in the level of SINAT oligomers was prevented in the atg7-3 mutant (Figure 7D), confirming that, like in yeast cells, SINAT protein oligomers are targeted to the autophagy machinery for turnover in Arabidopsis.
NEXT TO BRA1 GENE1 (NBR1) is a ubiquitin receptor in Arabidopsis and guides protein aggregates into autophagosomes (Zhou et al., 2013, 2014). The ubiquitin receptor DOMINANT SUPPRESSOR OF KAR2 (DSK2) also interacts with SINAT2 to target ubiquitinated BRI1-EMS-SUPPRESSOR1 (BES1) for autophagic degradation (Nolan et al., 2017). To further explore whether these two ubiquitin receptors are engaged in the differential degradation of SINAT proteins, we examined the interaction between SINAT proteins and NBR1 or DSK2. As shown in Supplemental Figure 10B, both FLAG-NBR1 and DSK2A-FLAG were associated with SINAT-HA proteins in a Co-IP assay. Notably, although the input samples contained less than 30% SINAT oligomers, the Co-IP samples contained roughly 70% SINAT oligomers in samples coimmunoprecipitated by FLAG-NBR1 (Supplemental Figure 10B). Similarly, samples coimmunoprecipitated by FLAG-FREE1 and FLAG-VPS23A contained ∼50% SINAT oligomers, compared to 20% in the total pools (Supplemental Figure 10A). Therefore, we suggest that SINAT oligomers may have higher affinities for the ubiquitin receptor NBR1 and their targets FREE1 and VPS23A, which likely contributes to the autophagic degradation of SINAT proteins and their target proteins.
To link the oligomerization-guided vacuolar degradation of SINAT proteins to ABA signaling, we further examined the levels of oligomers and monomers of SINAT2-HA upon ABA exposure for 0 and 12 h and at 3, 6, and 12 h during recovery post-ABA treatment. As expected, compared to untreated control (0 h), SINAT2-HA monomers and oligomers accumulated 12 h after ABA treatment (Figure 7E). However, SINAT2-HA oligomers were gradually degraded from 3 to 12 h of recovery after ABA treatment, which was inhibited by the application of ConA, but not by MG132 (Figure 7E). By contrast, SINAT2 monomers were relatively stable during the whole post-ABA treatment period compared to after 12 h of ABA treatment (Figure 7E). Moreover, the FREE1 protein showed a similar pattern to SINAT2 oligomers before and after ABA treatment at various time points post-ABA exposure (Figure 7E), indicating that during recovery stages post-ABA signaling, SINAT oligomers were codegraded with FREE1 by the autophagy-mediated vacuolar degradation pathway.
Given the translocation of FREE1 into the nucleus in response to ABA exposure (Li et al., 2019), we propose the elevation of FREE1 protein upon 12-h ABA treatment (Figure 7E) may due to its nuclear accumulation. To confirm this idea, we determined the abundances of SINAT2, FREE1, and PYL4 proteins in the cytosolic and nuclear fractions in response to ABA treatment at various time points (0, 3, 6, 12, 24, and after a 12-h recovery period) in the GFP-SINAT2-HA transgenic seedlings. As shown in Supplemental Figure 11A, the cytosolic fraction of FREE1 was degraded from 6 h after ABA treatment, while the nuclear FREE1 accumulated at various time points after ABA exposure and at 12-h recovery stage post-ABA exposure (Supplemental Figure 11A). Consistent with this, the PYL4 and/or PYR1 proteins detected by the PYL4-specific antibodies (Yu et al., 2016) showed mild accumulation from 12 h after ABA exposure in the cytosol fraction in the GFP-SINAT2-HA transgenic seedlings (Supplemental Figure 11A). Unlike the reduction of PYL4 and PYR1 protein levels in the wild-type seedlings in the presence of ABA (Figure 4F; García-León et al., 2019; Fernandez et al., 2020), these data confirmed that overexpression of SINAT2 enhanced the PYL4 and PYR1 protein stabilities by relieving them from vacuolar degradation. Together, these findings suggest that the SINAT proteins may predominately contribute to modulate protein stabilities of FREE1 in the cytoplasm.
To further establish the specific cellular compartments where each protein stabilization/destabilization process occurred, we performed confocal microscopy to determine the subcellular distributions of GFP-SINAT2 and GFP-FREE1 fusions upon treatment with ABA, ConA, or MG132, using stable transgenic seedlings expressing GFP-SINAT2 and GFP-FREE1. As shown in Supplemental Figure 11B, the puncta-localized GFP-SINAT2 and GFP-FREE1 did not change significantly in response to a 12-h ABA treatment. By contrast, a fraction of GFP-FREE1 translocated to the nucleus in response to ABA treatment (Supplemental Figure 11B), consistent with previous findings (Li et al., 2019). Interestingly, we observed that in response to ABA exposure, the GFP-SINAT2 signal was also detected in the nucleus, which was confirmed by fractionation and immunoblotting analyses (Supplemental Figure 11A). Moreover, MG132 application led to the accumulation of both GFP-SINAT2 and GFP-FREE1 in the nucleus (Supplemental Figure 11B), indicating that they are likely involved in modulation of ABA signaling in the nucleus. Furthermore, application of ConA, but not MG132, resulted in accumulation of both GFP-SINAT2 and GFP-FREE1 in puncta (Supplemental Figure 11B). Consistent with the immunoblot results (Figures 5A and 7B; Supplemental Figure 11A), these findings further support the idea that the turnover of SINAT and/or FREE1/VPS23A proteins is controlled via both proteasome and vacuole degradation systems.
Moreover, we examined the transcript levels of PYR1, PYL4, and ABI5 during ABA exposure for 6 and 12 h, followed by recovery for 6 and 12 h. PYR1 and PYL4 transcripts were downregulated upon ABA treatment and then returned to basal levels during recovery (Supplemental Figure 11C). As a control, the expression of ABI5 showed significant upregulation upon ABA exposure and returned to basal levels at 6 and 12 h after recovery. This observation suggests that during the recovery stages after ABA exposure, the clearance of ABA receptors may be primarily controlled by posttranslational modifications, among which vacuolar degradation is likely an important mechanism that downregulates ABA signaling for rapid cell recovery.
DISCUSSION
In plants, FREE1 and VPS23A are key components of ESCRT and function in regulating MVB biogenesis and the endosomal sorting of membrane proteins (Spitzer et al., 2006; Gao et al., 2014). During this process, ubiquitin and ubiquitin binding proteins, including FREE1 and VPS23A, are removed from the ubiquitinated cargoes before the cargo proteins are sorted into intraluminal vesicles (Fan et al., 2016; Gao et al., 2017; Isono and Kalinowska, 2017). FREE1 and VPS23A are involved in the modulation of ABA signaling, either by targeting the ABA receptors PYR1/PYL4 for vacuole-mediated turnover (Belda-Palazon et al., 2016; Yu et al., 2016) or by shuttling to the nucleus to suppress the transcriptional activity of ABA-responsive factors (Li et al., 2019). However, much less is known about how FREE1 and VPS23A protein abundance is regulated in plant cells.
In this study, we provide several lines of evidence to suggest that four SINAT family E3 ubiquitin ligases, SINAT1 to SINAT4, promote ABA signaling by controlling the ubiquitination and degradation of FREE1 and VPS23A. First, confocal microscopy indicated that GFP-SINAT1, GFP-SINAT2, GFP-SINAT3, and GFP-SINAT4 fusions localized to punctate structures in the cytoplasm, and these puncta colocalized with autophagosome- and MVB-labeled markers (Figure 1; Supplemental Figure 1). Second, these four SINAT proteins physically interact with FREE1 and VPS23A in vivo to promote their ubiquitination and degradation (Figures 2 and 3; Supplemental Figure 4). Consistent with this, genetic analyses revealed that similar to the vps23a mutant (Yu et al., 2016), SINAT-overexpression lines were hypersensitive to ABA and had upregulated transcripts of ABA-responsive genes and increased PYL4 protein levels (Figure 4; Supplemental Figure 6). Moreover, we observed that SINAT and FREE1 protein stabilities were affected by both proteasomal and vacuolar pathways, and the SINAT–FREE1 interaction resulted in their proteins being engulfed into the lumen of MVBs and cocolocalizing with the autophagosome marker mCh-ATG8a (Figure 5). Furthermore, during recovery post-ABA exposure, SINAT proteins formed homo- and hetero-oligomers in vivo, which were selectively degraded by the autophagy machinery (Figures 6 and 7). Thus, our findings reveal a novel mechanism for the interaction of proteasomal, endosomal, and autophagic degradation pathways in mediating the turnover of SINAT–FREE1/VSP23A complex proteins to promote ABA signaling.
Distinct from the ubiquitination of the mammalian ESCRT-I component Tsg101, which regulates the activities of the ESCRT-I complex in a proteasome-independent manner (Kim et al., 2007), the ubiquitination of Arabidopsis FREE1 and VPS23A proteins leads to the degradation of these two ubiquitin binding proteins via the UPS. FREE1 is a unique plant ESCRT component that specifically interacts with VPS23A in ESCRT-I (Gao et al., 2014); therefore, the ubiquitination of FREE1 and VPS23A by SINATs is probably a plant-specific mechanism that regulates particular physiological processes. To further confirm the direct interaction between SINATs and VPS23A, we performed an in vitro pull-down assay and observed that VPS23A could physically bind SINAT1/2 without FREE1 (Figure 2D), suggesting that the observed interaction between SINATs and VPS23A is not due to FREE1 forming a bridge between these proteins. In fact, both FREE1 and VPS23A are ubiquitin binding proteins of ESCRT-I (Spitzer et al., 2006; Gao et al., 2014), which might explain the observation that they bind to SINATs simultaneously.
Previous findings suggest that FREE1 plays distinct roles in the cytosol and nucleus in response to ABA (Li et al., 2019). Consistently, our results further revealed that the cytosolic FREE1 was degraded in response to ABA from 6 h after treatment (Supplemental Figure 11A). However, the total protein level of FREE1 was greatly enhanced by ABA treatment for 12 h compared to that in the untreated control (Figure 7E), possibly due to the accumulation of the nuclear FREE1 fraction (Supplemental Figure 11A). Alternatively, the increase of FREE1 might be a mechanism for clearance of ABA receptors (Belda-Palazon et al., 2016; Yu et al., 2016) and/or a feedback mechanism to repress ABA-induced transcription (Li et al., 2019) under prolonged ABA exposure (e.g., 12 h). By contrast, at 6 and 12 h of recovery following ABA treatment, FREE1 protein returned to basal levels, which was partially inhibited by application of ConA and MG132 (Figure 7E), indicating that both vacuolar and proteasomal degradation pathways contribute to the modulation of FREE1 stability. We suggested that SINAT2 oligomers mediated degradation of FREE1 in the cytosol through the autophagy pathway during the ABA recovery period (Figure 7E). We further revealed that a small fraction of SINAT2 protein presented in the nucleus after 12 h of ABA treatment (Supplemental Figures 11A and 11B). In particular, we observed that SINAT3 and SINAT6 also showed nuclear localizations under normal growth conditions (Figure 1A; Supplemental Figure 1A). It is therefore worth exploring the physiological significance of SINATs in the regulation of FREE1 in the nucleus.
Unlike the growth defects and impaired vacuole biogenesis of free1 and vps23a mutants (Spitzer et al., 2006; Gao et al., 2014, 2015; Yu et al., 2016), SINAT-overexpression lines showed few morphological differences from the controls (Figure 4), suggesting that SINATs specifically regulate FREE1 and VPS23A abundance in response to certain cues, such as increased levels of ABA. Under normal growth conditions, SINAT proteins are potentially maintained at relatively low levels due to auto-ubiquitination and UPS-mediated degradation, allowing FREE1 and VPS23A proteins to functionally regulate MVB biogenesis and plant growth and development.
Taken together, our findings show that (1) upon prolonged ABA treatment, SINAT proteins accumulate, mediate ubiquitination and destabilization of FREE1 and VPS23A, and modulate the homeostasis of ABA receptors to an appropriate cellular level; and (2) during recovery post-ABA exposure, the proteolytic degradation of FREE1 and VPS23A mediated by SINATs is necessary to terminate the vacuolar sorting of the PYR1/PYL4 ABA receptor proteins.
Compared to the hypersensitivity of the free1 and vps23a mutants to ABA treatment (Figure 4; Belda-Palazon et al., 2016; Yu et al., 2016), overexpression of FREE1 and VPS23A resulted in few phenotypic changes in response to ABA treatment (Supplemental Figure 6). Moreover, ESCRT-III–related protein LYST-INTERACTING PROTEIN5 (LIP5) positively regulates drought tolerance via ABA-mediated signaling (Xia et al., 2016), suggesting that the endomembrane system might play multiple roles in modulating ABA signaling. Given that we identified several other endomembrane trafficking proteins as potential SINAT1 interactors, including retromer components, coatomer subunits for endoplasmic reticulum–Golgi transport, exocyst elements, clathrin heavy chains, and AP-2 members (Table 1), it is possible that SINATs contribute to signaling in ESCRT-I–dependent and –independent manners. Further investigation of the roles of SINATs in modulating the stabilities of endomembrane trafficking proteins will deepen our understanding of SINAT functions and the molecular regulation of ABA signaling.
SINAT1 and SINAT2 target both ATG6 and ATG13 for degradation to modulate autophagosome biogenesis (Qi et al., 2017, 2020a). Many animal and plant E3 ligases recognize more than one target protein. For example, in mammalian cells, Siah proteins promote proteolysis of synaptophysin, DELETED IN COLORECTAL CANCER (DCC), and Carboxyl-terminal binding protein-interacting protein (Hu and Fearon, 1999; Germani et al., 2003). Moreover, the mammalian E3 ligase Cul3-KLHL20 controls UNC-51-LIKE KINASE1 (ULK1) and VPS34 stabilities to terminate autophagy (Liu et al., 2016). In the Arabidopsis Landsberg erecta ecotype, SINAT5 targets various proteins such as NAC DOMAIN CONTAINING PROTEIN1 (NAC1), LATE ELONGATED HYPOCOTYL (LHY), and FLOWERING LOCUS C (FLC) for ubiquitination and degradation (Xie et al., 2002; Park et al., 2007, 2010). Targeting of different proteins, particularly distinct components in the same biological pathway, by the same E3 ligase may be an efficient way for eukaryotic cells to control cellular signaling. Given that proteasomal and vacuolar degradation are controlled by different types of ubiquitination, it is possible that SINAT E3 ligases differentially ubiquitinate FREE1 and VPS23A by interacting with different E2 enzymes dependent on distinct physiological conditions. Although we did not obtain the specific E2 proteins in the SINAT1 interactome by IP-MS (Table 1), further identification of the interactors of SINATs will be needed to better understand the molecular mechanism of SINATs in the regulation of differential protein degradation in plant cells.
Exploiting the ability of the chemical inhibitors MG132, WM, and ConA to block proteasomal or vacuolar activity (Zhuang et al., 2013; Qi et al., 2017, 2020a), we demonstrated that the turnover of FREE1 and VPS23A proteins involves the proteasomal and vacuolar machineries (Figure 5A). Consistent with this, genetic analyses showed that the protein stability of FREE1 and VPS23A in the proteasomal mutant rpn10-1 and autophagic mutant atg7-3 was greatly enhanced (Figures 5B and 5C). Furthermore, we observed that the BiFC signals from interacting FREE1 and SINAT proteins were engulfed into the lumen of MVBs, whereas the signals from the interacting FREE1 and VPS23A proteins localized to the MVB membrane (Figure 5D), which clearly indicates the presence of two distinct functional patterns of FREE1. Conceivably, FREE1–VPS23A heterodimers in the MVB membrane might regulate MVB biogenesis, whereas the MVB-engulfed SINAT–FREE1 heterodimers are probably en route to vacuolar degradation. This phenomenon resembles that observed for the Arabidopsis autophagy-related proteins ATG1s, ATG13s, and ATG8s (Yoshimoto et al., 2004; Suttangkakul et al., 2011). In the autophagy pathway, the outer membrane of an autophagosome fuses with the tonoplast to release the inner membrane together with the luminal cargoes and ATG8 proteins into the vacuole for degradation (Yoshimoto et al., 2004; Marshall and Vierstra, 2018). Moreover, ATG1 and ATG13, two central kinase complex proteins that initiate autophagosome biogenesis, are degraded via the autophagy pathway (Suttangkakul et al., 2011), thus providing feedback control of autophagy. Because FREE1 plays dual roles in regulating vacuolar sorting and autophagy-mediated degradation (Gao et al., 2015, 2017), it probably acts as a regulator and as a target for the MVB and/or autophagy pathways.
Our findings suggested that SINATs are codegraded with FREE1 and VPS23A via the MVB- and autophagy-mediated vacuolar degradation pathways (Figure 5). Several lines of evidence suggest that these two pathways may act in coordinate and independent manners to degrade particular proteins including SINATs, FREE1, and VPS23A in plant cells. First, the MVB-mediated vacuolar sorting and autophagy pathways both engage the endomembrane system to sequester cargo proteins with ubiquitin as common initiation signals and the vacuole as final destination (Dikic, 2017; Gao et al., 2017; Cui et al., 2018). Second, several ESCRT components such as FREE1, VPS2.1, and CHMP1 are involved in autophagosome biogenesis or autophagy regulation (Katsiarimpa et al., 2013; Gao et al., 2015; Spitzer et al., 2015). Third, MVBs are constantly organized for vacuolar sorting and homeostasis of membrane or cytosolic proteins, whereas autophagy occurs at a basal level under normal growth conditions and is induced in response to developmental and environmental cues.
Increasing evidence suggests that protein quantity is controlled through either the proteasome or autophagy system. However, the mechanism underlying the selection of one of the two pathways remains largely unknown. In yeast cells, the oligomerization of ubiquitinated cargo proteins can guide degradation selectively via the proteasomal or selective autophagic systems (Lu et al. 2017). In particular, proteasomes generally cannot digest abnormal protein aggregates; the presence of these aggregates can trigger induction of autophagy to degrade the stress- or age-induced protein aggregates (Lu et al. 2017).
We report here that, similar to the situation in yeast cells, degradation of Arabidopsis SINAT protein monomers and oligomers is selectively performed via either the proteasomal or autophagic pathways. Using chemical inhibitors, we demonstrated that the degradation of SINAT monomers and oligomers was specifically inhibited by treatment with MG132 and ConA (Figure 7C), respectively, indicating that SINAT monomers are degraded by the proteasome system, whereas the oligomers are processed by the vacuolar pathway. We further confirmed that the half-lives of monomers and oligomers were much longer in the proteasome- and autophagy-deficient mutants rpn10-1 and atg7-3, respectively, compared with the wild type (Figure 7D). Given that the oligomers, but not monomers, of SINAT2 were degraded during recovery from ABA exposure (Figure 7E), our findings thus provide direct evidence to support the significance of the autophagy machinery in the clearance of SINAT oligomers post-ABA signaling, a process that might play essential roles in the termination of ABA responses. Notably, we observed that during the recovery period, the levels of FREE1 protein declined at 6 and 12 h after ABA treatment, and ConA application limited this decrease (Figure 7E). This suggests that a certain pool of FREE1 will undergo autophagy-mediated vacuolar degradation together with SINAT oligomers. However, compared to 95% degradation of SINAT2 oligomers at 12 h after recovery, only 60% of FREE1 was degraded under the same conditions (Figure 7E), which is probably connected with the role of FREE1 in attenuating transcription during ABA signaling (Li et al., 2019).
Although it remains unknown how autophagy targets SINAT oligomers for clearance, it has been suggested that the ubiquitin receptor DSK2 and the autophagy protein ATG8 interact with the E3 ligase SINAT2 and guide ubiquitinated BES1 for autophagic degradation (Nolan et al., 2017). In addition, the Arabidopsis ubiquitin receptor NBR1 guides protein aggregates into the autophagosomes (Zhou et al., 2013, 2014). By Co-IP analysis, we indeed observed that NBR1 preferentially associated with SINAT1 and SINAT2 oligomers (Supplemental Figure 10B), suggesting that NBR1 is involved in differentiation of SINAT oligomers for autophagic degradation. Future studies into the molecular mechanism by which NBR1 targets the SINAT oligomers for autophagy will deepen our understanding of which pathway is used for SINAT protein degradation in plant cells.
In mammalian cells, the TRAF domain is required for protein trimeric self-association and interactions with trimeric ligands, initiating downstream signaling (Park et al., 1999). This possibility cannot be excluded in the SINAT-mediated ABA signaling pathway given that the SINAT oligomers are visible before ABA treatment and enhanced at 12 h upon ABA exposure (Figure 7E). Thus, it is necessary to further determine the physiological significance of SINAT oligomerization in signal transduction in plant cells.
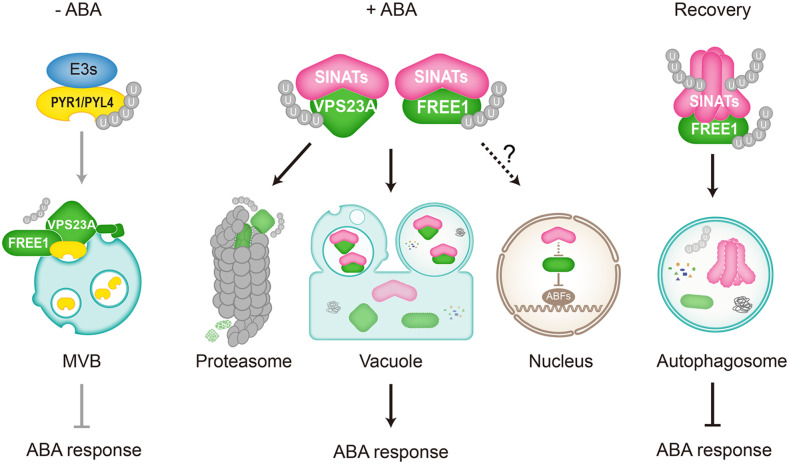
In conclusion, we elucidated the interplay between the UPS system and ESCRT machinery in regulating ABA signaling (Figure 8), revealing a role for E3 ubiquitin ligases in modulating the protein stability of ESCRT components. In the absence of ABA, the ESCRT-I components FREE1 and VPS23A interact with the ubiquitinated ABA receptors PYR1 and PYL4 for vacuolar degradation, thus suppressing ABA response (Figure 8, left). By contrast, the accumulation of ABA triggers SINAT proteins to target FREE1 and VPS23A for proteasomal and vacuolar degradation to activate the ABA response (Figure 8, middle). SINATs may also contribute to modulate FREE1 stability in the nucleus, although the precise molecular mechanism remains to be further elucidated. Moreover, we established a functional link for the autophagy pathway in the regulation of E3 ubiquitin ligase post-ABA signaling. During the recovery post-ABA application, SINAT proteins form homo- or hetero-oligomers in planta, which are predominantly degraded though the autophagy pathway, reactivating the ESCRT machine to remove excess PYR1 and PYL4 and thus terminate ABA signaling (Figure 8, right). Furthermore, SINAT oligomers may target excess FREE1 and/or VPS23A proteins for autophagic clearance to reset cellular homeostasis. Our findings therefore provide novel insight into the communication between different protein degradation systems and suggest that cooperation between distinct pathways contributes to ABA signal transduction in plant cells.
Figure 8.
Working Model for the Roles of SINATs in the Regulation of ABA Signaling by Modulating the Stabilities of FREE1 and VPS23A.
In the absence of ABA, the ESCRT-I components FREE1 and VPS23A interact with the ubiquitinated ABA receptors PYR1 and PYL4 for vacuolar degradation, thus suppressing ABA responses. By contrast, the accumulation of ABA triggers SINAT proteins to target FREE1 and VPS23A for both proteasomal and vacuolar degradation thus activating ABA responses. SINATs may also modulate FREE1 stability in the nucleus, while the precise mechanism remains to be further elucidated. During the post-ABA application recovery, however, SINAT proteins form homo- or hetero-oligomers in planta, which are predominantly degraded though the autophagy pathway, reactivating ESCRT to remove excess PYR1 and PYL4 proteins and terminate ABA signaling. Moreover, SINAT oligomers may target excess FREE1 and/or VPS23A proteins for autophagic clearance to reset cellular homeostasis.
METHODS
Plant Material, Growth Conditions, and Treatments
The seed pools of the Arabidopsis (Arabidopsis thaliana) T-DNA insertional mutants sinat1 (SALK_010417C), sinat2 (SALK_002174C), sinat3 (SALK-125517C), and sinat4 (CS415212) were obtained from The Arabidopsis Information Resource (http://www.arabidopsis.org). The vps23a mutant was described previously by Yu et al. (2016). The sinat single mutants were crossed to generate double, triple, or quadruple mutants. The primers used for genotyping are listed in Supplemental Data Set 1. All of the transgenic plants were generated by Agrobacterium-medium transformation via the floral dip method (Clough and Bent, 1998).
Surface-sterilized seeds were sown on MS medium plates (Sigma-Aldrich) containing 1% Suc (w/v) and 0.8% agar (w/v). Following cold treatment for 3 d, the plates were incubated at 22°C under a 16-h-light/8-h-dark cycle with a light intensity of 170 mmol/m2/s using fluorescent bulbs (catalog no. F17T8/TL841 17W; Philips).
Plasmid Construction
Most constructs were generated using the ClonExpress II One Step Cloning Kit (catalog no. C112; Vazyme) to avoid interference of restriction enzyme sites in genes. Primers for all constructs are listed in the Supplemental Table.
To generate constructs for transient expression, the vectors pHBT, pUC119, pUC120, and pUC121 were used. The pHBT vector contains a 35SPPDK promoter followed by nYFP or cYFP and was digested with BamHI for generating the SINAT-YN and SINAT-YC constructs. The pUC119 plasmid possesses a UBQ10-GFP-HA-nopaline synthase terminator (NOS) expression cassette. For GFP-SINAT-HA series constructs, pUC119 vectors were digested with StuI for SINAT-gene insertion between GFP and HA. The pUC120 plasmid, derived from pUC119, contains a UBQ10-FLAG/HA-FP(GFP/mCh/YN/YC)-GS linker-NOS expression cassette for molecular manipulation. Vector pUC121, derived from pUC119, contains a UBQ10-XTEN linker-FP(GFP/mCh/YN/YC)-FLAG/HA-NOS operation window. Using these two vectors, genes can be fused with fluorescent proteins and/or small epitope tags at the N or C terminus. Genes designed for N-terminally fused tags, such as FLAG-FREE1, FLAG-VPS23A, FLAG-mCh-FREE1, FLAG-mCh-VPS23A, HA-GFP-VPS23A, YN-FEE1, YN-VPS23A, YN-VPS28A, YN-VPS37A, and FLAG-NBR1 were cloned into pUC120 via BamHI and/or PstI digestion. Genes designed for C-terminally fused tags, such as SINATs-GFP-HA, SINATs-mCh-FLAG, PYL4-YC, SINATs-FLAG, and DSK2A-FLAG were cloned into pUC121 via BamHI and/or StuI digestion.
The UBQ10-NOS expression cassettes of three pUC vectors are flanked by two AscI sites and can be transferred to the binary vector pFGC-RCS or pCAMBIA1300 by digestion and ligation. For ProSINAT1:SINAT1-GFP and ProSINAT1:SINAT1-GFP construction, the genomic sequence of SINAT1 and SINAT6 including the 1.5-kb sequence upstream of ATG were cloned into pFGC-RCS.
To generate constructs for Y2H, the pGADT7 and pGBKT7 vectors were digested with EcoRI and BamHI to expose recombination arms. For GST-FREE1 and GST-VPS23A construction, the vector pGEX-6p-1 was digested with BamHI and XhoI. Generation of MBP-SINAT1 and MBP-SINAT2 was described by Qi et al. (2017).
Transient Expression Analyses and Microscopy
To establish SINAT subcellular localization, their colocalization with FREE1 or VPS23A, and BiFC assays, mesophyll protoplasts were prepared from the 4-week-old wild-type (Columbia-0) Arabidopsis rosettes. The plasmids were transfected as previously described by Yoo et al. (2007). Protoplasts were cultured overnight in the dark or light and were subsequently visualized by confocal microscopy. To determine the colocalization of SINATs and other endomembrane markers, protoplasts were prepared from PSB-D suspension cells and transient expression was performed as described by Miao and Jiang (2007). The colocalization ratio was calculated as described by Shen et al. (2018) by comparing the number of colocalization puncta with total GFP-SINAT puncta. The experiment was repeated three times, and average percentages of three independent experiments were used to calculate the colocalization ratio. For each experiment, four to six cells were collected for analysis.
IP-MS Assay
For the IP-MS assay, 10 g of material from 7-d-old transgenic seedlings expressing SINAT1-HA grown in liquid MS medium was ground in liquid nitrogen and homogenized in 10 mL of 2× IP buffer (100 mM Tris-HCl, pH 7.5, 200 mM NaCl, 2 mM EDTA, 20% glycerol, 0.4% Triton X-100, and protease inhibitor cocktail). After centrifugation at 4°C for 30 min at 12,000g, the supernatant was incubated with anti-GFP beads (catalog no. KTSM1301; KTSM-life) at 4°C overnight with gentle top-to-bottom rotation. Beads were then washed five times with IP buffer containing 0.1% Triton X-100 and were eluted with SDS sample buffer. The eluate was subjected to SDS-PAGE followed by silver staining. The dark- and light-stained bands were cut out separately and sent for liquid chromatography–tandem MS LTQ Orbitrap Elite analysis. Seedlings expressing free GFP protein were used as a blank control for identifying the nonspecific interactions.
Y2H Assay
A one-step-transformation method was used for Y2H assays as previously described by Chen et al. (1992). In brief, 300 μL of saturated yeast AH109 culture was centrifuged and resuspended in transformation buffer (10 μL of 2 M LiAc, 10 μL of 1 M DTT, 80 μL of 50% polyethylene glycol 3350, and 20 μg of salmon sperm DNA) with 200 ng of each AD or BD plasmid. After incubation at 45°C for 30 min, the total suspension was plated directly onto synthetic defined (SD)-Leu-Trp medium and incubated at 30°C for 48 h. Positive transformants were then transferred onto SD-Leu-Trp-His-Ade medium for another 48-h culture. The saturated yeast cultures were stored at 4°C for up to 1 month.
Co-IP and BiFC Assays
For Co-IP assays, total protein was extracted from the wild-type Arabidopsis protoplasts or from 7-d-old stable transgenic seedlings with IP buffer containing 0.2% Triton X-100. Approximately 10% of total lysate was reserved as input, and the rest was incubated with anti-HA magnetic beads (catalog no. HY-K0201; MCE) or anti-FLAG magnetic beads (catalog no. m8823; Sigma-Aldrich) for 4 h at 4°C with gentle rotation. Beads were then washed five times with IP buffer containing 0.1% Triton X-100 and were eluted with sample buffer at 95°C for 5 min, followed by SDS-PAGE.
For the BiFC assay, the split nYFP and cYFP were fused to the respective proteins and coexpressed in protoplasts overnight. The reconstituted YFP signals indicating a positive protein interaction were detected by confocal microscopy. For chemical treatments, 5 μM WM (catalog no. HY-10197; MCE) and 1 μM ConA (catalog no. C9705; Sigma-Aldrich) were applied to protoplast culture medium to visualize the enlarged MVBs and to stabilize the autophagosomes, respectively.
Pull-Down Assay
MBP-fused SINAT1 and SINAT2 proteins were purified from transgenic Escherichia coli as previously described by Qi et al. (2017). For GST-FREE1△446-664 and GST-VPS23A protein expression, cultures were incubated in a shaker at 20°C for 20 h, with 200 μM isopropyl β-d-1-thiogalactopyranoside and 2% Glc, followed by purification using Glutathione Sepharose 4B (catalog. no. 17-0756; GE Healthcare) according to the manufacturer’s instructions. Harvested fusion proteins were desalted and concentrated with IP buffer using Amicon Ultra-4 Centrifugal Filter Devices (catalog no. UFC803024; Millipore) and then the levels were examined with Coomassie Brilliant Blue R 250 staining using BSA as a standard.
For in vitro pull-down assays, 1 μg of GST-FREE1△446-664 or GST-VPS23A was mixed with 1 μg of MBP-SINAT1, MBP-SINAT2, or free MBP in IP buffer with 0.2% Triton X-100 and incubated at 16°C, with shaking at 1200 rpm for 2 h, followed by another 2 h with additional 10 μL of amylose resin. Resins were washed in IP buffer with 0.2% Triton X-100 five times and then eluted in sample buffer at 95°C for 5 min, followed by SDS-PAGE.
Protein Ubiquitination and Degradation Assays
For in vivo ubiquitination assays, total protein was extracted from protoplasts expressing FLAG-tagged FREE1 or VPS23A together with the HA-tagged SINATs in IP buffer containing 0.5% Triton X-100. FLAG-FREE1 and FLAG-VPS23A fusion proteins were then immunoprecipitated using anti-FLAG magnetic beads, and the ubiquitination levels were detected using anti-ubiquitin antibodies (catalog no. 12134-2-AP; Proteintech).
For protein degradation analysis, 7-d-old seedlings grown on solid MS medium were transferred to liquid MS medium with 500 μM CHX (catalog no. 2112; Cell Signaling Technology) for the indicated times, as previously described by Liu and Stone (2010), Nolan et al. (2017), and Yang et al. (2017). For inhibitor treatments, 50 μM MG132 (catalog no. A2585; APExBIO), 1 μM ConA, or 10 μM WM was added to block proteasome activity, vacuolar degradation, or vacuolar sorting, respectively. Total proteins were extracted for immunoblot analyses using specific antibodies.
Protein Extraction and Immunoblotting
For total protein extraction, Arabidopsis material was ground in liquid nitrogen and homogenized in ice-cold IP buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 1 mM EDTA, and 10% glycerol) with 0.5% Triton X-100 and protease inhibitor cocktail. After incubation on ice for 30 min and centrifugation at 4°C for 30 min at 12,000g, the supernatant was transferred to a new tube and denatured by boiling in SDS sample buffer. For protein oligomer detection, reductant (2-ME) was omitted from the SDS sample buffer.
For sulfhydrylation-reduction two-dimensional SDS-PAGE, SINAT1 proteins were extracted from 2 g of ProSINAT1:SINAT1-GFP-HA transgenic seedlings, enriched by anti-HA magnetic beads, and subjected to 1D SDS-PAGE without 2-ME in sample buffer. The strip from 1D SDS-PAGE was further reduced with SDS sample buffer adding 1% 2-ME at 95°C for 5 min and room temperature for 30 min, followed by the second-dimensional SDS-PAGE.
Cytosol and nuclear fractionation were performed as previously described by Li et al. (2019). In brief, 0.5 g of 7-d-old seedlings was grounded in liquid nitrogen and suspended in 1 mL of lysis buffer (20 mM Tris-HCl, pH 7.4, 20 mM KCl, 2.5 mM MgCl2, 2.0 mM EDTA, 25% glycerol, and 250 mM Suc) with protease inhibitor cocktail. The homogenate was filtered through two layers of Miracloth and centrifuged at 1500g for 10 min at 4°C. The supernatant was transferred to a new tube for another 10-min centrifuge at 14,000g at 4°C to obtain the cytosol fraction. The pellet containing crude nucleus extractions from the first centrifuge was gently resuspended in 1 mL of NRBT buffer (20 mM Tris-HCl, pH 7.4, 2.5 mM MgCl2, 25% glycerol, and 0.2% Triton X-100) and centrifuged at 1500g for 5 min at 4°C. The pellet was washed repeatedly with NRBT buffer until the green pellet became white. The nuclear pellet was resuspended in 40 μL of lysis buffer and 10 μL of 5× SDS-sample buffer.
Specific anti-HA (catalog no. H6533; Sigma-Aldrich), anti-FLAG (catalog no. A8592; Sigma-Aldrich), anti-ubiquitin (catalog no. 12134-2-AP; Proteintech), anti-PYL4 (Yu et al., 2016), anti-FREE1 (Gao et al., 2014), anti-FBPase (catalog no. AS04 043; Agrisera), and anti-HISTONE3 (ab179; Abcam) antibodies were used for protein blotting.
Leaf Senescence and Chlorophyll Content Measurement
Rosette leaves were detached from 4-week-old soil-grown plants and floated on water with or without 50 μM ABA for 3 d, followed by photography. For chlorophyll content measurement, fresh leaves were weighed and then immersed in absolute ethanol at 4°C for 2 d. The extracts were quantified spectrophotometrically at 664 and 647 nm. Chlorophyll content was calculated according to the equation (20.21 × OD645 + 8.02 × OD663)/1000 × V/W, where V indicates the volume of extraction buffer (in milliliters) and W indicates the fresh weight of leaves (in grams).
RNA Extraction and RT-qPCR Analysis
Total RNA was extracted from Arabidopsis seedlings using TRIzol reagent (Invitrogen) following the manufacturer’s instructions. The isolated RNA was reverse transcribed using HiScript II Q Select RT Super Mix with gDNA Eraser (Vazyme) followed by RT-qPCR analysis using ChamQ SYBR Color qPCR Master Mix (Vazyme) on a StepOnePlus Real-Time PCR System (Bio-Rad). Three technical replicates were performed per reaction with ACTIN2 as the reference gene. The gene-specific primers used for RT-qPCR are listed in Supplemental Data Set 1.
Statistical Analysis
The relative intensities of each band on immunoblots were quantified using ImageJ, with the first lanes set as 1 in each experiment. The fluorescence profiles of colocalized puncta in cells were measured and graphed with ImageJ. Data presented in this study are means ± sd of three independent experiments unless otherwise indicated. The significance of the differences between groups was determined by a two-tailed Student’s t test (Supplemental Data Set 2). P values < 0.05 or < 0.01 were considered significant.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: SINAT1 (At2g41980); SINAT2 (At3g58040); SINAT3 (At3g61790); SINAT4 (At4g27880); SINAT5 (At5g53360); SINAT6 (At3g13672); FREE1 (At1g20110); VPS23A (At3g12400); VPS23B (At5g13860); VPS28A (At4g21560); VPS28B (At4g05000); VPS37A (At3g53120); PYL4 (At2g38310); ABI5 (At2g36270); ABF2 (At1g45249); RD29A (At5g52310); NBR1 (AT4G24690); DSK2A (AT2G17200).
Supplemental Data
Supplemental Figure 1. Subcellular localization of SINAT proteins.
Supplemental Figure 2. Amino acid sequence alignment and domain structures of SINAT proteins.
Supplemental Figure 3. Interaction assays for SINATs, FREE1, and VPS23A proteins.
Supplemental Figure 4. In vivo ubiquitination of VPS23A and FREE1 by SINAT1 and SINAT2.
Supplemental Figure 5. Identification of SINAT-overexpression lines.
Supplemental Figure 6. SINAT transgenic lines show enhanced sensitivity to ABA.
Supplemental Figure 7. Co-localization of SINAT, FREE1, and VPS23A with MVB or autophagosome.
Supplemental Figure 8. Interaction between SINAT proteins.
Supplemental Figure 9. Degradation Pattern of SINAT proteins.
Supplemental Figure 10. ESCRT-I components and ubiquitin receptor NBR1 show higher affinity for SINAT oligomers.
Supplemental Figure 11. Expression pattern of SINAT2, FREE1, and PYL4 upon ABA treatment.
Supplemental Table. Transgenic plants used in this study.
Supplemental Data Set 1. Primers used in this study.
Supplemental Data Set 2. T.TEST analysis in this study.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank the Arabidopsis Biological Resources Center (www.arabidopsis.org) for providing sinat1, sinat2, sinat3, and sinat4 mutant seed pools. This work was supported by the National Natural Science Foundation of China (grants 31725004, 31670276, and 31461143001 to S.X.) and the Research Grants Council of Hong Kong (grants AoE/M-05/12 and C4002-17G to L.J.).
AUTHOR CONTRIBUTIONS
S.X. designed the research. F.-N.X., H.Q., H.-S.L., L.-J.X., L.-J.Y., and Q.-F.C carried out the experiments. S.X., J.-F.L., Y.-Q.C., B.Z., and L.J. analyzed the data. S.X. and F.-N.X. wrote the article.
References
- Barberon, M., Dubeaux, G., Kolb, C., Isono, E., Zelazny, E., Vert, G.(2014). Polarization of IRON-REGULATED TRANSPORTER 1 (IRT1) to the plant-soil interface plays crucial role in metal homeostasis. Proc. Natl. Acad. Sci. USA 111: 8293–8298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belda-Palazon, B., et al. (2016). FYVE1/FREE1 interacts with the PYL4 ABA receptor and mediates its delivery to the vacuolar degradation pathway. Plant Cell 28: 2291–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai, Y., Zhuang, X., Gao, C., Wang, X., Jiang, L.(2014). The Arabidopsis endosomal sorting complex required for transport III regulates internal vesicle formation of the prevacuolar compartment and is required for plant development. Plant Physiol. 165: 1328–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthew, R.W., Rubin, G.M.(1990). seven in absentia, a gene required for specification of R7 cell fate in the Drosophila eye. Cell 63: 561–577. [DOI] [PubMed] [Google Scholar]
- Chen, D.C., Yang, B.C., Kuo, T.T.(1992). One-step transformation of yeast in stationary phase. Curr. Genet. 21: 83–84. [DOI] [PubMed] [Google Scholar]
- Chen, L., et al. (2015). Autophagy contributes to regulation of the hypoxia response during submergence in Arabidopsis thaliana. Autophagy 11: 2233–2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., Bent, A.F.(1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Cui, Y., He, Y., Cao, W., Gao, J., Jiang, L.(2018). The multivesicular body and autophagosome pathways in plants. Front. Plant Sci. 9: 1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, Y., Shen, J., Gao, C., Zhuang, X., Wang, J., Jiang, L.(2016). Biogenesis of plant prevacuolar multivesicular bodies. Mol. Plant 9: 774–786. [DOI] [PubMed] [Google Scholar]
- Della, N.G., Senior, P.V., Bowtell, D.D.(1993). Isolation and characterisation of murine homologues of the Drosophila seven in absentia gene (sina). Development 117: 1333–1343. [DOI] [PubMed] [Google Scholar]
- Dikic, I.(2017). Proteasomal and autophagic degradation systems. Annu. Rev. Biochem. 86: 193–224. [DOI] [PubMed] [Google Scholar]
- Fan, T., Huang, Z., Chen, L., Wang, W., Zhang, B., Xu, Y., Pan, S., Mao, Z., Hu, H., Geng, Q.(2016). Associations between autophagy, the ubiquitin-proteasome system and endoplasmic reticulum stress in hypoxia-deoxygenation or ischemia-reperfusion. Eur. J. Pharmacol. 791: 157–167. [DOI] [PubMed] [Google Scholar]
- Fernandez, M.A., Belda-Palazon, B., Julian, J., Coego, A., Lozano-Juste, J., Iñigo, S., Rodriguez, L., Bueso, E., Goossens, A., Rodriguez, P.L.(2020). RBR-type E3 ligases and the ubiquitin-conjugating enzyme UBC26 regulate abscisic acid receptor levels and signaling. Plant Physiol. 182: 1723–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floyd, B.E., Morriss, S.C., Macintosh, G.C., Bassham, D.C.(2012). What to eat: Evidence for selective autophagy in plants. J. Integr. Plant Biol. 54: 907–920. [DOI] [PubMed] [Google Scholar]
- Gao, C., Luo, M., Zhao, Q., Yang, R., Cui, Y., Zeng, Y., Xia, J., Jiang, L.(2014). A unique plant ESCRT component, FREE1, regulates multivesicular body protein sorting and plant growth. Curr. Biol. 24: 2556–2563. [DOI] [PubMed] [Google Scholar]
- Gao, C., Zhuang, X., Cui, Y., Fu, X., He, Y., Zhao, Q., Zeng, Y., Shen, J., Luo, M., Jiang, L.(2015). Dual roles of an Arabidopsis ESCRT component FREE1 in regulating vacuolar protein transport and autophagic degradation. Proc. Natl. Acad. Sci. USA 112: 1886–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, C., Zhuang, X., Shen, J., Jiang, L.(2017). Plant ESCRT complexes: Moving beyond endosomal sorting. Trends Plant Sci. 22: 986–998. [DOI] [PubMed] [Google Scholar]
- Gao, S., Gao, J., Zhu, X., Song, Y., Li, Z., Ren, G., Zhou, X., Kuai, B.(2016). ABF2, ABF3, and ABF4 promote ABA-mediated chlorophyll degradation and leaf senescence by transcriptional activation of chlorophyll catabolic genes and senescence-associated genes in Arabidopsis. Mol. Plant 9: 1272–1285. [DOI] [PubMed] [Google Scholar]
- García-León, M., et al. (2019). Arabidopsis ALIX regulates stomatal aperture and turnover of abscisic acid receptors. Plant Cell 31: 2411–2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge, C., Gao, C., Chen, Q., Jiang, L., Zhao, Y.(2019). ESCRT-dependent vacuolar sorting and degradation of the auxin biosynthetic enzyme YUC1 flavin monooxygenase. J. Integr. Plant Biol. 61: 968–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germani, A., Prabel, A., Mourah, S., Podgorniak, M.P., Di Carlo, A., Ehrlich, R., Gisselbrecht, S., Varin-Blank, N., Calvo, F., Bruzzoni-Giovanelli, H.(2003). SIAH-1 interacts with CtIP and promotes its degradation by the proteasome pathway. Oncogene 22: 8845–8851. [DOI] [PubMed] [Google Scholar]
- Hu, G., Chung, Y.L., Glover, T., Valentine, V., Look, A.T., Fearon, E.R.(1997). Characterization of human homologs of the Drosophila seven in absentia (sina) gene. Genomics 46: 103–111. [DOI] [PubMed] [Google Scholar]
- Hu, G., Fearon, E.R.(1999). Siah-1 N-terminal RING domain is required for proteolysis function, and C-terminal sequences regulate oligomerization and binding to target proteins. Mol. Cell. Biol. 19: 724–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isono, E., Kalinowska, K.(2017). ESCRT-dependent degradation of ubiquitylated plasma membrane proteins in plants. Curr. Opin. Plant Biol. 40: 49–55. [DOI] [PubMed] [Google Scholar]
- Katsiarimpa, A., Kalinowska, K., Anzenberger, F., Weis, C., Ostertag, M., Tsutsumi, C., Schwechheimer, C., Brunner, F., Hückelhoven, R., Isono, E.(2013). The deubiquitinating enzyme AMSH1 and the ESCRT-III subunit VPS2.1 are required for autophagic degradation in Arabidopsis. Plant Cell 25: 2236–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, B.Y., Olzmann, J.A., Barsh, G.S., Chin, L.S., Li, L.(2007). Spongiform neurodegeneration-associated E3 ligase Mahogunin ubiquitylates TSG101 and regulates endosomal trafficking. Mol. Biol. Cell 18: 1129–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., et al. (2019). The plant ESCRT component FREE1 shuttles to the nucleus to attenuate abscisic acid signalling. Nat. Plants 5: 512–524. [DOI] [PubMed] [Google Scholar]
- Liu, C.C., Lin, Y.C., Chen, Y.H., Chen, C.M., Pang, L.Y., Chen, H.A., Wu, P.R., Lin, M.Y., Jiang, S.T., Tsai, T.F., Chen, R.H.(2016). Cul3-KLHL20 ubiquitin ligase governs the turnover of ULK1 and VPS34 complexes to control autophagy termination. Mol. Cell 61: 84–97. [DOI] [PubMed] [Google Scholar]
- Liu, H., Stone, S.L.(2010). Abscisic acid increases Arabidopsis ABI5 transcription factor levels by promoting KEG E3 ligase self-ubiquitination and proteasomal degradation. Plant Cell 22: 2630–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, K., den Brave, F., Jentsch, S.(2017). Receptor oligomerization guides pathway choice between proteasomal and autophagic degradation. Nat. Cell Biol. 19: 732–739. [DOI] [PubMed] [Google Scholar]
- Marshall, R.S., Vierstra, R.D.(2018). Autophagy: The master of bulk and selective recycling. Annu. Rev. Plant Biol. 69: 173–208. [DOI] [PubMed] [Google Scholar]
- Miao, Y., Jiang, L.(2007). Transient expression of fluorescent fusion proteins in protoplasts of suspension cultured cells. Nat. Protoc. 2: 2348–2353. [DOI] [PubMed] [Google Scholar]
- Morvan, J., Rinaldi, B., Friant, S.(2012). Pkh1/2-dependent phosphorylation of Vps27 regulates ESCRT-I recruitment to endosomes. Mol. Biol. Cell 23: 4054–4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan, T.M., Brennan, B., Yang, M., Chen, J., Zhang, M., Li, Z., Wang, X., Bassham, D.C., Walley, J., Yin, Y.(2017). Selective autophagy of BES1 mediated by DSK2 balances plant growth and survival. Dev. Cell 41: 33–46.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oelmüller, R., Peškan-Berghöfer, T., Shahollari, B., Trebicka, A., Sherameti, I., Varma, A.(2005). MATH domain proteins represent a novel protein family in Arabidopsis thaliana, and at least one member is modified in roots during the course of a plant–microbe interaction. Physiol. Plant. 124: 152–166. [Google Scholar]
- Paez Valencia, J., Goodman, K., Otegui, M.S.(2016). Endocytosis and endosomal trafficking in plants. Annu. Rev. Plant Biol. 67: 309–335. [DOI] [PubMed] [Google Scholar]
- Park, B.S., Eo, H.J., Jang, I.C., Kang, H.G., Song, J.T., Seo, H.S.(2010). Ubiquitination of LHY by SINAT5 regulates flowering time and is inhibited by DET1. Biochem. Biophys. Res. Commun. 398: 242–246. [DOI] [PubMed] [Google Scholar]
- Park, B.S., Sang, W.G., Yeu, S.Y., Do Choi, Y., Paek, N.C., Kim, M.C., Song, J.T., Seo, H.S.(2007). Post-translational regulation of FLC is mediated by an E3 ubiquitin ligase activity of SINAT5 in Arabidopsis. Plant Sci. 173: 269–275. [Google Scholar]
- Park, Y.C., Burkitt, V., Villa, A.R., Tong, L., Wu, H.(1999). Structural basis for self-association and receptor recognition of human TRAF2. Nature 398: 533–538. [DOI] [PubMed] [Google Scholar]
- Polekhina, G., House, C.M., Traficante, N., Mackay, J.P., Relaix, F., Sassoon, D.A., Parker, M.W., Bowtell, D.D.(2002). Siah ubiquitin ligase is structurally related to TRAF and modulates TNF-alpha signaling. Nat. Struct. Biol. 9: 68–75. [DOI] [PubMed] [Google Scholar]
- Qi, H., Li, J., Xia, F.N., Chen, J.Y., Lei, X., Han, M.Q., Xie, L.J., Zhou, Q.M., Xiao, S.(2020a). Arabidopsis SINAT proteins control autophagy by mediating ubiquitylation and degradation of ATG13. Plant Cell 32: 263–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, H., Xia, F.N., Xiao, S.(2020b). Autophagy in plants: Physiological roles and post-translational regulation. J. Integr. Plant Biol. (Published ahead of print). [DOI] [PubMed] [Google Scholar]
- Qi, H., Xia, F.N., Xie, L.J., Yu, L.J., Chen, Q.F., Zhuang, X.H., Wang, Q., Li, F., Jiang, L., Xie, Q., Xiao, S.(2017). TRAF-family proteins regulate autophagy dynamics by modulating AUTOPHAGY PROTEIN6 stability in Arabidopsis. Plant Cell 29: 890–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiborg, C., Stenmark, H.(2009). The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature 458: 445–452. [DOI] [PubMed] [Google Scholar]
- Reumann, S.(2004). Specification of the peroxisome targeting signals type 1 and type 2 of plant peroxisomes by bioinformatics analyses. Plant Physiol. 135: 783–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, J., Zhao, Q., Wang, X., Gao, C., Zhu, Y., Zeng, Y., Jiang, L.(2018). A plant Bro1 domain protein BRAF regulates multivesicular body biogenesis and membrane protein homeostasis. Nat. Commun. 9: 3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle, J., Kurepa, J., Yang, P., Emborg, T.J., Babiychuk, E., Kushnir, S., Vierstra, R.D.(2003). The pleiotropic role of the 26S proteasome subunit RPN10 in Arabidopsis growth and development supports a substrate-specific function in abscisic acid signaling. Plant Cell 15: 965–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spallek, T., Beck, M., Ben Khaled, S., Salomon, S., Bourdais, G., Schellmann, S., Robatzek, S.(2013). ESCRT-I mediates FLS2 endosomal sorting and plant immunity. PLoS Genet. 9: e1004035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer, C., Li, F., Buono, R., Roschzttardtz, H., Chung, T., Zhang, M., Osteryoung, K.W., Vierstra, R.D., Otegui, M.S.(2015). The endosomal protein CHARGED MULTIVESICULAR BODY PROTEIN1 regulates the autophagic turnover of plastids in Arabidopsis. Plant Cell 27: 391–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer, C., Reyes, F.C., Buono, R., Sliwinski, M.K., Haas, T.J., Otegui, M.S.(2009). The ESCRT-related CHMP1A and B proteins mediate multivesicular body sorting of auxin carriers in Arabidopsis and are required for plant development. Plant Cell 21: 749–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer, C., Schellmann, S., Sabovljevic, A., Shahriari, M., Keshavaiah, C., Bechtold, N., Herzog, M., Müller, S., Hanisch, F.G., Hülskamp, M.(2006). The Arabidopsis elch mutant reveals functions of an ESCRT component in cytokinesis. Development 133: 4679–4689. [DOI] [PubMed] [Google Scholar]
- Sun, S., Sun, L., Zhou, X., Wu, C., Wang, R., Lin, S.H., Kuang, J.(2016). Phosphorylation-dependent activation of the ESCRT function of ALIX in cytokinetic abscission and retroviral budding. Dev. Cell 36: 331–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunnerhagen, M., Pursglove, S., Fladvad, M.(2002). The new MATH: Homology suggests shared binding surfaces in meprin tetramers and TRAF trimers. FEBS Lett. 530: 1–3. [DOI] [PubMed] [Google Scholar]
- Suttangkakul, A., Li, F., Chung, T., Vierstra, R.D.(2011). The ATG1/ATG13 protein kinase complex is both a regulator and a target of autophagic recycling in Arabidopsis. Plant Cell 23: 3761–3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, M., Xie, Q.(2013). Non-26S proteasome proteolytic role of ubiquitin in plant endocytosis and endosomal trafficking(F). J. Integr. Plant Biol. 55: 54–63. [DOI] [PubMed] [Google Scholar]
- Vierstra, R.D.(2009). The ubiquitin-26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 10: 385–397. [DOI] [PubMed] [Google Scholar]
- Xia, Z., Huo, Y., Wei, Y., Chen, Q., Xu, Z., Zhang, W.(2016). The Arabidopsis LYST INTERACTING PROTEIN 5 acts in regulating abscisic acid signaling and drought response. Front. Plant Sci. 7: 758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Q., Guo, H.S., Dallman, G., Fang, S., Weissman, A.M., Chua, N.H.(2002). SINAT5 promotes ubiquitin-related degradation of NAC1 to attenuate auxin signals. Nature 419: 167–170. [DOI] [PubMed] [Google Scholar]
- Yang, M., Li, C., Cai, Z., Hu, Y., Nolan, T., Yu, F., Yin, Y., Xie, Q., Tang, G., Wang, X.(2017). SINAT E3 ligases control the light-mediated stability of the brassinosteroid-activated transcription factor BES1 in Arabidopsis. Dev. Cell 41: 47–58.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo, S.D., Cho, Y.H., Sheen, J.(2007). Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- Yoshimoto, K., Hanaoka, H., Sato, S., Kato, T., Tabata, S., Noda, T., Ohsumi, Y.(2004). Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 16: 2967–2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, F., Lou, L., Tian, M., Li, Q., Ding, Y., Cao, X., Wu, Y., Belda-Palazon, B., Rodriguez, P.L., Yang, S., Xie, Q.(2016). ESCRT-I component VPS23A affects ABA signaling by recognizing ABA receptors for endosomal degradation. Mol. Plant 9: 1570–1582. [DOI] [PubMed] [Google Scholar]
- Zhang, C., Hao, Z., Ning, Y., Wang, G.L.(2019). SINA E3 ubiquitin ligases: Versatile moderators of plant growth and stress response. Mol. Plant 12: 610–612. [DOI] [PubMed] [Google Scholar]
- Zhou, J., Wang, J., Cheng, Y., Chi, Y.J., Fan, B., Yu, J.Q., Chen, Z.(2013). NBR1-mediated selective autophagy targets insoluble ubiquitinated protein aggregates in plant stress responses. PLoS Genet. 9: e1003196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J., Zhang, Y., Qi, J., Chi, Y., Fan, B., Yu, J.Q., Chen, Z.(2014). E3 ubiquitin ligase CHIP and NBR1-mediated selective autophagy protect additively against proteotoxicity in plant stress responses. PLoS Genet. 10: e1004116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang, X., Wang, H., Lam, S.K., Gao, C., Wang, X., Cai, Y., Jiang, L.(2013). A BAR-domain protein SH3P2, which binds to phosphatidylinositol 3-phosphate and ATG8, regulates autophagosome formation in Arabidopsis. Plant Cell 25: 4596–4615. [DOI] [PMC free article] [PubMed] [Google Scholar]