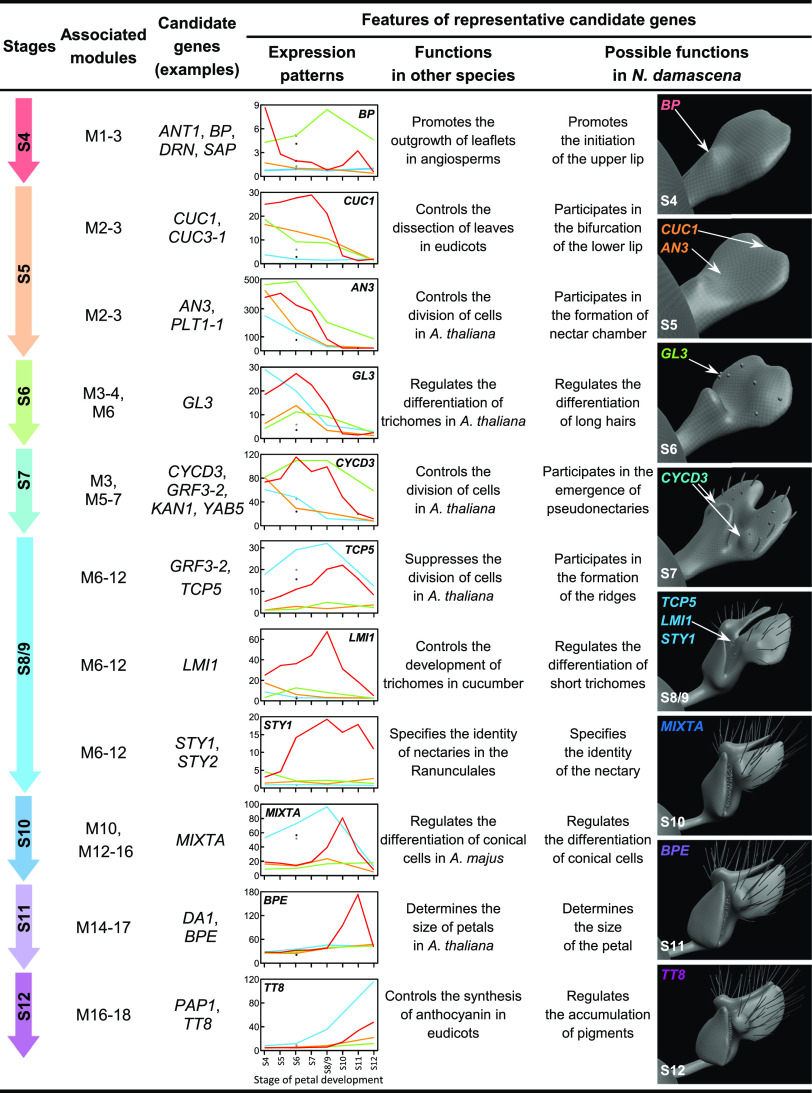
Extensive transcriptomic and functional analyses of elaborate petal development and specialized character formation in Nigella damascena point to the mechanisms underlying lateral organ diversification in plants.
Abstract
Petals can be simple or elaborate, depending on whether they have lobes, teeth, fringes, or appendages along their margins, or possess spurs, scales, or other types of modifications on their adaxial/abaxial side, or both. Elaborate petals have been recorded in 23 orders of angiosperms and are generally believed to have played key roles in the adaptive evolution of corresponding lineages. The mechanisms underlying the formation of elaborate petals, however, are largely unclear. Here, by performing extensive transcriptomic and functional studies on Nigella damascena (Ranunculaceae), we explore the mechanisms underlying elaborate petal development and specialized character formation. In addition to the identification of genes and programs that are specifically/preferentially expressed in petals, we found genes and programs that are required for elaborate rather than simple petal development. By correlating the changes in gene expression with those in petal development, we identified 30 genes that are responsible for the marginal/ventral elaboration of petals and the initiation of several highly specialized morphological characters (e.g., pseudonectaries, long hairs, and short trichomes). Expression and functional analyses further confirmed that a class I homeodomain-leucine zipper family transcription factor gene, Nigella damascena LATE MERISTEM IDENTITY1 (NidaLMI1), plays important roles in the development of short trichomes and bifurcation of the lower lip. Our results not only provide the first portrait of elaborate petal development but also pave the way to understanding the mechanisms underlying lateral organ diversification in plants.
INTRODUCTION
Petals are highly specialized lateral organs that lie between the outer sepals and inner stamens and/or carpels in a flower with a dimorphic perianth and play key roles in plant–animal interactions. Petals display spectacular arrays of diversity in size, shape, color, and function and are increasingly being used as a model system for the study of plant organogenesis and evolution (Irish, 2008; Huang and Irish, 2016). In Arabidopsis (Arabidopsis thaliana; Brassicaceae) and many other petalous taxa (such as snapdragon [Antirrhinum majus], petunia [Petunia hybrida], columbine [Aquilegia], peonies [Paeonia], roses [Rosa], lilies [Lilium], and orchids [Orchidaceae]), petals, like any other types of lateral organs of plants, are formed through at least three highly conserved developmental processes: initiation, growth, and maturation. Genes, pathways, and networks involved in the proliferation, expansion, and differentiation of cells, as well as those specifying the adaxial–abaxial, proximal–distal, and lateral–medial polarities of the entire organ, are therefore indispensable (Irish, 2008; Huang and Irish, 2016; Walcher-Chevillet and Kramer, 2016; Shan et al., 2019). Yet, unlike many other types of lateral organs, petals are usually brightly colored and/or unusually shaped, suggesting that, in addition to the common themes of lateral organs, there are developmental processes that are specific to petals (such as determination of petal identity; generation of the sometimes highly specialized, three-dimensional structures; and/or formation of intriguing color patterns; Irish, 2008, 2017; Huang and Irish, 2016; Moyroud and Glover, 2017). Hence, identifying the regulatory genes, pathways, and networks involved in petal morphogenesis is not only the prerequisite for elucidating how petals were made through development and evolution but also is helpful for understanding the mechanisms underlying lateral organ diversification.
Notably, almost all our knowledge about the mechanisms underlying petal organogenesis was gained from studying species with simple petals (such as those of Arabidopsis). In flowering plants, however, there are many species whose petals are highly specialized, either having lobes, teeth, fringes, or appendages along their margins, or possessing scales, spurs, pockets, or other types of modifications on their adaxial/abaxial sides, or both (Figures 1A to 1H; Endress and Matthews, 2006). These petals, also known as elaborate petals, have been recorded in hundreds of families of at least 23 orders of angiosperms and are generally believed to have played key roles in the adaptive evolution of the corresponding lineages (Endress and Matthews, 2006; Yao et al., 2019). Interestingly, in addition to the modification of their basic structures through ventral or marginal elaboration (Figure 1I), many elaborate petals also bear some newly gained characters (such as the labellum of the orchid Ophrys speculum, which mimics a female wasp [Colpa aurea]; Endress and Matthews, 2006; Yao et al., 2019). Uncovering the developmental mechanisms underlying the organogenesis of elaborate petals therefore will not only pave the way to elucidating the mechanisms underlying petal elaboration but also help in understanding the general patterns of new character origination and flowering plant evolution.
Figure 1.
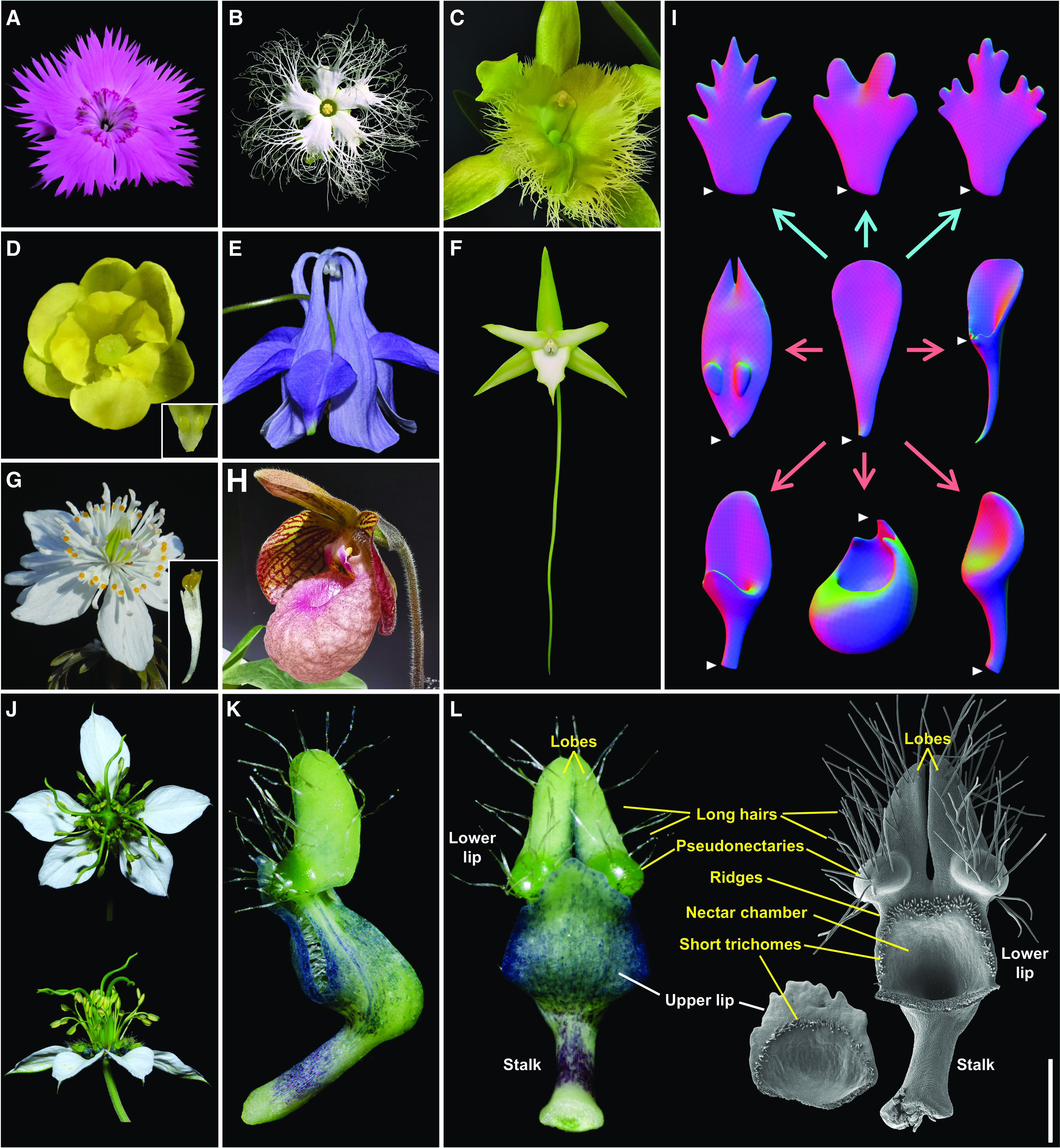
Elaborate Petals and Petal Elaboration.
(A) to (H) Representative species with elaborate petals. (A) Dianthus chinensis (Caryophyllaceae), with five elaborate petals with marginal teeth. (B) Trichosanthes kirilowii (Cucurbitaceae), with five elaborate petals with marginal fringes. (C) Rhyncholaelia digbyana (Orchidaceae), with one elaborate petal (i.e., labellum) with marginal fringes. (D) Berberis sp. (Berberidaceae), with five petals with each bearing two marginal lobes and two ventral lobes. Inset image shows the magnified two ventral lobes. (E) Aquilegia yabeana (Ranunculaceae), with five elaborate petals with curved spurs. (F) Angraecum sesquipedale (Orchidaceae), with one labellum with a long spur (∼30 cm). (G) Eranthis stellata (Ranunculaceae), with about 12 petals, each of which is tubular with a stalk and two yellowish pseudonectaries. Inset image shows the magnified elaborate petal. (H) Paphiopedilum micranthum (Orchidaceae), with one labellum with a pocket.
(I) Virtual clay models of elaborate petals and main ways of petal elaboration. White triangles indicate the positions from which the petals were connected to the receptacles. Blue and red arrows imply marginal and ventral elaborations, respectively.
(J) N. damascena, with eight highly elaborate petals (see [K] and [L]).
(K) and (L) Side (K) and front (L) views of a mature petal under stereomicroscope and scanning electron microscope. The three basic components and the highly specialized characters are shown in white and yellow, respectively. Bar = 1 mm.
The genus Nigella (Ranunculaceae) appears to be an excellent system for the study of petal elaboration and its underlying mechanisms, for four reasons. First, petals of several species in this genus have long been regarded as being elaborate, possessing not only a rod-like stalk, a laminar upper lip, and a bifurcate lower lip but also several highly specialized morphological characters, such as long hairs, short trichomes, a nectary, and a pair of pseudonectaries (Figures 1J to 1L; Strid, 1970; Tamura, 1995; Zhao et al., 2011). This attribute makes it possible to study both marginal/ventral elaboration and new character origination. Second, within the genus, the complexity of the elaborate petals shows considerable variation among species so that the evolutionary histories of many features can be inferred once a reliable phylogenetic tree is available (Zohary, 1983; Bittkau and Comes, 2005; Jaros et al., 2018; Yao et al., 2019). Third, in addition to elaborate petals, species of Nigella also possess petaloid sepals that are almost indistinguishable from the simple petals of many other species (including Arabidopsis) in both morphology and function (Figure 1J). This juxtaposition allows us to compare the elaborate petals with simple petals within the same flower. Fourth, almost all species of the genus are easy to collect, cultivate, and manipulate for which virus-induced gene silencing (VIGS) and many other technologies are applicable (Wang et al., 2015; Liao et al., 2020).
Recently, using species of Nigella as materials, some interesting progress has been made to understand the process, pattern, and mechanisms of petal elaboration. For example, it has been shown that, like all other types of lateral organs, the highly specialized elaborate petals of N. damascena have a very simple starting point—a dome-like primordium. During development, as the upper lip, long hairs, pseudonectaries, ridges, and short trichomes were initiated successively, the complexity of the petal increased gradually (Yao et al., 2019). In addition, when the ancestral states of important characters were reconstructed, it becomes evident that the evolutionary elaboration of Nigella petals was a gradual process, involving not only modifications of pre-existing structures but also originations of new characters (Yao et al., 2019). More interestingly, in N. damascena, genes required for petal identity determination, such as NidaAPETALA3-3 (NidaAP3-3), NidaPISTILLATA1 (NidaPI1), NidaPI2, NidaAGAMOUS-LIKE6 (NidaAGL6), and NidaSEPALLATA1 (NidaSEP1), NidaSEP2, and NidaSEP3 (Gonçalves et al., 2013; Zhang et al., 2013; Wang et al., 2015), as well as those involved in pseudonectary development, such as NidaYABBY5 (NidaYAB5; Liao et al., 2020), have been identified and functionally characterized. Genes controlling the formation of many other characters, however, remain largely unknown.
Here, using N. damascena as a model, we conducted a detailed time-course transcriptomic analysis of petals to identify the genes and programs that are potentially involved in elaborate petal development. By correlating the expression pattern of gene with the morphogenetic events during petal development, we successfully identified 30 candidate genes associated with the formation of key morphological characters. Functional analyses further confirmed that a class I homeodomain-leucine zipper (HD-Zip) family transcription factor gene, Nigella damascena LATE MERISTEM IDENTITY1 (NidaLMI1), plays important roles in the development of short trichomes and bifurcation of the lower lip. Our results not only elucidate the differences between simple and elaborate petals in molecular bases but also pave the way to a comprehensive understanding of the mechanisms underlying lateral organ diversification and the new character origination.
RESULTS
Reference Transcriptome and DGE Profiles
To understand how elaborate petals of N. damascena were made through development, we conducted a detailed time-course transcriptomic analysis. Before that, we first obtained a reference transcriptome using mixed RNAs of floral buds, bracts, and leaves at different developmental stages, in which a total of 38,310 protein-coding genes were predicted (Supplemental Figure 1). Second, we generated digital gene expression (DGE) profiles for petals at eight developmental stages: S4, S5, S6, S7, S8/9, S10, S11, and S12 (Figure 2A). For comparison, sepals, stamens, and carpels at four developmental stages (i.e., those in the flowers bearing S4, S6, S8/9, and S12 petals, respectively), as well as developing bracts and leaves (i.e., those that are just underneath the floral buds with S6 petals), were also included in the DGE profiles (Figure 2A). Because the reproducibility among the biological triplicates was very high (coefficient of determination [R2] > 0.85; Supplemental Figure 2), we used the average reads per kilobase of transcript per million mapped reads (RPKM) values of the triplicates as the expression level of the gene in the corresponding sample. As a result, a total of 30,790 genes were defined as expressed (RPKM ≥1.0) in at least one sample, of which 26,169 were expressed in petals (Supplemental Data Set 1).
Figure 2.
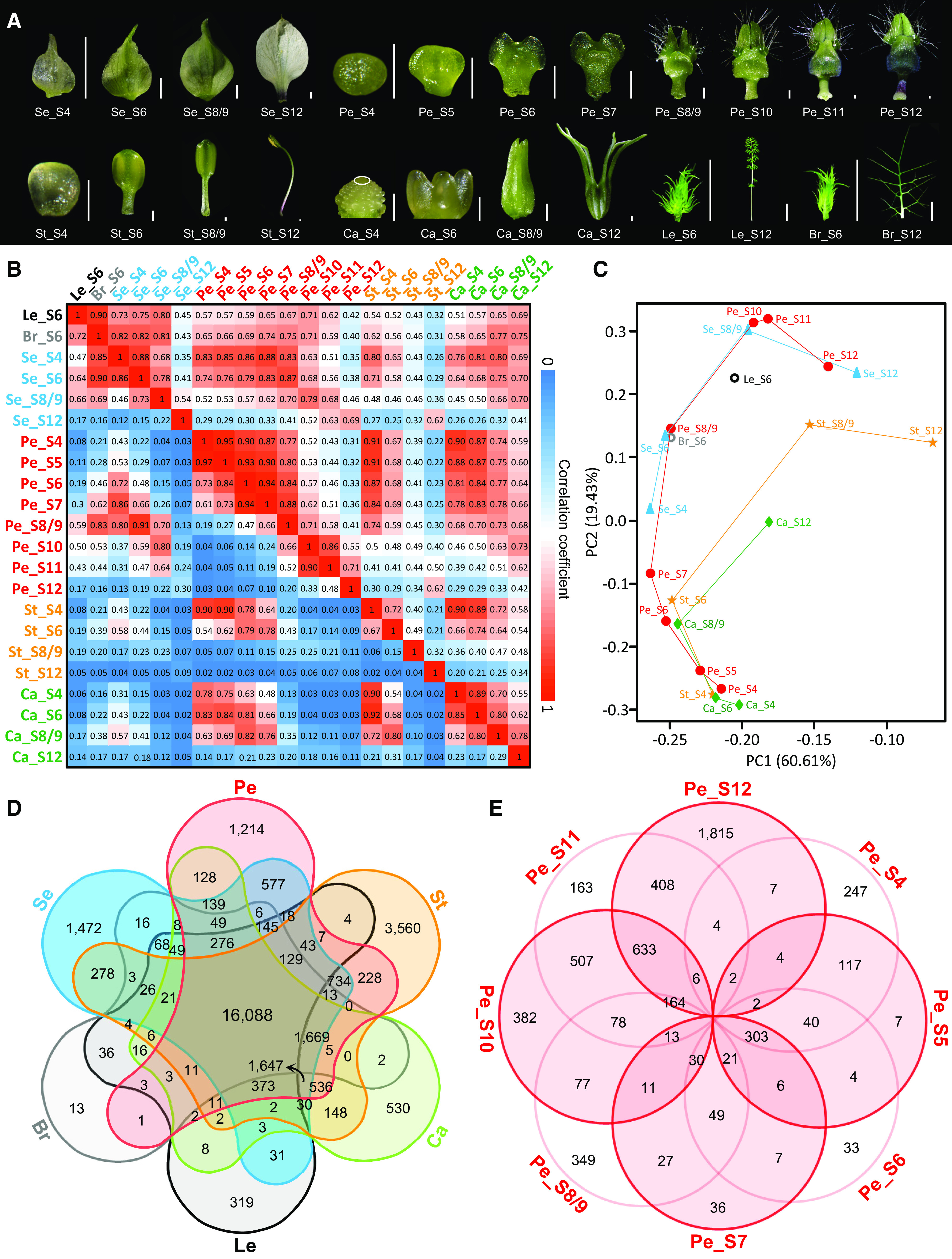
Landscape of DGE Profiles and Genes Preferentially Expressed in Different Organs and in Petals at Different Stages.
(A) DGE sampling for petals and other lateral organs at representative stages in N. damascena. Note that carpels at S4 are indicated by the white ellipse. All but leaves and bracts at S12 are sampled for DGE profiling. Bar for Se and Ca = 1 mm; bar for Pe and St = 200 μm; and bar for Br and Le = 1 cm. Br, bract; Ca, carpel; Le, leaf; Pe, petal; Se, sepal; St, stamen.
(B) PCC (bottom left) and SCC (top right) analyses of the 22 samples.
(C) PCA analysis of the 22 samples. PC1, principal component 1; PC2, principal component 2.
(D) and (E) Venn diagrams showing the numbers of genes preferentially expressed in different organs (D) and in petals at different stages (E), respectively.
To understand the general pattern of our data, we performed pairwise Pearson and Spearman correlation coefficient (PCC and SCC, respectively) analyses of the aforementioned 22 samples. We found that, for each type of organ, adjacent stages usually had higher correlation values than nonadjacent stages, suggestive of progressive changes at the transcriptomic level (Figure 2B). The PCC/SCC correlation value between petals at S4 and those at all other stages (i.e., S5 to S12), for example, was 0.97/0.95, 0.73/0.90, 0.61/0.87, 0.19/0.77, 0.04/0.52, 0.04/0.43, and 0.03/0.31, respectively. In addition, we found that the correlation values between different types of floral organs were sometimes larger than those between the same types of organs at different stages (Figure 2B). The PCC/SCC correlation values between S4 petals and S4 stamens, S5 petals and S4 stamens, and S4 stamens and S4 carpels, for instance, were all higher than 0.90, suggestive of very high similarity in the genes expressed. Similar phenomena were observed in a principal component analysis (PCA), although the relationships among the different stages of the same types of organs became more evident (Figure 2C). Taken together, these results suggest that at the early stages of flower development, different types of floral organs are actually not very different from each other in terms of the genes expressed; yet, during development, when different sets of genes were switched on and off, the differences become greater and greater.
Genes Specifically and Preferentially Expressed in Elaborate Petals
To understand the uniqueness of the petals, we compared the genes expressed in petals with those in other organs (Figure 2D; Supplemental Figure 3; Supplemental Data Sets 2 and 3). Of the genes expressed in petals, 918 and 1214 were specifically (i.e., a gene whose RPKM was ≥1.0 only in this organ but <1.0 in any other organs) and preferentially (i.e., a gene whose RPKM in this organ was at least twofold higher than those in any other organs) expressed in petals, respectively. The number of genes that were specifically/preferentially expressed in leaves, bracts, sepals, stamens, and carpels, for example, was 35/319, 23/13, 867/1472, 2037/3560, and 403/530, respectively. As expected, many of the petal-specific/preferential genes have already been reported to be involved in petal or other later organ sculpting (Supplemental Data Sets 2 and 3). Among them, the best known are the two petal identity genes NidaAP3-3 and NidaPI2 (Gonçalves et al., 2013; Zhang et al., 2013; Wang et al., 2015), as well as the orthologs/homologs (hereafter vaguely called “orthologs” for simplicity, although the exact relationships await to be determined by phylogenetic analyses) of the genes that control nectary development (e.g., STYLISH1 [STY1]; Min et al., 2019), petal size (e.g., BIGPETAL [BPE]; Szécsi et al., 2006), and compound or dissected leaf formation (e.g., KNOX ARABIDOPSIS THALIANA MEINOX [KNATM]; and LMI1; Magnani and Hake, 2008; Vlad et al., 2014; Andres et al., 2017) in other species. Interestingly, neither KNATM nor LMI1 has been shown to be expressed in petals in other species, suggesting that they may have new roles in N. damascena.
To understand the uniqueness of elaborate petals, we also investigated the expressed genes and programs that were shared or not shared with species with simple petals (such as Arabidopsis; Supplemental Data Sets 2 and 3). Of the shared programs, the best known were petal identity determination, represented by NidaAGL6, NidaAP3-1/NidaAP3-2/NidaAP3-3, NidaPI1/NidaPI2, and NidaSEP1/NidaSEP2/NidaSEP3 (Wang et al., 2015); cell proliferation and division, represented by the orthologs of AINTEGUMENTA (ANT; Mizukami and Fischer, 2000), ANGUSTIFOLIA3 (AN3; Kim and Kende, 2004), JAGGED (JAG; Dinneny et al., 2004; Ohno et al., 2004), and PETAL LOSS (PTL; Brewer et al., 2004); adaxial/abaxial polarity specification, represented by the orthologs of ASYMMETRIC LEAVES1 (AS1; Waites et al., 1998), PHABULOSA/PHAVOLUTA-related (PHX; McConnell and Barton, 1998; Prigge and Clark, 2006), and YAB2 (Siegfried et al., 1999); conical cell differentiation, represented by the ortholog of MIXTA (Noda et al., 1994); and anthocyanin biosynthesis, represented by the orthologs of PRODUCTION OF ANTHOCYANIN PIGMENT2 (PAP2; Borevitz et al., 2000) and TRANSPARENT TESTA GLABRA1 (TTG1; Walker et al., 1999). The programs specific to petals of N. damascena, by contrast, included meristem development, represented by the orthologs of BREVIPEDICELLUS (BP; Lincoln et al., 1994), CLAVATA1 (CLV1; Clark et al., 1993), and KNOTTED-LIKE FROM ARABIDOPSIS THALIANA2 (KNAT2; Dockx et al., 1995); stamen development and meiosis, represented by the orthologs of ABORTED MICROSPORES (AMS; Sorensen et al., 2003) and DISRUPTION OF MEIOTIC CONTROL1 (DMC1; Klimyuk and Jones, 1997); trichome differentiation, represented by the orthologs of GLABROUS1 (GL1; Marks and Feldmann, 1989), TRANSPARENT TESTA8 (TT8; Maes et al., 2008), and TTG2 (Johnson et al., 2002); and nectary development, represented by the orthologs of STY1, STY2, and LATERAL ROOT PRIMORDIUM (LRP; Min et al., 2019).
Genes Specifically/Preferentially Expressed in Petals at Different Developmental Stages
To identify the developmental programs involved in petal elaboration, we first searched for genes that were specifically/preferentially expressed in petals at different stages. Of the 26,169 genes expressed in petals, 584/247, 80/7, 137/33, 127/36, 726/349, 279/382, 113/163, and 702/1815 genes were specifically/preferentially expressed in petals at S4, S5, S6, S7, S8/9, S10, S11, and S12, respectively (Figure 2E; Supplemental Figure 3; Supplemental Data Sets 4 and 5). These stage-specific/preferential genes included known genes, or orthologs of known genes, that play key roles in petal development (Supplemental Figure 3; Supplemental Data Sets 4 and 5). At S4, for example, there were NidaSEP1 and NidaSEP2, two floral organ identity genes in N. damascena (Wang et al., 2015), and the orthologs of DORNRÖCHEN (DRN; Chandler and Werr, 2017) and CUP-SHAPED COTYLEDON3 (CUC3; Vroemen et al., 2003), which are involved in floral organ initiation. At S10, there were genes whose orthologs in other species are involved in cell wall biogenesis, such as GLUCURONIC ACID SUBSTITUTION OF XYLAN1 (GUX1) and GUX2 (Lee et al., 2012); lignin biosynthesis, such as LACCASE (LAC) and CINNAMYL ALCOHOL DEHYDROGENASE (CAD; Thévenin et al., 2011; Zhao et al., 2013); and starch biosynthesis, such as ADPGLC-PPASE LARGE SUBUNIT2 (APL2; Streb and Zeeman, 2012). At S12, there were genes whose orthologs regulate anthocyanin biosynthesis, for example, PAP1 (Borevitz et al., 2000). Interestingly, at S4 petals, there were genes not required in petal development in other species, such as the stamen and carpel identity gene NidaAGAMOUS1 (NidaAG1; Wang et al., 2015) and the class I KNOTTED1-LIKE HOMEOBOX (KNOXI) genes involved in meristem and compound leaf development, such as BP and SHOOT MERISTEMLESS (STM; Lincoln et al., 1994; Long et al., 1996; Hay and Tsiantis, 2010).
To gain more insight into the functions of the stage-specific/preferential genes, we performed Gene Ontology (GO) enrichment analyses (Supplemental Figure 3; Supplemental Data Set 6). We found that the GO terms enriched included “flower development,” “specification of floral organ identity,” “regulation of reproductive process,” “carpel development,” “gynoecium development,” “maintenance of meristem identity,” and “meristem development” at S4; “defense response” at S7; “cell wall biogenesis,” “xylan biosynthetic process,” “xylan metabolic process,” “starch metabolic process,” and “lignin biosynthetic process” at S10; and “flavonoid biosynthetic process,” “pigment biosynthetic process,” “response to hormone,” and “response to auxin” at S12.
Stage-Specific Coexpression Modules
To understand the dynamic changes of the genes and programs expressed during petal development, we performed weighted correlation network analysis (WGCNA) based on the 13,395 genes whose expression showed higher than 0.5 coefficient of variation across the eight stages. In total, 18 coexpression modules were identified, and the number of genes per module ranged from 26 (in module 4, M4) to 2735 (in M2; Figures 3A and 3B; Supplemental Data Set 7). Module-stage association analyses further revealed that six of these modules (i.e., M1, M4, M5, M9, M13, and M18) showed high correlations with one specific stage (PCC ≥0.85 for one stage and PCC < 0.30 for other stages), whereas other modules were correlated with two or more adjacent stages (PCC ≥0.30; Figure 3C). To understand the differences between the 18 modules in functional properties, we also performed GO enrichment analyses (Supplemental Figure 4; Supplemental Data Set 6). We found that, as expected, different GO categories were indeed enriched in different modules, although some overlap did exist. The GO categories enriched in M1, for example, included “flower development,” “specification of floral organ identity,” “floral organ development,” “phyllome development,” “meristem maintenance,” “meristem development,” and “microsporogenesis.” Interestingly, apart from the similar GO categories in M1, we found that “adaxial/abaxial axis specification” was enriched in M2 and “adaxial/abaxial axis specification,” “cell proliferation,” “cell division,” “petal development,” “male meiosis,” and “epidermis development” were enriched in M3. In addition, the representative GO categories enriched in other modules included “DNA biosynthetic process” (for M8 and M9); “photosynthesis” (for M10); “cell wall biogenesis,” “lignin metabolic process,” “starch metabolic process,” and “xylem development” (for M13) and “lignin metabolic process” and “pigment metabolic process” (for M18).
Figure 3.
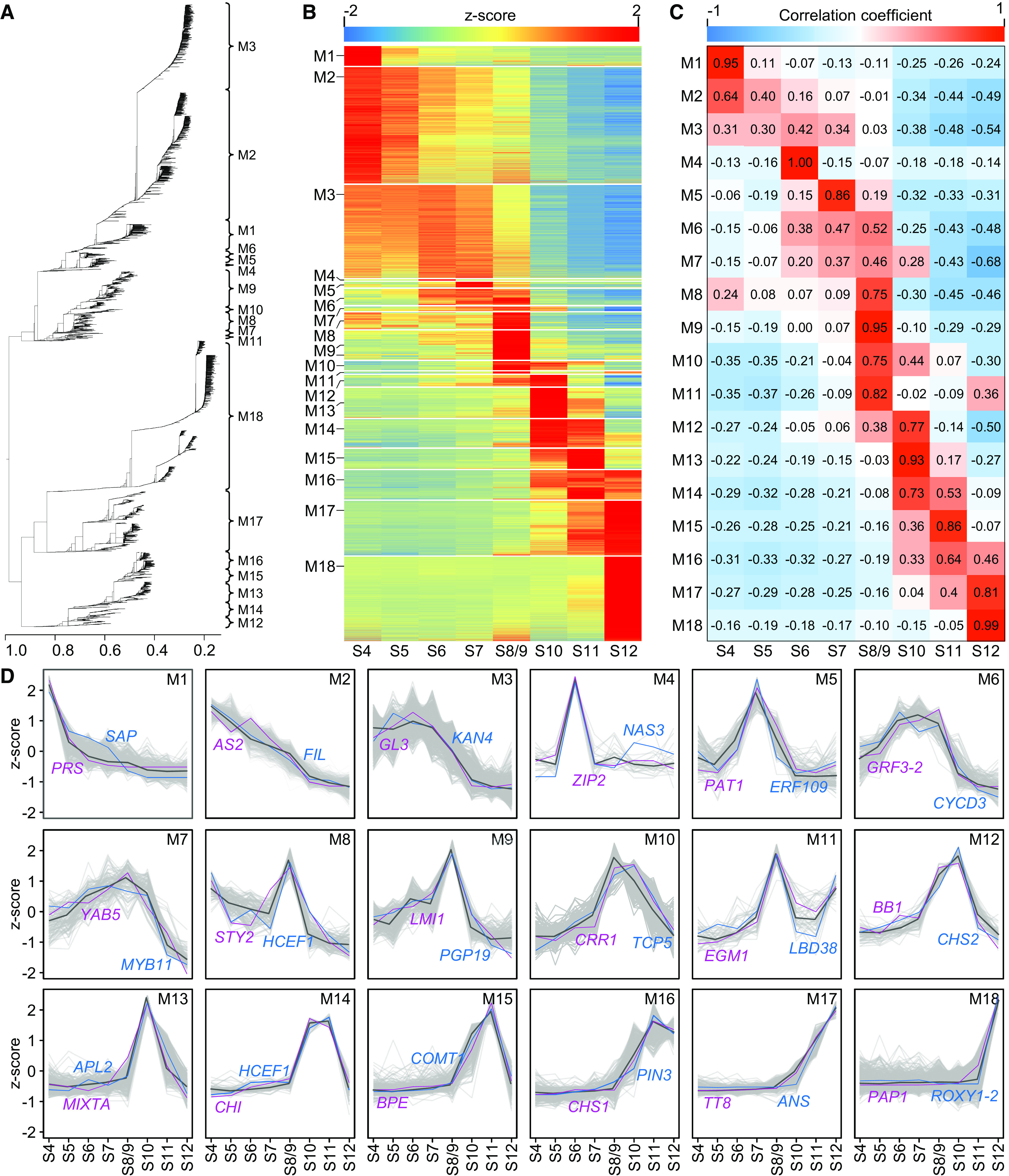
Gene Coexpression Modules Involved in the Development of Petals.
(A) WGCNA cluster dendrogram showing 18 coexpression modules.
(B) Heatmap showing relative expression of genes in 18 coexpression modules across different stages.
(C) Heatmap describing the correlations between coexpression modules and developmental stages.
(D) Expression profiles for the 18 coexpression modules. In each module, gray lines represent the expression profiles of all individual genes, the black line indicates the median expression profile of all genes, and the purple and blue lines denote the expression profiles of representative genes with known functions.
Among the GO categories enriched, there were also known genes (or orthologs of known genes in other species) that may control petal development (Figure 3D; Supplemental Data Set 7). In M1, for example, in addition to the aforementioned NidaSEP1, NidaSEP2, and NidaAG1, as well as the orthologs of DRN and CUC3, there was a gene whose ortholog in Arabidopsis, PRESSED FLOWER (PRS), regulates floral organ development (Nakata et al., 2012). In M2, there were NidaAP3-2, the floral organ identity gene in N. damascena (Wang et al., 2015), and orthologs of AUXIN RESPONSE FACTOR3 (ARF3; Pekker et al., 2005), AS2 (Semiarti et al., 2001), and FILAMENTOUS FLOWER (FIL; Sawa et al., 1999; Siegfried et al., 1999), which are all involved in adaxial/abaxial polarity specification. In M3, there were genes whose orthologs are involved in polarity specification, such as KANADI4 (KAN4; McAbee et al., 2006) and YAB2 (Siegfried et al., 1999); cell proliferation, such as GROWTH-REGULATING FACTOR5 (GRF5; Horiguchi et al., 2005); and trichome differentiation, such as GLABRA3 (GL3; Payne et al., 2000). In M10, there were genes whose orthologs are involved in photosynthesis, such as CHLORORESPIRATION REDUCTION1 (CRR1; Shimizu and Shikanai, 2007); cell division control, such as TEOSINTE BRANCHED1/CYCLOIDEA/PCF5 (TCP5; Koyama et al., 2010); and nectary determination, such as STY1 (Min et al., 2019). In M13, there was a gene whose ortholog, MIXTA, controls the differentiation of conical cells of petals (Noda et al., 1994). In M17 and M18, there were genes whose orthologs regulate anthocyanin biosynthesis, such as PAP1 (Borevitz et al., 2000), FLAVONOL SYNTHASE (FLS; Holton et al.., 1993), DIHYDROFLAVONOL REDUCTASE (DFR; O’Reilly et al., 1985; Holton and Cornish, 1995), and ANTHOCYANIDIN SYNTHASE (ANS; Menssen et al., 1990; Holton and Cornish, 1995); petal development, such as ROXY1 (Xing et al., 2005); and trichome differentiation, such as TT8 (Maes et al., 2008) and SCARECROW-LIKE8 (SCL8; Morohashi and Grotewold, 2009). Notably, in the modules with no enriched GO category, there were also genes whose orthologs in other species are involved in cell division regulation (in M6), such as GRF3 (Kim et al., 2003), TCP4 (Nag et al., 2009), and CYCLIN D3 (CYCD3; Riou-Khamlichi et al., 1999); polarity specification (in M7), such as YAB5 (Siegfried et al., 1999); and organ shape sculpting/trichome development (in M9), such as LMI1 (Hofer et al., 2009; Vlad et al., 2014; Zhao et al., 2015; Andres et al., 2017).
Candidate Genes Associated with Morphological Characters
To identify the genes responsible or indispensable for the formation of key morphological characters on N. damascena petals, we conducted more detailed investigations of the DGE profiles. We started with six essential and well-characterized developmental programs in lateral organ development: organ initiation, organ size determination, organ polarity specification, nectary development, trichome differentiation, and anthocyanin biosynthesis (Hay and Tsiantis, 2010; Yang and Ye, 2013; Albert et al., 2014; Fukushima and Hasebe, 2014; Huang and Irish, 2016; Min et al., 2019). We compared the expression patterns of the component genes in petals with those in sepals and stamens (Figure 4; Supplemental Figure 5). In all these programs, there were genes whose expression patterns in petals were largely consistent with those in sepals and stamens (Figure 4B), suggestive of functional conservativeness. A small number (23 of 66) of genes, however, showed obviously different expression patterns in petals, having either distinct peaks or much higher levels of expression in petals than in sepals or stamens (Figures 4B and 4C; Supplemental Figure 5), suggestive of possible functional diversification. Based on its expression pattern, and because its counterparts in Arabidopsis, PLETHORA1 (PLT1) and AINTEGUMENTA-LIKE6 (AIL6), function to promote cell proliferation in floral organs (Krizek, 2009; Wuest et al., 2012), we hypothesize that NidaPLT1-1 (peaked at S5) is required for the formation of the nectar chamber, where an increased rate of cell division was observed at S5. Similarly, because ectopic expression of polarity genes can sometimes lead to regional outgrowths (Fukushima and Hasebe, 2014), we propose that the polarity genes NidaKAN1 (peaked at S7) and NidaYAB5 (peaked at S8/9) are required for the formation of pseudonectaries. Consistent with this notion, a recent study has demonstrated that knockdown of NidaYAB5 can indeed led to the complete loss of pseudonectaries (Liao et al., 2020).
Figure 4.
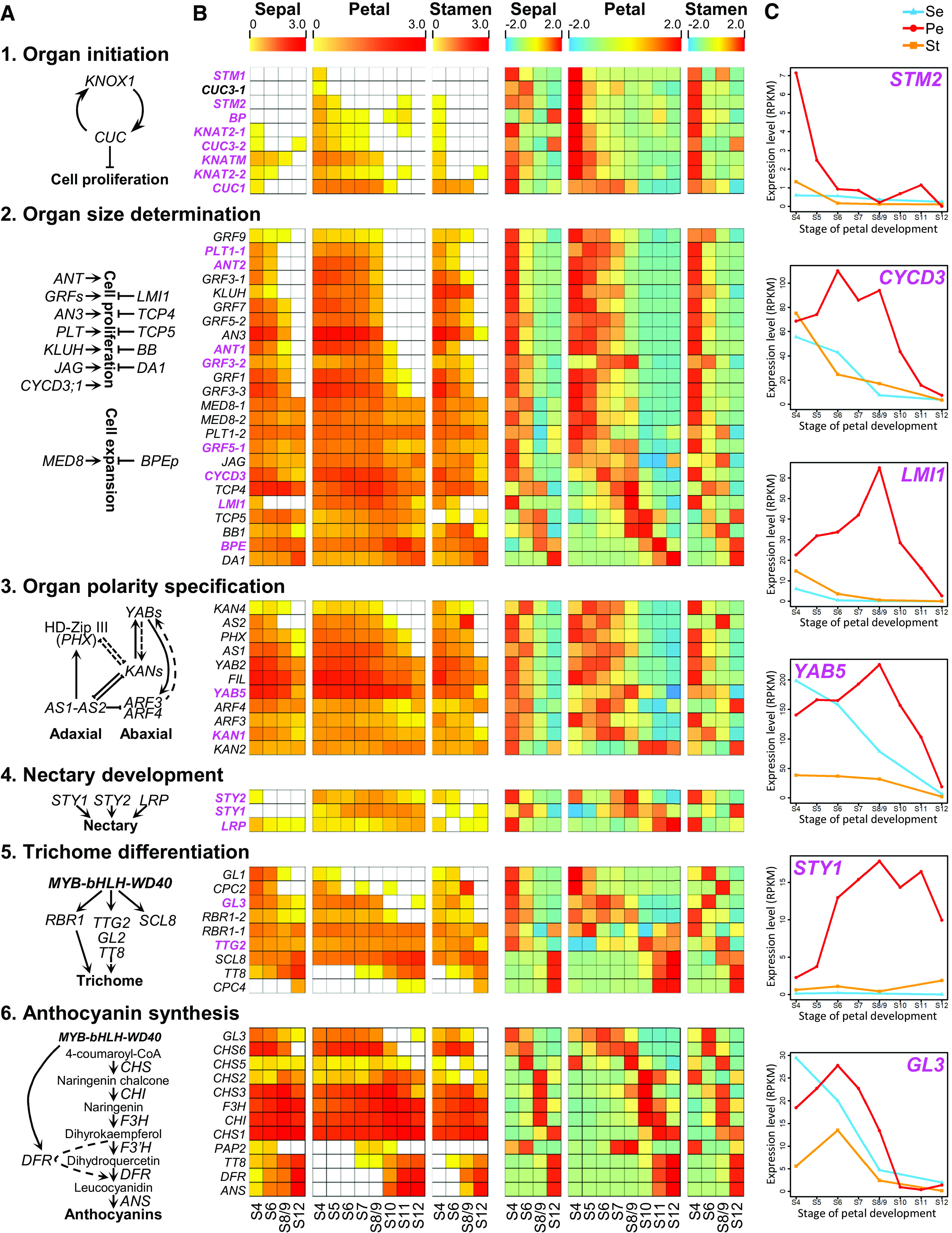
Expression Patterns of Genes in Six Well-Characterized Regulatory Programs.
(A) Six regulatory programs associated with different developmental events.
(B) Heatmaps showing the log10-transformed (left) and z-score transformed (right) gene expression values in sepals, petals, and stamens at different stages. The genes showing obviously different expression patterns in petals relative to sepals and stamens are highlighted with purple fonts.
(C) Expression profiles of the representative genes.
By using the same strategy and, more importantly, by correlating the expression patterns of genes with key morphological events in petal development, we identified many other candidate genes (Figure 5; Supplemental Figure 6). The bifurcation of the lower lip, for example, is very likely controlled by NidaCUC1 (peaked at S7), a NAM, ATAF1/2, and CUC2 (NAC) family member whose ortholog has been reported to control leaf dissection in several distantly related species (Blein et al., 2008). Likewise, the differentiation of long hairs and short trichomes is very likely regulated by NidaGL3 (peaked at S6) and NidaLMI1 (peaked at S8/9), respectively, because their orthologs are well-characterized regulators of trichome differentiation and development (Payne et al., 2000; Zhao et al., 2015). The differentiation of conical cells and the production of anthocyanin are highly likely regulated by two evolutionarily conserved transcription factor genes, NidaMIXTA (peaked at S10) and NidaTT8 (peaked at S12), respectively. In all, 30 candidate genes associated with the formation of different morphological characters were identified.
Figure 5.
Representative Candidate Genes Associated with the Formation of Different Characters.
Virtual clay models show the petals at different developmental stages. Arrows point to the characters that might be under the control of the corresponding genes.
NidaLMI1 as an Essential Regulator for Petal Elaborations
To test the reliability of our predictions, we conducted expression and functional experiments for one of the predicted candidate genes, NidaLMI1 (Supplemental Figure 7). It has been reported that, as a class I HD-Zip family transcription factor gene, LMI1 is involved in the formation of dissected leaves in Cardamine hirsuta and Gossypium hirsutum, tendrils in Pisum sativum, and trichomes in Cucumis sativus (Hofer et al., 2009; Vlad et al., 2014; Zhao et al., 2015; Andres et al., 2017). In N. damascena, NidaLMI1 was preferentially expressed in petals, with the highest expression being detected in S8/9 petals (Figure 5), suggesting that its expression has been expanded into petals. To understand the precise spatiotemporal expression of NidaLMI1, we conducted detailed in situ hybridization assays. The expression of NidaLMI1 was undetectable in shoot apical meristem, leaf primordia, or floral primordia before S6 of petal development (Supplemental Figure 8). At S6, the expression signals of NidaLMI1 were first detectable in the newly formed junction of the forked lamina (Supplemental Figure 8) and then spread to the area just above the developing nectar chamber, thereby bridging the two pseudonectaries at S7 (Figure 6A). Subsequently, the expression signals of NidaLMI1 were restricted to the regions where short trichomes would arise (at S8/9) and the developing short trichomes (at S11 and S12; Figures 6B and 6C). Clearly, the expression of NidaLMI1 not only coincides with the formation of short trichomes but also correlates with the bifurcation of the lower lip.
Figure 6.
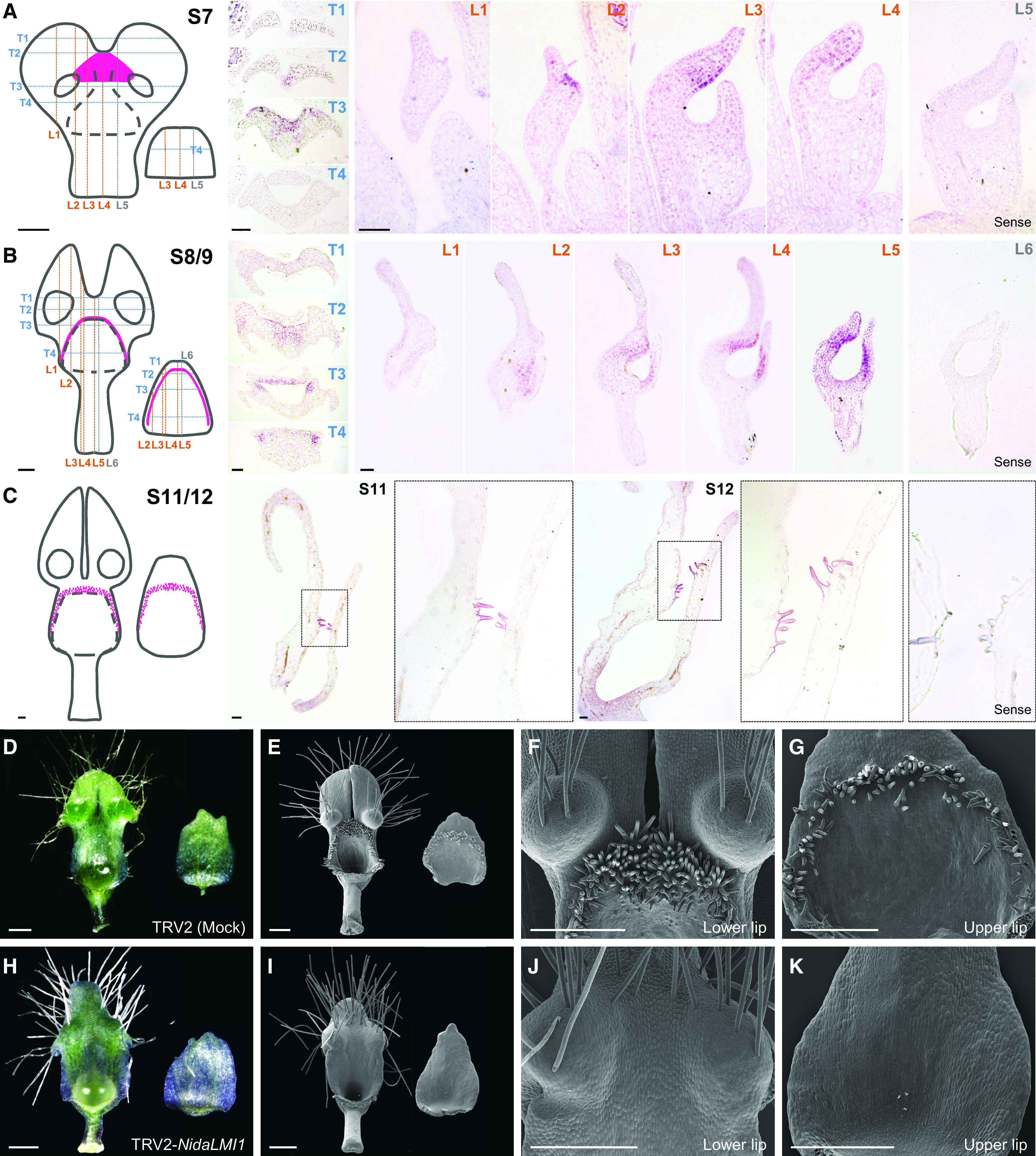
Expression and Function of NidaLMI1.
(A) to (C) Results of in situ hybridization of NidaLMI1 in petals at S7 (A), S8/9 (B), and S11/12 (C). For petals at S7 and S8/9, series transverse (T in blue) and longitudinal (L in orange or gray) sections are presented, with their relative positions being shown with dashed lines in the left cartoons (adaxial view for the lower lip and abaxial view for the upper lip). For petals at S11 and S12, only longitudinal sections are presented. In the cartoons, the black dashed lines indicate the edge of the nectar chamber and the pink regions represent the expression domains of NidaLMI1 in petals. Hybridization results of the sense probe are presented as negative controls. Bars = 100 μm.
(D) to (K) Phenotypes of petals in the mock (see [D] to [G]) and TRV2-NidaLMI1–treated plants (see [H] to [K]). Mature petals (adaxial view for the lower lip and abaxial view for the upper lip) captured by stereomicroscope and scanning electron microscope are shown in (D) and (H) and (E) and (I), respectively. The zoom-in views of lower lips and upper lips under scanning electron microscope are shown in (F) and (J) and (G) and (K), respectively. Bars = 500 μm.
To understand the function of NidaLMI1, we knocked down its expression using the VIGS technique with two Tobacco rattle virus (TRV) constructs, NidaLMI1_V1 and NidaLMI1_V2 (Supplemental Figure 9). Compared with the TRV2-treated plants (i.e., mock), the TRV2-NidaLMI1_V1– and TRV2-NidaLMI1_V2–treated plants with strong phenotypic changes were very similar to each other. Specifically, the two lobes of the lower lip were completely fused and the short trichomes disappeared from the chewing surface of the lower and upper lips, while leaves and other floral organs remained largely unaffected (Figures 6D to 6K; Supplemental Figure 9). In plants with moderate phenotypic changes, the two petal lobes were partially fused and the short trichomes were sparsely distributed (Supplemental Figure 9). In accordance with their morphological features, very low expression of NidaLMI1 was detected in flowers and petals with strong and moderate phenotypic changes (Supplemental Figure 9). These results not only confirmed the predication that NidaLMI1 is required for the differentiation of short trichomes but also suggest that it is involved in the bifurcation of the lower lip.
DISCUSSION
In this study, by conducting detailed transcriptomic and functional analyses on N. damascena, we explored the mechanisms underlying elaborate petal development and specialized character formation. We found that, in addition to the genes and programs indispensable for simple petal development (such as initiation, identity determination, growth, polarity specification, and maturation), there are genes and programs that are not normally expressed in simple petals (such as the formation of lobes, nectaries, and trichomes), suggestive of uniqueness of the N. damascena petals. By analyzing functionally important developmental programs and correlating the expression patterns of genes with morphological changes of petals, we also identified 30 candidate genes that likely play key roles in the development of elaborate petals (especially marginal/ventral elaboration) and the occurrence of morphological events. Our results not only provide a portrait of elaborate petal development but also elucidate the mechanisms underlying lateral organ diversification in plants.
Mechanisms Underlying Marginal and Ventral Elaboration
It has been proposed that elaborate petals can arise through marginal elaboration or ventral elaboration, or both, with the former being the formation of lobes, teeth, fringes, or appendages along the margin of the petal. In leaves, the formation of lobes, teeth, and leaflets has been generally believed to result from regional expression of cell division inhibitors, such as CUC-like genes of the NAC family, LMI1-like genes of the HD-Zip family, and CINCINNATA (CIN)-like genes of the TCP family (Crawford et al., 2004; Blein et al., 2008; Hofer et al., 2009; Koyama et al., 2011; Sicard et al., 2014; Vlad et al., 2014; Andres et al., 2017). In N. damascena, the bifurcate lobes of the lower petal lip are produced through marginal elaboration at S6. Coincidently, at S6, the expression of NidaLMI1 is restricted to the junction region of the forked lower lip. The fact that knockdown of NidaLMI1 led to the generation of intact rather than bifurcate lower petal lips further confirmed that NidaLMI1 is responsible for the bifurcation of the lower lip. In addition, we found that NidaCUC1 shows a relatively higher expression level in S4 to S7 petals relative to sepals and stamens (Supplemental Figure 5), suggestive of a possible role in marginal elaboration. Taken together, these results suggest that, similar to the formation of lobes, teeth, and leaflets in leaves, marginal elaboration of the N. damascena petals also requires the regional expression of cell division inhibitors.
Unlike marginal elaboration, ventral elaboration refers to the formation of lips, scales, spurs, or other protruded structures on the ventral (i.e., abaxial) side of the petal (Endress and Matthews, 2006; Yao et al., 2019). The mechanisms underlying ventral elaboration are still unclear; yet, the data suggest that it may be related to the reactivation of the meristem program or rearrangement of the expression of adaxial/abaxial genes, or both (Walcher-Chevillet and Kramer, 2016). In Antirrhinum majus and Linaria vulgaris (Plantaginaceae), reactivation of the KNOXI genes, a key regulator of meristem development and maintenance, can cause the production of spur-like outgrowths on petals (Golz et al., 2002; Box et al., 2011). The formation of the peltate leaves of Tropaeolum majus (Tropaeolaceae) and traps of the carnivorous plant Utricularia gibba (Lentibulariaceae), however, has likely been caused by the rearrangement of the expression of the adaxial/abaxial genes (Gleissberg et al., 2005; Whitewoods et al., 2020). In this study, we found that, at S4, when the upper lip starts to emerge through ventral elaboration, genes of the KNOXI-CUC module, such as NidaSTM1/2, NidaBP, and NidaCUC1, show elevated expression in petals compared to sepals and stamens, suggestive of possible reactivation of the program (Figure 4). Presumably, it was the reactivation of the KNOXI and CUC genes that led to the formation of the upper lip. Nevertheless, we still cannot exclude the possibility that expression rearrangement of adaxial/abaxial genes also contributed considerably to this process, because the abaxial gene NidaYAB5 is also expressed in the adaxial side of the upper lip (Yao et al., 2019; Liao et al., 2020). More studies are needed to elucidate the exact roles of these genes.
Molecular Bases of Other Morphological Events
In addition to ventral and marginal elaboration, there are other morphological events that play key roles in the making of elaborate petals. Some of these events have significantly changed the developmental trajectory of the petals and eventually led to the formation of new characters (such as nectaries, pseudonectaries, short trichomes, and long hairs). The exact mechanisms underlying the occurrences of these morphological characters are still unclear, except that a recent study has shown that ectopic expression of one of the abaxial genes, NidaYAB5, may be responsible for the formation of pseudonectaries through regional thickening; knockdown of NidaYAB5 led to the complete loss of pseudonectaries (Liao et al., 2020). In this study, we not only confirmed the importance of NidaYAB5 in petal development but also identified the genes tightly associated with the formation of other characters. The nectary, for example, is likely controlled by NidaSTY1, a STY family gene whose ortholog in Aquilegia coerulea is indispensable for nectary development (Min et al., 2019), whereas the formation of the ridges between pseudonectaries and nectar chamber may be regulated by the NidaTCP5 gene (Figure 5).
In addition to conical cells, petals of N. damascena also possess short trichomes and long hairs, both of which are unicellular structures. Long hairs, which start to emerge at S6, are ∼1200 μm in length and sparsely distributed on the lobes and pseudonectaries of the lower lip, whereas short trichomes, which start to initiate at S10, are ∼100 μm in length and densely distributed on the chewing surface of the upper and lower lips. Recently, it has been suggested that while long hairs on petals likely existed before the diversification of the genus Nigella, short trichomes evolved during the evolution of the genus (Yao et al., 2019). In this study, we proposed that NidaGL3 may be responsible for the differentiation of long hairs whereas NidaLMI1 is indispensable for the emergence of short trichomes. When the expression of NidaLMI1 was knocked down by using the VIGS technique, short trichomes disappeared, whereas long hairs remained unaffected. This differential effect confirms that long hairs and short trichomes are related but clearly different morphological characters that are possibly controlled by different regulators. Interestingly, it appears that, like conical cells, the initiation and development of short trichomes also require the proper functioning of NidaMIXTA, the ortholog of the well-known conical cell inducer MIXTA, for three reasons. First, during development, short trichomes and conical cells were not distinguishable until S10 (Yao et al., 2019). Second, like NidaLMI1, the expression of NidaMIXTA was very low at the early stages (before S7) of petal development but started to increase at S8/9, when both short trichomes and conical cells started to initiate (Figure 5). Third, ectopic expression of MIXTA into tobacco (Nicotiana tabacum) can trigger the formation of excess trichomes on leaves and floral organs (Glover et al., 1998). These results suggest that NidaLMI1 and NidaMIXTA have something in common in determining the differentiation of short trichomes, although the exact relationship between the two genes awaits to be tested.
Implications for the Origination of New Characters
It has been proposed that co-option is one of the most important mechanisms underlying the origination of new characters (Shubin et al., 2009; Arthur, 2011). By definition, co-option is the deployment of a gene or program with a well-established function to a new function over the course of evolution (Arthur, 2011). The origins of zygomorphic flowers in multiple flowering plant lineages and horns in beetles, for example, can be explained by the co-option of the CYCLOIDEA (CYC)-like genes and the limb program, respectively (Shubin et al., 2009; Hileman, 2014). In the genus Nigella, elaborate petals are made through both marginal/ventral elaboration and the successive originations of several new characters, such as the aforementioned nectary, pseudonectaries, long hairs, and short trichomes (Yao et al., 2019). In this study, we found that the originations of at least some new characters may have also been the results of co-option. The short trichomes, for instance, have likely evolved through co-option of the LMI1 gene, because the LMI1 orthologs in other species (including Arabidopsis) are usually not expressed in petals. In fact, a recent study has shown that the nectaries likely arose from co-option of the STY-like genes into the nectariferous petals before the divergence of the Ranunculaceae and Berberidaceae (Min et al., 2019). If these hypotheses are correct, it would be interesting to know how these genes and programs were co-opted into petals.
It has been suggested that the origination of one character sometimes can induce, or stimulate, the origination of other characters, as is the case for the feathers and wings of birds (Prum, 1999; Prum and Brush, 2002). If this prediction is correct, it implies that different characters may have connections in terms of the underlying mechanisms. Interestingly, in this study, we found that the NidaLMI1 gene has dual functions, that is, in both the bifurcation of the lower lip and the production of short trichomes; when the expression of NidaLMI1 was knocked down, short trichomes disappeared and the two lobes of the lower lip failed to bifurcate (Figure 6). In Nigella, the lobes on the lower lip and short trichomes are the new characters originated at different stages of evolution (Yao et al., 2019). It is therefore very likely that LMI1 has been co-opted twice during evolution, with the first being related to the bifurcation of the lower lip and the second being associated with the production of short trichomes. While this hypothesis awaits to be tested, there are already some data that support it. For example, it has been shown that the LMI1 orthologs are involved in the formation of dissected leaves, tendrils, and trichomes in Ca. hirsuta/G. hirsutum, P. sativum, and Cu. sativus, respectively (Hofer et al., 2009; Vlad et al., 2014; Zhao et al., 2015; Andres et al., 2017). Hence, it would not be surprising if it were co-opted in Nigella petals to control the formation of lip lobes and short trichomes. The relationship between the two new characters (i.e., whether the formation of the former is the prerequisite for the latter), however, remains unclear. A phylostratigraphic study on the DGE data and functional dissection of the key candidate genes will probably help elucidate the molecular mechanisms underlying, as well as the relationship between, these two successively originated new characters.
Problems, Challenges, and Future Directions
It should be mentioned that although the candidate genes identified in this study are very likely key regulators of petal development in N. damascena, it is still too early to say that the mechanisms underlying petal elaboration have been elucidated, for three reasons. First, except for NidaLMI1 and NidaYAB5, the expression patterns and functional properties of all other candidate genes have not yet been studied. Even for the genes with functional studies, the results were based on the VIGS rather than mutant, transgenic, or other more reliable analyses (Gonçalves et al., 2013; Wang et al., 2015; Liao et al., 2020; this study). Second, the candidate genes were identified because their expression patterns in petals match the occurrence of key morphological event and because their orthologs in other species have been reported to perform relevant functions (Figure 5; Supplemental Figure 6). Yet, because members of the same coexpression module tend to have the same or very similar functions, it is quite possible that other genes, especially those whose orthologs in other species have not yet been functionally characterized or do not perform relevant functions, actually play more important roles than the ones we identified. Third, and most importantly, the candidate genes were interesting because they have interesting expression patterns, yet the reasons why they have such interesting expression patterns are still unclear. In the future, while detailed expression and functional investigations could and should be conducted on the identified candidate genes, special attention should also be paid to the other genes of the corresponding coexpression modules and the factors that shape their expression patterns. It is hoped that, by doing this, we can not only identify more regulators of elaborated petal development but also elucidate the molecular bases of key morphological events.
In spite of these caveats, our study clearly demonstrates that detailed time-course transcriptomic analyses, followed by necessary expression (e.g., in situ hybridization) and functional (especially VIGS) investigations, may be a very promising strategy in elucidating the molecular basis of plant evolution. Functional studies on nonmodel systems can be extremely challenging; yet, many plants with interesting and/or important traits (including, but not limited to, the elaborate petals) are nonmodel species for which transgenic or even genetic manipulations are very difficult. A high-quality transcriptome, therefore, can act as an excellent starting point for studies when the reference genome of the species is lacking. Next, by conducting detailed DGE and bioinformatic analyses, genes, pathways, and networks responsible for, or at least tightly related to, the formation of the trait under consideration may be identified, and functional verification (if necessary) can be done by using RNA in situ hybridization, VIGS, and/or other technologies. More importantly, when this strategy is applied to multiple species, it will become possible to understand the commonness and peculiarity of a trait and elucidate the molecular underpinnings of its evolution.
METHODS
Plant Materials and Growth Conditions
Seeds of Nigella damascena were sown in nutrient soil and grown in a growth chamber under a 12-h-light (4280 Lux)/12-h-dark photoperiod at 24°C and 60% relative humidity.
Generation of the Reference Transcriptome and the DGE Profiles
Total RNAs were separately extracted from floral buds, bracts, and leaves at different developmental stages using the SV Total RNA Isolation System (Promega) and then equally mixed to generate a pool for library construction. Paired-end 100-bp-long reads were sequenced by Illumina HiSeq2000 (Novogene), and the clean reads were assembled into transcripts using Trinity (Grabherr et al., 2011) after the adaptors and the low-quality reads were removed. Next, an all-by-all BLASTN was performed to group transcripts with different alternative splice forms into unigenes. Subsequently, the unigenes were annotated by sequence similarity comparisons against the nonredundant protein database in National Center for Biotechnology Information and SwissProt using BLASTX (E value < 1e–5). If no best hit was found, the unigenes were annotated through ESTScan (Iseli et al., 1999), and only protein sequences longer than 100 amino acids were retained. Finally, the reference transcriptome of N. damascena was generated (Supplemental Figure 1).
For the generation of the DGE profiles, 22 samples, each with three biological replicates, were subjected to total RNA extraction as described above. In total, 66 libraries were constructed independently for single-end 100-bp-long reads sequencing on Illumina HiSeq2000. The clean reads of resultant 66 DGE profiles were separately mapped to the reference transcriptome by TopHat2 (Kim et al., 2013) and then the mapped reads were counted by HTSeq v0.5.4p3 (Anders et al., 2015) to calculate the RPKM values (Mortazavi et al., 2008). The quality of all the 66 DGE profiles was reflected by read utilization rates (82.40 to 87.49%) and gene coverage (74.84 to 90.35%; Supplemental Data Set 8). The reproducibility among the biological triplicates was evaluated by the pairwise coefficient of determination of the RPKM values, which was calculated in R using log10-transformed RPKM values [i.e., log10 (RPKM + 1)] as inputs.
PCC, SCC, and PCA Analyses
The overall comparisons of the expression profiles between samples were conducted by PCC, SCC, and PCA analyses. PCC and SCC were performed under the cor.test function in R using the methods of Pearson and Spearman, respectively. PCA was performed under the prcomp function in R using the original RPKM values as inputs.
Identification of Specifically and Preferentially Expressed Genes and GO Enrichment Analysis
A gene was defined as expressed in a specific type of organ if its highest RPKM is ≥1.0 in the sampled stages, and its expression level was determined in terms of the highest RPKM value. A gene was considered as specifically expressed in a specific organ if its RPKM is ≥1.0 only in this organ but <1.0 in any other organs. A gene was regarded as preferentially expressed in a specific organ if its RPKM in this organ is at least twofold higher than those in any other organs. The same criterion was used to identify the genes specifically and preferentially expressed in the petals at different stages. For the GO enrichment analysis of specifically and preferentially expressed genes, the protein sequences were BLAST against the Arabidopsis (Arabidopsis thaliana) nonredundant protein database with an E value cutoff <1e–10 (Ashburner et al., 2000). Next, the GO terms of each gene were adopted in terms of its best hit, and the GO enrichment of a cluster of genes was performed using the agriGO program (Tian et al., 2017) with the false discovery rate threshold of 0.05.
Gene Coexpression Analysis
The coexpression modules were identified by R package WGCNA (Zhang and Horvath, 2005; Langfelder and Horvath, 2008) based on the RPKM data of 13,395 genes whose expression showed coefficient of variation >0.5 across the eight stages. We first created a matrix of pairwise PCCs between all pairs of genes and constructed an adjacency matrix by raising the coexpression measure, 0.5 + 0.5 × PCC, to the power β = 14, which was interpreted as an optimal soft threshold of the correlation matrix. The resultant adjacency matrix was then used to calculate the topological overlap, based on which genes were hierarchically clustered. The Dynamic Hybrid Tree Cut algorithm was used to cut the hierarchical clustering tree. Clusters with fewer than 20 genes were merged into their closest larger neighbor cluster. Each module was summarized by the first principal component of the scaled gene expression profiles (referred to as module eigengene). Module-stage associations were qualified by PCC analysis, where each module was represented by its module eigengene, and each stage was represented with a numeric vector of “1” for a specific stage and “0” for all the other stages. The same strategy as described above was used for the GO enrichment analysis of the coexpression modules.
Phylogenetic Analysis
Coding sequences of the class I HD-Zip family genes were retrieved through BLAST searches against publicly available databases by using Arabidopsis genes as queries (Supplemental Table 1). Sequence alignment and adjustment were conducted as previously described by Yu et al. (2016). The alignable nucleotide matrix was used for phylogenetic reconstruction in PhyML 3.0 using the maximum-likelihood method (Guindon et al., 2010). The general time reversible (GTR) + I + Γ model was applied, and 1000 bootstrap replicates were performed. The matrix in a PHYLIP format and the tree topology in a Newick format can be found in Supplemental Data Sets 9 and 10, respectively.
Gene Expression and Functional Studies
To amplify the cDNA sequence of NidaLMI1, the total RNA was reverse-transcribed into cDNA using SuperScript III First-Strand Synthesis System (Life Technologies) and used as the template. Amplified fragments of expected length were purified and cloned into pEASY-T3 cloning vector (TransGen) for sequencing. The expression patterns of NidaLMI1 were revealed by mRNA in situ hybridization, following the procedures as described by Wang et al. (2015).
The function of NidaLMI1 was investigated through the VIGS technique. Two specific fragments, which spanned the 3′ and 5′ ends of NidaLMI1, respectively, were amplified, purified, and independently introduced into the TRV2-based PY156 vector to generate constructs NidaLMI1_V1 and NidaLMI1_V2 for VIGS experiments (Supplemental Figure 9). Wild-type plants were treated at least two rounds (Supplemental Table 2). Flowers showing phenotypic changes were photodocumented. The morphology and micromorphology of flowers with visible phenotypic changes were investigated as described by Yao et al. (2019). The silencing efficiency of VIGS treatments was determined by quantitative RT-PCR (qRT-PCR) as described by Wang et al. (2015). The cDNAs from individual floral buds or petals at S8/9 of the wild-type and VIGS-treated plants were used as templates. The primers used in this study are provided in Supplemental Table 3. The significant difference of the relative expression levels between the mock and the lines showing strong phenotypic changes was evaluated using the wilcox.test with the two-sided function in R. To avoid the off-target effect, the sequence specificities of the in situ probe, VIGS fragments, and qRT-PCR primers were evaluated by BLASTN against the reference transcriptome with an E value cutoff < 1e–10 (Supplemental Table 4).
Accession Numbers
The RNA-sequencing data have been deposited in National Center for Biotechnology Information Short Read Archive with the accession number PRJNA638226. Sequence data from this article can be found in the GenBank data library under the following accession number: NidaLMI1 (MT780182).
Supplemental Data
Supplemental Figure 1. Generation of the reference transcriptome.
Supplemental Figure 2. Assessment of the reproducibility of the DGE data among triplicates of each sample.
Supplemental Figure 3. Genes specifically expressed in different organs and in petals at different stages.
Supplemental Figure 4. Selected GO terms enriched in the 18 co-expression modules.
Supplemental Figure 5. Expression patterns of genes in the six regulatory programs as shown in Figure 4.
Supplemental Figure 6. Expression patterns of the candidate genes associated with morphological characters.
Supplemental Figure 7. Classification of NidaLMI1 and its sequence features.
Supplemental Figure 8. In situ hybridization of NidaLMI1.
Supplemental Figure 9. Functional characterization of NidaLMI1 by VIGS.
Supplemental Table 1. Gene information used for the phylogenetic analysis.
Supplemental Table 2. Information on VIGS experiments.
Supplemental Table 3. Primers used in this study.
Supplemental Table 4. Evaluation of sequence specificities of the in situ probe, VIGS fragments and qRT-PCR primers.
Supplemental Data Set 1. RPKM values of the 30,790 genes in 22 samples.
Supplemental Data Set 2. Genes specifically expressed in different organs.
Supplemental Data Set 3. Genes preferentially expressed in different organs.
Supplemental Data Set 4. Genes specifically expressed in petals at different stages.
Supplemental Data Set 5. Genes preferentially expressed in petals at different stages.
Supplemental Data Set 6. Information on the GO enrichment analyses.
Supplemental Data Set 7. Genes in the 18 co-expressed modules identified by WGCNA.
Supplemental Data Set 8. Quality evaluation of the DGE data.
Supplemental Data Set 9. Nucleotide alignment matrix of the class I HD-Zip family genes.
Supplemental Data Set 10. Tree file for the phylogeny of the class I HD-Zip family genes.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Shin-Han Shiu and Wengen Zhang for valuable comments and members of the Kong laboratory for helpful discussions. We also thank editors and three anonymous reviewers for valuable suggestions. This work was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (grant XDB27010304) and the Chinese Academy of Sciences (grant ZDBS-LY-SM022).
AUTHOR CONTRIBUTIONS
R.Z., H.S., and H.K. designed the research. R.Z. carried out the sample collection. P.W. performed the de novo assembly of the reference transcriptome of N. damascena. R.Z. and X.F. analyzed the DGE data with the help of G.X. R.Z. and C.Z. conducted mRNA in situ hybridization, VIGS, and qRT-PCR experiments with the help of H.L. X.Y., X.D., and Y.Y. performed the scanning electron microscopy experiment. J.C. made the virtual clay models. R.Z., X.F., H.S., E.M.K., and H.K. wrote the article.
Footnotes
Articles can be viewed without a subscription.
References
- Albert, N.W., Davies, K.M., Lewis, D.H., Zhang, H., Montefiori, M., Brendolise, C., Boase, M.R., Ngo, H., Jameson, P.E., Schwinn, K.E.(2014). A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots. Plant Cell 26: 962–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders, S., Pyl, P.T., Huber, W.(2015). HTSeq--A Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andres, R.J., et al. (2017). Modifications to a LATE MERISTEM IDENTITY1 gene are responsible for the major leaf shapes of Upland cotton (Gossypium hirsutum L.). Proc. Natl. Acad. Sci. USA 114: E57–E66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur, W.(2011). Evolution: A Developmental Approach. (Oxford: Wiley-Blackwell Press; ). [Google Scholar]
- Ashburner, M., et al. et al. ; The Gene Ontology Consortium (2000). Gene ontology: Tool for the unification of biology. Nat. Genet. 25: 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittkau, C., Comes, H.P.(2005). Evolutionary processes in a continental island system: Molecular phylogeography of the Aegean Nigella arvensis alliance (Ranunculaceae) inferred from chloroplast DNA. Mol. Ecol. 14: 4065–4083. [DOI] [PubMed] [Google Scholar]
- Blein, T., Pulido, A., Vialette-Guiraud, A., Nikovics, K., Morin, H., Hay, A., Johansen, I.E., Tsiantis, M., Laufs, P.(2008). A conserved molecular framework for compound leaf development. Science 322: 1835–1839. [DOI] [PubMed] [Google Scholar]
- Borevitz, J.O., Xia, Y., Blount, J., Dixon, R.A., Lamb, C.(2000). Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12: 2383–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Box, M.S., Dodsworth, S., Rudall, P.J., Bateman, R.M., Glover, B.J.(2011). Characterization of Linaria KNOX genes suggests a role in petal-spur development. Plant J. 68: 703–714. [DOI] [PubMed] [Google Scholar]
- Brewer, P.B., Howles, P.A., Dorian, K., Griffith, M.E., Ishida, T., Kaplan-Levy, R.N., Kilinc, A., Smyth, D.R.(2004). PETAL LOSS, a trihelix transcription factor gene, regulates perianth architecture in the Arabidopsis flower. Development 131: 4035–4045. [DOI] [PubMed] [Google Scholar]
- Chandler, J.W., Werr, W.(2017). DORNRÖSCHEN, DORNRÖSCHEN-LIKE, and PUCHI redundantly control floral meristem identity and organ initiation in Arabidopsis. J. Exp. Bot. 68: 3457–3472. [DOI] [PubMed] [Google Scholar]
- Clark, S.E., Running, M.P., Meyerowitz, E.M.(1993). CLAVATA1, a regulator of meristem and flower development in Arabidopsis. Development 119: 397–418. [DOI] [PubMed] [Google Scholar]
- Crawford, B.C., Nath, U., Carpenter, R., Coen, E.S.(2004). CINCINNATA controls both cell differentiation and growth in petal lobes and leaves of Antirrhinum. Plant Physiol. 135: 244–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinneny, J.R., Yadegari, R., Fischer, R.L., Yanofsky, M.F., Weigel, D.(2004). The role of JAGGED in shaping lateral organs. Development 131: 1101–1110. [DOI] [PubMed] [Google Scholar]
- Dockx, J., Quaedvlieg, N., Keultjes, G., Kock, P., Weisbeek, P., Smeekens, S.(1995). The homeobox gene ATK1 of Arabidopsis thaliana is expressed in the shoot apex of the seedling and in flowers and inflorescence stems of mature plants. Plant Mol. Biol. 28: 723–737. [DOI] [PubMed] [Google Scholar]
- Endress, P.K., Matthews, M.(2006). Elaborate petals and staminodes in eudicots: Diversity, function, and evolution. Org. Divers. Evol. 6: 257–293. [Google Scholar]
- Fukushima, K., Hasebe, M.(2014). Adaxial-abaxial polarity: The developmental basis of leaf shape diversity. Genesis 52: 1–18. [DOI] [PubMed] [Google Scholar]
- Gleissberg, S., Groot, E.P., Schmalz, M., Eichert, M., Kölsch, A., Hutter, S.(2005). Developmental events leading to peltate leaf structure in Tropaeolum majus (Tropaeolaceae) are associated with expression domain changes of a YABBY gene. Dev. Genes Evol. 215: 313–319. [DOI] [PubMed] [Google Scholar]
- Glover, B.J., Perez-Rodriguez, M., Martin, C.(1998). Development of several epidermal cell types can be specified by the same MYB-related plant transcription factor. Development 125: 3497–3508. [DOI] [PubMed] [Google Scholar]
- Golz, J.F., Keck, E.J., Hudson, A.(2002). Spontaneous mutations in KNOX genes give rise to a novel floral structure in Antirrhinum. Curr. Biol. 12: 515–522. [DOI] [PubMed] [Google Scholar]
- Gonçalves, B., Nougué, O., Jabbour, F., Ridel, C., Morin, H., Laufs, P., Manicacci, D., Damerval, C.(2013). An APETALA3 homolog controls both petal identity and floral meristem patterning in Nigella damascena L. (Ranunculaceae). Plant J. 76: 223–235. [DOI] [PubMed] [Google Scholar]
- Grabherr, M.G., et al. (2011). Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat. Biotechnol. 29: 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon, S., Dufayard, J.F., Lefort, V., Anisimova, M., Hordijk, W., Gascuel, O.(2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59: 307–321. [DOI] [PubMed] [Google Scholar]
- Hay, A., Tsiantis, M.(2010). KNOX genes: Versatile regulators of plant development and diversity. Development 137: 3153–3165. [DOI] [PubMed] [Google Scholar]
- Hileman, L.C.(2014). Trends in flower symmetry evolution revealed through phylogenetic and developmental genetic advances. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369: 20130348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer, J., Turner, L., Moreau, C., Ambrose, M., Isaac, P., Butcher, S., Weller, J., Dupin, A., Dalmais, M., Le Signor, C., Bendahmane, A., Ellis, N.(2009). Tendril-less regulates tendril formation in pea leaves. Plant Cell 21: 420–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holton, T.A., Brugliera, F., Tanaka, Y.(1993). Cloning and expression of flavonol synthase from Petunia hybrida. Plant J. 4: 1003–1010. [DOI] [PubMed] [Google Scholar]
- Holton, T.A., Cornish, E.C.(1995). Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 7: 1071–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiguchi, G., Kim, G.T., Tsukaya, H.(2005). The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. Plant J. 43: 68–78. [DOI] [PubMed] [Google Scholar]
- Huang, T., Irish, V.F.(2016). Gene networks controlling petal organogenesis. J. Exp. Bot. 67: 61–68. [DOI] [PubMed] [Google Scholar]
- Irish, V.F.(2008). The Arabidopsis petal: A model for plant organogenesis. Trends Plant Sci. 13: 430–436. [DOI] [PubMed] [Google Scholar]
- Irish, V.(2017). The ABC model of floral development. Curr. Biol. 27: R887–R890. [DOI] [PubMed] [Google Scholar]
- Iseli, C., Jongeneel, C.V., Bucher, P.(1999). ESTScan: A program for detecting, evaluating, and reconstructing potential coding regions in EST sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 138–148. [PubMed] [Google Scholar]
- Jaros, U., Tribsch, A., Comes, H.P.(2018). Diversification in continental island archipelagos: New evidence on the roles of fragmentation, colonization and gene flow on the genetic divergence of Aegean Nigella (Ranunculaceae). Ann. Bot. 121: 241–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, C.S., Kolevski, B., Smyth, D.R.(2002). TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell 14: 1359–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D., Pertea, G., Trapnell, C., Pimentel, H., Kelley, R., Salzberg, S.L.(2013). TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J.H., Choi, D., Kende, H.(2003). The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant J. 36: 94–104. [DOI] [PubMed] [Google Scholar]
- Kim, J.H., Kende, H.(2004). A transcriptional coactivator, AtGIF1, is involved in regulating leaf growth and morphology in Arabidopsis. Proc. Natl. Acad. Sci. USA 101: 13374–13379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimyuk, V.I., Jones, J.D.G.(1997). AtDMC1, the Arabidopsis homologue of the yeast DMC1 gene: Characterization, transposon-induced allelic variation and meiosis-associated expression. Plant J. 11: 1–14. [DOI] [PubMed] [Google Scholar]
- Koyama, T., Mitsuda, N., Seki, M., Shinozaki, K., Ohme-Takagi, M.(2010). TCP transcription factors regulate the activities of ASYMMETRIC LEAVES1 and miR164, as well as the auxin response, during differentiation of leaves in Arabidopsis. Plant Cell 22: 3574–3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama, T., Ohme-Takagi, M., Sato, F.(2011). Generation of serrated and wavy petals by inhibition of the activity of TCP transcription factors in Arabidopsis thaliana. Plant Signal. Behav. 6: 697–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek, B.(2009). AINTEGUMENTA and AINTEGUMENTA-LIKE6 act redundantly to regulate Arabidopsis floral growth and patterning. Plant Physiol. 150: 1916–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder, P., Horvath, S.(2008). WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics 9: 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, C., Teng, Q., Zhong, R., Ye, Z.H.(2012). Arabidopsis GUX proteins are glucuronyltransferases responsible for the addition of glucuronic acid side chains onto xylan. Plant Cell Physiol. 53: 1204–1216. [DOI] [PubMed] [Google Scholar]
- Liao, H., Fu, X., Zhao, H., Cheng, J., Zhang, R., Yao, X., Duan, X., Shan, H., Kong, H.(2020). The morphology, molecular development and ecological function of pseudonectaries on Nigella damascena (Ranunculaceae) petals. Nat. Commun. 11: 1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln, C., Long, J., Yamaguchi, J., Serikawa, K., Hake, S.(1994). A knotted1-like homeobox gene in Arabidopsis is expressed in the vegetative meristem and dramatically alters leaf morphology when overexpressed in transgenic plants. Plant Cell 6: 1859–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, J.A., Moan, E.I., Medford, J.I., Barton, M.K.(1996). A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379: 66–69. [DOI] [PubMed] [Google Scholar]
- Maes, L., Inzé, D., Goossens, A.(2008). Functional specialization of the TRANSPARENT TESTA GLABRA1 network allows differential hormonal control of laminal and marginal trichome initiation in Arabidopsis rosette leaves. Plant Physiol. 148: 1453–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnani, E., Hake, S.(2008). KNOX lost the OX: The Arabidopsis KNATM gene defines a novel class of KNOX transcriptional regulators missing the homeodomain. Plant Cell 20: 875–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks, M.D., Feldmann, K.A.(1989). Trichome development in Arabidopsis thaliana. I. T-DNA tagging of the GLABROUS1 gene. Plant Cell 1: 1043–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAbee, J.M., Hill, T.A., Skinner, D.J., Izhaki, A., Hauser, B.A., Meister, R.J., Venugopala Reddy, G., Meyerowitz, E.M., Bowman, J.L., Gasser, C.S.(2006). ABERRANT TESTA SHAPE encodes a KANADI family member, linking polarity determination to separation and growth of Arabidopsis ovule integuments. Plant J. 46: 522–531. [DOI] [PubMed] [Google Scholar]
- McConnell, J.R., Barton, M.K.(1998). Leaf polarity and meristem formation in Arabidopsis. Development 125: 2935–2942. [DOI] [PubMed] [Google Scholar]
- Menssen, A., Höhmann, S., Martin, W., Schnable, P.S., Peterson, P.A., Saedler, H., Gierl, A.(1990). The En/Spm transposable element of Zea mays contains splice sites at the termini generating a novel intron from a dSpm element in the A2 gene. EMBO J. 9: 3051–3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min, Y., Bunn, J.I., Kramer, E.M.(2019). Homologs of the STYLISH gene family control nectary development in Aquilegia. New Phytol. 221: 1090–1100. [DOI] [PubMed] [Google Scholar]
- Mizukami, Y., Fischer, R.L.(2000). Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc. Natl. Acad. Sci. USA 97: 942–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morohashi, K., Grotewold, E.(2009). A systems approach reveals regulatory circuitry for Arabidopsis trichome initiation by the GL3 and GL1 selectors. PLoS Genet. 5: e1000396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi, A., Williams, B.A., McCue, K., Schaeffer, L., Wold, B.(2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5: 621–628. [DOI] [PubMed] [Google Scholar]
- Moyroud, E., Glover, B.J.(2017). The evolution of diverse floral morphologies. Curr. Biol. 27: R941–R951. [DOI] [PubMed] [Google Scholar]
- Nag, A., King, S., Jack, T.(2009). miR319a targeting of TCP4 is critical for petal growth and development in Arabidopsis. Proc. Natl. Acad. Sci. USA 106: 22534–22539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata, M., Matsumoto, N., Tsugeki, R., Rikirsch, E., Laux, T., Okada, K.(2012). Roles of the middle domain-specific WUSCHEL-RELATED HOMEOBOX genes in early development of leaves in Arabidopsis. Plant Cell 24: 519–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda, K., Glover, B.J., Linstead, P., Martin, C.(1994). Flower colour intensity depends on specialized cell shape controlled by a Myb-related transcription factor. Nature 369: 661–664. [DOI] [PubMed] [Google Scholar]
- O’Reilly, C., Shepherd, N.S., Pereira, A., Schwarz-Sommer, Z., Bertram, I., Robertson, D.S., Peterson, P.A., Saedler, H.(1985). Molecular cloning of the a1 locus of Zea mays using the transposable elements En and Mu1. EMBO J. 4: 877–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno, C.K., Reddy, G.V., Heisler, M.G., Meyerowitz, E.M.(2004). The Arabidopsis JAGGED gene encodes a zinc finger protein that promotes leaf tissue development. Development 131: 1111–1122. [DOI] [PubMed] [Google Scholar]
- Payne, C.T., Zhang, F., Lloyd, A.M.(2000). GL3 encodes a bHLH protein that regulates trichome development in arabidopsis through interaction with GL1 and TTG1. Genetics 156: 1349–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekker, I., Alvarez, J.P., Eshed, Y.(2005). Auxin response factors mediate Arabidopsis organ asymmetry via modulation of KANADI activity. Plant Cell 17: 2899–2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigge, M.J., Clark, S.E.(2006). Evolution of the class III HD-Zip gene family in land plants. Evol. Dev. 8: 350–361. [DOI] [PubMed] [Google Scholar]
- Prum, R.O.(1999). Development and evolutionary origin of feathers. J. Exp. Zool. 285: 291–306. [PubMed] [Google Scholar]
- Prum, R.O., Brush, A.H.(2002). The evolutionary origin and diversification of feathers. Q. Rev. Biol. 77: 261–295. [DOI] [PubMed] [Google Scholar]
- Riou-Khamlichi, C., Huntley, R., Jacqmard, A., Murray, J.A.H.(1999). Cytokinin activation of Arabidopsis cell division through a D-type cyclin. Science 283: 1541–1544. [DOI] [PubMed] [Google Scholar]
- Sawa, S., Watanabe, K., Goto, K., Liu, Y.G., Shibata, D., Kanaya, E., Morita, E.H., Okada, K.(1999). FILAMENTOUS FLOWER, a meristem and organ identity gene of Arabidopsis, encodes a protein with a zinc finger and HMG-related domains. Genes Dev. 13: 1079–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semiarti, E., Ueno, Y., Tsukaya, H., Iwakawa, H., Machida, C., Machida, Y.(2001). The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development 128: 1771–1783. [DOI] [PubMed] [Google Scholar]
- Shan, H., Cheng, J., Zhang, R., Yao, X., Kong, H.(2019). Developmental mechanisms involved in the diversification of flowers. Nat. Plants 5: 917–923. [DOI] [PubMed] [Google Scholar]
- Shimizu, H., Shikanai, T.(2007). Dihydrodipicolinate reductase-like protein, CRR1, is essential for chloroplast NAD(P)H dehydrogenase in Arabidopsis. Plant J. 52: 539–547. [DOI] [PubMed] [Google Scholar]
- Shubin, N., Tabin, C., Carroll, S.(2009). Deep homology and the origins of evolutionary novelty. Nature 457: 818–823. [DOI] [PubMed] [Google Scholar]
- Sicard, A., Thamm, A., Marona, C., Lee, Y.W., Wahl, V., Stinchcombe, J.R., Wright, S.I., Kappel, C., Lenhard, M.(2014). Repeated evolutionary changes of leaf morphology caused by mutations to a homeobox gene. Curr. Biol. 24: 1880–1886. [DOI] [PubMed] [Google Scholar]
- Siegfried, K.R., Eshed, Y., Baum, S.F., Otsuga, D., Drews, G.N., Bowman, J.L.(1999). Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development 126: 4117–4128. [DOI] [PubMed] [Google Scholar]
- Sorensen, A.M., Kröber, S., Unte, U.S., Huijser, P., Dekker, K., Saedler, H.(2003). The Arabidopsis ABORTED MICROSPORES (AMS) gene encodes a MYC class transcription factor. Plant J. 33: 413–423. [DOI] [PubMed] [Google Scholar]
- Streb, S., Zeeman, S.C.(2012). Starch metabolism in Arabidopsis. Arabidopsis Book 10: e0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strid, A.(1970). Studies in the Aegean flora. XVI. Biosystematics of the Nigella arvensis complex. Opera Bot. 28: 1–169. [Google Scholar]
- Szécsi, J., Joly, C., Bordji, K., Varaud, E., Cock, J.M., Dumas, C., Bendahmane, M.(2006). BIGPETALp, a bHLH transcription factor is involved in the control of Arabidopsis petal size. EMBO J. 25: 3912–3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura, M.(1995). Ranunculaceae. In Bie Naturlichen Pflanzenfamilien, Engler A., Prantl K., and Hiepko P., eds (Berlin: Duncker & Humblot; ), pp. 1–555. [Google Scholar]
- Thévenin, J., Pollet, B., Letarnec, B., Saulnier, L., Gissot, L., Maia-Grondard, A., Lapierre, C., Jouanin, L.(2011). The simultaneous repression of CCR and CAD, two enzymes of the lignin biosynthetic pathway, results in sterility and dwarfism in Arabidopsis thaliana. Mol. Plant 4: 70–82. [DOI] [PubMed] [Google Scholar]
- Tian, T., Liu, Y., Yan, H., You, Q., Yi, X., Du, Z., Xu, W., Su, Z.(2017). agriGO v2.0: A GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 45 (W1): W122–W129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlad, D., et al. (2014). Leaf shape evolution through duplication, regulatory diversification, and loss of a homeobox gene. Science 343: 780–783. [DOI] [PubMed] [Google Scholar]
- Vroemen, C.W., Mordhorst, A.P., Albrecht, C., Kwaaitaal, M.A., de Vries, S.C.(2003). The CUP-SHAPED COTYLEDON3 gene is required for boundary and shoot meristem formation in Arabidopsis. Plant Cell 15: 1563–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waites, R., Selvadurai, H.R.N., Oliver, I.R., Hudson, A.(1998). The PHANTASTICA gene encodes a MYB transcription factor involved in growth and dorsoventrality of lateral organs in Antirrhinum. Cell 93: 779–789. [DOI] [PubMed] [Google Scholar]
- Walcher-Chevillet, C.L., Kramer, E.M.(2016). Breaking the mold: Understanding the evolution and development of lateral organs in diverse plant models. Curr. Opin. Genet. Dev. 39: 79–84. [DOI] [PubMed] [Google Scholar]
- Walker, A.R., Davison, P.A., Bolognesi-Winfield, A.C., James, C.M., Srinivasan, N., Blundell, T.L., Esch, J.J., Marks, M.D., Gray, J.C.(1999). The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 11: 1337–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, P., Liao, H., Zhang, W., Yu, X., Zhang, R., Shan, H., Duan, X., Yao, X., Kong, H.(2015). Flexibility in the structure of spiral flowers and its underlying mechanisms. Nat. Plants 2: 15188. [DOI] [PubMed] [Google Scholar]
- Whitewoods, C.D., Gonçalves, B., Cheng, J., Cui, M., Kennaway, R., Lee, K., Bushell, C., Yu, M., Piao, C., Coen, E.(2020). Evolution of carnivorous traps from planar leaves through simple shifts in gene expression. Science 367: 91–96. [DOI] [PubMed] [Google Scholar]
- Wuest, S.E., O’Maoileidigh, D.S., Rae, L., Kwasniewska, K., Raganelli, A., Hanczaryk, K., Lohan, A.J., Loftus, B., Graciet, E., Wellmer, F.(2012). Molecular basis for the specification of floral organs by APETALA3 and PISTILLATA. Proc. Natl. Acad. Sci. USA 109: 13452–13457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing, S., Rosso, M.G., Zachgo, S.(2005). ROXY1, a member of the plant glutaredoxin family, is required for petal development in Arabidopsis thaliana. Development 132: 1555–1565. [DOI] [PubMed] [Google Scholar]
- Yang, C., Ye, Z.(2013). Trichomes as models for studying plant cell differentiation. Cell. Mol. Life Sci. 70: 1937–1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, X., Zhang, W., Duan, X., Yuan, Y., Zhang, R., Shan, H., Kong, H.(2019). The making of elaborate petals in Nigella through developmental repatterning. New Phytol. 223: 385–396. [DOI] [PubMed] [Google Scholar]
- Yu, X., Duan, X., Zhang, R., Fu, X., Ye, L., Kong, H., Xu, G., Shan, H.(2016). Prevalent exon-intron structural changes in the APETALA1/FRUITFULL, SEPALLATA, AGAMOUS-LIKE6, and FLOWERING LOCUS C MADS-box gene subfamilies provide new insights into their evolution. Front. Plant Sci. 7: 598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, B., Horvath, S.(2005). A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 4: e17. [DOI] [PubMed] [Google Scholar]
- Zhang, R., et al. (2013). Disruption of the petal identity gene APETALA3-3 is highly correlated with loss of petals within the buttercup family (Ranunculaceae). Proc. Natl. Acad. Sci. USA 110: 5074–5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, J.L., Pan, J.S., Guan, Y., Zhang, W.W., Bie, B.B., Wang, Y.L., He, H.L., Lian, H.L., Cai, R.(2015). Micro-trichome as a class I homeodomain-leucine zipper gene regulates multicellular trichome development in Cucumis sativus. J. Integr. Plant Biol. 57: 925–935. [DOI] [PubMed] [Google Scholar]
- Zhao, L., Liu, P., Che, X.F., Wang, W., Ren, Y.(2011). Floral organogenesis of Helleborus thibetanus and Nigella damascena (Ranunculaceae) and its systematic significance. Bot. J. Linn. Soc. 166: 431–443. [Google Scholar]
- Zhao, Q., Nakashima, J., Chen, F., Yin, Y., Fu, C., Yun, J., Shao, H., Wang, X., Wang, Z.Y., Dixon, R.A.(2013). Laccase is necessary and nonredundant with peroxidase for lignin polymerization during vascular development in Arabidopsis. Plant Cell 25: 3976–3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zohary, M.(1983). The genus Nigella (Ranunculaceae)—A taxonomic revision. Plant Syst. Evol. 142: 71–105. [Google Scholar]