Paik and Farber demonstrate that lung-localized T cell–mediated protection to heterosubtypic influenza challenge requires priming of new effector T cells in the lung-draining lymph node, which is fortified by increased numbers of dendritic cells from prior influenza infection.
Abstract
Influenza infection generates tissue-resident memory T cells (TRMs) that are maintained in the lung and can mediate protective immunity to heterologous influenza strains, but the precise mechanisms of local T cell–mediated protection are not well understood. In a murine heterosubtypic influenza challenge model, we demonstrate that protective lung T cell responses derive from both in situ activation of TRMs and the enhanced generation of effector T cells from the local lung draining mediastinal lymph nodes (medLNs). Primary infection fortified the medLNs with an increased number of conventional dendritic cells (cDCs) that mediate enhanced priming of T cells, including those specific for newly encountered epitopes; cDC depletion during the recall response diminished medLN T cell generation and heterosubtypic immunity. Our study shows that during a protective recall response, cDCs in a fortified LN environment enhance the breadth, generation, and tissue migration of effector T cells to augment lung TRM responses.
Introduction
Diseases of the respiratory tract, triggered by diverse viral and bacterial pathogens that repeatedly infect individuals over their lifetimes, are a leading cause of morbidity and mortality worldwide. Respiratory viruses such as influenza and more recently, coronaviruses, can cause devastating worldwide pandemics due to their high transmissibility through respiration and targeted damage to the lung. In particular, influenza virus rapidly mutates to evade immune protection and continues to pose a major public health burden, causing up to 5 million global cases and 650,000 deaths annually, disproportionally affecting young children and the elderly (Paules et al., 2018). Vaccines for influenza promote the generation of strain-specific neutralizing antibodies but are poorly efficacious, resulting in seasonal outbreaks of unpredictable severity. T cells, however, can recognize invariant determinants of influenza virus to mediate efficacious cross-strain protection in mouse models (Liang et al., 1994). Moreover, T cell clones recognizing different influenza strains are readily detectable in human blood (Koutsakos et al., 2019; Pizzolla et al., 2018; Richards et al., 2010), indicating that promoting T cell–mediated immunity is a promising strategy for generating broad-based protection in the population.
Recent studies have shown that tissue localization is important for T cell–mediated protective immunity, particularly in the respiratory tract. In primary influenza infection, T cell responses are primed by dendritic cells (DCs) that migrate from the lung to the draining LN to present influenza antigens to naive T cells (Ho et al., 2011), resulting in generation of lung-homing CD4+ and CD8+ effector cells to mediate viral clearance. Heterogeneous subsets of memory T cells are generated from this initial infection and persist in multiple tissue sites, including noncirculating tissue-resident memory T cells (TRMs) within the lung (Marshall et al., 2001; Teijaro et al., 2011; Turner et al., 2014). TRMs comprise a distinct subset in diverse tissue sites in mice and humans and are transcriptionally and functionally distinct from circulating effector memory T cells (TEM; Mackay et al., 2016; Schenkel and Masopust, 2014; Szabo et al., 2019). Lung CD4+ and CD8+ TRMs mediate efficacious viral clearance and reduce morbidity to heterosubtypic influenza strains (McMaster et al., 2015; Teijaro et al., 2011; Turner et al., 2014; Wu et al., 2014; Zens et al., 2016). Functionally, lung TRMs can be activated in situ in the presence of inhibitors of lymphoid egress (Turner et al., 2014) and produce effector cytokines (McMaster et al., 2015). Although these studies suggest that lung TRMs may dominate secondary responses, the contributions of and requirements for peripheral T cell populations during the recall response to respiratory virus challenge are not clear.
Here, we demonstrate in a mouse model of heterosubtypic influenza challenge that lung T cells mount local protective responses independently of B cells and humoral immunity. This local recall response involves both in situ lung TRM proliferation and recruitment of effector T cells from the periphery. Using a T cell–tracking model, we demonstrate that in previously infected mice, priming of T cells in the mediastinal LN (medLN) DCs leads to rapid generation and trafficking of effector CD4+ and CD8+ T cells to the lung-resident niche, including those T cells specific for influenza antigens newly introduced during heterosubtypic challenge. We further demonstrate that this efficacious generation of lung-homing effector cells is due to increased numbers of conventional DCs (cDCs) in the local medLN from the prior infection. Preferential ablation of cDCs in the medLN during heterosubtypic challenge abrogated enhanced T cell priming and inhibited the protective response. Therefore, the lung T cell–mediated recall response is comprised of robust effector generation from fortified local LNs that coordinates with lung TRMs to mediate efficacious protection against future infectious challenges.
Results
Primary infection establishes lung-localized memory T cells that mediate heterosubtypic protection independent of B cells
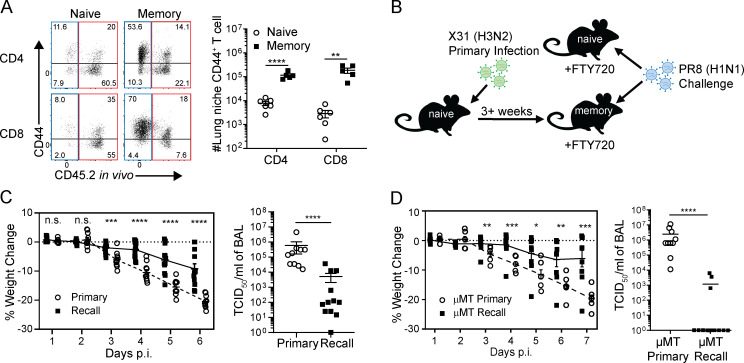
We assessed the tissue localization and differentiation state of T cells in the lung and draining medLNs from mice previously infected with influenza 3–4 wk prior (“memory”) mice compared with uninfected (“naive”) mice. Intravenous antibody labeling was used to distinguish between T cells in the circulation that are labeled by antibody from those cells in the tissues that are protected from labeling, as previously described (Turner et al., 2014). Memory mice had significantly increased frequencies and numbers of protected CD44hiCD4+ and CD8+ T cells in the lung tissue niche (Fig. 1 A), which were enriched for influenza-specific T cells shown by tetramer staining of CD8+ T cells (Fig. S1 A), consistent with our previous findings that lung niche T cells are a surrogate for antigen-specific responses (Turner et al., 2014). Lung niche CD44hi memory T cells expressed elevated levels of CD69, CXCR6, and CD103 (Fig. S1 B), consistent with a TRM phenotype (Kumar et al., 2017; Thome et al., 2014; Turner et al., 2014; Wein et al., 2019; Zens et al., 2016). These results show a robust, quantifiable generation of lung localized memory T cells following influenza infection.
Figure 1.
Influenza infection generates memory CD4+ and CD8+ T cells in the lung tissue niche and draining LN. T cells were isolated from the lung and lung-draining medLNs of uninfected naive mice and memory mice (infected with X31 influenza 3–4 wk previously) following i.v. administration of anti-CD45.2 antibody (see Materials and methods). (A) Analysis of lung T cells shown in representative flow cytometry plots of CD44 expression by circulating labeled (red) and lung tissue niche (blue) CD4+ and CD8+ T cells (left) and graphs of numbers of lung niche CD44+CD4+ and CD44+CD8+ T cells in naive and memory mice (right). Data were compiled from two groups; n = 5 or 6 mice/group. (B) Schematic diagram depicting heterosubtypic challenge model in which uninfected naive mice and memory mice, generated from prior X31 infection, are simultaneously challenged with PR8 (H1N1) virus with concurrent FTY720 treatment. (C) Heterosubtypic protection in FTY720-treated mice. Shown are weight loss morbidity plots at indicated days post-infection (p.i.) during primary and recall influenza challenge of WT C57BL/6 mice (left) and viral titers in the BAL assessed on day 6 after challenge (right). Data were compiled from two independent experiments; n = 10–12 mice/group. (D) Lung-localized protection is independent of B cells. Weight loss morbidity (left) and BAL viral titers determined at day 7 after infection (right) after PR8 challenge of µMT naive and memory mice. Data were compiled from two experiments; n = 10–14 mice/group. Significance was determined using Student’s unpaired t test; ****, P < 0.0001; ***, P ≤ 0.001; **, P < 0.01; *, P ≤ 0.05. All error bars show mean ± SEM. n.s., not significant.
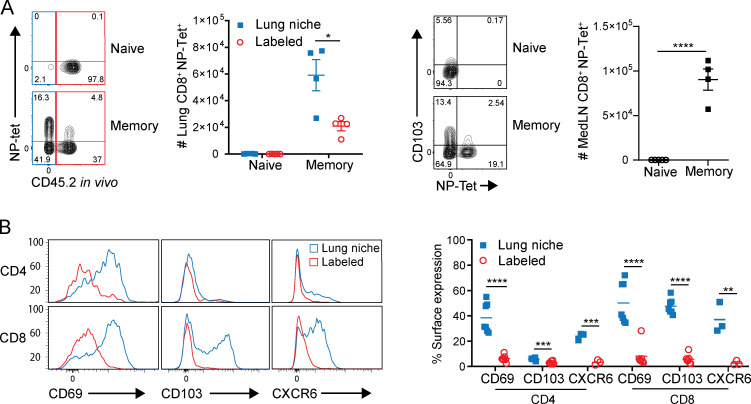
Figure S1.
Primary infection generates influenza-specific lung TRMs. (A) Lung cells isolated from mice infected 3–5 wk after primary infection. Left: Flow cytometry plots gated on lung CD44+CD8+ showing NP tetramer expression by lung niche (blue) and labeled (red) cells and graph showing number of lung CD44+CD8+NP-Tet+ cells. Right: Flow cytometry plots gated on medLN CD44+CD8+ showing CD103 and NP tetramer expression and graph depicting absolute number of medLN CD44+CD8+NP-Tet+ T cells. Data are representative of two experiments; n = 4 mice/group. (B) Lung niche T cells from memory mice express surface TRM markers. Surface expression of CD69, CD103, and CXCR6 by lung niche (blue) versus circulating (labeled; red) T cells shown as representative histograms (left) and graphs showing individual mice (right) treated with FTY720 for 2 d. Data were compiled from two independent experiments; n = 7 mice/group. Significance was determined by Student’s unpaired t test; ****, P ≤ 0.0001; ***, P ≤ 0.001; **, P ≤ 0.01; *, P ≤ 0.05. Error bars are mean ± SEM.
We used a modified mouse model of heterosubtypic immunity to assess local protective immunity to viral challenge in the context of a polyclonal T cell response. Naive and memory mice previously infected with the X31(H3N2) strain of influenza virus were treated with the sphingosine-1-phosphate receptor (S1PR1) agonist FTY720 before and during challenge with the heterosubtypic PR8 (H1N1) strain of influenza in order to minimize the contributions of circulating cells at the time of challenge (Fig. 1 B); protective immunity was measured at sequential time points after challenge compared with primary influenza infection of naive mice. In the presence of FTY720, which selectively reduces the number of circulating (labeled) T cells in the lung but maintains TRM in the lung niche (Turner et al., 2014; Zens et al., 2016), memory mice exhibited significantly reduced weight loss morbidity compared with primary infection of naive mice, and significantly lower lung viral load at 6 d after infection (Fig. 1 C)—the time point of peak viral load for primary infection (Teijaro et al., 2010). Notably, reduced morbidity and near-complete viral clearance (Fig. 1 D) were also observed in B cell–deficient (µMT) memory mice compared with primary infection of μMT naive mice, which lack mature B cells (Kitamura et al., 1991). These data provide evidence that local T cell memory directs protective immunity with the canonical features of an enhanced and rapid recall response, independent of humoral immunity.
Rapid accumulation and proliferation of T cells in the lung tissue–resident niche in recall responses to influenza challenge
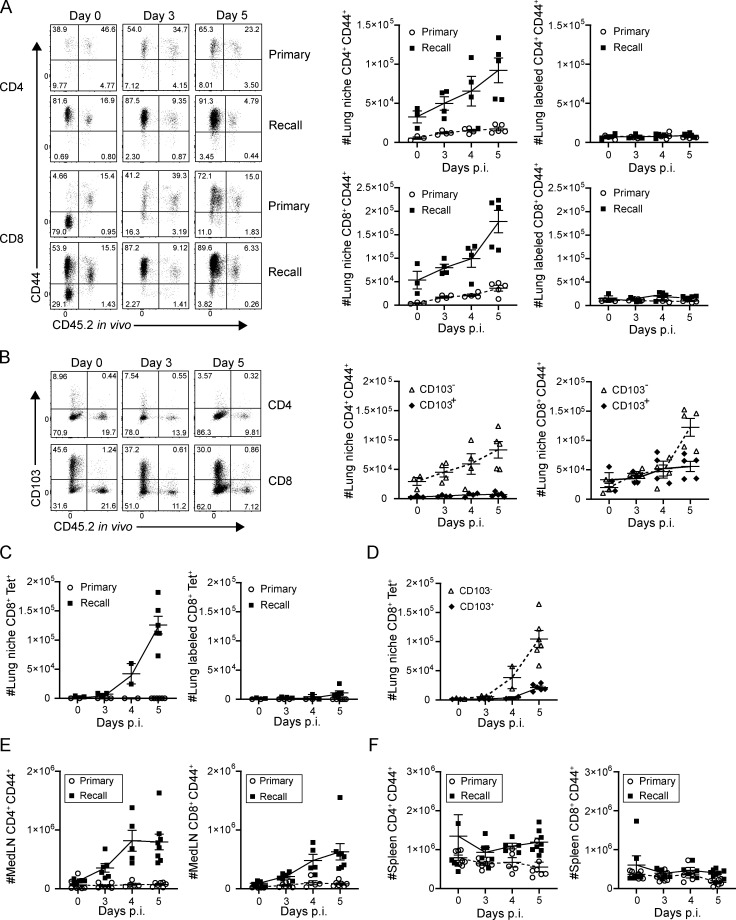
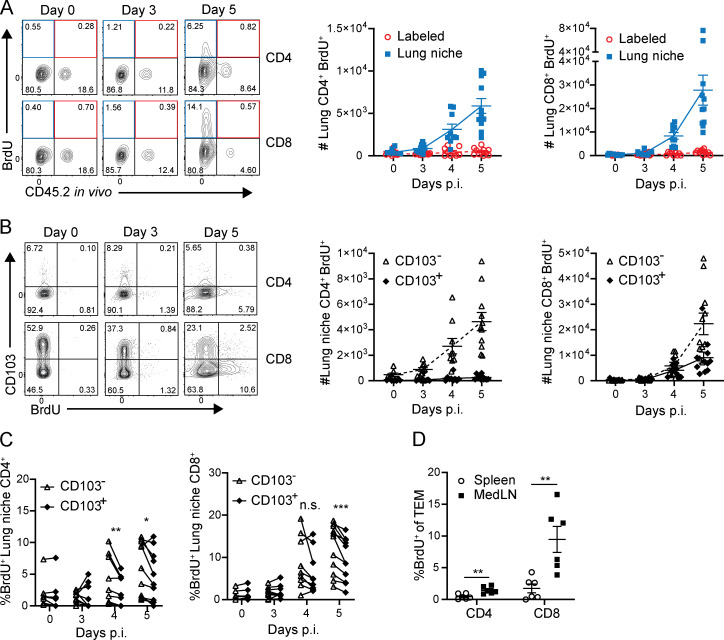
We further investigated the lung T cell dynamics that contribute to the protective recall response. After PR8 challenge, the numbers of CD4+ and CD8+CD44hi T cells within the lung-resident niche significantly increased at 4–5 d after infection in memory mice (recall) compared with the primary response (primary; Fig. 2 A, left). By contrast, circulating lung CD4+ and CD8+ T cells identified by in vivo antibody labeling did not increase in number after infection (Fig. 2 A, right), revealing that the enhanced accumulation of T cells during recall is specific to the lung tissue niche. This accumulation of lung niche T cells in the recall response was similar with or without FTY720 treatment (Fig. S2 A), suggesting that in situ expansion may be occurring. However, for both CD4+ and CD8+ T cells, the major subset accumulating in the lung-resident niche during the recall response was deficient in CD103, the TRM marker (Fig. 2 B), suggesting that non-TRM may contribute to T cell expansion. A similar pattern of rapid accumulation of lung niche CD103− cells was observed with influenza nucleoprotein (NP)-specific CD8+ T cells in the recall response (Fig. 2, C and D). In addition, there were increased numbers of memory CD4+ and CD8+CD44+ T cells in the recall (but not primary) response in the local medLN (Fig. 2 E) but not the spleen of memory mice (Fig. 2 F). Together, these results show robust, local, virus-driven expansion of T cells in the lung niche and associated LN in response to infectious challenge; however, the source of the expanding T cells required further investigation.
Figure 2.
Tissue-localized protection to heterosubtypic challenge is mediated by rapid lung niche T cell expansion. Lung T cells were isolated from naive and memory mice following in vivo antibody labeling at the indicated days post-infection (p.i.), as in Fig. 1. (A) Accumulation of CD4+CD44+ (upper) and CD8+CD44+ (lower) T cells in the lung following primary and recall challenge is shown in representative flow cytometry plots (left) and in graphs depicting absolute numbers of T cells in the lung niche (middle) and labeled (right) CD44+ T cells. (B) Accumulation of CD103− and CD103+CD4+CD44+ and CD8+CD44+ T cells in the lung during the recall response is shown in representative flow cytometry plots (left), and graphs show absolute numbers of lung niche CD4+CD44+ (middle) and CD8+CD44+ (right) T cells. Data from A and B are from one experiment with three to five mice per group, representative of three independent experiments. (C) Accumulation of lung niche (left) and lung labeled (right) influenza-specific tetramer+ (NP-Tet+) CD8+ T cells over the course of a primary or recall response. (D) Accumulation of CD103− and CD103+CD8+CD44+ NP-Tet+ lung niche T cells during the recall response. Data from C and D were compiled from two independent experiments; n = 2–6 mice/group. (E) Absolute numbers of medLN CD4+CD44+ (left) and CD8+CD44+ (right) T cells during primary and recall responses to PR8 challenge. (F) Absolute numbers of spleen CD4+CD44+ (left) and CD8+CD44+ (right) T cells during primary and recall responses to PR8 challenge. Data from E and F are compiled from three experiments; n = 6–8 mice/group. All error bars show mean ± SEM.
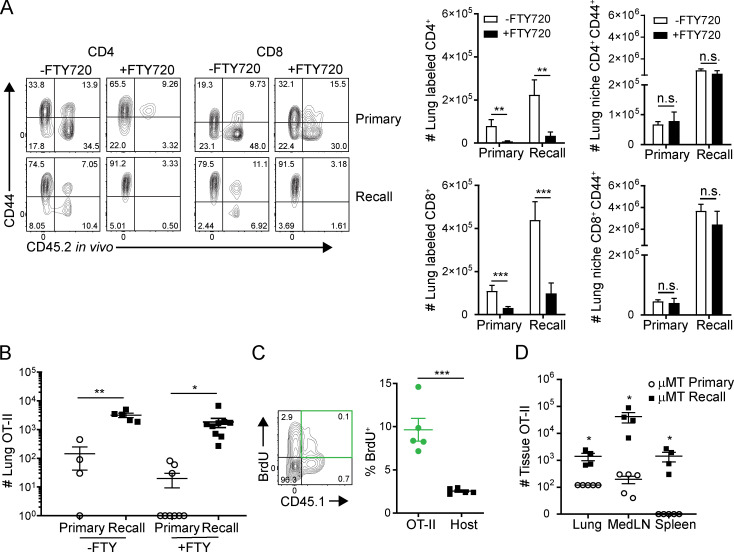
Figure S2.
Lung niche T cell migration during influenza infection and FTY720 treatment. (A) T cells were isolated from the lungs of memory and naive mice 5 d after PR8 challenge with or without daily FTY720 treatment. Left: Representative flow plot of lung CD4+ and CD8+ T cells showing CD44 expression versus in vivo administered CD45.2 antibody labeling. Right: Histograms depicting number of CD44+CD4+ or CD8+ T cells either labeled or protected by in vivo fluorescent antibody with or without FTY720 treatment. n = 4 or 5 mice per group. (B) CD44loCD45.1+ OT-II indicator population is transferred into both naive and memory mice and challenged with PR8-OVA (PR8-OTII). Lung cells were isolated 5 d after PR8-OVA challenge. Graph depicts number of lung CD4+CD45.1+ OT-II T cells during the primary and recall responses with or without FTY720 treatment. Data were compiled from two independent experiments; n = 4–9 mice/group. (C) BrdU incorporation by lung OT-II cells and CD45.1− host polyclonal cells 5 d after recall challenge with PR8-OVA. Data are representative of two independent experiments; n = 3–5 mice/group. (D) OT-II cells in the lung, medLN, and spleen of μMT hosts 4 d after primary or recall challenge shown in representative flow cytometry plots (left) and graphs depicting total numbers of OT-II cells in each tissue site in primary or recall responses. Data are representative of two independent experiments; n = 3–5 mice/group/experiment. Significance was determined by Student’s unpaired t test; ***, P ≤ 0.001; **, P ≤ 0.01; *, P ≤ 0.05. Error bars show mean ± SEM. n.s., not significant.
To determine the source of enhanced lung niche T cell expansion, in situ T cell proliferation was assessed by short-term in vivo BrdU administration. BrdU+CD4+ and CD8+ T cells were detectable 4–5 d after challenge predominantly in the lung-resident niche and not within circulating (labeled) cells (Fig. 3 A). For CD8+ T cells, BrDU+ cells were detected among the CD103+ as well as CD103− subsets, with a significant increase in BrdU+CD103− cells compared with CD103+ TRM counterparts by absolute numbers (Fig. 3 B) and frequencies (Fig. 3 C). (BrdU+ cells among lung CD4+ T cells are largely CD103−, consistent with the low frequency of CD103+CD4+ TRM.) The preferential BrdU incorporation by CD103− rather than CD103+ lung T cells even at early time points before accumulation suggests that TRM may not be the sole source of proliferating T cells in situ. Accordingly, BrdU incorporation during a recall response was significantly higher in medLN CD44hiCD62Llo (TEM) T cells than in the spleen (Fig. 3 D). These data suggest that local proliferation and expansion of T cells in the lung-draining LN may contribute to the proliferation and accumulation of lung niche T cells during the recall response.
Figure 3.
Heterosubtypic recall promotes rapid proliferation of CD103− T cells in the lung niche and medLN. Lung, medLN, and spleen T cells were isolated from naive and memory mice challenged as in Fig. 2 A following administration of BrdU i.p. and i.n. 2 h before tissue harvest. (A) BrdU incorporation by lung niche and labeled T cells is shown in representative flow cytometry plots (left) and graphs depicting absolute numbers of CD4+CD44+ (middle) and CD8+CD44+ lung niche (blue) and labeled (red) BrdU+ T cells. (B) BrdU incorporation of lung niche T cells as a function of CD103 expression depicted in representative flow cytometry plots (left) and graphs showing the number of BrdU+ lung niche CD103− and CD103+CD44+CD4+ and CD8+T cells (right). A and B were compiled from three independent experiments; n = 7–11 mice/group. (C) Graphs showing frequency of BrdU expression by CD103+ and CD103− lung niche CD44+CD62L−CD4+ (left) and CD8+ (right) T cells, with paired values shown for each mouse. Data were compiled from three independent experiments; n = 7–11 mice/group. Significance was determined by paired t test between CD103− and CD103+ groups. (D) Graph depicting medLN and spleen CD4+ and CD8+ TEM (CD44+CD62L−) BrdU incorporation during day 5 of the recall response. C and D were compiled from two independent experiments; n = 5–7 mice/group. Significance was determined by Student’s unpaired t test except for C. ***, P ≤ 0.001; **, P ≤ 0.01; *, P ≤ 0.05. All error bars show mean ± SEM. n.s., not significant.
Recall response promotes enhanced activation of new lung-homing effector cells in the medLN
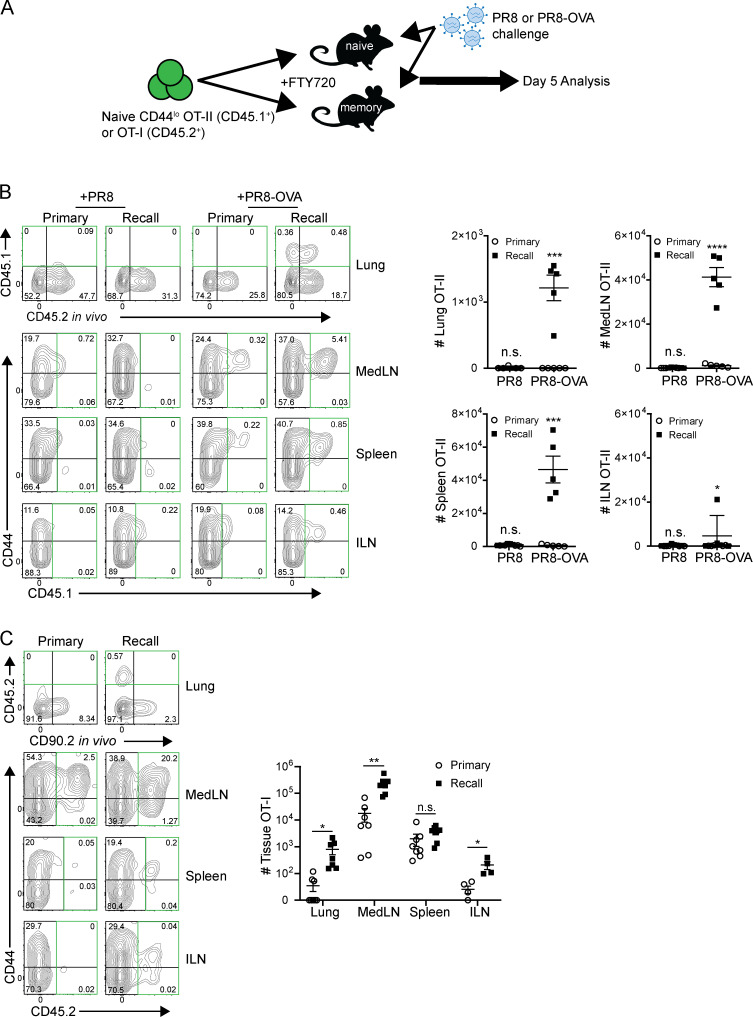
To assess the contribution of influenza-specific T cell activation and migration from the periphery to the lung-localized recall response, we adoptively transferred naive CD44lo OT-II (CD4+) cells expressing a transgene-encoded TCR specific for OVA peptide into naive and memory mice that were subsequently challenged with a recombinant PR8-OVA strain specific for the OT-II TCR, or PR8 virus as a control (Fig. 4 A). This experimental setup allowed us to address whether peripheral T cells were involved in the lung tissue–resident response and whether responses to newly introduced epitopes during the challenge were similar or different during recall versus primary infection.
Figure 4.
Recall response enhances local LN T cell priming and migration to the lung tissue niche. (A) Experimental schematic for tracking new effector and circulating T cell responses: CD44lo OT-II CD4+ or OT-I CD8+ transgenic T cells were transferred into naive and memory mouse hosts, which were subsequently challenged (in the presence of FTY720 treatment) with PR8 as a control and/or recombinant PR8-OVA expressing the OT-II or OT-I peptide epitope, respectively; tissues were harvested 4–5 d after infection following in vivo antibody labeling. (B) The presence of OT-II (CD45.1+) CD4+ T cells in the indicated sites of mice at day 5 of the primary or recall challenge with PR8 or PR8-OVA is shown in representative flow cytometry plots (left, OT-II–containing quadrants outlined in green) and in graphs (right) depicting total numbers of OT-II cells in each tissue site for each challenge condition in individual mice. FACS plots are gated on total CD4+ cells. Data are representative of two independent experiments with n = 3–5 mice/group. (C) The presence of OT-I (CD45.2+) CD8+ T cells in the indicated sites of mice at day 5 of the primary or recall challenge with PR8-OVA is shown in representative flow cytometry plots (left, OT-I–containing quadrants outlined in green) and in graphs (right) depicting total numbers of OT-I cells in each tissue site for individual mice. FACS plots are gated on total CD8+ T cells. Data are compiled from two independent experiments; n = 4–8 mice/group. Significance between primary and recall groups was determined by Student’s unpaired t test; ****, P ≤ 0.0001; ***, P ≤ 0.001; **, P ≤ 0.01; *, P ≤ 0.05. All error bars show mean ± SEM. n.s., not significant.
In response to PR8-OVA challenge, there were increased frequencies and numbers of OT-II cells in the lung, medLN, and spleen at 5 d after infection in memory mice compared with negligible to low numbers of OT-II cells in any site during primary infection (Fig. 4 B). Minimal numbers of OT-II cells were detected in the inguinal LN (ILN) distal to the infection site in both naive and memory hosts, indicating that priming of OT-II cells occurred local to the infection site. Like the broader polyclonal T cell response, OT-II expansion in the medLN and migration to the lung occurred independently of FTY720 treatment (Fig. S2 B) and proliferated in situ (Fig. S2 C). Moreover, OT-II cells were minimally present in tissues during primary or recall challenge with PR8 lacking the OVA epitope (Fig. 4 B), indicating that bystander migration was not sufficient to recruit and/or activate OT-II CD4+T cells.
Because B cells are a major APC, we investigated whether expansion and migration of T cells during the recall response also occurred in µMT memory mice. Similar to results in WT hosts, OT-II cells were present in the lung-resident niche, medLN, and spleen in significantly greater numbers in memory μMT mice than in naive μMT mice at 4 d after challenge with PR8-OVA (Fig. S2 D). These results show that local recall responses are associated with enhanced generation of effector T cells from the local draining LN independent of B cell–mediated priming, including T cells specific for newly encountered influenza epitopes.
We investigated whether this enhanced priming of CD4+ T cells for new epitopes introduced during the recall response also occurred for CD8+ T cells. OVA-specific OT-I TCR transgenic CD8+ T cells were transferred into mouse hosts subsequently challenged with a different recombinant PR8-OVA strain expressing the epitope recognized by the OT-I TCR (Fig. 4 A; see Materials and methods). In response to PR8-OVA challenge, OT-I cells were present in significantly increased numbers in the lung and medLN (but not in the distal ILN or spleen) in the recall response compared with a primary response (Fig. 4 D). These results show enhanced activation, proliferation, and lung-specific migration of newly primed, influenza-specific effector cells in the presence of an ongoing recall response to influenza challenge.
Prior influenza infection fortifies local LNs for T cell priming
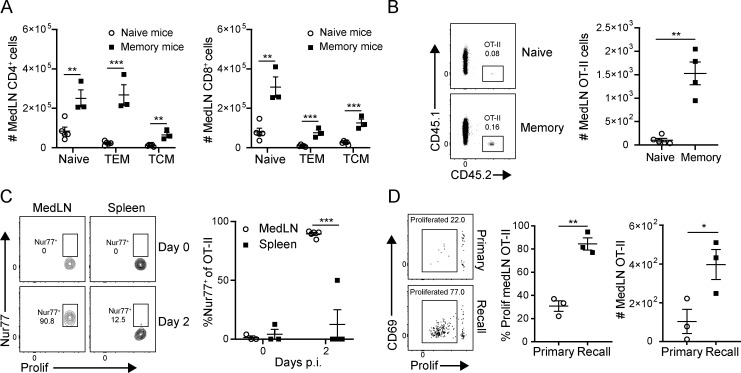
We hypothesized that the local LN environment in memory mice promoted activation and priming of new influenza-specific effector cells. Following previous influenza infection, there was a marked increase in T cell numbers—both naive and memory—even at 35 d after infection (Fig. 5 A), suggesting that local infection may confer preferential homing or retention of T cells in the medLN. Indeed, transfer of equal numbers of naive OT-II cells into the different hosts resulted in greater numbers of OT-II retained in the medLN of memory mice than in that of naive mice (Fig. 5 B), indicating that the environment of medLN in previously infected mice enabled increased retention of T cells compared with uninfected mice.
Figure 5.
Enhanced naive T cell recruitment, activation, and proliferation in the medLN during the recall response. (A) Prior influenza infection results in increased numbers of naive and memory T cells in the medLN. Graphs show numbers of indicated CD4+ and CD8+ T cell subsets (T-naive, CD44loCD62Lhi; TEM, CD44hiCD62Llo; central memory T cells [TCM], CD44intCD62Lhi) in the medLN of uninfected (naive) mice and memory mice 3–5 wk after infection. Data are representative of two experiments compiled from three to five mice per group. (B) Recruitment and/or retention of naive CD45.1+ OT-II CD4+ T cells in medLN 2 d following transfer of equal numbers (50,000 cells/mouse) into naive and memory congenic mice is shown in representative flow cytometry plots (left) and a graph (right) showing absolute numbers of OT-II cells, compiled from four or five mice per group. (C and D) Enhanced priming in the medLN of memory mice during recall. Proliferation dye–labeled (eFluor 450) OT-II/Nur77-GFP cells were transferred into naive and memory mouse hosts and subsequently challenged with PR8-OVA 1 d later. (C) Nur77-GFP expression by T cells isolated from medLN and spleen 2 d after challenge in the recall response is shown in representative flow cytometry plots gated on OT-II cells (left) and graphs (right) showing frequency of GFP+ cells. (D) Proliferation of OT-II T cells in the medLN 3 d after PR8-OVA challenge measured by proliferation dye dilution is shown in representative flow cytometry plots (left) and graphs depicting the percentage of OT-II cells divided (middle) and total number of OT-II cells (right). Data shown are representative of two independent experiments; n = 3–5 mice/group. Significance was determined by Student’s unpaired t test; ***, P ≤ 0.001; **, P ≤ 0.01; *, P ≤ 0.05. Error bars show mean ± SEM.
We then sought to clarify whether increased medLN T cell expansion during the recall response was due to enhanced activation and proliferation. At day 2 after challenge with PR8-OVA, nearly all of the OT-II cells in the medLN of memory mice expressed the early TCR activation marker Nur77 (known to be induced several hours after TCR signaling; Ashouri and Weiss, 2017; Cunningham et al., 2006), whereas OT-II cells in the spleen did not express Nur77 at this early time point (Fig. 5 C), demonstrating that the medLN is the initial site of early influenza-specific T cell activation. By day 3 after challenge, there was already extensive proliferation of medLN OT-II cells during the recall response compared with a significantly lower proliferation (and greatly reduced numbers) of medLN OT-II cells in the primary response (Fig. 5 D). These data demonstrate that prior influenza infection fortifies the local medLN to promote TCR-mediated signaling and proliferation during heterosubtypic challenge.
Increased numbers of cDCs persist in the medLN following influenza infection
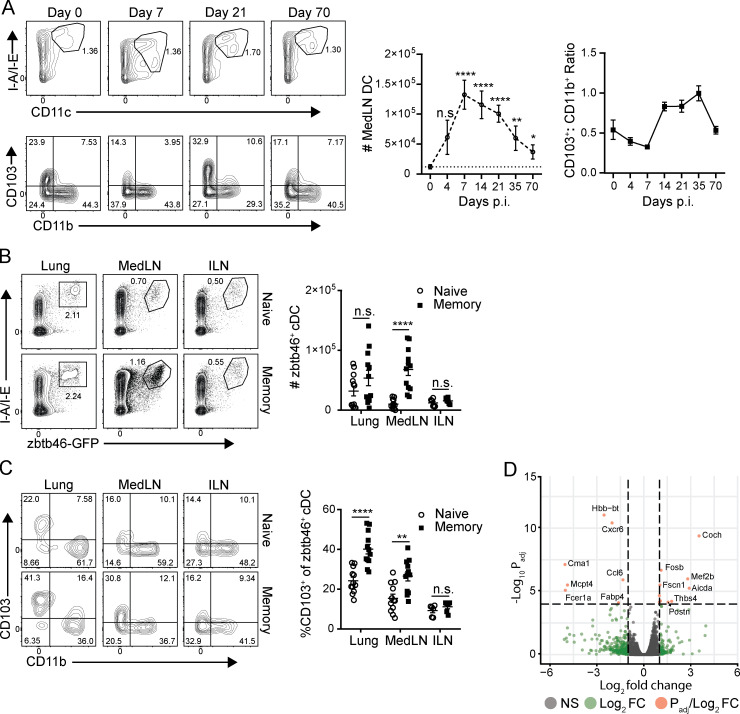
We examined the role of cDCs in promoting enhanced local T cell activation during the recall versus primary response. The number of medLN cDCs (CD45+CD11c+ I-A/I-Ehi) increased in the first week following primary infection, and this increase persisted up to 70 d after primary challenge (Fig. 6 A). The ratio of CD11b−CD103+ lung-migratory DCs to CD11b+ LN-resident DCs (CD103+CD11b+) increased significantly at 7–14 d after infection, which persisted for several weeks thereafter, reverting to baseline levels by day 70 after infection (Fig. 6 A). To better identify cDCs between sites, because CD11c can also be expressed by tissue macrophages (Helft et al., 2015), we assessed cDC numbers based on expression of the cDC-specific transcription factor zbtb46 (Meredith et al., 2012; Satpathy et al., 2012) using zbtb46-GFP reporter mice (Satpathy et al., 2012). Consistent with the DC results based on CD11c surface expression, zbtb46+ cDCs are present in greater numbers specifically within the medLN of memory mice than in that of naive (uninfected) mice, whereas cDC numbers in the nondraining ILN and lung are unchanged between naive and memory mice (Fig. 6 B). This quantifiable increase in cDC content induced from prior influenza infection is therefore specific to the local lung-draining LN and is not present at distal sites, nor were increased DC numbers found in the lung itself.
Figure 6.
Influenza infection promotes long-term quantitative increases of DCs in the medLN. (A) Persistence of lung-migratory DCs in the medLN following influenza infection. Left: Frequency of total DCs (CD11c+MHC class II+; top) and lung-migratory cDCs (CD11b−CD103+; bottom) in the medLN at indicated time points after primary X31 infection shown in representative flow cytometry plots (left) and in graphs depicting absolute numbers of medLN CD45+CD11c+ I-A/I-Ehi DCs (top) and ratio of CD11b−CD103+/CD11b+CD103− DCs (bottom) over time after infection. Data are compiled from two independent experiments; n = 4–8 mice per group. (B) Quantification of cDCs using zbtb46-GFP reporter mice. Left: Flow cytometry plots show zbtb46-GFP expression gated on CD45+ cells from indicated sites of naive or memory mice. Right: Graphs show absolute numbers of zbtb46+CD11chi I-A/I-Ehi cDCs in these sites compiled from four experiments; n = 6–12 mice/group. (C) CD103 expression is enhanced in cDCs from memory mice. CD103 and CD11b expression by zbtb46-GFP+ cDCs in indicated sites of naive and memory mice is shown in representative flow cytometry plots (left) and graphs displaying proportion of CD11b−CD103+zbtb46-GFP+ cDCs (right) compiled from four experiments; n = 6–12 mice/group. (D) Whole-transcriptome profiling by population RNA-seq of CD45+zbtb46-GFP+CD11c+ I-A/I-E+ cDCs sorted from medLNs of naive and memory mice as in B. Volcano plot depicts genes by the absolute value of log10Padj and the relative fold change comparing memory cDCs with naive cDCs. Genes with differential gene expression of at least twofold change are shown in green, and genes with an absolute value of log10Padj >4 are shown in red. For all experiments, significance between naive and memory mice was determined by Student’s unpaired t test; ****, P ≤ 0.001; **, P ≤ 0.01; *, P ≤ 0.05. Significance in A was determined by comparison with number of medLN DCs at day 0. Error bars show mean ± SEM. n.s., not significant.
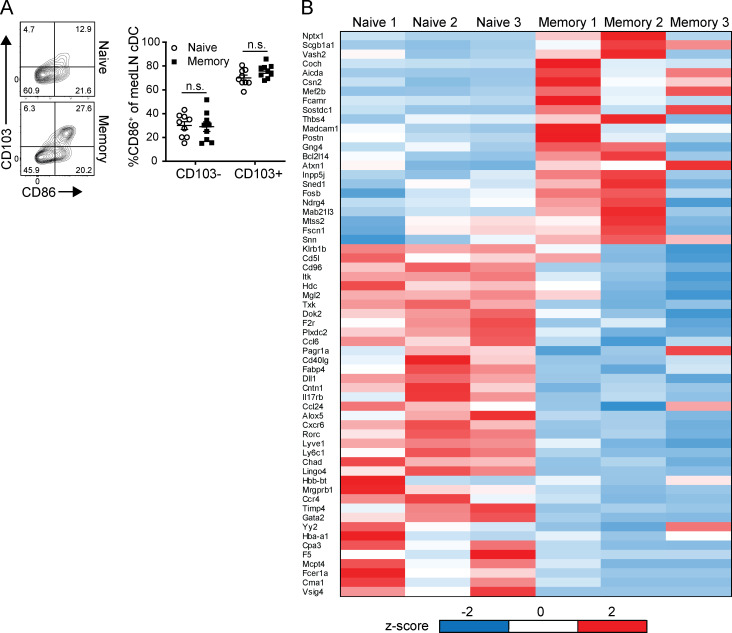
We next examined the augmented medLN cDC population for qualitative functional changes, including markers of migration and activation. Zbtb46+ cDCs from both the lung and medLN of memory mice contained higher proportions of CD11b−CD103+ migratory cells (Fig. 6 C), which also express higher levels of the costimulatory ligand and DC activation marker CD86 (Fig. S3). (There was no change in the level of CD86 expression for a given cDC subset between naive and memory mice.) In an unbiased approach to identifying potential changes in medLN cDCs between naive and memory mice, we analyzed the transcriptome profile of zbtb46+ cDCs from the medLN of naive and memory mice by RNA sequencing (RNA-seq; Fig. 6 D). Gene expression between the two cDC populations was highly similar, with only 61 genes differentially expressed (Fig. 6 D and Fig. S3 B), including the tissue residence marker Cxcr6 and chemokines Ccl6 and Ccl24, but without a clear signature of increased cDC activation or antigen presentation. These data show increased numbers and migration of zbtb46+ cDC populations in the medLN of memory mice that are transcriptionally similar to their naive counterparts, suggesting that the enhanced T cell priming during the recall response derives from greater numbers of cDCs.
Figure S3.
Qualitative analysis of cDCs in the medLN of memory compared with naive mice. (A) CD103+ lung migratory cDCs exhibit elevated CD86 expression. Analysis of CD86 expression by CD103-expressing cDC subsets in naive and memory mice is shown in representative flow cytometry plots of medLN zbtb46+ cDCs (left) and graph depicting percentage CD86 expression by CD11b−CD103+ and CD11b+CD103− medLN cDCs (right). Data were compiled from three experiments; n = 9 mice/group. (B) Whole-transcriptome profiling of medLN cDCs from Fig. 6 D. Heat map of top differentially expressed genes between naive and memory cDCs based on transcripts per million (TPM) and determined by Padj ≤ 0.05 and mean fold difference ≥2. z-Score is based on SD from mean TPM value per gene. Error bars show mean ± SEM. n.s., not significant.
LN DCs are required for protection and enhanced CD4+ T cell priming in the recall response
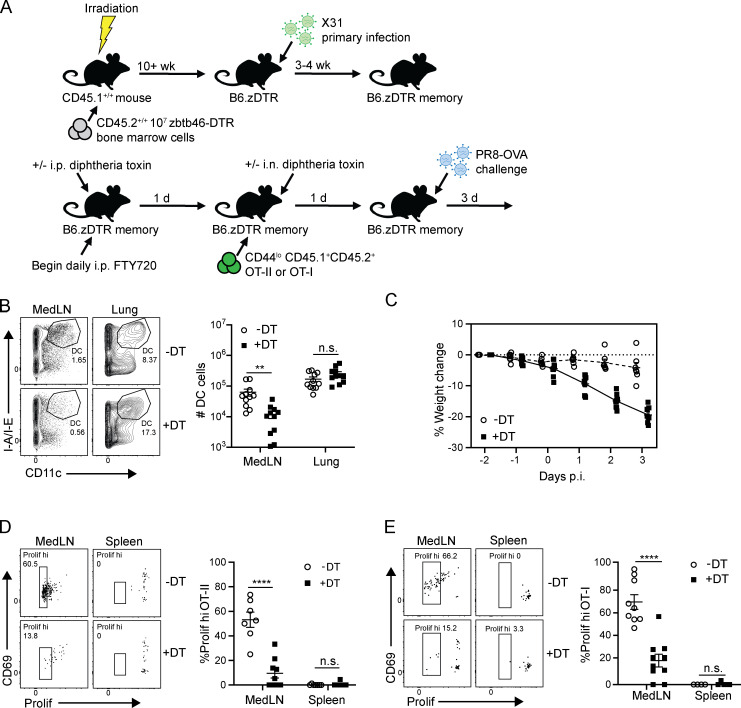
To assess the role of increased cDC numbers in key features of the recall response, including protective immunity and enhanced T cell priming within the medLN, we used mice expressing diphtheria toxin receptor (DTR) driven by the zbtb46 promoter (zbtb46-DTR) in order to deplete cDCs in vivo by diphtheria toxin (DT) administration. Due to broad expression of zbtb46 in nonhematopoietic cells (Satpathy et al., 2012), we generated bone marrow chimeras (B6.zDTR) using bone marrow cells derived from CD45.2+/+ zbtb46-DTR mice transferred into lethally irradiated CD45.1+/+ host mice (Fig. 7 A; see Materials and methods). B6.zDTR mice were infected with X31 to generate memory mice as shown previously, then treated with DT to deplete cDC; OT-I or OT-II cells were subsequently transferred into both DT-treated and untreated B6.zDTR memory mice before subsequent challenge with PR8-OVA (Fig. 7 A). In this way, we could assess the contribution of cDC numbers to enhanced priming and the overall recall response.
Figure 7.
medLN cDCs are required for enhanced T cell priming and protection during the recall response. (A) Schematic for cDC depletion experiments: B6 CD45.1+/+ mice were lethally irradiated and reconstituted with 107 CD45.2+/+ zbtb46-DTR bone marrow cells to create B6.zDTR bone marrow chimeras. After 10 wk, B6.zDTR chimeras were infected with X31 to generate B6.zDTR memory mice. B6.zDTR memory mice were treated with FTY720 and treated with either i.p. DT (40 µg/g body weight) or PBS as a vehicle control. 1 d later, mice were administered i.n. DT (20 µg/g body weight) or PBS vehicle control, and eFluor 450 proliferation dye–labeled CD45.1+CD45.2+ OT-II or OT-I cells were transferred into FTY720-treated B6.zDTR memory mice. 1 d later, mice were challenged with PR8-OVA (see Materials and methods), and recall protection and OT-II/OT-I priming was assessed 3 d after infection. (B) Preferential reduction in medLN cDCs following DT treatment in B6.zDTR memory mice depicted in representative flow cytometry plots (left) gated on CD45+ cells showing CD11c+ cDCs in the medLNs and lungs of PR8-OVA–challenged mice and in graphs of cDC numbers in each site (right). Data are representative of two independent experiments; n = 3 or 4 mice/group. (C) Weight loss morbidity of DT-treated and untreated memory mice at indicated times after PR8-OVA challenge. Data were compiled from two independent experiments; n = 6 mice/group. (D) DT treatment abrogates enhanced priming of medLN OT-II cells. Left: Flow cytometry plots show medLN and spleen OT-II proliferation and CD69 expression with or without DT treatment. Right: Graph showing proportion of medLN and spleen OT-II cells that express diluted proliferation dye. (E) Same as D but showing medLN OT-I cells. Data are compiled from two independent experiments; n = 7–9 mice/group in D and n = 4–10 mice/group in E. Significance was determined by Student’s unpaired t test; ****, P ≤ 0.0001; **, P ≤ 0.01. Error bars exhibit mean ± SEM. n.s., not significant.
Following influenza challenge, the frequency and numbers of cDCs were significantly reduced in the medLN of DT-treated compared with untreated mice; however, cDC numbers in the lung were comparable in infected hosts with or without DT (Fig. 7 B). This medLN-localized cDC depletion was associated with significant weight loss in DT-treated mice compared with the untreated group (Fig. 7 C), demonstrating a loss of T cell–mediated protection. The proliferation of both OT-II and OT-I cells in the medLN was significantly reduced in DT-treated compared with untreated mice (Fig. 7, D and E). At the same time, transgenic T cells did not proliferate in the spleen, demonstrating that the enhanced antigen-specific T cell priming is locally regulated (Fig. 7, D and E). These results demonstrate that cDCs in the medLN are required for enhanced priming and proliferative expansion of influenza-specific T cells during the recall response that are critical for optimal protective immunity in the lung.
Discussion
Influenza infection generates cross-strain protective immunity in the lung, but how systemic responses contribute to lung-resident immunity have not been defined. Here, we reveal that lung-localized T cell protection to heterosubtypic influenza challenge is augmented by enhanced generation and lung migration of peripheral effector T cells from the local LN. During secondary influenza challenge, increased numbers of T cells in the lung niche early after infection derive from in situ TRM proliferation along with the recruitment of effector cells generated in response to newly encountered influenza epitopes in the medLN. Importantly, prior influenza infection imprints the medLN with greater numbers of cDCs that persist for months after infection, resulting in more rapid priming of T cells, and that are required for optimal protection. Our results showing that the efficacious secondary response to respiratory infection in the lung is augmented by the fortified priming environment in the local lymphoid tissue demonstrate a new mechanism for cross-strain immunity for viruses, important for targeting in vaccines.
Prior influenza infection generates high frequencies of memory T cells that reside within the lung and local medLN that are enriched for reactivity to influenza-derived epitopes (Turner et al., 2014; Wu et al., 2014; Zens et al., 2016, 2017). Lung TRMs can be readily identified by their localization in the lung tissue niche that is inaccessible to circulation and protected by labeling with intravenously administered antibody. Heterosubtypic influenza challenge of memory mice led to protective responsessuch as reduced weight loss morbidity and more rapid viral clearance compared with the primary response, independently of mature B cells, consistent with previous studies showing that B cells were not required for memory T cell–directed protection to influenza challenge (McKinstry et al., 2012; Teijaro et al., 2010). This polyclonal T cell–directed response was marked by extensive accumulation of T cells in the lung-resident niche, consistent with TRM expansion in other tissue sites following recall challenge (Beura et al., 2018; Park et al., 2018). However, the expanding T cells in the lung-resident niche did not predominantly exhibit phenotypic features of TRMs such as CD103 expression, contrary to findings in skin and female reproductive tract (Beura et al., 2018; Park et al., 2018). The specific and distinct anatomical features of the lung, including extensive vascularization of the tissue and associated LNs, may influence the relative involvement of tissue and systemic responses in different sites.
Using a tracking model for monitoring activation and migration of peripheral naive, OVA-specific T cells, we demonstrate that the environment within the medLN of previously infected mice promotes superior T cell effector generation and lung migration after challenge for newly encountered influenza epitopes. These results indicate a broadening of the early T cell response during a recall challenge. A previously described effect of prior infection on subsequent immune responses is referred to as “original antigenic sin,” whereby antibody production in the recall response favors the original antigens, resulting in a narrowing of the antibody repertoire over repeated pathogen exposures (Francis, 1960; Kim et al., 2009; Zhang et al., 2019). The amplification of new T cell responses during recall identified here suggests that T cell immunity may be more robust than humoral immunity in promoting universal or broad-based protection, particularly related to viral strains for which neutralizing antibodies do not cross-react. The generation of T cell responses to SARS-CoV-2 in previously infected individuals (Grifoni et al., 2020) suggests that promoting T cell immunity could be a universal vaccine strategy for the current pandemic.
We identified that enhanced priming in the recall response occurred in local medLNs that exhibited a fortified state, maintaining larger numbers of T cells and increased numbers of lung-migratory cDCs. Recent studies have identified components of the innate immune response that become altered from previous infection and exhibit enhanced responses during recall—a phenomenon known as “trained immunity” (Mitroulis et al., 2018; Netea et al., 2016). In the lungs, infection with adenovirus induced long-term changes in alveolar macrophages with superior functional responses to viral challenge (Yao et al., 2018). Here, we describe durable alterations in the local medLN following influenza infection manifested by elevated numbers of migratory CD103+ cDCs that were specifically augmented in the medLN but not in the nondraining LN (Beauchamp et al., 2010; Ho et al., 2011; Kedl et al., 2017). These cDCs were essential for promoting enhanced T cell activation in the medLN and for efficacious protection. Interestingly, we found that numbers of CD103+ cDCs gradually declined to baseline levels several months after infection, which may also account for the observation that heterosubtypic immunity is lost over time (Slütter et al., 2017).
Our results demonstrating endogenous boosting properties of the local lymphoid environment have implications for generating protective immunity to diverse respiratory infections in vaccines. Protective lung TRM can be generated by diverse respiratory pathogens, including respiratory syncytial virus, tuberculosis, and pertussis (Morabito et al., 2017; Sakai et al., 2014; Wilk et al., 2017). I.n. routes of immunization can also generate lung TRMs and protective immunity to influenza and pertussis in mouse models (Wilk et al., 2017; Zens et al., 2016). In light of the emergence of several novel coronaviruses over the past decade, we propose that site-specific boosting in the respiratory tract can promote more robust responses to new or heterologous respiratory pathogens through targeting the local fortified LN and thus augment in situ T cell–mediated immunity.
Materials and methods
Mice
zbtb46-GFP, zbtb46-DTR (Satpathy et al., 2012), µMT (Kitamura et al., 1991), CD45.1, Nur77-GFP(Moran et al., 2011), OT-II, OT-I (The Jackson Laboratory), and C57BL/6 mice (Charles River) were housed and bred in specific pathogen–free Columbia University Medical Center (CUMC) animal facilities. Infections were performed in BSL-2 level biocontainment animal facilities. All animal studies were approved by the Columbia University Institutional Animal Care and Use Committee.
Influenza virus infection
C57BL/6 mice were anesthetized with isoflurane and infected i.n. with influenza virus diluted in 30 µl of PBS. To generate memory mice, uninfected naive mice were infected with X31 influenza virus at a 50% tissue-culture infective dose (TCID50) of 5,000. For heterosubtypic challenge, 4,000 TCID50 of PR8 was administered i.n. to mice. For experiments with transferred CD4+ OT-II T cells, mice were challenged with 2,000 TCID50 of a recombinant PR8-OVA strain expressing the OVA323–339 peptide (sequence ISQAVHAAHAEINEAGR; provided by Dr. Paul Thomas, St. Jude Children's Research Hospital, Memphis, TN; Thomas et al., 2010). Mouse hosts with transferred CD8+ OT-I T cells were challenged with 200–500 TCID50 of recombinant PR8-OVA expressing the OVA257–264 peptide (sequence SIINFEKL; provided by Dr. Adolfo Garcia-Sastre, Mount Sinai, New York, NY; Jenkins et al., 2006). For µMT mice, 2,000 TCID50 of X31 was administered for primary infection, and 1,000–2,000 TCID50 of PR8 was administered for heterosubtypic challenge, based on dose responses for this strain.
Viral titers
Bronchoalveolar lavage (BAL) was isolated from control and infected mice by flushing lungs with 1 ml of PBS. Viral titers were measured by hemagglutination of chicken RBCs from serial dilutions of BAL co-cultured with Madin-Darby canine kidney cells as previously described (Teijaro et al., 2010), and the Reed-Muench method was used to calculate TCID50.
FTY720 treatment
FTY720/Fingolimond (Sigma) was diluted in normal saline and injected i.p. daily into mice (200 µl/mouse) at a dose of 1 mg/kg body weight. FTY720 treatment was initiated 2 d before heterosubtypic challenge and maintained throughout secondary infection.
In vivo antibody labeling and flow cytometry
Mice were injected i.v. with 2.5 µg of Alexa Fluor 488– or Alexa Fluor 647–conjugated anti-CD45.2 (clone 104) or anti-Thy1.2 (clone 30-H12) antibody. After 5 min, lung lobes were collected and mechanically disrupted using gentleMACS tubes (Miltenyi Biotec) with a spleen 4.1 dissociation protocol. Lung tissue was incubated in Liberase TM (Roche) solution (20 µg/ml in DMEM) at 37°C for 1 h before dissociation with gentleMACS. LN cells were minced and incubated at 37°C in Liberase TM solution and mechanically dissociated by pipetting up and down. The spleen was minced and incubated at 37°C for 30 min in Liberase TM solution before being ground through a 70-µm filter. Cells were stained for 30 min at room temperature in PBS containing 2% FBS using the antibodies listed in Table S1. The CD8 tetramer was specific for influenza A NP linear epitope ASNENMETM (MBL International). For BrdU detection, cells were fixed and permeabilized using the Phase-Flow Alexa Fluor 647 BrdU kit (BioLegend). Cell numbers were determined using CountBright Absolute Counting Beads (BD Biosciences). All samples were acquired on a BD Fortessa, FACSCanto, or LSRII flow cytometer. Data were analyzed using FlowJo version 10 software (Tree Star).
BrdU administration
2 h before being killed, mice were anesthetized with isoflurane and i.n. administered 0.5 mg of BrdU in aqueous solution (BioLegend). At the same time, 2 mg of BrdU was administered by i.p. injection.
DT treatment
Unnicked DT (List Biological Laboratories) was reconstituted at 1 mg/1 ml in PBS and aliquoted and stored at −80°C. Mice were injected i.p. at a dose of 40 ng/g 2 d before secondary challenge and were administered i.n. 50 µl (12 ng/µl) 1 d before secondary challenge.
Generation of B6.zbtb46-DTR bone marrow chimeras
CD45.1+/+ C57BL/6 mice were irradiated with 1,100–1,200 rad by γ-irradiation in a secure CUMC facility. Bone marrow cells were isolated and filtered in HBSS from femurs and tibias of sex-matched CD45.2+/+zbtb46-DTR−/+ mice, and 107 cells were i.v. injected into each irradiated mouse. Mice were maintained on antibiotic-containing water (2 mg/ml neomycin) for 1 wk after irradiation and allowed to recover for 10 wk after irradiation before immune reconstitution.
RNA-seq and analysis
CD45+zbtb46+CD11c+ I-A/I-Ehi DCs were FACS sorted from the medLN cells of naive and memory mice, and RNA was isolated from cell pellets using the RNeasy Mini Kit (Qiagen) and quantitated using an Agilent 2100 Bioanalyzer (Agilent Technologies). Triplicate samples from memory mice are derived from cDC samples of three mice, whereas triplicate samples from naive mice were derived from three independent DC samples, each of which was pooled from three individual mice, due to low cDC numbers in naive mice. For low-input sequencing, RNA-seq libraries were prepared and sequenced by the CUMC Genome Center using the SMART-seq v4 Ultra Low Input RNA kit (Takara Bio USA) to create amplified cDNA and using the Nextera XT kit (Illumina) for library preparation. Libraries were sequenced on the Illumina NovaSeq 6000 system at the Columbia Genome Center. RNA-seq reads were mapped using Kallisto (version 0.44.0) with default parameters to the mouse reference genome build GRCh38. Non–protein-coding genes were filtered out, and then the bottom third of genes were filtered out by their gene counts. Differential gene expression analysis was performed with DESeq2 using variance-stabilized transformation to generate normalized gene expression data. Genes with Padj ≤ 0.05 and absolute value of fold change ≥2 were considered to be differentially expressed. Heat maps were generated from transcripts per million counts generated from Kallisto pseudoalignment. Data are available from the Gene Expression Omnibus database (GSE144755).
Statistical analysis
Statistical analyses were performed using Prism software (version 8.0.0; GraphPad Software). Significance tests as well as cutoffs used are specified in each figure legend.
Online supplemental material
Fig. S1 shows lung tissue–localized TRM generation after primary influenza infection. Fig. S2 shows that polyclonal and OT-II transgenic T cell expansion in the lung after heterosubtypic influenza challenge is FTY720 independent and that lung OT-II responses after heterosubtypic challenge are consistent with polyclonal responses. Fig. S3 shows that medLN cDCs found in naive or memory mice are similar in terms of CD86 expression and exhibit few qualitative transcriptional differences. Table S1 is provided online and shows flow cytometry antibodies used for this study, including manufacturer, fluorophore, and antibody clone.
Supplementary Material
lists the flow cytometry antibodies used in this study.
Acknowledgments
We thank Dr. Andrew Yates and Dr. Stuart P. Weisberg for reviewing the manuscript and for their helpful comments and discussion and Dr. Pranay Dogra for assistance with RNA-seq analysis and helpful comments and discussion.
This work was supported by National Institutes of Health grants HL116136 and AI100119 awarded to D.L. Farber. D.H. Paik was supported by National Institutes of Health grant NIH T32GM007367. These studies were performed in the Columbia Center for Translational Immunology Flow Cytometry Core funded in part by the Office of the Director, National Institutes of Health, under awards S10RR027050 and S10OD020056. These studies also used the resources of the CUMC Cancer Center Flow Core Facility funded in part through the National Institutes of Health center grant P30CA013696 as well as the CUMC Cancer Center Radiation Research Core Facility.
Author contributions: D.H. Paik designed and performed experiments, analyzed data, and wrote the manuscript; D.L. Farber planned experiments, analyzed data, and wrote the manuscript.
References
- Ashouri J.F., and Weiss A.. 2017. Endogenous Nur77 is a specific indicator of antigen receptor signaling in human T and B cells. J. Immunol. 198:657–668. 10.4049/jimmunol.1601301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauchamp N.M., Busick R.Y., and Alexander-Miller M.A.. 2010. Functional divergence among CD103+ dendritic cell subpopulations following pulmonary poxvirus infection. J. Virol. 84:10191–10199. 10.1128/JVI.00892-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beura L.K., Mitchell J.S., Thompson E.A., Schenkel J.M., Mohammed J., Wijeyesinghe S., Fonseca R., Burbach B.J., Hickman H.D., Vezys V., et al. 2018. Intravital mucosal imaging of CD8+ resident memory T cells shows tissue-autonomous recall responses that amplify secondary memory. Nat. Immunol. 19:173–182. 10.1038/s41590-017-0029-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham N.R., Artim S.C., Fornadel C.M., Sellars M.C., Edmonson S.G., Scott G., Albino F., Mathur A., and Punt J.A.. 2006. Immature CD4+CD8+ thymocytes and mature T cells regulate Nur77 distinctly in response to TCR stimulation. J. Immunol. 177:6660–6666. 10.4049/jimmunol.177.10.6660 [DOI] [PubMed] [Google Scholar]
- Francis T. 1960. On the doctrine of original antigenic sin. Proc. Am. Philos. Soc. 104:572–578. [Google Scholar]
- Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S., et al. 2020. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 181:1489–1501.e15. 10.1016/j.cell.2020.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helft J., Böttcher J., Chakravarty P., Zelenay S., Huotari J., Schraml B.U., Goubau D., and Reis e Sousa C.. 2015. GM-CSF mouse bone marrow cultures comprise a heterogeneous population of CD11c+MHCII+ macrophages and dendritic cells. Immunity. 42:1197–1211. 10.1016/j.immuni.2015.05.018 [DOI] [PubMed] [Google Scholar]
- Ho A.W., Prabhu N., Betts R.J., Ge M.Q., Dai X., Hutchinson P.E., Lew F.C., Wong K.L., Hanson B.J., Macary P.A., et al. 2011. Lung CD103+ dendritic cells efficiently transport influenza virus to the lymph node and load viral antigen onto MHC class I for presentation to CD8 T cells. J. Immunol. 187:6011–6021. 10.4049/jimmunol.1100987 [DOI] [PubMed] [Google Scholar]
- Jenkins M.R., Webby R., Doherty P.C., and Turner S.J.. 2006. Addition of a prominent epitope affects influenza A virus-specific CD8+ T cell immunodominance hierarchies when antigen is limiting. J. Immunol. 177:2917–2925. 10.4049/jimmunol.177.5.2917 [DOI] [PubMed] [Google Scholar]
- Kedl R.M., Lindsay R.S., Finlon J.M., Lucas E.D., Friedman R.S., and Tamburini B.A.J.. 2017. Migratory dendritic cells acquire and present lymphatic endothelial cell-archived antigens during lymph node contraction. Nat. Commun. 8:2034 10.1038/s41467-017-02247-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.H., Skountzou I., Compans R., and Jacob J.. 2009. Original antigenic sin responses to influenza viruses. J. Immunol. 183:3294–3301. 10.4049/jimmunol.0900398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura D., Roes J., Kühn R., and Rajewsky K.. 1991. A B cell-deficient mouse by targeted disruption of the membrane exon of the immunoglobulin mu chain gene. Nature. 350:423–426. 10.1038/350423a0 [DOI] [PubMed] [Google Scholar]
- Koutsakos M., Illing P.T., Nguyen T.H.O., Mifsud N.A., Crawford J.C., Rizzetto S., Eltahla A.A., Clemens E.B., Sant S., Chua B.Y., et al. 2019. Human CD8+ T cell cross-reactivity across influenza A, B and C viruses. Nat. Immunol. 20:613–625. 10.1038/s41590-019-0320-6 [DOI] [PubMed] [Google Scholar]
- Kumar B.V., Ma W., Miron M., Granot T., Guyer R.S., Carpenter D.J., Senda T., Sun X., Ho S.H., Lerner H., et al. 2017. Human tissue-resident memory T cells are defined by core transcriptional and functional signatures in lymphoid and mucosal sites. Cell Rep. 20:2921–2934. 10.1016/j.celrep.2017.08.078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang S., Mozdzanowska K., Palladino G., and Gerhard W.. 1994. Heterosubtypic immunity to influenza type A virus in mice. Effector mechanisms and their longevity. J. Immunol. 152:1653–1661. [PubMed] [Google Scholar]
- Mackay L.K., Minnich M., Kragten N.A., Liao Y., Nota B., Seillet C., Zaid A., Man K., Preston S., Freestone D., et al. 2016. Hobit and Blimp1 instruct a universal transcriptional program of tissue residency in lymphocytes. Science. 352:459–463. 10.1126/science.aad2035 [DOI] [PubMed] [Google Scholar]
- Marshall D.R., Turner S.J., Belz G.T., Wingo S., Andreansky S., Sangster M.Y., Riberdy J.M., Liu T., Tan M., and Doherty P.C.. 2001. Measuring the diaspora for virus-specific CD8+ T cells. Proc. Natl. Acad. Sci. USA. 98:6313–6318. 10.1073/pnas.101132698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinstry K.K., Strutt T.M., Kuang Y., Brown D.M., Sell S., Dutton R.W., and Swain S.L.. 2012. Memory CD4+ T cells protect against influenza through multiple synergizing mechanisms. J. Clin. Invest. 122:2847–2856. 10.1172/JCI63689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMaster S.R., Wilson J.J., Wang H., and Kohlmeier J.E.. 2015. Airway-resident memory CD8 T cells provide antigen-specific protection against respiratory virus challenge through rapid IFN-γ production. J. Immunol. 195:203–209. 10.4049/jimmunol.1402975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meredith M.M., Liu K., Darrasse-Jeze G., Kamphorst A.O., Schreiber H.A., Guermonprez P., Idoyaga J., Cheong C., Yao K.H., Niec R.E., et al. 2012. Expression of the zinc finger transcription factor zDC (Zbtb46, Btbd4) defines the classical dendritic cell lineage. J. Exp. Med. 209:1153–1165. 10.1084/jem.20112675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitroulis I., Ruppova K., Wang B., Chen L.S., Grzybek M., Grinenko T., Eugster A., Troullinaki M., Palladini A., Kourtzelis I., et al. 2018. Modulation of myelopoiesis progenitors is an integral component of trained immunity. Cell. 172:147–161.e12. 10.1016/j.cell.2017.11.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morabito K.M., Ruckwardt T.R., Redwood A.J., Moin S.M., Price D.A., and Graham B.S.. 2017. Intranasal administration of RSV antigen-expressing MCMV elicits robust tissue-resident effector and effector memory CD8+ T cells in the lung. Mucosal Immunol. 10:545–554. 10.1038/mi.2016.48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran A.E., Holzapfel K.L., Xing Y., Cunningham N.R., Maltzman J.S., Punt J., and Hogquist K.A.. 2011. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. J. Exp. Med. 208:1279–1289. 10.1084/jem.20110308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netea M.G., Joosten L.A.B., Latz E., Mills K.H.G., Natoli G., Stunnenberg H.G., O’Neill L.A.J., and Xavier R.J.. 2016. Trained immunity: A program of innate immune memory in health and disease. Science. 352 aaf1098 10.1126/science.aaf1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.L., Zaid A., Hor J.L., Christo S.N., Prier J.E., Davies B., Alexandre Y.O., Gregory J.L., Russell T.A., Gebhardt T., et al. 2018. Local proliferation maintains a stable pool of tissue-resident memory T cells after antiviral recall responses. Nat. Immunol. 19:183–191. 10.1038/s41590-017-0027-5 [DOI] [PubMed] [Google Scholar]
- Paules C.I., Sullivan S.G., Subbarao K., and Fauci A.S.. 2018. Chasing seasonal influenza — the need for a universal influenza vaccine. N. Engl. J. Med. 378:7–9. 10.1056/NEJMp1714916 [DOI] [PubMed] [Google Scholar]
- Pizzolla A., Nguyen T.H., Sant S., Jaffar J., Loudovaris T., Mannering S.I., Thomas P.G., Westall G.P., Kedzierska K., and Wakim L.M.. 2018. Influenza-specific lung-resident memory T cells are proliferative and polyfunctional and maintain diverse TCR profiles. J. Clin. Invest. 128:721–733. 10.1172/JCI96957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards K.A., Topham D., Chaves F.A., and Sant A.J.. 2010. Cutting edge: CD4 T cells generated from encounter with seasonal influenza viruses and vaccines have broad protein specificity and can directly recognize naturally generated epitopes derived from the live pandemic H1N1 virus. J. Immunol. 185:4998–5002. 10.4049/jimmunol.1001395 [DOI] [PubMed] [Google Scholar]
- Sakai S., Kauffman K.D., Schenkel J.M., McBerry C.C., Mayer-Barber K.D., Masopust D., and Barber D.L.. 2014. Cutting edge: control of Mycobacterium tuberculosis infection by a subset of lung parenchyma-homing CD4 T cells. J. Immunol. 192:2965–2969. 10.4049/jimmunol.1400019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satpathy A.T., Kc W., Albring J.C., Edelson B.T., Kretzer N.M., Bhattacharya D., Murphy T.L., and Murphy K.M.. 2012. Zbtb46 expression distinguishes classical dendritic cells and their committed progenitors from other immune lineages. J. Exp. Med. 209:1135–1152. 10.1084/jem.20120030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenkel J.M., and Masopust D.. 2014. Tissue-resident memory T cells. Immunity. 41:886–897. 10.1016/j.immuni.2014.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slütter B., Van Braeckel-Budimir N., Abboud G., Varga S.M., Salek-Ardakani S., and Harty J.T.. 2017. Dynamics of influenza-induced lung-resident memory T cells underlie waning heterosubtypic immunity. Sci. Immunol. 2 eaag2031 10.1126/sciimmunol.aag2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo P.A., Miron M., and Farber D.L.. 2019. Location, location, location: Tissue resident memory T cells in mice and humans. Sci. Immunol. 4 eaas9673 10.1126/sciimmunol.aas9673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teijaro J.R., Verhoeven D., Page C.A., Turner D., and Farber D.L.. 2010. Memory CD4 T cells direct protective responses to influenza virus in the lungs through helper-independent mechanisms. J. Virol. 84:9217–9226. 10.1128/JVI.01069-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teijaro J.R., Turner D., Pham Q., Wherry E.J., Lefrançois L., and Farber D.L.. 2011. Cutting edge: Tissue-retentive lung memory CD4 T cells mediate optimal protection to respiratory virus infection. J. Immunol. 187:5510–5514. 10.4049/jimmunol.1102243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P.G., Brown S.A., Morris M.Y., Yue W., So J., Reynolds C., Webby R.J., and Doherty P.C.. 2010. Physiological numbers of CD4+ T cells generate weak recall responses following influenza virus challenge. J. Immunol. 184:1721–1727. 10.4049/jimmunol.0901427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thome J.J., Yudanin N., Ohmura Y., Kubota M., Grinshpun B., Sathaliyawala T., Kato T., Lerner H., Shen Y., and Farber D.L.. 2014. Spatial map of human T cell compartmentalization and maintenance over decades of life. Cell. 159:814–828. 10.1016/j.cell.2014.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner D.L., Bickham K.L., Thome J.J., Kim C.Y., D’Ovidio F., Wherry E.J., and Farber D.L.. 2014. Lung niches for the generation and maintenance of tissue-resident memory T cells. Mucosal Immunol. 7:501–510. 10.1038/mi.2013.67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wein A.N., McMaster S.R., Takamura S., Dunbar P.R., Cartwright E.K., Hayward S.L., McManus D.T., Shimaoka T., Ueha S., Tsukui T., et al. 2019. CXCR6 regulates localization of tissue-resident memory CD8 T cells to the airways. J. Exp. Med. 216:2748–2762. 10.1084/jem.20181308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilk M.M., Misiak A., McManus R.M., Allen A.C., Lynch M.A., and Mills K.H.G.. 2017. Lung CD4 tissue-resident memory T cells mediate adaptive immunity induced by previous infection of mice with Bordetella pertussis. J. Immunol. 199:233–243. 10.4049/jimmunol.1602051 [DOI] [PubMed] [Google Scholar]
- Wu T., Hu Y., Lee Y.T., Bouchard K.R., Benechet A., Khanna K., and Cauley L.S.. 2014. Lung-resident memory CD8 T cells (TRM) are indispensable for optimal cross-protection against pulmonary virus infection. J. Leukoc. Biol. 95:215–224. 10.1189/jlb.0313180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y., Jeyanathan M., Haddadi S., Barra N.G., Vaseghi-Shanjani M., Damjanovic D., Lai R., Afkhami S., Chen Y., Dvorkin-Gheva A., et al. 2018. Induction of autonomous memory alveolar macrophages requires T cell help and is critical to trained immunity. Cell. 175:1634–1650.e17. 10.1016/j.cell.2018.09.042 [DOI] [PubMed] [Google Scholar]
- Zens K.D., Chen J.-K., and Farber D.L.. 2016. Vaccine-generated lung tissue-resident memory T cells provide heterosubtypic protection to influenza infection. JCI Insight. 1 e85832 10.1172/jci.insight.85832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zens K.D., Chen J.K., Guyer R.S., Wu F.L., Cvetkovski F., Miron M., and Farber D.L.. 2017. Reduced generation of lung tissue-resident memory T cells during infancy. J. Exp. Med. 214:2915–2932. 10.1084/jem.20170521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A., Stacey H.D., Mullarkey C.E., and Miller M.S.. 2019. Original antigenic sin: how first exposure shapes lifelong anti-influenza virus immune responses. J. Immunol. 202:335–340. 10.4049/jimmunol.1801149 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists the flow cytometry antibodies used in this study.