Abstract
The COVID-19 pandemic is an ongoing global challenge for public health systems. Ultrasensitive and early identification of infection is critical in preventing widespread COVID-19 infection by presymptomatic and asymptomatic individuals, especially in the community and in-home settings. We demonstrate a multiplexed, portable, wireless electrochemical platform for ultra-rapid detection of COVID-19: the SARS-CoV-2 RapidPlex. It detects viral antigen nucleocapsid protein, IgM and IgG antibodies, as well as the inflammatory biomarker C-reactive protein, based on our mass-producible laser-engraved graphene electrodes. We demonstrate ultrasensitive, highly selective, and rapid electrochemical detection in the physiologically relevant ranges. We successfully evaluated the applicability of our SARS-CoV-2 RapidPlex platform with COVID-19-positive and COVID-19-negative blood and saliva samples. Based on this pilot study, our multiplexed immunosensor platform may allow for high-frequency at-home testing for COVID-19 telemedicine diagnosis and monitoring.
Keywords: COVID-19, biosensors, telemedicine, graphene, antibody, antigen, C-reactive protein, SARS-CoV-2, electrochemical
Graphical Abstract
Highlights
-
•
Multiplexed detection of SARS-CoV-2 antigen, antibodies, and C-reactive protein
-
•
Low-cost and mass manufacturable platform using laser-engraved graphene
-
•
Wireless telemedicine sensing platform with rapid sample-to-answer turnaround
-
•
Successful device evaluation in both COVID-19 patient blood and saliva samples
Progress and Potential
The need for widespread testing to control the spread of COVID-19 has faced challenges due to testing backlogs, limited access to required equipment, and inaccurate assay results. To address this, we propose an ultrasensitive and low-cost telemedicine platform, the SARS-CoV-2 RapidPlex, based on target-specific immunoassays built off laser-engraved graphene for rapid and remote assessment of COVID-19 biomarkers (i.e., nucleocapsid protein, anti-spike protein IgG and IgM, and C-reactive protein). Multiplex sensing of these targets provides information on three key aspects of COVID-19 disease: viral infection, immune response, and disease severity. We successfully demonstrated the platform's applicability using COVID-19-positive and COVID-19-negative serum and saliva samples. The SARS-CoV-2 RapidPlex has the potential to quickly and effectively triage patients and track infection progression, allowing for the clear identification of individuals who are infectious, vulnerable, and/or immune.
The SARS-CoV-2 RapidPlex is developed on the basis of laser-engraved graphene immunosensors to electrochemically quantify SARS-CoV-2 nucleocapsid protein, IgG and IgM, and C-reactive protein (CRP). The platform is characterized by rapid detection and high molecular sensitivity and selectivity. Pilot study results demonstrate that it can successfully detect these biomarkers in COVID-19-positive patient serum and saliva samples. CRP results are well correlated with symptom severity, indicating the potential for this integrated system to be used as a diagnostic tool for telemedicine COVID-19 patient care.
Introduction
On March 11, 2020, the World Health Organization characterized the COVID-19 outbreak as a pandemic. Six months later, the global health crisis had continued with over 30 million confirmed cases of novel coronavirus globally, with over 22% of these being in the United States.1 It is estimated that 14%–20% of patients will develop severe illness requiring hospitalization.2 Initial efforts to mitigate the spread through state-mandated “stay-at-home” orders appeared effective; however, reopening of the United States economy resulted in renewed exponential spread of novel coronavirus, as predicted.3 It is estimated that the United States gross domestic product will suffer losses upward of $45.3 billion during a flu-like pandemic without available vaccines.4 Safe reopening of the economy, schools, and universities requires multiple approaches to mitigate the risks associated with COVID-19, including simple, affordable, and effective test-and-trace measures.
Containing the spread is difficult due to the challenges in identifying infectious individuals. Most COVID-19 community spread may occur in the absence of symptoms. Peak viremia may be at the end of the incubation period, allowing for a transmission-sufficient viral load 1–2 days prior to symptom onset.3 Additionally, due to the unknown duration and prevalence of asymptomatic cases, the true reproduction number may be underestimated.5 , 6 Reported incidence of asymptomatic patients ranges from 17.9% to 30.8%.7 , 8
Increased access to COVID-19 testing has allowed increased monitoring of the community spread, but several diagnostic challenges remain. Currently, the standard testing method is viral nucleic acid real-time PCR (RT-PCR), which is a slow process9 and requires expensive equipment and trained technicians for nasopharyngeal swab sample collection and analysis.9 In addition, the sheer volume of testing required is overwhelming the ability for healthcare systems to report RT-PCR results to patients, causing, in some states, delays of ∼7–10 days to inform positive10 findings and enact isolation and monitoring protocols. Despite the recent advances on point-of-care (POC) rapid RT-PCR testing,11, 12, 13, 14, 15 nucleic acid tests are also known to produce false negatives, which may limit containment strategies and access to treatment.16 An additional consideration for RT-PCR is that it only identifies active carriers of the virus. Identifying convalescent persons based on COVID-19 antibody presentation is equally important, as it may provide health officials with crucial information regarding the potential impact of reopening measures.17 Serologic assays detect circulating antibodies specific to SARS-CoV-2 antigens, including the nucleocapsid protein and the outer spike protein.9 , 18 However, it is not possible to differentiate between asymptomatic carriers and immune persons using antibody detection. Therefore, to effectively mitigate the risks of COVID-19 community spread, systems are required that determine simultaneously both the viral and serologic status of an individual. Moreover, recent studies show a correlation between circulating inflammatory biomarker concentration and COVID-19 severity.19 Increased C-reactive protein (CRP) concentration is found in patients diagnosed with COVID-19 pneumonia and is associated with increasing severity, suggesting a role in the diagnosis and prognosis of COVID-19 patients.20 , 21
There is a clear and urgent need for a highly sensitive, rapid, inexpensive, telemedicine COVID-19 test that can identify a patient's past and present infection status.22 There has been progress toward POC COVID-19 testing, but all commercially available test kits provide only qualitative results. Quantitative analysis of COVID-19 biomarkers using a telemedicine device may provide predictive information of disease severity and provide seroconversion information regarding time course of the disease. Electrochemical biosensors, in this regard, are advantageous due to their rapid detection efficacy and ease of use for POC applications.23, 24, 25, 26, 27 Simple, safe, and effective COVID-19 sample collection has proved challenging given current assay requirements. Saliva-compatible POC assays would be advantageous, since saliva contains rich information and can be easily and non-invasively collected by patients themselves for telemedicine testing.28
Here, we present a novel multiplexed, portable, wireless electrochemical platform for ultra-rapid detection of COVID-19: SARS-CoV-2 RapidPlex (Figure 1 ). This platform quantitatively detects biomarkers specific to COVID-19 in both blood and saliva including SARS-CoV-2 nucleocapsid protein (NP), specific immunoglobulins (Igs) against SARS-CoV-2 spike protein (S1) (S1-IgM and S1-IgG), and CRP, within physiologically relevant ranges. The platform uses capture antigens and antibodies immobilized on mass-producible, low-cost, laser-engraved graphene (LEG)29 , 30 electrodes. This multiplexed platform tracks the infection progression by diagnosing the stage of the disease, allowing for the clear identification of individuals who are infectious, vulnerable, and/or immune (Table 1 ). The main features of SARS-CoV-2 RapidPlex are high sensitivity, low cost, ultra-fast detection, wireless remote, and multiplexed sensing that provides information on three key aspects of COVID-19 disease: viral infection (NP),31 immune response (IgG and IgM),9 and disease severity (CRP).19, 20, 21
Figure 1.
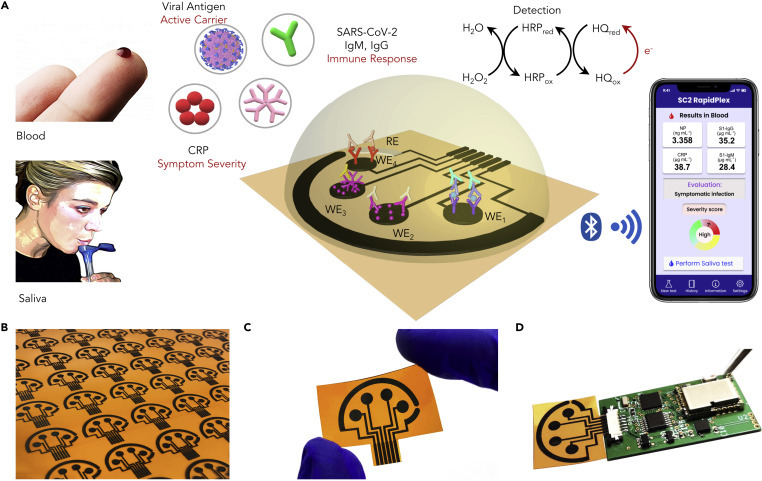
A Wireless Graphene-Based Telemedicine Platform (SARS-CoV-2 RapidPlex) for Rapid and Multiplex Electrochemical Detection of SARS-CoV-2 in Blood and Saliva
(A) Schematic illustration of the SARS-CoV-2 RapidPlex multisensor telemedicine platform for detection of SARS-CoV-2 viral proteins, antibodies (IgG and IgM), and inflammatory biomarker C-reactive protein (CRP). Data can be wirelessly transmitted to a mobile user interface. WE, working electrode; CE, counter electrode; RE, reference electrode.
(B) Mass-producible laser-engraved graphene sensor arrays.
(C) Photograph of a disposable and flexible graphene array.
(D) Image of a SARS-CoV-2 RapidPlex system with a graphene sensor array connected to a printed circuit board for signal processing and wireless communication.
Table 1.
Key Information on an Individual's COVID-19 Infection Status Provided by the SARS-CoV-2 RapidPlex
| Viral Antigen | IgM | IgG | CRP | Expected Outcome |
|---|---|---|---|---|
| – | – | – | – | healthy |
| + | + || − | – | – | infectious, presymptomatic |
| + | + || + | − | infectious, asymptomatic | |
| + | + || + | + | infectious, symptomatic | |
| – | + | + || − | + || − | recovered (recently) |
| – | – | + | – | recovered (long term) |
| – | – | – | + | inflammation/infection not due to COVID-19 |
+, higher than threshold; −, lower than threshold; ||, or.
Results and Discussion
Design of the SARS-CoV-2 RapidPlex Platform
As illustrated in Figure 1A, SARS-CoV-2 RapidPlex is composed of four graphene working electrodes (WEs), a Ag/AgCl reference electrode (RE), and a graphene counter electrode (CE), all of them patterned on a polyimide (PI) substrate via CO2 laser engraving, a fast, high-throughput, and cost-effective production method (Figures 1B and 1C). Our group has recently demonstrated the use of mesoporous graphene structure fabricated by laser engraving for high performance and low-cost biosensing.29 , 30 The materials cost for the unmodified RapidPlex platform is within $0.05; additional chemical and reagent costs for the multiplexed sensor preparation are at the level of dollars depending on the order sizes. Detection of selected target proteins (NP and CRP) and specific immunoglobulins (S1-IgG and S1-IgM) is achieved through sandwich- and indirect-based immunosensing strategies onto the LEG electrodes, respectively. The superior properties of graphene, in terms of high charge mobility and surface area together with the high sensitivity and selectivity of sensing strategies involving both capture and detector receptors, make our device (Figure 1D) a highly convenient tool for the rapid, accurate, and stage-specific detection of COVID-19 infection in blood as well as in non-invasive biofluid samples, such as saliva.
Electrochemical Characterization of SARS-CoV-2 RapidPlex Platform
Functionalization and modification steps carried out on the LEG surfaces for the covalent attachment of each of the specific receptors required for the development of our SARS-CoV-2 RapidPlex platform is schematized in Figure 2 A. 1-Pyrenebutyric acid (PBA) is used as the linker to anchor the required receptors to the graphene layer. Although attachment of functional groups directly on the sp2 carbon atom surface is one of the common ways to functionalize graphene, these methods are associated with the requirement of defects or edges in the sensor material, which could alter its specific physical properties.32 , 33 In contrast, introduction of functional groups on the sensing layer by means of pyrene derivatives is preferred here, as it does not disrupt the conjugation of the graphene sheets and improves its stability.34 , 35 PBA consisting of a pyrene group that contains π-electrons and a carboxylic group is used to functionalize graphene layers via π-stacking and hydrophobic interactions. The pyrene units of PBA strongly interact with graphene layers in the way that original structure and properties of the graphene are well maintained. The functional moieties contained in each PBA molecule allow the preparation of the affinity-based biosensing platform through the covalent coupling between the carboxylic groups on PBA units and the –NH2 groups of the respective capture receptors (specific antibodies or capture proteins). Blocking of unreacted sites with bovine serum albumin (BSA) impedes the non-specific adsorption of other molecules involved in each assay configuration or circulating in the sample of interest.
Figure 2.

Characterization of Electrochemical Graphene Biosensors Comprising the SARS-CoV-2 RapidPlex Platform
(A) Scheme detailing the methodology developed for the covalent attachment of the corresponding bioreceptor for the specific capture of the target analytes SARS-CoV-2 NP and CRP (left), and IgG and IgM isotypes against SARS-CoV-2 S1 protein (right). PBA, 1-pyrenebutyric acid; BSA, bovine serum albumin; CAb, capture antibody; PI, polyimide.
(B and C) Differential pulse voltammetry (B) and Nyquist plots (C) of a graphene electrode in 0.01 M PBS (pH 7.4) containing 2.0 mM K4Fe(CN)6/K3Fe(CN)6 (1:1) after each modification step (S1-IgG assay as representative example): bare graphene (Bare), functionalization with PBA (PBA), immobilization of SARS-CoV-2 S1 protein (Protein), blocking with BSA (BSA), recognition of specific S1-IgG (Target), and incubation with enzyme-tagged anti-human IgG antibody (DAb).
(D) Comparison of amperometric responses and overlaid signal-to-blank (S/B) ratio (black lines) for SARS-CoV-2-specific IgG and CRP detection using PBA and 1H-pyrrole-1-propionic acid (PPA) as linkers for the attachment of the corresponding capture bioreceptors. Data are presented as mean ± SD (n = 3).
(E) Amperometric responses and overlaid S/B ratio (black lines) observed for 0.0 and 500 pg mL−1 NP, 0.0 and 250 ng mL−1 SARS-CoV-2-specific IgG and IgM, and 0.0 and 50 ng mL−1 CRP, with 1-, 5-, and 10-min incubation. Data are presented as mean ± SD (n = 3).
Differential pulse voltammetry (DPV) and open-circuit potential-electrochemical impedance spectroscopy (OCP-EIS) techniques are employed to electrochemically characterize and prove the stepwise self-assembled processes in both assay configurations for the detection of selected target molecules. DPV readings reflect lower peak current intensity after each modification step related to S1-Ig assay due to the hindered diffusion of the redox label to the electrode surface derived from both the carboxyl groups and the attached proteins and biological macromolecules (Figure 2B). At the same time, resistance in the Nyquist plots from OCP-EIS is increased after each functionalization step (Figure 2C). The successful anchorage of PBA was also verified with scanning electron microscopy (SEM) (Figure S1). Electrochemical characterization of the sandwich assay-based sensor modification using CRP as a model molecule and the aforementioned techniques are presented in Figure S2.
To preserve the native structure and properties of the bound biomolecules, we chose PBA as a heterobifunctional linker, effectively preventing the direct interaction between large biomolecules and the graphene surface.33 To verify this selection, we constructed CRP- and SARS-CoV-2-specific IgG assay configurations on graphene electrodes functionalized with PBA and another common linker, 1H-pyrrole-1-propionic acid (PPA).30 Greater signal-to-blank (S/B) ratios were observed for both assays where PBA was used as a linker support (Figure 2D), mainly due to a significant decrease in the signals obtained in the absence of the corresponding target molecule when PBA was used instead of PPA. Together with an optimal blocking strategy, PBA can be used for the immobilization of specific biomolecular probes (e.g., antibodies, proteins) while avoiding non-specific adsorptions in the context of immunoassays.36
The orientation of modified antigenic proteins on solid surfaces is strongly associated with their activity and reactivity. Specific anti-His antibodies can be used to orient the immobilization of antigenic receptors containing histidine residues, but this implies an additional step compared with their direct attachment on the sensing layer, as schematized in Figure S3A. Our results show no significant differences in assay performance for IgG detection on PBA-graphene electrodes covalently functionalized with the specific coating protein (direct immobilization) and with anti-His antibodies for the previous capture of the polyhistidine-tag specific coating protein (oriented immobilization) (Figure S3B), proving that random protein orientation does not interfere with the epitope accessibility for effective recognition by specific target antibodies. This is in agreement with other reports confirming that His-tagged fusion antigens can be directly immobilized on different surfaces with protein orientations completely compatible with antibody recognition.37, 38, 39, 40 To simplify and reduce the cost and time of the assay, we carried out direct immobilization of S1 protein for specific Ig detection.
Considering that rapid target binding is essential to the successful implementation of our proposed platform as a POC system, we investigated how target (or sample) incubation time affects the response of each biosensor comprising our SARS-CoV-2 RapidPlex platform. Figure 2E summarizes the amperometric signals obtained for each of the four sensing units at different incubation times (1, 5 and 10 min) in the absence (blank, B) and presence (S) of 500 pg mL−1, 250 ng mL−1, and 50 ng mL−1 of NP, SARS-CoV-2-specific IgG and IgM isotypes, and CRP, respectively. It is important to note that although a 10-min incubation time was selected for most of the studies here in order to ensure the highest sensitivity for the determination of ultra-low levels of each target molecule, a significant difference between the absence and the presence of each of the corresponding targets is obtained with just 1-min incubation time. This provides one of the major advantages of our SARS-CoV-2 RapidPlex system as a rapid POC device for SARS-CoV-2 infection monitoring with the required sensitivity for both protein and Ig determination. ELISA,17 , 41, 42, 43, 44 nucleic acid amplification,45, 46, 47, 48, 49 mass spectrometry,50 or even combinations51 have been reported very recently for determination of the proposed SARS-CoV-2-specific target molecules, among others. However, most of these methods show crucial pitfalls, mainly in terms of sample preparation, complexity, and expensive and bulky equipment requirements, making them still highly difficult to be implemented as POC systems. Our device provides an attractive alternative to standard assays for protein determination, such as ELISA, because of its multiplexing capabilities, remote functionality, and short sample-to-answer time.
Evaluation of Analytical Performance of the SARS-CoV-2 RapidPlex
The performance of each biosensor contained in the SARS-CoV-2 RapidPlex was characterized in phosphate-buffered saline (PBS) solutions supplemented with 1.0% BSA by measuring the amperometric readout in the presence of increased concentrations of NP, S1-IgG, S1-IgM, and CRP (Figure 3 ). The selected strategies for NP viral antigen and CRP proteins are based on double-sandwich and sandwich configurations, respectively, as illustrated in Figure 3A. The sandwich-based immunoassays for antigen detection are, in general, highly sensitive due to the involvement of two different antibodies as capture and detector entities. According to the low levels that must be reached for NP and CRP in diluted serum and saliva (pg mL−1 to ng mL−1), we think these strategies are the most suitable to be implemented on our platform. Variation of cathodic currents with the concentration for NP and CRP in buffered solutions is presented in Figures 3B and 3C, respectively. S1-IgG and S1-IgM were detected based on indirect immunoassays (Figure 3D), which are considered highly suitable for detection of circulating macromolecules in antisera and other biofluids. Figures 3E and 3F show the calibration curves for S1-specific Ig determination (S1-IgG and S1-IgM, respectively) in buffered solutions. Reproducibility was demonstrated through the relative standard deviation (RSD) values obtained with different biosensors prepared in the same manner on different days. RSD values of 6.3%, 8.4%, 6.0%, and 7.6% for 20 ng mL−1 CRP, 250 ng mL−1 S1-IgG, 250 ng mL−1 S1-IgM, and 500 pg mL−1 NP antigen (n = 5) demonstrate good reproducibility in both device preparation and signal transduction. In addition, the sensors showed stable responses over a 5-day storage period at 4°C (Figure S4). We did not observe significant slope variations between data obtained in properly diluted human serum and in buffered solutions for the determination of each target analyte (for instance, the slope sensitivity value [16.28 nA mL ng−1] obtained for CRP as model analyte in PBS-buffered solutions is nearly the same as that in diluted serum samples from a healthy volunteer [16.64 nA mL ng−1]); therefore, accurate quantification of the proposed target analytes can be carried out by conducting a simple interpolation of the cathodic readings obtained for each sample tested in the corresponding calibration curve constructed in buffered solution.
Figure 3.

Evaluation of Analytical Sensor Performance for the Detection of Physiological Levels of Target COVID-19 Biomarkers
(A) Scheme of sensor preparation for detection of SARS-CoV-2 NP and CRP based on double-sandwich and sandwich assay configurations, respectively. CAb, capture antibody; DAb, detector antibody; DAb2, secondary detector antibody; HRP, horseradish peroxidase.
(B and C) Calibration curves constructed for NP (B) and CRP (C) detection in PBS (pH 7.4) supplemented with 1.0% BSA. Data are presented as mean ± SD (n = 3).
(D) Scheme of sensor preparation for detection of S1-IgG and S1-IgM isotypes based on direct assay configurations.
(E and F) Calibration curves constructed for S1-IgG (E) and S1-IgM (F) isotype detection in PBS (pH 7.4) supplemented with 1.0% BSA. Data are presented as mean ± SD (n = 3).
Since diagnostic sensitivity and specificity of seroprevalence studies can be improved by using a mixture of antigenic proteins instead of a single protein,52 , 53 we modified graphene with a mixture of SARS-CoV-2 related antigens, NP and S1, to capture specific immunoglobulin isotypes against both antigens in the same WE. A calibration curve for (NP + S1)-IgG detection is shown in Figure S5. Thus, this methodology can be tailored for detecting isotype-specific IgG (or IgM) or a combination of both Ig isotypes in the same sensing surface to better capture total Ig concentration and thus increase assay sensitivity across the patient population.
Investigation of the Selectivity and Multiplexed Performance of the SARS-CoV-2 RapidPlex
Human biofluids contain a complex and variable mixture of circulating molecules that could interfere with molecular sensing. In addition, negligible crosstalk between different working surfaces is an essential requirement for performing multiplexed detection readings accurately and meaningfully. Therefore, selectivity and crosstalk of the SARS-CoV-2 RapidPlex platform were evaluated. Amperometric readings obtained for each developed biosensor against non-target molecules are presented in Figure 4 A. We evaluated the specific binding for SARS-CoV-2 biomarkers in comparison with biomarkers of similar coronaviruses, including SARS-CoV and MERS-CoV. We observed no significant cross-reaction for NP, S1-IgG, S1-IgM, and CRP assays in the presence of each tested interferent, including SARS-CoV-2 S1, SARS-CoV S1, and CRP (for NP assay), SARS-CoV-2 NP-IgG, SARS-CoV IgG, MERS-CoV IgG, S1-IgG, and negative controls containing mixtures of IgG and IgM against both MERS-CoV and SARS-CoV (for S1-IgG and S1-IgM assays), and B-type natriuretic peptide (BNP), NP, SARS-CoV NP, and SARS-CoV S1 (for CRP assay), respectively. However, SARS-CoV NP viral antigen interferent provided a cathodic current corresponding to ∼80% of the raw current obtained for the detection of the specific NP antigen. Spike, envelope, and membrane SARS-CoV-2 proteins share 76%–95% sequence identity with those of SARS-CoV. This percentage homology is reduced to 30%–40% for MERS-CoV. Similarly, since SARS-CoV-2 NP is 90% identical to SARS-CoV NP,17 , 54, 55, 56 the interference observed from SARS-CoV NP antigen was expected. However, the lack of selectivity in this particular case is not a major concern due to the absence of new SARS-CoV cases detected recently; therefore, it can be inferred that this interference will not produce a barrier for selective SARS-CoV-2 NP determination in real samples. We further evaluated the amperometric-derived concentrations with absorbance-derived concentrations collected via ELISA. As presented in Figure 4B, the results from our functionalized electrochemical biosensor were linearly correlated (r = 0.955) with the results using the same reagents in a traditional ELISA protocol.
Figure 4.

Investigation of the Selectivity and Multiplexed Performance of the Wireless SARS-CoV-2 RapidPlex Platform
(A) Selective response of NP, S1-IgG and S1-IgM isotypes, and CRP sensors against different non-target circulating analytes. Interferential molecules were tested at 500 pg mL−1 (with an exception of 50 ng mL−1 for CRP), 250 ng mL−1, and 50 ng mL−1 for NP, S1-IgG and S1-IgM, and CRP assays, respectively. Data are presented as mean ± SD (n = 3).
(B) Validation of sample concentrations measured using the designed electrochemical sensor against sample concentrations measured using ELISA.
(C) Block diagram of the SARS-CoV-2 RapidPlex platform. UART, universal asynchronous receiver/transmitter; MCU, microcontroller unit; DAC, digital-to-analog converter; ADC, analog-to-digital converter.
(D) Schematic illustration of the graphene sensor array layout.
(E) Experimental readings obtained with the functionalized SARS-CoV-2 RapidPlex platform after incubation of the four WEs with PBS (pH 7.4) supplemented with 1.0% BSA containing 1.0 ng mL−1 NP (I), 250 ng mL−1 S1-IgG (II), 250 ng mL−1 S1-IgM (III), and 50 ng mL−1 CRP (IV).
Once the performance and selectivity of each constructed biosensor was individually and exhaustively evaluated, we demonstrate the multiplexing capabilities of our four-working-electrode (4WEs) graphene array device designed with a Ag/AgCl RE and a graphene CE. The block diagram showing the functional units that constitute the integrated electronic system is illustrated in Figures 4C and 4D. Amperometric readings from the four channels are concurrently taken and data are wirelessly transmitted to a user device over Bluetooth. The electronic system, including the printed circuit board (PCB) and a lithium-ion polymer battery, is 20 × 35 × 7.3 mm in dimension. The compact device can perform amperometric measurements continuously for over 5 h on a single charge.
With the objective of demonstrating the utility of our SARS-CoV-2 RapidPlex array for multiplexed and simultaneous quantification of selected target molecules, we evaluated the potential cross-reaction resulting from the diffusion of signal substances between adjacent immunosurfaces. To this end, each of the four conveniently functionalized working surfaces were incubated with buffered solutions containing significantly high concentration of each of the selected targets, followed by the corresponding detector receptors in each case. The absence of crosstalk between the adjacent WEs is verified from the experimental readings in buffered solutions containing 1.0 ng mL−1 NP antigen (I), 250 ng mL−1 S1-specific IgG (II) and IgM (III), and 50 ng mL−1 CRP (IV) (Figure 4E). As envisaged, significantly higher signal was obtained when each target was specifically captured and further labeled by its tracer antibody in the corresponding functionalized immunosurface. These results, in conjunction with those from Figure 4A, demonstrate the feasibility of the developed SARS-CoV-2 RapidPlex platform for fast, selective, and reliable determination of NP, S1-IgG, and S1-IgM isotypes, and CRP in one single experiment. It should be noted that since IgG and IgM have similar binding mechanisms to viral antigens and individual quantification of immunoglobulins requires no mixing of the specific detector labels, individual droplets were used on IgG and IgM sensing electrodes during modification and labeling.
Detection of SARS-CoV-2-Related Selected Targets in Human Biospecimens
To prove the utility of our device in a more complex and real scenario, we evaluated the multiplexed capabilities of SARS-CoV-2 RapidPlex in representative serum samples from COVID-19 RT-PCR-negative and -positive subjects. Sensor data from the serum samples of an RT-PCR-negative subject (Figure 5 A) and an RT-PCR-positive patient (Figure 5B) show minimal crosstalk in a real and complex sample matrix, indicating the efficient functionality of SARS-CoV-2 RapidPlex to simultaneously differentiate the overexpressed presence of SARS-CoV-2-related target reporters in COVID-19-positive specimens. Moreover, the SARS-CoV-2 RapidPlex device is able to provide significant positive readings for all targets after incubating the COVID-19-positive serum sample for just 1 min (Figures 5C and S6): The maintained high signal in positive patient samples demonstrates the great potential in future translation of the SARS-CoV-2 RapidPlex device as an ultra-fast POC remote diagnostic tool.
Figure 5.
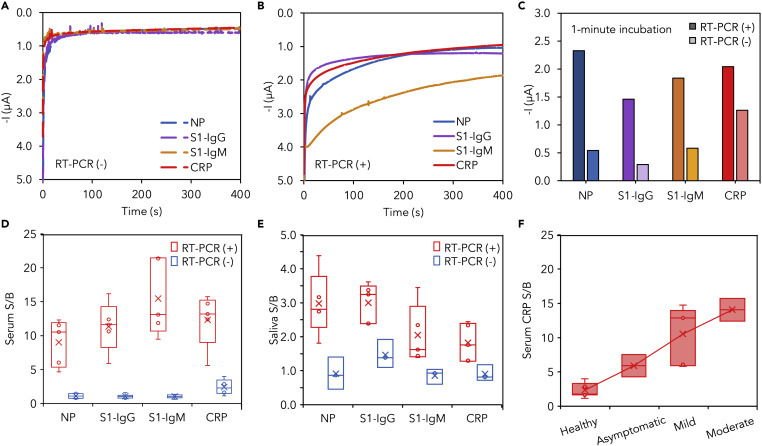
Application of SARS-CoV-2 RapidPlex in SARS-CoV-2 Detection in Blood and Saliva Samples from COVID-19-Positive and -Negative Subjects
(A and B) Experimental readings obtained with SARS-CoV-2 RapidPlex after 10-min incubation of the sensor array with serum samples from a representative COVID-19 RT-PCR-negative (A) and -positive (B) patient.
(C) Signal of individual sensor obtained after 1-min incubation with a serum sample from a COVID-19-positive patient (dark color) versus the signal obtained after 10-min incubation with a serum sample from a COVID-19-negative patient (light color).
(D) Box-and-whisker plot of measured signal-to-blank ratios (S/B) for NP, S1-IgG, S1-IgM, and CRP in RT-PCR-confirmed COVID-19-positive (n = 5) and -negative (n = 6) serum samples.
(E) Box-and-whisker plot of measured S/B for NP, S1-IgG, S1-IgM, and CRP in RT-PCR-confirmed COVID-19-positive (n = 5) and -negative (n = 3) saliva samples.
(F) CRP levels in diluted serum samples plotted against given COVID-19 symptom severity, with “Healthy” referring to COVID-19-negative patient samples (n = 7). Positive COVID-19 patients were classified according to disease severity as asymptomatic (n = 2), mild (n = 5), and moderate (n = 2).
To further investigate NP, S1-IgG, S1-IgM, and CRP response to SARS-CoV-2 infection using our LEG-based biosensors, we measured each target molecule in serum and saliva samples from RT-PCR-confirmed COVID-19-positive and -negative subjects. Obtained results were plotted as the ratio between the amperometric readings for each sample tested (S) and the respective blank (B) in each case to compare target detection in different concentration ranges. Using the graphene sensors, a total of 17 COVID-19 RT-PCR-tested serum samples (10 positive, 7 negative) were assayed, and a total of 8 COVID-19 RT-PCR-tested saliva samples (5 positive, 3 negative) were analyzed (Table S1).
Results from Figures 5D and 5E corroborate that, as expected, compared with RT-PCR-negative subjects, RT-PCR-positive COVID-19 patients show significantly elevated levels of the selected targets in both serum and saliva samples, with median S/B ratios of 10.53, 11.62, 10.67, and 12.39 in serum and 2.81, 3.24, 1.62, and 1.76 in saliva, for NP, S1-IgG, S1-IgM, and CRP, respectively. We observed a concentration of NP in the range of 0.1–0.8 μg mL−1 and 0.5–2.0 ng mL−1 in COVID-19 patient serum and saliva, respectively; S1-IgG in the range of 20–40 μg mL−1 and 0.2–0.5 μg mL−1 in COVID-19 patient serum and saliva, respectively; S1-IgM in the range of 20–50 μg mL−1 and 0.6–5.0 μg mL−1 in COVID-19 patient serum and saliva, respectively; and CRP in the range of 10–20 μg mL−1 and 0.1–0.5 μg mL−1 in COVID-19 patient serum and saliva, respectively. The fact that all the positive samples show much higher signals compared with negative samples proves the real utility for the accurate evaluation of the COVID-19 biomarkers in biofluids using our LEG-based biosensors. In particular, the observed significant presence of COVID-19 biomarkers in saliva demonstrates the exceptional utility of this biofluid as a valuable source for non-invasively diagnosing and monitoring SARS-CoV-2 infection.
With the aim of confirming the relationship between the levels of inflammatory biomarkers involved in the cytokine storm directly associated with disease progression, severity, and outcome in COVID-19,57, 58, 59, 60, 61, 62 we evaluated the variation of serum CRP levels in RT-PCR-negative subjects (n = 7) and RT-PCR-positive COVID-19 patients who were classified clinically according to disease severity as asymptomatic (n = 2), mild (n = 5), and moderate (n = 2). As shown in Figure 5F, we observed a positive association between CRP concentration and COVID-19 symptom severity grade, consistent with the recent literature reports.20 , 61 Future clinical testing using paired saliva and serum samples over the course of the infection is required to determine the relationship between saliva and serum concentrations and to validate the utility of our platform in identifying severity-specific COVID-19 (Table 1).
Conclusions
To address the increasing demands for effective diagnostic tools for simple COVID-19 detection with immediate sample-to-answer turnaround, we have developed and implemented the first multiplexed electrochemical graphene-based platform, SARS-CoV-2 RapidPlex, for sensitive, rapid, and selective simultaneous interrogation of NP viral antigen, S1-IgG and S1-IgM isotypes, and CRP in serum and saliva biofluids from healthy and RT-PCR-confirmed COVID-19-infected patients. The combination of the advantageous properties of graphene material with the high sensitivity and specificity of the immunosensing strategies makes our SARS-CoV-2 RapidPlex platform a promising diagnostic device for the accurate monitoring of COVID-19 infection in serum and non-invasively accessible body fluids free from complex sample pretreatment requirements. Due to the ease of use, saliva sample compatibility, and rapid time to results, the SARS-CoV-2 RapidPlex platform has high potential for implementation at POC for patient triage, as well as for at-home use for telemedicine care and remote monitoring.
Monitoring of selected targets in one single and fast experiment (target capture can be as low as 1 min) provides substantial information, not only regarding early COVID-19 infection through viral antigen and IgM isotype detection but also about disease severity by means of CRP evaluation and potential acquired immunity through IgG isotype quantification. The rapid on-site evaluation of disease severity enabled by our SARS-CoV-2 RapidPlex introduces the unparalleled advantage of immediate COVID-19 triaging. In future clinical applications, this could not only alert attending physicians of cases requiring major and careful medical attention but also facilitate the efficient allocation of precious medical resources, such as ventilators and ICU beds, in the event of resurging outbreaks to optimize patient outcomes under an overloading of local healthcare systems.
Our proposed methodologies based on simple yet well-established surface functionalization techniques and sensing principles allow the ease of translation to the detection of other highly informative SARS-CoV-2-related reporters by simply changing the coating capture receptor. Further technological improvement could be achieved by introducing a fully automated sample-handling process through a microfluidic module for telemedicine deployment. Modification of our platform design may allow for rapid viral antigen and antibody panel testing such that COVID-19 infection could be clearly distinguished from non-specific symptoms of seasonal respiratory infections such as influenza. Additionally, the wireless telemedicine diagnostic platform, when coupled with emerging wearable biosensors to continuously monitor vital signs and other chemical biomarkers, could provide comprehensive information on an individual's health status during the COVID-19 pandemic.63, 64, 65, 66, 67
Our platform pioneers multiplexed detection of stage-specific SARS-CoV-2-related biomarkers to provide a detailed and personalized snapshot of the COVID-19 infection. We firmly believe that our developed platform will be a high-utility testing method toward fighting this and future pandemics, helping to end one of the deepest global health, economic, and humanitarian crises in modern history.
Experimental Procedures
Resource Availability
Lead Contact
Further information and requests for materials should be directed to and will be fulfilled by the Lead Contact, Wei Gao, weigao@caltech.edu.
Materials Availability
The materials generated in this study are available from the corresponding author upon request.
Data and Code Availability
The data used to support the findings of this study are available from the corresponding author upon request.
Materials and Reagents
PBA (97%), PPA (97%), 1-ethyl-3-(3-dimethylamonipropyl)carbodiimide (EDC), N-hydroxysulfosuccinimide (sulfo-NHS), BSA, hydroquinone (HQ), 2-(N-morpholino)ethanesulfonic acid hydrate (MES), Tween 20, sodium thiosulfate, sodium bisulfite, and potassium ferrocyanide(II) were purchased from Sigma-Aldrich. Streptavidin-POD conjugate (streptavidin-HRP, 11089153001) was purchased from Roche. Sodium dihydrogen phosphate, potassium hydrogen phosphate, potassium chloride, hydrogen peroxide (30% [w/v]), sulfuric acid, and flat-bottom NUNC 96-well microplates were purchased from Fisher Scientific. Potassium ferricyanide(III) and silver nitrate, iron(III) chloride, and 0.1 M PBS (pH 7.4) were purchased from Acros Organics and Alfa Aesar, respectively. N,N-Dimethylformamide (DMF) and isopropyl alcohol (IPA) were purchased from Fisher Chemical and VWR Chemicals, respectively. Anti-CRP murine monoclonal antibody and human CRP standard were purchased from R&D Systems (DY1707). CRP polyclonal antibody labeled with horseradish peroxidase (HRP) (PA1-28329) and 3,3′,5,5′-tetramethylbenzidine (TMB) colorimetric substrate was purchased from Invitrogen. Mouse NP monoclonal antibody (mAb) (40143-MM05), SARS-CoV-2 NP antigen (40588-V08B), SARS-CoV/SARS-CoV-2 nucleocapsid antibody, rabbit mAb (40143-R001), SARS-CoV NP antigen (HCoV-OC43; 40643-V07E), SARS-CoV-2 Spike S1-His recombinant protein (HPLC-verified) (40591-V08H), and SARS-CoV Spike S1 protein (S1 subunit, His tag) (40150-V08B1) were purchased from Sino Biological. His-tagged SARS-CoV-2 Spike S1 protein (PNA002), His-tagged SARS-CoV-2 NP (PNA006), anti-Spike-RBD fully human mAb (IgG) (S1-IgG, AHA013), anti-NP mAb (IgG) (SARS-CoV-2 NP-IgG, AHA009), SARS-CoV antibody-80R (IgG) (CHA001), and MERS-CoV antibody-2E6 (IgG) (CHA002) were purchased from Sanyou Bio. Rabbit anti-human IgG H&L (HRP) (ab6759), recombinant human BNP (ab87200), rabbit anti-human IgM mu chain (HRP) (ab97210), goat anti-rabbit IgG H&L (HRP) (ab97051), and rabbit polyclonal anti-6× His-tag antibody (ab9108) were purchased from Abcam. Human Spike-SARS-CoV-2 IgM (S1-IgM, MBS2614311) were purchased from MyBiosource. MERS-CoV/SARS-CoV negative control (CI 2601-0101 Z) was purchased from Euroimmun. PI film (125 μm thick) was purchased from DuPont.
Fabrication of Multiplex Array Electrode
For four-channel graphene sensor fabrication, a PI film was attached onto a supporting substrate in a 50-W CO2 laser cutter (Universal Laser System VLS3.50). Selected laser-cutting parameters were: power 8.0%, speed 15%, points per inch 1,000, in raster mode and at focused height. Ag/AgCl REs were fabricated by electrodeposition in 40 μL of a mixture solution containing silver nitrate, sodium thiosulfate, and sodium bisulfite (final concentrations 250 mM, 750 mM, and 500 mM, respectively) for 100 s at −0.2 mA, followed by drop-casting a 20-μL aliquot of FeCl3 for 1 min.
Functionalization of the SARS-CoV-2 RapidPlex and Electrochemical Readout
A 10-μL aliquot of 5.0 mM PBA in DMF was drop-cast on the graphene surface and incubated for 2 h at room temperature in a humid chamber. After rinsing with DMF, IPA, and deionized (DI) water, and drying under air flow, electrodes were incubated with 10 μL of a mixture solution containing 0.4 M EDC and 0.1 M sulfo-NHS in 0.025 M MES (pH 6.5) for 35 min at room temperature under humid ambient conditions. Specific antibodies or coating protein were covalently attached onto activated surface by drop-casting 5.0 μL of the specific reagent (250 μg mL−1 for S1-IgG, S1-IgM, and CRP, or 50× dilution for NP, in 0.01 M PBS [pH 7.4]) and incubated for 3 h at room temperature, followed by a 90-min blocking step with 2.0% BSA prepared in 0.01 M PBS. Subsequently, 10 μL of the corresponding target analyte prepared in 0.01 M PBS containing 1.0% BSA was incubated for 1, 5, or 10 min at room temperature and, after one washing step with PBS, corresponding detector antibody (HRP-labeled or unlabeled) (250× dilution for NP, 2.0 μg mL−1 for S1-IgG and S1-IgM, and 1.0 μg mL−1 for CRP) in 0.01 M PBS containing 1.0% BSA was incubated for 5 min at room temperature. In the case of NP assay, after incubating detector antibody and performing the corresponding washing step with PBS, 10 μL of 1.0 μg mL−1 HRP-goat anti-rabbit IgG prepared in 0.01 M PBS containing 1.0% BSA was incubated for 5 min at room temperature. For each type of developed assay, amperometric readings were registered at −0.2 V (versus Ag/AgCl) in 0.05 M sodium phosphate buffer (pH 6.0) containing 2.0 mM HQ. The readout signal was obtained in the presence of 1.0 mM H2O2.
To characterize the morphology and material properties before and after surface modification with PBA, we obtained SEM images of graphene electrodes by focused ion beam SEM (FEI Nova 600 NanoLab).
Electrochemical Characterization of the SARS-CoV-2 RapidPlex
Amperometry, OCP-EIS, cyclic voltammetry, and DPV were carried out on a CHI820 electrochemical station. The electrochemical setup comprised laser-induced graphene electrodes as the WEs, a platinum wire as the CE, and a commercial Ag/AgCl electrode as the RE.
For each type of proposed assay, surface modification after each step was electrochemically characterized by DPV and OCP-EIS. Corresponding readings by means of each technique were carried out in 0.01 M PBS (pH 7.4) containing 2.0 mM K4Fe(CN)6/K3Fe(CN)6 (1:1) and under the following detailed conditions: potential range, −0.2 and 0.6 V; pulse width, 0.2 s; incremental potential, 4 mV; amplitude, 50 mV; frequency range, 0.1–106 Hz; amplitude, 5 mV. Graphene functionalization methods were evaluated for both CRP- and SARS-CoV-2-specific IgG assays, by comparing current responses obtained after developing each assay on both PBA and PPA-graphene, in the absence and in the presence of each of the corresponding target biomolecules (tested levels were 50 ng mL−1 for CRP and 500 ng mL−1 for SARS-CoV-2-specific IgG). Selectivity study was carried out by incubating corresponding interferential non-target molecules on the previously functionalized PBA-graphene. Concentration levels assayed for each interferent were the same as (or even higher than) the concentration of the target molecule in each case. Amperometric signals obtained for each interferent tested were compared with the current signals obtained in the absence and in the presence of the corresponding target analyte for each type of assay.
Design and Fabrication of Electronic System for the SARS-CoV-2 RapidPlex
The four-channel chronoamperometric measurements were performed by a custom PCB-based wireless potentiostat. An Arm Cortex-M4 microcontroller (STM32L432KC; STMicroelectronics), and a Bluetooth module (SPBT3.0DP2; STMicroelectronics) were used for potentiostat control and wireless communication. A single operational amplifier (AD8605; Analog Devices) was used as the control amplifier, and a quad operational amplifier (AD8608; Analog Devices) was used as a four-transimpedance amplifier to construct the potentiostat loop. A series voltage reference (ISL60002; Renesas Electronics) and the microcontroller unit’s (MCU) built-in digital-to-analog converter were used to generate the voltage bias across the RE and WEs. Four MCU built-in analog-to-digital converter channels were used to concurrently acquire the measurements.
Subjects and Procedures
In compliance with the protocols approved by the Institutional Review Board (no. 19-089417-292-A2) at the California Institute of Technology (Caltech), the performance of SARS-CoV-2 RapidPlex was evaluated in human serum and saliva samples from healthy and confirmed COVID-19-infected patients. Serum samples from ten RT-PCR and IgG/IgM serology-confirmed COVID-19 patients (age range 24–77 years) and seven healthy subjects (age range 18–65 years) were purchased from BioIVT and Ray Biotech. The severity information of the BioIVT samples was provided by the phlebotomists during sample collection. Saliva samples from five RT-PCR and IgG/IgM serology test-confirmed COVID-19 patients (age range 28–46 years) were purchased from BioIVT. Three healthy saliva samples were used from pre-existing stocks collected from volunteers prior to the pandemic (recommended tips before saliva collection include avoiding foods with high sugar and caffeine content, not eating a major meal within 60 min of sample collection, and rinsing the mouth with water prior to sample collection). After receiving, serum and saliva samples were stored at −80°C until required for its analysis. For the analysis of NP, CRP, S1-IgG, and S1-IgM, no sample treatment was required for both serum and saliva samples; a simple dilution with 0.01 M PBS containing 1.0% BSA was performed prior to analysis.
Determination of SARS-CoV-2-Related Selected Target Molecules in Serum and Saliva Samples
NP antigen, CRP, and S1-IgG and S1-IgM isotypes were analyzed in commercial serum and saliva samples from RT-PCR COVID-19-confirmed positive patients (nserum = 10; nsaliva = 5) and healthy subjects (nserum = 7; nsaliva = 3). After 100× and 5× dilution of corresponding serum and saliva samples in PBS with 1.0% BSA, respectively, a 10-μL aliquot was incubated in each WE for 10 min at room temperature (25× dilution was used for 1-min incubation study in serum for Figure 5C). After a washing step with PBS buffer, corresponding detector reagents were incubated in each WE for 5 min and subsequent detection was performed. Comparison of sensor performance in buffer and diluted body fluids from healthy subjects spiked with increasing concentrations of target molecule was performed using CRP as a model molecule.
Validation of Human Samples with the Gold-Standard ELISA
ELISA tests for NP, S1-IgG, and CRP (selected as model targets) were performed in an accuSkan FC Filter-Based Microplate Photometer at a detection wavelength of 450 nm according to the manufacturer's instructions. In brief, plates were coated for 3 h, shaking at 37°C, with 4.0 μg mL−1 of the corresponding capture receptor in each case and blocked with PBS containing 1.0% BSA for 2 h, shaking at 37°C, and standards (or properly diluted samples) were added to coated microtiter plate wells and incubated for 2 h, shaking at 37°C. Next, corresponding HRP-labeled detector antibody was incubated for 30 min at room temperature. Finally, 100 μL of TMB substrate was incubated for 15 min, and absorbance values were measured immediately after addition of 25 μL of 1 M H2SO4 in each well. Three washings with PBS containing 1.0% BSA were performed after each modification step.
Acknowledgments
This project was supported by the Merkin Institute for Translational Research at Caltech, Emergency COVID-19 Research Seed Funding and COVID-19 Continuation Funding from Tobacco-Related Disease Research Program (TRDRP), TRDRP High Impact Pilot Research Award, National Institutes of Health (5R21NR018271), and the Translational Research Institute through NASA NNX16AO69A. J.T. was supported by the National Science Scholarship from the Agency for Science, Technology and Research (A∗STAR), Singapore. We gratefully acknowledge critical support and infrastructure provided for this work by the Kavli Nanoscience Institute at Caltech.
Author Contributions
W.G., R.M.T.-R., and H.L. initiated the concept and designed the experiments; R.M.T.-R., H.L., and J.T. led the experiments and collected the overall data; Y.Y. performed electrode fabrication and characterization; J.M. performed the circuit design and platform test; C.X. contributed to sensor characterization; W.G., R.M.T.-R., and H.L. contributed the data analysis and co-wrote the paper. All authors provided the feedback on the manuscript.
Declaration of Interests
The authors declare no competing interests.
Published: October 1, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.matt.2020.09.027.
Supplemental Information
References
- 1.COVID-19 Map—Johns Hopkins Coronavirus Resource Center. https://coronavirus.jhu.edu/map.html
- 2.Zand M., Wang J. Potential mechanisms of age related severity of COVID-19 infection: implications for development of vaccines, convalescent serum, and antibody therapies. OSF Preprints. 2020 doi: 10.31219/osf.io/f3pze. [DOI] [Google Scholar]
- 3.Anderson R.M., Heesterbeek H., Klinkenberg D., Hollingsworth T.D. How will country-based mitigation measures influence the course of the COVID-19 epidemic? Lancet. 2020;395:931–934. doi: 10.1016/S0140-6736(20)30567-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prager F., Wei D., Rose A. Total economic consequences of an influenza outbreak in the United States. Risk Anal. 2017;37:4–19. doi: 10.1111/risa.12625. [DOI] [PubMed] [Google Scholar]
- 5.Park S.W., Cornforth D.M., Dushoff J., Weitz J.S. The time scale of asymptomatic transmission affects estimates of epidemic potential in the COVID-19 outbreak. Epidemics. 2020;31 doi: 10.1016/j.epidem.2020.100392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., Ren R., Leung K.S.M., Lau E.H.Y., Wong J.Y. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N. Engl. J. Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mizumoto K., Kagaya K., Zarebski A., Chowell G. Estimating the asymptomatic proportion of coronavirus disease 2019 (COVID-19) cases on board the Diamond Princess cruise ship, Yokohama, Japan, 2020. Eurosurveillance. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.10.2000180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nishiura H., Kobayashi T., Miyama T., Suzuki A., Jung S., Hayashi K., Kinoshita R., Yang Y., Yuan B., Akhmetzhanov A.R. Estimation of the asymptomatic ratio of novel coronavirus infections (COVID-19) Int. J. Infect. Dis. 2020;94:154–155. doi: 10.1016/j.ijid.2020.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Z., Yi Y., Luo X., Xiong N., Liu Y., Li S., Sun R., Wang Y., Hu B., Chen W. Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J. Med. Virol. 2020;92:1518–1524. doi: 10.1002/jmv.25727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.‘It’s like having no testing’: coronavirus test results are still delayed - The New York Times. https://www.nytimes.com/2020/08/04/us/virus-testing-delays.html
- 11.ID NOW COVID-19 | Rapid point of care diagnostics—Abbott. https://www.globalpointofcare.abbott/en/product-details/id-now-covid-19.html
- 12.van Dongen J.E., Berendsen J.T.W., Steenbergen R.D.M., Wolthuis R.M.F., Eijkel J.C.T., Segerink L.I. Point-of-care CRISPR/Cas nucleic acid detection: recent advances, challenges and opportunities. Biosens. Bioelectron. 2020;166:112445. doi: 10.1016/j.bios.2020.112445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang Z., Tian D., Liu Y., Lin Z., Lyon C.J., Lai W., Fusco D., Drouin A., Yin X., Hu T. Ultra-sensitive and high-throughput CRISPR-powered COVID-19 diagnosis. Biosens. Bioelectron. 2020;164:112316. doi: 10.1016/j.bios.2020.112316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo L., Sun X., Wang X., Liang C., Jiang H., Gao Q., Dai M., Qu B., Fang S., Mao Y. SARS-CoV-2 detection with CRISPR diagnostics. Cell Discov. 2020;6:4–7. doi: 10.1038/s41421-020-0174-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kellner M.J., Koob J.G., Gootenberg J.S., Abudayyeh O.O., Zhang F. SHERLOCK: nucleic acid detection with CRISPR nucleases. Nat. Protoc. 2019;14:2986–3012. doi: 10.1038/s41596-019-0210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang Y., Yang M., Shen C., Wang F., Yuan J., Li J., Zhang M., Wang Z., Xing L., Wei J. Evaluating the accuracy of different respiratory specimens in the laboratory diagnosis and monitoring the viral shedding of 2019-nCoV infections. medRxiv. 2020 doi: 10.1101/2020.02.11.20021493. [DOI] [Google Scholar]
- 17.Sethuraman N., Jeremiah S.S., Ryo A. Interpreting diagnostic tests for SARS-CoV-2. JAMA. 2020;323:2249–2251. doi: 10.1001/jama.2020.8259. [DOI] [PubMed] [Google Scholar]
- 18.Lisboa Bastos M., Tavaziva G., Abidi S.K., Campbell J.R., Haraoui L.P., Johnston J.C., Lan Z., Law S., MacLean E., Trajman A. Diagnostic accuracy of serological tests for covid-19: systematic review and meta-analysis. BMJ. 2020;370:m2516. doi: 10.1136/bmj.m2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gong J., Dong H., Xia S.Q., Huang Y.Z., Wang D., Zhao Y., Liu W., Tu S., Zhang M., Wang Q. Correlation analysis between disease severity and inflammation-related parameters in patients with COVID-19 pneumonia. medRxiv. 2020 doi: 10.1101/2020.02.25.20025643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu C., Chen X., Cai Y., Xia J., Zhou X., Xu S., Huang H., Zhang L., Zhou X., Du C. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern. Med. 2020;180:934–943. doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou B. Utility of ferritin, procalcitonin, and C-reactive protein in severe patients with 2019 novel coronavirus disease. Res. Square. 2019 doi: 10.21203/rs.3.rs-18079/v1. [DOI] [Google Scholar]
- 22.Morales-Narváez E., Dincer C. The impact of biosensing in a pandemic outbreak: COVID-19. Biosens. Bioelectron. 2020;163:112274. doi: 10.1016/j.bios.2020.112274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jalandra R., Yadav A.K., Verma D., Dalal N., Sharma M., Singh R., Kumar A., Solanki P.R. Strategies and perspectives to develop SARS-CoV-2 detection methods and diagnostics. Biomed. Pharmacother. 2020;129:110446. doi: 10.1016/j.biopha.2020.110446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seo G., Lee G., Kim M.J., Baek S.H., Choi M., Ku K.B., Lee C.S., Jun S., Park D., Kim H.G. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. 2020;14:5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- 25.Chandra P. Miniaturized label-free smartphone assisted electrochemical sensing approach for personalized COVID-19 diagnosis. Sens. Int. 2020;1:100019. doi: 10.1016/j.sintl.2020.100019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mahari S., Roberts A., Shahdeo D., Gandhi S. eCovSens-ultrasensitive novel in-house built printed circuit board based electrochemical device for rapid detection of nCovid-19. bioRxiv. 2020 doi: 10.1101/2020.04.24.059204. [DOI] [Google Scholar]
- 27.Tripathy S., Singh S.G. Label-free electrochemical detection of DNA hybridization: a method for COVID-19 diagnosis. Trans. Indian Natl. Acad. Eng. 2020 doi: 10.1007/s41403-020-00103-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pasomsub E., Watcharananan S.P., Boonyawat K., Janchompoo P., Wongtabtim G., Suksuwan W., Sungkanuparph S., Phuphuakrat A. Saliva sample as a non-invasive specimen for the diagnosis of coronavirus disease 2019: a cross-sectional study. medRxiv. 2020 doi: 10.1101/2020.04.17.20070045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang Y., Song Y., Bo X., Min J., Pak O.S., Zhu L., Wang M., Tu J., Kogan A., Zhang H. A laser-engraved wearable sensor for sensitive detection of uric acid and tyrosine in sweat. Nat. Biotechnol. 2020;38:217–224. doi: 10.1038/s41587-019-0321-x. [DOI] [PubMed] [Google Scholar]
- 30.Torrente-Rodríguez R.M., Tu J., Yang Y., Min J., Wang M., Song Y., Yu Y., Xu C., Ye C., IsHak W.W. Investigation of cortisol dynamics in human sweat using a graphene-based wireless mHealth system. Matter. 2020;2:921–937. doi: 10.1016/j.matt.2020.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Diao B., Wen K., Chen J., Liu Y., Yuan Z., Han C., Chen J., Pan Y., Chen L., Dan Y. Diagnosis of acute respiratory syndrome coronavirus 2 infection by detection of nucleocapsid protein. medRxiv. 2020 doi: 10.1101/2020.03.07.20032524. [DOI] [Google Scholar]
- 32.Hinnemo M., Zhao J., Ahlberg P., Hägglund C., Djurberg V., Scheicher R.H., Zhang S.L., Zhang Z.B. On monolayer formation of pyrenebutyric acid on graphene. Langmuir. 2017;33:3588–3593. doi: 10.1021/acs.langmuir.6b04237. [DOI] [PubMed] [Google Scholar]
- 33.Gao F., Zhang X., Gao F., Cai X., Zheng M., Jiang S., Wang Q. Application of graphene-pyrenebutyric acid nanocomposite as probe oligonucleotide immobilization platform in a DNA biosensor. Mater. Sci. Eng. C. 2013;33:3851–3857. doi: 10.1016/j.msec.2013.05.022. [DOI] [PubMed] [Google Scholar]
- 34.Yi E., Teo L., Yusoff M.M., Chong K.F. 1-Pyrenebutyric acid functionalized reduced graphene oxide (1-Pb-Rgo) energy storage. In: Hashim M.d.A., editor. ICGSCE 2014. 2015. pp. 185–193. [Google Scholar]
- 35.Karachevtsev V.A., Stepanian S.G., Glamazda A.Y., Karachevtsev M.V., Eremenko V.V., Lytvyn O.S., Adamowicz L. Noncovalent interaction of single-walled carbon nanotubes with 1-pyrenebutanoic acid succinimide ester and glucoseoxidase. J. Phys. Chem. C. 2011;115:21072–21082. [Google Scholar]
- 36.Fernandes E., Cabral P.D., Campos R., Machado G., Cerqueira M.F., Sousa C., Freitas P.P., Borme J., Petrovykh D.Y. Functionalization of single-layer graphene for immunoassays. Appl. Surf. Sci. 2020;480:709–716. [Google Scholar]
- 37.Ericsson E.M., Enander K., Bui L., Lundström I., Konradsson P., Liedberg B. Site-specific and covalent attachment of his-tagged proteins by chelation assisted photoimmobilization: a strategy for microarraying of protein ligands. Langmuir. 2013;29:11687–11694. doi: 10.1021/la4011778. [DOI] [PubMed] [Google Scholar]
- 38.Dobaño C., Vidal M., Santano R., Jiménez A., Chi J., Barrios D., Ruiz-Olalla G., Melero N.R., Carolis C., Parras D. Highly sensitive and specific multiplex antibody assays to quantify immunoglobulins M, A and G against SARS-CoV-2 antigens. bioRxiv. 2020 doi: 10.1101/2020.06.11.147363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qian C., Zhou M., Cheng F., Lin X., Gong Y., Xie X., Li P., Li Z., Zhang P., Liu Z. Development and multicenter performance evaluation of fully automated SARS-CoV-2 IgM and IgG immunoassays. Clin. Chem. Lab. Med. 2020;58:1601–1607. doi: 10.1515/cclm-2020-0548. [DOI] [PubMed] [Google Scholar]
- 40.Ma H., Zeng W., He H., Zhao D., Yang Y., Jiang D., Zhou P., Qi Y., He W., Zhao C. COVID-19 diagnosis and study of serum SARS-CoV-2 specific IgA, IgM and IgG by a quantitative and sensitive immunoassay. medRxiv. 2020 doi: 10.1101/2020.04.17.20064907. [DOI] [Google Scholar]
- 41.Ni L., Ye F., Cheng M.L., Feng Y., Deng Y.Q., Zhao H., Wei P., Ge J., Gou M., Li X. Detection of SARS-CoV-2-specific humoral and cellular immunity in COVID-19 convalescent individuals. Immunity. 2020;52:971–977. doi: 10.1016/j.immuni.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao R., Li M., Song H., Chen J., Ren W., Feng Y., Gao G.F., Song J., Peng Y., Su B. Early detection of severe acute respiratory syndrome coronavirus 2 antibodies as a serologic marker of infection in patients with coronavirus disease 2019. Clin. Infect. Dis. 2020:ciaa523. doi: 10.1093/cid/ciaa523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao J., Yuan Q., Wang H., Liu W., Liao X., Su Y., Wang X., Yuan J., Li T., Li J. Antibody responses to SARS-CoV-2 in patients with novel coronavirus disease 2019. Clin. Infect. Dis. 2020:ciaa344. doi: 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nicol T., Lefeuvre C., Serri O., Pivert A., Joubaud F., Dubée V., Kouatchet A., Ducancelle A., Lunel-Fabiani F., Le Guillou-Guillemette H. Assessment of SARS-CoV-2 serological tests for the diagnosis of COVID-19 through the evaluation of three immunoassays: two automated immunoassays (Euroimmun and Abbott) and one rapid lateral flow immunoassay (NG Biotech) J. Clin. Virol. 2020;129:104511. doi: 10.1016/j.jcv.2020.104511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W., Tao Q., Sun Z., Xia L. Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020;2019:200642. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chan J.F.W., Yip C.C.Y., To K.K.W., Tang T.H.C., Wong S.C.Y., Leung K.H., Fung A.Y.F., Ng A.C.K., Zou Z., Tsoi H.W. Improved molecular diagnosis of COVID-19 by the novel, highly sensitive and specific COVID-19-RdRp/Hel real-time reverse transcription-PCR assay validated in vitro and with clinical specimens. J. Clin. Microbiol. 2020;58:e00310–e00320. doi: 10.1128/JCM.00310-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yip C.C.Y., Ho C.C., Chan J.F.W., To K.K.W., Chan H.S.Y., Wong S.C.Y., Leung K.H., Fung A.Y.F., Ng A.C.K., Zou Z. Development of a novel, genome subtraction-derived, SARS-CoV-2-specific COVID-19-nsp2 real-time RT-PCR assay and its evaluation using clinical specimens. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21072574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Y., Odiwuor N., Xiong J., Sun L., Nyaruaba R.O., Wei H., Tanner N.A. Rapid molecular detection of SARS-CoV-2 (COVID-19) virus RNA using colorimetric LAMP. medRxiv. 2020 doi: 10.1101/2020.02.26.20028373. [DOI] [Google Scholar]
- 49.Teng F., Guo L., Cui T., Wang X.G., Xu K., Gao Q., Zhou Q., Li W. CDetection: CRISPR-Cas12b-based DNA detection with sub-attomolar sensitivity and single-base specificity. Genome Biol. 2019;20 doi: 10.1186/s13059-019-1742-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nachtigall, F.M., Pereira, A., Trofymchuk, O.S., and Santos, L.S. Detection of SARS-CoV-2 in nasal swabs using MALDI-MS. Nat. Biotechnol. 38 10.1038/s41587-020-0644-7. [DOI] [PubMed]
- 51.Wang P. Combination of serological total antibody and RT-PCR test for detection of SARS-COV-2 infections. J. Virol. Methods. 2020;283:113919. doi: 10.1016/j.jviromet.2020.113919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carvalho A.M.R.S., De Oliveira Mendes T.A., Coelho E.A.F., Duarte M.C., Menezes-Souza D. New antigens for the serological diagnosis of human visceral leishmaniasis identified by immunogenomic screening. PLoS One. 2018;13 doi: 10.1371/journal.pone.0209599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alizadeh S.A., Abdolahpour G., Pourmand M., Naserpour T., Najafipour R., Eshraghi S.S. Evaluation of new ELISA based on rLsa63-rLipL32 antigens for serodiagnosis of human leptospirosis. Iran. J. Microbiol. 2014;6:184–189. [PMC free article] [PubMed] [Google Scholar]
- 54.Feng W., Newbigging A., Le C., Pang B., Peng H., Cao Y., Wu J., Abbas G., Song J., Wang D.-B. Molecular diagnosis of COVID-19: challenges and research needs. Anal. Chem. 2020;92:10196–10209. doi: 10.1021/acs.analchem.0c02060. [DOI] [PubMed] [Google Scholar]
- 55.Grifoni A., Sidney J., Zhang Y., Scheuermann R.H., Peters B., Sette A. A sequence homology and bioinformatic approach can predict candidate targets for immune responses to SARS-CoV-2. Cell Host Microbe. 2020;27:671–680. doi: 10.1016/j.chom.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kannan S., Subbaram K., Ali S., Kannan H. Molecular characterization and amino acid homology of nucleocapsid (N) protein in SARS-CoV-1, SARS-CoV-2, MERS-CoV, and bat coronavirus. J. Pure Appl. Microbiol. 2020;14:757–763. [Google Scholar]
- 57.Liu F., Li L., Xu M., Wu J., Luo D., Zhu Y., Li B., Song X. Prognostic value of interleukin-6, C-reactive protein, and procalcitonin in patients with COVID-19. J. Clin. Virol. 2020;127:104370. doi: 10.1016/j.jcv.2020.104370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang I., Pranata R., Lim M.A., Oehadian A., Alisjahbana B. C-reactive protein, procalcitonin, D-dimer, and ferritin in severe coronavirus disease-2019: a meta-analysis. Ther. Adv. Respir. Dis. 2020;14 doi: 10.1177/1753466620937175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sun Y., Dong Y., Wang L., Xie H., Li B., Chang C., Wang F.S. Characteristics and prognostic factors of disease severity in patients with COVID-19: the Beijing experience. J. Autoimmun. 2020;112:102473. doi: 10.1016/j.jaut.2020.102473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shang W., Dong J., Ren Y., Tian M., Li W., Hu J., Li Y. The value of clinical parameters in predicting the severity of COVID-19. J. Med. Virol. 2020;92:2188–2192. doi: 10.1002/jmv.26031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tan C., Huang Y., Shi F., Tan K., Ma Q., Chen Y., Jiang X., Li X. C-reactive protein correlates with computed tomographic findings and predicts severe COVID-19 early. J. Med. Virol. 2020;92:856–862. doi: 10.1002/jmv.25871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen W., Zheng K.I., Liu S., Yan Z., Xu C., Qiao Z. Plasma CRP level is positively associated with the severity of COVID-19. Ann. Clin. Microbiol. Antimicrob. 2020;19 doi: 10.1186/s12941-020-00362-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xu C., Yang Y., Gao W. Skin-interfaced sensors in digital medicine: from materials to applications. Matter. 2020;2:1414–1445. doi: 10.1016/j.matt.2020.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang Y., Gao W. Wearable and flexible electronics for continuous molecular monitoring. Chem. Soc. Rev. 2019;48:1465–1491. doi: 10.1039/c7cs00730b. [DOI] [PubMed] [Google Scholar]
- 65.Adans-dester C.P., Bamberg S., Bertacchi F.P., Caulfield B., Chappie K., Demarchi D., Erb M.K., Estrada J., Fabara E.E., Freni M. Can mHealth technology help mitigate the effects of the COVID-19 pandemic? IEEE Eng. Med. Biol. 2020;1:242–248. doi: 10.1109/OJEMB.2020.3015141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jeong H., Adv S., Jeong H., Rogers J.A., Xu S. Continuous on-body sensing for the COVID-19 pandemic: gaps and opportunities. Sci. Adv. 2020;6:eabd4794. doi: 10.1126/sciadv.abd4794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim J., Campbell A.S., De Ávila B.E., Wang J. Wearable biosensors for healthcare monitoring. Nat. Biotechnol. 2019;37:389–406. doi: 10.1038/s41587-019-0045-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.


