Supplemental Digital Content is available in the text
Keywords: long non-coding RNA, breast cancer, overall survival, relapse free survival
Abstract
Breast cancer (BC) is a disease of high mortality rate because of high malignant, while early diagnosis and personal management may make a better prognosis possible. This study aimed to establish and validate lncRNAs signatures to improve the prognostic prediction for BC.
RNA sequencing data along with the corresponding clinical information of patients with BC were gained from The Cancer Genome Atlas (TCGA). Prognostic differentially expressed lncRNAs were obtained using differentially expressed lncRNAs analysis (P value <.01 and |fold change| > 2) and univariate cox regression (P value <.05). By applying least absolute shrinkage and selection operation (LASSO) Cox regression analysis along with 10-fold cross-validation, 2 lncRNA-based signatures were constructed in the training, test and whole set.
A 14-lncRNAs signature and a 10-lncRNAs signature were built for overall survival (OS) and relapse-free survival (RFS) respectively in the 3 sets. BC patients were divided into high-risk groups and low-risk groups depended on median risk score value. Significant differences were found for OS and RFS between 2 groups in the 3 sets. The time-dependent receiver operating characteristic (ROC) curves analysis demonstrated that our lncRNAs signatures had better predictive capacities of survival and recurrence for BC patients as well as enhancing the predictive ability of the tumor node metastasis (TNM) stage system.
These results indicate that the 2 lncRNAs signatures with the potential to be biomarkers to predict the prognosis of BC for OS and RFS.
1. Introduction
Breast cancer (BC) is one of the most common cancers and the leading cause of cancer death in females around the world, estimating that 24% of diagnosed cases and 15% of death cases among them.[1] There are various treatment strategies for BC patients, including surgery, chemotherapy, radiation therapy and hormone-blocking therapy,[2] but the prognosis of the patients remains dismal.[3] Therefore, it is particularly important to explore the potential molecular mechanisms in the development of BC and find new biomarkers and therapeutic targets to improve the prognosis of patients with BC. The outcome of BC varies depending on quite a few factors, such as age, stage and grade of cancer.[4] As is known to all, tumor node metastasis (TNM) stage system is a tool extensively used in predicting prognosis of patients and making treatment plants in clinical practice.[5] A single biomarker or a clinical characteristic often is lack of adequate accuracy for predicting outcome of patients, while integrating multiple biomarkers and clinical characteristics can significantly improve prognostic performance.[6,7] It is highly necessary that making a comprehensive approach of combining biomarkers and traditional clinicopathological factors to achieve more reliable prognostic assessment.
Long non-coding RNA (lncRNA), a type of RNA molecules with a length of more than 200 nucleotides, is a major class of ncRNA which can regulate gene expression at the levels of transcription, epigenetics and translation.[8,9,10] Increasing studies have found that lncRNAs exist abnormal expression and related to prognosis in BC.[11,12,13] However, there are few studies on the prognostic value of uniting biomarkers and other clinical indicators.
In the present study, by using least absolute shrinkage and selection operation (LASSO) Cox regression analysis and 10-fold cross-validation, a 14-lncRNAs signature for overall survival (OS) and a 10-lncRNAs signature for relapse-free survival (RFS) were built to appraise predictive value of lncRNAs for survival and recurrence of BC. Besides, Correlation analysis and Cox regression analysis between lncRNAs signatures and clinicopathological characteristics were performed. Moreover, a comparison of predictive ability between lncRNA signatures and TNM system was evaluated. Through this research, we hope to develop reliable prognostic predictors from lncRNAs signatures for BC patients.
2. Material and methods
2.1. Data collection
A flowchart showing the major steps in the study process (Fig. S1). The RNA sequencing datasets of BC patients and related clinical profiles were obtained from The Cancer Genome Atlas (TCGA) website (https://cancergenome.nih.gov/ , which contained 1109 BC tissue samples and 113 adjacent non-cancerous tissue samples. No ethical approval is required since all raw data came from public databases for this study.
2.2. Construction of survival-predicting models
The raw data were converted into an expression matrix, and lncRNAs names from the dataset were annotated by Ensembl (http://asia.ensembl.org/index.html). We utilized “edgeR” package in R studio to screen the differentially expressed lncRNAs (DElncRNAs) with P value <0.01 and |fold change| > 2 between BC tissues and adjacent non-cancerous tissues. Besides, we carried out univariate Cox regression analysis for lncRNAs in BC tissues and the lncRNAs with P value <.05 were identified to be prognostic lncRNAs. Prognostic DElncRNAs were obtained from the intersection between and DElncRNAs and prognostic lncRNAs for the next analysis. Tumor samples (the whole set) were randomly divided into the training set and the test set at a 2:1 ratio. Then, survival-predicting models were built by combining LASSO Cox regression analysis and 10-fold cross-validation in the 3 sets.[14,15]
2.3. Functional enrichment analyses of lncRNAs in the signatures
Expression correlation analysis between the lncRNAs and genes was used to find the putative genes of lncRNAs in the signatures. The absolute value of the Pearson correlation coefficient >0.3 and P value <.001 were set as the cutoff values. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis were performed by the R package “clusterProfiler”. classifiers
2.4. Statistical analysis
The lncRNAs prognosis risk score value was calculated by using the formula: risk score value = EXP1∗βlncRNA1 + EXP2∗β2… + EXPx∗βx, EXP presents the expression level of each lncRNA and β presents the regression coefficient from LASSO Cox selection method at 10-fold cross-validation. Among them, the median risk score value is the criterion for splitting BC patients into high-risk groups and low-risk groups. Pearsons Chi-Squared test was applied to investigate the relationship between the models and clinicopathologic characteristics. Prognostic factors were selected through univariate and multivariate Cox regression analysis in the above parameters. To further evaluate the prognostic value and clinical application value of the risk scores in each model and stage, we performed Kaplan–Meier survival analysis and time-dependent receiver operating characteristic (ROC) curves.[16,17]
3. Results
3.1. Derivation of the lncRNAs signatures for OS and RFS from BC patients
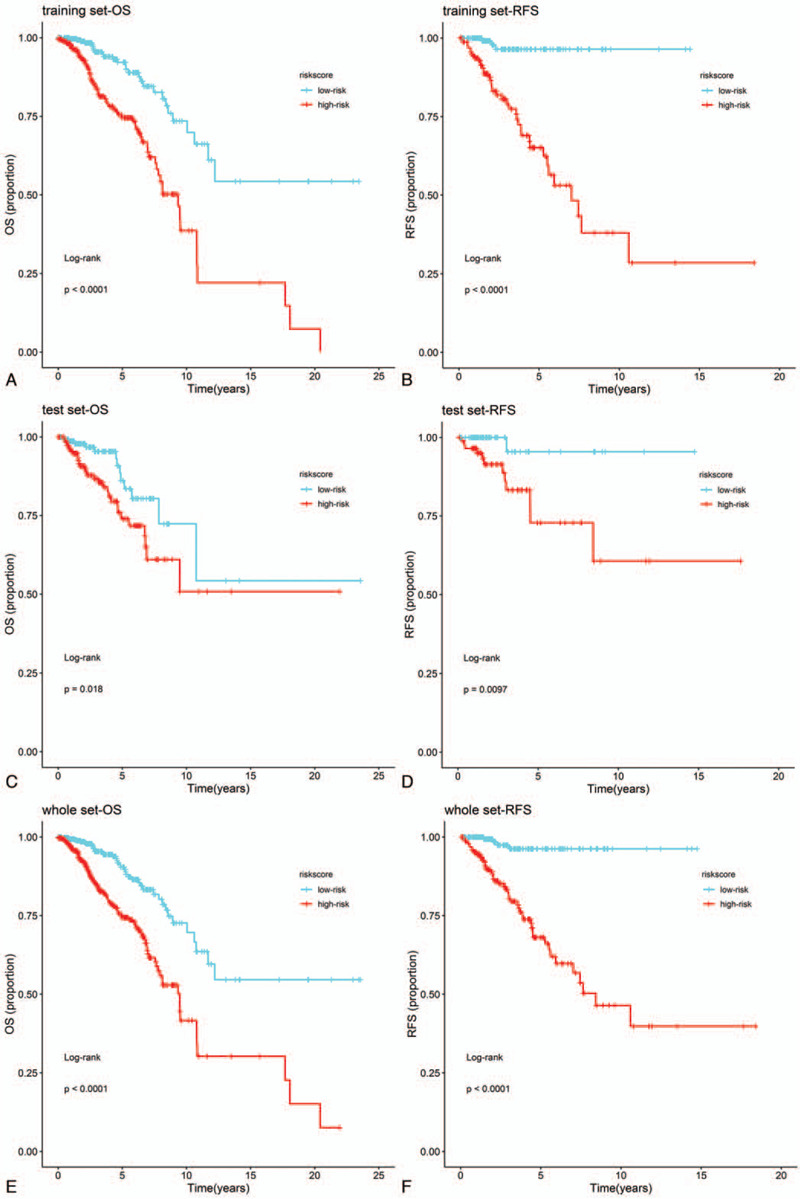
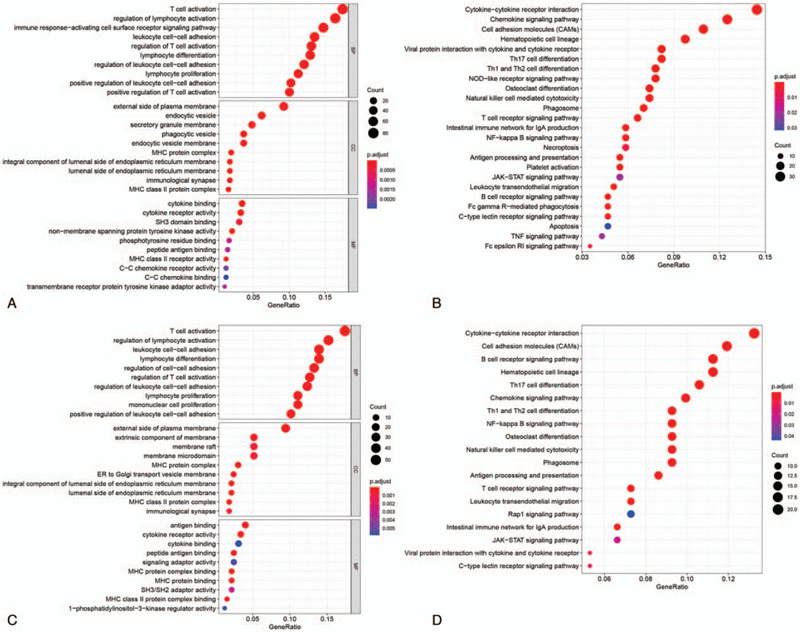
Firstly, there were 1042 DElncRNAs in BC tissue samples compare with normal tissue samples based on “edgeR” package at the threshold of P value <.01 and |fold change| > 2 (Fig. 1A). Moreover, lncRNAs with log-rank P < .05 were screened from all lncRNAs by univariate Cox regression analysis. Totally, 93 prognostic DElncRNAs (OS, Fig. 1B) and 87 prognostic DElncRNAs (RFS, Fig. 1C) were retained for subsequent analysis. For OS, BC samples (n = 1069) were randomly divided into training set (n = 712) and test set (n = 357) at a 2:1 ratio. Likewise, BC samples (n = 488) were randomly divided into training set (n = 325) and test set (n = 163) at the same ratio for OS. Survival-predicting models were built by combining LASSO Cox regression analysis and 10-fold cross-validation in the training sets (OS: Fig. 1D, 1E; RFS: Fig. 1F, 1G). As a result, a 14-lncRNAs signature for OS and a 10-lncRNAs signature for RFS were established. To confirm our results, the same signatures for OS and RFS were also constructed in the test set and the whole set. Detailed information about the lncRNAs of signatures as shown in Table 1S. BC patients were ranked on the basis of risk score value and split into high-risk groups or low-risk groups using the median risk score value of the sets as the cut-off point. Kaplan–Meier survival plots revealed that the low-risk group had better OS and RFS in the training sets (Fig. 2A and B), the test sets (Fig. 2C and D) and the whole sets (Fig. 2E and F). GO and KEGG analyses showed that the lncRNAs might involve in immune cells activation, formation of membrane and vesicle, cytokine activity and immune-related pathways for OS (Fig. 3A and B). According to the same analysis, the lncRNAs were enriched in immune cells activation, membrane formation, antigen binding and immune-related pathways for RFS (Fig. 3C and D).
Figure 1.
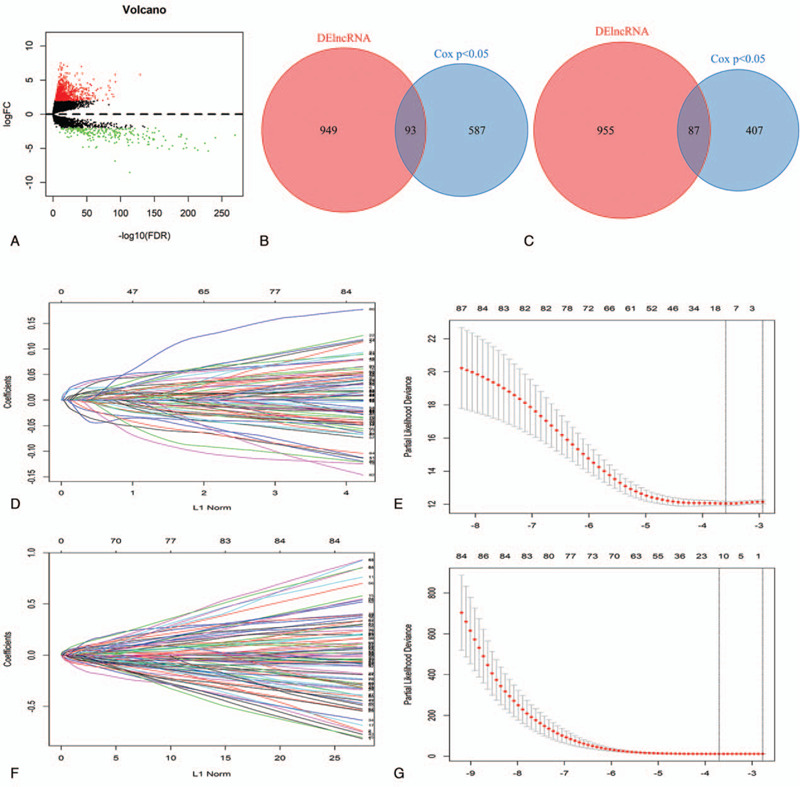
Construction of the lncRNAs-based models for OS and RFS. (A) Volcano plot of DElncRNAs (P value <.01 and |fold change| > 2). (B and C) Venn diagram of prognostic DElncRNAs in prognostic lncRNAs for OS and RFS (univariate cox P value <.05). (D and F) 10-fold cross-validation for tuning parameter selection in the LASSO model for OS and RFS. (E and G) LASSO coefficient profiles of prognostic DElncRNAs for OS and RFS. lncRNA = long non-coding RNA, OS = overall survival, RFS = relapse free survival, LASSO = least absolute shrinkage and selection operation, DElncRNAs = differentially expressed long non-coding RNAs.
Figure 2.
Functional enrichment analyses for lncRNAs of the 2 signatures. (A and B) GO and KEGG analysis of lncRNAs in 14-lncRNA-based classifier. (C and D) GO and KEGG analysis of lncRNAs in 10-lncRNA-based classifier. lncRNA = long non-coding RNA, GO = Gene Ontology, KEGG = Kyoto Encyclopedia of Genes and Genomes.
Figure 3.
Kaplan–Meier survival analysis between 2 groups in the 3 sets. (A, C, and E): Survival curves of BC patients from high-risk groups and low-risk groups in training, test and whole sets based on 14-lncRNA-based classifier risk score level for OS. (B, D, and F): Survival curves of BC patients from high-risk groups and low-risk groups in training, test and whole sets based on 10-lncRNA-based classifier risk score level for RFS. lncRNA = long non-coding RNA, OS = overall survival, RFS = relapse free survival.
3.2. LncRNAs signatures associated with clinical and pathologic features
To assess the association between lncRNAs signatures and clinical and pathologic features, we performed Pearsons Chi-Squared test for the 3 sets independently. As shown in Table 1 , 2 groups existed significant differences in 4 characteristics (subtype, Her2, ER, and PR) in the 3 sets for OS. Besides, 2 groups existed significant differences on only 1 characteristic (ER) in 3 sets for RFS (Table 2 ). We also analyzed associations between risk scores and clinical characteristics. Patients with Her2 positive, distant metastases, ER negative, PR negative tended more towards high-risk score (Fig. 4).
Table 1.
Correlations of risk score of the 14-lncRNA-based classifier for OS with clinicopathological characteristics in training set, test set, and whole set.
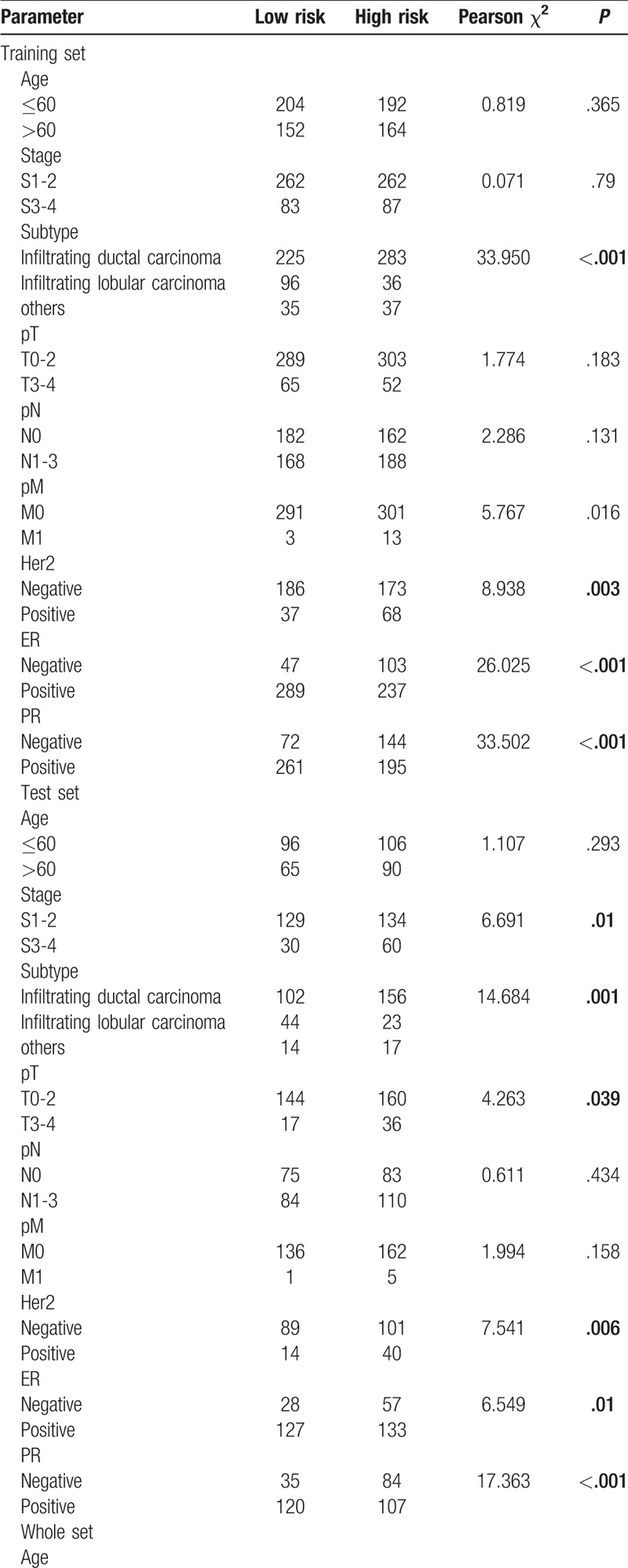
Table 1 (Continued).
Correlations of risk score of the 14-lncRNA-based classifier for OS with clinicopathological characteristics in training set, test set, and whole set.
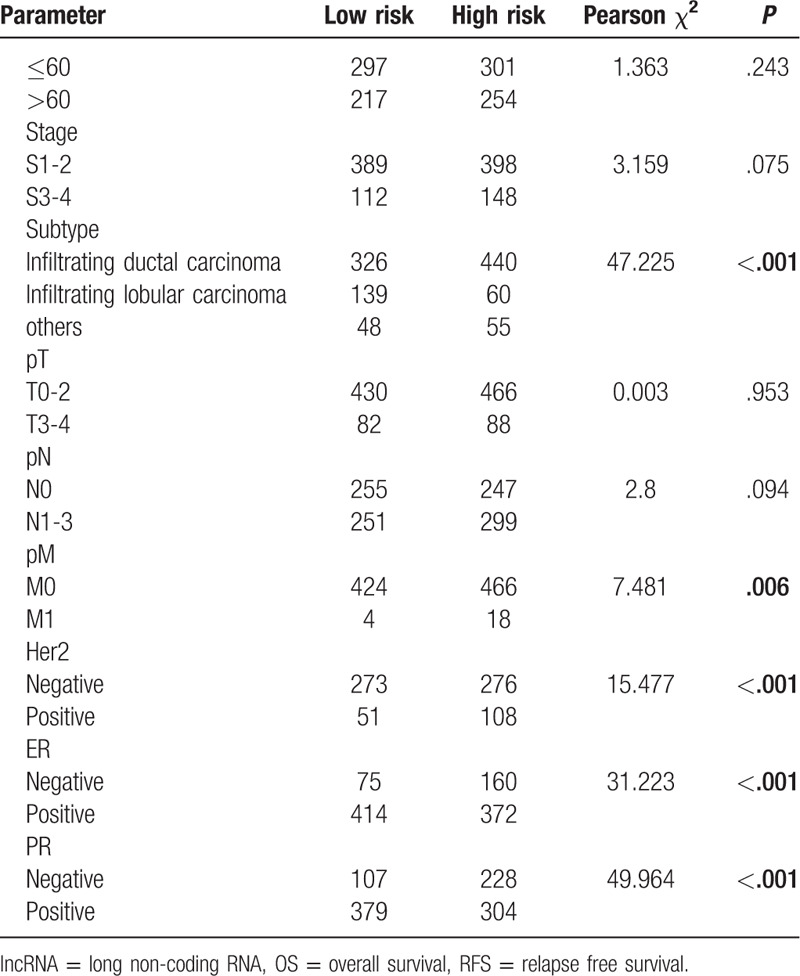
Figure 4.
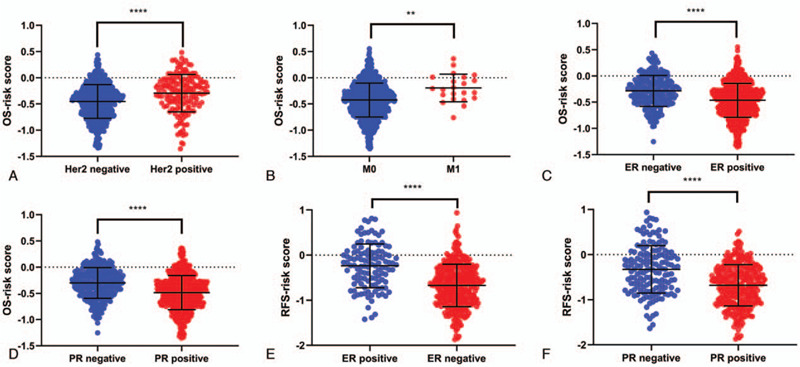
Correlation between risk score and clinical and pathologic features in the whole set. Boxplot of risk scores in BC patients with Her2 (A, OS), pM (B, OS), ER (C, OS; E, RFS) and PR (D, OS; F, RFS). ∗ P < .05, ∗∗ P < .01, ∗∗∗ P < .001 and ∗∗∗∗ P < .0001. OS = overall survival, RFS = relapse free survival.
3.3. Prognostic value of lncRNAs signatures for BC
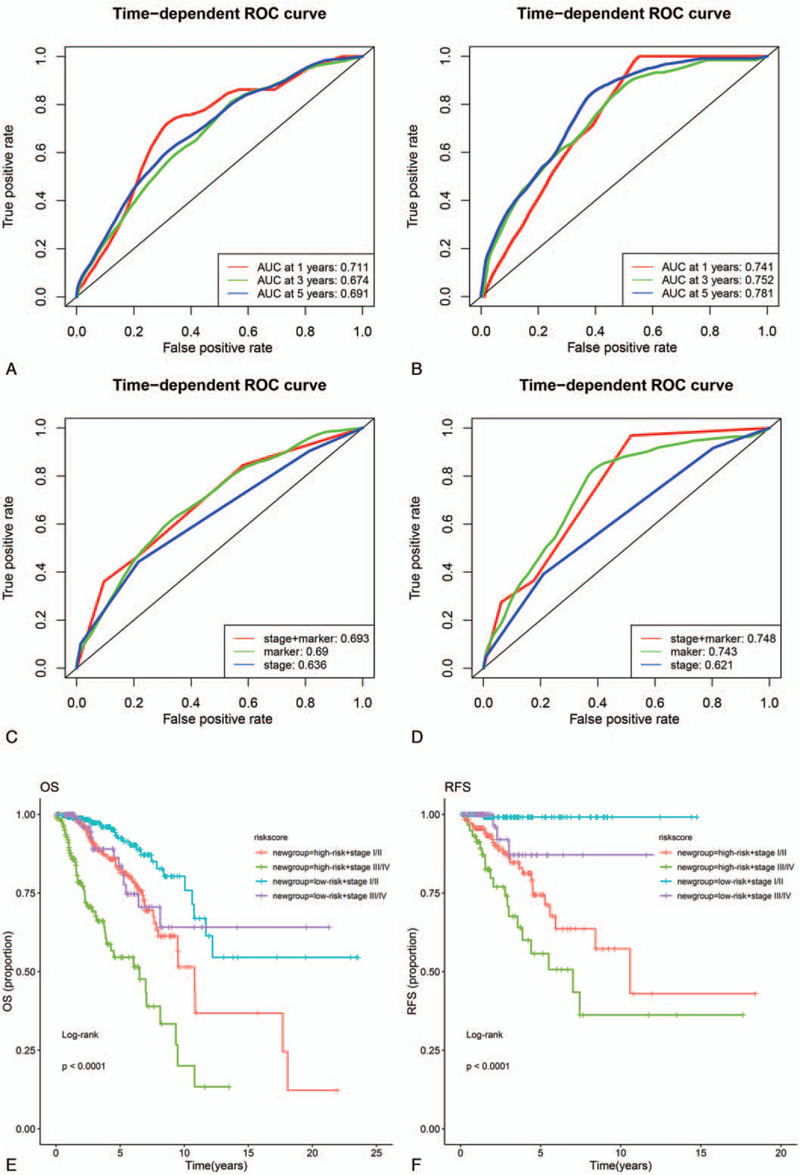
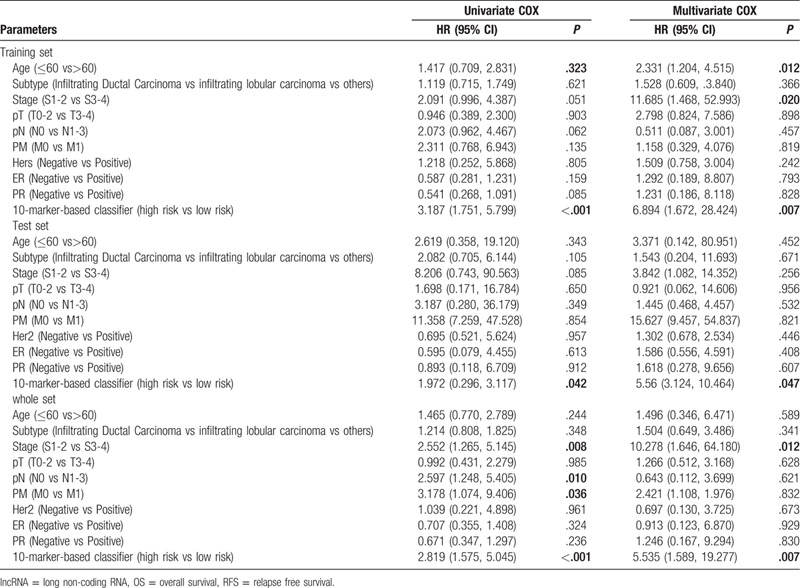
Cox regression analysis was applied to investigate the independent predictive power of lncRNAs signatures and other clinicopathological data. Univariate Cox regression analysis found that age, stage, pM, and the 14-lncRNAs signature were obviously related to OS in the 3 sets (Table 3). But age, stage and the 14-lncRNAs signature were remarkably related to OS after multivariate Cox regression analysis of BC in the 3 sets (Table 3). However, in both univariate and multivariate Cox regression analyses, only the 10-lncRNAs signature remained an independent predictor for RFS in the 3 sets (Table 4). In addition, time-dependent ROC curves demonstrated prediction power of the 14-lncRNAs signature for OS (Fig. 5A) in the 1 year, 3 years and 5 years achieved area under curves (AUCs) were 0.711, 0.674, and 0.691 separately. As for RFS, the AUCs based on the 10-lncRNAs signature in the 1 year, 3 years and 5 years were 0.741, 0.752, and 0.781 separately (Fig. 5B). The curves indicate that 2 signatures had a considerable predictive performance for BC survival and recurrence. TNM stage system is the most frequently used tool to distinguish poor prognosis and good prognosis for cancer patients in clinical practice. Therefore, we compared predicting prognostic ability of the lncRNAs signatures, TNM stage and their combination. The multi-index time-dependent ROC cures analysis revealed that the 14-lncRNAs signature and the 10-lncRNAs signature had better predictive power than TNM stage for OS and RFS. What is more, combining the lncRNAs signatures and TNM stage may increase predictive accuracy for survival and recurrence (Fig. 5C and D). Kaplan–Meier curves and log-rank tests demonstrated that differences between groups were significant after separating BC patients into 4 groups according to lncRNAs risk scores and TNM stage (Fig. 5E and F).
Table 2.
Correlations of risk score of the 10-lncRNA-based classifier for RFS with clinicopathological characteristics in training set, test set, and whole set.
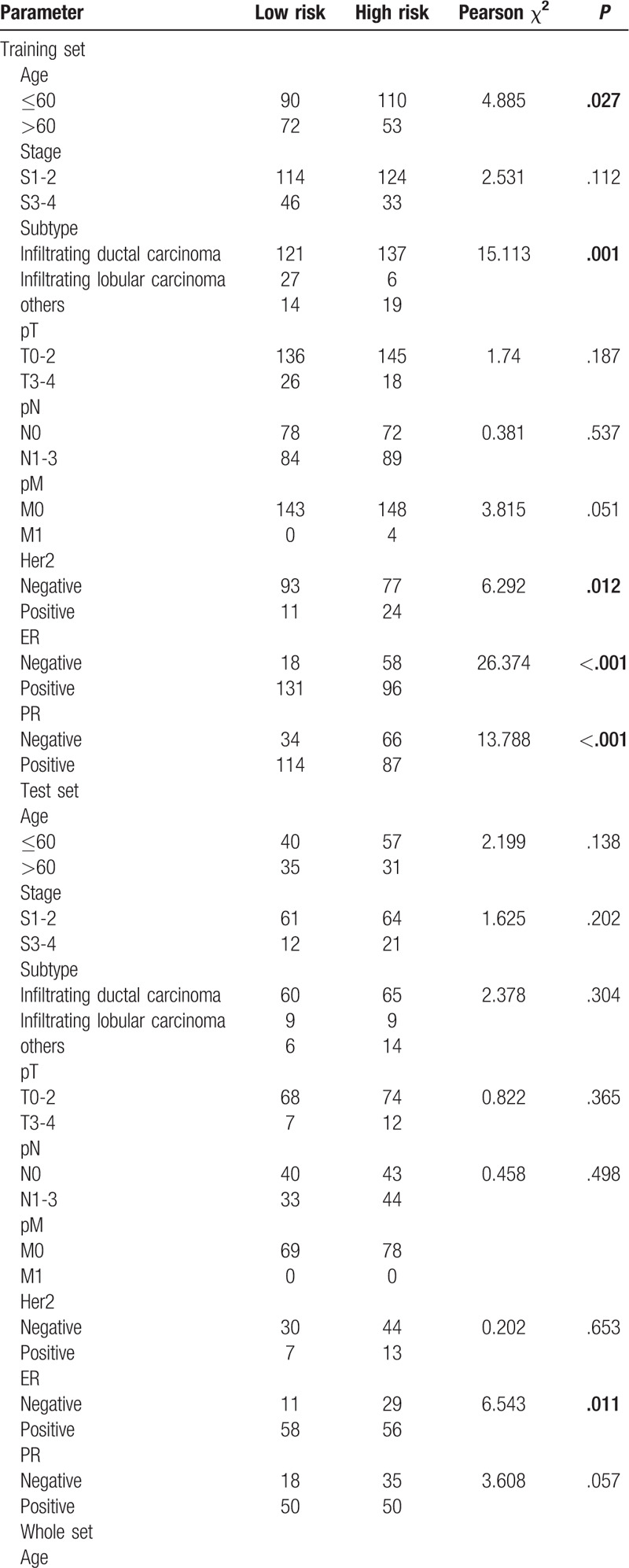
Table 2 (Continued).
Correlations of risk score of the 10-lncRNA-based classifier for RFS with clinicopathological characteristics in training set, test set, and whole set.
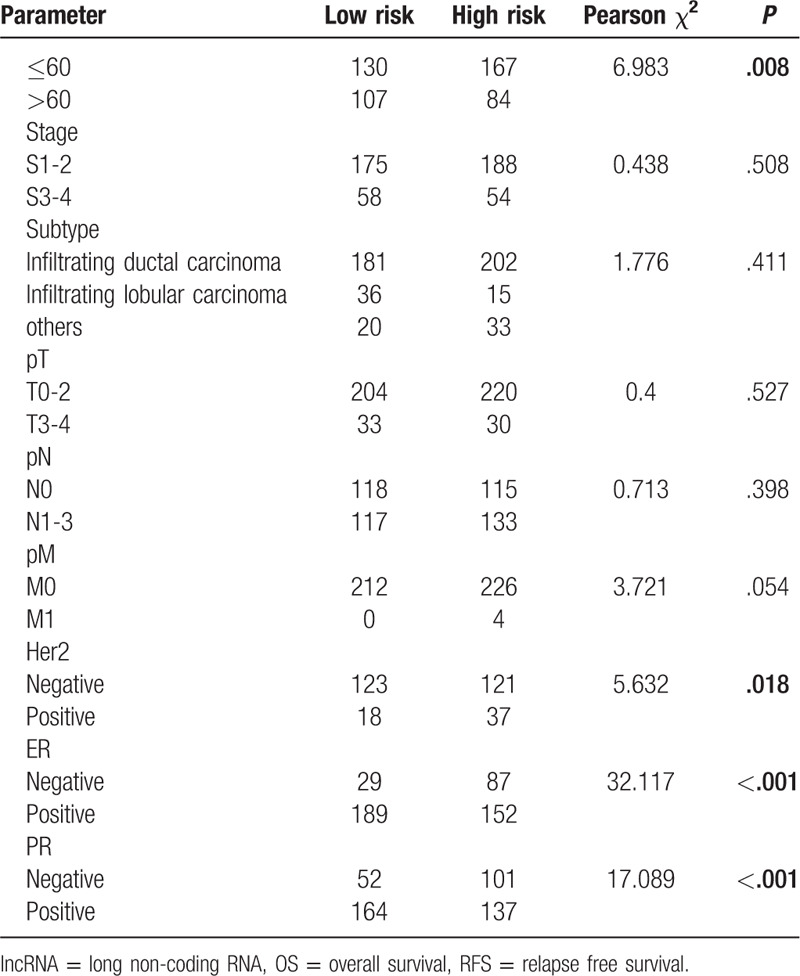
Figure 5.
Evaluating the predicting prognostic ability for models and stage in the whole set. (A and B) Time-dependent ROC curves of lncRNA-based classifiers at 1, 3 and 5 years for OS and RFS. (C and D) Time-dependent ROC curves of lncRNA-based classifiers, stage, and combinations of them for OS and RFS. (E and F) Survival curves of BC patients with combinations of lncRNA-based classifiers and stage for OS and RFS. lncRNA = long non-coding RNA, OS = overall survival, RFS = relapse free survival.
Table 3.
Cox regression analysis of the 14-lncRNA-based classifier with OS in training set, test set, and whole set.
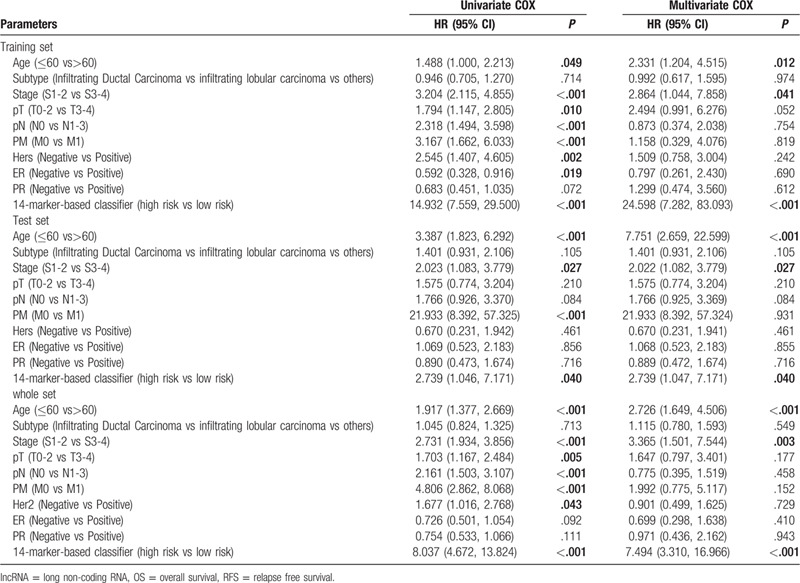
Table 4.
Cox regression analysis of the 10-lncRNA-based classifier with RFS in training set, test set and whole set.
4. Discussion
There is a large population with cancer in China, and the incidence and mortality have been increasing at a rapid rate.[18] BC is the first cause of new cancer diagnoses at the same time the most commonly diagnosed cancer at young and middle age in females.[19] Improvement in the clinical management of breast cancer has alleviated mortality rate in recent years, but conventional diagnostic criteria and treatment means are far from mainly satisfactory because of tumor heterogeneity in molecular level.[20,21] Fortunately, the rapid development of molecular biology makes it possible to discover highly specific markers of diagnosis and treatment for patients with BC. Cumulative studies have indicated that lncRNAs are aberrantly expressed in numerous cancers and play significant roles in tumorigenesis, cancer recurrence and metastasis.[22,23] For BC, emerging investigations have emphasized the vital function of lncRNAs in tumorigenesis and development.[24,25,26] For example, UASR1 has a high expression in BC tissues and promotes cancer cell growth, proliferation, wound healing and migration through the AKT/mTOR signaling pathway. [27] MALAT1 is considered an oncogenic regulator of BC, which accelerates angiogenesis through miR-145 in MCF-7 cells to enhance aggressiveness of BC.[28] Its worth noting that some lncRNAs have been regarded as potential prognosis markers for BC, according to the previous researches.[29,30] But they have limited prognostic value and insufficient credibility because of small sample sizes and deficiency of verification from multiple data sets. Therefore, we explored and validated lncRNAs signatures to improve predictive power of prognosis in BC based on a large sample size from TCGA cohort. LASSO Cox regression model not only has advantages to solve the common multicollinearity problem but also holds some traits such as numerical stability and interpretability.[31] Besides, LASSO Cox regression model combines with 10-fold cross-validation can contribute to deal with the “curse-of-dimensionalit” in high-throughput biological data. Hence, we established a 14-lncRNAs signature for OS and a 10-lncRNAs signature for RFS by way of LASSO Cox regression model along with 10-fold cross-validation, which may have better predictive power for BC. BC patients could be split into high-risk groups and low-risk groups with remarkable differences through 2 signatures for OS and RFS in the training sets. What is more, 2 signatures could distinguish 2 groups in the test sets and the whole sets, further confirming the robustness and reliability of the 2 lncRNAs signatures in predicting BC prognosis. Interestingly, GO and KEGG analyses show that the lncRNAs in 2 signatures were both involved in some similar biological processes and pathways. Next, Pearsons Chi-Squared test was performed to evaluate correlations between risk scores and clinicopathological characteristics. The results had shown that risk scores of the 14-lncRNAs signature correlated with subtype, Her2, ER, and PR for OS, while risk scores of the 10-lncRNAs signature only correlated with ER. Afterward, multivariate Cox regression analyses revealed age, stage and the 14-lncRNAs signature were creditable independent predictors for OS and only the 10-lncRNAs signature was a creditable independent predictor for RFS. Based on the time-dependent ROC curves analysis, the 14-lncRNAs signature and the 10-lncRNAs signature had an excellent and effective performance for survival and recurrence prediction, respectively. It is widely agreed that TNM stage is the most commonly used classified tool for cancer. For this reason, we explored the relationship between the lncRNAs signatures and TNM stage. Compared to TNM stage, the lncRNAs signatures had a perceptibly better predictive power than TNM stage. Importantly, combing 2 indexes may improve the performance of predicting prognosis for survival and recurrence. As shown in Fig. 3E and F, the prognosis differences between BC patients divided by both the lncRNAs risk scores and TNM stage were evident.
In the present study, the 14-lncRNAs signature and the 10-lncRNAs signature has been proven to be significantly connected with the OS and RFS of BC. The functions of a majority of the lncRNAs in the signatures have not been totally expounded up to now. But there are some reports about several lncRNAs in our signatures from the former researches. FEZF1-AS1 has been identified as a nuclear-restricted lncRNA. The expression levels of FEZF1-AS1 are downregulated in human prostate cancer tissues, and it paly tumor-promotive roles in prostate cancer via Notch signaling pathway.[32] LINC00536 is a newly identified lncRNA and named by Nakajima that located at chromosome 8q23.3 in 2014. It could induce malignant phenotypes to promote BC cell proliferation, migration, and invasion.[33] Gong et al found that LINC01224 regulated the expression levels of CHEK1 by competitively binding to miR-330-5p, thus inhibiting hepatocellular carcinoma progression.[34] LINC00668 is overexpressed in BC tissues and contributes to the progression of BC by accelerating cell cycle progression and inhibiting cell apoptosis.[35] In view of the outstanding performance of prognostic prediction, the roles of these lncRNAs in cancers should be investigated in the future, especially for BC.
However, some shortcomings in this study should not be ignored. On the 1 hand, this study was a retrospective nature because all the data came from the public database which lacked further test in a prospective clinical trial. Besides, TCGA cohort could not provide some helpful clinicopathologic characteristics, such as treatments. On the other hand, the identified functions and potential mechanisms of the lncRNAs in our signatures are still veiled. Therefore, further studies are required to complete the clinical applications and explore the roles of lncRNAs, as this may help us to understand the signification of them in BC occurrence and development. Although these deficiencies are inevitable, our results remain to provide reliable lncRNAs signatures to predict BC survival and recurrence.
In summary, we assessed the genome-wide lncRNA expression profiles from TCGA database and constructed a 14-lncRNAs signature and a 10-lncRNAs signature by the LASSO Cox regression analysis and 10-fold cross-validation. The 2 lncRNAs signatures with the potential to be biomarkers to predict the prognosis of BC for OS and RFS. In addition, our study could also complement clinical and clinicopathologic characteristics analysis in an attempt to facilitate the personalized management of patients with BC.
Acknowledgments
We thank all the researchers involved in the consolidation and submission of the data from the TCGA database, which may provide convenience and possibility of tumors studies in a large cohort.
Author contributions
All authors read and approved the final manuscript.
Conceptualization: Yi Zhang, Tianhua Jiang.
Data curation: Yuzhi Wang.
Formal analysis: Yuzhi Wang, Gang Tian, Yi Zhang.
Funding acquisition: Gang Tian.
Investigation: Yi Zhang, Yuzhi Wang, Gang Tian.
Methodology: Yi Zhang, Gang Tian, Tianhua Jiang.
Project administration: Yi Zhang, Tianhua Jiang.
Resources: Yi Zhang, Tianhua Jiang.
Software: Yuzhi Wang, Gang Tian.
Supervision: Gang Tian, Yi Zhang.
Validation: Yuzhi Wang, Tianhua Jiang.
Visualization: Yuzhi Wang, Gang Tian.
Writing – original draft: Yi Zhang.
Writing – review & editing: Gang Tian, Tianhua Jiang.
Supplementary Material
Supplementary Material
Footnotes
Abbreviations: AUCs = area under curves, BC = breast cancer, DElncRNAs = differentially expressed long non-coding RNAs, ER = estrogen receptor, GO = Gene Ontology, Her2 = Human epidermal growth factor receptor 2, KEGG = Kyoto Encyclopedia of Genes and Genomes, lncRNA = long non-coding RNA, LASSO = least absolute shrinkage and selection operation, OS = overall survival, PR = progesterone receptor, RFS = relapse free survival, ROC = receiver operating characteristic, TCGA = The Cancer Genome Atlas, TNM = tumor node metastasis.
How to cite this article: Zhang Y, Wang Y, Tian G, Jiang T. Long non-coding RNA-based signatures to improve prognostic prediction of breast cancer. Medicine. 2020;99:40(e22203).
YZ and YW contributed equally to the study.
This study was supported by the doctor of medicine start-up capital of the Affiliated Hospital of Southwest Medical University (Grant number 18057) and the Luzhou-Southwest Medical University applied basic research project (Grant number 2018LZXNYD-ZK30).
The authors report no conflicts of interest.
The datasets generated during and/or analyzed during the current study are publicly available.
References
- [1].Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA 2018;68:394–424. [DOI] [PubMed] [Google Scholar]
- [2].Radice D, Redaelli A. Breast cancer management: quality-of-life and cost considerations. Pharmacoeconomics 2003;21:383–96. [DOI] [PubMed] [Google Scholar]
- [3].Pelosi A, Careccia S, Sagrestani G, et al. Dual promoter usage as regulatory mechanism of let-7c expression in leukemic and solid tumors. Mol Cancer Res 2014;12:878–89. [DOI] [PubMed] [Google Scholar]
- [4].Jiralerspong S, Goodwin PJ. Obesity and Breast Cancer Prognosis: Evidence, Challenges, and Opportunities. J Clin Oncol 2016;34:4203–16. [DOI] [PubMed] [Google Scholar]
- [5].Ni YB, Tsang JY, Chan SK, et al. A novel morphologic-molecular recurrence predictive model refines traditional prognostic tools for invasive breast carcinoma. Ann Surg Oncol 2014;21:2928–33. [DOI] [PubMed] [Google Scholar]
- [6].Qi L, Yao Y, Zhang T, et al. A four-mRNA model to improve the prediction of breast cancer prognosis. Gene 2019;721:144100. [DOI] [PubMed] [Google Scholar]
- [7].Li G, Hu J, Hu G. Biomarker studies in early detection and prognosis of breast cancer. Adv Exp Med Biol 2017;1026:27–39. [DOI] [PubMed] [Google Scholar]
- [8].Ezkurdia I, Juan D, Rodriguez JM, et al. Multiple evidence strands suggest that there may be as few as 19,000 human protein-coding genes. Hum Mol Genet 2014;23:5866–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Yang S, Sun Z, Zhou Q, et al. MicroRNAs, long noncoding RNAs, and circular RNAs: potential tumor biomarkers and targets for colorectal cancer. Cancer Manag Res 2018;10:2249–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell 2016;29:452–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Yao F, Wang Q, Wu Q. The prognostic value and mechanisms of lncRNA UCA1 in human cancer. Cancer Manag Res 2019;11:7685–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Keshavarz M, Asadi MH. Upregulation of pluripotent long noncoding RNA ES3 in HER2-positive breast cancer. J Cell Biochem 2019;120:18398–405. [DOI] [PubMed] [Google Scholar]
- [13].Zheng T, Pang Z, Zhao Z. A gene signature predicts response to neoadjuvant chemotherapy in triple-negative breast cancer patients. Biosci Rep 2019;39: [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Tibshirani R. Regression shrinkage and selection via the lasso. J R Stat Soc 1996;58:267–88. [Google Scholar]
- [15].Tibshirani R. The lasso method for variable selection in the Cox model. Stat Med 1997;16:385–95. [DOI] [PubMed] [Google Scholar]
- [16].Jia W, Sheng-Kai H, Mei Z, et al. Identification of a circulating microRNA signature for colorectal cancer detection. Plos One 2014;9:e87451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Yakushiji S, Tateishi U, Nagai S, et al. Computed tomographic findings and prognosis in thymic epithelial tumor patients. J Comput Assist Tomogr 2008;32:799–805. [DOI] [PubMed] [Google Scholar]
- [18].Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA 2016;66:115–32. [DOI] [PubMed] [Google Scholar]
- [19].Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018;68:7–30. [DOI] [PubMed] [Google Scholar]
- [20].Bertoli G, Cava C, Castiglioni I, et al. New biomarkers for diagnosis, prognosis, therapy prediction and therapeutic tools for breast cancer. Theranostics 2015;5:1122–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Charles M. Comprehensive molecular portraits of human breast tumors. Nature 2012;490:61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell 2013;154:26–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Luka B, Metka R-G, Damjan G. Long nncoding RNAs as biomarkers in cancer. Dis Markers 2017;1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Rodriguez Bautista R, Ortega Gomez A. Long non-coding RNAs: implications in targeted diagnoses, prognosis, and improved therapeutic strategies in human non- and triple-negative breast cancer. Clin Epigenetics 2018;10:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Dong L, Qian J, Chen F, et al. LINC00461 promotes cell migration and invasion in breast cancer through miR-30a-5p/integrin beta3 axis. J Cell Biochem 2019;120:4851–62. [DOI] [PubMed] [Google Scholar]
- [26].Qiao E, Chen D, Li Q, et al. Long noncoding RNA TALNEC2 plays an oncogenic role in breast cancer by binding to EZH2 to target p57(KIP2) and involving in p-p38 MAPK and NF-kappaB pathways. J Cell Biochem 2019;120:3978–88. [DOI] [PubMed] [Google Scholar]
- [27].Cao Z, Wu P, Su M, et al. Long non-coding RNA UASR1 promotes proliferation and migration of breast cancer cells through the AKT/mTOR pathway. J Cancer 2019;10:2025–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Huang XJ, Xia Y, He GF, et al. MALAT1 promotes angiogenesis of breast cancer. Oncol Rep 2018;40:2683–9. [DOI] [PubMed] [Google Scholar]
- [29].Tu C, Ren X, He J, et al. The value of LncRNA BCAR4 as a prognostic biomarker on clinical outcomes in human cancers. J Cancer 2019;10:5992–6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Wang Y, Zhang G, Han J. HIF1A-AS2 predicts poor prognosis and regulates cell migration and invasion in triple-negative breast cancer. J Cell Biochem 2019;120:10513–8. [DOI] [PubMed] [Google Scholar]
- [31].Mao Y, Fu Z, Zhang Y, et al. A six-microRNA risk score model predicts prognosis in esophageal squamous cell carcinoma. J Cell Physiol 2019;234:6810–9. [DOI] [PubMed] [Google Scholar]
- [32].Zhu LF, Song LD, Xu Q, et al. Highly expressed long non-coding RNA FEZF1-AS1 promotes cells proliferation and metastasis through Notch signaling in prostate cancer. Eur Rev Med Pharmacol Sci 2019;23:5122–32. [DOI] [PubMed] [Google Scholar]
- [33].Li R, Zhang L, Qin Z, et al. High LINC00536 expression promotes tumor progression and poor prognosis in bladder cancer. Exp Cell Res 2019;378:32–40. [DOI] [PubMed] [Google Scholar]
- [34].Gong D, Feng PC, Ke XF, et al. Silencing Long Non-coding RNA LINC01224 inhibits hepatocellular carcinoma progression via MicroRNA-330-5p-induced inhibition of CHEK1. Mol Ther Nucleic Acids 2019;19:482–97. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [35].Qiu X, Dong J, Zhao Z, et al. LncRNA LINC00668 promotes the progression of breast cancer by inhibiting apoptosis and accelerating cell cycle. Onco Targets Ther 2019;12:5615–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


