OsRDR6 plays a crucial role in meiosis that might be related to its function in mediating small RNA genesis.
Abstract
RNA-dependent RNA polymerase 6 (RDR6) is a core component of the small RNA biogenesis pathway, but its function in meiosis is unclear. Here, we report a new allele of OsRDR6 (Osrdr6-meiosis [Osrdr6-mei]), which causes meiosis-specific phenotypes in rice (Oryza sativa). In Osrdr6-mei, meiotic double-strand break (DSB) formation is partially blocked. We created a biallelic mutant with more severe phenotypes, Osrdr6-bi, by crossing Osrdr6-mei with a knockout mutant, Osrdr6-edit. In Osrdr6-bi meiocytes, 24 univalents were observed, and no histone H2AX phosphorylation foci were detected. Compared with the wild type, the number of 21-nucleotide small RNAs in Osrdr6-mei was dramatically lower, while the number of 24-nucleotide small RNAs was significantly higher. Thousands of differentially methylated regions (DMRs) were discovered in Osrdr6-mei, implying that OsRDR6 plays an important role in DNA methylation. There were 457 genes downregulated in Osrdr6-mei, including three genes, CENTRAL REGION COMPONENT1, P31comet, and O. sativa SOLO DANCERS, related to DSB formation. Interestingly, the downregulated genes were associated with a high level of 24-nucleotide small RNAs but less strongly associated with DMRs. Therefore, we speculate that the alteration in expression of small RNAs in Osrdr6 mutants leads to the defects in DSB formation during meiosis, which might not be directly dependent on RNA-directed DNA methylation.
INTRODUCTION
In plants, RNA-dependent RNA polymerases (RDRs) have primarily been studied for their role in antiviral defense, and it is becoming increasingly apparent that they also have important endogenous functions, including the control of chromatin structure and the regulation of cellular gene expression (Voinnet, 2008). The effects of plant RDRs are intimately linked to the DICER-like and Argonaute (AGO) functions with which they are associated. Under the action of particular trigger RNAs, RDRs synthesize double-stranded RNAs (dsRNAs) using single-stranded RNA as a template. The dsRNAs are then processed into ∼20- to 25-nucleotide RNA duplexes by Dicer. One strand of the small RNA duplex preferentially binds to an AGO protein to form an RNA-induced silencing complex that silences the target transcripts based on sequence complementarity (Song et al., 2012a).
RNA-dependent RNA polymerase 6 (RDR6) is a critical member of the RDR family, members of which are necessary for the synthesis of dsRNAs in sense-transgene–mediated silencing, for the synthesis of trans-acting small interfering RNA (siRNA), and for antivirus silencing pathways (Kumakura et al., 2009). In Arabidopsis (Arabidopsis thaliana), RDR6 acts together with TEX1, a component of the THO/TREX complex, to regulate megaspore mother cell specification (Su et al., 2017). It also plays a central role in the temporal control of shoot development (Peragine et al., 2004). Arabidopsis RDR6 also participates in regulating transposon expression mediated by Serrate (Ma et al., 2018). In rice (Oryza sativa), loss of function of OsRDR6 often leads to serious defects in embryonic organ development, such as the absence of a shoot apical meristem (SAM; Satoh et al., 2003; Nagasaki et al., 2007). By studying the phenotype of a mutant carrying a weak OsRDR6 allele, researchers were able to document the function of OsRDR6 in regulating reproductive organ development (Song et al., 2012a). However, the role of OsRDR6 in meiosis remains unclear.
Small RNAs are important types of noncoding RNAs that cause RNA-mediated silencing (Castel and Martienssen, 2013). They are commonly grouped into three classes: microRNAs (miRNAs), siRNAs, and PIWI-interacting RNAs. The processing of miRNAs and siRNAs is Dicer dependent, but the processing of PIWI-interacting RNAs is not (Valencia-Sanchez et al., 2006; Aravin et al., 2007). miRNAs are hairpin-derived RNAs with imperfect complementarity to targets and cause translational repression; siRNAs have perfect complementarity to targets and cause transcript degradation (Castel and Martienssen, 2013). Phased siRNA (phasiRNA) is the general term for a class of siRNAs that are arranged regularly in 21- or 24-nucleotide phased intervals in plant genomes (Komiya et al., 2014). During the reproductive stage in rice, large quantities of 21- and 24-nucleotide phasiRNAs are specifically produced by RDRs and Dicers (Johnson et al., 2009; Song et al., 2012b). miRNAs and 21-nucleotide siRNAs usually silence target mRNAs by forming an RNA-induced silencing complex with AGOs and DICER-like proteins. This pathway is named posttranscription gene silencing (PTGS; Cuerda-Gil and Slotkin, 2016). By contrast, 24-nucleotide small RNAs generally participate in RNA-directed DNA methylation (RdDM) to modify gene transcription, resulting in transcriptional gene silencing (Matzke and Mosher, 2014).
Meiosis consists of one round of DNA replication and two rounds of cell division. The first round of cell division is meiosis I in which recombination of homologous chromosomes occurs. Homologous recombination is initiated by programmed DNA double-strand break (DSB) formation, which is catalyzed by Spo11, a protein homologous to subunit A of an archaeal topoisomerase (TopoVIA; Bergerat et al., 1997; Keeney et al., 1997). The conserved protein TopoVIBL, which shares strong structural similarity to the TopoVIB subunit of TopoVI DNA topoisomerase, was reported to be required for meiotic DSB formation in mice (Robert et al., 2016).
In plants, in addition to functionally conserved proteins such as SPO11 and meiotic topoisomerase VIB-like (MTOPVIB; Stacey et al., 2006; Yu et al., 2010; Vrielynck et al., 2016; Xue et al., 2016), proteins with plant-specific roles in DSB formation have been identified. In rice, CENTRAL REGION COMPONENT1 (CRC1), the homolog of PCH2 in Caenorhabditis elegans, has been reported to be essential for DSB formation (Miao et al., 2013). P31comet, the human mitotic arrest-deficient 2 (Mad2) binding protein, participates in the spindle checkpoint and coordinates cell cycle events in mitosis (Xia et al., 2004). However, its ortholog in rice plays a key role in DSB formation by associating with CRC1 (Ji et al., 2016). In Arabidopsis, the meiosis-specific cyclin SOLO DANCERS (SDS) plays a specific role in regulating synapsis in prophase I (Azumi et al., 2002). But its rice ortholog, OsSDS, was reported to be necessary for meiotic DSB formation (Wu et al., 2015).
In this study, we report that OsRDR6 might function in meiotic DSB formation in rice. Mutation of OsRDR6 caused alterations in small RNA levels and ultimately led to the downregulation of genes related to DSB formation.
RESULTS
Characterization of a Sterile Mutant in Rice
A sterile mutant induced by 60Co γ-ray irradiation was obtained from the indica rice var Zhongxian 3037. The mutant showed normal vegetative growth but was completely sterile (Figure 1A). The anthers of the mutant appeared similar to those of the wild type. However, they were pale yellow and failed to produce viable pollen (Figure 1A). The female gametes of the mutant were also defective; no functional embryonic sac was produced (Figure 1A), and no seed was set when the mutant pistil was pollinated with the wild-type pollen grains. By examining the progeny of a self-fertilized heterozygous plant, we found that the segregation ratio of fertile plants to sterile plants was ∼3:1 (172 fertile and 56 sterile plants), implying that the sterile phenotype is controlled by a single recessive nuclear gene (χ2 = 0.0234, P > 0.05).
Figure 1.
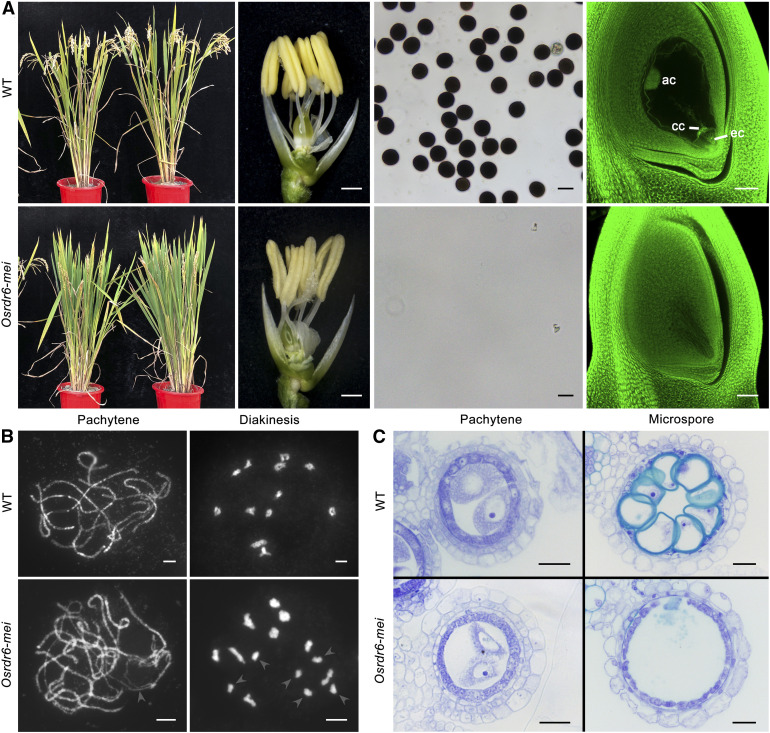
Phenotypic Analysis of Osrdr6-mei.
(A) From left to right: wild-type (WT; top) and Osrdr6-mei (bottom) plants, spikelets with the lemma and palea removed, pollen grains, and embryo sac. ac, antipodal cell; cc, central cell; ec, egg cell. Bars = 500 μm for spikelets; bars = 50 μm for pollen and embryo sacs.
(B) Meiotic chromosome behavior of meiocytes from wild-type (WT) and Osrdr6-mei plants. In Osrdr6-mei meiocytes, arrowheads indicate unpaired chromosomes at pachytene and univalents at diakinesis. Bars = 5 μm.
(C) Transverse section analysis of anthers from the wild type (WT) and Osrdr6-mei. The images are cross sections of a single locule. Bars = 20 μm.
The target gene was mapped to the long arm of chromosome 1, and its position was further delimited to a region of 110 kb (Supplemental Figure 1A). Within this region, one candidate gene (LOC_Os01g34350) was found to have a single base substitution at the nucleotide position 1862 (G to T) of the cDNA, resulting in the change of Arg576 to Leu (Supplemental Figure 1A). LOC_Os01g34350 encodes the RDR6 in rice. We named the mutant Osrdr6-meiosis (Osrdr6-mei) based on its abnormal chromosome behavior in meiosis. No mutation in any other gene was found in the fine-mapping region. To confirm that the sterility of the mutant plants indeed results from the mutation of OsRDR6, a complementation experiment was performed. The sterility phenotype was successfully rescued by introducing the wild-type genomic sequence of OsRDR6 into the homozygous mutant (Supplemental Figure 1B), further verifying that the defects were caused by the mutation of OsRDR6.
OsRDR6 Is Required for DSB Formation in Rice
We investigated the meiotic chromosome behavior of meiocytes from the wild-type and Osrdr6-mei plants. In the wild type, synaptonemal complex (SC) formation was complete at pachytene, and chromosomes were present as thick threads. Twelve bivalents became highly condensed at diakinesis. However, in Osrdr6-mei, some homologous chromosomes did not pair at pachytene and univalents were clearly detected at diakinesis (Figure 1B).
To monitor homologous pairing, a fluorescence in situ hybridization (FISH) assay was performed using a probe specific to 5S rDNA. The 5S rDNA probe signals were well paired at pachytene in the 142 wild-type meiocytes examined (Supplemental Figure 2A). However, in 33.5% of Osrdr6-mei meiocytes (n = 173) at pachytene, the probe signals were separated (Supplemental Figures 2B and 2C). These results confirmed that homologous pairing of some homologous chromosomes did not proceed normally in Osrdr6-mei.
In addition, we found that the meiotic process was blocked in Osrdr6-mei meiocytes, most of which exhibited variable degrees of meiotic arrest. In particular, about half of the meiocytes were arrested at pachytene (Supplemental Figure 3; Supplemental Table 1). We observed that the anthers from the wild type and Osrdr6-mei did not show significant differences in development at the three-layer stage and preleptotene (Supplemental Figure 4). Even when entering pachytene, all somatic cell layers were well developed (Figure 1C). However, the defective meiocytes of Osrdr6-mei gradually began to disintegrate and no microspores were observed in anthers after meiosis (Figure 1C), indicating that the meiocytes might have undergone apoptosis.
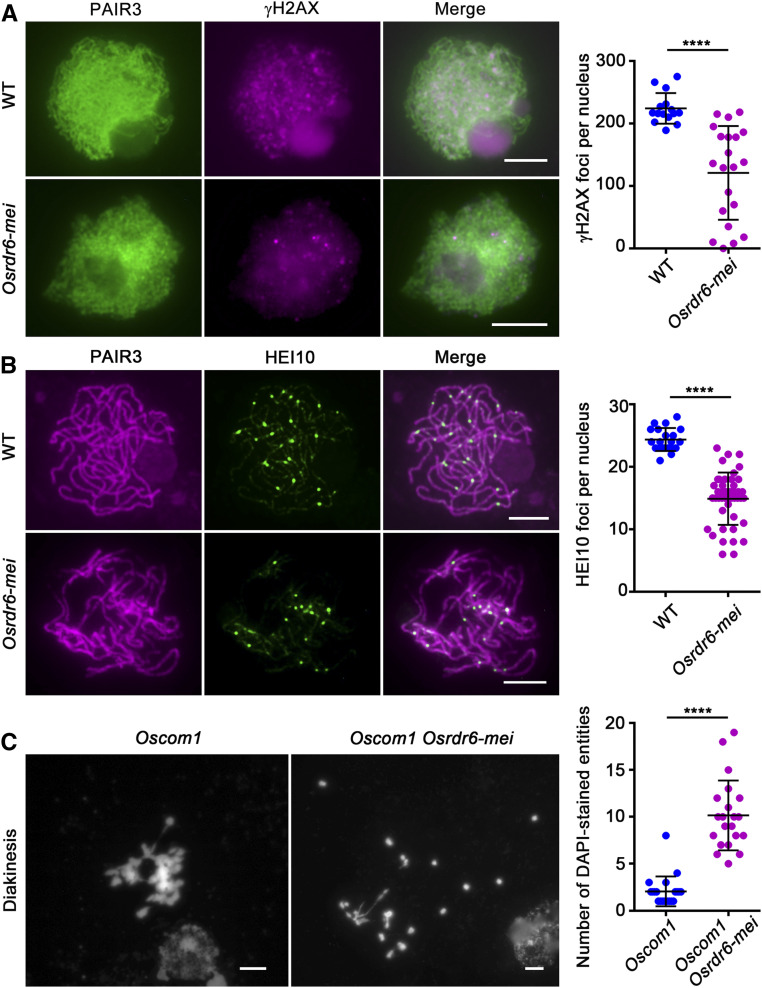
DSB formation is necessary for homologous chromosome pairing during meiosis in most eukaryotes, including rice. The inability of some homologous chromosomes to pair suggests that DSB formation may be blocked in Osrdr6-mei. Phosphorylation of histone H2AX from leptotene to early zygotene is induced by meiotic DSB formation; therefore, immunostaining with antibodies specifically recognizing the phosphorylated form of rice histone H2AX (γH2AX) was performed to monitor DSB formation. Dot-like γH2AX signals were observed at zygotene in both the wild-type and Osrdr6-mei meiocytes. However, the average number of γH2AX foci in Osrdr6-mei (120.8 ± 16.4; range, 0 to 218; n = 21) was significantly lower than that in the wild type (223.9 ± 6.3; range, 189 to 275; n = 15), indicating that the number of DSBs was decreased (Figure 2A). Since DSB formation is necessary for the generation of crossovers, we next performed an immunostaining assay to visualize HEI10, a marker of class I crossovers (Wang et al., 2012), in wild-type and Osrdr6-mei meiocytes. At late pachytene, the number of bright HEI10 foci in Osrdr6-mei meiocytes (14.9 ± 0.69; range, 6 to 23; n = 44) was lower than that in wild-type meiocytes (24.4 ± 0.4; range, 21 to 28; n = 20; Figure 2B), indicating that the maturation of class I crossovers in Osrdr6-mei was affected.
Figure 2.
Cytological Analysis of Osrdr6-mei.
(A) Immunofluorescence imaging of γH2AX signals on meiotic chromosome spreads of wild-type (WT) and Osrdr6-mei meiocytes. ****P < 0.0001, two-tailed Student’s t test.
(B) Dual immunolocalization of PAIR3 (red) and HEI10 (green) in wild-type (WT) and Osrdr6-mei meiocytes. PAIR3 was used to visualize the meiotic chromosomes. ****P < 0.0001, two-tailed Student’s t test.
(C) Meiotic chromosome behavior in Oscom1 and Oscom1Osrdr6-mei. Chromosomes were stained with DAPI. Numbers of DAPI-stained entities per cell at diakinesis in Oscom1 and Oscom1Osrdr6-mei plants were analyzed. ****P < 0.0001, two-tailed Student’s t test. Bars = 5 μm.
To further confirm the role of OsRDR6 in meiotic DSB formation, the Oscom1 Osrdr6-mei double mutant was constructed. In rice, loss of function of OsCOM1 leads to nonhomologous chromosome entanglements (Ji et al., 2012). In Oscom1 Osrdr6-mei double mutants, the formation of abnormal chromosome associations in Oscom1 was partially repressed; more free-univalents were observed (Figure 2C). This result provided direct evidence demonstrating that fewer DSBs were formed in Osrdr6-mei.
Analysis of a Biallelic Mutant, Osrdr6-bi, Verified That OsRDR6 Is Required for DSB Formation
Different from other Osrdr6 alleles identified to date, Osrdr6-mei shows an obvious meiotic phenotype. To further investigate the role of OsRDR6 in rice meiosis, we created a knockout mutant allele, Osrdr6-edit (Osrdr6-edi), using the CRISPR-Cas9 system. However, we could not obtain a plant homozygous for this allele. We thus crossed Osrdr6-edi/+ and Osrdr6-mei/+ to create the biallelic mutant Osrdr6-edi/Osrdr6-mei (Osrdr6-bi; Figure 3A). Like Osrdr6-mei plants, the Osrdr6-bi plants grew normally but were sterile. Osrdr6-bi anthers were also similar to those of the wild type at early stages of development (Supplemental Figure 4).
Figure 3.
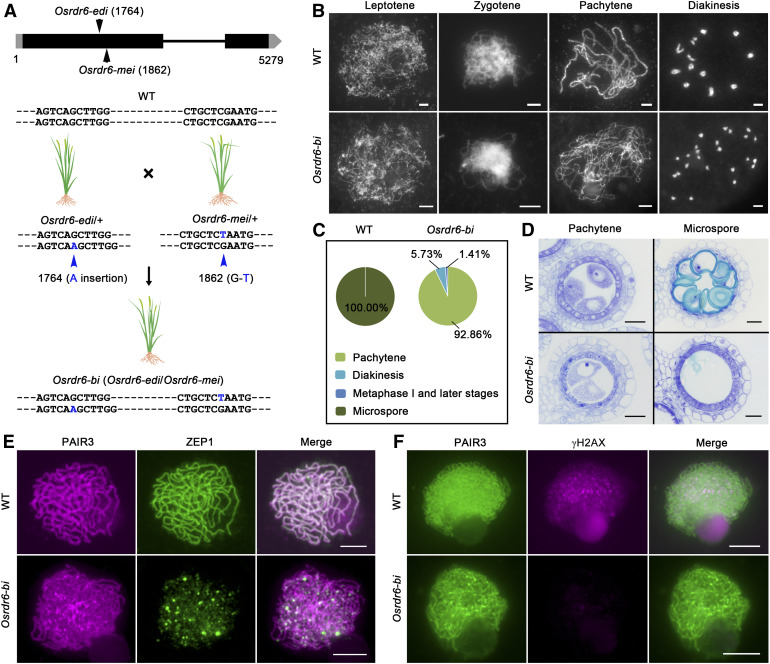
Genetic and Cytological Analysis of Osrdr6-bi.
(A) Mutation sites of Osrdr6-mei, Osrdr6-edi, and Osrdr6-bi.
(B) Meiotic chromosome behavior of meiocytes from the wild-type (WT) and Osrdr6-bi plants. Bars = 5 μm.
(C) Proportion of meiocytes at different stages in 8-mm anthers from the wild type (WT) and Osrdr6-bi.
(D) Transverse sections of the anthers from the wild type (WT) and Osrdr6-bi. Images are cross sections of a single locule. Bars = 20 μm.
(E) Dual immunolocalization of PAIR3 (red) and ZEP1 (green) in wild-type (WT) and Osrdr6-bi meiocytes. PAIR3 was used to visualize the meiotic chromosomes. Bars = 5 μm.
(F) Dual immunolocalization of PAIR3 (green) and γH2AX (red) in WT and Osrdr6-bi meiocytes. Bars = 5 μm.
We then investigated the meiotic chromosome behavior of meiocytes from Osrdr6-bi in detail (Figure 3B). In the wild type, meiotic chromosomes condensed as thin threads at leptotene. The chromosomes continued to condense at subsequent stages of meiosis, and the thin threads began to synapse at zygotene. In Osrdr6-bi, no obvious difference from the wild type was observed at leptotene and zygotene, but at pachytene, neither homologous chromosome pairing nor synapsis was observed. Subsequently, 24 univalents were clearly detected at diakinesis. We also found that the meiotic process was blocked in Osrdr6-bi meiocytes, more than 90% of which were arrested at pachytene (Figure 3C; Supplemental Table 2). In addition, the defective meiocytes of Osrdr6-bi might undergo apoptosis (Figure 3D).
ZEP1, the homolog of Saccharomyces cerevisiae ZIP1, is regarded as a central component of SC in rice (Wang et al., 2010). Thus, immunostaining for ZEP1 was conducted in the wild-type and Osrdr6-bi meiocytes (Figure 3E). In wild-type meiocytes, ZEP1 signals aligned perfectly along the entire lengths of the paired chromosomes at pachytene. By contrast, in Osrdr6-bi meiocytes, ZEP1 signals appeared as punctate foci on the chromosomes, indicating that the SC did not form normally in Osrdr6-bi. The process of DSB formation was also investigated in Osrdr6-bi by performing γH2AX immunostaining. No obvious γH2AX signal was observed in Osrdr6-bi, indicating that DSB formation was completely blocked (Figure 3F).
In the FISH assay, the probe signals of 5S rDNA were well paired at pachytene in the wild type but separated in Osrdr6-bi (Figure 4A). Two bulked oligonucleotide probes, specific to the short (11S) and long (11L) arms of chromosome 11, were also prepared and used in the FISH assay. In Osrdr6-bi, separated signals of these two probes were observed at pachytene, whereas in the wild type, the probe signal arrays were well paired (Figure 4A). Both assays indicated that homologous pairing was aberrant in Osrdr6-bi, which might be due to the blocking of DSB formation.
Figure 4.
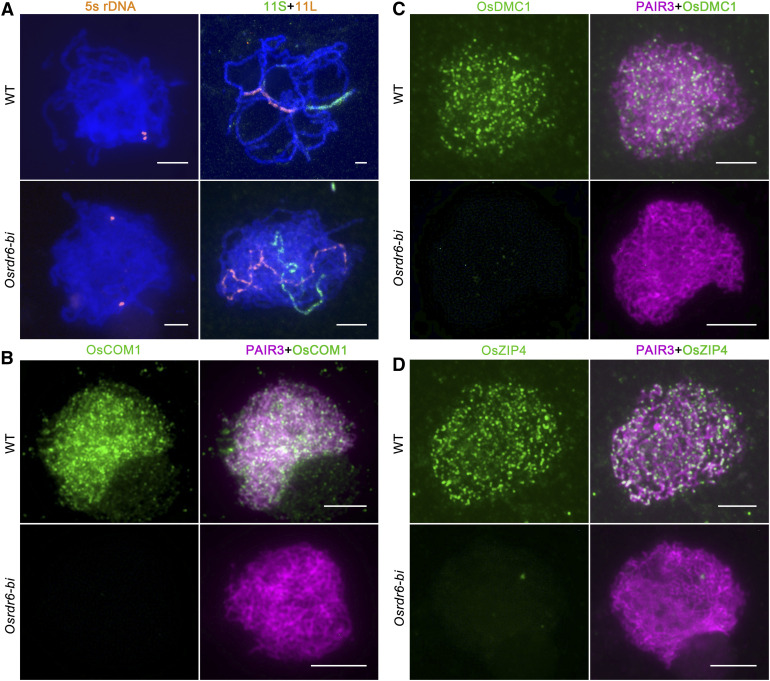
FISH and Immunolocalization Assays in the Wild-Type and Osrdr6-bi Meiocytes.
(A) Chromosome pairing in the wild type (WT) and Osrdr6-bi at pachytene analyzed by FISH using 5S rDNA (orange), bulked oligonucleotide probes specific to the 11S (green) and 11L (orange) of chromosome 11. Meiotic chromosomes are stained with DAPI (blue). The images are pseudocolored. Bar = 5 μm.
(B) Dual immunolocalization of PAIR3 (red) and OsCOM1 (green) in wild-type (WT) and Osrdr6-bi meiocytes. Bar = 5 μm.
(C) Dual immunolocalization of PAIR3 (red) and OsDMC1 (green) in wild-type (WT) and Osrdr6-bi meiocytes. Bar = 5 μm.
(D) Dual immunolocalization of PAIR3 (red) and OsZIP4 (green) in wild-type (WT) and Osrdr6-bi meiocytes. Bar = 5 μm.
To further confirm the absence of DSB formation in Osrdr6-bi, immunostaining of the wild-type and Osrdr6-bi meiocytes was conducted using antibodies against OsCOM1, OsDMC1, and OsZIP4, all of which are essential for rice DSB repair. OsCOM1 is essential for DSB processing (Ji et al., 2012); OsDMC1 mediates single-strand invasion during meiotic recombination (Wang et al., 2016); and OsZIP4, which is a component of the ZMM complex (a protein group required for crossover formation, including ZIP1, ZIP2, ZIP3, ZIP4, MSH4, MSH5, and MER3), is essential for interference-sensitive crossover formation in rice (Shen et al., 2012). In the wild-type meiocytes, OsCOM1, OsDMC1, and OsZIP4 signals were observed as punctuate foci on chromosomes at zygotene, but in Osrdr6-bi, no signals were observed (Figures 4B to 4D). These results indicate that OsRDR6 is essential for meiotic DSB formation.
Small RNA Biosynthesis Is Affected in the Osrdr6-mei Mutants
Although only reproductive cells undergo meiosis, other anther cells are important for the normal progression of meiosis. Ono et al. (2018) reported that ETERNAL TAPETUM1 (EAT1), which is localized in tapetal cells, activates meiotic small RNA biogenesis in the anther tapetum. They concluded that a subset of 24-nucleotide phasiRNAs triggered by EAT1 could move from tapetal cells to reproductive cells to support meiosis in rice anthers. Therefore, to gain a comprehensive view of the small RNAs expressed during rice meiosis in the wild type and Osrdr6-mei, we isolated whole anthers at the leptotene-like stage. Six small RNA libraries, three from the wild type and three from Osrdr6-mei, were created and sequenced. The small RNA expression levels were highly correlated between biological replicates, with correlation coefficients greater than 0.9 (Supplemental Figure 5).
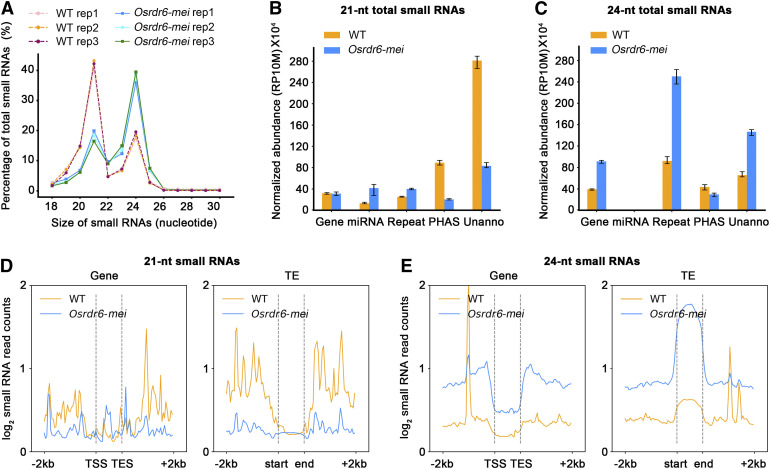
In rice, most small RNAs range from 21 to 24 nucleotides in length (Song et al., 2012a). Our results revealed a major peak in the lengths of small RNAs in meiotic anthers at 21 nucleotides and a smaller peak at 24 nucleotides (Figure 5A). Therefore, we paid the most attention to RNAs with lengths of 21 and 24 nucleotides. During the reproductive stage in rice, large quantities of 21- and 24-nucleotide phasiRNAs are specifically produced (Song et al., 2012b). Here, when the phase score was set to the gold standard (≥30), 234 21-nucleotide phasiRNA loci (phased loci) and 199 24-nucleotide phased loci were identified in the wild-type meiotic anthers. These phased loci were randomly distributed on all 12 chromosomes. However, the numbers of 21- and 24-nucleotide phased loci in Osrdr6-mei were only 122 and 98, respectively (Supplemental Figure 6). Moreover, the locations of the phased loci were different in Osrdr6-mei; some new loci were identified, while some loci disappeared (Supplemental Figure 6; Supplemental Data Sets 1 and 2). This is consistent with the previous finding that OsRDR6 is required for the biogenesis of both 21- and 24-nucleotide phasiRNAs (Song et al., 2012a).
Figure 5.
Size Distribution, Annotation, and Spatial Distribution of Small RNAs from High-Throughput Sequencing Data.
(A) Size distribution of small RNAs in the wild type (WT) and Osrdr6-mei. The abundance of small RNAs in each size class is shown as a percentage of the total abundance of small RNAs.
(B) Annotation of 21-nucleotide (nt) small RNAs in the wild type (WT) and Osrdr6-mei. RP10M, reads per 10 million.
(C) Annotation of 24-nucleotide (nt) small RNAs in the wild type (WT) and Osrdr6-mei.
(D) Distribution of 21-nucleotide (nt) small RNAs around genes (left) and TEs (right). Profiles of average abundance in the wild type (WT) and Osrdr6-mei are shown.
(E) Distribution of 24-nucleotide (nt) small RNAs around genes (left) and TEs (right). Profiles of average abundance in the wild type (WT) and Osrdr6-mei are shown.
In Osrdr6-mei, the number of 21-nucleotide small RNAs was dramatically lower than in the wild type, while the number of 24-nucleotide small RNAs was significantly higher (Figure 5A). Further analysis showed that the 21-nucleotide small RNAs that exhibited the most significant reduction in abundance in Osrdr6-mei were preferentially derived from phased loci and unannotated regions in the genome (Figure 5B), whereas the 24-nucleotide small RNAs that were significantly more abundant in number in Osrdr6-mei were derived from gene loci, repeat-associated regions, and unannotated regions (Figure 5C). Although small RNAs have been artificially grouped into different classes (miRNA, siRNA, etc.), we know very little about the function of each class in meiosis. For the purpose of this study, we only divided the small RNAs by size.
Small RNAs regulate gene and transposable element (TE) expression through the PTGS and RdDM pathways; therefore, we analyzed the small RNAs located around gene and TE loci. At gene loci, the number of 21-nucleotide small RNAs mapping to the 5′ and 3′ regulatory regions of Osrdr6-mei was lower compared with that in the wild type, while the number mapping to the gene body was higher (Figure 5D). At TE loci, the number of 21-nucleotide small RNAs mapping to the 5′ and 3′ regulatory regions was also dramatically lower in Osrdr6-mei, but no significant difference was found in the TE body (Figure 5D). For 24-nucleotide small RNAs, the number mapping to gene and TE loci was higher in Osrdr6-mei compared with that in the wild type (Figure 5E).
Small RNAs can usually be mapped to the genome as clusters. We found that Osrdr6-mei had fewer 21-nucleotide clusters but more 24-nucleotide clusters compared with the wild type (Supplemental Figure 7A). Many 24-nucleotide clusters specific to the mutant were identified (Supplemental Figure 7B). In addition, we selected clusters shared among replicates and identified clusters showing differential expression between the wild type and Osrdr6-mei. We found 748 (90% of 828) 21-nucleotide and 38,036 (82% of 46,583) 24-nucleotide small RNA clusters with differential expression (Supplemental Table 3; Supplemental Data Set 3). Taking these results together, we speculate that OsRDR6 modulates the small RNA levels in meiotic anthers.
Increased DNA Methylation in Osrdr6-mei
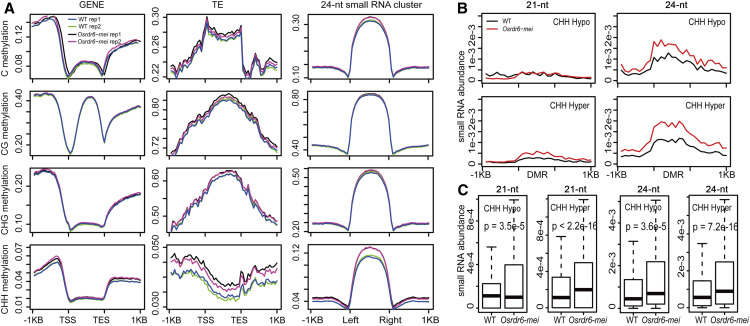
To investigate the changes in the methylome caused by OsRDR6 malfunction, we performed whole genome bisulfite sequencing (WGBS) of the wild-type and Osrdr6-mei anthers at the same stage as those used for small RNA sequencing. The quality of the methylome data was high (Supplemental Table 4). We found similar levels of CG and CHG methylation at non-TE genes and adjacent regions, but substantially higher CHH methylation at TE-surrounding regions and regions 1 kb upstream or downstream of non-TE genes in Osrdr6-mei (Figure 6A). This is consistent with the increased abundance of 24-nucleotide small RNA in Osrdr6-mei. We next identified differentially methylated regions (DMRs) between the wild type and Osrdr6-mei (Supplemental Data Set 4). More CHH hyper-DMRs were found in Osrdr6-mei as expected (Supplemental Table 5). In addition, more hyper-DMRs of CG and CHG were also found, implying that OsRDR6 might play a negative role in DNA methylation.
Figure 6.
Relationship between DNA Methylation and Small RNA.
(A) DNA methylation level surrounding non-TE genes (left), TE-related genes (middle), and 24-nucleotide (nt) small RNA clusters (right) with significant differential expression calculated as the ratio of normalized methylated C to total C. Left and right indicate left border and right border of small RNA clusters, respectively. TES, transcription end site; TSS, transcription start site.
(B) 21-nucleotide (nt; left) and 24-nt (right) small RNA abundance (count per million) surrounding DMRs. Hyper, significantly higher DNA methylation of DMR in Osrdr6-mei.
(C) Boxplots of small RNA abundance and statistical tests of differences between the wild type (WT) and Osrdr6-mei.
To elucidate the relationship between DNA methylation and changes in the abundance of small RNAs related to OsRDR6, we first investigated the overall levels of DNA methylation at 24-nucleotide small RNA clusters with differential expression. The 21-nucleotide small RNA clusters were not included in this analysis because they were too few in number (only 748). We found substantially higher CHH methylation in the regions of differentially expressed 24-nucleotide small RNA clusters in Osrdr6-mei (Figure 6A). We also observed significantly higher abundance of 24-nucleotide small RNAs and 21-nucleotide small RNAs in CHH hyper-DMRs in the Osrdr6-mei background (Figures 6B and 6C; Supplemental Figure 8). Moreover, 87% of 24-nucleotide small RNA clusters overlapping with CHH DMR regions were differentially expressed (Supplemental Table 5). Interestingly, 24-nucleotide small RNA abundance was also significantly higher in CHH hypo-DMRs (Figures 6B and 6C), implying an intricate relationship between 24-nucleotide small RNAs and DNA methylation, which is consistent with the finding in a previous study in Arabidopsis that DNA methylation still occurs even if all Dicers are mutated (Yang et al., 2016).
Expression Levels of CRC1, P31comet, and OsSDS Were Lower in Osrdr6 Mutants
We next asked whether the change in small RNA levels affected gene expression. Four (two replicates of the wild type and two of Osrdr6-mei) of the six samples used for small RNA sequencing were further used for transcriptome sequencing (RNA sequencing). The correlation coefficient for each pair of biological replicates was greater than 0.9 (Supplemental Figure 9). Compared with the wild type, 778 differentially expressed genes (DEGs) were identified in Osrdr6-mei; 321 genes were upregulated and 457 genes were downregulated (Figure 7A; Supplemental Data Set 5). In addition, 422 (54%) of 778 DEGs were associated with differentially expressed 24-nucleotide small RNA clusters located in the regions from 2 kb upstream to 2 kb downstream of genes (Supplemental Data Set 3). Previously, 24-nucleotide siRNAs were reported to be predominantly associated with miniature inverted repeat transposable elements (MITEs; Wei et al., 2014). In our study, we found that the small RNA clusters mapping to the majority (264 of 422) of differentially expressed small RNA cluster–associated DEG-adjacent regions were related to repeat sequences (Supplemental Data Set 5). All these results implied a possible comprehensive effect of small RNAs related to OsRDR6 on gene expression.
Figure 7.
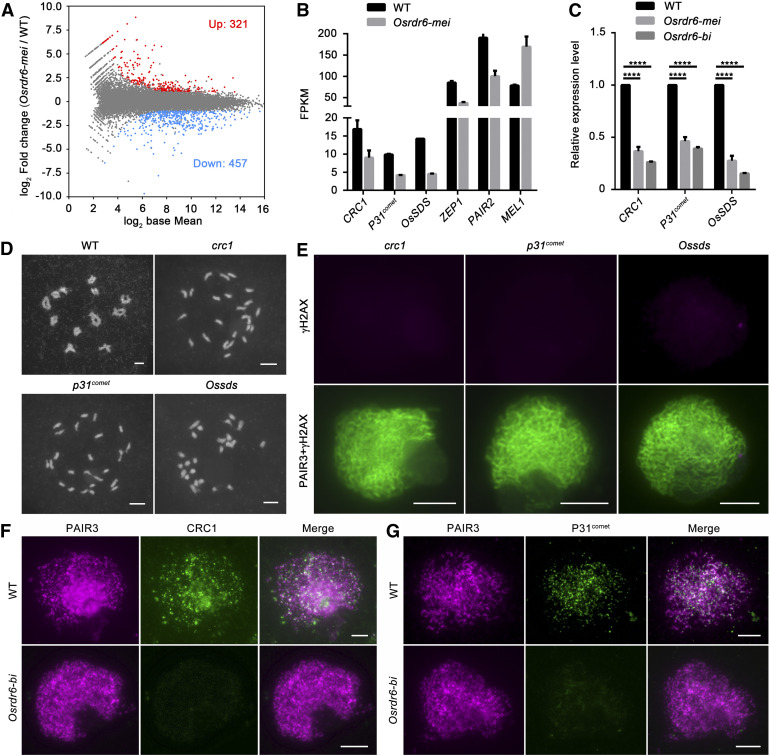
CRC1, P31comet, and OsSDS Are Downregulated in the Osrdr6 Mutants.
(A) M-versus-A plot showing the expression changes between wild type (WT) and Osrdr6-mei; 321 up- and 457 downregulated DEGs compared with the WT were detected in Osrdr6-mei.
(B) Fragments per kilobase of transcript per million fragments mapped (FPKM) values for CRC1, P31comet, OsSDS, ZEP1, PAIR2, and MEL1 in the wild type (WT) and Osrdr6-mei.
(C) Expression levels of CRC1, P31comet, and OsSDS in the wild type (WT), Osrdr6-mei, and Osrdr6-bi confirmed by real-time PCR. ****P < 0.0001, two-tailed Student’s t test. The expression level in WT was normalized to 1.
(D) Meiotic chromosome behavior in meiocytes from the wild-type (WT), crc1, p31comet, and Ossds plants. Bar = 5 μm.
(E) Immunodetection of γH2AX on meiotic chromosome spreads of crc1, p31comet and Ossds meiocytes. PAIR3 signals showing meiotic chromosomes (green); γH2AX signals are in red. Bar = 5 μm.
(F) Dual immunolocalization of PAIR3 (red) and CRC1 (green) in the wild-type (WT) and Osrdr6-bi meiocytes. Bar = 5 μm.
(G) Dual immunolocalization of PAIR3 (red) and P31comet (green) in the wild-type (WT) and Osrdr6-bi meiocytes. Bar = 5 μm.
To date, dozens of genes have been reported to be involved in the regulation of rice meiosis. We searched for meiosis-related genes in our RNA sequencing data and found that the expression levels of multiple genes were altered, such as CRC1, P31comet, OsSDS, PAIR2, MEL1, and ZEP1 (Figure 7B). The downregulation of CRC1, P31comet, and OsSDS caught our attention; previously, we showed that these genes are all essential for meiotic DSB formation in rice (Figures 7D and 7E; Miao et al., 2013; Wu et al., 2015; Ji et al., 2016). Real-time PCR results also showed that the expression levels of CRC1, P31comet, and OsSDS were dramatically lower in Osrdr6-mei than in the wild type; what is more interesting is that the expression levels of these genes were even lower in Osrdr6-bi (Figure 7C).
We next determined the localization of CRC1 and P31comet in wild-type and Osrdr6-bi chromosomes. Signals of both proteins could be clearly observed as punctuate foci at leptotene in the wild-type chromosomes, but no signal could be detected in Osrdr6-bi chromosomes (Figures 7F and 7G). We speculated that OsRDR6 modulates meiotic DSB formation by mediating the expression levels of CRC1, P31comet, and OsSDS in rice.
Genes Downregulated and Upregulated in Osrdr6-mei Are Regulated by Different Pathways
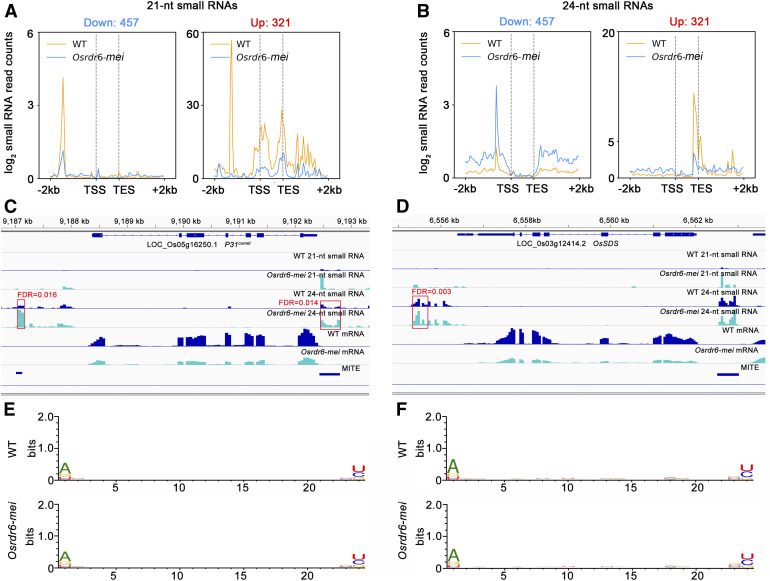
The relationship between the alteration in gene expression levels and small RNA levels in Osrdr6-mei was further investigated. We first analyzed the changes in small RNA abundance around DEGs. For downregulated genes, we did not find significant differences in the numbers of 21-nucleotide small RNAs mapping to the regulatory regions or gene bodies between Osrdr6-mei and the wild type (Figure 8A). However, a significantly higher number of 24-nucleotide small RNAs mapped to the promoters and 3′ regulatory regions in Osrdr6-mei compared with the wild type (Figure 8B). For upregulated genes, the number of 21-nucleotide small RNAs in Osrdr6-mei mapping to the regulatory regions and gene bodies was dramatically lower than that in the wild type (Figure 8A), while the number of 24-nucleotide small RNAs mapping to gene bodies in Osrdr6-mei was lower but the number mapping to regulatory regions was higher (Figure 8B). We speculate that the increased 24-nucleotide small RNA levels at the promoter and 3′ regulatory regions might be associated with gene silencing (downregulation) in Osrdr6-mei and that the upregulation of genes might be the result of differential expression of both 21- and 24-nucleotide small RNAs. We found that differentially expressed 24-nucleotide small RNA clusters were located in the regions adjacent to P31comet and OsSDS (Figures 8C and 8D). However, no differentially expressed small RNA clusters were found around CRC1 (Supplemental Figure 10A), indicating that the mechanism modulating CRC1 expression might be more complicated than that modulating P31comet and OsSDS expression. Interestingly, the differentially expressed 24-nucleotide small RNA clusters in the region adjacent to P31comet were associated with MITEs (Figure 8C; Supplemental Data Set 5).
Figure 8.
Association between Small RNA Level and Gene Expression.
(A) Distribution of 21-nucleotide (nt) small RNAs surrounding genes downregulated (left) and upregulated (right) in Osrdr6-mei. Profiles of average abundance in the wild type (WT) and Osrdr6-mei are shown.
(B) Distribution of 24-nucleotide (nt) small RNAs surrounding genes downregulated (left) and upregulated (right) in Osrdr6-mei. Profiles of average abundance in the wild type (WT) and Osrdr6-mei are shown.
(C) Small RNA abundance and MITE loci surrounding P31comet. Red box indicating the small RNA clusters with significant differential expression between the wild type (WT) and Osrdr6-mei. FDR, false discovery rate.
(D) Small RNA abundance and MITE loci surrounding OsSDS. Red box indicating the small RNA cluster with significant differential expression between the wild type (WT) and Osrdr6-mei. FDR, false discovery rate.
(E) Relative nucleotide bias at each position of the 24-nucleotide (nt) small RNAs mapping to the promoter regions of downregulated genes.
(F) Relative nucleotide bias at each position of the 24-nucleotide (nt) small RNAs mapping to the 3′ regulatory regions of downregulated genes.
According to previous studies, 21-nucleotide small RNAs can modulate gene expression via the PTGS pathway, and 24-nucleotide small RNAs exert their functions through the RdDM pathway (Matzke and Mosher, 2014; Cuerda-Gil and Slotkin, 2016). Small RNAs need to be assembled with AGO proteins to be functional. Sorting of small RNAs into AGO proteins in plants is directed by the 5′ terminal nucleotide (Mi et al., 2008; Wu et al., 2009). We analyzed the relative nucleotide bias at each position of the 24-nucleotide small RNAs mapping to regions around the downregulated genes. The 5′ terminal nucleotides of the 24-nucleotide small RNAs that mapped to the promoter and 3′ regulatory regions of the downregulated genes were both biased toward adenine (A; Figures 8E and 8F); thus, most of these small RNAs are predicted to be sorted to AGO2 or AGO4. AGO2 and AGO4 have been reported to function in RdDM (Matzke and Mosher, 2014; Cuerda-Gil and Slotkin, 2016).
We next checked the effect of DMRs on differential gene expression between the wild type and Osrdr6-mei. We found that 106 (14%) DEGs were associated with DMRs (Supplemental Data Set 5). However, CRC1, P31comet, and OsSDS were not in this list, indicating that small RNAs might not modulate the expression of these genes by directly changing the level of DNA methylation. Furthermore, the percentage of DEGs associated with DMRs was significantly lower than that (54%) of DEGs related to 24-nucleotide small RNA clusters (chi-square test, P < 2.2e–16), indicating that 24-nucleotide small RNAs possibly play many other important roles in modulating gene expression in Osrdr6-mei other than RdDM.
Judging from the distribution of small RNAs in regions around the upregulated genes, their regulatory mechanism is more complicated than that of the downregulated genes. We selected a few typical regions surrounding upregulated genes to analyze the relative nucleotide bias of small RNAs. The 5′ terminal nucleotides of 21-nucleotide small RNAs mapping to gene bodies were biased toward uracil (Supplemental Figure 10B). Therefore, most of these small RNAs are predicted to be sorted to AGO1. Meanwhile, the 5′ terminal nucleotides of 24-nucleotide small RNAs mapping to the promoter and 3′ regulatory regions were biased toward A (Supplemental Figures 10C and 10D), which indicates that most of these small RNAs are sorted to AGO2 or AGO4. We speculate that 21- and 24-nucleotide small RNAs might modulate the expression of upregulated genes by binding to different AGOs.
DISCUSSION
OsRDR6 is essential for small RNA biogenesis in rice. Based on its core role in small RNA processing, it plays an important role in plant development and immunity (Satoh et al., 2003; Jiang et al., 2012). To date, multiple alleles of OsRDR6 have been isolated, and increasingly sophisticated regulation networks associated with it have been drawn. More than 10 years ago, OsRDR6 was verified to be crucial for SAM development; mutants with a strong OsRDR6 allele lack a complete SAM in the embryo (Nagasaki et al., 2007). In the following years, multiple other alleles of OsRDR6 were identified. Toriba et al. (2010) showed that OsRDR6 is essential for stamen development. Interestingly, a temperature-sensitive mutant of OsRDR6, Osrdr6-1, was identified by Song et al. (2012a). Osrdr6-1 produces a rod-like lemma when grown at high temperature but produces a normal lemma when grown at low temperature. However, it is still unclear how OsRDR6 functions in rice meiosis.
In this study, we isolated a new allele of OsRDR6 (Osrdr6-mei), which causes a meiosis-specific phenotype. By introducing the wild-type genomic sequence of OsRDR6 into the homozygous Osrdr6-mei mutant, we confirmed that sterility was due to the OsRDR6 mutation. We also created a stronger biallelic mutant, Osrdr6-bi, and showed that OsRDR6 is indispensable for meiotic DSB formation. The disruption of small RNA levels and defective meiotic phenotypes in Osrdr6-mei suggest that regulation of rice meiosis is RDR6 dependent. High-throughput sequencing of small RNAs showed that OsRDR6 is important for regulating small RNA levels in meiotic anthers. In addition, we found that the regulation of small RNA levels by OsRDR6 is important for whole genome DNA methylation. In Osrdr6-mei, altered small RNA levels result in the downregulation of three genes related to DSB formation, which may lead to abnormal DSB formation in meiocytes.
OsRDR6 Is Important for Maintaining Both Small RNA Levels and DNA Methylation in Meiotic Anthers
Small RNA sequencing, transcriptome sequencing, and WGBS were performed for both the wild type and Osrdr6-mei. In order to exclude the possibility of interference from other mutated genes, the plant materials (the wild type and Osrdr6-mei) used for these assays were obtained from the offspring of Osrdr6-mei/+. Based on the transcriptome data, we found only two genes with sequence differences among all the detected expressed genes (Supplemental Table 6). One gene is OsRDR6, and the other gene is annotated as a multidrug resistance–like ATP‐binding cassette transporter in the Rice Genome Annotation Project and has not been reported to be related to small RNAs or DNA methylation. Therefore, we are confident that the changes in small RNA and DNA methylation in the mutant are caused by the mutation of OsRDR6.
In Osrdr6-mei, small RNA levels were dramatically altered. Levels of both 21- and 24-nucleotide phasiRNAs were lower in the mutant, revealing that the biogenesis of phasiRNAs during rice meiosis is dependent on OsRDR6. However, we were surprised to find that the mutant had more 24-nucleotide small RNAs than the wild type on average. The reason for the increased levels of these small RNAs is still unclear. It might be that OsRDR6-dependent small RNAs repress the biogenesis of abnormal small RNAs, but we cannot rule out the possibility that this might be a compensation mechanism for the loss of function of OsRDR6. The increased 24-nucleotide small RNA levels might result in a change in genome-wide DNA methylation level. We found that 24-nucleotide small RNA abundance was also significantly higher in hypo-DMRs of CHH, indicating the intricate impact of small RNAs on DNA methylation. Similar phenomena were reported in a previous study by Yang et al. (2016).
MEIOSIS ARRESTED AT LEPTOTENE1 (MEL1) is a rice AGO that specifically functions in the development of premeiotic germ cells and the progression of meiosis by binding small RNAs (Nonomura et al., 2007). Komiya et al. (2014) predicted that the synthesis of small RNAs bound by MEL1 is dependent on OsRDR6. In this study, we experimentally verified that OsRDR6 plays an essential role in regulating small RNA levels, DNA methylation, and meiotic DSB formation; however, more in-depth investigation is needed to determine the detailed function of OsRDR6 in modulating meiotic small RNA levels.
OsRDR6 Contributes to Cell Cycle Transition during Rice Meiosis
In yeast, hop2, dmc1, pch2, and zip1 are well-characterized mutants that arrest at the pachytene stage (Leu and Roeder, 1999; San-Segundo and Roeder, 1999). In animals, the mutation of homologs of these genes, such as HOP2 and PCH2 in mouse, also leads to meiocyte arrest at the pachytene stage (Petukhova et al., 2003; Li and Schimenti, 2007). However, in Arabidopsis, the mutation of AtDMC1 causes abnormal chromosome segregation, but not meiotic arrest (Couteau et al., 1999). HOP2 was reported to be involved in meiotic DSB repair in Arabidopsis, but meiosis can be completed in the hop2 mutant (Uanschou et al., 2013). Depletion of Arabidopsis ZYP1 results in a delay, but not arrest, in progression through meiotic prophase I (Higgins et al., 2005). No meiotic arrest was observed in the Atpch2 mutant either (Lambing et al., 2015). Previously, we identified mutants of HOP2, DMC1, PCH2 (CRC1), and ZIP1 (ZEP1) in rice and found that the meiotic process could be completed in all of these mutants (Wang et al., 2010, 2016; Miao et al., 2013; Shi et al., 2019). This suggests that the functions of genes involved in the cell cycle transition in yeast and animals may not be conserved in plants. To date, no checkpoint protein has been reported to participate in plant meiosis.
As to how OsRDR6 functions in cell cycle regulation during rice meiosis, we have two hypotheses. First, small RNAs are reported to be associated with DNA methylation, which is important for maintaining chromosome structure. Because the small RNA levels were dramatically altered in Osrdr6 mutants and thousands of DMRs were identified, we speculate that the chromosome structure might be affected. Changes in chromosome structure lead to an abnormal meiotic process. Second, although no checkpoint protein involved in cell cycle transition during rice meiosis has been identified, we cannot exclude the possibility that such a protein(s) exists. OsRDR6 might be required for the expression of the potential checkpoint protein. Further studies need to be done before we can fully understand the role of OsRDR6 in cell cycle regulation during rice meiosis.
OsRDR6 Is Essential for Rice Meiotic DSB Formation
In Arabidopsis, the small RNAs in meiocytes were different from those of leaves in terms of genome-wide distribution and regions where they work. The meiocyte-specific small RNAs have a significant positive correlation with higher expressed genes, indicating that they might be the outcome of the upregulation of meiotic genes that need to be subsequently suppressed in the next developmental stage. The genesis of a series of small RNAs in meiocytes is dependent on AtSPO11-1, and AtSPO11-1-dependent small RNAs tend to be associated with open chromatin structure, which might favor meiotic recombination hotspots (Huang et al., 2019). The relationship between small RNAs and meiosis has rarely been reported in rice. In this study, we preliminarily established the relationship between small RNAs and programed DSB formation in meiocytes.
Previously, we reported that both CRC1 and P31comet are essential for meiotic DSB formation in rice. Although a direct interaction between P31comet and HOMOLOGOUS PAIRING ABERRATION IN RICE MEIOSIS1 (PAIR1) was not detected, P31comet can interact with CRC1, and CRC1 can interact with PAIR1. Excitingly, CRC1 and P31comet both have stellate distributions in early prophase I of meiosis (Miao et al., 2013; Ji et al., 2016). These findings imply an intimate relationship among these three proteins in which CRC1/P31comet may coordinate with PAIR1 to promote DSB formation. OsSDS is also required for DSB formation during rice meiosis, in contrast to the Arabidopsis SDS ortholog (Azumi et al., 2002; Wu et al., 2015). Therefore, we conclude that CRC1, P31comet, and OsSDS play specific roles in DSB formation in rice meiosis. Previously, we had no knowledge about the regulation of CRC1, P31comet, and OsSDS expression. Here, we speculate that the OsRDR6-mediated small RNA pathway might be involved in regulating the expression of these genes.
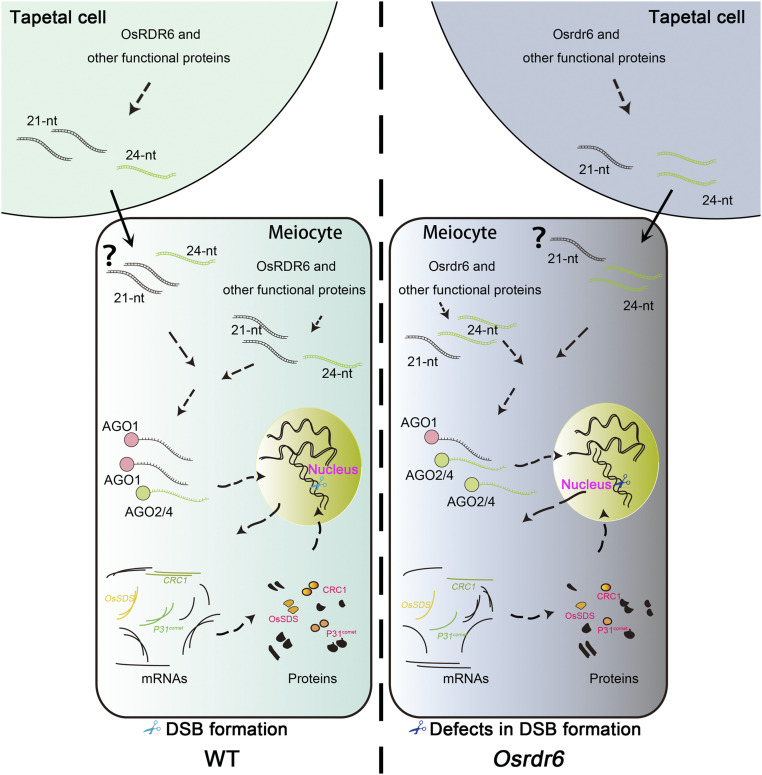
Based on our results, we propose a potential working model for the role of OsRDR6 in regulating meiotic DSB formation (Figure 9). In the wild type, OsRDR6 works in concert with other small RNA processing pathways to maintain a balance in small RNA levels, both in tapetal cells and meiocytes. A previous study found that some 24-nucleotide small RNAs are transferred from tapetal cells to meiocytes (Ono et al., 2018), where an appropriate level of small RNAs precisely regulates the expression of multiple genes by binding to AGO proteins. In Osrdr6-mei, abnormal function of OsRDR6 results in higher 24-nucleotide small RNA levels, especially at the promoter and 3′ regulatory regions of downregulated genes. Moreover, these 24-nucleotide small RNAs are more likely to be assembled into AGO2/4 proteins to repress the expression of these genes. Among these downregulated genes, OsSDS, P31comet, and CRC1 are known to be required for meiotic DSB formation.
Figure 9.
A Proposed Working Model of OsRDR6-Mediated DSB Formation in Rice Meiosis.
In Osrdr6 mutants, increased 24-nucleotide (nt) small RNA levels lead to downregulation of 457 genes through binding to AGO2/AGO4. Three genes associated with meiotic DSB formation, OsSDS, P31comet, and CRC1, are among the downregulated genes. As a result, meiotic DSB formation is defective in Osrdr6 mutants.
In the regions adjacent to OsSDS and P31comet, significantly differentially expressed 24-nucleotide small RNA clusters were identified; these clusters might induce the downregulation of these genes. In rice, gene expression has been shown to be regulated by nearby MITEs. Furthermore, 24-nucleotide siRNAs are tightly associated with MITEs or other TEs and to some extent negatively regulate the expression of nearby genes (Wei et al., 2014). Interestingly, we found two 24-nucleotide small RNA clusters with significantly higher expression in Osrdr6-mei that are associated with MITE loci nearby P31comet. We speculate that these MITEs might be a reason for the downregulation of P31comet. For CRC1, the regulation of its expression may be more complicated, since a direct relationship between small RNA level and its expression level was not found.
AGO2 and AGO4 usually mediate the RdDM pathway in rice (Matzke and Mosher, 2014). Given that the 5′ terminal nucleotides of the 24-nucleotide small RNAs that mapped to the promoter and 3′ regulatory regions of genes downregulated in Osrdr6-mei were biased toward A, we hypothesized that the RdDM pathway might be responsible for the downregulation of most genes in the mutant. However, according to our results, not many downregulated genes are associated with DMRs, indicating that the 24-nucleotide small RNAs with increased abundance may function in some undiscovered pathway(s) other than RdDM to repress the expression of genes in the mutant. In addition, small RNA-mediated gene silencing may also not function through direct binding of the AGO/small RNA complex to the regulatory regions of some downregulated genes. However, the specific reason still needs to be further investigated.
As a major member of the small RNA biogenesis pathway, RDR6 has been reported to be involved in many biological processes. In this study, we found that RDR6 is also essential for meiosis in rice.
METHODS
Plant Materials and Growth Conditions
Osrdr6-mei was derived from the indica rice (Oryza sativa) cv Zhongxian 3037, which was exposed to 60Co γ-ray radiation. Osrdr6-edi was derived from the japonica rice cv Yandao 8 using the CRISPR-Cas9 system. Osrdr6-bi was obtained by crossing Osrdr6-mei/+ with Osrdr6-edi/+. The normal plants identified in the offspring of Osrdr6-mei/+ were used as the wild type. Plants were grown in paddy fields during the growing season in Beijing (40.22°N, 116.20°E) or Hainan (18.51°N, 110.04°E), China.
Map-Based Cloning, Complementation, and CRISPR-Cas9 Targeting of OsRDR6
Mapping populations were generated by crossing Osrdr6-mei/+ mutants with Yandao 8. Using 106 sterile plants identified in the F2 segregating population, the locus was first mapped to the long arm of chromosome 1. Fine mapping was performed using 318 sterile plants identified in the F3 segregating population. Markers were developed based on genomic sequence differences between the reference genomes of Nipponbare and 9311, using the sequences from the National Center for Biotechnology Information (NCBI) nucleotide database (http://www.ncbi.nlm.nih.gov/). Primer sequences used for map-based cloning are listed in Supplemental Table 7. The cloning vector SK-gRNA and the CRISPR-Cas9 binary vector pC1300-cas9 were used in CRISPR/Cas9-mediated editing to knock out OsRDR6 in Yandao 8. The target sequence was 5'-AGTTAAGGGACAAGTCAGCT-3'. Transformation was mediated by the Agrobacterium tumefaciens strain EHA105. The primer pair GDF and GDR was used for genotyping.
A 10.7-kb genomic DNA fragment containing the entire OsRDR6 coding region, the 2.1-kb upstream sequence, and the 3.8-kb downstream sequence was amplified from a bacterial artificial chromosome clone and subcloned into the SacI site of the binary vector pCAMBIA1300. The plasmid was transformed into EHA105 and then into embryonic calli of Osrdr6-mei/+ plants. The primer pair CF and CR was used for genotyping of mutant plants and then the primer pair VF and GR was used to verify whether the construct containing the insertion segment was successfully transformed into the homozygous mutant. We obtained six independent homozygous plants that were complemented by the transgene. Primer sequences used for genotyping are listed in Supplemental Table 7.
Small RNA Sequencing and Data Analyses
The wild-type and Osrdr6-mei anthers at the leptotene-like stage, determined according to floret length, were collected and frozen in liquid nitrogen. The plant materials (the wild type and Osrdr6-mei) used for the assays were obtained from the offspring of Osrdr6-mei/+. Anthers with the wild-type genotype of OsRDR6 were used as the control group, while anthers with the Osrdr6-mei genotype were used as the mutant group. Each sample contained fresh anthers from three independent plants, and three biological repeats were performed for both groups. In this condition, the repeatability between samples was good. The same sample collection method was used for transcriptome sequencing and WGBS. Small RNA library construction and sequencing were performed by Oebiotech (http://www.oebiotech.com/).
Low-quality reads and adapter sequences in the raw data were removed using Cutadapt (Martin, 2011). tRNAs (http://rice.plantbiology.msu.edu/analyses_search_tRNA.shtml), rRNAs (Rice Genome Annotation MSU7.0), and small nucleolar RNAs and small nuclear RNAs (https://www.arb-silva.de/no_cache/download/archive/release_132/Exports/) were also removed. Clean reads were mapped to the rice genome to identify small RNA clusters using the default parameters of Shortstack (Axtell, 2013); a phased score of ∼30 or more indicates a well-phased siRNA locus (phased locus). Annotation and distribution analysis of small RNAs were performed using deepTools2 (Ramírez et al., 2016) and BEDtools (Quinlan and Hall, 2010). Sequence logos were created by Weblogo (Crooks et al., 2004). For differential expression analysis of small RNA clusters, the regions of clusters identified in all three replicates of the wild type or Osrdr6-mei were kept and normalized reads counts (count per million) of the clusters were calculated. We used Student’s t test to perform statistical tests of the differentially expressed small RNAs with multiple comparison correction using R (false discovery rate less than 0.05). We used RepeatMasker (http://repeatmasker.org/) to search the O. sativa Repeat Database to identify the MITE repeats overlapping with small RNA clusters.
Transcriptome Sequencing and Data Analyses
Total RNA samples used for small RNA sequencing were also used for transcriptional sequencing. Library construction and sequencing were performed by Oebiotech.
Low-quality reads and adapter sequences in the raw data were removed using Cutadapt (Martin, 2011). Clean reads were mapped to the rice genome using the default parameters of Hisat2 (Kim et al., 2015). Feature Counts (Liao et al., 2014) and DESeq2 (Love et al., 2014) were used to identify differentially expressed genes based on the criteria: q-value ≤ 0.01 and log2 fold change ≥ 1.
WGBS and Data Analyses
The wild-type and Osrdr6-mei anthers at the leptotene-like stage (the same stage as that used for small RNA sequencing), which was determined according to floret length, were collected and frozen in liquid nitrogen. Library construction and sequencing were performed by Oebiotech.
We used Bismark software (Krueger and Andrews, 2011) to map bisulfite sequencing data to the rice reference genome with default parameters and methylKit (Akalin et al., 2012) to identify DMRs (window size = 100 bp, step size = 50 bp, meth.diff = 25, q-value = 0.01).
Semithin Sections
Semithin sectioning experiments were performed as previously described by Liu et al. (2018). In brief, samples were fixed in 2.5% (v/v) glutaraldehyde at 4°C for 10 h and then dehydrated in ethanol. Samples were embedded in Technovit_7100 for sectioning.
Ovule Clearing
Ovule clearing was performed as previously described by Liu et al. (2018). Mature ovules of the wild type and Osrdr6-mei were fixed for 10 h in formaldehyde-acetic acid-ethanol stationary liquid. Samples were gradually rehydrated in 70, 50, and 30% (v/v) ethanol and distilled water. Fixed ovules were treated with a 2% (w/v) aluminum potassium sulfate solution for 20 min and stained in a solution of eosin B dissolved in 4% (w/v) Suc at a concentration of 10 mg/L for 10 h. After dehydration with ethanol, the ovules were transferred into a mixture of ethanol and methyl salicylate (1:1 [v/v]) for 1 h and cleared in pure methyl salicylate overnight.
Meiotic Chromosome Preparation
Young panicles of both the wild-type and mutant lines were fixed in Carnoy’s solution (ethanol:glacial acetic acid, 3:1 [v/v]). Anthers at different meiotic stages were squashed in an acetocarmine solution on glass slides and washed with 45% (v/v) acetic acid. The cover slips were removed after freezing the slides in liquid nitrogen. Next, the slides were dehydrated through an ethanol gradient (70, 90, and 100% [v/v]). Chromosome spreads were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) in an antifade solution (Vector Laboratories). Cytological analysis was performed under an A2 fluorescence microscope (Zeiss).
FISH Assay
FISH analysis was conducted as previously described by Ji et al. (2016) using pTa794 containing 5S rRNA genes from wheat (Triticum aestivum) as the probe (Cuadrado and Jouve, 1994). The bulked oligonucleotide probes (11S and 11L) were developed based on the procedures described in a previous report by Han et al. (2015), and procedures were performed as previously described by Shi et al. (2019). The secondary antibodies used in these assays were the fluorescein anti-biotin antibody (catalog no. SP-3040; Vectors) and the anti–digoxigenin-rhodamine Fab fragments antibody (catalog no. 11207750910; Roche).
Immunofluorescence
Fresh young panicles were fixed in 4% (w/v) paraformaldehyde for 30 to 90 min at room temperature. Anthers at the proper stage were squashed in PBS solution on glass slides and covered with a cover slip. The slides were then frozen in liquid nitrogen. After removing cover slips, slides were dehydrated using an ethanol gradient (70, 90, and 100% [v/v]). Next, slides were incubated in a humid chamber at 37°C for 4 h with different combinations of primary antibodies, all diluted 1:500 (v/v) in TNB buffer (0.1 M Tris-HCl, pH 7.5, 0.15 M NaCl, and 0.5% [w/v] blocking reagent). After three rounds of washing in PBS, the appropriate secondary antibodies (1:500 [v/v]) were added to the slides. Next, slides were incubated for 30 min. After another three rounds of washing in PBS, chromosomes were counterstained with DAPI. The primary antibodies against γH2AX, PAIR3, CRC1, P31comet, OsDMC1, OsCOM1, OsZIP4, and HEI10 were previously described by Miao et al. (2013) and Ji et al. (2016). The secondary antibodies include the fluorescein-conjugated goat anti-mouse antibody (cat. no. 1010-02; Southern Biotech), rhodamine-conjugated goat anti-rabbit antibody (cat. no. 4010-03; Southern Biotech), and DyLight 594-conjugated goat anti-guinea pig IgG antibody (cat. no. GtxGp-003-D594NHSX; ImmunoReagents).
Real-Time PCR
Samples were ground in liquid nitrogen, and total RNA was extracted using a HiPure Plant RNA Mini kit (Magen). Roughly 2 μg of total RNA was treated with RNase-free DNase I (Invitrogen). In vitro RT was performed using SuperScript III Reverse Transcriptase (Invitrogen). Real-time PCR analysis was performed on a Bio-Rad CFX96 instrument using the Fast Evagreen qPCR master mix (Biotium). Primers are listed in Supplemental Table 7.
Accession Numbers
Sequence data used in this article can be found in the NCBI databases under the following accession numbers: OsRDR6 (AB353923); OsSDS (XP_015628868); P31comet (XM_015783459); CRC1 (KF245924). All large-scale sequencing data have been deposited in the NCBI Gene Expression Omnibus database under accession number GSE151871.
Supplemental Data
Supplemental Figure 1. Map-based cloning of OsRDR6 and complementation of Osrdr6-mei.
Supplemental Figure 2. Homologous pairing was affected in Osrdr6-mei meiocytes.
Supplemental Figure 3. Proportion of meiocytes at different stages in wild-type (WT) and Osrdr6-mei spikelets of various lengths.
Supplemental Figure 4. Semi thin slices of wild type (WT), Osrdr6-mei and Osrdr6-bi anthers at early developmental stages.
Supplemental Figure 5. The correlation coefficient for each pair of biological replicates used in small RNA sequencing.
Supplemental Figure 6. Phased loci analysis in wild type (WT) and Osrdr6-mei.
Supplemental Figure 7. Small RNA clusters in wild type (WT) and Osrdr6-mei.
Supplemental Figure 8. The abundance of 21-nt and 24-nt small RNAs (CPM) surrounding DMRs with different types of DNA methylation.
Supplemental Figure 9. The correlation between each pair of biological replicates used in transcriptome sequencing.
Supplemental Figure 10. Supplemental information about the association between small RNA level and gene expression.
Supplemental Table 1. Proportion of meiocytes at different stages in wild-type (WT) and Osrdr6-mei spikelets of various lengths.
Supplemental Table 2. Proportion of meiocytes at different stages in 8-mm wild-type (WT) and Osrdr6-bi spikelets.
Supplemental Table 3. Statistical information for small RNA clusters.
Supplemental Table 4. Statistical information for WGBS.
Supplemental Table 5. Number of differentially methylated regions.
Supplemental Table 6. Mutations in Osrdr6-mei revealed by transcriptome sequencing.
Supplemental Table 7. Primers used in this study.
Supplemental Data Set 1. PhaseLoci_21 nt.
Supplemental Data Set 2. PhaseLoci_24 nt.
Supplemental Data Set 3. The overlap between differentially expressed small RNA clusters and DEGs.
Supplemental Data Set 4. List of DMRs overlapped with small RNA clusters.
Supplemental Data Set 5. DEGs, asso small RNAs, asso DMRs, and asso TEs.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
This work was supported by the National Natural Science Foundation of China (grants 31930018 and 31971912) and the Strategic Priority Research Program of the Chinese Academy of Sciences (grant XDA 24010302).
AUTHOR CONTRIBUTIONS
Z.C., C.L., and Y.S. conceived the research; C.L., B.Q., and W.S. performed the majority of biological experiments; Y.W., H.W., J.C., and F.M. did the informatics analysis; C.L., Y.W., G.D., Y.L., and D.T. analyzed the data; C.L. wrote the article; and Y.W., Y.S., and Z.C. supervised and completed the writing.
References
- Akalin, A., Kormaksson, M., Li, S., Garrett-Bakelman, F.E., Figueroa, M.E., Melnick, A., Mason, C.E.(2012). methylKit: A comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 13: R87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin, A.A., Hannon, G.J., Brennecke, J.(2007). The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science 318: 761–764. [DOI] [PubMed] [Google Scholar]
- Axtell, M.J.(2013). ShortStack: Comprehensive annotation and quantification of small RNA genes. RNA 19: 740–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azumi, Y., Liu, D., Zhao, D., Li, W., Wang, G., Hu, Y., Ma, H.(2002). Homolog interaction during meiotic prophase I in Arabidopsis requires the SOLO DANCERS gene encoding a novel cyclin-like protein. EMBO J. 21: 3081–3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergerat, A., de Massy, B., Gadelle, D., Varoutas, P.C., Nicolas, A., Forterre, P.(1997). An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature 386: 414–417. [DOI] [PubMed] [Google Scholar]
- Castel, S.E., Martienssen, R.A.(2013). RNA interference in the nucleus: Roles for small RNAs in transcription, epigenetics and beyond. Nat. Rev. Genet. 14: 100–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couteau, F., Belzile, F., Horlow, C., Grandjean, O., Vezon, D., Doutriaux, M.P.(1999). Random chromosome segregation without meiotic arrest in both male and female meiocytes of a dmc1 mutant of Arabidopsis. Plant Cell 11: 1623–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks, G.E., Hon, G., Chandonia, J.-M., Brenner, S.E.(2004). WebLogo: A sequence logo generator. Genome Res. 14: 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuadrado, A., Jouve, N.(1994). Mapping and organization of highly-repeated DNA sequences by means of simultaneous and sequential FISH and C-banding in 6x-triticale. Chromosome Res. 2: 331–338. [DOI] [PubMed] [Google Scholar]
- Cuerda-Gil, D., Slotkin, R.K.(2016). Non-canonical RNA-directed DNA methylation. Nat. Plants 2: 16163. [DOI] [PubMed] [Google Scholar]
- Han, Y., Zhang, T., Thammapichai, P., Weng, Y., Jiang, J.(2015). Chromosome-specific painting in Cucumis species using bulked oligonucleotides. Genetics 200: 771–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins, J.D., Sanchez-Moran, E., Armstrong, S.J., Jones, G.H., Franklin, F.C.(2005). The Arabidopsis synaptonemal complex protein ZYP1 is required for chromosome synapsis and normal fidelity of crossing over. Genes Dev. 19: 2488–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, J., Wang, C., Wang, H., Lu, P., Zheng, B., Ma, H., Copenhaver, G.P., Wang, Y.(2019). Meiocyte-specific and AtSPO11–1-dependent small RNAs and their association with meiotic gene expression and recombination. Plant Cell 31: 444–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji, J., Tang, D., Shen, Y., Xue, Z., Wang, H., Shi, W., Zhang, C., Du, G., Li, Y., Cheng, Z.(2016). P31comet, a member of the synaptonemal complex, participates in meiotic DSB formation in rice. Proc. Natl. Acad. Sci. USA 113: 10577–10582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji, J., Tang, D., Wang, K., Wang, M., Che, L., Li, M., Cheng, Z.(2012). The role of OsCOM1 in homologous chromosome synapsis and recombination in rice meiosis. Plant J. 72: 18–30. [DOI] [PubMed] [Google Scholar]
- Jiang, L., Qian, D., Zheng, H., Meng, L.Y., Chen, J., Le, W.J., Zhou, T., Zhou, Y.J., Wei, C.H., Li, Y.(2012). RNA-dependent RNA polymerase 6 of rice (Oryza sativa) plays role in host defense against negative-strand RNA virus, Rice stripe virus. Virus Res. 163: 512–519. [DOI] [PubMed] [Google Scholar]
- Johnson, C., Kasprzewska, A., Tennessen, K., Fernandes, J., Nan, G.L., Walbot, V., Sundaresan, V., Vance, V., Bowman, L.H.(2009). Clusters and superclusters of phased small RNAs in the developing inflorescence of rice. Genome Res. 19: 1429–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney, S., Giroux, C.N., Kleckner, N.(1997). Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88: 375–384. [DOI] [PubMed] [Google Scholar]
- Kim, D., Langmead, B., Salzberg, S.L.(2015). HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 12: 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiya, R., Ohyanagi, H., Niihama, M., Watanabe, T., Nakano, M., Kurata, N., Nonomura, K.(2014). Rice germline-specific Argonaute MEL1 protein binds to phasiRNAs generated from more than 700 lincRNAs. Plant J. 78: 385–397. [DOI] [PubMed] [Google Scholar]
- Krueger, F., Andrews, S.R.(2011). Bismark: A flexible aligner and methylation caller for bisulfite-seq applications. Bioinformatics 27: 1571–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumakura, N., Takeda, A., Fujioka, Y., Motose, H., Takano, R., Watanabe, Y.(2009). SGS3 and RDR6 interact and colocalize in cytoplasmic SGS3/RDR6-bodies. FEBS Lett. 583: 1261–1266. [DOI] [PubMed] [Google Scholar]
- Lambing, C., Osman, K., Nuntasoontorn, K., West, A., Higgins, J.D., Copenhaver, G.P., Yang, J., Armstrong, S.J., Mechtler, K., Roitinger, E., Franklin, F.C.(2015). Arabidopsis PCH2 mediates meiotic chromosome remodeling and maturation of crossovers. PLoS Genet. 11: e1005372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leu, J.Y., Roeder, G.S.(1999). The pachytene checkpoint in S. cerevisiae depends on Swe1-mediated phosphorylation of the cyclin-dependent kinase Cdc28. Mol. Cell 4: 805–814. [DOI] [PubMed] [Google Scholar]
- Li, X.C., Schimenti, J.C.(2007). Mouse pachytene checkpoint 2 (trip13) is required for completing meiotic recombination but not synapsis. PLoS Genet. 3: e130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, Y., Smyth, G.K., Shi, W.(2014). featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930. [DOI] [PubMed] [Google Scholar]
- Liu, C., Xue, Z., Tang, D., Shen, Y., Shi, W., Ren, L., Du, G., Li, Y., Cheng, Z.(2018). Ornithine δ-aminotransferase is critical for floret development and seed setting through mediating nitrogen reutilization in rice. Plant J. 96: 842–854. [DOI] [PubMed] [Google Scholar]
- Love, M.I., Huber, W., Anders, S.(2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, Z., Castillo-Gonzalez, C., Wang, Z., Sun, D., Hu, X., Shen, X., Potok, M.E., Zhang, X.(2018). Arabidopsis serrate coordinates histone methyltransferases ATXR5/6 and RNA processing factor RDR6 to regulate transposon expression. Dev. Cell 45: 769–784. [DOI] [PubMed] [Google Scholar]
- Martin, M.(2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17: 10–12. [Google Scholar]
- Matzke, M.A., Mosher, R.A.(2014). RNA-directed DNA methylation: An epigenetic pathway of increasing complexity. Nat. Rev. Genet. 15: 394–408. [DOI] [PubMed] [Google Scholar]
- Mi, S., et al. (2008). Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell 133: 116–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao, C., Tang, D., Zhang, H., Wang, M., Li, Y., Tang, S., Yu, H., Gu, M., Cheng, Z.(2013). Central region component1, a novel synaptonemal complex component, is essential for meiotic recombination initiation in rice. Plant Cell 25: 2998–3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasaki, H., Itoh, J., Hayashi, K., Hibara, K., Satoh-Nagasawa, N., Nosaka, M., Mukouhata, M., Ashikari, M., Kitano, H., Matsuoka, M., Nagato, Y., Sato, Y.(2007). The small interfering RNA production pathway is required for shoot meristem initiation in rice. Proc. Natl. Acad. Sci. USA 104: 14867–14871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonomura, K., Morohoshi, A., Nakano, M., Eiguchi, M., Miyao, A., Hirochika, H., Kurata, N.(2007). A germ cell specific gene of the ARGONAUTE family is essential for the progression of premeiotic mitosis and meiosis during sporogenesis in rice. Plant Cell 19: 2583–2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono, S., Liu, H., Tsuda, K., Fukai, E., Tanaka, K., Sasaki, T., Nonomura, K.I.(2018). EAT1 transcription factor, a non-cell-autonomous regulator of pollen production, activates meiotic small RNA biogenesis in rice anther tapetum. PLoS Genet. 14: e1007238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peragine, A., Yoshikawa, M., Wu, G., Albrecht, H.L., Poethig, R.S.(2004). SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 18: 2368–2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petukhova, G.V., Romanienko, P.J., Camerini-Otero, R.D.(2003). The Hop2 protein has a direct role in promoting interhomolog interactions during mouse meiosis. Dev. Cell 5: 927–936. [DOI] [PubMed] [Google Scholar]
- Quinlan, A.R., Hall, I.M.(2010). BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez, F., Ryan, D.P., Grüning, B., Bhardwaj, V., Kilpert, F., Richter, A.S., Heyne, S., Dündar, F., Manke, T.(2016). deepTools2: A next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44 (W1): W160–W165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert, T., Nore, A., Brun, C., Maffre, C., Crimi, B., Bourbon, H.M., de Massy, B., Massy, B.(2016). The TopoVIB-Like protein family is required for meiotic DNA double-strand break formation. Science 351: 943–949. [DOI] [PubMed] [Google Scholar]
- San-Segundo, P.A., Roeder, G.S.(1999). Pch2 links chromatin silencing to meiotic checkpoint control. Cell 97: 313–324. [DOI] [PubMed] [Google Scholar]
- Satoh, N., Itoh, J., Nagato, Y.(2003). The SHOOTLESS2 and SHOOTLESS1 genes are involved in both initiation and maintenance of the shoot apical meristem through regulating the number of indeterminate cells. Genetics 164: 335–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, Y., Tang, D., Wang, K., Wang, M., Huang, J., Luo, W., Luo, Q., Hong, L., Li, M., Cheng, Z.(2012). ZIP4 in homologous chromosome synapsis and crossover formation in rice meiosis. J. Cell Sci. 125: 2581–2591. [DOI] [PubMed] [Google Scholar]
- Shi, W., Tang, D., Shen, Y., Xue, Z., Zhang, F., Zhang, C., Ren, L., Liu, C., Du, G., Li, Y., Yan, C., Cheng, Z.(2019). OsHOP2 regulates the maturation of crossovers by promoting homologous pairing and synapsis in rice meiosis. New Phytol. 222: 805–819. [DOI] [PubMed] [Google Scholar]
- Song, X., Wang, D., Ma, L., Chen, Z., Li, P., Cui, X., Liu, C., Cao, S., Chu, C., Tao, Y., Cao, X.(2012a). Rice RNA-dependent RNA polymerase 6 acts in small RNA biogenesis and spikelet development. Plant J. 71: 378–389. [DOI] [PubMed] [Google Scholar]
- Song, X., et al. (2012b). Roles of DCL4 and DCL3b in rice phased small RNA biogenesis. Plant J. 69: 462–474. [DOI] [PubMed] [Google Scholar]
- Stacey, N.J., Kuromori, T., Azumi, Y., Roberts, G., Breuer, C., Wada, T., Maxwell, A., Roberts, K., Sugimoto-Shirasu, K.(2006). Arabidopsis SPO11-2 functions with SPO11-1 in meiotic recombination. Plant J. 48: 206–216. [DOI] [PubMed] [Google Scholar]
- Su, Z., Zhao, L., Zhao, Y., Li, S., Won, S., Cai, H., Wang, L., Li, Z., Chen, P., Qin, Y., Chen, X.(2017). The THO complex non-cell-autonomously represses female germline specification through the TAS3–ARF3 module. Curr. Biol. 27: 1597–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toriba, T., Suzaki, T., Yamaguchi, T., Ohmori, Y., Tsukaya, H., Hirano, H.Y.(2010). Distinct regulation of adaxial-abaxial polarity in anther patterning in rice. Plant Cell 22: 1452–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uanschou, C., Ronceret, A., Von Harder, M., De Muyt, A., Vezon, D., Pereira, L., Chelysheva, L., Kobayashi, W., Kurumizaka, H., Schlögelhofer, P., Grelon, M.(2013). Sufficient amounts of functional HOP2/MND1 complex promote interhomolog DNA repair but are dispensable for intersister DNA repair during meiosis in Arabidopsis. Plant Cell 25: 4924–4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia-Sanchez, M.A., Liu, J., Hannon, G.J., Parker, R.(2006). Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 20: 515–524. [DOI] [PubMed] [Google Scholar]
- Voinnet, O.(2008). Use, tolerance and avoidance of amplified RNA silencing by plants. Trends Plant Sci. 13: 317–328. [DOI] [PubMed] [Google Scholar]
- Vrielynck, N., Chambon, A., Vezon, D., Pereira, L., Chelysheva, L., De Muyt, A., Mézard, C., Mayer, C., Grelon, M.(2016). A DNA topoisomerase VI-like complex initiates meiotic recombination. Science 351: 939–943. [DOI] [PubMed] [Google Scholar]
- Wang, H., Hu, Q., Tang, D., Liu, X., Du, G., Shen, Y., Li, Y., Cheng, Z.(2016). OsDMC1 is not required for homologous pairing in rice meiosis. Plant Physiol. 171: 230–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, K., Wang, M., Tang, D., Shen, Y., Miao, C., Hu, Q., Lu, T., Cheng, Z.(2012). The role of rice HEI10 in the formation of meiotic crossovers. PLoS Genet. 8: e1002809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, M., Wang, K., Tang, D., Wei, C., Li, M., Shen, Y., Chi, Z., Gu, M., Cheng, Z.(2010). The central element protein ZEP1 of the synaptonemal complex regulates the number of crossovers during meiosis in rice. Plant Cell 22: 417–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, L., Gu, L., Song, X., Cui, X., Lu, Z., Zhou, M., Wang, L., Hu, F., Zhai, J., Meyers, B.C., Cao, X.(2014). Dicer-like 3 produces transposable element-associated 24-nt siRNAs that control agricultural traits in rice. Proc. Natl. Acad. Sci. USA 111: 3877–3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, L., Zhang, Q., Zhou, H., Ni, F., Wu, X., Qi, Y.(2009). Rice MicroRNA effector complexes and targets. Plant Cell 21: 3421–3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Z., Ji, J., Tang, D., Wang, H., Shen, Y., Shi, W., Li, Y., Tan, X., Cheng, Z., Luo, Q.(2015). OsSDS is essential for DSB formation in rice meiosis. Front. Plant Sci. 6: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, G., Luo, X., Habu, T., Rizo, J., Matsumoto, T., Yu, H.(2004). Conformation-specific binding of p31(comet) antagonizes the function of Mad2 in the spindle checkpoint. EMBO J. 23: 3133–3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue, Z., Li, Y., Zhang, L., Shi, W., Zhang, C., Feng, M., Zhang, F., Tang, D., Yu, H., Gu, M., Cheng, Z.(2016). OsMTOPVIB promotes meiotic DNA double-strand break formation in rice. Mol. Plant 9: 1535–1538. [DOI] [PubMed] [Google Scholar]
- Yang, D.L., Zhang, G., Tang, K., Li, J., Yang, L., Huang, H., Zhang, H., Zhu, J.K.(2016). Dicer-independent RNA-directed DNA methylation in Arabidopsis. Cell Res. 26: 66–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, H., Wang, M., Tang, D., Wang, K., Chen, F., Gong, Z., Gu, M., Cheng, Z.(2010). OsSPO11-1 is essential for both homologous chromosome pairing and crossover formation in rice. Chromosoma 119: 625–636. [DOI] [PubMed] [Google Scholar]