PhasiRNAs are increasingly well-described features of plant genomes, with multifaceted roles in processes such as stress responses and anther biology.
Abstract
Phased secondary small interfering RNAs (phasiRNAs) constitute a major category of small RNAs in plants, but most of their functions are still poorly defined. Some phasiRNAs, known as trans-acting siRNAs, are known to target complementary mRNAs for degradation and to function in development. However, the targets or biological roles of other phasiRNAs remain speculative. New insights into phasiRNA biogenesis, their conservation, and their variation across the flowering plants continue to emerge due to the increased availability of plant genomic sequences, deeper and more sophisticated sequencing approaches, and improvements in computational biology and biochemical/molecular/genetic analyses. In this review, we survey recent progress in phasiRNA biology, with a particular focus on two classes associated with male reproduction: 21-nucleotide (accumulate early in anther ontogeny) and 24-nucloetide (produced in somatic cells during meiosis) phasiRNAs. We describe phasiRNA biogenesis, function, and evolution and define the unanswered questions that represent topics for future research.
INTRODUCTION
Small RNAs (sRNAs) play central roles in regulating many plant developmental and physiological processes. These activities typically occur via transcriptional gene silencing or posttranscriptional gene silencing (PTGS; Borges and Martienssen, 2015). Among the classes of sRNAs, microRNAs (miRNAs) are the best studied and are relevant here because they trigger phased secondary small interfering RNA (phasiRNA) production. Typically, plant miRNAs are generated from a noncoding product of RNA Polymerase II (Pol II) that forms a stem-loop secondary structure; the stem-loop is recognized and processed by the RNase III enzyme DICER-LIKE1 (DCL1; Rogers and Chen, 2013; Bologna and Voinnet, 2014). After sequential cleavage steps by DCL1 (Bologna et al., 2009; Zhang et al., 2017), a duplex RNA of ∼21 nucleotides is released, containing the mature miRNA and the complementary strand called miRNA* (pronounced miRNA-star). The mature miRNA is then incorporated into an ARGONAUTE protein, typically ARGONAUTE1 (AGO1), to form an RNA-induced silencing complex (RISC) that activates the function of the miRNA in PTGS, thereby guiding the silencing of target mRNAs via complementary nucleotide base pairing between the miRNA and its mRNA “target” site (Henderson et al., 2006; Bologna and Voinnet, 2014; Borges and Martienssen, 2015). The miRNA* is the unneeded complement or “passenger” strand that is typically degraded. In cases when the stem-loop precursor includes an asymmetric bulge within the paired miRNA-miRNA* region, DCL1 cleavage will generate a 22-nucleotide/21-nucleotide duplex. The AGO1 RISC loaded with the 22-nucleotide miRNA can trigger the biogenesis of secondary small interfering RNAs (siRNAs) that have a distinctive, phased configuration. Specifically, cleavage generates regularly spaced siRNAs (see below), which is evident when these siRNAs are mapped back to a precursor transcript. These siRNAs are phasiRNAs, and the loci that generate them are known as PHAS loci. The PHAS precursor RNAs may be protein-coding mRNAs or long, noncoding RNA (lncRNAs); lncRNAs are generally recognized as RNAs lacking an open reading frame encoding a protein of at least 100 amino acids. The 21-nucleotide secondary siRNAs (i.e., phasiRNAs) negatively regulate target transcripts, such as during plant development (Chen et al., 2010; Cuperus et al., 2010a).
Plant PHAS loci can be subdivided into two major groups based on their genomic source: PHAS loci found within noncoding regions that produce lncRNAs, and those located within protein-coding genes (Fei et al., 2013). A subset of the first group of PHAS loci, the TAS loci, encode lncRNAs that generate trans-acting siRNAs (tasiRNAs); these loci were described ∼15 years ago in Arabidopsis (Arabidopsis thaliana). The name tasiRNAs was derived from the experimentally validated activity of these phasiRNAs to silence transcripts from other loci (Fei et al., 2013). The 21-nucleotide tasiRNAs derived from TAS loci negatively regulate their target transcripts, triggering their selective degradation (Chen et al., 2010; Cuperus et al., 2010a). There are still fewer than 10 known TAS loci or families of loci, making it the smallest family of known PHAS loci. More recently, another subclass of reproductive phasiRNAs was discovered that is also derived from lncRNAs. The targets of the reproductive phasiRNAs are unknown, but they appear to be involved in reproductive development (e.g., they are highly enriched in anther tissue) and in some cases have been shown to be essential for male fertility (Johnson et al., 2009; Zhai et al., 2015; Fei et al., 2016a; Xia et al., 2019).
PHAS loci within protein-coding genes encode a much larger subgroup of phasiRNAs. These PHAS loci include nucleotide binding leucine-rich repeat (NLR) genes (Zhai et al., 2011; Fei et al., 2015; Xia et al., 2015a), arguably the largest subgroup when data are compared across many plant genomes, as well as PENTATRICOPEPTIDE REPEAT (PPR) genes (Howell et al., 2007; Xia et al., 2013, 2015b), MYB transcription factor (TF) genes (Xia et al., 2013, 2015a), AUXIN RESPONSE FACTOR (ARF) genes (Xia et al., 2013, 2017), and NAC TF genes (Liu et al., 2017; Xie et al., 2017; Ma et al., 2018), among a long list of diverse genes yielding phasiRNAs. These loci, and thus presumably the resulting phasiRNAs, function as negative regulators in many biological processes, such as disease resistance, plant vegetative and reproductive development, seed germination, and plant parasitism (Marin et al., 2010; Yifhar et al., 2012; Zhou, 2013; Cabrera et al., 2016; Hobecker et al., 2017; Guo, 2018; Shahid et al., 2018).
We reviewed phasiRNAs in 2013 (Fei et al., 2013), focusing on protein-coding genes, and in particular on genes encoding NLR proteins. Here, we update what is known about phasiRNAs, focusing on those derived from lncRNAs, with the majority of new information concerning the reproductive phasiRNAs. We describe recent progress in understanding phasiRNA biogenesis, evolution, mobility, and function.
BIOGENESIS OF phasiRNAs
The biogenesis of phasiRNAs occurs after cleavage of the target mRNA or lncRNAs, typically (but not exclusively) by a 22-nucleotide miRNA. After cleavage, the 5′ fragment of the target mRNA is rapidly degraded by a 3′→5′ exonucleolytic complex (e.g., the SKI2-3-8 complex; Figure 1A; Branscheid et al., 2015). The 3′ fragment is converted to double-stranded RNA (dsRNA) via the activity of RNA-DEPENDENT RNA POLYMERASE6 (RDR6), which may be recruited by AGO1-RISC or AGO7-RISC and assisted by SUPPRESSOR OF GENE SILENCING3 (SGS3), which in turn may prevent the degradation of the 3′ fragment from a 5′→3′ exoribonuclease (e.g., XRN4; Souret et al., 2004). The resulting dsRNA is iteratively cleaved by a Dicer protein from the 5′ end of the “top” (Pol II-derived) strand containing the cleavage site, yielding duplexes of phasiRNAs (Figure 1A). There are likely at least three different Dicer family members capable of producing phasiRNAs, as described in detail below. The function of the Dicer family member DCL4 requires the assistance of a DOUBLE-STRANDED RNA BINDING FACTOR (DRB) protein to produce 21-nucleotide phasiRNAs (Vazquez et al., 2004; Adenot et al., 2006; Fukudome et al., 2011; Song et al., 2012). Similar to miRNA duplexes, tasiRNA duplexes are sorted during loading into AGO proteins via a process known to be dependent on the 5′ nucleotide (Mi et al., 2008), but this process is otherwise not well described. The tasiRNA-containing RISC subsequently interacts with target RNAs in a homology-dependent manner, as with miRNAs. The production of RISCs loaded with other phasiRNAs, like the reproductive phasiRNAs described below, presumably follows a similar process. Interestingly, only some of the phasiRNAs produced from a precursor accumulate and are detectable; the rest of the phasiRNAs are likely degraded or not even loaded in AGO in the first place, which is common for phasiRNAs other than tasiRNAs.
Figure 1.
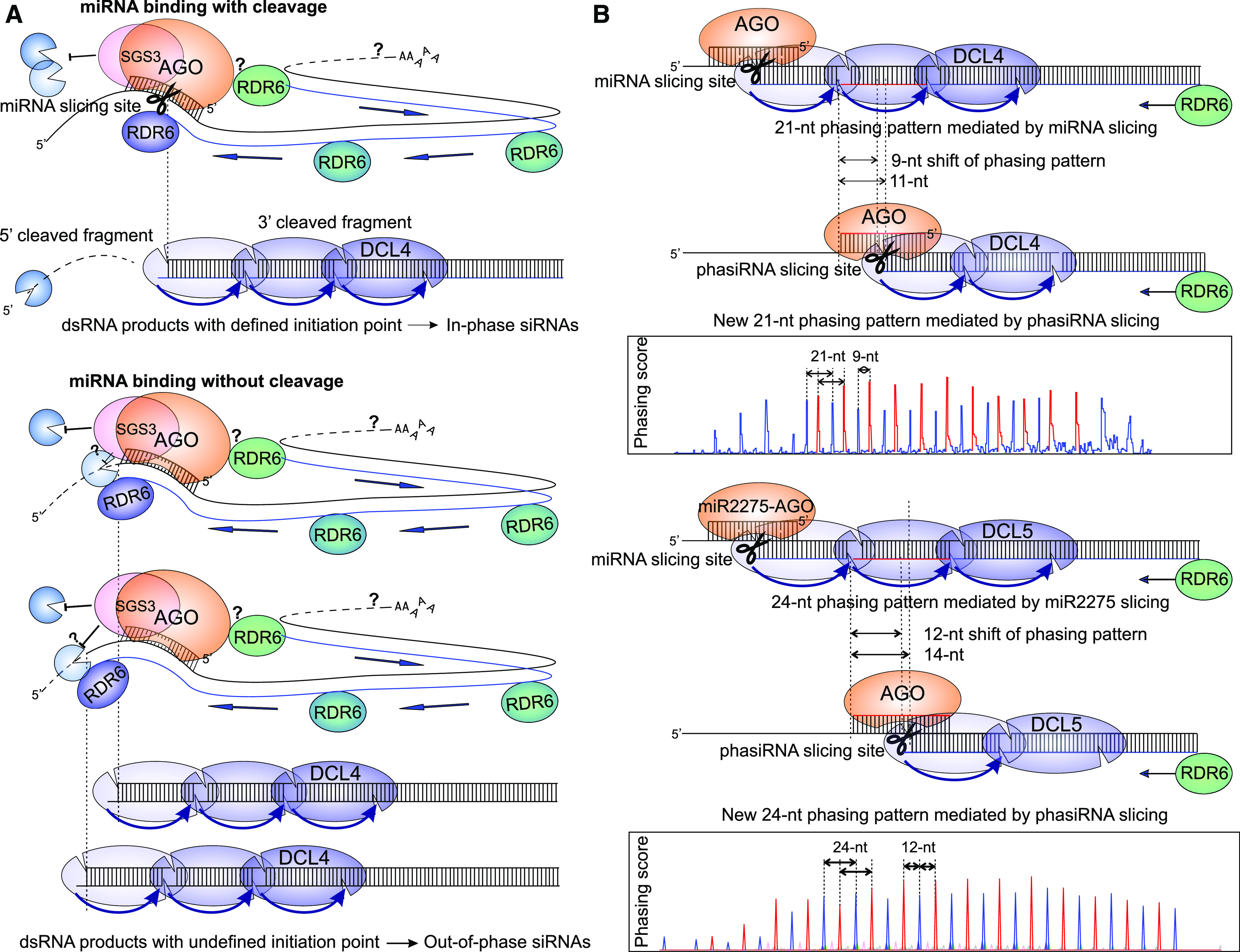
Mechanisms That Generate the Phased Patterns of PhasiRNAs.
(A) Cleavage is responsible for establishing the phasing pattern. During miRNA-mediated secondary siRNA biogenesis, RDR6, recruited by AGO (with the assistance of SGS3), converts the mRNA substrate into dsRNA, followed by processing by DCL4 or DCL5. Cleavage by the miRNA-AGO complex at a consistent nucleotide position marks the defined initiation point of the resulting siRNAs, establishing the phasing pattern. Conversely, the absence of precise cleavage on the mRNA substrate yields dsRNAs with undefined initiation points, producing out-of-phase siRNAs. The dark-blue “Pac-Man” represents the SKI2-3-8 complex that directs 3′→5′ exonucleolysis, while the light-blue Pac-Man represents a 5′→3′-exoribonuclease (e.g., XRN4). The question mark indicates possible degradation.
(B) Alternative phasing patterns. At a locus with a primarily 21-nucleotide (nt) phasing pattern (blue phasing signal in the line plot), the resulting phasiRNAs can target and slice the PHAS/TAS precursor in cis, giving rise to a new round of phasiRNA production, with a phasing pattern (red phasing signal in the line plot) shifted 9 nt relative to the original miRNA-mediated phasing pattern (Tamim et al., 2018). Superposition of two phasing patterns creates a new 12-nt/9-nt phasing pattern. Similarly, from a locus generating a primary 24-nt phasing pattern, phasiRNAs targeting a precursor in cis yield a new 12-nt phasing pattern (Xia et al., 2019).
PhasiRNA Production Triggered by One versus Two “Hits”
The critical first steps in initiating phasiRNA production require (1) cleavage of the precursor RNA with single-nucleotide precision to define the 5′ end and phasing “register,” and (2) a mechanism to make the cleaved RNA double stranded and thus a substrate for Dicer processing. In most cases, (1) results from miRNA activity and (2) results from RDR6 activity. Yet, as is often the case with biology, evolution has demonstrated that these are not inviolable rules, as exceptions exist to almost every mechanistic generalization about phasiRNA biogenesis.
The pathway for phasiRNA production likely originated early in plant evolution, as analyses of phasiRNAs have demonstrated that the TAS3 (tasiRNA) locus is present in one of the earliest diverged land plants, a liverwort (Marchantia polymorpha), and phasiRNA-generating loci (including TAS3) are found in all angiosperm genomes analyzed to date (Xia et al., 2017). While TAS3 has distinct attributes, understanding this locus is essential, since it may well be the progenitor of all plant PHAS loci.
tasiRNA production from most TAS3 loci is described by the two-hit model. This model proposes that the 21-nucleotide miRNA trigger, miR390, is loaded into its specialized protein partner AGO7 and targets or “hits” the TAS3 lncRNA precursor at two different positions to trigger tasiRNA biogenesis (Axtell et al., 2006; Montgomery et al., 2008; Endo et al., 2013). Just one or two of the resulting tasiRNAs, the tasiARFs, target transcripts from AUXIN RESPONSIVE FACTOR (ARF) genes to suppress ARF activity (Axtell et al., 2006; Montgomery et al., 2008). There are two types of TAS3 loci in vascular plants, the longer and shorter variants (TAS3L and TAS3S, respectively), but the difference in the biological roles of these two types of loci is unclear (Xia et al., 2017). Typically, the 5′ proximal miR390 target site is not cleaved but is required (Cuperus et al., 2010b), while the 3′ proximal target site is cleaved, suggesting that a pair of miR390-loaded RISCs may be important, perhaps for recruiting RDR6. The miR390:TAS3 pairing and cleavage patterns are generally conserved across thousands of plant species (Xia et al., 2017). The cleavage occurring only at the 3′ proximal target site triggers tasiRNA biogenesis from that end, with processing occurring in the 3′→5′ direction. Yet, there is rich diversity in TAS3 configurations: in numerous gymnosperms (for TAS3L) and eudicots (for TAS3S), the 5′ miR390 target sites are cleavable, likely generating tasiARFs in the 5′→3′ direction, which is consistent with a bidirectional processing mechanism not found in Arabidopsis (Xia et al., 2017). This notion was validated in work using artificial TAS constructs; with cleavage at both ends, the phasing of the sRNAs was poor, due to out-of-phase superposition of tasiRNAs initiated from each of the two ends (de Felippes et al., 2017). The canonical configuration of TAS3 may provide insight into the mechanism of tasiRNA production: the first step is cleavage to remove the poly(A) tail of the lncRNAs precursor, which may be helpful for RDR6 recruitment, a second step leading to dsRNA conversion (Baeg et al., 2017). Thus, this “two-hit, one-cleavage” configuration for tasiRNA production is likely efficient, precise, and accurate, as reflected in its conservation over hundreds of millions of years.
The one-hit model, in which one miRNA target is present in a transcript, is a much more prevalent pathway for producing phasiRNAs in plants. This pathway uses a trigger miRNA (generally 22-nucleotides long) with a 5′ uridine (U; Chen et al., 2010). The initial “U” directs miRNA loading to AGO1, the canonical AGO in phasiRNA generation (apart from AGO7, which loads miR390 for tasiARF generation; Mi et al., 2008). Loading of a 22-nucleotide, but not 21-nucleotide, miRNA presumably activates the recruitment of RDR6 and SGS3 following miRNA-directed cleavage (Chen et al., 2010), perhaps via a change in the structural conformation of the RISC. Recent work has started to describe the process of dsRNA precursor production. Baeg et al. (2017) demonstrated that RDR6 prefers deadenylated mRNAs over canonical polyadenylated mRNAs as templates during the initiation step of complementary strand synthesis. Other studies showed that miRNA-directed cleavage and secondary siRNA generation can be uncoupled. Arribas-Hernández et al. (2016) analyzed a slicer-deficient ago1 mutant in Arabidopsis and identified abundant secondary siRNAs from TAS/PHAS precursors whose production was dependent on RDR6 and SGS3 but lacked a clearly phased pattern (i.e., the siRNAs were out of phase). de Felippes et al. (2017) obtained similar results from the analysis of TAS3: if a single, 5′-proximate miR390 target site was noncleavable, out-of-phase siRNAs were still generated. These results indicate that miRNA-mediated cleavage is not essential to recruit RDR6/SGS3 for dsRNA synthesis and subsequent siRNA production, but it is important for the formation of phased siRNAs, as it sets the initiation point for subsequent Dicer processing. In the absence of slicing, multiple dsRNA products with different 5′ ends result from a combination of factors, yielding out-of-phase siRNAs (Figure 1A).
How and where on the precursor RNA does RDR6 initiate? The observations described in the previous paragraph suggest that RDR6 recruitment without cleavage requires a noncleaved miRNA target site, which is consistent with AGO-mediated recruitment of RDR6 for dsRNA conversion. However, if so, it is not known whether this is a direct or indirect interaction, or precisely how RDR6 is recruited. In the two-hit pathway of TAS3, tasiRNAs are generated from the region between two miR390 target sites, with the cleaved 3′ site setting the phase of tasiRNAs and the noncleaved 5′ site defining the boundary of dsRNA synthesis by RDR6 (Rajeswaran and Pooggin, 2012). Removal of the poly(A) tail by 3′ target site cleavage likely makes the cleaved TAS3 transcript more suitable as an RDR6 substrate. Yet, the mRNA or lncRNA PHAS precursors cleaved at a single, 22-nucleotide miRNA target site should retain a poly(A) tail on the fragment converted to siRNAs. Perhaps deadenylation or a shortening of the poly(A) tail on this fragment assists in the recruitment of RDR6. For example, once phasiRNA production starts, the phasiRNAs may act in cis to accelerate or amplify the production of poly(A)-minus substrates for RDR6. In support of this hypothesis, a TAS1c-derived tasiRNA in Arabidopsis directs cleavage of TAS1a/b/c and TAS2 transcripts (i.e., in cis and in trans), helping restrict siRNA production within a region between the miRNA target sites (miR173) and the TAS1c tasiRNA target site (Rajeswaran et al., 2012). Alternatively, the initial set of phasiRNAs produced may subsequently function in cis as primers for RDR6 activity, possibly explaining why the abundance of phasiRNAs is greater at sites closer to the target site of the miRNA trigger (Tamim et al., 2018), as each phasiRNA could prime production of short substrates from a target Pol II transcript.
Another unknown is how the direction of tasiRNA generation is regulated. It is unclear why, in the two-hit mode, the cleaved fragment 3′ of the 3′ miR390 site is not the substrate for RDR6-mediated dsRNA synthesis and subsequent phasiRNA production, as observed in the one-hit model, even though a single miR390 target site at a TAS3 locus is sufficient for tasiRNA production (de Felippes et al., 2017). In other words, why is the downstream fragment (relative to the miRNA cleavage site) converted to phasiRNAs with one hit but the upstream fragment converted for the two-hit precursors? A possible explanation is the distinct pairing pattern of the 3′ miR390 site in TAS3 genes, which has a consistently matched middle region, with the last four nucleotides (the 3′ end of miR390) always unpaired (Xia et al., 2017). The 3′ unpaired region may dictate the direction of production of TAS3 tasiRNAs, perhaps via recruitment of RDR6 (Xia et al., 2017). However, earlier work in Medicago truncatula described a similar two-hit PHAS locus with a cleaved miR172 target site lacking this unpaired region, coupled with an uncleaved miR156 region (Zhai et al., 2011). As mentioned above, perhaps the proximity of two RISCs (dimerizing?) at the two-hit target sites causes RDR6 recruitment to the 5′ fragment after cleavage, whereas for one-hit loci, the single AGO could recruit RDR6 to the 3′ fragment initially quite poorly. However, after a few initial rounds of phasiRNA production, the phasiRNA-RISC could function in cis, interacting with the miRNA-RISC in a feed-forward cycle of phasiRNA biogenesis.
22-Nucleotide miRNAs as Triggers of PhasiRNAs
A number of researchers have investigated how 22-nucleotide miRNAs are generated and what triggers phasiRNA production at most PHAS loci. An early observation was that 22-nucleotide miRNAs are derived from miRNA/miRNA* duplexes containing an asymmetric bulge on the miRNA strand (Chen et al., 2010); DCL1 cleavage of this structure yields a 21/22-nucleotide duplex. Manavella et al. (2012) argued that it is this asymmetric structure of the duplex that activates the process of phasiRNA generation from target transcripts. They showed that a 21-nucleotide miRNA from such a mismatched duplex can also trigger phasiRNA biogenesis. However, contrasting observations were made in Arabidopsis and Phaseoleae species. To understand those observations, it is useful to know that in plants, miRNAs are normally 2′-O-methylated at the 3′ terminus by HUA ENHANCER1 (HEN1) after DCL1 processing; the absence of HEN1-directed methylation leads to the addition of 3′ Us, changing the length of the sRNA. In an Arabidopsis hen1 mutant, miR171 (a 21-nucleotide mature miRNA at biogenesis) is monouridylated from 21 nucleotides to 22 nucleotides in length, and it then triggers phasiRNA production from Scarecrow-like (SCL) target transcripts (Zhai et al., 2013). In most Phaseoleae plants, the miR1510 precursor has no asymmetric bulge, yielding a 21-nucleotide miRNA. However, miR1510 is incompletely 2′-O-methylated at the 3′ terminus due to a unique structure in the miR1510/miR1510* duplex (Fei et al., 2018). Unmethylated 21-nucleotide miR1510 is thereafter monouridylated, leading to the accumulation of a 22-nucleotide isoform of miR1510, again triggering phasiRNA production from the targeted NLR transcripts (Fei et al., 2018). In addition, 22-nucleotide miRNAs can be generated via a DCL2-dependent pathway from symmetric miRNA/miRNA* duplexes capable of initiating the biogenesis of phasiRNAs (Wang et al., 2018). These observations suggest that no matter how the 22-nucleotide miRNA is produced, its length is a key determinant of phasiRNA generation from target RNAs.
Patterns of PhasiRNAs Indicative of Low-Frequency Cis Cleavage
The distinctive head-to-tail pattern of siRNAs, the key attribute of phasiRNAs, is generated by progressive processing of DCL proteins from the miRNA cleavage site on PHAS transcripts. These patterns are 21 nucleotides long or 24 nucleotides long, depending on the Dicer. However, other patterns have been observed that are reminiscent of overlapped patterns shifted by a few nucleotides; these may result from the cis cleavage activity of phasiRNAs. For the 21-PHAS loci, phasiRNAs loaded into a cleavage-competent AGO may target transcripts in cis, that is, from the cognate PHAS loci, resulting in a new round of phasiRNA biogenesis and forming a 21-nucleotide phasing pattern offset from the primary pattern (Tamim et al., 2018). This offset was calculated and observed as a 9-nucleotide shift, with the superposition of two different phasing patterns generating a phasing pattern offset by a 12-nucleotide and 9-nucleotide shift relative to the primary PHAS pattern (Figure 1B; Tamim et al., 2018). Similarly, a transcript yielding 24-nucleotide phasiRNAs that could act in cis would also generate phasiRNAs with an offset, in this case apparently yielding a 12-nucleotide shift between phasing patterns superpositioned at the same locus (Figure 1B; Xia et al., 2019). However, in each case in which these offset patterns indicative of cis activity have been observed, the shifted phasiRNAs are present at low abundance, suggesting that cis-directed cleavage is infrequent.
Variability of PhasiRNA Biogenesis across Species
As mentioned above, for every “rule” of phasiRNA biogenesis, there are observations inconsistent with that rule. In most cases, these confounding observations come from studies using species widely diverged from Arabidopsis. One striking case is from our work in nongrass monocots, including garden asparagus (Asparagus officinalis) and lily (Lilium maculatum). In these species, the analysis of phasiRNAs identified 24-nucleotide reproductive phasiRNAs that corresponded to only one strand of the genomic DNA—that is, these are apparently produced independently of RDR activity (Kakrana et al., 2018). This is possible because the PHAS loci yield lncRNA precursors that contain lengthy inverted repeats, such that the foldback could be processed by a Dicer directly to yield phasiRNAs. Confusingly, most of these loci also lacked the canonical miR2275 target site, or in fact, any evidence of a miRNA trigger, suggesting that (1) these phased siRNAs are not secondary but rather primary siRNAs, and (2) there are as-yet unknown triggering mechanisms for phasiRNAs capable of yielding phasing without an obvious initiation site (Kakrana et al., 2018). The production of 24-nucleotide phasiRNAs without an apparent miRNA trigger was also observed in Solanaceous species, but in that case, phasiRNAs matched to both strands in the genome, which is consistent with RDR activity (Xia et al., 2019). While we and others have focused on the roles of these reproductive phasiRNAs in male organs, reproductive phasiRNAs are also present in the female flowers or organs of some plants (Xia et al., 2015a; Kakrana et al., 2018), suggesting they play a role in female reproduction. Thus, from the context of phasiRNA biogenesis, there is much left to be learned both in terms of elucidating currently unclear steps in the molecular process and from analyses across a broader set of plant species and organs.
Subcellular Localization of PhasiRNA Biogenesis
Where in the cell does the biogenesis of phasiRNAs occur? SGS3 and RDR6 interact and colocalize in membrane-associated granules called “siRNA bodies” (Kumakura et al., 2009). Arabidopsis AGO7 was later found to accumulate in siRNA bodies, which is consistent with the observation that the biogenesis of tasiRNAs from TAS3 occurs in these cytoplasmic membrane structures (Jouannet et al., 2012). Two studies have implicated the membrane system of the endoplasmic reticulum (ER) as the site of phasiRNA biogenesis. Li et al. (2016) combined genomic approaches with cellular fractionation experiments to compare sRNA sequencing data from whole Arabidopsis cells, total cellular polysomes (TPs), and membrane-bound polysomes (MBPs). They found that among 22-nucleotide sRNAs, the proportion of 22-nucleotide miRNAs was greater in MBPs than in TPs. Also, the 21- and 22-nucleotide sRNAs enriched in MBPs relative to TPs overlapped with MIR (miRNA precursor) and TAS genes; the authors concluded that 22-nucleotide miRNAs that trigger the biogenesis of 21-nucleotide tasiRNAs are localized to the MBP (Li et al., 2016). The data are consistent with the notion that AGO1 associates with membranes, partly in an RNA-independent manner, to recruit miRNA triggers and direct the cleavage of target transcripts (Li et al., 2016). Interestingly, analysis of ribosome-protected transcripts from MBPs revealed an association with TAS transcripts, which are thought—as lncRNAs—to lack protein-coding capacity. The authors thus proposed that ribosomes function to specify or expose the phasiRNA-generating precursor to the biogenesis machinery via the binding of PHAS precursor transcripts (Li et al., 2016). These conclusions were mirrored in a global analysis of ribosome-protected mRNA fragments (Hou et al., 2016). This work found that TAS3 is bound by ribosomes and that miR390-directed binding of AGO7 to TAS3 deters ribosome movement; these observations support the notion that tasiRNA biogenesis from TAS3 occurs on the membrane system of the ER, to which ribosomes are attached (Hou et al., 2016).
Mobility of PhasiRNAs
Plant RNAs are capable of both cell-to-cell short-distance movement via plasmodesmata or exosome-like vesicles and long-distance migration through the phloem to function in plant development, stress responses, and many other physiological processes (Liu and Chen, 2018). sRNAs, including both miRNAs and siRNAs, are also mobile (Liu and Chen, 2018; Shahid et al., 2018; Tsikou et al., 2018). miRNAs such as miR165/166 and miR394 move from the cells that produce them to neighboring cells and function in a dose-dependent or signaling gradient manner (Carlsbecker et al., 2010; Knauer et al., 2013). The diffusion of tasiARFs, the functional tasiRNAs produced from the miR390-TAS3 module, is essential for the establishment of adaxial-abaxial leaf polarity in Arabidopsis (Chitwood et al., 2009). The authors demonstrated that miR390 is expressed in both the adaxial and abaxial sides of Arabidopsis leaves, whereas tasiARFs are produced only in the adaxial sides of leaves. A gradient of tasiARFs was observed by in situ hybridization, reflecting diffusion from the adaxial to abaxial side of the leaf, suggesting that movement of the tasiARFs functions as a non-cell-autonomous silencing signal (Chitwood et al., 2009). Skopelitis et al. (2017) found that concentration gradients generated by the non-cell-autonomous movement of miR166 and tasiARF create sharply defined domains of target gene expression, which contribute to the formation of robust developmental boundaries.
More recent experiments have uncovered a mechanism of sRNA mobility. Artificial miRNAs (“miRGFP”) targeting GFP-encoding transcripts driven by different tissue-specific promoters were transformed into Arabidopsis constitutively expressing GFP (Skopelitis et al., 2018). By detecting the spread of GFP silencing, the authors found that miRGFP movement was directional at defined cell–cell interfaces, a so-called “gating mechanism.” This polarized gating mechanism restricted long-distance movement of the miRGFP by limiting miRNA transport into phloem companion cells, reflecting domain-autonomous behaviors within stem cell niches (Skopelitis et al., 2018). Whether this gating mechanism functions for phasiRNAs or other sRNAs remains unclear. In general, siRNAs are thought to be more mobile than miRNAs, as miRNA functions may be restricted to the cells in which they are produced (Liu and Chen, 2018). de Felippes et al. (2011) set out to compare differences in mobility between tasiRNAs and miRNAs using either a miRNA or TAS precursor designed to yield the same sRNA; this was expressed in phloem companion cells to target a reporter gene (CH42, a SUL homolog) in leaf mesophyll cells to produce a bleached phenotype. Silencing caused by tasiRNAs led to greater bleaching than silencing caused by miRNAs, suggesting greater movement of the tasiRNAs (de Felippes et al., 2011). One possible explanation is that tasiRNA biogenesis occurs on membrane-bound polysomes (see above); this may facilitate tasiRNA delivery to adjacent cells through plasmodesmata, which are an extension of the cell membrane system.
sRNAs can also move across species boundaries, although it is not clear whether phasiRNAs are more mobile than miRNAs in this regard. This notion is discussed in more detail below concerning their role in biotic stress responses. Cai et al. (2018) observed that Arabidopsis delivers sRNAs, including tasiRNAs and miRNAs, to the pathogen Botrytis cinerea via exosome-like vesicles. There is evidence for specific enrichment of some tasiRNA sequences in vesicles relative to their abundance in host cells, which is consistent with selective loading (Cai et al., 2018). However, independent work also utilizing Arabidopsis found no evidence of specificity in vesicle-localized sRNAs for phasiRNAs (Baldrich et al., 2019), perhaps due to the use of different vesicle isolation protocols. However, a subset of miRNAs demonstrated differential accumulation in vesicles, perhaps also supporting specificity in vesicle loading (Baldrich et al., 2019). In another study, PPR-derived phasiRNAs were transported from Arabidopsis to Phytophthora via vesicles, guiding gene silencing in the pathogen to confer resistance (described in more detail below; Hou et al., 2019). Work on the parasitic plant dodder (Cuscuta campestris) demonstrated that miRNAs can move from the parasite to trigger phasiRNA production from targets in the host cells (Shahid et al., 2018). Therefore, data from a number of sources and systems demonstrate varying degrees of mobility for both phasiRNAs and their miRNA triggers between adjacent cells and over long distances.
EVOLUTION OF phasiRNAs AND THEIR miRNA TRIGGERS
PhasiRNAs, their biogenesis components, and the proteins needed for their function apparently evolved with the emergence of land plants. While PHAS loci that primarily generate 21-nucleotide phasiRNAs are reportedly present in the single-cell green alga Chlamydomonas reinhardtii (Zheng et al., 2015), our own analyses indicated that these are likely artifacts derived from mis-scoring dense tracts of repeat-derived siRNAs. Additionally, no PHAS loci (or miRNAs) are conserved between algae and land plants, or at least none that have been confidently described. PIWI-associated RNAs (piRNAs) in animals show a low degree of phasing and lack precision in length (Han et al., 2015), unlike the exactly 21-nucleotide or 24-nucleotide sRNAs generated from the trigger-directed mechanism of plant phasiRNA biogenesis.
Among all PHAS loci identified to date in land plants, the miR390-TAS3-ARF pathway is the most conserved and archetypal. The three components, miR390, TAS3, and the ARF targets, are present in one of the oldest known land plants, a liverwort (Figure 2; Xia et al., 2017). TAS3 is arguably the most conserved lncRNA in plants. In vascular plants, the tasiARF of TAS3 genes likely originated from a miR390 target site (Xia et al., 2017). miR390 is somewhat unusual as a 21-nucleotide trigger of phasiRNAs, with conserved features including a hairpin structure and a 5′ adenine in the mature miRNA (Xia et al., 2017). The AGO partner, AGO7, evolved only in seed plants (Xia et al., 2017). Evolutionary analysis identified a major change in the functional tasiRNAs that occurred after the split of gymnosperms from their common ancestor with mosses (Xia et al., 2017). The tasiARF target sites in ARF2/3/4 genes are under strong selection for conservation, reflecting their functional importance (Xia et al., 2017).
Figure 2.
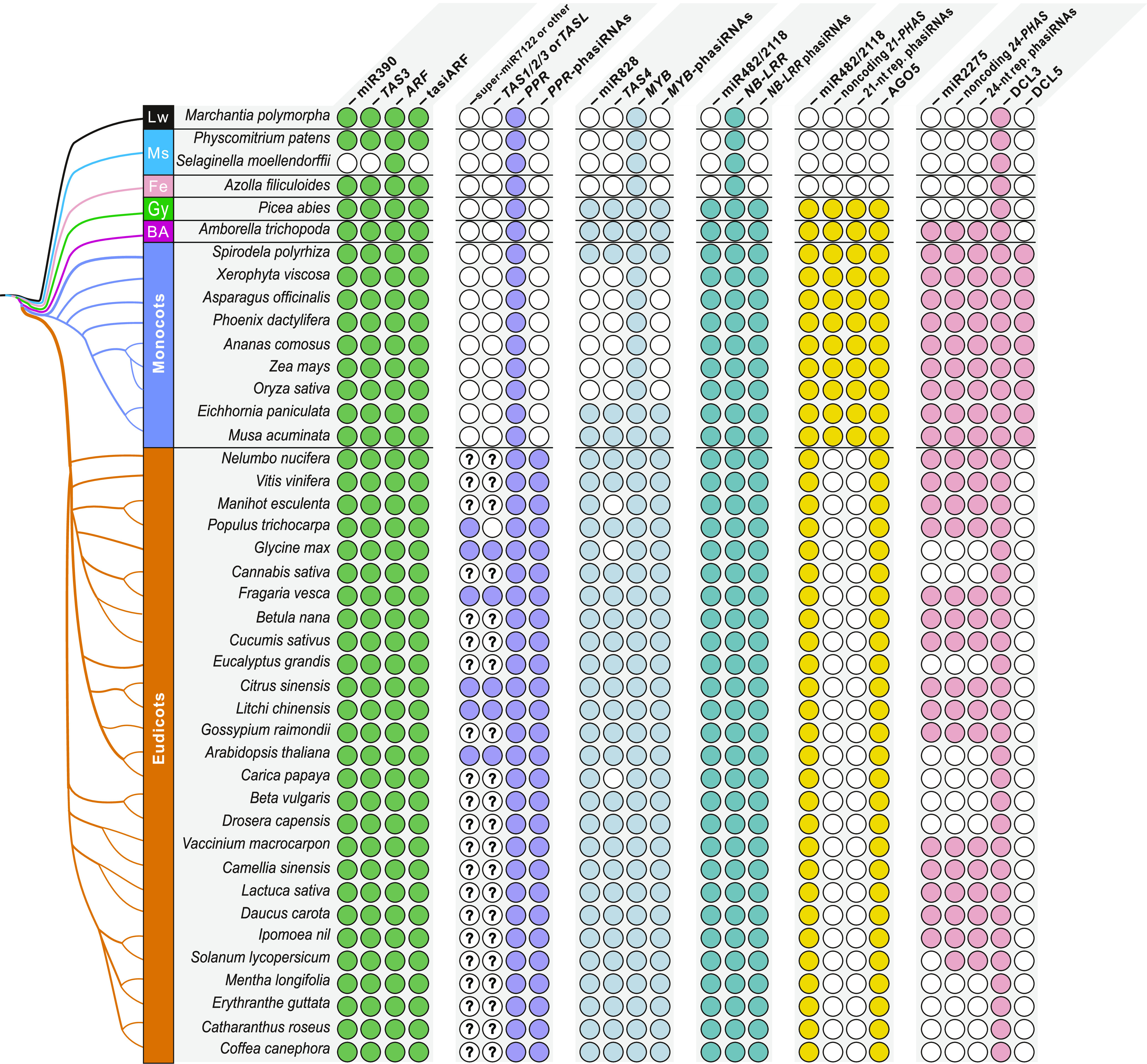
Conservation of PhasiRNA Pathways in Land Plants.
A phylogenetic tree of representative plant species is shown on the left; major groups in the top left are Lw, liverworts; Ms, mosses; Fe, ferns; Gy, gymnosperms; and BA, basal angiosperms. Components of phasiRNA pathways, including trigger miRNAs, PHAS loci, phasiRNAs, and relevant proteins are listed across the top, grouped by pathways. Below these headings, colored circles indicate that the component is present in the species, while empty circles indicate their absence. A circle with a question mark indicates that the triggering miRNA is still unknown.
PPR genes in plants produce profuse phasiRNAs, especially in eudicots (Figure 2). The pathway for the generation of PPR phasiRNAs includes numerous, diverse trigger miRNAs. The miRNA superfamily super-miR7122 was identified as a major group of triggers, including miR173 in Arabidopsis, miR7122 in the Rosaceae, and miR1509 in legumes (Xia et al., 2013). In addition, some PPR phasiRNAs are tertiary siRNAs whose production is activated by tasiRNA triggers from noncoding TAS or TAS-like (TASL) genes. For instance, miR7122 is able to target PPR transcripts directly or via phasiRNAs from TASL genes, and miR1509 can trigger PPR phasiRNA production via two layers of TASL-tasiRNA interactions (Xia et al., 2013). Interestingly, super-miR7122 miRNAs share a common origin with miR390 and with miRNAs of the miR4376 superfamily, which initiate phasiRNA generation from Ca2+-ATPase genes (Xia et al., 2013). Other unrelated miRNAs were also identified as triggers of PPR phasiRNAs (Xia et al., 2015a). This great diversity of miRNA triggers might be due to the high sequence divergence of PPR genes in plants.
Another relatively highly conserved phasiRNA pathway is the miR828-(TAS4)-MYB module. This regulatory circuit is widely present in seed plants but was likely lost in a few monocot lineages, including grasses (Figure 2). TAS4 loci might have evolved from MIR828 or MYB pseudogenes via neofunctionalization, and they are apparently missing in certain eudicot species (Rock, 2013). Unlike most other 22-nucleotide miRNAs, miR828 is produced from a precursor without an asymmetric bulge in the miRNA/miRNA duplex, implying that its biogenesis occurs via a mechanism other than that based on precursor structure or via a noncanonical DCL protein, such as DCL2 (Wang et al., 2018).
Analyses across diverse plant genomes have shown that NLR genes comprise the largest gene family that produces phasiRNAs (Fei et al., 2013). One group of primary trigger miRNAs, the miR482/2118 superfamily, apparently emerged in gymnosperms via a mechanism involving the tandem duplication of their target NLR genes (Xia et al., 2015a). The miR482-NLR-phasiRNA pathway is widely conserved in seed plants, although to date, the phenomenon of phasiRNA production from NLR genes appears to be more widespread in eudicot than monocot genomes. Many other miRNAs evolved independently to target NLR and trigger phasiRNA production (Zhang et al., 2016), notably in Norway spruce (Picea abies; Xia et al., 2015a). Interestingly, the diversification of NLR genes drives the evolution of the trigger miR482/2118, likely because the encoded P-loop region targeted by miR482/2118 is required for pathogen defense (Zhang et al., 2016).
The miR482/2118 superfamily is unusual because in addition to its widespread role in targeting NLR genes for phasiRNA production, in many species but particularly grasses, miR482/2118 preferentially targets lncRNA transcripts from (in many grass genomes) hundreds to thousands of genomic loci, instigating 21-nucleotide “reproductive phasiRNA” production (Johnson et al., 2009; Song et al., 2012; Zhai et al., 2015). The 21-nucleotide reproductive phasiRNAs are principally enriched in early-stage anthers and are thus known as premeiotic reproductive phasiRNAs (Zhai et al., 2015). A clue to their origin might be found in Norway spruce, a gymnosperm in which miR428/2118 initiates phasiRNA production from both NLR genes and noncoding transcripts in reproductive tissues (male or female cones; Xia et al., 2015a). Therefore, perhaps these dual functions of the miR482/2118 superfamily evolved concurrently but were differentially preserved in many angiosperm species (Xia et al., 2015a).
In addition to the 21-nucleotide reproductive phasiRNAs, Johnson et al. (2009) described a second, 24-nucleotide class of reproductive phasiRNAs. These 24-nucleotide phasiRNAs are also highly enriched in anthers, coincident with meiosis (and are therefore known as “meiotic phasiRNAs”; Zhai et al., 2015). The biogenesis of 24-nucleotide phasiRNAs typically requires miR2275 as a trigger that also primarily targets lncRNA precursors. Both miR2275 and PHAS transcripts are thought to have emerged only in monocots (possibly only in grasses), as 24-nucleotide phasiRNAs are absent in a number of well-studied eudicots, including Arabidopsis and soybean (Glycine max; Zhai et al., 2015). We recently found that the 24-nucleotide reproductive phasiRNA pathway is widely present in eudicots, although it is absent in many eudicot plant families (Figure 2; Xia et al., 2019), and miR2275 was reported in eudicots (Polydore et al., 2018). In contrast to monocots, in which a Dicer protein (DCL5, a DCL3 homolog) evolved to be responsible for the biogenesis of 24-nucleotide phasiRNAs, eudicots apparently lack DCL5, suggesting that DCL3 may perform this activity (Xia et al., 2019), in addition to its role in producing heterochromatic or “Pol IV” siRNAs. DCL5 is absent in basal angiosperm genomes including Amborella, and thus the appearance of DCL5 in monocots is consistent with subfunctionalization after gene duplication of DCL3. There is apparently substantial variation in how 24-nucleotide reproductive phasiRNAs are generated, including via hairpin transcript precursors that lack miR2275-directed cleavage in asparagus (Kakrana et al., 2018) and from precursors that seem to lack miRNA target sites in tomato (Solanum lycopersicum; Xia et al., 2019).
FUNCTIONS AND ROLES OF phasiRNAs
PhasiRNA Pathways in Plant Development
Plant phasiRNAs are known or predicted to function in diverse biological processes (Table 1). Yet, given the large number of PHAS loci and populations of phasiRNAs in many species, their precise functions are surprisingly poorly described. The miR390-TAS3-ARF pathway is an exception, with known roles in plant development, including leaf morphogenesis, developmental timing and patterning, lateral root growth, and somatic embryogenesis (Adenot et al., 2006; (Fahlgren et al., 2006; Marin et al., 2010; Cho et al., 2012; Yifhar et al., 2012; Zhou et al., 2013; Lin et al., 2015; Hobecker et al., 2017). Its function in leaf morphogenesis is conserved in plants, with diverse phenotypic variations observed when the pathway is disordered. For example, loss of function of AGO7 or DCL4 in tomato blocked the biogenesis of tasiARFs and increased the levels of ARF transcripts, causing a wiry leaf syndrome, whereas overexpressing these ARFs in Arabidopsis, tobacco, and potato (Solanum tuberosum) failed to produce similar wiry leaves (Yifhar et al., 2012). An ago7 M. truncatula mutant displays lobed leaf margins (Zhou et al., 2013), and the maize (Zea mays) ragged seedling2 mutant (an ago7 mutant) displays cylindrical leaves with dorsiventral polarity (Douglas et al., 2010). These observations are consistent with the notion that the tasiARF pathway functions with many endogenous variants to confer developmental plasticity across species (Figure 3). In rice (Oryza sativa) and maize, tasiARFs and miR166 work together and are thought to be involved in shoot meristem initiation and the maintenance of leaf polarity (Nagasaki et al., 2007; Nogueira et al., 2007; Petsch et al., 2015). miR167 also targets ARFs and triggers secondary phasiRNA biogenesis (Kakrana et al., 2018; Ma et al., 2018). This regulatory module is involved in pollen release through various hormonal pathways (Ru et al., 2006; Wu et al., 2006), but the function and regulatory mechanism of ARF-derived phasiRNAs remain unknown. Another relatively highly conserved phasiRNA pathway is the one in which miR828 targets TAS4 or MYB transcripts and directs the production of phasiRNAs that reinforce the regulation of MYBs (Xia et al., 2012). These MYB TFs are associated with anthocyanin and lignin biosynthesis pathways, bioflavonoid biosynthesis, and fruit development (Xia et al., 2012; Rock, 2013). In addition, the targeting of GhMYB2 in cotton (Gossypium hirsutum) by miR828 and miR858 plays an important role in fiber development (Guan et al., 2014). Thus, even conserved phasiRNA pathways contribute to phenotypic diversity during development (Figure 3).
Table 1. Summary of Plant Loci Generating PhasiRNAs.
| Biogenesis Mode | Trigger miRNA | Trigger Length | Trigger-AGO | Target Gene (PHAS Loci) | DCL and phasiRNA Length | Secondary Target Gene | Biological Role | Plant Lineage | Reference(s) |
|---|---|---|---|---|---|---|---|---|---|
| One-hit | miR173 | 22-nt | AGO1 | TAS1 | DCL4; 21-nt | HTT | Thermotolerance | Arabidopsis | (Li et al., 2014) |
| One-hit | miR173/miR161a | 22-nt | AGO1 | TAS1/TAS2/PPRs | DCL4; 21-nt | Pathogen genes | Biotic resistance | Arabidopsis | (Cai et al., 2018; Hou et al., 2019) |
| Two-hit | miR390 | 21-nt | AGO7 | TAS3 | DCL4; 21-nt | ARF | Auxin signaling | Land plants | (Xia et al., 2017) |
| One-hit | miR828 | 22-nt | AGO1 | TAS4, MYB | DCL4; 21-nt | MYB | Trichome development; secondary metabolism; seed development | Eudicots | (Xia et al., 2012; Guan et al., 2014; Shuai et al., 2016) |
| One-hit | miR828 | 22-nt | AGO1 | MYB | DCL4; 21-nt | FAR1, PPR | Light signal | Populus | (Shuai et al., 2016) |
| One-hit | miR828 | 22-nt | AGO1 | MYB/TLD | DCL4; 21-nt | MYB/TLD | Wounding response | Populus | (Shuai et al., 2016) |
| Two-hit | miR156 and miR529b | 21-nt | AGO1 | TAS6 | DCL4; 21-nt | Zinc finger domain transcripts | Developmental timing | Moss | (Arif et al., 2012; Cho et al., 2012) |
| One-hit | miR173/miR7122/miR1509a | 22-nt | AGO1 | PPR, TASL | DCL4; 21-nt | PPR | Unknown | Eudicots | (Xia et al., 2013) |
| One-hit | miR482/miR1507/miR2109/miR2118/miR9863a | 22-nt | AGO1 | NLR/TAS5 | DCL4; 21-nt | NLR | Disease resistance | Angiosperms | (Zhai et al., 2011; Li et al., 2012; Liu et al., 2014; Wu et al., 2015) |
| One-hit | miR482/miR1507/miR1510a | 22-nt | AGO1 | NLR | DCL4; 21-nt | NLR | Nodule development | Soybean | (Zhai et al., 2011) |
| One-hit | miR2275 | 22-nt | AGO1 | Noncoding RNA | DCL3(?) and DCL5; 24-nt | Unknown | Reproductive growth | Angiosperms | Zhai et al., 2015, Teng et al., 2020, Xia et al., 2019) |
| One-hit | miR2118 | 22-nt | AGO1 | Noncoding RNA | DCL4; 21-nt | Unknown | Reproductive growth | Monocots or angiosperms | (Zhai et al., 2015; Fan et al., 2016) |
| One-hit | miR4392 | 22-nt | AGO1 | Noncoding RNA | DCL4; 21-nt | LTR retrotransposons | Reproductive growth | Soybean | (Arikit et al., 2014) |
| One-hit | miR4376 | 22-nt | AGO1 | ACA10 (Ca2+-ATPase) | DCL4; 21-nt | Unknown | Reproductive growth | Solanaceae | (Wang et al., 2011) |
| One-hit | miR1514 | 22-nt | AGO1 | NAC | DCL4; 21-nt | NAC | Drought response | Legumes | (Sosa-Valencia et al., 2017) |
| One-hit | miR6445 | 22-nt | AGO1 | PtPHAS18, NAC | DCL4; 21-nt | NAC | Drought response | Populus | (Shuai et al., 2016; Xie et al., 2017) |
| One-hit | miR3954 | 22-nt | AGO1 | Noncoding RNA, NAC | DCL4; 21-nt | NAC | Flowering time | Citrus | (Liu et al., 2017) |
| One-hit | miRFBX cluster | 22-nt | AGO1 | F-BOX | DCL4; 21-nt | F-BOX | Fruit shape | Strawberry | (Xia et al., 2015b) |
| One-hit | miR9470-3p | 22-nt | AGO1 | Sly-TAS9 | DCL4; 21-nt | Unknown | Chilling response | Solanaceae | (Zuo et al., 2017) |
| One-hit | miR9678 | 22-nt | AGO1 | WSGAR | DCL4; 21-nt | Germination genes | Seed germination | Wheat | (Guo et al., 2018) |
| One-hit | miR12480 | 22-nt | AGO1 | SEOR1 | DCL4; 21-nt | Host genes | Parasitism | Cuscuta | (Shahid et al., 2018) |
| One-hit | miR2118 | 22-nt | AGO1 | SGS3 | DCL4; 21-nt | SGS3 | sRNA biogenesis | Medicago, soybean | (Zhai et al., 2011; Arikit et al., 2014) |
| One-hit | miR1507/miR1515a | 22-nt | AGO1 | DCL2 | DCL4; 21-nt | DCL2 | sRNA biogenesis | Medicago | (Zhai et al., 2011) |
| One-hit | miR1885 | 22-nt | AGO1 | TAS-like (BraTIR1) | DCL4; 21-nt | BraCP24 | Flowering | Brassica | (Cui et al., 2020) |
| One-hit | miR1885 | 22-nt | AGO1 | TIR-NBS-LRR | DCL4; 21-nt | TIR-NBS-LRR | Disease resistance | Brassica | (Cui et al., 2020) |
Note that AGO interactions, Dicers, biological roles, and species distributions are in some cases inferred rather than definitely proven. nt, nucleotide. Brassica (Brassica rapa); Populus (Populus trichocarpa); Strawberry (Fragaria × ananassa).
These PHAS loci could be triggered by different miRNAs.
This PHAS locus is targeted by miR156 and miR529 in two target sites.
Figure 3.
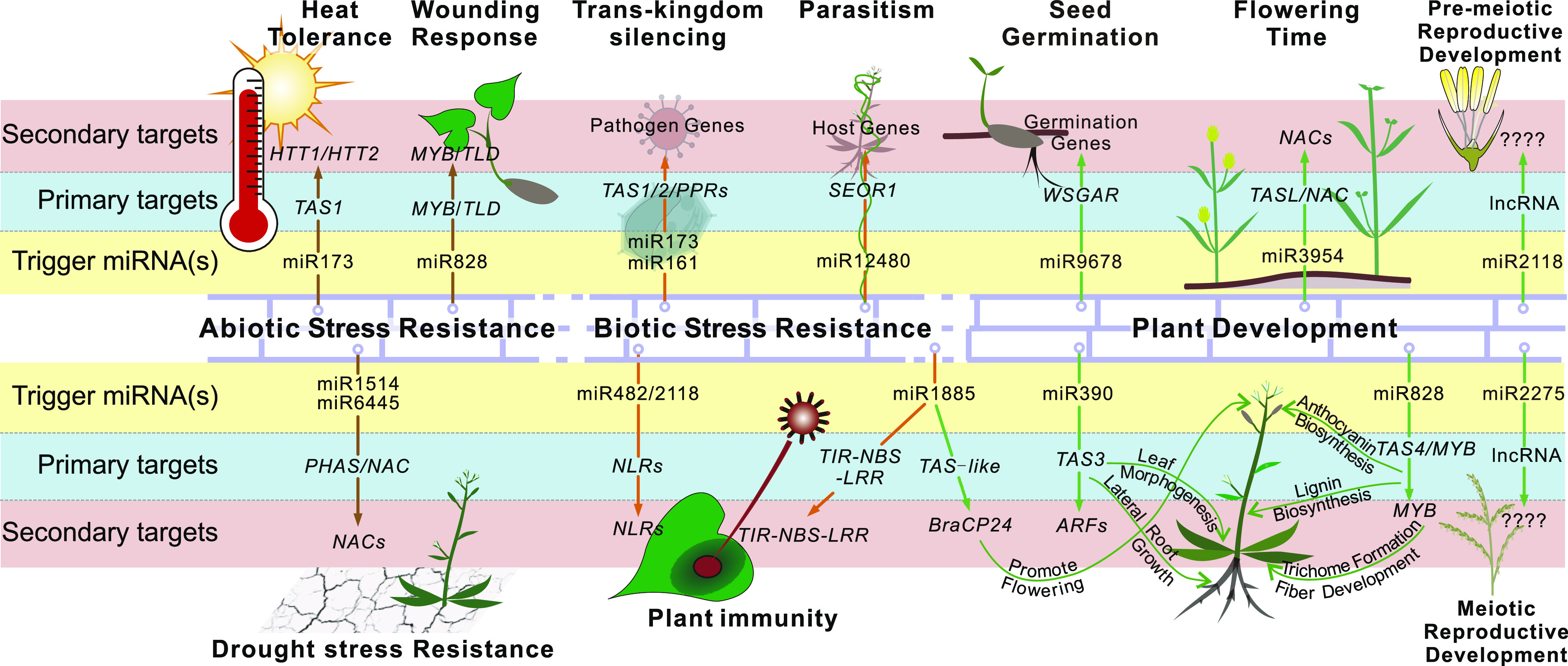
Roles of PhasiRNAs in Plant Stress Responses and Development.
Regulatory cascades mediated by phasiRNAs, largely described in this review, are represented as three layers for each pathway, in which the yellow, blue, and pink highlighting indicate the trigger miRNA(s), the primary target transcripts that are the precursors generating phasiRNAs, and the secondary target genes that are regulated by phasiRNAs, respectively.
Nonconserved or lineage-specific phasiRNA pathways play other roles in plant development. miR3954 triggers phasiRNAs from NAC genes or lncRNAs and regulate a set of NAC genes in citrus (Citrus sinensis) and litchi (Litchi chinensis; Liu et al., 2017; Ma et al., 2018). Overexpressing miR3954 increased the abundance of phasiRNAs and reduced the expression of NAC genes, resulting in early flowering in citrus (Liu et al., 2017). miR9678, a wheat (Triticum aestivum)–specific miRNA that is expressed only in the scutellum of developing and germinating seeds, targets an lncRNA (WSGAR) to trigger phasiRNA production (Guo et al., 2018). Overexpressing miR9678 decreased the level of WSGAR, thereby delaying immature embryo generation, whereas silencing miR9678 increased germination (Figure 3; Guo et al., 2018). Guo et al. (2018) found that abscisic acid signaling proteins activate the expression of miR9678, which affects the expression levels of genes associated with gibberellic acid homeostasis, suggesting that miR9678 influences the regulation of germination via abscisic acid–gibberellic acid crosstalk. However, the downstream target genes of WSGAR-derived phasiRNAs are still unknown, leaving unclear the mechanism by which these phasiRNAs function.
PhasiRNA Pathways in Biotic Resistance
We previously reviewed the roles of phasiRNAs from NLR genes and their potential roles in plant immunity (Fei et al., 2013). However, the past 7+ years have brought new insights. NLR genes possess a common target site for miRNAs from the miR482/2118 superfamily in the region encoding the functionally important P-loop motif (Shivaprasad et al., 2012; Fei et al., 2015; Xia et al., 2015a). This targeting relationship between miR482/2118 and NLR genes is conserved in seed plants (Xia et al., 2015a) and is likely an important component in plant immunity (Fei et al., 2016b, 2013); yet, studies supporting this notion have only been performed in Arabidopsis (Boccara et al., 2014).
It is now clear that over evolutionary time, there has been constant generation of miRNA triggers of phasiRNAs that target disease resistance genes (R genes), supporting the importance of this function. These miRNAs include miR1507 and miR1510 in legumes (Zhai et al., 2011; Fei et al., 2015; Zhao et al., 2015) and miR6019 and miR6027 in Solanaceae species (Li et al., 2012). miR9863, identified in barley (Hordeum vulgare) and wheat, regulates MLA genes that encode a coiled-coil–type NLR protein; miR9863 triggers the production of phasiRNAs that are critical for the regulation of MLA (Liu et al., 2014). In Norway spruce, the miR482/2118 family includes 23 members, and as many as 18 novel 22-nucleotide miRNAs target NLR genes to generate phasiRNAs (Xia et al., 2015a). Thus, miRNAs and secondary phasiRNAs constitute key components in plant immunity (Fei et al., 2013, 2016b).
Plant NLR genes direct effector-triggered immunity by inducing programmed cell death (PCD) in the host plant (Cui et al., 2015). In the absence of a pathogen, these trigger miRNAs and the resulting NLR-derived phasiRNAs may keep NLR transcripts and thus NLR proteins at low levels to minimize auto-activation and undesired PCD. During infection, the levels of miR482/2118 (and subsequently phasiRNAs) may drop to release the expression of NLR genes and enhance effector-triggered immunity (Fei et al., 2016b). In tomato, using transgenic lines expressing short tandem target mimic RNAs against miR482/2118, Canto-Pastor et al. (2019) demonstrated that miR482 targets a conserved motif in NLR transcripts, while miR2118 directs the cleavage of a noncoding transcript, triggering the biogenesis of phasiRNAs. phasiRNAs derived from both cascades and miR482/2118 regulate NLRs, but tomato short tandem target mimic lines showed enhanced resistance to pathogens (Figure 3; Canto-Pastor et al., 2019) In addition, Cui et al. (2020) found that 22-nucleotide miR1885 from Brassica targets an R gene (BraTNL1) encoding a TIR-NBS-LRR disease-resistance protein and transcripts of a TAS gene (BraTIR1) that potentially encodes only a TIR domain. The sRNA phasiR130-4 derived from BraTIR1 targets the photosynthesis and flowering-related gene BraCP24. Under natural conditions, low levels of miR1885 maintain immunity and vegetative growth, but the abundance of miR1885 increases during the floral transition to promote flowering (Cui et al., 2020). Upon viral infection, miR1885, BraTNL1, and BraTIR1 transcript levels all increase, thereby increasing the levels of phasiR130-4 and leading to the repression of BraCP24 and early flowering. Viral-induced BraTNL1 levels overwhelm miR1885-mediated repression, resulting in increased immunity (Figure 3). These recent observations provide insights into the novel regulatory roles of NLR-associated miRNAs and phasiRNAs.
Recent studies in Arabidopsis demonstrated that phasiRNAs from PPR genes are also involved in biotic resistance (Figure 3; Cai et al., 2018; Hou et al., 2019). Cai et al. (2018) found that when Arabidopsis is infected by the fungal pathogen B. cinerea, tasiRNAs from TAS1c and TAS2 are transported from the plant host to the pathogen via exosome-like extracellular vesicles to induce the silencing of fungal pathogenic genes. tasiRNA overexpression enhanced plant resistance to the pathogen, whereas lines with reduced tasiRNA levels showed hypersusceptibility to B. cinerea (Cai et al., 2018). Similar anti-pathogen roles of PPR-derived phasiRNAs were demonstrated against the oomycete pathogen Phytophthora capsici. Constitutive expression of the Phytophthora suppressors of RNAi in transgenic Arabidopsis reduced the levels of almost all PPR-phasiRNAs (Hou et al., 2019). miR161 contributes to plant defenses against P. capsica by triggering the production of PPR-siRNAs; plants with increased levels of miR161-triggered phasiRNAs showed enhanced resistance, while plants with reduced levels showed hypersusceptibility to this pathogen (Hou et al., 2019). Both of these studies describe roles of PPR-derived phasiRNAs in defense, but how do they achieve such a broad effect on diverse pathogens? Hou et al. (2019) speculated that phasiRNAs generated from noncoding genes or genes within large families (such as PPRs) have high sequence complexity and transcript abundances, which could potentially overcome the more rapid, selection-driven diversification of pathogen target genes. PPRs are indeed one of the largest and most diverse plant gene families (Schmitz-Linneweber and Small, 2008). However, much work remains to be done, as only a subset of these PPRs generate phasiRNAs, and the pathogen target genes of PPR phasiRNAs have not been thoroughly examined.
miRNAs and phasiRNAs are also involved in plant parasitism. Shahid et al. (2018) found that a parasitic plant (Cuscuta campestris) uses trans-species silencing to repress transcript production in the host plant, thereby facilitating its parasitism. A group of 22-nucleotide novel miRNAs from Cuscuta accumulate to high levels in haustoria while parasitizing Arabidopsis. These miRNAs direct the cleavage of target transcripts in the host (Arabidopsis), thereby triggering phasiRNA production. These miRNAs have no endogenous C. campestris targets, which is consistent with a role only in pathogenic, trans-species gene silencing (Shahid et al., 2018). The mutation of two of these target genes in Arabidopsis promoted the growth of C. campestris (Figure 3; Shahid et al., 2018). However, there was no significant difference in C. campestris growth on dcl4 or sgs mutants compared to the wild type, implying that the absence of phasiRNAs triggered by C. campestris miRNAs does not affect parasitism, or perhaps other factors are involved in this process (Shahid et al., 2018). Therefore, the biological relevance of these phasiRNAs requires further investigation.
miRNAs and phasiRNAs from host plants appear to work synergistically to optimize plant health and/or counter pathogen infection, either by tuning the endogenous immune system (target NLR genes) or via export to pathogens to silence their genes. Additionally, this system of silencing is hijacked by parasitic plants to facilitate their parasitism. In every case, numerous questions remain about how, where, and to what effect these sRNA act.
PhasiRNA Pathways in Abiotic Resistance
PhasiRNAs also play roles in abiotic stress resistance (Figure 3). PhasiRNAs triggered by miR173 from Arabidopsis TAS1 genes target two genes involved in heat tolerance, HEAT-INDUCED TAS1 TARGET1 (HTT1) and HTT2. Overexpressing TAS1 boosted the accumulation of the tasiRNAs, resulting in lower thermotolerance (Li et al., 2014). In Populus, phasiRNAs triggered by miR482, miR828, and miR6445 are responsive to drought stress (Shuai et al., 2016). Among these, 22-nucleotide miR6445 is Populus specific and targets NAC genes, yielding phasiRNAs that target additional NAC genes responsive to drought stress (Shuai et al., 2016; Xie et al., 2017). Similarly, legume-specific miR1514a triggers the production of phasiRNAs from NAC 700 during drought stress (Sosa-Valencia et al., 2017). Under well-watered conditions, the NAC 700 TF regulates downstream genes that participate in normal metabolism and growth. However, under water deficit, miR1514a represses the expression of NAC 700, and the NAC 700–derived phasiRNAs NAC 700 3′D2(+), which is conserved among different NAC genes in common bean (Phaseolus vulgaris), is recruited into AGO1 to amplify the silencing of NAC homologs, thereby regulating drought-stress responses (Sosa-Valencia et al., 2017). In sweet potato (Ipomoea batatas), miR828 accumulates in wounded leaves, triggering the production of phasiRNAs from its targets IbMYB and IbTLD that then function in cis to enhance their silencing. Consequently, the repression of IbMYB and IbTLD increases lignin and H2O2 contents to help protect the plant against damage (Lin et al., 2012).
REPRODUCTIVE phasiRNAs
As mentioned above, work over the last decade by our laboratories and others has demonstrated that abundant phasiRNAs are produced in anthers, although their functions are still somewhat unknown. In brief, reproductive phasiRNAs were first described over a decade ago from work focused on rice inflorescences. Limited maize data confirmed the presence of two pathways that produce 21-nucleotide and 24-nucleotide phasiRNAs from large numbers of loci (Johnson et al., 2009). This work came after the heyday of the discovery of the biogenesis pathways of plant sRNAs in the mid-2000s, and since the reproductive phasiRNAs were apparently absent in Arabidopsis, the importance or broader relevance of these sRNAs was arguably overlooked. In fact, for much of the last decade, our conclusion, and perhaps that of others, was that these sRNAs might be a grass-specific aberration—an incorrect idea that shaped our thinking during this time. Based on the analyses of Johnson et al. (2009), we knew that large numbers of loci for lncRNA precursors of 21- and 24-nucleotide reproductive phasiRNAs are found in grass genomes; the authors identified clusters and singleton PHAS loci that were widely distributed in the rice genome without obvious overlaps with protein-coding genes or transposons. Two families of miRNAs were described as the triggers: miR2118 for 21-nucleotide phasiRNAs and miR2275 for 24-nucleotide phasiRNAs (Johnson et al., 2009). Subsequent work from a number of laboratories have led to a better understanding of the biogenesis, evolution, spatiotemporal accumulation and regulation, and functional relevance of these pathways.
Because of the increasingly accessible genomes of diverse seed and flowering plants, it is now clear that reproductive phasiRNAs are widely, but not comprehensively, found in flowering plants (Kakrana et al., 2018; Xia et al., 2019). These data suggest that reproductive phasiRNAs emerged either in conjunction with flowering (i.e., the developmental process and structure) or perhaps earlier (Xia et al., 2015a). Their functional importance to reproductive success is also now evident (see below). Maize anthers are a useful model system, as the all-male tassel is easily staged to obtain specific developmental time points (Kelliher and Walbot, 2011; Kelliher et al., 2014). Unlike most plant sRNAs, the prediction of targets of reproductive phasiRNAs has been relatively uninformative (Song et al., 2012), with targets poorly validated by the parallel analysis of RNA ends. Gene Ontology terms are also poorly enriched among the predicted targets, which is consistent with the notion that reproductive phasiRNAs have distinct activities or roles relative to miRNAs, tasiRNAs, or Pol IV siRNAs (i.e., heterochromatic siRNAs, 24-nucleotide long; Zhai et al., 2015). Our application of machine learning methods to classify sRNAs has demonstrated that reproductive phasiRNAs have distinct features relative to miRNAs and Pol IV siRNAs, features that are conserved across flowering plants (Patel et al., 2018). For example, the 21-nucleotide phasiRNAs have an enriched U/cytidine (C) and depleted guanosine (G) in the 5′ end, with internal positions biased differently from miRNAs; similarly, 24-nucleotide phasiRNAs have a distinctive sequence composition (Patel et al., 2018, Xia et al., 2019). These characteristics may influence the AGO-phasiRNA interaction or reflect interactions with targets.
We and others (Zhai et al., 2015; Komiya, 2017) assert that the plant reproductive phasiRNAs are analogous to metazoan PIWI-associated RNAs. Despite the fact that piRNA biogenesis utilizes an entirely unrelated process (Ozata et al., 2019), animal pachytene piRNAs and plant reproductive phasiRNAs share a striking number of features. These shared features include their enrichment in male reproductive organs and function in male fertility, the presence of both premeiotic and meiotic pathways, phasing, and other less prominent characteristics. piRNAs are now known to accumulate in other tissues and play roles beyond those in reproductive tissues (Liu et al., 2019). In addition, insect and meiotic metazoan piRNAs function in the suppression of transposons (Han et al., 2015), a role seemingly unnecessary in plants given the specialization of the plant-specific Pol IV siRNA pathway for this activity. However, given that phasiRNAs evolved in plants, there is no evidence to support a common origin of phasiRNAs and piRNAs. Until we have more clarity on the specific roles of reproductive phasiRNAs in plants, the selective pressures that drove the emergence of this case of apparent convergent evolution will remain unknown.
Similar to phasiRNAs, 21/22-nucleotide epigenetically activated siRNAs (easiRNAs) are secondary siRNAs that are generated in germ cells from activated transposable elements. The biogenesis of easiRNAs is triggered by miR845b and is dependent on DCL2 and DCL4 during or right after meiosis (Borges et al., 2018; Wang et al., 2020). Interestingly, the depletion of paternal easiRNAs in the Arabidopsis Pol IV mutant nrpd1 bypasses the triploid block in the omission of second division (osd1) mutant, which forms unreduced (2n) male gametes (Martinez et al., 2018). This suggests that easiRNAs could be a quantitative signal for paternal chromosome number and that their presence is required for post-fertilization genome stability and seed viability (Martinez et al., 2018). Even though both phasiRNAs and easiRNAs are secondary siRNAs important for plant reproduction, there is no evidence suggesting they are closely related classes of sRNAs.
In the following sections, we focus on insights from recent progress on the biogenesis, function, and evolution of 21- and 24-nucleotide reproductive phasiRNAs. Table 2 shows some of the major characteristics of plant reproductive phasiRNAs along with key references, as covered in more detail below.
Table 2. Key Characteristics of Reproductive phasiRNAs.
| Features | 21-nt PhasiRNA | References | 24-nt PhasiRNA | Reference(s) |
|---|---|---|---|---|
| Species distribution | Originated with angiosperms or possibly gymnosperms (such as Norway spruce) | ( Xia et al., 2015a; Chen et al., 2019) | Originated in and widespread in angiosperms | (Kakrana et al., 2018; Xia et al., 2019) |
| Tissue specificity and enrichment | Premeiotic anthers | (Zhai et al., 2015; Araki et al., 2020) | Meiotic anthers; also premeiotic anthers in wheat and barley | (Zhai et al., 2015; Xia et al., 2019; Bélanger et al., 2020) |
| miRNA trigger | miR2118 in monocots | ( Xia et al., 2015a; Araki et al., 2020) | miR2275 in most monocots; other angiosperms have diverse modes of biogenesis, with or without miR2275 as a trigger, and with or without RDR6 (i.e., from inverted repeats) | (Kakrana et al., 2018; Xia et al., 2019) |
| miRNA trigger localization | Epidermis | (Zhai et al., 2015; Araki et al., 2020; Huang et al., 2020b) | Tapetum and other layers | (Zhai et al., 2015; Kakrana et al., 2018; Huang et al., 2020b) |
| Dicer for biogenesis and its localization | DCL4, likely in all cell types | (Song et al., 2012) | In grasses, DCL5 in the tapetum; in eudicots, the absence of DCL5 but presence of both 24-nt phasiRNAs and 24-nt Pol IV siRNAs suggests a dual role for DCL3 | (Huang et al., 2020a) |
| PHAS precursor loci | 21-PHAS loci are mostly lncRNAs | (Xia et al., 2015a; Zhai et al., 2015; Araki et al., 2020) | 24-PHAS loci are mostly lncRNAs | (Zhai et al., 2015; Kakrana et al., 2018; Ono et al., 2018; Xia et al., 2019; Bélanger et al., 2020) |
| phasiRNA localization | Tapetum and germ cells | (Zhai et al., 2015; Huang et al., 2020b) | Tapetum and pollen mother cells, meiocytes | (Zhai et al., 2015; Huang et al., 2020b) |
| phasiRNA function | Largely unknown; at least two 21-PHAS loci (PMS1 and PMS3) in rice are required for full male fertility under long-day conditions in N58S rice, rescued with short days; molecular mechanism still unknown, possibly PTGS or transcriptional gene silencing | (Ding et al., 2012; Fan et al., 2016) | Largely unknown; loss of 24-nt phasiRNAs in rice eat1 and maize dcl5 mutants impacts tapetal development and male fertility; dcl5 phenotype rescued under slow-growth environmental conditions; molecular mechanism still unknown | (Ono et al., 2018; Teng et al., 2020) |
nt, nucleotide.
Premeiotic 21-Nucleotide PhasiRNAs
A number of components of the biogenesis pathway for 21-nucleotide reproductive phasiRNAs have now been described, with varying degrees of confirmation and validation. OUTER CELL LAYERS4 (OCL4) is an HD-ZIP IV TF that is expressed in the epidermis of many tissues in maize, including premeiotic anthers, in which OCL4 transcripts are restricted to epidermal cells (Figures 4A and 4B; Vernoud et al., 2009). Anthers of a homozygous ocl4 null mutant have an extra subepidermal layer with endothecium characteristics that results in substantially reduced male fertility (Vernoud et al., 2009). Interestingly, miR2118, 21-nucleotide phasiRNAs, and their long noncoding precursors are almost completely absent in premeiotic anthers of ocl4 (Zhai et al., 2015), suggesting that OCL4 may transcriptionally activate these components in the epidermis. Given the defect in the endothecium of ocl4, these phasiRNAs might function in the specification of somatic cell fate.
Figure 4.
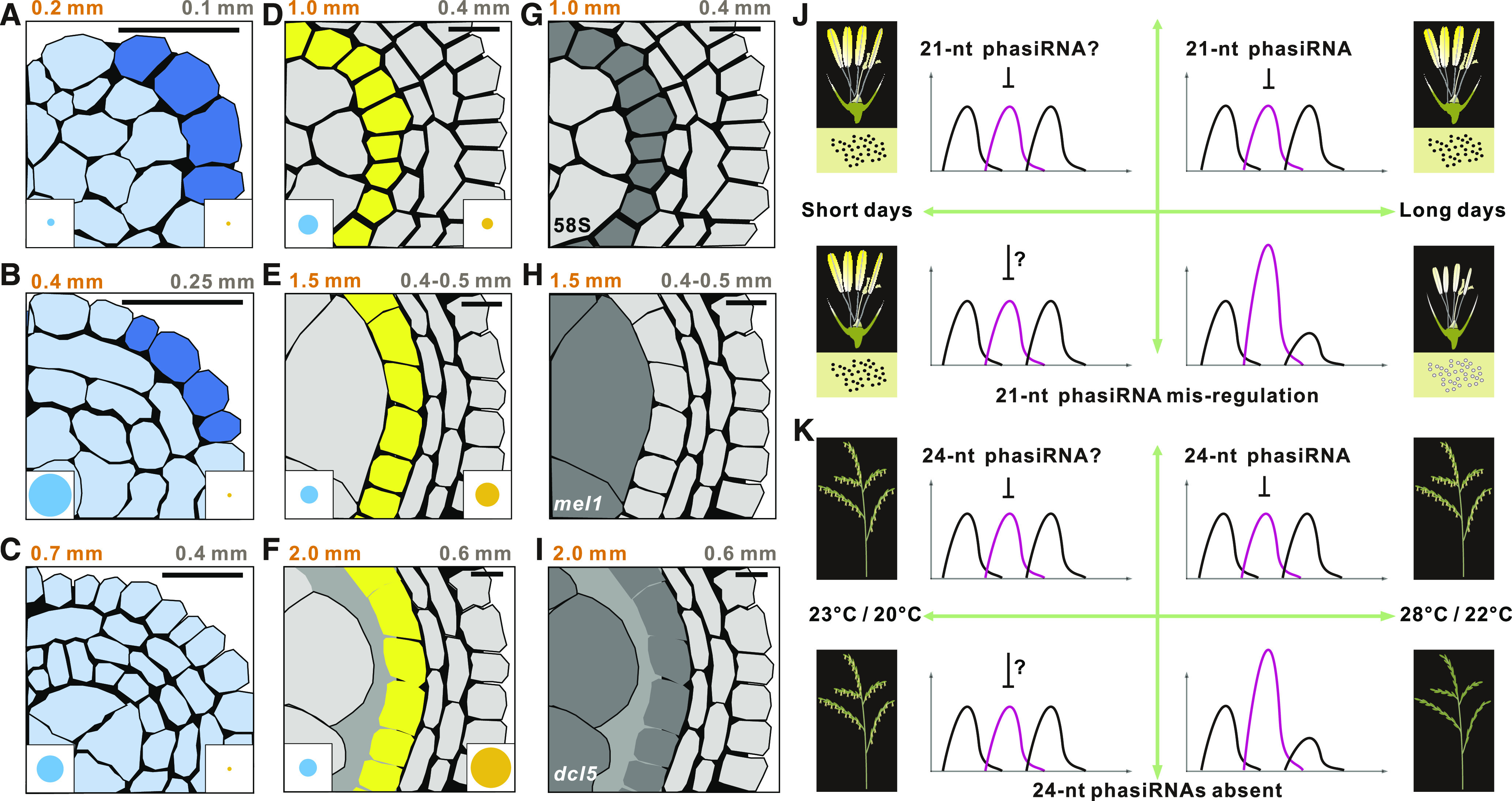
Schematic Representation of the Accumulation and Roles of Reproductive PhasiRNAs.
(A) to (C) Premeiotic 21-nucleotide (nt) phasiRNAs are dependent on a functional epidermis; OCL4, an epidermis-constrained basic leucine zipper–type transcription factor, and the miR2118 triggers are expressed only in epidermal cells (indicated in dark blue); the 21-nt phasiRNAs are enriched in all cell types (indicated in light blue). The lengths of maize (orange) and rice (brown) anthers at the corresponding stages are shown above each panel. In rice and maize, these cells are among those in three-cell-layer (A) and four-cell-layer (B) anthers. The blue circles in the bottom left corner of each panel indicate the abundance of 21-nt phasiRNAs at each stage, with peak levels in all premeiotic stages (see [B] to [D]); data from Zhai et al. (2015).
(D) to (F) Meiotic, 24-nucleotide (nt) phasiRNAs are dependent on a functional tapetal layer (indicated in yellow). These phasiRNAs accumulate during these later stages (abundance indicated by the size of the gold circles in the bottom right corner) and peak in (F); data from Zhai et al. (2015). The miR2275 triggers for 24-nt phasiRNA production are most abundant in the tapetum of premeiotic (1.0 mm in maize) anthers (D) and drop slightly in early meiotic (1.5 mm in maize) anthers (E). The bHLH-type TFs, including MS23 in maize and EAT1 in rice, are expressed specifically in the tapetal cells of early meiotic anthers (E). The 24-PHAS precursors and DCL5 transcripts accumulate in tapetal cells at the same stage (E), with DCL5 protein detected slightly later during this stage (2.0 mm in maize), and are likely constrained to the tapetum (F) when the cell wall of the tapetal cells facing the meiocytes degenerates.
(G) to (K) Under male-sterile-inducing conditions, rice line Nongken 58S shows altered mitochondria, ER, and premature PCD in the tapetum during premeiotic to early meiotic stages (G). The absence of MEL1/AGO5 in rice stops meiotic progression before pachytene (H). The absence of DCL5 causes defective tapetal development and male sterility (I). The defective tapetum of 58S and dcl5, and the defective meiocytes of mel1 and dcl5, are indicated in dark gray. Bar in each image = 10 μm. (J) and (K) represent environmentally induced male sterility associated with perturbed reproductive phasiRNAs; in each of the four quadrants in (J) and (K), the x axis indicates the stages of anther development and the y axis indicates the transcript abundance of direct or indirect phasiRNA targets; in both cases, longer days or higher temperatures (to the right) yield male sterility in the absence of regulation by phasiRNAs (bottom right). These phenotypes are rescued when phasiRNAs are unperturbed (top quadrants) or in a slower growth environment (i.e., shorter days or lower temperatures, left quadrants), perhaps due to the suppressive activity of phasiRNAs or in their absence, a factor related to growth conditions.
(J) In Nongken 58N rice (fertile), optimal (long-day) and permissive (short-day) growth conditions may cause different rates of metabolism/development/growth with no impact on fertility; SNPs in two 21-PHAS loci in 58S cause male sterility under the same conditions.
(K) In a maize dcl5 mutant, typical temperatures or photoperiod conditions for maize growth cause male sterility; lower temperatures or shorter days yield male-fertile mutant plants.
Ken-ichi Nonomura and colleagues (National Institute of Genetics, Japan) characterized an AGO5 family member in rice (MEL1) that functions by loading 21-nucleotide reproductive phasiRNAs. MEL1 binds mostly to 21-nucleotide phasiRNAs in the cytoplasm of the male germ cells (the pollen mother cells [PMCs] or meiocytes) and in the corresponding stage of the female germ cells (Komiya et al., 2014). The maize ortholog of MEL1, AGO5c, is highly expressed in 0.7-mm anthers, just after the peak abundance of 21-nucleotide, premeiotic phasiRNAs (Figure 4C; Zhai et al., 2015), which is consistent with a role as their binding partner. The absence of MEL1/AGO5 in a rice mutant did not affect the production of the 21-nucleotide phasiRNAs, yet markers of meiotic, double-stranded breaks were lost, including H3K9 tri-methylation and the recruitment of histone H2A variant γH2AX (Nonomura et al., 2007). This likely reflects the failure of meiosis to progress into pachytene. Nonomura and colleagues concluded that the 21-nucleotide premeiotic phasiRNAs are produced and bound by MEL1/AGO5 in the cytoplasm of PMCs at the initiation of meiosis, affecting meiotic chromosomal programming, recombination, and synapsis during meiosis (Nonomura et al., 2007; Komiya et al., 2014; Liu and Nonomura, 2016; Ono et al., 2018). However, we wonder why the peak of abundance of the 21-nucleotide phasiRNAs occurs substantially earlier than expected from their role interpreted from the MEL1/AGO5 work. In our studies of the spatiotemporal accumulation of phasiRNAs in maize anthers (Figures 4A to 4F), the 21-nucleotide phasiRNAs accumulate in the just-differentiated tapetum (Zhai et al., 2015). Perhaps the perceived impact on meiosis reflects an indirect and downstream consequence of earlier anther failure, rather than a direct function of the 21-nucleotide phasiRNAs.
In any case, the biogenesis of 21-nucleotide premeiotic phasiRNAs begins with the transcriptional activation of miR2118 and their precursors (from “21-PHAS” loci), apparently directly or indirectly by OCL4 in the epidermis. Like all miRNAs, 22-nucleotide miR2118 is bound by an AGO protein, probably AGO1. Recent work showed that the deletion of many copies of miR2118 in rice destabilizes AGO1b and AGO1d, suggesting a role in the biogenesis or loading of 21-nucleotide premeiotic phasiRNAs (Araki et al., 2020). The 21-PHAS precursors are cleaved, converted by RDR6 to dsRNA substrates, and diced into 21-mers by DCL4 (Song et al., 2012). Analysis of these phasiRNAs demonstrated a highly disproportionate strand specificity, with phasiRNAs from the Pol II strand overrepresented relative to the RDR6-strand phasiRNAs, perhaps reflecting differential stability due to posttranscriptional modifications on the Pol II strand (Tamim et al., 2018). Genetic and in situ data demonstrate that in maize, a differentiated anther epidermis is both necessary and sufficient for 21-nucleotide phasiRNA biogenesis, yet the phasiRNAs accumulate in the tapetum (Zhai et al., 2015). Immunoprecipitation data from rice demonstrate that MEL1/AGO5 (predominantly loaded with 21-nucleotide phasiRNAs) is exclusively localized in the PMCs, which is consistent with the translocation of at least the phasiRNAs (Komiya et al., 2014). MEL1/AGO5 mostly (82%) binds 21-nucleotide phasiRNAs with C at the 5′ end (Komiya et al., 2014), which is indeed one characteristic of these sRNAs (Tamim et al., 2018). How the sRNAs move across cell layers is as-yet unclear (i.e., loaded on AGO, naked, or via plasmodesmata).
Once in the tapetum, the 21-nucleotide phasiRNAs presumably do what other 21-nucleotide sRNAs do in plants: interact with a target RNA to direct cleavage. In fact, there are data supporting the notion that the 21-nucleotide premeiotic phasiRNAs can direct cleavage; analysis of phasing and cleaved mRNA data demonstrate that they act in cis to cleave their own precursors, albeit at a low level (Tamim et al., 2018). While these data demonstrate that 21-nucleotide phasiRNAs can function in cis, since the precursors are made in the epidermis and the phasiRNAs are translocated to the tapetum, presumably they have other trans targets in the tapetal cells, or perhaps in the PMCs. Why haven’t these been identified by bioinformatics or validated by cleavage assays? There may be several reasons: (1) Target identification is confounded because there are thousands of distinct phasiRNAs, and with moderate degeneracy in the target interactions, there may be tens of thousands of possible targets. Perhaps this is the function of these phasiRNAs: to massively disrupt most-to-all transcripts at a particular stage and location during anther development? Even extensive target analysis of these 21-nucleotide phasiRNAs has been largely inconclusive, although demonstrating that if they target genes, this interaction is likely at a lower levels of complementarity to their targets than miRNAs, and they are more likely to target genes than transposons (Patel et al., 2018). (2) Target validation by parallel analysis of RNA ends/degradome analysis is also likely challenging because tapetal cells may comprise only a small portion of all cells in the anther (i.e., one of only five cell layers per lobe, and there is a substantial number of connective cells between lobes), and thus libraries of cleaved RNA targets must be made from isolated tapetal cells, which is currently a major technical challenge. (3) Like pachytene piRNAs in animals (Wu et al., 2020), perhaps only a small number of phasiRNAs in the total population are guides that induce cleavage of target transcripts to promote reproduction. Wu et al. (2020) concluded that the large diversity of piRNAs might selfishly reinforce their own production rather exist to regulate large numbers of mRNAs. Likewise, few individual 21-nucleotide phasiRNAs have been shown to affect male sterility in plants.
The direct function of 21-nucleotide reproductive phasiRNAs thus remains largely speculative. However, a consideration of the features of these sRNAs may help guide speculation. For example, there is substantial variation in the number of loci, indicating no correlation with genome size (or the related characteristic, transposon composition), ranging from just three reproductive-enriched 21-PHAS loci in asparagus to several thousand in rice (Fei et al., 2016a; Kakrana et al., 2018). There is a lack of conservation of phasiRNA sequences across lineages, and even within a single genome most PHAS loci are single copy (Zhai et al., 2015). Maybe this reflects neutral-to-positive selection to diverge rapidly, such that each single phasiRNA has almost insignificant utility and they instead function en masse, as a group. Unlike heterochromatic siRNAs, the sequences of 21-nucleotide reproductive phasiRNAs are generally single copy, with no apparent relationship to transposons (Song et al., 2012; Zhai et al., 2015), which could suggest a genic role. In addition, the presence of the Pol IV pathway might mitigate the need for a secondary pathway for transposon control.
One intriguing exception to the absence of functional data for 21-nucleotide reproductive phasiRNAs is from environmentally sensitive male sterile rice lines, in which male sterility or fertility is modulated by altered photoperiod or temperature during sensitive stages of development (Fan and Zhang, 2018). Two loci have been cloned from the rice line ‘Nongken 58S,’ PMS1 and PMS3, which is used for hybrid rice production via photoperiod-induced male sterility. These two loci generate lncRNA precursors and are targets of miR2118 to yield 21-nucleotide phasiRNAs (Ding et al., 2012; Zhou et al., 2012; Fan et al., 2016). Both loci were mapped and cloned based on the male sterile phenotype, with the relevant polymorphism found to be a single-nucleotide polymorphism (SNP) within one of the 21-nucleotide phasiRNAs. With two SNPs downstream of the miR2118 targeting sites, the production of 21-nucleotide phasiRNAs was altered or better processed in the 58S line, causing male sterility under long-day growth conditions. The SNP at PMS1 is semidominant, as male fertility in the heterozygous plants is only half that of plants with the wild type locus (Fan et al., 2016). This 21-nucleotide premeiotic phasiRNA from the PMS1 locus in 58S may act at unknown downstream targets to reduce male fertility (Figures 4G and 4J; Fan et al., 2016). What do these studies tell us about the possible roles of 21-nucleotide premeiotic phasiRNAs? It is clear that just a single phasiRNA (or the duplex) can have a major impact on male fertility, perhaps due to the target transcripts from genes required for cellular development. Do all phasiRNAs likely have such functions? Given the sheer number of distinct 21-nucleotide reproductive phasiRNAs in rice, this seems highly unlikely, given that environmentally sensitive male sterile lines have not been widely found or described. It is likely that most 21-nucleotide reproductive phasiRNAs target genes with varying degrees of functional importance, such that the phenotypic consequences of the loss of a single phasiRNAs, are unnoticeable.
The most accurate genetic analysis of a broad-scale role of 21-nucleotide premeiotic phasiRNAs will likely await the generation of CRISPR-mediated deletion of the triggers of 21-nucleotide reproductive phasiRNAs, such as miR2118 in grasses. Since other biogenesis components (DCL4, RDR6, presumably SGS3, and likely a DRB protein) also function in tasiRNA production (i.e., from TAS3) important for normal development, other options for clean genetic deletion of all 21-nucleotide reproductive phasiRNAs are limited. Even if OCL4 is a master regulator of precursor expression, it has redundant roles that led to its initial characterization (Vernoud et al., 2009). The miR2118 family of miRNAs is arguably the largest family of miRNAs found in a single plant genome, with 25+ members in rice (Xia et al., 2015a). Yet, these miRNAs are produced from two major clusters of precursor loci on different chromosomes, and thus complete deletions could be generated relatively easily in rice and maize for functional analysis of 21-nucleotide reproductive phasiRNAs. One large cluster of miR2118 copies in rice (cv Nipponbare) was recently knocked out, leading to daylength-sensitive male sterility (Araki et al., 2020). Analysis of the mutant revealed that the developmental alteration likely initiated early and in the epidermis during the leptotene–zygotene stage of the germ cells, following a secondary alteration to the tapetum during or after meiosis. This is different from the phenotype of mel1/ago5, which displays altered meiotic progression rather than anther wall development. This difference might be partially explained by the different groups of 21-nucleotide phasiRNAs that were altered in these mutation events. The partial knockout of miR2118 blocked the processing of some of the 21-PHAS precursors and the biogenesis of 21-nucleotide phasiRNAs enriched with 5′-U in the first step of the 21-nucleotide phasiRNA pathway (Araki et al., 2020). By contrast, the knockout of mel1/ago5 blocked the function of 21-nucleotide phasiRNAs enriched with 5′-C bound to MEL1/AGO5; initial phasiRNA biogenesis in the mel1/ago5 mutant is mostly normal. The mutants described in Araki et al. (2020) retained four miR2118 copies, and the PMS1 locus was largely unaffected, suggesting redundancy of the miR2118 members. The mel1 mutant provides additional insights in rice (Figure 4H; Komiya et al., 2014), and complete deletion lines of mir2118 may well precisely phenocopy this mutant, although MEL1 does bind sRNAs other than 21-nucleotide phasiRNAs, and there are other closely related AGO proteins that might provide some redundancy. Comparative analysis of mir2118 complete deletion lines across several grass species would provide the most robust insight into 21-nucleotide reproductive phasiRNA function, at least in the Poaceae in which they are highly numerous and abundant. Therefore, there are exciting new discoveries yet to be made about the functions of these sRNAs.
Finally, can we infer anything about function or conservation of function (or lack thereof) based on when this pathway emerged during evolution and how it is comprised in different plant lineages? The short answer is that it is too early to say, and much more data are needed. It is still unclear when 21-nucleotide reproductive phasiRNAs emerged. As mentioned above, they may date to at least the gymnosperms (Xia et al., 2015a), and there is substantial variation in the number of loci within the monocots (Kakrana et al., 2018). What about the eudicots? Observations in Norway spruce suggested that the 21-nucleotide reproductive phasiRNAs predate the monocot–eudicot split; yet, extensive analyses by our group and others have not turned up this class of sRNAs in Arabidopsis or soybean. Perhaps, like the 24-nucleotide reproductive phasiRNAs (see below; Xia et al., 2019), there is lineage-specific loss of the pathway in eudicots. Premeiotic anthers are difficult to isolate, so sRNAs in these organs have not been well characterized for most species with sequenced genomes, including most eudicots. While miR2118 is widely present in eudicots, like its sister group miR482, it largely seems to function in the regulation of NLR transcripts, a role it has mostly transitioned away from in grasses.
Meiotic 24-Nucleotide PhasiRNAs
The pathway for 24-nucleotide phasiRNA production is highly, and apparently specifically, upregulated just prior to the initiation of meiosis in the tapetum of the anther in many angiosperm species. The molecular components required for the biogenesis of these phasiRNAs include those described earlier in this review: miR2275 as the typical trigger, the (mostly) lncRNA precursors (from “24-PHAS” loci), and either DCL5 (in most monocots) or DCL3 in eudicots. Other biogenesis components may be shared with the miRNA and 21-nucleotide phasiRNA pathways, including DCL1 (to make the miRNA trigger), AGO1 (to load the trigger), RDR6, and presumably proteins that function with the RISC (orthologs of SGS3 and a DRB protein). The 24-PHAS precursors are Pol II products with a poly(A) tail, as evidenced by their presence in RNA-sequencing libraries (Zhai et al., 2015; Teng et al., 2020), and our best guess, absent any experimental data, is that 24-nucleotide phasiRNAs are processed like tasiRNAs in Arabidopsis, associated with cytoplasmic, ER-bound ribosomes (Li et al., 2016).
How are these components regulated within the anther? Basic-helix-loop-helix (bHLH) TFs are a recurring theme in anther development and the regulation of these components. For example, in the rice tapetum during early meiosis, the bHLH TF ETERNAL TAPETUM1 (EAT1) binds to an “E-box” motif (a CANNTG sequence) in the promoters of most rice 24-PHAS loci (>100 loci; Ono et al., 2018). The eat1-4 mutant is male sterile, and in this mutant, the 24-PHAS loci are not transcribed and most 24-nucleotide phasiRNAs are missing, which is consistent with a role for EAT1 as a master regulator of 24-PHAS precursor production (Ono et al., 2018). EAT1 also regulates DCL5 expression by binding to its promoter-localized motifs (Ono et al., 2018). However, the precursors of the one examined member of the miR2275 family were unaffected in eat1-4, suggesting they are regulated by a different TF or TFs (Ono et al., 2018). In the same article, TIP2, another bHLH transcriptional factor, was reported to interact with EAT1, suggesting it plays a role in meiotic 24-nucleotide phasiRNA biogenesis, while UNDEVELOPED TAPETUM1 (UDT1; yet another bHLH) is a potential interacting partner of both EAT1 and TIP2. In maize male sterile mutants of the MS23 bHLH TF, 24-nucleotide phasiRNAs and DCL5 are mostly missing, and miR2275 families are altered as well (Nan et al., 2017). MS23 directly interacts with bHLH122, the maize ortholog of rice EAT1 (Nan et al., 2017). Other bHLH TFs have been implicated in anther development, but they are not described here, as they have not yet been connected to phasiRNAs (see Walbot and Egger, 2016), and bHLH TFs commonly function as heterodimers. Therefore, network and interaction analyses are likely to identify partners of EAT1, MS23, and orthologs that function in the production of 24-nucleotide meiotic phasiRNAs and their various biogenesis components.
Numerous studies now point to the tapetum as a cell layer of primary importance for 24-nucleotide phasiRNA activity. The EAT1 and MS23 TFs that drive the expression of the 24-PHAS precursors localize to the tapetum, and mutants in these TFs display defective tapetal cells (Nan et al., 2017; Ono et al., 2018). Earlier work on the eat1 mutant demonstrated that the tapetum fails to progress into a stage of PCD during later stages when microspores have formed (Niu et al., 2013). Perhaps the failure of PCD in the eat1 tapetum is a consequence of the loss of 24-nucleotide phasiRNAs, resulting indirectly (via “non-cell-autonomous signaling or some nutrient delivery,” as suggested by Ono et al. [2018]) in a block in meiotic progression due to a failure of the tapetum to support meiocytes development. The rice msp1 and ostdl1a mutants, which are defective in genes important for tapetal differentiation but without direct roles in phasiRNA production, have defective tapetal layers, with the levels of miR2275, 24-PHAS transcripts, and consequently 24-nucleotide phasiRNAs all severely reduced (Fei et al., 2016a). A similar analysis of maize mutants defective in anther cell layers other than the tapetum also demonstrated that the production of 24-PHAS precursors and 24-nucleotide phasiRNAs depends solely on the presence of a normal tapetum (Zhai et al., 2015). DCL5 is specifically expressed in early, meiotic-stage anthers (Teng et al., 2020), with transcripts (as measured by single-molecule fluorescent in situ hybridization) highly restricted to the tapetum (Huang et al., 2020a). Finally, single-cell transcriptional analysis of isolated, developing meiocytes from maize failed to find 24-PHAS precursors (Nelms and Walbot, 2019). Combining all of these observations, it is clear that 24-nucleotide phasiRNAs are made in tapetal cells, but is this where they end up? Fluorescent in situ hybridization data demonstrate that 24-nucleotide phasiRNAs are strongly localized to both the tapetum and PMCs (Zhai et al., 2015; Xia et al., 2019), with much lower levels in the outer layers of meiotic anthers (Huang et al., 2020b). These observations suggest that 24-nucleotide phasiRNAs are translocated from the tapetum to the PMCs; this is perhaps not unexpected, as tapetal cells act as nurse cells, providing sustenance to PMCs. While a role in tapetal development is our favored hypothesis, absent information on the bona fide targets of 24-nucleotide phasiRNAs, we cannot exclude the possibility that these phasiRNAs function in the PMCs.
The AGO protein that is loaded with the 24-nucleotide phasiRNAs is unknown, but it seems likely that a specialized AGO binds to the 24-nucleotide phasiRNAs and acts on their targets. An intriguing and perhaps related observation is that despite the “standard” 22-nucleotide length of miR2275 as a trigger of phasiRNAs, cleavage directed by miR2275 leads to the recruitment of DCL5 rather than DCL4, as DCL4 appears to be present in the tapetal cells (Huang et al., 2020a). One possible explanation is that there is a specialized copy of an AGO that binds to miR2275, perhaps a paralog of AGO1, as there are multiple paralogs of AGO1, at least in the grass genomes (Zhang et al., 2015); this AGO could specifically load miR2275 much like AGO10 specializes in loading miR165/166 or AGO7 with miR390. While the question of DCL5 recruitment remains intriguing and unsolved, analyses of RNA-sequencing data from developing anthers or isolated meiocytes have yielded several candidate AGO genes that are strongly upregulated coincident with 24-nucleotide phasiRNAs. Multiple studies identified maize AGO18b as highly enriched in anthers, including localization in tapetal and meiotic cells (Zhai et al., 2014), and enrichment of AGO18 transcripts is coincident with 24-nucleotide phasiRNAs (Zhai et al., 2015) and in isolated maize meiocytes (Nelms and Walbot, 2019). In rice, OsAGO2b, OsAGO1d, and OsAGO18 transcripts were identified as anther enriched, with the latter two AGO levels validated by mRNA in situ analysis, demonstrating a spatiotemporal pattern of expression that correlates with 24-nucleotide phasiRNA accumulation (Fei et al., 2016a). The enrichment of AGO1d is interesting—perhaps it is the miR2275-specialized partner that recruits DCL5 instead of DCL4? The multiple studies pointing to AGO18 are likewise intriguing, yet AGO18 appears to be specific to grasses (Zhang et al., 2015), so which AGO could bind 24-nucleotide phasiRNAs in angiosperms outside of the grasses? A relative of AGO4 is a possibility (in the AGO4/6/8/9 clade, as named based on Arabidopsis), since AGO4 binds 24-nucleotide Pol IV siRNAs, and other members of this clade function in the specification of cell fate in female flowers (Olmedo-Monfil et al., 2010; Singh et al., 2011; Zhang et al., 2015). Given the role of AGO4 in directing epigenetic modifications, perhaps 24-nucleotide phasiRNA activity yields a similar outcome. However, what would distinguish the 24-nucleotide phasiRNAs from loading into AGO4 itself to function in RNA-directed DNA methylation? As mentioned above, the subcellular location of 24-nucleotide phasiRNA biogenesis is likely distinct from that of 24-nucleotide Pol IV siRNAs (cytoplasmic versus nuclear); plus, 24-nucleotide phasiRNAs have shown distinct sequences compared to all other types of plant sRNAs, demonstrating an overall adenosine (A)/U enrichment (Patel et al., 2018). Finally, if we knew which AGO loads the 24-nucleotide phasiRNAs, finding the targets would be easier, as bioinformatics analysis have yielded relatively few insights. Like the 21-nucleotide reproductive phasiRNAs, the 24-nucleotide phasiRNAs are generally single copy, and extensive target analysis has found low homology to transposons and a greater match to genic sequences (Patel et al., 2018). Since the 24-nucleotide phasiRNAs move from their site of production in the tapetum to PMCs, are the targets located in either or both of these layers? Answers to all of these questions may await the discovery of the AGO (or AGOs) that binds these sRNAs, coupled with biochemical, molecular, and genomic analysis.
What do we know about the broader roles of 24-nucleotide phasiRNAs, even without knowing their targets? Data from isolated meiocytes revealed elevated CHG and CHH methylation at 24-PHAS loci concurrent with the peak abundance of 24-nucleotide phasiRNAs at the time of zygotene, suggesting that 24-nucleotide phasiRNAs play a role in chromatin remodeling during meiosis (Dukowic-Schulze et al., 2016). On the other hand, the sequences of 24-nucleotide phasiRNAs are relatively distinct within a genome (Zhai et al., 2015), similar to the premeiotic 21-nucleotide phasiRNAs. However, unlike the PMS1 and PMS3 loci in rice (yielding 21-nucleotide phasiRNAs), there are no reports of mutants of single 24-nucleotide phasiRNAs that produce a phenotype. Since tens to hundreds of 24-PHAS precursor loci are distributed across genomes (Johnson et al., 2009; Xia et al., 2019), the best opportunity for functional analysis is their complete elimination, perhaps by CRISPR-mediated deletion of the miR2275 loci or by mutation of DCL5. We have identified and characterized dcl5 mutant alleles in maize (Figure 4I; Teng et al., 2020). sRNA analysis of dcl5 alleles revealed essentially a complete loss of 24-nucleotide phasiRNAs, indicating no redundancy with DCL3 (Teng et al., 2020). The 21-nucleotide and 24-nucleotide reproductive phasiRNAs share biogenesis factors such as DCL1, AGO1, RDR6, and so on, making the loss of DCL5 particularly informative. The dcl5 null mutants are male sterile yet apparently fully female fertile, which is arguably inconsistent with a major role for phasiRNAs in female flowers. Curiously, these mutants demonstrate different degrees of male sterility depending on the temperature during meiosis; that is, normal temperatures yield sterility, while low temperatures rescue fertility, perhaps reflecting rescue by slower growth conditions (Figure 4K; Teng et al., 2020). Unlike the previous finding in Arabidopsis that miRNA biogenesis could be rescued without DCL1 cofactors (HYL1 and SE) under ambient temperature conditions (Ré et al., 2019), DCL3 or DCL4 did not substitute for DCL5 under ambient temperature conditions in the maize dcl5 mutant (Teng et al., 2020). This phenotype of conditional male sterility is uncommon in maize. The basis of the sterility phenotype is a slowly developing or defective tapetum, which is consistent with rice eat1 and maize ms23. Since the loss of DCL5 blocks processing of the dsRNA precursors, the tapetal defects described above are presumably due to mis-regulation of the targets of 24-nucleotide phasiRNAs—assuming that they, like all other sRNAs, interact with other, longer RNAs with a suppressive effect.
Does the evolutionary emergence and presence/absence of this pathway across plant lineages provide clues about its function? The short answer is that much more data are needed from more species. The longer, more speculative answer stems from the observation that the 24-nucleotide reproductive phasiRNAs appear to have emerged coincident with angiosperms. There is substantial variation in the number of loci within the monocots (Kakrana et al., 2018) and presence/absence within the eudicots (Xia et al., 2019). The presence of 24-nucleotide reproductive phasiRNAs in many eudicots, in the absence of DCL5, suggests that DCL3 has a dual role, generating Pol IV siRNAs in the nucleus and possibly reproductive 24-nucleotide phasiRNAs in the cytoplasm of tapetal cells, while in some monocots, DCL5 emerged and specialized to generate 24-nucleotide reproductive phasiRNAs. Our phylogenetic analysis demonstrated that DCL5 likely emerged in the monocots after the divergence of the Acorales/Alismatales and before the Dioscoreales emerged, more than110 million years ago (Patel et al., 2020). miR2275 is present in earlier diverged flowering plants, such as Nymphaea colorata, a basal angiosperm (Patel et al., 2020). It may be informative to study miR2275 diversity in early-diverged angiosperms, including Magnoliids, to assess its origins; together with its lncRNA 24-PHAS targets, miR2275 was perhaps one of the first-evolved components unique to this pathway. A related observation that intrigues us is the origin and functional implications of the tandem repeat, or dual hairpin structure of miR2275 conserved from eudicots to monocots (Xia et al., 2019). What selective pressures maintain this precursor structure? Does the tandem structure play a role in specifying loading of miR2275 into an AGO that can recruit DCL5? There is also much work to be done in studies of 24-nucleotide reproductive phasiRNAs.
Major Unanswered Questions about Reproductive PhasiRNAs
At this point in the review, our readers should understand that numerous questions fuel our interest in and speculation about the function, evolution, and biogenesis of reproductive phasiRNAs. Since so much is unknown about the biology of reproductive phasiRNAs, we list here what we consider to be the major unanswered questions, hopefully to facilitate further research in the coming years. We feel that the most pertinent questions are as follows:
What is the molecular basis of the environmental sensitivity in grasses with perturbed reproductive phasiRNAs?
Are photoperiod and temperature effects in phasiRNA mutants connected or separable?
How does the rescue of male sterility phenotypes occur under slow-growth conditions?
Do reproductive phasiRNAs normally function to modulate environmental variation—or is the environmental sensitivity observed when certain phasiRNAs are perturbed an indirect phenotype?
Reproductive phasiRNAs are abundant in anthers for longer than 1 week—does this reflect stability and longevity, or constant turnover and replacement?
How is it that DCL5 (or DCL3 in eudicots), instead of DCL4, is recruited by the activity of a 22-nucleotide miRNA?
Is there functional significance to the miR2275 tandem duplication/polycistronic precursor?
Do 24-nucleotide phasiRNAs function in RNA-directed DNA methylation, or do they have other functions? What are the targets at which they are directing these activities?
Do 21-nucleotide phasiRNAs function in trans to direct cleavage of genic targets?
As no obvious endogenous targets have yet been found for reproductive phasiRNAs, could they have a function in regulating exogenous RNAs?
What role do the 21- and 24-nucleotide phasiRNAs play in the development of the tapetum (or other anther layers)?
What is the AGO partner of 24-nucleotide phasiRNAs? Is it AGO18, and if so, which AGO is this partner in species that lack AGO18, that is, outside of the grasses?
Is there a role for phasiRNAs in female organs, or outside of reproductive tissues, perhaps similar to that of animal piRNAs?
When and how did the 21- and 24-nucleotide phasiRNA pathways emerge in plants?
Why is there such variation in the number of reproductive phasiRNA loci across species?
Are there common selective pressures that led to the apparent parallels in plant reproductive phasiRNAs and animal piRNAs?
Is miR2275 the only miRNA that triggers 24-nucleotide phasiRNA production? If so, what makes miR2275 unique in this respect—perhaps a partnership with a specialized AGO?
Can miR2275 only trigger 24-nucleotide phasiRNA production? Does it only target lncRNAs, or did it emerge during evolution as a “normal” miRNA that triggers 21-nucleotide phasiRNA production from protein-coding genes?
Why are the lncRNA precursors and thus the reproductive phasiRNAs themselves so divergent in sequence when compared across species? Does this reflect something about their function or targets?
If we review phasiRNAs in another 7 years (the time since our last phasiRNA review in these pages; Fei et al., 2013), perhaps most of these questions will have been addressed and replaced by a new set of questions that will continue to drive research into these fascinating sRNAs. Beyond basic insights into RNA biology, the greatest translational impact of answers to these questions would be a system for the production of hybrid crops. In many crops, particularly grasses such as rice, wheat, and barley, there is a strong need for environmentally controlled male sterility, an application that could result from mastering the phenotype of conditional male sterility that is consistently associated with reproductive phasiRNAs.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Joanna Friesner, Rachel Egger, and Virginia Walbot for helpful comments and input on earlier versions of the article. Support for this work was provided by the National Key Research and Developmental Program of China (grant 2018YFD1000104 to R.X.), the Special Support Program of Guangdong Province (grant 2019TX05N193 to R.X.), the National Science Foundation of China (grant 31872063 to R.X.) and the National Science Foundation Plant Genome Research Program (grant 1754097 to B.C.M.).
AUTHOR CONTRIBUTIONS
Y.L., C.T., R.X., and B.C.M. outlined, wrote, and edited this review.
References
- Adenot, X., Elmayan, T., Lauressergues, D., Boutet, S., Bouché, N., Gasciolli, V., Vaucheret, H.(2006). DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr. Biol. 16: 927–932. [DOI] [PubMed] [Google Scholar]
- Araki, S., Le, N.T., Koizumi, K., Villar-Briones, A., Nonomura, K.I., Endo, M., Inoue, H., Saze, H., Komiya, R.(2020). miR2118-dependent U-rich phasiRNA production in rice anther wall development. Nat. Commun. 11: 3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arif, M.A., Fattash, I., Ma, Z., Cho, S.H., Beike, A.K., Reski, R., Axtell, M.J., Frank, W.(2012). DICER-LIKE3 activity in Physcomitrella patens DICER-LIKE4 mutants causes severe developmental dysfunction and sterility. Mol. Plant 5: 1281–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arikit, S., Xia, R., Kakrana, A., Huang, K., Zhai, J., Yan, Z., Valdés-López, O., Prince, S., Musket, T.A., Nguyen, H.T., Stacey, G., Meyers, B.C.(2014). An atlas of soybean small RNAs identifies phased siRNAs from hundreds of coding genes. Plant Cell 26: 4584–4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arribas-Hernández, L., Marchais, A., Poulsen, C., Haase, B., Hauptmann, J., Benes, V., Meister, G., Brodersen, P.(2016). The slicer activity of ARGONAUTE1 is required specifically for the phasing, not production, of trans-acting short interfering RNAs in Arabidopsis. Plant Cell 28: 1563–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell, M.J., Jan, C., Rajagopalan, R., Bartel, D.P.(2006). A two-hit trigger for siRNA biogenesis in plants. Cell 127: 565–577. [DOI] [PubMed] [Google Scholar]
- Baeg, K., Iwakawa, H.O., Tomari, Y.(2017). The poly(A) tail blocks RDR6 from converting self mRNAs into substrates for gene silencing. Nat. Plants 3: 17036. [DOI] [PubMed] [Google Scholar]
- Baldrich, P., Rutter, B.D., Karimi, H.Z., Podicheti, R., Meyers, B.C., Innes, R.W.(2019). Plant extracellular vesicles contain diverse small RNA species and are enriched in 10- to 17-nucleotide “tiny” RNAs. Plant Cell 31: 315–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bélanger, S., Pokhrel, S., Czymmek, K., Meyers, B.C.(2020). Pre-meiotic, 24-nt reproductive phasiRNAs are abundant in anthers of wheat and barley but not rice and maize. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boccara, M., Sarazin, A., Thiébeauld, O., Jay, F., Voinnet, O., Navarro, L., Colot, V.(2014). The Arabidopsis miR472-RDR6 silencing pathway modulates PAMP- and effector-triggered immunity through the post-transcriptional control of disease resistance genes. PLoS Pathog. 10: e1003883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bologna, N.G., Mateos, J.L., Bresso, E.G., Palatnik, J.F.(2009). A loop-to-base processing mechanism underlies the biogenesis of plant microRNAs miR319 and miR159. EMBO J. 28: 3646–3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bologna, N.G., Voinnet, O.(2014). The diversity, biogenesis, and activities of endogenous silencing small RNAs in Arabidopsis. Annu. Rev. Plant Biol. 65: 473–503. [DOI] [PubMed] [Google Scholar]
- Borges, F., Martienssen, R.A.(2015). The expanding world of small RNAs in plants. Nat. Rev. Mol. Cell Biol. 16: 727–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges, F., Parent, J.-S., van Ex, F., Wolff, P., Martínez, G., Köhler, C., Martienssen, R.A.(2018). Transposon-derived small RNAs triggered by miR845 mediate genome dosage response in Arabidopsis. Nat. Genet. 50: 186–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branscheid, A., Marchais, A., Schott, G., Lange, H., Gagliardi, D., Andersen, S.U., Voinnet, O., Brodersen, P.(2015). SKI2 mediates degradation of RISC 5′-cleavage fragments and prevents secondary siRNA production from miRNA targets in Arabidopsis. Nucleic Acids Res. 43: 10975–10988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera, J., Barcala, M., García, A., Rio-Machín, A., Medina, C., Jaubert-Possamai, S., Favery, B., Maizel, A., Ruiz-Ferrer, V., Fenoll, C., Escobar, C.(2016). Differentially expressed small RNAs in Arabidopsis galls formed by Meloidogyne javanica: A functional role for miR390 and its TAS3-derived tasiRNAs. New Phytol. 209: 1625–1640. [DOI] [PubMed] [Google Scholar]
- Cai, Q., Qiao, L., Wang, M., He, B., Lin, F.M., Palmquist, J., Huang, S.D., Jin, H.(2018). Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science 360: 1126–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto-Pastor, A., Santos, B.A.M.C., Valli, A.A., Summers, W., Schornack, S., Baulcombe, D.C.(2019). Enhanced resistance to bacterial and oomycete pathogens by short tandem target mimic RNAs in tomato. Proc. Natl. Acad. Sci. USA 116: 2755–2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsbecker, A., et al. (2010). Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature 465: 316–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C., Feng, J., Liu, B., Li, J., Feng, L., Yu, X., Zhai, J., Meyers, B.C., Xia, R.(2019). sRNAanno—A database repository of uniformly-annotated small RNAs in plants. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H.M., Chen, L.T., Patel, K., Li, Y.H., Baulcombe, D.C., Wu, S.H.(2010). 22-Nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proc. Natl. Acad. Sci. USA 107: 15269–15274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, S.H., Coruh, C., Axtell, M.J.(2012). miR156 and miR390 regulate tasiRNA accumulation and developmental timing in Physcomitrella patens. Plant Cell 24: 4837–4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, C., Wang, J.J., Zhao, J.H., Fang, Y.Y., He, X.F., Guo, H.S., Duan, C.G.(2020). A Brassica miRNA regulates plant growth and immunity through distinct modes of action. Mol. Plant 13: 231–245. [DOI] [PubMed] [Google Scholar]
- Cui, H., Tsuda, K., Parker, J.E.(2015). Effector-triggered immunity: From pathogen perception to robust defense. Annu. Rev. Plant Biol. 66: 487–511. [DOI] [PubMed] [Google Scholar]
- Cuperus, J.T., Carbonell, A., Fahlgren, N., Garcia-Ruiz, H., Burke, R.T., Takeda, A., Sullivan, C.M., Gilbert, S.D., Montgomery, T.A., Carrington, J.C.(2010a). Unique functionality of 22-nt miRNAs in triggering RDR6-dependent siRNA biogenesis from target transcripts in Arabidopsis. Nat. Struct. Mol. Biol. 17: 997–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuperus, J.T., Montgomery, T.A., Fahlgren, N., Burke, R.T., Townsend, T., Sullivan, C.M., Carrington, J.C.(2010b). Identification of MIR390a precursor processing-defective mutants in Arabidopsis by direct genome sequencing. Proc. Natl. Acad. Sci. USA 107: 466–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitwood, D.H., Nogueira, F.T., Howell, M.D., Montgomery, T.A., Carrington, J.C., Timmermans, M.C.(2009). Pattern formation via small RNA mobility. Genes Dev. 23: 549–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Felippes, F.F., Marchais, A., Sarazin, A., Oberlin, S., Voinnet, O.(2017). A single miR390 targeting event is sufficient for triggering TAS3-tasiRNA biogenesis in Arabidopsis. Nucleic Acids Res. 45: 5539–5554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Felippes, F.F., Ott, F., Weigel, D.(2011). Comparative analysis of non-autonomous effects of tasiRNAs and miRNAs in Arabidopsis thaliana. Nucleic Acids Res. 39: 2880–2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, J., Lu, Q., Ouyang, Y., Mao, H., Zhang, P., Yao, J., Xu, C., Li, X., Xiao, J., Zhang, Q.(2012). A long noncoding RNA regulates photoperiod-sensitive male sterility, an essential component of hybrid rice. Proc. Natl. Acad. Sci. USA 109: 2654–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas, R.N., Wiley, D., Sarkar, A., Springer, N., Timmermans, M.C.P., Scanlon, M.J.(2010). Ragged seedling2 encodes an ARGONAUTE7-like protein required for mediolateral expansion, but not dorsiventrality, of maize leaves. Plant Cell 22: 1441–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dukowic-Schulze, S., Sundararajan, A., Ramaraj, T., Kianian, S., Pawlowski, W.P., Mudge, J., Chen, C.(2016). Novel meiotic miRNAs and indications for a role of phasiRNAs in meiosis. Front. Plant Sci. 7: 762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo, Y., Iwakawa, H.O., Tomari, Y.(2013). Arabidopsis ARGONAUTE7 selects miR390 through multiple checkpoints during RISC assembly. EMBO Rep. 14: 652–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren, N., Montgomery, T.A., Howell, M.D., Allen, E., Dvorak, S.K., Alexander, A.L., Carrington, J.C.(2006). Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr. Biol. 16: 939–944. [DOI] [PubMed] [Google Scholar]
- Fan, Y., et al. (2016). PMS1T, producing phased small-interfering RNAs, regulates photoperiod-sensitive male sterility in rice. Proc. Natl. Acad. Sci. USA 113: 15144–15149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, Y., Zhang, Q.(2018). Genetic and molecular characterization of photoperiod and thermo-sensitive male sterility in rice. Plant Reprod. 31: 3–14. [DOI] [PubMed] [Google Scholar]
- Fei, Q., Li, P., Teng, C., Meyers, B.C.(2015). Secondary siRNAs from Medicago NB-LRRs modulated via miRNA-target interactions and their abundances. Plant J. 83: 451–465. [DOI] [PubMed] [Google Scholar]
- Fei, Q., Xia, R., Meyers, B.C.(2013). Phased, secondary, small interfering RNAs in posttranscriptional regulatory networks. Plant Cell 25: 2400–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei, Q., Yang, L., Liang, W., Zhang, D., Meyers, B.C.(2016a). Dynamic changes of small RNAs in rice spikelet development reveal specialized reproductive phasiRNA pathways. J. Exp. Bot. 67: 6037–6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei, Q., Yu, Y., Liu, L., Zhang, Y., Baldrich, P., Dai, Q., Chen, X., Meyers, B.C.(2018). Biogenesis of a 22-nt microRNA in Phaseoleae species by precursor-programmed uridylation. Proc. Natl. Acad. Sci. USA 115: 8037–8042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei, Q., Zhang, Y., Xia, R., Meyers, B.C.(2016b). Small RNAs add zing to the Zig-Zag-Zig model of plant defenses. Mol. Plant Microbe Interact. 29: 165–169. [DOI] [PubMed] [Google Scholar]
- Fukudome, A., Kanaya, A., Egami, M., Nakazawa, Y., Hiraguri, A., Moriyama, H., Fukuhara, T.(2011). Specific requirement of DRB4, a dsRNA-binding protein, for the in vitro dsRNA-cleaving activity of Arabidopsis Dicer-like 4. RNA 17: 750–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan, X., Pang, M., Nah, G., Shi, X., Ye, W., Stelly, D.M., Chen, Z.J.(2014). miR828 and miR858 regulate homoeologous MYB2 gene functions in Arabidopsis trichome and cotton fibre development. Nat. Commun. 5: 3050. [DOI] [PubMed] [Google Scholar]
- Guo, G., et al. (2018). Wheat miR9678 affects seed germination by generating phased siRNAs and modulating abscisic acid/gibberellin signaling. Plant Cell 30: 796–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, B.W., Wang, W., Li, C., Weng, Z., Zamore, P.D.(2015). Noncoding RNA. piRNA-guided transposon cleavage initiates zucchini-dependent, phased piRNA production. Science 348: 817–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson, I.R., Zhang, X., Lu, C., Johnson, L., Meyers, B.C., Green, P.J., Jacobsen, S.E.(2006). Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat. Genet. 38: 721–725. [DOI] [PubMed] [Google Scholar]
- Hobecker, K.V., Reynoso, M.A., Bustos-Sanmamed, P., Wen, J., Mysore, K.S., Crespi, M., Blanco, F.A., Zanetti, M.E.(2017). The microRNA390/TAS3 pathway mediates symbiotic nodulation and lateral root growth. Plant Physiol. 174: 2469–2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, Y., et al. (2019). A Phytophthora effector suppresses trans-kingdom RNAi to promote disease susceptibility. Cell Host Microbe 25: 153–165.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, C.Y., Lee, W.C., Chou, H.C., Chen, A.P., Chou, S.J., Chen, H.M.(2016). Global analysis of truncated RNA ends reveals new insights into ribosome stalling in plants. Plant Cell 28: 2398–2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell, M.D., Fahlgren, N., Chapman, E.J., Cumbie, J.S., Sullivan, C.M., Givan, S.A., Kasschau, K.D., Carrington, J.C.(2007). Genome-wide analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 pathway in Arabidopsis reveals dependency on miRNA- and tasiRNA-directed targeting. Plant Cell 19: 926–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, K., Batish, M., Teng, C., Harkess, A., Meyers, B.C., Caplan, J.L.(2020a). Quantitative fluorescence in situ hybridization detection of plant mRNAs with single-molecule resolution. In RNA Tagging: Methods and Protocols, Heinlein M., ed, Volume. Vol. 2166 (New York: Springer; ). [DOI] [PubMed] [Google Scholar]
- Huang, K., Huang, K., Demirci, F., Meyers, B.C., Caplan, J.L.(2020b). Quantitative, super-resolution localization of small RNAs with sRNA-PAINT. Nucleic Acids Res. 716696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, C., Kasprzewska, A., Tennessen, K., Fernandes, J., Nan, G.-L., Walbot, V., Sundaresan, V., Vance, V., Bowman, L.H.(2009). Clusters and superclusters of phased small RNAs in the developing inflorescence of rice. Genome Res. 19: 1429–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouannet, V., Moreno, A.B., Elmayan, T., Vaucheret, H., Crespi, M.D., Maizel, A.(2012). Cytoplasmic Arabidopsis AGO7 accumulates in membrane-associated siRNA bodies and is required for ta-siRNA biogenesis. EMBO J. 31: 1704–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakrana, A., et al. (2018). Plant 24-nt reproductive phasiRNAs from intramolecular duplex mRNAs in diverse monocots. Genome Res. 28: 1333–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelliher, T., Egger, R.L., Zhang, H., Walbot, V.(2014). Unresolved issues in pre-meiotic anther development. Front. Plant Sci. 5: 347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelliher, T., Walbot, V.(2011). Emergence and patterning of the five cell types of the Zea mays anther locule. Dev. Biol. 350: 32–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knauer, S., Holt, A.L., Rubio-Somoza, I., Tucker, E.J., Hinze, A., Pisch, M., Javelle, M., Timmermans, M.C., Tucker, M.R., Laux, T.(2013). A protodermal miR394 signal defines a region of stem cell competence in the Arabidopsis shoot meristem. Dev. Cell 24: 125–132. [DOI] [PubMed] [Google Scholar]
- Komiya, R.(2017). Biogenesis of diverse plant phasiRNAs involves an miRNA-trigger and Dicer-processing. J. Plant Res. 130: 17–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiya, R., Ohyanagi, H., Niihama, M., Watanabe, T., Nakano, M., Kurata, N., Nonomura, K.(2014). Rice germline-specific Argonaute MEL1 protein binds to phasiRNAs generated from more than 700 lincRNAs. Plant J. 78: 385–397. [DOI] [PubMed] [Google Scholar]
- Kumakura, N., Takeda, A., Fujioka, Y., Motose, H., Takano, R., Watanabe, Y.(2009). SGS3 and RDR6 interact and colocalize in cytoplasmic SGS3/RDR6-bodies. FEBS Lett. 583: 1261–1266. [DOI] [PubMed] [Google Scholar]
- Li, F., Pignatta, D., Bendix, C., Brunkard, J.O., Cohn, M.M., Tung, J., Sun, H., Kumar, P., Baker, B.(2012). MicroRNA regulation of plant innate immune receptors. Proc. Natl. Acad. Sci. USA 109: 1790–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S., et al. (2016). Biogenesis of phased siRNAs on membrane-bound polysomes in Arabidopsis. eLife 5: e22750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S., Liu, J., Liu, Z., Li, X., Wu, F., He, Y.(2014). HEAT-INDUCED TAS1 TARGET1 mediates thermotolerance via HEAT STRESS TRANSCRIPTION FACTOR A1a-directed pathways in Arabidopsis. Plant Cell 26: 1764–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, J.S., Lin, C.C., Lin, H.H., Chen, Y.C., Jeng, S.T.(2012). MicroR828 regulates lignin and H2O2 accumulation in sweet potato on wounding. New Phytol. 196: 427–440. [DOI] [PubMed] [Google Scholar]
- Lin, Y., Lin, L., Lai, R., Liu, W., Chen, Y., Zhang, Z., XuHan, X., Lai, Z.(2015). microRNA390-directed TAS3 cleavage leads to the production of tasiRNA-ARF3/4 during somatic embryogenesis in Dimocarpus longan Lour. Front. Plant Sci. 6: 1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H., Nonomura, K.I.(2016). A wide reprogramming of histone H3 modifications during male meiosis I in rice is dependent on the Argonaute protein MEL1. J. Cell Sci. 129: 3553–3561. [DOI] [PubMed] [Google Scholar]
- Liu, J., Cheng, X., Liu, D., Xu, W., Wise, R., Shen, Q.H.(2014). The miR9863 family regulates distinct Mla alleles in barley to attenuate NLR receptor-triggered disease resistance and cell-death signaling. PLoS Genet. 10: e1004755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, L., Chen, X.(2018). Intercellular and systemic trafficking of RNAs in plants. Nat. Plants 4: 869–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y., Dou, M., Song, X., Dong, Y., Liu, S., Liu, H., Tao, J., Li, W., Yin, X., Xu, W.(2019). The emerging role of the piRNA/piwi complex in cancer. Mol. Cancer 18: 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y., Ke, L., Wu, G., Xu, Y., Wu, X., Xia, R., Deng, X., Xu, Q.(2017). miR3954 is a trigger of phasiRNAs that affects flowering time in citrus. Plant J. 92: 263–275. [DOI] [PubMed] [Google Scholar]
- Ma, W., Chen, C., Liu, Y., Zeng, M., Meyers, B.C., Li, J., Xia, R.(2018). Coupling of microRNA-directed phased small interfering RNA generation from long noncoding genes with alternative splicing and alternative polyadenylation in small RNA-mediated gene silencing. New Phytol. 217: 1535–1550. [DOI] [PubMed] [Google Scholar]
- Manavella, P.A., Koenig, D., Weigel, D.(2012). Plant secondary siRNA production determined by microRNA-duplex structure. Proc. Natl. Acad. Sci. USA 109: 2461–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin, E., Jouannet, V., Herz, A., Lokerse, A.S., Weijers, D., Vaucheret, H., Nussaume, L., Crespi, M.D., Maizel, A.(2010). miR390, Arabidopsis TAS3 tasiRNAs, and their AUXIN RESPONSE FACTOR targets define an autoregulatory network quantitatively regulating lateral root growth. Plant Cell 22: 1104–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez, G., Wolff, P., Wang, Z., Moreno-Romero, J., Santos-González, J., Conze, L.L., DeFraia, C., Slotkin, R.K., Köhler, C.(2018). Paternal easiRNAs regulate parental genome dosage in Arabidopsis. Nat. Genet. 50: 193–198. [DOI] [PubMed] [Google Scholar]
- Mi, S., et al. (2008). Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell 133: 116–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery, T.A., Howell, M.D., Cuperus, J.T., Li, D., Hansen, J.E., Alexander, A.L., Chapman, E.J., Fahlgren, N., Allen, E., Carrington, J.C.(2008). Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell 133: 128–141. [DOI] [PubMed] [Google Scholar]
- Nagasaki, H., Itoh, J., Hayashi, K., Hibara, K., Satoh-Nagasawa, N., Nosaka, M., Mukouhata, M., Ashikari, M., Kitano, H., Matsuoka, M., Nagato, Y., Sato, Y.(2007). The small interfering RNA production pathway is required for shoot meristem initiation in rice. Proc. Natl. Acad. Sci. USA 104: 14867–14871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan, G.L., Zhai, J., Arikit, S., Morrow, D., Fernandes, J., Mai, L., Nguyen, N., Meyers, B.C., Walbot, V.(2017). MS23, a master basic helix-loop-helix factor, regulates the specification and development of the tapetum in maize. Development 144: 163–172. [DOI] [PubMed] [Google Scholar]
- Nelms, B., Walbot, V.(2019). Defining the developmental program leading to meiosis in maize. Science 364: 52–56. [DOI] [PubMed] [Google Scholar]
- Nogueira, F.T.S., Madi, S., Chitwood, D.H., Juarez, M.T., Timmermans, M.C.P.(2007). Two small regulatory RNAs establish opposing fates of a developmental axis. Genes Dev. 21: 750–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu, N., Liang, W., Yang, X., Jin, W., Wilson, Z.A., Hu, J., Zhang, D.(2013). EAT1 promotes tapetal cell death by regulating aspartic proteases during male reproductive development in rice. Nat. Commun. 4: 1445. [DOI] [PubMed] [Google Scholar]
- Nonomura, K., Morohoshi, A., Nakano, M., Eiguchi, M., Miyao, A., Hirochika, H., Kurata, N.(2007). A germ cell specific gene of the ARGONAUTE family is essential for the progression of premeiotic mitosis and meiosis during sporogenesis in rice. Plant Cell 19: 2583–2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmedo-Monfil, V., Durán-Figueroa, N., Arteaga-Vázquez, M., Demesa-Arévalo, E., Autran, D., Grimanelli, D., Slotkin, R.K., Martienssen, R.A., Vielle-Calzada, J.P.(2010). Control of female gamete formation by a small RNA pathway in Arabidopsis. Nature 464: 628–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono, S., Liu, H., Tsuda, K., Fukai, E., Tanaka, K., Sasaki, T., Nonomura, K.I.(2018). EAT1 transcription factor, a non-cell-autonomous regulator of pollen production, activates meiotic small RNA biogenesis in rice anther tapetum. PLoS Genet. 14: e1007238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozata, D.M., Gainetdinov, I., Zoch, A., O’Carroll, D., Zamore, P.D.(2019). PIWI-interacting RNAs: Small RNAs with big functions. Nat. Rev. Genet. 20: 89–108. [DOI] [PubMed] [Google Scholar]
- Patel, P., Mathioni, S., Kakrana, A., Shatkay, H., Meyers, B.C.(2018). Reproductive phasiRNAs in grasses are compositionally distinct from other classes of small RNAs. New Phytol. 220: 851–864. [DOI] [PubMed] [Google Scholar]
- Patel, P., Mathioni, S.M., Hammond, R., Harkess, A.E., Kakrana, A., Arikit, S., Dusia, A., Meyers, B.C.(2020). Reproductive phasiRNA loci and DICER-LIKE5, but not microRNA loci, diversified in monocotyledonous plants. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petsch, K., Manzotti, P.S., Tam, O.H., Meeley, R., Hammell, M., Consonni, G., Timmermans, M.C.P.(2015). Novel DICER-LIKE1 siRNAs bypass the requirement for DICER-LIKE4 in maize development. Plant Cell 27: 2163–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polydore, S., Lunardon, A., Axtell, M.J.(2018). Several phased siRNA annotation methods can frequently misidentify 24 nucleotide siRNA-dominated PHAS loci. Plant Direct 2: e00101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajeswaran, R., Aregger, M., Zvereva, A.S., Borah, B.K., Gubaeva, E.G., Pooggin, M.M.(2012). Sequencing of RDR6-dependent double-stranded RNAs reveals novel features of plant siRNA biogenesis. Nucleic Acids Res. 40: 6241–6254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajeswaran, R., Pooggin, M.M.(2012). RDR6-mediated synthesis of complementary RNA is terminated by miRNA stably bound to template RNA. Nucleic Acids Res. 40: 594–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ré, D.A., Lang, P.L.M., Yones, C., Arce, A.L., Stegmayer, G., Milone, D., Manavella, P.A.(2019). Alternative use of miRNA-biogenesis co-factors in plants at low temperatures. Development 146: dev172932. [DOI] [PubMed] [Google Scholar]
- Rock, C.D.(2013). Trans-acting small interfering RNA4: Key to nutraceutical synthesis in grape development? Trends Plant Sci. 18: 601–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers, K., Chen, X.(2013). Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell 25: 2383–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ru, P., Xu, L., Ma, H., Huang, H.(2006). Plant fertility defects induced by the enhanced expression of microRNA167. Cell Res. 16: 457–465. [DOI] [PubMed] [Google Scholar]
- Schmitz-Linneweber, C., Small, I.(2008). Pentatricopeptide repeat proteins: A socket set for organelle gene expression. Trends Plant Sci. 13: 663–670. [DOI] [PubMed] [Google Scholar]
- Shahid, S., Kim, G., Johnson, N.R., Wafula, E., Wang, F., Coruh, C., Bernal-Galeano, V., Phifer, T., dePamphilis, C.W., Westwood, J.H., Axtell, M.J.(2018). MicroRNAs from the parasitic plant Cuscuta campestris target host messenger RNAs. Nature 553: 82–85. [DOI] [PubMed] [Google Scholar]
- Shivaprasad, P.V., Chen, H.M., Patel, K., Bond, D.M., Santos, B.A.C.M., Baulcombe, D.C.(2012). A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs. Plant Cell 24: 859–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuai, P., Su, Y., Liang, D., Zhang, Z., Xia, X., Yin, W.(2016). Identification of phasiRNAs and their drought-responsiveness in Populus trichocarpa. FEBS Lett. 590: 3616–3627. [DOI] [PubMed] [Google Scholar]
- Singh, M., Goel, S., Meeley, R.B., Dantec, C., Parrinello, H., Michaud, C., Leblanc, O., Grimanelli, D.(2011). Production of viable gametes without meiosis in maize deficient for an ARGONAUTE protein. Plant Cell 23: 443–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skopelitis, D.S., Benkovics, A.H., Husbands, A.Y., Timmermans, M.C.P.(2017). Boundary formation through a direct threshold-based readout of mobile small RNA gradients. Dev. Cell 43: 265–273.e6. [DOI] [PubMed] [Google Scholar]
- Skopelitis, D.S., Hill, K., Klesen, S., Marco, C.F., von Born, P., Chitwood, D.H., Timmermans, M.C.P.(2018). Gating of miRNA movement at defined cell-cell interfaces governs their impact as positional signals. Nat. Commun. 9: 3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, X., et al. (2012). Roles of DCL4 and DCL3b in rice phased small RNA biogenesis. Plant J. 69: 462–474. [DOI] [PubMed] [Google Scholar]
- Sosa-Valencia, G., Palomar, M., Covarrubias, A.A., Reyes, J.L.(2017). The legume miR1514a modulates a NAC transcription factor transcript to trigger phasiRNA formation in response to drought. J. Exp. Bot. 68: 2013–2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souret, F.F., Kastenmayer, J.P., Green, P.J.(2004). AtXRN4 degrades mRNA in Arabidopsis and its substrates include selected miRNA targets. Mol. Cell 15: 173–183. [DOI] [PubMed] [Google Scholar]
- Tamim, S., Cai, Z., Mathioni, S.M., Zhai, J., Teng, C., Zhang, Q., Meyers, B.C.(2018). Cis-directed cleavage and nonstoichiometric abundances of 21-nucleotide reproductive phased small interfering RNAs in grasses. New Phytol. 220: 865–877. [DOI] [PubMed] [Google Scholar]
- Teng, C., Zhang, H., Hammond, R., Huang, K., Meyers, B.C., Walbot, V.(2020). Dicer-like 5 deficiency confers temperature-sensitive male sterility in maize. Nat. Commun. 11: 2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsikou, D., Yan, Z., Holt, D.B., Abel, N.B., Reid, D.E., Madsen, L.H., Bhasin, H., Sexauer, M., Stougaard, J., Markmann, K.(2018). Systemic control of legume susceptibility to rhizobial infection by a mobile microRNA. Science 362: 233–236. [DOI] [PubMed] [Google Scholar]
- Vazquez, F., Vaucheret, H., Rajagopalan, R., Lepers, C., Gasciolli, V., Mallory, A.C., Hilbert, J.L., Bartel, D.P., Crété, P.(2004). Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol. Cell 16: 69–79. [DOI] [PubMed] [Google Scholar]
- Vernoud, V., Laigle, G., Rozier, F., Meeley, R.B., Perez, P., Rogowsky, P.M.(2009). The HD-ZIP IV transcription factor OCL4 is necessary for trichome patterning and anther development in maize. Plant J. 59: 883–894. [DOI] [PubMed] [Google Scholar]
- Walbot, V., Egger, R.L.(2016). Pre-meiotic anther development: Cell fate specification and differentiation. Annu. Rev. Plant Biol. 67: 365–395. [DOI] [PubMed] [Google Scholar]
- Wang, Y., Itaya, A., Zhong, X., Wu, Y., Zhang, J., van der Knaap, E., Olmstead, R., Qi, Y., Ding, B.(2011). Function and evolution of a MicroRNA that regulates a Ca2+-ATPase and triggers the formation of phased small interfering RNAs in tomato reproductive growth. Plant Cell 23: 3185–3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z., Butel, N., Santos-González, J., Borges, F., Yi, J., Martienssen, R.A., Martinez, G., Köhler, C.(2020). Polymerase IV plays a crucial role in pollen development in Capsella. Plant Cell 32: 950–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z., Hardcastle, T.J., Canto Pastor, A., Yip, W.H., Tang, S., Baulcombe, D.C.(2018). A novel DCL2-dependent miRNA pathway in tomato affects susceptibility to RNA viruses. Genes Dev. 32: 1155–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, M.F., Tian, Q., Reed, J.W.(2006). Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development 133: 4211–4218. [DOI] [PubMed] [Google Scholar]
- Wu, P.H., Fu, Y., Cecchini, K., Özata, D.M., Arif, A., Yu, T., Colpan, C., Gainetdinov, I., Weng, Z., Zamore, P.D.(2020). The evolutionarily conserved piRNA-producing locus pi6 is required for male mouse fertility. Nat. Genet. 52: 728–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, X.M., Kou, S.J., Liu, Y.L., Fang, Y.N., Xu, Q., Guo, W.W.(2015). Genomewide analysis of small RNAs in nonembryogenic and embryogenic tissues of citrus: microRNA- and siRNA-mediated transcript cleavage involved in somatic embryogenesis. Plant Biotechnol. J. 13: 383–394. [DOI] [PubMed] [Google Scholar]
- Xia, R., Chen, C., Pokhrel, S., Ma, W., Huang, K., Patel, P., Wang, F., Xu, J., Liu, Z., Li, J., Meyers, B.C.(2019). 24-nt reproductive phasiRNAs are broadly present in angiosperms. Nat. Commun. 10: 627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, R., Meyers, B.C., Liu, Z., Beers, E.P., Ye, S., Liu, Z.(2013). MicroRNA superfamilies descended from miR390 and their roles in secondary small interfering RNA biogenesis in Eudicots. Plant Cell 25: 1555–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, R., Xu, J., Arikit, S., Meyers, B.C.(2015a). Extensive families of miRNAs and PHAS loci in Norway spruce demonstrate the origins of complex phasiRNA networks in seed plants. Mol. Biol. Evol. 32: 2905–2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, R., Xu, J., Meyers, B.C.(2017). The emergence, evolution, and diversification of the miR390-TAS3-ARF pathway in land plants. Plant Cell 29: 1232–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, R., Ye, S., Liu, Z., Meyers, B.C., Liu, Z.(2015b). Novel and recently evolved microRNA clusters regulate expansive F-BOX gene networks through phased small interfering RNAs in wild diploid strawberry. Plant Physiol. 169: 594–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, R., Zhu, H., An, Y.-Q., Beers, E.P., Liu, Z.(2012). Apple miRNAs and tasiRNAs with novel regulatory networks. Genome Biol. 13: R47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, J., Yang, X., Song, Y., Du, Q., Li, Y., Chen, J., Zhang, D.(2017). Adaptive evolution and functional innovation of Populus-specific recently evolved microRNAs. New Phytol. 213: 206–219. [DOI] [PubMed] [Google Scholar]
- Yifhar, T., Pekker, I., Peled, D., Friedlander, G., Pistunov, A., Sabban, M., Wachsman, G., Alvarez, J.P., Amsellem, Z., Eshed, Y.(2012). Failure of the tomato trans-acting short interfering RNA program to regulate AUXIN RESPONSE FACTOR3 and ARF4 underlies the wiry leaf syndrome. Plant Cell 24: 3575–3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai, J., et al. (2011). MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev. 25: 2540–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai, J., Zhang, H., Arikit, S., Huang, K., Nan, G.L., Walbot, V., Meyers, B.C.(2015). Spatiotemporally dynamic, cell-type-dependent premeiotic and meiotic phasiRNAs in maize anthers. Proc. Natl. Acad. Sci. USA 112: 3146–3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai, J., Zhao, Y., Simon, S.A., Huang, S., Petsch, K., Arikit, S., Pillay, M., Ji, L., Xie, M., Cao, X., Yu, B., Timmermans, M., et al. (2013). Plant microRNAs display differential 3′ truncation and tailing modifications that are ARGONAUTE1 dependent and conserved across species. Plant Cell 25: 2417–2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai, L., Sun, W., Zhang, K., Jia, H., Liu, L., Liu, Z., Teng, F., Zhang, Z.(2014). Identification and characterization of Argonaute gene family and meiosis-enriched Argonaute during sporogenesis in maize. J. Integr. Plant Biol. 56: 1042–1052. [DOI] [PubMed] [Google Scholar]
- Zhang, H., Xia, R., Meyers, B.C., Walbot, V.(2015). Evolution, functions, and mysteries of plant ARGONAUTE proteins. Curr. Opin. Plant Biol. 27: 84–90. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Xia, R., Kuang, H., Meyers, B.C.(2016). The diversification of plant NBS-LRR defense genes directs the evolution of microRNAs that target them. Mol. Biol. Evol. 33: 2692–2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z., Guo, X., Ge, C., Ma, Z., Jiang, M., Li, T., Koiwa, H., Yang, S.W., Zhang, X.(2017). KETCH1 imports HYL1 to nucleus for miRNA biogenesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 114: 4011–4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, M., Meyers, B.C., Cai, C., Xu, W., Ma, J.(2015). Evolutionary patterns and coevolutionary consequences of MIRNA genes and microRNA targets triggered by multiple mechanisms of genomic duplications in soybean. Plant Cell 27: 546–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, Y., Wang, Y., Wu, J., Ding, B., Fei, Z.(2015). A dynamic evolutionary and functional landscape of plant phased small interfering RNAs. BMC Biol. 13: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, C., et al. (2013). The trans-acting short interfering RNA3 pathway and no apical meristem antagonistically regulate leaf margin development and lateral organ separation, as revealed by analysis of an argonaute7/lobed leaflet1 mutant in Medicago truncatula. Plant Cell 25: 4845–4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, H., et al. (2012). Photoperiod- and thermo-sensitive genic male sterility in rice are caused by a point mutation in a novel noncoding RNA that produces a small RNA. Cell Res. 22: 649–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo, J., Wang, Q., Han, C., Ju, Z., Cao, D., Zhu, B., Luo, Y., Gao, L.(2017). SRNAome and degradome sequencing analysis reveals specific regulation of sRNA in response to chilling injury in tomato fruit. Physiol. Plant. 160: 142–154. [DOI] [PubMed] [Google Scholar]


