The induction of polyploid divisions in the tomato gbp1 mutant reveals the role of the guanylate-binding protein in maintaining fruit tissue differentiation.
Abstract
Cell fate maintenance is an integral part of plant cell differentiation and the production of functional cells, tissues, and organs. Fleshy fruit development is characterized by the accumulation of water and solutes in the enlarging cells of parenchymatous tissues. In tomato (Solanum lycopersicum), this process is associated with endoreduplication in mesocarp cells. The mechanisms that preserve this developmental program, once initiated, remain unknown. We show here that analysis of a previously identified tomato ethyl methanesulfonate-induced mutant that exhibits abnormal mesocarp cell differentiation could help elucidate determinants of fruit cell fate maintenance. We identified and validated the causal locus through mapping-by-sequencing and gene editing, respectively, and performed metabolic, cellular, and transcriptomic analyses of the mutant phenotype. The data indicate that disruption of the SlGBP1 gene, encoding GUANYLATE BINDING PROTEIN1, induces early termination of endoreduplication followed by late divisions of polyploid mesocarp cells, which consequently acquire the characteristics of young proliferative cells. This study reveals a crucial role of plant GBPs in the control of cell cycle genes, and thus, in cell fate maintenance. We propose that SlGBP1 acts as an inhibitor of cell division, a function conserved with the human hGBP-1 protein.
INTRODUCTION
The growth dynamics of fruit tissues typically involve asynchronous evolution of cells into different developmental states: some still dividing and others differentiating and expanding, despite being in the same cell layers. In most fruit tissues, growth starts immediately after fertilization with frequent cell divisions (early growth) and continues through a cell enlargement phase (late growth) that lasts until the fruit ripening phase (Gillaspy et al., 1993; Renaudin et al., 2017). In most fleshy fruits, cells in the mesocarp tissues undergo massive enlargement driven by vacuolar expansion, which provides the major force that drives cell growth. In many fruit species, such as tomato (Solanum lycopersicum), this vacuolar expansion is coupled with endoreduplication, which allows cytoplasmic volume to increase rapidly according to the nuclear DNA content, a concept consistent with the karyoplasmic ratio theory (Chevalier et al., 2014). Endoreduplication results from a shift between the mitotic cell cycle and the endocycle, which consists of repeated DNA replication rounds alternating between the G and S phases and a bypass of mitosis and cytokinesis to produce a single endopolyploid nucleus (for review, see Orr-Weaver, 2015). Even if a positive correlation exists between cell ploidy and cell expansion, the exact role of endoreduplication in cell growth is still unclear. Recently, Bhosale et al. (2019) proposed that endopolyploidy controls the transcription of cell wall-modifying genes to satisfy the high demand for new cell wall material during extremely rapid cell expansion. Endoreduplication has also been proposed to enhance metabolic activity in cells undergoing differentiation (Lee et al., 2009; Chevalier et al., 2011). In tomato fruit, endocycling marks the onset of cell differentiation of mesocarp cells into an expanding cell type, and this has an obvious impact on transcriptional activities (Lemaire-Chamley et al., 2005; Pirrello et al., 2018).
The progression through the mitotic cycle and endocycle requires the activity of cyclin-dependent kinase (CDK)–cyclin complexes (CYCs; Inzé and de Veylder, 2006). High CDK/CYC activity is essential for the initiation of mitosis, while low CDK/CYC activity drives endoreduplication (de Veylder et al., 2007). Several mechanisms account for the control of CDK/CYC activity, including control at the transcriptional level by regulating the availability of components of the CDK/CYC complexes and at the post-translational level via the binding of CDK-specific inhibitors or the specific degradation of the cyclin regulatory subunit (Chevalier et al., 2014). This latter mechanism involves CCS52 proteins, which are known as activators of the anaphase-promoting complex/cyclosome that mediates degradation of mitotic cyclins and consequently triggers endoreduplication (Baloban et al., 2013). For instance, the downregulation of SlCCS52A in tomato resulted in smaller fruits with lower nuclear ploidy level and reduced cell size (Mathieu-Rivet et al., 2010). Multiple functional gene studies in tomato revealed that cell cycle regulators act early during fruit growth, thus affecting the onset of cell differentiation in the mesocarp by governing the timing of the transition from the mitotic cycle to the endocycle (for a review, Chevalier et al., 2014). Key determinants of fruit growth and morphology identified to date based on the natural diversity of tomato are also related to early events that affect meristem architecture and cell division patterning within the ovary before anthesis or in the developing fruit early after anthesis (for review, see van der Knaap et al., 2014; van der Knaap and Østergaard, 2018). However, the subsequent events that control the maintenance or arrest of differentiation programs in mesocarp cells in tomato remain unknown. CELL SIZE REGULATOR is thought to act after the cell division phase by functioning in cell differentiation of the vasculature during the later stages of fruit development, with an indirect effect on ploidy and cell size (Mu et al., 2017).
In a previous study of tomato ethyl methanesulfonate (EMS) mutants with abnormal fruit size and pericarp thickness (Musseau et al., 2017), we identified several mutants in which the number of cell layers in the fruit was not altered, indicating that late events but not early cell division and fruit growth were affected in these mutants. This study also revealed the existence of an isotropic cell expansion module that appears to affect the growth of all fruit tissues and an anisotropic cell expansion module that only affects pericarp growth. One of the mutants presenting anisotropic cell expansion, named p3d3, displayed a striking reduction in fruit pericarp thickness. Detailed observations revealed an extreme disorganization of the pericarp tissue that was accompanied by abnormal cell size and shape in this mutant. Here, we identified the mutation underlying the p3d3 mutant phenotype and found that it is associated with a poorly studied guanylate binding protein (GBP) gene. We confirmed by clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 gene editing that GBP disruption is indeed responsible for the observed cellular alterations in this mutant. Further characterization of the mutant further revealed that the polyploid cells in the mesocarp, which are normally committed to an enlargement process, instead enter into a cell division mode and acquire or maintain characteristics of young proliferative cells. These results highlight the crucial role of a plant GBP in the commitment to undergo cell differentiation and endocycling.
RESULTS
Mapping-by-Sequencing Reveals that a Mutation in the GBP Gene SlGBP1 Is Responsible for the Tomato p3d3 Phenotype
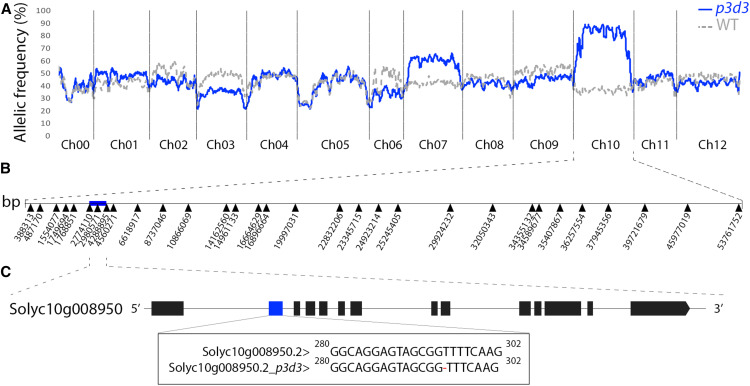
The p3d3 mutant, initially identified from our EMS mutant collection based on its thin pericarp phenotype (Musseau et al., 2017), exhibits cellular disorganization in the pericarp (Figure 1). Segregation analysis of pericarp thickness in the BC1F2 population indicated the presence of a single recessive mutation. To identify the mutation responsible for this phenotype, we used a mapping-by-sequencing strategy that was previously developed to discover causal mutations in tomato EMS mutants (Garcia et al., 2016). After whole genome sequencing of F2 mutant- and wild-type–like bulks, an analysis of the allelic frequency distribution of EMS mutations throughout the tomato genome revealed a major association of the p3d3 phenotype with chromosome 10 (Figure 2A). A subsequent recombination analysis allowed us to reduce the candidate region to 1.51 Mb, encompassing 131 genes, between the single nucleotide polymorphisms at positions 2,774,110 and 4,288,895 (Figure 2B). Only one single nucleotide polymorphism was detected at position 2,980,571 in this region, and this was located in the coding region of the Solyc10g008950 gene, encoding a large GTPase GBP, hereafter named SlGBP1 (Figure 2C). The mutation corresponded to a deletion of a thymine in the second exon of the gene causing a frameshift in the open reading frame and a premature termination codon at position 120 (Figure 2C; Supplemental Figure 1A).
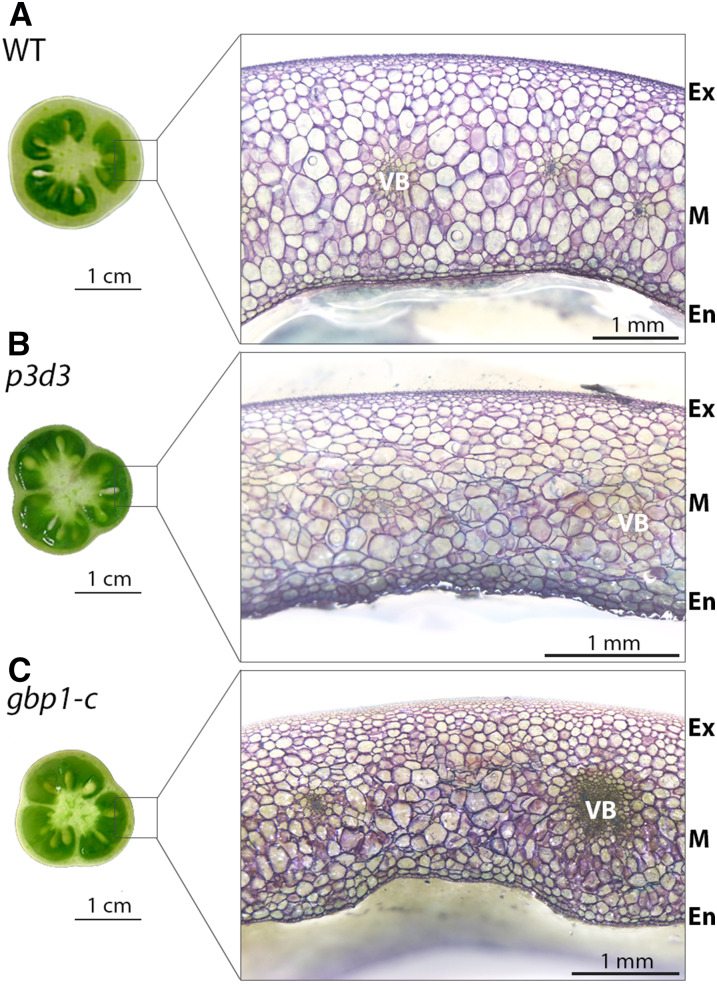
Figure 1.
Pericarp Phenotypes of the gbp1-c Mutants.
(A) to (C) Equatorial and transverse sections of fruit pericarp at the breaker stage in the wild type (A), p3d3 EMS mutant (B), and CRISPR gbp1-c mutant (C).
Pericarp sections were stained with toluidine blue. En, endocarp; Ex, exocarp; M, mesocarp; WT, wild type.
Figure 2.
Identification of the p3d3 Causal Mutation through Mapping-by-Sequencing.
(A) Distribution of the allelic frequencies of EMS mutations in the wild-type (WT)–like and mutant-like bulks are represented along the tomato genome in sliding windows of 30 consecutive mutations. In the case of a recessive mutation, allelic frequency of the causal mutation/region should be 1 and 0.33 in the mutant-like and wild-type–like bulks, respectively. Ch, chromosome.
(B) The mutations used as markers for recombination analysis are indicated along Ch10. The blue box represents the causal 1.51-Mb region encompassing 131 genes.
(C) Solyc10g008950 is the only gene containing an EMS-induced mutation in the causal region. The mutation corresponds to a T deletion in exon 2 (blue box) of Solyc10g008950. Exons are represented by boxes.
Disruption of the SlGBP1 Locus Using CRISPR-Cas9 Gene Editing Phenocopies the p3d3 Phenotype
Because the genetic background of EMS mutants is complex, we generated an allelic series of CRISPR mutants (here referred to as gbp1-c) in the Micro-Tom background to validate the causal mutation in p3d3. The first exon of the SlGBP1 gene was targeted by Cas9, causing premature termination of the protein between positions 117 and 120 (Supplemental Figure 1A). Quantitative expression analysis revealed that SlGBP1 mRNA abundance was highly reduced in both the p3d3 and CRISPR gbp1-c mutants (Supplemental Figures 1B and 1C). This reduction was consistent with the action of the Nonsense-Mediated Decay pathway, which eliminates mRNA transcripts containing premature termination codons (He and Jacobson, 2015). As expected for a recessive mutation, the heterozygous CRISPR mutants displayed a wild-type phenotype while homozygous gene edited mutants phenocopied the EMS mutant cellular phenotype, thus confirming the identity of the causal mutations in the SlGBP1 locus (Figure 1C).
SlGBP1 belongs to the GBP family, comprising large GTPases, whose function was first described in humans in response to interferon (Prakash et al., 2000). In tomato, a second gene closely homologous to SlGBP1 is located on chromosome 2 (Solyc02g077600, hereafter named SlGBP2), and these genes encode proteins with >80% sequence identity. Both tomato genes are expressed in vegetative and reproductive tomato organs, including in fruit tissues, with major expression in mature seeds (Supplemental Figures 2 and 3). SlGBP1 is expressed at higher levels than SlGBP2, and more than 2-fold in the fruit (Supplemental Figures 2 and 3). Interestingly, SlGBP2 was expressed at higher levels in the gpb1-c lines at the breaker stage than during other stages (Supplemental Figures 2 and 3).
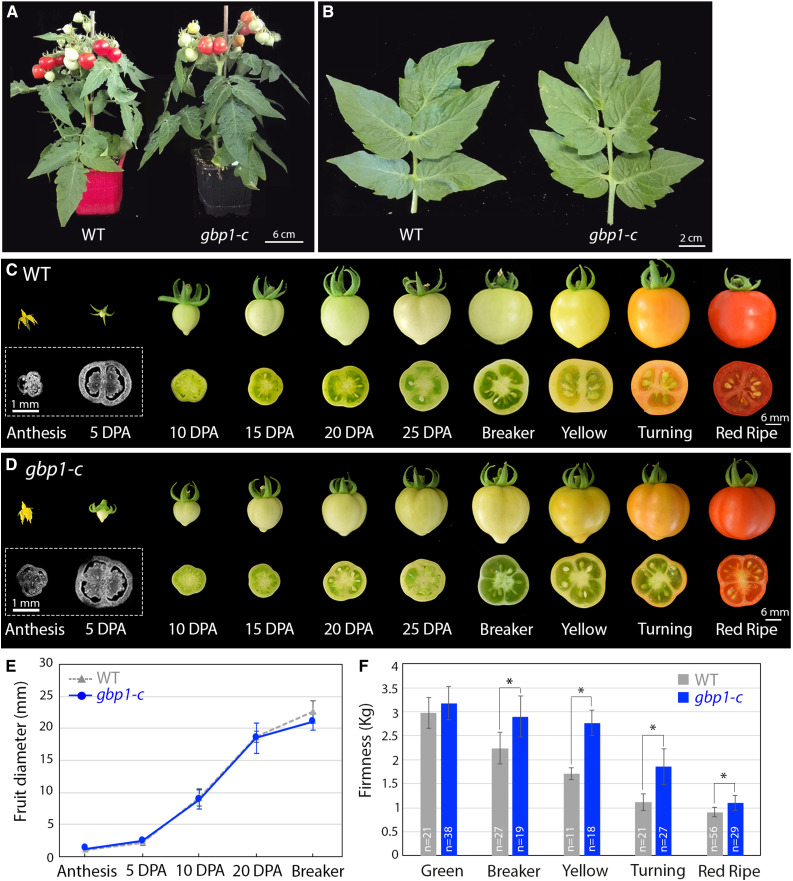
Alterations in Pericarp Cell Size Appear during Late Stages of Fruit Expansion in the gbp1 -c Mutant
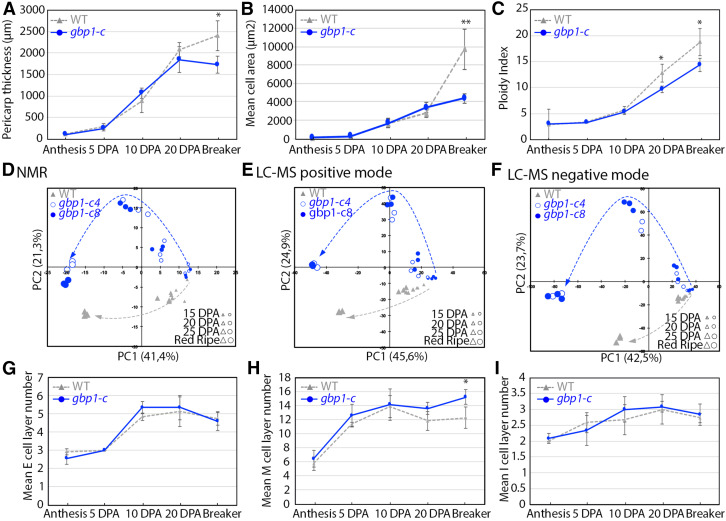
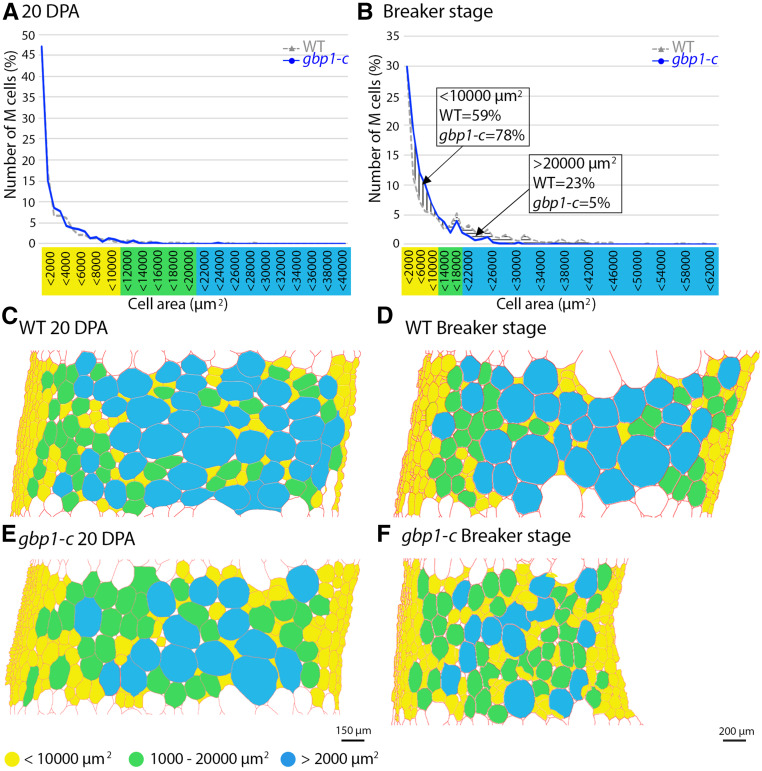
The gpb1-c mutant was comparable to the wild type in terms of vegetative growth, and the only phenotypic differences that were noted were related to fruit development, including seed number (Figures 3A and 3B; Musseau et al., 2017). We examined the fruit morphology and pericarp histology from anthesis to the breaker stage (the onset of ripening) to determine the time course of the appearance of abnormalities. All subsequent analyses were performed using the Micro-Tom CRISPR mutants to exclude any effect on fruit phenotype derived from other EMS mutations present in the p3d3 mutant. No significant differences in fruit size were observed between the mutant and the wild type (Figures 3C to 3E), while differences in pericarp thickness and cell area were observed beginning at 20 days post anthesis (DPA; Figures 4A and 4B). In wild-type pericarp, the mean cell size increased by >3.5-fold (×3.53) between 20 DPA and the breaker stage, while only a slight increase (×1.28) occurred in the mutant (Figure 4B). Analysis of the distribution of mesocarp cell area suggested that this difference was due to a higher proportion of small cells (+19% more cells <10,000 μm2) and a reduction in the number of large cells (−18% fewer cells >20,000 µm2) in the mutant compared with the wild type at the breaker stage, while no difference was apparent at 20 DPA (Figures 5A and 5B). During fruit growth, a typical gradient of cell size is established across the pericarp, spanning large round cells in the inner cell layers and smaller cells in the outer cell layers (Figures 1A and 5C to 5E; Renaudin et al., 2017). This gradient of cell size was substantially different after 20 DPA in the gbp1-c mutants compared with wild type, comprising a highly heterogeneous cell size and shape distribution resulting in a partial loss of tissue structure (Figures 1B, 1C, and 5F). The presence of a higher proportion of small cells in the inner mesocarp at the breaker stage suggests that the increase in cell expansion rate that normally takes place after 20 DPA did not occur in the mutant.
Figure 3.
Plant and Fruit Development in the Wild Type and gbp1-c.
(A) Plant phenotypes of the wild type (WT) and gbp1-c.
(B) Leaf phenotypes of the wild type and gbp1-c.
(C) and (D) Whole fruits and half transverse sections of the wild type (WT) and gbp1-c during fruit development. For anthesis and 5DPA, equatorial sections were stained with Calcofluor white.
(E) Fruit diameter during the growth period. Values represent means ± sd (n = 9 to 16 fruits). Mutant samples include gbp1c-8 and gbp1-c4 fruits.
(F) Fruit firmness along development in wild type and mutant. Values represent means ± sd (n = 11 to 56 fruits). Mutant samples include gbp1c-8, gbp1-c10, and gbp1-c4.
In (E) and (F), significant differences between the mutant and the wild type: Wilcoxon test, P-value < 0.05 with FC >1.2 and > 2 are indicated by * and **, respectively.
Figure 4.
Cellular Parameters and Composition during Pericarp Development.
(A) Pericarp thickness during pericarp development (n = 4 to 6 fruits).
(B) Mean cell area during pericarp development (n = 4 to 5 fruits).
(C) Ploidy index corresponding to the mean C-level of a pericarp cell (Bertin et al., 2009) during pericarp development (n = 5 to 8 fruits).
(D) to (F) Principal component analyses of metabolomics data in two independent gbp1-c lines and wild-type (WT) fruit. 1H-NMR data includes 329 spectra regions. LC-MS under positive ionization includes 1,523 metabolite features. LC-MS under negative ionization includes 3,915 metabolite features. Changes in the metabolic patterns along fruit development are represented by an arrow for each genotype.
(G) to (I) Exocarp (E), mesocarp (M), and endocarp (I) cell layer number during pericarp development (n = 3 to 6 fruits).
In (A) to (C) and (G) to (I), values represent means ± sd. Mutant samples include gbp1c-8 and gbp1-c4 fruits. Significant differences between the mutant and the wild type: Wilcoxon test, P-value < 0.05 with FC >1.2 and > 2 are indicated by * and **, respectively.
Figure 5.
Mesocarp Cell Area Distribution at 20 DPA and the Breaker Stage.
(A) and (B) Proportion (%) of mesocarp (M) cells in successive cell area categories in μm2. Values represent means of four wild-type and mutant (gbp1c-8 and gbp1-c4) fruits.
(C) to (F) Schematic representation of the spatial distribution of M cells in mutant and wild-type (WT) sections. Segmented cells walls obtained using the program CellSeT (Pound et al., 2012) are represented in red. A color code is given according to cell area.
Changes in Nuclear Ploidy Precede Decelerated Cell Expansion in the gbp1 -c Mutant
Because endoreduplication generally correlates with cell size variations in the tomato pericarp (Chevalier et al., 2011; Musseau et al., 2017; Renaudin et al., 2017), we investigated the nuclear ploidy levels of pericarp cells in the gbp1-c mutant. The ploidy index at 20 DPA onwards was much lower in the mutant compared with wild type (Figure 4C). This difference was due to a marked increase in 4C nuclei, corresponding to the first endocycle and dividing cells at the G2 phase of the cell cycle (Supplemental Figure 4B), and to a 13% and 7% reduction in polyploid nuclei (8C to 128C) at 20 DPA and the breaker stage, respectively (Supplemental Figure 4C). In particular, 64C and 128C DNA ploidy levels were much lower in the mutant than in the wild type (Supplemental Figures 4G and 4H). As nuclear populations with 16C, 32C, 64C, or 128C were detected at a similar developmental stage in the mutant and wild type (Supplemental Figure 4), the timing of endoreduplication was unaffected but the process appeared to be less efficient in the gbp1-c mutant. In the mutant, a high proportion of cells did not proceed to the highest DNA ploidy levels; instead, they stopped endocycling while still at a low ploidy state, including the first tetraploid (4C) state.
A Metabolic Shift Occurs in the gbp1 -c Pericarp during the Late Expansion Stage
We further investigated the development of mesocarp cells, which in the wild type are normally large and accumulate water, solutes, and minerals (Mounet et al., 2007), by examining the composition of primary and specialized metabolites in the wild-type and gbp1-c fruit pericarp using proton nuclear magnetic resonance (1H-NMR) and liquid chromatography-mass spectrometry (LC-MS) profiling, respectively. Principal component analysis (PCA) revealed that the metabolite composition of the pericarp followed divergent trajectories during fruit development in the gbp1-c mutant and wild type (Figures 4D to 4F). Regardless of the analytical method used (PCA or univariate analyses in the form of Volcano plots), most of the shift in metabolite composition in fruit occurred at 25 DPA, when most of the significant differences in metabolite signatures were evident between the mutant and wild-type pericarp (Figures 4D to 4F; Supplemental Figure 5). Glc, which normally accumulates during ripening, stayed at a low level throughout fruit development in gbp1-c fruit, and many amino acids, including glutamine, accumulated at lower levels in the mutant (Table 1). Conversely, the levels of several major green fruit-associated metabolites or classes of metabolites that normally exhibit substantial reductions during ripening remained high in gbp1-c (Table 1). These included Suc, which is normally hydrolyzed into Glc and Fru, and γ-amino butyric acid (GABA), which usually accumulates until the onset of ripening before being catabolized (Akihiro et al., 2008). Other examples included glycoalkaloids, which in the Solanaceae family are antinutritional compounds that deter animals from consuming unripe fruit (Itkin et al., 2013) and hydroxycinnamic acid derivatives, which confer a bitter or astringent taste (Verdu et al., 2014) and accumulate in unripe fruit. Higher levels of lactic acid and ethanol were detected in gbp1-c fruit tissues than the wild type (Table 1), which may reflect the onset of anaerobic metabolism. Taken together, these results all mark the arrest of many processes executed during the normal developmental program of polyploid mesocarp cells after 25 DPA, resulting in a ripe gbp1-c fruit that is analogous in several regards to an unripe growing fruit.
Table 1. Metabolic Profiles of the gbp1 Mutant.
| Biochemical Family | Metabolite | Fruit Developmental Stage | Analytical Method | MSI Levela | NMR or MS Signalsb | |||
|---|---|---|---|---|---|---|---|---|
| 15 DPA | 20 DPA | 25 DPA | Red Ripe | |||||
| Amino Acids | Alanine | — | — | WT* | — | NMR | MSI1 | B1.4762; B1.4907 |
| Asparagine | — | — | WT** | — | NMR | MSI1 | B2.7978; B2.8135; B2.8461; B2.8525; B2.9398 | |
| Phenylalanine | — | — | WT** | — | NMR | MSI1 | B7.3342; B7.3395 | |
| MS(pos) | MSI1 | X41 | ||||||
| Valine | — | WT** | WT** | - | NMR | MSI1 | B0.9991; B1.0115; B1.0503 | |
| Glutamine | — | WT** | WT** | WT* | NMR | MSI1 | B2.4369;B2.4451;B2.4515;B2.4569;B2.4623;B2.4689;B2.4778 | |
| Aspartic acid | — | WT* | WT* | gbp1* | NMR | MSI1 | B2.6126;B2.6484;B2.6642;B2.7853;B2.7931;B2.8214;B2.8274 | |
| Isoleucine | — | WT* | WT* | gbp1* | NMR | MSI1 | B1.0175;B1.0304 | |
| Glutamic acid | — | gbp1* | gbp1* | - | NMR | MSI1 | B2.0372;B2.0520 | |
| GABA | — | gbp1* | gbp1** | gbp1** | NMR | MSI1 | B1.8691;B1.8879;B1.9021;B1.9168;B1.9311;B2.2887;B2.3051;B2.3202;B3.0264 | |
| Tyrosine | — | — | gbp1** | gbp1** | NMR | MSI1 | B6.8424;B6.8598;B7.1731;B7.1915 | |
| Organic Acids | Fumaric acid | — | — | gbp1** | gbp1** | NMR | MSI1 | B6.5291 |
| Lactic acid | — | — | — | gbp1** | NMR | MSI3 | B1.3608 | |
| Isobutyric acid | — | — | — | gbp1* | NMR | MSI3 | B1.0840; B1.0964 | |
| Pantothenic acid hexose | — | — | — | gbp1* | MS(neg) | MSI3 | X852 | |
| Alcohols | Ethanol | — | — | — | gbp1** | NMR | MSI1 | B1.1753 |
| Sugars | Glc | — | WT* | WT* | WT* | NMR | MSI1 | B3.1862; B3.2052; B3.6791; B3.7049; B3.7252; B3.7354; B3.8619;B4.5769; B4.5985; B5.1805; B5.1996 |
| Fru | — | — | gbp1* | WT* | NMR | MSI1 | B3.7692; B3.9612 | |
| Gal | — | — | gbp1* | WT* | NMR | MSI1 | B4.5262; B5.2358 | |
| Suc | — | — | — | gbp1** | NMR | MSI1 | B4.1662; B4.1847; B5.3988; B5.4131 | |
| Cyclitols and derivatives | Inositol | — | — | gbp1* | — | NMR | MSI1 | B3.2454; B3.2621 |
| Alkaloids | Trigonelline | — | — | — | gbp1* | NMR | MSI1 | B4.4496; B8.8670; B9.1435 |
| Glycoalkaloids | Deglucosylated lycoperoside F/deglucosylated esculeoside A | — | — | — | gbp1** | MS(pos) | MSI3 | X1667 |
| Lycoperoside C | — | — | gbp1** | MS(pos) | MSI3 | X1661 | ||
| Lycoperoside F-G/esculeoside A isomer 2 | — | — | — | gbp1* | MS(pos) | MSI3 | X1692 | |
| Alpha-Tomatine | — | — | — | gbp1* | MS(pos) | MSI3 | X1638 | |
| Dehydrotomatine | — | — | — | gbp1** | MS(pos) | MSI3 | X1634 | |
| Hydroxycinnamic acid derivatives | Chlorogenic acid (3CQA) | — | gbp1** | gbp1** | gbp1** | NMR | MSI1 | B1.9856;B2.0063;B4.1933;B4.1990;B4.2059;B4.2125;B6.3635;B6.3963;B6.8889;B7.0665;B7.0737;B7.084;B7.0912;B7.1545;B7.1620;B7.6106 |
| — | — | gbp1** | — | MS(pos) | MSI2 | X492 | ||
| — | — | — | gbp1** | MS(neg) | MSI2 | X661 | ||
| Cryptochlorogenic acid (4CQA) | — | — | — | gbp1* | MS(neg) | MSI2 | X663 | |
| Dicaffeoylquinic acid isomer 2 | — | — | — | gbp1** | MS(pos) | MSI3 | X982 | |
| MS(neg) | MSI3 | X1893 | ||||||
| Dicaffeoylquinic acid isomer 1 | — | — | — | gbp1** | MS(neg) | MSI3 | X1895 | |
| Tricaffeoylquinic acid | — | — | — | gbp1** | MS(neg) | MSI3 | X2798 | |
| — | — | — | MS(pos) | MSI3 | X1397 | |||
| Feruloylputrescine isomer 2 | — | — | — | gbp1** | MS(pos) | MSI3 | X214 | |
| Feruloylquinic acid | — | — | — | gbp1** | MS(neg) | MSI3 | X771 | |
| gbp1* | MS(pos) | MSI3 | X534 | |||||
| Feruloyltyramine | — | — | — | gbp1** | MS(neg) | MSI3 | X424 | |
| gbp1** | MS(pos) | MSI2 | X367 | |||||
| Coumaroylquinic acid isomer 1 | — | — | — | gbp1** | MS(neg) | MSI3 | X549 | |
| gbp1** | MS(pos) | MSI3 | X443 | |||||
| Coumaroylquinic acid isomer 2 | — | — | — | gbp1** | MS(neg) | MSI3 | X548 | |
| Flavonoids | Rutin | — | WT* | WT** | gbp1* | NMR | MSI3 | B1.3288;B1.3416,B6.5544;B6.5599;B6.9886;B7.0071;B7.6804;B7.6869 |
| Xanthosine | — | — | — | gbp1** | MS(neg) | MSI2 | X291 | |
| Inosine | — | — | — | gbp1** | MS(neg) | MSI3 | X225 | |
| Uridine monophosphate | — | — | — | WT* | MS(neg) | MSI2 | X472 | |
Annotated metabolites with significant differences (Wilcoxon test, FDR-corrected P-value < 0.05) in levels between the mutant and the wild type (WT) at one or several stages of fruit development are shown. The genotype (wild type or gbp1-c) with higher content in the pericarp is indicated (FC > 1.2 and > 2 are indicated by * and **, respectively). Only metabolites that are consistent between the two CRISPR lines are shown. Corresponding analytical methods and signals of detection are shown. Dashes indicate no significant difference.
Groups of the Metabolomics Standards Initiative (MSI) level according to Sumner et al. (2007). For NMR MSI1, 1D and 2D 1H and/or 13C confirmation + internal NMR database of reference compounds acquired in the same conditions of solvent and pH or spiking have been performed; for NMR MSI3, as only 1D 1H detection + external NMR database of reference compounds or literature data have been performed, annotations are putative.
NMR signals correspond to integrated spectral region or bucket centered at a chemical shift expressed in ppm (B1.4762, where “B” indicates Bucket and 1.4762 is the center of the bucket in ppm); MS signals correspond to software MS-DIAL variable IDs.
Changes in Cell Morphology in the gbp1 -c Mutant Are Associated with Modifications in Cell Wall Pectins
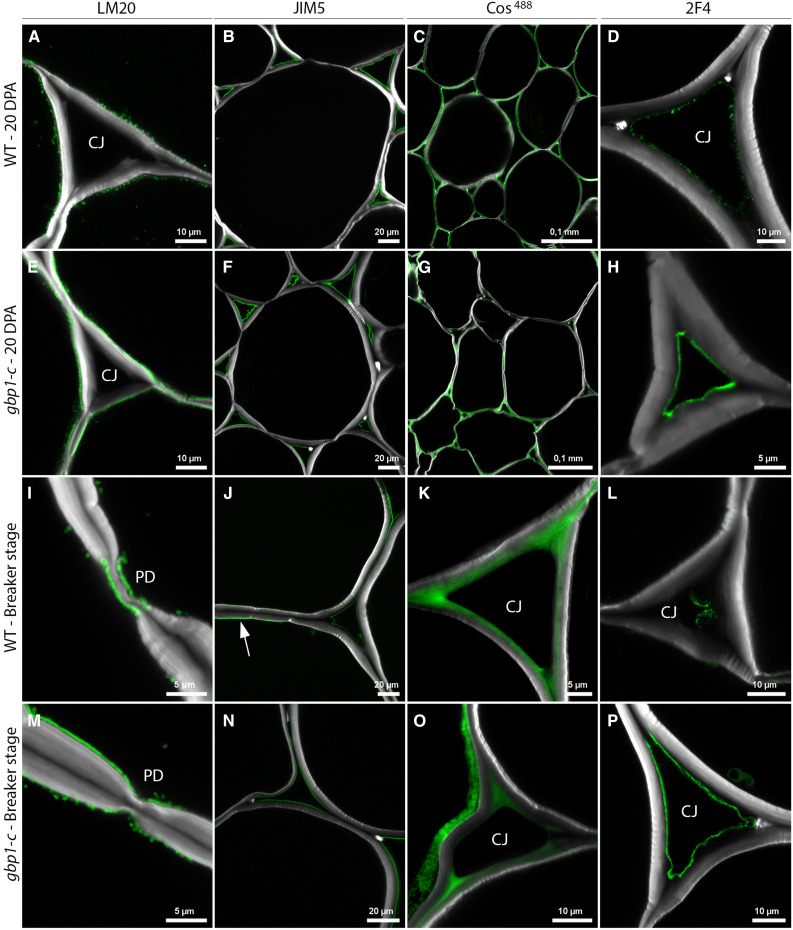
Plant cell walls play crucial roles in determining cell size and shape during growth. The pectin component homogalacturonan (HG) is secreted into the wall in a highly methyl-esterified form and is subsequently demethylesterified during cell growth, where it contributes to the balance between wall rigidity and flexibility (Willats et al., 2001; Daher and Braybrook, 2015). Given their importance as major matrix components of the middle lamella and primary walls of dicotyledonous plants, including fleshy fruit (Prasanna et al., 2007), we examined the distribution and composition of pectins at 10, 15, and 20 DPA, and the breaker stage using antibodies and probes that distinguish between different degrees of esterification of HG. Of these four stages, differences in the distribution patterns of esterified and non-esterified pectins between the mutant and the wild type were observed only at 20 DPA and the breaker stage (Figure 6). Cells from gbp1-c showed stronger immunolabeling in the primary wall adjacent to the plasma membrane than did wild-type cells using the LM20 monoclonal antibody, which recognizes methylesterified pectins with a higher degree of esterification. In wild-type cells, the fluorescent signal had a punctate pattern at 20 DPA and remained only around the plasmodesmata at the breaker stage, whereas it was more contiguous in the mutant at 20 DPA and showed a punctate distribution at the breaker stage (Figures 6A, 6E, 6I, and 6M), suggesting delayed demethylesterification in gbp1-c at the onset of ripening. Conversely, JIM5, a monoclonal antibody that recognizes HG with a low degree of esterification (Knox et al., 1990), showed strong labeling of the wall close to the plasma membrane in wild-type fruit at the breaker stage (Figure 6J), but no such labeling in mutant fruit at the breaker stage (Figure 6N) or in younger fruit of either genotype (Figures 6B and 6F). This again indicated delayed pectin demethylesterification in gbp1-c. Notably, there were no differences between the wild type and gbp1-c in JIM5 labeling of pectins in the middle lamella and in the corners of cell junctions (Figures 6B, 6F, 6J, and 6N), which are regions thought to be important for cell–cell adhesion (Wang et al., 2018).
Figure 6.
Cell Wall Alterations in the gbp1-c Mutant.
(A) to (P) Indirect immunofluorescence microscopy of the wild type (WT) and gbp1-c at 20 DPA or at the breaker stage.
Indirect immunofluorescence microscopy was performed using antibodies and probes that indicate different degrees of esterification of the HG component of pectin, from highly esterified (LM20 antibody) to partially esterified (JIM5 antibody, COS488), including no-esterified forms that are able to bind to cations such as calcium (Ca2+) and form gels in the middle lamella (2F4 antibody). Green represents specific antibody or probe signal. Arrow indicates JIM5 signal close to the plasma membrane. The white signal shows Calcofluor white staining of cell walls (β-linked glucans: callose and cellulose). CJ, cell junctions; PD, plasmodesmata.
However, differences in the middle lamella and cellular junctions between gbp1-c and wild type were revealed by staining with the fluorescent dye COS488, which recognizes HG epitopes with a seven-residue stretch of unesterified HG (Mravec et al., 2014), as well as labeling with the monoclonal antibody 2F4, which recognizes demethylesterified HG associated through calcium ions (Figures 6C, 6D, 6G, and 6H; Liners et al., 1989). A homogeneous signal was observed with COS488 at 20 DPA and the breaker stage in the intercellular junctions and middle lamella regions of the wild-type mesocarp, but the signal was patchy in the gbp1-c mutant, which is consistent with less de-methylesterification. By contrast, we observed consistently high labeling with 2F4 in the mutant throughout development, whereas there was minimal labeling in the wild type. A decrease in residues with calcium binding sites in intercellular pectins has been associated with the fruit softening that normally occurs at late stages of fruit growth (Ng et al., 2013). These results suggest that the degree of calcium-mediated pectin complexation is higher in a subset of pectins in the intercellular junctions of the gbp1-c mutant versus the wild type, and they highlight differences in the labeling of pectins by COS488 and 2F4. Taken together, these results suggest that the demethylesterification of primary wall HG that typically occurs during tomato fruit growth and dissolution of the middle lamella during ripening did not progress normally in the mutant mesocarp. In agreement with the cell wall alterations, gbp1-c fruit was ∼30% firmer than wild-type fruit beginning at the breaker stage and softening during ripening was delayed in the mutant (Figure 3F).
Identification of Additional Cell Walls in Large Mesocarp Cells of the gbp1 -c Mutant
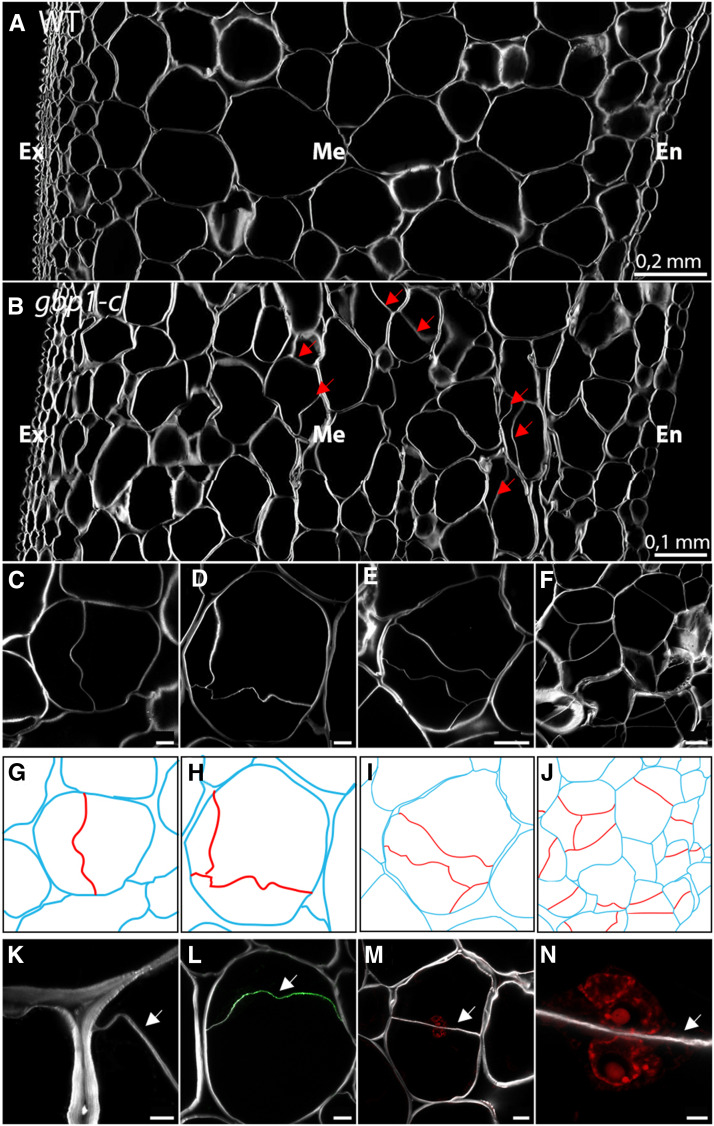
We observed unusual patterns of cell wall deposition in ∼5% of mesocarp cells at the breaker stage (Figures 7A and 7B), while <0.5% of such abnormalities were present at 20 DPA. These newly formed cell walls were either wrinkled (61%, e.g., Figure 7C) or flat (39%, e.g., Figure 7M) and divided the parental cell areas symmetrically (21%, e.g., Figure 7M) or asymmetrically (79%, e.g., Figure 7H). In most cases, a single additional cell wall was noticed in each parental cell, but up to 15% of parental cells had two to three additional cell walls that were deposited in various orientations and anchor positions at the parental cell wall, as well as at newly formed cell walls (Figures 7D, 7E, 7H, and 7I). The latter situation resulted in daughter cells with unexpected geometric shapes, while the overall initial form of parental cells was conserved. In some cases, many adjacent cells were affected by these additional cell walls, creating patchy regions of disordered cellular organization in the mesocarp (Figures 7F and 7J). Parental cells with additional cell walls had an average area of 13,000 µm2 and could reach up to 33,000 µm2, corresponding to medium and large mesocarp cells of the wild type. Labeling with Calcofluor white revealed that these new walls were thinner than those from the parental cells (Figure 7K). Because most of the additional cell walls were reminiscent of immature cell plates, which have a wavy appearance (Assaad, 2001), we immunolabeled pericarp sections with callose anti-β-(1,3)-d-glucan antibodies to detect the cell plate. In many instances, the callose epitopes were recognized on the additional cell walls, suggesting that some of them were immature and probably recently formed (Figure 7L). Although it was technically difficult to image the entire volume of large mesocarp cells by extended depth stacking, it was possible to detect nuclei in the different daughter cells by propidium iodide staining (Figures 7M and 7N; Supplemental Figure 6). These observations suggest that additional cell walls were newly formed after 20 DPA in the gbp1-c mutant due to cell division activity within the large cells of the mesocarp. Consistently, we noticed an increase in mesocarp cell layer number in the mutant at the breaker stage compared to the wild type (Figures 4G to 4I).
Figure 7.
Newly Formed Cell Walls in Large Mesocarp Cells of the gbp1-c Mutant at the Breaker Stage.
(A) and (B) Pericarp sections in the wild type (WT) and gbp1-c at the breaker stage.
(C) to (F) Additional cell walls inside mesocarp cells in the gbp1-c mutant.
(G) to (J) Schematic representations of cells shown in (A) to (D). The parental cell walls are shown in blue and additional cell walls are shown in red.
(K) Close-up of fusion site between parental and new cell wall.
(L) Indirect immunofluorescence with an anti-β-1,3-glucan antibody (green signal) indicating callose deposition.
(M) and (N) Newly divided parental mesocarp cell showing a nucleus in both daughter cells. Nuclei in red are labeled with propidium iodide.
Scale bars represent 5 µm in (K) and (N), 20 µm in (C), (D), (L), and (M) and 50 µm in (E) and (F).
The white signal shows Calcofluor white staining of cell walls (β-linked glucans: callose and cellulose). Arrows indicate new cell walls. Ex, exocarp; M, mesocarp; En, endocarp.
Cell Cycle and Endocycle Regulators Are Mis-Expressed during Late Stages of Fruit Development in the gbp1 -c Mutant
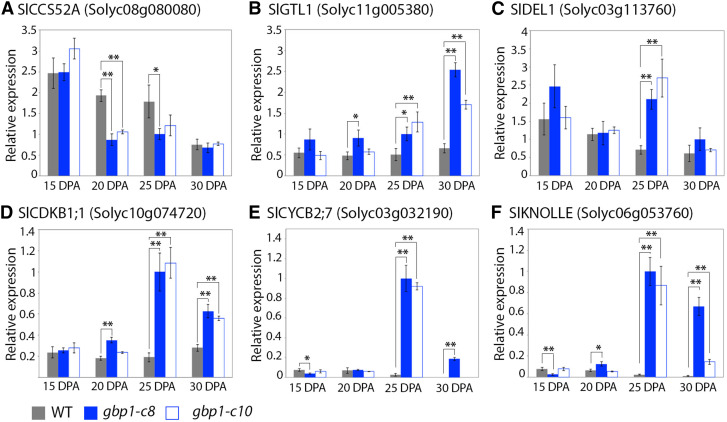
Finally, because endoreduplication is reduced (Figure 4C) and new divisions likely arise (Figures 4H and 7) in the gbp1-c mutant, we investigated the expression of genes involved in regulating the endocycle and the mitotic cycle. SlCCS52A was used as a positive effector of endoreduplication (Mathieu-Rivet et al., 2010). GTL1 and DEL1 encode direct transcriptional repressors of CCS52A in Arabidopsis (Arabidopsis thaliana); the closest tomato homologs, SlGTL1 and SlDEL1, were used as negative effectors of endoreduplication (Lammens et al., 2008; Breuer et al., 2012). SlCDKB1;1 and SlCYCB2;7 were used as gene markers for the G2-to-M transition (Inzé and de Veylder, 2006). SlKNOLLE was used as a gene marker associated with cytokinesis (Figure 8; Lauber et al., 1997). SlCCS52A was downregulated in the gbp1-c mutant at 20 DPA compared with the wild type while SlGTL1 and SlDEL1 were highly upregulated in the mutant beginning at 20 DPA, in accordance with its lower ploidy status. The expression of genes associated with mitosis and cytokinesis peaked at 25 DPA in the gbp1-c mutant. These results are in agreement with the notion that cell division activity resumes at a late stage of fruit growth at the expense of endoreduplication when the expression of SlGBP1 is disrupted.
Figure 8.
Expression Patterns of Cell Cycle and Endocycle Regulators in the gbp1-c Mutant.
(A) SlCCS52A gene expression.
(B) SlGTL1 gene expression.
(C) SlDEL1 gene expression.
(D) SlCDKB1;1 gene expression.
(E) SlCYCB2;7 gene expression.
(F) SlKNOLLE gene expression.
Normalized relative expression of cell cycle genes in wild type (WT) and two gbp1-c mutant lines is given in arbitrary units. Values represent means ± sd for technical triplicates. Significant differences (Student's t test) between the mutants and the wild type are indicated by *P-value < 0.05 and **P-value < 0.01.
DISCUSSION
Here, we identified the key role of SlGBP1, a gene from a family that is poorly functionally characterized in plants, in maintaining the normal developmental program of large polyploid cells in the growing fruit pericarp. Slgbp1 loss-of-function mutants exhibit a reprogramming at late stages of fruit growth from a specialized cell type characteristic of fleshy fruit tissues (Lemaire-Chamley et al., 2005) to a dividing cell type that is a feature of proliferative tissues. We showed that maintenance of the differentiation program in pericarp cells, and hence fleshy fruit characteristics, depends on the repression of the divisions of polyploid cells by SlGBP1. Such a role highlights the importance of SlGBP1 in cell-fate maintenance to preserve cell specialization and tissue function. A recent report described the human GBP1 homolog as a negative regulator of cell proliferation (Unterer et al., 2018), suggesting a conserved function for GBPs in plants and animals. Our study reveals an important regulatory pathway that determines cell fate and maintenance in plants.
SlGBP1 Acts Late in Fruit Development to Maintain the Differentiation Program of Polyploid Cells in the Mesocarp
The fleshy fruit trait arose independently in many fruit species, including examples in the Solanaceae family (Knapp, 2002), and is characterized by the development of a juicy tissue that eventually ripens, making the fruit appealing for frugivores. The tomato pericarp acquires its fleshy properties during fruit growth as a result of the enlargement of mesocarp cells, which can reach up to 400 µm in diameter (Cheniclet et al., 2005). The mesocarp cells enlarge due to the progressive accumulation of solutes and water that sustain vacuolar expansion (Lemaire-Chamley et al., 2005). This process involves endoreduplication (Lee et al., 2009; Bourdon et al., 2012), which starts before anthesis in young tomato ovaries and accounts for the rapid cell growth after fruit set (Renaudin et al., 2017). It is generally accepted that once a cell enters the endocycle, it is no longer able to divide and starts to differentiate. The gbp1 mutant deviates from this normal developmental program in that cell divisions occur in polyploid mesocarp cells at a late stage of fruit growth, thereby revealing a function for SlGBP1 in maintaining mesocarp cell differentiation. Mesocarp from the gbp1 mutant shows reduced cell size, lower ploidy status, and untimely cell division patterns. The mutant retains the ability to produce high C-value nuclei, but in a lower proportion than the wild type, which is likely insufficient to support the boost of cell expansion that typically occurs after 20 DPA. In tomato, changes in ploidy levels are mostly related to variations in the duration of the cell division phase and the onset of endoreduplication (Bertin, 2005), which appears to be unaffected in the gbp1 mutant, as few differences are observed before 10 DPA. While the initiation of endoreduplication is well documented in plants and has been associated with the action of CCS52A in promoting the endocycle (Breuer et al., 2010), less is known about the maintenance and termination of endoreduplication. In plants, E2Fe/DEL1 and the trihelix transcription factor GTL1 are key regulators of the progression of, and exit from, the endocycle by directly repressing CCS52A expression (Breuer et al., 2009, 2012; Heyman et al., 2017). Compared to the wild type, the expression of both homologs from tomato, SlGTL1 and SlDEL1, is upregulated, while that of SlCCS52A is downregulated in gbp1-c fruit from 20 DPA onwards, suggesting an early termination of endoreduplication. This is counterbalanced by the elevated expression of mitotic promoting factors (CDKB and Cyclin), which likely creates favorable conditions to resume cell divisions in the mutant mesocarp cells after 20 DPA. Such divisions are both cytoplasmic and nuclear, as suggested by the occurrence of the upregulation of the cytokinesis gene marker SlKNOLLE and a nucleus in the different daughter cells in the gbp1-c mutant. The compensation of the SlGBP1 knock-out by the increase in SlGBP2 expression, which may affect cell division at late stages of fruit development, cannot be ruled out.
Based on their size and position across the pericarp, the late-dividing cells in the gbp1-c mesocarp are likely highly polyploid. Interestingly, cells from the inner mesocarp are not involved in early proliferative activity, unlike those in the exocarp and endocarp (Renaudin et al., 2017). In this context, the consequence of the disruption of SlGBP1 in the mutant should not be viewed as a return to a cell division mode for a given cell, but rather as a reprogramming of cell fate causing the conversion of an expanding cell into a novel proliferating cell type. This new developmental program is associated with several cell wall and metabolic modifications. The gbp1-c mutant maintains high levels of methylesterified pectins after 20 DPA, which is indicative of cell proliferation, with de-esterified pectin-rich cell walls found in differentiating cells (Bárány et al., 2010; Pérez-Pérez et al., 2019). Other major ripening-associated modifications are affected, including the reduced accumulation of soluble sugars, which are required for cell enlargement and fruit growth, and the maintenance of high levels of organic acids, GABA, and steroidal glycoalkaloids, the degradation of which is necessary to render the fruit edible and appealing. The metabolic status of the gbp1 mesocarp cells also likely reflects their new proliferative state, which is characterized in young dividing fruit by a high Suc demand (Biais et al., 2014) and substantial glycolytic flux (Carrari et al., 2006), to sustain rapid fruit growth at this stage.
SlGBP1 Inhibits Polyploid Divisions in the Tomato Mesocarp
Endopolyploidy is generally regarded as a determinant of terminal differentiation (Orr-Weaver, 2015). Hence, the occurrence of mitotic divisions in large endopolyploid cells appears counterintuitive. However, even if the developmentally programmed endopolyploidization is mainly considered as an irreversible differentiated cell status, several observations from the animal kingdom have shown that polyploid cells can resume mitosis. For example, in vitro cell cultures of human megakaryocytes and mouse hepatocytes under a normal developmental program can revert to complete mitotic cycles (Duncan et al., 2010; Leysi-Derilou et al., 2010). Polyploid divisions have long been reported to occur in plants. For instance, mitotic divisions in polyploid cells occur spontaneously in roots and during root nodule formation in pea (Pisum sativum; Wipf and Cooper, 1938) and after hormone treatment or responses to wounding in various organs and tissues (Bradley and Crane, 1955; Matthysse and Torrey, 1967; Torrey and Fosket, 1970). These latter examples relate to the occurrence of endopolyploidization resulting from endomitosis (a process that differs from endoreduplication, as mitosis is initiated during endomitosis, leading to an increased number of chromosomes, but cytokinesis is aborted). Drosophila melanogaster (fruit fly) and Culex pipiens (mosquito) rectal papillar cells are rare examples of polyploid cells that are able to re-enter the mitotic cell cycle after endoreduplication (Fox et al., 2010; Schoenfelder et al., 2014). Stormo and Fox (2016) found existence of a pre-anaphase mechanism that eliminates polyteny in these endoreduplicated cells to allow proper segregation of pairs of sister chromatids during mitosis. In A. thaliana leaves, endoreduplicated trichome-neighboring cells overexpressing the CDK inhibitor KRP1 were also shown to reenter a mitotic cycle (Weinl et al., 2005). In addition, these trichome-neighboring cells formed new cell walls.
Taken together, these findings suggest that cell cycle/endocycle control is far more plastic than is generally stated. Although the exact cytological mechanism involved remains unknown, polyploid cells in the gbp1 mesocarp can divide without hormonal treatments or wounding and generate new cell walls. According to the studies cited above, nuclei of high polyploidy categories appear to be less susceptible to division. The only example of division of a 64C nucleus (endomitotic) that we have identified in the literature was in apricot (Prunus armeniaca) fruit (Bradley and Crane, 1955). Further investigation would be required to determine the ploidy levels of cells that are still able to resume mitosis in the gbp1 mutant, as tomato mesocarp cells can reach 128C and in some cases 512C (Cheniclet et al., 2005). The results reported here provide evidence for a role of plant GBPs in repressing the division of polyploid cells to maintain the endoreduplication status.
GBPs Are Master Genes Controlling the Expression of Cell Cycle Genes
GBPs are large GTPases belonging to the dynamin protein superfamily, although they can be distinguished from other dynamin-related proteins due to low sequence homology (Praefcke and McMahon, 2004). In addition, unlike dynamins, GBPs can hydrolyze GTP not only to GDP, but also to GMP (Schwemmle and Staeheli, 1994). While dynamins are involved in a broad range of activities, such as endocytosis, actin dynamics, cytokinesis, and viral resistance (Praefcke and McMahon, 2004; Konopka et al., 2006; Roux et al., 2006), GBPs have mostly been described in humans and animals as major interferon-gamma (IFN-γ)–stimulated factors upon pathogen infection, inflammatory diseases, and cancer (Vestal and Jeyaratnam, 2011; Britzen-Laurent et al., 2016; Praefcke, 2018). The best-studied human GBP, hGBP-1, interacts directly with actin via its indispensable GTPase activity (Ostler et al., 2014), inducing rearrangements of the cytoskeleton at the origin of the IFN-γ–dependent anti-viral function (Zou et al., 2017). Recently, Unterer et al. (2018) demonstrated that hGBP-1 mediates the anti-proliferative and anti-tumorigenic effects of IFN-γ via its C-terminal helical domain by regulating the Hippo pathway (Unterer et al., 2018). Via a direct interaction, hGBP-1 deactivates the Hippo-signaling TEA-Domain transcription factor (Unterer et al., 2018), which controls cell cycle gene expression downstream (Santucci et al., 2015). No such role for GBPs in plants has been described to date. Although TEA-Domain homologs have not yet been identified in plants, homologs for Hippo kinases that act upstream of hGBP-1 in regulating the Hippo pathway and control cell proliferation and cytokinesis (Pinosa et al., 2013; Xiong et al., 2016) are indeed present in plants. Given the tomato gbp1 mutant phenotype, the anti-proliferative functions may be conserved in plant GBPs, likely via the indirect control of the expression of cell cycle genes, but the pathway remains to be elucidated.
The roles of GBPs in plants are not well understood. By analogy with their recently described functions in animal immunity, it has been hypothesized that plant GBPs could also play a cell-autonomous role in plant immunity (Huang et al., 2019). Our results show that SlGBP1 is central to cell proliferation and cell differentiation control, suggesting a possible role in regulating the balance between plant growth and defense. Further studies are required to decipher the mechanisms by which GBP exerts its effects, especially studies focused on uncovering the regulatory events that fine-tune the extent of endoreduplication during fruit growth.
METHODS
Plant Materials and Growth Conditions
The tomato p3d3 mutant was identified in a highly mutagenized EMS mutant collection of plants in the miniature cv Micro-Tom (Solanum lycopersicum) background (Just et al., 2013; Musseau et al., 2017). Plants were grown in a greenhouse at the Institut National de Recherche pour l'Agriculture, l'Alimentation, et l'Environnement (Bordeaux, France) under standard conditions as described by Rothan et al. (2016) from 2015 to 2019. A description of Micro-Tom fruit developmental and phenological stages is provided in Figure 3, with the breaker stage corresponding to the final fruit size.
Mapping-by-Sequencing and Recombination Analysis
Mapping-by-sequencing and recombination analysis were performed as described by Garcia et al. (2016) and Petit et al. (2016). The BC1F2 population consisted of 180 plants obtained by crossing the p3d3 mutant with the wild type. Bulks were produced by pooling 26 plants with the wild-type–like or the mutant thin pericarp phenotype. Library preparation of bulked DNA and sequencing using a HiSeq 2500 sequencer (Illumina) was done at the INRA-GeT-PlaGe-GENOTOUL platform, operating in a 100-bp paired-end run mode. Sequence analyses were performed as previously described by Garcia et al. (2016), and version SL2.50 of the reference tomato genome was used for read mapping (ftp://ftp.solgenomics.net/genomes/Solanum_lycopersicum/Heinz1706/assembly/build_2.50/). EMS variants with a read depth between 10 and 100 were considered for allelic frequency analysis. Recombination analysis of the BC1F2 individuals was performed using EMS mutations as markers in a Kompetitive allele-specific PCR assay (Smith and Maughan, 2015). Subsequent genotyping of the causal p3d3 mutation was done through Sanger sequencing of PCR products. All primer sequences are listed in Supplemental Data Set 1.
Cloning and Plant Transformation
The pEn-Chimera (single guide RNA) entry vector and pDe-CAS9 (Streptococcus pyogenes nuclease) destination vector including the plant Kanamycin resistance cassette were kindly provided by Holger Puchta (Karlsruhe Institute of Technology, Germany). Gateway-compatible cloning for single sgRNAs was performed according to the protocol available at http://www.botanik.kit.edu/molbio/940.php (Schiml et al., 2016) using the P3D3-CP-F and P3D3-CP-R oligos to target SlGBP1 (primers in Supplemental Data Set 1). Agrobacterium-mediated tomato transformation of Micro-Tom cotyledons was performed as described in Cortina and Culiáñez-Macià (2004), and 15 independent transformation events with expected mutations in SlGBP1 were validated by Sanger sequencing (primers in Supplemental Data Set 1). Three independent CRISPR lines were used for this study: gbp1-c4, c8, and c10 in the T2 and T3 generations (Supplemental Figure 1). For illustrations or when data corresponded to means of different CRISPR lines, samples were called gbp1-c. The absence of off-targets in the homologous SlGBP2 gene was verified by Sanger sequencing. We confirmed by RT-qPCR that SlGBP2 expression was not reduced in the gbp1-c mutants compared with SlGBP1 expression (Supplemental Figure 1D).
Histological Analyses of Pericarp Tissue
Pericarp thickness, cell number, and cell areas were measured in pericarp cross sections. Sample preparations and equatorial transverse sections of fruit pericarps were performed as described by Musseau et al. (2017). Nuclei were labeled using a 10-µg/mL propidium iodide solution (Sigma Aldrich). Samples were observed using an epifluorescence microscope (Axiophot; Zeiss) or a confocal microscope (LSM880; Zeiss) at the Bordeaux Imaging Center (http://www.bic.u-bordeaux.fr/). Parameters, including cross-sectional cell number and area other than those of the vasculature tissue, were estimated as described by Musseau et al. (2017). For each genotype and stage, the biological replicates included a minimum of three pericarp sections from three fruits from different plants. For the mutant samples, these included fruits from two different transgenic lines. For each pericarp section, three measurements of pericarp thickness and of number of cell layers were made. Pericarp cell layers (exocarp, mesocarp, and endocarp) were defined according to Renaudin et al. (2017). Mean cell area was estimated based on all cells of the cross sections excluding pericarp regions with vasculature tissue (between 100 and 550 cells were measured in each section).
Ploidy Analysis
Cell ploidy of equatorial pericarp samples was determined by flow cytometry (CyFlow Space, Partec; Sysmex) as described by Cheniclet et al. (2005). Supplemental Figure 7 shows cell sorter chromatograms that we obtained for the mutant and the wild type. The 2C nucleus value at the breaker stage was set to zero in both the mutant and the wild type, as the corresponding peaks were not discernable above background noise (Supplemental Figure 7). For each stage and genotype, a minimum of five fruits from different plants was analyzed as biological replicates. The mutant samples included fruits from two different transgenic lines.
Cell Wall Labeling
Experiments were performed on fruits at 10 DPA, 15 DPA, 20 DPA, and the breaker stage from wild type and the CRISPR mutant lines (n = 2 to 4 fruits per genotype and stage as biological replicates). The mutant sample replicates included fruits from at least two different CRISPR lines among gbp1-c8, gbp1-c10, or gbp1-c4. Callose and pectin labeling was performed on formaldehyde alcohol acetic acid-fixed equatorial portions of tomato equatorial pericarp, using 100-µm to 150-µm–thick sections cut with a vibratome microtome (model no. Microm HM 650 V; Thermo Fisher Scientific). Sections were incubated with 5% (w/v) milk/phosphate-buffered saline (PBS) at pH 7.2 for immunolabeling by JIM5, LM20, and anti-β-1,3 glucan or 5% (w/v) milk/TCaS (20 mM Tris–HCl at pH 8.2, 0.5 mM of CaCl2, and 150 mM of NaCl) for immunolabeling by 2F4 for 30 min with gentle shaking. Fresh buffer without 5% milk was then added, and the samples were incubated with JIM5, LM20, 2F4 (cat. nos. JIM5, LM20, and 2F4; Plantprobes), and anti-β-1,3 glucan (cat. no. 400-2; Biosupplies) antibodies overnight at 4°C in dilutions of 0.05 or 0.004 milk/PBS or TCaS. The sections were washed three times for 5 min with PBS and incubated in a 0.01 dilution of goat anti-rat (JIM5, LM20) or anti-mouse (2F4, anti-β-1,3 glucan) IgG coupled to fluorescein isothiocyanate (FITC; cat. nos. F6258, lot SLBN3827V and cat. no. F0257, lot SLBT0812, respectively; Sigma Aldrich) in milk/PBS for 90 min in the dark at room temperature under slow agitation. Control sections omitting the primary antibody were also made. COS488, kindly provided by William G.T. Willats (Newcastle University, United Kingdom) was diluted 0.001 in MES buffer, pH 5.7, as described by Mravec et al. (2014). Sections were washed three times for 5 min with MES buffer and incubated with Calcofluor white (Sigma Aldrich) for 1 min and rinsed in the corresponding buffer. Sections were mounted with Citifluor (Electron Microscopy Science) for observations under a confocal microscope (model no. LSM880; Zeiss) at the Bordeaux Imaging Center. No signals were observed in the controls.
Tomato Fruit Firmness
Fruit firmness was measured by penetrometry at the fruit equator excluding septum position, using a Fruit Texture Analyzer (GüSS) equipped with a 5-mm diameter probe and using the following parameters: trigger threshold: 0.05 kg, forward speed: 10 mm/s, reverse speed: 20 mm/s, measure speed: 5 mm/s, measure distance: 3.5 mm, reverse increment: 20 mm. For each stage and genotype, a minimum of 11 fruits from at least five different plants were analyzed as biological replicates. The mutant samples included fruits from the three different transgenic lines (gbp1-c8, gbp1-c10, or gbp1-c4).
Metabolomic Analyses by NMR and LC-MS Profiling
Metabolomic analyses of 15 DPA, 20 DPA, 25 DPA, and Red Ripe fruit from the wild type, gbp1-c4, and gbp1-c8 independent mutant lines were performed at the Bordeaux Metabolome Facility of MetaboHUB. Three biological replicates were made for each stage and genotype (wild type, gbp1-c8, and gbp1-c4), each replicate corresponding to whole fruit pericarp pooled from three fruits from different plants. Freeze-dried pericarp was used for extraction. 1H-NMR metabolomic profiling of semi-polar extracts was performed as described by Deborde et al. (2019). Annotations of resonances based on the literature and an in-house database are presented in Supplemental Data Set 2. LC-MS–based metabolomic profiling of extracts was performed as previously described by Lamari et al. (2018). Blank extracts from the same procedure, but without sample, were also analyzed. A quality-control sample was produced by pooling 50 μL of each sample extract. The resulting methanolic extracts were analyzed using an LC-MS (Ultimate 3000 LTQ-Orbitrap Elite; Thermo Fisher Scientific). The sample injection order was randomized. The quality-control sample was injected every 10 samples to correct for mass spectrometer signal drift. Data were processed using the software MS-DIAL v3.66 (Tsugawa et al., 2015). Variables detected in blank extracts were filtered out. Intensity drift was corrected using lowest regression, retention time correction was performed using known metabolites, and zero intensities were replaced by 10% of the minimum height. Finally, intensities were normalized according to the sample powder mass used for extraction. Annotation of intense ions, shown in Supplemental Data Set 3, was performed using RT, accurate m/z, and MS/MS fragments.
RT-qPCR Gene Expression Analysis
Gene expression was analyzed in the wild type and two gbp1-c lines for SlGBP1, SlGBP2, and cell cycle genes corresponding to the closest Arabidopsis (Arabidopsis thaliana) homologous genes. Accession numbers and primer sequences and efficiencies are listed in Supplemental Data Set 1. Whole fruit pericarp from different plants was pooled (n = 3 to 6) for each genotype (wild type, gbp1-c8, gbpc1-c10-gbp1-c4, and p3d3) and each stage. RNA extraction and reverse transcription were performed as described in Mounet et al. (2012). RT-qPCR was performed in a 10-μL final volume using GoTaq qPCR Master Mix (Promega) according to the manufacturer's instructions, on a CFX-96 instrument (Bio-Rad). Five housekeeping genes were included, and the two genes with the most consistent expression patterns (UBI and EIF4a) were used for ∆∆CT normalization of gene expression (Supplemental Data Set 1).
Statistical Analyses
Multivariate and univariate statistical analyses were performed using the BioStatFlow web application based on R scripts (biostatflow, v2.9; http://biostatflow.org/). PCAs of metabolome data were performed on data mean-centered and scaled to unit variance. Significant variations of metabolite signatures in a gbp1-c mutant compared with wild type at each stage of development were identified using a Wilcoxon test with false-discovery rate (FDR) correction (Benjamini and Hochberg, 1995), and the ratios between means were visualized on Volcano plots (Wilcoxon test corrected P < 0.05, ratio between means threshold of 1.2). For mean comparisons of the fruit cellular data (Wilcoxon test, P < 0.05) and expression data (Student's t test, P < 0.05); no FDR correction was applied. P-values of statistic tests are shown in Supplemental Data Sets 4 and 5.
Accession Numbers
Accession numbers from this article are listed in Supplemental Data Set 1. NMR and LC-MS data and metadata have been deposited into the Institut National de la Recherche Agronomique Dataverse repository (https://data.inrae.fr/dataverse/Sl-gbp1).
Supplemental Data
Supplemental Figure 1. Effect of SlGBP1 mutations on the predicted protein sequence and mRNA abundance.
Supplemental Figure 2. SlGBP1 expression in tomato available in databases.
Supplemental Figure 3. SlGBP2 expression in tomato available in databases.
Supplemental Figure 4. DNA ploidy levels in pericarp during fruit growth in wild type and the gbp1-c mutant.
Supplemental Figure 5. Volcano plots showing the statistical significance of differences in primary and specialized metabolite levels against fold change (FC) between the gbp1-c mutant lines and wild type.
Supplemental Figure 6. Newly divided mesocarp cells exhibiting a nucleus in daughter cells in the gbp1-c mutant.
Supplemental Figure 7. Flow cytometry for estimating DNA ploidy levels in wild type and the gbp1-c mutant.
Supplemental Data Set 1. List of primer sequences and accession numbers.
Supplemental Data Set 2. List of metabolites detected in the 1H-NMR spectra (500 MHz) of semi-polar extracts of tomato fruit pericarp.
Supplemental Data Set 3. List of compounds detected in the LC-MS spectra of semi-polar extracts of tomato fruit pericarp.
Supplemental Data Set 4. Statistical analyses of cellular and molecular data.
Supplemental Data Set 5. Statistical analyses of MS- and NMR-based metabolomic data.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Supplementary Material
Acknowledgments
The authors express their deepest thanks to Fabienne Wong for her help with bioinformatic analysis; Catherine Cheniclet, Nathalie Frangne, and Jean Pierre Renaudin for helpful discussions; Lysiane Brocard from the Bordeaux Imaging Center for plant imaging assistance; and Sabine Mueller and Nathalie Gonzalez for critical reading of the article. This work was supported by MetaboHUB (grant ANR-11-INBS-0010) and the Ministère de l’Enseignement Supérieur et de la Recherche (Ph.D. for C.M.).
AUTHOR CONTRIBUTIONS
L.F.-L., F.G., C.R., and C.M. designed the research; I.A. contributed to plant material production; J.-P.M. performed the genotyping; C.M., C.D., S.B., and A.M. performed the metabolomic analysis; I.S. and J.K.C.R. contributed to cell wall study; C.M., J.J., S.G., and L.F.-L. performed all other research; C.M. and L.F.-L. performed data analysis and wrote the original draft of the article; C.R., C.C., I.S., J.K.C.R., M.L.-C., and A.M. reviewed and edited the article; F.G. and L.F.-L. supervised C.M.'s PhD.
References
- Akihiro, T. , et al. (2008). Biochemical mechanism on GABA accumulation during fruit development in tomato. Plant Cell Physiol. 49: 1378–1389. [DOI] [PubMed] [Google Scholar]
- Assaad, F.F. (2001). Plant cytokinesis. Exploring the links. Plant Physiol. 126: 509–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baloban, M. , Vanstraelen, M. , Tarayre, S. , Reuzeau, C. , Cultrone, A. , Mergaert, P. , Kondorosi, E. (2013). Complementary and dose-dependent action of AtCCS52A isoforms in endoreduplication and plant size control. New Phytol. 198: 1049–1059. [DOI] [PubMed] [Google Scholar]
- Bárány, I. , Fadón, B. , Risueño, M.C. , Testillano, P.S. (2010). Microspore reprogramming to embryogenesis induces changes in cell wall and starch accumulation dynamics associated with proliferation and differentiation events. Plant Signal. Behav. 5: 341–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini, Y. , Hochberg, Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57: 289–300. [Google Scholar]
- Bertin, N. (2005). Analysis of the tomato fruit growth response to temperature and plant fruit load in relation to cell division, cell expansion and DNA endoreduplication. Ann. Bot. 95: 439–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertin, N. , Causse, M. , Brunel, B. , Tricon, D. , Génard, M. (2009). Identification of growth processes involved in QTLs for tomato fruit size and composition. J. Exp. Bot. 60: 237–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhosale, R. , Maere, S. , de Veylder, L. (2019). Endoreplication as a potential driver of cell wall modifications. Curr. Opin. Plant Biol. 51: 58–65. [DOI] [PubMed] [Google Scholar]
- Biais, B. , et al. (2014). Remarkable reproducibility of enzyme activity profiles in tomato fruits grown under contrasting environments provides a roadmap for studies of fruit metabolism. Plant Physiol. 164: 1204–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourdon, M. , Pirrello, J. , Cheniclet, C. , Coriton, O. , Bourge, M. , Brown, S. , Moïse, A. , Peypelut, M. , Rouyère, V. , Renaudin, J.-P. , Chevalier, C. , Frangne, N. (2012). Evidence for karyoplasmic homeostasis during endoreduplication and a ploidy-dependent increase in gene transcription during tomato fruit growth. Development 139: 3817–3826. [DOI] [PubMed] [Google Scholar]
- Bradley, M. , Crane, J. (1955). The effect of 2,4,5-trichlorophenoxyacetic acid on cell and nuclear size and endopolyploidy in parenchyma of apricot fruits on JSTOR. Am. J. Bot. 42: 273–281. [Google Scholar]
- Breuer, C. , Ishida, T. , Sugimoto, K. (2010). Developmental control of endocycles and cell growth in plants. Curr. Opin. Plant Biol. 13: 654–660. [DOI] [PubMed] [Google Scholar]
- Breuer, C. , Kawamura, A. , Ichikawa, T. , Tominaga-Wada, R. , Wada, T. , Kondou, Y. , Muto, S. , Matsui, M. , Sugimoto, K. (2009). The trihelix transcription factor GTL1 regulates ploidy-dependent cell growth in the Arabidopsis trichome. Plant Cell 21: 2307–2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuer, C. , Morohashi, K. , Kawamura, A. , Takahashi, N. , Ishida, T. , Umeda, M. , Grotewold, E. , Sugimoto, K. (2012). Transcriptional repression of the APC/C activator CCS52A1 promotes active termination of cell growth. EMBO J. 31: 4488–4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britzen-Laurent, N. , Herrmann, C. , Naschberger, E. , Croner, R.S. , Stürzl, M. (2016). Pathophysiological role of guanylate-binding proteins in gastrointestinal diseases. World J. Gastroenterol. 22: 6434–6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrari, F. , Baxter, C. , Usadel, B. , Urbanczyk-Wochniak, E. , Zanor, M.I. , Nunes-Nesi, A. , Nikiforova, V. , Centero, D. , Ratzka, A. , Pauly, M. , Sweetlove, L.J. , Fernie, A.R. (2006). Integrated analysis of metabolite and transcript levels reveals the metabolic shifts that underlie tomato fruit development and highlight regulatory aspects of metabolic network behavior. Plant Physiol. 142: 1380–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheniclet, C. , Rong, W.Y. , Causse, M. , Frangne, N. , Bolling, L. , Carde, J.P. , Renaudin, J.P. (2005). Cell expansion and endoreduplication show a large genetic variability in pericarp and contribute strongly to tomato fruit growth. Plant Physiol. 139: 1984–1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier, C. , Bourdon, M. , Pirrello, J. , Cheniclet, C. , Gévaudant, F. , Frangne, N. (2014). Endoreduplication and fruit growth in tomato: evidence in favour of the karyoplasmic ratio theory. J. Exp. Bot. 65: 2731–2746. [DOI] [PubMed] [Google Scholar]
- Chevalier, C. , Nafati, M. , Mathieu-Rivet, E. , Bourdon, M. , Frangne, N. , Cheniclet, C. , Renaudin, J.P. , Gévaudant, F. , Hernould, M. (2011). Elucidating the functional role of endoreduplication in tomato fruit development. Ann. Bot. 107: 1159–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortina, C. , Culiáñez-Macià, F.A. (2004). Tomato transformation and transgenic plant production. Plant Cell Tissue Organ Cult. 76: 269–275. [Google Scholar]
- Daher, F.B. , Braybrook, S.A. (2015). How to let go: Pectin and plant cell adhesion. Front Plant Sci 6: 523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deborde, C. , Fontaine, J.X. , Jacob, D. , Botana, A. , Nicaise, V. , Richard-Forget, F. , Lecomte, S. , Decourtil, C. , Hamade, K. , Mesnard, F. , Moing, A. , Molinié, R. (2019). Optimizing 1D 1H-NMR profiling of plant samples for high throughput analysis: extract preparation, standardization, automation and spectra processing. Metabolomics 15: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Veylder, L. , Beeckman, T. , Inzé, D. (2007). The ins and outs of the plant cell cycle. Nat. Rev. Mol. Cell Biol. 8: 655–665. [DOI] [PubMed] [Google Scholar]
- Duncan, A.W. , Taylor, M.H. , Hickey, R.D. , Hanlon Newell, A.E. , Lenzi, M.L. , Olson, S.B. , Finegold, M.J. , Grompe, M. (2010). The ploidy conveyor of mature hepatocytes as a source of genetic variation. Nature 467: 707–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox, D.T. , Gall, J.G. , Spradling, A.C. (2010). Error-prone polyploid mitosis during normal Drosophila development. Genes Dev. 24: 2294–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia, V. , et al. (2016). Rapid identification of causal mutations in tomato EMS populations via mapping-by-sequencing. Nat. Protoc. 11: 2401–2418. [DOI] [PubMed] [Google Scholar]
- Gillaspy, G. , Ben-David, H. , Gruissem, W. (1993). Fruits: A developmental perspective. Plant Cell 5: 1439–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, F. , Jacobson, A. (2015). Nonsense-mediated mRNA decay: Degradation of defective transcripts is only part of the story. Annu. Rev. Genet. 49: 339–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyman, J. , Polyn, S. , Eekhout, T. , de Veylder, L. (2017). Tissue-specific control of the endocycle by the anaphase promoting complex/cyclosome inhibitors UVI4 and DEL1. Plant Physiol. 175: 303–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, S. , Meng, Q. , Maminska, A. , MacMicking, J.D. (2019). Cell-autonomous immunity by IFN-induced GBPs in animals and plants. Curr. Opin. Immunol. 60: 71–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inzé, D. , De Veylder, L. (2006). Cell cycle regulation in plant development. Annu. Rev. Genet. 40: 77–105. [DOI] [PubMed] [Google Scholar]
- Itkin, M. , et al. (2013). Biosynthesis of antinutritional alkaloids in solanaceous crops is mediated by clustered genes. Science 341: 175–179. [DOI] [PubMed] [Google Scholar]
- Just, D. , et al. (2013). Micro-Tom mutants for functional analysis of target genes and discovery of new alleles in tomato. Plant Biotechnol. 30: 225–231. [Google Scholar]
- van der Knaap, E. , Chakrabarti, M. , Chu, Y.H. , Clevenger, J.P. , Illa-Berenguer, E. , Huang, Z. , Keyhaninejad, N. , Mu, Q. , Sun, L. , Wang, Y. , Wu, S. (2014). What lies beyond the eye: The molecular mechanisms regulating tomato fruit weight and shape. Front Plant Sci 5: 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp, S. (2002). Tobacco to tomatoes: A phylogenetic perspective on fruit diversity in the Solanaceae. J. Exp. Bot. 53: 2001–2022. [DOI] [PubMed] [Google Scholar]
- Knox, J.P. , Linstead, P.J. , King, J. , Cooper, C. , Roberts, K. (1990). Pectin esterification is spatially regulated both within cell walls and between developing tissues of root apices. Planta 181: 512–521. [DOI] [PubMed] [Google Scholar]
- Konopka, C.A. , Schleede, J.B. , Skop, A.R. , Bednarek, S.Y. (2006). Dynamin and cytokinesis. Traffic 7: 239–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamari, N. , et al. (2018). Metabotyping of 30 maize hybrids under early-sowing conditions reveals potential marker-metabolites for breeding. Metabolomics 14: 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammens, T. , Boudolf, V. , Kheibarshekan, L. , Zalmas, L.P. , Gaamouche, T. , Maes, S. , Vanstraelen, M. , Kondorosi, E. , La Thangue, N.B. , Govaerts, W. , Inzé, D. , de Veylder, L. (2008). Atypical E2F activity restrains APC/CCCS52A2 function obligatory for endocycle onset. Proc. Natl. Acad. Sci. USA 105: 14721–14726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber, M.H. , Waizenegger, I. , Steinmann, T. , Schwarz, H. , Mayer, U. , Hwang, I. , Lukowitz, W. , Jürgens, G. (1997). The Arabidopsis KNOLLE protein is a cytokinesis-specific syntaxin. J. Cell Biol. 139: 1485–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, H.O. , Davidson, J.M. , Duronio, R.J. (2009). Endoreplication: Polyploidy with purpose. Genes Dev. 23: 2461–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire-Chamley, M. , Petit, J. , Garcia, V. , Just, D. , Baldet, P. , Germain, V. , Fagard, M. , Mouassite, M. , Cheniclet, C. , Rothan, C. (2005). Changes in transcriptional profiles are associated with early fruit tissue specialization in tomato. Plant Physiol. 139: 750–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leysi-Derilou, Y. , Robert, A. , Duchesne, C. , Garnier, A. , Boyer, L. , Pineault, N. (2010). Polyploid megakaryocytes can complete cytokinesis. Cell Cycle 9: 2589–2599. [DOI] [PubMed] [Google Scholar]
- Liners, F. , Letesson, J.-J. , Didembourg, C. , van Cutsem, P. (1989). Monoclonal antibodies against pectin: Recognition of a conformation induced by calcium. Plant Physiol. 91: 1419–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu-Rivet, E. , Gévaudant, F. , Cheniclet, C. , Hernould, M. , Chevalier, C. (2010). The Anaphase Promoting Complex activator CCS52A, a key factor for fruit growth and endoreduplication in Tomato. Plant Signal. Behav. 5: 985–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthysse, A.G. , Torrey, J.G. (1967). DNA synthesis in relation to polyploid mitoses in excised pea root segments cultured in vitro. Exp. Cell Res. 48: 484–498. [DOI] [PubMed] [Google Scholar]
- Mounet, F. , et al. (2007). Quantitative metabolic profiles of tomato flesh and seeds during fruit development: Complementary analysis with ANN and PCA. Metabolomics 3: 273–288. [Google Scholar]
- Mounet, F. , et al. (2012). Down-regulation of a single auxin efflux transport protein in tomato induces precocious fruit development. J. Exp. Bot. 63: 4901–4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mravec, J. , et al. (2014). Tracking developmentally regulated post-synthetic processing of homogalacturonan and chitin using reciprocal oligosaccharide probes. Development 141: 4841–4850. [DOI] [PubMed] [Google Scholar]
- Mu, Q. , Huang, Z. , Chakrabarti, M. , Illa-Berenguer, E. , Liu, X. , Wang, Y. , Ramos, A. , van der Knaap, E. (2017). Fruit weight is controlled by Cell Size Regulator encoding a novel protein that is expressed in maturing tomato fruits. PLoS Genet. 13: e1006930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musseau, C. , Just, D. , Jorly, J. , Gévaudant, F. , Moing, A. , Chevalier, C. , Lemaire-Chamley, M. , Rothan, C. , Fernandez, L. (2017). Identification of two new mechanisms that regulate fruit growth by cell expansion in tomato. Front Plant Sci 8: 988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, J.K.T. , Schröder, R. , Sutherland, P.W. , Hallett, I.C. , Hall, M.I. , Prakash, R. , Smith, B.G. , Melton, L.D. , Johnston, J.W. (2013). Cell wall structures leading to cultivar differences in softening rates develop early during apple (Malus × domestica) fruit growth. BMC Plant Biol. 13: 183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver, T.L. (2015). When bigger is better: The role of polyploidy in organogenesis. Trends Genet. 31: 307–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostler, N. , et al. (2014). Gamma interferon-induced guanylate binding protein 1 is a novel actin cytoskeleton remodeling factor. Mol. Cell. Biol. 34: 196–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Pérez, Y. , Carneros, E. , Berenguer, E. , Solís, M.T. , Bárány, I. , Pintos, B. , Gómez-Garay, A. , Risueño, M.C. , Testillano, P.S. (2019). Pectin de-methylesterification and AGP increase promote cell wall remodeling and are required during somatic embryogenesis of Quercus suber. Front Plant Sci 9: 1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit, J. , Bres, C. , Mauxion, J.P. , Tai, F.W.J. , Martin, L.B.B. , Fich, E.A. , Joubès, J. , Rose, J.K.C. , Domergue, F. , Rothan, C. (2016). The glycerol-3-phosphate acyltransferase GPAT6 from tomato plays a central role in fruit cutin biosynthesis. Plant Physiol. 171: 894–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinosa, F. , Begheldo, M. , Pasternak, T. , Zermiani, M. , Paponov, I.A. , Dovzhenko, A. , Barcaccia, G. , Ruperti, B. , Palme, K. (2013). The Arabidopsis thaliana Mob1A gene is required for organ growth and correct tissue patterning of the root tip. Ann. Bot. 112: 1803–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirrello, J. , et al. (2018). Transcriptome profiling of sorted endoreduplicated nuclei from tomato fruits: How the global shift in expression ascribed to DNA ploidy influences RNA-seq data normalization and interpretation. Plant J. 93: 387–398. [DOI] [PubMed] [Google Scholar]
- Pound, M.P. , French, A.P. , Wells, D.M. , Bennett, M.J. , Pridmore, T.P. (2012). CellSeT: novel software to extract and analyze structured networks of plant cells from confocal images. Plant Cell 24: 1353–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praefcke, G.J.K. (2018). Regulation of innate immune functions by guanylate-binding proteins. Int. J. Med. Microbiol. 308: 237–245. [DOI] [PubMed] [Google Scholar]
- Praefcke, G.J.K. , McMahon, H.T. (2004). The dynamin superfamily: Universal membrane tubulation and fission molecules? Nat. Rev. Mol. Cell Biol. 5: 133–147. [DOI] [PubMed] [Google Scholar]
- Prakash, B. , Praefcke, G.J.K. , Renault, L. , Wittinghofer, A. , Herrmann, C. (2000). Structure of human guanylate-binding protein 1 representing a unique class of GTP-binding proteins. Nature 403: 567–571. [DOI] [PubMed] [Google Scholar]
- Prasanna, V. , Prabha, T.N. , Tharanathan, R.N. (2007). Fruit ripening phenomena--an overview. Crit. Rev. Food Sci. Nutr. 47: 1–19. [DOI] [PubMed] [Google Scholar]
- Renaudin, J.-P. , Deluche, C. , Cheniclet, C. , Chevalier, C. , Frangne, N. (2017). Cell layer-specific patterns of cell division and cell expansion during fruit set and fruit growth in tomato pericarp. J. Exp. Bot. 68: 1613–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothan, C. , Just, D. , Fernandez, L. , Atienza, I. , Ballias, P. , Lemaire-Chamley, M. (2016). Culture of the Tomato Micro-Tom Cultivar in Greenhouse. (New York, NY: Humana Press; ), pp. 57–64. [DOI] [PubMed] [Google Scholar]
- Roux, A. , Uyhazi, K. , Frost, A. , de Camilli, P. (2006). GTP-dependent twisting of dynamin implicates constriction and tension in membrane fission. Nature 441: 528–531. [DOI] [PubMed] [Google Scholar]
- Santucci, M. , Vignudelli, T. , Ferrari, S. , Mor, M. , Scalvini, L. , Bolognesi, M.L. , Uliassi, E. , Costi, M.P. (2015). The Hippo pathway and YAP/TAZ-TEAD protein-protein interaction as targets for regenerative medicine and cancer treatment. J. Med. Chem. 58: 4857–4873. [DOI] [PubMed] [Google Scholar]
- Schiml, S. , Fauser, F. , Puchta, H. (2016). CRISPR/Cas-mediated site-specific mutagenesis in Arabidopsis thaliana using Cas9 nucleases and paired nickases. Methods Mol. Biol. 1469: 111–122. [DOI] [PubMed] [Google Scholar]
- Schoenfelder, K.P. , Montague, R.A. , Paramore, S.V. , Lennox, A.L. , Mahowald, A.P. , Fox, D.T. (2014). Indispensable pre-mitotic endocycles promote aneuploidy in the Drosophila rectum. Development 141: 3551–3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwemmle, M. , Staeheli, P. (1994). The interferon-induced 67-kDa guanylate-binding protein (hGBP1) is a GTPase that converts GTP to GMP. J. Biol. Chem. 269: 11299–11305. [PubMed] [Google Scholar]
- Smith, S.M. , Maughan, P.J. (2015). SNP genotyping using KASPar assays. Methods Mol. Biol. 1245: 243–256. [DOI] [PubMed] [Google Scholar]
- Stormo, B.M. , Fox, D.T. (2016). Distinct responses to reduplicated chromosomes require distinct Mad2 responses. eLife 5: 1–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumner, L.W. , et al. (2007). Proposed minimum reporting standards for chemical analysis. Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 3: 211–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrey, J.G. , Fosket, D.E. (1970). Cell division in relation to cytodifferentiation in cultured pea root segments. Am. J. Bot. 57: 1072–1080. [Google Scholar]
- Tsugawa, H. , Cajka, T. , Kind, T. , Ma, Y. , Higgins, B. , Ikeda, K. , Kanazawa, M. , VanderGheynst, J. , Fiehn, O. , Arita, M. (2015). MS-DIAL: Data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods 12: 523–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterer, B. , Wiesmann, V. , Gunasekaran, M. , Sticht, H. , Tenkerian, C. , Behrens, J. , Leone, M. , Engel, F.B. , Britzen-Laurent, N. , Naschberger, E. , Wittenberg, T. , Stürzl, M. (2018). IFN-γ-response mediator GBP-1 represses human cell proliferation by inhibiting the Hippo signaling transcription factor TEAD. Biochem. J. 475: 2955–2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Knaap, E. , Østergaard, L. (2018). Shaping a fruit: Developmental pathways that impact growth patterns. Semin. Cell Dev. Biol. 79: 27–36. [DOI] [PubMed] [Google Scholar]
- Verdu, C.F. , Childebrand, N. , Marnet, N. , Lebail, G. , Dupuis, F. , Laurens, F. , Guilet, D. , Guyot, S. (2014). Polyphenol variability in the fruits and juices of a cider apple progeny. J. Sci. Food Agric. 94: 1305–1314. [DOI] [PubMed] [Google Scholar]
- Vestal, D.J. , Jeyaratnam, J.A. (2011). The guanylate-binding proteins: emerging insights into the biochemical properties and functions of this family of large interferon-induced guanosine triphosphatase. J. Interferon Cytokine Res. 31: 89–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, D. , Yeats, T.H. , Uluisik, S. , Rose, J.K.C. , Seymour, G.B. (2018). Fruit softening: Revisiting the role of pectin. Trends Plant Sci. 23: 302–310. [DOI] [PubMed] [Google Scholar]
- Weinl, C. , Marquardt, S. , Kuijt, S.J.H. , Nowack, M.K. , Jakoby, M.J. , Hülskamp, M. , Schnittger, A. (2005). Novel functions of plant cyclin-dependent kinase inhibitors, ICK1/KRP1, can act non-cell-autonomously and inhibit entry into mitosis. Plant Cell 17: 1704–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willats, W.G.T. , Orfila, C. , Limberg, G. , Buchholt, H.C. , van Alebeek, G.J.W.M. , Voragen, A.G.J. , Marcus, S.E. , Christensen, T.M.I.E. , Mikkelsen, J.D. , Murray, B.S. , Knox, J.P. (2001). Modulation of the degree and pattern of methyl-esterification of pectic homogalacturonan in plant cell walls. Implications for pectin methyl esterase action, matrix properties, and cell adhesion. J. Biol. Chem. 276: 19404–19413. [DOI] [PubMed] [Google Scholar]
- Wipf, L. , Cooper, D.C. (1938). Chromosome numbers in nodules and roots of red clover, common vetch and garden pea. Proc. Natl. Acad. Sci. USA 24: 87–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, J. , Cui, X. , Yuan, X. , Yu, X. , Sun, J. , Gong, Q. (2016). The Hippo/STE20 homolog SIK1 interacts with MOB1 to regulate cell proliferation and cell expansion in Arabidopsis. J. Exp. Bot. 67: 1461–1475. [DOI] [PubMed] [Google Scholar]
- Zou, Z. , Meng, Z. , Ma, C. , Liang, D. , Sun, R. , Lan, K. (2017). Guanylate-binding protein 1 inhibits nuclear delivery of Kaposi's sarcoma-associated herpesvirus virions by disrupting formation of actin filament. J. Virol. 91: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.