Abstract
Cancer metastasis accounts for the major cause of cancer-related deaths. How disseminated cancer cells cope with hostile microenvironments in secondary site for full-blown metastasis is largely unknown. Here, we show that AMPK (AMP-activated protein kinase), activated in mouse metastasis models, drives pyruvate dehydrogenase complex (PDHc) activation to maintain TCA cycle (tricarboxylic acid cycle) and promotes cancer metastasis by adapting cancer cells to metabolic and oxidative stresses. This AMPK-PDHc axis is activated in advanced breast cancer and predicts poor metastasis-free survival. Mechanistically, AMPK localizes in the mitochondrial matrix and phosphorylates the catalytic alpha subunit of PDHc (PDHA) on two residues S295 and S314, which activates the enzymatic activity of PDHc and alleviates an inhibitory phosphorylation by PDHKs, respectively. Importantly, these phosphorylation events mediate PDHc function in cancer metastasis. Our study reveals that AMPK-mediated PDHA phosphorylation drives PDHc activation and TCA cycle to empower cancer cells adaptation to metastatic microenvironments for metastasis.
Keywords: AMPK, PDHA, TCA cycle, cancer metastasis, metabolic stress, breast cancer
Graphical Abstract

Highlights
-
•
AMPKα facilitates TCA cycle by maintaining PDH complex activity
-
•
AMPKα phosphorylates PDHA subunit on Ser295 and Ser314 to activate PDH complex
-
•
Activation of AMPKα-PDHA axis promotes tumor lung metastasis
-
•
AMPKα-PDHA axis predicts poor metastasis-free survival in breast cancer patients
Cai et al. demonstrate that phosphorylation of the catalytic subunit PDHA on Ser295 and S314 by AMPKα is essential for the maintenance of pyruvate dehydrogenase complex activity and TCA cycle. Activation of AMPKα-PDHA axis predicts poor metastasis-free survival in breast cancer patients and facilitates tumor lung metastasis.
Introduction
Cancer-related death is largely attributed to metastasis, which involves multiple steps of biological processes (Steeg, 2016; Wirtz et al., 2011). Although metastasis is life threatening, it is extremely challenging for disseminated cancer cells to successfully seed metastases from their primary site (Chambers et al., 2002). During this risky process, cancer cells encounter tremendous crisis, including anoikis and shear stress-induced apoptosis in systemic circulation (Douma et al., 2004; Reymond et al., 2013), hyper-oxidative stress when colonizing at the distant organ (Piskounova et al., 2015), as well as metabolic stress when exposing to new inhospitable environments (Kim et al., 2017; Senft and Ronai, 2016b). Only less than 0.02% of metastatic cancer cells, once getting into the circulation, could survive in the secondary site (Chambers et al., 2002; Luzzi et al., 1998). It has been proposed that metabolic reprogramming confers metastatic cancer cells to adapt to hostile environments with metabolic stress (Celià-Terrassa and Kang, 2016) and oxidative stress (Wu et al., 2014).
The role of cancer metabolism in cancer progression and metastasis starts to emerge. Accumulating evidence reveals that cancer cells display distinct metabolic states with respect to their normal counterparts. They often utilize aerobic glycolysis known as Warburg effect to generate energy, building blocks, and NADPH to maintain redox balance for their survival and proliferation. Such metabolic reprogramming, which is believed to play important roles in cancer progression and metastasis, could be partly due to the defect in mitochondria TCA cycle and/or mitochondria functions resulting from the mutations of TCA cycle enzymes (Mullen and DeBerardinis, 2012). Therefore, it is thought that defect in mitochondria activity may be a common event associated with Warburg effect and cancer progression. However, such concept has been challenged and may need to be revisited, because recent studies demonstrated that mitochondria activity is fully functional in cancers on the basis of 13C-labeled metabolomic analysis (Corbet and Feron, 2017) and that PGC1α (peroxisome proliferator-activated receptor gamma coactivator 1-alpha), which could induce mitochondrial biogenesis and oxidative phosphorylation, promotes metastasis (LeBleu et al., 2014). Mitochondria TCA cycle also generates energy and building blocks for cells to maintain their survival, but hyperactivation of TCA cycle was previously considered to produce excess reaction oxygen species (ROS) that is otherwise toxic to cells; however, more recent reports also suggest the essential role of certain TCA intermediates, such as oxaloacetate (OAA) and α-ketoglutarate (α-KG) in detoxification of ROS (Sawa et al., 2017). It is currently unclear about what exact roles the mitochondria TCA cycle plays in cancer progression and metastasis.
AMPK is a classical energy sensor activated under diverse stresses, such as metabolic and oxidative stresses (Hardie et al., 2012). It activates ATP-producing pathways but limits ATP-consuming pathways in response to metabolic stress. As such, its function is tightly regulated, and the defect in its function is associated with aging process and multiple metabolic disorders (Burkewitz et al., 2014; Ruderman et al., 2013). AMPK is thought to serve as a tumor suppressor because of its role in suppressing oncogenic mTORC1 activation (Shackelford and Shaw, 2009), and the fact that numerous anti-cancer agents, such as metformin (Sośnicki et al., 2016), phenformin (Appleyard et al., 2012), and resveratrol (Puissant et al., 2010), could all activate AMPK. However, some studies revealed that AMPK may promote tumorigenesis by maintaining redox balance and inducing Akt activation (Han et al., 2018; Jeon et al., 2012). Therefore, the role of AMPK in cancer regulation is controversial. Interestingly, AMPK deficiency renders cancer cells more vulnerable to stresses, including metabolic stress and cell detachment, which are critical barriers needed to be overcome during cancer metastasis, although the underlying mechanisms are not well understood (Ng et al., 2012; Saito et al., 2015; Svensson and Shaw, 2012).
Pyruvate dehydrogenase complex (PDHc), as a rate-limiting enzyme complex for maintaining TCA cycle, catalyzes pyruvate to acetyl-coenzyme A (Ac-CoA) and links glycolysis to oxidative phosphorylation (Sun et al., 2015). This trimeric complex consists of rate-limiting E1 (pyruvate dehydrogenase, composed of catalytic α [PDHA] and regulatory β [PDHB] subunits), E2 (dihydrolipoyl transacetylase [DLAT]), and E3 (dihydrolipoyl dehydrogenase [DLD]), and the integrity of this complex is critical for PDHc activity (Zhou et al., 2001). How PDHc activity is maintained is currently not well understood. It was previously shown that PDHA S293 phosphorylation serves as a negative signal for PDHc activation, and the removal of PDHA S293 phosphorylation is necessary for PDHc activation (Patel et al., 2014). However, the molecular basis of how PDHA S293 phosphorylation serves as a barrier for PDHc activation is unclear. Moreover, little is known about what physiological cues may prevent and/or antagonize PDHA S293 phosphorylation.
Although dysregulation of PDHc leads to multiple metabolic disorders and neurodegeneration (Brown, 2012), its role in cancer progression and cancer metastasis is still elusive. Previous study reported the PDHK1, which phosphorylates PDHA and suppresses PDHc activity upon hypoxia, displays oncogenic activity in certain cancer contexts, such as lung cancer (Hitosugi et al., 2011), implying tumor suppressor function of PDHc. However, more recent publication indicated that depletion of PDHc E1 β subunit (PDHB) resembles Warburg effect with impaired PDHc activity but compromises breast cancer growth (Yonashiro et al., 2018). Therefore, the role of PDHc in cancer regulation appears to be controversial. In this study, we aimed to dissect the role of PDHA, the critical rate-limiting catalytic subunit of PDHc, in cancer metastasis and its underlying mechanisms.
Results
AMPKα1 Critically Regulates Cancer Cell Survival under Stress to Maintain Metastasis
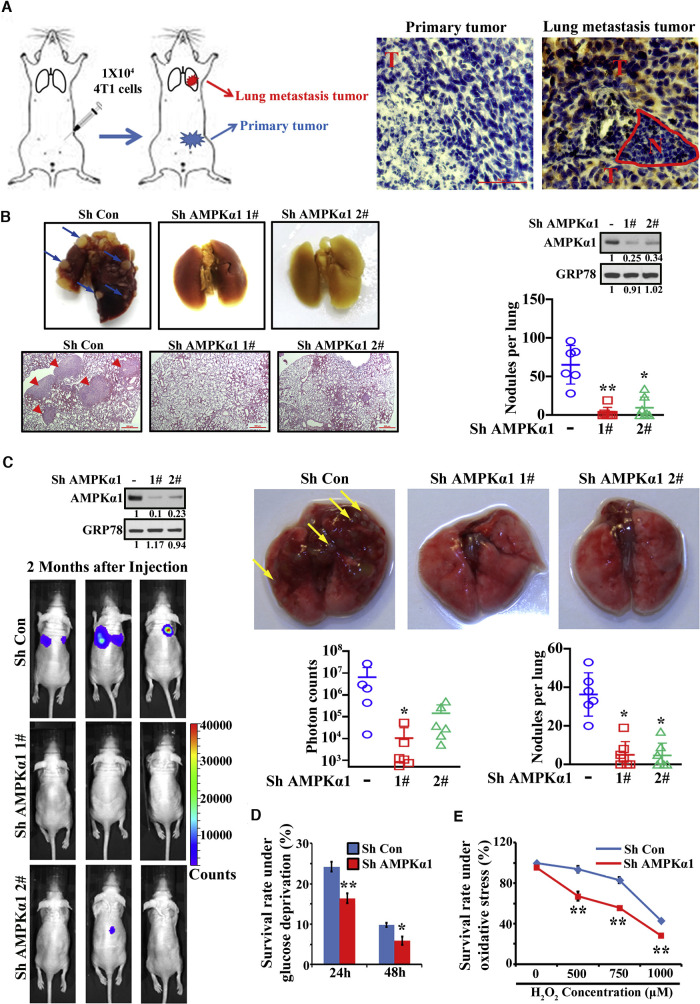
The TCA cycle maintenance is affected by mitochondria homeostasis, which is dynamically regulated by mitochondria biogenesis and mitophagy (Palikaras et al., 2015; Senft and Ronai, 2016a). Interestingly, mitochondria biogenesis and mitophagy are known to be regulated by AMPK (Marin et al., 2017; Pei et al., 2018). AMPK is widely believed as a tumor suppressor for its role in repressing mTOR activation (Shackelford and Shaw, 2009), although oncogenic activity of AMPK in certain tumor contexts is also reported (Han et al., 2018; Jeon et al., 2012). Surprisingly, overexpression of AMPK was associated with shorter metastasis-free survival in breast cancer cohort (Figure S1A). Because AMPK regulates cancer cell survival under metabolic stresses (Saito et al., 2015; Svensson and Shaw, 2012), which are critical barriers needed to be overcome during cancer metastasis, we hypothesized that AMPK may be a critical player for cancer metastasis by empowering cancer cells to adapt to diverse stresses in the metastatic microenvironments. To address this question, we generated 4T1 orthotopic model by inoculating 4T1 breast cancer cells into mammary fat pads that develop spontaneous metastasis to lung and harvested both primary tumor and metastatic tumor tissues for immunohistochemistry (IHC) staining (Figure 1 A). Notably, by staining p-AMPK (T172), AMPK activity was higher in lung metastatic tumor than primary tumor (Figure 1A), indicating that AMPK activation may be essential for metastatic adaptation of metastatic processes. In support of this notion, impairment of AMPK activation by AMPKα1 depletion (Figure S1B) abolished cancer lung metastasis using 4T1 orthotopic metastasis models (Figure 1B). However, the primary tumor growth was not significantly affected upon AMPKα1 deficiency in 4T1 cells (Figure S1C).
Figure 1.
AMPKα1 Is Essential for Tumor Metastasis
(A) AMPK activity by immunostaining of p-AMPKα(T172) in primary tumor and lung metastatic tumor was shown. N, normal; T, tumor. Scale bars indicate 50 μm.
(B) Orthotopic metastasis model from AMPKα1 knockdown 4T1 cells was performed. Representative lung tissues, H&E images, and average number of nodules per lung were shown. Arrows indicate metastatic nodules. Scale bars indicate 500 μm.
(C) Tail vein injection metastasis model from AMPKα1 knockdown 231 cells was performed. Representative bioluminescence images, lung tissues, and average photon counts and number of nodules per lung were shown. Arrows indicate metastatic nodules.
(D) Survival rate of control and AMPKα1 knockdown cells upon glucose deprivation was determined.
(E) Survival rate of control and AMPKα1 knockdown cells treated with H2O2 was determined.
Data are means ± SEM from 5–7 mice for metastasis assays and means ± SD from 3 independent experiments for other assays. ∗p < 0.05 and ∗∗p < 0.01. See also Figure S1.
By using tail vein injection metastasis model, AMPKα1 depletion also impaired lung metastasis in multiple cell models, including 4T1, 231, and Hep3B (Figures 1C, S1D, and S1E). Because AMPK activity is elevated in metastatic tissues where metastatic cancer cells frequently encounter metabolic stresses, we determined whether the impaired metastasis upon AMPK deficiency resulted from the failure of cancer cells to adapt to stressful conditions during the colonization process. Indeed, AMPKα1 knockdown rendered cancer cells more vulnerable to metabolic stress (Figure 1D) and enhanced apoptosis upon glucose deprivation (Figures S1F and S1G). Oxidative stress is another critical barrier preventing disseminated tumor cells from surviving in distant organ (Piskounova et al., 2015). AMPKα1 knockdown also conferred cancer cells more sensitive to oxidative stress (Figures 1E, S1H, and S1I). Thus, AMPKα1 is critical for cancer cell survival under diverse stresses, thereby protecting metastatic cells from stress-induced cell death and maintaining cancer metastasis.
TCA Cycle Is Crucially Maintained by AMPKα1
Although glycolysis via glucose utilization for the generation of energy and building blocks is considered as a major strategy for cancer cell proliferation and survival, this mechanism is likely impaired during cancer metastasis processes where metastatic cancer cells constantly experience metabolic stress and/or oxidative stress in the secondary site (Piskounova et al., 2015; Senft and Ronai, 2016b). It is conceivable that metastatic cancer cells will need to rewire their metabolic demands from glycolysis to TCA cycle for the production of energy and building blocks, thereby allowing metastatic cancer cells to survive in the distant site (Pascual et al., 2018).
Because activation of AMPK renders cancer cells to survive better under glucose deprivation, where TCA cycle may become a key strategy for cancer cell survival, we speculated that AMPK may be crucial for maintaining TCA cycle. Indeed, metabolomics study revealed that several key intermediates in TCA cycle, such as α-KG, fumarate, and oxaloacetate, were markedly decreased in AMPKα1 knockdown cells (Figure 2 A). Impaired α-KG level was also observed in multiple AMPKα1 knockdown cells and AMPKα1 −/− α2 −/− mouse embryonic fibroblasts (MEFs) (Figure 2B). Moreover, ATP level and oxygen consumption rates (OCRs) were declined in AMPKα1-deficient cells (Figures 2C and 2D). Thus, AMPKα1 is critical for TCA cycle maintenance.
Figure 2.
TCA Cycle Is Critically Regulated by AMPKα1
(A) Metabolomics analysis in control and AMPKα1 knockdown Hep3B cells was performed. Relative expression levels of three representative TCA intermediates were shown.
(B) Relative α-KG level in AMPKα1 knockdown cells and AMPKα1−/−α2−/− MEFs was shown.
(C) Relative ATP level in AMPKα1 knockdown cells and AMPKα1−/−α2−/− MEFs was shown.
(D) Basal and maximal oxygen consumption rate (OCR) was determined in control and AMPKα1 knockdown cells.
(E) The enrichment of isotope derived from 13C3-labeled pyruvate or 13C5-labeled glutamine in TCA intermediates and derivative (glutamate) from TCA cycle was determined by mass spectrometry analysis. Green circle: 13C-labeled carbon atom derived from 13C3-labeled pyruvate; yellow circle: 13C-labeled carbon atom derived from 13C3-labeled glutamine; blank circle: no isotope labeled carbon atom.
Data are means ± SD from 3 independent experiments. ∗p < 0.05 and ∗∗p < 0.01. See also Figure S2.
However, the expression level of TCA cycle enzymes and other enzymes regulating the connection between TCA cycle and other metabolic pathways were not affected upon AMPK deficiency (Figures S2A and S2B). We also ruled out the possibility that AMPK regulates TCA cycle acting through its known targets, such as PGC1α functioning in mitochondria biogenesis (Jäger et al., 2007) and ACC (acetyl-CoA carboxylase) involved in lipogenesis, a process squeezing acetyl-CoA needed for TCA cycle (Minokoshi et al., 2002), as depletion of PGC1α or ACC failed to compromise TCA cycle, as indicated by α-KG and ATP level (Figures S2C and S2D). We then speculated that AMPKα1 may maintain TCA cycle partly through orchestrating glucose uptake (Wu et al., 2013), because glucose is one of the major nutritional sources providing pyruvate for TCA cycle. AMPKα1 knockdown reduced glucose uptake and multiple metabolites from glycolysis, including pyruvate (Figures S2E and S2F). However, the level of lactate, as final product of glycolysis, remained unchanged (Figure S2G), indicating AMPK may favor pyruvate shunting through TCA cycle instead of converting it to lactate.
To address whether AMPK maintains TCA cycle through directly facilitating pyruvate metabolism, we performed 13C-labeled pyruvate- and glutamine-tracing experiments. Notably, the level of TCA cycle metabolites (citrate, fumarate, malate, and succinate) incorporated from 13C-labeled pyruvate was reduced in AMPKα1 knockdown cells (Figure 2E). However, the level of corresponding TCA metabolites derived from 13C-labeled glutamine was comparable between control and AMPKα1 knockdown cells (Figure 2E). Collectively, our data suggest AMPKα1 is an essential regulator for TCA cycle maintenance through controlling pyruvate metabolism.
PDHc Activation and TCA Cycle by AMPKα1 Are Required for Cancer Metastasis
Pyruvate is located at the crossroads of glycolysis and TCA cycle, and its metabolic fate is mainly controlled by lactate dehydrogenase (LDH) and PDHc. The former facilitates glycolysis by converting pyruvate to lactate, although the latter maintains TCA cycle by catalyzing pyruvate to Ac-CoA (Figure S3A). LDH activity remained unchanged upon AMPKα1 knockdown (Figure S3B), but PDHc activity was impaired in AMPKα1 −/− α2 −/− MEFs and AMPKα1 knockdown cells (Figure 3 A). As a consequence, the level of Ac-CoA, a direct product of PDHc from pyruvate, in both whole-cell lysates and mitochondria was markedly reduced in AMPKα1-deficient cells (Figure 3B). Thus, AMPKα1 is essential for maintaining PDHc activity.
Figure 3.
Disruption of PDHc Activity by PDHA Depletion Abolishes Tumor Metastasis
(A) PDHc activity (kinetic curve and quantification based on time-dependent absorption change) in control and AMPKα1 knockdown cells and AMPKα1−/−α2−/− MEFs was shown.
(B) Relative whole cell and mitochondria Ac-CoA level in control and AMPKα1 knockdown cells was shown.
(C) Orthotopic metastasis model from PDHA knockdown 4T1 cells was performed. Representative lung tissues, H&E images, and average number of nodules per lung were shown. Arrows indicate metastatic nodules.
(D) Tail vein injection metastasis model from PDHA knockdown MDA-MB-231 cells was performed. Representative lung tissues, H&E images, and average number of nodules per lung were shown. Arrows indicate metastatic nodules.
Data are means ± SEM from 5–7 mice for metastasis assays and means ± SD from 3 independent experiments for other assays. ∗p < 0.05 and ∗∗p < 0.01. Scale bars indicate 500 μm. See also Figure S3.
Similar to AMPKα1 depletion, knockdown of PDHA, the catalytic subunit of PDHc, specifically impaired cancer metastasis to lung, but not primary tumor growth, using 4T1 orthotopic metastasis model (Figures 3C and S3C) and 231 tail vein injection metastasis model (Figure 3D), underscoring the fundamental role of PDHc in tumor metastasis. Remarkably, introduction of constitutively active PDHA (PDHA S293A), which restores PDHc activity in AMPKα1-deficient cells, but not wild-type (WT) PDHA, fully rescued the defects in TCA cycle, indicated by ATP and α-KG (Figure 4 A), and tumor metastasis (Figures 4B, 4C, S3D, and S3E). Thus, AMPK acts through PDHc activation to facilitate cancer metastasis.
Figure 4.
Maintenance of PDHc Activity and TCA Cycle by AMPKα1 Regulates Cell Survival under Diverse Stresses and Tumor Metastasis
(A) Relative PDHc activity, ATP, and α-KG level in AMPKα1 knockdown 4T1 cells with restoration of WT or constitutively active PDHA (S293A) were shown.
(B) Orthotopic metastasis model from AMPKα1 knockdown 4T1 cells with restoration of WT or PDHA (S293A) was performed. Representative lung tissues, H&E images, and average number of nodules per lung were shown. Arrows indicate metastatic nodules.
(C) Tail vein injection metastasis model from AMPKα1 knockdown 231 cells with restoration of WT or PDHA (S293A) was performed. Representative bioluminescence images and average photon counts per lung were shown.
(D) Survival rate of control cells and PDHA knockdown cells supplied with vehicle, pyruvate (5 mM), or α-KG (10 mM) under glucose deprivation condition was shown.
(E) Survival rate of control cells, AMPKα1 knockdown cells, and AMPKα1 knockdown cells with restoration of PDHA (S293A) supplied with vehicle, pyruvate (5 mM), or α-KG (10 mM) under glucose deprivation was shown.
(F) Survival rate upon H2O2 treatment in AMPKα1 knockdown cells with restoration of PDHA (S293A) was shown.
Data are means ± SEM from 5–7 mice for metastasis assays and means ± SD from 3 independent experiments for other assays. ∗p < 0.05 and ∗∗p < 0.01. Scale bars indicate 500 μm. See also Figure S3.
Activation of PDHc by AMPKα1 Empowers Tumor Cells to Survive under Various Stresses
PDHA depletion also heightened the sensitivity of cancer cells to metabolic stress (Figure 4D), similar to AMPKα1 loss. Introducing active PDHA in AMPKα1 knockdown cells rescued cancer cell survival under metabolic and oxidative stresses (Figures 4E and 4F). Interestingly, add-back pyruvate or α-KG largely rescued cancer cell survival under metabolic stress in control cells, but not in AMPKα1 or PDHA knockdown cancer cells where TCA cycle is compromised (Figures 4D and 4E). Remarkably, introduction of active PDHA in AMPKα1 knockdown cells enabled pyruvate or α-KG to protect cancer cells from death under metabolic stress (Figure 4E). Collectively, TCA maintenance ensured by AMPK-mediated PDHc activation is essential for cancer cell survival under diverse stresses, thereby facilitating cancer metastasis.
AMPK-PDHc Axis Is Activated in Advanced Breast Cancer Patients and Predicts Poor Metastasis-free Survival
Although PDHc is a crucial regulator for pyruvate metabolism and TCA cycle, how it is activated to participate in TCA cycle remains largely unclear. Earlier studies revealed that PDHA S293 phosphorylation by PDHKs serves as a negative mechanism to limit PDHc activation (Kolobova et al., 2001). Interestingly, elevated PDHA S293 phosphorylation and enhanced interaction between PDHA and PDHK1 were observed in AMPKα1 knockdown cells (Figures 5A and 5C), accounting for the impairment of PDHc activity. Conversely, activation of AMPK upon either specific activator A-769662 or glucose deprivation resulted in dissociation between PDHA and PDHK1 and reduced PDHA S293 phosphorylation, which were reversed by AMPKα1 depletion or AMPK inhibitor compound C treatment (Figures 5B, 5D, and S4A–S4D). Impaired interaction between PDHA and other PDHKs (PDHK2 and PDHK3) was also observed upon AMPK activation by glucose deprivation or overexpression of constitutively active AMPKα1 (AMPK-CA) (Figure S4E).
Figure 5.
AMPK-PDHc Axis Is Activated in Advanced Breast Cancer Patients, and Its Activation Predicts Poor Metastasis-free Survival
(A) PDHA S293 phosphorylation was determined in AMPKα1 knockdown cells.
(B) PDHA S293 phosphorylation was determined in control and AMPKα1 knockdown cells with or without A-769662 (100 μM) treatment.
(C) The interaction between PDHA and PDHK1 was determined in control and AMPKα1 knockdown cells.
(D) The interaction between endogenous PDHA and PDHK1 was determined in control and AMPKα1 knockdown cells upon A-769662 treatment.
(E) Immunostaining of p-AMPK (T172) and p-PDHA (S293) in breast cancer tissues with different stages were shown.
(F) Scatterplot of pAMPK expression versus pPDHA expression in breast cancer tissues was shown.
(G) Kaplan-Meier plots showed high expression of pAMPK and low expression of pPDHA significantly predicted metastasis-free survival.
Scale bars indicate 100 μm. See also Figure S4 and Tables S1–S3.
By IHC staining, comparing with corresponding primary tumor, we found elevated activity of PDHc, as indicated by lower expression of p-PDHA (S293), in metastatic tumor, correlated with enhanced activity of AMPK, as determined by p-AMPK (T172; Figure S4F). However, both control and AMPKα1 knockdown primary 4T1 tumors displayed low AMPK and low PDHc activity. Of note, AMPKα1 knockdown in 4T1 cells, which impairs lung metastasis, remarkably reduced PDHc activity as indicated by enhanced p-PDHA level (Figure S4F), consistent with our in vitro results. To further support the pathophysiological link between AMPK and PDHc, we detected p-AMPK (T172) and p-PDHA (S293) in our in-house 184 breast cancer samples with different tumor stages and metastasis statuses by IHC staining. Notably, p-AMPK level was positively correlated with higher tumor stage, although p-PDHA level was negatively correlated with higher tumor stage (Figure S4G; Table S1). Moreover, the p-AMPK expression was negatively correlated with p-PDHA expression level (Figures 5E and 5F). Importantly, high p-AMPK or low p-PDHA level predicted worse metastasis-free survival (Figure 5G; Tables S2 and S3), highlighting the importance of AMPK-PDHc axis in breast cancer progression and metastasis.
PDHA Phosphorylation by AMPK Maintains PDHc Activity
To gain further insight into how AMPK regulates PDHA S293 phosphorylation and PDHc activation, we determined whether AMPK can be localized in the mitochondrial matrix together with PDHc. To address this question, we performed mitochondrial isolation and mitochondrial subfractionation as previously described (Nishimura and Yano, 2014). AMPKα could indeed localize in mitochondrial matrix with PDHA (Figures 6 A and S5A–S5C). Further, AMPKα was found to interact with PDHA, but not with PDHK1 (Figure S5D). Moreover, active AMPKα co-localized with both PDHA and Tomm20 in mitochondria, as indicated by immunofluorescence staining (Figures 6B and S5E). Thus, AMPK is localized in mitochondria matrix along with PDHc.
Figure 6.
PDHA Phosphorylation by AMPK Maintains PDHc Activity
(A) Mitochondrial isolation and sub-fractionation were performed, and AMPK and PDHA expression in mitochondrial matrix was determined. Tomm20 is marker for mitochondrial outer membrane, COX4 is marker for mitochondrial inner membrane, and CS is marker for mitochondrial matrix. Mito, mitochondria; WCL, whole cell lysis.
(B) p-AMPK (T172) and PDHA co-localization was determined by immunofluorescence in cells upon A-769662 treatment.
(C) Phosphorylation level on PDHA or Mff was determined by anti-phosphor-Ser/Thr antibody after in vitro kinase assay. L.E., long exposure; S.E., short exposure.
(D) In vitro γ-32P ATP incorporation was determined by incubating recombinant PDHA and active AMPK complex with γ-32P ATP.
(E) Phosphorylation level on PDHA was determined by anti-phosphor-Ser/Thr antibody after in vitro kinase assay (left). All the samples were subjected to SDS-PAGE, and PDHA bands were cut for mass spectrometry analysis. Phosphorylation statuses on each sample were shown (right). Relative phosphorylation intensity was indicated by the number of .
(F) Venn diagram was used to integrate phosphorylation site detected by mass spectrometry and phosphorylation site predicted by GPS 3.0 (group-based prediction system; http://gps.biocuckoo.org/).
(G) S295 and S314 phosphorylation on WT PDHA and mutant PDHA after in vitro kinase assay was determined by PDHA-specific phospho-antibodies.
(H) p-PDHA (S295) and p-PDHA (S314) were determined in control and AMPKα1 knockdown cells treated with glucose deprivation (left) or A-769662 (right).
(I) Relative PDHc activity in control and AMPKα1 knockdown cells with or without A-769662 treatment was shown.
(J) Relative PDHc activity was determined in purified PDHc with or without in vitro kinase reaction with active AMPK complex.
(K) Relative PDHc activity in AMPK knockdown cells with restoration of PDHA WT and mutants was shown.
(L) The interaction between PDHK1 and PDHA WT or PDHA mutants was determined.
(M) S293 phosphorylation levels of PDHA WT or PDHA mutants were determined.
(N) Relative level of pyruvate trapped in purified PDHc with PDHA WT, S295D, or S295A was shown.
(O) PDHc was purified from control, AMPK knockdown cells, and AMPK knockdown cells with PDHA S295D or S295A restoration. Relative pyruvate trapped in PDHc was shown.
Data are means ± SD from 3 independent experiments. ∗∗p < 0.01. Scale bars indicate 20 μm. See also Figures S5 and S6.
In light of these findings, we hypothesized that mitochondria-localized AMPK could directly induce PDHA phosphorylation, thereby affecting PDHc activity. In vitro kinase assay revealed that PDHA could be readily phosphorylated by active AMPK complex in a dose-dependent manner (Figure 6C). AMPK complex could induce phosphorylation of Mff, a well-known AMPK substrate (Toyama et al., 2016), but the overall phosphorylation of Mff was much lower than that of PDHA (Figure 6C). Additionally, γ-32P ATP incorporation kinase assays were conducted by incubating recombinant PDHA with different concentrations of active AMPK complex and γ-32P ATP to study the phosphorylation stoichiometry. The results indicate that AMPK gradually enhanced γ-32P ATP incorporation on PDHA in a time-dependent and dose-dependent manner. These results further demonstrate that AMPK serves as a direct kinase for PDHA (Figure 6D).
In vitro kinase assay followed by subsequent mass spectrometry analysis and combined bio-informatic phosphorylation site prediction indicated that AMPK induced PDHA phosphorylation at S295 and S314, which were highly conserved among various species (Figures 6E, 6F, and S5F–S5H). We then developed and characterized phospho-PDHA antibodies at S295 and S314 (Figures S5I and S5J). Using these phospho-PDHA antibodies, we validated that AMPK could specifically induce in vitro PDHA phosphorylation on S295 and S314 in a dose-dependent manner, which was abolished upon phospho-dead mutation on the corresponding site (Figures 6G, S5K, and S5L). AMPK activation by either AMPK activator A-769662 treatment or glucose deprivation induced in vivo PDHA phosphorylation on serine 295 and 314, which was compromised upon AMPK deficiency (Figure 6H), indicating that AMPK activation facilitates PHDA S295 and S314 phosphorylation under physiological conditions. By performing IHC staining in lung metastatic tumor tissues, enhanced p-AMPK level was positively correlated with elevated expression of p-PDHA (S295) and p-PDHA (S314) but inversely correlated with p-PDHA (S293) in control group, although impaired expression of p-PDHA (S295) and p-PDHA (S314) and enhanced expression of p-PDHA (S293) were observed in AMPKα1 knockdown group (Figure S6A).
In vivo S295 phosphorylation on PDHA was also validated by mass spectrometry analysis in cells either upon overexpression of constitutively active AMPK or glucose deprivation (Figures S6B and S6C). Combined treatment of AMPK inhibitor compound C could abolish glucose-deprivation-induced S295 phosphorylation on PDHA (Figure S6C), suggesting that PDHA S295 is one of the major phosphorylation sites in vivo upon AMPK activation. Consistent with IHC staining results, S293 phosphorylation and S295 phosphorylation inversely expressed (Figures S6B and S6C), indicative of the potential negative regulation between each other.
To investigate the role of AMPK-dependent phosphorylation on PDHA in PDHc activation, we activated AMPK by A-769662 treatment in both control and AMPKα1-deficient cells. A-769662 treatment enhanced activity of PDHc in control cells, but not in AMPKα1-deficient cells (Figure 6I). Moreover, we purified PDHc by pulling down PDHA and boosted PDHA phosphorylation by incubating it with active AMPK complex. Elevated PDHA phosphorylation after in vitro kinase reaction facilitated activation of PDHc (Figure 6J). Collectively, these results indicate that PDHA phosphorylation by active AMPK enhances the enzymatic activity of PDHc. Notably, we found PDHA S295D or S314D, but not WT PDHA, PDHA S295A, or S314A, could rescue the defect of PDHc activity in AMPKα1-deficient cells (Figure 6K), indicative of the crucial role of AMPK-mediated PDHA S295 and S314 phosphorylation in PDHc activation.
PDHA S314 Phosphorylation Prevents It from Binding to Its Negative Regulator, and PDHA S295 Serves as an Intrinsic Catalytic Site
To gain insight into how AMPK-mediated PDHA S295 and S314 phosphorylation maintains PDHc activity, we determined whether S295 and S314 phosphorylation affects PDHA-PDHKs interaction and PDHA S293 phosphorylation. Of note, PDHA S314D, but not WT PDHA or PDHA S314A, displayed impaired interaction with PDHK1 (Figure 6L), indicative of the suppressive role of PDHA S314 phosphorylation in PDHA and PDHK1 interaction. As a result, PDHA S293 phosphorylation was markedly reduced in PDHA S314D compared with WT PDHA and PDHA S314A (Figure 6M). Surprisingly, both PDHA S295D and PDHA S295A displayed attenuated interaction with PDHK1 and decreased S293 phosphorylation when comparing with WT PDHA (Figures 6L and 6M). We rationalize that this result is likely due to the steric effect of S295 on S293, because these two sites are physically close with each other. Indeed, PDHA S295C mutant also exhibited impaired S293 phosphorylation (Figure S6D), suggesting that any substitution of amino acid on S295 would destroy the docking site of PDHKs, leading to reduced S293 phosphorylation. Although both PDHA S295D and S295A mutants exhibit compromised S293 phosphorylation, only PDHA S295D, but not PDHA S295A, displays activated enzymatic activity of PDH complex, suggesting S295 phosphorylation is sufficient for determining the activity of PDHc. This result also suggests that, although loss of PDHA S293 phosphorylation elicits PDHc activation, the failure in S295 phosphorylation will drive PDHc inactivation, even when PDHA S293 phosphorylation is lost (e.g., PDHA S295A). Collectively, AMPK-mediated S314 phosphorylation, but not S295 phosphorylation, blocks the interaction of PDHA with PDHK1 and subsequent PDHA S293 phosphorylation.
We then assessed how PDHA S295 phosphorylation maintains PDHc activity. We rationalized that PDHA S295 phosphorylation may either regulate the integrity of the PDHc or serve as an intrinsic active site essential for PDHc activation. We ruled out the first possibility, as there was no difference between PDHA S295D and PDHA S295A in its interaction with other PDHc subunits (Figure S6E). To test the second possibility, we purified PDHc with WT PDHA, PDHA S295D, and PDHA S295A from cells and incubated them with C14-labeled pyruvate to determine the amounts of C14-labeled pyruvate trapped in PDHc. Because the active PDHc could quickly and efficiently catalyze C14-labeled pyruvate, it is not expected to observe C14-labeled pyruvate trapped on the active PDHc. Of note, elevated C14-labeled pyruvate was detected in PDHc with PDHA S295A but decreased C14-labeled pyruvate in PDHc with S295D PDHA (Figure 6N). Similarly, enhanced C14-labeled pyruvate trapped on the PDHc was also observed in AMPK-deficient cells, which could be rescued by re-introduction of PDHA S295D, but not of PDHA S295A (Figure 6O). Collectively, PDHA S295 phosphorylation is an intrinsic catalytic site critical for PDHc activation and pyruvate metabolism.
We determined whether PDHA S293 phosphorylation interferes with PDHA S295 phosphorylation and thus blocks intrinsic PDHc activity. Remarkably, S293 phospho-mimic mutant (PDHA S293D) abrogated PDHA S295 phosphorylation compared with the WT PDHA or S293 phospho-deficient mutant (PDHA S293A) (Figure S7A). Similarly, inhibition of PDHA S293 phosphorylation by DCA (dichloroacetate)-mediated PDHKs inactivation induced PDHA S295 phosphorylation (Figure S7B). PDHA S314A with enhanced S293 phosphorylation displayed compromised S295 phosphorylation, although PDHA S314D with impaired phosphorylation on S293 conversely exhibited increased S295 phosphorylation (Figure S7C). Although PDHA S314 phosphorylation is required for PDHA S295 phosphorylation, loss of PDHA S295 phosphorylation did not affect PDHA S314 phosphorylation (Figure S7D). Thus, PDHKs-mediated PDHA S293 phosphorylation serves as a barrier to prevent PDHA S295 phosphorylation, although AMPK-mediated PDHA S314 phosphorylation relieves the inhibitory effect of PDHA S293 phosphorylation on PDHA S295 phosphorylation, allowing for PDHc activation.
Phosphorylation of S295 and S314 by AMPK Is Essential for PDHA Function in TCA Cycle and Cancer Metastasis
We investigated whether PDHA S295 and S314 phosphorylation is essential for the maintenance of TCA cycle. Only PDHA S295D or S314D, but not WT PDHA, PDHA S295A, or S314A, could rescue the defects in TCA cycle, as indicated by restored α-KG level and ATP level (Figure 7 A), suggesting AMPK-mediated PDHA S295 and S314 phosphorylation maintains TCA cycle through activating PDHc.
Figure 7.
PDHA Phosphorylation by AMPK Is Essential for TCA Cycle and Cancer Metastasis
(A) Relative ATP and α-KG level in control, AMPKα1 knockdown alone, and AMPKα1 knockdown cells with restoration of WT PDHA or mutants were shown.
(B and C) Orthotopic metastasis model from PDHA knockdown 4T1 cells with WT PDHA or mutants knockin was performed (B). Orthotopic metastasis model from AMPKα1 knockdown 4T1 cells with restoration of WT PDHA or mutants was performed (C). Representative lung tissues, H&E staining, and average number of nodules per lung were shown. Arrows indicate metastatic nodules.
(D) Tail vein injection metastasis model from AMPKα1 knockdown 231 cells with restoration of WT PDHA or mutants was performed. Average photon counts per lung were shown.
(E) A schematic model is presented to dissect the role of AMPK-PDHc axis in cancer metastasis.
Data are means ± SEM from 5–7 mice for metastasis assays and means ± SD from 3 independent experiments for other assays. ∗p < 0.05 and ∗∗p < 0.01. Scale bars indicate 500 μm. See also Figure S7.
To directly investigate whether PDHA S295 and S314 phosphorylation is critical for PDHA-mediated metastasis, we knocked in human WT PDHA and its mutants resistant to mouse PDHA short hairpin RNA (shRNA) to PDHA knockdown 4T1 cells and found that the introduction of PDHA S295D and S314D, but not PDHA S295A and S314A, rescued impaired lung cancer metastasis upon PDHA knockdown, albeit their expression level was still lower than endogenous mouse PDHA level (Figure 7B). However, WT PDHA only partially rescued impaired lung metastasis upon PDHA depletion, likely due to partial restoration of WT PDHA expression in PDHA knockdown cells (Figure 7B). PDHA mutants knockin did not affect primary tumor growth (Figure S7E). Remarkably, the impairment of tumor metastasis in AMPKα1-deficient cells was also rescued by expressing PDHA S295D or PDHA S314D, but not WT PDHA, PDHA S295A, or PDHA S314A, in both 4T1 orthotopic metastasis model and 231 tail vein injection model (Figures 7C, 7D, and S7F), although the primary tumor was not affected by these treatment conditions (Figure S7G). Collectively, AMPK promotes PDHc activation, TCA cycle, and cancer cell metastasis through facilitating PDHA S295 and S314 phosphorylation.
Discussion
Cancer cells are considered to bypass the TCA cycle and primarily utilize glycolysis based on the early dogma; however, emerging evidence indicates that certain cancer cells depend heavily on the TCA cycle for energy production and macromolecule synthesis (Anderson et al., 2018). Thus, investigation of the exact function of TCA cycle and its regulation in various cellular contexts and different cancer progression stages may yield great insights into cancer metabolism and potential strategies for tackling advanced cancer. PDHc is a rate-limiting enzyme directly controlling pyruvate influx for the maintenance of TCA cycle, but its function in cancer progression is still under debate (Chen et al., 2018; Kaplon et al., 2013). However, in our orthotopic breast cancer models, impaired PDHc activity by PDHA deficiency fails to affect primary tumor growth but dramatically impairs tumor metastasis, suggesting that PDHA is a previously unrecognized, oncogenic player that promotes tumor metastasis at least in breast cancer models. In further support of this notion, PDHc activity is significantly upregulated in our advanced breast cancer patient cohorts, and its activation correlates with poor metastasis-free survival of breast cancer patients. Thus, our study not only identifies PDHA as a biomarker for the prediction of tumor metastasis but also provides the evidence that targeting PDHc is a promising strategy for tackling breast cancer metastasis. Future study aiming to develop PDHc inhibitors, which are expected to achieve a promising efficacy for breast cancer metastasis, should be warranted.
How PDHc activity is regulated remains largely unknown. We provide a detailed model to explain how AMPK activation induced by metabolic stress inhibits PDHA S293 phosphorylation and maintains PDHc activity. AMPK triggers phosphorylation of PDHA on S314 and S295, which is required for PDHc activation. PDHA S295 phosphorylation is an intrinsic catalytic site required for PDHc activation and pyruvate metabolism, whereas PDHA S314 phosphorylation is a primed site required for subsequent PDHA S295 phosphorylation through preventing PDHA-PDHKs interaction and subsequent PDHA S293 phosphorylation, which serves as a barrier for PDHA S295 phosphorylation. The finding that AMPK-mediated PDHA S295 and S314 phosphorylation serves as a crucial step for PDHc activation provides the strategy of how PDHc can be pharmacologically targeted for the intervention of breast cancer metastasis.
AMPK selects its substrate with the consensus motif of AMPK and prefers hydrophobic residues at −5 and +4 positions of actual phosphorylation site (Schaffer et al., 2015). We identified PDHA S295 and S314 as two sites phosphorylated by AMPK. Consistent with optimal AMPK consensus motif, PDHA contains hydrophobic residues valine (V) and methionine (M) at +4 position for S295 and S314 sites, respectively, but histidine (H) for S295 and glutamic acid (E) for S314 at −5 position do not fit perfect AMPK consensus motif. However, there are also known AMPK substrates without this optimal consensus motif, such as H2B (Bungard et al., 2010) with arginine (R) at −5 position and PGC1α (Jäger et al., 2007) with asparagine (N) and phenylalanine (F) at −5 position for two phosphorylation sites, respectively, indicating the complicity and flexibility of AMPK consensus motif. Moreover, basic residues at −3 position and serine (S) at −2 position are considered additional AMPK consensus motif. PDHA also contains histidine (H) at −3 position, serine (S) at −2 position for S295 and arginine (R) at −3 position, serine (S) at −2 position for S314. Using site-specific phospho-antibodies, PDHA S295 and S314 phosphorylation indeed occurs in vivo either upon A-769662 treatment or under glucose deprivation in an AMPK-dependent manner. Moreover, mass spectrometry analysis and site-specific phospho-antibodies from in vitro kinase assay reveal that AMPK phosphorylates PDHA on S295 and S314. Thus, PDHA is a bona fide substrate of AMPK.
Supplying α-KG could protect cancer cells with intact PDHc from metabolic stress, but not for cancer cells with compromised PDHc activation. Apart from its generation by TCA cycle enzyme IDH (isocitrate dehydrogenase), α-KG can be also produced through glutaminolysis (Villar et al., 2015), which is considered as a key metabolic reprogramming for metastasis (Elia et al., 2018). We rationalize that accumulating α-KG in disseminated cancer cells during metastasis may not only promote energy production and building blocks through TCA cycle maintenance but also orchestrate epigenetic reprogramming to empower cancer cell adaptation to hostile microenvironment through activating histone and/or DNA demethylases (Rinaldi et al., 2018). α-KG also functions in regulating pro-survival signal, such as mTOR pathway (Durán et al., 2012), which regulates cancer cells survival under oxidative stress (Malone et al., 2017). Thus, our study defines the critical role of AMPK-PDHc-TCA cycle-α-KG axis in cancer cell survival under diverse stresses, allowing for subsequent cancer metastasis.
In summary, our study defines the crucial role of PDHc and TCA cycle maintained by AMPK in empowering cancer cell survival in malicious environments for cancer metastasis (Figure 7E). Moreover, we also reveal that activation of PDHc is maintained by AMPK through phosphorylating its catalytic subunit PDHA. The strategy through PDHA phosphorylation and PDHc activation by AMPK allows for breast cancer cell adaption to the hostile microenvironments for developing full-blown metastatic cancer. Our study advances our current understanding of how PDHc activity and TCA cycle are maintained and offers potential strategies to tackle cancer metastasis.
Limitations of Study
We demonstrated that AMPK induces PDHA phosphorylation in vivo under physiological conditions using multiple approaches. Importantly, we generated site-specific antibodies to validate PDHA S295 and S314 phosphorylation in cells upon AMPK activation. However, currently we are only able to detect in vivo S295 phosphorylation in cells using mass spectrometry analysis, which occurs in around 10%–20% of PDHA based on the number of S295 phosphopeptides versus total peptides (Figures S6B and S6C), although in vitro S314 phosphorylation was detected by mass spectrometry analysis (Figure S5F). We rationalize that this result may be due to the low abundance of PDHA protein with S314 phosphorylation in vivo condition and/or the condition for our mass spectrometry analysis that may not be fully optimized. Unfortunately, given the current situation with COVID-19, it is not possible for us to pursue further the mass spectrometry analysis. Thus, our future work will dissect how exactly the AMPK-dependent phosphorylation events regulate PDHc function.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| P-AMPK (T172) | Cell Signaling Technology | Cat# 2535S; RRID:AB_331250 |
| AMPK | Cell Signaling Technology | Cat# 2532L; RRID:AB_330331 |
| P-ACC (S79) | Cell Signaling Technology | Cat# 3661S; RRID:AB_330337 |
| ACC | Cell Signaling Technology | Cat# 3662S; RRID:AB_2219400 |
| PDHK1 | Cell Signaling Technology | Cat# 3820S; RRID:AB_1904078 |
| P-ULK1 (S555) | Cell Signaling Technology | Cat# 5869S; RRID:AB_10707365 |
| ULK1 | Cell Signaling Technology | Cat# 8054S; RRID:AB_11178668 |
| HSP60 | Cell Signaling Technology | Cat# 4870S; RRID:AB_2295614 |
| Cleaved Caspase-3 (Asp175) | Cell Signaling Technology | Cat# 9664S; RRID:AB_2070042 |
| P-PDHA (S293) | abcam | Cat# Ab92696; RRID:AB_10711672 |
| P-Ser/Thr | abcam | Cat# Ab17464; RRID:AB_443891 |
| Tomm20 | abcam | Cat# Ab56783; RRID:AB_945896 |
| PDHK2 | Sigma | Cat# SAB4502155; RRID:AB_10747420 |
| PDHK3 | Sigma | Cat# SAB2103934; RRID:AB_10667235 |
| Flag | Sigma | Cat# F1804; RRID:AB_262044 |
| Actin | Sigma | Cat# A5441; RRID:AB_476744 |
| GRP78 | Santa Cruz | Cat# sc-13539; RRID:AB_627698 |
| AMPKα1 | Santa Cruz | Cat# sc-398861 |
| SDHA | Santa Cruz | Cat# sc-390381 |
| CS | Santa Cruz | Cat# sc-390693; RRID:AB_2813783 |
| IDH1 | Santa Cruz | Cat# sc-515396; RRID:AB_2827767 |
| IDH2 | Santa Cruz | Cat# sc-374476; RRID:AB_10986415 |
| FH | Santa Cruz | Cat# sc-393992 |
| SUCLG2 | Santa Cruz | Cat# sc-390818 |
| PDHA | Santa Cruz | Cat# sc-377092; RRID:AB_2716767 |
| DLD | Santa Cruz | Cat# sc-365977; RRID:AB_10917587 |
| DLAT | Santa Cruz | Cat# sc-271534; RRID:AB_10649809 |
| COX4 | Santa Cruz | Cat# sc-376731 |
| PDHB | Proteintech | Cat# 14744-1-AP; RRID:AB_2162941 |
| HA | Covance | Cat# MMS-101R; RRID:AB_291262 |
| P-PDHA S295 | This paper | N/A |
| P-PDHA S314 | This paper | N/A |
| Bacterial and Virus Strains | ||
| Top10 | Hui-Kuan Lin lab | N/A |
| DH5α | Hui-Kuan Lin lab | N/A |
| Stabl3 | Hui-Kuan Lin lab | N/A |
| BL21 | Hui-Kuan Lin lab | N/A |
| Biological Samples | ||
| Human breast cancer tissues | Chi Mei Medical Center from 1998 to 2004. IRB: 10210004 | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| AMPK (α1, β1, γ1) Protein, active | millipore | 14-840 |
| PDHA | Sigma | SRP5238 |
| Pyruvate | Thermo | 10569010 |
| Permeable α-ketoglutarate (Dimethyl 2-oxoglutarate) | Sigma | 349631 |
| ATP, [γ-32P]- 6000Ci/mmol 10mCi/ml | PerkinElmer | BLU002Z250UC |
| Pyruvic acid [3-14C] SODIUM SALT | Amrican radiolabeled chemicals | ARC0220 |
| 2-NBDG | CAYMAN CHEMICAL | 11046 |
| D-Luciferin, Potassium Salt | Gold biotechnology | LUCK-100 |
| ProLong Gold Antifade Mountant | Fisher | P36930 |
| protein A/G beads | Santa Cruz | sc-2003 |
| Critical Commercial Assays | ||
| Pyruvate Dehydrogenase (PDH) Activity Colorimetric Assay Kit | Biovision | K679 |
| Alpha-Ketoglutarate Colorimetric/Fluorometric Assay Kit | Biovision | K677 |
| Lactate Dehydrogenase Activity Colorimetric Assay Kit | Biovision | K726 |
| ATP Determination Kit | Fisher | A22066 |
| lactate test strip | nova biomedical | 40813 |
| DAB Substrate Kit | Abcam | ab64238 |
| VECTASTAIN® Elite® ABC HRP Kit | Vector | PK-7200 |
| Deposited Data | ||
| Unprocessed immunoblotting images | Mendeley datasets | https://dx.doi.org/10.17632/7vjm5bsyyt.1 |
| Experimental Models: Cell Lines | ||
| 4T1 | ATCC | Cat# CRL-2539; RRID:CVCL_0125 |
| Hep3B | Hui-Kuan Lin lab | N/A |
| MDA-MB-231 (MDA-MB-231-Luci) | MD Anderson Cancer Center | N/A |
| 293T | Hui-Kuan Lin lab | N/A |
| AMPK knockout MEF | Hui-Kuan Lin lab | N/A |
| Experimental Models: Organisms/Strains | ||
| BALB/c (NCI BALB-cAnNCr) mice | Charles Rivers | Code 555 |
| Nude (Crl:NU(NCr)-Foxn1nu) mice | Charles Rivers | Code 490 |
| Oligonucleotides | ||
| AMPKα1 (Human) Sh RNA-1#: GTGACCTCACTTGACTCTTCT | Sigma | N/A |
| AMPKα1 (Human) Sh RNA-2#: GAAGGTTGTAAACCCATATTA | Sigma | N/A |
| PDHA (Human) Sh RNA-1#: CGAGAAATTCTCGCAGAGCTT | Sigma | N/A |
| PDHA (Human) Sh RNA-2#: GCCAATCAGTGGATCAAGTTT | Sigma | N/A |
| AMPKα1 (Mouse) Sh RNA-1#: CGTAGTATTGATGATGAGATT | Sigma | N/A |
| AMPKα1 (Mouse) Sh RNA-2#: GAATCCTCATAGACCTTATTA | Sigma | N/A |
| PDHA (Mouse) Sh RNA-1#: GCTCAAGTACTACAGGATGAT | Sigma | N/A |
| PDHA (Mouse) Sh RNA-2#: GCTCAAGTACTACAGGATGAT | Sigma | N/A |
| ACC (Human) Sh RNA: TATGAGGTGGATCGGAGATTT | Sigma | N/A |
| PGC1α (Human) Sh RNA: TCGTGTTCCCGATCACCATAT | Sigma | N/A |
| See Table S4 for a detailed primer list | This paper | N/A |
| Recombinant DNA | ||
| pcDNA3-HA-AMPKα1 | Hui-Kuan Lin lab | Lee et al., 2015 |
| pcDNA3-Flag-PDHA | From Dr. Zhimin Lu (The University of Texas MD Anderson Cancer Center) | N/A |
| pBabe-Flag-PDHA S293A | From Dr. Amit Maity (Perelman School of Medicine at the University of Pennsylvania) | Cerniglia et al., 2015 |
| pCDH-3xFlag vector | From Dr. Xiaobing Shi (Van Andel Research Institute) | N/A |
| pCag-3xFlag vector | From Dr. Xiaobing Shi | N/A |
| pMD2G | From Dr. Xiaobing Shi | N/A |
| pPAX | From Dr. Xiaobing Shi | N/A |
| VSVG | Hui-Kuan Lin lab | N/A |
| Gal-Pol | Hui-Kuan Lin lab | N/A |
| pGEX-PDHA WT | This paper | N/A |
| pGEX-PDHA S295A | This paper | N/A |
| pGEX-PDHA S314A | This paper | N/A |
| pCDH-PDHA WT-Flag | This paper | N/A |
| pCDH-PDHA S295 A/D-Flag | This paper | N/A |
| pCDH-PDHA S314 A/D-Flag | This paper | N/A |
| pCag-PDHA WT-Flag | This paper | N/A |
| pCag-PDHA S295A/D-Flag | This paper | N/A |
| pCag-PDHA S314A/D-Flag | This paper | N/A |
| Software and Algorithms | ||
| GPS 3.0 (phosphorylation site prediction) | http://gps.biocuckoo.org/ | N/A |
| PROGgeneV2 online prognostic database | https://watson.compbio.iupui.edu/chirayu/proggene/database/index.php | Goswami and Nakshatri, 2013, 2014 |
| ImageQuant TL | GE healthcare | N/A |
| Excel | Microsoft | N/A |
| SPSS | IBM | N/A |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dr. Hui-Kuan Lin (hulin@wakehealth.edu).
Materials Availability
The plasmids, stable cell lines and antibodies generated in this study have not been deposited to any repositories yet. These materials will be available upon request. MTA may apply.
Data and Code Availability
The unprocessed data of immunoblotting have been deposited to Mendeley Data: https://dx.doi.org/10.17632/7vjm5bsyyt.1
Published datasets included in this study are available through PROGgeneV2 online prognostic database.
(https://watson.compbio.iupui.edu/chirayu/proggene/database/index.php)
Experimental Model and Subject Details
Animal Studies
The animals used in this study include BALB/c (NCI BALB-cAnNCr) mice (Female, 6 weeks) for orthotopic metastasis model and nude (Crl:NU(NCr)-Foxn1 nu) mice (Female, 6 weeks) for tail vein injection metastasis model. All the mice were purchased from Charles River Laboratories and group housed in a specific-pathogen-free facility under standard condition. All the related protocols were approved by Institutional Animal Care and Use Committee of Wake Forest School of Medicine.
Cell Culture
4T1, Hep3B, MDA-MB-231, 293T, AMPKα1 −/− α2 −/− MEFs were cultured in DMEM supplied with 10% Fetal Bovine Serum (FBS) and 1% Penicillin/Streptomycin at 37°C under 5% CO2. All the cell lines were tested to confirm no mycoplasma contamination.
Human Tumor Samples
All the patient tumor samples included in this study were retrieved from biobank that collected from female patients who underwent surgical resection in Chi Mei Medical Center from 1998 to 2004. The age of patients ranged from 27 to 85, with a mean of 51.5 and a medium of 50. As a rule, informed consent was obtained from all subjects and all the samples are anonymized. This study was approved by the Institutional Review Board (IRB) of the Chi Mei Medical Center (10210004).
Method Details
Transfection and Viral Infection
Calcium phosphate transfection method was used to transfect plasmids into 293T cells for virus package. Briefly, for lentiviral shRNA infection, 293T cells were co-transfected with pLKO.1-shRNA-puro constructs (Sigma) and packaging plasmids (pMD2G and pPAX). After 48h, virus containing supernatants were harvested to infect target cells. All stably transfected cells were selected by 3 μg/ml puromycin for 5 days.
For PDHA S293A, S925D, S295A, S314D, S314A restoration in AMPK deficient cells, 293T were co-transfected with pBabe-PDHA S293A-neo or pCDH-PDHA S295D, S295A, S314D, S314A-blasticidin and package plasmids (Gag-pol/VSVG or pPAX/pMD2G). After 48h, virus containing supernatants were harvested to infect AMPK deficient cells. All stably transfected cells were selected by 1 mg/ml neomycin or 5 μg/ml blasticidin for 7 days.
RNA Isolation and q-PCR
Cells were lysed by TRIZON reagent (Invitrogen) and total RNA were extracted according to the manufacture instruction. 2μg RNA was immediately processed to cDNA synthesis using PrimeScript RT Master Mix Kit (Clontech). q-PCR was performed in 20ul system containing 5μl H2O, 1μl P1 (10 μM), 1μl P2 (10 μM), 3μl diluted cDNA template and 10μl iTaq Universal SYBR® Green Supermix (Bio-Rad). The reaction was performed on ABI7300 Real-time PCR system. Melting curve analysis was used to guarantee the specificity of primers. β-Actin (ACTB) was used as an internal control and for normalization. ΔΔCt method was used to indicate the relative expression level of corresponding genes. See Table S4 for a detailed primer list:
Metabolomics analysis and 13C labeled Pyruvate/Glutamine tracing
Methanol extraction was used to prepare metabolomics samples. Briefly, 2x106 cells were seeded on 10cm dish and refreshed with complete medium for 24h. After quickly aspirating cell culture medium, gently rinse the cells with 37°C PBS. Then immediately add 1 mL 8:2 methanol:H2O (−75°C, on dry ice) into the plates placed on dry ice and incubate for 30min at −75°C to quench metabolism and perform extraction. Scrape all the cells from dishes at −75°C, and transfer them into tubes. Add 0.7 mL 8:2 methanol:H2O (−75°C, on dry ice) to perform second extraction, scrape and transfer all the cell contents to tubes. Spin the mixture at 13,000 rpm for 5 min at 0-4°C. Remove all the soluble extract into a vial and completely dry samples using the Speedvac at 30°C. For 13C labeled Pyruvate/Glutamine tracing assay, before extraction, the cells were treated with 13C3 labeled Pyruvate or 13C5 labeled Glutamine for 6h. Global metabolomics analysis was performed using Globally Optimized Targeted–MS (GOT-MS) by Northwest Metabolomics Research Center (NW-MRC)(Gu et al., 2015).
α-KG, ATP and Ac-CoA Level Detection
The expression levels of α-KG, ATP and Ac-CoA level were determined by using Alpha-Ketoglutarate Colorimetric/Fluorometric Assay Kit (Biovision), ATP Determination Kit (Invitrogen) and Acetyl-Coenzyme A Assay Kit (Sigma) respectively. Briefly, 2x106 cells were lysed by corresponding assay buffer. Then deproteinize samples by using 10 kDa molecular weight cut off spin columns (Biovision). Perform specific reaction according to manufacture instruction and measure the absorption/luminescence/fluorescence using spectrophotometer.
Oxygen Consumption Rate (OCR) Measurement
Oxygen consumption rate was determined using a Seahorse Bioscience XF24 Extracellular Flux Analyzer (Seahorse Bioscience). 0.8x104 Hep3B and 1.5x104 MDA-MB-231 cells were seeded in specialized V7 Seahorse tissue culture plates for overnight. Before the experiment, cells were washed and changed to seahorse assay medium (supplement with 10 mM Glucose, 1 mM Pyruvate, 2 mM Glutamine, adjusted to pH 7.4), then incubated in non-CO2 incubator for 1h. After measuring basal OCR level, Oligomycin, FCCP and Antimycin/Rotenone were sequentially injected into the cell chamber according to Seahorse standard protocol. Basal OCR level was calculated based on the AUC (area under the curve) before the injection of oligomycin. Maximal OCR level was calculated based on the AUC between the injection of FCCP and Antimycin/Rotenone.
PDHc Activity Detection
Activity of PDHc was determined by using Pyruvate Dehydrogenase Activity Colorimetric Assay Kit (Biovision). Briefly, 2x106 cells were homogenized with ice cold assay buffer and incubate on ice for 10min. Spin down at 10,000 x g for 5 min and transfer supernatant to fresh tube. Perform 100μl reaction (40μl sample, 10μl assay buffer, 50μl reaction mix) in 96 well plate. Measure absorption immediately at 450 nm in kinetic mode for 0-100 min at 37°C. According to manufacture instruction, calculate the PDHc activity based on the alteration of absorption (ΔOD) during specific time frame (ΔT). Apply ΔOD to NADH Standard Curve to obtain B nmol of NADH generated. PDHc Activity = B/(ΔT x 0.04) = nmol/min/ml = mU/ml.
In Vitro Kinase Assay and γ-32P ATP Incorporation Assay
Recombinant PDHA was purchased from Sigma, and active AMPK complex (α1β1γ1) was purchased from Millipore. 10X kinase assay buffer was purchased from Hyclone. Substrate PDHA was incubated with active AMPK complex for 30 min at 30°C in 20 μL reaction system (2ul 10X kinase assay buffer, 0.5 mM ATP, 250ng substrate, 500ng kinase). The reaction was terminated by adding SDS-loading buffer and all the samples were subjected to SDS-PAGE directly. For γ-32P ATP incorporation assay, GST-tagged PDHA was purified from bacteria and conjugated with glutathione beads. After performing in vitro kinase assay through incubating recombinant PDHA with different concentrations of active AMPK complex (250ng, 500ng) and γ-32P ATP for 0, 5, 20, 30 min, we washed out un-incorporated γ-32P ATP and determined γ-32P ATP incorporation on PDHA by liquid scintillation counter.
Immunoprecipitation and Immunoblotting
For immunoblotting assay, cell lysis buffer (50 mM Tris-HCl pH7.4, 250 mM NaCl, 0.5% Triton X-100, 10% Glycerol, 1 mM DTT) containing a protease inhibitor cocktail (Roche) was used to lyse cells. For immunoprecipitation (IP) assay, E1A lysis buffer (250 mM NaCl, 50 mM HEPES pH 7.5, 0.1% NP40, 5 mM EDTA) containing a protease inhibitor cocktail was used to lyse cells. After sonication for 10 s for three times, cell debris were removed by spin down at 13,000 rpm for 15 min. Clarified cell lysates were then incubated overnight with magnetic beads pre-incubated with indicated antibody. The beads were washed with PBST for 6 times before eluting with SDS sample buffer and subjected to immunoblotting analysis. All the blots were quantified by ImageQuant TL software. The main antibodies included in the study were listed below: anti-p-AMPK (T172) (1:1000), AMPK (1:1000), anti-p-ACC (S79) (1:1000), anti-ACC (1:1000), anti-PDHK1 (1:1000) from Cell Signaling Technology; anti-PDHK2 (1:1000), anti-PDHK3 (1:1000), anti-Flag (1:3000) from Sigma; anti-HA (1:5000) from Covance; anti-GRP78 (1:5000) from BD Transduction Laboratories; anti-phosphor-Ser/Thr antibody (1:1000), anti-p-PDHA(S293) (1:1000) from Abcam; anti-PDHA (1:1000) from Santa Cruz. anti-p-PDHA(S295), anti-p-PDHA(S314) were generated by Proteintech Inc.
Immunohistochemistry (IHC)
Primary tumor and lung metastatic tumor were subjected to paraffin embedding after fixation with 10% formaldehyde overnight. After dewaxing, the sections were heated at 95°C in pH 6.0 citric acid solution for epitope retrieval, quenched in 3% H2O2 to abolish endogenous peroxidase. Then standard procedure was applied to determine the expression of p-AMPK (T172), p-PDHA (S293), p-PDHA (S295) and p-PDHA (S314) in tissue samples.
Immunofluorescence (IF)
Cells were seeded on 8-well chamber slide and treated with AA769662 (100 μM) for 4h at 80% confluence to activate AMPK. After wash with ice cold PBS, the cells were fixed with 4% Paraformaldehyde under RT for 1h, and then incubated with 0.4% Triton X-100 at 4°C for 30min. After wash with PBS twice, the cells were blocked with 2% BSA under RT for 30min, followed by incubation with the primary antibody (p-AMPK, 1:100, PDHA 1:200, Tomm20 1:100) at 4°C overnight. After wash with PBS three times to remove the primary antibody, the cells were incubated with fluorescence conjugated secondary antibody (1:1000) under RT for 45min. After wash with PBS three times, the cells were incubated with ProLong® Gold Antifade Reagent with DAPI to stain nucleus. Images were documented by confocal microscope (Olympus FV1200 SPECTRAL Laser scanning Confocal Microscope).
Glucose Uptake Assay
Cells were seeded in 6 well plate and treated with glucose deprivation overnight before the assay. After incubation with 50μM 2-NBDG for 1h, glucose uptake was determined by FACS analysis. Using cells without 2-NBDG treatment as a blank, the shift of the FITC peak indicated the relative glucose uptake level.
Lactate Level Detection
Equal number of the cells was seeded in 6 well plate and refreshed with medium for 24h. Lactate level was determined by lactate test strips and Accutrend Lactate analyzer (Accutrend Lactate, Roche). Then the total cell number was counted, and the final concentration of lactate was presented as mM per 1x106 cells.
LDH Activity Assay
Activity of LDH was determined by using Lactate Dehydrogenase Activity Colorimetric Assay Kit (Biovision). 1x106 cells were homogenized with ice cold assay buffer and incubated on ice for 10min. Spin down at 10,000 x g for 15 min and transfer supernatant to fresh tube. Perform 100μl reaction (10μl sample, 40μl assay buffer, 50μl reaction mix) in 96 well plate. Measure absorption immediately at 450 nm in kinetic mode for 0-20 min at 37°C. According to manufacture instruction, calculate the LDH activity based on the alteration of absorption (ΔOD) during specific time frame (ΔT). Apply ΔOD to NADH Standard Curve to obtain B nmol of NADH generated. LDH Activity = B/(ΔT x 0.01) = nmol/min/ml = mU/ml
Cell Survival under Metabolic or Oxidative Stress
2x104 cells were seeded in 24 well plate. After culturing overnight, the cells were treated with either glucose deprivation (for metabolic stress) or H2O2 (for oxidative stress) with the supplement of vehicle, pyruvate (5 mM), α-KG (10 mM) for indicated 48h. Rinsing the well with PBS to remove the dead cells and stain the remaining cells with crystal violet. After dissolving in 2% SDS, absorption at 600nm was measured to calculate the relative survival rate for each group. Multiple wells were calculated, and the data were presented as means ± s.d.
Mitochondrial Isolation and Subfractionation
Mitochondrial isolation and Mitochondrial subfractionation were performed as previously described (Nishimura and Yano, 2014). Briefly, 1x108 cells were harvested after PBS wash and suspended in ice cold MTiso-buffer (3 mM HEPES (pH 7.4), 210 mM mannitol, 70 mM sucrose, 0.2 mM EGTA supplemented with protease inhibitor), then subjected to gently homogenize in a Dounce glass homogenizer using a glass “B”-type pestle for 50-80 strokes. Pile up the homogenate on an equal volume of 340 mM sucrose and centrifuge it at 500 x g for 10min at 4 °C to remove nuclei and unbroken cells as pellet. Collect the supernatant and then centrifuge it at 3,000 x g for 10min at 4 °C to isolate mitochondria as pellet. To ensure the purity of the mitochondria, 10% mitochondria pellet was subjected to SDS-PAGE after boiling with SDS loading buffer. The expression levels of various sub-cellular biomarkers between whole cell lysates and intact mitochondrial lysates were determined by immunoblotting. For mitochondrial subfractionation, re-suspend the isolated mitochondria with 0.5ml of MTiso-buffer with 5 mg/ml digitonin, and mix the suspension intensely for 15min by vortex. Centrifuge the suspension at 10,000 x g for 10min at 4°C to isolate the pellet containing mitoplast (inner membrane plus matrix) and the supernatant containing solubilized outer membrane (OM) and inter-membrane space (IMS) proteins. Re-suspend the pellet (mitoplast) in 100 μL of MTiso-buffer, and perform sonication to disrupt mitoplast, and then centrifuge it at 100,000 x g for 30min at 4°C, resulting in inner membrane (IM) fraction as pellet and matrix fraction as supernatant.
Pyruvate Trapped in PDHc Assay
Cells were transfected with Flag tagged WT or mutant PDHA. Fresh PDHc was purified through immunoprecipitation with Flag antibody and conjugated with protein A/G beads. Then the complex was directly incubated with C14-labeled pyruvate for 1h at room temperature. After washing with PBS for three times, pyruvate trapped in PDHc was determined by scintillation counter based on C14 level.
In vivo Tumor Growth and Metastasis Assay
2x106 (1x104 for 4T1) cells with AMPKα1 knockdown, PDHA knockdown or AMPKα1 knockdown restored with constitutive active form of PDHA were injected into 7-week old female nude mice (n = 5-7 for each group) through tail veins. Two months later (35 days for 4T1), mice were sacrificed, and lung tissues were analyzed for the incidence of metastasis. All lung tissues were subjected to 10% formaldehyde fixation and paraffin embedding, followed by H&E (Hematoxylin and Eosin) staining, and metastatic nodules were further confirmed by microscope. For bioluminescence imaging, mice were provided with 150 mg/g D-luciferin through intraperitoneal injection. After 5 min, IVIS imaging system was used to detect bioluminescence. For spontaneous metastasis model, 1x104 4T1 cells were injected into the 4th mammary fat pad of 7-week old female BALB/c mice (n = 5-7 for each group). Primary tumor was measured by Caliper for length and width every three to four days, and growth curve was documented based on the volume of tumor (V = (Length x Width x Width)/2). 35-40 days later, mice were sacrificed and lung tissues were analyzed for the incidence of metastasis. If no clear metastatic nodules were found, lung tissues were fixed with 10% formaldehyde and paraffin embedding, followed by H&E staining.
Quantification and Statistical Analysis
Statistical analyses
Unless otherwise noted, all data were quantified based on 3 independent results and presented as means ± s.d. Two-tailed Student`s t test was performed to calculate P value between different assay groups. For metastasis assay, the data were presented as means ± SEM from 5∼7 mice and Wilcoxon rank sum test (non-parametric two sample t test) (Coffelt et al., 2015) was performed to calculate P value. For all analyses, p < 0.05 regarded as statistically significant was indicated by asterisk (∗) and p < 0.01 regarded as statistically highly significant was indicated by two asterisks (∗∗).
Acknowledgments
We thank Drs. Z. Lu and Amit Maity for providing reagents. We acknowledge the support of Wake Forest Baptist Comprehensive Cancer Center Cell & Flow Cytometry Shared Resource, supported by the National Cancer Institute’s Cancer Center Support Grant (P30CA012197), United States. This work was supported by start-up funds from Wake Forest School of Medicine, endowed funds from Anderson Discovery Professor for Cancer Research, NIH grants (R01CA136787, R01CA182424, and R01CA193813), PPG developmental grant from Wake Forest Cancer Center, United States (to H.-K.L.), Breast Cancer Pilot Award from Wake Forest Cancer Center, United States (to Z.C.), and Health and welfare surcharge of tobacco products (MOHW109-TDU-B-212-134014), Taiwan (to C.-F.L.).
Author Contributions
Z.C. and H.-K.L. designed the experiments. Z.C., C.-F.L., X.Z., C.-C.H., A.-H.R., D.P., G.J., F.H., C.-Y.W., A.Z., S.-Y.W., and B.-S.P. performed the experiments. F.-C.H. and R.B.D. performed statistical analysis. Y.-H.W., S.-C.Y., G.W., W.Z., C.M.F., G.L.K., J.S.P., F.H.C., C.-Y.H., F.-J.T., B.P., and K.W. provided technical support, scientific insights, and suggestions. Z.C., D.P., and H.-K.L. wrote the manuscript. All authors discussed and read the manuscript.
Declaration of Interests
The authors declare no competing interests.
Published: October 5, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.molcel.2020.09.018.
Supplemental Information
References
- Anderson N.M., Mucka P., Kern J.G., Feng H. The emerging role and targetability of the TCA cycle in cancer metabolism. Protein Cell. 2018;9:216–237. doi: 10.1007/s13238-017-0451-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appleyard M.V., Murray K.E., Coates P.J., Wullschleger S., Bray S.E., Kernohan N.M., Fleming S., Alessi D.R., Thompson A.M. Phenformin as prophylaxis and therapy in breast cancer xenografts. Br. J. Cancer. 2012;106:1117–1122. doi: 10.1038/bjc.2012.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown G. Pyruvate dehydrogenase deficiency and the brain. Dev. Med. Child Neurol. 2012;54:395–396. doi: 10.1111/j.1469-8749.2012.04266.x. [DOI] [PubMed] [Google Scholar]
- Bungard D., Fuerth B.J., Zeng P.Y., Faubert B., Maas N.L., Viollet B., Carling D., Thompson C.B., Jones R.G., Berger S.L. Signaling kinase AMPK activates stress-promoted transcription via histone H2B phosphorylation. Science. 2010;329:1201–1205. doi: 10.1126/science.1191241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkewitz K., Zhang Y., Mair W.B. AMPK at the nexus of energetics and aging. Cell Metab. 2014;20:10–25. doi: 10.1016/j.cmet.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celià-Terrassa T., Kang Y. Distinctive properties of metastasis-initiating cells. Genes Dev. 2016;30:892–908. doi: 10.1101/gad.277681.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerniglia G.J., Dey S., Gallagher-Colombo S.M., Daurio N.A., Tuttle S., Busch T.M., Lin A., Sun R., Esipova T.V., Vinogradov S.A. The PI3K/Akt pathway regulates oxygen metabolism via pyruvate dehydrogenase (PDH)-E1α phosphorylation. Mol. Cancer Ther. 2015;14:1928–1938. doi: 10.1158/1535-7163.MCT-14-0888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers A.F., Groom A.C., MacDonald I.C. Dissemination and growth of cancer cells in metastatic sites. Nat. Rev. Cancer. 2002;2:563–572. doi: 10.1038/nrc865. [DOI] [PubMed] [Google Scholar]
- Chen J., Guccini I., Di Mitri D., Brina D., Revandkar A., Sarti M., Pasquini E., Alajati A., Pinton S., Losa M. Compartmentalized activities of the pyruvate dehydrogenase complex sustain lipogenesis in prostate cancer. Nat. Genet. 2018;50:219–228. doi: 10.1038/s41588-017-0026-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffelt S.B., Kersten K., Doornebal C.W., Weiden J., Vrijland K., Hau C.S., Verstegen N.J.M., Ciampricotti M., Hawinkels L.J.A.C., Jonkers J., de Visser K.E. IL-17-producing γδ T cells and neutrophils conspire to promote breast cancer metastasis. Nature. 2015;522:345–348. doi: 10.1038/nature14282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbet C., Feron O. Cancer cell metabolism and mitochondria: nutrient plasticity for TCA cycle fueling. Biochim. Biophys. Acta Rev. Cancer. 2017;1868:7–15. doi: 10.1016/j.bbcan.2017.01.002. [DOI] [PubMed] [Google Scholar]
- Douma S., Van Laar T., Zevenhoven J., Meuwissen R., Van Garderen E., Peeper D.S. Suppression of anoikis and induction of metastasis by the neurotrophic receptor TrkB. Nature. 2004;430:1034–1039. doi: 10.1038/nature02765. [DOI] [PubMed] [Google Scholar]
- Durán R.V., Oppliger W., Robitaille A.M., Heiserich L., Skendaj R., Gottlieb E., Hall M.N. Glutaminolysis activates Rag-mTORC1 signaling. Mol. Cell. 2012;47:349–358. doi: 10.1016/j.molcel.2012.05.043. [DOI] [PubMed] [Google Scholar]
- Elia I., Doglioni G., Fendt S.M. Metabolic hallmarks of metastasis formation. Trends Cell Biol. 2018;28:673–684. doi: 10.1016/j.tcb.2018.04.002. [DOI] [PubMed] [Google Scholar]
- Goswami C.P., Nakshatri H. PROGgene: gene expression based survival analysis web application for multiple cancers. J. Clin. Bioinforma. 2013;3:22. doi: 10.1186/2043-9113-3-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goswami C.P., Nakshatri H. PROGgeneV2: enhancements on the existing database. BMC Cancer. 2014;14:970. doi: 10.1186/1471-2407-14-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H., Zhang P., Zhu J., Raftery D. Globally optimized targeted mass spectrometry: reliable metabolomics analysis with broad coverage. Anal. Chem. 2015;87:12355–12362. doi: 10.1021/acs.analchem.5b03812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han F., Li C.F., Cai Z., Zhang X., Jin G., Zhang W.N., Xu C., Wang C.Y., Morrow J., Zhang S. The critical role of AMPK in driving Akt activation under stress, tumorigenesis and drug resistance. Nat. Commun. 2018;9:4728. doi: 10.1038/s41467-018-07188-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie D.G., Ross F.A., Hawley S.A. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat. Rev. Mol. Cell Biol. 2012;13:251–262. doi: 10.1038/nrm3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitosugi T., Fan J., Chung T.W., Lythgoe K., Wang X., Xie J., Ge Q., Gu T.L., Polakiewicz R.D., Roesel J.L. Tyrosine phosphorylation of mitochondrial pyruvate dehydrogenase kinase 1 is important for cancer metabolism. Mol. Cell. 2011;44:864–877. doi: 10.1016/j.molcel.2011.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäger S., Handschin C., St-Pierre J., Spiegelman B.M. AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1alpha. Proc. Natl. Acad. Sci. USA. 2007;104:12017–12022. doi: 10.1073/pnas.0705070104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon S.M., Chandel N.S., Hay N. AMPK regulates NADPH homeostasis to promote tumour cell survival during energy stress. Nature. 2012;485:661–665. doi: 10.1038/nature11066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplon J., Zheng L., Meissl K., Chaneton B., Selivanov V.A., Mackay G., van der Burg S.H., Verdegaal E.M., Cascante M., Shlomi T. A key role for mitochondrial gatekeeper pyruvate dehydrogenase in oncogene-induced senescence. Nature. 2013;498:109–112. doi: 10.1038/nature12154. [DOI] [PubMed] [Google Scholar]
- Kim N.H., Cha Y.H., Lee J., Lee S.H., Yang J.H., Yun J.S., Cho E.S., Zhang X., Nam M., Kim N. Snail reprograms glucose metabolism by repressing phosphofructokinase PFKP allowing cancer cell survival under metabolic stress. Nat. Commun. 2017;8:14374. doi: 10.1038/ncomms14374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolobova E., Tuganova A., Boulatnikov I., Popov K.M. Regulation of pyruvate dehydrogenase activity through phosphorylation at multiple sites. Biochem. J. 2001;358:69–77. doi: 10.1042/0264-6021:3580069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBleu V.S., O’Connell J.T., Gonzalez Herrera K.N., Wikman-Kocher H., Pantel K., Haigis M.C., de Carvalho F.M., Damascena A., Domingos Chinen L.T., Rocha R.M. PGC-1α mediates mitochondrial biogenesis and oxidative phosphorylation to promote metastasis. Nat. Cell Biol. 2014;16:992–1003. doi: 10.1038/ncb3039. 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.W., Li C.F., Jin G., Cai Z., Han F., Chan C.H., Yang W.L., Li B.K., Rezaeian A.H., Li H.Y. Skp2-dependent ubiquitination and activation of LKB1 is essential for cancer cell survival under energy stress. Mol. Cell. 2015;57:1022–1033. doi: 10.1016/j.molcel.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luzzi K.J., MacDonald I.C., Schmidt E.E., Kerkvliet N., Morris V.L., Chambers A.F., Groom A.C. Multistep nature of metastatic inefficiency: dormancy of solitary cells after successful extravasation and limited survival of early micrometastases. Am. J. Pathol. 1998;153:865–873. doi: 10.1016/S0002-9440(10)65628-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone C.F., Emerson C., Ingraham R., Barbosa W., Guerra S., Yoon H., Liu L.L., Michor F., Haigis M., Macleod K.F. mTOR and HDAC inhibitors converge on the TXNIP/thioredoxin pathway to cause catastrophic oxidative stress and regression of RAS-driven tumors. Cancer Discov. 2017;7:1450–1463. doi: 10.1158/2159-8290.CD-17-0177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin T.L., Gongol B., Zhang F., Martin M., Johnson D.A., Xiao H., Wang Y., Subramaniam S., Chien S., Shyy J.Y. AMPK promotes mitochondrial biogenesis and function by phosphorylating the epigenetic factors DNMT1, RBBP7, and HAT1. Sci. Signal. 2017;10:eaaf7478. doi: 10.1126/scisignal.aaf7478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minokoshi Y., Kim Y.B., Peroni O.D., Fryer L.G., Müller C., Carling D., Kahn B.B. Leptin stimulates fatty-acid oxidation by activating AMP-activated protein kinase. Nature. 2002;415:339–343. doi: 10.1038/415339a. [DOI] [PubMed] [Google Scholar]
- Mullen A.R., DeBerardinis R.J. Genetically-defined metabolic reprogramming in cancer. Trends Endocrinol. Metab. 2012;23:552–559. doi: 10.1016/j.tem.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng T.L., Leprivier G., Robertson M.D., Chow C., Martin M.J., Laderoute K.R., Davicioni E., Triche T.J., Sorensen P.H. The AMPK stress response pathway mediates anoikis resistance through inhibition of mTOR and suppression of protein synthesis. Cell Death Differ. 2012;19:501–510. doi: 10.1038/cdd.2011.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura N., Yano M. Separation of the inner and outer mitochondrial membrane in HeLa cells. Bio-protocol. 2014;4:e1299. [Google Scholar]
- Palikaras K., Lionaki E., Tavernarakis N. Coordination of mitophagy and mitochondrial biogenesis during ageing in C. elegans. Nature. 2015;521:525–528. doi: 10.1038/nature14300. [DOI] [PubMed] [Google Scholar]
- Pascual G., Domínguez D., Benitah S.A. The contributions of cancer cell metabolism to metastasis. Dis. Model. Mech. 2018;11 doi: 10.1242/dmm.032920. dmm032920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel M.S., Nemeria N.S., Furey W., Jordan F. The pyruvate dehydrogenase complexes: structure-based function and regulation. J. Biol. Chem. 2014;289:16615–16623. doi: 10.1074/jbc.R114.563148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei S., Minhajuddin M., Adane B., Khan N., Stevens B.M., Mack S.C., Lai S., Rich J.N., Inguva A., Shannon K.M. AMPK/FIS1-mediated mitophagy is required for self-renewal of human AML stem cells. Cell Stem Cell. 2018;23:86–100.e6. doi: 10.1016/j.stem.2018.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskounova E., Agathocleous M., Murphy M.M., Hu Z., Huddlestun S.E., Zhao Z., Leitch A.M., Johnson T.M., DeBerardinis R.J., Morrison S.J. Oxidative stress inhibits distant metastasis by human melanoma cells. Nature. 2015;527:186–191. doi: 10.1038/nature15726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puissant A., Robert G., Fenouille N., Luciano F., Cassuto J.P., Raynaud S., Auberger P. Resveratrol promotes autophagic cell death in chronic myelogenous leukemia cells via JNK-mediated p62/SQSTM1 expression and AMPK activation. Cancer Res. 2010;70:1042–1052. doi: 10.1158/0008-5472.CAN-09-3537. [DOI] [PubMed] [Google Scholar]
- Reymond N., d’Água B.B., Ridley A.J. Crossing the endothelial barrier during metastasis. Nat. Rev. Cancer. 2013;13:858–870. doi: 10.1038/nrc3628. [DOI] [PubMed] [Google Scholar]
- Rinaldi G., Rossi M., Fendt S.M. Metabolic interactions in cancer: cellular metabolism at the interface between the microenvironment, the cancer cell phenotype and the epigenetic landscape. Wiley Interdiscip. Rev. Syst. Biol. Med. 2018;10:e1397. doi: 10.1002/wsbm.1397. [DOI] [PubMed] [Google Scholar]
- Ruderman N.B., Carling D., Prentki M., Cacicedo J.M. AMPK, insulin resistance, and the metabolic syndrome. J. Clin. Invest. 2013;123:2764–2772. doi: 10.1172/JCI67227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y., Chapple R.H., Lin A., Kitano A., Nakada D. AMPK protects leukemia-initiating cells in myeloid leukemias from metabolic stress in the bone marrow. Cell Stem Cell. 2015;17:585–596. doi: 10.1016/j.stem.2015.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa K., Uematsu T., Korenaga Y., Hirasawa R., Kikuchi M., Murata K., Zhang J., Gai X., Sakamoto K., Koyama T., Satoh T. Krebs cycle intermediates protective against oxidative stress by modulating the level of reactive oxygen species in neuronal HT22 cells. Antioxidants. 2017;6:21. doi: 10.3390/antiox6010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer B.E., Levin R.S., Hertz N.T., Maures T.J., Schoof M.L., Hollstein P.E., Benayoun B.A., Banko M.R., Shaw R.J., Shokat K.M., Brunet A. Identification of AMPK phosphorylation sites reveals a network of proteins involved in cell invasion and facilitates large-scale substrate prediction. Cell Metab. 2015;22:907–921. doi: 10.1016/j.cmet.2015.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senft D., Ronai Z.A. Regulators of mitochondrial dynamics in cancer. Curr. Opin. Cell Biol. 2016;39:43–52. doi: 10.1016/j.ceb.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senft D., Ronai Z.E.A. Adaptive stress responses during tumor metastasis and dormancy. Trends Cancer. 2016;2:429–442. doi: 10.1016/j.trecan.2016.06.004. [DOI] [PubMed] [Google Scholar]
- Shackelford D.B., Shaw R.J. The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nat. Rev. Cancer. 2009;9:563–575. doi: 10.1038/nrc2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sośnicki S., Kapral M., Węglarz L. Molecular targets of metformin antitumor action. Pharmacol. Rep. 2016;68:918–925. doi: 10.1016/j.pharep.2016.04.021. [DOI] [PubMed] [Google Scholar]
- Steeg P.S. Targeting metastasis. Nat. Rev. Cancer. 2016;16:201–218. doi: 10.1038/nrc.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W., Liu Q., Leng J., Zheng Y., Li J. The role of pyruvate dehydrogenase complex in cardiovascular diseases. Life Sci. 2015;121:97–103. doi: 10.1016/j.lfs.2014.11.030. [DOI] [PubMed] [Google Scholar]
- Svensson R.U., Shaw R.J. Cancer metabolism: tumour friend or foe. Nature. 2012;485:590–591. doi: 10.1038/485590a. [DOI] [PubMed] [Google Scholar]
- Toyama E.Q., Herzig S., Courchet J., Lewis T.L., Jr., Losón O.C., Hellberg K., Young N.P., Chen H., Polleux F., Chan D.C., Shaw R.J. Metabolism. AMP-activated protein kinase mediates mitochondrial fission in response to energy stress. Science. 2016;351:275–281. doi: 10.1126/science.aab4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villar V.H., Merhi F., Djavaheri-Mergny M., Durán R.V. Glutaminolysis and autophagy in cancer. Autophagy. 2015;11:1198–1208. doi: 10.1080/15548627.2015.1053680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtz D., Konstantopoulos K., Searson P.C. The physics of cancer: the role of physical interactions and mechanical forces in metastasis. Nat. Rev. Cancer. 2011;11:512–522. doi: 10.1038/nrc3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu N., Zheng B., Shaywitz A., Dagon Y., Tower C., Bellinger G., Shen C.H., Wen J., Asara J., McGraw T.E. AMPK-dependent degradation of TXNIP upon energy stress leads to enhanced glucose uptake via GLUT1. Mol. Cell. 2013;49:1167–1175. doi: 10.1016/j.molcel.2013.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y.T., Wu S.B., Wei Y.H. Metabolic reprogramming of human cells in response to oxidative stress: implications in the pathophysiology and therapy of mitochondrial diseases. Curr. Pharm. Des. 2014;20:5510–5526. doi: 10.2174/1381612820666140306103401. [DOI] [PubMed] [Google Scholar]
- Yonashiro R., Eguchi K., Wake M., Takeda N., Nakayama K. Pyruvate dehydrogenase PDH-E1β controls tumor progression by altering the metabolic status of cancer cells. Cancer Res. 2018;78:1592–1603. doi: 10.1158/0008-5472.CAN-17-1751. [DOI] [PubMed] [Google Scholar]
- Zhou Z.H., McCarthy D.B., O’Connor C.M., Reed L.J., Stoops J.K. The remarkable structural and functional organization of the eukaryotic pyruvate dehydrogenase complexes. Proc. Natl. Acad. Sci. USA. 2001;98:14802–14807. doi: 10.1073/pnas.011597698. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The unprocessed data of immunoblotting have been deposited to Mendeley Data: https://dx.doi.org/10.17632/7vjm5bsyyt.1
Published datasets included in this study are available through PROGgeneV2 online prognostic database.
(https://watson.compbio.iupui.edu/chirayu/proggene/database/index.php)