Abstract
Background:
Chronic exposure to arsenic (As), a human toxicant and carcinogen, remains a global public health problem. Health risks persist after As exposure has ended, suggesting epigenetic dysregulation as a mechanistic link between exposure and health outcomes.
Objectives:
We investigated the association between total urinary As and locus-specific DNA methylation in the Strong Heart Study, a cohort of American Indian adults with low-to-moderate As exposure [total urinary As, creatinine: 11.7 (10.6)].
Methods:
DNA methylation was measured in 2,325 participants using the Illumina MethylationEPIC array. We implemented linear models to test differentially methylated positions (DMPs) and the DMRcate method to identify regions (DMRs) and conducted gene ontology enrichment analysis. Models were adjusted for estimated cell type proportions, age, sex, body mass index, smoking, education, estimated glomerular filtration rate, and study center. Arsenic was measured in urine as the sum of inorganic and methylated species.
Results:
In adjusted models, methylation at 20 CpGs was associated with urinary As after false discovery rate (FDR) correction (). After Bonferroni correction, 5 CpGs remained associated with total urinary As (), located in SLC7A11, ANKS3, LINGO3, CSNK1D, ADAMTSL4. We identified one DMR on chromosome 11 (chr11:2,322,050-2,323,247), annotated to C11orf2; TSPAN32 genes.
Discussion:
This is one of the first epigenome-wide association studies to investigate As exposure and locus-specific DNA methylation using the Illumina MethylationEPIC array and the largest epigenome-wide study of As exposure. The top DMP was located in SLC7A11A, a gene involved in cystine/glutamate transport and the biosynthesis of glutathione, an antioxidant that may protect against As-induced oxidative stress. Additional DMPs were located in genes associated with tumor development and glucose metabolism. Further research is needed, including research in more diverse populations, to investigate whether As-related DNA methylation signatures are associated with gene expression or may serve as biomarkers of disease development. https://doi.org/10.1289/EHP6263
Introduction
Arsenic (As) exposure through drinking water is a global public health concern affecting at least 140 million people (World Health Organization 2012). In 2001, the U.S. Environmental Protection Agency (U.S. EPA) lowered maximum contaminant level (MCL) from 50 to . With the implementation of this new standard, the number of people served by public water systems with water As has declined from 13 million in 2001 to 296,000 in 2018 (U.S. EPA 2018). An estimated 2.1 million individuals, however, remain exposed to As in drinking water from private wells (Ayotte et al. 2017), which are not regulated under the MCL.
Arsenic is a human toxicant and group 1 carcinogen (World Health Organization 2011). Chronic As exposure increases the risk of numerous health conditions, including skin lesions, impaired intellectual function, cardiovascular disease, diabetes, inflammation, and cancers including bladder, lung, kidney, liver, skin, and possibly prostate (IARC Working Group 2009; Moon et al. 2017; National Research Council 2013). Elevated risk of cancer mortality (Roh et al. 2018; Smith et al. 2018) and lung disease (Steinmaus et al. 2016) following early life exposure persists decades after exposure has been reduced. Epigenetic dysregulation, including changes in DNA methylation patterns, may provide a mechanistic link between As exposure and health outcomes with prolonged latency periods (Bailey et al. 2016). DNA methylation can influence gene expression by inhibiting transcription factor binding in promoter regions and recruiting DNA binding proteins, and DNA methylation is involved in maintaining chromosomal stability (Robertson 2005).
In vitro, animal, and human evidence supports the findings that alterations in the epigenome are involved in the etiology of As-induced health outcomes and carcinogenesis (Bailey et al. 2016; Bailey and Fry 2014; Carlin et al. 2015). In epidemiological studies, As exposure has been associated with global DNA methylation levels (Intarasunanont et al. 2012; Kile et al. 2012; Niedzwiecki et al. 2015, 2013; Pilsner et al. 2007, 2009). Loci-specific DNA methylation has also been assessed in epigenome-wide association studies (EWAS) using the Illumina Infinium HumanMethylation BeadChip (450K), which interrogates DNA methylation at loci (Argos 2015). Studies of in utero As exposure and studies of As exposure in adults have identified significant associations with DNA methylation at individual CpG sites; however, the identity and number of CpGs identified (ranging from 0 to 4,771 after adjusting for multiple comparisons) differ between studies (Table S1).
Inconsistent results among epidemiological studies of the association between As exposure and epigenetic dysregulation may be due to differences between populations studied (e.g., age, sex, genetic structure), levels of As exposure, differential residual confounding, and methods for quantifying DNA methylation (Argos 2015). Previous EWAS of As exposure have included birth cohorts in the United States (Green et al. 2016; Koestler et al. 2013), Bangladesh (Broberg et al. 2014; Cardenas et al. 2015; Gliga et al. 2018; Kile et al. 2012), Mexico (Rojas et al. 2015), and Taiwan (Kaushal et al. 2017), and studies of adults in the United States (Liu et al. 2014), Bangladesh (Argos et al. 2015; Demanelis et al. 2019), Argentina (Ameer et al. 2017), and China (Guo et al. 2018). In addition, the samples sizes of previous EWAS have ranged from , resulting in limited statistical, particularly when interrogating a large number of CpG sites (Argos 2015).
The objective of this study was to investigate the association between total urinary As and locus-specific DNA methylation in the Strong Heart Study (SHS), a population-based prospective cohort of American Indian adults with low-to-moderate levels of As exposure primarily through drinking water (Navas-Acien et al. 2009; Nigra et al. 2019). Exposure levels were consistent in the 10-y period following recruitment (1989–1999) due primarily to contaminated drinking water (Navas-Acien et al. 2009; Nigra et al. 2019). In the SHS, As exposure has been associated with cardiovascular disease incidence and mortality (Moon et al. 2013); reduced lung function (Powers et al. 2019); diabetes incidence (Grau-Perez et al. 2017); incident chronic kidney disease (Zheng et al. 2015); and lung, prostate, and pancreatic cancer mortality (García-Esquinas et al. 2013). In the current analyses, we analyzed associations between total urinary As levels and DNA methylation measured using the Infinium MethylationEPIC BeadChip.
Methods
Study Population
The SHS has been described in detail (Lee et al. 1990). Briefly, from July 1989 to June 1991, SHS participants were recruited from 13 tribes in Arizona, Oklahoma, and North and South Dakota. All noninstitutionalized tribal members age 45–74 y were eligible for enrollment. Eligible persons were identified from tribal rolls and invited to participate in person or by mail. Cluster sampling was used in North and South Dakota. A total of 4,549 men and women age 45–75 y accepted to participate in the period 1989–1991 (62% participation rate). Most participants were born in their communities and had lived there for their entire lives. In 2009, As was measured in the urine of 3,973 participants with sufficient urine available. For this study, one of the tribes declined to participate, leaving 3,516 potential participants. We further excluded 251 participants with prevalent cardiovascular disease, 534 participants missing data on other cardiovascular risk factors, and 380 lacking sufficient DNA samples, leaving 2,351 participants for blood DNA analyses (Figure S1). Participants included in analyses of DNA methylation were similar to all eligible participants (Table S2).
Ethics
The study protocols were approved by institutional review boards from the participating research institutions, the Indian Health Service, and the tribal communities. Participating communities approved this manuscript and received a lay summary of findings as part of the SHS community-based research approach that included partnership with local communities.
Data Collection
During baseline visits, a clinical exam was conducted, and biospecimens were collected by trained and certified nurses and medical examiners.
Urinary as Concentration
Arsenic concentrations were measured in all urine samples collected at baseline (1989–1991) with a sufficient volume available. Analytical methods and quality control for urine As measurement in urine samples collected at baseline has been described in detail (Scheer et al. 2012). In summary, baseline spot urine samples were stored in polypropylene tubes and frozen samples were shipped on dry ice to the MedStar Health Research Institute (Washington, DC, USA), where they were stored at . In the period 2009–2010, samples were thawed and an aliquot up to was transported on dry ice to the Trace Element Laboratory, Graz University (Austria), where they were stored at until analysis for total urinary As concentrations and As metabolite concentrations.
Urinary As concentrations were calculated as the sum of the concentrations of inorganic (iAs), monomethyl- (MMA), and dimethyl- (DMA) As species and are referred to herein as total urinary As. iAs, MMA, and DMA concentrations were measured using high-performance liquid chromatography coupled to inductively coupled plasma mass spectrometry (Agilent z HPLC and Agilent 7700x ICP-MS; Agilent Technologies) (Scheer et al. 2012). Interassay coefficients of variation were: 6.0%, 6.5%, and 5.9% for iAs, MMA, and DMA, respectively. The limits of detection (LOD) for iAs (), MMA, and DMA was (Scheer et al. 2012). Samples with As species concentrations beneath the LOD were replaced with (iAs: , 5.5%; MMA: , 0.7%; DMA: ). Concentrations of arsenobetaine, a nontoxic As species found in seafood (Joint FAO/WHO Export Committee on Food Additives 1989), were low (median creatinine), reflecting little seafood intake in the study population.
Urinary creatinine was measured at the National Institute of Diabetes and Digestive and Kidney Diseases Epidemiology and Clinical Research Branch laboratory (Phoenix, Arizona, USA) using an automated alkaline picrate methodology run on a rapid flow analyzer (Lee et al. 1990). Total urinary As concentrations () were divided by urinary creatinine (g/L) to account for urine dilution. Serum creatinine was measured in fasting blood samples on a Hitachi 717 platform (Hitachi Ltd.) using an automated alkaline-picrate rate method (Roche Diagnostics). Urine specific gravity was measured using a refractometer (Leica TS 400 Refractometer; Leica Microsystems Inc.). Serum creatinine, age, and sex were used to calculate estimated glomerular filtration rate (eGFR) using the Chronic Kidney Disease Epidemiology Collaboration (CKD-EPI) equation (Levey et al. 2009; Shara et al. 2012).
Epigenome-Wide DNA Methylation Assessment and Quality Control
DNA methylation was measured in blood samples collected at baseline. Blood samples were collected in EDTA tubes. DNA from white blood cells was isolated using organic solvents at the MedStar Health Research Institute or at the Texas Biomed Research Institute and stored at . All epigenetic analyses were performed at the Texas Biomedical Research Institute. Genomic DNA was bisulfite-converted and eluted in buffer. DNA methylation was measured in all available samples (Figure S1). DNA methylation was measured using the Infinium MethylationEPIC BeadChip (850K) (Illumina, Inc.) according to the manufacturer’s instructions. The MethylationEPIC BeadChip provides a measure of DNA methylation at a single nucleotide resolution at methylation sites, including of loci measured by the 450K microarray (Illumina 2015). Samples were randomized within and across plates to minimize potential batch artifacts. A 0% methylation and 100% methylation control were included on each plate. In addition, two control samples were included on each plate and standard quality control checks (e.g., performance of Illumina controls, call rate for detected CpGs) using the Illumina GenomeStudio software (Illumina, Inc.). Samples with low performance were also identified using GenomeStudio and rerun using a different DNA source if available.
Raw methylation image files were processed using the minfi package (Aryee et al. 2014) in R (R Core Development Team). Density plots were generated to analyze the distribution of Beta-values, and 18 samples that did not have classical bimodal distributions were excluded. Normalization was performed using single sample Noob (Fortin et al. 2017). An additional eight samples were excluded with low median intensity of methylated and unmethylated channels []. Probes determined to be technical failures () were removed. Batch effects for plate and row were corrected using the ComBat function in the sva package (version 3.36.0), which employs an empirical Bayesian framework (Leek et al. 2012). Clustering by batch and row before and after applying ComBat was visually assessed using the first two principal components. The proportions of T cells, T cells, natural killer (NK) cells, B cells, monocytes, and granulocytes in each sample were estimated using the Houseman projection method (Houseman et al. 2012). In addition, probes located in X and Y chromosomes, probes associated with single-nucleotide polymorphisms with a minor allele frequency in an admixed American population, and probes previously identified as cross-reactive were removed prior to analysis (McCartney et al. 2016; Pidsley et al. 2016). After exclusion of probes and samples, DNA methylation data measured at 788,753 loci in 2,325 samples were available for analysis.
For sensitivity analyses, to correct for potential probe-type bias, Representative Concentration Pathway (RCP) normalization was performed (Niu et al. 2016). We also conducted sensitivity analyses using a reference-free approach to correct for cell type distribution. We implemented ReFACTor, a method based on sparse principal component analysis (Rahmani et al. 2016), using our processed Beta-value data set to estimate ReFACTor components of six cell types.
Other Variables
Height and weight were measured for body mass index (BMI) calculation. A trained interviewer collected data on sociodemographics and health-related behaviors (e.g., age, sex, history of smoking) (Lee et al. 1990). Diabetes status was determined according to the American Diabetes Association classification using fasting glucose , 2-h post-load plasma glucose , hemoglobin A1c , or taking diabetes medication (American Diabetes Association 2014).
Statistical Analysis
Descriptive statistics [means and standard deviations (SDs) for continuous variables, frequencies for categorical variables] were calculated for all covariates. To adjust for between-individual differences in urine dilution, total urinary As concentrations were divided by urinary creatinine concentrations and expressed as creatinine. Adjusted urinary As levels were right-skewed and natural log-transformed to reduce the influence of extreme values. Associations between estimated cell type proportions and ln(total urinary As) were evaluated using linear models adjusted for age, sex, BMI, and smoking status (never smoker, former smoker, current smoker).
Differentially methylated positions (DMPs) were identified using linear regression models implemented in the limma package in R with empirical Bayes smoothing of standard errors (Ritchie et al. 2015). Beta-values were logit transformed to M-values—i.e., to meet linear regression model assumptions (Du et al. 2010). Models were adjusted for sex, age, BMI, self-reported smoking status, education (, high school graduate or GED, ), study center (Arizona, Oklahoma, North and South Dakota), eGFR, and cell type proportion estimates. Potential systematic biases were evaluated using Q-Q plots and the genomic inflation factor (). For the fully adjusted models, , suggesting that our analyses were not affected by genomic inflation (Figure S2). Multiple comparisons were accounted for using the Benjamini and Hochberg method for false discovery rates (FDR) (Benjamini and Hochberg 1995) and the Bonferroni correction with a level of significance of . Using the p.adjust function in R, FDR-adjusted -values were calculated as the number of expected divided by the number of observed [i.e., for N-ordered -values through ], and Bonferroni-adjusted -values were calculated as multiplied by the number of tests (i.e., ).
Sensitivity analyses were performed for the potential effect modifiers of sex, smoking status, study center, and diabetes status. For participants without diabetes, specific gravity, rather than urinary creatinine, was used to correct total urinary As concentrations () for between-individual differences in urine dilution by multiplying the sample urinary As concentration by the ratio (mean specific gravity – 1)/(participant’s specific gravity – 1) (Miller et al. 2004). In addition, limma analyses were performed for each stratum. Due to reduced sample size with stratification, significance was assessed at a nominal . For loci that did not achieve significance at in stratified analyses, we assessed effect modification using linear models which included an interaction term. Sensitivity analyses were also performed for the effects of adjusting for probe-type bias due to differences between Infinium I and Infinium II probes on the MethylationEPIC BeadChip. Overall limma analyses were performed on data processed using RCP normalization in addition to single sample Noob normalization. We also tested for the effect of using a reference-free method to adjust for cell type proportion by including ReFACTor components in adjusted limma models.
Recognizing the limitations of nonpenalized linear regression for accommodating high-dimensional data or highly correlated predictors, we also used GLMnet penalized linear regression to identify DMPs (elastic-net, R package glmnet, version 4.0-2). The elastic-net framework allows us to include all CpGs in the same model, thus being able to account for the complex interrelationships across CpGs and leverage the rich EWAS information of the microarray. Elastic-net approach is a mix between Ridge and Lasso regression (Friedman et al. 2010) that can successfully model high-dimensional DNA methylation (Benton et al. 2017). The algorithm fits a generalized linear model using penalized maximum likelihood with penalty controlled by the parameter; we selected to ensure that our model was more flexible for correlated data. The regularization path is computed for the penalty at values as specified by the regularization parameter so that the minimum mean squared error is achieved. was selected using 10-fold cross-validation in our study. We fit a linear elastic-net using ln(total urinary As) as the outcome and methylation levels as predictors, adjusting for sex, age, BMI, self-reported smoking status, education, study center, eGFR, and cell type proportion estimates.
Differentially methylated regions (DMRs) were tested using the DMRcate package in R, which uses a Gaussian kernel smoothing function to grouping of significant probes identified by limma (Peters et al. 2015). DMRcate was applied using a Gaussian kernel bandwidth with a smoothing factor . The bandwidth value defines the maximum distance in nucleotides used to group methylated loci.
Gene ontology (GO) enrichment analysis was conducted using the gometh (Geeleher et al., 2013) function implemented in the R package missMethyl (Phipson et al. 2016). The gometh algorithm identifies GO terms that contain an overrepresentation of genes with DMPs while accounting for differences in the a priori probabilities of genes to include DMPs based on representation among probes included on the MethylationEPIC BeadChip. All probes tested in limma analyses (representing 25,715 genes) and probes identified as significantly associated with urinary As levels at were used as input for the gometh function.
Evaluation of Previously Identified Signals
To identify previous studies investigating the association between As exposure and loci-specific DNA methylation, we searched PubMed for the terms “arsenic” and “DNA methylation.” Loci associated with As exposure reported by those studies measuring DNA methylation using the Illumina HumanMethylation BeadChip or the Infinium MethylationEPIC BeadChip were included in a lookup approach. Loci with a nominal in the limma analysis described above were considered statistically significant.
All analyses were performed using R 3.4.3 (R Core Development Team).
Results
Participant Characteristics
The mean (SD) age of participants was 56.2 (8.1) y, 58.5% of participants were female, and 41.6% of participants had diabetes (Table 1). Total urinary As levels ranged from creatinine, with mean (±SD) creatinine and median [interquartile range (IQR)] creatinine. Total urinary As was associated with the imputed proportions of NK cells and B cells: on average, with every increase in one log-unit of As (), there was an increase of 4% in the proportion of NK cells (), and a decrease of 7% in the proportion of B cells () (Table S3). Total urinary As was not significantly associated with the imputed proportions of monocytes (), T cells (), T cells (), granulocytes ().
Table 1.
Participant characteristics ().
| N | (%) | |
|---|---|---|
| Sex | ||
| Female | 1,361 | (58.5) |
| Male | 964 | (41.5) |
| Age [mean years (SD)] | 56.2 | (8.1) |
| Study center | ||
| Arizona | 312 | (13.4) |
| Oklahoma | 981 | (42.2) |
| North and South Dakota | 1,032 | (44.4) |
| Total urinary As [mean creatinine (SD)] | 11.7 | (10.6) |
| creatinine | 533 | (22.9) |
| creatinine | 1,252 | (53.8) |
| creatinine | 540 | (23.2) |
| Total urinary As by study center [mean creatinine (SD)] | ||
| Arizona | 18.9 | (12.14) |
| Oklahoma | 6.9 | (5.51) |
| North and South Dakota | 14.1 | (11.75) |
| Education | ||
| diploma | 963 | (41.4) |
| High school diploma or GED | 658 | (28.3) |
| diploma | 704 | (30.3) |
| Smoking | ||
| Never smoker | 684 | (29.4) |
| Former smoker | 748 | (32.2) |
| Current smoker | 893 | (38.4) |
| BMI [mean (SD)] | 30.3 | (6.1) |
| Diabetes statusa | ||
| Diabetes diagnosis | 968 | (41.6) |
| Not diagnosed with diabetes | 1,357 | (58.4) |
| eGFR [mean mL/min (SD)] | 987.4 | (16.8) |
Note: BMI, body mass index; eGFR, Estimated glomerular filtration rate; GED, General Education Diploma.
Fasting glucose (), 2-h post-load plasma glucose (), HbA1c (), or taking diabetes medication. Data are complete for all variables.
Differentially Methylated Positions
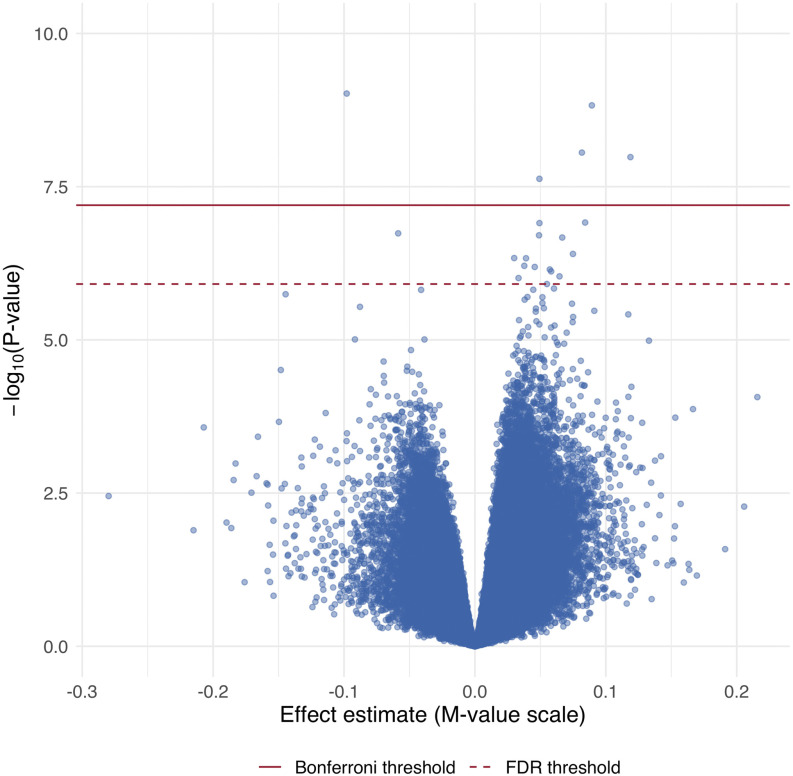
In locus-specific analyses, 788,753 methylated positions were tested for associations with ln(total urinary As), of which 39,857 (5.1%) were significantly associated with As at a nominal . Nominally significant loci are listed in Excel Table S1. After adjusting for multiple comparisons using an FDR and Bonferroni approach, 20 () and five () loci remained significantly associated with ln(total urinary As), respectively (Figure 1). Table 2 summarizes the loci significant at the FDR threshold, and includes effect size estimates from models of Beta-values for interpretation of results. Most significant loci () were located within gene bodies. Two genes, leucine rich repeat and Ig domain containing 3 (LINGO3) and casein kinase 1 delta (CSNK1D) contained two FDR-significant CpGs located within the same genomic feature (in LINGO3, cg22294740 and cg08059112 are located 74 nucleotides apart; in CSNK1D, cg20493718 and cg21369801 are located 18 nucleotides apart). Of the 20 FDR-significant loci, the association between urinary As levels and DNA methylation was positive at 18 loci and inverse at two loci (cg06690548 and cg00500428). Among all probes, urinary As levels also appeared to be related to hypermethylation: 87% of the top 100 probes ranked by -values were positively associated with As, and 58% of all probes were positively associated with As (Figure 2).
Figure 1.
Manhattan plot for the epigenome-wide association of log(total urinary arsenic levels) and DNA methylation levels. limma models adjusted for age, sex, BMI, self-reported smoking status, education (, high school graduate or GED, ), study center (Arizona, Oklahoma, North and South Dakota), estimated glomerular filtration rate, and cell-type proportion estimates. The solid line represents the Bonferroni threshold, and the dashed line represents the FDR threshold for significance. Note: BMI, body mass index; FDR, false discovery rate; GED, General Education Diploma.
Table 2.
Differentially methylated CpG sites associated with ln(total urinary arsenic levels) at , sorted by chromosome and position.
| CpG | Chromosome | Positiona | Gene | Feature category | -Value | Median methylation (%)b | Mean difference in % methylation (95% CI)c | ||
|---|---|---|---|---|---|---|---|---|---|
| cg12106731 | 1 | 36023279 | NCDN; KIAA0319L | TSS200; TSS1500 | 0.014 | 0.098 | 0.60 | 0.04 (0.02, 0.05) | |
| cg07317306 | 1 | 110314824 | Intergenic | — | 0.041 | 0.773 | 17.75 | 0.50 (0.31, 0.69) | |
| cg18616702 | 1 | 150523808 | ADAMTSL4; MIR4257 | 5′UTR; TSS1500 | 0.004 | 0.019 | 22.45 | 0.80 (0.50, 1.10) | |
| cg04940901 | 3 | 52418250 | DNAH1 | Body | 0.017 | 0.155 | 71.58 | 0.97 (0.60, 1.33) | |
| cg06690548 | 4 | 139162808 | SLC7A11 | Body | 0.001 | 0.001 | 96.24 | (, ) | |
| cg01538969 | 6 | 30624636 | DHX16 | Body | 0.035 | 0.559 | 57.05 | 1.31 (0.80, 1.82) | |
| cg14827056 | 8 | 141550539 | EIF2C2 | Body | 0.017 | 0.169 | 88.76 | 0.58 (0.35, 0.81) | |
| cg09280971 | 9 | 73036509 | Intergenic | — | 0.028 | 0.365 | 31.54 | 0.64 (0.40, 0.89) | |
| cg14595618 | 10 | 71135446 | HK1 | Body | 0.035 | 0.602 | 39.52 | 1.29 (0.76, 1.82) | |
| cg27178850 | 11 | 9039431 | Intergenic | — | 0.028 | 0.367 | 69.43 | 0.82 (0.51, 1.13) | |
| cg03036214 | 15 | 63640658 | CA12 | Body | 0.048 | 0.967 | 56.18 | 1.30 (0.78, 1.82) | |
| cg00500428 | 16 | 3074502 | HCFC1R1; THOC6 | TSS1500; Body | 0.017 | 0.144 | 5.61 | (, ) | |
| cg03497652 | 16 | 4751569 | ANKS3 | Body | 0.001 | 0.001 | 74.34 | 1.57 (1.08, 2.06) | |
| cg07021906 | 16 | 87866833 | SLC7A5 | Body | 0.040 | 0.725 | 82.02 | 0.85 (0.51, 1.20) | |
| cg12116137 | 17 | 1576449 | PRPF8 | Body | 0.014 | 0.096 | 77.19 | 1.33 (0.83, 1.84) | |
| cg21369801 | 17 | 80202961 | CSNK1D | Body; 3′UTR | 0.028 | 0.312 | 77.81 | 1.17 (0.73, 1.62) | |
| cg20493718 | 17 | 80202979 | CSNK1D | Body; 3′UTR | 0.002 | 0.008 | 97.47 | 0.35 (0.20, 0.50) | |
| cg08059112 | 19 | 2294887 | LINGO3 | 5′UTR | 0.034 | 0.488 | 56.18 | 0.89 (0.54, 1.24) | |
| cg22294740 | 19 | 2294961 | LINGO3 | 5′UTR | 0.002 | 0.007 | 55.92 | 1.84 (1.20, 2.48) | |
| cg20509831 | 19 | 58866362 | A1BG-AS1; A1BG; ZNF497 | Body, TSS1500; 3′UTR | 0.034 | 0.509 | 28.10 | 0.89 (0.53, 1.25) |
Note: —, no data; BMI, body mass index; CI, confidence interval; GED, General Education Diploma.
GRCh37/hg19 assembly.
Percent methylation calculated .
Effect size estimate from limma models of Beta-values [i.e., mean difference in percent methylation for each unit change in ln(total urinary arsenic levels)] adjusted for age, sex, BMI, self-reported smoking status, education (, high school graduate or GED, ), study center (Arizona, Oklahoma, North and South Dakota), estimated glomerular filtration rate, and cell-type proportion estimates.
Figure 2.
Volcano plot for the epigenome-wide association of log(total urinary arsenic levels) and DNA methylation levels. Limma models adjusted for age, sex, BMI, self-reported smoking status, education (, high school graduate or GED, ), study center (Arizona, Oklahoma, North and South Dakota), estimated glomerular filtration rate, and cell-type proportion estimates. The solid line represents the Bonferroni threshold and the dashed line represents the FDR threshold for significance. Note: BMI, body mass index; FDR, false discovery rate; GED, General Education Diploma.
Sensitivity Analyses
Results were consistent in stratified analyses by sex (Table S4), smoking status (Table S5), study center (Table S6), and diabetes status (Table S7). By sex and smoking status, all 20 FDR-significant sites achieved significance at a nominal in stratified analyses. In the smallest stratum representing Arizona (), 4 loci were not associated with urinary As (cg20509831 located in A1BG-AS1; A1BG; ZNF497: nominal ; cg18616702 located in ADAMTSL4; MIR4257: nominal ; cg09280971: nominal ; cg07317306: nominal ), although the observed directions of association were consistent with unstratified analyses. All FDR-significant loci achieved nominal significance among participants located in Oklahoma and North and South Dakota. By diabetes status, all FDR-significant loci achieved nominal significance in both strata, with the exception of one CpG among participants without diabetes when using urinary creatinine concentrations to correct for urine dilution [cg14595618 located in hexokinase 1 (HK1): nominal ], and two CpGs among participants without diabetes when using specific gravity to correct for urine dilution (cg14595618 located in HK1: nominal , and cg14827056 located in EIF2C2: nominal ). We assessed possible interaction between As exposure and diabetes status on DNA methylation at cg14595618 using a linear model including the interaction term status and adjusted age, sex, BMI, smoking status, education, study center, eGFR, and cell type proportion estimates. Interaction between As levels and diabetes status was statistically significant () (Table S8). Across strata of sex, smoking status, study center, and diabetes status, the effect estimates were in the same direction.
Results of analyses of probe-type normalized data were similar to those of our primary analyses. The Q-Q plot for adjusted limma models is provided in Figure S3 (). Seventeen of the 20 FDR-significant sites achieved significance at (Table S9). Although three CpGs failed to achieve FDR significance (cg12106731: ; cg07317306: ; cg03036214: ), they were nominally significant at , and the observed magnitudes and directions of association were consistent with our primary analyses.
We also conducted sensitivity analyses using a reference-free approach to control for cell type proportions by adjusting limma models for six ReFACTor components. The Q-Q plot is provided in Figure S4 (). Sixteen of the 20 FDR-significant sites achieved significance at (Table S10). The four CpGs that did not achieve FDR significance (cg14827056: ; cg27178850: ; cg20509831: ; cg07317306: ) were nominally significant at and were consistent with our primary analyses in the magnitude and direction of associations.
Elastic-Net Analysis
We also used the elastic-net framework to evaluate all CpGs in the same model. The elastic-net model selected 315 CpGs associated with As levels (Excel Table S2). Eight of these CpGs were among those that achieved FDR-significance in linear models implemented in limma models (cg06690548, annotated to SLC7A11; cg03497652, ANKS3; cg22294740, LINGO3; cg20493718, CSNK1D; cg18616702, ADAMTSL4 and MIR4257; cg12106731, NCDN and KIAA0319L; cg00500428, HCFC1R1 and THOC6; and cg14827056, EIF2C2).
Evaluation of Previously Identified Differentially Methylated Positions
Out of 396 PubMed results containing the terms “arsenic” and “DNA methylation,” 14 studies reported EWAS of the association between As exposure and DNA methylation; 13 measured DNA methylation using the 450K microarray, and one measured DNA methylation using the 850K microarray (Table S1). Across these studies, 5,801 unique CpGs were associated with As exposure after adjustment for multiple comparisons (criteria used in each manuscript to determine significance are summarized in Table S1). A total of 4,595 DMPs were evaluated in the current study, including 17 DMPs identified in two studies, of which 191 achieved nominal significance in the current study () (Excel Table S3; the number and proportion of replicated CpGs for each study is shown is Table S1). Among 168 unique DMPs identified in studies of adults, 12 were nominally significant in our analyses. These CpGs included cg06121226 (located in SLC4A4), identified as significantly associated with total blood and urinary As levels among adults in Bangladesh () (Argos et al. 2015), and cg05428706, cg19534475 (ATP1B3), and cg06466147 (GBAP1) identified as significantly associated with urinary or water As levels among a separate cohort of adults in Bangladesh (); cg14718533 was found to be significantly associated with urinary As levels in a meta-analysis, including both Bangladeshi cohorts (Demanelis et al. 2019). In addition, cg15019001 (HLA-DPB2), cg22809683 (LAMC1), cg07466788 (SLC16A3), cg19504605 (ZFP41), cg22143856 (ZNF389), cg13251666, and cg13844779, identified as differentially methylated between As-exposed and control families in China () (Guo et al. 2018), were nominally significant in our analyses. Among 4,427 CpGs previously associated with in utero As exposure (Cardenas et al. 2015; Gliga et al. 2018; Green et al. 2016; Kaushal et al. 2017; Kile et al. 2014; Rojas et al. 2015) 179 were nominally significant in the current study. Demanelis et al. also reported all nominally significant loci, allowing us to check for significance of our FDR-significant CpGs. Furthermore, cg04940901 (DNAH1) and cg09280971 were associated in urinary or water As levels among the Bangladeshi cohort, and cg00500428 (HCFC1R1; THOC6) was associated with urinary As levels in the meta-analysis reported by Damanelis et al. at .
Differentially Methylated Regions
One DMR was identified including 20 CpGs located on chromosome 11 (chr11:2,322,050-2,323,247) (Figure 3). Table 3 lists the 20 CpGs ordered by genomic coordinates. This region spans the body, first exon, and TSS200 of C11orf21, and the first exon, TSS200, TSS1500, and 5′UTR of TSPAN32. Overall, the DNA methylation in this region was low, ranging from less than 1% to 12%. Fifteen of the 20 loci located within this region were positively and significantly associated with total urinary As levels at a nominal . With the exception of cg02537342, all CpGs were positively correlated with each other ().
Figure 3.
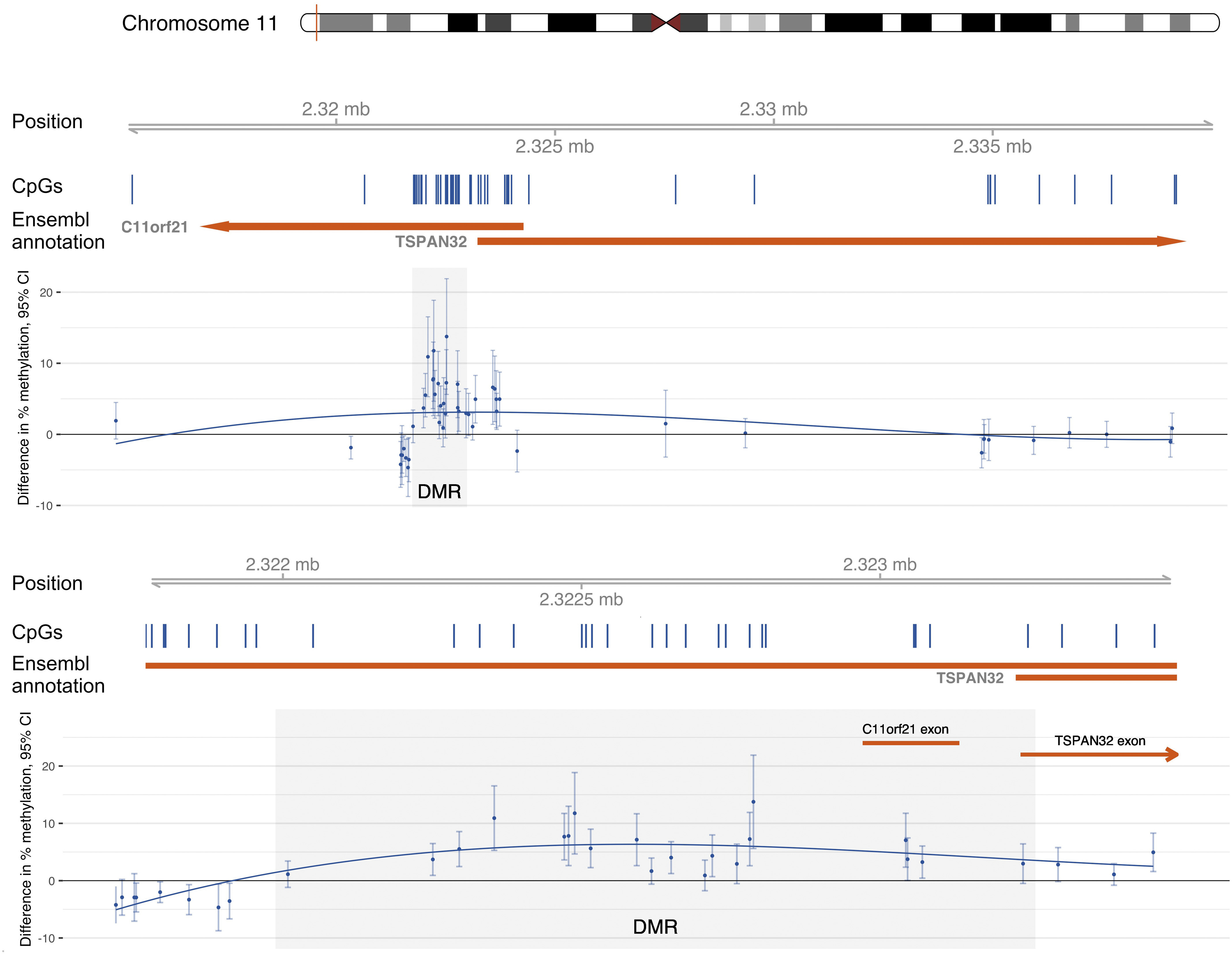
Epigenome-wide association results in the genomic region containing the differentially methylated region (DMR) chr11:2,322,050-2,323,247 (shaded area). The first panel shows the region from 7,000 nucleotides upstream of the DMR to 17,000 nucleotides downstream of the DMR; the second panel shows the region from 300 nucleotides upstream of the DMR to 300 nucleotides downstream of the DMR. Difference in % methylation from limma models of Beta-values [i.e., mean difference in percent methylation for each unit change in ln(total urinary arsenic levels)] adjusted for age, sex, BMI, self-reported smoking status, education (, high school graduate or GED, ), study center (Arizona, Oklahoma, North and South Dakota), estimated glomerular filtration rate, and cell type proportion estimates. Trend in odds of methylation related to As levels is indicated by a solid blue line fitted using polynomial splines. Note: BMI, body mass index; GED, General Education Diploma.
Table 3.
CpGs Located in the differentially methylated region chr11:2,322,050-2,323,247.
| CpG | Positiona | Gene | Feature category | -Value | Median % methylationb | Mean difference in % methylation (95% CI)c |
|---|---|---|---|---|---|---|
| cg25961099 | 2322050 | C11orf21; TSPAN32 | Body; TSS1500 | 0.019 | 9.02 | 0.08 (, 0.26) |
| cg01211906 | 2322286 | C11orf21; TSPAN32 | Body; TSS1500 | 0.002 | 2.91 | 0.10 (0.02, 0.18) |
| cg09114153 | 2322329 | C11orf21; TSPAN32 | Body; TSS1500 | 0.009 | 9.09 | 0.40 (0.15, 0.64) |
| cg01612681 | 2322386 | C11orf21; TSPAN32 | Body; TSS1500 | 1.12 | 0.17 (0.06, 0.29) | |
| cg13592872 | 2322500 | C11orf21; TSPAN32 | Body; TSS1500 | 0.502 | 4.20 | 0.28 (0.12, 0.45) |
| cg21027517 | 2322507 | C11orf21; TSPAN32 | Body; TSS1500 | 0.150 | 3.80 | 0.27 (0.08, 0.46) |
| cg05509777 | 2322517 | C11orf21; TSPAN32 | Body; TSS1500 | 0.096 | 3.75 | 0.37 (0.14, 0.60) |
| cg10782575 | 2322543 | C11orf21; TSPAN32 | Body; TSS1500 | 0.001 | 10.77 | 0.49 (0.18, 0.81) |
| cg00502099 | 2322618 | C11orf21; TSPAN32 | Body; TSS1500 | 0.092 | 1.18 | 0.09 (, 0.20) |
| cg03494648 | 2322642 | C11orf21; TSPAN32 | Body; TSS1500 | 1.06 | 0.02 (, 0.05) | |
| cg15924868 | 2322674 | C11orf21; TSPAN32 | Body; TSS1500 | 0.024 | 0.57 | 0.03 (0.00, 0.05) |
| cg02537342 | 2322729 | C11orf21; TSPAN32 | Body; TSS1500 | 0.001 | 6.96 | 0.04 (, 0.22) |
| cg00041575 | 2322741 | C11orf21; TSPAN32 | Body; TSS1500 | 1.61 | 0.06 (, 0.15) | |
| cg05403469 | 2322781 | C11orf21; TSPAN32 | Body; TSS1500 | 0.048 | 1.34 | (, 0.12) |
| cg19766471 | 2322802 | C11orf21; TSPAN32 | Body; TSS1500 | 0.005 | 11.75 | 0.72 (0.29, 1.16) |
| cg22210337 | 2322808 | C11orf21; TSPAN32 | Body; TSS1500 | 0.002 | 11.46 | 1.05 (0.40, 1.70) |
| cg21201830 | 2323056 | C11orf21; TSPAN32 | Body, 1stExon; TSS200 | 0.003 | 1.80 | 0.13 (0.01, 0.25) |
| cg15579389 | 2323059 | C11orf21; TSPAN32 | Body, 1stExon; TSS200 | 0.003 | 0.89 | 0.04 (, 0.09) |
| cg09358071 | 2323083 | C11orf21; TSPAN32 | Body, 1stExon; TSS200 | 0.92 | 0.04 (0.01, 0.07) | |
| cg05572370 | 2323247 | C11orf21; TSPAN32 | TSS200, 1stExon; 5′UTR | 0.337 | 2.22 | 0.08 (, 0.17) |
Note: BMI, body mass index; CI, confidence interval; GED, General Education Diploma: .
GRCh37/hg19 assembly.
Percent methylation calculated .
Effect size estimate from limma models of Beta-values [i.e., mean difference in percent methylation for each unit change in ln(total urinary arsenic levels)] adjusted for age, sex, BMI, self-reported smoking status, education (, high school graduate or GED, ), study center (Arizona, Oklahoma, North and South Dakota), estimated glomerular filtration rate, and cell type proportion estimates.
GO Analysis
GO analysis identified 190 GO terms overrepresented among genes containing FDR-significant DMPs and consisting of at least two genes (molecular function: 52 terms; biological process: 111 terms; cellular component: 27 terms); however, none of these terms achieved statistical significance after accounting for multiple comparisons. Table 4 presents the top 10 GO terms in each ontology ranked by -value. This list includes several sets of related terms associated with the transport of cysteine. For example, amino acid transmembrane transporter activity (GO: 0015171; 2 of 76 genes differentially methylated; ) is a parent of neutral amino acid transmembrane transporter activity (GO: 0015175; 2 of 33 genes differentially methylated; ), L-amino acid transmembrane transporter activity (GO: 0015179; 2 of 56 genes differentially methylated; ), and sulfur amino acid transmembrane transporter activity (GO: 0000099; 1 of 7 genes differentially methylated; ). We excluded the GO term cystine:glutamate antiporter activity (GO: 0015327), an offspring of GO: 0015171 representing the single gene solute carrier family 7 member 11 (SLC7A11), which was identified as overrepresented among differential methylated genes (). However, cg06690548, the top DMP, is annotated to SLC7A11, suggesting the importance of differential methylation within this pathway.
Table 4.
Top gene ontology (GO) terms overrepresented among genes containing DMPs ordered by -value and stratified by ontology. The top 10 significant GO terms represented by two or more genes in each ontology are shown. All probes tested in limma analyses (representing 25,715 genes) and probes identified as significantly associated with urinary as levels at were used as input for GO analysis.
| Accession number | Term | Genes in GO term | Differentially methylated genes | -Value |
|---|---|---|---|---|
| Molecular function | ||||
| GO: 0015175 | Neutral amino acid transmembrane transporter activity | 33 | 2 | 0.001 |
| GO: 0015179 | L-amino acid transmembrane transporter activity | 56 | 2 | 0.002 |
| GO: 0000386 | Second spliceosomal transesterification activity | 3 | 1 | 0.003 |
| GO: 0098808 | mRNA cap binding | 2 | 1 | 0.004 |
| GO: 0015297 | Antiporter activity | 77 | 2 | 0.004 |
| GO: 0015171 | Amino acid transmembrane transporter activity | 76 | 2 | 0.005 |
| GO: 0140098 | Catalytic activity, acting on RNA | 361 | 3 | 0.006 |
| GO: 0030620 | U2 snRNA binding | 6 | 1 | 0.006 |
| GO: 0097157 | Pre-mRNA intronic binding | 7 | 1 | 0.008 |
| GO: 0000099 | Sulfur amino acid transmembrane transporter activity | 7 | 1 | 0.008 |
| Biological process | ||||
| GO: 1902475 | L-alpha-amino acid transmembrane transport | 44 | 2 | 0.001 |
| GO: 0006396 | RNA processing | 902 | 5 | 0.002 |
| GO: 0015807 | L-amino acid transport | 62 | 2 | 0.003 |
| GO: 1905424 | Regulation of Wnt-mediated midbrain dopaminergic neuron differentiation | 2 | 1 | 0.003 |
| GO: 1905426 | Positive regulation of Wnt-mediated midbrain dopaminergic neuron differentiation | 2 | 1 | 0.003 |
| GO: 0035279 | mRNA cleavage involved in gene silencing by miRNA | 2 | 1 | 0.004 |
| GO: 0098795 | mRNA cleavage involved in gene silencing | 2 | 1 | 0.004 |
| GO: 0003333 | Amino acid transmembrane transport | 74 | 2 | 0.004 |
| GO: 1904958 | Positive regulation of midbrain dopaminergic neuron differentiation | 3 | 1 | 0.004 |
| GO: 1905616 | Regulation of miRNA mediated inhibition of translation | 2 | 1 | 0.004 |
| Cellular component | ||||
| GO: 0036126 | Sperm flagellum | 81 | 2 | 0.003 |
| GO: 0097729 | motile cilium | 81 | 2 | 0.003 |
| GO: 0000347 | THO complex | 5 | 1 | 0.003 |
| GO: 0000445 | THO complex part of transcription export complex | 5 | 1 | 0.003 |
| GO: 0036156 | Inner dynein arm | 4 | 1 | 0.008 |
| GO: 0031514 | Motile cilium | 152 | 2 | 0.010 |
| GO: 0070578 | RISC-loading complex | 9 | 1 | 0.010 |
| GO: 0035068 | Micro-ribonucleoprotein complex | 9 | 1 | 0.012 |
| GO: 0097223 | Sperm part | 186 | 2 | 0.012 |
| GO: 0000346 | Transcription export complex | 12 | 1 | 0.013 |
Discussion
This EWAS investigated the relationship between low-to-moderate levels of As exposure and loci-specific DNA methylation in a population-based prospective cohort study of American Indian adults. Twenty novel CpGs were associated with total urinary As levels (); methylation levels of 18 CpGs were positively associated with As. In addition, one DMR (chr11:2,322,050-2,323,247) was identified.
The most significantly associated CpG, cg06690548, is located in the gene body of solute carrier family 7 member 11 (SLC7A11). SLC7A11 is a protein coding gene for a subunit of the amino-acid transporter cystine:glutamate antiporter system , which exchanges cystine for glutamate within cells (Lim and Donaldson 2011), providing cysteine for glutathione (GSH) biosynthesis (Conrad and Sato, 2012). GSH is an endogenous antioxidant (Forman et al. 2009) and may be protective against As-induced oxidative stress. In a cross-sectional study of As-exposed adults in Bangladesh, water As concentrations were negatively associated with GSH concentrations in blood (Hall et al. 2013). Arsenic may deplete GSH through the induction of reactive oxygen species, or it may affect GSH biosynthesis through an epigenetic mechanism. A paralog of SLC7A11, solute carrier family 7 member 5 (SLC7A5) was also identified as containing a differentially methylated CpG (cg07021906). SLC7A5 is involved in the transport of amino acids including glutamine (Pochini et al. 2014), which may be deaminated to glutamate and used for GSH biosynthesis (Y. Liu et al. 2014).
Additional DMPs may be biologically responsive to As exposure. Urinary As levels were associated with DNA methylation of cg22294740 and cg08059112, located in LINGO3, a transmembrane protein expressed in the nervous system (Haines and Rigby, 2008). LINGO3 has also been identified as a gene hub in metastatic melanoma tumors (Wang et al. 2018). Urinary As levels were also associated with DNA methylation of cg20493718 and cg21369801, located in the gene body of CSNK1D. The casein kinase 1 (CK1) protein family is involved in a broad range of cellular processes, including control of DNA replication and repair, apoptosis, and circadian rhythm (Schittek and Sinnberg 2014) and has been association with regulation of the tumor suppressor protein p53 (Schittek and Sinnberg 2014). Prenatal As exposure has been associated with differential expression (Fry et al. 2007; Rojas et al. 2015) and DNA methylation (Rojas et al. 2015) of CSNK1D in cord blood.
Seven of the 20 CpGs associated with total urinary As levels were novel to the 850K (cg07317306, cg27178850, cg09280971, cg14595618, cg04940901, cg18616702, cg20509831), all of which were located in ENCODE DNase hypersensitive sites. The ENCODE project has mapped DNase sites, associated with accessible chromatin regions (The ENCODE Project Consortium 2012). The three significant CpGs not annotated to a RefSeq gene (cg07317306, cg27178850, cg09280971) were, however, annotated to DNase hypersensitive sites, suggesting that they may have functional roles.
Overall, results were consistent in analyses of data normalized for probe type and using a reference-free approach to adjust for cell type proportions. In addition, results were robust in sensitivity analyses stratifying by sex, smoking status, study center, and diabetes status. This study includes American Indian participants recruited from three genetically, environmentally, and culturally distinct study centers in Arizona, Oklahoma, and North and South Dakota. Consistency across study centers provide internal validity to our analysis and suggest that factors associated with study center do not modify the observed relationship between As exposure and DNA methylation. In addition, analyses stratified by diabetes status were consistent. Due to the effect of uncontrolled diabetes on urine osmolality (Voinescu et al. 2002), in overall analyses, we corrected for between-individual differences in urine dilution by dividing total urinary As concentrations by urinary creatinine concentrations. In analyses of participants without diabetes, total urinary As concentrations were corrected for both urinary creatinine and specific gravity. All FDR-significant CpGs achieved significance at a nominal among participants with diabetes. Among those without diabetes cg14595618 located in HK1, a protein coding gene involved in glucose metabolism, was not significantly associated with total urinary As concentration corrected for urine dilution using urinary creatinine (nominal ) or specific gravity (nominal ). In a linear model we observed significant interaction between As levels and diabetes status on cg14595618 methylation (). Due to the role of HK1 in glucose metabolism, further research is needed to determine the relationship between As exposure, diabetes status, and HK1 DNA methylation.
We found overlap between results from our elastic-net model, which is suitable for modeling high-dimensional data with correlated predictors and results from linear models implemented in limma. Among 315 CpG selected by the elastic-net model as associated with urinary As levels, 8 were identified as FDR-significant, and 280 achieved nominal significance () in limma analyses. Replication of our top CpGs using an elastic-net approach suggests that our results are robust to limitations of performing nonpenalized linear regression on high-dimensional EWAS data.
The one identified DMR (chr11:2,322,050-2,323,247) included the open reading frame C11orf21 and spanned several functional regions of tetraspanin 32 (TSPAN32), including the TSS1500, TSS22, 5′UTR, and first exon. C11orf21/TSPAN32 is located in a genomic region containing a cluster of imprinted genes (Smith et al. 2007), and alterations of this region have been associated with Beckwith–Wiedemann syndrome, a condition associated with abnormal growth and tumors in childhood (Koufos et al. 1989). Genetic variation in C11orf21/TSPAN32 has been associated with chronic lymphocytic leukemia in a genome-wide association study (Berndt et al. 2013).
Previous EWAS among adults have found conflicting results. Studies of adult populations have included cohorts in Bangladesh, identifying 4 CpGs associated with As measured in urine or blood at (; 3 CpGs overlapped for both exposure measures) (Argos et al. 2015) and 50 CpGs associated with As measured in urine or drinking water at (; 8 CpGs overlapped for both exposure measures) (Demanelis et al. 2019). In addition, in a study of women in Argentina (), differential methylation was found at 6 loci (Ameer et al. 2017), and in a study of families in China (adults and children; ), differential methylation was found at 85 loci (Guo et al. 2018). To understand the overlap between loci previously identified as differentially methylated with As exposure, we used a lookup approach of studies analyzing the association between As exposure and DNA methylation measured using the 450K or 850K microarrays. CpGs previously associated with As exposure were evaluated in the current study. Among 4,595 CpGs previously identified as associated with As exposure after adjustment for multiple comparisons, 191 achieved nominal significance in our analyses (). This overlap between significant loci may be due to differences in population, time of exposure, tissue evaluated, and analytical methods (Table S1). Of the 14 studies identified in our lookup approach, 9 evaluated the association between prenatal As exposure and DNA methylation (8 measured DNA methylation in cord blood or placental samples, and one measured DNA methylation in blood mononuclear cells collected at 9 y). Only one study of adults reported results of DNA methylation measured using the 850K microarray (Demanelis et al. 2019). In addition, levels of As exposure differed markedly between population studies; for example, among studies reporting urinary As concentrations adjusted for urinary creatinine, mean concentrations ranged from creatinine (Kaushal et al. 2017) to creatinine (Argos et al. 2015). Although speculative, limited replication across EWAS may also be due in part to broad, nonspecific epigenetic dysregulation, as suggested by the range of health outcomes that have been associated with As exposure.
We also identified significant associations between urinary As levels and imputed leukocyte proportions, specifically a positive association with the proportion of NK cells (; ) and a negative association with the proportion of B cells (; ). Associations between maternal drinking water As levels and imputed leukocyte proportions in cord blood have previously been reported by Kile et al. (2014); however, As levels were positively associated with the proportion of T cells and negatively associated with the proportion of T cells. Although our study found relationships between urinary As levels and the proportions of T and T in the same directions, these associations were not significant ( T-cell proportion: and ; T-cell proportion: and ).
This study was limited by measuring DNA methylation in peripheral blood, and observed associations may not be present in other tissues, although As exposure is known to affect a broad range of tissues (Naujokas et al. 2013). Peripheral blood leukocytes, moreover, consist of a mixture of cell types, including T cells, B cells, NK cells, monocytes, and granulocytes. Due to epigenetic control of cellular differentiation (Khavari et al. 2010), DNA methylation patterns differ among cell types, and blood cell composition may be associated with environmental exposures (Lurà et al. 2018; Stiegel et al. 2016). Therefore, EWAS using mixtures of cell types may be subject to confounding. In the current study, the Houseman regression calibration method (Houseman et al. 2012) was implemented to estimate leukocyte composition, and the proportions of six cell types were controlled for in all models. This method is generally accepted to remove confounding by cell type, although EWAS results may be influenced by variation in cell subtype proportions not characterized by the six main cell types (Bauer et al. 2015).
This study was also limited in measuring DNA methylation with standard bisulfite conversion, which is not able to distinguish between 5-methylcytosine and 5-hydroxymethylcytosine, an intermediate of active demethylation (Klug et al. 2013). Although 5-hydroxymethylcytosine is less abundant in the human genome, in a study of adults in Bangladesh As exposure was found to have sex-specific associations with global levels of 5-hydroxymethylcytosine (Niedzwiecki et al. 2015). Our observed associations may be due in part to As-associated variation in 5-hydroxymethylcytosine levels. We were also limited by lack of data on gene expression. Although CpGs located in biologically relevant genes were identified, it is not known if alterations in these epigenetic markers are associated with functional changes in gene expression. In addition, this study included American Indian adults with low-to-moderate As exposure through drinking water. It is not known if the association between As exposure and DNA methylation may differ among populations.
The strengths of this study include the large sample size in a population with low-to-moderate levels of As exposure, use of the 850K microarray to measure DNA methylation, and robustness of the findings in sensitivity analyses. This study had a large sample size () in comparison with previous EWAS of As exposure, in which ranged from (Table S1). Because power in EWAS depends on sample size, effect size, and correction for multiple testing, large sample size is particularly advantageous for 850K studies to allow for the detection of small effect sizes. Few EWAS of adults have focused on populations with low-to-moderate levels of As exposure through drinking water. In our study, low concentrations of arsenobetaine in urine (median creatinine) reflect that seafood intake is not a major contributor to urinary As species including DMA, in contrast with the general U.S. population in which DMA often reflects exposure to seafood arsenicals rather than to inorganic As (Navas-Acien et al. 2011). In addition, previous EWAS of As exposure have predominantly measured DNA methylation using the 450K microarray, which interrogates CpGs; the 850K microarray, however, interrogates CpGs, including of 450K loci and increased coverage of regulatory elements including ENCODE DNase hypersensitive sites and FANTOM5 enhancers (Pidsley et al. 2016).
Conclusions
To our knowledge, this is the largest study to investigate the association between chronic As exposure, mostly through drinking water (Navas-Acien et al. 2009), and epigenome-wide DNA methylation in blood, and one of the first using the 850K microarray. In a cohort of American Indian adults with low-to-moderate levels of exposure, significant associations between total urinary As levels and DNA methylation were observed at 20 novel CpGs, including loci located in genes involved in As-related mechanistic pathways and health outcomes. Further investigation is necessary to determine whether As-related DNA methylation signatures are common across populations and levels of exposure, and whether observed differences in DNA methylation are associated with gene expression or may serve as biomarkers of disease development.
Supplementary Material
Acknowledgments
The authors would like to thank all Strong Heart Study participants and staff who have made this work possible. This study was supported by the National Institute of Environmental Health Sciences (NIEHS) grants T32ES007322, F31ES029019, R01ES025216, P42ES010349, and P30ES009089; the National Center for Advancing Translational Sciences (NCATS) grant TL1TR001875; and the National Heart, Lung, and Blood Institute grants 75N92019D00027, 75N92019D00028, 75N92019D00029, and 75N92019D00030.
References
- Ameer SS, Engström K, Hossain MB, Concha G, Vahter M, Broberg K. 2017. Arsenic exposure from drinking water is associated with decreased gene expression and increased DNA methylation in peripheral blood. Toxicol Appl Pharmacol 321:57–66, PMID: 28242323, 10.1016/j.taap.2017.02.019. [DOI] [PubMed] [Google Scholar]
- American Diabetes Association. 2014. Diagnosis and classification of diabetes mellitus. Diabetes Care 37 (suppl 1):S81–S90, 10.2337/dc14-S081. [DOI] [PubMed] [Google Scholar]
- Argos M. 2015. Arsenic exposure and epigenetic alterations: recent findings based on the Illumina 450K DNA Methylation Array. Curr Environ Health Rep 2(2):137–144, PMID: 26231363, 10.1007/s40572-015-0052-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argos M, Chen L, Jasmine F, Tong L, Pierce BL, Roy S, et al. 2015. Gene-specific differential DNA methylation and chronic arsenic exposure in an epigenome-wide association study of adults in Bangladesh. Environ Health Perspect 123(1):64–71, PMID: 25325195, 10.1289/ehp.1307884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aryee M, Jaffe A, Corrada-Bravo H, Ladd-Acosta C, Feinberg A, Hansen K, et al. 2014. Minfi: A flexible and comprehensive Bioconductor package for the analysis of Infinium DNA Methylation microarrays. Bioinformatics 30(10):1363–1369, PMID: 24478339, 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayotte JD, Medalie L, Qi SL, Backer LC, Nolan BT. 2017. Estimating the high-arsenic domestic-well population in the conterminous United States. Environ Sci Technol 51(21):12443–12454, PMID: 29043784, 10.1021/acs.est.7b02881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey KA, Fry RC. 2014. Arsenic-associated changes to the epigenome: what are the functional consequences? Curr Environ Health Rep 1(1):22–34, PMID: 24860721, 10.1007/s40572-013-0002-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey KA, Smith AH, Tokar EJ, Graziano JH, Kim K-W, Navasumrit P, et al. 2016. Mechanisms underlying latent disease risk associated with early-life arsenic exposure: current research trends and scientific gaps. Environ Health Perspect 124(2):170–175, PMID: 26115410, 10.1289/ehp.1409360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer M, Linsel G, Fink B, Offenberg K, Hahn AM, Sack U, et al. 2015. A varying T cell subtype explains apparent tobacco smoking induced single CpG hypomethylation in whole blood. Clin Epigenetics 7(1):81, PMID: 26246861, 10.1186/s13148-015-0113-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc 57(1):289–300, 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- Benton MC, Sutherland HG, Macartney-Coxson D, Haupt LM, Lea RA, Griffiths LR. 2017. Methylome-wide association study of whole blood DNA in the Norfolk Island isolate identifies robust loci associated with age. Aging (Albany NY). 9(3):753–768, PMID: 28255110, 10.18632/aging.101187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berndt SI, Skibola CF, Joseph V, Camp NJ, Nieters A, Wang Z, et al. 2013. Genome-wide association study identifies multiple risk loci for chronic lymphocytic leukemia. Nat Genet 45(8):868–876, PMID: 23770605, 10.1038/ng.2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broberg K, Ahmed S, Engström K, Hossain MB, Jurkovic Mlakar S, Bottai M, et al. 2014. Arsenic exposure in early pregnancy alters genome-wide DNA methylation in cord blood, particularly in boys. J Dev Orig Health Dis 5(4):288–298, PMID: 24965135, 10.1017/S2040174414000221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas A, Houseman EA, Baccarelli AA, Quamruzzaman Q, Rahman M, Mostofa G, et al. 2015. In utero arsenic exposure and epigenome-wide associations in placenta, umbilical artery, and human umbilical vein endothelial cells. Epigenetics 10(11):1054–1063, PMID: 26646901, 10.1080/15592294.2015.1105424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlin DJ, Naujokas MF, Bradham KD, Cowden J, Heacock M, Henry HF, et al. 2015. Arsenic and environmental health: state of the science and future research opportunities. Environ Health Perspect, 10.1289/ehp.1510209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad M, Sato H. 2012. The oxidative stress-inducible cystine/glutamate antiporter, system xc: cystine supplier and beyond. Amino Acids 42(1):231–246, PMID: 21409388, 10.1007/s00726-011-0867-5. [DOI] [PubMed] [Google Scholar]
- Demanelis K, Argos M, Tong L, Shinkle J, Sabarinathan M, Rakibuz-Zaman M, et al. 2019. Association of arsenic exposure with whole blood DNA methylation: an epigenome-wide study of Bangladeshi adults. Environ Health Perspect 127, 10.1289/EHP3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du P, Zhang X, Huang C-C, Jafari N, Kibbe WA, Hou L, et al. 2010. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics 11:587, PMID: 21118553, 10.1186/1471-2105-11-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forman HJ, Zhang H, Rinna A. 2009. Glutathione: overview of its protective roles, measurement, and biosynthesis. Mol Aspects Med 30(1–2):1–12, PMID: 18796312, 10.1016/j.mam.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortin J-P, Triche TJ, Hansen KD, Hansen KD. 2017. Preprocessing, normalization and integration of the Illumina HumanMethylationEPIC array with minfi. Bioinformatics 33(4):558–560, PMID: 28035024, 10.1093/bioinformatics/btw691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J, Hastie T, Tibshirani R. 2010. Regularization paths for generalized linear models via coordinate descent. J Stat Softw 33(1):1–22, PMID: 20808728. [PMC free article] [PubMed] [Google Scholar]
- Fry RC, Navasumrit P, Valiathan C, Svensson JP, Hogan BJ, Luo M, et al. 2007. Activation of inflammation/NF-κB signaling in infants born to arsenic-exposed mothers. PLoS Genet 3(11):e207, PMID: 18039032, 10.1371/journal.pgen.0030207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Esquinas E, Pollán M, Umans JG, Francesconi KA, Goessler W, Guallar E, et al. 2013. Arsenic exposure and cancer mortality in a US-based prospective cohort: the Strong Heart Study. Cancer Epidemiol Biomarkers Prev 22(11):1944–1953, PMID: 23800676, 10.1158/1055-9965.EPI-13-0234-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geeleher P, Hartnett L, Egan LJ, Golden A, Raja Ali RA, Seoighe C. 2013. Gene-set analysis is severely biased when applied to genome-wide methylation data. Bioinformatics 29(15):1851–1857, PMID: 23732277, 10.1093/bioinformatics/btt311. [DOI] [PubMed] [Google Scholar]
- Gliga AR, Engström K, Kippler M, Skröder H, Ahmed S, Vahter M, et al. 2018. Prenatal arsenic exposure is associated with increased plasma IGFBP3 concentrations in 9-year-old children partly via changes in DNA methylation. Arch Toxicol 92(8):2487–2500, PMID: 29947889, 10.1007/s00204-018-2239-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grau-Perez M, Kuo C-C, Gribble MO, Balakrishnan P, Jones Spratlen M, Vaidya D, et al. 2017. Association of low-moderate arsenic exposure and arsenic metabolism with incident diabetes and insulin resistance in the Strong Heart Family Study. Environ Health Perspect 125(12):127004, PMID: 29373862, 10.1289/EHP2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green BB, Karagas MR, Punshon T, Jackson BP, Robbins DJ, Houseman EA, et al. 2016. Epigenome-wide assessment of DNA methylation in the placenta and arsenic exposure in the New Hampshire Birth Cohort Study. Environ Health Perspect 124(8):1253–1260, PMID: 26771251, 10.1289/ehp.1510437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Chen X, Wang J, Liu Z, Gaile D, Wu H, et al. 2018. Multi-generational impacts of arsenic exposure on genome-wide DNA methylation and the implications for arsenic-induced skin lesions. Environ Int 119:250–263, PMID: 29982128, 10.1016/j.envint.2018.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haines BP, Rigby PWJ. 2008. Expression of the Lingo/LERN gene family during mouse embryogenesis. Gene Expr Patterns 8(2):79–86, PMID: 18297755, 10.1016/j.modgep.2007.10.003. [DOI] [PubMed] [Google Scholar]
- Hall MN, Niedzwiecki M, Liu X, Harper KN, Alam S, Slavkovich V, et al. 2013. Chronic arsenic exposure and blood glutathione and glutathione disulfide concentrations in Bangladeshi adults. Environ Health Perspect 121(9):1068–1074, PMID: 23792557, 10.1289/ehp.1205727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, et al. 2012. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics 13:86, PMID: 22568884, 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IARC Working Group. 2009. Arsenic, metals, fibres, and dusts. IARC Monogr Eval Carcinog Risks Hum 100:11–465, PMID: 23189751. [PMC free article] [PubMed] [Google Scholar]
- Illumina. 2015. Infinium MethylationEPIC BeadChip Data Sheet.
- Intarasunanont P, Navasumrit P, Waraprasit S, Chaisatra K, Suk WA, Mahidol C, et al. 2012. Effects of arsenic exposure on DNA methylation in cord blood samples from newborn babies and in a human lymphoblast cell line. Environ Heal 11:31, 10.1186/1476-069X-11-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joint FAO/WHO Export Committee on Food Additives. 1989. Toxicological Evaluation of Certain Food Additives and Contaminants. Geneva, Switzerland: Cambridge University Press. [Google Scholar]
- Kaushal A, Zhang H, Karmaus WJJ, Everson TM, Marsit CJ, Karagas MR, et al. 2017. Genome-wide DNA methylation at birth in relation to in utero arsenic exposure and the associated health in later life. Environ Health 16(1):50, PMID: 28558807, 10.1186/s12940-017-0262-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khavari DA, Sen GL, Rinn JL. 2010. DNA methylation and epigenetic control of cellular differentiation. Cell Cycle 9(19):3880–3883, PMID: 20890116, 10.4161/cc.9.19.13385. [DOI] [PubMed] [Google Scholar]
- Kile ML, Baccarelli A, Hoffman E, Tarantini L, Quamruzzaman Q, Rahman M, et al. 2012. Prenatal arsenic exposure and DNA methylation in maternal and umbilical cord blood leukocytes. Environ Health Perspect 120(7):1061–1066, PMID: 22466225, 10.1289/ehp.1104173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kile ML, Houseman EA, Baccarelli AA, Quamruzzaman Q, Rahman M, Mostofa G, et al. 2014. Effect of prenatal arsenic exposure on DNA methylation and leukocyte subpopulations in cord blood. Epigenetics 9(5):774–782, PMID: 24525453, 10.4161/epi.28153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klug M, Schmidhofer S, Gebhard C, Andreesen R, Rehli M. 2013. 5-Hydroxymethylcytosine is an essential intermediate of active DNA demethylation processes in primary human monocytes. Genome Biol 14(5):R46, PMID: 23705593, 10.1186/gb-2013-14-5-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koestler DC, Avissar-Whiting M, Houseman EA, Karagas MR, Marsit CJ. 2013. Differential DNA methylation in umbilical cord blood of infants exposed to low levels of arsenic in utero. Environ Health Perspect 121(8):971–977, PMID: 23757598, 10.1289/ehp.1205925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koufos A, Grundy P, Morgan K, Aleck KA, Hadro T, Lampkin BC, et al. 1989. Familial Wiedemann-Beckwith syndrome and a second Wilms tumor locus both map to 11p15.5. Am J Hum Genet 44:711–719, PMID: 2539717. [PMC free article] [PubMed] [Google Scholar]
- Lee ET, Welty TK, Fabsitz R, Cowan LD, Le NA, Oopik AJ, et al. 1990. The Strong Heart Study. A study of cardiovascular disease in American Indians: design and methods. Am J Epidemiol 132(6):1141–1155, PMID: 2260546, 10.1093/oxfordjournals.aje.a115757. [DOI] [PubMed] [Google Scholar]
- Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. 2012. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28(6):882–883, PMID: 22257669, 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levey AS, Stevens LA, Schmid CH, Zhang Y(L), Castro AF, Feldman HI, et al. 2009. A new equation to estimate glomerular filtration rate. Ann Intern Med 150(9):604, PMID: 19414839, 10.7326/0003-4819-150-9-200905050-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim JC, Donaldson PJ. 2011. Focus on molecules: the cystine/glutamate exchanger (System x(c)(−)). Exp Eye Res 92(3):162–163, PMID: 20488177, 10.1016/j.exer.2010.05.007. [DOI] [PubMed] [Google Scholar]
- Liu Y, Hyde AS, Simpson MA, Barycki JJ. 2014. Emerging regulatory paradigms in glutathione metabolism. Adv Cancer Res 122:69–101, PMID: 24974179, 10.1016/B978-0-12-420117-0.00002-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Zheng Y, Zhang W, Zhang X, Lloyd-Jones DM, Baccarelli AA, et al. 2014. Blood methylomics in response to arsenic exposure in a low-exposed US population. J Expo Sci Environ Epidemiol 24(2):145–149, PMID: 24368509, 10.1038/jes.2013.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurà MP, Gorlanova O, Müller L, Proietti E, Vienneau D, Reppucci D, et al. 2018. Response of cord blood cells to environmental, hereditary and perinatal factors: a prospective birth cohort study. PLoS One 13(7):e0200236, PMID: 29979752, 10.1371/journal.pone.0200236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney DL, Walker RM, Morris SW, McIntosh AM, Porteous DJ, Evans KL. 2016. Identification of polymorphic and off-target probe binding sites on the Illumina Infinium MethylationEPIC BeadChip. Genom Data 9:22–24, PMID: 27330998, 10.1016/j.gdata.2016.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RC, Brindle E, Holman DJ, Shofer J, Klein NA, Soules MR, et al. 2004. Comparison of specific gravity and creatinine for normalizing urinary reproductive hormone concentrations. Clin Chem 50(5):924–932, PMID: 15105350, 10.1373/clinchem.2004.032292. [DOI] [PubMed] [Google Scholar]
- Moon KA, Guallar E, Umans JG, Devereux RB, Best LG, Francesconi KA, et al. 2013. Association between exposure to low to moderate arsenic levels and incident cardiovascular disease. Ann Intern Med 159:649–659, PMID: 24061511, 10.7326/0003-4819-159-10-201311190-00719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon KA, Oberoi S, Barchowsky A, Chen Y, Guallar E, Nachman KE, et al. 2017. A dose–response meta-analysis of chronic arsenic exposure and incident cardiovascular disease. Int J Epidemiol 46(6):1924–1939, PMID: 29040626, 10.1093/ije/dyx202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Research Council. 2013. Critical Aspects of EPA’s and IRIS Assessment of Inorganic Arsenic, Interim Report. Washington, DC: The National Academies of Sciences, Engineering, and Medicine. [Google Scholar]
- Naujokas MF, Anderson B, Ahsan H, Vasken Aposhian H, Graziano JH, Thompson C, et al. 2013. The broad scope of health effects from chronic arsenic exposure: update on a worldwide public health problem. Environ Health Perspect 121(3):295–302, PMID: 23458756, 10.1289/ehp.1205875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navas-Acien A, Francesconi KA, Silbergeld EK, Guallar E. 2011. Seafood intake and urine concentrations of total arsenic, dimethylarsinate and arsenobetaine in the US population. Environ Res 111(1):110–118, PMID: 21093857, 10.1016/j.envres.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navas-Acien A, Umans JG, Howard BV, Goessler W, Francesconi KA, Crainiceanu CM, et al. 2009. Urine arsenic concentrations and species excretion patterns in American Indian communities over a 10-year period: The Strong Heart Study. Environ Health Perspect 117(9):1428–1433, PMID: 19750109, 10.1289/ehp.0800509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedzwiecki MM, Hall MN, Liu X, Oka J, Harper KN, Slavkovich V, et al. 2013. A dose-response study of arsenic exposure and global methylation of peripheral blood mononuclear cell DNA in Bangladeshi adults. Environ Health Perspect 121(11–12):1306–1312, 10.1289/ehp.1206421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedzwiecki MM, Liu X, Hall MN, Thomas T, Slavkovich V, Ilievski V, et al. 2015. Sex-specific associations of arsenic exposure with global DNA methylation and hydroxymethylation in leukocytes: results from two studies in Bangladesh. Cancer Epidemiol Biomarkers Prev 24(11):1748–1757, 10.1158/1055-9965.EPI-15-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigra AE, Olmedo P, Grau-Perez M, O’Leary R, O’Leary M, Fretts AM, et al. 2019. Dietary determinants of inorganic arsenic exposure in the Strong Heart Family Study. Environ Res 177:108616, PMID: 31442790, 10.1016/j.envres.2019.108616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu L, Xu Z, Taylor JA. 2016. RCP: a novel probe design bias correction method for Illumina Methylation BeadChip. Bioinformatics 32(17):2659–2663, PMID: 27153672, 10.1093/bioinformatics/btw285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters TJ, Buckley MJ, Statham AL, Pidsley R, Samaras K, Lord RV, et al. 2015. De novo identification of differentially methylated regions in the human genome. Epigenetics Chromatin 8(8):6, PMID: 25972926, 10.1186/1756-8935-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phipson B, Maksimovic J, Oshlack A. 2016. missMethyl: an R package for analyzing data from Illumina’s HumanMethylation450 platform. Bioinformatics 32(2):286–288, PMID: 26424855, 10.1093/bioinformatics/btv560. [DOI] [PubMed] [Google Scholar]
- Pidsley R, Zotenko E, Peters TJ, Lawrence MG, Risbridger GP, Molloy P, et al. 2016. Critical evaluation of the Illumina MethylationEPIC BeadChip microarray for whole-genome DNA methylation profiling. Genome Biol 17(1):208, PMID: 27717381, 10.1186/s13059-016-1066-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilsner JR, Liu X, Ahsan H, Ilievski V, Slavkovich V, Levy D, et al. 2009. Folate deficiency, hyperhomocysteinemia, low urinary creatinine, and hypomethylation of leukocyte DNA are risk factors for arsenic-induced skin lesions. Environ Health Perspect 117(2):254–260, PMID: 19270796, 10.1289/ehp.11872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilsner JR, Liu X, Ahsan H, Ilievski V, Slavkovich V, Levy D, et al. 2007. Genomic methylation of peripheral blood leukocyte DNA: influences of arsenic and folate in Bangladeshi adults. Am J Clin Nutr 86(4):1179–1186, PMID: 17921400, 10.1093/ajcn/86.4.1179. [DOI] [PubMed] [Google Scholar]
- Pochini L, Scalise M, Galluccio M, Indiveri C. 2014. Membrane transporters for the special amino acid glutamine: structure/function relationships and relevance to human health. Front Chem 2:61, PMID: 25157349, 10.3389/fchem.2014.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers M, Sanchez TR, Grau-Perez M, Yeh F, Francesconi KA, Goessler W, et al. 2019. Low-moderate arsenic exposure and respiratory in American Indian communities in the Strong Heart Study. Environ Heal 18:104, PMID: 31779614, 10.1186/s12940-019-0539-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmani E, Zaitlen N, Baran Y, Eng C, Hu D, Galanter J, et al. 2016. Sparse PCA corrects for cell type heterogeneity in epigenome-wide association studies. Nat Methods 13(5):443–445, PMID: 27018579, 10.1038/nmeth.3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie M, Phipson B, Wu D, Hu Y, Law C, Shi W, et al. 2015. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43(7):e47, PMID: 25605792, 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson KD. 2005. DNA methylation and human disease. Nat Rev Genet 6(8):597–610, PMID: 16136652, 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- Roh T, Steinmaus C, Marshall G, Ferreccio C, Liaw J, Smith AH. 2018. Age at exposure to arsenic in water and mortality 30–40 years after exposure cessation. Am J Epidemiol 187(11):2297–2305, PMID: 30084889, 10.1093/aje/kwy159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas D, Rager JE, Smeester L, Bailey KA, Drobná Z, Rubio-Andrade M, et al. 2015. Prenatal arsenic exposure and the epigenome: identifying sites of 5-methylcytosine alterations that predict functional changes in gene expression in newborn cord blood and subsequent birth outcomes. Toxicol Sci 143(1):97–106, PMID: 25304211, 10.1093/toxsci/kfu210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer J, Findenig S, Goessler W, Francesconi KA, Howard B, Umans JG, et al. 2012. Arsenic species and selected metals in human urine: validation of HPLC/ICPMS and ICPMS procedures for a long-term population-based epidemiological study. Anal Methods 4(2):406–413, PMID: 22685491, 10.1039/C2AY05638K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schittek B, Sinnberg T. 2014. Biological functions of casein kinase 1 isoforms and putative roles in tumorigenesis. Mol Cancer 13:231, PMID: 25306547, 10.1186/1476-4598-13-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shara NM, Wang H, Mete M, Al-Balha YR, Azalddin N, Lee ET, et al. 2012. Estimated GFR and incident cardiovascular disease events in American Indians: The Strong Heart Study. Am J Kidney Dis 60(5):795–803, PMID: 22841159, 10.1053/j.ajkd.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AC, Choufani S, Ferreira JC, Weksberg R. 2007. Growth regulation, imprinted genes, and chromosome 11p15.5. Pediatr Res 61(5 pt 2):43R–47R, PMID: 17413842, 10.1203/pdr.0b013e3180457660. [DOI] [PubMed] [Google Scholar]
- Smith AH, Marshall G, Roh T, Ferreccio C, Liaw J, Steinmaus C. 2018. Lung, bladder, and kidney cancer mortality 40 years after arsenic exposure reduction. J Natl Cancer Inst 110(3):241–249, PMID: 29069505, 10.1093/jnci/djx201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmaus C, Ferreccio C, Acevedo J, Balmes JR, Liaw J, Troncoso P, et al. 2016. High risks of lung disease associated with early-life and moderate lifetime arsenic exposure in northern Chile. Toxicol Appl Pharmacol 313:10–15, PMID: 27725189, 10.1016/j.taap.2016.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiegel MA, Pleil JD, Sobus JR, Madden MC. 2016. Inflammatory cytokines and white blood cell counts response to environmental levels of diesel exhaust and ozone inhalation exposures. PLoS One 11(4):e0152458, PMID: 27058360, 10.1371/journal.pone.0152458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489:57–74, PMID: 22955616, 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. EPA (U.S. Environmental Protection Agency). 2018. Government Performance and Results Act (GPRA) Tool-Submission Year Quarter 2018Q3, Violation Report. https://obipublic.epa.gov/analytics/saw.dll?PortalPages&PortalPath=/shared/SFDW/_portal/Public&Page=Violation [accessed 10 January 2019].
- Voinescu GC, Shoemaker M, Moore H, Khanna R, Nolph KD. 2002. The relationship between urine osmolality and specific gravity. Am J Med Sci 323(1):39–42, PMID: 11814141, 10.1097/00000441-200201000-00007. [DOI] [PubMed] [Google Scholar]
- Wang L-X, Li Y, Chen G-Z. 2018. Network-based co-expression analysis for exploring the potential diagnostic biomarkers of metastatic melanoma. PLoS One 13(1):e0190447, PMID: 29377892, 10.1371/journal.pone.0190447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. 2011. Arsenic in Drinking-Water: Background Document for the Development of WHO Guidelines for Drinking-Water Quality, Geneva, Switzerland: World Health Organization, 10.1016/j.kjms.2011.05.002, https://www.who.int/water_sanitation_health/dwq/chemicals/arsenic.pdf. [DOI] [Google Scholar]
- World Health Organization. 2012. Arsenic. WHO Fact Sheets. http://www.who.int/mediacentre/factsheets/fs372/en/ [accessed 16 May 2016].
- Zheng LY, Umans JG, Yeh F, Francesconi KA, Goessler W, Silbergeld EK, et al. 2015. The association of urine arsenic with prevalent and incident chronic kidney disease. Epidemiology 26(4):601–612, PMID: 25929811, 10.1097/EDE.0000000000000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.