Abstract
Background
We screened long non-coding RNAs (lncRNAs) specifically expressed in the serum of cervical squamous carcinoma (CESC) patient samples and investigated the role of these specific lncRNAs in the diagnosis of CESC and cervical intraepithelial neoplasia (CIN).
Methods
The expression levels of the lncRNAs CCAT2, LINC01133, and LINC00511 in the serum of normal controls and patient with CESC and CIN were measured using reverse transcription–quantitative polymerase chain reaction (RT-qPCR). Next, we analyzed the correlation between the serum lncRNAs levels and the clinical characteristics of CESC. Thereafter, we estimated their combined diagnostic value by receiver operating characteristic (ROC) curve analysis.
Results
The results showed that CCAT2, LINC01133, and LINC00511 were highly expressed in the serum of patients with CESC. When these lncRNAs and squamous cell carcinoma (SCC) antigen were combined, the area under the ROC curve (AUC) value reached 0.94. We also found that the AUC value of the diagnostic model combining CCAT2 and LINC01133 reached 0.894.
Conclusion
The serum lncRNAs (CCAT2, LINC01133, and LINC00511) and SCC may be new non-invasive biomarkers for the diagnosis of CESC.
Keywords: CCAT2, LINC01133, LINC00511, SCC, CESC, diagnosis
Introduction
Cervical cancer is the fourth most common malignant tumor in women and a disease that seriously threatens women’s physical and mental health.1 According to the 2015 annual report on cancer registration in China, cervical cancer was ranked as the 9th most common malignancy among women in mainland China, with an incidence becoming more prevalent at younger ages.2 Early cervical cancer often has no detectable signs; thus, patients have mostly developed to the late stage when symptoms appear, contributing to the high mortality rate. In recent years, cervical cancer mortality has dropped significantly with the continuous improvement of cervical screening techniques to identify high-risk cervical cancer populations. However, most patients do not comply with completion of the entire process of cervical cancer screening owing to cumbersome screening steps, time-consuming tasks, costly healthcare. Thus they miss the timing of early diagnosis and early treatment.3 A long precancerous stage exists before cervical cancer occurs, called cervical intraepithelial neoplasia (CIN). With the early detection and early treatment of precancerous lesions, the occurrence of cervical cancer can be completely prevented.4 Therefore, to reduce mortality from cervical cancer, screening methods for early tumor detection are highly demanded.
Not only cervical smear cytological examination but also pathological evidence of malignant cells typically require invasive strategies, such as vaginoscopy and cervical biopsy, or a loop electrosurgical excisional procedure (LEEP). Common clinical serum tumor biomarkers, such as squamous cell carcinoma (SCC) antigen and carbohydrate antigen 125 (CA125), have limited clinical applications owing to their poor sensitivity and specificity. Novel, reliable, effective and noninvasive tumor biomarkers are required for the early detection of cervical cancer to reduce the incidence and improve prognosis.
During tumorigenesis, nucleic acid molecules can be released into the blood circulation through the apoptosis and necrosis of tumor cells. Active tumor cells can also release nucleic acid molecules into the blood circulation, resulting in the changes in DNA, mRNA, microRNA and other nucleic acid molecules in the blood. Free nucleic acid molecules in the blood circulation can detect preclinical tumor and are more ideal candidate tumor markers.5 In recent years, studies have found that long non-coding RNAs (lncRNAs) are stably present in serum or plasma, reflecting the patient’s pathophysiological changes.6 With the identification of numerous tumor-related lncRNAs, lncRNAs may have an important role in non-invasive diagnosis of tumors.7 Colon cancer-associated transcript 2 (CCAT2), a 1752-bp lncRNA that maps to chromosome 8q24.21, was originally detected as being highly expressed in colorectal cancer.8 The long intergenic non-coding RNA 1133 (LINC01133) is a 1154-bp lncRNA on chromosome 1q23.2. The long intergenic non-coding RNA 511 (LINC00511) is a 2265-bp lncRNA on chromosome17q24.3.
Among cervical cancers, cervical squamous carcinoma (CESC) accounts for the majority, with the proportion reaching 80%–85%. In this study, we investigated serum samples from a large cohort of patients with CESC, patients with CIN and healthy control subjects. After step-wise screening and validation, we established a panel comprising CCAT2, LINC01133, LINC00511 and SCC for early CESC detection. The expression levels of these lncRNAs in the serum of patients with CESC were analyzed to estimate the association between the expression levels and clinical features. Receiver operating characteristic (ROC) curve analysis was used to estimate the panel in the diagnosis of CESC.
Methods
Ethics Statement and Patients
This study was approved by the ethical committee of Cancer Hospital, Chinese Academy of Medical Sciences, in accordance with the Declaration of Helsinki. Serum samples from 115 cases of CESC, 79 cases of CIN and 101 healthy controls were collected. The patients were recruited at the National Cancer Center/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College between January 2016 and November 2017. All the patients with CESC and CIN were confirmed according to their histopathological examination results. The tumors were staged following the International Federation of Gynecology and Obstetrics (FIGO) Surgical staging system. Venous blood samples were collected from patients before surgery or chemoradiotherapy. Healthy controls were age- and gender-matched to the patients and were free of any known malignancy or active inflammatory condition. Written informed consents were obtained from all patients and healthy volunteers before the study. The demographics and clinical features of the study cohort are listed in Table 1.
Table 1.
Clinical Features of the CESC and CIN Patients and Healthy Controls
| Characteristics | Healthy Controls | CIN | CESC |
|---|---|---|---|
| Age (years) | |||
| ≤49 | 54 | 38 | 64 |
| >49 | 47 | 41 | 51 |
| Menopause age (years) | |||
| <49 | – | – | 55 |
| ≥49 | – | – | 60 |
| FIGO stage | |||
| Ι | – | – | 62 |
| ΙΙ+ΙΙΙ+ΙV | – | – | 53 |
| Tumor size | |||
| <3.5 cm | – | – | 53 |
| ≥3.5 cm | – | – | 62 |
| Lymphatic metastasis | |||
| N0 | – | – | 73 |
| N1 or above | – | – | 42 |
| The grade of tumor differentiation | |||
| Poor | – | – | 56 |
| Moderate and Well | – | – | 59 |
| SCC* | |||
| Positive | – | – | 84 |
| Negative | – | – | 31 |
Notes: *SCC reference range is 0–1.5ng/mL. SCC≥1.5ng/mL is defined as positive. Negative:<1.5ng/mL.
Serum Collection
Venous blood samples (<5 mL) were collected into serum collection tubes for clinical routine biochemistry tests. After analysis, 250 µL of the serum samples were incubated at room temperature for approximately 1h before being centrifuged at 820 g for 10 min at 4°C. The resulting serum samples were transferred into new tubes, followed by further centrifugation at 16,000 g for 10 min at 4°C to completely remove any cell debris. Thereafter, the serum samples were transferred to the new RNase- and DNase-free tubes and stored at −80°C until total RNA extraction.
RNA Isolation
Total RNA was isolated from 250 µL of serum using Trizol LS Reagent (Invitrogen, Carlsbad, CA, USA) and a co-purification technique. For each 250 µL of serum, phase separation was performed by the addition of 750 µL of Trizol LS. Next, 200 µL of trichloromethane was added to augment the RNA phase separation process. Total RNA was precipitated using isopropanol and washed with 75% ethanol, and then 26 µL of RNase-free water was added for solubilization.
RT–qPCR Assay
First-strand cDNA was synthesized using a reverse transcriptase M-MLV kit (Takara, Japan) according to the manufacturer’s protocol. The reverse transcribed products were diluted to one fifth with RNase-free water and used as templates for further PCR analysis. Next, quantitative PCR was performed using the SYBR Premix Ex Taq II kit (Takara, Japan) in the ViiA 7 PCR System (Applied Biosystems, Foster City, CA, USA). Each reaction was performed in a 20-µL volume system containing 2 µL of cDNA, 0.8 µL of each primer (10 mM), 0.4 µL of ROX reference dye (506), 6 µL of sterile distilled water, and 10 µL of SYBR Premix Ex Taq II. The PCR program was as follows: denaturation at 95°C for 30 seconds, followed by 40 cycles of denaturation for 10 s at 95°C, and extension for 30 s at 60°C. Each experiment was performed in triplicate and repeated three times. The specificity of each PCR reaction was confirmed by melting curve analysis. The expression of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was detected as the endogenous control. All of the samples were normalized to GAPDH according to the 2−ΔΔCT method. The primers used in the current study are shown in Table 2.
Table 2.
The Primers Used for qRT-PCR Analysis
| lncRNA | Primers (5ʹ-3ʹ) |
|---|---|
| CCAT2 | Forward: CCCTGGTCAAATTGCTTAACCT |
| Reverse: TTATTCGTCCCTCTGTTTTATGGAT | |
| LINC00511 | Forward: TCCTCACAGGGGGTAGTAGG |
| Reverse: CCCTTCTCCCTCGGTCATTT | |
| LINC01133 | Forward: GCTGTGGTGGAGAGAATGGA |
| Reverse: CCCCAGCTTTCCAGATCCAAA | |
| GAPDH | Forward: CCTGGTATGACAACGAATTTG |
| Reverse: CAGTGAGGGTCTCTCTCTTCC |
Statistical Analysis
Statistical analysis was performed using the computer software SPSS (Statistical Package for Social Sciences, version 22.0) and GraphPad Prism (version 6.01). A comparison of the demographics and clinical features among the groups was determined by chi-squared test, Student’s t-test, and the nonparametric test. The significance of the lncRNA expression level was determined by the nonparametric test. Multivariate logistic regression analysis were performed to establish the panel (CCAT2, LINC00511, LINC01133, SCC) model for ROC analysis. ROC curves and areas under the curve (AUC) values for each lncRNA were constructed and calculated with 95% confidence intervals (95% CIs) to assess the performance of lncRNA detection in identifying patients with cancer. The optimal cut-off point, diagnostic sensitivity and specificity, positive predictive value, and negative predictive value from ROC curves were determined by a commonly used method with 95% CIs. All P values were two-sided, and P<0.05 was considered statistically significant.
Results
Differential Expression of Serum lncRNAs in Normal Controls and Patients with CIN and CESC
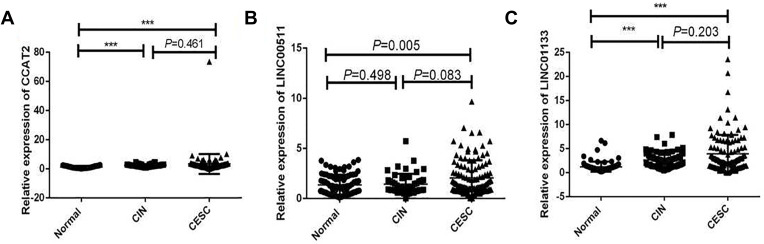
The expression levels of CCAT2, LINC00511 and LINC01133 were significantly different between normal control and patients with CESC. Compared with those in the normal control group, the CCAT2, LINC00511, and LINC01133 levels were significantly elevated in the serum of CESC patients: the average fold increases in CCAT2, LINC00511 and LINC01133 were 2.70, 1.57 and 3.15 times, respectively (Table 3, Figure 1).
Table 3.
Differential Expression of Three lncRNAs in the Serum of the Patients with CESC and Normal Controls
| lncRNA | Control | CESC | Average Fold Change | P-value |
|---|---|---|---|---|
| CCAT2 | 1.234±0.077 | 3.336±0.639 | 2.70 | P =0.000 |
| LINC00511 | 1.374±0.1044 | 2.062±0.164 | 1.57 | P =0.005 |
| LINC01133 | 1.247±0.1086 | 3.931±0.367 | 3.15 | P =0.000 |
Figure 1.
Differential expression of lncRNAs in the serum of the normal controls, patients with CIN and CESC. (A) CCAT2, (B) LINC00511, (C) LINC01133. *** P<0.001.
We also selected 79 serum samples from patients with CIN. The results showed that the expression of LINC01133 was significantly different between normal control and patients with CIN (P<0.001), but not between the CIN and CESC groups (P=0.461). In addition, no significant difference in expression of LINC00511 was found between normal control and patients with CIN (P=0.498), as well as between the CIN and CESC groups (P=0.083). CCAT2 expression was significantly different between normal controls and patients with CIN (P<0.001), but not between the CIN and CESC groups (P=0.203) (Figure 1).
ROC Analysis of CESC Using the Three lncRNAs and SCC
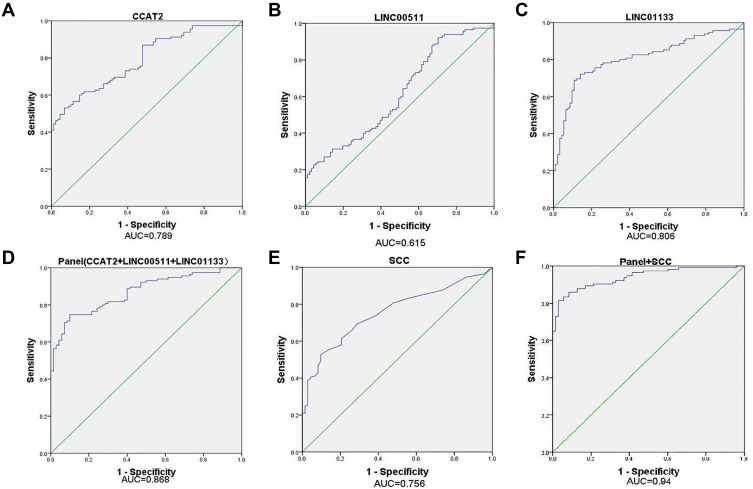
We analyzed the efficiency of each IncRNA in the CESC diagnosis and found that the AUC values of CCAT2, LINC00511, and LINC01133 were 0.789, 0.615, and 0.806, respectively. The ROC curve was generated by combining the three lncRNAs, and the sensitivity and specificity were 74.8% and 90.0%, respectively. Additionally, the AUC value reached 0.868 (95% CI: 0.819–0.918). While the ROC curve was generated by combining three lncRNAs with SCC, the sensitivity and specificity were 81.6% and 97.1%, respectively. Additionally, the AUC value reached 0.940 (95% CI: 0.907–0.972). We also analyzed the AUC value of SCC, a serum marker commonly used in the clinical diagnosis of CESC. The AUC value was 0.756 (95% CI: 0.688–0.824), and the sensitivity and specificity were 52.6% and 90.4%, respectively. Although the specificity of SCC as a diagnosis marker is relatively high, the sensitivity is poor, especially for the diagnosis of early CESC. The results showed that the combination of multiple lncRNAs and SCC was more efficient, sensitive, and specific than using the single SCC. Therefore, the diagnostic value of a diagnostic model based on the panel (CCAT2, LINC00511, LINC01133, SCC) is higher than that of SCC in the diagnosis of CESC (Table 4, Figure 2).
Table 4.
Comparison of the Sensitivity and Specificity of the Panel and SCC
| Method | AUC | 95% CI | Sensitivity | Specificity |
|---|---|---|---|---|
| Panel* | 0.868 | 0.819–0.918 | 74.8% | 90.0% |
| Panel*+SCC | 0.940 | 0.907–0.972 | 81.6% | 97.1% |
| SCC | 0.756 | 0.688–0.824 | 52.6% | 90.4% |
Note: *Panel: CCAT2+LINC01133+LINC00511.
Figure 2.
ROC curve analysis for the detection of lncRNAs and SCC. (A) CCAT2, (B) LINC00511, (C) LINC01133, (D) Panel (CCAT2+LINC00511+LINC01133), (E) SCC, (F) Panel +SCC.
Correlation Analysis of Serum lncRNAs and SCC and the Clinical Characteristics of CESC
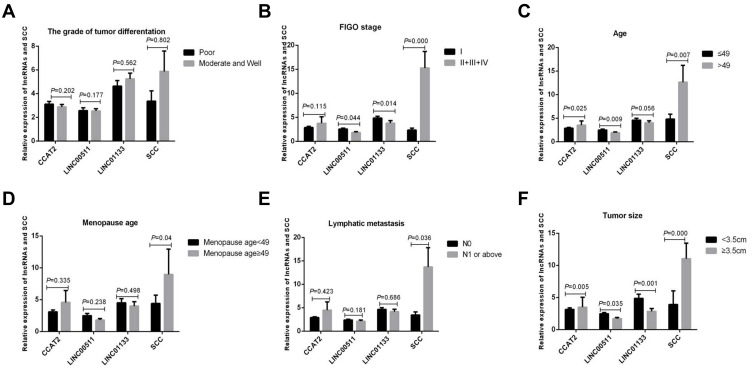
To further verify the correlation among the three types of lncRNAs and SCC and the clinical features of CESC, we analyzed the relationship between CCAT2, LINC00511, LINC01133 and SCC and age, lymph node metastasis, FIGO stage, menopause age, tumor size, and the grade of tissue differentiation. The analysis results showed that the expression of CCAT2 showed a significant upward trend with the increase in tumor size and age. No statistical difference was found between the expression of CCAT2 and FIGO stage, the grade of tissue differentiation, menopause age, and lymph node metastasis. The expression of LINC00511 was significantly associated with FIGO stage, tumor size, and age, but no significant relationship was found with the grade of tissue differentiation and menopause age. The expression of LINC01133 showed a remarkably decreased trend with the increase in tumor size and FIGO stage, but no significant relationship was found with age, the grade of tissue differentiation, menopause age, and lymph node metastasis. Furthermore, The expression of SCC was significantly associated with FIGO stage, tumor size, age, and menopause age, but no significant relationship was found with the grade of tissue differentiation (Figure 3).
Figure 3.
Correlation between relative expression of lncRNAs and SCC and the clinical features of CESC. (A) The grade of tissue differentiation, (B) FIGO stage, (C) Age, (D) Menopause age,(E) Lymph node metastasis, (F) Tumor size.
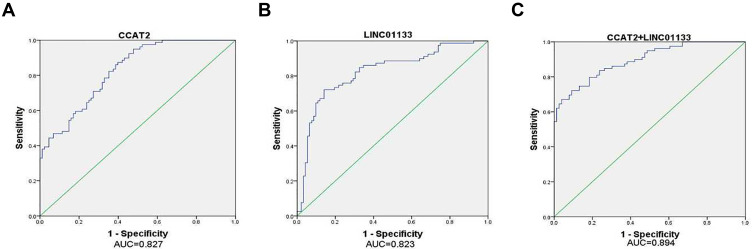
ROC Analysis of CIN Using CCAT2 and LINC01133
The above experimental results showed a significant difference in the expression of CCAT2 and LINC01133 between the normal control group and CIN group. Therefore, we used this diagnostic model for ROC analysis to observe its possible diagnostic efficacy in CIN. The AUC values of CCAT2 and LINC01133 were 0.827 and 0.823, respectively (Figure 4). The AUC value of the diagnostic model combining these two lncRNAs reached 0.894 (95% CI: 0.846–0.942), and the sensitivity and specificity were 67.1% and 96.1%, respectively. The diagnostic model comprising CCAT2 and LINC01133 showed high diagnostic value for identification in healthy humans and patients with CIN, with high specificity. However, the sensitivity of 67% was too low for screening, so the negative predictive value was much more important than the positive predictive value.
Figure 4.
ROC curve analysis for the detection of CIN using lncRNA. (A) CCAT2, (B) LINC01133, (C) Panel (CCAT2+LINC01133).
Discussion
Cervical cancer is a major disease that seriously threatens the life and health of women. Early intervention can be achieved through effective early screening and early prevention. Epidemiological data have shown that screening can significantly reduce the incidence and mortality of cervical cancer. The prognosis of patients with cervical cancer is closely related to the clinical stage of the tumor. Therefore, early diagnosis and early treatment of cervical cancer are of utmost importance. Presently, thin-layer liquid-based cytology (LBC), HPV DNA detection (second-generation hybrid capture technique, HC2), colposcopy and cervical biopsy are used for clinical “three-step ladder” screening methods. Among them, cytology has high positive predictive value and high specificity, but low sensitivity, and requires high professional skills of pathologists. HPV screening has high sensitivity, but the positive predictive value is low, and some patients present with transient infections. The biopsy of cervical tissue under colposcopy is a traumatic examination; the patients’ acceptance was limited and the method was difficult to promote among the population.9 SCC, a tumor marker closely related to CESC, is associated with the clinical diagnosis, curative effect, prognosis, and recurrence of squamous cell carcinoma.10 However, because of its low specificity and sensitivity, the value of SCC in assisted diagnosis is limited. Therefore, better inspection methods are needed with higher efficiency, sensitivity, and specificity. In recent years, studies have found that blood non-coding RNA is a non-invasive and inexpensive type of screening tool that is conducive to the screening of large-scale high-risk groups. Therefore, we urgently need to identify new targets to improve the survival rate of cervical cancer patients.
In recent years, many studies have shown that lncRNAs have many important biological functions. They are differentially expressed during tumorigenesis and are closely related to cancer invasion, metastasis, and prognosis.11–13 Previous studies have found that lncRNA CCAT2 is up-regulated in cervical squamous cell cancer tissues compared with that in the adjacent non-tumor tissues.14,15 LncRNA CCAT2 promotes the proliferation and survival of cervical cancer cells.16 We found that the expression levels of LINC01133 and LINC00511 are up-regulated in CESC tissues compared with those in normal tissues in the TCGA and GTEx database from GEPIA. Feng et al17 revealed that LINC01133 promotes the progression of cervical cancer by sponging miR-4784 to up-regulate the AT-hook DNA binding motif containing 1(AHDC1).18,19 Shi et al20 suggested that LINC00511 might promote the occurrence of cervical cancer by upregulating phospholipase D1 (PLD1) expression via recruiting transcription factor retinoic X receptor alpha (RXRA). Their use as new tumor markers has received increasing attention owing to their stability in blood circulation and differential expression of specificity. In this study, we screened three lncRNAs (CCAT2, LINC01133, and LINC00511) that showed a consistent difference in the serum of patients with CESC and healthy human subjects and these lncRNAs all had a higher AUC for the diagnosis of CESC. With the combined use of these three lncRNAs with SCC, the AUC for the diagnosis of CESC can reach 0.94 with high sensitivity and specificity. Additionally, we directly compared the diagnostic efficiency of the three lncRNA diagnosis models with the traditional CESC marker SCC in the diagnosis of CESC. The results showed that the diagnostic efficiency of lncRNAs with SCC in CESC was higher than that of SCC, and its sensitivity is significantly higher than SCC especially in the diagnosis of early CESC. Based on the above results, we believe that this model has a high clinical value for the early diagnosis of CESC. Additionally, we verified the correlation between the three types of lncRNAs and the clinical features of CESC. We found that the expression levels of LINC01133 and LINC00511 were significantly associated with the FIGO stage, and these three lncRNAs all showed significant differences with the increase in tumor size.
CIN represents a group of cervical lesions that are closely related to the occurrence of invasive cervical cancer. Most of the low-grade CIN lesions can be regressed, but the high-grade CIN has the potential to become cancerous and can develop into invasive cancer, which is considered as a precancerous lesion. Clinically, CIN lesions are found through cervical screening. Patients with CIN were followed up regularly and the lesions were treated promptly. Therefore, the use of simple and effective methods to identify healthy people and patients with CIN is essential to reduce the incidence of cervical cancer.
In this study, the results showed that CCAT2 and LINC01133 were differentially expressed in the normal control and CIN groups. We also tried to use this model to observe its possible diagnostic efficacy in CIN and found that the AUC value of the model combining CCAT2 and LINC01133 reached 0.894 (95% CI: 0.846–0.942), and the sensitivity and specificity were 67.1% and 96.1%, respectively.
In conclusion, we have found that the serum levels of CCAT2, LINC01133, LINC00511, and SCC may serve as novel non-invasive biomarkers for the diagnosis of CESC, and the diagnostic model comprising CCAT2 and LINC01133 showed high diagnostic value for the identification in healthy humans and patients with CIN.
Acknowledgments
We thank Nicole Okoh, PhD, from Edanz Group (https://en-author-services.edanzgroup.com/) for editing a draft of this manuscript. We thank Sheng-kai Huang, PhD, Department of Clinical Laboratory, National Cancer Center/National Clinical Research Center for Cancer/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College for contributing to the paper. This work was supported by the National Natural Science Foundation of China (Grant no. 81872038) (Grant no. 81902503), CAMS Innovation Fund for Medical Sciences (CIFMS) (Grant no.2016-I2M-1-001) and the Fundamental Research Funds for the Central Universities (Grant no. 3332019056).
Disclosure
The authors declare no potential competing interests regarding this manuscript.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–386. doi: 10.1002/ijc.29210 [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CACancer J Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338 [DOI] [PubMed] [Google Scholar]
- 3.Leufkens AM, van den Bosch MA, van Leeuwen MS, Siersema PD. Diagnostic accuracy of computed tomography for colon cancer staging: a systematic review. Scand J Gastroenterol. 2011;46(7–8):887–894. [DOI] [PubMed] [Google Scholar]
- 4.Olson B, Gribble B, Dias J, et al. Cervical cancer screening programs and guidelines in low- and middle-income countries. Int J Gynaecol Obstet. 2016;134(3):239–246. [DOI] [PubMed] [Google Scholar]
- 5.Lin XJ, Chong Y, Guo ZW, et al. A serum microRNA classifier for early detection of hepatocellular carcinoma: a multicentre, retrospective, longitudinal biomarker identification study with a nested case-control study. Lancet Oncol. 2015;16(7):804–815. [DOI] [PubMed] [Google Scholar]
- 6.Tong YS, Wang XW, Zhou XL, et al. Identification of the long non-coding RNA POU3F3 in plasma as a novel biomarker for diagnosis of esophageal squamous cell carcinoma. Mol Cancer. 2015;14(1):3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zeng S, Xiao YF, Tang B, et al. Long noncoding RNA in digestive tract cancers: function, mechanism, and potential biomarker. Oncologist. 2015;20(8):898–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ling H, Spizzo R, Atlasi Y, et al. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res. 2013;23(9):1446–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.P.D.Q. Screening, B. Prevention Editorial. Cervical Cancer Screening (PDQ(R)): Health Professional Version, PDQ Cancer Information Summaries. Bethesda, National Cancer Institute (US); 2002. [Google Scholar]
- 10.Salvatici M, Achilarre MT, Sandri MT, Boveri S, Vanna Z, Landoni F. Squamous cell carcinoma antigen (SCC-Ag) during follow-up of cervical cancer patients: role in the early diagnosis of recurrence. Gynecol Oncol. 2016;142(1):115–119. doi: 10.1016/j.ygyno.2016.04.029 [DOI] [PubMed] [Google Scholar]
- 11.Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–1076. doi: 10.1038/nature08975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vitiello M, Tuccoli A, Poliseno L. Long non-coding RNAs in cancer: implications for personalized therapy. Cell Oncol (Dordr). 2015;38(1):17–28. doi: 10.1007/s13402-014-0180-x [DOI] [PubMed] [Google Scholar]
- 13.Fu M, Zou C, Pan L, et al. Long noncoding RNAs in digestive system cancers: functional roles, molecular mechanisms, and clinical implications (Review). Oncol Rep. 2016;36(3):1207–1218. doi: 10.3892/or.2016.4929 [DOI] [PubMed] [Google Scholar]
- 14.Chen X, Liu L, Zhu W. Up-regulation of long non-coding RNA CCAT2 correlates with tumor metastasis and poor prognosis in cervical squamous cell cancer patients. Int J Clin Exp Pathol. 2015;8(10):13261–13266. [PMC free article] [PubMed] [Google Scholar]
- 15.Lazniak S, Lutkowska A, Warenczak-Florczak Z, et al. The association of CCAT2 rs6983267 SNP with MYC expression and progression of uterine cervical cancer in the polish population. Arch Gynecol Obstet. 2018;297(5):1285–1292. doi: 10.1007/s00404-018-4740-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu L, Jin L, Zhang W, Zhang L. Roles of long non-coding RNA CCAT2 in cervical cancer cell growth and apoptosis. Med Sci Monit. 2016;22:875–879. doi: 10.12659/MSM.897754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feng Y, Qu L, Wang X, Liu C. LINC01133 promotes the progression of cervical cancer by sponging miR-4784 to up-regulate AHDC1. Cancer Biol Ther. 2019;20(12):1453–1461. doi: 10.1080/15384047.2019.1647058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu C-L, Xu X-L, Yuan F. LINC00511 is associated with the malignant status and promotes cell proliferation and motility in cervical cancer. Biosci Rep. 2019;39(9):BSR20190903. doi: 10.1042/BSR20190903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mao B-DI, Xu P, Xu P, Zhong Y, Ding W-W, Meng Q-Z. LINC00511 knockdown prevents cervical cancer cell proliferation and reduces resistance to paclitaxel. J Biosci. 2019;44(2):44. doi: 10.1007/s12038-019-9851-0 [DOI] [PubMed] [Google Scholar]
- 20.Shi Y, Liu M, Huang Y, Zhang J, Yin L. Promotion of cell autophagy and apoptosis in cervical cancer by inhibition of long noncoding RNA LINC00511 via transcription factor RXRA-regulated PLD1. J Cell Physiol. 2020. doi: 10.1002/jcp.29529 [DOI] [PubMed] [Google Scholar]