A protein engineering approach allows the constitutive activation of LRR membrane receptor kinase signaling pathways in plants.
Abstract
Receptor kinases with extracellular leucine-rich repeat domains (LRR-RKs) form the largest group of membrane signaling proteins in plants. LRR-RKs can sense small molecule, peptide, or protein ligands and may be activated by ligand-induced interaction with a shape complementary SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE (SERK) coreceptor kinase. We have previously shown that SERKs can also form constitutive, ligand-independent complexes with the LRR ectodomains of BAK1-INTERACTING RECEPTOR-LIKE KINASE3 (BIR3) receptor pseudokinases, negative regulators of LRR-RK signaling. Here, we report that receptor chimera in which the extracellular LRR domain of BIR3 is fused to the cytoplasmic kinase domains of the SERK-dependent LRR-RKs BRASSINOSTEROID INSENSITIVE1, HAESA and ERECTA form tight complexes with endogenous SERK coreceptors in the absence of ligand stimulus. Expression of these chimeras under the control of the endogenous promoter of the respective LRR-RK leads to strong gain-of-function brassinosteroid, floral abscission, and stomatal patterning phenotypes, respectively. Importantly, a BIR3-GASSHO1 (GSO1)/SCHENGEN3 (SGN3) chimera can partially complement sgn3 Casparian strip formation phenotypes, suggesting that SERK proteins also mediate GSO1/SGN3 receptor activation. Collectively, our protein engineering approach may be used to elucidate the physiological functions of orphan LRR-RKs and to identify their receptor activation mechanism in single transgenic lines.
INTRODUCTION
Plant-unique membrane receptor kinases characterized by an extracellular domain, a single membrane-spanning helix, and a cytoplasmic dual-specificity kinase domain control many aspects of plant growth and development. They form the first layer of the plant immune system and mediate symbiotic interactions (Hohmann et al., 2017). These leucine-rich repeat receptor kinases (LRR-RKs) constitute the largest class of receptor kinases known in plants (Shiu and Bleecker, 2001). Members of the family have been shown to sense small molecule (Wang et al., 2001), peptide (Gómez-Gómez and Boller, 2000; Matsubayashi, 2014; Santiago et al., 2016), and protein (Huang et al., 2016; Lin et al., 2017; Zhang et al., 2017) ligands.
Brassinosteroids, whose biosynthesis involves the steroid 5α reductase DE-ETIOLATED2 (DET2; Chory et al., 1991; Noguchi et al., 1999), are a class of phytohormones that are sensed by the ectodomain of the LRR-RK BRASSINOSTEROID INSENSITIVE1 (BRI1) with nanomolar affinity (Wang et al., 2001; Hothorn et al., 2011; Hohmann et al., 2018b). Brassinosteroid binding to the BRI1 ectodomain triggers BRI1 interaction with the LRR domain of a SOMATIC EMBRYOGENESIS RECEPTOR LIKE KINASE (SERK) coreceptor (Hothorn et al., 2011; She et al., 2011; Santiago et al., 2013; Sun et al., 2013; Hohmann et al., 2018b). The formation of this heterodimeric complex at the cell surface promotes interaction and trans-phosphorylation of the receptor and coreceptor kinase domains inside the cell (Wang et al., 2008; Bojar et al., 2014; Hohmann et al., 2018b; Perraki et al., 2018). BRI1 receptor activation initiates a cytoplasmic signaling cascade, which ultimately results in the dephosphorylation and activation of a family of basic helix-loop-helix transcription factors, including the Arabidopsis (Arabidopsis thaliana) proteins BRASSINAZOLE-RESISTANT1 (BZR1) and BRI1-EMS-SUPPRESSOR1 (BES1; Wang et al., 2002; Yin et al., 2002; Vert and Chory, 2006; Nosaki et al., 2018). In bes1-D plants, BES1 Pro-233 is replaced by a Leu residue, which leads to constitutive brassinosteroid signaling responses by enhancing protein phosphatase 2A–mediated dephosphorylation (Yin et al., 2002; Tang et al., 2011).
The plant-unique mechanism of SERK coreceptor-dependent activation is conserved among many LRR-RKs (Hohmann et al., 2017), for example, the LRR-RK HAESA (HAE), whose functions include the control of floral organ abscission in Arabidopsis by interacting with the peptide hormone INFLORESCENCE DEFICIENT IN ABSCISSION (IDA; Jinn et al., 2000; Meng et al., 2016; Santiago et al., 2016; Hohmann et al., 2018b). A SERK-dependent mitogen-activated protein kinase (MAPK) signaling pathway (Meng et al., 2015) involves the LRR-RK ERECTA (ER) and its paralogues ERECTA-LIKE1 (ERL1) and ERL2 (Torii et al., 1996; Shpak, 2013) and plays diverse roles in plant development. ERECTA, ERL1, and ERL2 together control stomata development and their correct spacing on the leaf surface (Shpak et al., 2005). Cys-rich EPIDERMAL PATTERNING FACTOR (EPF) peptides bind to the ectodomains of ERECTA, ERL1, and ERL2, which form constitutive complexes with the ectodomain of the receptor-like protein (RLP) TOO MANY MOUTH (TMM; Yang and Sack, 1995; Nadeau and Sack, 2002; Lee et al., 2012, 2015; Lin et al., 2017). Binding of EPF peptides to these LRR-RK/LRR-RLP complexes triggers their interaction with SERK coreceptor kinases (Meng et al., 2015; Lin et al., 2017) that in turn leads to the initiation of a MAPK signaling pathway that includes the MAPK kinase kinase YODA (Bergmann et al., 2004). Stimulation of the ERECTA pathway negatively regulates stomata formation (Lampard et al., 2009).
The determination of complex structures and quantitative biochemical comparisons of different ligand-activated LRR-RK–SERK complexes have revealed a structurally and functionally conserved activation mechanism, relying on the interaction of the ligand-bound receptor LRR ectodomain with the shape-complementary ectodomain of the SERK coreceptor (Santiago et al., 2013, 2016; Wang et al., 2015; Hohmann et al., 2017, 2018b; Lin et al., 2017). The ligand binding specificity of plant LRR-RKs is encoded in their LRR ectodomains (Hohmann et al., 2017; Okuda et al., 2020). The kinase domain of the receptor, not of the SERK coreceptor, confers cytoplasmic signaling specificity (Hohmann et al., 2018b; Chen et al., 2019; Zheng et al., 2019). Recent genetic, biochemical, and structural evidence suggests that not all plant LRR-RKs rely on SERKs as essential coreceptor kinases (Zhang et al., 2017; Anne et al., 2018; Cui et al., 2018; Hu et al., 2018; Smakowska-Luzan et al., 2018).
Protein engineering approaches have previously been used to dissect LRR-RK receptor activation in planta: a fusion protein combining the extracellular and transmembrane domains of BRI1 (outerBRI1 [oBRI1]) with the cytoplasmic kinase domain of the rice (Oryza sativa) immune receptor XA21 (innerXA21 [iXA21]) triggered an immune response in rice cells upon stimulation with brassinosteroids (He et al., 2000). We now know that both BRI1 and XA21 rely on SERK coreceptor kinases for receptor activation (Li et al., 2002; Nam and Li, 2002; Santiago et al., 2013; Chen et al., 2014; Hohmann et al., 2018b). The heteromeric nature of LRR-RK–SERK complexes has been validated in planta using similar protein engineering approaches. Coexpression of a chimeric construct between the immune receptor FLAGELLIN SENSING2 (FLS2) and its coreceptor SERK3 (oFLS2-iSERK3) with an oSERK3-iFLS2 construct led to immune signaling after stimulation with the FLS2 ligand flg22 in a transient expression system (Albert et al., 2013). Stable transgenic lines coexpressing oBRI1-iSERK3 and oSERK3-iBRI1 constructs partially rescued the BRI1 weak loss-of-function mutant bri1-301 (Hohmann et al., 2018b).
The signaling specificity of the cytoplasmic kinase domain of LRR-RKs has been dissected using an oBRI-iHAESA chimera, which rescued the floral abscission phenotypes when expressed under the control of the HAESA promoter in the haesa haesa-like2 (hsl2) double mutant (Hohmann et al., 2018b). A similar approach recently demonstrated that the LRR-RKs BRI1 and EXCESS MICROSPOROCYTES1 (EMS1) share a common cytoplasmic signaling cascade (Zheng et al., 2019). However, these approaches all rely on ligand stimulus.
Recently, two studies reported a constitutive, ligand-independent interaction between the LRR ectodomains of SERKs and of BAK1-INTERACTING RECEPTOR-LIKE KINASEs (BIRs; Ma et al., 2017; Hohmann et al., 2018a). While BIR1 appears to have a catalytically active cytoplasmic kinase domain, BIR2 to BIR4 are receptor pseudokinases (Gao et al., 2009; Wang et al., 2011; Blaum et al., 2014). Different BIRs have been characterized as negative regulators of plant immunity, floral abscission, and brassinosteroid signaling (Gao et al., 2009; Leslie et al., 2010; Halter et al., 2014; Imkampe et al., 2017). Structural and biochemical analyses now implicate BIR proteins as general negative regulators of SERK coreceptor-mediated LRR-RK signaling pathways (Moussu and Santiago, 2019). The ectodomains of BIR1 to BIR4 bind to SERK ectodomains with dissociation constants in the low micromolar range and target a surface area in SERKs normally required for the interaction with ligand-bound LRR-RKs (Ma et al., 2017; Hohmann et al., 2018a, 2018b). Thus, BIRs can efficiently compete with LRR-RKs for SERK binding, negatively regulating LRR-RK signaling pathways. In line with this observation, the elongated (elg) allele of SERK3, which weakens the interaction with BIRs, but not with BRI1, results in a brassinosteroid-specific gain-of-function signaling phenotype, as BRI1 can more efficiently compete with BIRs for coreceptor binding (Jaillais et al., 2011; Hohmann et al., 2018a). Structure-guided mutations in the BIR-SERK ectodomain complex interface (BIR3 residues Phe-146-Ala/Arg-170-Ala) efficiently disrupt BIR-SERK signaling complexes in vitro and in planta (Hohmann et al., 2018a). Here, we present protein fusions of the BIR3 LRR ectodomain and transmembrane helix (oBIR3) with the cytoplasmic domains of different SERK-dependent LRR-RKs (iBRI1, iHAESA, iER, and iFLS2). Expressing these chimeric constructs under the control of endogenous/context-specific promoters, we obtain strong gain-of-function phenotypes for different developmental signaling pathways triggered by LRR-RKs. In addition, an oBIR3-iGSO1/SGN3 chimera supports a SERK-dependent activation mechanism for the LRR-RK GASSHO1 (GSO1; also called SCHENGEN3 [SGN3]) in Casparian strip formation (Pfister et al., 2014; Okuda et al., 2020). Our strategy allows for the identification of gain-of-function phenotypes of orphan LRR-RKs whose ligands are unknown and enables the elucidation of their receptor activation mechanism.
RESULTS
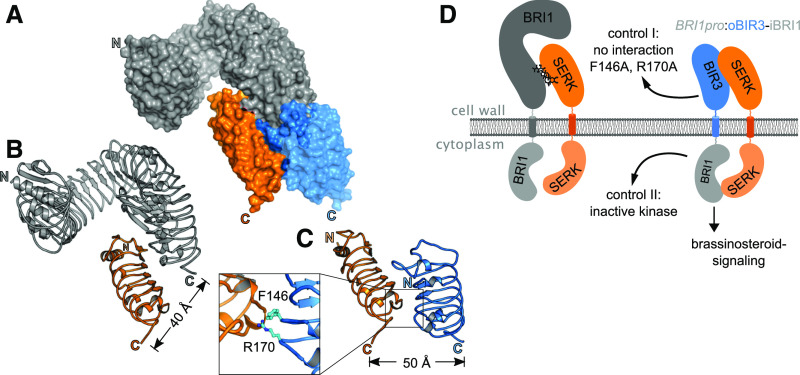
We compared the structure of a previously reported BRI1-brassinolide-SERK1 complex (Protein Data Bank [PDB] ID: 4LSX; http://rcsb.org) with the recently reported structure of a BIR3-SERK1 complex (PDB-ID: 6FG8; Santiago et al., 2013; Hohmann et al., 2018a). The BRI1 and BIR3 ectodomains bind SERK1 using overlapping, but nonidentical, binding surfaces (Figure 1A). As in the BRI1-SERK1 complex, the C termini of BIR3 and SERK1 are in close proximity in the complex structure (Figures 1B and 1C). Based on their structural similarities, we generated an oBIR3-iBRI1 chimera, in which the BIR3 ectodomain and trans-membrane helix are connected to the cytoplasmic domain of BRI1 (see Methods; Figure 1D).
Figure 1.
Structural Overview of the BRI1-SERK and BIR3-SERK Complexes.
(A) Surface view of a structural superposition of the BRI1-SERK1 (ectodomains shown in gray and orange, respectively; PDB ID: 4LSX; http://www.rcsb.org/) and SERK1-BIR3 (orange and blue; PDB ID: 6FG8) complexes. The two structures are aligned on SERK1 (root mean square deviation ∼0.3 Å comparing 143 corresponding SERK1 Cɑ atoms).
(B) and (C) Ribbon diagrams of the BRI1-SERK1 (B) and BIR3-SERK1 (C) complexes, with SERK1 shown in the same orientation. The distances between the respective C termini are indicated (colors as in [A]). Inset: close-up view of the BIR3-SERK1 complex interface, with the interface residues Phe-146 and Arg-170 highlighted in bonds representation. Mutation of both residues to Ala disrupts the BIR3-SERK1 complex in vitro and in vivo (Hohmann et al., 2018a).
(D) Schematic overview of an entire BRI1-brassinolide-SERK signaling complex and the envisioned oBIR3-iBRI1-SERK interaction.
We introduced the oBIR3-iBRI1 chimeric construct, driven by the BRI1 promoter and with a C-terminal mCitrine fluorescent protein tag, in a previously characterized bri1 null mutant (Jaillais et al., 2011). We used chimeric constructs encoding oBIR3F146A/R170A-iBRI1 and oBIR3-iBRI1D1027N as controls, as they block BIR-SERK complex formation (Hohmann et al., 2018a) and BRI1 kinase activity (Bojar et al., 2014; Hohmann et al., 2018b), respectively. Independent oBIR3-iBRI1 transgenic lines, but none of the control lines, displayed the wavy hypocotyl phenotype characteristic of gain-of-function brassinosteroid mutants (Figure 2A). Importantly, we also observed the wavy hypocotyl phenotype in oBIR3-iBRI1 lines in plants grown in the presence of the brassinosteroid biosynthesis inhibitor brassinazole (BRZ; Figure 2A; Asami et al., 2000). This suggests that oBIR3-iBRI1–triggered brassinosteroid signaling does not depend on endogenous brassinosteroids (Figure 2A). To confirm this hypothesis, we introduced the oBIR3-iBRI1 chimera into the det2-1 background (Chory et al., 1991), characterized by reduced brassinosteroid levels (Fujioka et al., 1997): all oBIR3-iBRI1 det2-1 lines, but none of the controls, exhibited a constitutively active phenotype (Figure 2A). Quantification of three independent oBIR3-iBRI1 T3 lines revealed strong gain-of-function phenotypes, which were even more pronounced than the previously reported phenotype of the constitutively active bes1-1D mutant (Figure 2A; Supplemental Figure 1B to 1D; Supplemental Data Set; Yin et al., 2002). Introduction of oBIR3-iBRI1 into the bri1 null or det2-1 mutants complemented their dwarf phenotype and resulted in extremely elongated petioles, another hallmark of enhanced brassinosteroid signaling (Supplemental Figure 1A). Consistent with a constitutive activation of brassinosteroid signaling, BES1 was dephosphorylated in oBIR3-iBRI1 lines, but not in the control lines (Figure 2B; Supplemental Figure 2). We also detected dephosphorylated BES1 in oBIR3-iBRI1 det2-1 lines (Figure 2C; Supplemental Figure 2). We next performed coimmunoprecipitation (co-IP) experiments in our stable lines and determined that oBIR3-iBRI1 and oBIR3-iBRI1D1027N efficiently interacted with the endogenous SERK3 coreceptor in vivo, whereas the oBIR3F146A/R170A-iBRI1 control, which disrupts the interaction of the isolated BIR3 and SERK1/3 ectodomain in vitro (Hohmann et al., 2018a), could no longer bind SERK3 in planta (Figure 2D; Supplemental Figure 3). Taken together, the BIR3 ectodomain can promote a brassinosteroid-independent interaction with SERK3, and possibly other SERKs in vivo, resulting in a constitutive activation of the brassinosteroid signaling pathway. The control lines further suggest that this signaling complex is formed and stabilized by the ectodomains of BIR3 and SERK3 and requires the catalytic activity of the BRI1 kinase domain for signaling (Figure 2A).
Figure 2.
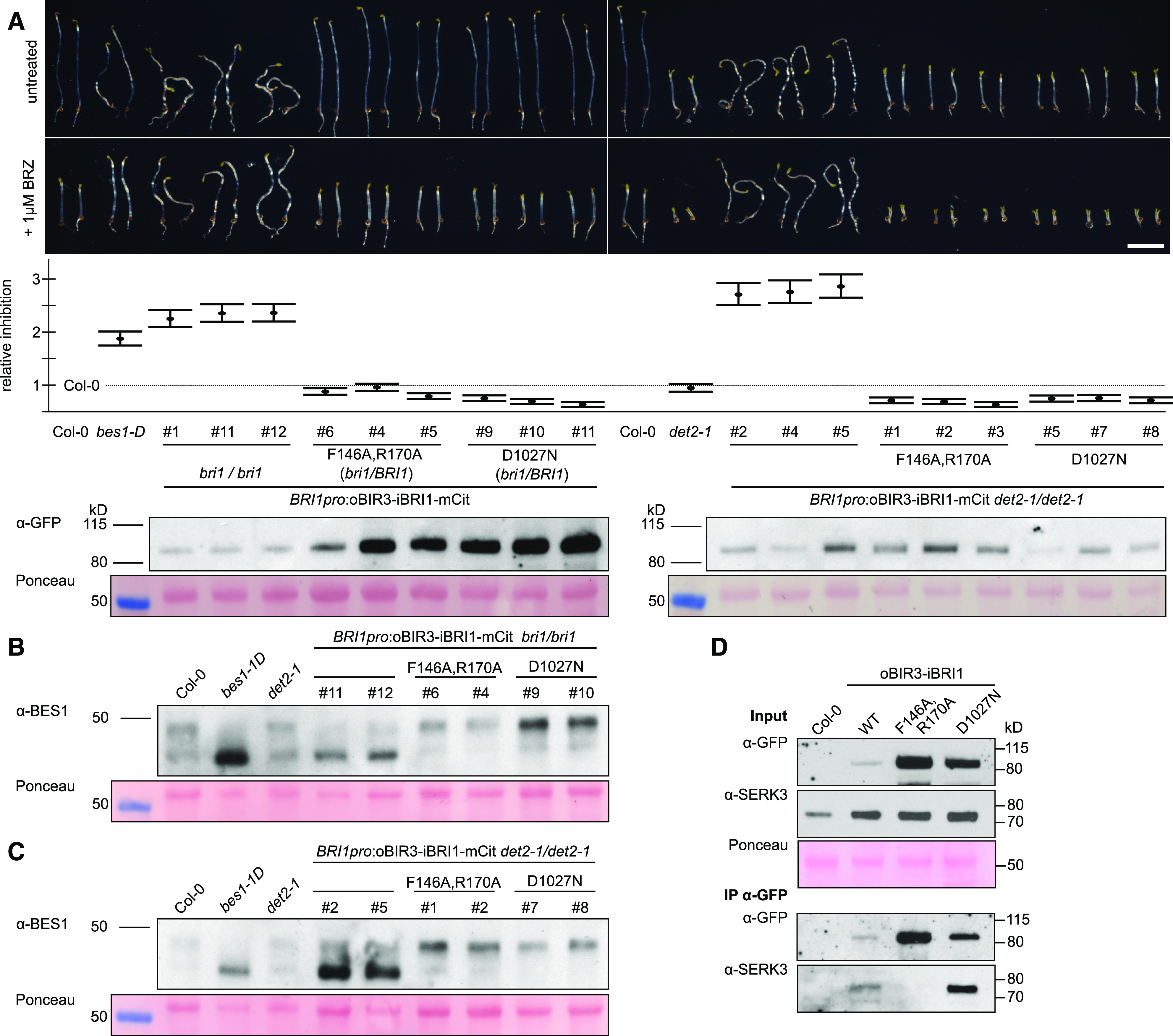
oBIR3-iBRI1 Chimeras Constitutively Activate Brassinosteroid Signaling.
(A) Hypocotyl growth assay of dark-grown seedlings in the presence and absence of the BR biosynthesis inhibitor BRZ. Representative seedlings are shown in the top panel, with the quantification of the data (relative inhibition of hypocotyl growth in the presence of BRZ plotted together with lower and upper confidence intervals) below. For each sample n = 50 hypocotyls from five different half-strength MS plates were measured. The # numbers indicate independent lines. Steady state protein levels were quantified by immunoblot with an anti-GFP antibody (detecting the mCitrine tag present in each chimera); the Ponceau-stained membrane is shown as loading control. Homozygous bri1-null plants could be obtained only upon expression of oBIR3-iBRI1, but not of the control chimeras. Bar = 0.5 cm.
(B) and (C) Anti-BES1 immunoblot on oBIR3-iBRI chimeras in the bri1-null (B) and det2 (C) backgrounds, with the corresponding Ponceau-stained membranes.
(D) co-IP experiment of oBIR3-iBRI1 chimera and SERK3. Shown alongside are the input immunoblots and the Ponceau-stained membrane. WT, wild type.
We next tested whether BIR3-based protein chimeras would also activate a functionally distinct LRR-RK signaling pathway. The LRR-RK HAE shares the same overall structure and activation mechanism as BRI1 (Santiago et al., 2013, 2016; Hohmann et al., 2018b), but the two receptors control very different developmental processes (Li and Chory, 1997; Jinn et al., 2000). We introduced an oBIR3-iHAE fusion construct (Figure 3A) with a C-terminal mCitrine tag driven by the HAE promoter into the hae hsl2 mutant, which displays delayed floral organ abscission (Stenvik et al., 2008). We observed that only expression of the oBIR3-iHAE chimera, but not that of the control constructs bearing point mutations in the BIR3 or HAE ectodomains, rescued the floral abscission phenotype of the hae hsl2 mutant (Figures 3B and 3C). In agreement with these results, oBIR3-iHAE and oBIR3-iHAED1027N interacted with SERK3 in co-IP assays, but not oBIR3F146A/R170A-iHAE (Figure 3D).
Figure 3.
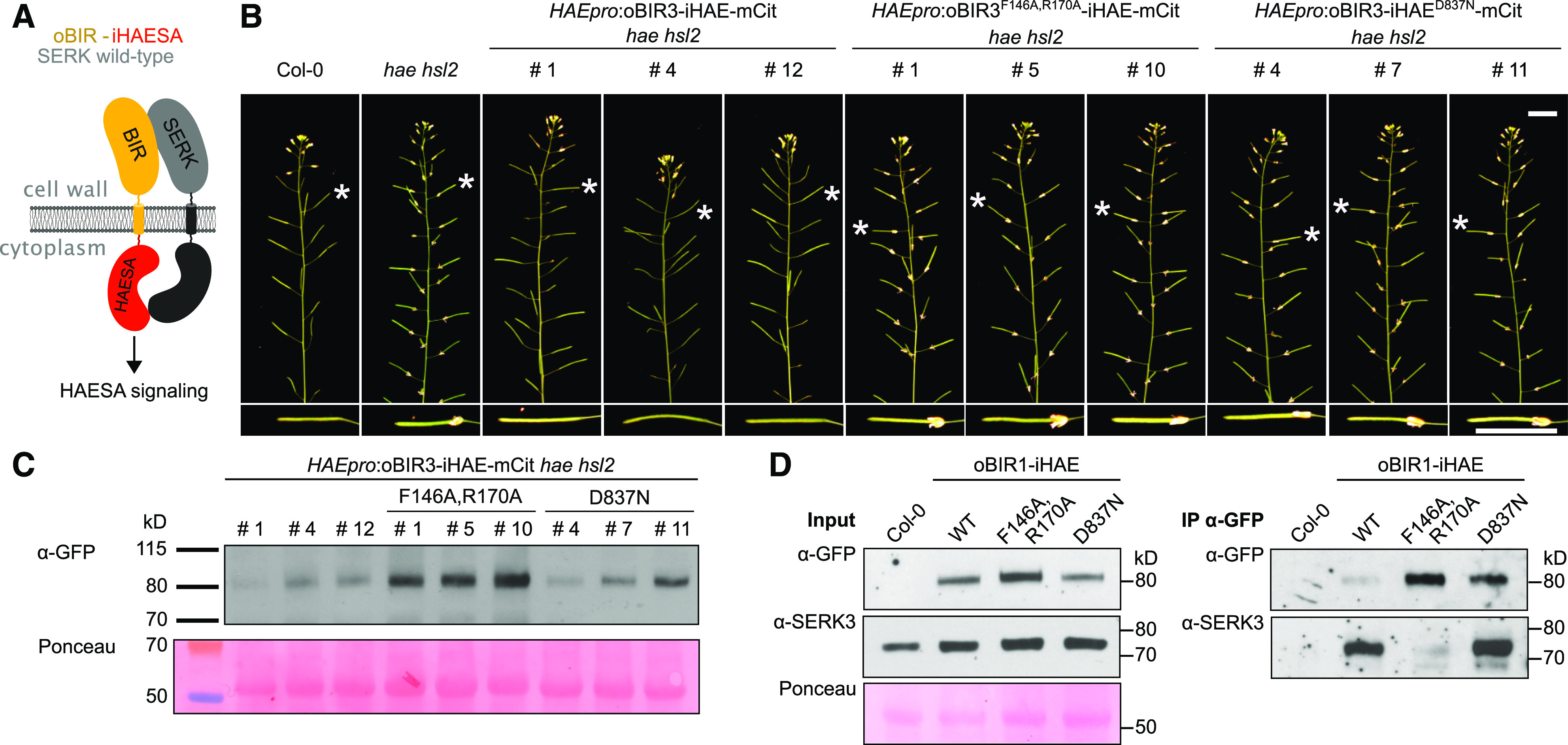
oBIR3-iHAE Chimeras Restore Floral Organ Shedding in hae hsl2 Mutant Plants.
(A) Cartoon representation of the oBIR3-iHAE chimera.
(B) Representative inflorescences of ∼9-week-old Arabidopsis Col-0, hae hsl2, and oBIR3-iHAE chimera, with one silique (indicated with a white star) shown magnified below. The # numbers indicate independent lines. Bars = 2 cm.
(C) Steady state protein levels are visualized by immunoblot with an anti-GFP antibody (detecting the mCitrine tag present in each chimera). The Ponceau-stained membrane is shown as loading control.
(D) co-IP experiment of oBIR3-iHAE chimera and SERK3. Shown alongside are the input immunoblots and the Ponceau-stained membrane from input samples (left). WT, wild type.
SERK proteins were previously shown to allow for receptor activation of the ERECTA family of receptor kinases during protoderm formation and stomatal patterning (Meng et al., 2015). ERECTA forms constitutive complexes with the LRR-RLP TMM to sense EPF peptides in stomatal patterning (Yang and Sack, 1995; Nadeau and Sack, 2002; Lee et al., 2012, 2015; Lin et al., 2017). However, it is not understood at the mechanistic level how SERK coreceptor kinases allow for receptor activation of this LRR-RK/LRR-RLP signaling complex (Lin et al., 2017). To test for the conservation of the receptor activation mechanism between BRI1, HAESA, and ERECTA, we expressed a chimeric oBIR3-iER construct with a C-terminal yellow fluorescent protein for energy transfer (YPet) specifically in the stomata lineage by using the meristemoid-specific MUTE promoter (Figure 4A; Pillitteri et al., 2007). Previous experiments demonstrated that constitutive activation of the ERECTA pathway in differentiating meristemoids leads to developmental arrest of guard mother cells (GMCs; Lampard et al., 2009). To validate the signaling specificity of our oBIR3-iER chimera, we also expressed a chimeric fusion of the innate immunity receptor FLS2 (Gómez-Gómez and Boller, 2000) driven by the MUTE promoter (oBIR3-iFLS2-YPet; Figure 4A; Supplemental Data Set).
Figure 4.
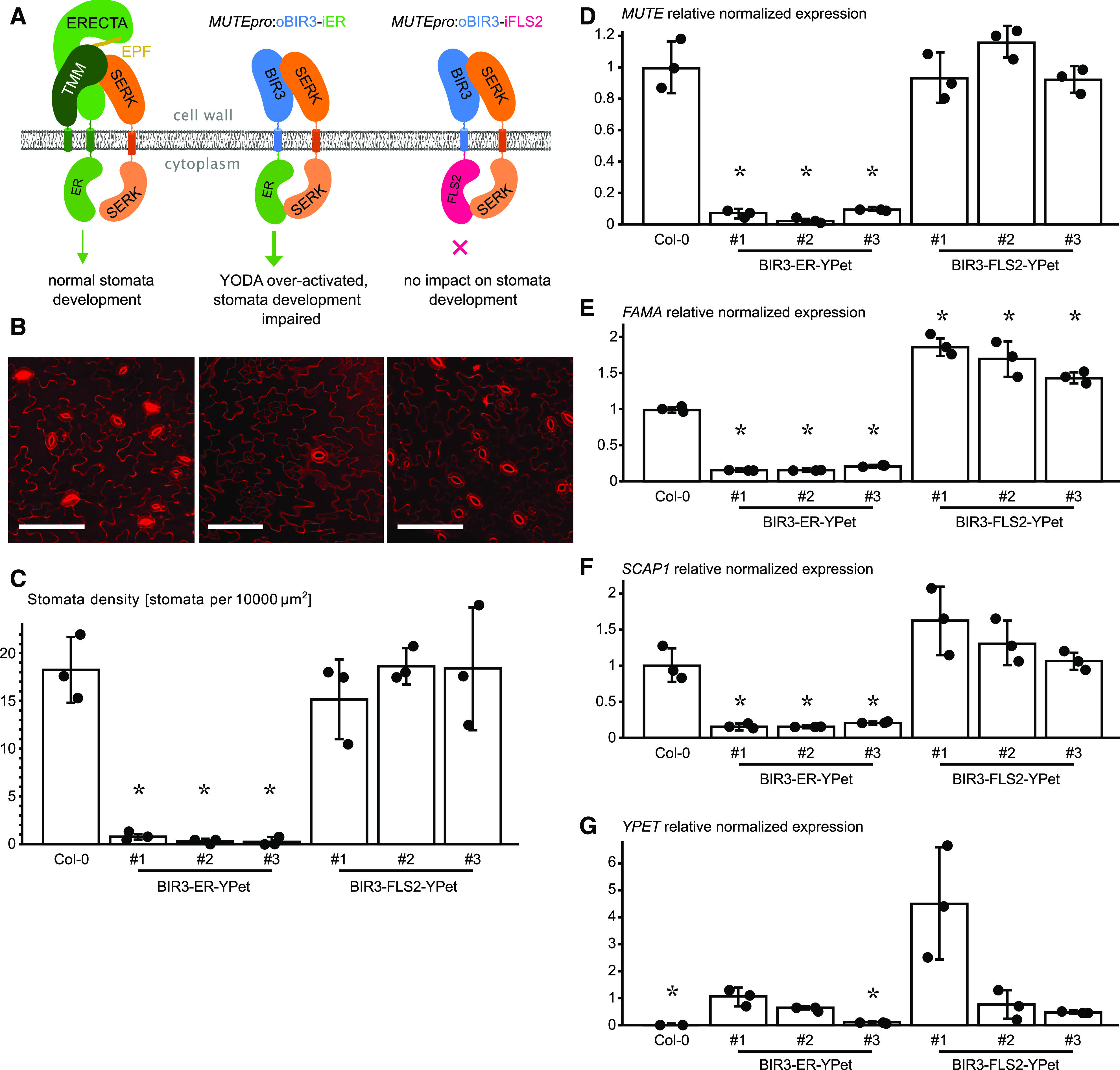
BIR3 chimeras Reveal a Conserved Receptor Activation Mechanism in the LRR-RK ERECTA.
(A) Schematic overview of the ectopically expressed BIR chimera. The receptor kinase ERECTA (ER) interacts with SERK-coreceptor kinases upon ligand (EPF) binding and regulates stomata development (left). Expression of an oBIR3-iER chimera in the epidermis under the MUTE promoter (MUTEpro) leads to pathway over-activation and the loss of stomata (middle), while the expression of an oBIR3-iFLS2 chimera has no effect on stomata development.
(B) Confocal microscopy images of PI-stained epidermis of the indicated genotype. Representative images of Col-0 (left), BIR3-ER-YPet (center), and BIR3-FLS2-YPet (right) are shown. Bar = 100 µm.
(C) Abaxial stomata density of cotyledons (# numbers indicate independent lines). The average value of stomata density for three individual plants of each transgenic line is shown. Error bars depict sds. Individual data points are shown as dots. Significant differences to the wild type are indicated by an asterisk (t test; P < 0.05).
(D) Expression level of the respective transgenes detected by RT-qPCR of MUTE. Data are shown as means ± sd (n = 3). Individual data points are shown as dots. Expression in the wild type was arbitrarily set to 1. Significant differences to the wild-type levels are indicated by an asterisk (t test; P < 0.05).
(E) Relative normalized expression of FAMA. Normalized expression values of FAMA determined by RT-qPCR are shown as means ± sd (n = 3). Individual data points are shown as dots. Expression in the wild type was arbitrarily set to 1. Significant differences to the wild-type levels are indicated by an asterisk (t test; P < 0.05).
(F) Relative normalized expression of SCAP1. Normalized expression values determined by RT-qPCR are shown as means ± sd (n = 3). Individual data points are shown as dots. Expression in the wild type was arbitrarily set to 1. Significant differences to the wild-type levels are indicated by an asterisk (t test; P < 0.05).
(G) Expression level of the respective transgenes detected by RT-qPCR of YPet. Data are shown as means ± sd (n = 3). Individual data points are shown as dots. Expression in the oBIR3-iER-YPET line #1 was arbitrarily set to 1. Significant differences in transgene expression to line #1 is indicated by an asterisk (t test; P < 0.05).
Because of the low abundance of our oBIR3-iER and oBIR3-iFLS2 chimeric fusions in meristemoids, we did not perform immunoblot analyses. We did however confirm that all chimeric constructs were expressed and that the fusion proteins localized to the plasma membrane in meristemoids (Supplemental Figure 4). We selected three representative lines according to their YPet fluorescence. The oBIR3-iER lines showed a drastic reduction in mature stomata and an increase in meristemoid-like cells at the leaf surface (Figures 4B and 4C). Consistent with these observations, the oBIR3-iER chimeras downregulated MUTE expression (Figure 4D). By contrast, none of the oBIR3-iFLS2 lines displayed any significant deviation from the wild-type stomata phenotype, even though the transgenes were expressed at a similar or higher level than the BIR3-ER chimeric constructs (Figure 4G).
To analyze the observed phenotype at a molecular level, we determined the transcript levels of the GMC-specific transcription factor FAMA (Ohashi-Ito and Bergmann, 2006) and the guard cell–specific Dof-type transcription factor STOMATAL CARPENTER1 (SCAP1; Negi et al., 2013). The three independent BIR3-ER lines displayed a strong reduction in FAMA and SCAP1 expression (Figures 4E and 4F), suggesting that the abnormal epidermal cells were arrested at the meristemoid stage and did not express GMC-specific or guard cell–specific genes. None of the oBIR3-iFLS2 lines showed a reduction in FAMA or SCAP1 expression. While SCAP1 transcript levels did not differ significantly from the wild type, FAMA expression was significantly upregulated in these lines relative to the wild type (Figures 4E and 4F).
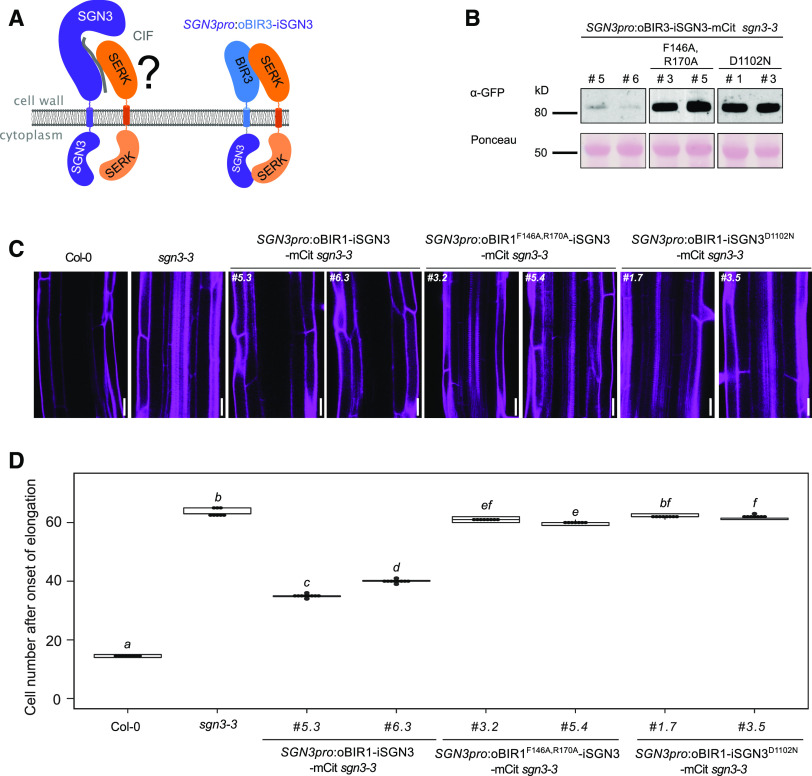
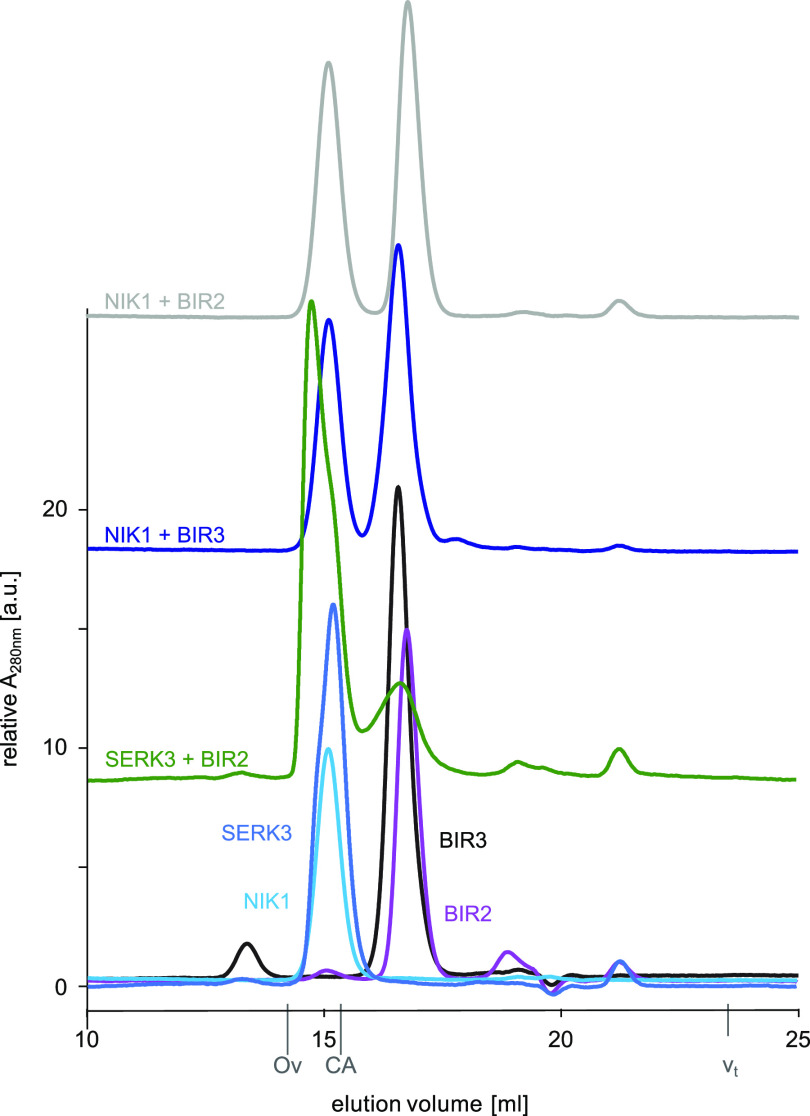
Finally, we tested whether a fusion between the BIR3 ectodomain and the LRR-RK GSO1/SGN3 (Tsuwamoto et al., 2008; Pfister et al., 2014) would restore the apoplastic barrier defects of the sgn3-3 mutant (Pfister et al., 2014). GSO1/SGN3 directly senses the peptide ligands CASPARIAN STRIP INTEGRITY FACTOR1 (CIF1) and CIF2 to ensure proper formation of the Casparian strip, an endodermal diffusion barrier enabling selective nutrient uptake in the root (Pfister et al., 2014; Doblas et al., 2017; Nakayama et al., 2017; Okuda et al., 2020). A biochemical interaction screen recently identified SERK proteins as putative coreceptor kinases for GSO1/SGN3 (Okuda et al., 2020), but it is presently unclear whether SERKs mediate GSO1/SGN3 receptor activation in vivo (Figure 5A). We introduced chimeric constructs, driven by the SGN3 promoter and encoding the chimeric proteins oBIR3-iSGN3, oBIR3-iSGN3F146A,R170A, and oBIR3-iSGN3D1102N into the sgn3-3 mutant background (Figure 5B). As previously described, the sgn3-3 mutant has a nonfunctional apoplastic barrier that can be visualized and quantified by visualizing the uptake of the apoplastic tracer propidium iodide (PI) along the root and its access to the central vasculature (Figure 5C). We established that the oBIR3-iSGN3 chimera, but none of the point mutants, partially rescued the sgn3-3 apoplastic defects (Figures 5C and 5D; Supplemental Data Set), indicating a SERK-mediated GSO1/SGN3 receptor activation mechanism in Casparian strip formation. Notably, BIR ectodomains specifically bind the ectodomains of SERKs (Ma et al., 2017; Hohmann et al., 2018a), while not forming complexes with the LRR ectodomain of the sequence-related NSP-INTERACTING KINASE1 (NIK1; Figure 6). This result suggests that SERK coreceptor kinases may have redundant functions in SGN3/GSO1 signaling in the endodermis (Pfister et al., 2014; Okuda et al., 2020).
Figure 5.
oBIR3-iSGN3 Chimeras Suggest a Role for SERK Proteins in Casparian Strip Formation.
(A) Schematic overview of a biochemically defined SGN3-CIF-SERK signaling complex. The oBIR3-iSGN3 chimera is shown alongside.
(B) Steady state protein levels are visualized by immunoblot with an anti-GFP antibody (detecting the mCitrine tag present in each chimera). The Ponceau-stained membrane is shown as loading control.
(C) Complementation of the sgn3-3 endodermal barrier defect by the chimeric construct SGN3pro:oBIR3-iSGN3. Visualization of endodermal defects with the apoplastic tracer PI, which can reach the stele in barrier-defective plants but is blocked at the endodermis of plants with functional barriers. Pictures were taken around the 50th endodermal cell from the onset of elongation. Bar = 20 µm.
(D) Quantification of the PI block, measured as the number of endodermal cells after the onset of elongation where the PI block is observed. Data are presented as box plots with dot plots overlaid (n ≥ 7). For multiple comparisons between genotypes, Kruskal-Wallis test was performed and nonparametric Tukey’s test was subsequently used as a multiple comparison procedure. Different letters indicate significant difference (P < 0.05).
Figure 6.
LRR Ectodomains of BIRs and NIK1 Do Not Interact in Vitro.
Analytical size-exclusion chromatography binding experiments using the NIK1, BIR2, and BIR3 ectodomains. BIR2 (gray absorption trace) and BIR3 (in dark blue) do not form a complex with NIK1, as their respective elution volumes correspond to that of the isolated protein (BIR2 in magenta, BIR3 in black, NIK1 in light blue). By contrast, SERK3 and BIR2 form a complex (green absorption trace), resulting in a peak elution volume distinct from isolated SERK3 or BIR2 (SERK3 in medium blue). The NIK1 LRR domain shares 49% protein sequence identity with the SERK1 ectodomain. The total volume (vt) is shown together with elution volumes for molecular mass standards (Ov, ovalbumin, 44 kD; CA, carbonic anhydrase, 29 kD). a.u., arbitrary units.
DISCUSSION
The identification of a constitutive, ligand-independent interaction between the LRR ectodomains of two plant membrane signaling proteins prompted us to investigate whether protein chimeras between the BIR3 ectodomain and the cytoplasmic domain of various receptor kinases might lead to constitutively active signaling complexes. Despite the significant structural differences between the LRR-RK-SERK and BIR-SERK complexes, our data demonstrate that a wide range of oBIR3-iLRR-RK chimeras are functional in planta.
Expression of the oBIR3-iBRI1 chimera resulted in a strong, constitutive activation of the brassinosteroid signaling pathway. The gain-of-function effect was more pronounced than in the previously described BRI1sud1 and SERK3elongated alleles (Jaillais et al., 2011; Belkhadir et al., 2012; Hohmann et al., 2018a) and was comparable to the constitutive activation of BES1 (Figure 2A; Yin et al., 2002). The constitutive signaling activity of the oBIR3-iBRI1 chimera depends on (1) the ability of the BIR3 ectodomain to bind SERK ectodomains and (2) the kinase activity of the BRI1 cytosolic segment (Figure 2). These results reinforce the notion that formation of the heterodimeric extracellular signaling complex drives LRR-RK receptor activation and that signaling specificity is encoded in the kinase domain of the receptor, not the coreceptor (Bojar et al., 2014; Hohmann et al., 2018b; Zheng et al., 2019). The similar phenotypes seen in oBIR3-iBRI1 lines and bes1-1D plants suggest that little signal amplification occurs throughout the brassinosteroid signaling pathway (Figure 2).
We noted that different active signaling chimeras accumulated to low protein levels, whereas their corresponding noninteracting or kinase-dead controls accumulated to higher levels (Figures 2A, 3C, and 5B). We speculate that expression, protein accumulation, and/or protein stability of the constitutively active chimeras may be negatively regulated to dampen their signaling capacity. Such regulation may be achieved in part by known processes regulating LRR-RK internalization and degradation in plants (Russinova et al., 2004; Robatzek et al., 2006; Geldner et al., 2007; Beck et al., 2012; Irani et al., 2012; Doblas et al., 2017; Zhou et al., 2018).
Analysis of the oBIR3-iHAE chimeric receptor revealed a strongly conserved activation mechanism between different SERK-dependent LRR-RK signaling pathways, as previously suggested (Figure 3; Hohmann et al., 2018b). In addition, our experiments imply that BIR ectodomains can interact with SERK proteins in the abscission zone and thus that BIR proteins may act as negative regulators of HAESA- and HSL2-mediated signaling cascades in the wild-type plants (Figure 3). In this respect, it is worth noting that the bir1 suppressor SOBIR1/EVERSHED was previously characterized as a genetic component of the floral abscission signaling pathway (Leslie et al., 2010).
The ERECTA family of kinases require SERK coreceptor kinases to control stomatal patterning and immune responses (Meng et al., 2015; Jordá et al., 2016). Our functional oBIR3-iER chimera now suggests that, despite the requirement for TMM, EPF-bound ER signaling complexes are activated by SERK proteins in very similar ways as those previously reported for other LRR-RKs (Figure 4; Hohmann et al., 2017). Expression of the oBIR3-iER chimera in meristemoid cells led to a similar phenotype as that described for the expression of constitutively active versions of YODA, MAPPK KINASE4 (MKK4), and MKK5 (Lampard et al., 2009). This similarity in phenotypes therefore indicates that the oBIR3-iER chimera displays constitutive, ligand-independent signaling activity. The specificity of signal transduction appears to be largely maintained, as expression of oBIR3-iFLS2 led to the wild-type–like stomatal development. At the molecular level, we observed a significant increase in FAMA expression for all tested oBIR3-iFLS2 lines and a decrease in MUTE expression in oBIR3-iER lines. These results are consistent with an antagonistic regulation of these two pathways (Sun et al., 2018). The upregulation of FAMA expression, however, did not significantly alter stomata density, likely because the transcriptional activation of the oBIR3-iFLS2 construct in this experiment was restricted to meristemoid cells by the use of the MUTE promoter and might be compensated by posttranscriptional regulation.
Expression of an oBIR3-iSGN3 chimera partially rescued the Casparian strip phenotype of sgn3-3 plants (Figure 5). BIR ectodomains specifically interacted with SERKs, but not with related LRR-RKs, in vitro (Figure 6). This result suggests that SGN3/GSO1 requires SERKs for receptor activation.
Taken together, our simple, Lego-style assembly of BIR3 chimeras (Figure 7) and the availability of suitable control lines now allow for the genetic characterization of orphan LRR-RKs with unknown/unclear loss-of-function phenotypes and the dissection of their potential activation mechanism. BIR3 protein chimeras may also be of use for biochemical or genetic interaction screens in which a constitutively active form of the receptor is desirable.
Figure 7.
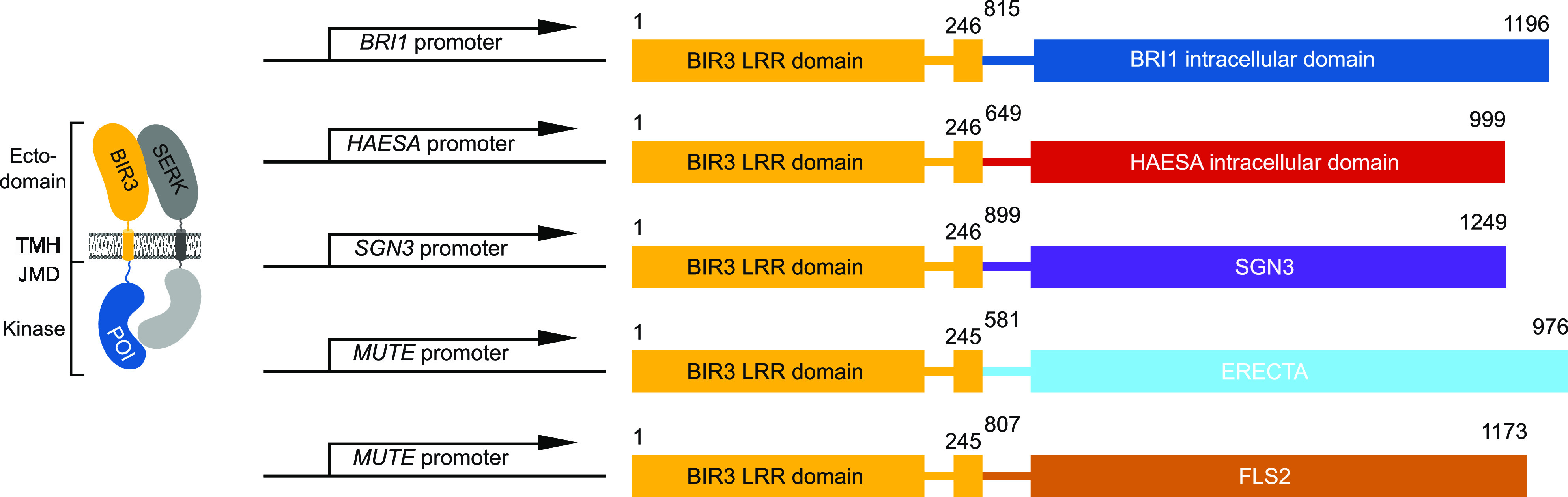
Design Principles of BIR Chimeras.
Schematic overview of selected BIR3 chimeras used in this study. Chimeric constructs are expressed under the endogenous promoter of the respective receptor gene. JMD, juxtamembrane domain; POI, protein of interest; TMH, transmembrane helix.
METHODS
Plant Materials, Growth Conditions, and Generation of Transgenic Lines
To design chimeric receptor kinases, we predicted the transmembrane helix of all LRR-RKs using TMHMM version 2.0 (https://services.healthtech.dtu.dk/service.php?TMHMM-2.0; Krogh et al., 2001). We fused the native signal peptide, extracellular domain, and the transmembrane helix from Arabidopsis BIR3 (residues 1 to 246) to the juxtamembrane and kinase domains of the respective receptor (BRI1 residues 815 to 1196, HAE residues 649 to 999, and SGN3 residues 899 to 1249). We added no additional linker sequences (Figure 7). We PCR amplified all fragments from Arabidopsis (accession Columbia-0 [Col-0]) genomic or cDNA and cloned the resulting PCR products into pDONR221 (Thermo Fisher Scientific) using Gibson-cloning technology. We introduced mutations through site-directed mutagenesis (Supplemental Table 1). We assembled binary vectors via multi-site Gateway technology into the binary vector pB7m34GW, conferring Basta resistance gene (Thermo Fisher Scientific). We introduced all constructs into Agrobacterium (Agrobacterium tumefaciens) strain C58C1 harboring the pGV2260 plasmid and transformed Arabidopsis (Arabidopsis thaliana) plants using the floral dip method (Clough and Bent, 1998). For each construct, we selected 12 primary transformants. We confirmed the presence of a single insertion by segregation in T2 lines of which we selected three independent lines based on protein accumulation for subsequent analysis. We analyzed all lines in the T3 generation. All transgenic lines generated in the course of this study are listed in Supplemental Table 2.
We used the bri1 null allele GABI_134E10 (Jaillais et al., 2011), bes1-1D (ABRC CS65988; Yin et al., 2002), and det2-1 (ABRC CS6159; Chory et al., 1991). The mutants hae hsl2 and sgn3-3 were previously reported by Stenvik et al. (2008) and Pfister et al. (2014). All plants were grown in soil (Einheitserde Classic, ref. CL Ton Kokos mix containing white peat moss, coco fiber, and clay with 30% [v/v] perlite added) under 50% humidity at 21°C and a 16-h-light/8-h dark cycle (photosynthetic active radiation was ∼150 μmol m−2 s−1 originating from Sylvania–T8 luxline plus, half F58W/T8/840 bulbs [4000 K/5200 lumen] and half F58W/T8/830 [3000 K/5200 lumen] light bulbs).
To generate the chimeric MUTEpro:oBIR3-iFLS2-YPet and MUTEpro:oBIR3-iER-YPet constructs, we synthesized (Baseclear) a 1946-bp DNA fragment encoding the N-terminal extracellular domain of BIR3 (residues 1 to 245), followed by a short multiple cloning site, the coding sequence of YPet, and a 411-bp terminator sequence from the Arabidopsis UBQ10 gene. We inserted the synthetic DNA fragment in the T-DNA of a modified pCambia3300 binary vector. We PCR amplified a 2432-bp promoter region from the Arabidopsis MUTE gene from Col-0 genomic DNA and inserted the resulting PCR product directly upstream of the synthetic BIR3 fusion construct by in-fusion cloning (Clontech). We PCR amplified the coding regions for the intracellular domains of ER (residues 581 to 976) and FLS2 (residues 807 to 1173) from cDNAs derived from Arabidopsis seedlings and inserted in frame between the coding region of the BIR3 extracellular domain and the YPet coding region (Figure 7). All constructs were confirmed by Sanger sequencing.
Hypocotyl Growth Assay
Seeds were surface sterilized, stratified at 4°C for 2 d, and plated on half-strength Murashige and Skoog (MS) medium containing 0.8% (w/v) agar and supplemented with 1 μM BRZ from a 10 mM stock solution in 100% DMSO (Tokyo Chemical Industry) or, for the controls, with 0.1% (v/v) DMSO. Following a 1-h light exposure to induce germination, we wrapped the plates in aluminum foil and incubated them in the dark at 22°C for 5 d. We then scanned the plates at 600 dots per inch resolution on a regular flatbed scanner (CanoScan 9000F; Canon), measured hypocotyl lengths using Fiji (Schindelin et al., 2012), and analyzed the results in R version 3.6.1 (R Core Team, 2014) using the packages mratios (Kitsche and Hothorn, 2014) and multcomp (Hothorn et al., 2008). Rather than P-values, we report unadjusted 95% confidence limits for fold-changes. We used a mixed-effects model for the ratio of a given line to the wild-type Col-0, allowing for heterogeneous variances, to analyze log-transformed end point hypocotyl lengths. To evaluate treatment-by-mutant interactions, we calculated the 95% two-sided confidence intervals for the relative inhibition (Col-0: untreated versus BRZ-treated hypocotyl length)/(any genotype: untreated versus BRZ-treated hypocotyl length) for the log-transformed length.
Plant Protein Extraction and Immunoprecipitation
We sowed surface-sterilized seeds on half-strength MS medium with 0.8% (w/v) agar and allowed seedlings to grow for ∼14 d after release from stratification. We harvested seedlings, padded them dry carefully on paper towels, snap-froze them in liquid nitrogen, and ground them to a fine powder using a pre-cooled mortar and pestle. We resuspended 1 g of powder per sample in 3 mL of ice-cold extraction buffer (50 mM Bis-Tris, pH 7.0, 150 mM NaCl, 10% [v/v] glycerol, 1% [v/v] Triton X-100, 5 mM DTT, and protease inhibitor cocktail [P9599; Sigma-Aldrich]) and agitated gently at 4°C for 1 h. Subsequently, we centrifuged samples (30 min at 16,000g and 4°C), transferred the supernatant to a fresh tube, and estimated protein concentration by Bradford assay against a BSA standard curve.
For each co-IP, we incubated 20 mg of total protein in a volume of 5 mL with 50 μL of antiGFP superparamagnetic MicroBeads (Miltenyi Biotec) for 1 h at 4°C with gentle agitation. Using a magnetic rack and μMACS columns (washed once with extraction buffer; Miltenyi Biotec), we collected the beads and then washed them four times with 1 mL of ice-cold extraction buffer. We then eluted bound proteins twice in 20 μL of extraction buffer pre-heated to 95°C. We separated samples on 10% SDS-PAGE gels and analyzed resolved proteins by standard immunoblot using the following antibodies: anti-GFP antibody coupled to horseradish peroxidase (anti-GFP-HRP, 130-091-833; Miltenyi Biotec) at 1:2000 dilution to detect mCitrine; anti-SERK3 (Bojar et al., 2014) at 1:5000 dilution in conjunction with a secondary anti-rabbit HRP antibody (1:10,000, no. 401353; Calbiochem) to detect SERK3.
Immunoblot for BES1
For each sample, we harvested ∼100 µg of 7-d-old seedlings grown on half-strength MS medium with 0.8% (w/v) agar, froze the tissue in liquid nitrogen, and ground it to a fine powder using a bead mill (MM400; Retsch). We resuspended samples in ∼200 μL of ice-cold extraction buffer (25 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% (w/v) SDS, 10 mM DTT, and protease inhibitor cocktail [P9599; Sigma-Aldrich]), and incubated them with gentle agitation for 1 h at 4°C before centrifugation for 30 min at 4°C and 16,000g. We transferred the supernatant to a fresh tube and assessed their protein concentration by Bradford assay. We separated 80 µg of total protein on a 12% SDS-PAGE gel and analyzed the resolved proteins by immunoblot (primary antibody: anti-BES1, 1:2000 [Yin et al., 2002]; secondary antibody: anti-rabbit HRP [1:10,000, no. 40135; Calbiochem).
Stomata Density Measurements and Microscopy
We used 7-d-old T2 seedlings resistant to Basta to determine stomata density. For confocal imaging, we incubated seedlings in a 10 mg/L PI solution for 30 min and then washed them with water. We imaged the abaxial epidermal regions of cotyledons using a confocal LSM 780 nonlinear optical microscope (Zeiss) equipped with a Plan-Apochromat 25×/0.8 Imm Corr differential interference contrast objective. We visualized PI staining with an excitation wavelength of 514 nm and recorded emission between 566 nm and 643 nm. We counted mature stomata over a 0.5 mm by 0.5 mm epidermal area for three seedlings per line. For analysis of fluorescent protein accumulation, we stained 2-d-old seedlings with PI solution and imaged them by confocal microscopy as described above. In addition, we recorded YPet fluorescence by excitation at 514 nm and recorded emission between 517 nm and 544 nm.
Gene Expression Analysis
We used 7-d-old T2 seedlings to analyze transcript levels for the transgene and the endogenous genes. For each independent line, we extracted RNA from 24 pooled T2 seedlings using the RNase Plant Mini Kit (Qiagen). We synthesized first-strand cDNAs with the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific). We measured the relative abundance of the endogenous FAMA and SCAP1 transcripts as well as chimeric YPet-containing BIR3 transcripts by RT-qPCR; program: 1. 50°C for 10 min, 2. 95°C for 5 min, 3. 95°C for 10 s, 4. 60°C for 30 s, Plate Read; repeat step 3 – 4 40 times; 5. 95°C for 10 s, 6. ramp 65°C to 95 and increase 0.5°C every 5 s, Plate Read). We used the expression levels of endogenous ACTIN2 for normalization.
PI Permeability Assay and Confocal Microscopy of the Wild-Type and Complemented sgn3-3 Plants
We performed PI permeability assays on 5-d-old seedlings. Briefly, we stained the seedlings in the dark for 10 min in 10 µg/mL PI, rinsed them twice in water, and quantified the staining as previously described by Naseer et al. (2012). We counted endodermal cells using an epifluorescence microscope (Leica). We acquired representative confocal images with an SP8 microscope (Leica), with excitation and detection windows set as follows for PI: excitation, 488 nm; emission, 500 to 550 nm. We processed and analyzed confocal images using ImageJ (Schindelin et al., 2012). We performed all statistical analyses in R (R Core Team, 2014). For multiple comparisons between genotypes, we performed the Kruskal-Wallis test and nonparametric Tukey’s test as a multiple comparison procedure. Different letters indicate significant differences (P < 0.05). Data are presented as box plots overlaid with dot plots.
Protein Expression, Purification, and Size-Exclusion Chromatography
We PCR amplified the coding regions of Arabidopsis NIK132-248 and SERK31-220 from Arabidopsis Col-0 cDNAs, and PCR-amplified Arabidopsis BIR21–222 and BIR31–213 from Arabidopsis Col-0 genomic DNA, before cloning all PCR products into a modified pFastBac vector (Geneva Biotech), providing a tobacco etch virus protease–cleavable C-terminal StrepII-9xHis tag. We fused NIK1 to an N-terminal azurocidin secretion peptide. We expressed proteins by infection of cabbage looper (Trichoplusia ni) cells (strain Tnao38; Hashimoto et al., 2010) with 15 mL of Autographa californica nucleopolyhedrovirus in 250 mL of cells at a density of ∼2 × 106 cells mL−1 (multiplicity of infection was ∼3), followed by incubation for 26 h at 28°C and 110 rpm shaking and then for another 48 h at 22°C and 110 rpm. We purified the secreted proteins from the supernatant by sequential Ni2+ (HisTrap excel; GE Healthcare; equilibrated in 25 mM KPi pH 7.8, 500 mM NaCl) and StrepII (Strep-Tactin XT; IBA; equilibrated in 25 mM Tris-HCl, pH 8.0, 250 mM NaCl, and 1 mM EDTA) affinity chromatography followed by size-exclusion chromatography on a HiLoad 16/600 Superdex 200-pg column (GE Healthcare), equilibrated in 20 mM sodium citrate, pH 5.0, and 250 mM NaCl. The theoretical molecular weight of the purified ectodomains is 23.6 kD for NIK132-248, 24.6 kD for SERK326-220, 23.4 kD for BIR21–222, and 24.0 kD for BIR325–213.
For analytical size-exclusion chromatography experiments, we pre-equilibrated a Superdex 200 increase 10/300 GL column (GE Healthcare) in 20 mM sodium citrate, pH 5.0, and 250 mM NaCl. For each run, we injected 40 μg of the individual NIK1, SERK3, BIR2, or BIR3 ectodomains in a volume of 100 μL and monitored elution at a rate of 0.75 mL min−1 by UV light absorbance at 280 nm. To probe interactions between NIK1, SERK3, BIR2, and BIR3, we mixed 40 μg of the respective proteins in a total volume of 100 μL and incubated the mixture on ice for 30 min before analysis as outlined above.
Accession Numbers
Sequence data from this article can be found in the National Center for Biotechnology Information and The Arabidopsis Information Resource databases under the following accession numbers: BRI1 (At4g39400); BIR2 (At3g28450); BIR3 (At1g27190); SERK1 (At1g71830); SERK3 (At4g33430); BES1 (At1g19350); HAE (At4g28490); HSL2 (At5g65710); ER (At2g26330); FLS2 (At5g46330); MUTE (At3g06120); UBQ10 (At4g05320); FAMA (At3g24140); SCAP (At5g65590); ACTIN2 (At3g18780); GSO1/SGN3 (At4g20140).
Supplemental Data
Supplemental Figure 1. Rosette phenotypes of plants expressing oBIR3-iBRI1 chimeras and raw data for the hypocotyl growth assays (supports Figure 2A).
Supplemental Figure 2. Full immunoblot films and Ponceau-stained membranes (supports Figures 2A to 2C).
Supplemental Figure 3. Full immunoblot films and Ponceau-stained membranes (supports Figures 2D, 3C, and D3D).
Supplemental Figure 4. Accumulation and subcellular localization of oBIR3-iER and oBIR3-iFLS2 chimeras (supports Figure 4).
Supplemental Table 1. Primers used in this study.
Supplemental Table 2. Transgenic lines created in this study.
Supplemental Data Set. Summary of statistical analyses in this study.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Jenny Russinova for providing us with the BES1 antibody. This work was supported by the Swiss National Science Foundation (grant 31003A_176237 to M.H., grant 31CP30_180213 to M.H., and grant 3003A_179159 to M. Barberon), the Howard Hughes Medical Institute (International Research Scholar Award to M.H.), the Max Planck Society, and the Deutsche Forschungsgemeinschaft (grant SFB 1101/B01 to M. Bayer).
AUTHOR CONTRIBUTIONS
U.H., M. Bayer, and M.H. designed the project and the experiments. U.H., P.R., K.W., L.L.-O., J.N., and A.H. performed the experiments. U.H., P.R., K.W., M. Barberon, M. Bayer, and M.H. analyzed the data and prepared the figures. Funding was secured by M. Barberon, M. Bayer, and M.H.
Footnotes
Articles can be viewed without a subscription.
References
- Albert, M., Jehle, A.K., Fürst, U., Chinchilla, D., Boller, T., Felix, G.(2013). A two-hybrid-receptor assay demonstrates heteromer formation as switch-on for plant immune receptors. Plant Physiol. 163: 1504–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anne, P., Amiguet-Vercher, A., Brandt, B., Kalmbach, L., Geldner, N., Hothorn, M., Hardtke, C.S.(2018). CLERK is a novel receptor kinase required for sensing of root-active CLE peptides in Arabidopsis. Development 145: dev162354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asami, T., Min, Y.K., Nagata, N., Yamagishi, K., Takatsuto, S., Fujioka, S., Murofushi, N., Yamaguchi, I., Yoshida, S.(2000). Characterization of brassinazole, a triazole-type brassinosteroid biosynthesis inhibitor. Plant Physiol. 123: 93–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck, M., Zhou, J., Faulkner, C., MacLean, D., Robatzek, S.(2012). Spatio-temporal cellular dynamics of the Arabidopsis flagellin receptor reveal activation status-dependent endosomal sorting. Plant Cell 24: 4205–4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkhadir, Y., Jaillais, Y., Epple, P., Balsemão-Pires, E., Dangl, J.L., Chory, J.(2012). Brassinosteroids modulate the efficiency of plant immune responses to microbe-associated molecular patterns. Proc. Natl. Acad. Sci. USA 109: 297–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann, D.C., Lukowitz, W., Somerville, C.R.(2004). Stomatal development and pattern controlled by a MAPKK kinase. Science 304: 1494–1497. [DOI] [PubMed] [Google Scholar]
- Blaum, B.S., Mazzotta, S., Nöldeke, E.R., Halter, T., Madlung, J., Kemmerling, B., Stehle, T.(2014). Structure of the pseudokinase domain of BIR2, a regulator of BAK1-mediated immune signaling in Arabidopsis. J. Struct. Biol. 186: 112–121. [DOI] [PubMed] [Google Scholar]
- Bojar, D., Martinez, J., Santiago, J., Rybin, V., Bayliss, R., Hothorn, M.(2014). Crystal structures of the phosphorylated BRI1 kinase domain and implications for brassinosteroid signal initiation. Plant J. 78: 31–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, W., Lv, M., Wang, Y., Wang, P.-A., Cui, Y., Li, M., Wang, R., Gou, X., Li, J.(2019). BES1 is activated by EMS1-TPD1-SERK1/2-mediated signaling to control tapetum development in Arabidopsis thaliana. Nat. Commun. 10: 4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X., Zuo, S., Schwessinger, B., Chern, M., Canlas, P.E., Ruan, D., Zhou, X., Wang, J., Daudi, A., Petzold, C.J., Heazlewood, J.L., Ronald, P.C.(2014). An XA21-associated kinase (OsSERK2) regulates immunity mediated by the XA21 and XA3 immune receptors. Mol. Plant 7: 874–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chory, J., Nagpal, P., Peto, C.A.(1991). Phenotypic and genetic analysis of det2, a new mutant that affects light-regulated seedling development in Arabidopsis. Plant Cell 3: 445–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., Bent, A.F.(1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Cui, Y., et al. (2018). CIK receptor kinases determine cell fate specification during early anther development in Arabidopsis. Plant Cell 30: 2383–2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doblas, V.G., Smakowska-Luzan, E., Fujita, S., Alassimone, J., Barberon, M., Madalinski, M., Belkhadir, Y., Geldner, N.(2017). Root diffusion barrier control by a vasculature-derived peptide binding to the SGN3 receptor. Science 355: 280–284. [DOI] [PubMed] [Google Scholar]
- Fujioka, S., Li, J., Choi, Y.H., Seto, H., Takatsuto, S., Noguchi, T., Watanabe, T., Kuriyama, H., Yokota, T., Chory, J., Sakurai, A.(1997). The Arabidopsis deetiolated2 mutant is blocked early in brassinosteroid biosynthesis. Plant Cell 9: 1951–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, M., Wang, X., Wang, D., Xu, F., Ding, X., Zhang, Z., Bi, D., Cheng, Y.T., Chen, S., Li, X., Zhang, Y.(2009). Regulation of cell death and innate immunity by two receptor-like kinases in Arabidopsis. Cell Host Microbe 6: 34–44. [DOI] [PubMed] [Google Scholar]
- Geldner, N., Hyman, D.L., Wang, X., Schumacher, K., Chory, J.(2007). Endosomal signaling of plant steroid receptor kinase BRI1. Genes Dev. 21: 1598–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Gómez, L., Boller, T.(2000). FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5: 1003–1011. [DOI] [PubMed] [Google Scholar]
- Halter, T., et al. (2014). The leucine-rich repeat receptor kinase BIR2 is a negative regulator of BAK1 in plant immunity. Curr. Biol. 24: 134–143. [DOI] [PubMed] [Google Scholar]
- Hashimoto, Y., Zhang, S., Blissard, G.W.(2010). Ao38, a new cell line from eggs of the black witch moth, Ascalapha odorata (Lepidoptera: Noctuidae), is permissive for AcMNPV infection and produces high levels of recombinant proteins. BMC Biotechnol. 10: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, Z., Wang, Z.Y., Li, J., Zhu, Q., Lamb, C., Ronald, P., Chory, J.(2000). Perception of brassinosteroids by the extracellular domain of the receptor kinase BRI1. Science 288: 2360–2363. [DOI] [PubMed] [Google Scholar]
- Hohmann, U., Lau, K., Hothorn, M.(2017). The structural basis of ligand perception and signal activation by receptor kinases. Annu. Rev. Plant Biol. 68: 109–137. [DOI] [PubMed] [Google Scholar]
- Hohmann, U., Nicolet, J., Moretti, A., Hothorn, L.A., Hothorn, M.(2018a). The SERK3 elongated allele defines a role for BIR ectodomains in brassinosteroid signalling. Nat. Plants 4: 345–351. [DOI] [PubMed] [Google Scholar]
- Hohmann, U., Santiago, J., Nicolet, J., Olsson, V., Spiga, F.M., Hothorn, L.A., Butenko, M.A., Hothorn, M.(2018b). Mechanistic basis for the activation of plant membrane receptor kinases by SERK-family coreceptors. Proc. Natl. Acad. Sci. USA 115: 3488–3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hothorn, M., Belkhadir, Y., Dreux, M., Dabi, T., Noel, J.P., Wilson, I.A., Chory, J.(2011). Structural basis of steroid hormone perception by the receptor kinase BRI1. Nature 474: 467–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hothorn, T., Bretz, F., Westfall, P.(2008). Simultaneous inference in general parametric models. Biom. J. 50: 346–363. [DOI] [PubMed] [Google Scholar]
- Hu, C., et al. (2018). A group of receptor kinases are essential for CLAVATA signalling to maintain stem cell homeostasis. Nat. Plants 4: 205–211. [DOI] [PubMed] [Google Scholar]
- Huang, J., Zhang, T., Linstroth, L., Tillman, Z., Otegui, M.S., Owen, H.A., Zhao, D.(2016). Control of anther cell differentiation by the small protein ligand TPD1 and its receptor EMS1 in Arabidopsis. PLoS Genet. 12: e1006147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imkampe, J., et al. (2017). The Arabidopsis leucine-rich repeat receptor kinase BIR3 negatively regulates BAK1 receptor complex formation and stabilizes BAK1. Plant Cell 29: 2285–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irani, N.G., et al. (2012). Fluorescent castasterone reveals BRI1 signaling from the plasma membrane. Nat. Chem. Biol. 8: 583–589. [DOI] [PubMed] [Google Scholar]
- Jaillais, Y., Belkhadir, Y., Balsemão-Pires, E., Dangl, J.L., Chory, J.(2011). Extracellular leucine-rich repeats as a platform for receptor/coreceptor complex formation. Proc. Natl. Acad. Sci. USA 108: 8503–8507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinn, T.L., Stone, J.M., Walker, J.C.(2000). HAESA, an Arabidopsis leucine-rich repeat receptor kinase, controls floral organ abscission. Genes Dev. 14: 108–117. [PMC free article] [PubMed] [Google Scholar]
- Jordá, L., Sopeña-Torres, S., Escudero, V., Nuñez-Corcuera, B., Delgado-Cerezo, M., Torii, K.U., Molina, A.(2016). ERECTA and BAK1 receptor like kinases interact to regulate immune responses in Arabidopsis. Front. Plant Sci. 7: 897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitsche, A., Hothorn, L.A.(2014). Testing for qualitative interaction using ratios of treatment differences. Stat. Med. 33: 1477–1489. [DOI] [PubMed] [Google Scholar]
- Krogh, A., Larsson, B., von Heijne, G., Sonnhammer, E.L.(2001). Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305: 567–580. [DOI] [PubMed] [Google Scholar]
- Lampard, G.R., Lukowitz, W., Ellis, B.E., Bergmann, D.C.(2009). Novel and expanded roles for MAPK signaling in Arabidopsis stomatal cell fate revealed by cell type-specific manipulations. Plant Cell 21: 3506–3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.S., Hnilova, M., Maes, M., Lin, Y.-C.L., Putarjunan, A., Han, S.-K., Avila, J., Torii, K.U.(2015). Competitive binding of antagonistic peptides fine-tunes stomatal patterning. Nature 522: 439–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.S., Kuroha, T., Hnilova, M., Khatayevich, D., Kanaoka, M.M., McAbee, J.M., Sarikaya, M., Tamerler, C., Torii, K.U.(2012). Direct interaction of ligand-receptor pairs specifying stomatal patterning. Genes Dev. 26: 126–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie, M.E., Lewis, M.W., Youn, J.-Y., Daniels, M.J., Liljegren, S.J.(2010). The EVERSHED receptor-like kinase modulates floral organ shedding in Arabidopsis. Development 137: 467–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., Chory, J.(1997). A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90: 929–938. [DOI] [PubMed] [Google Scholar]
- Li, J., Wen, J., Lease, K.A., Doke, J.T., Tax, F.E., Walker, J.C.(2002). BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110: 213–222. [DOI] [PubMed] [Google Scholar]
- Lin, G., Zhang, L., Han, Z., Yang, X., Liu, W., Li, E., Chang, J., Qi, Y., Shpak, E.D., Chai, J.(2017). A receptor-like protein acts as a specificity switch for the regulation of stomatal development. Genes Dev. 31: 927–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, C., Liu, Y., Bai, B., Han, Z., Tang, J., Zhang, H., Yaghmaiean, H., Zhang, Y., Chai, J.(2017). Structural basis for BIR1-mediated negative regulation of plant immunity. Cell Res. 27: 1521–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubayashi, Y.(2014). Posttranslationally modified small-peptide signals in plants. Annu. Rev. Plant Biol. 65: 385–413. [DOI] [PubMed] [Google Scholar]
- Meng, X., Chen, X., Mang, H., Liu, C., Yu, X., Gao, X., Torii, K.U., He, P., Shan, L.(2015). Differential function of arabidopsis SERK family receptor-like kinases in stomatal patterning. Curr. Biol. 25: 2361–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng, X., Zhou, J., Tang, J., Li, B., de Oliveira, M.V.V., Chai, J., He, P., Shan, L.(2016). Ligand-induced receptor-like kinase complex regulates floral organ abscission in Arabidopsis. Cell Rep. 14: 1330–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussu, S., Santiago, J.(2019). Structural biology of cell surface receptor-ligand interactions. Curr. Opin. Plant Biol. 52: 38–45. [DOI] [PubMed] [Google Scholar]
- Nadeau, J.A., Sack, F.D.(2002). Control of stomatal distribution on the Arabidopsis leaf surface. Science 296: 1697–1700. [DOI] [PubMed] [Google Scholar]
- Nakayama, T., Shinohara, H., Tanaka, M., Baba, K., Ogawa-Ohnishi, M., Matsubayashi, Y.(2017). A peptide hormone required for Casparian strip diffusion barrier formation in Arabidopsis roots. Science 355: 284–286. [DOI] [PubMed] [Google Scholar]
- Nam, K.H., Li, J.(2002). BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110: 203–212. [DOI] [PubMed] [Google Scholar]
- Naseer, S., Lee, Y., Lapierre, C., Franke, R., Nawrath, C., Geldner, N.(2012). Casparian strip diffusion barrier in Arabidopsis is made of a lignin polymer without suberin. Proc. Natl. Acad. Sci. USA 109: 10101–10106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negi, J., Moriwaki, K., Konishi, M., Yokoyama, R., Nakano, T., Kusumi, K., Hashimoto-Sugimoto, M., Schroeder, J.I., Nishitani, K., Yanagisawa, S., Iba, K.(2013). A Dof transcription factor, SCAP1, is essential for the development of functional stomata in Arabidopsis. Curr. Biol. 23: 479–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi, T., Fujioka, S., Takatsuto, S., Sakurai, A., Yoshida, S., Li, J., Chory, J.(1999). Arabidopsis det2 is defective in the conversion of (24R)-24-methylcholest-4-En-3-one to (24R)-24-methyl-5alpha-cholestan-3-one in brassinosteroid biosynthesis. Plant Physiol. 120: 833–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosaki, S., Miyakawa, T., Xu, Y., Nakamura, A., Hirabayashi, K., Asami, T., Nakano, T., Tanokura, M.(2018). Structural basis for brassinosteroid response by BIL1/BZR1. Nat. Plants 4: 771–776. [DOI] [PubMed] [Google Scholar]
- Ohashi-Ito, K., Bergmann, D.C.(2006). Arabidopsis FAMA controls the final proliferation/differentiation switch during stomatal development. Plant Cell 18: 2493–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda, S., Fujita, S., Moretti, A., Hohmann, U., Doblas, V.G., Ma, Y., Pfister, A., Brandt, B., Geldner, N., Hothorn, M.(2020). Molecular mechanism for the recognition of sequence-divergent CIF peptides by the plant receptor kinases GSO1/SGN3 and GSO2. Proc. Natl. Acad. Sci. USA 117: 2693–2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perraki, A., et al. (2018). Phosphocode-dependent functional dichotomy of a common co-receptor in plant signalling. Nature 561: 248–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister, A., et al. (2014). A receptor-like kinase mutant with absent endodermal diffusion barrier displays selective nutrient homeostasis defects. eLife 3: e03115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillitteri, L.J., Sloan, D.B., Bogenschutz, N.L., Torii, K.U.(2007). Termination of asymmetric cell division and differentiation of stomata. Nature 445: 501–505. [DOI] [PubMed] [Google Scholar]
- R Core Team (2014). R: A language and environment for statistical computing.. (Vienna: R Foundation for Statistical Computing; ). [Google Scholar]
- Robatzek, S., Chinchilla, D., Boller, T.(2006). Ligand-induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis. Genes Dev. 20: 537–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russinova, E., Borst, J.-W., Kwaaitaal, M., Caño-Delgado, A., Yin, Y., Chory, J., de Vries, S.C.(2004). Heterodimerization and endocytosis of Arabidopsis brassinosteroid receptors BRI1 and AtSERK3 (BAK1). Plant Cell 16: 3216–3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago, J., Brandt, B., Wildhagen, M., Hohmann, U., Hothorn, L.A., Butenko, M.A., Hothorn, M.(2016). Mechanistic insight into a peptide hormone signaling complex mediating floral organ abscission. eLife 5: e15075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago, J., Henzler, C., Hothorn, M.(2013). Molecular mechanism for plant steroid receptor activation by somatic embryogenesis co-receptor kinases. Science 341: 889–892. [DOI] [PubMed] [Google Scholar]
- Schindelin, J., et al. (2012). Fiji: An open-source platform for biological-image analysis. Nat. Methods 9: 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She, J., Han, Z., Kim, T.-W., Wang, J., Cheng, W., Chang, J., Shi, S., Wang, J., Yang, M., Wang, Z.-Y., Chai, J.(2011). Structural insight into brassinosteroid perception by BRI1. Nature 474: 472–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu, S.H., Bleecker, A.B.(2001). Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl. Acad. Sci. USA 98: 10763–10768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpak, E.D.(2013). Diverse roles of ERECTA family genes in plant development. J. Integr. Plant Biol. 55: 1238–1250. [DOI] [PubMed] [Google Scholar]
- Shpak, E.D., McAbee, J.M., Pillitteri, L.J., Torii, K.U.(2005). Stomatal patterning and differentiation by synergistic interactions of receptor kinases. Science 309: 290–293. [DOI] [PubMed] [Google Scholar]
- Smakowska-Luzan, E., et al. (2018). An extracellular network of Arabidopsis leucine-rich repeat receptor kinases. Nature 553: 342–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenvik, G.-E., Tandstad, N.M., Guo, Y., Shi, C.-L., Kristiansen, W., Holmgren, A., Clark, S.E., Aalen, R.B., Butenko, M.A.(2008). The EPIP peptide of INFLORESCENCE DEFICIENT IN ABSCISSION is sufficient to induce abscission in arabidopsis through the receptor-like kinases HAESA and HAESA-LIKE2. Plant Cell 20: 1805–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, T., Nitta, Y., Zhang, Q., Wu, D., Tian, H., Lee, J.S., Zhang, Y.(2018). Antagonistic interactions between two MAP kinase cascades in plant development and immune signaling. EMBO Rep. 19: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Y., Han, Z., Tang, J., Hu, Z., Chai, C., Zhou, B., Chai, J.(2013). Structure reveals that BAK1 as a co-receptor recognizes the BRI1-bound brassinolide. Cell Res. 23: 1326–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, W., et al. (2011). PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat. Cell Biol. 13: 124–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torii, K.U., Mitsukawa, N., Oosumi, T., Matsuura, Y., Yokoyama, R., Whittier, R.F., Komeda, Y.(1996). The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. Plant Cell 8: 735–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuwamoto, R., Fukuoka, H., Takahata, Y.(2008). GASSHO1 and GASSHO2 encoding a putative leucine-rich repeat transmembrane-type receptor kinase are essential for the normal development of the epidermal surface in Arabidopsis embryos. Plant J. 54: 30–42. [DOI] [PubMed] [Google Scholar]
- Vert, G., Chory, J.(2006). Downstream nuclear events in brassinosteroid signalling. Nature 441: 96–100. [DOI] [PubMed] [Google Scholar]
- Wang, J., Li, H., Han, Z., Zhang, H., Wang, T., Lin, G., Chang, J., Yang, W., Chai, J.(2015). Allosteric receptor activation by the plant peptide hormone phytosulfokine. Nature 525: 265–268. [DOI] [PubMed] [Google Scholar]
- Wang, X., Kota, U., He, K., Blackburn, K., Li, J., Goshe, M.B., Huber, S.C., Clouse, S.D.(2008). Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Dev. Cell 15: 220–235. [DOI] [PubMed] [Google Scholar]
- Wang, Z., Meng, P., Zhang, X., Ren, D., Yang, S.(2011). BON1 interacts with the protein kinases BIR1 and BAK1 in modulation of temperature-dependent plant growth and cell death in Arabidopsis. Plant J. 67: 1081–1093. [DOI] [PubMed] [Google Scholar]
- Wang, Z.Y., Nakano, T., Gendron, J., He, J., Chen, M., Vafeados, D., Yang, Y., Fujioka, S., Yoshida, S., Asami, T., Chory, J.(2002). Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell 2: 505–513. [DOI] [PubMed] [Google Scholar]
- Wang, Z.Y., Seto, H., Fujioka, S., Yoshida, S., Chory, J.(2001). BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 410: 380–383. [DOI] [PubMed] [Google Scholar]
- Yang, M., Sack, F.D.(1995). The too many mouths and four lips mutations affect stomatal production in Arabidopsis. Plant Cell 7: 2227–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, Y., Wang, Z.Y., Mora-Garcia, S., Li, J., Yoshida, S., Asami, T., Chory, J.(2002). BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109: 181–191. [DOI] [PubMed] [Google Scholar]
- Zhang, X., Liu, W., Nagae, T.T., Takeuchi, H., Zhang, H., Han, Z., Higashiyama, T., Chai, J.(2017). Structural basis for receptor recognition of pollen tube attraction peptides. Nat. Commun. 8: 1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, B., Bai, Q., Wu, L., Liu, H., Liu, Y., Xu, W., Li, G., Ren, H., She, X., Wu, G.(2019). EMS1 and BRI1 control separate biological processes via extracellular domain diversity and intracellular domain conservation. Nat. Commun. 10: 4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J., et al. (2018). Regulation of Arabidopsis brassinosteroid receptor BRI1 endocytosis and degradation by plant U-box PUB12/PUB13-mediated ubiquitination. Proc. Natl. Acad. Sci. USA 115: E1906–E1915. [DOI] [PMC free article] [PubMed] [Google Scholar]