Abstract
Naïve pluripotency can be established in human pluripotent stem cells (hPSCs) by manipulation of transcription factors, signaling pathways, or a combination thereof. However, differences exist in the molecular and functional properties of naïve hPSCs generated by different protocols, which include varying similarities with pre‐implantation human embryos, differentiation potential, and maintenance of genomic integrity. We show here that short treatment with two chemical agonists (2a) of nuclear receptors, liver receptor homologue‐1 (LRH‐1) and retinoic acid receptor gamma (RAR‐γ), along with 2i/LIF (2a2iL) induces naïve‐like pluripotency in human cells during reprogramming of fibroblasts, conversion of pre‐established hPSCs, and generation of new cell lines from blastocysts. 2a2iL‐hPSCs match several defined criteria of naïve‐like pluripotency and contribute to human–mouse interspecies chimeras. Activation of TGF‐β signaling is instrumental for acquisition of naïve‐like pluripotency by the 2a2iL induction procedure, and transient activation of TGF‐β signaling substitutes for 2a to generate naïve‐like hPSCs. We reason that 2a2iL‐hPSCs are an easily attainable system to evaluate properties of naïve‐like hPSCs and for various applications.
Keywords: chimera formation, human naïve pluripotency, nuclear receptors, TGF‐β signaling pathway
Subject Categories: Regenerative Medicine
Short‐term exposure of nuclear receptors LRH‐1 and RARɣ with chemical agonists (2a) together with 2iL induces human naïve‐like pluripotency in fibroblasts, hPSC and blastocysts.
Introduction
Mammalian pluripotency is defined as the ability of a cell to differentiate into all the cell types of an adult organism. The epiblast of an early embryo is considered to contain bona fide in vivo pluripotent cells. In vitro‐derived pluripotent stem cells (PSCs), including embryonic stem cells (ESCs) and induced PSCs (iPSCs), recapitulate numerous properties of epiblast cells (Evans & Kaufman, 1981; Martin, 1981; Thomson et al, 1998; Takahashi & Yamanaka, 2006). In mice, both ESCs and iPSCs are the genuine counterparts of pre‐implantation epiblast cells. Naïve PSCs can participate in embryo development to form chimeric organisms. However, human PSCs (hPSCs) are in a primed state of pluripotency, similar to mouse post‐implantation epiblast‐derived stem cells (EpiSCs). Both hPSCs and EpiSCs lack the ability to form chimeras (Brons et al, 2007; Tesar et al, 2007; Nichols & Smith, 2009). Although naïve and primed pluripotent states have some common features, there are considerable differences at the cellular and molecular levels. Naïve PSCs show a dome‐shaped morphology, two active X‐chromosomes in female cells, rapid growth rates, robust single‐cell survival, and amenability to genetic manipulation. In contrast, primed PSCs show flattened morphology, an inactive X‐chromosome in female cells, transcriptional leakages of a number of early lineage differentiation genes, and sensitivity to single‐cell dissociation (Hassani et al, 2014, 2019).
Several studies have attempted to convert primed hPSCs into naïve cells by overexpressing naïve‐related transcriptional regulators and/or impressing naïve‐related signaling pathways, which might overcome primed characteristics that lead to inconsistent lineage differentiation protocols, limiting the use of hPSCs in regenerative medicine (Li et al, 2009; Buecker et al, 2010; Hanna et al, 2010; Chan et al, 2013; Gafni et al, 2013; Takashima et al, 2014; Theunissen et al, 2014; Ware et al, 2014; Chen et al, 2015; Duggal et al, 2015; Guo et al, 2016, 2017; Qin et al, 2016; Zimmerlin et al, 2016; Liu et al, 2017; Di Stefano et al, 2018). Both molecular and functional properties of naïve hPSCs generated by various protocols differ in terms of gene regulatory network (GRN), in vitro and in vivo differentiation potential, similarity to pre‐implantation human embryos, and maintenance of genomic integrity (Theunissen et al, 2016). Therefore, further research is needed to establish authentic hPSCs with naïve‐specific features.
In this study, we introduce an easy and efficient approach to obtain functional naïve‐like hPSCs without the need for ectopic gene expression. We took advantage of the power of two chemical agonists (2a) specifically activating two nuclear receptors, liver receptor homologue‐1 (LRH‐1 or NR5A2) and retinoic acid receptor gamma (RAR‐γ). 2a in conjunction with GSK3 and MEK1/2 chemical inhibitors (2i) and LIF (hereafter called 2a2iL) reprograms human cells into the naïve‐like state during fibroblast reprogramming, conversion of conventional primed human ESCs (hESCs), and hESC derivation from blastocysts. Our study builds on previous reports demonstrating that enhanced expression of RAR‐γ and LRH‐1 leads to rapid and efficient reprogramming of mouse embryonic fibroblasts (MEF), EpiSCs, and human fibroblasts toward ground state‐like pluripotency (Wang et al, 2011; Yang et al, 2015). Our study reveals that a chemical approach based on 2a2iL treatment induces naïve pluripotency in human cells and an efficient contribution to interspecies chimeras. Since transient activation of the TGF‐β pathway is able to substitute for 2a treatment, we conclude that 2a2iL‐hPSCs depends on TGF‐β signal transduction for derivation and maintenance of the naïve‐like state.
Results
Chemical agonists of LRH‐1 and RAR‐γ induce naïve‐like pluripotency during reprogramming of human dermal fibroblasts
In order to induce naïve pluripotency during reprogramming of human dermal fibroblasts (HDFs), we used three episomal vectors expressing OCT4, SOX2, KLF4, LIN28, L‐MYC, and p53 shRNA (Okita et al, 2011) along with RJW101 and CD437, two chemical agonists (2a) of LRH‐1 and RAR‐γ, respectively. We added 2a on the first day after electroporation. The duration of 2a treatment was tested for different time periods that ranged from at least 4 days to a maximum of 10 days. Small colonies with naive‐like morphology appeared 20–24 days after electroporation (Fig EV1A and B). We found that 5 days of treatment with 2a followed by addition of 2iL in serum‐free medium led to the appearance of colonies with a naïve‐like morphology (Fig EV1B). Extension of treatment period had adverse effects and seemed to promote exit from pluripotency.
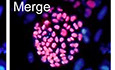
Figure EV1. (related to Fig 1). Generation of naïve‐like hiPSCs by 2a2iL induction.
-
ASchematic drawing of the strategy used to produce safe, transgene‐free hiPSCs. After electroporation of the three episomal vectors that contained OCT4, SOX2, KLf4, LIN28, L‐MYC, and p53 shRNA into human dermal fibroblasts (HDF), we added two chemical agonists (2a), RJW101 and CD437, of nuclear receptors liver receptor homologue‐1 (LRH‐1) and retinoic acid receptor gamma (RARγ), respectively, to fibroblast medium for 5 days. The cells were re‐plated on a mouse embryonic fibroblast (MEF)‐coated plate on day 6 with serum‐free medium that contained 2i and human LIF (2iL). Small colonies that had a distinct morphology from primed colonies were observed around day 20. After trypsinization and passage onto a feeder‐coated plate, dome‐shaped colonies were clearly visible. RT–PCR analysis showed the status of episomal expression in nRepi‐iPSC1.
-
BBright‐field images of nRepi‐iPSC1 colonies on day 20 after reprogramming and at passage 18 (scale bar: 200 μm).
-
CnRepi‐iPSC1 showed alkaline phosphatase (ALP) activity (scale bar: 100 μm).
-
DnRepi‐iPSC1 displayed a normal karyotype.
-
EImmunostaining showed the expressions of pluripotency markers OCT4, NANOG, TRA‐1-81, and SSEA4 in addition to nuclear localization of STAT3 and TFE3 in nRepi‐iPSC1 (scale bar: 50 μm).
-
FThe teratoma formation assay showed the ability of nRepi‐iPSC1 to differentiate into the derivatives of three embryonic germ cell layers as seen by: (i) rosette structures (ectoderm), (ii) cartilage (mesoderm), and (iii) primitive gut (endoderm; scale bar: 50 μm).
-
GCloning efficiency assay of pRepi‐iPSC4 and nRepi‐iPSC1 in the absence of ROCK inhibitor (ROCKi) and Y27632. **t‐test: P < 0.01. Error bar indicates SD (n = 3). nRepi‐iPSC1, Naïve‐like Royan episomal iPSC1; pRepi‐iPSC4, Primed Royan episomal iPSC4.
Evaluation of one of the resulting lines, nRepi‐iPSC1, revealed the appearance of dome‐shaped colonies during repetitive passaging by TrypLE without the need for ROCK inhibitor (ROCKi; Y27632) treatment (Fig EV1B). Further assessment also showed alkaline phosphatase (ALP) enzyme activity and a stable karyotype after 20 passages (Fig EV1C and D). Analysis of pluripotency markers revealed strong expressions of OCT4, NANOG, TRA‐1‐81, and SSEA4 along with nuclear enrichment of STAT3 and TFE3 by immunofluorescence staining (Fig EV1E). The differentiation potential of nRepi‐iPSC1 was examined by subcutaneous injection into immunodeficient nude mice. We observed teratoma formation in these mice after 2 months. Histological analysis confirmed the generation of derivatives of all three embryonic germ layers, including rosette structures, cartilage, and primitive gut, which indicated the presence of ectoderm, mesoderm, and endoderm derivatives, respectively, in nRepi‐iPSC1‐derived teratomas (Fig EV1F). In comparison to primed human iPSCs (hiPSCs; pRepi‐iPSC4), derived in parallel, nRepi‐iPSC1 showed significantly higher single‐cell cloning efficiency in the absence of ROCKi (Fig EV1G). These results indicated that 2a substitutes for previously reported transgene‐dependent overexpressions of LRH‐1 and RAR‐γ to induce a naïve‐like pluripotent state during hiPSC generation (Wang et al, 2011).
2a2iL treatment induces naïve‐like pluripotency in primed hPSCs
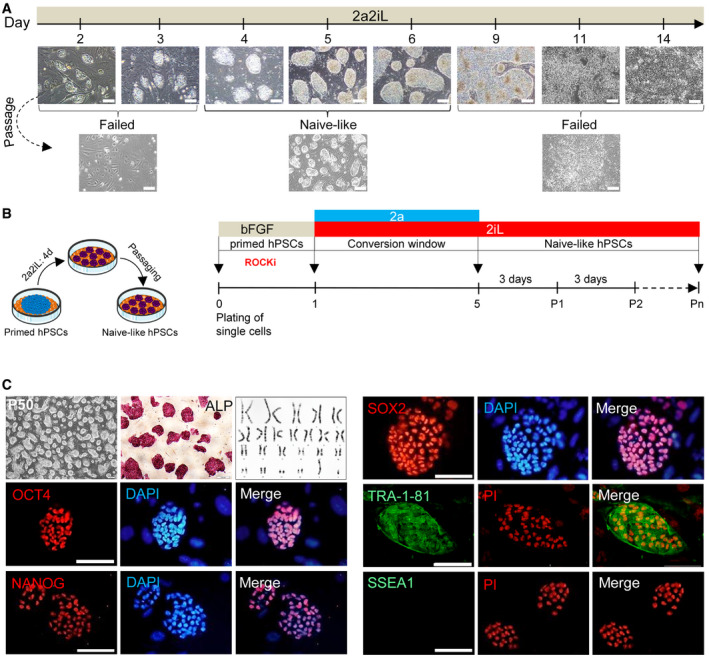
We next analyzed whether 2a converts pre‐established primed hPSC lines into the naïve‐like state. A primed hESC line RH6 (pRH6) was cultured on a feeder layer in serum‐free medium supplemented with 2a2iL from 2 to 14 days in order to determine the optimal exposure time (Fig 1A). Compacted dome‐shaped colonies were first observed on day three and became widespread on days four to six (Fig 1A), while extended exposure with 2a2iL promoted differentiation in hPSCs (Figs 1A and EV2A). To avoid such complications, we developed a protocol for conversion of primed to naïve‐like pluripotency where the primed hPSCs were cultured as single cells for 1 day in conventional primed hPSC medium containing ROCKi, followed by treatment with 2a2iL for 4 days in the absence of ROCKi, and further subsequent passage as single cells only under 2iL conditions (Fig 1B). The converted hPSCs were passaged every 3–4 days and maintained the naïve‐like morphology for over 50 passages in 2iL medium (Fig 1C). The application of 2iL medium as a naïve‐maintenance medium for 2a2iL‐induced hPSCs (hereafter named 2a2iL‐hPSCs) was intriguing, since we were unable to maintain naïve‐like hPSCs derived by other published protocols (Gafni et al, 2013; Theunissen et al, 2014). The resultant naïve‐like 2a2iL‐RH6 had ALP activity and normal karyotype in 2iL medium (Fig 1C). Moreover, 2a2iL‐RH6 expressed the pluripotency markers OCT4, NANOG, SOX2, and TRA‐1‐81, but were negative for SSEA1 (Fig 1C).
Figure 1. 2a2iL induces conversion of primed hPSCs into a naïve‐like state.
-
ADetermination of a time window for 2a2iL treatment needed to induce a naïve‐like pluripotency state in primed hPSCs. One day of treatment with 2a followed by the addition of 2a2iL for up to 14 days. Scale bars: 200 μm.
-
BSchematic illustration that outlines induction of the naïve‐like state from primed hPSCs. Primed hPSCs were first cultured as single cells in conventional hPSC medium in the presence of ROCK inhibitor (ROCKi) on day 0. The medium was replaced by serum‐free medium plus 2a2iL on day 1 without ROCKi. Compact dome‐shaped colonies began to appear on day 3. The resultant naïve‐like cells were passaged as single cells in the absence of ROCKi on day 5 in 2iL medium. 2a2iL‐hPSCs were passaged every 3–4 days in 2iL, and kept typical naïve‐like morphology for over 50 passages.
-
CCharacteristics of 2a2iL‐RH6 cells represented by dome‐shaped colonies, ALP activity, and a normal male karyotype (46, XY). 2a2iL‐RH6 expressed the common pluripotency markers OCT4, NANOG, SOX2, and TRA‐1-81, but not SSEA1. Nuclei were stained with 4′, 6‐diamidino‐2-phenylindole (DAPI) and propidium iodide (PI). Scale bars: 200, 100, and 50 μm in the inset.
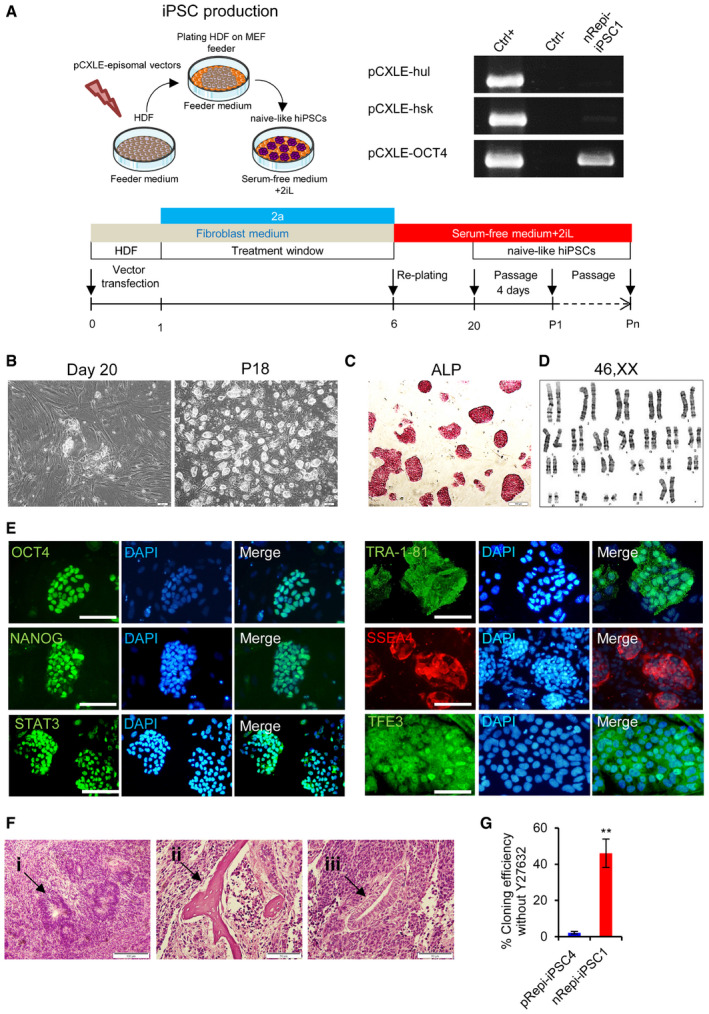
Figure EV2. (related to Fig 1). Conversion of primed hPSCs into the naïve‐like state by 2a2iL induction.
-
AMorphological change of pRH6 during 14 days of induction with 2a2iL.
-
BNaïve‐like colony morphology, alkaline phosphatase (ALP) staining, and immunostaining against pluripotency markers for 2a2iL‐RiPSC1 and 2a2iL‐RH5.
-
CA modified method for induction of naïve‐like pluripotency by 2a2iL in some lines with impurity of mesenchymal‐like cells (MLCs). (i) The hPSC line (pRH5) began to differentiate into mesenchymal‐like cells (MLCs) after passage 4 or 5 (black and white arrows show naïve‐like colonies and MLCs, respectively). (ii) Naïve‐like cells could be separated from MLCs by differential adherence (iii) or by two rounds of single‐cell passaging in the primed state before induction into the naïve‐like state.
To determine whether the 2a2iL induction protocol is applicable to other pre‐established primed hPSC lines, we investigated various different hPSC lines. RH5, RiPSC1, RH2, and Bom‐phiPSC11 responded to 2a2iL induction and were converted into a naïve‐like cells that expressed ALP, OCT4, NANOG, and TRA‐1‐81. Figure EV2B shows the characteristics of RH5 and RiPSC1. Careful morphological analysis revealed the appearance of mesenchymal‐like cells (MLCs) in some lines during early passages. However, such cells were easily eliminated by a differential adhesion culture during subsequent passages (Fig EV2C).
Next, we sought to determine whether 2a2iL supports feeder‐independent conversion of primed hPSCs into the naïve‐like state. We found that the lack of feeder cells did not support long‐term expansion (> 24 passages) of naïve‐like cells in the 2iL condition, although 2a2iL‐hPSCs maintained a naïve‐like morphology and expressed naïve‐specific markers in the feeder‐free culture during serial passaging (Fig EV3A). Finally, we investigated whether 2a2iL‐hPSCs are able to return to the primed state. Transfer of naïve‐like single cells onto the MEF‐coated plates containing basic fibroblast growth factor (bFGF) supplemented medium caused a switch back to the primed state, which required mechanical passaging (Fig EV3B). Overall, these data indicated that the 2a2iL induction protocol supports acquisition of naïve‐like pluripotency and keep the ability to return to the primed state under proper conditions.
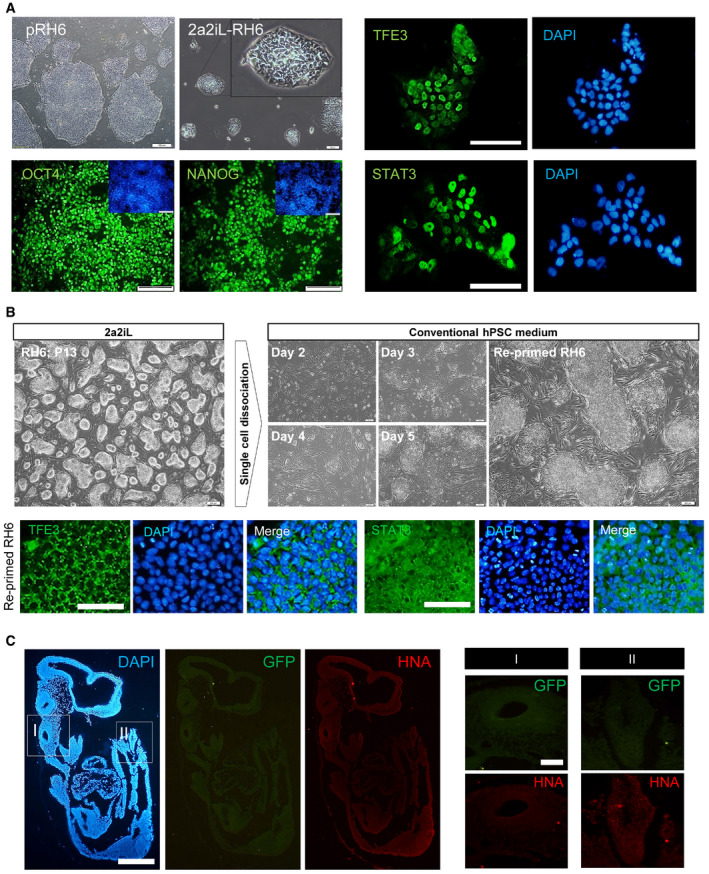
Figure EV3. (related to Figs 1 and 3). Conversion of pRH6 into the naïve‐like state by 2a2iL under feeder‐free conditions.
-
AThe same protocol as Fig 1B was used for conversion of primed hESCs into naïve‐like cells without a feeder layer. Phase contrast morphology of 2a2iL‐RH6 and immunostaining for OCT4 and NANOG in nRH6 (left panel) and TFE3 and STAT3 indicated nuclear localization (right panel) scale bar: 50 μM.
-
BReversion of 2a2iL‐RH6 into the primed state (re‐primed RH6) with cytoplasmic localization of the TFE3 and STAT3 transcription factors.
-
CFluorescence images of tissue sections of non‐injected mouse in E10.5 (negative control group) showed no green or red signals representing GFP and human nuclear antigen (HNA) in different parts of the embryo.
2a2iL‐hPSCs exhibit various hallmarks of naïve pluripotency
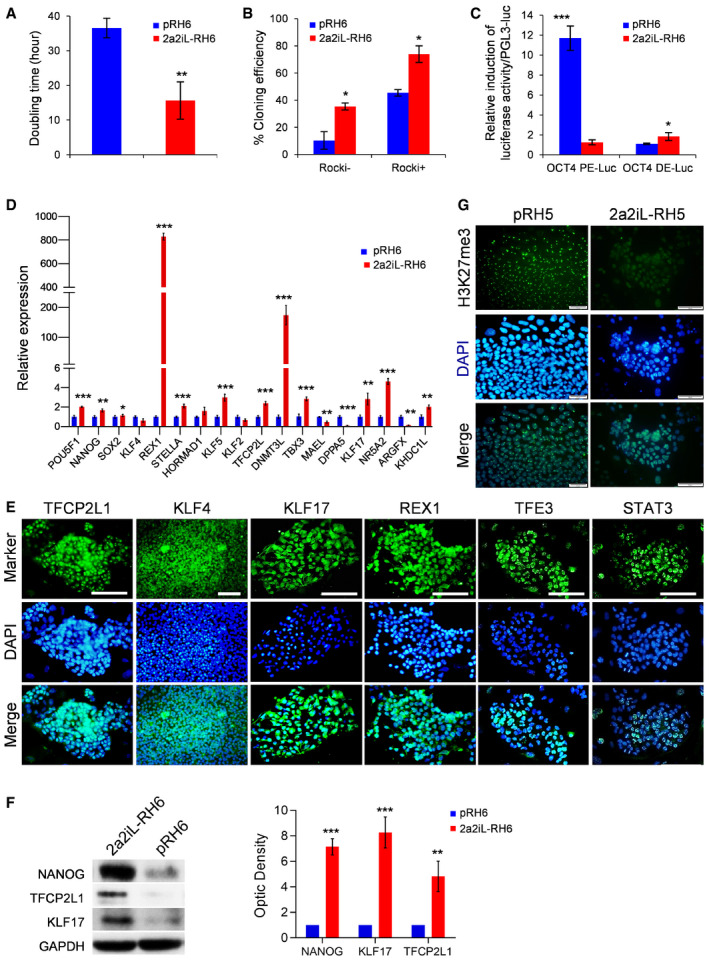
2a2iL‐hPSCs showed a reduction in doubling time to ~ 16 h compared to primed hPSCs with ~ 36 h (Fig 2A). 2a2iL‐hPSCs cultured as single cells without ROCKi had a 40% increase in cloning efficiency compared to primed hPSCs, which further increased to 80% increase when 2a2iL‐hPSCs were cultured as single cells in the presence of ROCKi (Fig 2B). To monitor OCT4 distal enhancer activity, pRH6 and the corresponding 2a2iL‐RH6 were transfected with Luciferase/Renilla reporter constructs, which are under the control of distal or proximal enhancer sequences of the OCT4 gene (Gafni et al, 2013). Our results indicated that OCT4 transcription depends on the distal enhancer in 2a2iL‐RH6 cells in comparison to proximal enhancer dependency in primed parental cells (Fig 2C). Blastocyst‐ and naïve‐specific markers TFCP2L, KLF17, REX1, STELLA, DNMT3L, TBX3, and TET1 were upregulated in 2a2iL‐hPSCs at the transcript and protein levels (Fig 2D–F). Nuclear enrichment of KLF17, KLF4, TFE3, and STAT3 transcription factors was observed in 2a2iL‐hPSCs (Fig 2E). Analysis of X‐chromosome activity in female 2a2iL‐hPSCs showed almost complete absence of H3K27me3 nuclear foci in female cell lines compared to their primed counterparts (Fig 2G). Taken together, our analysis demonstrated that 2a2iL‐hPSCs exhibited various properties of naïve pluripotency.
Figure 2. 2a2iL‐hPSCs show characteristics of naïve pluripotency.
-
ADoubling time in pRH6 versus 2a2iL‐RH6. **P < 0.01 (t‐test). Data presented as mean ± SD (n = 3).
-
BCloning efficiency in the presence (+) and absence (−) of ROCK inhibitor (ROCKi) in pRH6 versus 2a2iL‐RH6. ROCKi (−) *P < 0.05, ROCKi (+) *P < 0.01 (t‐test), data presented as mean ± SD (n = 3).
-
CFirefly luciferase expression of constructs that contain distal and proximal enhancers of OCT4 after transfection of pRh6 and 2a2iL‐RH6. The ratio of firefly luciferase expression to Renilla luciferase expression (expressed from co‐transfected plasmid) measured 48 h after transfection showed proximal enhancer activity in pRH6 and distal enhancer activation in 2a2iL‐RH6. OCT4 PE‐Luc ***P < 0.001, OCT4 DE‐Luc *P < 0.05 (t‐test). Data presented as mean ± SD (n = 3).
-
DqRT–PCR analysis of pluripotency‐related genes in pRH6 and 2a2iL‐RH6. *P < 0.05, **P < 0.01, ***P < 0.001 (t‐test). Data presented as mean ± SD (n = 3).
-
EImmunofluorescence staining for several naïve‐specific markers in 2a2iL‐RH6. Scale bar: 100 μm.
-
FWestern blot analysis of naïve‐related pluripotency markers in pRH6 and 2a2iL‐RH6. **P < 0.01, ***P < 0.001 (t‐test). Data presented as mean ± SD (n = 3).
-
GImmunofluorescence staining against pRH5 and 2a2iL‐RH5 (46, XX) for H3K27me3. Nuclei were stained with 4′, 6‐diamidino‐2-phenylindole (DAPI). Reactivation of inactive X‐chromosome in female 2a2iL‐RH5 was confirmed by the lack of H3K27me3 nuclear foci. Scale bar: 50 μm.
2a2iL‐hPSCs contribute to interspecies chimeras
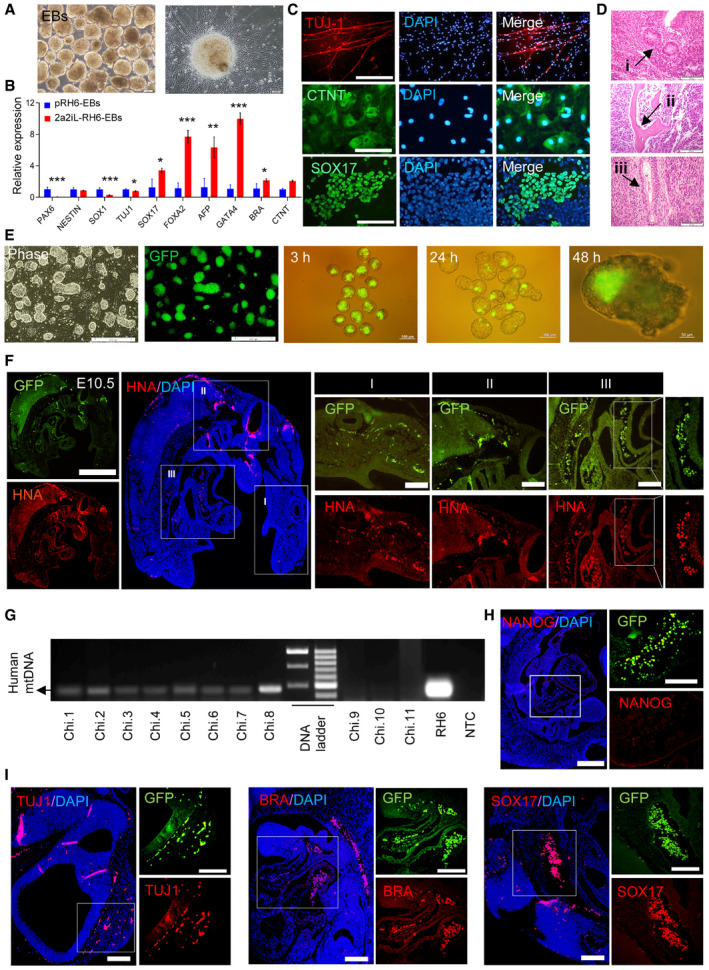
Next, we evaluated the developmental potential of 2a2iL‐hPSCs by identifying derivatives of three embryonic germ layers. 2a2iL‐hPSCs efficiently formed embryoid bodies (EBs) (Fig 3A). qRT–PCR analysis of 25‐day EBs derived from 2a2iL‐RH6 indicated expression of different lineage markers that showed different expression levels compared to their parental primed cells (Fig 3B). Directed differentiation into the three germ lineages demonstrated the potential of 2a2iL‐RH6 to differentiate into neuroectodermal, mesodermal, and endodermal cells, which were visualized by immunofluorescence staining (Fig 3C). Moreover, subcutaneous injection of 2a2iL‐RH6 cells into nude mice resulted in teratoma formation that contained derivatives of all three embryonic germ layers (Fig 3D).
Figure 3. Functional pluripotency assessment of 2a2iL‐hPSCs.
-
APhase contrast images of embryoid bodies (EBs) from 2a2iL‐RH6 cells that differentiated into neurons. Scale bars: 200 and 100 μm in the inset.
-
BqRT–PCR analysis of 20‐day EBs from pRH6 and 2a2iL‐RH6 for markers of different embryonic germ layer derivatives. *P < 0.05, **P < 0.01, ***P < 0.001 (t‐test). Data presented as mean ± SD (n = 3).
-
CImmunofluorescence images of differentiated 2a2iL‐RH6 cells that depict expression of markers for neuron‐like cells (TUJ‐1), cardiac‐muscle‐like cells (CTNT), and hepatocyte‐like cells (SOX17). Nuclei were stained with 4′, 6‐diamidino‐2-phenylindole (DAPI). Scale bar: 100 μm.
-
D2a2iL‐RH6 cells form teratomas that consist of three embryonic germ layer‐derived tissues. (i) Rosette structures (ectoderm), (ii) cartilage (mesoderm), and (iii) primitive gut (endoderm; scale bar 50 μm).
-
EFrom left to right: Phase contrast and GFP representation in 2a2iL‐RH6-eGFP; presence of injected 2a2iL‐RH6-eGFP in the mouse blastocyst and integration into the inner cell mass (ICM) after 3‐, 24-, and 48‐h incubation periods. Scale bars: 200, 100, and 50 μm in the inset.
-
FFluorescence images of tissue sections from human–mouse chimeras in E10.5 showed cells positive for GFP (top) and human nuclear antigen (HNA) (bottom) in different parts of the embryos, including the head (i), trunk (ii), and tail (iii) with the higher magnification in the right panels. Scale bars: 200 and 100 μm in the inset.
-
GGenomic PCR analysis for a human‐specific mitochondrial sequence (391 bp) in human–mouse chimeras.
-
HImmunofluorescence staining against NANOG shows lack of expression in E10.5 human–mouse chimera tissue sections. Scale bars: 200 and 100 μm in the inset.
-
IImmunofluorescence staining against TUJ1, BRACHYURY (BRA), and SOX17 in E10.5 human–mouse chimera tissue sections represents differentiation derivatives of 2a2iL‐RH6. Scale bars: 200 and 100 μm in the inset.
Formation of chimeras is the gold standard to assess pluripotency. Hence, we evaluated the contribution of 2a2iL‐hPSCs to human–mouse interspecies chimeras. To this end, 2a2iL‐RH6 was engineered to express the pCAG‐eGFP transgene allowing cell tracing (Fig 3E). Ten 2a2iL‐RH6‐eGFP cells were injected into the blastocoel of mouse blastocysts at embryonic day (E) 3.5 and analyzed for chimera formation within 2 days of in vitro culture (Fig 3E). Most of the 2a2iL‐RH6‐eGFP cells survived and integrated into the inner cell mass (ICM) 48 h after the injection, which indicated compatibility between 2a2iL‐hPSCs and mouse ICM cells (Fig 3E). To determine the contribution of 2a2iL‐RH6‐eGFP to post‐implantation development, 431 blastocysts were injected with 2a2iL‐RH6‐eGFP cells and transferred into the uteri of foster mothers. Sixty injected embryos implanted successfully and developed normally until E10.5, when the embryos were harvested (nearly 14% implantation efficiency; Table EV1). All 60 fetuses were sectioned and evaluated for the presence of GFP signals to assess for chimera formation. We then selected eight embryos that showed higher levels of detectable GFP signal for further analysis (Fig 3F and G). To validate the presence of 2a2iL‐RH6‐eGFP cells, we stained with an antibody detecting human nuclear antigen (HNA), which recapitulated the results obtained by GFP‐based cell tracing (Fig 3F). Furthermore, PCR analysis for a human‐specific mitochondrial sequence in embryos with high numbers of GFP‐positive cells in comparison to low number confirmed the presence of human cells in the chimeras with high numbers of GFP‐positive cells (Fig 3G). To obtain a more detailed view of the fate of 2a2iL‐RH6 in chimeric embryos, we performed immunofluorescence analysis against pluripotency‐ and lineage‐specific markers. The results showed nearly complete loss of NANOG expression in randomly selected tissue sections from eight chimeras (Fig 3H). In addition, antibodies against human TUJ1, SOX17, and BRACHYURY (BRA) confirmed the presence of differentiated derivatives of 2a2iL‐RH6 in chimeric tissue sections (Fig 3I). We also checked green and red signals in non‐injected mouse (E10.5, negative control) that had been stained with HNA (negative control) to exclude cellular auto‐fluorescents (Fig EV3C). These results demonstrated the high developmental potential of 2a2iL‐hPSCs by various in vitro and in vivo differentiation assays and confirmed the functionality of 2a2iL‐hPSCs by successful formation of human–mouse interspecies chimeras.
2a2iL permits direct generation of naïve‐like hESCs from blastocysts
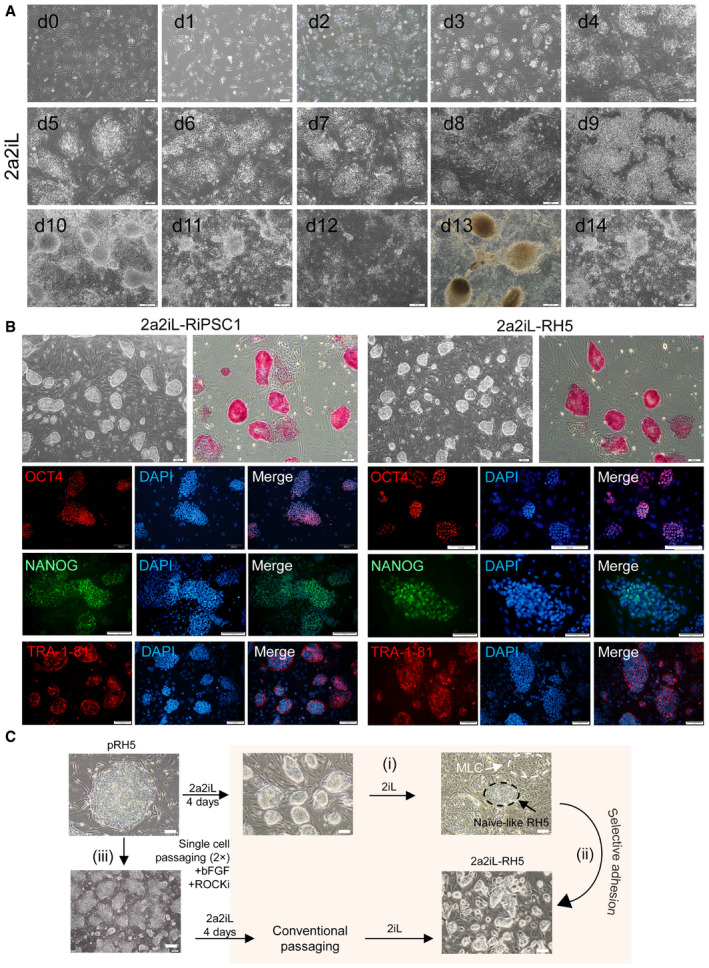
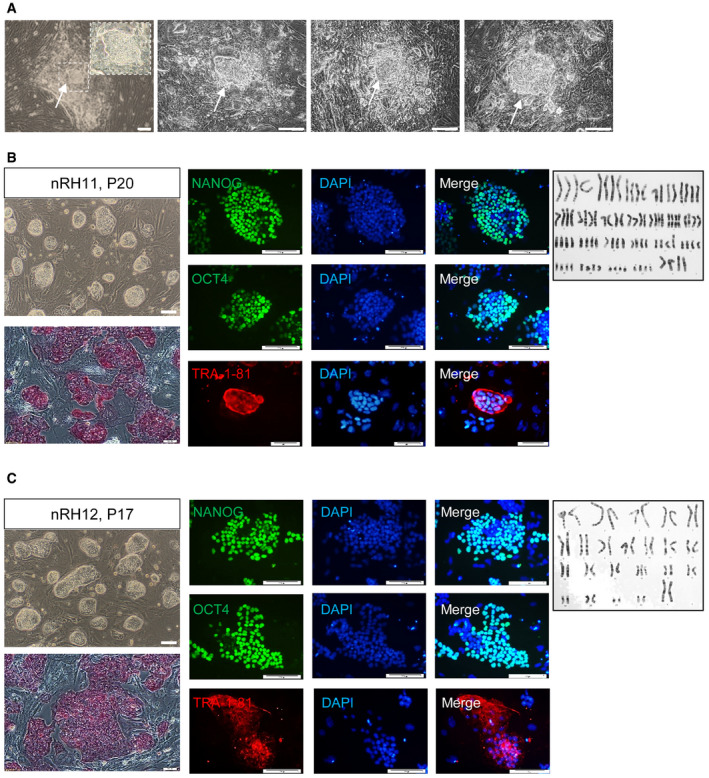
Next, we determined whether 2a2iL allows direct derivation of naïve‐like hESC lines from blastocysts. Initially, we cultured a number of whole zona‐free human blastocysts in 2a2iL‐supplemented medium on MEF‐coated plates. We observed ICM outgrowths that were phenotypically similar to naïve‐like colonies. However, no pluripotent‐like cells were observed after enzymatic passaging (Fig EV4A). We assumed that trophectoderm (TE) cells which cover the ICM outgrowths probably prompted exit from pluripotency. Thus, we removed TE cells by microsurgery and proceeded using two slightly different protocols (Fig 4A). In protocol 1, human blastocysts were cultured in conventional bFGF‐containing hPSC medium for 4 days and further cultivated in 2a2iL until day 10, when the first enzymatically passage was performed (Fig 4A and B). Using this approach, we generated three naïve hESC lines that had similar morphologies to mESCs from 19 ICM outgrowths (~ 16% efficiency of line derivation; Fig 4C). One of the three hESC lines was lost at passage 8 because of a technical problem. Of the remaining two lines, nRH11 was propagated up to passage 20 and nRH12 was propagated up to passage 17 before storage (Fig EV4B and C). nRH11 and nRH12 readily dissociated into single cells in the absence of ROCKi, responded to 2iL medium for continuous passaging, and expressed a panel of pluripotency markers (Fig EV4B and C).
Figure EV4. (related to Fig 4). Characterization of 2a2iL‐hESCs that were derived directly from human blastocysts.
-
AWhole blastocyst outgrowths after 5 days of culture in the 2a2iL condition. Despite the formation of naïve‐like ICM outgrowths, there were no cell lines established by this protocol, which was most likely due to the presence of trophectoderm (TE) cells. More than 100 early, poor quality human embryos were used in this study.
-
B, CColony morphology, ALP staining, immunostaining against pluripotency markers, and karyotype in the 2a2iL‐derived nRH11 and nRH12 cell lines.
Figure 4. 2a2iL allows derivation of naïve‐like hESCs from human blastocysts.
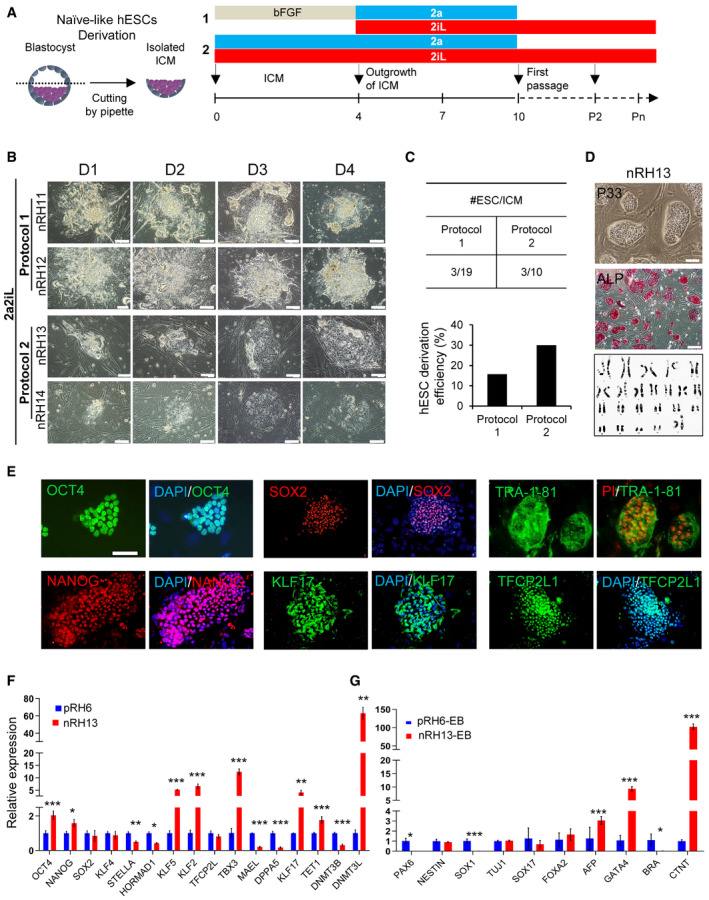
-
ASchematic overview of protocols used to derive naïve‐like hESC lines by 2a2iL from human blastocysts. Two protocols were used to generate naïve hESC lines. In protocol 1, isolated ICMs were cultured in hESC medium that contained basic fibroblast growth factor (bFGF) for 4 days followed by 2a2iL‐supplemented medium until day 10. Dome‐shaped colonies representative of the naïve state formed after single‐cell dissociation. In protocol 2, ICMs were exposed to 2a2iL‐supplemented medium at the beginning of seeding.
-
BMorphology of ICM outgrowths by protocols 1 and 2 during the first 4 days after seeding. nRH11‐14 (naïve Royan H11, 12, 13, and 14): naïve hESC lines produced from human embryos using 2a2iL condition. Scale bar: 100 μm.
-
CComparison of the efficiency of naïve‐like hESCs derivation from human blastocysts by two protocols.
-
DRepresentative example of naïve‐like hESCs (nRH13) at passage 33. Dome‐shaped colonies were stained for ALP activity and karyotyped. Scale bars: 200 and 100 μm in the inset.
-
EExpression of the common pluripotency markers OCT4, SOX2, and TRA-1‐81, and naïve‐related pluripotency markers NANOG, KLF17, and TFCP2L1 in nRH13. Nuclei were stained with 4′, 6‐diamidino‐2-phenylindole (DAPI). Scale bar: 100 μm.
-
FqRT–PCR analysis for pluripotency markers in nRH13 in comparison to a primed hESC line, pRH6. *P < 0.05, **P < 0.01, ***P < 0.001 (t‐test). Data presented as mean ± SD (n = 3).
-
GExpression of lineage markers after embryoid body (EB) formation and spontaneous differentiation in nRH13 cells. *P < 0.05, ***P < 0.001 (t‐test). Data presented as mean ± SD (n = 3).
In protocol 2, we treated the isolated ICMs with a 2a2iL cocktail for 10 days (Fig 4A and B). The ICM outgrowths were dissociated as single cells on day 10 and cultured in the absence of ROCKi in 2iL medium (Fig 4A). After the first passage, a naïve‐like morphology appeared in three of 10 trypsinized ICM outgrowths (~ 30% line derivation efficiency; Fig 4C). One line was lost before passage 10, and the two surviving lines, nRH13 and nRH14, were propagated for further analysis. Importantly, karyotyping of nRH11, nRH12, and nRH13 by G‐banding at passages 16 or 17 revealed that the nRH13 cells had a normal 46XX karyotype (Fig 4D), but nRH11 and nRH12 cells showed tetraploidy and trisomy for chromosome 16, respectively. Because of the quality of human embryos as the source for hESC derivation was not optimal, we could not distinguish whether the observed abnormality was attributed to genomic instability in the embryos or was the result of single‐cell dissociation during repetitive passaging (Nguyen et al, 2013). nRH13 was used for further analysis.
nRH13 expressed the common pluripotency markers ALP (Fig 4D), OCT4, SOX2 and TRA‐1‐81, and naïve‐related pluripotency markers NANOG, KLF17, and TFCP2L1 (Fig 4E). Expression analysis of mRNAs related to naïve‐associated markers revealed elevated KLF5, KLF2, TBX3, DNMT3L, and KLF17 gene expressions in nRH13 compared to primed pRH6 cells (Fig 4F). Moreover, nRH13 readily formed EBs and showed spontaneous differentiation as shown by qRT–PCR analysis for differentiation‐associated markers on day 20 (Fig 4G). In summary, the data indicated that the highly efficient 2a2iL regimen allows derivation of naïve‐like hESCs directly from human blastocysts.
Transcriptional profiling of 2a2iL‐hPSCs
We performed a whole‐transcriptome RNA‐sequencing (RNA‐seq) analysis for 2a2iL‐hPSCs and the parental primed cells for two cell lines, RH5 and RH6. The pairwise Pearson correlation coefficient heatmap showed a high degree of correlation among primed and 2a2iL‐hPSCs (Fig 5A). We also found a clear separation of primed versus 2a2iL‐hPSCs (adjusted P < 0.01; fold change: > 2) by principal component analysis (PCA; Fig 5B). Bioinformatics analysis revealed a set of 1,728 genes that was differentially expressed between primed and 2a2iL‐hPSCs, including 948 and 780 upregulated transcripts, respectively (Benjamini–Hochberg adjusted P < 0.05; log2 fold change: > 1; Fig 5C). Several pluripotency markers were similarly expressed in the two cell types; however, the 2a2iL‐hPSCs showed significantly higher expressions of TBX3, DNMT3L, GDF3, BMP4, FGF4, MYC, NODAL, KLF5, and WNT3 (Dataset EV1).
Figure 5. Transcriptome analysis of 2a2iL‐hPSCs.
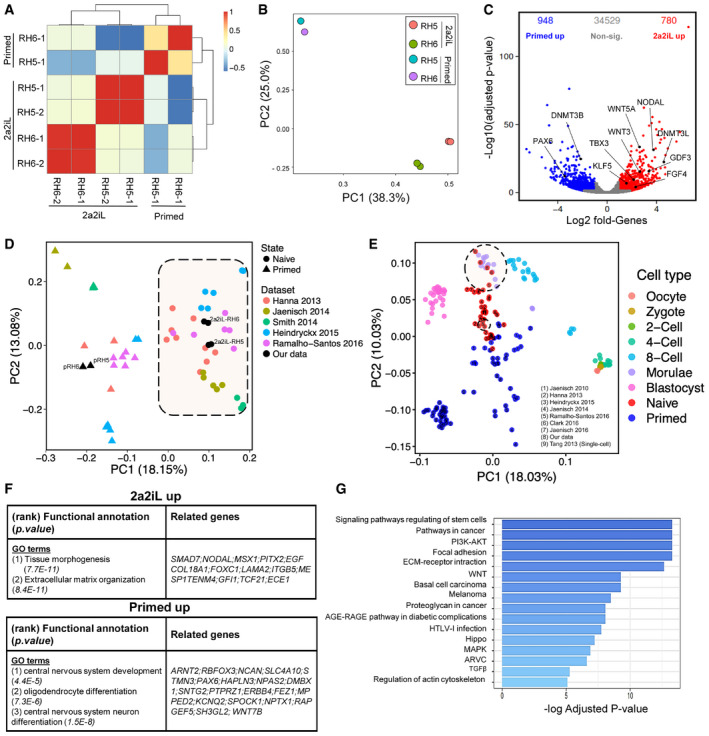
-
APairwise Pearson correlation coefficient heatmap of transcriptome profiles (RNA‐seq) from 2a2iL‐, pRH5, and pRH6 cell lines. Each cell of the heatmap shows the pairwise Pearson correlation coefficient heatmap of genes with the largest expression variances in a pair of samples.
-
BPrincipal component analysis (PCA) of the mean‐centered transcriptome profiles. The share of variation captured by each principal component is mentioned in the axis labels.
-
CThe volcano plot shows a differential expression between 2a2iL‐ versus primed hPSCs lines. X‐axis shows the log2 fold change, and the y‐axis shows the negative log10 scaled adjusted P‐value (Benjamini–Hochberg). Red and blue points represent upregulated genes in 2a2iL‐ and primed cells, respectively (absolute log2 fold change > 1, adjusted P‐value < 0.05). Gray points indicate non‐significant genes between these states.
-
DIntegrated analysis of six different naïve‐primed datasets. Circular and triangular points in the PCA plot represent naïve and primed samples, respectively. Each color indicates one dataset. Black points represent data generated in this study. Each dataset is indicated by the name of the corresponding author and the year of publication (Dataset EV2).
-
EIntegrated analysis of transcriptome profiles of primed and naïve hPSCs, including primary human embryonic cells. Each point represents a biological sample; the colors represent the cell type. Naïve and primed samples are depicted in red and blue, respectively. Samples from pre‐implantation embryo are shown in different colors. The number inside each circle indicates the study from which the data are obtained. Large dash circle indicates morula stage of human embryo which is almost in line with our data (small dash circle). All empty points are from dataset (9) Tang 2013 (Yan et al, 2013).
-
FFunctional annotation of 2a2iL‐ and primed upregulated genes. The most significant over‐represented Gene Ontology (GO) terms and their significance level, along with related genes, are presented in each row. Adjusted P < 0.05 (adjusted P‐value was obtained using the Benjamini and Hochberg correction to determine the false discovery rate with a log2 fold change > 1).
-
GBar plot showing KEGG signaling pathway analysis of genes significantly upregulated in 2a2iL‐hPSCs (Benjamini–Hochberg adjusted P < 0.05 and log2 fold change > 1). X‐axis shows −log10 of adjusted P‐value.
Next, we compared the transcriptome of 2a2iL‐hPSCs to previously published datasets to determine how they relate to each other (Gafni et al, 2013; Theunissen et al, 2014, 2016; Duggal et al, 2015; Qin et al, 2016). We included expression profiles from five different studies of 49 samples (29 naïve and 20 primed; Dataset EV2) along with our samples (two primed and four 2a2iL‐hPSCs). PCA readily discriminated 2a2iL‐hPSCs from primed hPSCs and indicated that 2a2iL‐hPSCs clustered with other reported naïve hPSCs (Fig 5D).
Interestingly, the elevated expression of FGF4, GDF3, NODAL, LEFTY2, TDGF1, and WNT3 in 2a2iL‐hPSCs resembled the transcriptional signature found in human epiblasts of pre‐implantation embryos (Stirparo et al, 2018) and 2a2iL‐hPSCs are closely to early human pre‐implantation embryos (Yan et al, 2013) together with other naïve hPSCs in the PCA (Huang et al, 2014; Pastor et al, 2016; Theunissen et al, 2016) (Fig 5E). While PC1 showed that 2a2iL‐hPSCs are in line with morula and closer to primed cells, they have been entirely separated from their parental primed cells. Functional annotation of genes upregulated in the naïve‐like state identified pathways related to tissue morphogenesis and extracellular matrix (Fig 5F), whereas primed hESCs typically activate pathways related to nervous system development (Fig 5F). We observed that transcripts for various signal molecules such as FGFs, WNTs, and growth factors that belonged to the TGF‐β superfamily were enriched in 2a2iL‐hPSCs. KEGG pathway analysis of the RNA‐seq data revealed significant upregulation of 16 pathways, of which pluripotency regulation had the highest rank (Fig 5G). Similar to the naïve state in rodents, the PI3K‐AKT, WNT, Hippo, and MAPK pathways were strongly enriched in naïve‐like human cells, which suggests that activation of these pathways is necessary to establish human naïve pluripotency.
Moreover, we observed upregulation of the components of the TGF‐β signaling pathway in 2a2iL‐hPSCs. This was of particular interest since growth factors or chemicals that directly modulate TGF‐β signaling were absent in our culture media (Fig 5G). The role of the TGF‐β pathway in human naïve pluripotency has been poorly explored, despite the fact that a number of reported human naïve cells require supplementation with growth factors such as TGF‐β and Activin A (Gafni et al, 2013; Theunissen et al, 2014) or endogenous activation of these pathways (Qin et al, 2016). This prominent upregulation of TGF‐β pathway components in the 2a2iL‐hPSCs caught our attention for further experiments.
The TGF‐β pathway is critical for sustaining naïve‐like pluripotency in 2a2iL‐hPSCs
A comparison of the transcriptional profiles focusing on TGF‐β pathway‐related molecules, extracted from MSigDB and KEGG databases, showed increased expressions of NODAL, LEFTY1, LEFTY2, SMAD7, and PITX2 (Fig 6A). qRT–PCR analysis confirmed the higher expressions of various TGF‐β pathway‐related molecules in 2a2iL‐hPSCs in comparison to primed cells (Fig 6B). In order to analyze the role of TGF‐β signaling, 2a2iL‐RH6 was cultured in the presence of a TGF‐β signaling chemical inhibitor, SB431542 (SB43). The number of naïve‐like cells significantly declined after 24‐h treatment with SB43, and the cells began to differentiate into MLCs in subsequent days (Fig 6C). After one passage, most naïve‐like cells disappeared and MLCs covered the culture dish, which clearly indicated the dependency of 2a2iL‐hPSCs on TGF‐β signaling. This finding was also validated by another TGF‐β inhibitor, A83‐01 (A83) (Fig 6C). We examined whether activation of TGF‐β signaling was related to the 2a protocol by focusing on the target genes of the RA‐ and LRH‐1 signaling pathways (Balmer & Blomhoff, 2002; Rhinn & Dolle, 2012; Tsankov et al, 2015). Twenty‐seven putative, upregulated target genes were detected for RA‐related signaling pathways and 16 target genes for the LRH‐1‐related signaling pathways in 2a2iL‐hPSCs (Fig 6D and E). Upregulation of the putative target genes correlated with upregulation of TGF‐β signaling (Fig 6F), suggesting that the 2a treatment stimulates TGF‐β signaling. We concluded that induction of naïve pluripotency in 2a2iL‐hPSCs might be dependent on activation of intrinsic TGF‐β signaling.
Figure 6. TGF‐β pathway is critical for maintenance of 2a2iL‐hPSCs.
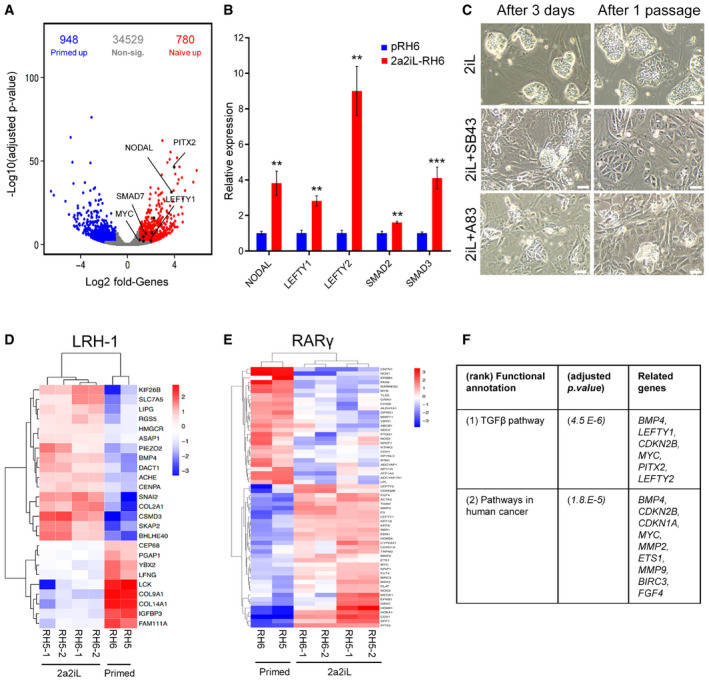
-
APosition of TGF‐β family members in the volcano plot of differentially expressed genes in 2a2iL‐ and primed hPSCs. X‐axis shows log2 fold change, and y‐axis shows negative log10 adjusted P‐value < 0.05 (adjusted P‐value was obtained using the Benjamini and Hochberg correction to determine the false discovery rate). Red and blue points represent upregulated genes in 2a2iL‐ and primed hPSCs, respectively. Genes with no significant expression change are shown in gray.
-
BqRT–PCR analysis of several TGF‐β‐related genes in pRH6 and 2a2iL‐RH6. **P < 0.01, ***P < 0.001 (t‐test). Data presented as mean ± SD (n = 3).
-
CInhibition of the TGF‐β pathway by two small molecules, SB43 and A83. Left panel: morphology of naïve‐like cells 3 days after treatment. Right panel: morphology of treated cells after passaging. Scale bar: 100 μm.
-
DHeatmap shows expression of putative LRH‐1 target genes in this study's transcriptome profiles. Colors indicate log2 fold change of marker genes in the naïve versus primed hPSCs, with a range from dark blue for lower expression to dark red for higher expression.
-
EHeatmap shows expression of putative RARγ target genes in this study's transcriptome profiles. Colors indicate log2 fold change of marker genes in the naïve versus primed hPSCs, with a range from dark blue for lower expression to dark red for higher expression.
-
FKEGG analysis of putative LRH‐1 and RARγ target genes is upregulated in 2a2iL‐hPSCs. Adjusted P < 0.05 (adjusted P‐value was obtained using the Benjamini and Hochberg correction to determine the false discovery rate with a log2 fold change > 1).
To further confirm the functional role of TGF‐β signaling in 2a2iL‐hPSCs, we focused on the time window for conversion of primed hPSCs into naïve‐like by the 2a2iL induction protocol. Our experiments showed increased expressions of NODAL and LEFTY1/2 during the conversion window after exposure with 2a for 1, 3, and 5 days in comparison to primed‐ and 2iL‐treated hPSCs (Fig 7A). Next, primed hPSCs were treated with SB43 for 5 days in conjunction with 2a2iL during the conversion window. The results showed while primed hPSCs started to form dome‐shaped colonies with a sharp border during the first 3 days, this morphology became gradually disturbed and high numbers of non‐viable MLCs emerged during the next 2 days (Fig 7B).
Figure 7. TGFβ‐2iL converts primed hPSCs to the naïve state.
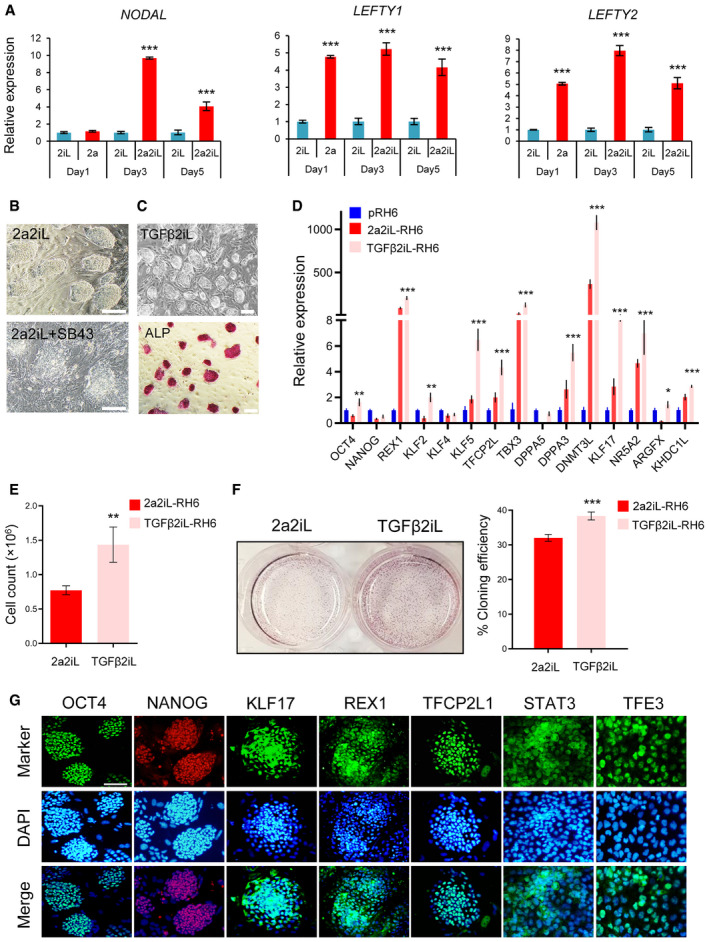
-
AqRT–PCR analysis of ligands associated with TGF‐β signaling during conversion of primed to naïve‐like pluripotency by 2a2iL induction protocol at days 1, 3, and 5. 2iL was selected as a control group. ***P < 0.0001. One‐way ANOVA. Data presented as mean ± SD (n = 3).
-
BInhibition of TGF‐β pathway using SB43 during conversion of primed to naïve pluripotency. Scale bar: 100 μm.
-
CMorphology of TGFβ2iL‐treated RH6 cells along with ALP activity. Scale bar: 200 μm.
-
DqRT–PCR analysis of naïve‐related genes in pRH6, 2a2iL‐, and TGFβ2iL‐RH6. *P < 0.05, **P < 0.01, ***P < 0.001. One‐way ANOVA. Data presented as mean ± SD (n = 3).
-
EBar plot representing cell proliferation rates of 2a2iL‐ and TGFβ2iL‐RH6 by cell counts. **P < 0.01 (t‐test). Data presented as mean ± SD (n = 3).
-
FBar plot of the cloning efficiency of 2a2iL and TGFβ2iL‐RH6 based on the number of ALP positive colonies. ***P < 0.001 (t‐test). Data presented as mean ± SD (n = 3).
-
GImmunofluorescence images show the expression of naïve pluripotency markers in TGF‐β2iL‐RH6. Nuclei were stained with 4′, 6‐diamidino‐2-phenylindole (DAPI). Scale bar: 100 μm.
To investigate whether activation of the TGF‐β pathway mimics effects of 2a during induction of the naïve‐like state, we added TGF‐β to 2iL medium (TGFβ2iL) for 5 days. This treatment promoted the appearance of naïve‐like hPSC colonies, which could be dissociated into single cells in 2iL medium without overt differentiation or cell death (Fig 7C). The TGF2iL‐hPSCs showed substantially increased expressions of REX1, KLF2, KLF5, TFCP2L, TBX3, DPPA3, DNM3L, NR5A2, ARGFX, KHDC1L, and KLF17 compared to primed cells (Fig 7D). TGFβ2iL‐hPSCs had a proliferation rate and clonogenicity comparable to naïve‐like 2a2iL‐hPSCs (Fig 7E and F). They also expressed OCT4, NANOG, KLF17, REX1, and TFCP2L1, and showed nuclear localization of TFE3 and STAT3 (Fig 7G). Taken together, the results demonstrated that TGF‐β is able to substitute for 2a to induce naïve‐like pluripotency in 2iL culture medium. Our findings emphasize the importance of the TGF‐β pathway for both generation and maintenance of naïve‐like 2a2iL‐hPSCs.
Discussion
The main motivation for development of an efficient protocol to establish human naïve pluripotency is the production of cells that have a high growth rate, are resistant to single‐cell dissociation, and show the capability to differentiate into three germ layers while maintaining genome integrity. Various approaches have been tried, including forced expression of naïve‐related transcription factors (Li et al, 2009; Buecker et al, 2010; Hanna et al, 2010; Takashima et al, 2014; Chen et al, 2015), manipulation of different signaling pathways with small molecules (Chan et al, 2013; Ware et al, 2014; Duggal et al, 2015; Qin et al, 2016), or targeting of numerous protein kinases such as PKC, p38, JNK, BRAF, SRC, CDK, and ROCK (Gafni et al, 2013; Theunissen et al, 2014; Guo et al, 2016, 2017; Zimmerlin et al, 2016; Szczerbinska et al, 2019) to induce the naïve pluripotent state in human cells. These different culture conditions induce different levels of naivety in PSCs. Although most of these protocols lead to the generation of similar cellular phenotype, their gene profiling shows a spectrum of naïve pluripotency levels that can be typically clustered into two separate groups; bona fide and intermediate naïve pluripotency (Taei et al, 2020). According to this clustering, naïve‐like cells derived from studies including Chan et al (2013), Duggal et al (2015), Chen et al (2015), Qin et al (2016), Zimmerlin et al (2016), Gafni et al (2013), and Ware et al (2014) are in intermediate state, while 5i/LAF‐ and t2iLGö‐derived cells related to studies of Theunissen et al (2014) and Takashima et al (2014) are in bona fide state of naïve pluripotency (Taei et al, 2020).
Here, we report that synthetic small molecule ligands specific to the nuclear receptors LRH‐1 and RAR‐γ in combination with 2i and LIF (2a2iL) induce naïve‐like pluripotency in human cells during (i) reprogramming of fibroblasts, (ii) conversion of existing primed hPSCs, and (iii) derivation of hESCs from human blastocysts. We found that 2a2iL‐hPSCs own the majority of key criteria of naïve pluripotency: (i) dome‐shaped morphology of the colonies, (ii) single‐cell passaging and high cloning efficiency without the need for ROCKi treatment, (iii) short doubling time that resembled mouse PSCs, (iv) activation of luciferase reporter constructs under the DE of OCT4, (v) nuclear localization of naïve‐related markers such as STAT3 and TFE3, and (vi) normal karyotype after long‐term passaging. The expression of a panel of naïve‐related markers including REX1, KLF17, and TFCP2L1 indicated upregulation in 2a2i‐PSCs versus primed cells at the transcript and protein levels. Our transcriptome analysis also indicated that 2a2iL‐hPSCs are molecularly in the intermediate state of naïve pluripotency (Fig EV5; Taei et al, 2020).
Figure EV5. (related to discussion). Dendrogram clustering.
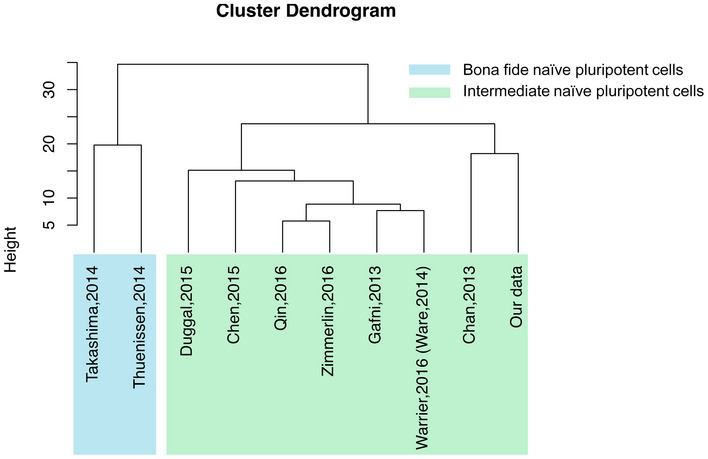
Hierarchical clustering of naïve, primed, and lineage specific‐associated marker genes in 10 studies. The color blue indicates bona fide and green indicates intermediate states of human naïve pluripotency. This clustering shows our study in the intermediate state of human naivety.
We rigorously tested the properties of 2a2iL‐hPSCs by assessing their contribution to human–mouse chimeras. In contrast to previous reports that described a rather inefficient integration of naïve hPSCs into hosts (Theunissen et al, 2014, 2016; Wu et al, 2017), we observed considerable contribution of 2a2iL‐hPSCs to different parts of the host embryos indicating that intermediate naïve hPSCs have higher potency in generation of chimera (Taei et al, 2020). The successful integration of 2a2iL‐hPSCs into post‐implantation mouse embryos suggests a potential stage matching between 2a2iL‐hPSCs and pre‐implantation epiblast of mouse ICM. Therefore, it seems that the transient induction with 2a2iL might position hPSCs in a state that maintains self‐renewal ability in 2iL‐supplemented medium. We suppose that long‐term proliferation of 2a2iL‐hPSCs in 2iL medium without the need for ROCKi for single‐cell dissociation will lead to generation of mPSC‐like hPSCs that can be matched with mouse ICMs to participate into post‐implantation tissue development. Results from other researchers have shown that previously established naïve hPSCs lacked the ability to proliferate in 2iL alone and other supplements such as different kinase inhibitors were needed (Gafni et al, 2013; Takashima et al, 2014; Theunissen et al, 2014). Many of these studies have used ROCKi in naïve hPSC‐maintenance medium.
The ability of these naïve hPSCs to form interspecies chimeras has been subject to an intense debate. Some studies have claimed that established naïve hPSCs have the ability to form chimeras (Gafni et al, 2013); however, others challenged the reproducibility of those experiments (Theunissen et al, 2014). Various potential reasons were raised to explain the barriers that prevent effective formation of interspecies chimeras with hPSCs, including the proliferation rate of donor cells (Masaki et al, 2016), evolutionary distance, and developmental matching between donor cells and host embryos (Wu et al, 2017). For example, Wu et al (2017) have reported that the use of an intermediate type of pluripotency, which is a type of pluripotency that is molecularly between the primed and naïve states, exhibits a high degree of chimerism in post‐implantation pig embryos (Taei et al, 2020). In addition, Yang et al (2017) have reported that human extended pluripotent stem (EPS) cells, which can self‐renew in LCDM medium containing hLIF and the small molecules CHIR99021, DiM, and MiH, can colonize both embryonic and extraembryonic tissues in mouse conceptuses. Since the molecular state of the cells in both mentioned studies is different from other published naïve hPSCs ((Wu et al, 2017; Yang et al, 2017) and our unpublished data), we assume that the ability of chimera formation in 2a2iL‐hPSCs is related to the specific molecular state of the cells that are functionally matched to pre‐implantation mouse embryos.
Our study emphasizes the importance of nuclear receptors to achieve functional pluripotency. In recent years, nuclear receptors have found a special position among the other regulators of pluripotency and reprogramming (Gonzales & Ng, 2013). Nuclear receptor acts as transcription factors but more than half of them are considered to be orphan receptors. It was suggested that deorphanization might provide novel targets for pharmaceuticals (Whitby et al, 2011). The foremost members include Esrrb (Feng et al, 2009; Martello et al, 2012), LRH‐1 (Gu et al, 2005; Guo & Smith, 2010; Heng et al, 2010), Nr5a1 (Barnea & Bergman, 2000), Dax1 (Niakan et al, 2006), RARs (Pikarsky et al, 1994), and Ncoa3 (Wu et al, 2012). However, in‐depth knowledge about the function of nuclear receptors in hPSCs is critically missing. Transcriptome analysis has shown expressions of several nuclear receptors, including ESRRB, RAR‐γ, LRH‐1, and NCOA3 in early pre‐implantation human embryos (Stirparo et al, 2018). It has been reported that expression of ESRRB and RAR‐γ increases from zygotes and 2‐, 4‐, and 8‐cell embryos to the morula developmental stage, peak at the early stage of ICM development, and decreases thereafter (Stirparo et al, 2018). Additionally, LRH‐1 is strongly expressed during all stages of pre‐implantation human embryos, with a peak at the 8‐cell stage. LRH‐1 serves as a marker for the human naïve state (Zimmerlin et al, 2016). Nr5a2 can be substituted for OCT4 in the reprogramming process and modulates expression of NANOG (Heng et al, 2010). On the other hand, short‐term activation of retinoic acid (RA) signaling prevents differentiation of hPSCs (De Angelis et al, 2018), although other studies have suggested that RAR‐γ promotes lineage differentiation (Goncalves et al, 2009; Gely‐Pernot et al, 2012; Rhinn & Dolle, 2012; Zhang et al, 2015). RAR‐γ is indispensable for reprogramming of mouse fibroblasts and EpiSCs into mESC‐like cells through activation of β‐catenin signaling (Yang et al, 2015). In addition, the heterodimer of LRH‐1 and RAR‐γ activates OCT4 expression by binding to RA‐responsive elements (RAREoct) and subsequent recruitment of the transcriptional co‐activators (Gonzales & Ng, 2013).
Transcriptome analysis in this study showed upregulation of a number of ligands, including ligands related to TGF‐β signaling in 2a2iL‐hPSCs. In this regard, evaluation of LRH‐1 and RAR‐γ target genes and gain‐ and loss‐of‐function studies suggested that 2a might activate TGF‐β signaling. We observed that short‐term treatment of cells with 2a during conversion from primed to naïve‐like pluripotency led to upregulation of NODAL and LEFTY1/2. We also found that the transient activation of TGF‐β signaling substitutes for 2a induction to stimulate naïve‐like pluripotency in human cells. Overall, our data revealed that the TGF‐β signaling pathway is critical both for induction and maintenance of naïve‐like pluripotency. Several reports also used TGF‐β/Activin A to establish naïve pluripotency in hPSCs (Qin et al, 2016). Consistent with this, it has been reported that inhibition of TGF‐β signaling in bona fide naïve pluripotent cells can generate extraembryonic trophectoderm indicating the dependency of human naïve pluripotency to TGF‐β signaling (Dong et al, 2020; preprint: Guo et al, 2020). Compared to primed hPSCs in which continuous supplementation of TGF‐β/Activin A is necessary to protect pluripotency, we found that transient stimulation of TGF‐β signaling is sufficient to induce and maintain the naïve‐like state in hPSCs. Further experiments need to determine the molecular mechanisms of TGF‐β signaling pathway in human naïve pluripotency regulation.
Materials and Methods
Ethical issues
Early human embryos were obtained from the Assisted Reproductive Technology (ART) Clinic of Royan Institute (Tehran, Iran) in compliance with the approval statute of the Institutional Review Board and Ethical Committee. Informed consent forms for embryo donation were signed by infertile couple who referred for treatment at Royan. Royan Institutional Review Board approved all procedures conducted with mice and their embryos. In this study, mice were maintained on a 12‐h light/dark schedule. Royan ethical committee is composed of members with scientific, legal, clergyman, and bioethical expertise.
Primed and naïve‐like hPSC culture
Serum‐free medium used in this study consisted of DMEM/F12 (Invitrogen), 20% knockout serum replacement (KOSR, Invitrogen), 2 mM L‐glutamine (Invitrogen), 1% non‐essential amino acids (NEAA, Invitrogen), 1% insulin‐transferrin‐selenium (ITS, Invitrogen), 1% penicillin–streptomycin (Invitrogen), and 0.1 mM β‐mercaptoethanol (β‐ME, Sigma‐Aldrich). Primed hPSC medium contained serum‐free medium supplemented with 12 ng/ml basic fibroblast growth factor (bFGF, Royan Institute).
Previously established primed hPSCs, including hESCs Royan H5 (RH5), RH6 (Baharvand et al, 2006), human iPSCs Royan iPSC1 (RiPSC1) (Totonchi et al, 2010), and the newly derived primed iPSCs in this study, pRepi‐iPSC4, were maintained on a mitomycin C‐inactivated MEF feeder layer in primed hPSCs medium. The primed cells were passaged every 5–7 days using dissociation solution composed of 2.5% trypsin solution (10 ml), 20% KOSR (20 ml), 100 mM CaCl2 (1 ml, Sigma‐Aldrich), and PBS without Ca2+ and Mg2+ (69 ml, Invitrogen). For single‐cell passaging, primed hPSCs were treated with TrypLE (1X; Invitrogen) and cultured in primed hPSC medium supplemented with 10 μM Y27632 (Sigma‐Aldrich) as a ROCK inhibitor for 24 h to prevent cell death.
For induction of the naïve‐like state of pluripotency, we used serum‐free medium supplemented with 2a2iL. 2a2iL consisted of two chemical agonists of nuclear receptors (2a), RJW101 (10 μg/ml; kindly gifted by Dr. Richard J. Withby) and CD437 (0.1 μg/ml, Invitrogen) that were related to LRH‐1 and RARγ, respectively, and 2iL, which included PD0325901 (1 μM; Sigma‐Aldrich), CHIR99021 (3 μM; Stemgent), and human leukemia inhibitory factor (hLIF; 10 ng/ml). For conversion of primed hPSCs into the naïve‐like state, primed hPSCs were dissociated using TrypLE (Invitrogen) and seeded at a density of 2 × 105 cell per 3.5 cm2 on MEF‐coated plates that contained primed hPSC medium in the presence of 10 μM ROCK inhibitor (ROCKi, Y27632) (day 0). On day 1, the medium was refreshed with serum‐free medium supplemented with 2a2iL. The cells were treated with this medium for 4 days in the absence of ROCKi. The medium was refreshed every day. On day five, naïve‐like colonies were dissociated as single cells by TrypLE and re‐plated on MEF‐coated dishes in serum‐free medium plus 2iL. Passaging of naïve‐like hPSCs was carried out by TrypLE every three to 4 days, and the cells were seeded on inactivated MEF in the presence of 2iL. Induction of naïve‐like pluripotency under feeder‐free culture conditions was performed similar to the feeder condition, but on Matrigel (Sigma‐Aldrich)‐coated plates. Naïve‐like hPSCs were also induced with TGF‐β (1 ng/ml; Fitzgerald) in 2iL medium. Inhibition of TGF‐β signaling was performed using SB431542 (2 μM, Cayman Chemical Company) or A83‐01 (Sigma‐Aldrich).
Generation of naïve‐like hiPSCs
Human dermal fibroblasts were obtained from the Royan Stem Cell Bank (Royan Institute) and cultured in feeder medium that included DMEM supplemented with 10% fetal bovine serum (FBS), 1% NEAAs, 2 mM L‐glutamine, 1% penicillin/streptomycin, and 0.1 mM β‐ME. Reprogramming was carried out as previously described (Okita et al, 2011). Briefly, 40 μg of expression plasmid mixtures (Addgene plasmid, 15 μg of each) that included pCXLE‐hOCT3/4‐shp53 (OCT4, P53 shRNA), pCXLE‐hSK (SOX2, KLF4), and pCXLE‐hUL (L‐MYC, LIN28) were transfected into 1 × 10⁶ HDF cells via electroporation (Bio‐Rad) at conditions of 250 mV, 500 μF with a one‐time pulse. The transfected cells were plated on 100 mm dishes covered with gelatin (0.01%, Sigma‐Aldrich). For induction of naïve‐like pluripotency, the culture medium was refreshed with fibroblast medium supplemented with 2a on day one post‐electroporation. The cells were treated with TrypLE on day 6 and re‐plated on MEF‐coated 60 mm dishes in serum‐free medium supplemented with 2iL. After 20–24 days, there were dome‐shaped colonies. Induction of the primed state hiPSCs was performed as previously described (Okita et al, 2011). For further evaluation, naïve‐like hiPSCs were passaged by TrypLE to single cells every 3 or 3 days, and cultured in 2iL medium on MEF‐ or gelatin‐coated plates.
Derivation of naïve‐like ESCs from human blastocysts
We generated hESC in a naïve‐like state as follows. The zona pellucida of 6‐ to 7‐day‐old blastocysts was removed by brief incubation in acid Tyrode's solution. Inner cell masses (ICM) of the denuded blastocysts were isolated mechanically by a microsurgical approach (Meng et al, 2010) and cultured on MEF feeder layer in 2a2iL medium. Initial outgrowths of ICM were harvested on day 10 using TrypLE and then cultured in 2iL medium. Subsequent passaging of the naïve‐like hESC lines was performed for storage and further analysis.
Spontaneous differentiation
In vitro differentiation of 2a2iL‐hPSCs was performed via embryoid body (EB) formation. Dissociated 2a2iL‐hPSCs were separated from MEF by the differential adhesion method and cultured as suspensions in bacterial dishes that contained 2iL medium. After 2 days, the medium was replaced by differentiation medium that included DMEM/F12 with 15% FBS. The medium was refreshed every other day. After 8 days, EBs were plated onto gelatin‐coated plates. The EBs were evaluated for lineage‐specific markers ≤ 21 days later by immunofluorescence staining and qRT–PCR analysis.
Directed differentiation into cardiomyocytes
Induction of differentiation into beating cardiomyocytes was performed as previously described (Fonoudi et al, 2015). Briefly, individual 2a2iL‐hPSCs were forced to form EBs at a density of 2 × 105 cells per ml in 2iL medium for 2 days. Then, the medium was replaced by basal cardiomyocyte differentiation medium that included RPMI 1640 (Invitrogen) supplemented with 2% B27 (Life Technology) without RA or insulin, 1% NEAA, 2 mM L‐glutamine, and 0.1 mM β‐ME. For the first 24 h, we added 12 μM of CHIR99021. The following day, CHIR99021 was removed and the EBs were cultured for an additional 24 h in basal cardiomyocyte differentiation medium. Subsequently, the cells were treated with 5 μM purmorphamine (Stemgent, sonic hedgehog agonist), 5 μM IWP2 (Tocris Bioscience, WNT antagonist), and 5 μM SB431542 (Sigma‐Aldrich, TGF‐β receptor inhibitor) for 48 h. EBs mostly began contractions on day 10.
Directed differentiation into hepatocytes
Embryoid body formation was performed by culturing 2 × 105 cells per ml of 2a2iL‐hPSCs in 2iL medium for 2 days. Then, the medium was replaced by RPMI 1640 supplemented with 2% B27 without RA or insulin, 1% NEAA, 2 mM L‐glutamine, 0.1 mM β‐ME, and 12 μM CHIR for 24 h. The next day, the EBs were plated on Matrigel‐coated plates in RPMI/B27 supplemented with 10 ng/ml Activin A.
Teratoma formation
Teratomas were induced as follows. 2a2iL‐hPSCs were dissociated using TrypLE. Approximately 2–3 × 106 cells were suspended in 100 μl Matrigel and injected subcutaneously into the flanks of 7‐ to 8‐week‐old nude male mice. The resulting teratomas were isolated 30 days after cell transplantation and fixed in 4% paraformaldehyde (PFA), embedded in paraffin, sectioned into 5–7 μm sections, and stained with hematoxylin and eosin. The stained sections were studied under a bright‐field microscope for derivatives of all three germ layers.
Mouse‐human chimera formation
For in vitro and in vivo interspecies chimera generation, E3.5 mouse blastocysts were injected with 10–15 single 2a2iL‐RH6‐eGFP. For in vitro evaluation, chimeric embryos were cultured in 2iL medium for 48 h. For in vivo chimera formation, the chimeric embryos were cultured in 2iL medium for 2 h and a number of the 10–12 embryos were transferred into each of the 2.5‐day post‐coitus pseudo‐pregnant foster mothers. The implanted embryos were harvested from the uteri of the foster mothers on E10.5. The contribution of 2a2iL‐hPSCs in chimeric embryos was analyzed via immunohistochemistry and PCR for human‐specific mitochondrial sequences (Table 1) (Cooper et al, 2007).
Table 1.
Primer sequences and reaction conditions of qRT–PCR or PCR
| Gene name | Primer (5′–3′) | Accession no. | Annealing temperature (°C) |
|---|---|---|---|
| qRT–PCR | |||
| GAPDH | F: ctc att tcc tgg tat gac aac ga | NM_001289746.1 | 60 |
| R: ctt cct ctt gtg ctc ttg ct | |||
| OCT4 | F: ctg ggt tga tcc tcg gac ct | NM_002701.4 | 60 |
| R: cac aga act cat acg gcg gg | |||
| NANOG | F: aaa gaa tct tca cct atg cc | NM_024865.2 | 60 |
| R: gaa gga aga gga gag aca gt | |||
| SOX2 | F: ggg aaa tgg aag ggg tgc aaa aga gg | NM_003106.3 | 60 |
| R: ttg cgt gag tgt gga tgg gat tgg tg | |||
| KLF4 | F: att acc aag agc tca tgc ca | NM_004235.4 | 60 |
| R:cct tga gat ggg aac tct ttg | |||
| REX1 | F: ttt acg ttt ggg agg agg | NM_174900.3 | 60 |
| R: gtg gtc agc tat tca gga g | |||
| DPPA3 | F: aaa gct tcc gat aga ggg ga | NM_199286.3 | 60 |
| R: tgc tca ccg aag aaa att cc | |||
| TBX3 | F: ttt gaa gac cat gga gcc cg | NM_005996.3 | 60 |
| R: aca ttc gcc ttc ccg act tg | |||
| KLF2 | F: ggc aag acc tac acc aag ag | NM_016270 | 60 |
| R: cac aga tgg cac tgg aat gg | |||
| KLF5 | F: cct ggt cca gac aag atg tga | NM_001730 | 60 |
| R: gaa ctg gtc tac gac tga ggc | |||
| DPPA5 | F: gtc gtg gtt tac ggc tcc tat | NM_001025290 | 60 |
| R: ggc aag ttt gag cat ccc tc | |||
| MAEL | F: ttg ctg atg cca tcc ctt act | NM_032858 | 60 |
| R: agctgacatatctggaggtgaa | |||
| HORMAD1 | F: gcc cag ttg cag agg act c | NM_001199829 | 60 |
| R: tct tgt tcc ata agc gca ttc t | |||
| TFCP2L1 | F: cag ccc gag cac tac aac c | NM_014553 | 60 |
| R: ctc cca gct tcc gat tct cc | |||
| SOX1 | F: cac aac tcg gag atc agc aa | NM_005986.2 | 60 |
| R: ggt act tgt aat ccg ggt gc | |||
| PAX6 | F: gtc cat ctt tgc ttg gga aa | NM_001127612 | 60 |
| R: tag cca ggt tgc gaa gaa ct | |||
| TUBB3 | F: gta tcc cga ccg cat cat | NM_006086 | 60 |
| R: tct cat ccg tgt tct cca | |||
| NESTIN | F: tcc agg aac gga aaa tca ag | NM_006617.1 | 60 |
| R: gcc tcc tca tcc cct act tc | |||
| GATA4 | F: cct gtc atc tca cta cgg | NM_002052.3 | 60 |
| R: gct gtt cca aga gtc ctg | |||
| BRA | F: aat tgg tcc agc ctt gga at | NM_003181.2 | 60 |
| R: cgt tgc tca cag acc aca | |||
| CTNT | F: atg atg cat ttt ggg ggt ta | NM_000364.2 | 60 |
| R: cag cac ctt cct cct ctc ag | |||
| AFP | F: aaa tgc gtt tct cgt tgc tt | NM_001134.1 | 60 |
| R: gcc aca ggc caa tag ttt gt | |||
| FOXA2 | F: gga gcg gtg aag atg gaa | NM_021784.4 | 60 |
| R: tac gtg ttc atg ccg ttc at | |||
| SOX17 | F: ctc tgc ctc ctc cac gaa | NM_022454.3 | 60 |
| R: cag aat cca gac ctg cac aa | |||
| DNMT3β | F: tcc cag ctc tta cct tac ca | NM_001207056.1 | 60 |
| F: aaa ctc ctt ccc atc ctg ata ctc | |||
| TET1 | F: cca acc tta ggg agt aac act g | NM_030625.2 | 60 |
| R: ggg agt gct gct tct ttc tg | |||
| DNMT3L | F: atg aag tca agg cta acc agc | NM_175867 | 60 |
| R: cgt cat cgt cgt aca gga aga g | |||
| KLF17 | F: gtctttcttccgttctgatgag | NM_173484.3 | 60 |
| R: ttcttcattcacctaaggacca | |||
| SMAD2 | F: aactatctcctactactctttccc | NM_005901.6 | 60 |
| R: cactcctcttcctatatgcct | |||
| SMAD3 | F: agacaccagttctacctcct | NM_005902.4 | 60 |
| R: ggtctctggaatattgctctg | |||
| NODAL | F: gcg tac atg ctg agc ctc ta | NM_018055 | 60 |
| R: ggt gac ctg gga caa agt g | |||
| NR5A2 | F: ctttgtcccgtgtgtggagat | NM_205860.3 | 60 |
| R: gtcggcccttacagcttcta | |||
| ARGFX | F: gaacgtacttctttcacccac | NM_001012659 | 60 |
| R: ttcctgaaccaaacctttactg | |||
| KHDC1L | F: gacttgatgacacgtaccttcg | NM_001126063.3 | 60 |
| R: agcgtgacacttggagtcct | |||
| LEFTY‐1 | F: tgctacaggtgtcggtgcagagg | NM_020997.4 | 60 |
| R: agaaacggacacttgaaggccagg | |||
| LEFTY‐2 | F: gggaattgggatacctggat | NM_003240.5 | 60 |
| R: ctaaatatgcacgggcaagg | |||
| PCR | |||
| Mitochondrial cytochrome C oxidase subunit I (COI)_Homo Sapiens | F: tagacatcgtactacacgacacg | ‐ | 58 |
| R: tccaggtttatggagggttc | |||
BRA, brachyury.
Immunocytofluorescence staining
For immunocytofluorescence, naïve‐like hPSCs or differentiated cells were fixed in 4% PFA (Sigma‐Aldrich) for 20 min at 37°C and permeabilized with 0.2% Triton X‐100 for 10 min at room temperature for the intracellular markers. Non‐specific antibody binding sites were blocked with 10% host serum of the secondary antibody for 15 min at 37°C. The cells were exposed to the primary antibodies diluted in PBS that contained 0.1% bovine serum albumin (BSA, Sigma‐Aldrich) overnight at 4°C, then rinsed three times with PBS without Ca2+ and Mg2+ supplemented with 0.05% Tween‐20 (PBS/Tween), and incubated with secondary antibodies for 1 h at 37°C. The nuclei were counterstained with 1 mg/ml 4′, 6‐diamidino‐2‐phenylindole (DAPI; Sigma‐Aldrich). An Olympus fluorescent microscope (Olympus BX51) was used to visualize the cells. The primary and secondary antibodies used in this study are listed in Table 2.
Table 2.
Primary and secondary antibodies used for immunofluorescence staining
| Antigen | Company | Cat no. | Dilution/concentration |
|---|---|---|---|
| OCT4 | Santa Cruz | Sc5279 | 1:200 |
| NANOG | Sigma‐Aldrich | N3038 | 1:500 |
| SOX2 | Santa Cruz | Sc20088 | 1:200 |
| TRA1‐81 | Invitrogen | 41‐1100 | 1:100 |
| SSEA1 | R&D System | MAB2155 | 1:250 |
| KLF4 | R&D System | Af3158 | 1:200 |
| TFE3 | Sigma‐Aldrich | HPA023881 | 1:200 |
| STAT3 | Cell Signaling | 4904S | 1:200 |
| TFCP2L1 | Sigma‐Aldrich | HPA029708 | 1:100 |
| KLF17 | Sigma‐Aldrich | HPA024629 | 1:100 |
| REX1 | Santa Cruz | Sc50671 | 1:100 |
| H3K27me3 | Abcam | Ab6002 | 1:500 |
| TUJ‐1 | Sigma‐Aldrich | T8660 | 1:200 |
| CTNT | Abcam | Ab64623 | 1:200 |
| SOX17 | R&D System | AF1924 | 1:100 |
| TBRA | R&D System | AF2085 | 1:100 |
| HNA | AbD Serotec | ab6970 | 1:200 |
| GAPDH | Proteintech | 60004‐1‐Ig | 1:30,000 |
| Anti‐mouse IgG‐Atto488 | Sigma‐Aldrich | 62197 | 1:500 |
| Anti‐mouse IgM‐Texas Red | Invitrogen | Sc2983 | 1:200 |
| Anti‐rabbit IgG‐Alexa Fluor 488 | Santa Cruz | A11008 | 1:500 |
| Anti‐goat IgG‐Alexa Fluor 488 | Invitrogen | A11055 | 1:500 |
| Anti‐rabbit IgG‐Alexa Fluor 546 | Invitrogen | A10040 | 1:500 |
| Anti‐mouse IgG‐Alexa 546 | Invitrogen | A10036 | 1:500 |
| Anti‐rabbit IgG‐peroxidase | Sigma‐Aldrich | A0545 | 1:1,000 |
| Anti‐goat TgG‐peroxidase | Sigma‐Aldrich | A5420 | 1:20,000 |
| Anti‐mouse IgG‐peroxidase | Sigma‐Aldrich | A2554 | 1:50,000 |
HNA, human nuclear antigen.
Immunohistofluorescence staining
We sought to validate the contribution of 2a2iL‐RH6‐eGFP in mouse embryos. Initially, the embryos were isolated from sacrificed mouse at E10.5. The embryos were fixed for 24 h in 10% neutral buffered formalin solution (Merck) in PBS (pH 7.4) at 25◦C and gradually dehydrated using serial dilutions of ethanol and xylol (Merck). The embryos were embedded with melted paraffin (Merck) and sectioned in 6 μm thicknesses by microtome (SLEE). For immunostaining, the sections were placed on slides and the sections were blocked by non‐specific antibodies with 10% goat serum for 1 h at 37°C. The sections were incubated overnight with the selected primary antibodies at 4°C. Subsequently, they were incubated with the specific secondary antibodies for 1 h at room temperature and then studied under a fluorescent microscope (Olympus BX5L). Before and after each antibody stage, the slides were washed with PBS‐tween. The primary and secondary antibodies used in this study are listed in Table 2.
Cell population doubling time
Cell population doubling time was assessed according to a protocol mentioned by Ware et al (2014). Briefly, naïve‐like and primed hPSCs were treated with TrypLE and the single cells were seeded on 3.5‐cm2 MEF‐coated plates at a density of 80,000 cells. Cell counts were performed at two time points, after 48 h (baseline) and 96 h (intervening growth). Three biological replicates were used for the experiments.
Cloning efficiency assay
We determined clonogenicity of naïve‐like versus primed hPSCs by plating the individualized cells on MEF‐coated petri dishes at a density of 10 and 100 cells per 3.5 cm2 plate and 1,000 cells per 6 cm2 plate. Colony counting was performed at day 6 after ALP staining. The clonogenic capacity was calculated as the percent of total colony numbers per the initial number of seeded cells.
Karyotyping
In order to assess genetic integrity at the chromosome level, G‐banding staining was performed according to a standard protocol in the Cytogenetics Laboratory of Royan Institute (Tehran, Iran). Briefly, 2a2iL‐hPSCs were treated with 0.66 mM thymidine (Sigma‐Aldrich) for 16 h at 37°C for cell cycle synchronization. Next, the cells were refreshed with 2iL medium and allowed to enter the M phase for 5 h at 37°C. We added 0.15 mg/ml colcemid (Life Technology) for 45 min at 37°C to arrest the cells in metaphase. Subsequently, 2a2iL‐hPSCs were harvested using TrypLE and allowed to expand by the addition of a hypotonic solution, 0.075 M KCl (Merck), for 10 min at 37°C. The cells were fixed with a 3:1 ratio of ice‐cold methanol:acetic acid (Merck) at three sequential times and dropped on chilled slides. The chromosomes were visualized using G‐banding staining, and we evaluated at least 30 spreads to assess for genetic stability.
Western blotting
Total proteins were extracted from primed and 2a2iL‐hPSCs using a Qproteome Mammalian Protein Prep Kit (Qiagen, cat no. 37901). We added 5× Laemmli buffer (0.25 M Tris–HCl, pH 6.8, 50% glycerol, 10% SDS, 0.01% bromophenol blue, 50 mM DTT) to 20 μg of total protein and the solution was allowed to boil for 5 min at 95°C. Then, the proteins were separated by 12% SDS–PAGE electrophoresis at 100 V for 1.5 h using a Mini‐PROTEAN electrophoresis cell (Bio‐Rad) and transferred to a GE‐PVDF membrane by wet blotting at 15 V for 16 h using a Trans‐Blot (Bio‐Rad). The membranes were blocked for 1 h with 2% milk and incubated overnight at 4°C with the following primary antibodies: anti‐NANOG, TFCP2L1, KLF17, and GAPDH. Next, the membranes were washed with tris‐buffered saline tween (TBST) and incubated with secondary antibodies. Visualization of the blots was performed using ECL substrate (Amersham, RPN2232) with a Uvitec Alliance Q9 Advanced imaging system. Quantification was performed by ImageJ software. Information related to the primary and secondary antibodies is listed in Table 2.
Evaluation of OCT4 distal and proximal enhancer activity
Activation of OCT4 enhancer was evaluated by luciferase activity as described by Chen et al (2015). Briefly, 1 × 106 hPSCs were separately cultured on a 6 cm2 MEF‐coated petri dish. The following day, the cells were co‐transfected with 2.5 μg of each of the PGL3 reporter plasmids: pGL3‐human OCT4 DESV40‐Luc (Addgene), pGL3‐human OCT4 PE‐SV40‐Luc (Addgene), and pGL3‐empty vector (Promega) with the internal control, 2.5 μg of psiCHECK™‐1 Vector (Promega), which expresses Renilla by 5 μl lipofection (Lipofectamine 3000; Invitrogen). After 8 h, the medium was replaced by 2iL medium. At 48 h post‐transfection, the cells were lysed and we measured the activities of both firefly and Renilla luciferases by a Berthold detection system (Promega). Relative luciferase was calculated by normalizing firefly luciferase activity (reporter) to Renilla luciferase activity (internal control).
RNA isolation, reverse transcription and quantitative real‐time PCR
Total RNA was extracted using a RNeasy mini Kit (Qiagen). The isolated RNA was evaluated for purity and concentration using a UV/visible spectrophotometer (WPA, Biowave II). Integrity and quality were checked by 1% agarose gel electrophoresis. Subsequently, 2 μg of total RNA was converted to cDNA using a Revert Aid First‐strand Kit and random hexamer primer (Thermo Scientific) in a final reaction volume of 20 μl. qRT–PCR was performed with a mixture of 10 μl SYBR Premix Ex Taq II (Takara), 0.4 μl ROX reference dye, 1 μl of 5 pmol for each forward and reverse primer, and 2 μl sample (12.5 ng/μl) in a final reaction volume of 20 μl by using Applied Biosystems StepOnePlus (ABI). All experiments were carried out in three biological and two technical replicates. The expression levels of the target genes were normalized to GAPDH as the housekeeping gene and quantified based on the ∆∆Ct method. The sequence primers are listed in Table 1.
RNA‐sequencing and pre‐processing of the data
RNA was isolated from the samples by using a PureLink miRNA Isolation Kit (Ambion) in combination with a PureLink and RNA Mini Kit to isolate RNA > 150 nucleotides. Large‐sized RNA fractions were treated with on‐column DNase digestion (RNase‐Free DNase Set, Qiagen) to avoid contamination by genomic DNA. RNA and library preparation integrity were verified by a BioAnalyzer 2100 (Agilent) or LabChip Gx Touch 24 (Perkin Elmer). We used 1 μg of total RNA as the input for the SMARTer Stranded Total RNA Sample Prep Kit—HI Mammalian (Clontech). Sequencing was performed on a NextSeq500 instrument (Illumina) using v2 chemistry, which resulted in a minimum of 30M reads per library with a 2 × 75 bp paired end setup. The resultant raw reads were assessed for quality, adapter content, and duplication rates with FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc), a quality control tool for high‐throughput sequence data. Reaper version 13–100 was employed to trim reads after a quality drop below a mean of Q20 in a window of 10 nucleotides (Davis et al, 2013). Only reads between 30 and 150 nucleotides were cleared for further analyses. Trimmed and filtered reads were aligned versus the Ensembl human genome version hg19 (GRCh37.p5) using STAR 2.4.0a with the parameter “–outFilterMismatchNoverLmax 0.1” to increase the maximum ratio of mismatches to mapped length to 10% (Dobin et al, 2013). The number of reads that aligned to the genes was counted with featureCounts 1.4.5‐p1 tool from the Subread package (Liao et al, 2014), which is an efficient general purpose program for assigning sequence reads to genomic features. Only reads that mapped at least partially inside the exons were admitted and aggregated per gene. Reads that overlapped multiple genes or aligned to multiple regions were excluded.
Bioinformatics analysis
Analysis of the data was performed using R/Bioconductor. Differentially expressed genes were identified using DESeq2 version 1.62 (Love et al, 2014). Only genes with a minimum absolute log2 fold change of +1 a maximum Benjamini–Hochberg corrected P‐value of 0.05 were deemed to be significantly differentially expressed. The Ensemble annotation was enriched with UniProt data (release 06.06.2014) based on Ensembl gene identifiers (Activities at the Universal Protein Resource [UniProt]).
Author contributions
AT, TK, and ZT contributed to the establishment of the naïve protocol and performed most of the experiments. SM contributed to the study and experimental design, cell culture, and discussions. AS performed qRT–PCR and analyzed the results. SM and KK contributed to the cell culture. AS‐Z and RK contributed to bioinformatics and the results analysis, and prepared the diagrams. SGperformed RNA‐sequencing. BAA assisted with chimera formation. MN‐A and AA contributed to histological assessments. AT and S‐NH wrote the manuscript. AT, SNH, TB, and HB conceived and designed the study, designed the experiments, and analyzed and interpreted the data. TB supported RNA‐seq analysis, interpreted the data, and edited the manuscript. HB and SNH provided financial and administrative support, designed and analyzed the experiments, interpreted and discussed the results, and approved the manuscript. All authors reviewed and confirmed the manuscript before submission.
Conflict of interest
The authors declare that they have no conflict of interest
Supporting information
Expanded View Figures PDF
Table EV1
Dataset EV1
Dataset EV2
Review Process File
Acknowledgements
This work was supported by a grant from Royan Institute, the Iranian Council of Stem Cell Research and Technology, the Iran National Science Foundation (INSF) to H.B. T.B. was supported by grants from the DFG (Excellence Cluster Cardio‐Pulmonary System (ECCPS), SFB TRR81 TP A02 and SFB 1213, TP A02 and B02) and the European Research Area Network on Cardiovascular Diseases (grant number CLARIFY). We are grateful to Aboulfazl Khameh for his work with teratoma formation; Poya Tavakol for mouse manipulations; Hassan Ansari, Zahra Farzaneh, and Ebrahim Shahbazi for their assistance with directed differentiation; Mehdi Totonchi for discussions about the study; Elham Yektadost for Western blotting; and Paniz Rasooli for dendrogram cluster analysis.
EMBO Reports (2020) 21: e47533
Contributor Information
Seyedeh‐Nafiseh Hassani, Email: snafiseh.hassani@royaninstitute.org.
Hossein Baharvand, Email: baharvand@royaninstitute.org.
Data availability
The datasets and computer code produced in this study are available in the following database: RNA‐seq data: Gene Expression Omnibus GSE116501 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE116501).
References
- Baharvand H, Ashtiani SK, Taee A, Massumi M, Valojerdi MR, Yazdi PE, Moradi SZ, Farrokhi A (2006) Generation of new human embryonic stem cell lines with diploid and triploid karyotypes. Dev Growth Differ 48: 117–128 [DOI] [PubMed] [Google Scholar]
- Balmer JE, Blomhoff R (2002) Gene expression regulation by retinoic acid. J Lipid Res 43: 1773–1808 [DOI] [PubMed] [Google Scholar]
- Barnea E, Bergman Y (2000) Synergy of SF1 and RAR in activation of Oct‐3/4 promoter. J Biol Chem 275: 6608–6619 [DOI] [PubMed] [Google Scholar]
- Brons IG, Smithers LE, Trotter MW, Rugg‐Gunn P, Sun B, Chuva de Sousa Lopes SM, Howlett SK, Clarkson A, Ahrlund‐Richter L, Pedersen RA et al (2007) Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature 448: 191–195 [DOI] [PubMed] [Google Scholar]
- Buecker C, Chen HH, Polo JM, Daheron L, Bu L, Barakat TS, Okwieka P, Porter A, Gribnau J, Hochedlinger K et al (2010) A murine ESC‐like state facilitates transgenesis and homologous recombination in human pluripotent stem cells. Cell Stem Cell 6: 535–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan YS, Goke J, Ng JH, Lu X, Gonzales KA, Tan CP, Tng WQ, Hong ZZ, Lim YS, Ng HH (2013) Induction of a human pluripotent state with distinct regulatory circuitry that resembles preimplantation epiblast. Cell Stem Cell 13: 663–675 [DOI] [PubMed] [Google Scholar]
- Chen H, Aksoy I, Gonnot F, Osteil P, Aubry M, Hamela C, Rognard C, Hochard A, Voisin S, Fontaine E et al (2015) Reinforcement of STAT3 activity reprogrammes human embryonic stem cells to naive‐like pluripotency. Nat Commun 6: 7095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JK, Sykes G, King S, Cottrill K, Ivanova NV, Hanner R, Ikonomi P (2007) Species identification in cell culture: a two‐pronged molecular approach. In Vitro Cell Dev Biol Anim 43: 344–351 [DOI] [PubMed] [Google Scholar]
- Davis M, Dongen SV, Abreu‐Gooder C, Bartonicek N, Enright A (2013) Kraken: a set of tools for quality control and analysis of high‐throughput sequence data. Methods 63: 41–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Angelis MT, Parrotta EI, Santamaria G, Cuda G (2018) Short‐term retinoic acid treatment sustains pluripotency and suppresses differentiation of human induced pluripotent stem cells. Cell Death Dis 9: 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Stefano B, Ueda M, Sabri S, Brumbaugh J, Huebner AJ, Sahakyan A, Clement K, Clowers KJ, Erickson AR, Shioda K et al (2018) Reduced MEK inhibition preserves genomic stability in naive human embryonic stem cells. Nat Methods 15: 732–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR (2013) STAR: ultrafast universal RNA‐seq aligner. Bioinformatics 29: 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C, Beltcheva M, Gontarz P, Zhang B, Popli P, Fischer LA, Khan SA, Park KM, Yoon EJ, Xing X et al (2020) Derivation of trophoblast stem cells from naive human pluripotent stem cells. Elife 9: e52504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggal G, Warrier S, Ghimire S, Broekaert D, Van der Jeught M, Lierman S, Deroo T, Peelman L, Van Soom A, Cornelissen R et al (2015) Alternative routes to induce naive pluripotency in human embryonic stem cells. Stem Cells 33: 2686–2698 [DOI] [PubMed] [Google Scholar]
- Evans MJ, Kaufman MH (1981) Establishment in culture of pluripotential cells from mouse embryos. Nature 292: 154–156 [DOI] [PubMed] [Google Scholar]
- Feng B, Jiang J, Kraus P, Ng JH, Heng JC, Chan YS, Yaw LP, Zhang W, Loh YH, Han J et al (2009) Reprogramming of fibroblasts into induced pluripotent stem cells with orphan nuclear receptor Esrrb. Nat Cell Biol 11: 197–203 [DOI] [PubMed] [Google Scholar]
- Fonoudi H, Ansari H, Abbasalizadeh S, Larijani MR, Kiani S, Hashemizadeh S, Zarchi AS, Bosman A, Blue GM, Pahlavan S et al (2015) A universal and robust integrated platform for the scalable production of human cardiomyocytes from pluripotent stem cells. Stem Cells Transl Med 4: 1482–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gafni O, Weinberger L, Mansour AA, Manor YS, Chomsky E, Ben‐Yosef D, Kalma Y, Viukov S, Maza I, Zviran A et al (2013) Derivation of novel human ground state naive pluripotent stem cells. Nature 504: 282–286 [DOI] [PubMed] [Google Scholar]
- Gely‐Pernot A, Raverdeau M, Celebi C, Dennefeld C, Feret B, Klopfenstein M, Yoshida S, Ghyselinck NB, Mark M (2012) Spermatogonia differentiation requires retinoic acid receptor gamma. Endocrinology 153: 438–449 [DOI] [PubMed] [Google Scholar]
- Goncalves MB, Agudo M, Connor S, McMahon S, Minger SL, Maden M, Corcoran JP (2009) Sequential RARbeta and alpha signalling in vivo can induce adult forebrain neural progenitor cells to differentiate into neurons through Shh and FGF signalling pathways. Dev Biol 326: 305–313 [DOI] [PubMed] [Google Scholar]
- Gonzales KA, Ng HH (2013) Driving pluripotency and reprogramming: nuclear receptors at the helm. Semin Cell Dev Biol 24: 670–678 [DOI] [PubMed] [Google Scholar]
- Gu P, Goodwin B, Chung AC, Xu X, Wheeler DA, Price RR, Galardi C, Peng L, Latour AM, Koller BH et al (2005) Orphan nuclear receptor LRH‐1 is required to maintain Oct4 expression at the epiblast stage of embryonic development. Mol Cell Biol 25: 3492–3505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G, Smith A (2010) A genome‐wide screen in EpiSCs identifies Nr5a nuclear receptors as potent inducers of ground state pluripotency. Development 137: 3185–3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G, von Meyenn F, Santos F, Chen Y, Reik W, Bertone P, Smith A, Nichols J (2016) Naive pluripotent stem cells derived directly from isolated cells of the human inner cell mass. Stem Cell Reports 6: 437–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G, von Meyenn F, Rostovskaya M, Clarke J, Dietmann S, Baker D, Sahakyan A, Myers S, Bertone P, Reik W et al (2017) Epigenetic resetting of human pluripotency. Development 144: 2748–2763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G, Stirparo GG, Strawbridge S, Yang J, Clarke J, Li MA, Myers S, Ozel BN, Nichols J, Smith A (2020) Trophectoderm potency is retained exclusively in human naïve cells. bioRxiv 10.1101/2020.02.04.933812v1 [PREPRINT] [DOI] [Google Scholar]
- Hanna J, Cheng AW, Saha K, Kim J, Lengner CJ, Soldner F, Cassady JP, Muffat J, Carey BW, Jaenisch R (2010) Human embryonic stem cells with biological and epigenetic characteristics similar to those of mouse ESCs. Proc Natl Acad Sci USA 107: 9222–9227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassani SN, Totonchi M, Gourabi H, Scholer HR, Baharvand H (2014) Signaling roadmap modulating naive and primed pluripotency. Stem Cells Dev 23: 193–208 [DOI] [PubMed] [Google Scholar]
- Hassani SN, Moradi S, Taleahmad S, Braun T, Baharvand H (2019) Transition of inner cell mass to embryonic stem cells: mechanisms, facts, and hypotheses. Cell Mol Life Sci 76: 873–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng JC, Feng B, Han J, Jiang J, Kraus P, Ng JH, Orlov YL, Huss M, Yang L, Lufkin T et al (2010) The nuclear receptor Nr5a2 can replace Oct4 in the reprogramming of murine somatic cells to pluripotent cells. Cell Stem Cell 6: 167–174 [DOI] [PubMed] [Google Scholar]
- Huang K, Maruyama T, Fan G (2014) The naive state of human pluripotent stem cells: a synthesis of stem cell and preimplantation embryo transcriptome analyses. Cell Stem Cell 15: 410–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Wei W, Zhu S, Zhu J, Shi Y, Lin T, Hao E, Hayek A, Deng H, Ding S (2009) Generation of rat and human induced pluripotent stem cells by combining genetic reprogramming and chemical inhibitors. Cell Stem Cell 4: 16–19 [DOI] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W (2014) FeatureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923‐930 [DOI] [PubMed] [Google Scholar]
- Liu X, Nefzger CM, Rossello FJ, Chen J, Knaupp AS, Firas J, Ford E, Pflueger J, Paynter JM, Chy HS et al (2017) Comprehensive characterization of distinct states of human naive pluripotency generated by reprogramming. Nat Methods 14: 1055–1062 [DOI] [PubMed] [Google Scholar]
- Love M, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA‐Seq data with DESeq2. Genome Biol 15: 550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martello G, Sugimoto T, Diamanti E, Joshi A, Hannah R, Ohtsuka S, Gottgens B, Niwa H, Smith A (2012) Esrrb is a pivotal target of the Gsk3/Tcf3 axis regulating embryonic stem cell self‐renewal. Cell Stem Cell 11: 491–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GR (1981) Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci USA 78: 7634–7638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masaki H, Kato‐Itoh M, Takahashi Y, Umino A, Sato H, Ito K, Yanagida A, Nishimura T, Yamaguchi T, Hirabayashi M et al (2016) Inhibition of apoptosis overcomes stage‐related compatibility barriers to chimera formation in mouse embryos. Cell Stem Cell 19: 587–592 [DOI] [PubMed] [Google Scholar]
- Meng G, Liu S, Li X, Krawetz R, Rancourt DE (2010) Derivation of human embryonic stem cell lines after blastocyst microsurgery. Biochem Cell Biol 88: 479–490 [DOI] [PubMed] [Google Scholar]
- Nguyen HT, Geens M, Spits C (2013) Genetic and epigenetic instability in human pluripotent stem cells. Human Reprod Update 19: 187–205 [DOI] [PubMed] [Google Scholar]
- Niakan KK, Davis EC, Clipsham RC, Jiang M, Dehart DB, Sulik KK, McCabe ER (2006) Novel role for the orphan nuclear receptor Dax1 in embryogenesis, different from steroidogenesis. Mol Genet Metab 88: 261–271 [DOI] [PubMed] [Google Scholar]
- Nichols J, Smith A (2009) Naive and primed pluripotent states. Cell Stem Cell 4: 487–492 [DOI] [PubMed] [Google Scholar]
- Okita K, Matsumura Y, Sato Y, Okada A, Morizane A, Okamoto S, Hong H, Nakagawa M, Tanabe K, Tezuka K et al (2011) A more efficient method to generate integration‐free human iPS cells. Nat Methods 8: 409–412 [DOI] [PubMed] [Google Scholar]
- Pastor WA, Chen D, Liu W, Kim R, Sahakyan A, Lukianchikov A, Plath K, Jacobsen SE, Clark AT (2016) Naive human pluripotent cells feature a methylation landscape devoid of blastocyst or germline memory. Cell Stem Cell 18: 323–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikarsky E, Sharir H, Ben‐Shushan E, Bergman Y (1994) Retinoic acid represses Oct‐3/4 gene expression through several retinoic acid‐responsive elements located in the promoter‐enhancer region. Mol Cell Biol 14: 1026–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H, Hejna M, Liu Y, Percharde M, Wossidlo M, Blouin L, Durruthy‐Durruthy J, Wong P, Qi Z, Yu J et al (2016) YAP induces human naive pluripotency. Cell Rep 14: 2301–2312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhinn M, Dolle P (2012) Retinoic acid signalling during development. Development 139: 843–858 [DOI] [PubMed] [Google Scholar]
- Stirparo GG, Boroviak T, Guo G, Nichols J, Smith A, Bertone P (2018) Integrated analysis of single‐cell embryo data yields a unified transcriptome signature for the human pre‐implantation epiblast. Development 145: dev158501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczerbinska I, Gonzales KAU, Cukuroglu E, Ramli MNB, Lee BPG, Tan CP, Wong CK, Rancati GI, Liang H, Goke J et al (2019) A chemically defined feeder‐free system for the establishment and maintenance of the human naive pluripotent state. Stem Cell Reports 13: 612–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taei A, Rasooli P, Braun T, Hassani SN, Baharvand H (2020) Signal regulators of human naive pluripotency. Exp Cell Res 389: 111924 [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126: 663–676 [DOI] [PubMed] [Google Scholar]
- Takashima Y, Guo G, Loos R, Nichols J, Ficz G, Krueger F, Oxley D, Santos F, Clarke J, Mansfield W et al (2014) Resetting transcription factor control circuitry toward ground‐state pluripotency in human. Cell 158: 1254–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, McKay RD (2007) New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature 448: 196–199 [DOI] [PubMed] [Google Scholar]
- Theunissen TW, Powell BE, Wang H, Mitalipova M, Faddah DA, Reddy J, Fan ZP, Maetzel D, Ganz K, Shi L et al (2014) Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell Stem Cell 15: 471–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theunissen TW, Friedli M, He Y, Planet E, O'Neil RC, Markoulaki S, Pontis J, Wang H, Iouranova A, Imbeault M et al (2016) Molecular criteria for defining the naive human pluripotent state. Cell Stem Cell 19: 502–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz‐Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM (1998) Embryonic stem cell lines derived from human blastocysts. Science 282: 1145–1147 [DOI] [PubMed] [Google Scholar]
- Totonchi M, Taei A, Seifinejad A, Tabebordbar M, Rassouli H, Farrokhi A, Gourabi H, Aghdami N, Hosseini‐Salekdeh G, Baharvand H (2010) Feeder‐ and serum‐free establishment and expansion of human induced pluripotent stem cells. Int J Dev Biol 54: 877–886 [DOI] [PubMed] [Google Scholar]
- Tsankov AM, Gu H, Akopian V, Ziller MJ, Donaghey J, Amit I, Gnirke A, Meissner A (2015) Transcription factor binding dynamics during human ES cell differentiation. Nature 518: 344–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Yang J, Liu H, Lu D, Chen X, Zenonos Z, Campos LS, Rad R, Guo G, Zhang S et al (2011) Rapid and efficient reprogramming of somatic cells to induced pluripotent stem cells by retinoic acid receptor gamma and liver receptor homolog 1. Proc Natl Acad Sci USA 108: 18283–18288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware CB, Nelson AM, Mecham B, Hesson J, Zhou W, Jonlin EC, Jimenez‐Caliani AJ, Deng X, Cavanaugh C, Cook S et al (2014) Derivation of naive human embryonic stem cells. Proc Natl Acad Sci USA 111: 4484–4489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitby RJ, Stec J, Blind RD, Dixon S, Leesnitzer LM, Orband‐Miller LA, Williams SP, Willson TM, Xu R, Zuercher WJ et al (2011) Small molecule agonists of the orphan nuclear receptors steroidogenic factor‐1 (SF‐1, NR5A1) and liver receptor homologue‐1 (LRH‐1, NR5A2). J Med Chem 54: 2266–2281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Yang M, Liu H, Guo H, Wang Y, Cheng H, Chen L (2012) Role of nuclear receptor coactivator 3 (ncoa3) in pluripotency maintenance. J Biol Chem 287: 38295–38304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Platero‐Luengo A, Sakurai M, Sugawara A, Gil MA, Yamauchi T, Suzuki K, Bogliotti YS, Cuello C, Morales Valencia M et al (2017) Interspecies chimerism with mammalian pluripotent stem cells. Cell 168: 473–486.e415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Yang M, Guo H, Yang L, Wu J, Li R, Liu P, Lian Y, Zheng X, Yan J et al (2013) Single‐cell RNA‐Seq profiling of human preimplantation embryos and embryonic stem cells. Nat Struct Mol Biol 20: 1131–1139 [DOI] [PubMed] [Google Scholar]
- Yang J, Wang W, Ooi J, Campos LS, Lu L, Liu P (2015) Signalling through retinoic acid receptors is required for reprogramming of both mouse embryonic fibroblast cells and epiblast stem cells to induced pluripotent stem cells. Stem Cells 33: 1390–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Liu B, Xu J, Wang J, Wu J, Shi C, Xu Y, Dong J, Wang C, Lai W et al (2017) Derivation of pluripotent stem cells with in vivo embryonic and extraembryonic potency. Cell 169: 243–257.e225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Gao Y, Yu M, Wu H, Ai Z, Wu Y, Liu H, Du J, Guo Z, Zhang Y (2015) Retinoic acid induces embryonic stem cell differentiation by altering both encoding RNA and microRNA expression. PLoS ONE 10: e0132566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerlin L, Park TS, Huo JS, Verma K, Pather SR, Talbot CC Jr, Agarwal J, Steppan D, Zhang YW, Considine M et al (2016) Tankyrase inhibition promotes a stable human naive pluripotent state with improved functionality. Development 143: 4368–4380 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expanded View Figures PDF
Table EV1
Dataset EV1
Dataset EV2
Review Process File
Data Availability Statement
The datasets and computer code produced in this study are available in the following database: RNA‐seq data: Gene Expression Omnibus GSE116501 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE116501).


