Abstract
BACKGROUND
Relationships between microbiota composition and clinical outcomes after allogeneic hematopoietic-cell transplantation have been described in single-center studies. Geographic variations in the composition of human microbial communities and differences in clinical practices across institutions raise the question of whether these associations are generalizable.
METHODS
The microbiota composition of fecal samples obtained from patients who were undergoing allogeneic hematopoietic-cell transplantation at four centers was profiled by means of 16S ribosomal RNA gene sequencing. In an observational study, we examined associations between microbiota diversity and mortality using Cox proportional-hazards analysis. For stratification of the cohorts into higher- and lower-diversity groups, the median diversity value that was observed at the study center in New York was used. In the analysis of independent cohorts, the New York center was cohort 1, and three centers in Germany, Japan, and North Carolina composed cohort 2. Cohort 1 and subgroups within it were analyzed for additional outcomes, including transplantation-related death.
RESULTS
We profiled 8767 fecal samples obtained from 1362 patients undergoing allogeneic hematopoietic-cell transplantation at the four centers. We observed patterns of microbiota disruption characterized by loss of diversity and domination by single taxa. Higher diversity of intestinal microbiota was associated with a lower risk of death in independent cohorts (cohort 1: 104 deaths among 354 patients in the higher-diversity group vs. 136 deaths among 350 patients in the lower-diversity group; adjusted hazard ratio, 0.71; 95% confidence interval [CI], 0.55 to 0.92; cohort 2: 18 deaths among 87 patients in the higher-diversity group vs. 35 deaths among 92 patients in the lower-diversity group; adjusted hazard ratio, 0.49; 95% CI, 0.27 to 0.90). Subgroup analyses identified an association between lower intestinal diversity and higher risks of transplantation-related death and death attributable to graft-versus-host disease. Baseline samples obtained before transplantation already showed evidence of microbiome disruption, and lower diversity before transplantation was associated with poor survival.
CONCLUSIONS
Patterns of microbiota disruption during allogeneic hematopoietic-cell transplantation were similar across transplantation centers and geographic locations; patterns were characterized by loss of diversity and domination by single taxa. Higher diversity of intestinal microbiota at the time of neutrophil engraftment was associated with lower mortality. (Funded by the National Cancer Institute and others.)
ALLOGENEIC HEMATOPOIETIC-CELL transplantation is a curative therapy for hematologic cancers in which a patient receives a cytotoxic conditioning regimen followed by infusion of hematopoietic precursor cells from a donor matched for major histocompatibility antigens. Complications such as graft-versus-host disease (GVHD) remain a major cause of illness and death, limiting the broader applicability of allogeneic hematopoietic-cell transplantation.
The intestinal microbiota play a role in host physiology.1 We and others have reported that patients undergoing allogeneic hematopoietic-cell transplantation have microbiota disruption that is characterized by expansions of potentially pathogenic bacteria and loss of alpha diversity — a variable that reflects the number of unique bacterial taxa present and their relative frequencies.2–5 Diversity of the intestinal microbiota has previously been linked with inflammatory bowel disease and response to cancer immunotherapy.6,7 The major adverse outcomes after allogeneic hematopoietic-cell transplantation are relapse, GVHD, infection, and toxic effects to organs. Each of these outcomes, as well as overall survival, has been associated in single-center studies with features of the intestinal microbiota in the period after hematopoietic-cell transplantation.2–4,8–15 For example, domination of intestinal communities by a single bacterial taxon leads to an increased risk of bloodstream infection by a bacterium of that same taxon,3 and exposure to certain antibiotic agents at specific times is associated with transplantation-related mortality and GVHD-related mortality.10,16 Preclinical models have shown that commensal bacteria influence the pathophysiology of GVHD.10,17–20
These findings in small, single-center studies raise questions regarding the development of clinical strategies to manipulate the microbiota with the goal of improving outcomes of allogeneic hematopoietic-cell transplantation.21–24 However, it is unclear whether the relationships between microbiota composition and outcomes of allogeneic hematopoietic-cell transplantation are generalizable. Practice patterns vary across transplantation centers, particularly with regard to antibiotics and nutrition — two critical determinants of microbiota injury.25–27 Moreover, geographic variations in the composition of intestinal microbiota have been described and implicated in the development of autoimmunity.28,29 We studied patterns of microbiota disruption and their associations with clinical outcomes in recipients of allogeneic hematopoietic-cell transplantation at four institutions on three continents.
METHODS
STUDY DESIGN
We conducted this study at four centers: Memorial Sloan Kettering Cancer Center (MSK) in New York; Duke University Medical Center in Durham, North Carolina; the University Medical Center, University Hospital Regensburg, in Regensburg, Germany; and Hokkaido University Hospital in Sapporo, Japan. The study center at MSK was defined as cohort 1, and the other three centers made up cohort 2.
Stool samples were obtained prospectively at each center with the use of similar procedures. The study protocol (available with the full text of this article at NEJM.org) was approved by the institutional review board at MSK. Samples were obtained at the other centers with approval from the local institutional review board. Written informed consent was obtained from all the participants, including recipients of allogeneic hematopoietic-cell transplantation and healthy volunteers. DNA extraction, polymerase-chain-reaction amplification of genomic 16S ribosomal RNA V4–V5 regions, and sequencing were performed in a central laboratory as described in the Supplementary Appendix, available at NEJM.org.8,9
The hematopoietic-cell transplantation comorbidity index (HCT-CI) is a tool for assessing the risk of death after hematopoietic-cell transplantation.30 Weighted scores for individual organ dysfunctions before transplantation are summed; scores range from 0 to a theoretical maximum of 26, with higher scores indicating a higher risk of death after transplantation.
OUTCOMES
The primary outcome was overall survival. Transplantation-related death, relapse (defined here as relapse or progression of disease), and GVHD-related death were also analyzed. The variable of alpha diversity, which we calculated using the inverse Simpson index, is a single value that summarizes a microbiome community according to the count of unique species and how evenly their frequencies are distributed. However, this value does not provide any information about the actual species present.
STATISTICAL ANALYSIS
For the primary analysis of clinical outcomes, patients were stratified into higher-diversity and lower-diversity groups according to the median diversity value observed in the periengraftment samples obtained at MSK. When diversity was additionally analyzed as a continuous variable, hazard ratios refer to the risk of the outcome per 1 log10 change in inverse Simpson values. Analyses were adjusted for multiple comparisons with the use of the Benjamini–Hochberg procedure.31
Associations between microbial diversity and outcomes were assessed with overall and cause-specific Cox proportional-hazards multivariable regression models. In cohort 1 and exploratory subgroups within it, the cumulative incidences of transplantation-related death, GVHD-related death, and relapse were estimated in analyses that accounted for respective competing outcomes. Subgroups within cohort 1 were defined according to allograft manipulation (T-cell depletion) and analyzed for overall survival and cause-specific outcomes. Analyses of clinical outcomes are presented as hazard ratios with 95% confidence intervals. We used a Wilcoxon rank-sum test to compare continuous microbiota features between groups and Fisher’s exact test to compare categorical microbiota features. Microbiota composition was visualized with the use of the t-distributed stochastic neighbor embedding (t-SNE) algorithm (see the Supplementary Appendix).
RESULTS
CHARACTERISTICS OF THE PATIENTS
Stool samples were obtained prospectively at the four institutions: at MSK; Duke University Medical Center; University Medical Center, University Hospital Regensburg; and Hokkaido University Hospital (Table 1). Samples were requested weekly, which resulted in a median of four samples per patient. A cohort of 1362 participants was identified, each of whom had at least one sample that could be evaluated that had been obtained no more than 30 days before the first allogeneic hematopoietic-cell transplantation.
Table 1.
Clinical Characteristics of the Patients.*
| Characteristic | Overall (N = 1362) |
MSK (N = 1076) |
Regensburg (N = 78) |
Duke (N = 142) |
Hokkaido (N = 66) |
|---|---|---|---|---|---|
| Age at HCT — yr | 52.9±13.0 | 53.6±12.8 | 51.3±11.7 | 50.6±13.2 | 49.1±14.5 |
| Male sex — no. (%) | 833 (61) | 650 (60) | 47 (60) | 99 (70) | 37 (56) |
| Disease — no. (%) | |||||
| Acute myeloid leukemia | 490 (36) | 373 (35) | 43 (55) | 42 (30) | 32 (48) |
| Myelodysplastic syndrome or myeloproliferative neoplasm | 264 (19) | 208 (19) | 8 (10) | 41 (29) | 7 (11) |
| Non-Hodgkin’s lymphoma | 229 (17) | 187 (17) | 12 (15) | 25 (18) | 5 (8) |
| Acute lymphoid leukemia | 128 (9) | 94 (9) | 6 (8) | 13 (9) | 15 (23) |
| Myeloma | 111 (8) | 97 (9) | 3 (4) | 10 (7) | 1 (2) |
| Chronic lymphocytic leukemia | 33 (2) | 30 (3) | 3 (4) | 0 | 0 |
| Hodgkin’s lymphoma | 31 (2) | 27 (3) | 0 | 3 (2) | 1 (2) |
| Chronic myeloid leukemia | 29 (2) | 23 (2) | 1 (1) | 4 (3) | 1 (2) |
| Aplastic anemia | 10 (1) | 5 (<1) | 2 (3) | 2 (1) | 1 (2) |
| Other† | 37 (3) | 32 (3) | 0 | 2 (1) | 3 (5) |
| Graft type — no. (%) | |||||
| Unmodified bone marrow | 113 (8) | 83 (8) | 11 (14) | 13 (9) | 6 (9) |
| Cord blood | 211 (15) | 178 (17) | 0 | 19 (13) | 14 (21) |
| T-cell–depleted PBSCs | 447 (33) | 447 (42) | 0 | 0 | 0 |
| Unmodified PBSCs | 591 (43) | 368 (34) | 67 (86) | 110 (77) | 46 (70) |
| Intensity of conditioning regimen — no. (%) | |||||
| Ablative | 771 (57) | 598 (56) | 10 (13) | 116 (82) | 47 (71) |
| Reduced intensity | 468 (34) | 367 (34) | 68 (87) | 14 (10) | 19 (29) |
| Nonmyeloablative | 123 (9) | 111 (10) | 0 | 12 (8) | 0 |
| HCT-CI‡ | 2.6±2.0 | 2.6±2.1 | 2.2±1.7 | 3.4±.7 | 1.6±1.7 |
| Follow-up of survivors — mo | |||||
| Median | 25.2 | 34.2 | 32.5 | 15.0 | 8.3 |
| Interquartile range | 12.8–49.9 | 15.5–56.5 | 26.0–44.4 | 5.8–23.5 | 4.5–12.9 |
A total of 79% of the patients in the overall study population were enrolled at Memorial Sloan Kettering Cancer Center (MSK), New York; 6% at University Medical Center, University Hospital Regensburg, Regensburg, Germany; 10% at Duke University Medical Center, Durham, North Carolina; and 5% at Hokkaido University Hospital, Sapporo, Japan. Additional details are provided in Tables S1 and S2 and in Figure S1. HCT denotes hematopoietic-cell transplantation, and PBSC peripheral-blood stem cell.
Diseases categorized as “other” included biphenotypic acute leukemia, natural killer–cell large granular lymphocyte leukemia, plasmacytoid dendritic-cell neoplasms, and nonmalignant hematologic disorders, including familial hemophagocytic lymphohistiocytosis, X-linked lympho-proliferative disease, and paroxysmal nocturnal hemoglobinuria.
In the hematopoietic-cell transplantation comorbidity index (HCT-CI), weighted scores for individual organ dysfunctions before transplantation are summed; scores range from 0 to a theoretical maximum of 26, with higher scores indicating a higher risk of death after transplantation.30
Acute leukemia was the most common among a range of indications for transplantation. Various intensities of conditioning regimens were used before hematopoietic-cell transplantation. Unmodified peripheral-blood stem cells were the most common graft type (in 43% of patients). At MSK, infused grafts were ex vivo T-cell–depleted in 42% of patients. Practices regarding the use of prophylactic and empirical antibiotic therapies varied across centers (Table S3 in the Supplementary Appendix). The mean (±SD) HCT-CI score was 2.6±2.0 (range 0 to 11).
ASSOCIATION BETWEEN SURVIVAL AND DIVERSITY IN THE PERIENGRAFTMENT PERIOD
To characterize patterns of microbiota injury across different geographic locations, we used 16S ribosomal RNA gene sequencing to analyze 8767 stool samples that had been obtained from 1362 patients across all four centers. A loss of diversity was observed in patients at all four centers during the course of the transplantation period (P<0.001 for each center) (Fig. 1A).
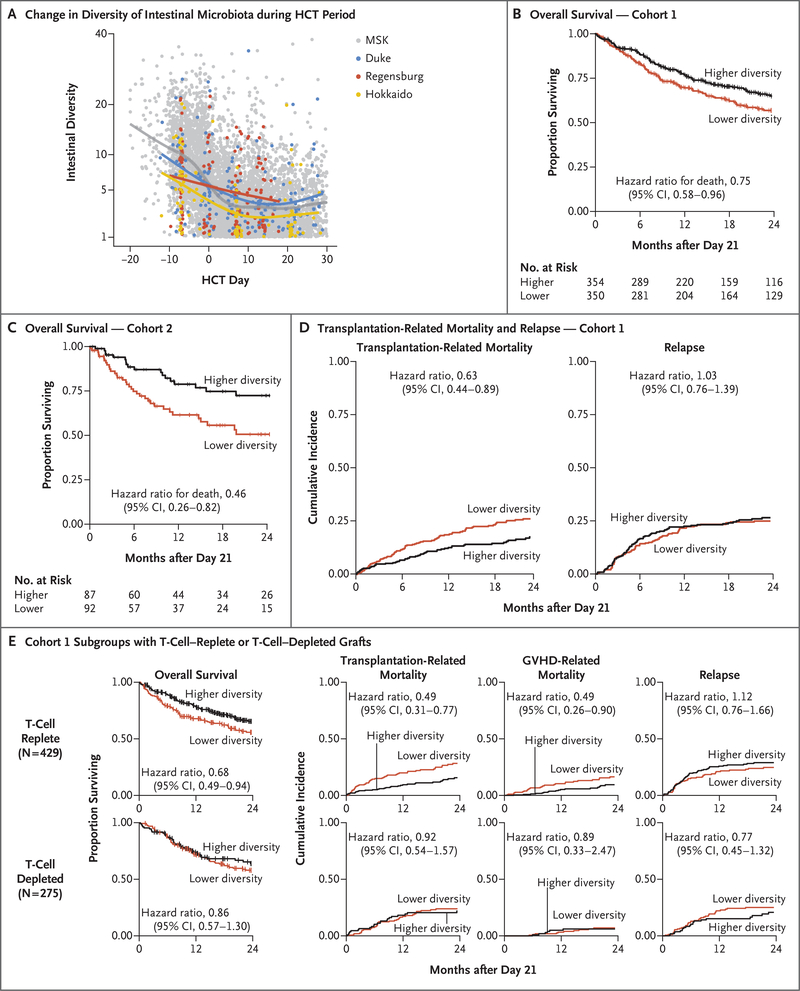
Figure 1 (facing page). Intestinal Diversity during Transplantation Period and Association with Overall Survival.
Intestinal microbiota diversity, as measured by 16S ribosomal RNA gene sequencing and the inverse Simpson index, declined similarly during the course of allogeneic hematopoietic-cell transplantation (HCT) at all four institutions (Panel A). Each point represents a stool sample, color coded according to institution. Curves are loess-smoothed averages. Between each patient’s baseline sample (earliest sample obtained on day −30 to −6) and the periengraftment period (median value of samples obtained from day 7 to 21), the median diversity decreased by a factor of 4.3 at Memorial Sloan Kettering Cancer Center (MSK); by a factor of 3.3 at Duke University Medical Center, Durham, North Carolina; by a factor of 1.7 at University Medical Center, University Hospital Regensburg, Regensburg, Germany; and by a factor of 2.5 at Hokkaido University Hospital, Sapporo, Japan (Fig. S2). Overall survival was longer among patients with higher intestinal diversity in periengraftment samples, both in cohort 1, which comprised patients from MSK (Panel B), and in cohort 2, which comprised patients from the other three sites (Panel C). Tick marks indicate censored data. The median diversity value in cohort 1 (2.64) was used as the stratification cutoff in these landmark analyses (day 21). In cohort 1, there were 104 deaths among 354 patients in the higher-diversity group and 136 deaths among 350 patients in the lower-diversity group; in cohort 2, there were 18 deaths among 87 patients in the higher-diversity group and 35 deaths among 92 patients in the lower-diversity group. CI denotes confidence interval. The cumulative incidences of transplantation-related death and of relapse or progression of disease are shown for cohort 1 (Panel D). There were 52 transplantation-related deaths in 354 patients in the higher-diversity group and 82 such deaths in 349 patients in the lower-diversity group. There were 84 relapse events in 354 patients in the higher-diversity group and 81 relapse events in 349 patients in the lower-diversity group. Panel E shows the subgroup analysis involving recipients of T-cell–replete (unmodified) grafts or T-cell–depleted grafts in cohort 1 (Fig. S4). The numbers of patients at risk in the analyses shown in Panels D and E are provided in Table S4. Subsets of the data plotted in Panels A and D have been reported previously.12,21
To evaluate the association between diversity and overall survival in a multicenter fashion, we considered patients who had stool samples that could be evaluated that had been obtained between days 7 and 21 and who had survived to day 21. For patients who had more than one stool specimen in the sampling window, the median diversity value of those samples was used. Among patients at MSK (i.e., in cohort 1), the median diversity value was 2.64; higher diversity (>2.64) was associated with a lower risk of death than lower diversity (≤2.64) (104 deaths among 354 patients in the higher-diversity group vs. 136 deaths among 350 patients in the lower-diversity group; hazard ratio, 0.75; 95% confidence interval [CI], 0.58 to 0.96) (Fig. 1B and Table 2). This association was also observed after multivariable adjustment for age, intensity of the conditioning regimen, graft source, and the HCT-CI (hazard ratio, 0.71; 95% CI, 0.55 to 0.92), as well as in an analysis in which diversity was considered as a continuous variable (240 deaths among 704 patients; adjusted hazard ratio, 0.50; 95% CI, 0.31 to 0.80).
Table 2.
Association of Intestinal Microbiota Diversity with Survival after Allogeneic HCT, According to Study-Site Cohort.*
| Variable | No. of Deaths | No. of Patients | Univariate Analysis | Continuous Multivariable Analysis | Binary Multivariable Analysis |
|---|---|---|---|---|---|
| hazard ratio (95% CI) | |||||
| Cohort 1: MSK | |||||
| Continuous | 240 | 704 | 0.58 (0.37–0.91) | 0.50 (0.31–0.80) | — |
| Binary | |||||
| Lower diversity | 136 | 350 | Reference | — | — |
| Higher diversity | 104 | 354 | 0.75 (0.58–0.96) | — | 0.71 (0.55–0.92) |
| Cohort 2: Regensburg, Duke, and Hokkaido | |||||
| Continuous | 53 | 179 | 0.33 (0.13–0.84) | 0.38 (0.14–1.04) | — |
| Binary | |||||
| Lower diversity | 35 | 92 | Reference | — | — |
| Higher diversity | 18 | 87 | 0.46 (0.26–0.82) | — | 0.49 (0.27–0.90) |
Shown are multivariable Cox proportional-hazards analyses of the association of diversity of intestinal microbiota during the periengraftment period (median of samples obtained during days 7 to 21) with overall survival. The multivariable models were adjusted for age, intensity of conditioning regimen, graft source, and HCT-CI and were stratified according to institution. Intestinal diversity was measured by the inverse Simpson index and is considered here separately as either a log10-transformed continuous variable or as a median-stratified binary variable. The median diversity value in the MSK cohort (2.64) was used as the cutoff in both cohorts 1 and 2; the lower-diversity group was defined as diversity of 2.64 or less, and the higher-diversity group as diversity of more than 2.64.
The same association between higher intestinal diversity in the periengraftment period (days 7 to 21) and longer survival was observed in the combined cohort of the other three centers (i.e., cohort 2). Patients with higher intestinal diversity had a lower risk of death than those with lower diversity both in the univariate analysis (18 deaths among 87 patients in the higher-diversity group vs. 35 deaths among 92 patients in the lower-diversity group; hazard ratio, 0.46; 95% CI, 0.26 to 0.82) and in the multivariable analysis (hazard ratio, 0.49; 95% CI, 0.27 to 0.90) (Fig. 1C and Table 2). In cohort 2, diversity considered as a continuous variable was predictive of mortality only in the univariate analysis (53 deaths among 179 patients; hazard ratio, 0.33; 95% CI, 0.13 to 0.84) but not in the multivariable model. These data showed that higher diversity of intestinal microbiota during the periengraftment period was associated with a lower risk of death after allogeneic hematopoietic-cell transplantation.
In cohort 1, higher intestinal diversity was associated with a lower risk of transplantation-related death than lower diversity (52 deaths among 354 patients in the higher-diversity group vs. 82 deaths among 349 patients in the lower-diversity group; hazard ratio, 0.63; 95% CI, 0.44 to 0.89) and was not associated with a higher risk of relapse (84 relapse events among 354 patients in the higher-diversity group vs. 81 relapse events among 349 patients in the lower-diversity group; hazard ratio, 1.03; 95% CI, 0.76 to 1.39) (Fig. 1D). Because some recipients in cohort 1 received T-cell–depleted grafts, we had the opportunity to explore the influence of graft composition on the association between diversity and clinical outcomes. Although diversity declined similarly in recipients of unmodified (T-cell replete) grafts and recipients of T-cell–depleted grafts (Fig. S3), the association of microbiota diversity with survival and transplantation-related mortality was observed among recipients of unmodified grafts (30 transplantation-related deaths among 244 patients in the higher-diversity group vs. 46 such deaths among 184 patients in the lower-diversity group; hazard ratio for transplantation-related death, 0.49; 95% CI, 0.31 to 0.77) and not among recipients of T-cell–depleted grafts (Fig. 1E). Among recipients of unmodified grafts, the risk of GVHD-related death was lower among patients with higher intestinal diversity than among those with lower diversity (17 GVHD-related deaths among 244 patients in the higher-diversity group vs. 26 such deaths among 184 patients in the lower-diversity group; hazard ratio, 0.49; 95% CI, 0.26 to 0.90).
In further exploratory analysis, the association between microbiota diversity and survival was also observed in cohort 1 when multivariable models were modified to include exposure to two antibiotic agents (piperacillin–tazobactam and meropenem) that we identified as being associated with a decline in diversity during hematopoietic-cell transplantation (Fig. S5 and Tables S5 and S6). This finding was consistent with previous reports that antibiotics are an important determinant of microbiota composition, including in patients undergoing allogeneic hematopoietic-cell transplantation.3,4,8,10,16,21,32
SPECTRUM OF MICROBIOTA DISRUPTION IN TRANSPLANTATION
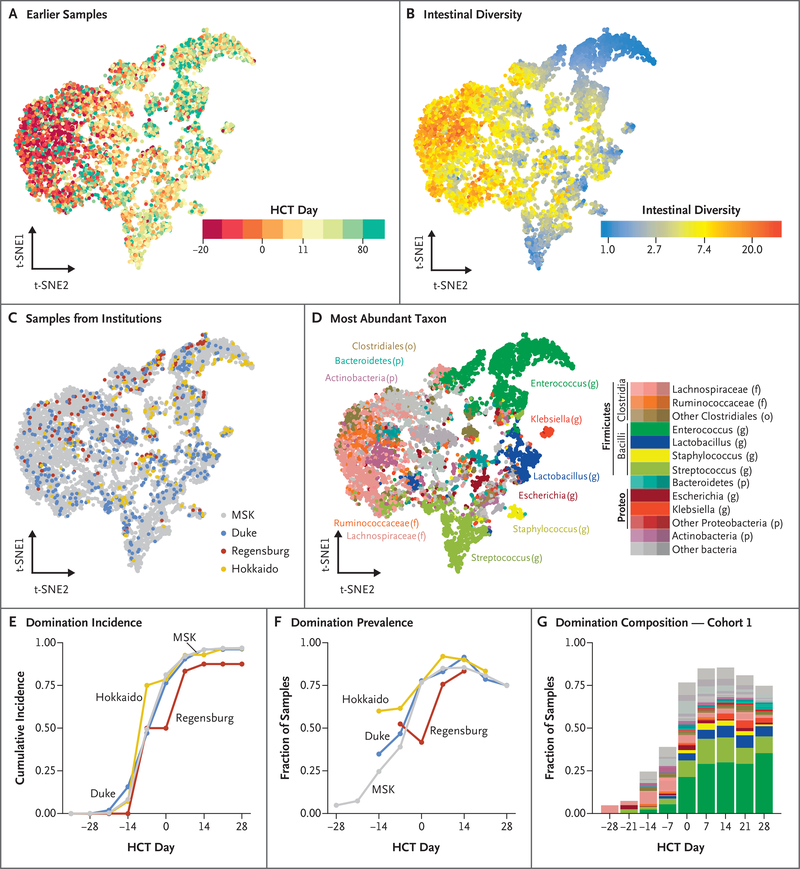
Differences in microbiota composition among samples can be visualized by means of the t-SNE algorithm, such that each point represents a single stool sample. Similar samples are located relatively close to each other, and clusters of distinct microbiota compositions can be appreciated (Fig. 2). Color-coding a t-SNE projection according to the time that the sample was obtained showed a large cluster of early samples and several clusters of samples obtained mostly at later time points (Fig. 2A). As expected from the known decline in microbiota diversity over time (Fig. 1A), the samples in the early cluster tended to be highly diverse, whereas samples from later time points were enriched for several distinct lower-diversity compositions (Fig. 2B).
Figure 2 (facing page). Global Spectrum of Microbiota Disruption in Patients Undergoing Allogeneic HCT.
The microbiota composition of 8767 samples from 1362 patients from all four institutions are shown according to the t-distributed stochastic neighbor embedding (t-SNE) algorithm. Each point represents a single stool sample, and the axes (t-SNE1 and t-SNE2) have arbitrary units. The more similar the samples are, the closer together they appear on the t-SNE plot. Earlier samples (Panel A, left side) are enriched for higher-diversity configurations (Panel B, left side), whereas later samples are enriched for lower-diversity configurations (Panel B, right side). Samples from all four institutions were well distributed across t-SNE space (Panel C). A quantitative comparison of beta-diversity distances between samples from different institutions is shown in Figure S6A. Color coding according to the most abundant taxon in each sample shows that the early cluster was characterized by Clostridia (brown, orange, and pink), Bacteroidetes (teal), and Actinobacteria (purple) (Panel D). Some low-diversity states were characterized by domination with the genera enterococcus (dark green), streptococcus (light green), klebsiella (bright red), and escherichia (dark red). Among patients who had at least one sample obtained both before and after hematopoietic-cell transplantation, the fraction of patients who had had at least one instance of domination each week, defined as a relative abundance of at least 30% for any taxonomic unit, is shown in Panel E. The fraction of samples with domination in each 7-day sliding window is shown in Panel F. The odds over time of a sample being dominated was similar in three of the cohorts and was slightly higher at Hokkaido (Fig. S6B). The taxa that contributed to domination events in cohort 1 are shown in Panel G. Domination was defined at the level of operational taxonomic units; color coding is at higher taxonomic ranks (see color legend in Panel D).
Samples from all four centers spread across the different composition clusters, revealing no obvious transplantation-center–specific effect (Fig. 2C). To quantitatively assess microbiota differences between institutions (over geography) versus within institutions (over time) we compared samples that had been obtained before and after hematopoietic-cell transplantation at each institution. The variation among institutions in microbiota composition before hematopoietic-cell transplantation was smaller than the changes that occurred over the course of transplantation, as measured by Bray–Curtis beta-diversity distances (adjusted P<0.005) (Fig. S6A).33
Color-coding the t-SNE according to the most abundant taxon (Fig. 2D) showed that several of the lower-diversity compositions were characterized by an abundance of enterococcus, klebsiella, escherichia, staphylococcus, and streptococcus. Enterococcus domination, a lower-diversity state that was previously shown to confer an increased risk of vancomycin-resistant enterococcal bacteremia3 as well as a higher risk of GVHD,2,20 occurred at all four centers.
We previously described a pattern of intestinal microbiota domination characterized by dramatic expansions of single taxonomic units that is common in patients undergoing allogeneic hematopoietic-cell transplantation.3 To better understand these microbiota disruption patterns across geographic locations, we quantified the occurrence of domination during the transplantation period at each institution, defined as a relative-abundance threshold of any single taxonomic unit of at least 30%.3 The cumulative incidence of domination by any taxonomic unit rose similarly at the four institutions; the fraction of samples with domination peaked 1 week after transplantation and decreased moderately thereafter (Fig. 2E and 2F). A variety of taxonomic groups contributed to domination events, the most common belonging to the genera enterococcus and streptococcus, at both MSK (Fig. 2G) and the other study sites. Taken together, these data show that microbiota disruption frequently accompanies allogeneic hematopoietic-cell transplantation and that patterns of microbiota injury are observed consistently across transplantation centers, although not in all patients.
MICROBIOTA DISRUPTION BEFORE HEMATOPOIETIC-CELL TRANSPLANTATION AND SURVIVAL
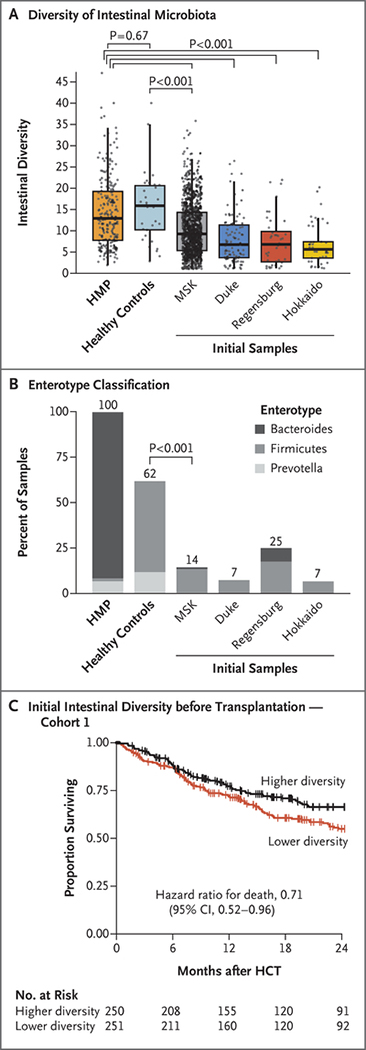
Given the link between intestinal microbiota integrity and outcomes after allogeneic hematopoietic-cell transplantation, we asked whether this association was evident earlier than the periengraftment period. To characterize the microbiota composition of fecal samples obtained from patients arriving at transplantation units for allogeneic hematopoietic-cell transplantation, we compared the composition of the first sample obtained from patients (within a sampling window from day −30 to −6) with those from two sources of healthy volunteers: 313 samples obtained from 212 participants in the Human Microbiome Project,34 whose publicly available raw sequences were processed with the use of our computational pipeline; and stool samples from 34 healthy adult volunteers that were obtained and sequenced at MSK.
The initial samples from 606 patients at all four institutions had lower diversity than did those of the healthy volunteers (P<0.001), and the diversity between the two cohorts of healthy volunteers did not differ significantly (Fig. 3A). We also evaluated the extent to which initial microbiota compositions could be described according to enterotype, a reference classifier of healthy human intestinal communities (see the Supplementary Appendix).35 The initial compositions of stool samples from patients were distinct from those of healthy volunteers, as assessed according to the fraction of samples that could be categorized to an enterotype (adjusted P = 0.002 for Regensburg; adjusted P<0.001 for all other study sites) (Fig. 3B). This finding indicates that patients arrived for transplantation with microbiota compositions that were already distinct from those of healthy volunteers.
Figure 3. Microbiota Injury before HCT and Association with Transplantation Outcomes.
The diversity of the initial pretransplantation samples from patients (obtained on day −30 to −6) is lower than the diversity of 313 samples from 212 participants of the Human Microbiome Project (HMP) and 1 sample obtained from each of 34 healthy adult volunteers at MSK (healthy controls) (Panel A). A total of 993 patients were included in the analysis at MSK, 95 at Duke, 40 at Regensburg, and 45 at Hokkaido. The horizontal line in each box represents the median, the lower and upper boundaries of the boxes the interquartile range, the ends of the whisker lines the minimum and maximum values within 1.5 times the interquartile range, and the dots the individual data points. The proportions of samples from healthy volunteers and initial samples from patients that could be classified according to the enterotype scheme are plotted (Panel B). Samples obtained before HCT were less likely to be classified as belonging to an enterotype than were samples obtained from healthy volunteers. In cohort 1, overall survival was longer among patients with higher intestinal diversity in the initial samples than among those with lower diversity (Panel C). There were 72 deaths among 250 patients in the higher-diversity group and 101 deaths among 251 patients in the lower-diversity group (Table S7). Tick marks indicate censored data. Combined analyses of diversity before transplantation and during the periengraftment period are shown in Table S8 and Figure S7.
In the MSK cohort, higher diversity in the period before hematopoietic-cell transplantation was associated with a lower risk of death (173 deaths among 501 patients; adjusted hazard ratio with diversity as a continuous variable, 0.41, 95% CI, 0.24 to 0.71; two-group analyses are shown in Fig. 3C) and a lower risk of transplantation-related death (103 deaths among 501 patients; adjusted hazard ratio with diversity as a continuous variable, 0.44; 95% CI, 0.22 to 0.87). We observed a weak correlation between diversity values measured at baseline and those measured during the periengraftment period (Pearson’s r = 0.22; 95% CI, 0.12 to 0.31). The association between microbiota diversity before transplantation and survival was not observed at the other three sites (Duke, Regensburg, and Hokkaido), either individually or combined; however, 15 or fewer deaths had occurred among the patients with available baseline samples at each of those centers.
PREDICTIVE MICROBIOTA SIGNATURE
Since alpha-diversity metrics provide no information about which specific species are present, we sought to identify a signature of bacterial abundances in the periengraftment period that could predict mortality. From the set of taxonomic levels (species through phylum), we selected as candidates those taxa that were present in at least 10% of samples with a minimum relative abundance of 10−4. We also removed features that were correlated according to a Pearson’s r greater than 0.75 with a hierarchically lower taxonomic rank, which yielded an input set of 172 candidate taxonomic features. Regularized Cox regression with cross-validation was performed in cohort 1 to derive a risk score (see the Supplemental Methods section). We found that higher values of the score were associated with a higher risk of death in cohort 2 (53 deaths among 178 patients; adjusted hazard ratio, 1.39; 95% CI, 1.02 to 1.91) (Fig. S8), which indicates that not only a diversity metric but also a signature of specific bacterial abundances were informative about the risk of death after hematopoietic-cell transplantation across institutions.
DISCUSSION
We report the results of a multicenter study of longitudinal profiling of intestinal microbiota in patients undergoing allogeneic hematopoietic-cell transplantation. We observed similar patterns of microbiota disruption and associations with clinical outcomes across transplantation centers and geographic locations. Microbiota compositions before transplantation were relatively similar across geographic locations but were distinct from those of healthy persons. Profound microbiota injuries — namely, loss of diversity and domination by single taxa — are common events that occur with strikingly convergent kinetics worldwide. These microbiota disruptions are of interest because lower diversity at the time of neutrophil engraftment predicts poor overall survival, particularly among recipients of T-cell–replete grafts. The results of this international study extend previous observations that have been made in smaller, single-center cohorts.3,4,8,36
The association of lower microbiota diversity with poor survival was explained in part by higher transplantation-related mortality and was most prominently observed in recipients of T-cell–replete (unmodified) grafts, even though the diversity of intestinal microbiota declined similarly in recipients of T-cell–depleted grafts. Among recipients of unmodified grafts, higher diversity of the intestinal microbiome was associated with lower GVHD-related mortality, a result consistent with previous observations.4,9 This finding suggests a hypothesis that the correlation between microbiota injury and mortality requires early alloreactivity (e.g., that driven by mature T cells in the graft); however, it may also reflect other clinical variables that differ in these two patient groups, which include indications for transplantation, coexisting conditions, antimicrobial exposures, intensity of the conditioning regimen, and GVHD prophylaxis.
We found that by the day of cell infusion, many patients already had an intestinal community that differed markedly from those in healthy volunteers and that was characterized by loss of diversity and domination by single taxa. This finding is consistent with previous work that showed that the risk of bloodstream infections could be predicted by microbiota composition before hematopoietic-cell transplantation37 and that the risk of transplantation-related death could be predicted by the timing of exposure to antibiotics and by colonization with antibiotic-resistant bacteria before transplantation.16,38–40 Taken together, these results highlight two specific times relative to transplantation at which strategies to remediate or prevent microbiota injury could be evaluated in clinical trials — before transplantation and during the periengraftment period.
Our results extend beyond microbial diversity to provide multicenter evidence that microbiota compositions — specifically the relative abundances of bacterial taxa — may offer relevant information about outcomes of allogeneic hematopoietic-cell transplantation. Microbiota-based classification algorithms that distinguish cases from controls and that can be extrapolated across geographic regions have been described previously for inflammatory bowel disease and colorectal cancer.41,42 However, these studies did not consider clinical outcomes beyond diagnosis, and microbiome variation across geographic regions has limited other attempts to apply classifiers across populations.43 Despite the clinical heterogeneity of the participants in this study, we found that a microbiota-composition risk score that was trained in one cohort (cohort 1) could predict survival in an independent international cohort (cohort 2). It may be of interest to integrate microbiota classifiers into prospective trials of GVHD-predictive biomarkers.
Strengths of this study include its international design, longitudinal serial sampling, and the uniform tracking of standardized clinical outcomes by transplantation centers. Many larger-scale observational studies of the human microbiome have detailed the interactions between microbial communities and their human hosts,44 but few have examined an outcome as important as mortality. The central analysis of samples in this study was another important element because both wet-laboratory processing and computational pipelines are critical factors in the reproducibility and quality of microbiota data.45
The diversity of clinical practices across institutions and differences in underlying diseases, conditioning regimens, antibiotic exposures, and graft sources imposed considerable heterogeneity on the study population; nevertheless, we observed parallel patterns of microbiota injury and associations with clinical outcomes. Another limitation of this study is that samples were analyzed by targeted sequencing of genes encoding the 16S ribosomal RNA across all bacteria, which allows reliable genus-level annotation, or in some cases species information, and limits the scope of the analysis to bacteria. The alternative of metagenomic shotgun sequencing would expand the scope to viruses and fungi and allow the identification of encoded metabolic pathways but with the support of less well annotated reference databases. In addition, samples were not obtained at uniform time points relative to transplantation owing to the inherently unscheduled nature of obtaining stool samples. A key limitation of an observational study such as this is that it can show only correlations and not causative relationships. However, our findings are consistent with preclinical models of allogeneic transplantation and GVHD, which have provided mechanisms by which microbial communities can modulate alloreactivity.10,17–20
This study defines opportunities for rational interventions to restore integrity to the intestinal microbiota, such as with fecal microbiota replacement or other strategies,21–24 which could also be evaluated in clinical settings beyond allogeneic hematopoietic-cell transplantation.7,46 The similarities we observed in patterns of microbiota injury and their associations with clinical outcomes highlight the important interactions that occur between microbial communities and their hosts. In patients undergoing allogeneic hematopoietic-cell transplantation, these associations were observed consistently across graft sources, conditioning regimens, and around the world, despite local variations in microbiota composition and clinical practice.
Supplementary Material
Acknowledgments
Supported by grants from the National Cancer Institute (R01-CA228308, to Dr. van den Brink; R01 CA203950-01, to Drs. Sung and Chao; R01CA203950-01, to Dr. Sung, Dr. Chao, Ms. Bush, and Ms. Bohannon; P30 CA008748 [Cancer Center Core Grant]; and P01-CA023766, to Dr. van den Brink), by grants from the National Heart, Lung, and Blood Institute (K08HL143189-01A1, to Dr. Peled; and 1R01HL124112, to Drs. Peled, Sung, and Jenq), by grants from the National Institute of Allergy and Infectious Diseases (U01 AI124275, to Drs. Xavier and van den Brink; R01 AI032135 and AI095706, to Dr. Pamer; R01 AI137269-01, to Drs. Taur and Xavier; and 9R01AI100288, to Drs. Peled, Shono, and van den Brink), by a grant from the National Institutes of Health (KL2 TR001115-03 [National Center for Advancing Translational Sciences Clinical and Translational Science Award], to Dr. Sung), by a grant from the Claude D. Pepper Older Americans Independence Center of the National Institute on Aging (2P30AG028716-11, to Dr. Sung), by the Lymphoma Foundation (to Dr. Khan), by the Parker Institute for Cancer Immunotherapy at MSK (to Drs. Peled and van den Brink), by the Sawiris Foundation (to Dr. Peled), by the Society of MSK (to Dr. Peled), by the Empire Clinical Research Investigator Program (to Dr. Peled), by Seres Therapeutics (to Dr. Peled, Dr. Gomes, Mr. Slingerland, Ms. Clurman and Dr. van den Brink, and to Dr. Sung and Ms. Slingerland), by grants from the Japan Society for the Promotion of Science Grants-in-Aid for Scientific Research (KAKENHI) (17H04206, to Dr. Teshima; and 17K09945 to Dr. Hashimoto), by the Center of Innovation Program from Japan Science and Technology Agency (to Dr. Teshima), by grants from the Mochida Memorial Foundation for Medical and Pharmaceutical Research (to Dr. Hashimoto), by the Conquer Cancer Foundation Young Investigator Award and Gilead Sciences (to Dr. Khan), by the Alfonso Martin Escudero Foundation (to Dr. Castillo Flores), by grants from MSK Cycle for Survival (to Dr. Peled), and by funding from the Collaborative Research Center Transregio 221 (to Drs. Weber, Gessner, and Holler).
APPENDIX
The authors’ full names and academic degrees are as follows: Jonathan U. Peled, M.D., Ph.D., Antonio L.C. Gomes, Ph.D., Sean M. Devlin, Ph.D., Eric R. Littmann, B.A., Ying Taur, M.D., Anthony D. Sung, M.D., Daniela Weber, M.D., Daigo Hashimoto, M.D., Ph.D., Ann E. Slingerland, B.S., John B. Slingerland, B.S., Molly Maloy, M.S., Annelie G. Clurman, B.A., Christoph K. Stein-Thoeringer, M.D., Kate A. Markey, M.B., B.S., Ph.D., Melissa D. Docampo, B.S., Marina Burgos da Silva, Ph.D., Niloufer Khan, M.D., André Gessner, M.D., Ph.D., Julia A. Messina, M.D., Kristi Romero, B.S., Meagan V. Lew, B.S., Amy Bush, B.A., Lauren Bohannon, B.S., Daniel G. Brereton, B.A., Emily Fontana, B.A., Luigi A. Amoretti, B.S., Roberta J. Wright, M.S., M.B.S., Gabriel K. Armijo, B.S., Yusuke Shono, M.D., Ph.D., Míriam Sanchez-Escamilla, M.D., Nerea Castillo Flores, M.D., Ph.D., Ana Alarcon Tomas, M.D., Richard J. Lin, M.D., Ph.D., Lucrecia Yáñez San Segundo, M.D., Ph.D., Gunjan L. Shah, M.D., Christina Cho, M.D., Michael Scordo, M.D., Ioannis Politikos, M.D., Kasumi Hayasaka, Yuta Hasegawa, M.D., Boglarka Gyurkocza, M.D., Doris M. Ponce, M.D., Juliet N. Barker, M.B., B.S., Miguel-Angel Perales, M.D., Sergio A. Giralt, M.D., Robert R. Jenq, M.D., Takanori Teshima, M.D., Ph.D., Nelson J. Chao, M.D., Ernst Holler, M.D., Joao B. Xavier, Ph.D., Eric G. Pamer, M.D., and Marcel R.M. van den Brink, M.D., Ph.D.
Footnotes
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
Contributor Information
J.U. Peled, Adult Bone Marrow Transplantation Service, Memorial Sloan Kettering Cancer Center, New York Department of Medicine, Weill Cornell Medical College, New York.
A.L.C. Gomes, Department of Immunology, Sloan Kettering Institute, Memorial Sloan Kettering Cancer Center, New York
S.M. Devlin, Department of Medicine, the Department of Epidemiology and Biostatistics, New York
E.R. Littmann, Department of Immunology, Sloan Kettering Institute, Memorial Sloan Kettering Cancer Center, New York Duchossois Family Institute of the University of Chicago, Chicago.
Y. Taur, Infectious Disease Service, Memorial Sloan Kettering Cancer Center, New York Department of Medicine, Weill Cornell Medical College, New York.
A.D. Sung, Division of Hematologic Malignancies and Cellular Therapy, Department of Medicine, Duke University Medical Center, Durham, NC
D. Weber, Department of Hematology and Oncology, Internal Medicine III, University Medical Center, Regensburg, Germany Collaborative Research Center Transregio 221, Regensburg, Germany.
D. Hashimoto, Department of Hematology, Hokkaido University Faculty of Medicine, Sapporo, Japan
A.E. Slingerland, Department of Immunology, Sloan Kettering Institute, Memorial Sloan Kettering Cancer Center, New York
J.B. Slingerland, Department of Immunology, Sloan Kettering Institute, Memorial Sloan Kettering Cancer Center, New York
M. Maloy, Adult Bone Marrow Transplantation Service, Memorial Sloan Kettering Cancer Center, New York
A.G. Clurman, Adult Bone Marrow Transplantation Service, Memorial Sloan Kettering Cancer Center, New York
C.K. Stein-Thoeringer, Department of Immunology, Sloan Kettering Institute, Memorial Sloan Kettering Cancer Center, New York
K.A. Markey, Adult Bone Marrow Transplantation Service, Memorial Sloan Kettering Cancer Center, New York Department of Medicine, Weill Cornell Medical College, New York.
M.D. Docampo, Department of Immunology, Sloan Kettering Institute, Memorial Sloan Kettering Cancer Center, New York Department of Medicine, Weill Cornell Medical College, New York.
M. Burgos da Silva, Department of Immunology, Sloan Kettering Institute, Memorial Sloan Kettering Cancer Center, New York
N. Khan, Adult Bone Marrow Transplantation Service, Memorial Sloan Kettering Cancer Center, New York
A. Gessner, Institute of Clinical Microbiology and Hygiene, University Hospital Regensburg, Regensburg, Germany Collaborative Research Center Transregio 221, Regensburg, Germany.
J.A. Messina, Division of Infectious Diseases, Department of Medicine, Duke University, Durham, NC
K. Romero, Duke Office of Clinical Research, Duke University School of Medicine, Durham, NC
M.V. Lew, Division of Hematologic Malignancies and Cellular Therapy, Department of Medicine, Duke University Medical Center, Durham, NC
A. Bush, Division of Hematologic Malignancies and Cellular Therapy, Department of Medicine, Duke University Medical Center, Durham, NC
L. Bohannon, Division of Hematologic Malignancies and Cellular Therapy, Department of Medicine, Duke University Medical Center, Durham, NC
D.G. Brereton, Adult Bone Marrow Transplantation Service, Memorial Sloan Kettering Cancer Center, New York
E. Fontana, Infectious Disease Service, Memorial Sloan Kettering Cancer Center, New York
L.A. Amoretti, Infectious Disease Service, Memorial Sloan Kettering Cancer Center, New York
R.J. Wright, Infectious Disease Service, Memorial Sloan Kettering Cancer Center, New York
G.K. Armijo, Department of Immunology, Sloan Kettering Institute, Memorial Sloan Kettering Cancer Center, New York
Y. Shono, Department of Immunology, Sloan Kettering Institute, Memorial Sloan Kettering Cancer Center, New York
M. Sanchez-Escamilla, Adult Bone Marrow Transplantation Service, Memorial Sloan Kettering Cancer Center, New York Research Institute Marqués de Valdecilla-IDIVAL, Spain.
N. Castillo Flores, Adult Bone Marrow Transplantation Service, Memorial Sloan Kettering Cancer Center, New York
A. Alarcon Tomas, Adult Bone Marrow Transplantation Service, Memorial Sloan Kettering Cancer Center, New York Santander, and Hospital Universitario Puerta de Hierro, Madrid, Spain.
R.J. Lin, Adult Bone Marrow Transplantation Service, Memorial Sloan Kettering Cancer Center, New York Department of Medicine, Weill Cornell Medical College, New York.
L. Yáñez San Segundo, Adult Bone Marrow Transplantation Service, Memorial Sloan Kettering Cancer Center, New York Department of Hematology, Hospital Universitario Marqués de Valdecilla–IDIVAL, University of Cantabria, Spain.
G.L. Shah, Adult Bone Marrow Transplantation Service, Memorial Sloan Kettering Cancer Center, New York Department of Medicine, Weill Cornell Medical College, New York.
C. Cho, Adult Bone Marrow Transplantation Service, Memorial Sloan Kettering Cancer Center, New York Department of Medicine, Weill Cornell Medical College, New York.
M. Scordo, Adult Bone Marrow Transplantation Service, Memorial Sloan Kettering Cancer Center, New York Department of Medicine, Weill Cornell Medical College, New York.
I. Politikos, Adult Bone Marrow Transplantation Service, Memorial Sloan Kettering Cancer Center, New York Department of Medicine, Weill Cornell Medical College, New York.
K. Hayasaka, Division of Laboratory and Transfusion Medicine, Hokkaido University Hospital, Sapporo, Japan
Y. Hasegawa, Department of Hematology, Hokkaido University Faculty of Medicine, Sapporo, Japan
B. Gyurkocza, Adult Bone Marrow Transplantation Service, Memorial Sloan Kettering Cancer Center, New York Department of Medicine, Weill Cornell Medical College, New York.
D.M. Ponce, Adult Bone Marrow Transplantation Service, Memorial Sloan Kettering Cancer Center, New York Department of Medicine, Weill Cornell Medical College, New York.
J.N. Barker, Adult Bone Marrow Transplantation Service, Memorial Sloan Kettering Cancer Center, New York Department of Medicine, Weill Cornell Medical College, New York.
M.-A. Perales, Adult Bone Marrow Transplantation Service, Memorial Sloan Kettering Cancer Center, New York Department of Medicine, Weill Cornell Medical College, New York.
S.A. Giralt, Adult Bone Marrow Transplantation Service, Memorial Sloan Kettering Cancer Center, New York Department of Medicine, Weill Cornell Medical College, New York.
R.R. Jenq, Departments of Genomic Medicine and Stem Cell Transplantation Cellular Therapy, Division of Cancer Medicine, University of Texas M.D. Anderson Cancer Center, Houston
T. Teshima, Division of Laboratory and Transfusion Medicine, Hokkaido University Hospital, Sapporo, Japan Department of Hematology, Hokkaido University Faculty of Medicine, Sapporo, Japan.
N.J. Chao, Division of Hematologic Malignancies and Cellular Therapy, Department of Medicine, Duke University Medical Center, Durham, NC
E. Holler, Department of Hematology and Oncology, Internal Medicine III, University Medical Center, Regensburg, Germany Collaborative Research Center Transregio 221, Regensburg, Germany.
J.B. Xavier, Program for Computational and Systems Biology, Memorial Sloan Kettering Cancer Center, New York
E.G. Pamer, Infectious Disease Service, Memorial Sloan Kettering Cancer Center, New York Duchossois Family Institute of the University of Chicago, Chicago.
M.R.M. van den Brink, Adult Bone Marrow Transplantation Service, Memorial Sloan Kettering Cancer Center, New York Department of Immunology, Sloan Kettering Institute, Memorial Sloan Kettering Cancer Center, New York; Department of Medicine, Weill Cornell Medical College, New York.
REFERENCES
- 1.Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell 2014;157:121–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holler E, Butzhammer P, Schmid K, et al. Metagenomic analysis of the stool microbiome in patients receiving allogeneic stem cell transplantation: loss of diversity is associated with use of systemic antibiotics and more pronounced in gastrointestinal graft-versus-host disease. Biol Blood Marrow Transplant 2014;20:640–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taur Y, Xavier JB, Lipuma L, et al. Intestinal domination and the risk of bacteremia in patients undergoing allogeneic hematopoietic stem cell transplantation. Clin Infect Dis 2012;55:905–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Golob JL, Pergam SA, Srinivasan S, et al. Stool microbiota at neutrophil recovery is predictive for severe acute graft vs host disease after hematopoietic cell transplantation. Clin Infect Dis 2017;65: 1984–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stoma I, Littmann ER, Peled JU, et al. Compositional flux within the intestinal microbiota and risk for bloodstream infection with gram-negative bacteria. Clin Infect Dis 2020. January 24 (Epub ahead of print). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gevers D, Kugathasan S, Denson LA, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe 2014;15:382–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gopalakrishnan V, Spencer CN, Nezi L, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 2018;359:97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taur Y, Jenq RR, Perales MA, et al. The effects of intestinal tract bacterial diversity on mortality following allogeneic hematopoietic stem cell transplantation. Blood 2014;124:1174–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jenq RR, Taur Y, Devlin SM, et al. Intestinal blautia is associated with reduced death from graft-versus-host disease. Biol Blood Marrow Transplant 2015;21:1373–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shono Y, Docampo MD, Peled JU, et al. Increased GVHD-related mortality with broad-spectrum antibiotic use after allogeneic hematopoietic stem cell transplantation in human patients and mice. Sci Transl Med 2016;8:339ra71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hidaka D, Hayase E, Shiratori S, et al. The association between the incidence of intestinal graft-vs-host disease and antibiotic use after allogeneic hematopoietic stem cell transplantation. Clin Transplant 2018;32(9):e13361. [DOI] [PubMed] [Google Scholar]
- 12.Peled JU, Devlin SM, Staffas A, et al. Intestinal microbiota and relapse after hematopoietic-cell transplantation. J Clin Oncol 2017;35:1650–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ubeda C, Taur Y, Jenq RR, et al. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J Clin Invest 2010;120:4332–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haak BW, Littmann ER, Chaubard JL, et al. Impact of gut colonization with butyrate-producing microbiota on respiratory viral infection following allo-HCT. Blood 2018;131:2978–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harris B, Morjaria SM, Littmann ER, et al. Gut microbiota predict pulmonary infiltrates after allogeneic hematopoietic cell transplantation. Am J Respir Crit Care Med 2016;194:450–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weber D, Jenq RR, Peled JU, et al. Microbiota disruption induced by early use of broad-spectrum antibiotics is an independent risk factor of outcome after allogeneic stem cell transplantation. Biol Blood Marrow Transplant 2017;23:845–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Bekkum DW, Roodenburg J, Heidt PJ, van der Waaij D. Mitigation of secondary disease of allogeneic mouse radiation chimeras by modification of the intestinal microflora. J Natl Cancer Inst 1974;52: 401–4. [DOI] [PubMed] [Google Scholar]
- 18.Mathewson ND, Jenq R, Mathew AV, et al. Gut microbiome-derived metabolites modulate intestinal epithelial cell damage and mitigate graft-versus-host disease. Nat Immunol 2016;17:505–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jenq RR, Ubeda C, Taur Y, et al. Regulation of intestinal inflammation by microbiota following allogeneic bone marrow transplantation. J Exp Med 2012;209: 903–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stein-Thoeringer CK, Nichols KB, Lazrak A, et al. Lactose drives Enterococcus expansion to promote graft-versus-host disease. Science 2019;366:1143–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taur Y, Coyte K, Schluter J, et al. Reconstitution of the gut microbiota of antibiotic-treated patients by autologous fecal microbiota transplant. Sci Transl Med 2018;10(460):eaap9489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peled JU, Jenq RR, Holler E, van den Brink MR. Role of gut flora after bone marrow transplantation. Nat Microbiol 2016;1:16036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DeFilipp Z, Peled JU, Li S, et al. Third-party fecal microbiota transplantation following allo-HCT reconstitutes microbiome diversity. Blood Adv 2018;2:745–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kakihana K, Fujioka Y, Suda W, et al. Fecal microbiota transplantation for patients with steroid-resistant acute graft-versus-host disease of the gut. Blood 2016; 128:2083–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.David LA, Maurice CF, Carmody RN, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014; 505:559–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rashidi A, Wangjam T, Bhatt AS, Weisdorf DJ, Holtan SG. Antibiotic practice patterns in hematopoietic cell transplantation: a survey of blood and marrow transplant clinical trials network centers. Am J Hematol 2018;93:E348–E350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peric Z, Botti S, Stringer J, et al. Variability of nutritional practices in peritransplant period after allogeneic hematopoietic stem cell transplantation: a survey by the Complications and Quality of Life Working Party of the EBMT. Bone Marrow Transplant 2018;53:1030–7. [DOI] [PubMed] [Google Scholar]
- 28.Yatsunenko T, Rey FE, Manary MJ, et al. Human gut microbiome viewed across age and geography. Nature 2012; 486:222–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vatanen T, Kostic AD, d’Hennezel E, et al. Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell 2016;165:842–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sorror ML, Maris MB, Storb R, et al. Hematopoietic cell transplantation (HCT)-specific comorbidity index: a new tool for risk assessment before allogeneic HCT. Blood 2005;106:2912–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc B 1995;57:289–300. [Google Scholar]
- 32.Weber D, Hiergeist A, Weber M, et al. Detrimental effect of broad-spectrum antibiotics on intestinal microbiome diversity in patients after allogeneic stem cell transplantation: lack of commensal sparing antibiotics. Clin Infect Dis 2019; 68:1303–10. [DOI] [PubMed] [Google Scholar]
- 33.Legendre P, Legendre L. Numerical ecology. 3rd ed. Amsterdam: Elsevier, 2012. [Google Scholar]
- 34.Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012;486:207–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Costea PI, Hildebrand F, Arumugam M, et al. Enterotypes in the landscape of gut microbial community composition. Nat Microbiol 2018;3:8–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weber D, Oefner PJ, Hiergeist A, et al. Low urinary indoxyl sulfate levels early after transplantation reflect a disrupted microbiome and are associated with poor outcome. Blood 2015;126:1723–8. [DOI] [PubMed] [Google Scholar]
- 37.Montassier E, Al-Ghalith GA, Ward T, et al. Pretreatment gut microbiome predicts chemotherapy-related bloodstream infection. Genome Med 2016;8:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zirakzadeh A, Gastineau DA, Mandrekar JN, Burke JP, Johnston PB, Patel R. Vancomycin-resistant enterococcal colonization appears associated with increased mortality among allogeneic hematopoietic stem cell transplant recipients. Bone Marrow Transplant 2008;41:385–92. [DOI] [PubMed] [Google Scholar]
- 39.Bilinski J, Robak K, Peric Z, et al. Impact of gut colonization by antibiotic-resistant bacteria on the outcomes of allogeneic hematopoietic stem cell transplantation: a retrospective, single-center study. Biol Blood Marrow Transplant 2016; 22:1087–93. [DOI] [PubMed] [Google Scholar]
- 40.Rashidi A, Ebadi M, Shields-Cutler RR, et al. Pretransplant gut colonization with intrinsically vancomycin-resistant enterococci (E. gallinarum and E. casseliflavus) and outcomes of allogeneic hematopoietic cell transplantation. Biol Blood Marrow Transplant 2018;24:1260–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou Y, Xu ZZ, He Y, et al. Gut microbiota offers universal biomarkers across ethnicity in inflammatory bowel disease diagnosis and infliximab response prediction. mSystems 2018;3(1):e00188–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu J, Feng Q, Wong SH, et al. Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer. Gut 2017; 66:70–8. [DOI] [PubMed] [Google Scholar]
- 43.He Y, Wu W, Zheng HM, et al. Regional variation limits applications of healthy gut microbiome reference ranges and disease models. Nat Med 2018;24:1532–5. [DOI] [PubMed] [Google Scholar]
- 44.Lynch SV, Pedersen O. The human intestinal microbiome in health and disease. N Engl J Med 2016;375:2369–79. [DOI] [PubMed] [Google Scholar]
- 45.Sinha R, Abu-Ali G, Vogtmann E, et al. Assessment of variation in microbial community amplicon sequencing by the Microbiome Quality Control (MBQC) project consortium. Nat Biotechnol 2017;35: 1077–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tang WH, Wang Z, Levison BS, et al. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N Engl J Med 2013;368:1575–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.