OBJECTIVE
Cervical cancer remains the most common gynecological malignancy internationally, and nearly one third of patients succumb to this disease within 5 years from diagnosis.1,2 While patients with malignancy who have limited life expectancy benefit from less aggressive treatment intervention,3 there is currently little evidence to guide the prediction of the length of life expectancy. The aims of the study were (1) to examine predictors for survival by utilizing clinicolaboratory variables among women with recurrent cervical cancer and (2) to examine the utility of a new analytic approach using a deep-learning neural networks model.
STUDY DESIGN
This institutional review board—approved retrospective study evaluated 157 women who developed recurrent cervical cancer among 431 women with cervical cancer diagnosed between January 2008 and December 2014. Prediction of 3 and 6 month survival after recurrence was compared between the historical approach (linear regression model) and experimental approach (deep-learning neural networks model) as described in our previous study.4,5 Model inputs included 13 unique variables (clinical: age, body habitus change, pain score, blood pressure [systolic and diastolic], and heart rate, and laboratory: white blood cell, hemoglobin, platelet, bicarbonate, blood urea nitrogen, creatinine, and albumin) from all outpatient visits following recurrence (5421 total data points).
RESULTS
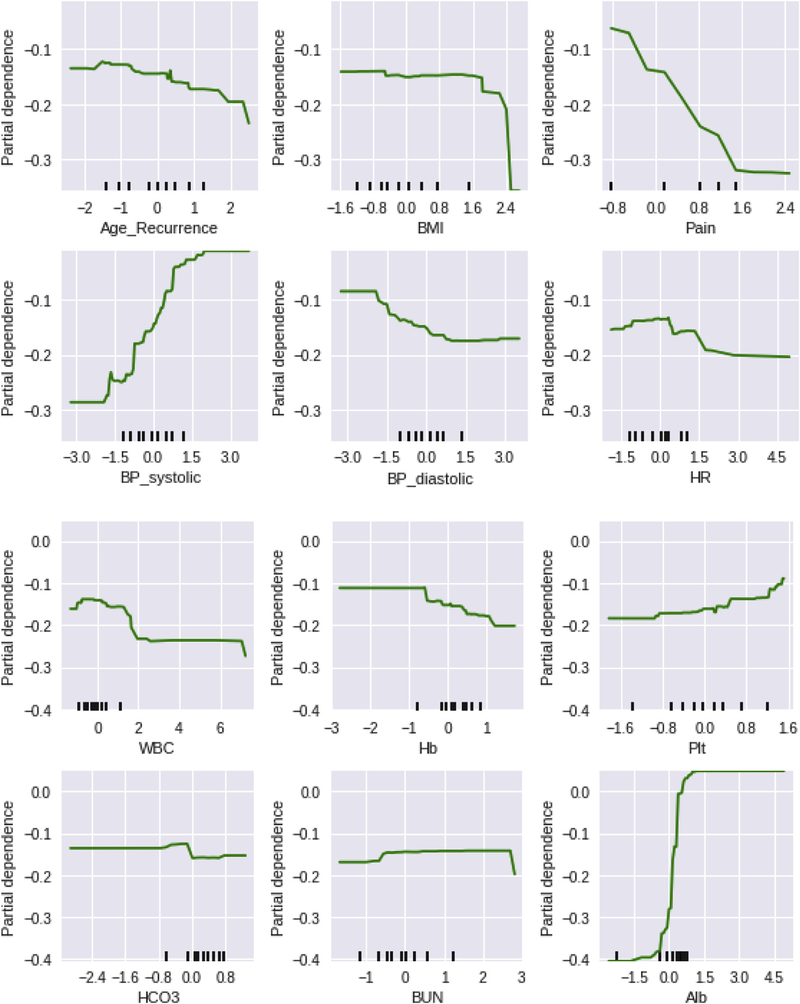
There were 102 women who died of cervical cancer (65.0%), with a median time to death after recurrence being 7.7 months. The deep-learning model had a significantly better prediction for 3 month (area under the curve, 0.747 vs 0.652, P < .0001) and 6 month (0.724 vs 0.685, P < .0001) survival compared with the linear regression model. In the deep-learning model, decreased 3 month survival was associated with older age, decreasing albumin level, decreasing body mass index, increasing pain score, decreasing systolic blood pressure, decreasing white blood cell count, increasing platelet counts, and decreasing hemoglobin levels determined as the interval changes from the baseline values at the recurrence (all, P < .05; Figure 1). Similar findings were seen in the 6 month survival prediction model.
FIGURE 1. Partial dependency plots for 3 month survival prediction.
Deep-learning model was used for the analysis. The x-axis represents the interval change of each (z-score normalization) clinicolaboratory factor from the first recurrence. Normalization was used to eliminate the effects of different feature ranges. The y-axis represents the interval change in survival chance (partial dependency). Partial dependency plots are graphical visualizations of the marginal effect of a given variable (or multiple variables) on an outcome. Intuitively, we can interpret the partial dependence as the expected outcome as a function of the target variables. The y-axis values are negative because the expected outcome decreases for our target variables. The lower the partial dependence value, the less chance of 3 month survival. Similar results were also seen in 6 month survival predictors. The tick marks on the x-axis represent the deciles of the feature values in the training data.
CONCLUSION
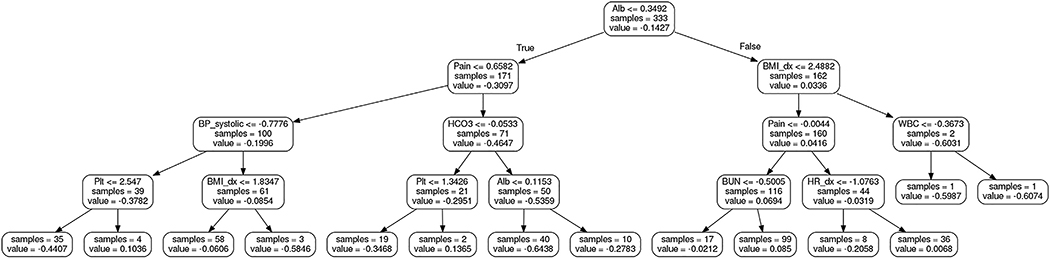
Our study suggests that deep learning by evaluating certain clinicolaboratory parameters commonly used in daily practice may be a useful approach to predict limited life expectancy in women with recurrent cervical cancer that can then translate into patient counseling regarding palliative care. Deep-learning models can be converted to a set of decision trees,5 whose rules may be more interpretable to the clinician (Figure 2). In this model, interval change of albumin was the most important factor to triage the patient. Then pain score and body habitus change need to be factored based on the albumin level. Based on the interval changes in these values, additional factors need to be evaluated for 3 month survival prediction.
FIGURE 2. First decision tree model for 3 month survival prediction.
First decision tree for 3 month survival prediction was obtained by mimicking the performance of deep neural networks.5 In the decisio- tree plots, the thresholds on the feature nodes are for normalized features. Samples include the number of samples to that node. The value of a node is the prediction score of a sample from the corresponding decision rules.
There is recent mounting evidence demonstrating the utility of a deep-learning in decision making for clinical medicine.5 Because early discussion for palliative care is beneficial to reduce aggressive care,3 objective assessment of survival time will add more concrete information in patient counseling. Given the complexity and difficulty in predicting actual survival time as opposed to proportional survival chance, the utility of deep-learning models merits further study and validation in oncology practice.
Acknowledgments
This study was supported by Ensign Endowment for Gynecologic Cancer Research (to K.M.).
Footnotes
The authors report no conflict of interest.
Contributor Information
Koji Matsuo, Division of Gynecologic Oncology, Department of Obstetrics and Gynecology, University of Southern California, Los Angeles, CA; Norris Comprehensive Cancer Center, University of Southern California, Los Angeles, CA.
Sanjay Purushotham, Department of Computer Science, University of Southern California, Los Angeles, CA.
Aida Moeini, Division of Gynecologic Oncology, Department of Obstetrics and Gynecology, University of Southern California, Los Angeles, CA.
Guangyu Li, Department of Computer Science, University of Southern California, Los Angeles, CA.
Hiroko Machida, Division of Gynecologic Oncology, Department of Obstetrics and Gynecology, University of Southern California, Los Angeles, CA.
Yan Liu, Department of Computer Science, University of Southern California, Los Angeles, CA.
Lynda D. Roman, Division of Gynecologic Oncology, Department of Obstetrics and Gynecology, University of Southern California, Los Angeles, CA; Norris Comprehensive Cancer Center, University of Southern California, Los Angeles, CA.
REFERENCES
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87–108. [DOI] [PubMed] [Google Scholar]
- 2.National Cancer Institute, Surveillance, Epidemiology, and End Results Program. Available at: https://seer.cancer.gov/statfacts/html/cervix.html. Accessed June 20, 2017.
- 3.Mack JW, Cronin A, Keating NL, et al. Associations between end-oflife discussion characteristics and care received near death: a prospective cohort study. J Clin Oncol 2012;30:4387–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature 2015;521: 436–44. [DOI] [PubMed] [Google Scholar]
- 5.Che Z, Purushotham S, Khemani R, Liu Y. Interpretable deep models for ICU outcome prediction. AMIA Annu Symp Proc 2017;2016:371–80. [PMC free article] [PubMed] [Google Scholar]