Geminivirus infection induces the expression of a plant imprinted E3 ligase gene, leading to the proteosomal degradation of DNA methyltransferases and epigenetic activation of viral gene transcription.
Abstract
Flowering plants and mammals contain imprinted genes that are primarily expressed in the endosperm and placenta in a parent-of-origin manner. In this study, we show that early activation of the geminivirus genes C2 and C3 in Arabidopsis (Arabidopsis thaliana) plants, encoding a viral suppressor of RNA interference and a replication enhancer protein, respectively, is correlated with the transient vegetative expression of VARIANT IN METHYLATION5 (VIM5), an endosperm imprinted gene that is conserved in diverse plant species. VIM5 is a ubiquitin E3 ligase that directly targets the DNA methyltransferases MET1 and CMT3 for degradation by the ubiquitin-26S proteasome proteolytic pathway. Infection with Beet severe curly top virus induced VIM5 expression in rosette leaf tissues, possibly via the expression of the viral replication initiator protein, leading to the early activation of C2 and C3 coupled with reduced symmetric methylation in the C2-3 promoter and the onset of disease symptoms. These findings demonstrate how this small DNA virus recruits a host imprinted gene for the epigenetic activation of viral gene transcription. Our findings reveal a distinct strategy used by plant pathogens to exploit the host machinery in order to inhibit methylation-mediated defense responses when establishing infection.
INTRODUCTION
Geminiviruses (family Geminiviridae) belong to one of the largest and most important families of plant viruses, with genomes comprising one or two single-stranded, 2.5- to 3.0-kb circular DNAs (Raja et al., 2010; Hanley-Bowdoin et al., 2013; Zhou, 2013). Curtoviruses (genus Curtovirus) and begomoviruses (genus Begomovirus) are two well-studied genera in the family Geminiviridae (Fontes et al., 1992; Sunter and Bisaro, 1992; Hormuzdi and Bisaro, 1995). The monopartite geminiviruses often contain positionally conserved open reading frames from complementary-sense strand designated as C1 (also known as Replication initiator protein [Rep], equivalent to L1 or AC1 in begomoviruses), C2 (L2 or AC2, known as transcriptional activator protein [TrAP] in begomoviruses, but not curtoviruses), C3 (L3 or AC3, also known as replication enhancer protein [REn]), and C4 (L4 or AC4), while the open reading frames encoded in the virion-sense strand are named V1 (coat protein [CP]), V2, and V3 (moment protein [MP]; Raja et al., 2010).
Geminiviruses do not encode DNA or RNA polymerases and thus depend on viral proteins to redirect the host machinery and processes to DNA replication and gene expression (Raja et al., 2010; Hanley-Bowdoin et al., 2013). Transcription in geminiviruses occurs bidirectionally from an intergenic region (IR) containing oppositely oriented promoters separated by the origin of replication (Raja et al., 2010). Extensive characterization of the transcription programs of begomoviruses suggested that a single mRNA is produced from the virion-sense strand (Hanley-Bowdoin et al., 1989). By contrast, multiple virion-sense mRNAs have been detected for curtoviruses (Frischmuth et al., 1993; Mullineaux et al., 1993). For the transcription from the complementary-sense strand in both begomoviruses and curtoviruses, several reports support the production of a distinct bicistronic mRNA driven by one promoter to control the expression of both the C2/AC2 and C3/AC3 genes (Frischmuth et al., 1991; Shivaprasad et al., 2005; Shung et al., 2006; Jeske, 2009).
DNA virus infection in plants triggers both transcriptional and posttranscriptional gene silencing antiviral mechanisms. As a counter-defense strategy, geminiviruses have evolved multiple viral suppressors of RNA silencing (VSRs; Glick et al., 2008; Raja et al., 2010; Zhang et al., 2011b; Aregger et al., 2012; Hanley-Bowdoin et al., 2013; Yang et al., 2013; Jackel et al., 2015; Ramesh et al., 2017; Guo et al., 2018; Rosas-Diaz et al., 2018). For instance, geminivirus Rep protein reduces the mRNA levels of cytosine methyltransferase genes METHYLTRANSFERASE1 (MET1) and CHROMOMETHYLASE3 (CMT3; Rodríguez-Negrete et al., 2013). The curtovirus protein C2/L2 interferes with the host’s methyl cycle maintenance by inactivating adenosine kinase or impairing the ubiquitination of the S-adenosylmethionine decarboxylase SAMDC1 (Trinks et al., 2005; Wang et al., 2005; Buchmann et al., 2009; Zhang et al., 2011b). The AC4/C4 protein of geminiviruses interacts with long/short RNA and single-stranded/double-stranded (ss/ds) RNAs and blocks the transport of small interfering RNAs (Vanitharani et al., 2004; Chellappan et al., 2005; Amin et al., 2011; Deom and Mills-Lujan, 2015; Carluccio et al., 2018; Li et al., 2018). Finally, the expression of V2 protein of the monopartite geminiviruses in transgenic plants reduced genomic methylation levels and reversed transcriptional gene silencing (Zrachya et al., 2007; Zhang et al., 2012). Another important strategy used by geminiviruses to evade gene silencing is specialized replication mechanisms (Bian et al., 2006; Paprotka et al., 2011; Pooggin, 2013). Rep-dependent rolling circle replication and recombination-dependent replication generate abundant unmethylated ssDNAs to resurrect their dsDNA forms from repressive cytosine methylation (Hanley-Bowdoin et al., 2000; Jeske et al., 2001).
DNA viruses coordinate replication and virion assembly by regulating the timing of their gene expression (Broyles, 2003; Lee et al., 2016). Simian Virus 40, one of the most thoroughly studied mammalian DNA viruses, contains a circular dsDNA genome divided into early and late transcription regions (Elder et al., 1981). The general strategy in which the products of the early-class genes activate the expression of the late-class genes may also be used by geminiviruses (Elder et al., 1981; Borah et al., 2016; Saribas et al., 2019). The geminivirus genes (C1, C2, C3, and C4) encoded by the complementary-sense strand are transcribed early during infection to promote viral replication and counter-defense responses, whereas the virion-sense strand-encoded genes (V1, V2, and V3) are usually expressed after DNA replication and participate in virion assembly and movement (Hanley-Bowdoin et al., 2013; Borah et al., 2016; Ramesh et al., 2017). Much is known about how early-class proteins regulate late-class gene expression. For instance, TrAP/AC2 in Tomato golden mosaic virus (TGMV) activates AV1/CP expression through the essential cis-elements TATA box and the conserved late element motif within the CP promoter in coordination with the host PEAPOD2 protein (Lacatus and Sunter, 2009; Berger and Sunter, 2013; Liu et al., 2014). However, less is known about the regulation of early genes and the host factors involved in this process.
Beet severe curly top virus (BSCTV) is a curtovirus in the family Geminiviridae whose circular DNA genome is 2927 nucleotides long and encodes seven genes. In the current study, we characterized independent lines of Arabidopsis (Arabidopsis thaliana, Columbia [Col-0] ecotype) plants expressing a construct harboring an inducible BSCTV Rep gene, which also harbors the C4 coding region, the C2-3 promoter, and C2 N-terminal sequence (C2N; Figure 1A). By serendipity, we observed constitutive transcription of C2N in one of the five lines, pER-Rep-5. Characterization of pER-Rep-5 led to the surprising finding that the activation of the endosperm imprinted gene VARIANT IN METHYLATION5 (VIM5) in vegetative tissues was responsible for the transcriptional induction of C2N. BSCTV infection also transiently induced VIM5 expression, leading to elevated C2 and C3 transcription coupled with reduced symmetric methylation at the C2-3 promoter. We further showed that VIM5 functions as an E3 ligase that directly targets the DNA methyltransferases MET1 and CMT3 for ubiquitination and proteasomal degradation in planta. Viral DNA replication was delayed and DNA methylation was enhanced in the C2-3 promoter of Arabidopsis vim5 plants. These mutant phenotypes were restored via complementation with the 35S-VIM5 transgene. These findings reveal that a virus-activated host E3 ligase participates in posttranslational regulation of DNA methyltransferases MET1 and CMT3 to facilitate the expression of the early-class C2 and C3 genes of a plant-infecting DNA virus.
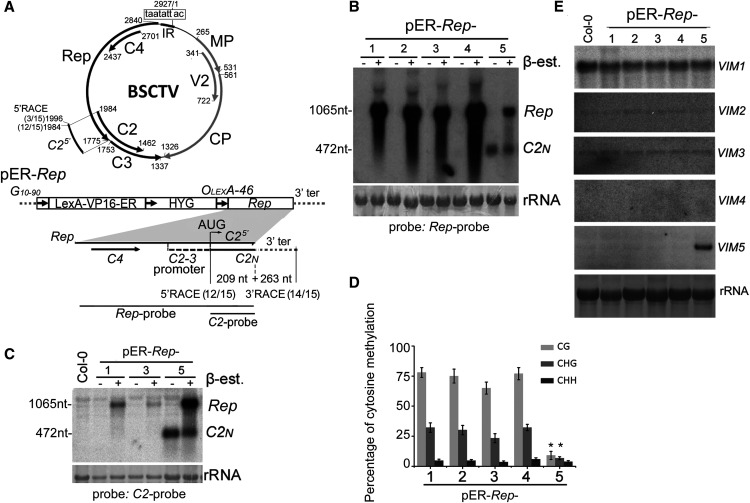
Figure 1.
Detection and Characterization of the C2N Short Transcript.
(A) Schematic diagram of the monopartite geminivirus BSCTV genome (top). BSCTV contains an IR with an invariant nonanucleotide (boxed) to direct the bidirectional transcription of viral mRNAs encoding Rep (also known as C1), C4, C2, and C3 from the complementary strand and CP (or V1), V2, and MP (or V3) from the virion strand, which are shown as thick arrows with the nucleotide positions indicated. The inducible transgene construct pER-Rep (bottom) consists of a strong synthetic constitutive promoter (G10-90); a chimeric transactivator (LexA-VP16-ER) containing the regulatory domain of an estrogen receptor (Zuo et al., 2000); a hygromycin-resistance marker (HYG); eight copies of the LexA DNA binding site fused to the -46 cauliflower mosaic virus 35S promoter (OLexA-46); and the Rep coding sequence of BSCTV, which also encodes the full-length C4, the C2-3 promoter, and a portion of the C2 in overlapping reading frame, C25′. C2N, a short C2 transcript of 472 nucleotides (nt; 209 nt C25′ + 263 nt 3′ terminal [ter] from vector) detected by 5′ and 3′ RACE. The total number of transcripts and the number sequenced from pER-Rep-5 plants at the indicated sites are shown in brackets.
(B) and (C) Detection of Rep mRNA (1065 nucleotides [nt]) and C2N (472 nt) from pER-Rep plants and Col-0. Total RNA was extracted from the rosette leaves of 2-week-old seedlings of different pER-Rep transgenic lines (60 individual seedlings per line) that were grown on MS medium with hygromycin, transferred onto MS medium with (+) or without (–) β-estradiol (β-est) for 16 h of induction, and analyzed by RNA blotting using a DNA probe specific to Rep (B) or C2 fragment (C). rRNA was stained by methylene blue as a loading control.
(D) Analysis of DNA methylation in the C2-3 promoter pER-Rep lines by bisulfite sequencing analysis. Samples were collected as described in (C). The original data are shown in Supplemental Figure 1. The statistical analysis was performed using OriginPro 8 (http://www.originlab.com). Values are means ± se, and asterisks indicate statistically significant differences in pER-Rep-5 compared with the other lines (n = 20; one-way ANOVA, P < 0.05).
(E) Detection of the expression of VIM family genes by RNA gel blotting. Rosette leaves of the 4-week-old wild-type Col-0 and transgenic pER-Rep plants were harvested for RNA isolation. VIM5 was highly induced in the pER-Rep-5 line.
Note that the seedlings analyzed in (B) were from the T2 generation, and the seedlings in (C) to (E) were the respective pER-Rep transgenic plants from the T6 generation.
RESULTS
Transcription of the C2N mRNA within the Rep Transgene in Arabidopsis Is Associated with Upstream Hypomethylation
Aiming to study the function of Rep, we generated and characterized independent lines of Arabidopsis (Col-0) carrying a BSCTV Rep transgene (pER-Rep; Figure 1A) under the control of a β-estradiol–inducible promoter (Zuo et al., 2000), as constitutive expression of C4 in Arabidopsis caused callus-like developmental defects in transgenic seedlings (Lai et al., 2009). As expected, RNA gel blot analysis detected accumulation of Rep mRNA in all five lines of pER-Rep transgenic plants grown on medium containing β-estradiol (Figure 1B). Interestingly, we detected an additional shorter RNA species in one transgenic plant line with or without β-estradiol treatment (Figure 1B). We hypothesized that the short RNA corresponded to transcript from the C2-3 promoter that was only activated in pER-Rep-5 plants. Both 5′ and 3′ rapid amplification of cDNA ends (RACE; Figure 1A) and RNA gel blot analysis using a C2 mRNA-specific probe (Figure 1C) demonstrated that the short RNA was indeed the predicted transcripts from the C2-3 gene promoter, designated C2N mRNA.
Bisulfite sequencing revealed significantly reduced cytosine methylation in the CG and CHG contexts, but not in the CHH context, in the C2-3 promoter region of the Rep transgene DNA integrated in pER-Rep-5 plants compared with the four transgenic plant lines in which C2N mRNA was undetectable (Figure 1D; Supplemental Figure 1). Based on the known regulatory mechanisms of plant gene expression (Zhang et al., 2018), these findings suggest that hypomethylation in the promoter region leads to the transcriptional activation of the viral C2 gene from the integrated transgene. The protein product of C2N mRNA lacked the C-terminal 103 amino acids of the full-length C2 and lost VSR activity to suppress transgene RNA silencing (Supplemental Figure 2), demonstrating that the C2N product was not responsible for hypomethylation in the promoter region. The C2-3 promoter was methylated normally in lines pER-Rep-1, pER-Rep-2, pER-Rep-3, and pER-Rep-4; thus, pER-Rep-5 provided a unique genetic material to explore the plant factor(s) involved in hypomethylation of the C2-3 promoter and the BSCTV–host plant interaction.
Vegetative Induction of VIM5 Is Responsible for the Hypomethylation of the Transgenic C2-3 Promoter in Arabidopsis
A genome-wide transcriptome comparison between the wild-type Arabidopsis and pER-Rep-5 plants (Supplemental Data Set 1) revealed that the most significant difference was the accumulation of Arabidopsis mRNAs from the group of methylation-related genes (Stroud et al., 2013) and the mRNA of VIM5 (Supplemental Figure 3A), a paternally imprinted gene conserved in Arabidopsis, rice (Oryza sativa), and maize (Zea mays; Gehring et al., 2011; Hsieh et al., 2011; Luo et al., 2011; Zhang et al., 2011a). RNA gel blot analysis detected VIM5 mRNA only in pER-Rep-5 plants (Figure 1E). By contrast, no obvious changes were found in the accumulation of mRNAs encoding MET1, CMT3, or DOMAINS REARRANGED METHYLTRANSFERASE2 (DRM2; Stroud et al., 2013; Zhang et al., 2018) between the wild-type and pER-Rep transgenic plants (Supplemental Figure 3B). pER-Rep-5 plants contained a single-copy insertion of the pER-Rep transgene in the first exon of ARABIDOPSIS HISTIDINE KINASE4 (AHK4, AT2g01830), and activation of VIM5 transcription in leaves was observed in AHK4 knockout mutant plants (Supplemental Figures 3C to 3E). To determine whether increases in the expression of VIM5 lead to C2N transcription, we introduced 35S-VIM5 into the pER-Rep-3 line, in which the C2N was not detectable. The constitutive expression of VIM5 from the transgene reduced symmetric methylation (∼67 to 30% for CG; 24 to 15% for CHG) in the C2N promoter and induced C2N transcription in these plants (Supplemental Figure 4). These findings suggest that the expression of the imprinted gene may be also responsible for the transcriptional activation of the viral C2-3 promoter in transgenic line pER-Rep-5.
BSCTV Infection Induces VIM5 Expression Early and Transiently in Systemically Infected Leaves
VIM5 transcription was transiently induced in systemically infected rosette leaves of the wild-type plants 2 days after inoculation (DAI) with BSCTV (Figure 2A), that, however, did not induce decreases in AHK4 expression (Supplemental Figure 5A). Transient expression of Rep also greatly activated the expression of VIM5 driven by its native promoter in Nicotiana benthamiana leaves (Supplemental Figure 5B). Similarly, pER-Rep-1 and pER-Rep-3 transgenic plants grown on medium containing β-estradiol for 7 d exhibited activated expression of VIM5 and C2N coupled with reduced CG and CHG methylation levels in the C2-3 promoter (Supplemental Figures 5C to 5E), suggesting a role for the expression of the viral Rep protein in BSCTV-induced VIM5 expression. To determine the effect of the aberrantly induced VIM5 expression on BSCTV replication and viral gene expression, we subjected Arabidopsis vim5 plants (SALK_071899) to BSCTV infection; these plants did not accumulate VIM5 transcripts in the endosperm-containing siliques and exhibited no developmental defects (Supplemental Figure 6A). Consistent with previous studies on other geminiviruses (Raja et al., 2010; Hanley-Bowdoin et al., 2013; Zhou, 2013) and unlike the delayed expression of the three virion-sense strand genes (CP, V2, and MP), all four complementary-sense strand genes (Rep, C2, C3, and C4) were transcriptionally active at 2 DAI in the systemically infected rosette leaves of wild-type plants (Figure 2B; Supplemental Figure 6B). In contrast to Rep and C4, however, C2 and C3 transcripts were undetectable in vim5 plants at 2 DAI (Figure 2B). Interestingly, abundant transcripts from all of the virion- and complementary-sense strand genes including C2 and C3 accumulated in vim5 plants at both 6 and 11 DAI (Figure 2B; Supplemental Figure 6B), indicating that VIM5 specifically contributes to the early expression of C2 and C3 from the infecting viral DNA genome. Our finding is consistent with the current view for curtoviruses that the expression of C2 and C3 is driven by a single C2-3 promoter (Jeske, 2009).
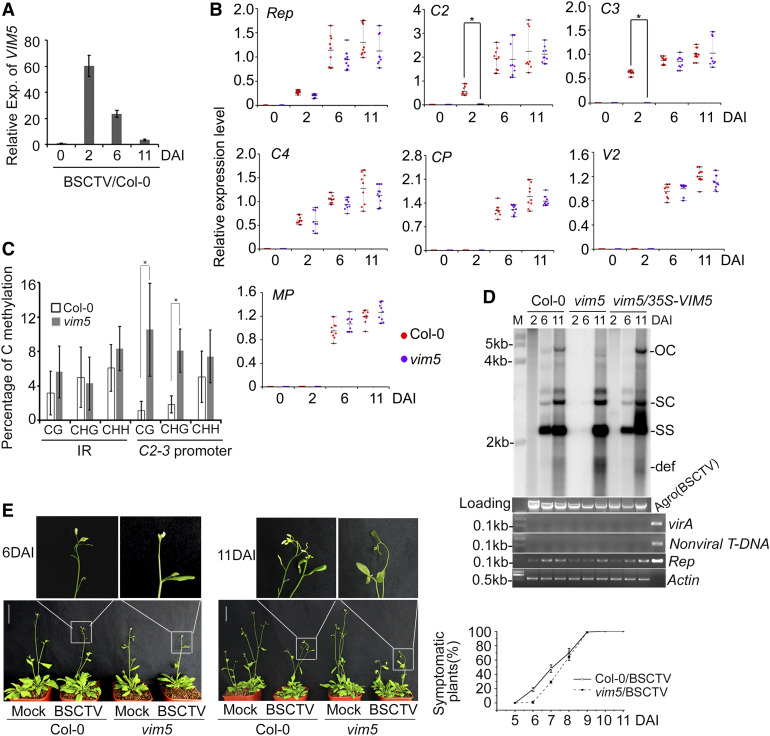
Figure 2.
Analysis of VIM5 and Viral Gene Expression, Viral DNA Accumulation, and Methylation.
(A) Detection of VIM5 expression (Exp.) in BSCTV-infected wild-type Col-0 plants by RT-qPCR. Data were normalized to the expression level of ACTIN (as the internal standard). The value at 0 DAI was arbitrarily designated as 1. Error bars represent the sd for three biological replicates. In each of three independent experiments, noninoculated rosette leaves of BSCTV-infected wild-type Col-0 plants in a pool of eight plants for each time points were harvested for RNA extraction and RT-qPCR.
(B) Detection of transcription of BSCTV-encoded early-class genes Rep, C2, C3, and C4 and late-class genes CP, V2, and MP by RT-qPCR and RT-PCR (see Supplemental Figure 6B). Data were normalized to the expression level of ACTIN (as the internal standard) and calculated by the 2–ΔCt method. The asterisk indicates significant difference in C2 and C3 expression of infected Col-0 wild-type plants versus vim5 plants at 2 DAI (Student’s t test, *P < 0.05; mean ± sd, n = 9 biologically independent samples). Three independent experiments were performed with pools of 32 plants per genotype (eight plants for each time point), and noninoculated rosette leaves were collected for total RNA isolation.
(C) DNA methylation analysis of the IR and C2-3 promoter regions of viral DNA in the BSCTV-infected wild-type Col-0 and vim5 plants at 2 DAI. Samples were collected as described in (B). The statistical analysis was performed using OriginPro 8. Values are means ± se, and asterisks indicate statistically significant differences in CG and CHG methylation levels of 2 DAI BSCTV-infected vim5 plants compared to wild-type Col-0 plants (n = 20; Student’s t test, *P < 0.05). The original data are shown in Supplemental Figure 7A.
(D) Detection of viral DNA accumulation by DNA gel blotting. Noninoculated leaves of 4-week-old BSCTV-infected plants were collected for DNA extraction as indicted in the Methods. The positions of open circle (OC), supercoiled (SC), and single-stranded (SS) DNAs and a population of defective interfering (def) DNAs are indicated. Rep-specific, isotope-labeled DNA sequence was used as a probe. Ethidium bromide–stained genomic DNA served as the loading/running control. Both the Agrobacterium virA gene (Wang et al., 2020) and a nonviral sequence within the T-DNA of the infectious pCambia-BSCTV1.8 plasmid served as controls in PCR (with 200 ng of respective total DNAs) to exclude any inoculated residue in extracted total DNAs. The Rep sequence and ACTIN gene was amplified as viral and loading control, respectively. Agro(BSCTV), representing Agrobacterium colonies containing the BSCTV infectious plasmids, served as a positive control. The sizes of virA, nonviral T-DNA, Rep, and ACTIN sequences are indicated.
(E) BSCTV disease symptoms. Photograph was taken at 6 and 11 DAI. Quantification of symptomatic plants (right) was performed by counting all symptomatic plants versus total number of plants at each time point for each genotype. Three independent experiments were performed. Values are means ± sd. For each independent experiment, 32 individual plants per genotype were used for observation and calculation. Bar = 5 cm.
We then compared cytosine methylation in the C2-3 promoter and the divergent promoters in the IR in the infected wild-type Col-0 and vim5 plants at 2 DAI. Total cytosine methylation of the viral genomic DNA was considerably lower than that of the integrated pER-Rep transgene DNA (5.75 versus 14.6%; Figure 2C, compare to Figure 1D). Nevertheless, whereas the viral IR DNA exhibited similar levels of methylation in the CG, CHG, and CHH contexts in both Col-0 and vim5 plants, the viral C2-3 promoter DNA was methylated at significantly higher levels in the CG and CHG contexts in vim5 versus Col-0 plants at 2 DAI (Figure 2C; Supplemental Figure 7A). Thus, transcriptional activation of the C2-3 promoter in both pER-Rep-5 plants and BSCTV-infected Col-0 plants (Figure 1) was correlated with significantly reduced methylation at CG and CHG, but not CHH, sites. These findings suggest that the aberrant vegetative expression of VIM5 contributes to the epigenetic activation of the viral C2-3 promoter during early stages of infection.
In support of a physiological function for the early activation of the C2 and C3 genes, the accumulation of various forms of virus DNA was readily detectable by DNA gel blot analysis at 6 DAI in systemically infected rosette leaves of wild-type plants and vim5 plants constitutively expressing VIM5 from a transgene (vim5/35S-VIM5), but not vim5 plants (Figure 2D; Supplemental Figures 6C and 6D). Examination of agrobacterial genes and sequences from the BSCTV infectious clone ruled out any DNA contamination from the inoculated leaves (Figure 2D; Supplemental Figure 6C). The wild-type plants also exhibited an earlier onset of symptoms than vim5 plants following BSCTV infection (Figure 2E). Both the wild-type and vim5 plants supported robust virus replication and developed severe disease symptoms at 11 DAI (Figures 2D and 2E), indicating that VIM5 is not essential for BSCTV infection but may contribute to viral C2 and C3 expression and the onset of disease symptoms.
VIM5 Is an Active E3 Ubiquitin Ligase That Directly Interacts with MET1 and CMT3 in Vitro and in Vivo
The Arabidopsis genome encodes four closely related proteins (VIM1 to VIM4) with sequence homology to VIM5 (also known as ORTH3), all of which contain a plant homeodomain, two Really Interesting New Gene (RING)-type zinc finger domains, and a Suvar Enhancer of Zeste Trithorax and RING Associated domain (Liu et al., 2007; Woo et al., 2007, 2008; Kraft et al., 2008; Yao et al., 2012; Kim et al., 2014; Shook and Richards, 2014). VIM1 (ORTH2), VIM2 (ORTH5), and VIM3 (ORTH1) are expressed in vegetative tissues, cooperate with MET1 to maintain global CG methylation and epigenetic silencing, and exhibit ubiquitin E3 ligase activity in vitro (Kraft et al., 2008; Woo et al., 2008; Kim et al., 2014; Shook and Richards, 2014). We found that VIM5 expressed as a fusion protein with maltose binding protein (MBP) from Escherichia coli was able to self-ubiquitinate in the presence of E1, E2, and ubiquitin (Figure 3A). However, the self-ubiquitination activity of VIM5 was abolished when three amino acid substitutions known to impair the intact RING structure and the enzyme activity of E3 ligases (Xie et al., 2002; Deng et al., 2016) were introduced into this protein (Supplemental Figures 8A and 8B). These results demonstrate that VIM5 has E3 ligase activity, as predicted previously (Kraft et al., 2008). Thus, we hypothesized that VIM5 might induce DNA hypomethylation, leading to C2 and C3 transcriptional activation by targeting one or more methyltransferases for degradation by the ubiquitin-26S proteasome proteolytic pathway.
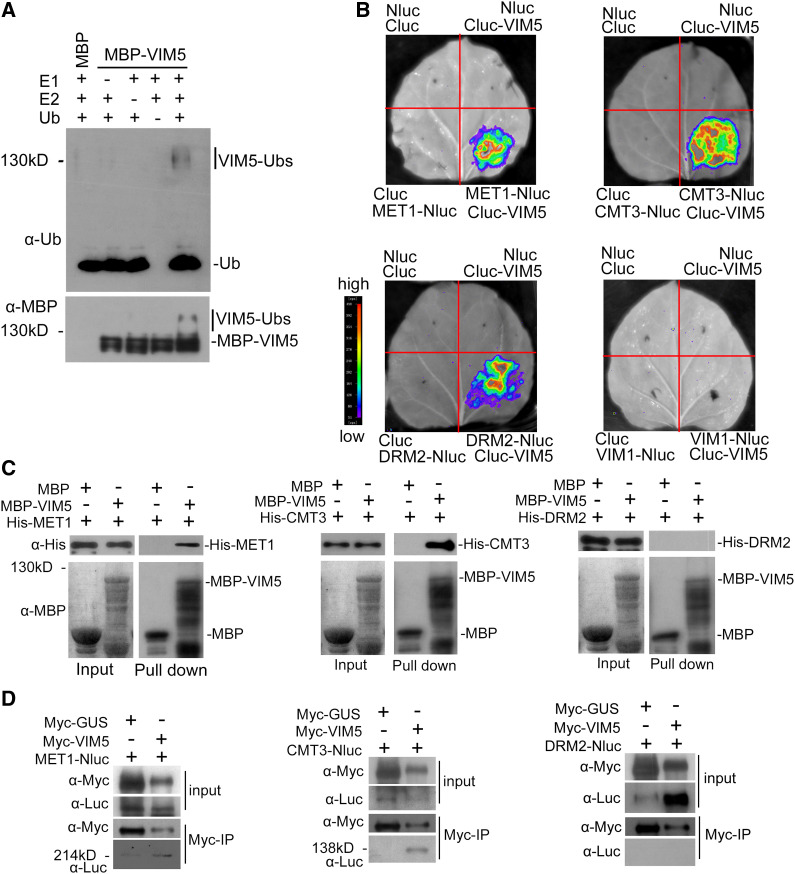
Figure 3.
Analysis of the E3 Ligase Activity of VIM5 and Interactions with DNA Methyltransferases.
(A) VIM5 exhibits E3 ubiquitin ligase activity. Purified MBP-VIM5 fusion protein was assayed for E3 activity in the presence of E1, E2, and ubiquitin (Ub). Ubiquitinated protein was detected by immunoblotting using anti-ubiquitin (α-Ub) and anti-MBP (α-MBP) antibodies, respectively.
(B) Detection of protein–protein interactions in planta by a luciferase complementation imaging assay. VIM5 interacts with MET1, CMT3, and DRM2, but not with VIM1, in N. benthamiana leaf cells. Fluorescence signal intensity is indicated by the pseudocolor bar.
(C) Detection of direct protein–protein interactions by an in vitro pull-down assay. Fusion proteins expressed in E. coli were purified. MBP-VIM5 pulled down His-MET1 and His-CMT3, but not His-DRM2. MBP was used as a negative control. Fusion proteins were detected by immunoblotting using anti-His (α-His) and anti-MBP (α-MBP) antibodies as appropriate.
(D) Detection of protein–protein interaction by an in vivo coimmunoprecipitation assay. Fusion protein MET1-Nluc, CMT3-Nluc, or DRM2-Nluc was transiently coexpressed in N. benthamiana with myc-VIM5. Myc-VIM5 coimmunoprecipitated MET1-Nluc or CMT3-Nluc, but not DRM2-Nluc. Myc-GUS was served as a negative control. Fusion proteins were detected by immunoblotting using anti-Myc (α-Myc) or anti-Luc (α-Luc) antibodies as indicated.
To test the hypothesis, we performed a firefly luciferase complementation imaging assay to determine whether VIM5 interacts in vivo with MET1, CMT3, or DRM2 (Chen et al., 2008). As shown in Figure 3B, fluorescent signals were detected when VIM5 fused with the C-terminal half of luciferase (Cluc) was coexpressed with MET1, CMT3, or DRM2 fused with the N-terminal half of luciferase (Nluc), but not with VIM1-Nluc. In an in vitro pull-down assay, MBP-VIM5 fusion protein pulled down both MET1 and CMT3, but not DRM2 (Figure 3C). The interaction of VIM5 with both MET1 and CMT3, but not DRM2, was also demonstrated by a coimmunoprecipitation assay in N. benthamiana leaves (Figure 3D). These findings suggest that VIM5 interacts directly with both MET1 and CMT3, but not DRM2, in vitro and in vivo.
MET1 and CMT3 Are Substrates of VIM5 for Proteasomal Degradation
To determine whether VIM5 expression would induce the degradation of its interacting methyltransferases in vivo, we coexpressed MET1-Nluc, CMT3-Nluc, or DRM2-Nluc with Myc-tagged VIM5 or β-glucuronidase (GUS) in N. benthamiana leaves by Agrobacterium tumefaciens (Agrobacterium)–mediated coinfiltration. No clear differences were observed in the accumulation of GFP) expressed from the coinfiltrated internal control transgene after coexpression with VIM5 or GUS (Figure 4A). However, both MET1-Nluc and CMT3-Nluc, but not DRM2-Nluc, accumulated to markedly lower levels following coexpression with VIM5 than GUS (Figure 4A). By contrast, coexpression of the mutant VIM5 defective in its E3 ligase activity for self-ubiquitination (Supplemental Figure 8B) did not induce the in vivo degradation of either MET1-Nluc or CMT3-Nluc (Figure 4B). Moreover, the in vivo degradation of MET1-Nluc and CMT3-Nluc was detected in N. benthamiana leaves following BSCTV-induced expression of VIM5, but not mutant VIM5 (Supplemental Figure 8D). Similar to the pER-Rep transgenic plants (Supplemental Figure 3B), we detected no significant differences in the accumulation of MET1 or CMT3 mRNA between the wild-type and vim5 plants after BSCTV infection (Supplemental Figure 8E). Together, these findings indicate that VIM5 targets the methyltransferases MET1 and CMT3 for degradation via a mechanism that depends on its E3 ligase activity.
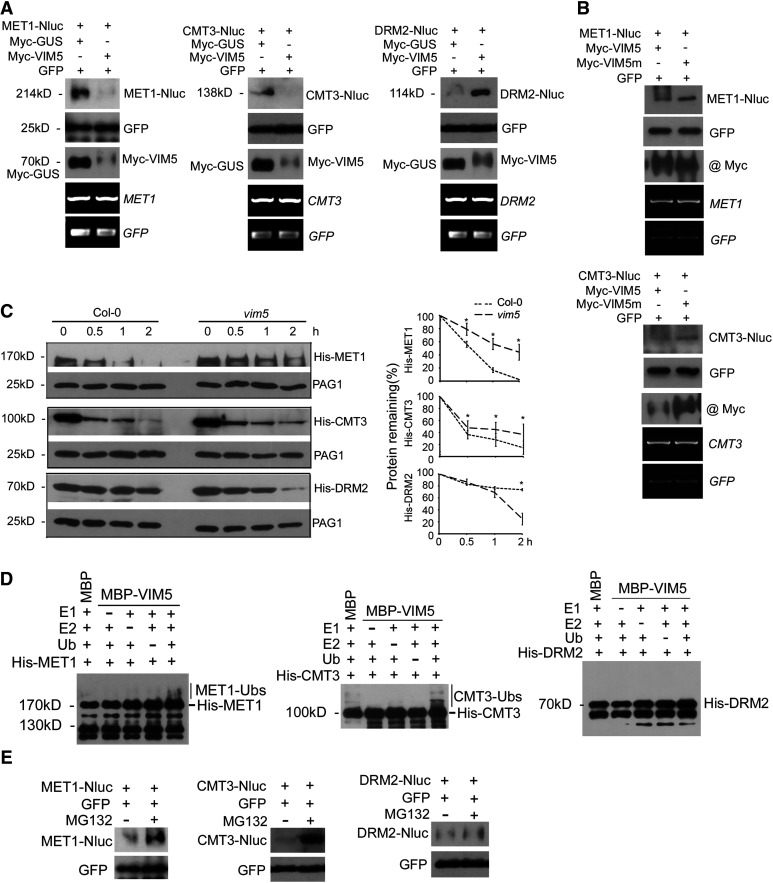
Figure 4.
VIM5 Targets the DNA Methyltransferases MET1 and CMT3 for Degradation by the 26S Proteasome Pathway.
(A) and (B) Agrobacterium strains directing transient expression of the E3 ligase VIM5 or the mutant VIM5 (VIM5m) with its candidate substrate MET1, CMT3, or DRM2 were coinfiltrated into N. benthamiana leaves, and total proteins/RNA were extracted 3 DAI. Also, coinfiltrated as controls were those for the expression of GUS and GFP. The accumulation of the expressed proteins in the leaves was analyzed by immunoblotting using antibodies specific to GFP; Myc tag fused with VIM5, VIM5m, and GUS, or the luciferase tag Nluc fused with MET1, CMT3, and DRM2. The accumulation of MET1, CMT3, DRM2, and GFP mRNA was examined by RT-PCR, as shown in the bottom two panels.
(C) Equal amounts of recombinant His-tagged MET1, CMT3, or DRM2 purified from E. coli were incubated with total protein extracts from the siliques of Col-0 and vim5 and 0, 0.5, 1, or 2 h later, the incubation was terminated and the accumulation level of the recombinant proteins was analyzed by immunoblotting using anti-His antibody. The accumulation of the endogenous 20S proteasome α subunit G-1 (PAG1) during each time-course incubation analysis was also examined using PAG1-specific antibodies as an internal control. Right panels show quantitative analysis of recombinant protein accumulation in three independent experiments using ImageJ software. Values are means ± sd. Asterisks indicate that statistically significant differences (Student’s t test, *P < 0.05).
(D) Detection of VIM5 substrates by in vitro ubiquitination assay. In the presence of E1, E2, ubiquitin (Ub), and MBP-VIM5, ubiquitination of MET1 and CMT3, but not DRM2, was detected. Samples were processed for immunoblotting using an anti-His antibody.
(E) Agroinfiltration and immunoblotting were as described above, except that 8 h before harvesting the sample for protein extraction, the leaves were infiltrated with MG132 (50 µM) or DMSO.
We further investigated the VIM5-mediated degradation of these methyltransferases in vitro. We observed rapid degradation of recombinant His-tagged MET1 and CMT3 proteins purified from E. coli following incubation with fresh total protein extracts from siliques of the wild-type plants compared with incubation with vim5 plant protein extracts (Figure 4C). Interestingly, incubation with vim5 extracts appeared to accelerate the turnover of DRM2 compared with incubation with the wild-type extracts (Figure 4C). Moreover, the recombinant MET1 and CMT3, but not DRM2, were ubiquitinated in vitro by MBP-VIM5 in the presence of E1, E2, and ubiquitin (Figure 4D), thereby identifying both MET1 and CMT3 as substrates of VIM5. Consistent with a role for the E3 ligase catalytic activity of VIM5 in the degradation of MET1 and CMT3 (Figures 3A and 4A), treatment with MG132, a 26S proteasome inhibitor (Smalle and Vierstra, 2004), enhanced the accumulation of Nluc-tagged MET1 and CMT3, but not DRM2 or the control GFP, in coinfiltrated N. benthamiana leaves (Figure 4E). These findings provide independent lines of evidence demonstrating that MET1 and CMT3, but not DRM2, are targeted by VIM5 for degradation by the ubiquitin-26S proteasome proteolytic pathway.
Methylation of C2-3 Promoter DNA at CG and CHG Sites Suppresses BSCTV Accumulation during the Early Stages of Systemic Infection and Disease Development
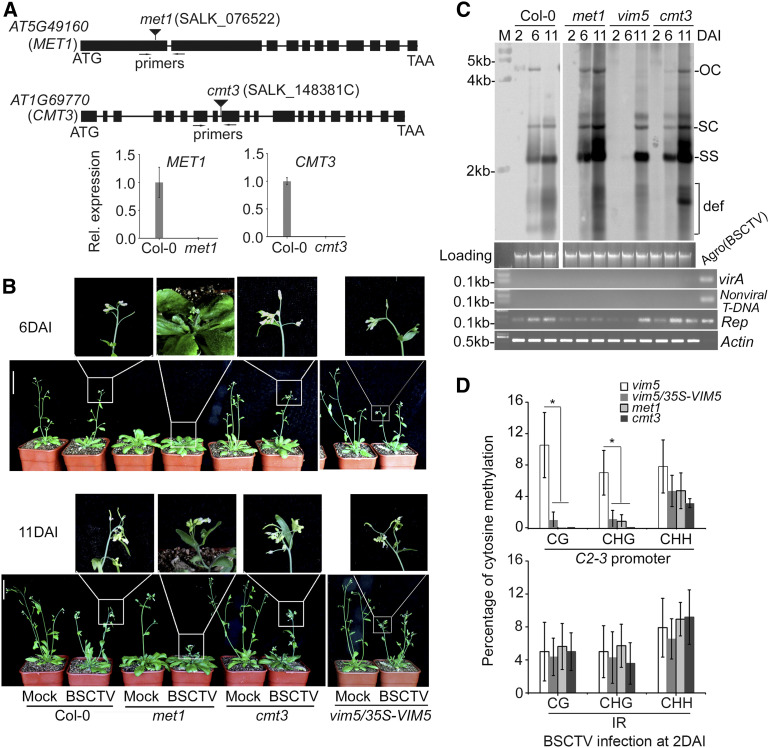
We further investigated the role of DNA methylation at the CG and CHG sites of the C2-3 promoter during BSCTV infection using two approaches. In Arabidopsis, the maintenance of CG and CHG methylation is performed by MET1 and CMT3, respectively, whereas DRM2 catalyzes de novo DNA methylation in the CG and CHG as well as CHH contexts (Zhang et al., 2018). Thus, we first compared BSCTV infection among Col-0 wild-type and met1, cmt3, and vim5 knockout mutant plants (Figure 5A) as well as 35S-VIM5–complemented vim5 plants. Col-0 and vim5/35S-VIM5 plants exhibited similar levels of susceptibility to BSCTV (Figure 5B, compare to Figure 2E), and as expected, the C2-3 promoter DNA in the infected vim5/35S-VIM5 plants did not exhibit the enhanced methylation at CG and CHG sites observed in vim5 plants (Figure 5D). In agreement with the findings for begomovirus infection (Raja et al., 2008), both met1 and cmt3 plants developed more severe disease symptoms than Col-0 plants after BSCTV infection (Figure 5B, compare to Figure 2E). In contrast to vim5 plants, however, met1 and cmt3 plants as well as vim5/35S-VIM5 plants maintained much lower levels of methylation at CG, CHG, and CHH sites in the C2-3 promoter at 2 DAI, and BSCTV accumulation was readily detectable at 6 DAI (Figures 2D and 5C), whereas the IR methylation status was comparable in all plants examined (Figures 2C and 5D).
Figure 5.
Characterization of met1 and cmt3 Plants and Analysis of Viral DNA Accumulation and Methylation in BSCTV-Infected Mutant Plants.
(A) Relative (Rel.) expression of MET1 or CMT3 was detected in the rosette leaves of the 4-week-old wild-type Col-0 plants, but not in met1 or cmt3 plants, by RT-qPCR. Values are means ± sd (n = 9). Primers used for RT-qPCR are denoted in the schematic diagrams at the top.
(B) BSCTV disease symptoms. Photograph was taken at 6 and 11 DAI. Three independent experiments were performed. For each independent experiment, 32 individual plants per genotype were used for observation. Compared with Col-0 plants, cmt3 knockout plants or transgene-complemented vim5 plants, met1 knockout plants developed earlier and showed more severe disease symptoms after BSCTV infection. Bar = 5 cm.
(C) Detection of viral DNA accumulation by DNA gel blotting. The positions of open circle (OC), supercoiled (SC), and single-stranded (SS) DNAs and a population of defective interfering (def) DNAs are indicated. Rep-specific, isotope-labeled DNA sequence was used as a probe. Ethidium bromide–stained genomic DNA served as the loading/running control, and the samples were examined by PCR as described in Figure 2D. Signals for viral accumulation were readily detected at 6 DAI in the infected Col-0 wild-type, met1, and cmt3 plants, but not vim5 plants.
(D) DNA methylation analysis of the C2-3 promoter region and IR of viral DNA in BSCTV-infected vim5, met1, and cmt3 plants and vim5/35S-VIM5 complemented plants at 2 DAI. The statistical analysis was performed using OriginPro 8. Values are means ± se, and asterisks indicate statistically significant differences in CG and CHG methylation levels in 2 DAI BSCTV-infected vim5 plants compared with other plants (n = 20; one-way ANOVA, P < 0.05). The original data are shown in Supplemental Figure 7C. In vim5/35S-VIM5 complemented plants, met1 plants, or cmt3 plants, the methylation levels of CG and CHG, but not CHH, in the C2-3 promoter were significantly lower than that in vim5 plants.
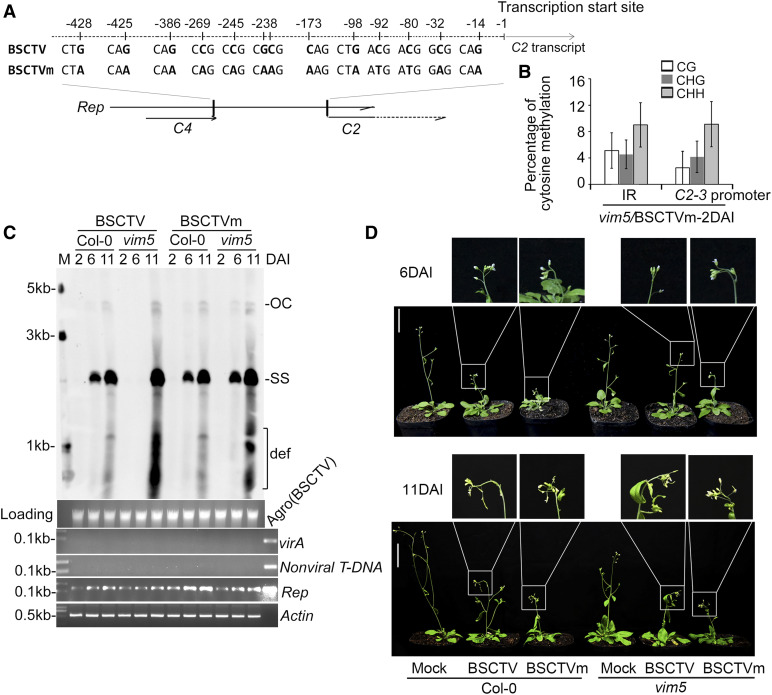
Finally, we created a mutant BSCTV protein (BSCTVm) containing 13 nucleotide substitutions in the CG and CHG sites of the C2-3 promoter to prevent DNA methylation by MET1 and CMT3 without altering the encoded Rep protein sequence (Figure 6A). As expected, the IR DNA of BSCTVm (Figure 6B) in vim5 plants exhibited similar levels of methylation in the CG, CHG, and CHH contexts to BSCTV in the Col-0 wild-type and vim5 plants (Figure 2C). Notably, BSCTVm accumulated to levels readily detectable by DNA gel blot analysis at 6 DAI in both Col-0 and vim5 plants and appeared more virulent than BSCTV (Figures 6C and 6D; Supplemental Figure 8G). These results demonstrate that BSCTV accumulation was inhibited during the early stages of systemic infection and disease development by methylation of C2-3 promoter DNA at both the CG and CHG sites. Together, our findings demonstrate that VIM5-mediated degradation of both MET1 and CMT3 promotes BSCTV infection by activating C2-C3 expression by preventing DNA methylation in the CG and CHG, but not the CHH, context in the C2-3 promoter.
Figure 6.
Characterization of the Modified BSCTV Infectious Clone and Its Viral DNA Accumulation and Methylation in vim5 Plants.
(A) Schematic diagram of the modified BSCTV infectious DNA clone, designated as BSCTVm. In BSCTVm, a total of 13 nucleotide substitutions were introduced into the CG and CHG sites of the BSCTV C2-3 promoter sequence, designed to prevent DNA methylation by MET1 and CMT3, ranging from the stop codon of C4 to the transcription start site of C2, as indicated in the schematic diagram.
(B) DNA methylation analysis of the C2-3 promoter regions and IR of viral DNA in BSCTVm-infected vim5 plants at 2 DAI. The statistical analysis was performed using OriginPro 8. Values are means ± se (n ≥ 20). The original data are shown in Supplemental Figure 7D.
(C) Detection of viral DNA accumulation by DNA gel blotting. The positions of open circle (OC) and ssDNAs (SS) and a population of defective interfering (def) DNAs are indicated. Rep-specific, biotin-labeled DNA sequence was used as a probe. StarGreen (DNA dye)–stained genomic DNA served as the loading/running control, and the samples were examined by PCR as described in Figure 2D. Signals for viral accumulation were readily detected at 6 DAI in the BSCTV- or BSCTVm-infected Col-0 wild-type plants and BSCTVm-infected vim5 plants, but not in BSCTV-infected vim5 plants.
(D) BSCTV disease symptoms. Photographs were taken at 6 and 11 DAI. Curly top symptoms were readily observed at 6 DAI for vim5 plants infected by BSCTVm plants, but not by wild-type BSCTV plants. Three independent experiments were performed. For each independent experiment, 32 individual plants per virus were used for observation. Bar = 5 cm.
DISCUSSION
In this study, we showed that the imprinted gene VIM5 encodes a ubiquitin E3 ligase that mediates 26S proteasome–dependent degradation of the DNA methyltransferases MET1 and CMT3, but not DRM2. We also showed that BSCTV infection induces transient expression of VIM5 in vegetative tissues, which is associated with hypomethylation at the CG and CHG sites of the C2-C3 promoter and the early activation of these viral genes (Supplemental Figure 9).
Local or systemic infection with several begomoviruses (e.g., Tomato yellow leaf curl Sardinia virus, African cassava mosaic virus, or TGMV) or the less virulent curtovirus Beet curly top virus reduced the mRNA levels of both MET1 and CMT3, but not DRM2, in both N. benthamiana and Arabidopsis plants at ∼3 DAI (Rodríguez-Negrete et al., 2013). In the current study, we showed that BSCTV infection of the Col-0 wild-type or vim5 plants induced no significant changes in MET1 or CMT3 mRNA accumulation from 2 to 11 DAI (Supplemental Figure 8E). Although why BSCTV, a virulent species of curtovirus (Zhang et al., 2011b), uses a host E3 ligase to degrade MET1 and CMT3 instead of downregulating their transcription is not yet known, the findings of both studies strongly suggest a universal strategy used by geminiviruses to modulate the expression of cytosine methyltransferases. Moreover, the finding (from both studies) that MET1 and CMT3, rather than DRM2, were modulated at either the transcriptional or posttranslational level suggests that geminiviruses tend to suppress the maintenance rather than the establishment of cytosine methylation. Total DNA methylation in the C2-3 promoter of the replicating BSCTV DNA was lower (Figure 2C) than that of the same region in the stably integrated pER-Rep transgene (Figure 1D), possibly as a result of continuing viral DNA replication. Indeed, the continuous rounds of replication in geminiviruses displacing newly synthesized ssDNA, and the removal of nucleosomes during the round of rolling circle replication or recombination-dependent replication prevents the access of hemimethylated dsDNA binding proteins required for the recruitment of methyltransferases (Pooggin, 2013).
VIM family proteins share highly structural similarities, including a plant homeodomain, two RING finger domains, and a Suvar Enhancer of Zeste Trithorax and RING Associated domain. While VIM5 is specifically expressed in endosperm, VIM1 to VIM3 are expressed in vegetative tissues, which is consistent with their redundant role in maintaining cytosine methylation in collaboration with MET1 (Liu et al., 2007; Kraft et al., 2008; Woo et al., 2008; Yao et al., 2012; Kim et al., 2014; Shook and Richards, 2014). Substantial work has pinpointed the in vitro E3 ligase activities associated with E2 (UBC8 family members) on VIM1, VIM2, and VIM3 (Kraft et al., 2008). However, little is known about the E3 ligase–associated functions of VIM family proteins. In the current study, we found that in contrast to VIM1-3, VIM5 mediates the 26S proteasome–dependent degradation of MET1 and CMT3. The divergent properties of VIMs may be attributed to the differences in their coding sequences in that VIM5 is distant from the VIM2, VIM3, VIM4 (62%), and VIM1 (73%) clades in the phylogenetic tree (Kraft et al., 2008).
How BSCTV induces VIM5 expression is an interesting issue. Both geminiviral Rep and C4 are early-class genes whose early expression is VIM5 independent, at least for BSCTV (Figure 2B). Multiple studies have suggested that Rep and C4 function in modulating the host cell cycles in a manner corresponding to host re-replication or even plant endoreduplication (Nagar et al., 2002; Kittelmann et al., 2009; Lai et al., 2009; Hipp et al., 2014). The incoming continuous re-establishment of epigenetic markers might cause the transient expression of VIM5 in vegetative tissues during the early stages of infection. In agreement with this notion, transient or induced expression of BSCTV-encoded Rep strongly activated the expression of VIM5 in both N. benthamiana and Arabidopsis plants (Supplemental Figures 5B and 5C), supporting the function of Rep in VIM5 activation.
In Arabidopsis, AHK proteins perceive and initiate cytokinin-associated phosphorelay signaling, which ultimately reaches and activates Arabidopsis response regulators; these proteins serve as DNA binding transcriptional activators or transcriptional repressors of their downstream target genes (Kakimoto, 2003; Müller and Sheen, 2007; Argueso et al., 2010). Although knocking out AHK4 led to the induction of VIM5 (Supplemental Figures 3C and 3D), BSCTV infection did not result in decreased AHK4 expression (Supplemental Figure 5A). It remains possible that BSCTV infection inhibits the activity of AHK4, leading to the induction of VIM5. Alternatively, BSCTV infection might target Arabidopsis response regulators to induce VIM5 expression. Indeed, our data suggest that the components and molecular mechanism(s) of BSCTV-induced expression of VIM5 are complex and warrant further investigation in the future.
Interestingly, we recently found that BSCTV infection of tomato (Solanum lycopersicum) plants at an early time point also induced transient expression of the imprinted locus ORTHRUS2, a VIM5 homolog (Supplemental Figure 10). Thus, activating an imprinted E3 ligase gene to inhibit viral DNA methylation may represent a conserved strategy used by BSCTV in diverse plant species to induce the expression of the viral RNA interference suppressor C2 and the replication enhancer C3 immediately after the expression of the early gene Rep. Previous studies provided clues about the role of VIM5 in embryo development (Zhang et al., 2018). The current findings reinforce the importance of a geminiviral counter-defense strategy involving suppressing the maintenance of DNA methylation of this gene via a specific posttranslational mechanism.
METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Col-0 was used as the wild type and the vim5 (salk_071899), met1 (salk_076522), cmt3 (salk_148381C), and ahk4 (salk_048970C) mutants (in the Columbia background) are T-DNA insertion lines obtained from the Arabidopsis Biological Resource Center (Ohio State University). The Arabidopsis seeds were germinated on Murashige and Skoog (MS) medium and stratified at 4°C for 48 h before being transferred to a 21°C growth chamber for 7 d. Seedlings with three expanded leaves were transplanted into soil and grown at 22°C under a 16-h-light/8-h-dark cycles with a white-light intensity of 120 µmol photons m–2 s–1. In the estradiol (stock solution in DMSO, E2758-250MG; Sigma-Aldrich) induction assays, Arabidopsis seedlings with (+) or without (−) induction indicate growth on MS medium containing 10 μM estradiol (+) or 10 μM DMSO (−). Nicotiana benthamiana and the GFP transgenic line 16c (Fabián E and Louise, 2002) were germinated and grown in soil at 25°C under a 16-h-light/8-h-dark cycle with the same light conditions as for Arabidopsis. Tomato seeds (Solanum lycopersicum cv Micro-Tom) provided by the Tomato Genetics Resources Center at the University of California, Davis (Li et al., 2004), were placed on moistened filter paper for 48 h for germination. Tomato seedlings were transferred to growth chambers and maintained under a long-day photoperiod (16-h-light at 25°C/8-h-dark at 18°C cycles with a white-light intensity of 200 mmol photons m–2 s–1).
Cloning and Plasmids
The following plasmids were constructed by recombination cloning using ClonExpress II or a ClonExpress MultiS kit (Vazyme) according to the manufacturer’s instructions. All oligonucleotide primers used are listed in Supplemental Data Set 2.
The full-length coding sequence (1065 bp) of BSCTV Rep was amplified from the BSCTV infectious DNA clone pCambia-BSCTV1.8 plasmid (Chen et al., 2010) and cloned into SpeI/XhoI-linearized pER-C4 vector (Lai et al., 2009). In pER-Rep, Rep was expressed under the control of the LexA operator-46 and the minimal 35S promoter of Cauliflower mosaic virus.
The 472-bp fragment including 209 bp from the start codon of the BSCTV C2 gene to the end of Rep and the 263 bp vector-derived 3A-terminator was amplified from pER-Rep and cloned into SpeI/BamHI-linearized vector pCambia1300-221. In 35S-C2N, C2N expression was under the control of the 35S promoter.
pMBP-VIM5 and 35S-VIM5 were generated by cloning the coding sequence (1944 bp) of VIM5 amplified from Col-0 plants into BamHI/PstI-linearized vector pMalC2 (Zhao et al., 2013) for translational fusion with MBP and SmaI/XbaI-linearized vector pCanG-3Flag (Liu et al., 2010) for transcriptional control by the 35S promoter.
35S-myc-VIM5 and 35S-myc-VIM5m constructs were obtained by cloning the wild-type and mutant (H158Y, H173Y, and H532Y) coding sequence of VIM5 into SmaI/BamHI-linearized vector pBI121-Myc.
The PVIM5-myc-VIM5 and PVIM5-myc-VIM5m constructs were generated by cloning the 2000-bp promoter sequence of VIM5 amplified from the genomic DNA of Col-0 plants into PacI/XbaI-linearized 35S-myc-VIM5 and 35S-myc-VIM5m, respectively, to replace the 35S promoter.
The His-MET1, His-CMT3, and His-DRM2 constructs used in the pull-down assay were generated by cloning the coding sequence of MET1, CMT3, and DRM2, respectively, into SacI/XhoI-linearized vector pET-28a (Zhao et al., 2013).
Mutant BSCTV (BSCTVm) contained a total of 13 nucleotide substitutions introduced into the CG and CHG sites of the BSCTV C2-3 promoter sequence in the infectious DNA clone pCambia-BSCTV1.8 (Chen et al., 2010), designed to prevent DNA methylation by MET1 and CMT3.
Derivatives of pCAMBIA1300-C-Luc and pCambia1300-N-Luc that, respectively, encode Cluc and Nluc driven by the 35S promoter: Cluc-VIM5 (35S-VIM5), Cluc-VIM5m (35S-myc-VIM5m), Cluc-GUS (pBI121-GUS), and the viral protein-Cluc constructs (pCambia-BSCTV1.8) Cluc-Rep, Cluc-C2, Cluc-C3, and Cluc-C4 were obtained by cloning the respective coding sequences from the parental plasmids (listed in parentheses) into KpnI/SalI-linearized pCAMBIA1300-C-Luc. To directly express Rep and C4 under the control of the 35S promoter as in Supplemental Figure 5B (right), the respective coding sequences were integrated into SacI/SalI-linearized pCAMBIA1300-C-Luc (the Cluc sequence was thus removed in both constructs). Similarly, the coding sequences of MET1, CMT3, DRM2, and VIM1 were each amplified from Col-0 plants and cloned into SacI/SalI-linearized pCambia1300-N-Luc to yield MET1-Nluc, CMT3-Nluc, DRM2-Nluc, and VIM1-Nluc, respectively.
Luciferase Complementation Imaging Assay
The luciferase complementation imaging assays, in which the N- and C-terminal halves of the firefly luciferase reconstitute active luciferase enzyme only when fused to two interacting proteins, was performed as described previously (Chen et al., 2008). The assay was used to examine the interaction between the E3 ligase VIM5 (or VIM5m) fused with Cluc and its substrate proteins MET1, CMT3, and DRM2 fused with Nluc. Equal volumes of Agrobacterium tumefaciens harboring pCambia-NLuc, pCambia-CLuc (or their derivative constructs) were mixed to a final concentration of OD600 = 1.5 with A. tumefaciens carrying 35S-P19, which encodes the tombusviral silencing suppressor p19 to ensure efficient expression of the fusion proteins. Different combinations of A. tumefaciens were infiltrated into four positions of the same N. benthamiana leaf. A low-light cooled charge-coupled device imaging apparatus (NightOWL II LB983 with indiGO software) was used to capture the luciferase images of the infiltrated leaves at 3 DAI.
In Vitro Pull-Down Assay
His-MET1, His-CMT3, His-DRM2, MBP-VIM5, and MBP were expressed in Escherichia coli (BL21 strain) and purified. Approximately 4 μg of purified MBP and MBP-VIM5 was used to bind to MBP Amylose resin beads for 2 h at 4°C. MBP Amylose resin beads were washed three times using 1× PBS and blocked by 5% (w/v) BSA for 1 h at 4°C. The beads were washed again three times with 1× PBS and incubated with 2 μg of purified His-MET1, His-CMT3, or His-DRM2 for 2 h at 4°C. Finally, the beads were washed six times with 1× PBS containing 150 mM NaCl and analyzed by SDS-PAGE.
In Vitro Cell-Free 26S Proteasomal Degradation Assay
Fresh total proteins were extracted from siliques of 6-week-old Col-0 and vim5 plants in protein extraction buffer NB1 (50 mM Tris-MES, pH 8.0, 0.5 M Suc, 1 mM MgCl2, 10 mM EDTA, 10 mM phenylmethylsulfonyl fluoride [PMSF], 1 mM DTT, and protease inhibitor cocktail Complete Mini tablets [Roche]) at 4°C. Equal amounts of His-MET1, His-CMT3, or His-DRM2 purified from E. coli were incubated with equal amounts of Col-0 or vim5 protein extracts with 5 mM ATP in each mixture at 30°C for 2 h. Samples were collected at 0, 0.5, 1, and 2 h and subjected to immunoblot analysis as described previously by Liu et al. (2011).
In Vitro Ubiquitination Assay
For the in vitro ubiquitination assay of the E3 ligase VIM5, His-tagged recombinant wheat (Triticum aestivum) E1 (GI: 136632; 50 ng), human E2 (UBCH5b; 100 ng), and Arabidopsis ubiquitin (UBQ14; At4g02890; 2 µg) purified from E. coli were incubated with MBP-tagged VIM5 or VIM5m (500 ng) as described previously by Xie et al. (2002). However, glutathione S-transferase–tagged E1, E2, and ubiquitin proteins were prepared for the in vitro ubiquitination of the candidate substrate proteins MET1, CMT3, and DRM2 (100 ng for each lane), all of which were His-tagged. MBP was used as the negative control.
Immunoblotting and Antibodies
Protein gel blot analysis was performed as described previously by Liu et al. (2011) with primary anti-LUC (1:5000 diluted, lot no. BE3612), anti-Myc (1:5000 diluted, lot no. ab127421), anti-MBP (1:5000 diluted, lot no. BE2021), anti-His (1:5000 diluted, lot no. 6200203), anti-PAG1 antibody (1:10,000 diluted, ab98962; all antibodies were purchased from EASY-BIO), anti-GFP (1:1500 diluted, lot no. 11814460001; Roche), or anti-ubiquitin (1:3000 diluted, produced by our laboratory, using ubiquitin protein expressed from the Arabidopsis UBQ14 gene as antigen; Zhao et al., 2013), followed by secondary goat anti-mouse (SA00001-1; Proteintech) or goat anti-rabbit (SA00001-2; Proteintech) antibody conjugated to horseradish peroxidase, and visualized using ECL solution (Amersham Pharmacia). Protein gel blot images were scanned, and the intensity of the bands was quantified by ImageJ (National Institutes of Health).
Agrobacterium Coinfiltration Assay, Plant Transformation, and BSCTV Inoculation by A. tumefaciens Infiltration
For the coinfiltration assays, A. tumefaciens strain EHA105 cultures carrying different plasmids were mixed together in equal volumes after OD600 values were adjusted and infiltrated into ∼5-week-old N. benthamiana leaves as previously described by Zhang et al. (2011b), with the following minor modifications. Samples were harvested at 3 DAI for analysis. The GFP transgenic line 16c of N. benthamiana was used in coinfiltration to examine the VSR activity of C2 and C2N proteins. For in planta protein degradation assays, the wild-type N. benthamiana leaves were coinfiltrated with Agrobacterium strains directing transient coexpression of myc-tagged E3 ligase VIM5 and its candidate substrate protein MET1-Nluc, CMT3-Nluc, or DRM2-Nluc as well as 35S-P19. Myc-tagged GUS and GFP constructs driven by the 35S promoter were included as controls. Wild-type N. benthamiana leaves were coinfiltrated with Agrobacterium strains harboring PVIM5-myc-VIM5 (or PVIM5-myc-VIM5m) and MET1-Nluc or CMT3-Nluc as well as those carrying 35S-GFP and 35S-P19. Total proteins were extracted at 3 DAI and subjected to immunoblotting with different antibodies as indicated. The mRNA levels of MET1, CMT3, and DRM2 were examined by RT-PCR using GFP mRNA as the internal control. Sixty-four hours after coinfiltration with Agrobacterium strains harboring 35S-GFP and 35S-P19 as well as MET1-Nluc, CMT3-Nluc, or DRM2-Nluc, 50 µM MG132 (stock solution in DMSO) suspended in 10 mM MgCl2 was infiltrated into the coinfiltrated leaves, and protein samples were harvested for immunoblotting 8 h after infiltration. Samples infiltrated with equal amounts of DMSO suspended in 10 mM MgCl2 solution served as a negative control. Stable transformation of Arabidopsis plants was performed using the vacuum infiltration method as previously described by Bechtold and Pelletier (1998).
Before BSCTV agro-inoculation, all bolts of 4-week-old Arabidopsis were removed. An approximately equal amount of a fresh 2-d culture of A. tumefaciens carrying pCambia1300-BSCTV1.8 on Luria-Bertani plates was picked up with a syringe needle to paint on the midvein of each of three rosette leaves per seedling before scratching three times to create mild wounding. Eight Arabidopsis seedlings per genotype were agro-inoculated for each time point of tissue harvest, and seedlings inoculated with A. tumefaciens carrying pCambia1300 empty vector served as the mock inoculation control. Photographs were taken at 6 and 11 DAI. After removing inoculated leaves, the remaining noninoculated rosette leaves were harvested at various time points for each genotype for further analysis. Three independent experiments were performed.
In Vivo Coimmunoprecipitation
Samples for coinfiltrated combinations between myc-VIM5 or myc-GUS and MET1-Nluc, CMT3-Nluc, or DRM2-Nluc were harvested as described above. Approximately 0.3 g of fresh total protein extracts in buffer (50 mM Tris-MES, pH 8.0, 0.5 M Suc, 1 mM MgCl2, 10 mM EDTA, 10 mM PMSF, 1 mM DTT, 50 µM MG132, and protease inhibitor cocktail Complete Mini tablets) was prepared for each sample as described for the cell-free degradation assay. The fresh total protein extracts were bound to protein-G myc beads (code no. MO47-10; MBL) for 2 h at 4°C. The antigen–antibody complexes were washed three times (5 min each time) in 1× PBS-NaCl (1 mM PMSF and 200 mM NaCl), diluted in 40 µL of 1× PBS, and analyzed by SDS-PAGE.
Nucleic Acid Gel Blotting
For DNA gel blotting, total DNA was extracted using cetyltrimethylammonium ammonium bromide (CTAB) buffer as described by Chen et al. (2010), and 15 µg total DNA was loaded and stained with ethidium bromide or DNA dye (StarGreen DNA Dye 10,000×, E107-10; GenStar) to monitor loading, separated by electrophoresis on a 0.9% (w/v) agarose gel, and transferred to Hybond N+ nylon membranes after depurination and neutralization as described by Zhang et al. (2011b).)
For RNA gel blotting, total RNA was extracted using the hot-phenol method as previously described by Fernández et al. (1997). Twenty micrograms of total RNA was separated on 2.4% (w/v) agarose gels containing 6% (v/v) formaldehyde, transferred to nylon N+ membranes, and stained with methylene blue to monitor loading.
For gel blotting using an isotope-labeling system, probe sequences were amplified and labeled with [α-32P]dCTP using the Rediprime II system according to the manufacturer’s manual. Membranes were hybridized at 65°C overnight as described by Chen et al. (2010) and washed once with 2× SSC and 0.2% (w/v) SDS at 65°C for 20 min and once with 0.2× SSC and 0.2% (w/v) SDS at 65°C for 10 min. The signal was visualized on a Typhoon FLA 7000 system (GE Healthcare).
For DNA gel blotting conducted by a biotin-labeling system (Qi et al., 2017), probe sequences were labeled with Biotin-dUTP by a LA-Taq–based PCR process (20-µL PCR volume) in addition to 1.35 µL of Biotin-11-dUTP (1 mM, catalog no. R0081; Thermo Fisher Scientific) according to the LA-Taq PCR manual (no. DRR002A; Takara). Membranes were hybridized at 65°C overnight with PerfectHyb Plus hybridization buffer (no. 3180216; Sigma-Aldrich) using the same procedure as described for the isotope-labeling system (Chen et al., 2010) and washed with a Chemiluminescent Nucleic Acid Detection Module kit (no. 89880; Thermo Fisher Scientific) according to the manufacturer’s instructions. The signal was visualized using the Azure c600 imaging system (Azure Biosystems).
qPCR and RT-PCR Analysis
After removing residual genome DNA using gDNA wiper mix (Vazyme), reverse transcription was performed using HiScript II qRT Supermix (Vazyme). Quantitative PCR was performed using ChamQ SYBR qPCR MasterMix (Vazyme) with the CFX96 real-time system (Bio-Rad). The transcript levels of the target genes were quantified relative to Arabidopsis ACTIN as an internal control. The primers used for quantitative PCR are listed in Supplemental Data Set 2. Three technical replicates within one experiment for each sample were performed, and three independent experiments were conducted for biological replicates.
RT-PCR was performed as previously described (Duan et al., 2012). cDNAs were subjected to qPCR to amplify fragments with 29 or 33 thermal cycles, as necessary. Three biological replicates were performed. The primers are listed in Supplemental Data Set 2. To measure mRNA levels in N. benthamiana inoculation samples, GFP expression served as the internal control. To detect BSCTV-encoded gene expression and verify VIM5 expression in pER-Rep-3/VIM5 and vim5/35S-VIM5 lines, ACTIN expression served as the internal control.
DNA Bisulfite Sequencing Analysis, 5′ and 3′ RACE Assays, and Thermal Asymmetric Interlaced PCR
Two micrograms of Arabidopsis genomic DNA was subjected to bisulfite treatment using an EpiTect bisulfite kit (Qiagen) according to the manufacturer’s handbook. The purified bisulfite-treated DNA was amplified using specific primer pairs designed as previously described by Zilberman et al. (2003; Supplemental Data Set 2). The PCR products were ligated into the pMD18-T vector. Approximately 10 colonies were used for sequencing, and cytosine methylation was analyzed using the CyMATE program (http://cymate.org/cymate.html). Sequences with the same cytosine methylation status were counted as one clone. At least three technical replicates of bisulfite treatment were performed for each viral infection experiment. The numbers of individual clones obtained from three independent viral infection experiments and used for statistical analysis in each figure are listed in the Supplemental Table. To determining the bisulfite conversion efficiency, each bisulfite-treated sample was also examined for the methylation status of an endogenous Arabidopsis DNA locus (At5g66750) as the internal control, which was previously shown not to contain any methylated cytosine (Hetzl et al., 2007).
The 5′ and 3′ RACE assays were performed using a First Choice RLM-RACE kit (Ambion) according to the manufacturer’s manuals. Both the 5′ RACE and 3′ RACE PCR products were ligated into the pMD18-T vector, and no fewer than 10 individual clones for each RACE assay were sequenced.
Thermal asymmetric interlaced PCR was conducted to characterize the pER-Rep insertion site for pER-Rep-5 plants using a Genome Walking kit (code no. 6108; Takara) according to the manufacturer’s instructions. Primers for the left border and right border are listed in Supplemental Data Set 2.
Statistical Analysis
ANOVA tables are provided in Supplemental File 1. Student’s t test tables are provided in Supplemental File 2.
Accession Numbers
The gene information in this article can be found in the TAIR10 or GenBank/EMBL under the following accession numbers: VIM5 (AT1G57800); MET1 (AT5G49160); CMT3 (AT1G69770); DRM2 (AT5G14620); VIM1 (AT1G57820); VIM2 (AT1G66050); VIM3 (AT5G39550); VIM4 (AT1G66040); AHK4 (AT2G01830); ACTIN2 (AT3G18780); ORTHRUS2 (XM_004239523.4); Tom52 (NM_001345841.1); N. benthamiana ACTIN (JQ256516.1); Beet severe curly top virus (U02311.1).
Supplemental Data
Supplemental Figure 1. Bisulfite sequencing analysis of methylated cytosine at the C2-3 promoter in pER-Rep transgenic lines.
Supplemental Figure 2. C2N lacks transgene RNA-silencing suppression activity.
Supplemental Figure 3. Analysis of DNA methylation-related genes in transgenic pER-Rep plants and characterization of pER-Rep-5 plants.
Supplemental Figure 4. C2N transcription and analysis of methylated cytosine at the C2-3 promoter in pER-Rep-3/VIM5 transgenic plants.
Supplemental Figure 5. Analysis of VIM5-inducing factors.
Supplemental Figure 6. Characterization of vim5 and vim5/35S-VIM5 plants and analysis of viral DNA accumulation, viral gene expression, and methylation in BSCTV-infected plants.
Supplemental Figure 7. Bisulfite sequencing analysis of methylated cytosine at the C2-3 promoter, intergenic region, and At5g66750 for BSCTV/BSCTVm-infected plants and transgenic plants harboring pER-Rep cassettes.
Supplemental Figure 8. Characterization of VIM5 mutant protein and its interactions with DNA methyltransferases.
Supplemental Figure 9. Graphical model of the role of BSCTV-induced transient expression of VIM5 in degrading methyltransferases to ensure viral DNA accumulation during the early infection period.
Supplemental Figure 10. Analysis of viral DNA accumulation and the expression of the VIM5 homolog in BSCTV-infected tomato plants.
Supplemental Table. Number of individual independent clones from three independent BSCTV infection experiments used for statistical analysis for each plant genotype.
Supplemental Data Set 1. Transcriptome data of genes involved in DNA methylation pathway.
Supplemental Data Set 2. Primers used in this study.
Supplemental File 1. ANOVA tables.
Supplemental File 2. Student’s t test tables.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Jifeng Fei for technical support and David Baulcombe for the GFP 16c seeds. This study was supported by the National Natural Science Foundation of China (grants 31730078 and 31390423).
AUTHOR CONTRIBUTIONS
Z.-Q.C., Q.X., and H.-S.G. designed the experiments. Z.-Q.C., Q.C., Z.-H.Z., and J.L. performed the experiments. Z.-Q.C., J.-H.Z., and Z.-X.G. analyzed the transcriptome and bisulfite sequencing data. Z.-Q.C., Q.X., S.-W.D., and H.-S.G. discussed the results and wrote the article.
References
- Amin, I., Hussain, K., Akbergenov, R., Yadav, J.S., Qazi, J., Mansoor, S., Hohn, T., Fauquet, C.M., Briddon, R.W.(2011). Suppressors of RNA silencing encoded by the components of the cotton leaf curl begomovirus-betasatellite complex. Mol. Plant Microbe Interact. 24: 973–983. [DOI] [PubMed] [Google Scholar]
- Aregger, M., Borah, B.K., Seguin, J., Rajeswaran, R., Gubaeva, E.G., Zvereva, A.S., Windels, D., Vazquez, F., Blevins, T., Farinelli, L., Pooggin, M.M.(2012). Primary and secondary siRNAs in geminivirus-induced gene silencing. PLoS Pathog. 8: e1002941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argueso, C.T., Raines, T., Kieber, J.J.(2010). Cytokinin signaling and transcriptional networks. Curr. Opin. Plant Biol. 13: 533–539. [DOI] [PubMed] [Google Scholar]
- Bechtold, N., Pelletier, G.(1998). In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol. Biol. 82: 259–266. [DOI] [PubMed] [Google Scholar]
- Berger, M.R., Sunter, G.(2013). Identification of sequences required for AL2-mediated activation of the tomato golden mosaic virus-yellow vein BR1 promoter. J. Gen. Virol. 94: 1398–1406. [DOI] [PubMed] [Google Scholar]
- Bian, X.Y., Rasheed, M.S., Seemanpillai, M.J., Ali Rezaian, M.(2006). Analysis of silencing escape of tomato leaf curl virus: An evaluation of the role of DNA methylation. Mol. Plant Microbe Interact. 19: 614–624. [DOI] [PubMed] [Google Scholar]
- Borah, B.K., Zarreen, F., Baruah, G., Dasgupta, I.(2016). Insights into the control of geminiviral promoters. Virology 495: 101–111. [DOI] [PubMed] [Google Scholar]
- Broyles, S.S.(2003). Vaccinia virus transcription. J. Gen. Virol. 84: 2293–2303. [DOI] [PubMed] [Google Scholar]
- Buchmann, R.C., Asad, S., Wolf, J.N., Mohannath, G., Bisaro, D.M.(2009). Geminivirus AL2 and L2 proteins suppress transcriptional gene silencing and cause genome-wide reductions in cytosine methylation. J. Virol. 83: 5005–5013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carluccio, A.V., Prigigallo, M.I., Rosas-Diaz, T.(2018). S-Acylation mediates Mungbean yellow mosaic virus AC4 localization to the plasma membrane and in turns gene silencing suppression. PLoS Pathog. 14: e1007207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chellappan, P., Vanitharani, R., Fauquet, C.M.(2005). MicroRNA-binding viral protein interferes with Arabidopsis development. Proc. Natl. Acad. Sci. USA 102: 10381–10386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H., Zhang, Z., Teng, K., Lai, J., Zhang, Y., Huang, Y., Li, Y., Liang, L., Wang, Y., Chu, C., Guo, H., Xie, Q.(2010). Up-regulation of LSB1/GDU3 affects geminivirus infection by activating the salicylic acid pathway. Plant J. 62: 12–23. [DOI] [PubMed] [Google Scholar]
- Chen, H., Zou, Y., Shang, Y., Lin, H., Wang, Y., Cai, R., Tang, X., Zhou, J.M.(2008). Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 146: 368–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, S., Jang, I.C., Su, L., Xu, J., Chua, N.H.(2016). JMJ24 targets CHROMOMETHYLASE3 for proteasomal degradation in Arabidopsis. Genes Dev. 30: 251–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deom, C.M., Mills-Lujan, K.(2015). Toward understanding the molecular mechanism of a geminivirus C4 protein. Plant Signal. Behav. 10: e1109758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan, C.G., Fang, Y.Y., Zhou, B.J., Zhao, J.H., Hou, W.N., Zhu, H., Ding, S.W., Guo, H.S.(2012). Suppression of Arabidopsis ARGONAUTE1-mediated slicing, transgene-induced RNA silencing, and DNA methylation by distinct domains of the Cucumber mosaic virus 2b protein. Plant Cell 24: 259–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elder, J.T., Spritz, R.A., Weissman, S.M.(1981). Simian virus 40 as a eukaryotic cloning vehicle. Annu. Rev. Genet. 15: 295–340. [DOI] [PubMed] [Google Scholar]
- Fabián E, Vaistij, Louise, Jones; David C Baulcombe (2002). Spreading of RNA targeting and DNA methylation in RNA silencing requires transcription of the target gene and a putative RNA-dependent RNA polymerase. Plant Cell 14: 857–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández, A., Guo, H.S., Sáenz, P., Simón-Buela, L., Gómez de Cedrón, M., García, J.A.(1997). The motif V of plum pox potyvirus CI RNA helicase is involved in NTP hydrolysis and is essential for virus RNA replication. Nucleic Acids Res. 25: 4474–4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontes, E.P., Luckow, V.A., Hanley-Bowdoin, L.(1992). A geminivirus replication protein is a sequence-specific DNA binding protein. Plant Cell 4: 597–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frischmuth, S., Frischmuth, T., Jeske, H.(1991). Transcript mapping of Abutilon mosaic virus, a geminivirus. Virology 185: 596–604. [DOI] [PubMed] [Google Scholar]
- Frischmuth, S., Frischmuth, T., Latham, J.R., Stanley, J.(1993). Transcriptional analysis of the virion-sense genes of the geminivirus beet curly top virus. Virology 197: 312–319. [DOI] [PubMed] [Google Scholar]
- Gehring, M., Missirian, V., Henikoff, S.(2011). Genomic analysis of parent-of-origin allelic expression in Arabidopsis thaliana seeds. PLoS One 6: e23687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick, E., Zrachya, A., Levy, Y., Mett, A., Gidoni, D., Belausov, E., Citovsky, V., Gafni, Y.(2008). Interaction with host SGS3 is required for suppression of RNA silencing by tomato yellow leaf curl virus V2 protein. Proc. Natl. Acad. Sci. USA 105: 157–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, Z., Li, Y., Ding, S.W.(2018). Small RNA-based antimicrobial immunity. Nat. Rev. Immunol. 19: 31–44. [DOI] [PubMed] [Google Scholar]
- Hanley-Bowdoin, L., Bejarano, E.R., Robertson, D., Mansoor, S.(2013). Geminiviruses: Masters at redirecting and reprogramming plant processes. Nat. Rev. Microbiol. 11: 777–788. [DOI] [PubMed] [Google Scholar]
- Hanley-Bowdoin, L., Elmer, J.S., Rogers, S.G.(1989). Functional expression of the leftward open reading frames of the A component of tomato golden mosaic virus in transgenic tobacco plants. Plant Cell 1: 1057–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanley-Bowdoin, L., Settlage, S.B., Orozco, B.M., Nagar, S., Robertson, D.(2000). Geminiviruses: Models for plant DNA replication, transcription, and cell cycle regulation. Crit. Rev. Biochem. Mol. Biol. 35: 105–140. [PubMed] [Google Scholar]
- Hetzl, J., Foerster, A.M., Raidl, G., Mittelsten Scheid, O.(2007). CyMATE: A new tool for methylation analysis of plant genomic DNA after bisulphite sequencing. Plant J. 51: 526–536. [DOI] [PubMed] [Google Scholar]
- Hipp, K., Rau, P., Schäfer, B., Gronenborn, B., Jeske, H.(2014). The RXL motif of the African cassava mosaic virus Rep protein is necessary for rereplication of yeast DNA and viral infection in plants. Virology 462-463: 189–198. [DOI] [PubMed] [Google Scholar]
- Hormuzdi, S.G., Bisaro, D.M.(1995). Genetic analysis of beet curly top virus: Examination of the roles of L2 and L3 genes in viral pathogenesis. Virology 206: 1044–1054. [DOI] [PubMed] [Google Scholar]
- Hsieh, T.F., Shin, J., Uzawa, R., Silva, P., Cohen, S., Bauer, M.J., Hashimoto, M., Kirkbride, R.C., Harada, J.J., Zilberman, D., Fischer, R.L.(2011). Regulation of imprinted gene expression in Arabidopsis endosperm. Proc. Natl. Acad. Sci. USA 108: 1755–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackel, J.N., Buchmann, R.C., Singhal, U., Bisaro, D.M.(2015). Analysis of geminivirus AL2 and L2 proteins reveals a novel AL2 silencing suppressor activity. J. Virol. 89: 3176–3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeske, H.(2009). Geminiviruses. Curr. Top. Microbiol. Immunol. 331: 185–226. [DOI] [PubMed] [Google Scholar]
- Jeske, H., Lütgemeier, M., Preiss, W.(2001). DNA forms indicate rolling circle and recombination-dependent replication of Abutilon mosaic virus. EMBO J. 20: 6158–6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakimoto, T.(2003). Perception and signal transduction of cytokinins. Annu. Rev. Plant Biol. 54: 605–627. [DOI] [PubMed] [Google Scholar]
- Kim, J., Kim, J.H., Richards, E.J., Chung, K.M., Woo, H.R.(2014). Arabidopsis VIM proteins regulate epigenetic silencing by modulating DNA methylation and histone modification in cooperation with MET1. Mol. Plant 7: 1470–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittelmann, K., Rau, P., Gronenborn, B., Jeske, H.(2009). Plant geminivirus rep protein induces rereplication in fission yeast. J. Virol. 83: 6769–6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft, E., Bostick, M., Jacobsen, S.E., Callis, J.(2008). ORTH/VIM proteins that regulate DNA methylation are functional ubiquitin E3 ligases. Plant J. 56: 704–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacatus, G., Sunter, G.(2009). The Arabidopsis PEAPOD2 transcription factor interacts with geminivirus AL2 protein and the coat protein promoter. Virology 392: 196–202. [DOI] [PubMed] [Google Scholar]
- Lai, J., Chen, H., Teng, K., Zhao, Q., Zhang, Z., Li, Y., Liang, L., Xia, R., Wu, Y., Guo, H., Xie, Q.(2009). RKP, a RING finger E3 ligase induced by BSCTV C4 protein, affects geminivirus infection by regulation of the plant cell cycle. Plant J. 57: 905–917. [DOI] [PubMed] [Google Scholar]
- Lee, J.S., Raja, P., Knipe, D.M.(2016). Herpesviral ICP0 protein promotes two waves of heterochromatin removal on an early viral promoter during lytic infection. MBio 7: e02007–e02015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., Zeng, R., Chen, Z., Liu, X., Cao, Z., Xie, Q., Yang, C., Lai, J.(2018). S-Acylation of a geminivirus C4 protein is essential for regulating the CLAVATA pathway in symptom determination. J. Exp. Bot. 69: 4459–4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L., Zhao, Y., McCaig, B.C., Wingerd, B.A., Wang, J., Whalon, M.E., Pichersky, E., Howe, G.A.(2004). The tomato homolog of CORONATINE-INSENSITIVE1 is required for the maternal control of seed maturation, jasmonate-signaled defense responses, and glandular trichome development. Plant Cell 16: 126–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, L., Chung, H.Y., Lacatus, G., Baliji, S., Ruan, J., Sunter, G.(2014). Altered expression of Arabidopsis genes in response to a multifunctional geminivirus pathogenicity protein. BMC Plant Biol. 14: 302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, L., Cui, F., Li, Q., Yin, B., Zhang, H., Lin, B., Wu, Y., Xia, R., Tang, S., Xie, Q.(2011). The endoplasmic reticulum-associated degradation is necessary for plant salt tolerance. Cell Res. 21: 957–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, L., Zhang, Y., Tang, S., Zhao, Q., Zhang, Z., Zhang, H., Dong, L., Guo, H., Xie, Q.(2010). An efficient system to detect protein ubiquitination by agroinfiltration in Nicotiana benthamiana. Plant J. 61: 893–903. [DOI] [PubMed] [Google Scholar]
- Liu, S., Yu, Y., Ruan, Y., Meyer, D., Wolff, M., Xu, L., Wang, N., Steinmetz, A., Shen, W.H.(2007). Plant SET- and RING-associated domain proteins in heterochromatinization. Plant J. 52: 914–926. [DOI] [PubMed] [Google Scholar]
- Luo, M., Taylor, J.M., Spriggs, A., Zhang, H., Wu, X., Russell, S., Singh, M., Koltunow, A.(2011). A genome-wide survey of imprinted genes in rice seeds reveals imprinting primarily occurs in the endosperm. PLoS Genet. 7: e1002125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, B., Sheen, J.(2007). Arabidopsis cytokinin signaling pathway. Sci. STKE 2007: cm5. [DOI] [PubMed] [Google Scholar]
- Mullineaux, P.M., Rigden, J.E., Dry, I.B., Krake, L.R., Rezaian, M.A.(1993). Mapping of the polycistronic RNAs of tomato leaf curl geminivirus. Virology 193: 414–423. [DOI] [PubMed] [Google Scholar]
- Nagar, S., Hanley-Bowdoin, L., Robertson, D.(2002). Host DNA replication is induced by geminivirus infection of differentiated plant cells. Plant Cell 14: 2995–3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paprotka, T., Deuschle, K., Metzler, V., Jeske, H.(2011). Conformation-selective methylation of geminivirus DNA. J. Virol. 85: 12001–12012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pooggin, M.M.(2013). How can plant DNA viruses evade siRNA-directed DNA methylation and silencing? Int. J. Mol. Sci. 14: 15233–15259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, L., Yue, L., Feng, D., Qi, F., Li, J., Dong, X.(2017). Genome-wide mRNA processing in methanogenic archaea reveals post-transcriptional regulation of ribosomal protein synthesis. Nucleic Acids Res. 45: 7285–7298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raja, P., Sanville, B.C., Buchmann, R.C., Bisaro, D.M.(2008). Viral genome methylation as an epigenetic defense against geminiviruses. J. Virol. 82: 8997–9007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raja, P., Wolf, J.N., Bisaro, D.M.(2010). RNA silencing directed against geminiviruses: Post-transcriptional and epigenetic components. Biochim. Biophys. Acta 1799: 337–351. [DOI] [PubMed] [Google Scholar]
- Ramesh, S.V., Sahu, P.P., Prasad, M., Praveen, S., Pappu, H.R.(2017). Geminiviruses and plant hosts: A closer examination of the molecular arms race. Viruses 9: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Negrete, E., Lozano-Durán, R., Piedra-Aguilera, A., Cruzado, L., Bejarano, E.R., Castillo, A.G.(2013). Geminivirus Rep protein interferes with the plant DNA methylation machinery and suppresses transcriptional gene silencing. New Phytol. 199: 464–475. [DOI] [PubMed] [Google Scholar]
- Rosas-Diaz, T., et al. (2018). A virus-targeted plant receptor-like kinase promotes cell-to-cell spread of RNAi. Proc. Natl. Acad. Sci. USA 115: 1388–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saribas, A.S., Coric, P., Bouaziz, S., Safak, M.(2019). Expression of novel proteins by polyomaviruses and recent advances in the structural and functional features of agnoprotein of JC virus, BK virus, and simian virus 40. J. Cell. Physiol. 234: 8295–8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaprasad, P.V., Akbergenov, R., Trinks, D., Rajeswaran, R., Veluthambi, K., Hohn, T., Pooggin, M.M.(2005). Promoters, transcripts, and regulatory proteins of Mungbean yellow mosaic geminivirus. J. Virol. 79: 8149–8163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shook, M.S., Richards, E.J.(2014). VIM proteins regulate transcription exclusively through the MET1 cytosine methylation pathway. Epigenetics 9: 980–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shung, C.-Y., Sunter, J., Sirasanagandla, S.S., Sunter, G.(2006). Distinct viral sequence elements are necessary for expression of Tomato golden mosaic virus complementary sense transcripts that direct AL2 and AL3 gene expression. Mol. Plant Microbe Interact. 19: 1394–1405. [DOI] [PubMed] [Google Scholar]
- Smalle, J., Vierstra, R.D.(2004). The ubiquitin 26S proteasome proteolytic pathway. Annu. Rev. Plant Biol. 55: 555–590. [DOI] [PubMed] [Google Scholar]
- Stroud, H., Greenberg, M.V., Feng, S., Bernatavichute, Y.V., Jacobsen, S.E.(2013). Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell 152: 352–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunter, G., Bisaro, D.M.(1992). Transactivation of geminivirus AR1 and BR1 gene expression by the viral AL2 gene product occurs at the level of transcription. Plant Cell 4: 1321–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinks, D., Rajeswaran, R., Shivaprasad, P.V., Akbergenov, R., Oakeley, E.J., Veluthambi, K., Hohn, T., Pooggin, M.M.(2005). Suppression of RNA silencing by a geminivirus nuclear protein, AC2, correlates with transactivation of host genes. J. Virol. 79: 2517–2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanitharani, R., Chellappan, P., Pita, J.S., Fauquet, C.M.(2004). Differential roles of AC2 and AC4 of cassava geminiviruses in mediating synergism and suppression of posttranscriptional gene silencing. J. Virol. 78: 9487–9498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., Buckley, K.J., Yang, X., Buchmann, R.C., Bisaro, D.M.(2005). Adenosine kinase inhibition and suppression of RNA silencing by geminivirus AL2 and L2 proteins. J. Virol. 79: 7410–7418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S., Chen, H., Wang, Y., Pan, C., Tang, X., Zhang, H., Chen, W., Chen, Y.Q.(2020). Effects of Agrobacterium tumefaciens strain types on the Agrobacterium-mediated transformation efficiency of filamentous fungus Mortierella alpina. Lett. Appl. Microbiol. 70: 388–393. [DOI] [PubMed] [Google Scholar]
- Woo, H.R., Dittmer, T.A., Richards, E.J.(2008). Three SRA-domain methylcytosine-binding proteins cooperate to maintain global CpG methylation and epigenetic silencing in Arabidopsis. PLoS Genet. 4: e1000156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo, H.R., Pontes, O., Pikaard, C.S., Richards, E.J.(2007). VIM1, a methylcytosine-binding protein required for centromeric heterochromatinization. Genes Dev. 21: 267–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Q., Guo, H.S., Dallman, G., Fang, S., Weissman, A.M., Chua, N.H.(2002). SINAT5 promotes ubiquitin-related degradation of NAC1 to attenuate auxin signals. Nature 419: 167–170. [DOI] [PubMed] [Google Scholar]
- Yang, L.P., Fang, Y.Y., An, C.P., Dong, L., Zhang, Z.H., Chen, H., Xie, Q., Guo, H.S.(2013). C2-mediated decrease in DNA methylation, accumulation of siRNAs, and increase in expression for genes involved in defense pathways in plants infected with beet severe curly top virus. Plant J. 73: 910–917. [DOI] [PubMed] [Google Scholar]
- Yao, Q., Song, C.X., He, C., Kumaran, D., Dunn, J.J.(2012). Heterologous expression and purification of Arabidopsis thaliana VIM1 protein: In vitro evidence for its inability to recognize hydroxymethylcytosine, a rare base in Arabidopsis DNA. Protein Expr. Purif. 83: 104–111. [DOI] [PubMed] [Google Scholar]
- Zhang, H., Lang, Z., Zhu, J.K.(2018). Dynamics and function of DNA methylation in plants. Nat. Rev. Mol. Cell Biol. 19: 489–506. [DOI] [PubMed] [Google Scholar]
- Zhang, J., Dong, J., Xu, Y., Wu, J.(2012). V2 protein encoded by Tomato yellow leaf curl China virus is an RNA silencing suppressor. Virus Res. 163: 51–58. [DOI] [PubMed] [Google Scholar]
- Zhang, M., Zhao, H., Xie, S., Chen, J., Xu, Y., Wang, K., Zhao, H., Guan, H., Hu, X., Jiao, Y., Song, W., Lai, J.(2011a). Extensive, clustered parental imprinting of protein-coding and noncoding RNAs in developing maize endosperm. Proc. Natl. Acad. Sci. USA 108: 20042–20047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z., et al. (2011b). BSCTV C2 attenuates the degradation of SAMDC1 to suppress DNA methylation-mediated gene silencing in Arabidopsis. Plant Cell 23: 273–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Q., Tian, M., Li, Q., Cui, F., Liu, L., Yin, B., Xie, Q.(2013). A plant-specific in vitro ubiquitination analysis system. Plant J. 74: 524–533. [DOI] [PubMed] [Google Scholar]
- Zhou, X.(2013). Advances in understanding begomovirus satellites. Annu. Rev. Phytopathol. 51: 357–381. [DOI] [PubMed] [Google Scholar]
- Zilberman, D., Cao, X., Jacobsen, S.E.(2003). ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299: 716–719. [DOI] [PubMed] [Google Scholar]
- Zrachya, A., Glick, E., Levy, Y., Arazi, T., Citovsky, V., Gafni, Y.(2007). Suppressor of RNA silencing encoded by Tomato yellow leaf curl virus-Israel. Virology 358: 159–165. [DOI] [PubMed] [Google Scholar]
- Zuo, J., Niu, Q.W., Chua, N.H.(2000). Technical advance: An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 24: 265–273. [DOI] [PubMed] [Google Scholar]