Abstract
Coronavirus disease 2019 (COVID-19) has become a severe threat to human health worldwide. Early etiological diagnosis plays a critical role in controlling COVID-19 pandemic. However, etiological diagnosis has been largely compromised by high “false-negative” rates of viral nucleic acid testing, resulting from limited sampling efficiency using conventional oropharyngeal swabs. Here, we engineer regular swabs by using a microneedle (MN) patch to significantly improve the quality and quantity of virus collection. The combination of MNs with different crosslinking levels endows the patches with dual capability of mucus penetration and virus extraction. Moreover, the antibody (Ab) against viral spike protein was integrated into the patch, conferring MNs with an active virus capture potential. By taking advantage of the biological and engineered species, we believe that the designed MN/Ab swabs could serve as a promising tool to improve current sampling efficiency with fewer false negatives, contributing to the containment of the COVID-19 pandemic.
Material Advancement Progression: MAP6: Development
Graphical Abstract
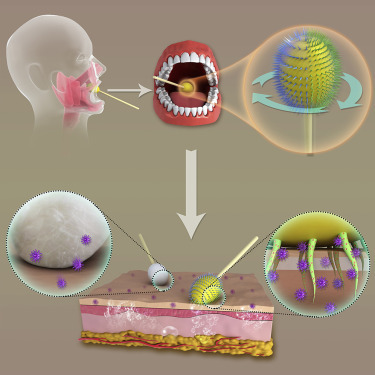
Highlights
-
•
Well-designed microneedles were introduced onto the oropharyngeal swab
-
•
Different crosslinked microneedles indicated distinct functions
-
•
Virus-specific antibody integration in the swab facilitated virus capture
-
•
Highly effective sampling significantly reduced “false negatives”
Progress and Potential
The epidemic of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has spread to most countries of the world, causing severe respiratory illness (coronavirus disease 2019). To combat the disease, early tests on viral nucleic acid act as the standard method, but this results in a high rate of “false negatives,” which may mislead the doctor's response. Aiming to reduce these false negatives, we fabricated a chemically engineered oropharyngeal swab by introducing a microneedle structure and integrating virus-specific antibodies. Importantly, the modified swab could provide effective penetration and active virus capture, achieving high-efficiency sampling by synergizing the advantages of biological and engineered species, which widen the difference between positive and negative samples. With current advances, we believe that the designed swab could serve as a promising candidate for other oral or respiratory disease diagnostics, suggesting the generalizability of the strategy.
A well-designed microneedle patch was invented to modify the heads of regular oropharyngeal swabs to improve sampling efficiency during COVID-19 detection. The microneedle patch consists of two different crosslinked clusters capable of both tissue penetration and virus capture. In combination with virus-specific antibodies, the swab achieved high-efficiency sampling by synergizing the advantages of biological and engineered species, facilitating accurate virus detection to significantly reduce “false negatives” during COVID-19 diagnostics.
Introduction
An outbreak of coronavirus disease 2019 (COVID-19), a severe respiratory illness, has become one of the most concerning threats to human health globally,1 affecting 215 countries (including territory or area), and causing over 30 million infected cases and more than 950,000 deaths by September 19, 2020.2 Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has been identified as an etiological cause of the disease.3 , 4 To effectively contain this pandemic, early detection of SARS-CoV-2 is critical, as it would facilitate timely diagnosis and treatment, and implementation of public health measures.5 Among the current diagnostic approaches, viral tests and antibody detection are two main methods that have been broadly applied.6 As serological tests (blood antibody detection) mainly reflect the infection at the mid-to-late stage of the illness,7 viral nucleic acid polymerase-chain-reaction (PCR) testing on specimens obtained through oropharyngeal (OP) swab sampling is a gold-standard and widely used method for early diagnosis, due to its practical convenience, simple equipment requirement, low cost, and negligible influence on sample integrity.8, 9, 10 However, early studies demonstrated that such tests using OP samples for detecting COVID-19 might produce up to 30% “false negatives.”11, 12, 13, 14 This is thought to be largely ascribed to ineffective sample collection when using regular swabs because they could only provide limited physical adsorption from and nearly no infiltration into mucosal tissues, with which they have weak interactions, or show poor recovery for low-volume samples with low transfer efficiency.12 , 13 , 15 Moreover, regular swabs can only collect superficial tissues from pharyngeal mucosal surface, which is readily contaminated or diluted by food and drink.6 In this regard, a more effective and precise viral sampling method is urgently needed to improve the current COVID-19 testing in the hope of avoiding false negatives, which can be highly essential to block the COVID-19 pandemic.
Notably, sampling by OP swabs, a process of local specimen collection within a short period of time, is similar to birds' intake of food and drink with the help of their tongues.16 , 17 For instance, the rainbow lorikeet (Trichoglossus haematodus) possesses the capability of effective nectar and pollen intake from flowers, largely owing to small brush structures (extended papillae) on the tip of its tongue,18 highlighting the functional importance of physical structure. Moreover, some animals frequently secrete sticky mucus on tongues to facilitate foodstuff collection via strong adhesion,19 which is derived from intense interactions (e.g., hydrogen bonds, covalent bonds) between mucus and molecules in foods, revealing a key role of molecular interactions. Inspired by “brush structures” and molecular interactions, we hypothesized that a highly efficient and precise sample collection from OP sites might be achieved by introducing chemically modified micropapillae/microneedles designed to allow deep-tissue penetration, effective virus extraction, and long sample retention.20
Here, we took advantage of soft-tissue penetration capability of microneedles (MNs) to improve the depth of sampling.21, 22, 23, 24, 25, 26, 27, 28 Meanwhile, to achieve effective and specific virus collection, the antibody targeting spike protein of SARS-CoV-2 was integrated into the MNs, with the aim of chemically binding to the virus through antigen-antibody interactions.29, 30, 31 Notably, the MN patch was fabricated by using alginate polymers under two different crosslinking conditions in order to provide variable strengths required by mucus insertion and virus recruitment.32 After optimization for base fabrication, a soft flexible patch was attached to an OP swab using biomedical device adhesives, thus conferring the regular swab with the abilities of mucus penetration and active virus extraction (Figure 1 ). Thanks to chemically engineered MN swabs, the diagnostics of COVID-19 has been remarkably improved with a decrease in false negatives, bringing great hopes in controlling COVID-19.
Figure 1.
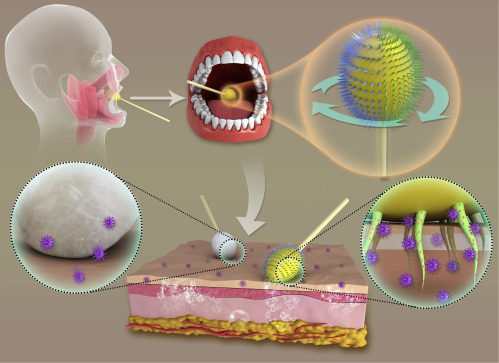
Scheme of an MN/Ab Swab Synergistically Providing Mucus Penetration and Virus Extraction for Highly Efficient COVID-19 Detection
After inserting into the oral cavity and contacting with posterior pharynx and tonsillar areas, the MN/Ab swab was rotated to guarantee differently crosslinked MNs touching the tissue for 5–10 s. SARS-CoV-2 would self-accumulate around and in the MNs as a result of antigen-antibody interactions. In such a way, a more specific and effective sampling was achieved.
Results and Discussion
To fabricate microneedle patches, we printed a well-designed master mold following the standard protocol (Figures 2A and 2B). A poly(dimethylsiloxane) (PDMS) mold (negative) was constructed from the master mold (Figure 2C) by mixing two-part resin systems containing vinyl groups and hydrosiloxane groups. To capture the SARS-CoV-2, we selected the antibody targeting the spike protein (Figures 2D and S1) on the virus surface as the source for virus acquirement.29, 30, 31 After being loaded into the PDMS mold, alginate polymer was cast to form the MNs via distinct crosslinking processes (Figure 2E). The high-level crosslinking (hard) was expected to provide extensive tissue penetration capability, while low-crosslinking level (soft) was expected to offer potent virus extraction and ease of release.33 Fluorescein isothiocyanate (FITC) and rhodamine dyes were respectively used to label the MN patches with different crosslinking levels (Figure S2). These patches were combined by biomedical device adhesives (Figure 2F). Scanning electron microscopy (SEM) images exhibited a recognizable distinction between the high- and low-crosslinked MNs (Figure 2G), consistent with the observation from fluorescence microscopy (Figure 2H). The obvious boundary between the MNs could be readily identified by fluorescence distribution (Figure 2I), suggesting the successful fabrication of the dually functionalized MN patches. The combination of MNs with diverse strengths complemented each other's deficiencies by preventing low virus migration (high-crosslinked MNs) and poor penetration (low-crosslinked MNs), providing a synergetic and interdependent profile. With this smart design, a dually functionalized MN patch was invented, holding great promise to improve SARS-CoV-2 collection.
Figure 2.
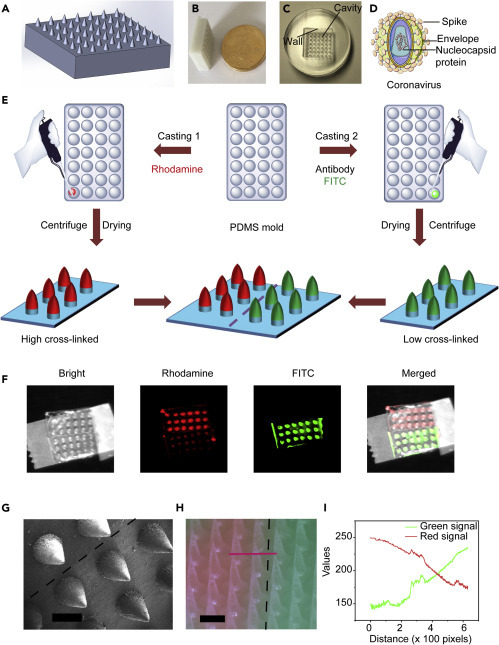
Fabrication of Dually Functionalized MN Patches
(A) Image of the designed master mold.
(B) Image of the printed master mold.
(C) Fabricated PDMS mold based on the master mold.
(D) Structure of SARS-CoV-2.
(E) Schematic of dually fictionalized MN patch fabrication.
(F) Fluorescence images of the MN arrays with two dyes distributed in distinct segments.
(G) SEM image of dually functionalized MNs. Scale bar, 500 μm.
(H and I) (H) Fluorescence microscopy image of dually functionalized MNs (scale bar, 500 μm) and (I) fluorescence distribution according to the red line.
After MN formation, the assembly of the patches onto the regular swab was optimized by adjusting the base intensity. Alginate (1% ) crosslinked by 50 mM CaCl2 was applied to construct the base, which would provide sufficient strength and toughness to allow flexible bending in a controllable fashion (Figures 3A–3C). This bendable property is indispensable for the MN patch to fit swab heads because brittle or hard materials would hinder shape alteration as well as the subsequent attachment. Notably, the transparent patch with a thin base (1 mm) could be effortlessly curled by hand and perfectly wrapped on the head of a regular swab (Figures 3D–3F). When using the swab, a rotated scratch was suggested, owing to the interlaced distribution of dually functionalized MNs on the patch. In such a way, the high-crosslinked MNs ones would penetrate the mucus, introducing numerous microchannels (slightly bent, Figure 3G), while low-crosslinked ones would facilitate virus infiltration and release in virus preservation solution (VPS) by controllable dissolution (Figure 3H). The difference in strength (Figure 3I) between the patches and controlled dissolving behaviors (Figure S3) of the patches verified the observations, further suggesting a tunable and precisely manageable sampling process.
Figure 3.
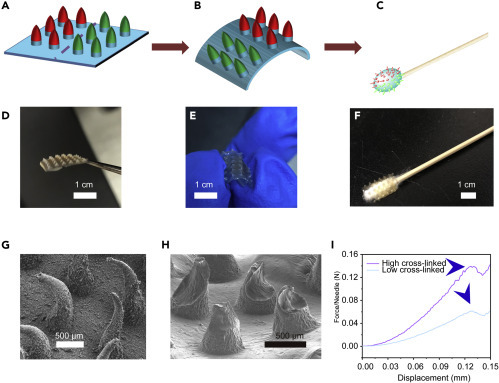
Attachment of Dually Functionalized MN Patches onto a Regular Swab
(A–C) Schematic of MN patch assembly on a regular swab.
(D–F) Images of MN patch assembly onto a regular swab.
(G) SEM image of MNs treated in rat throat for 10 s.
(H) SEM image of MNs treated by virus preservation solution for 5 min.
(I) Mechanical behaviors of MNs with high and low cross-linked levels.
To evaluate the sampling efficiency of engineered MN swabs, we introduced FITC-labeled streptavidin into the mouth of rats to act as a drug model for the in vivo test (Figure 4 A). After biotin was loaded into the MNs, dually functionalized MN (high- and low-crosslinked) patches were combined and attached to regular swabs. Similar to the treatment of patients, the swabs were inserted into the oral cavity of rats and rotated for a few seconds to collect samples (Figure 4B), then transferred to VPS, followed by fluorescence measurement. We observed that the MN/biotin swab turned green, indicating a successful capture of FITC-streptavidin (Figure 4C). Importantly, compared with empty MNs and regular swabs, a marked increase in FITC fluorescence intensity was observed after MN/biotin swab treatment (Figure 4D), revealing a synergistic effect of MN insertion and antigen-antibody interactions in sampling. After MN treatment, microscaled channels were generated, as evidenced by tissue staining (Figure 4E), suggesting an in-depth tissue sampling (the regular swab could not penetrate tissues). It should be emphasized that MN treatment is minimally invasive while the microchannels induced by MNs could be recovered spontaneously within 24 h (Figure S4). Due to the dually functionalized segments of the patch, an uneven fluorescence distribution was detected using an in vivo imaging system (In-Vivo FX PRO, Bruker) (Figure 4F). It should be noted that either a lower or higher ratio of two types of crosslinked MNs would result in decreased sampling efficiency while the ratio ranging from 0.4 to 0.6 produced the optimal sampling effect (Figure 4G). This was mainly because within this range, the advantages of tissue insertion and active binding could both be maximized, synergizing sampling efficiency. Similar to momentary sampling using regular swabs, this process with engineered MN swabs was also swift (less than 10 s, Figure 4H), thus retaining convenience while guaranteeing effectiveness. In such a way, this well-designed MN swab might rapidly substitute regular swabs given its superior sampling efficiency and negligible side effects.
Figure 4.
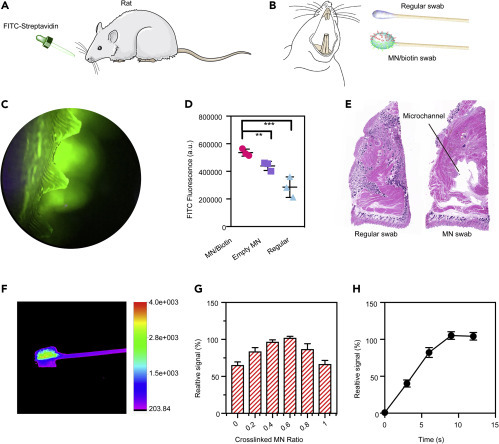
Evaluation of Sampling Efficiency of Different Swabs in a Rat Model
(A) The rat was fed with FITC-labeled streptavidin to mimic an infection model.
(B) The MN swab was loaded with biotin to capture FITC-labeled streptavidin. A regular swab was applied as a control.
(C) Fluorescence image of MNs after collecting samples from the oral rat cavity.
(D) Comparisons of sampling efficiency of different swabs. Mean ± SD (n = 3). ∗∗p < 0.01, ∗∗∗p < 0.001.
(E) Histology analysis indicated that MNs penetrated the oral epithelium to induce microscaled channels compared with the regular swab.
(F) Fluorescence image of MN/biotin swabs indicated separated signal distribution.
(G) The impacts of the ratios of high-crosslinked MNs to low-crosslinked ones on sampling efficiency of MN/biotin swabs.
(H) The fluorescence-time profile showed that the whole sampling process could be completed within 10 s, similar to that using regular swabs.
Facing the challenge of high false-negative rates during COVID-19 detection, we applied the MN-based oropharyngeal swabs on a COVID-19 rat model to evaluate their sampling efficiency. In brief, a COVID-19 pseudovirus developed by the Yeasen Company was introduced into the oral cavity of the rat model, followed by sampling using different swabs (Figure 5 A). After treatment in VPS, the samples were tested within 24 h by RT-PCR (Figure 5B). Three typical targets for COVID-19 testing, nucleocaspid (N), envelope (E), and RNA-dependent RNA polymerase (ORF1ab/RdRp) genes, were analyzed by using real-time RT-PCR assays. As shown in Figure 5C, regular swab sampling needed a higher cycle threshold (Ct) value (>30) to gain an obvious signal for the detection, indicating a low virus payload. Interestingly, an MN swab (without antibody) slightly enhanced the sampling by reducing the Ct value, ascribed to the MN penetration to mucus tissue for more virus collection. Notably, after antibody loading, the MN with antibody (MN/Ab) swab exhibited a superior sampling potential by showing a Ct value lower than 30 (Figure S5). In our test, Ct ≤ 30 was evaluated to be more significant compared with the value that was larger than 30, which was suggested based on the protocol of the detection kit. Besides, to verify the clinical potential of the MN-based swabs, a laboratory-confirmed COVID-19 patient with typical computed tomography (CT) imaging characteristics participated in the test (Figures 5D and S6). After swab sampling in patients or volunteers (COVID-19-negative) by well-trained nurses (Figure 5E), degree of pain, itching, nausea, saliva, cough, sputum, and dyspnea were evaluated by using a questionnaire, which was completed immediately after the tests. Four levels were provided (score 4, very serious; score 3, serious; score 2, mild; score 1, very mild) and the data from each patient/volunteer were presented. Generally, the regular swab did not induce intense discomfort and only one volunteer felt severe nausea (Figure 5F). Importantly, with or without antibody loading, no patient claimed severe discomfort from MN-based swabs, comparable with the regular swab, suggesting minor or negligible adverse effects (Figure 5F). During the test on the positive patient, the best candidate (the ratio of highly crosslinked MN was 0.5 and antibody amount 25 μg/patch) was applied for comparison with the clinically used ones. As expected, significantly reduced Ct values were found for all three genes after the MN/Ab swab treatment compared with a regular swab on the same patient (Figures 5G and S7), indicating a higher amount of virus on the engineered swab. To normalize the results, we divided the Ct values of target genes by that of the reference (added after sampling), whereby MN/Ab swab treatment still presented remarkably lower Ct values relative to regular ones (Figure 5H). It should be emphasized that this enhancement is essential for the COVID-19 test, as the Ct values from regular swabs were close to the detection limit of the method, which could easily mislead clinicians. Evidently, the compelling improvement in the detection allowed the collection of more viruses and widened the gap between Ct values of positive patients and those of negative patients. In this regard, the MN/Ab swab could serve as a useful tool for COVID-19 sampling compared with currently used ones. In negative patients, the MN/Ab swabs would not induce undesired “false positives” as did regular swabs (Figure S8), suggesting feasible and reliable detection profiles.
Figure 5.
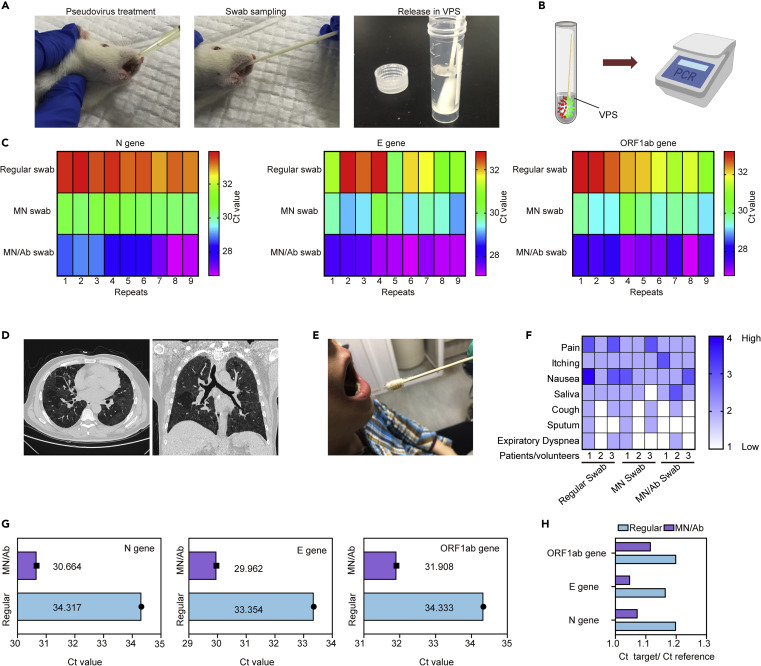
Assessment of Sampling Efficiency of MN/Ab Swabs on COVID-19 Models and Patients
(A) The rats were treated by the COVID-19 pseudovirus, which contains nuclear acid similar to that of SARS-CoV-2 but without high infection, followed by swab sampling and storage in VPS.
(B) The swabs were immersed in VPS within 24 h, followed by qRT-PCR testing.
(C) N gene, E gene, and ORF1ab gene were tested, indicating low Ct values for the MN/Ab group compared with others.
(D) The MN/Ab swab was applied to a patient, in similar manner to application of a regular swab.
(E) CT scanning verified the COVID-19-positive patient.
(F) Side-effect comparison between regular swabs, MN/swab, and MN/Ab swab.
(G) Comparison of absolute Ct value for three genes.
(H) Comparison of relative Ct value for three genes.
Conclusions
In summary, aiming to reduce the “false negatives” toward accurate COVID-19 etiological testing, we invented a well-designed MN patch to modify the heads of regular swabs. The MN patch consisted of two different clusters capable of both tissue penetration and virus collection. The high-level crosslinked MNs possessed sufficient strength for insertion into mucus layers in a minimally invasive manner, while low-level crosslinked MNs with loaded virus spike protein-targeting antibody facilitated virus capture. Through a rotating motion during sampling, the MN/Ab swab could provide effective penetration and active virus capture, achieving high-efficiency sampling by synergizing the advantages of biological and engineered species. Moreover, the higher amount of collected virus offered ease of distinction for positive from the negative patients, bringing great hopes of reducing “false negatives” in COVID-19 detection. Thus, the MN/Ab swab could not only be employed for COVID-19 testing but also act as a promising candidate for detection of other disease, including diverse oral or respiratory diseases, highlighting MNs' functional generalizability for various biomedical applications.
Experimental Procedures
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Xiaoyuan Chen (chen9647@gmail.com).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
This study did not generate/analyze any datasets/code.
Fabrication of MNs
Three-dimensional master molds were designed in Solidworks (Dassault Systèmes, Vèlizy-Villacoublay, France) and printed with white resin on a Lite 600 HD printer (Uniontech, Shanghai, China). PDMS mold (negative) was fabricated from the master mold by mixing two-part resin systems (Sylgard 184 (Dow, Midland, MI, USA)) containing vinyl groups (part A) and hydrosiloxane groups (part B), and casting the material around the master. 2019-nCoV Spike/receptor binding domain antibody (Sino Biological, China) was diluted in PBS with the concentration of 125 μg/mL. FITC (2%) was added as the indicator. After weighing the molds, 200 μL of solution was added to the mold, followed by 10 min of vacuum (2.5 kPa) and 10 min of centrifugation (2,000 rpm). It should be mentioned that all of the solution could be added to the PDMS mold; thus, each patch contained around 25 μg antibody. Next, 2 mL of 2% alginate solution was introduced to the PDMS molds, which were placed in an oven overnight (37°C). After cooling to room temperature, the patches were carefully peeled off from the PDMS molds by using tweezers. All patches were stored at 4°C until use.
Crosslinking of MNs
The fabricated patches were treated by different concentrations of CaCl2 solution to induce distinct crosslinked levels. In brief, for high-crosslinked patches, the samples were immersed in 1 M CaCl2 for 3 min, followed by wiping with a tissue paper. Meanwhile, low-crosslinked samples were generated by using 20 mM CaCl2 instead, and the treatment time was controlled at 5 s. High-crosslinked samples were labeled by rhodamine while low-crosslinked ones were indicated by FITC.
Attachment of MN Patches to the Regular Swab
The patches with different crosslinked levels were combined by using biomedical device adhesives (Loctite 4061, Henkel, Germany). The patches were then curved with the help of a rod-like fixture, followed by the assembly to the head of regular swab (Munkfoam, China) with the help of biomedical device adhesives. All of the samples were stored at 4°C until use.
Mechanical Strength Test
The mechanical strength of the MN arrays with high- or low-crosslinked levels was tested by using an MTS 30 G tensile testing machine. The MNs were pressed against a stainless-steel fixture. The initial gauge was set as 2 mm between the MN tips and the stainless-steel fixture, with 10.00 N as the load cell capacity. The speed of the top stainless-steel plate movement toward the MN arrays was set as 0.1 mm s−1. The failure force of MNs was recorded as the needle began to buckle.
In Vivo Studies Using Sprague-Dawley Rats
Male Sprague-Dawley rats (8–10 weeks) were obtained from the Experimental Animal Center of Tongji Medical College (Wuhan, China). The animals were housed with free access to food and water under 12-h light/12-h dark cycle. All animal procedures were approved by the Animal Care and Use Committee at the Huazhong University of Science and Technology and performed in compliance with the rules of the International Guiding Principles for Biomedical Research Involving Animals.
After anesthesia by chloral hydrate, the rats were treated with 5 μg of FITC-streptavidin solution (20 μL) through the mouth, followed by immediate sampling by using regular swab, MN swab, and MN/biotin swab (200 μL, 1.5 mg/mL biotin was loaded in the patch). The treatment time was adjusted within seconds. The swabs were immersed in VPS and the samples were measured within 24 h by using an EnSpire Multimode Plate Reader (PerkinElmer, USA). In addition, the ratio of high-crosslinked and low-crosslinked patches was optimized by using the same method.
Besides the FITC-streptavidin detection, a COVID-19 rat model was established by introducing COVID-19 pseudovirus (Yeasen, China) to the oral mucus layer in the rats. After COVID-19 pseudovirus (20 μL) treatment, different swabs (regular swab, MN swab, and MN/Ab swab) were applied in the oral cavity of the rat for 15 s, then transferred to VPS. Each group contained three rats and each rat was tested three times.
RNA was isolated from the samples with the use of a commercial kit (TianLong Ex DNA/RNA Extraction Kit) on a GeneRotex96 automated nucleic acid extraction system (TianLong, Xi'an, China), according to the procedure recommended by the manufacture. An initial volume of 200 μL was used for the nucleic acid extraction, and the final elution volume was 100 μL. Real-time RT-PCR was performed on a QuantStudio 5 system (Applied Biosystem, USA) with the use of a 2019-nCoV real-time multiplex RT-PCR kit (Shanghai ZJ Bio-Tech, China). A 25-μL reaction contained 5 μL of RNA, 19 μL of reaction buffer, and 1 μL of reverse transcriptase/Taq polymerase mixture from the kit. Thermal cycling was performed at 45°C for 10 min for reverse transcription, followed by 95°C for 3 min, then 45 cycles of 95°C for 15 s and 58°C for 30 s.
Three genes were tested, ORF1ab gene (channel A), N gene (channel B), and E gene (channel C).
Target 1 (ORF1a/b gene, RdRp region):
Forward, 5′-GTG ARA TGG TCA TGT GTG GCG G-3′
Reverse, 5′-CAR ATG TTA AAS ACA CTA TTA GCA TA-3′
Probe, 5′-FAM-CAG GTG GAA CCT CAT CAG GAG ATG C-BHQ1-3′
Target 2 (N gene):
Forward, 5′-CAC ATT GGC ACC CGC AAT C-3′
Reverse, 5′-GAG GAA CGA GAA GAG GCT TG-3′
Probe, 5′-HEX-ACT TCC TCA AGG AAC AAC ATT GCC A-BHQ1-3′
Target 3 (E gene):
Forward, 5′-ACA GGT ACG TTA ATA GTT AAT AGC GT-3′
Reverse, 5′-ATA TTG CAG CAG TAC GCA CAC A-3′
Probe, 5′-610-ACA CTA GCC ATC CTT ACT GCG CTT CG-BHQ2-3′
The references, primers, and probes were provided in the 2019-nCoV real-time multiplex RT-PCR kit (Shanghai ZJ Bio-Tech, China). After qRT-PCR test, the Ct values of ORF1ab gene, N gene, and E gene were divided by that of reference genes.
Patient Test
All the experiments followed the policy approved by the Ethics Committee of Wuhan Union Hospital. After sampling in COVID-19 patients/volunteers, regular and MN/Ab swabs (the ratio of high-crosslinked MN was 0.5) were stored in VPS. A questionnaire was completed by each patient/volunteer (one positive patient, one negative patient [recovery from positive to negative], and seven volunteers participated the study), which included their feelings about pain, itching, nausea, saliva, cough, sputum and dyspnea. A quantitative analysis was conducted based on questionnaire (score 4, very serious; score 3, serious; score 2, mild; score 1, very mild). The clinical samples were analyzed by using the method mentioned above.
Statistical Analysis
All experiments were repeated at least three times and the data presented as mean ± SD. SPSS statistical software (SPSS 16.0) was applied for statistical analyses. Any significant differences among mean values were evaluated by the two-tailed Student's t test or ANOVA (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001).
Acknowledgments
We thank the medical staff for their assistance in sampling and all healthcare workers in the frontline in Wuhan for their tremendous efforts and sacrifice made to fight against COVID-19 during this pandemic. This work was supported in part by the National Natural Science Foundation of China and the Intramural Research Program, National Institute of Biomedical Imaging and Bioengineering, National Institutes of Health (ZIA EB000073).
Author Contributions
Conceptualization, W.C., Z.W., L.W., and X.C.; Methodology, W.C., B.C., Z.G., and F.C.; Validation, Z.G. and F.C.; Formal Analysis, W.C., Z.W., and L.W.; Investigation, Z.W., L.W., and X.C.; Resources, Z.W. and L.W.; Writing – Original Draft, W.C.; Writing – Review & Editing, Z.W., L.W., and X.C.; Funding Acquisition, Z.W., L.W., and X.C.
Declaration of Interests
The authors declare no competing interests.
Published: October 11, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.matt.2020.09.021.
Supplemental Information
References
- 1.Lipsitch M., Swerdlow D.L., Finelli L. Defining the epidemiology of Covid-19—studies needed. N. Engl. J. Med. 2020;382:1194–1196. doi: 10.1056/NEJMp2002125. [DOI] [PubMed] [Google Scholar]
- 2.Coronavirus disease (COVID-19) World Health Organization; 2020. Situation Report. [Google Scholar]
- 3.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rothe C., Schunk M., Sothmann P., Bretzel G., Froeschl G., Wallrauch C., Zimmer T., Thiel V., Janke C., Guggemos W. Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N. Engl. J. Med. 2020;382:970–971. doi: 10.1056/NEJMc2001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo L., Ren L., Yang S., Xiao M., Chang D., Yang F., Dela Cruz C.S., Wang Y., Wu C., Xiao Y. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19) Clin. Infect. Dis. 2020;71:778–785. doi: 10.1093/cid/ciaa310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tang Y., Schmitz J.E., Persing D.H., Stratton C.W. The laboratory diagnosis of COVID-19 infection: current issues and challenges. J. Clin. Microbiol. 2020;58:e00512–e00520. doi: 10.1128/JCM.00512-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiang F., Wang X., He X., Peng Z., Yang B., Zhang J., Zhou Q., Ye H., Ma Y., Li H. Antibody detection and dynamic characteristics in patients with COVID-19. Clin. Infect. Dis. 2020:ciaa461. doi: 10.1093/cid/ciaa461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petruzzi G., De Virgilio A., Pichi B., Mazzola F., Zocchi J., Mercante G., Spriano G., Pellini R. COVID-19: nasal and oropharyngeal swab. Head Neck. 2020;42:1303–1304. doi: 10.1002/hed.26212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lan L., Xu D., Ye G., Xia C., Wang S., Li Y., Xu H. Positive RT-PCR test results in patients recovered from COVID-19. JAMA. 2020;323:1502–1503. doi: 10.1001/jama.2020.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laboratory testing for coronavirus disease 2019 (COVID-19) in suspected human cases: interim guidance (2020). (World Health Organization).
- 11.Xie X., Zhong Z., Zhao W., Zheng C., Wang F., Liu J. Chest CT for typical 2019-nCoV pneumonia: relationship to negative RT-PCR testing. Radiology. 2020;296:E41–E45. doi: 10.1148/radiol.2020200343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fang Y., Zhang H., Xie J., Lin M., Ying L., Pang P., Ji W. Sensitivity of chest CT for COVID-19: comparison to RT-PCR. Radiology. 2020;296:E115–E117. doi: 10.1148/radiol.2020200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Winichakoon P., Chaiwarith R., Liwsrisakun C., Salee P., Goonna A., Limsukon A., Kaewpoowat Q. Negative nasopharyngeal and oropharyngeal swabs do not rule out COVID-19. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.00297-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hase R., Kurita T., Muranaka E., Sasazawa H., Mito H., Yano Y. A case of imported COVID-19 diagnosed by PCR-positive lower respiratory specimen but with PCR-negative throat swabs. Infect. Dis. 2020;52:423–426. doi: 10.1080/23744235.2020.1744711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Panpradist N., Toley B.J., Zhang X., Byrnes S., Buser J.R., Englund J.A., Lutz B.R. Swab sample transfer for point-of-care diagnostics: characterization of swab types and manual agitation methods. PLoS One. 2014;9:e105786. doi: 10.1371/journal.pone.0105786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim W., Peaudecerf F., Baldwin M.W., Bush J.W. The hummingbird's tongue: a self-assembling capillary syphon. Proc. R. Soc. B Biol. Sci. 2012;279:4990–4996. doi: 10.1098/rspb.2012.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Al-Zahaby S.A., Elsheikh E. Ultramorphological and histological studies on the tongue of the common kingfisher in relation to its feeding habit. J. Basic Appl. Zool. 2014;67:91–99. [Google Scholar]
- 18.Emura S., Okumura T., Chen H. Scanning electron microscopic study of the tongue in the rainbow lorikeet (Trichoglossus haematodus) Okajimas Folia Anat. Jpn. 2011;88:17–21. doi: 10.2535/ofaj.88.17. [DOI] [PubMed] [Google Scholar]
- 19.Kleinteich T., Gorb S.N. Tongue adhesion in the horned frog Ceratophrys sp. Sci. Rep. 2014;4:5225. doi: 10.1038/srep05225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y., Feng P., Yu J., Yang J., Zhao J., Wang J., Shen Q., Gu Z. Ros-responsive microneedle patch for acne vulgaris treatment. Adv. Ther. 2018;1:1800035. [Google Scholar]
- 21.Blicharz T.M., Gong P., Bunner B.M., Chu L.L., Leonard K.M., Wakefield J.A., Williams R.E., Dadgar M., Tagliabue C.A., El Khaja R. Microneedle-based device for the one-step painless collection of capillary blood samples. Nat. Biomed. Eng. 2018;2:151–157. doi: 10.1038/s41551-018-0194-1. [DOI] [PubMed] [Google Scholar]
- 22.Lee H., Choi T.K., Lee Y.B., Cho H.R., Ghaffari R., Wang L., Choi H.J., Chung T.D., Lu N., Hyeon T. A graphene-based electrochemical device with thermoresponsive microneedles for diabetes monitoring and therapy. Nat. Nanotechnol. 2016;11:566–572. doi: 10.1038/nnano.2016.38. [DOI] [PubMed] [Google Scholar]
- 23.Coffey J.W., Meliga S.C., Corrie S.R., Kendall M.A. Dynamic application of microprojection arrays to skin induces circulating protein extravasation for enhanced biomarker capture and detection. Biomaterials. 2016;84:130–143. doi: 10.1016/j.biomaterials.2016.01.015. [DOI] [PubMed] [Google Scholar]
- 24.Paul R., Saville A.C., Hansel J.C., Ye Y., Ball C., Williams A., Chang X., Chen G., Gu Z., Ristaino J.B. Extraction of plant DNA by microneedle patch for rapid detection of plant diseases. ACS Nano. 2019;13:6540–6549. doi: 10.1021/acsnano.9b00193. [DOI] [PubMed] [Google Scholar]
- 25.Lee K.T., Muller D.A., Coffey J.W., Robinson K.J., McCarthy J.S., Kendall M.A., Corrie S.R. Capture of the circulating plasmodium falciparum biomarker HRP2 in a multiplexed format, via a wearable skin patch. Anal. Chem. 2014;86:10474–10483. doi: 10.1021/ac5031682. [DOI] [PubMed] [Google Scholar]
- 26.Coffey J.W., Corrie S.R., Kendall M.A. Early circulating biomarker detection using a wearable microprojection array skin patch. Biomaterials. 2013;34:9572–9583. doi: 10.1016/j.biomaterials.2013.08.078. [DOI] [PubMed] [Google Scholar]
- 27.Yeow B., Coffey J.W., Muller D.A., Grøndahl L., Kendall M.A., Corrie S.R. Surface modification and characterization of polycarbonate microdevices for capture of circulating biomarkers, both in vitro and in vivo. Anal. Chem. 2013;85:10196–10204. doi: 10.1021/ac402942x. [DOI] [PubMed] [Google Scholar]
- 28.Yu J., Zhang Y., Ye Y., DiSanto R., Sun W., Ranson D., Ligler F.S., Buse J.B., Gu Z. Microneedle-array patches loaded with hypoxia-sensitive vesicles provide fast glucose-responsive insulin delivery. Proc. Natl. Acad. Sci. U S A. 2015;112:8260–8265. doi: 10.1073/pnas.1505405112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang C., Li W., Drabek D., Okba N.M., van Haperen R., Osterhaus A.D., van Kuppeveld F.J., Haagmans B.L., Grosveld F., Bosch B.-J. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat. Commun. 2020;11:2251. doi: 10.1038/s41467-020-16256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 32.Chen W., Tian R., Xu C., Yung B.C., Wang G., Liu Y., Ni Q., Zhang F., Zhou Z., Wang J. Microneedle-array patches loaded with dual mineralized protein/peptide particles for type 2 diabetes therapy. Nat. Commun. 2017;8:1777. doi: 10.1038/s41467-017-01764-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nonoyama T., Wada S., Kiyama R., Kitamura N., Mredha M.T., Zhang X., Kurokawa T., Nakajima T., Takagi Y., Yasuda K. Double-network hydrogels strongly bondable to bones by spontaneous osteogenesis penetration. Adv. Mater. 2016;28:6740–6745. doi: 10.1002/adma.201601030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate/analyze any datasets/code.


