Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) disease, which started in Wuhan, Chin, has now become a public health challenge in most countries around the world. Proper preventive measures are necessary to prevent the spread of the virus to help control the pandemic. Because, SARS-CoV-2 is new, its transmission route has not been fully understood. In this study, we aimed to investigate the presence of SARS‐CoV‐2 in the sweat secretion of COVID‐19 patients. Sweat specimens of 25 COVID- 19 patients were collected and tested for SARS‐CoV‐2 RNA by Real‐time Polymerase Chain Reaction (RT-PCR) method. After RNA extraction and cDNA amplification, all samples were examined for the presence of ORF-1ab and N genes related to COVID-19. Results annotated by Realtime PCR machines software based on Dynamic algorithm. The results of this study showed the absence of SARS-CoV-2 in the sweat samples taken from the foreheads of infected people. Therefore, it can be concluded that the sweat of patients with COVID- 19 cannot transmit SARS-CoV-2. However they can be easily contaminated with other body liquids.
Keywords: Sweat, COVID- 19, Sars-Cov-2, Detection, Transmission, Contamination
1. Introduction
On December 31, 2019, a cluster of atypical pneumonia cases appeared in China [1]. Rapidly, investigations identified a novel family B betacoronavirus, now officially named as SARS‐CoV‐2 by the International Committee on Taxonomy of Viruses (ICTV), and introduced as the etiological factor responsible for this infection [2,3]. Acute fever, cough, myalgia, dyspnea, and radiological evidence of bilateral pneumonia with ground-glass opacity of lung have been observed in most patients with coronavirus disease 2019 (COVID-19), or SARS-CoV-2 infection [4,5]. Although, mildly symptomatic or asymptomatic cases have also been reported [[6], [7], [8]]. According to the findings of epidemiologic and genetic research, COVID-19 disease is transmitted by sustained human-to-human spread. It is now believed that transmission between individuals occurs mainly through respiratory droplets and contact [9]. On the other hand, with the discovery of SARS-CoV-2 in the feces of patients in the United States and China, it has been announced that there may be a risk of transmission of the virus through the feces of patients [10,11]. However, our knowledge of other possible ways of transmitting the virus, including vertical transmission (from infected mother to fetus), transmission through sperm and sweat, is not yet complete. In a study of 212 people infected with SARS-CoV-2, 114 reported “profuse sweating” and 102 of them reported “night sweats”. This indicates a high volume of substance for infection, if the sweat of these people has infectious SARS-CoV-2 [12]. On other hand, SARS-CoV-2 has similarities with acute respiratory coronavirus syndrome (SARS-CoV) and Middle East respiratory syndrome (MERS) in terms of symptoms and biological mechanisms, although they differ in terms of infectivity and mortality [13,14]. Given the novelty of this virus, the use of research findings on MERS and SARS can help us understand the behavior of this virus. Based on previous findings from related human betacoronaviruses, one of the potential ways of transmitting the virus can be through the sweat secretions of patients with coronavirus infection [15]. According to previous studies on the presence of betacoronaviruses in the sweat glands and the ability to transmit through the sweat of an infected person and the high rate of sweating in patients with SARS-CoV-2 and the high transmissibility of this virus, it is better not to ignore the possibility of transmission through contact with sweat Infected person [12,[16], [17], [18]]. Therefore, to clarify this issue, we have studied the sweat samples of 25 patients with COVID-19.
2. Materials and methods
2.1. Patients and clinical specimens
Twenty-five patients that were from the health staff were contributed in the testing of the presence of SARS-CoV-2 in sweat secretions. All patients were diagnosed based on clinical symptoms and contact history with further confirmation by positive results of qualitative real-time reverse transcriptase-polymerase chain reaction assay (qRT-PCR) targeting the ORF1ab and N gene (Sunsure, China) in respiratory specimens according to WHO interim guidance [19]. This study was approved by the Ethics Committee of Tabriz University of Medical Sciences with reference number IR.TBZMED.REC.1399.632. First, the patients' foreheads were disinfected with 70% ethanol and they were asked to go up and down the stairs (and do a little exercise) to make their bodies sweat. Sweat samples were taken from their foreheads by the Dacron swabs which were placed into viral transport media (VTM) and transferred to the laboratory by preserving the strict cold chain.
2.2. RNA extraction and real-time PCR
Viral RNA of SARS-CoV-2 was extracted using the viral RNA mini kit (Vazyme Biotech Co., Ltd., Nanjing, China) according to the manufacturer's protocol. In general, 200 μl of each sweat specimen were subjected to extraction with an elution volume of 50 μl. All samples were kept in −80 for any further study to prevent hydrolysis of RNA. For the best results, examination was done in the same day as RNA of viruses were extracted.
2.3. Real-time PCR
Real-Time PCR was performed to synthesize cDNA using Novel Coronavirus (2019-nCoV) Nucleic Acid Diagnostic Kit (PCR-Fluorescence Probing) (Sunsure Biotech. China). The kit contains PCR buffer, primers and probes, dNTPs, MgCl2, Rnasin, RT Enzyme, and Taq Enzyme. The detectors used are FAM (ORF-1ab region), ROX (N gene), and Internal Control (Cy5). Taqman RT- PCR was performed using the MIC PCR thermocycler (Applied Biomolecular system, Australia) at the following cycling conditions: 1 cycle at 50 °C for 20 min, 1 cycle at 95 °C for 1 min and 45 cycles at 95 °C for 15 s, 60 °C for 35 s then 72.0 °C for 15 s. For analysis of the final results dynamic algorithm was used by cycling analysis in green and yellow and red fluorescence and cut off for fluorescence was 5%. Cut off for detection of virus according to the company report was above 200 copy number per ml.
3. Results
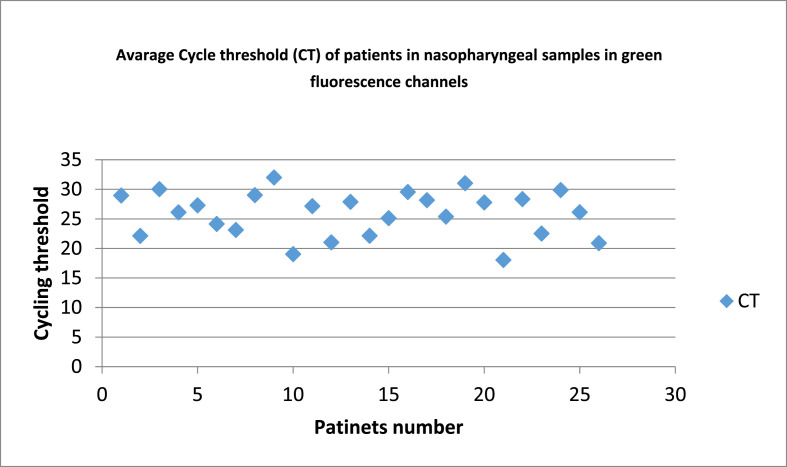
The 25 patients consisted of 15 males and 10 females. Their median age was 35 years (range 24–48 years). All of them had clinical features compatible with high viral load such as fatigue, fever, and low blood oxygenation levels and also positive result of real-time PCR of nasopharyngeal sample (19.2 ≤ Ct ≤ 32) (Fig. 1 ).
Fig. 1.
Average Cycle threshold (Ct) of patients in nasopharyngeal samples. These results indicate patient's nasopharyngeal contamination with Sars-Cov-2.
Out of twenty-five patients, twenty of them had CRP positive and blood changes such as lymphocyte depletion were observed in 18 of them.
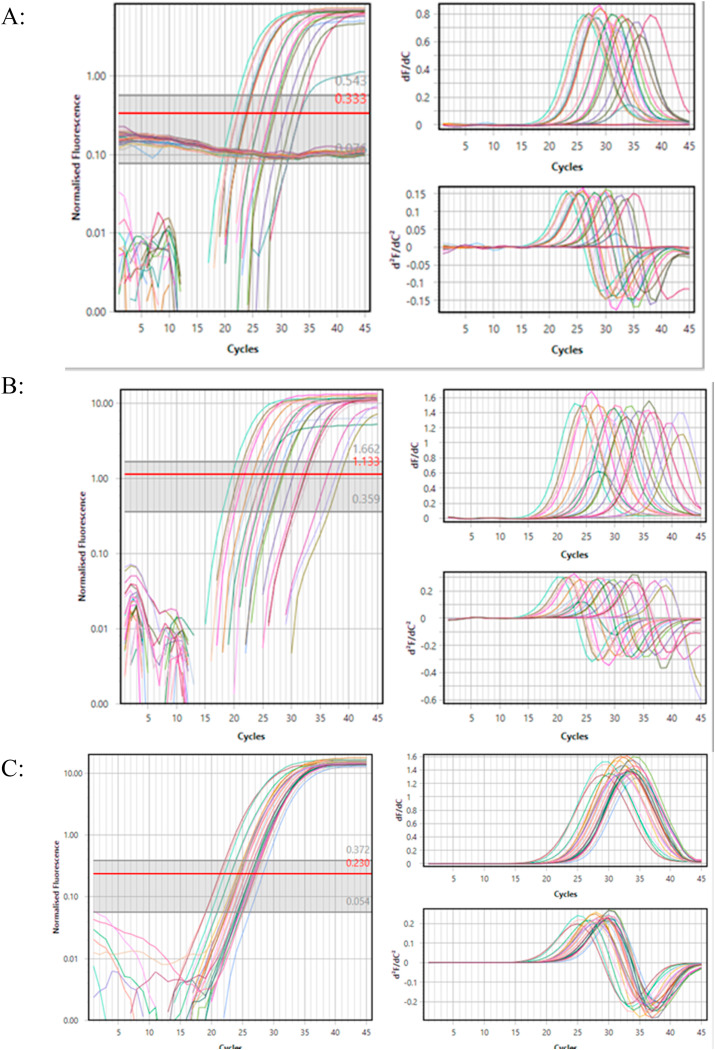
The result of Real-time PCR test for sweat samples taken from the forehead of these patients was that all samples were negative for the presence of SARS-CoV-2 that has been shown in Fig. 2 and only two positive cases were observed (Ct: 32.4 and Ct:28.17).
Fig. 2.
Amplification curve of the Sar-Cov-2 identification by A) ORF1-ab (in Green channel), B) N gene (in Yellow channel) and C) Internal control (In Red Channel). These curves show accuracy and sensitivity of the identification of target genes. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
Findings of the present study indicate sweat of the patients are not infective and is safe. SARS-CoV-2 is known as the seventh and newest coronavirus, which started spreading in December 2019 from Wuhan, China, and quickly spread around the world, becoming the biggest global health concern [20]. Clinical signs of COVID-19 disease range from mild upper respiratory tract infection to a multisystemic disease with the inflammatory and thrombotic immunological response [21]. The main reason for the wide spread of this virus is its transmission power and contagion, and evidence shows that asymptomatic people can transmit the disease [22,23]. Development of practical and effective strategies to protect high-risk individuals and prevent the spread of the virus is urgently needed. However, the pathogenicity of SARS-CoV2 infection and the pathways of virus transmission have not yet been clearly identified. In addition to proven routes of transmission of SARS-CoV-2, such as respiratory secretions and feces, the possibility of transmission through sweat is also considered. In this study, twenty-five infected patients, all of whom had clinical symptoms and high viral load in respiratory samples, were examined for the presence of SARS-CoV-2 in sweat samples. All of them were negative and only two positive cases were found. By asking patients who tested positive, we found that they had touched their foreheads during the test and that their hands may have been contaminated with oral secretions or mucosa, so the positive result was actually due to contamination of the sampling site with the secretions of other parts. Unfortunately, very little study has been done on the possible role of sweat in the transmission of any of the betacoronaviruses, especially with regard to other deadly viruses that can be transmitted through the sweat of an infected person [24]. In 2015, a report was presented that a Korean healthcare worker infected with the MERS coronavirus during cardiopulmonary resuscitation (CPR) of a MERS patient. According to the report, during CPR, a large amount of fluid was splashed and the nurse becomes infected by touching the mask and wiping the sweat from her face. Although this is not certain, one of the possible ways that this person became infected was through mucosal exposure to sweat contaminated with MERS coronavirus [17]. The presence of the SARS-CoV-2 in eccrine glands indicates that sweat can be a source of transmission [25]. In a 2004 study of four SARS-CoV patients, Ding and his colleagues found SARS-CoV RNA and nucleoprotein in the sweat glands, kidney, and intestine by in situ hybridization and immunohistochemistry. Accordingly, there were speculations that SARS-CoV could be transmitted through sweat, feces and urine [16]. One of the ways of transmission by sweat is through skin contact with mucus, similar to what has been said about the Korean healthcare worker [17]. From a theoretical point of view, one way of transmission can be path skin-object-mucosa so that contaminated sweat may remain on objects, which in turn may cause infection if touched by non-infected people [26]. The receptor for SARS-CoV-2 to enter human cells is human angiotensin converting enzyme 2 (ACE2), and it is thought that organs with high levels of this receptor will be able to accept more viruses and therefore be more likely to become infected [27]. ACE2 is itself exist in the skin, eccrine glands and smooth muscles around the sebaceous glands as well as within these glands [28]. The fact that the number of ACE2 receptors in the skin can absorb the SARS-CoV-2 raises concerns about how the SARS-CoV-2 is transmitted. Therefore, it is possible that not only inhaling or touching the oral and respiratory secretions and mucosa alone does not cause infection, but also simpler routes such as touching the sweat-soaked objects of the infected person can cause the infection. However, the results of this study refute the hypotheses about the possibility of transmitting SARS-CoV-2 through sweat, but still more research in this case with more samples and sweat sampling of other parts of the body are recommended to ensure this result.
CRediT authorship contribution statement
Hadis Fathizadeh: Writing - original draft, study design, Data collection, Data interpretation, manuscript preparation. Sepehr Taghizadeh: data interpretation, manuscript preparation. Rohollah Safari: Writing - original draft, Data collection, manuscript preparation. Saeid Shabestari Khiabani: Writing - original draft, data interpretation, manuscript preparation. Bayaz Babak: Writing - original draft, Data collection, manuscript preparation. Fatemeh Hamzavi: Writing - original draft, Data collection, manuscript preparation. Khudaverdi Ganbarov: Writing - original draft, Formal analysis, Data analysis, Data interpretation, manuscript preparation. Silvano Esposito: Writing - original draft, manuscript preparation. Elham Zeinalzadeh: Writing - original draft, Literature search, manuscript preparation. Sounkalo Dao: Writing - original draft, Formal analysis, Statistical analysis, manuscript preparation. Şükran Köse: Writing - original draft, Formal analysis, Statistical analysis, manuscript preparation. Hossein Samadi Kafil: Writing - original draft, Study design, Supervision, Funding acquisition, manuscript preparation.
Declaration of competing interest
None to declare.
Acknowledgment
This study was supported by Tabriz University of Medical Sciences with grant number 66041 and was approved in ethic committee with reference number IR.TBZMED.REC.1399.632. All participants filled consent form and were health workers who worked in Imam Reza Hospital and their participation was completely voluntarily and according to Helsinki declaration.
References
- 1.Ozma M.A., Maroufi P., Khodadadi E., Köse Ş., Esposito I., Ganbarov K., et al. Clinical manifestation, diagnosis, prevention and control of SARS-CoV-2 (Covid-19) during the outbreak period. Infez Med. 2020;28:153–165. [PubMed] [Google Scholar]
- 2.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fathizadeh H., Maroufi P., Momen-Heravi M., Dao S., Köse Ş., Ganbarov K., et al. Protection and disinfection policies against SARS-CoV-2 (COVID-19) Infez Med. 2020;28:185–191. [PubMed] [Google Scholar]
- 4.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khodadadi E., Maroufi P., Khodadadi E., Esposito I., Ganbarov K., Espsoito S., et al. Study of combining virtual screening and antiviral treatments of the Sars-CoV-2 (Covid-19) Microb. Pathog. 2020:145. doi: 10.1016/j.micpath.2020.104241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wei M., Yuan J., Liu Y., Fu T., Yu X., Zhang Z.J. Novel coronavirus infection in hospitalized infants under 1 Year of age in China. J. Am. Med. Assoc. 2020;323:1313–1314. doi: 10.1001/jama.2020.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arunmanee W., Ecoy G.A.U., Khine H.E.E., Duangkaew M., Prompetchara E., Chanvorachote P., et al. Colicin N mediates apoptosis and suppresses integrin-modulated survival in human lung cancer cells. Molecules. 2020;25:816. doi: 10.3390/molecules25040816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Del Rio C., Malani P.N. 2019 novel coronavirus-important information for clinicians. J. Am. Med. Assoc. 2020;323(22):1039–1040. doi: 10.1001/jama.2020.1490. [DOI] [PubMed] [Google Scholar]
- 10.Holshue M.L., DeBolt C., Lindquist S., Lofy K.H., Wiesman J., Bruce H., et al. First case of 2019 novel coronavirus in the United States. N. Engl. J. Med. 2020;382:929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gholizadeh P., Safari R., Marofi P., Zeinalzadeh E., Pagliano P., Ganbarov K., et al. Alteration of liver biomarkers in patients with SARS-CoV-2 (COVID-19) J. Inflamm. Res. 2020:13. doi: 10.2147/JIR.S257078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y.L.Z., Wang B., Cang J., Ma Y. Gastrointestinal tract symptoms in coronavirus disease 2019: analysis of clinical symptoms in adult patients. medRxiv Preprint. 2020 [Google Scholar]
- 13.da Costa V.G., Moreli M.L., Saivish M.V. The emergence of SARS, MERS and novel SARS-2 coronaviruses in the 21st century. Arch. Virol. 2020;165:1517–1526. doi: 10.1007/s00705-020-04628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Najafi K., Maroufi P., Khodadadi E., Zeinalzadeh E., Ganbarov K., Asgharzadeh M., et al. SARS-CoV-2 receptor ACE2 and molecular pathway to entre target cells during infection. Rev. Med. Microbiol. 2020;4:1–12. [Google Scholar]
- 15.Propper R.E. Is sweat a possible route of transmission of SARS-CoV-2? Exp. Biol. Med. 2020;245:997–998. doi: 10.1177/1535370220935409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ding Y., He L., Zhang Q., Huang Z., Che X., Hou J., et al. Organ distribution of severe acute respiratory syndrome (SARS) associated coronavirus (SARS-CoV) in SARS patients: implications for pathogenesis and virus transmission pathways. J. Pathol. 2004;203:622–630. doi: 10.1002/path.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nam H.S., Yeon M.Y., Park J.W., Hong J.Y., Son J.W. Healthcare worker infected with Middle East Respiratory Syndrome during cardiopulmonary resuscitation in Korea. Epidemiol. health. 2017;39 doi: 10.4178/epih.e2017052. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanche S., Lin Y.T., Xu C., Romero-Severson E., Hengartner N., Ke R. High contagiousness and rapid spread of severe acute respiratory syndrome coronavirus 2. Emerg. Infect. Dis. 2020;26:1470–1477. doi: 10.3201/eid2607.200282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gholizadeh P., Aghazadeh M., Asgharzadeh M., Kafil H.S. Suppressing the CRISPR/Cas adaptive immune system in bacterial infections. Eur. J. Clin. Microbiol. Infect. Dis. 2017;11 doi: 10.1007/s10096-017-3036-2. 017-3036. [DOI] [PubMed] [Google Scholar]
- 20.Deng Z., Hu Y., Yang P., Zheng P., Peng W., Ren B., et al. Diagnosis and treatment of an acute severe pneumonia patient with COVID-19: case report. J. Med. Virol. 2020 doi: 10.1002/jmv.25802. [DOI] [PubMed] [Google Scholar]
- 21.Hoehl S., Rabenau H., Berger A., Kortenbusch M., Cinatl J., Bojkova D., et al. Evidence of SARS-CoV-2 infection in returning travelers from Wuhan, China. N. Engl. J. Med. 2020;382:1278–1280. doi: 10.1056/NEJMc2001899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., et al. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N. Engl. J. Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bai Y., Yao L., Wei T., Tian F., Jin D.Y., Chen L., et al. Presumed asymptomatic carrier transmission of COVID-19. J. Am. Med. Assoc. 2020;323:1406–1407. doi: 10.1001/jama.2020.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vetter P., Fischer W.A., 2nd, Schibler M., Jacobs M., Bausch D.G., Kaiser L. Ebola virus shedding and transmission: review of current evidence. J. Infect. Dis. 2016;214 doi: 10.1093/infdis/jiw254. S177-s84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Santonja C., Heras F., Núñez L., Requena L. COVID-19 chilblain-like lesion: immunohistochemical demonstration of SARS-CoV-2 spike protein in blood vessel endothelium and sweat gland epithelium in a polymerase chain reaction-negative patient. Br. J. Dermatol. 2020 doi: 10.1111/bjd.19338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Doremalen N., Bushmaker T., Morris D.H., Holbrook M.G., Gamble A., Williamson B.N., et al. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N. Engl. J. Med. 2020;382:1564–1567. doi: 10.1056/NEJMc2004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020 doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hamming I., Timens W., Bulthuis M., Lely A., Gv Navis, van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]