Abstract
Dysregulated Th17 cell differentiation is associated with autoimmune diseases such as multiple sclerosis, which has no curative treatment. Understanding the molecular mechanisms of regulating Th17 cell differentiation will help find a novel therapeutic target for treating Th17 cells-mediated diseases. Here, we investigated the cell-intrinsic processes by which RNA-binding protein HuR orchestrates Th17 cell fate decisions by post-transcriptionally regulating transcription factor Irf4 and Runx1 and receptor Il12rb1 expression, in turn promoting Th17 cell and Th1-like Th17 cell differentiation in C57BL/6J mice. Knockout of HuR altered the transcriptome of Th17 cells characterized by reducing the levels of RORγt, IRF4, RUNX1, and T-bet, thereby reducing the number of pathogenic IL-17+IFN-γ+CD4+ T cells in the spleen during experimental autoimmune encephalomyelitis (EAE). In keeping with the fact that HuR increased the abundance of adhesion molecules VLA-4 on Th17 cells, knockout of HuR impaired splenic Th17 cell migration to the central nervous system and abolished the disease. Accordingly, targeting HuR by its inhibitor DHTS inhibited splenic Th17 cell differentiation and reduced EAE severity. In sum, we uncovered the molecular mechanism of HuR regulating Th17 cell functions, underscoring the therapeutic value of HuR for treatment of autoimmune neuroinflammation.
INTRODUCTION
Multiple sclerosis (MS) is an autoimmune inflammatory disease of the central nervous system (CNS) (1). Experimental autoimmune encephalomyelitis (EAE) is the animal model most widely used to investigate MS pathology and potential treatment. Accumulating evidence has demonstrated that both Th17 cells and Th1 cells are able to induce pathogenesis of EAE, albeit through different mechanisms (2-5). Currently, there is no curative treatment for MS. Further understanding the molecular mechanism underlying Th17 cell differentiation will help find a novel therapeutic target for MS.
Transcriptional gene regulation of Th17 and Th1 cell differentiation and function are well studied. During the cytokine-mediated Th17 cell differentiation, the two orphan nuclear receptors, RORγt (RORC) and RORα (RORA) and transcription factor STAT3, jointly regulate Th17 cell differentiation (6-8). In addition, several other transcriptional factors also participate in Th17 cell differentiation, including IRF4 (9). RUNX1 influences Th17 cell differentiation by inducing RORγt expression and by jointly driving IL-17 (IL-17A) transcription (10). A more recent report revealed that the key transcription factor TBX21 (T-bet) in Th1 cells is required for the ontogeny of pathogenic interferon-γ-producing Th17 cells in autoimmune encephalomyelitis (11).
In the immune system, T cell responses following activation are driven by the rapid induction of cytokines and chemokines involving both transcriptional and post-transcriptional regulation (12). However, it remains unknown how Th17 cell differentiation is post-transcriptionally regulated by RNA-binding proteins in autoimmune diseases. Considering the importance that post-transcriptional regulation modulates gene expression for quick responses to environmental stimuli, and that the abundance of mRNA is determined by two rates: transcription rate and decoy rate, there has been a strong interest in the post-transcriptional gene regulation of immune cell responses (12-17). HuR (ELAVL1) expressing ubiquitously in all tissues, is a critical post-transcriptional regulator of gene expression in cancer and immune cells (14,18-25). HuR binds to target mRNAs that contain U- and AU-rich sequences in the 3’ untranslated regions (3’UTRs) to prolong their lives, such as il17, Csf2, and Ccr6 in Th17 cells(18,26-28). Here, we have investigated that HuR influenced Th17 cell fate by controlling its transcripts of transcription factors and receptors. Mechanistically, HuR stabilized Irf4 and Runx1 mRNAs and prolonged their half-lives, therefore enhanced their expression, which in turn promoted the expression of RORγt and facilitated Th17 cell differentiation (11). Furthermore, HuR directly and indirectly regulated IL-12Rβ1 and T-bet expression, respectively, as well as VLA-4 expression. Accordingly, genetic ablation of HuR impaired pathogenic Th17 and Th1-like Th17 cell differentiation and migration to CNS, abrogated the severity of EAE. Finally, targeting HuR by its inhibitor DHTS was effective for delaying the onset and reducing EAE severity. These results support the notion that HuR might be a potential target for treatment of MS.
MATERIALS AND METHODS
Animals
HuRflox/flox mice were kindly provided by Dr. Ulus Atasoy (University of Missouri-Columbia). Eight to twelve week-old control (HuRflox/flox) mice and HuR conditional knockout mice (OX40-Cre HuRflox/flox) were used. OX40-Cre and Rag1−/− mice were purchased from Jackson Laboratory. All mice are on the C57BL/6J background and were breed at the animal facility of Thomas Jefferson University. Animal experiments were approved by the Institutional Animal Care and Use Committee and performed following federal and institutional guidelines. Both male and female mice were used in the experiments.
Actively induced EAE
Eight to twelve week-old WT were immunized with MOG35-55 (150 μg) and CFA, followed by Pertussis toxin (PTX) injection (300 ng/mouse) by i.p. at day 0 and 2 post-immunization. EAE score was monitored as previously described (29). For Dihydrotanshinone-I (DHTS) treatment, starting at 5 or 15 days after MOG immunization, mice received injection of DHTS (Sigma, D0947) (10mg/kg) dissolved in PBS or vehicle control every 48 hrs as literature reported (30). At the end of experiments, mice were euthanized and spinal cords were harvested for histopathology assay or CNS inflammatory cell isolation.
Adoptive transfer of EAE
Splenocytes and lymph nodes from 8- to 12-week old WT (HuRf/f) and HuR KO (OX40-Cre.HuRf/f) female mice were collected 11 days later following MOG35-55(150 μg) in CFA immunization. Cells from spleens and lymph nodes were stimulated with MOG (20 μg/ml) and murine IL-23 (20 ng/ml) for 3 days for further Th17 cell polarization(3,26). For further Th1 cell polarization, MOG (20 μg/ml), murine IL-12 (5 ng/ml), and IFN-γ (2 ng/ml)(2) were used. CD4+ T cells were then isolated by a CD4+ T cell isolation kit (Miltenyi Biotech), and 6 to 7x106 CD4+ T cells were transferred to 10 to 12 week old Rag1−/− mice by i.v. injection. The recipients were given 300 ng pertussis toxin i.p. on days 0 and 2 post-transfer. Clinical disease was monitored daily on a scale of 0-5 as follows: no disease, 0; partially limp tail, 1; limp tail and weakness of hind legs, 2; limp tail, complete paralysis of hind legs, 3; limp tail complete hind leg and one front leg paralysis, or paralysis of trunk, 4; moribund state or death, 5. Mean clinical scores were compared using Student’s t test.
Isolation and differentiation of CD4+ T cells in vitro
Naïve CD4+ T cells were purified from splenocytes using naïve CD4 isolation kits (#19765, STEMCELL Tech., Canada or Miltenyi Biotech.) following the manufacturer’s protocol. Purified naïve CD4+ T Cells were activated with plate-bound 5 μg/ml anti-CD3 (17A2, Thermo Fisher Scientific) and 2 μg/ml anti-CD28 (37.51, Thermo Fisher Scientific). For Th17 polarization, 0.5 ng/ml TGF-β (Thermo Fisher Scientific), 20 ng/ml IL-6 (Peprotech), 20 ng/ml IL-23 (Thermo Fisher Scientific), 10 μg/ml anti-IFN-γ (Thermo Fisher Scientific)(XMG1.2, Bio-X-cell) and 10 μg/ml anti-IL-4 (11B11,Thermo Fisher Scientific)(Bio-X-cell) were used additionally. For mRNA stability measurement, 3 μg/ml actinomycin D (MilliporeSigma) was added to the Th17 cell culture to stop nascent mRNA transcription on day 5 following activation. Cells were collected at 0 to 5 h after actinomycin D treatment, and the RNA was isolated from the cells by using TRIzol extraction. RT-qPCR was performed as described previously (26,28). The amount of RNA at 0 h was set to 100%.
RNA isolation and Quantitative real time-PCR
Cells were collected and total RNA was extracted using TRIzol (Invitrogen, Thermo Fisher Scientific). Five hundred ng of RNA was reverse-transcribed (RT) into cDNA using SuperScript III Kit (Invitrogen, Thermo Fisher Scientific) following the manufacturer’s protocols. The resulting cDNA template was subjected to real-time quantitative (q)PCR analysis using CFX96 Real-Time PCR Detection system (Bio-RAD) with SYBR Green reagent Kit (Invitrogen, Thermo Fisher Scientific) following the manufacturer’s protocols. The levels of specific mRNAs were normalized to levels of Gapdh mRNA in each sample. Forward and reverse primers for specific murine target genes were published (26-28).
RNA-seq DE
One hundred ng of total RNA was used to prepare libraries using TruSeq Stranded Total RNA kit (Illumina, CA, USA) following the manufacturer’s protocol. The final libraries at a concentration of 4 nM were sequenced on NextSeq 500 using 75-bp paired-end chemistry. Raw FASTQ sequencing reads were mapped against the reference genome of mouse Ensembl Version GRCm38 utilizing further information from the gene transfer format (.gtf) annotation from GENCODE version GRCM18 using RSEM. Total read counts and normalized Transcripts Per Million (TPM) were obtained using RSEM’s calculate-expression function. Before determining differential expression levels, batch effects and sample heterogeneity were tested using iSeqQC (https://github.com/gkumar09/iSeqQC). Differential gene expression was tested between KO and WT samples using the DESeq2 package in R/Bioconductor. Genes were considered differentially expressed (DE) if they had adjusted p ≤ 0.05 and absolute fold change ≥ 2. All the plots were constructed using R/Bioconductor.
Gene Set Enrichment Analysis (GSEA) was performed to evaluate Gene Ontology Biological Process (GOBP) terms and markers of cell migration in the resulting differential expression lists. The DESeq2 test statistic was used as a ranking metric to perform GSEA in pre-ranked mode, with genes having zero base mean or “NA” test statistic values filtered out to avoid providing numerous duplicate values to GSEA. GSEA pre-ranked analysis was performed using the “weighted” enrichment statistic. Cytoscape analysis was performed on the selected genes to examine their network patterns using Reactome functional interaction network.
Western blotting
Whole-cell lysates were prepared and Western blot analysis performed as described (26,28). The concentration of protein was determined with BCA protein assay reagent (Thermo Scientific Pierce, Rockford, IL, USA). Protein samples were size-separated by electrophoresis through 10% sodium dodecyl sulphate-polyacrylamide gels (SDS-PAGE) and transferred to nitrocellulose membranes (Bi-Rad, Hercules, CA, USA). Membranes were blocked and blotted with antibodies recognizing HuR (clone 3A2) (SC-5261, Santa Cruz Biotech), RORγt (ab138043, Abcam), β-Actin (SC-47778, Santa Cruz Biotech), IRF4 (SC-130921, Santa Cruz Biotech), RUNX1 (SC-365644, Santa Cruz Biotech), IL12Rβ1(ab97813, Abcam), T-bet (644802, Biolegend), or GAPDH (SC-32233, Santa Cruz Biotech), followed by detection using goat anti-mouse or goat anti-rabbit Ig conjugated with HRP secondary antibody (Jackson ImmunoResearch Lab. Inc., West Grove, PA 19390). Membranes were developed by Signal West Pico Chemiluminescent Substrate (Thermo Scientific Pierce, Rockford, IL, USA).
Intracellular cytokine staining and flow cytometry
Cells obtained from in vitro culture or isolated from CNS of mice with EAE were incubated for 4 h with 50 ng/ml PMA (Sigma), 500 ng/ml ionomycin (MilliporeSigma), and GolgiPlug (1 μg per 1x106 cells, BD Bioscience). Cells were stained for surface markers, then fixed in Fix & Perm Medium A (Thermo Fisher Scientific), Fix & Medium B (Thermo Fisher Scienctifc), and stained for intracellular cytokines. The following antibodies were from Biolegend, FITC or APC-anti-CD4 (GK1.5), APC or PE-anti-IL17A (TC11-18H10.1), FITC or PE-anti-IFN-γ (XMG1.2), PE-anti-RORγt (AFKJS-9), PE-anti-RUNX1 (RXDMC), PE-anti-VLA-4 (9c10), APC/Cy7-anti-CD45 (30-F11), PE-anti-IL-2 (JES6-5H4), PE-anti-IL-10 (JES5-16E3). Percp/Cy5-anti-GM-CSF (MP1-22E9), APC-anti-Foxp3 (FJK-16s), and PE-anti-Ki67(SolA15) were purchased from Thermo Fisher Scientific. PE-IL-12Rb1(551974) was from BD Biosciences. All antibodies were used at dilution of 1:50 to 1:100 as instructed by the manufacturers. Data were collected with BD FACSARIA fusion flow cytometry (BD Biosciences) and analyzed by using Flowjo software (v10, TreeStar).
Immunoprecipitation of endogenous messenger RNP complexes (RIP)
RIP was performed according to an established protocol (26). Briefly, Th17-polarized cells were lysed using polysome lysis buffer. Beads (50 μl) were coated by adding 30 μg of either IgG1 (BD Biosciences) as control or anti-HuR antibody (3A2), and incubated overnight at 4°C. After extensive washes, 100 μl of pre-cleared lysate was added and incubated for 4 h at 4°C with additives, and then 0.5 mg/ml of proteinase K was added and incubated for 30 min at 55°C to digest protein. After extraction, RNA was reverse-transcribed (RT) and amplified by real-time quantitative (q) PCR analysis to assess the presence of specific target mRNAs.
Polysome fraction Assay
WT and HuR KO Th17 cell pellets were lysed using polysome extraction buffer (20 mmol/L Tris-HCl, pH 7.5, 100 mmol/L KCl, 5 mmol/L MgCl2, 0.3% Igepal CA-630, protease inhibitors, and 0.1 mg/mL cycloheximide). After centrifugation to remove insoluble material, the lysate was overlaid on a 10% to 50% sucrose gradient. After ultracentrifugation at 39,000 rpm at 4°C, 1-ml fractions were obtained on a density gradient fractionation system. Absorbance (A254) was measured during the entire fractionation process. Total RNA was extracted from each fraction using TRIzol (Invitrogen, Thermo Fisher Scientific) and analyzed by RT-qPCR analysis.
Human naïve CD4+ T cells purification
Blood samples were obtained from the Biological Specialty Corporation, PA. Mononuclear cells were prepared from the buffy coats from healthy adult donors on Ficoll-paque plus gradients. Naïve CD4+ T cells were further isolated by using EasySep Human naïve CD4+ T cell isolation kit II (Cat. # 17555, Stemcell Technologies) according to the manufacture’s instruction. The purity of naïve CD4+ T cells (CD3+CD4+CD45RA+CD45RO−) was around 95%.
Human CD4+ T Cell culture and lentiviral transduction
Cells were cultivated with completed T cell medium (RPMI1640 medium supplemented with 10% FBS (Hyclone). For transduction experiments, human naïve CD4+ T cell were seeded on day 0 at a density of 1x106 cell per ml in 24-well plates with polybrene (10μg/ml). Lentiviral HuR shRNA or scranmble shRNA (Sigma-Aldrich) were added at a multiplicity of infection of 1-10. Cells were washed on day 1. Puromycin (2μg/ml) was added on day 2. For Th17 cell polarization experiments, human CD4+ T cells were stimulated with plated coated anti-CD3 (5μg/ml) plus soluble anti-CD28 (1μg/ml), IL-1β(20ng/ml), IL-6 (20ng/ml), TGF-β (1ng/ml), IL-23(20ng/ml) , anti-IL-4(10μg/ml), and anti-IFN-γ(10μg/ml). Cells were collected on day 6 to 10 for RT-qPCR and Western blots.
Statistical analysis
Student’s t-test was used to analyze differences between two groups. The data are expressed as the mean ± standard error of the means (SEM). A p-value < 0.05 was considered statistically significant.
Data availability
All the RNA-seq data have been deposited in the BioProject database under the accession ID PRJNA589592 (https://www.ncbi.nlm.nih.gov/bioproject/589592). All data generated or analyzed during this study are included in this published article and its supplementary information files.
RESULTS
HuR KO CD4+ T cells are defective in triggering autoimmune encephalomyelitis
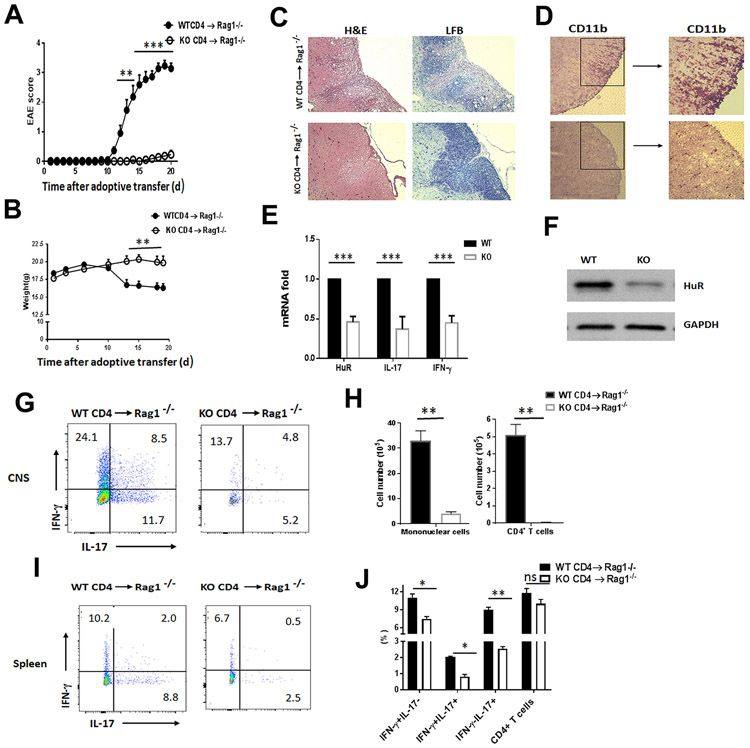
HuR plays an important role in immune cells (14,22,31) and is required for the initiation of EAE (26). Because OX40 expressed on CD4+ T cells after activation, HuR was not completely deleted in CD4+ T cells of OX40-Cre+/−.HuRf/f mice, which still play a partial role in actively induced EAE (26). To further understand the role of HuR in CD4+ T cells for inducing EAE, we did adoptive transfer of MOG-activated CD4+ T cells to Rag1−/− mice, which do not have endogenous T or B lymphocytes, keeping away from recipient endogenous lymphocytes contributing to pathogenesis of EAE (26,32). Splenocytes taken from MOG-immunized WT (HuRf/f) and HuR KO (OX40-Cre+/−.HuRf/f) mice at day 11 post-immunization were further activated with MOG and IL-23 under conditions of Th17 polarization (2,3,26). Three days later, purified MOG-activated CD4+ T cells (Th17 cells) were transferred into Rag1−/− mice. The clinical score of EAE was monitored daily according to reported criteria (4,26,33,34). All recipients that received WT Th17 cells developed very severe EAE early after transfer, whereas, only 3 out of 11 Rag1−/− recipients injected with HuR KO Th17 cells developed very mild EAE (0.5 to 1.5 score) at later times (Fig. 1A). The body weights of Rag1−/− recipients receiving WT Th17 cells were also significantly lower than those receiving HuR KO Th17 cells (Fig. 1B). Histopathological analysis showed that there were significantly decreased infiltrating inflammatory cells and reduced demyelination (Luxol fast blue (LFB) staining) in spinal cords of Rag1−/− mice receiving HuR KO Th17 cells, but not those receiving WT Th17 cells (Fig. 1C, 1D), suggesting that HuR KO CD4+ T cells cannot induce autoimmune neuroinflammation.
Figure 1. Genetic deletion of HuR in CD4+ T cells impairs their capacity to induce adoptive transfer of EAE.
(A) Mean clinical score of EAE in Rag1−/− mice that received MOG-activated WT and HuR KO (KO) Th17 cells (7 × 106/per mouse) (n=11 mice per group). (B) Body weight of Rag1−/− recipients with WT or HuR KO Th17 cells. (C, D) Hematoxylin and eosin (H&E), Luxol fast blue (LFB), and immunohistochemical staining (anti-CD11b) of spinal cords from Rag1−/− recipients of WT and HuR KO Th17 cells at the peak of disease at day 19. (E) Reverse transcription followed by quantitative PCR (RT-qPCR) analysis of Il17 and Ifng mRNA expression levels in MOG-activated HuR KO CD4+ T cells comparing with WT CD4+ T cells before transfer to Rag1−/− recipients. (F) Western blot analysis of the levels of HuR in MOG-activated WT and HuR KO CD4+ T cells before transfer to Rag1−/− recipients. (G-J) Analysis of proinflammatory cytokine produced by infiltrating CD4+ T cells in spinal cords (G) and spleens (I) of Rag1−/− recipients at the peak of disease at day 19 (n=5 mice) using flow cytometry; data summarizing three independent experiments of CNS infiltrating inflammatory cells (H) and CD4+ T cells (J) in the spleens of Rag1−/− recipients. *, p<0.05, ** p<0.01, *** p<0.001.
To understand why HuR KO CD4+ T cells were less functional in inducing EAE than WT CD4+ T cells, we characterized the proinflammatory cytokine expression by MOG-activated CD4+ T cells before cell transfer to Rag1−/− mice. Flow cytometry analysis indicated that the frequency of IFN-γ+CD4+ and IL-17+CD4+ T cells in HuR KO splenocytes was much lower than in WT splenocytes (fig. S1A, S1B). The same was true for the frequency of GM-CSF+CD4+ T cells (fig. S1C) and when analyzing inguinal lymph nodes (Data not shown). Given that IFN-γ+IL-17+CD4+ T cells are pathogenic inflammatory cells in EAE(11), the frequency of IFN-γ+IL-17+ CD4+ T cells was significantly reduced in the spleen of MOG-immunized HuR KO mice compared with WT mice (fig. S1A, S1B). Reverse transcription (RT) followed by quantitative (q) PCR analysis showed that the expression of Ifng and il17 mRNAs was much lower in HuR KO CD4+ T cells than WT CD4+ T cells (Fig. 1E). Western blot analysis confirmed the production of HuR was reduced in MOG-activated HuR KO CD4+ T cells before transfer to Rag1−/− mice (Fig. 1F).
To characterize the proinflammatory cytokines in CNS during EAE, we isolated the infiltrating inflammatory cells from the CNS in Rag1−/− recipients at the peak of disease around day 19. As expected, the number of monocytes was significantly reduced in the CNS of recipients injected with HuR KO CD4+ T cells than that of WT CD4+ T cells, so did the number and frequency of IFN-γ+, IFN-γ+IL-17+, and IL-17+CD4+ T cells in the CNS (Fig. 1G, 1H), which was consistent with the histopathological analysis of mice with EAE (Fig. 1C, 1D). In the spleen of Rag1−/− recipients, although the numbers of WT and KO CD4+ T cells were comparable, the percentage of CD4+ T cells secreting IFN-γ and IL-17 in WT CD4+ T cells was much higher than in HuR KO CD4+ T cells (Fig. 1I, 1J). Further comparison of WT and HuR KO CD4+ T cell proliferation in the spleen of Rag1−/− recipients by flow cytometry showed that knockout of HuR only reduced CD4+ T cell proliferation slightly relative to WT CD4+ T cells (fig. S1D, S1E). Thus, these results indicated that HuR deficient CD4+ T cells have impaired production of IL-17 and IFN-γ. Collectively, these data suggest that HuR is required for induction of autoimmune neuroinflammation by CD4+ T cells.
Knockout of HuR alters the transcriptome in Th17 cells
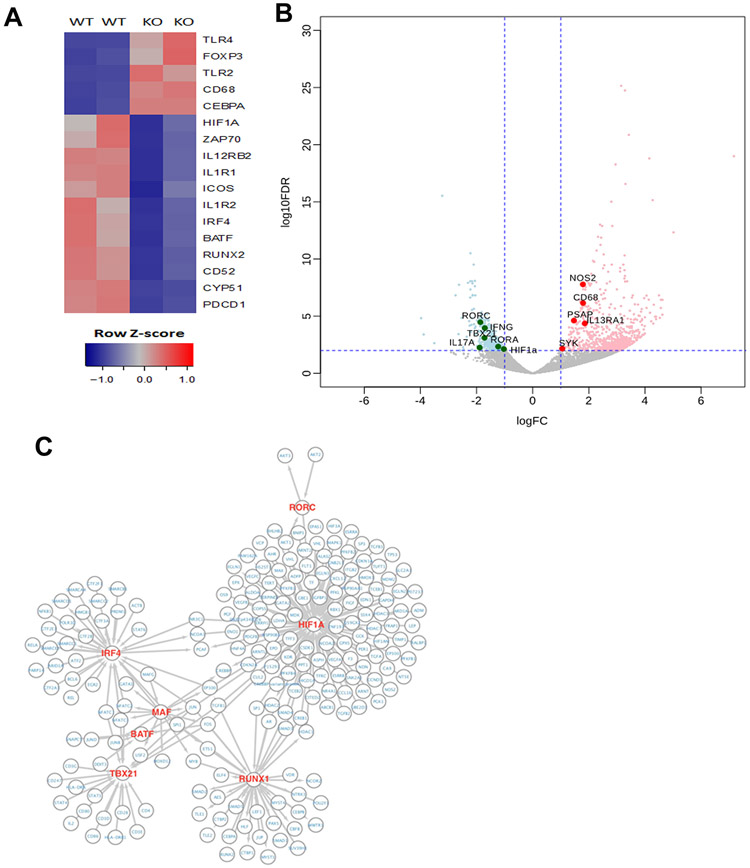
To investigate the post-transcriptional regulation of Th17 cell differentiation by HuR in EAE, we performed RNA-sequencing (RNA-seq) analysis to compare gene expression programs between MOG-activated WT and HuR KO Th17 cells (Fig. 2A, fig. S2). We found that 576 mRNAs were less abundant and 847 mRNAs were more abundant in HuR KO Th17 cells compared with WT Th17 cells (absolute log fold change ≥2; adjusted p-value ≤ 0.05) (Fig. 2B). Several transcripts encoding Th17 cell-related proteins, such as Il17, Ifng, Rora, Rorc, Tbx21, Cyp51 and Runx2 mRNAs were among those downregulated (Fig. 2A, 2B, fig. S2), while Il13r1, Tlr4, Nos2, and Foxp3 mRNAs were among those upregulated. By comparing the RNA-seq data of HuR KO relative to WT Th17 cells, we identified a network of transcription factors regulated by HuR (Fig. 2C). In line with the function of HuR KO CD4+ T cells in autoimmune neuroinflammation and flow cytometry data (Fig. 1G, 1I, fig. S1A, S1B), the abundance of Ifng and Il17 mRNAs was significantly reduced in HuR KO Th17 cells compared with WT Th17 cells. In sum, knockout of HuR severely impaired differentiation-related gene expression in Th17 cells.
Figure 2. HuR deficiency disrupts Th17 cell differentiation.
(A) Heat map of RNA-seq analysis of mRNAs that were more (red) or less (blue) abundantly expressed in MOG-reactivated HuR KO and WT Th17 cells before transfer to Rag1−/− recipients. (B) Volcano plot of mRNAs elevated (red) or reduced (green) in HuR KO Th17 cells compared with WT Th17 cells. (C) Network of transcription factors regulated directly or indirectly by HuR in Th17 cells derived from RNA-seq analysis.
HuR increases the levels of transcription factors RORγt and IRF4
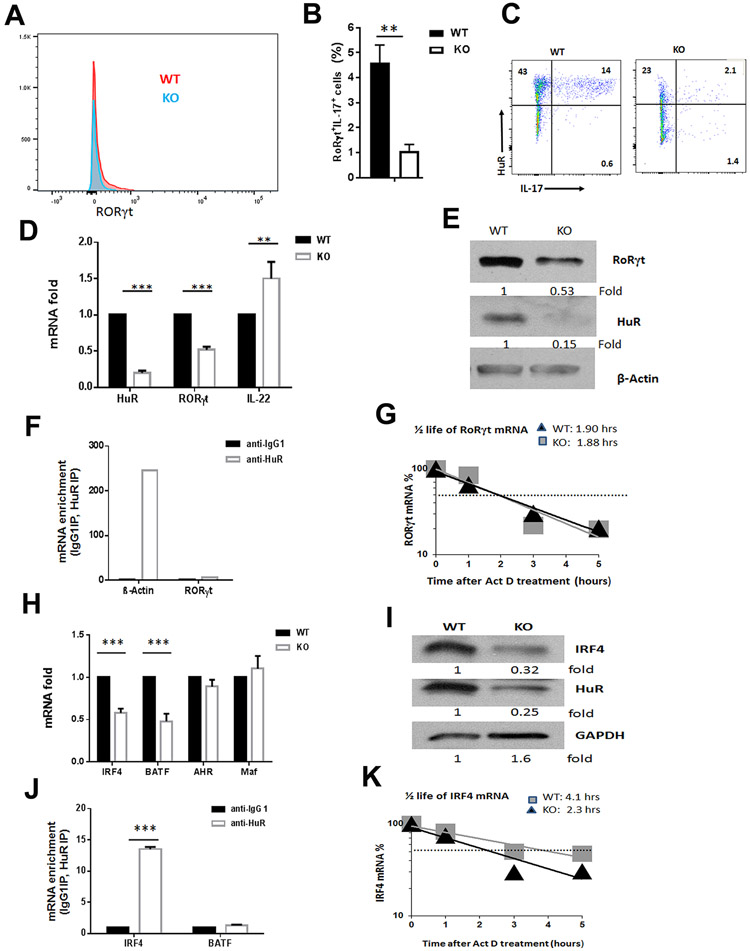
RORγt is a key transcription factor that orchestrates the differentiation of naïve CD4+ T cells into Th17 cells (6). Accordingly, targeting RORγt by its synthetic ligand suppresses Th17 cell differentiation and autoimmunity (35). Since cytokines including IL-17, IFN-γ and GM-CSF were significantly reduced in activated HuR KO CD4+ T cells compared with WT CD4+ T cells (Fig. 1G-J, fig. S1A-C), we sought to dissect the mechanisms by which HuR regulates Th17 cell function. To test whether HuR regulated the expression of transcription factor RORγt in Th17 cells, we assessed RORγt protein levels in MOG-activated Th17 cells. Flow cytometry analysis revealed that the level of RORγt in CD4+ T cells declined dramatically in the spleens of HuR KO mice compared with WT mice (Fig. 3A, 3B), implicating that HuR promoted RORγt expression. The data also showed that most HuR-negative CD4+ T cells in MOG-activated HuR KO mice did not produce IL-17 (Fig. 3C), which is consistent with our previous report that HuR directly modulates IL-17 expression (26).
Figure 3. Knockout of HuR reduces RORγt and IRF4 expression in MOG activated Th17 cells.
(A to C) Flow cytometric analysis of RORγt in MOG-activated WT and HuR KO Th17 cells before transferring to Rag1−/− mice. (D, E) The expression of Rorγt mRNA and RORγt protein in WT and HuR KO CD4+ T cells cultured under Th17-cell polarizing condition were examined by RT-qPCR (D) and Western blot (E) analyses, respectively. (F) RNP immunoprecipitation (RIP) followed by RT-qPCR analysis was performed to examine the relative amount of mRNA associated with HuR protein. B-actin mRNA that associated with HuR protein served as the positive control of RIP assay. (G) The relative amount of Rorγt mRNA in WT and HuR KO Th17 cells treated with actomycin D were evaluated by RT-qPCR analysis. (H, I) The relative of Irf4 mRNA (H) and IRF4 protein (I) in MOG-activated WT and HuR KO CD4+ T cells were examined by RT-qPCR and Western blots, respectively. (J) RIP assay showed that Irf4 mRNA was associated with HuR protein in WT Th17 cells. (K) The relative amount of Irf4 mRNA in WT and HuR KO Th17 cells treated with actomycin D were evaluated by RT-qPCR analysis. ** p<0.01, *** p<0.001.
To test the hypothesis that HuR might directly regulate RORγt expression in Th17 cells, naïve CD4+ T cells were isolated from the spleens of WT and HuR KO mice and cultured under Th17 cell-polarizing conditions (36). The abundance of Rorγt mRNA and RORγt protein was measured by RT-qPCR and western blot analyses, respectively (Fig. 3D, 3E). Interestingly, knockout of HuR increased Il22 mRNA (Fig. 3D). Although HuR knockout led to reductions in the levels of Rorγt mRNA and RORγt protein, HuR did not appear to bind to Rorγt mRNA, as revealed from two different experiments. Ribonucleoprotein (RNP) immunoprecipitation (RIP) analysis indicated that HuR did not bind to Rorγt mRNA; HuR binding to b-actin mRNA is the positive control for RIP assay (Fig. 3F). Actinomycin D treatment to quantity mRNA decay rate further revealed that the half-life of Rorγt mRNA in HuR KO Th17 cells was comparable to that of WT Th17 cells (Fig. 3G), suggesting that HuR indirectly modulate Rorγt expression in CD4+ T cells to promote Th17 cell differentiation.
The transcription factor IRF4 is required for the induction of Th17 lineage-specific regulators RORγt and RORα upon exposure to either TGF-β and IL-6 or TGF-β and IL-21(9); thus, Irf4−/− mice are completely resistant to EAE (9). Interestingly, knockout of HuR also reduced the expressions of Irf4 mRNA and protein levels in MOG-activated CD4+ T cells (Fig. 3H, 3I), and in in vitro Th17-polarized cells (fig. S3A). RIP assay showed that Irf4 mRNA, but not Batf mRNA, associated with HuR protein in WT Th17 cells (Fig. 3J). In addition, the half-life of Irf4 mRNA was shorter in HuR KO Th17 cells (2.3 h) than in WT Th17 cells (4.1 h) (Fig. 3K), suggesting that HuR directly binds to and stabilizes Irf4 mRNA. Given that IRF4 plays an essential role in inducing Rorγt mRNA levels during Th17 cell differentiation (9), our data indicate that HuR might modulate Rorγt mRNA levels by enhancing IRF4 production to promote Th17 cell differentiation.
Runx1 mRNA is a direct target of HuR in Th17 cells
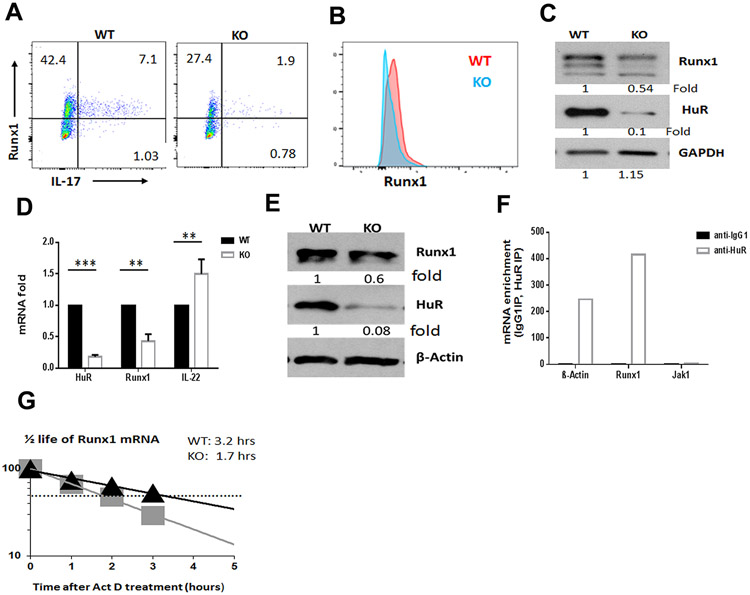
RUNX1 and RUNX3 are important transcription factors that upregulate Th17 cell differentiation (10,11). We performed RT-qPCR analysis to measure the levels of Runx1, Runx2, and Runx3 mRNAs in more than 3 pairs of WT and HuR KO Th17 cells. Given that Runx1 but not Runx2 and Runx3 expression was significantly decreased in MOG-activated HuR KO Th17 cells (fig. S3B), we hypothesized that HuR upregulates RUNX1 expression to facilitate Th17 cell differentiation. Flow cytometry and Western blot analyses indicated that there were fewer Runx1+IL-17+ cells in MOG-activated HuR KO CD4+ T cells in comparison with WT CD4+ T cells (Fig. 4A to C). Similar findings were observed in cultured WT and HuR KO Th17 cells (Fig. 4D, 4E). In addition, knockdown of HuR by lentivrial HuR shRNA also decreased the RUNX1 expression in human Th17 cells as determined by Western blot analysis (data not shown).
Figure 4. HuR promotes RUNX1 expression in Th17 cells.
(A to D). The expression of RUNX1 in MOG-activated WT and HuR KO CD4+ T cells was evaluated by flow cytometry (A, B) and Western blot analysis (C). (D, E) Levels of Runx1 mRNA (D) and RUNX1 protein (E) in HuR KO CD4+ T cells compared with WT CD4+ T cells under Th17 cell-polarizing condition. (F) RIP assay was used to measure the enrichment in Runx1 mRNA in HuR IP complexes relative to IgG complexes. (G) Actinomycin D treatments were used to assess the half-life of Runx1 mRNA in WT and HuR KO Th17 cells. ** p<0.01, *** p<0.001.
To further understand the underlying mechanism whereby HuR modulated RUNX1 abundance, we performed RIP assay with both mouse and human Th17 cells. As expected, Runx1 mRNA was highly enriched in HuR IP complexes compared with control IgG IP complexes, which indicated that Runx1 mRNA associated with HuR in both mouse (Fig. 4F) and human Th17 cells (data not shown). Furthermore, actinomycin D treatment of mouse Th17 cells revealed that the half-life of Runx1 mRNA was much shorter in HuR KO Th17 cells (1.7 h) than in WT Th17 cells (3.2 h) (Fig 4G). These results indicate that HuR enhances Runx1 mRNA levels by binding to and protecting it from decay, in turn increasing Th17 cell differentiation.
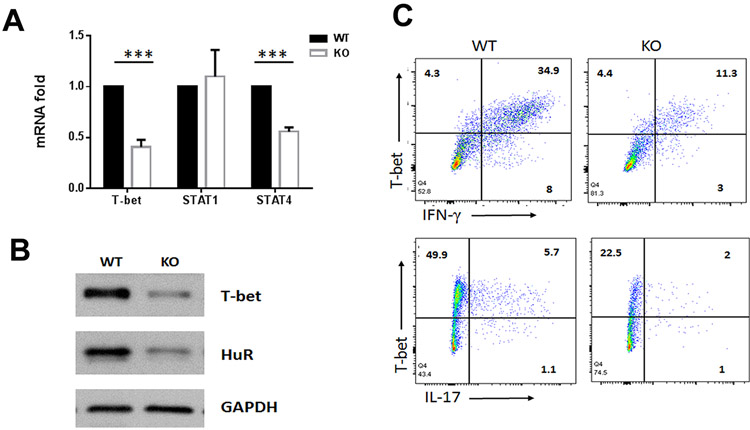
HuR deficiency in CD4+ T cells reduces the expression of T-bet
Our study showed that compared with WT mice, HuR KO mice had reduced percentages of IL-17+CD4+ T cells (4.2% vs. 6.8%), IFN-γ+CD4+ T cells (10.2% vs. 20%) and IFN-γ+IL-17+CD4+ T cells (1.2% vs. 2.8%) (fig. S1A, S1B). It is well known that transcription factor T-bet critically promotes Ifng gene expression (37,38), and that T-bet and RUNX1 are essential for the development of pathogenic IFN-γ+IL-17+CD4+ T cells (11). We thus examined whether HuR modulated T-bet abundance during MOG-activated Th17 cell differentiation. As shown, the expression levels of T-bet mRNA and protein were significantly reduced in MOG-activated HuR KO CD4+ T cells compared with WT CD4+ T cells (Fig. 5A to C), consistent with the flow cytometry data showing decreased frequencies of IFN-γ+IL-17+ CD4+ T cells in the spleens of mice with HuR KO CD4+ T cells (fig. S1A, S1B). However, Stat1 but not Stat4 mRNA was not reduced in HuR KO Th17 cells compared to WT Th17 cells (Fig. 5A).
Figure 5. HuR modulates T-bet expression in MOG-activated CD4+ T cells contributing to EAE.
(A to C) The expression of T-bet in MOG-activated WT and HuR KO CD4+ T cells was evaluated by RT-qPCR analysis (A), Western blot analysis (B), and flow cytometry assay (C). *** p<0.001. Data in panel A are summary of three independent experiments (Mean ± SEM). Data in panel B and C are representative data of three individual experiments.
Previous studies showed that Th1 cells also play a critical role in the pathogenesis of EAE (2), we assessed the pathogenic role of HuR in MOG-activated CD4+ T cells under Th1 cell polarizations (MOG+IL-12+IFN-γ) (2). By adoptive transfer EAE, similar to what was done with Th17 cells (Fig. 1A), Rag1−/− recipients of WT Th1 cells developed much more severe EAE while Rag1−/− recipients of HuR KO Th1 cells developed minimal EAE (fig. S4A). The results of histological analysis were also consistent with clinical EAE scores of Rag1−/− recipients receiving WT or HuR KO Th1 cells (fig. S4B). In addition, flow cytometry analysis showed that the T-bet expression in MOG-activated Th1 cells was significantly reduced in HuR KO mice compared with WT mice (fig. S4C to S4F). However, RIP assay and actinomycin D treatment suggested that HuR did not directly modulate T-bet expression in CD4+ T cells (data not shown).
HuR increases IL-12Rβ1 levels, contributing to Th17 and Th1-like Th17 cell differentiation
RNA-seq and RT-qPCR analyses showed that Il12rb1, Il12rb2 and Tyk2 but not gp130 mRNAs were dramatically decreased in MOG-activated HuR KO Th17 cells compared with WT Th17 cells (Fig. S2A and Fig. S5A). Since IL-12Rβ1 is the common receptor for IL-23 and IL-12 in Th17 and Th1 cells (39), and IL-23 causes Th17 cell pathogenicity (4,40), we examined whether HuR directly modulated IL-12Rβ1 expression; as shown, HuR knockout significantly decreased the frequency of IL-12Rβ1+IFN-γ+ T cells in MOG-activated KO Th17 cells compared with WT Th17 cells (Fig. S5B, S5C). Similar results were observed after in vitro culturing HuR KO Th17 cells (Fig. S5D). In addition, RIP assay showed that HuR bound to Il12rb1 mRNA in mouse (Fig. S5E) and human Th17 cells and stabilized Il12rb1 mRNA (data not shown). Given that IL-12Rβ1 is the upstream of T-bet in Th1 cells (41), our data suggested that HuR upregulates T-bet expressions in MOG-activated CD4+ T cells by increasing IL-12Rβ1 expression, which may endow the plasticity among Th17 and Th1 cells caused by the local microenvironment in vivo.
Abrogation of HuR impairs MOG-activated CD4+ T cell migration to the CNS
We found fewer CD4+ T cell infiltrating into CNS in Rag1−/− mice receiving HuR KO CD4+ T cells than WT CD4+ T cells (Fig. 1H), although the numbers of WT and HuR KO CD4+ T cells were comparable in the spleens of Rag1−/− recipients (Fig. 1J). We hypothesized that HuR might increase the expression of adhesion molecules in MOG-activated CD4+ T cells, promoting their migration to the CNS during EAE. To test this possibility, we analyzed RNA-seq data for expression of adhesion molecules related with T cell migration in WT and HuR KO Th17 cells. As shown, the abundance of mRNAs encoding proteins with functions in cell migration were significantly lower in HuR KO CD4+ T cells than in WT CD4+ T cells (fig. S6A, S6B). The levels of some of these transcripts, including Vla-4 (Itga4) mRNA, were validated by RT-qPCR analysis (fig. S6C). Consistent with the RNA-seq data, flow cytometric analysis also showed that knockout of HuR decreased the expression of VLA-4 in MOG-activated CD4+ T cells and in in vitro cultured Th17 cells (fig. S6D, S6E). Natalizumab (anti-VLA-4) has been approved by the FDA for treatment of multiple sclerosis (42). HuR promotes VLA-4 expression may contribute T cell migration to CNS.
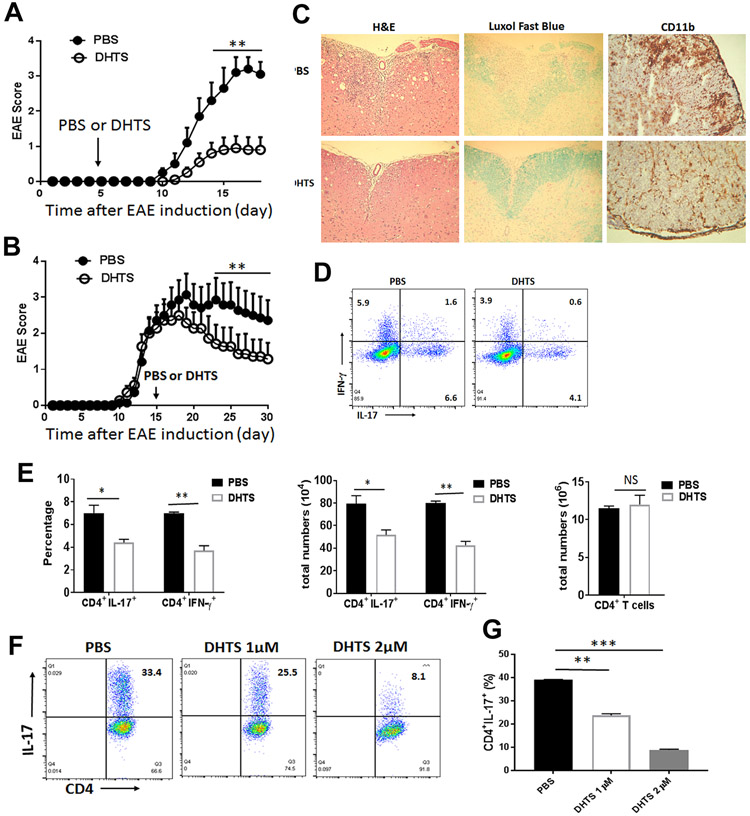
Targeting HuR by its inhibitor ameliorates chronic EAE
Dihydrotanshinone-I (DHTS) is a natural compound extracted from Salvia miltiorrhiza and recently identified as HuR inhibitor, which functions to block HuR binding to its target mRNAs (30). To investigate the impact of HuR inhibitor on EAE, we immunized WT mice with MOG and CFA for EAE, then treated them with a vehicle control or DHTS by i.p. starting at day 5 after immunization. The injection was repeated every other day. Targeting HuR by DHTS not only delayed the onset, but also reduced the severity of EAE progression (Fig. 6A). Interestingly, administration of DHTS starting at day 15 post-immunization when mice with 2+ score of EAE also significantly ameliorated the severity of EAE (Fig. 6B). The DHTS treatment reduced CNS inflammatory cell infiltration as determined by H&E staining and demyelination as determined by Luxol fast blue staining (Fig. 6C). Importantly, the CD11b positive myeloid cell infiltrating the spinal cords in DHTS-treated mice was reduced by more than 2 folds as compared to PBS-treated mice (Fig. 6C). Flow cytometric analysis showed that the frequency and number of IL-17- and IFN-γ-producing CD4+ T cells was significantly reduced in the spleen of DHTS-treated mice (Fig. 6D, E). However, DHTS treatment had no effect on the frequencies of CD4, CD8 T cells, B cells, and macrophages in the spleen in comparison with PBS-treated mice (fig. S7A, S7B), suggesting that administration of DHTS for treatment was safe to mice without systemic toxicity in vivo. Consistent with the reduced EAE severity, the proinflammatory cytokine ( Il17, Ifn-g, Csf2 ) and adhesion molecule (Vla-4) mRNA abundance in CNS inflammatory cells was also reduced in MOG-immunized mice treated with DHTS (data not shown). To determine if targeting HuR by DHTS down-regulates Th17 cell differentiation, we compared naïve CD4+ T cells differentiated into Th17 cells with or without DHTS. The results showed that the frequency of CD4+IL-17+ cells was dose-dependently decreased in Th17 cells cultured with DHTS in comparison with control group (Fig. 6F, G), suggesting that targeting HuR by DHTS impairs Th17 cell differentiation and functions. Furthermore, DHTS had the capacity to inhibit the cytokine and transcriptional factor mRNA expression in activated human Jurkat T cells (data not shown), suggesting that DHTS might have a similar effect on human MS T cells.
Figure 6. Targeting HuR by DHTS ameliorates chronic EAE.
EAE disease course in WT mice that were injected i.p. with either DHTS or PBS every other day starting from day 5 (A) or 15 (B) after disease induction with MOG in complete Freund’s adjuvant (CFA. (C) Representative histopathology of spinal cords of EAE mice treated with PBS or DHTS were stained by H&E, Luxol fast blue and anti-CD11b staining. (D) Representative CD4+IL-17 and CD4+ IFN-γ producing cells in the splenocytes of EAE mice treated with PBS or DHTS as described in panel B at the end of experiment (day 30) were shown. (E) Summary of the flow cytometry data of CD4+IL-17 and CD4+ IFN-γ-producing cells in the spleen of EAE mice treated with PBS or DHTS at the end of experiment (day 30). (F) Naïve CD4+ T cells were cultured under Th17 cell polarization condition with or without different dosage of DHTS (1 to 2μM/ml) for 72 hours. Representative flow cytometry data was shown. (G) Summary of three independent experiments were shown for frequency of CD4+IL-17+ T cells measured by flow cytometry assay (mean ± SEM). *, p<0.05; **, p<0.01; *** p<0.001. Data in panel A are summary of two experiments (N=10 mice/per group). Data in panel B represent one of two independent experiments (N=7 mice/per group).
In sum, by either using OX40-Cre+/−HuRf/f mice to delete HuR after T cell activation or using HuR inhibitor-DHTS to block HuR function in EAE, we report that downregulation of HuR abolishes Th17 and Th1-like Th17 cell differentiation and pathogenesis of autoimmune neuroinflammation, and targeting HuR could be a novel therapeutic approach for patients with multiple sclerosis.
DISCUSSION
Multiple sclerosis is a chronic inflammatory disease of the CNS. Although FDA approves several clinical treatments, there were no curative treatments for this disease, emphasizing the significance to develop a novel approach for treatment of MS.
Understanding post-transcriptional regulation of pathogenic Th17 cell differentiation will open a novel avenue to develop drugs to target Th17 cells-mediated autoimmune neuroinflammation. Here, we provide molecular evidence that HuR post-transcriptionally upregulated the transcription factor IRF4, RUNX1, and receptor IL-12Rβ1 expression, in turn upregulating RORγt and T-bet abundance to enhance Th17 cell and Th1-like Th17 cell differentiation, which was different from the mechanism of HuR in regulating Th2 cell differentiation (25,43). These findings highlight the complex regulatory program whereby HuR modulates transcription factors and receptor to regulate CD4+ T cell differentiation and to promote autoimmune disease.
HuR directly modulated the expression of transcription factors IRF4 and RUNX1 by stabilizing their respective mRNAs in Th17 cells, thereby controlling Th17 cell differentiation, given that IRF4 and RUNX1 critically promote Th17 cell differentiation (11). Although HuR did not directly bind nor stabilize Rorγt mRNA, HuR promoted Rorγt expression indirectly by enhancing the production of IRF4 and RUNX1, two proteins that cooperatively induce Rorγt expression in Th17 cells (10,44). We further observed that knockout of HuR not only reduced the frequency of IL-17+CD4+ T cells, but also decreased the frequency of IFN-γ+CD4+ T cells and IFN-γ+IL-17+CD4+ T cells, consistent with the fact that HuR deficiency decreased the expression of transcription factor T-bet. Although HuR did not directly modulate T-bet expression, HuR was found to promote the expression of upstream receptor IL-12Rβ1 by binding to and stabilizing its mRNA (Fig. 6). It is well known that IL-12 binding to IL12Rβ1 and IL-12Rβ2 complex activates STAT1 and STAT4, and to promote IFN-γ production, and IFN-γ in turn promotes expression of T-bet and IL-12Rb2 in Th1 cells (45). Like IL-12, IL-23 binds to the IL-12Rb1 and IL-23R complex, and activates Stat3 in Th17 cells (46); therefore, knockout of IL-12Rβ1 impairs both IL-12- and IL-23-mediated effects. Previous reports showed that IL-12Rβ1 or T-bet deficient mice are resistant to EAE (38,47,48). Our current study suggests that HuR promotes IFN-γ+IL-17+ Th1-like Th17 cell differentiation.
In line with our previous findings that HuR binds to and stabilizes Csf2 mRNA in Th17 cells (27), knockout of HuR reduced GM-CSF expression in MOG-activated CD4+ T cells with impaired function to induce EAE (fig. S1). These findings are consistent with reports that GM-CSF is a critical cytokine for the pathogenicity of Th17 cells in EAE (4,34,40), and extend our previous report to better understand the mechanism by which HuR plays a critical role in Th17 cells.
RNA-seq analysis showed that the expression of Foxp3 mRNA increased in MOG-activated HuR KO Th17 cells compared to WT counterparts (Fig. 2A). However, there were no significant differences on the number and percentage of T regulatory cells (CD25+Foxp3+) in the spleen and lymph node of HuR KO and WT mice at day 11 after MOG and CFA immunization as examined by flow cytometry assay (10.1% ± 0.46% vs 10.5% ± 0.63%). Which excluded the possibility that HuR KO CD4+ T cells defective for induction of EAE were due to the increased T regulatory cells, supporting the notion that knockout of HuR impaired Th17 cell differentiation and its pathogenicity for EAE.
Hypoxia-inducible factor 1(HIF-1) has been reported to enhance Th17 cell differentiation by directly transactivating Rorγt gene expression, and Hif-1α−/− mice are resistant to EAE (49). It is well known that HuR promotes the translation of HIF-1α in human cervical carcinoma Hela cell line and human lung carcinoma A549 cells (50). In this study, knockout of HuR reduced the expression of HIF-1α in Th17 cells, and RIP assay showed that Hif-1α mRNA was associated with HuR protein (data not shown), suggesting that HuR may further facilitate Th17 differentiation by regulating HIF-1α expression in CD4+ T cells. In addition, we previously showed that HuR promoted CCR6 expression on Th17 cells, and knockout of HuR reduced CCR6 expression on Th17 cells and impaired their migration to its ligand CCL20(28). The expression levels of several adhesion molecule such as VLA-4 were greatly reduced in HuR KO CD4+ T cells, supporting the notion that HuR promotes pathogenic Th17 cell migration to the site of inflammation in the CNS. Our results were consistent with the fact that administration of anti-VLA4 reduced the development of EAE (51) and benefited some patients with multiple sclerosis.
HuR not only modulates the expressions of Th17 cell transcription factors, but also regulates the expression of metabolism enzyme Cyp51 (cytochrome P450, family 51), whose product is the ligand to activate RORγt and Th17 cell differentiation (52). In line with the function of HuR in promoting Th17 cell differentiation, knockout of HuR significantly impaired the gene expression in other pathways (data not shown).
The expression of NOS2 (nitric oxide synthase, an enzyme that promotes NO production) was dramatically increased in MOG-activated HuR KO Th17 cells compared with WT Th17 cells (Fig. 2B). Interestingly, Nos2−/− mice developed exacerbated EAE (53). However, it remains to be studied if the increased NOS2 expression rendered HuR KO Th17 cells less capable of inducing EAE than WT Th17 cells.
HuR is highly expressed in many tumors(19), implicating that HuR could be a therapeutic target for cancer treatment, and many investigators developed HuR inhibitors aiming to blocking tumor growth and metastasis(30,54). However, so far there are no reports on utilizing HuR inhibitors to treat autoimmune diseases in mouse models. Based on our results HuR upregulating pathogenic Th17 cell differentiation, we administrated HuR inhibitor DHTS to mice with EAE. Clearly, DHTS treatment starting at the onset of or middle of the disease significantly reduced the severity of EAE compared to mice treated with vehicle control, but not impaired the T and B cell populations in the spleen, which suggested that administration of DHTS is safe and without system toxic to mice in vivo. We also provided evidence that DHTS inhibited in vitro cultured Th17 cell differentiation at a dose-dependent manner. Furthermore, our study showed that DHTS was able to reduce the proinflammatory cytokine production by human Jurkat T cells (data not shown), which provided a clue to test its effect on human MS T cells in future study.
In closing, our collective studies extend our previous reports on the function of HuR in stabilizing Il17, Csf2, and Ccr6 mRNAs (28), and identify a new function of HuR in upregulating the expression of transcription factor, receptor, and adhesion molecules, synergistically promoting Th17 cell differentiation and migration to contributing to autoimmune neuroinflammation. Our study provides further understanding of HuR as a potential therapeutic target in autoimmune diseases such as multiple sclerosis.
Supplementary Material
KEY POINTS:
HuR regulates Th17 cell and Th1-like Th17 cell differentiation
HuR directly and indirectly modulates RUNX1 and RORγt expression, respectively
Targeting HuR by its inhibitor reduces severity of autoimmune neuroinflammation
ACKNOWLEDGEMENTS
We thank Dr. Ulus Atasoy for kindly providing us the HuRf/f mice. We also thank Neuroimmunology Research group for comments and suggestion.
This work was supported by NIH grant (R01AI119135) and Thomas Jefferson University faculty startup package fund. JLM, KA and MG were supported by the NIA-IRP, NIH
Abbreviation used in this article:
- MS
multiple sclerosis
- EAE
experimental autoimmune encephalomyelitis
- MOG
myelin oligodendrocyte glycoprotein
- DHTS
Dihydrotanshinone-I
- Cyp51
cytochrome P450 family 51
- GSEA
Gene Set Enrichment Analysis
- MS
multiple sclerosis
- CNS
Central nervous system
Footnotes
Disclosure: The authors have no financial conflicts of interest.
References
- 1.Dendrou CA, Fugger L, and Friese MA (2015) Immunopathology of multiple sclerosis. Nature reviews. Immunology 15, 545–558 PMID: 26250739. [DOI] [PubMed] [Google Scholar]
- 2.Kroenke MA, Carlson TJ, Andjelkovic AV, and Segal BM (2008) IL-12- and IL-23-modulated T cells induce distinct types of EAE based on histology, CNS chemokine profile, and response to cytokine inhibition. The Journal of experimental medicine 205, 1535–1541 PMC2442630 PMID: 18573909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martin BN, Wang C, Zhang CJ, Kang Z, Gulen MF, Zepp JA, Zhao J, Bian G, Do JS, Min B, Pavicic PG Jr., El-Sanadi C, Fox PL, Akitsu A, Iwakura Y, Sarkar A, Wewers MD, Kaiser WJ, Mocarski ES, Rothenberg ME, Hise AG, Dubyak GR, Ransohoff RM, and Li X (2016) T cell-intrinsic ASC critically promotes T(H)17-mediated experimental autoimmune encephalomyelitis. Nature immunology 17, 583–592 PMC5385929 PMID: 26998763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.El-Behi M, Ciric B, Dai H, Yan Y, Cullimore M, Safavi F, Zhang GX, Dittel BN, and Rostami A (2011) The encephalitogenicity of T(H)17 cells is dependent on IL-1- and IL-23-induced production of the cytokine GM-CSF. Nature immunology 12, 568–575 PMC3116521 PMID: 21516111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stromnes IM, Cerretti LM, Liggitt D, Harris RA, and Goverman JM (2008) Differential regulation of central nervous system autoimmunity by T(H)1 and T(H)17 cells. Nature medicine 14, 337–342 PMC2813727 PMID: 18278054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, Cua DJ, and Littman DR (2006) The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell 126, 1121–1133 PMID: 16990136. [DOI] [PubMed] [Google Scholar]
- 7.Yang XO, Pappu BP, Nurieva R, Akimzhanov A, Kang HS, Chung Y, Ma L, Shah B, Panopoulos AD, Schluns KS, Watowich SS, Tian Q, Jetten AM, and Dong C (2008) T helper 17 lineage differentiation is programmed by orphan nuclear receptors ROR alpha and ROR gamma. Immunity 28, 29–39 PMC2587175 PMID: 18164222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laurence A, Tato CM, Davidson TS, Kanno Y, Chen Z, Yao Z, Blank RB, Meylan F, Siegel R, Hennighausen L, Shevach EM, and O'Shea J J (2007) Interleukin-2 signaling via STAT5 constrains T helper 17 cell generation. Immunity 26, 371–381 PMID: 17363300. [DOI] [PubMed] [Google Scholar]
- 9.Brustle A, Heink S, Huber M, Rosenplanter C, Stadelmann C, Yu P, Arpaia E, Mak TW, Kamradt T, and Lohoff M (2007) The development of inflammatory T(H)-17 cells requires interferon-regulatory factor 4. Nature immunology 8, 958–966 PMID: 17676043. [DOI] [PubMed] [Google Scholar]
- 10.Zhang F, Meng G, and Strober W (2008) Interactions among the transcription factors Runx1, RORgammat and Foxp3 regulate the differentiation of interleukin 17-producing T cells. Nature immunology 9, 1297–1306 PMC4778724 PMID: 18849990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y, Godec J, Ben-Aissa K, Cui K, Zhao K, Pucsek AB, Lee YK, Weaver CT, Yagi R, and Lazarevic V (2014) The transcription factors T-bet and Runx are required for the ontogeny of pathogenic interferon-gamma-producing T helper 17 cells. Immunity 40, 355–366 PMC3965587 PMID: 24530058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blackinton JG, and Keene JD (2016) Functional coordination and HuR-mediated regulation of mRNA stability during T cell activation. Nucleic acids research 44, 426–436 PMC4705648 PMID: 26490963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kafasla P, Skliris A, and Kontoyiannis DL (2014) Post-transcriptional coordination of immunological responses by RNA-binding proteins. Nature immunology 15, 492–502 PMID: 24840980. [DOI] [PubMed] [Google Scholar]
- 14.Diaz-Munoz MD, Bell SE, Fairfax K, Monzon-Casanova E, Cunningham AF, Gonzalez-Porta M, Andrews SR, Bunik VI, Zarnack K, Curk T, Heggermont WA, Heymans S, Gibson GE, Kontoyiannis DL, Ule J, and Turner M (2015) The RNA-binding protein HuR is essential for the B cell antibody response. Nature immunology 16, 415–425 PMC4479220 PMID: 25706746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y, Wang X, Zhang X, Wang J, Ma Y, Zhang L, and Cao X (2019) RNA-binding protein YTHDF3 suppresses interferon-dependent antiviral responses by promoting FOXO3 translation. Proceedings of the National Academy of Sciences of the United States of America 116, 976–981 PMC6338863 PMID: 30591559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Z, Yin W, Zhu L, Li J, Yao Y, Chen F, Sun M, Zhang J, Shen N, Song Y, and Chang X (2018) Iron Drives T Helper Cell Pathogenicity by Promoting RNA-Binding Protein PCBP1-Mediated Proinflammatory Cytokine Production. Immunity 49, 80–92.e87 PMID: 29958803. [DOI] [PubMed] [Google Scholar]
- 17.Garg AV, Amatya N, Chen K, Cruz JA, Grover P, Whibley N, Conti HR, Hernandez Mir G, Sirakova T, Childs EC, Smithgall TE, Biswas PS, Kolls JK, McGeachy MJ, Kolattukudy PE, and Gaffen SL (2015) MCPIP1 Endoribonuclease Activity Negatively Regulates Interleukin-17-Mediated Signaling and Inflammation. Immunity 43, 475–487 PMC4575280 PMID: 26320658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lopez de Silanes I, Zhan M, Lal A, Yang X, and Gorospe M (2004) Identification of a target RNA motif for RNA-binding protein HuR. Proceedings of the National Academy of Sciences of the United States of America 101, 2987–2992 PMC365732 PMID: 14981256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abdelmohsen K, and Gorospe M (2010) Posttranscriptional regulation of cancer traits by HuR. Wiley interdisciplinary reviews. RNA 1, 214–229 PMC3808850 PMID: 21935886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Techasintana P, Ellis JS, Glascock J, Gubin MM, Ridenhour SE, Magee JD, Hart ML, Yao P, Zhou H, Whitney MS, Franklin CL, Martindale JL, Gorospe M, Davis WJ, Fox PL, Li X, and Atasoy U (2017) The RNA-Binding Protein HuR Posttranscriptionally Regulates IL-2 Homeostasis and CD4(+) Th2 Differentiation. Immunohorizons 1, 109–123 PMC6052877 PMID: 30035254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stellato C, Gubin MM, Magee JD, Fang X, Fan J, Tartar DM, Chen J, Dahm GM, Calaluce R, Mori F, Jackson GA, Casolaro V, Franklin CL, and Atasoy U (2011) Coordinate regulation of GATA-3 and Th2 cytokine gene expression by the RNA-binding protein HuR. Journal of immunology (Baltimore, Md. : 1950) 187, 441–449 PMC5801757 PMID: 21613615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Turner M, and Diaz-Munoz MD (2018) RNA-binding proteins control gene expression and cell fate in the immune system. Nature immunology 19, 120–129 PMID: 29348497. [DOI] [PubMed] [Google Scholar]
- 23.Huang W, Thomas B, Flynn RA, Gavzy SJ, Wu L, Kim SV, Hall JA, Miraldi ER, Ng CP, Rigo F, Meadows S, Montoya NR, Herrera NG, Domingos AI, Rastinejad F, Myers RM, Fuller-Pace FV, Bonneau R, Chang HY, Acuto O, and Littman DR (2018) Retraction Note: DDX5 and its associated lncRNA Rmrp modulate TH17 cell effector functions. Nature 562, 150 PMC6172141 PMID: 29973715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghosh M, Aguila HL, Michaud J, Ai Y, Wu MT, Hemmes A, Ristimaki A, Guo C, Furneaux H, and Hla T (2009) Essential role of the RNA-binding protein HuR in progenitor cell survival in mice. The Journal of clinical investigation 119, 3530–3543 PMC2786787 PMID: 19884656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Techasintana P, Davis JW, Gubin MM, Magee JD, and Atasoy U (2015) Transcriptomic-Wide Discovery of Direct and Indirect HuR RNA Targets in Activated CD4+ T Cells. PloS one 10, e0129321 PMC4498740 PMID: 26162078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen J, Cascio J, Magee JD, Techasintana P, Gubin MM, Dahm GM, Calaluce R, Yu S, and Atasoy U (2013) Posttranscriptional gene regulation of IL-17 by the RNA-binding protein HuR is required for initiation of experimental autoimmune encephalomyelitis. Journal of immunology (Baltimore, Md. : 1950) 191, 5441–5450 PMC3831112 PMID: 24166976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen J, Adamiak W, Huang G, Atasoy U, Rostami A, and Yu S (2017) Interaction of RNA-binding protein HuR and miR-466i regulates GM-CSF expression. Scientific reports 7, 17233 PMC5722853 PMID: 29222492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen J, Martindale JL, Cramer C, Gorospe M, Atasoy U, Drew PD, and Yu S (2017) The RNA-binding protein HuR contributes to neuroinflammation by promoting C-C chemokine receptor 6 (CCR6) expression on Th17 cells. The Journal of biological chemistry 292, 14532–14543 PMC5582845 PMID: 28684423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xie L, Chen J, McMickle A, Awar N, Nady S, Sredni B, Drew PD, and Yu S (2014) The immunomodulator AS101 suppresses production of inflammatory cytokines and ameliorates the pathogenesis of experimental autoimmune encephalomyelitis. Journal of neuroimmunology 273, 31–41 PMC4113726 PMID: 24975323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lal P, Cerofolini L, D'Agostino VG, Zucal C, Fuccio C, Bonomo I, Dassi E, Giuntini S, Di Maio D, Vishwakarma V, Preet R, Williams SN, Fairlamb MS, Munk R, Lehrmann E, Abdelmohsen K, Elezgarai SR, Luchinat C, Novellino E, Quattrone A, Biasini E, Manzoni L, Gorospe M, Dixon DA, Seneci P, Marinelli L, Fragai M, and Provenzani A (2017) Regulation of HuR structure and function by dihydrotanshinone-I. Nucleic acids research 45, 9514–9527 PMC5766160 PMID: 28934484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DeMicco A, Naradikian MS, Sindhava VJ, Yoon JH, Gorospe M, Wertheim GB, Cancro MP, and Bassing CH (2015) B Cell-Intrinsic Expression of the HuR RNA-Binding Protein Is Required for the T Cell-Dependent Immune Response In Vivo. Journal of immunology (Baltimore, Md. : 1950) 195, 3449–3462 PMC4575876 PMID: 26320247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bai XF, Li O, Zhou Q, Zhang H, Joshi PS, Zheng X, Liu Y, Wang Y, Zheng P, and Liu Y (2004) CD24 controls expansion and persistence of autoreactive T cells in the central nervous system during experimental autoimmune encephalomyelitis. The Journal of experimental medicine 200, 447–458 PMC2211938 PMID: 15314074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fitzgerald DC, Zhang GX, El-Behi M, Fonseca-Kelly Z, Li H, Yu S, Saris CJ, Gran B, Ciric B, and Rostami A (2007) Suppression of autoimmune inflammation of the central nervous system by interleukin 10 secreted by interleukin 27-stimulated T cells. Nature immunology 8, 1372–1379 PMID: 17994023. [DOI] [PubMed] [Google Scholar]
- 34.Codarri L, Gyulveszi G, Tosevski V, Hesske L, Fontana A, Magnenat L, Suter T, and Becher B (2011) RORgammat drives production of the cytokine GM-CSF in helper T cells, which is essential for the effector phase of autoimmune neuroinflammation. Nature immunology 12, 560–567 PMID: 21516112. [DOI] [PubMed] [Google Scholar]
- 35.Solt LA, Kumar N, Nuhant P, Wang Y, Lauer JL, Liu J, Istrate MA, Kamenecka TM, Roush WR, Vidovic D, Schurer SC, Xu J, Wagoner G, Drew PD, Griffin PR, and Burris TP (2011) Suppression of TH17 differentiation and autoimmunity by a synthetic ROR ligand. Nature 472, 491–494 PMC3148894 PMID: 21499262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park H, Li Z, Yang XO, Chang SH, Nurieva R, Wang YH, Wang Y, Hood L, Zhu Z, Tian Q, and Dong C (2005) A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nature immunology 6, 1133–1141 PMC1618871 PMID: 16200068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Szabo SJ, Sullivan BM, Stemmann C, Satoskar AR, Sleckman BP, and Glimcher LH (2002) Distinct effects of T-bet in TH1 lineage commitment and IFN-gamma production in CD4 and CD8 T cells. Science (New York, N.Y.) 295, 338–342 PMID: 11786644. [DOI] [PubMed] [Google Scholar]
- 38.Yang Y, Weiner J, Liu Y, Smith AJ, Huss DJ, Winger R, Peng H, Cravens PD, Racke MK, and Lovett-Racke AE (2009) T-bet is essential for encephalitogenicity of both Th1 and Th17 cells. The Journal of experimental medicine 206, 1549–1564 PMC2715092 PMID: 19546248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vignali DA, and Kuchroo VK (2012) IL-12 family cytokines: immunological playmakers. Nature immunology 13, 722–728 PMC4158817 PMID: 22814351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Komuczki J, Tuzlak S, Friebel E, Hartwig T, Spath S, Rosenstiel P, Waisman A, Opitz L, Oukka M, Schreiner B, Pelczar P, and Becher B (2019) Fate-Mapping of GM-CSF Expression Identifies a Discrete Subset of Inflammation-Driving T Helper Cells Regulated by Cytokines IL-23 and IL-1beta. Immunity PMID: 31079916. [DOI] [PubMed]
- 41.Afkarian M, Sedy JR, Yang J, Jacobson NG, Cereb N, Yang SY, Murphy TL, and Murphy KM (2002) T-bet is a STAT1-induced regulator of IL-12R expression in naive CD4+ T cells. Nature immunology 3, 549–557 PMID: 12006974. [DOI] [PubMed] [Google Scholar]
- 42.Ransohoff RM (2007) Natalizumab for multiple sclerosis. The New England journal of medicine 356, 2622–2629 PMID: 17582072. [DOI] [PubMed] [Google Scholar]
- 43.Gubin MM, Techasintana P, Magee JD, Dahm GM, Calaluce R, Martindale JL, Whitney MS, Franklin CL, Besch-Williford C, Hollingsworth JW, Abdelmohsen K, Gorospe M, and Atasoy U (2014) Conditional knockout of the RNA-binding protein HuR in CD4(+) T cells reveals a gene dosage effect on cytokine production. Molecular medicine (Cambridge, Mass.) 20, 93–108 PMC3960399 PMID: 24477678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lazarevic V, Chen X, Shim JH, Hwang ES, Jang E, Bolm AN, Oukka M, Kuchroo VK, and Glimcher LH (2011) T-bet represses T(H)17 differentiation by preventing Runx1-mediated activation of the gene encoding RORgammat. Nature immunology 12, 96–104 PMC3077962 PMID: 21151104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu J, Cao S, Kim S, Chung EY, Homma Y, Guan X, Jimenez V, and Ma X (2005) Interleukin-12: an update on its immunological activities, signaling and regulation of gene expression. Current immunology reviews 1, 119–137 PMC2965603 PMID: 21037949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parham C, Chirica M, Timans J, Vaisberg E, Travis M, Cheung J, Pflanz S, Zhang R, Singh KP, Vega F, To W, Wagner J, O'Farrell AM, McClanahan T, Zurawski S, Hannum C, Gorman D, Rennick DM, Kastelein RA, de Waal Malefyt R, and Moore KW (2002) A receptor for the heterodimeric cytokine IL-23 is composed of IL-12Rbeta1 and a novel cytokine receptor subunit, IL-23R. Journal of immunology (Baltimore, Md. : 1950) 168, 5699–5708 PMID: 12023369. [DOI] [PubMed] [Google Scholar]
- 47.Zhang GX, Yu S, Gran B, Li J, Siglienti I, Chen X, Calida D, Ventura E, Kamoun M, and Rostami A (2003) Role of IL-12 receptor beta 1 in regulation of T cell response by APC in experimental autoimmune encephalomyelitis. Journal of immunology (Baltimore, Md. : 1950) 171, 4485–4492 PMID: 14568921. [DOI] [PubMed] [Google Scholar]
- 48.Bettelli E, Sullivan B, Szabo SJ, Sobel RA, Glimcher LH, and Kuchroo VK (2004) Loss of T-bet, but not STAT1, prevents the development of experimental autoimmune encephalomyelitis. The Journal of experimental medicine 200, 79–87 PMC2213316 PMID: 15238607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dang EV, Barbi J, Yang HY, Jinasena D, Yu H, Zheng Y, Bordman Z, Fu J, Kim Y, Yen HR, Luo W, Zeller K, Shimoda L, Topalian SL, Semenza GL, Dang CV, Pardoll DM, and Pan F (2011) Control of T(H)17/T(reg) balance by hypoxia-inducible factor 1. Cell 146, 772–784 PMC3387678 PMID: 21871655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Galban S, Kuwano Y, Pullmann R Jr., Martindale JL, Kim HH, Lal A, Abdelmohsen K, Yang X, Dang Y, Liu JO, Lewis SM, Holcik M, and Gorospe M (2008) RNA-binding proteins HuR and PTB promote the translation of hypoxia-inducible factor 1alpha. Molecular and cellular biology 28, 93–107 PMC2223304 PMID: 17967866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mindur JE, Ito N, Dhib-Jalbut S, and Ito K (2014) Early treatment with anti-VLA-4 mAb can prevent the infiltration and/or development of pathogenic CD11b+CD4+ T cells in the CNS during progressive EAE. PloS one 9, e99068 PMC4045930 PMID: 24896098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Santori FR, Huang P, van de Pavert SA, Douglass EF Jr., Leaver DJ, Haubrich BA, Keber R, Lorbek G, Konijn T, Rosales BN, Rozman D, Horvat S, Rahier A, Mebius RE, Rastinejad F, Nes WD, and Littman DR (2015) Identification of natural RORgamma ligands that regulate the development of lymphoid cells. Cell metabolism 21, 286–298 PMC4317570 PMID: 25651181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fenyk-Melody JE, Garrison AE, Brunnert SR, Weidner JR, Shen F, Shelton BA, and Mudgett JS (1998) Experimental autoimmune encephalomyelitis is exacerbated in mice lacking the NOS2 gene. Journal of immunology (Baltimore, Md. : 1950) 160, 2940–2946 PMID: 9510198. [PubMed] [Google Scholar]
- 54.Lang M, Berry D, Passecker K, Mesteri I, Bhuju S, Ebner F, Sedlyarov V, Evstatiev R, Dammann K, Loy A, Kuzyk O, Kovarik P, Khare V, Beibel M, Roma G, Meisner-Kober N, and Gasche C (2017) HuR Small-Molecule Inhibitor Elicits Differential Effects in Adenomatosis Polyposis and Colorectal Carcinogenesis. Cancer research 77, 2424–2438 PMID: 28428272. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the RNA-seq data have been deposited in the BioProject database under the accession ID PRJNA589592 (https://www.ncbi.nlm.nih.gov/bioproject/589592). All data generated or analyzed during this study are included in this published article and its supplementary information files.