Graphical abstract
Keywords: Adenosine analogue, Nucleoside, Methanocarba, Enterovirus, Antiviral, Structure activity relationship
Highlights
-
•
An in-house library of (N)-methanocarba nucleosides was screened against Enteroviruses.
-
•
A hit against Enterovirus A71, adenosine derivative 17, was structurally expanded providing an initial SAR.
-
•
The rigid bicyclo[3.1.0]hexane substitution of ribose was needed for antiviral activity.
-
•
A narrow SAR was found, not resembling other EV-A71 antiviral nucleosides.
-
•
Several novel nucleosides, including a 4′-fluoromethyl analogue MRS7704 48, had selectivity index >10 at EV-A71.
Abstract
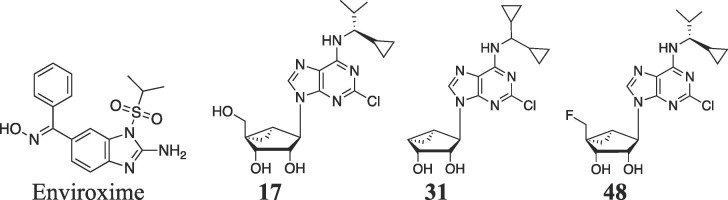
Various (North)-methanocarba adenosine derivatives, containing rigid bicyclo[3.1.0]hexane ribose substitution, were screened for activity against representative viruses, and inhibition was observed after treatment of Enterovirus A71 with a 2-chloro-N6-1-cyclopropyl-2-methylpropan-1-yl derivative (17). µM activity was also seen when testing 17 against other enteroviruses in the Picornaviridae family. Based on this hit, structural congeners of 17, containing other N6-alkyl groups and 5′ modifications, were synthesized and tested. The structure activity relationship is relatively narrow, with most modifications of the adenine or the methanocarba ring reducing or abolishing the inhibitory potency. 4′-Truncated 31 (MRS5474), 4′-fluoromethyl 48 (MRS7704) and 4′-chloromethyl 49 nucleosides displayed EC50 ~3–4 µM, and 31 and 48 achieved SI ≥10. However, methanocarba analogues of ribavirin and N6-benzyladenosine, shown previously to have anti-EV-A71 activity, were inactive. Thus, we identified methanocarba nucleosides as a new scaffold for enterovirus inhibition with a narrow structure activity relationship and no similarity to previously published anti-enteroviral nucleosides.
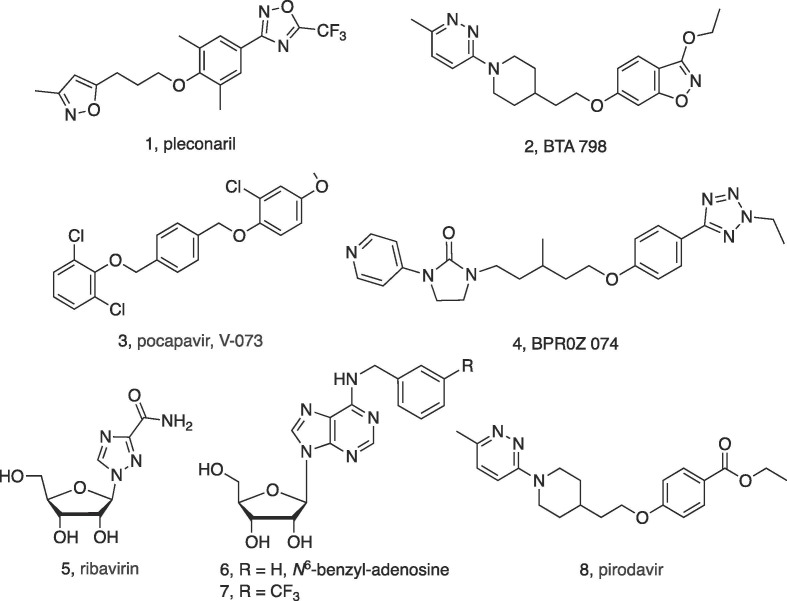
Human infection by members of the Picornaviridae family of enteroviruses is a continuing public health threat, leading to rashes and skin lesions (hand-foot-mouth disease) as well as neurological and respiratory symptoms that are often fatal.1 There have been periodic outbreaks caused by Enterovirus A71 (EV-A71) and Enterovirus D68, especially in Eastern Asia but also in Western countries. Recently, acute flaccid myelitis (AFM) associated with Enterovirus 68 has affected hundreds of children in the US.2 Poliovirus, which does have a vaccine, is a member of the same enterovirus family and causes poliomyelitis. The Picornaviridae are non-enveloped, positive-stranded RNA viruses. These viruses are usually spread by the fecal-to-mouth route and survive in the acidic gastric fluid before invading the lymphatic system. There are no approved potent antiviral small molecules for the enteroviruses.3, 4 Nevertheless, pleconaril 1 (Chart 1 , Phase 3 halted), BTA-798 2 (Phase 2b completed) and V-073 3 (Phase 2b) that block viral entry through the viral capsid have shown promise in clinical trials. An epidithio-diketopiperazine, KCN-21, potently inhibited several members of the Picornaviridae family.5, 6 Also, viral enzyme inhibitors, e.g. 3C protease and RNA-dependent RNA polymerase 3Dpol, and viral entry inhibitors are being developed.4, 7, 8
Chart 1.
EV-A71 antiviral compounds.
The crystal structure of EV-A71 revealed details of a lipidic pocket factor for regulating the uncoating process bound in solvent-accessible hydrophobic canyon, and this canyon might serve as a target region for small molecule inhibitors.9 P-selectin glycoprotein ligand-1 (PSGL-1) and scavenger receptor class B, member 2 (SCARB2) are putative cellular receptors on T cells that are associated with EV-A71 infection.10 There is no known effective treatment for EV-A71 infection,4 but a vaccine has shown efficacy in clinical trials.11 Small molecules that inhibit EV-A71 replication, such as pyridyl imidazolidinones (oxadiazoles),12 have been reported but not yet approved for clinical use.3, 13, 14, 15 Among a series of oxadiazoles, BPR0Z 074 4, inhibited the EV-A71 capsid protein with an IC50 of 0.8 nM.15 The known antiviral drug ribavirin 5, a nucleoside, is a weak inhibitor of EV-A71, of which resistant mutants have been identified, EV-D68 and the novel coronavirus SAR-CoV-2.4, 13, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18 Other synthetic nucleoside derivatives were found to interact with the EV-A71 2C peptide to inhibit viral infection more potently,19, 20, 21 but some of these nucleosides, such as N 6-benzyl-adenosine 6 (EC50 5 μM), are associated with a small selectivity index (SI) for antiviral activity compared to cell toxicity. Modification of the benzyl group, as in N 6-(3-trifluoromethyl)benzyl-adenosine 7 (EC50 0.068 μM), increased both the potency and SI.21 Other N 6- and C2′-derivatized adenine nucleosides showed moderate potency in inhibiting EV-A71.22, 23 None of the EV-A71 antivirals or vaccines are approved clinically, and although drug repurposing efforts are underway, novel antiviral agents for the Picornaviridae family are needed.24, 25
We have utilized a rigid bicyclic mimic of the ribose moiety for the design of nucleosides and nucleotides that bind to various G protein-coupled receptors (GPCRs) and transporters.26, 27, 28, 29, 30, 31 The methanocarba modification substitutes a bicyclo[3.1.0]hexane ring in place of the tetrahydrofuryl moiety of ribose. This modification was originally introduced by Marquez and colleagues for antiviral and anticancer therapeutics.32 One methanocarba isomer maintains the North (N)-conformation of the pseudoribose ring, and an alternate placement of the fused cyclopropyl ring maintains a South (S)-conformation. (N)-Methanocarba nucleosides are often associated with enhanced affinity at various target proteins, compared to the corresponding ribosides.26
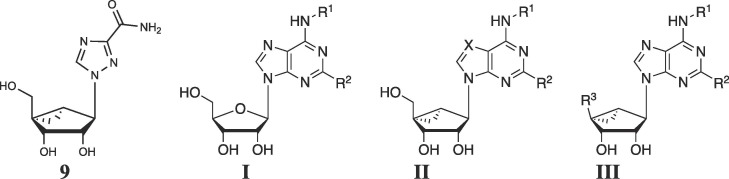
We screened adenosine derivatives from our in-house library to evaluate the applicability of the (N)-methanocarba nucleosides to viral inhibition including EV-A71 and other Picornaviridae. Based on the initial hit, the compounds in this study were either synthesized for this work, or samples were obtained from previous studies in our lab (Table 1 ).15, 16, 17, 18, 19 Several parallel series were compared depending on the substitution at the 4′ position: adenosine-like CH2OH (13 – 30), 4′-truncated (31 – 43), 4′-carbonyl (44 – 47) and substituted CH2OH (48 – 52) derivatives. Additional methanocarba nucleoside derivatives (53 – 59), including 7-deazaadenosine derivatives and (S)-methanocarba-uridine, that were found to be inactive EV-A71 and other viruses are listed in Table S1 (Supplementary data). We first examined the in vitro inhibition of the replication of EV-A71 in a cell-based assay and compared the potency (EC50) in inhibiting the Tainin/4643/98 strain with the CC50 (compound concentration that reduces cell viability by 50%) in Vero 76 (monkey kidney epithelial) cells.6, 33 The detailed procedures for these assays are described in Smee et al.5 Two antiviral parameters were measured: visual (virus yield reduction) and neutral red (cytopathy effect). The SI was calculated as the ratio of the toxic concentration, as CC50 divided by the viral inhibitory concentration, as EC50 (Table 1). Pirodavir 8 was used as a known standard antiviral drug and gave a SI ≥30.33
Table 1.
| Compound | R1 = | R2 =, other | Visual (µM)a |
Neutral red (µM)a |
||||
|---|---|---|---|---|---|---|---|---|
| EC50 | CC50 | SI50 | EC50 | CC50 | SI50 | |||
| 8 | [Pirodavir]c | – | 0.86 | >27 | >31 | 0.57 | 17 | 30 |
| 931 | – | – | >50 | >50 | b | >50 | >50 | b |
| formula I | ||||||||
| 1031 |  |
H | >50 | >50 | b | >50 | >50 | b |
| 1131 | 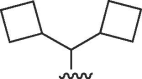 |
H | >100 | >100 | b | >100 | >100 | b |
| 1231 |  |
Cl | >32 | 32 | b | >27 | 27 | b |
| formula II (R2 = Cl, unless noted) | ||||||||
| 1329, 31 | H | Cl | >50 | >50 | b | >36 | 36 | b |
| 14 | 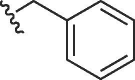 |
Cl | >100 | >100 | b | >100 | >100 | b |
| 15 | 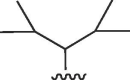 |
Cl | 32 | >100 | >3.1 | 32 | >100 | >3.1 |
| 1629, 31 | 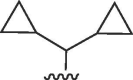 |
Cl | 10 | >100 | >10 | 3.2 | 68 | 21 |
| 1729 |  |
Cl | 7.7 EC90 | 24 | 3.1SI90 | 9.1 | 24 | 2.6 |
| 18 |  |
Cl, X = CH | 32 | >100 | >3.1 | 24 | 86 | 3.6 |
| 19 |  |
I | 18 | 100 | 5.6 | 20 | 46 | 2.3 |
| 20 |
 (R) (R) |
SCH3 | >100 | >100 | b | 66 | >100 | >1.5 |
| 2129 |
 (S) (S) |
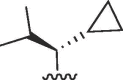
Cl | 12 | >50 | >4.2 | 2.3 | >50 | >22 |
| 22 |
 (S) (S) |
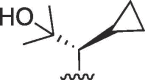
Cl | >100 | >100 | b | >100 | >100 | b |
| 23 |
 (S) (S) |
Cl, X = CH | >100 | >100 | b | >63 | 63 | b |
| 24 |
 (S) (S) |
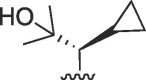
SCH3, X = CH | >100 | >100 | b | >100 | >100 | b |
| 25 |
 (R) (R) |
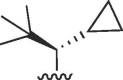
Cl | 32 | >100 | >3.1 | 31 | >100 | >3.2 |
| 2629 |
 (R) (R) |
Cl | 16 | >50 | >3.1 | 2.3 | >50 | >22 |
| 2729 |
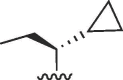 (S) (S) |
Cl | 16 | >50 | >3.1 | 5.3 | >50 | >9.4 |
| 2829 |
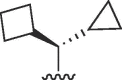 (R) (R) |
Cl | 16 | >50 | >3.1 | 8 | 13 | 1.6 |
| 2929 |
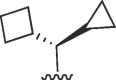 (S) (S) |
Cl | 16 | >50 | >3.1 | 9.3 | 14 | 1.5 |
| 3031 | 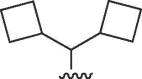 |
Cl | 16 | 50 | 3.1 | >28 | 28 | b |
| formula III, R3 = H | ||||||||
| 3127, 28, 31 | 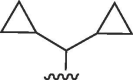 |
Cl | 3.6 | >100 | >28 | 3.4 | 34 | 10 |
| 3228 | 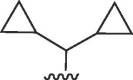 |
H | >50 | >50 | b | >50 | >50 | b |
| 3328 | 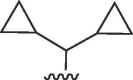 |
I | >50 | >50 | b | >50 | 50 | b |
| 3430 | 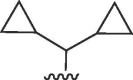 |
I,X = CH | >100 | 100 | b | 32 | 44 | 1.4 |
| 3528 | 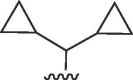 |
NHNH2 | >50 | >50 | b | >50 | 50 | b |
| 3628 | 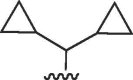 |
 |
>50 | >50 | b | >50 | 50 | b |
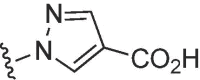
| 3728 | 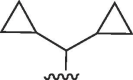 |
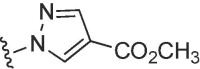 |
>50 | >50 | b | >50 | 50 | b |
| 3828 |
 (R) (R) |
Cl | 16 | >50 | >3.1 | 12 | >50 | >4.2 |
| 3928 |
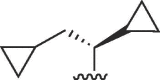 (R) (R) |
Cl | 21 | >50 | >2.4 | 8.1 | 9.9 | 1.2 |
| 4028 |
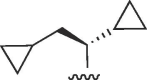 (S) (S) |
Cl | 18 | >50 | >2.8 | >7.6 | 7.6 | b |
| 4128 | 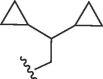 |
Cl | >50 | >50 | b | >10 | 10 | b |
| 4231 | 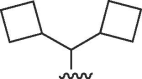 |
Cl | >16 | 16 | b | >17 | 17 | b |
| 4327 | 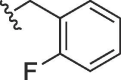 |
Cl | >50 | >50 | b | >23 | 23 | b |
| formula III, R3 = H | ||||||||
| 4429 | 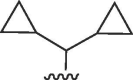 |
Cl, R3 = CO2CH2CH3 | >50 | >50 | b | >1.5 | 1.5 | b |
| 4529 | 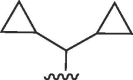 |
Cl, R3 = CO2H | >50 | >50 | b | >50 | 50 | b |
| 4629 | 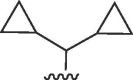 |
Cl, R3 = CONH(CH2)2NH2 | >50 | >50 | b | >50 | 50 | b |
| 4729 | 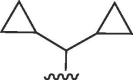 |
Cl, R3 = CONH(CH2)3NH2 | >50 | >50 | b | >50 | 50 | b |
| 48 |
 (R) (R) |
Cl, R3 = CH2F | 3.6 | 36 | 10 | 3.0 | 33 | 11 |
| 49 |
 (R) (R) |
Cl, R3 = CH2Cl | 4.3 | 32 | 7.4 | 3.9 | 27 | 6.9 |
| 50 |
 (R) (R) |
Cl, R3 = CH2SCH3 | >32 | 32 | b | 8.4 | 32 | 3.8 |
| 51 |
 (R) (R) |
Cl, R3 = CH2N3 | 5.3 | 32 | 6.0 | 4.8 | 32 | 6.7 |
| 52 |
 (R) (R) |
Cl, R3 = CH2NH2 | >32 | 32 | b | >32 | 32 | b |
Footnotes:
a - EC50 (or EC90 when indicated) - compound concentration that reduces viral replication by 50% (or 90%). CC50 ‐ compound concentration that reduces cell viability by 50%. SI50 (selectivity index) = CC50/EC50.
b - undetermined low value.
c - EC50 values of pirodavir were in the range of 0.036–0.32 (visual) and 0.056–0.22 (neutral red) µg/mL.
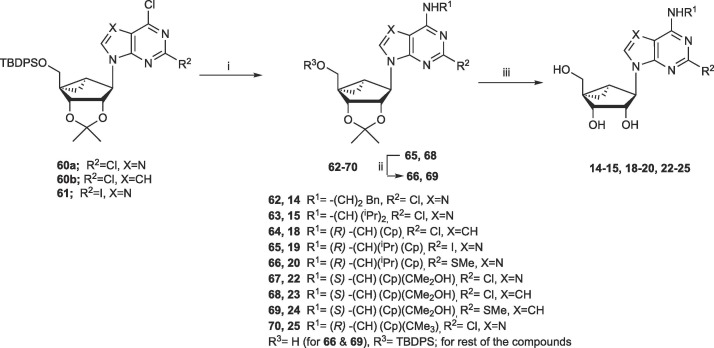
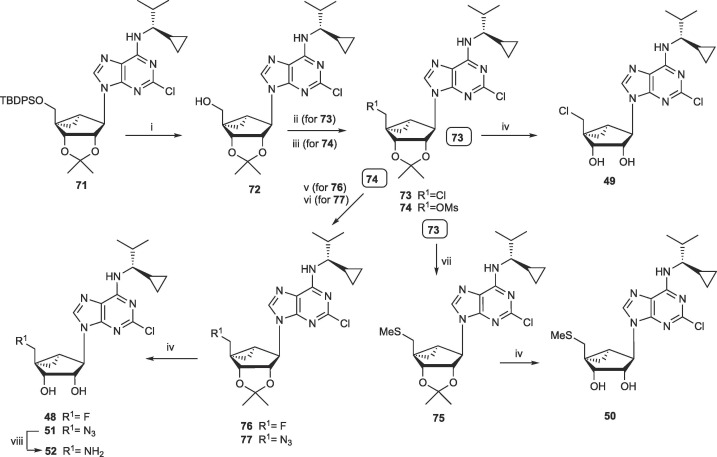
The synthetic routes to the novel (N)-methanocarba nucleoside analogues are shown in Scheme 1, Scheme 2 with procedures in Supplementary data. The syntheses of other analogues are found in structure activity relationship (SAR) studies of adenosine receptors or 5HT2B/5HT2C serotonin receptors as indicated in Table 1.27, 28, 29, 31 Additional compounds were either reported or utilized intermediates found in our earlier publications on opioid receptors or adenosine kinase inhibitors.30, 34 2′,3′-Isopropylidene-protected (N)-methanocarba intermediates 60, 61 and 71 were prepared as reported.28, 29, 30 For 5′-hydroxy nucleosides (with adenine or 7-deazaadenine as nucleobase), nucleophilic substitution of 6-chloro with various amines was performed, and a 2-chloro or 2-iodo was optionally substituted with a 2-methylthio group. For adenine nucleosides having groups other than OH at the 5′ position, functional group substitution was accomplished by nucleophilic attack on a 5′-chloro 73 or 5′-mesylate 74 intermediate.
Scheme 1.
Synthesis of 5′-hydroxy nucleosides (adenine and 7-deazaadenine). Reagents and Conditions: (i) R1NH2, DIPEA, 2-propanol, 70 ˚C (for X = N) or 140 ˚C, µwave (for X = CH); (ii) NaSMe, DMF, 90 ˚C; (iii) 10%TFA, MeOH, 70 ˚C.
Scheme 2.
Synthesis of 5′–non-hydroxy nucleosides. Reagents and Conditions: (i) TBAF, THF, rt; (ii) SOCl2, pyridine, CH3CN, −5 ˚C to rt; (iii) MsCl, pyridine, rt; (iv) 10% TFA, MeOH, 70 ˚C; (v) TBAF, THF, 70 ˚C; (vi) NaN3, DMF, 60 ˚C; (vii) NaSMe, DMF, 0 ˚C; (viii) PMe3, THF.
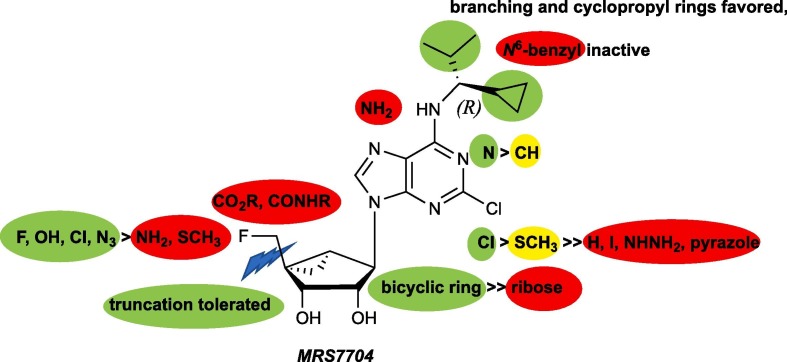
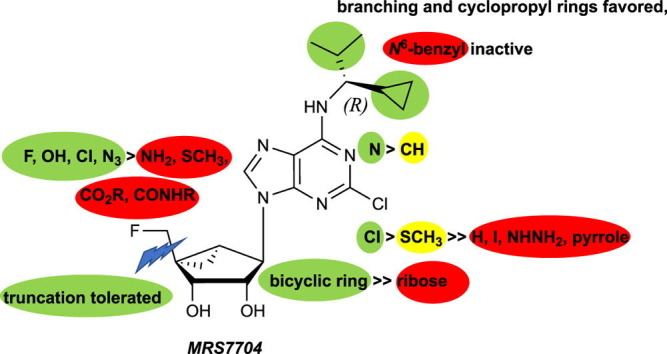
The SAR of (N)-methanocarba-adenosine derivatives as antiviral agents against Enterovirus A71 is summarized in Fig. 1 . Although ribavirin 5 weakly inhibited EV-71A viral yield in infected cells (IC50 of 65 µg/mL, 266 µM),17 the (N)-methanocarba analogue 9 of ribavirin31 was inactive at 50 µM and higher concentrations were not tested. However, various analogues (15 – 19, 21, and 25 – 29) in the (N)-methanocarba adenosine series containing a 5′–CH2OH displayed measurable potency in inhibition of EV-A71 replication, in two assays of the cytopathic effect of the virus. In the 4′–CH2OH series, compound 17 with a N 6-(R)-1-cyclopropyl-2-methylpropan-1-yl group inhibited viral replication with moderate potency as determined visually (Table 1, EC90 = 7.7 µM), and the corresponding (S) diastereoisomer 21 displayed EC50 values of 12 and 2.3 µM in the visual and neutral red assays, respectively. Other subtle N 6 group modifications of 17 produced EC50 values in the range of 2 – 31 µM (neutral red assay). Other potent nucleoside inhibitors with Formula II were compounds 16, 17 and 21. Slight enlargement of the rings of N 6-dicyclopropylmethyl analogue 16 to N 6-dicyclobutylmethyl in 30 prevented inhibition. Other analogues of 17 modified at the 5′ carbon indicated that fluoro 48, chloro 49, and azido 51 substitution of the hydroxyl group led to intermediate potency, but 5′-methylthio 50 and 5′-amino 52 substitution reduced inhibition. An N 6-benzyl analogue 14 with C2-Cl was inactive at 100 µM in both assays. Thus, the SAR surrounding hydrophobic alkyl or aryl groups at the N 6 position had distinct steric requirements.
Fig. 1.
SAR pattern of adenosine derivatives in antiviral activity against EV-A71, relative to 4′-fluoromethyl derivative 48.
The 4′-truncated (N)-methanocarba-adenosine analogues were originally synthesized as adenosine receptor ligands.27, 28 N 6-Dicyclopropylmethyl analogue 31 (MRS5474) was moderately active in inhibiting viral replication (neutral red EC50 = 3.4 µM), but replacement of its 2-Cl with H prevented this activity. Thus, we examined other N 6 modified analogues in both the truncated and the 4′–CH2OH series. The oxidized analogue 44 containing an ethyl ester group at the 4′ position was inactive. Homologation of 31 to the corresponding truncated N 6-dicyclobutylmethyl analogue 4 11 prevented activity, but the substitution with an N 6-(R)-1-cyclopropyl-2-methylpropan-1-yl group in 38 retained inhibition but with 4-fold less potency than the parent truncated compound 31.
Several ribose-containing 2-H analogues 10 and 11 were inactive against EV-A71. However, there was indication in the 4′-truncated series that a 2-Cl substituent enhances EV-A71 inhibition (31 vs. 32). Therefore, a direct comparison of (N)-methanocarba 2-Cl-adenosine analogue 30 and its ribose equivalent 12 indicated that the methanocarba ring of 30 promotes the antiviral activity.
Lead compound 17 was tested against three Picornaviridae viruses related to EV-A71, coxsackie virus B3, enterovirus 68 and poliovirus 1, and the EC50 values indicated moderate inhibition (Table 2 ). EC50 values were in the range of 16 – 32 µM. In addition, compounds 31 and 48 showed EC50/90 values of 8 – 16 µM against enterovirus 68, demonstrating activity as indicated by an SI >10. Also, more structurally diverse (N)- or (S)-methanocarba nucleosides (53 – 59) were evaluated in inhibition of EV-A71 replication (Table S1, Supplementary data), but all were inactive. Several nucleosides showed very weak activity (Table S2) against respiratory syncytial virus (31), Rift Valley fever virus (59) and Tacaribe virus (16).
Table 2.
| Compound | Visual (µM) |
Neutral red (µM) |
||||
|---|---|---|---|---|---|---|
| EC50, 90 | CC50 | SI50 | EC50 | CC50 | SI50 | |
| Coxsackie virus B3 (HA 201,933 strain, Vero76 cells) | ||||||
| Enviroxime | <0.01 | >10 | >1000 | <0.01 | 4.5 | >450 |
| 17 | 16 | >50 | >3.1 | 16 | >50 | >3.1 |
| Enterovirus 68 (US/KY/14–18953 strain, RD cells) | ||||||
| Enviroxime | 0.032 | 4.2 | 130 | 0.032 | 1.5 | 47 |
| 17 | 32 | >100 | >3.1 | 32 | >100 | >3.1 |
| 31 | 15 (EC90) | 41 | 2.7 | 9.3 | 41 | 4.4 |
| 48 | 7.7 (EC90) | 42 | 5.4 | 16 | 42 | 2.6 |
| Poliovirus 1 (Mahoney strain, Vero76 cells) | ||||||
| Enviroxime | 0.015 | >10 | >670 | <0.01 | 4.9 | >490 |
| 17 | 28 | >100 | >3.6 | 27 | >100 | >3.7 |
Off-target activities of these nucleosides are present, principally binding to adenosine and 5HT2B/C serotonin receptors, with affinities often in a similar range or greater than their antiviral potencies. Some of these other activities might be tolerable in antiviral agents. We previously reported that 2-Cl-adenine nucleoside 17 binds as an agonist to A1, A2A and A3 adenosine receptors (Ki values 103, 2200 and 1240 nM, respectively), and as an antagonist to the 5HT2B and 5HT2C receptors (Ki values 163 and 190 nM, respectively).29 Other adenosine receptor affinities, for 13, 16, 21, 26–29 and 44,29 for 12,31 and for 31–33, 35–41 and 43,28 were reported to be variable, but generally Ki > 0.1 µM. Truncated compound 31 was characterized as a moderately selective A1 agonist,28, 31 and compounds 11 and 12 were potent at the A1 receptor (Ki < 10 nM).31 Thus, there was no correlation between adenosine or serotonin receptor affinity of the reported compounds and their antiviral activity. The novel nucleosides were also tested systematically for off-target interactions at 45 different receptors, channels and transporters by the Psychoactive Drug Screening Program (PDSP, Supplementary data).35 Representative hits were 2-iodo 19 and 2-methylthio 20 analogues, which bound to the 5HT2B and 5HT2C receptors with Ki values in the range of 0.17 – 0.43 µM. Compound 15, which is a bond-opened form of 17, the 7-deaza analogue 18, and 5′-fluoro derivative 48 bound to the 5HT2B receptor with Ki values of 552 ± 167, 1310 ± 260 and 934 ± 66 nM, respectively.
In conclusion, we have discovered inhibition of EV-A71 by 17 and shown that other enteroviruses in the Picornaviridae family were similarly inhibited. We do not know the mode of interaction with the virus, and the approach must currently be through empirical SAR probing. The bicyclic ribose substitution and 2-Cl promote the viral inhibition. Thus, we have identified rigid (N)-methanocarba nucleosides as a new scaffold for enterovirus inhibition with a relatively narrow SAR. There was not a strong dependence of the viral inhibition on the N 6 group’s stereochemistry. Analogues that displayed more potent inhibition than the initial hit compound 17 include 4′-truncated 31, 4′-fluoromethyl 48 and 4′-chloromethyl 49 nucleosides that displayed EC50 values of ~3–4 µM. Compounds 31 and 48 achieved an SI of ≥10 by both criteria (visual and neutral red methods). Intermediate potency as was also associated with closely related analogues 16, 21, and 51. Larger N 6 groups, such as benzyl, and C2 substitution other than chloro resulted in inactivity. Thus, we have identified methanocarba nucleosides as a new scaffold for enterovirus inhibition and defined its initial SAR, which did not resemble previously published anti-enteroviral nucleosides, suggesting a site of action, or mode of binding that is different than reported nucleosides. Future studies will be needed to enhance both the antiviral potency and the selectivity of these nucleosides.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank the NIDDK Intramural Research Program (ZIADK31117) for support. We thank NIAID, Antiviral Discovery, Division of Microbiology and Infectious Diseases for providing antiviral testing under contract 75N93010D00021/Task Oder B01. We thank Bryan L. Roth, National Institute of Mental Health's Psychoactive Drug Screening Program (Univ. North Carolina at Chapel Hill, Contract # HHSN-271-2008-00025-C) for screening data.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bmcl.2020.127599.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Jones E., Pillay T.D., Liu F. Eur J Paediatr Neurol. 2018;22(5):763–773. doi: 10.1016/j.ejpn.2018.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Messacar K., Asturias E.J., Hixon A.M. Lancet Infectious Dis. 2018;18(8):e239–e247. doi: 10.1016/S1473-3099(18)30094-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benschop K.S., van der Avoort H.G., Duizer E., Koopmans M.P. Antivir Ther. 2015;20:121–130. doi: 10.3851/IMP2939. [DOI] [PubMed] [Google Scholar]
- 4.Bauer L., Lyoo H., van der Schaar H.M., Strating J.R.P.M., van Kuppeveld F.J.M. Curr Opin Virol. 2017;24:1–8. doi: 10.1016/j.coviro.2017.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smee D.F., Evans W.J., Nicolaou K.C., Tarbet E.B., Day C.W. Antivir Res. 2016;131:61–65. doi: 10.1016/j.antiviral.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nicolaou K.C., Lu M., Totokotsopoulos S. J Am Chem Soc. 2012;134:17320–17332. doi: 10.1021/ja308429f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van der Linden L., Wolthers K.C., van Kuppeveld F.J. Viruses. 2015;7:4529–4562. doi: 10.3390/v7082832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martínez-Gualda B., Sun L., Martí-Marí O. J Med Chem. 2020;63(1):349–368. doi: 10.1021/acs.jmedchem.9b01737. [DOI] [PubMed] [Google Scholar]
- 9.Plevka P., Perera R., Cardosa J., Kuhn R.J., Rossmann M.G. Science. 2012;336(6086):1274. doi: 10.1126/science.1218713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nishimura Y., Shimizu H. Front Microbiol. 2012;3:105. doi: 10.3389/fmicb.2012.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu F., Xu W., Xia J. New Engl J Med. 2014;370(9):818–828. doi: 10.1056/NEJMoa1304923. [DOI] [PubMed] [Google Scholar]
- 12.Shia K.-S., Li W.-T., Chang C.-M. J Med Chem. 2002;45(8):1644–1655. doi: 10.1021/jm010536a. [DOI] [PubMed] [Google Scholar]
- 13.H.J. Thibaut A.M. De Palma J. Neyts Biochem Pharmacol. 83 2 2012 185 192 10.1016/j.bcp.2011.08.016. [DOI] [PubMed]
- 14.Pourianfar HR, Grollo L. J Microbiol Immunol Infect. 2015;48(1):1–8. doi: 10.1016/j.jmii.2013.11.011. [DOI] [PubMed] [Google Scholar]
- 15.Shang L., Xu M., Yin Z. Antivir. Res. 2013;97(2):183–194. doi: 10.1016/j.antiviral.2012.12.005. [DOI] [PubMed] [Google Scholar]
- 16.Sadeghipour S., Bek E.J., McMinn P.C. J Virol. 2013;87(3):1759–1769. doi: 10.1128/JVI.02139-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Z.H., Li C.M., Ling P. J Infect Dis. 2008;197:854e7. doi: 10.1086/527326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khalili J.S., Zhu H., Mak N.S.A., Yan Y., Zhu Y. J Med Virol. 2020;92:740–746. doi: 10.1002/jmv.25798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drenichev M.S., Oslovsky V.E., Sun L. Eur J Med Chem. 2016;111:84–94. doi: 10.1016/j.ejmech.2016.01.036. [DOI] [PubMed] [Google Scholar]
- 20.Oslovsky V.E., Drenichev M.S., Sun L. Molecules. 2017;22:1219. doi: 10.3390/molecules22071219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arita M., Wakita T., Shimizu H. J Gen Virol. 2008;89(10):2518–2530. doi: 10.1099/vir.0.2008/002915-0. [DOI] [PubMed] [Google Scholar]
- 22.Tararov V.I., Tijsma A., Kolyachkina S.V. Eur J Med Chem. 2015;90:406–413. doi: 10.1016/j.ejmech.2014.11.048. [DOI] [PubMed] [Google Scholar]
- 23.Shang L., Wang Y., Qing J. Antivir Res. 2014;112:47–58. doi: 10.1016/j.antiviral.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 24.Sun J., Yogarajah T., Lee R.C.H. Sci Rep. 2020;10:8159. doi: 10.1038/s41598-020-65152-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin J., Kung Y., Shih S. J Biomed Sci. 2019;26:65. doi: 10.1186/s12929-019-0560-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacobson K.A., Tosh D.K., Toti K.S., Ciancetta A. Drug Disc Today. 2017;22:1782–1791. doi: 10.1016/j.drudis.2017.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tosh D.K., Phan K., Deflorian F., Wei Q., Gao Z.G., Jacobson K.A. ACS Med Chem Lett. 2011;2:626–631. doi: 10.1021/ml200114q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tosh D.K., Paoletta S., Deflorian F. J Med Chem. 2012;55:8075–8090. doi: 10.1021/jm300965a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tosh D.K., Ciancetta A., Warnick E., Crane S., Gao Z.G., Jacobson K.A. J Med Chem. 2016;59:11006–11026. doi: 10.1021/acs.jmedchem.6b01183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tosh D.K., Ciancetta A., Mannes P. ACS Omega. 2018;3:12658–12678. doi: 10.1021/acsomega.8b01237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tosh D.K., Rao H., Bitant A. J Med Chem. 2019;62:1502–1522. doi: 10.1021/acs.jmedchem.8b01662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marquez VE. The properties of locked methanocarba nucleosides in biochemistry, biotechnology and medicine. In Modified Nucleosides in Biochemistry, Biotechnology and Medicine (Herdewijn, P., ed.), 2008;307–341, Wiley-VCH.
- 33.Barnard D.L., Hubbard V.D., Smee D.F. Antimicrob Agents Chemother. 2004;48:1766–1772. doi: 10.1128/AAC.48.5.1766-1772.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Toti K.S., Osborne D., Ciancetta A., Boison D., Jacobson K.A. J Med Chem. 2016;59:6860–6877. doi: 10.1021/acs.jmedchem.6b00689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Besnard J., Ruda G.F., Setola V. Nature. 2012;492:215–220. doi: 10.1038/nature11691. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.