Abstract
The ADP-ribosylhydrolase ARH3 plays a key role in DNA damage repair, digesting poly(ADP-ribose) and removing ADP-ribose from serine residues of the substrates. Specific inhibitors that selectively target ARH3 would be a useful tool to examine DNA damage repair, as well as a possible strategy for tumor suppression. However, efforts to date have not identified any suitable compounds. Here, we used in silico and biochemistry screening to search for ARH3 inhibitors. We discovered a small molecule compound named ARH3 inhibitor 26 (AI26) as, to our knowledge, the first ARH3 inhibitor. AI26 binds to the catalytic pocket of ARH3 and inhibits the enzymatic activity of ARH3 with an estimated IC50 of ∼2.41 μm in vitro. Moreover, hydrolysis of DNA damage–induced ADP-ribosylation was clearly inhibited when cells were pretreated with AI26, leading to defects in DNA damage repair. In addition, tumor cells with DNA damage repair defects were hypersensitive to AI26 treatment, as well as combinations of AI26 and other DNA-damaging agents such as camptothecin and doxorubicin. Collectively, these results reveal not only a chemical probe to study ARH3-mediated DNA damage repair but also a chemotherapeutic strategy for tumor suppression.
Keywords: ADP-ribosylation, dePARylation, DNA damage response, cancer therapy, ARH3, inhibitor, DNA repair
Genomic DNA can be damaged by numerous internal and external hazards. However, during evolution, cells have developed sophisticated systems to sense and repair DNA lesions. One of the earliest DNA damage responses is protein ADP-ribosylation.
DNA damage–induced ADP-ribosylation is mainly catalyzed by poly(ADP-ribose) polymerases (PARPs), a group of enzymes transferring ADP-ribose (ADPR) residue from NAD+ to amino acid residues (1, 2). To date, 17 members have been identified in PARP family enzymes (3). Among these enzymes, PARP1, PARP2, PARP5a, and PARP5b catalyze poly(ADP-ribosyl)ation (also called PARylation) by mediating the glycosidic bond formation between each ADPR unit, whereas PARP9 and PARP13 are enzymatically inactive proteins because they lack key catalytic residues (4–6). The remaining 11 PARPs catalyze mono(ADP-ribosyl)ation (MARylation) (2). In response to DNA damage, both PARylation and MARylation occur to mediate DNA damage response and repair (7). Because each ADPR unit has two phosphate moieties, PARylation or multiple MARylation bring a huge amount of negative charges to the chromatin close to DNA lesions (7, 8). Because DNA is also negatively charged, the charge repulsion may induce chromatin relaxation, which facilitates DNA damage repair (7, 8). Over the past 15 years, accumulated evidence suggests that ADP-ribosylation acts as an early wave of signal at DNA lesions and is recognized by PARylation and/or MARylation binding motifs (7, 9, 10). These interactions mediate the recruitment of DNA damage repair factors, containing those ADPR-binding motifs, to the sites of DNA lesions within a very short period for early-phase DNA repair (7).
The acceptor residues for ADP-ribosylation have been identified on aspartic acid, glutamic acid, arginine, lysine, cysteine, serine, and tyrosine (7, 11). Among these residues, aspartic acid and glutamic acid can be linked with poly(ADP-ribose) (PAR) or mono(ADP-ribose) by PARPs (12–15). ADP-ribosylation on arginine is catalyzed by specific arginine ADP-ribosyltransferases (also called ATRCs) or bacterial toxins and may be not associated with DNA damage repair (16). More recently, serine was shown to be linked with ADPR, which is catalyzed by PARP1 following DNA damage (17–19). The detailed molecular mechanism of ADP-ribosylation on other residues remains elusive.
It has been shown that PARP1-mediated serine ADP-ribosylation is one of the major types of ADP-ribosylation that facilitates DNA damage repair (19). HPF1, a co-factor and functional partner of PARP1, interacts with the catalytic domain of PARP1 for promoting ADP-ribosylation on serine (17). Moreover, ARH3 was recently identified as an ADP-ribosylhydrolase that removes ADP-ribosylation from serine residue (20, 21). Interestingly, in addition to removing the last ADPR from serine, ARH3 is able to digest the glycosidic bond between ADPR units, thus hydrolyzing PAR chain, suggesting that ARH3 may specifically remove PARylation and/or MARylation on serine residue (20–22). Accumulated evidence suggests that like PARylation/MARylation, dePARylation and deMARylation may play equally important roles in DNA damage repair (7, 23, 24). It is likely that transient ADP-ribosylation mediates the recruitment of DNA damage repair factors to the proximity of DNA lesion (7). DePARylation and deMARylation may act as immediate downstream events following transient ADP-ribosylation and facilitate loading DNA damage repair factors onto the sites of DNA lesions (24). Otherwise, those ADPR-binding DNA repair factors may be trapped by any prolonged ADP-ribosylation (7, 23). Thus, it has been well-known that suppression of dePARylation severely inhibits DNA damage repair (24–28).
Targeting PARylation-dependent DNA damage repair by PARP inhibitors selectively kills tumor cells with other DNA repair defects, such as homologous recombination repair defects (29–31). Similar to PARP inhibitor, emerging evidence suggests that targeting dePARylation has similar effects on the suppression of DNA damage repair, as well as tumor cell growth (23, 24). Thus, searching for dePARylation/deMARylation inhibitors may have a huge impact on cancer research. However, to date, the specific inhibitors targeting ARH3 have not been identified yet. Here, we discovered a unique small molecule compound that suppresses the enzymatic activity of ARH3 and ARH3-dependent DNA damage repair.
Results
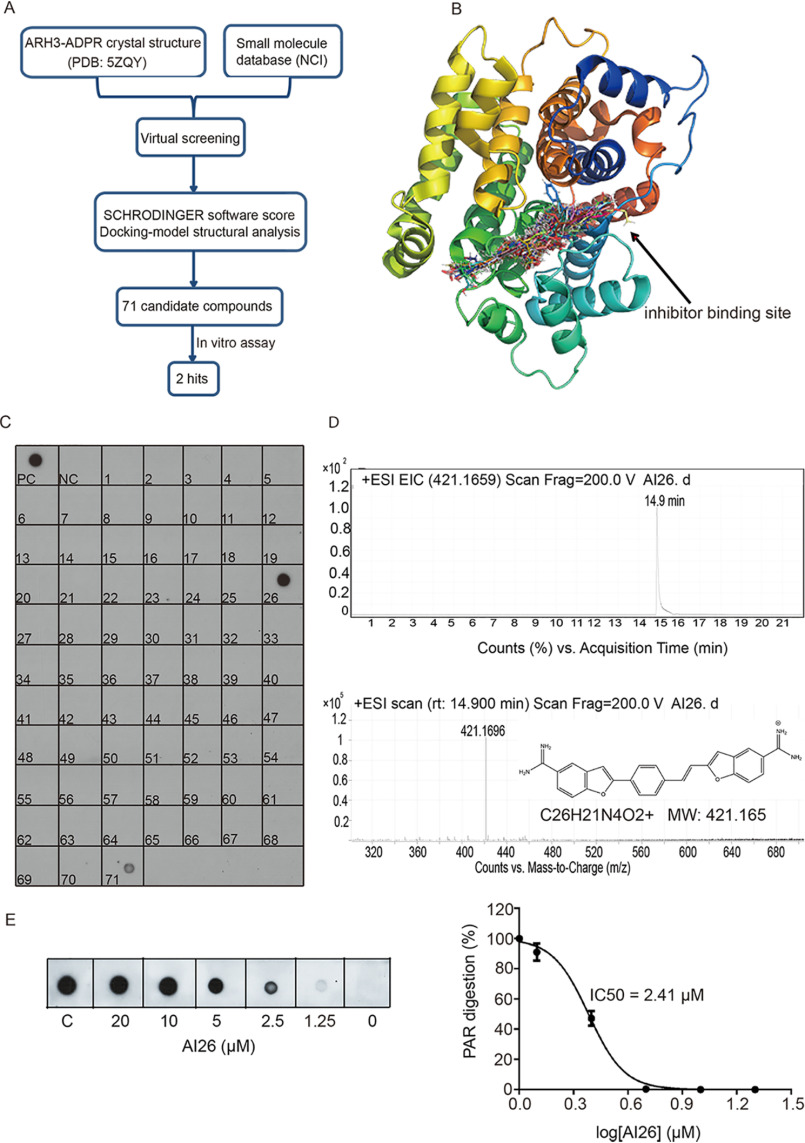
Recently, we and others solved the crystal structure of ARH3 and ADPR complex (PDB code 5ZQY), in which ADPR forms multiple contacts within the catalytic pocket of ARH3 (32, 33). Based on the structure of the complex and small molecule compound library of the National Cancer Institute (NCI), we designed a strategy that combines both virtual screening and biochemical screening to search for specific small molecules targeting ARH3 (Fig. 1A). Using an in silico approach, we first screened more than 260,000 compounds from NCI small molecule database and modeled the compounds that could be inserted into the catalytic pocket of ARH3 and suppress its activity (Fig. 1B). Based on the docking scores, chemical structures of the compounds, and their availability from the NCI, we selected 71 candidates for the secondary biochemical screening using dot-blotting assays with anti-PAR antibody. We generated recombinant ARH3 and synthesized PAR. With mock treatment, PAR can be detected by anti-PAR antibody in the dot-blotting assay as a positive control. However, when PAR was incubated with recombinant ARH3, PAR was digested by ARH3 and could not be detected in the dot-blotting assay, which served as a negative control. Next, we included each candidate compound (100 μm) in the reaction mixture. Interestingly, compound 26 completely suppressed ARH3-mediated PAR digestion, whereas compound 71 mildly inhibited the enzymatic activity of ARH3 (Fig. 1C).
Figure 1.
Identification of ARH3 small-molecule inhibitors. A, a diagram of screening approach for the ARH3 inhibitors. B, the docking models of small molecules fitting into the catalytic pocket of ARH3. The structure of ARH3 is displayed in rainbow cartoon, and the small molecule compounds are shown in stick. C, biochemical screening of the ARH3 inhibitor. 71 candidates from NCI library were examined. ARH3 (1 μm) was incubated with PAR (10 μm) for 30 min at room temperature in the presence of each compound (100 μm). PAR digestion results were measured by dot-blotting assays with anti-PAR antibody. PC and NC indicate the positive control and negative control, respectively. D, AI26 was examined by LC–MS. From top to bottom, HPLC chromatogram, extracted ion chromatogram, and chemical structure of AI26. EIC, extracted ion chromatogram, ESI, electron spray ionization, MW, molecular weight. E, the estimated IC50 of AI26 was calculated from the in vitro PAR digestion assay (n = 3 independent experiments). AI26 with the indicated concentration was incubated with ARH3 (0.5 μm) and PAR (10 μm). The dot-blotting assays were performed with anti-PAR antibody to examine the in vitro PAR digestion. The IC50 value was determined using GraphPad Prism 7 software and the equation: log (inhibitor) versus normalized response – variable slope.
To further explore the small molecule inhibitors of ARH3, we focused on compound 26 and named it ARH3 inhibitor 26 (AI26). We examined the compound using LC–MS and found the single retention peak in HPLC analysis, indicating the single molecule purity. The following quadrupole TOF–MS shows that the molecule weight of the compound is 421.1696, confirming the identity of the compound (Fig. 1D). Next, we diluted the concentration of AI26 in the in vitro ARH3-mediated PAR digestion assays and measured the estimated IC50 of AI26 as 2.41 μm in this in vitro assay (Fig. 1E).
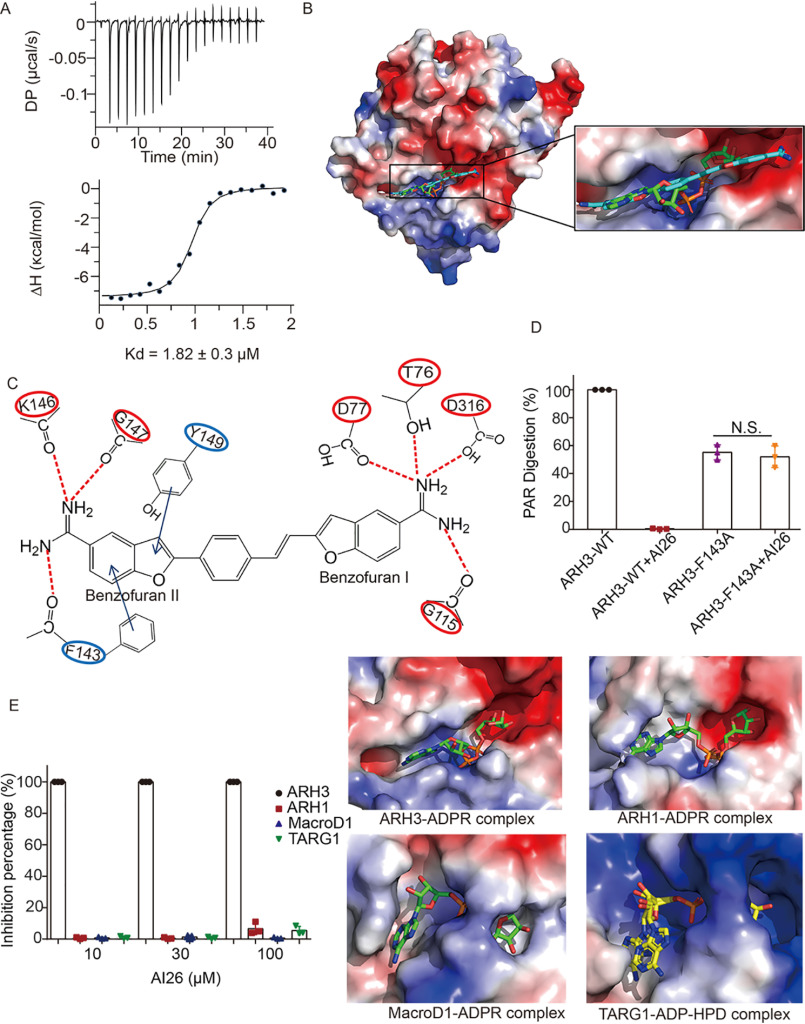
To further characterize the biochemical feature of this ARH3 inhibitor, we performed isothermal titration calorimetry (ITC) assay to measure the binding affinity between recombinant ARH3 and AI26. The average dissociation constant (Kd) obtained from three independent experiments is 1.82 ± 0.3 μm with 1:1 stoichiometric binding (Fig. 2A and Fig. S1). Moreover, computational modeling suggests that AI26 fits into the prolonged and linear catalytic groove, which may not only occupy the catalytic site but also extend the contact with other adjacent residues (Fig. 2B). Detailed analysis indicates that two amino groups in the benzofuran I of AI26 may form hydrogen bonds with catalytic residues such as Thr76, Asp77, and Asp316, which directly mediates the hydrolysis of the ester bond between ADPR and serine (serine deMARylation), as well as the glycosidic bond between two ADPR units (dePARylation). Also, another hydrogen bond occurs in the Gly115 and one amino group in the benzofuran I. Interestingly, two amino groups on the benzofuran II of AI26 may form hydrogen bonds with residues Phe143, Lys146, and Gly147 that are in the outside of the catalytic site. Additionally, Tyr149 and Phe143 may also interact with AI26 benzofuran II through π–π stacking interactions, which may not be involved in catalysis (Fig. 2C). Structural analyses show that the carbonyl group on the main chain of Phe143 forms hydrogen bonds with amino groups on the main chain of Gly145 and Lys146, and the side chain of Phe143 points to protein surface of ARH3 and does not have any interaction with other ARH3 residues (Fig. S2A). Thus, we generated a Phe143-to-Ala (F143A) mutation and hypothesized that this mutation does not abolish the tertiary structure of ARH3 but disrupts the interaction with AI26. To test the hypothesis, we performed thermal shift assay using the WT ARH3 and the F143A mutant proteins and found that the F143A mutant had very similar melting temperature with WT ARH3 (Fig. S2B), indicating that this mutation may not affect the overall folding of ARH3. Instead, the F143A mutation abolished the interaction between ARH3 and AI26 (Fig. S3A). However, because the side chain of Phe143 extends toward the outside of the catalytic groove, the F143A mutation still largely retained the enzymatic activity (Fig. S3B). Interestingly, because the mutation abolished the interaction with AI26, AI26 could not suppress the enzymatic activity of F143A (Fig. 2D). These results validate that AI26 occupied the catalytic pocket and surrounding areas of ARH3. In addition to ARH3, other enzymes such as MacroD1 and TARG1 also digest the ester bond between ADPR and amino acid residues. Moreover, like ARH3, MacroD1 can digest the glycosidic bond between two ADPRs (34). However, the catalytic pockets of both MacroD1 and TARG1 are L-shaped folds, whereas that of ARH3 adopts prolonged and linear conformation, which is more extended compared with those of TARG1 and MacroD1 (Fig. 2E). Thus, AI26 is unlikely to fit into the catalytic pockets of TARG1 and MacroD1. Moreover, because ADPR is twisted in the catalytic pocket of ARH1, this ADPR-recognition pocket is much smaller than that of ARH3, indicating there is no enough space for accommodating the benzofuran II of AI26. Consequently, AI26 did not inhibit the enzymatic activities of these ADPR hydrolases (Fig. 2E). Collectively, these results suggest that AI26 specifically suppresses the enzymatic activity of ARH3 by binding to its catalytic pocket.
Figure 2.
AI26 occupies the catalytic pocket of ARH3. A, the binding affinity between AI26 and recombinant ARH3 was measured using ITC. The Kd value was the average of three independent experiments shown in Fig. S1. B, a computer modeling of AI26 in the catalytic pocket of ARH3. The ARH3 is shown in electrostatic potential map, the AI26 is in cyan stick, and the ADPR is shown in green stick. C, schematic representation of the interaction between AI26 and ARH3. The dashed lines represent the predicted hydrogen bonds. The arrows represent the predicted π–π stacking interaction. D, AI26 does not suppress the enzymatic activity of the F143A mutant. AI26 (100 μm) was incubated with ARH3 (1 μm) or the F143A mutant (1 μm) and PAR (10 μm). The PAR digestion was measured by dot-blotting assays with anti-PAR antibody (n = 3 independent experiments). N.S., nonsignificant. E, AI26 specifically suppresses ARH3 but not other ADPR hydrolases. The relative ADPR digestion inhibition by AI26 treatment on each ADPR hydrolases was examined. The detailed approaches are included under “Experimental procedures.” Three independent experiments were performed on each inhibition assay, and the results are shown in a histogram (left panel). The catalytic pockets of ADPR hydrolases are shown in an electrostatic potential map. The ARH3–ADPR complex (PDB code 5ZQY), ARH1–ADPR complex (PDB code 6IUX), MacroD1–ADPR complex (PDB code 6LH4), and TARG1–ADP–HPD complex (PDB code 4J5R) are included in the right panels. ADPR is shown in green stick. The ADPR analog ADP-HPD is in yellow stick, and its adenine base exhibits two alternate conformations in the binding pocket.
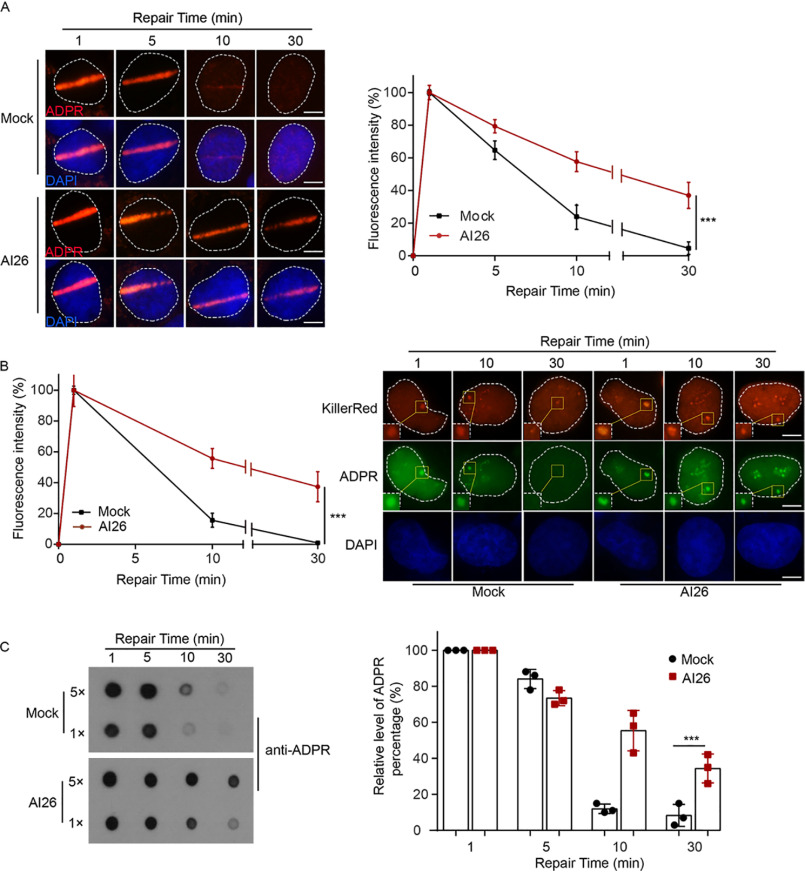
Next, we asked whether AI26 was able to suppress ARH3-mediated ADP-ribosylation hydrolysis in cells. We used laser microirradiation to induce DNA damage in the nucleus of U2OS cell. Using immunofluorescent staining with anti-ADPR antibody, we examined the kinetics of DNA damage–induced ADP-ribosylation in the time course assays. We found that ADP-ribosylation occurred within 1 min following laser microirradiation and was hydrolyzed within 30 min. However, when the cells were pretreated with AI26, the DNA damage–induced ADP-ribosylation was largely prolonged (Fig. 3A), which is in agreement with an earlier study that serine ADP-ribosylation is one of the major types of ADP-ribosylation during DNA damage repair (20, 21). To further validate the inhibition effect of AI26 treatment, we used the KillerRed system to induce oxidative damage in U2OS cells. Again, ADP-ribosylation was occurred quickly in the mock treated cells and was removed within 30 min following DNA damage. However, AI26 treatment clearly suppressed hydrolysis of ADP-ribosylation at the sites of DNA damage (Fig. 3B). Moreover, we treated U2OS cells with methyl methanesulfonate (MMS) to induce global ADP-ribosylation. Using dot-blotting assays, we examined the removal kinetics of ADP-ribosylation. Compared with the mock treatment, AI26 treatment was able to suppress the removal of global DNA damage–induced ADP-ribosylation (Fig. 3C).
Figure 3.
AI26 suppresses hydrolysis of ADP-ribosylation in cells. A, AI26 treatment suppresses the degradation of ADPR at laser strips. U2OS cells were with or without 10 μm AI26 followed by laser microirradition. ADP-ribosylation at DNA lesions was examined by IF with anti-ADPR antibody. The accumulation kinetics of ADP-ribosylation at DNA lesions was examined in 50 cells (n = 3 independent experiments). The results were summarized as means ± S.D. ***, P < 0.001. The scale bar represents 5 μm. B, AI26 treatment suppresses the digestion of ADPR at oxidative lesions. U2OS cells expressing KillerRed were treated with or without 10 μm AI26 for 1 h, following white light treatment at 25 °C for 10 min. The location of DNA lesions were indicated by IF with anti-KillerRed antibody. The kinetics of ADP-ribosylation at DNA lesions was examined by IF with anti-ADPR antibody. The results are summarized from 50 cells (n = 3 independent experiments) and shown as means ± S.D. ***, P < 0.001. The scale bar represents 5 μm. C, 293T cells were pretreated with or without 10 μm AI26 for 1 h, followed by 0.5 mm H2O2 treatment for 5 min at 37 °C. The cellular levels of ADP-ribosylation were examined by dot-blotting assays with anti-ADPR antibody (left panel). The histograms represent the time course results from three independent experiments (right panel). The results were summarized as means ± S.D. ***, P < 0.001. DAPI, 4[prime],6[prime]-diamino-2-phenylindole.
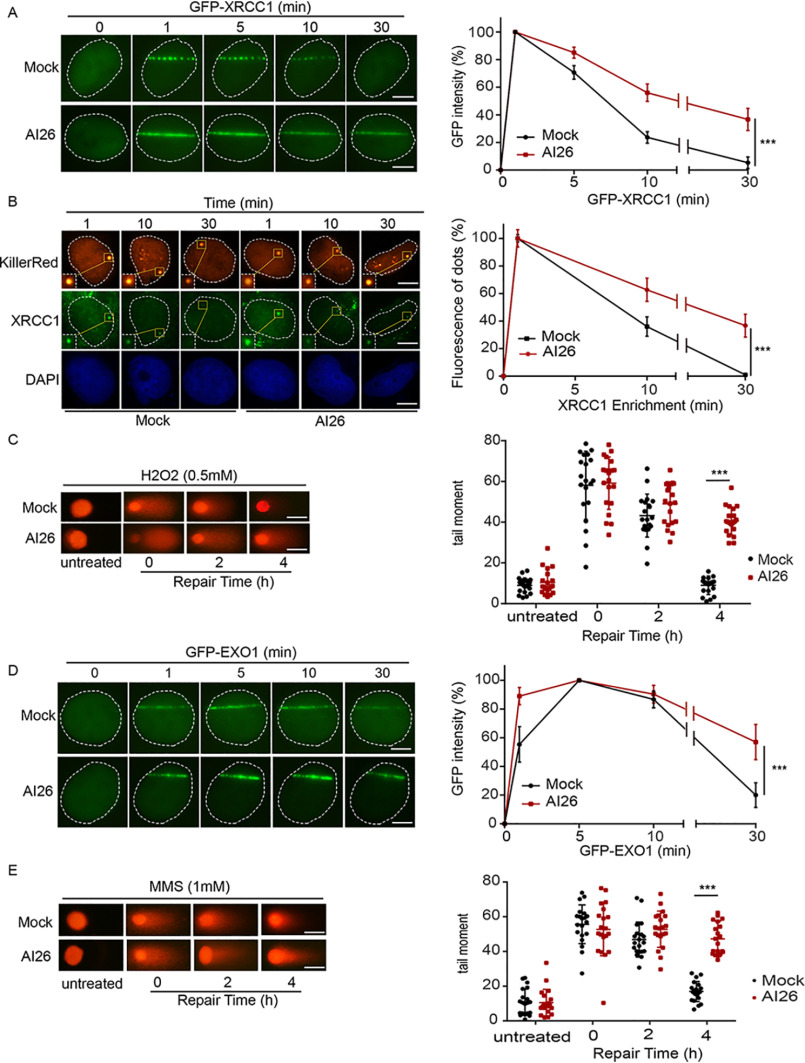
We and others have shown that ADP-ribosylation mediates the recruitment of DNA damage repair factors to DNA lesions (7). The prolonged ADP-ribosylation may trap these DNA damage repair factors at DNA lesions for prolonged time. To examine this possibility, we studied the recruitment of XRCC1, which is a bona fide ADPR-binding DNA repair factor that is involved in DNA single-strand break repair (SSBR) (35). Similar to the kinetics of DNA damage–induced ADP-ribosylation, XRCC1 was retained at laser strip as well as KillerRed sites for prolonged time when cells were pretreated with AI26 (Fig. 4, A and B), suggesting that suppression of ARH3-mediated ADPR hydrolysis traps DNA damage repair factors at DNA lesions. Because trapping ADPR-binding DNA repair factors may impair DNA damage repair, we performed comet assays with alkaline condition to measure the kinetics of SSBR and found that suppression of ARH3-mediated ADPR hydrolysis by AI26 remarkably impaired SSBR (Fig. 4C).
Figure 4.
AI26 treatment impairs DNA damage repair. A and B, AI26 treatment traps XRCC1 at DNA lesions. U2OS cells expressing GFP-XRCC1 were pretreated with or without 10 μm AI26 for 1 h, and the retention of XRCC1 at laser strip was examined with live cell imaging (A). U2OS cells expressing KillerRed were pretreated with or without 10 μm AI26 for 1 h, and the endogenous XRCC1 at the oxidative damage sites was examined by IF with anti-XRCC1 antibodies. The scale bar represents 5 μm. B, results are displayed as means ± S.D. from 50 cells (n = 3 independent experiments). ***, P < 0.001. The scale bar represents 5 μm. C, AI26 treatment suppresses SSBR. U2OS cells were pretreated with or without 10 μm AI26 for 1 h, followed by 0.5 mm H2O2 for 5 min. Alkaline comet assays were performed to examine the rate of SSBR in time course experiments. The tail moments were determined from at least 50 cells at each time point in each experiment, and three independent experiments were carried out. ***, P < 0.001. The scale bar represents 30 μm. D, AI26 treatment traps EXO1 at DNA lesions. U2OS cells expressing GFP-EXO1 were pretreated with or without 10 μm AI26 for 1 h, and the retention of EXO1 at laser strip was examined with live cell imaging. Three independent experiments were carried out. ***, P < 0.001. The scale bar represents 5 μm. E, AI26 treatment suppresses DSBR. U2OS cells were pretreated with or without 10 μm AI26 for 1 h, followed by 1 mm MMS for 30 min. Neutral comet assays were performed to examine the rate of DSBR in time course experiments. The scale bar represents 30 μm.
In addition to SSBR, ADP-ribosylation participates in DNA double-strand break repair (DSBR) (36), We and others have shown that EXO1, a double-strand break end-processing enzyme, is recruited to DNA lesions by ADP-ribosylation (37, 38). Here, we examined and found that EXO1 was also trapped at DNA lesions following AI26 treatment (Fig. 4D). Moreover, we performed comet assays in neutral condition to examine the kinetics of DNA DSBR. Similar to SSBR, DSBR was impaired as well (Fig. 4E). In addition, the cells treated with AI26 were sensitive to the MMS treatment. However, AI26 treatment did not affect the viability of ARH3-deficient cells under the similar DNA-damaging conditions (Fig. S4). Collectively, these results demonstrate that AI26 treatment suppresses DNA damage repair by trapping repair factors at DNA lesions.
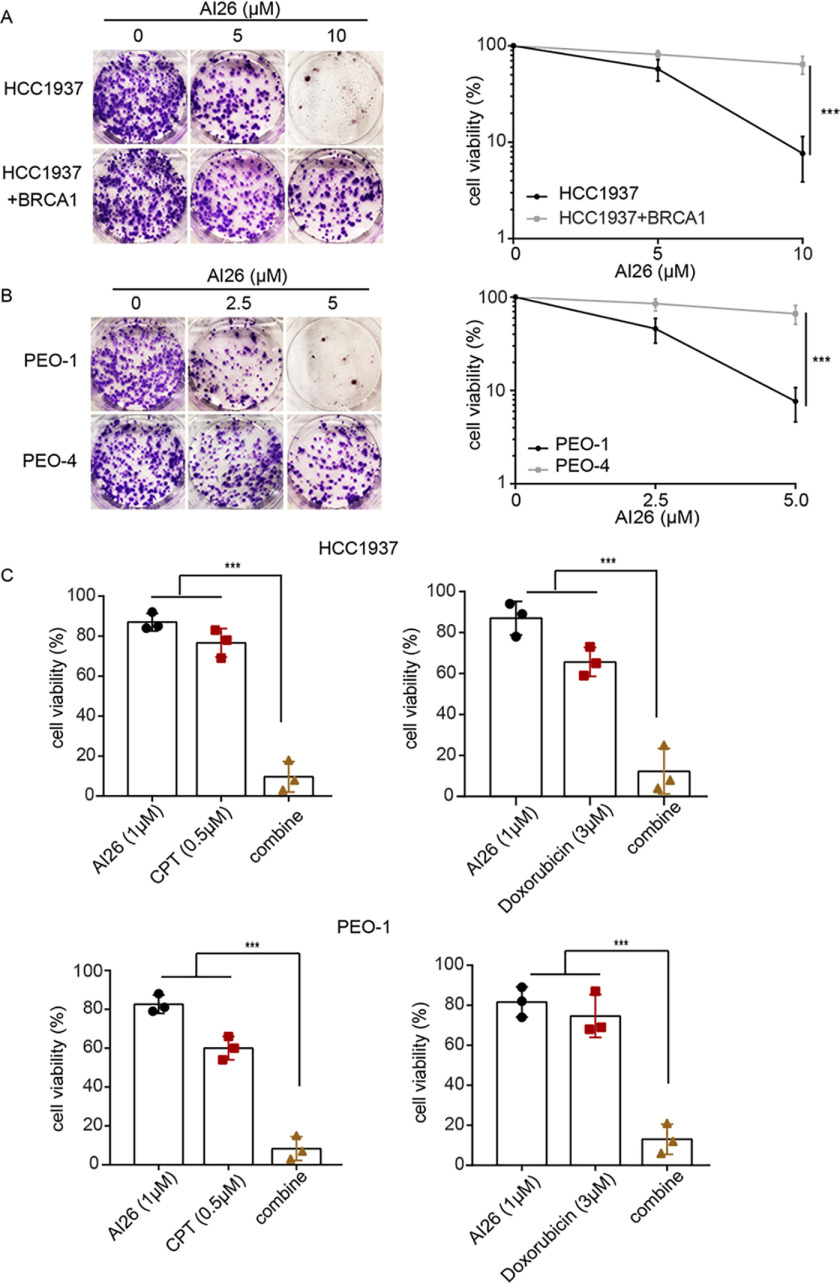
Because additional DNA damage repair suppression may sensitize tumor cells that already have repair defects, we asked whether AI26 was able to selectively kill tumor cells with repair defects, such as tumor cells with BRCA1/2 mutations. We examined HCC1937, a BRCA1-null cell line and derived from a triple-negative breast cancer patient. Of note, HCC1937 is notoriously insensitive to PARP inhibitor treatment because of unsolved molecular mechanism (39). Interestingly, AI26 was able to selectively suppress HCC1937 cell growth but not HCC1937 cell reconstituted with WT BRCA1 at the concentration of 10 μm (Fig. 5A). Moreover, AI26 was able to suppress the growth of BRCA2-deficient ovarian cancer cell PEO-1, but not BRCA2-proficient cell PEO4 at the concentration of 5 μm (Fig. 5B). In addition, we used low dose of AI26 and found that it sensitized these BRCA mutant tumor cells to other DNA-damaging agents such camptothecin and doxorubicin (Fig. 5C). Taken together, these results indicate that targeting ARH3 may be an effective strategy for tumor suppression.
Figure 5.
AI26 selectively kills tumor cells with DNA damage repair defects. A and B, AI26 suppresses the growth of tumor cells with BRCA1 or BRCA2 mutations. HCC1937, a BRCA1-null triple-negative breast cancer cell line (A), and PEO-1, a BRCA2-deificent ovarian cancer cell line (B), were treated with the indicated doses of AI26. BRCA1-reconstituted HCC1937 (HCC1937 + BRCA1) and PEO-4 (BRCA2-proficient ovarian cancer cell line) were used as controls, respectively. The cells were stained using crystal violet after 14 days of culture. Cell viability is displayed as means ± S.D. from three independent experiments. ***, P < 0.001. C, AI26 sensitizes tumor cells to camptothecin or doxorubicin. HCC1937 or PEO-1 cells were treated with the indicated dose of compounds. Cell viabilities from three independent experiments are displayed in the histograms. ***, P < 0.001. CPT, camptothecin.
Discussion
In this study, we have discovered the first-in-class ARH3 inhibitor that specifically occupies the catalytic pocket of ARH3. This enzymatic pocket is quite different from that in other ADPR hydrolases. To date, three major classes of ADPR hydrolases have been identified, namely macrodomain ADPR hydrolases, ADP-ribosylhydrolase family enzymes, and pyrophosphatases (23). Because of the conformational diversity of catalytic pocket, AI26 cannot fit into macrodomain ADPR hydrolases and pyrophosphatases (Fig. 2E and Fig. S5) and thus does not block their enzymatic activities. In the ADP-ribosylhydrolase family enzymes, ARH1 and ARH3 are active ADPR hydrolases (40–42). However, the catalytic pocket of ARH1 is quite different from that of ARH3. Thus, AI26 does not inhibit the enzymatic activity of ARH1 either. Moreover, because the F143A mutation of ARH3 disrupts the interaction between ARH3 and AI26, AI26 cannot inhibit the enzymatic activity of the F143A mutant, further suggesting that AI26 selectively suppresses the enzymatic activity of ARH3. The HillSlope along with the estimated IC50 is −1.6 with an R2 value of 0.99. Because these results were from dot-blotting assays, a type of semiquantitative assay, quantitative calculation of AI26 inhibition activity may need in future.
Although other ADPR hydrolases play important roles in ADPR metabolism, it has been shown that serine ADP-ribosylation is one of the major types of ADP-ribosylation during DNA damage repair (19). Moreover, ARH3 is the only known ADPR hydrolase to remove ADPR from serine residue (20, 21). Thus, inhibition of the enzymatic activity of ARH3 by AI26 abolishes ARH3-dependent DNA damage repair function, mainly through prolonged ADP-ribosylation and trapping DNA damage repair factors on ADPR at DNA lesions. Here, we found that XRCC1 was trapped at DNA lesions by AI26 treatment. XRCC1 contains C-terminal tandem BRCA1 C-terminal (BRCT) repeats, which recognize ADPR (43). Thus, loss of ARH3, the BRCTs domain of XRCC1 recognizes the prolonged ADP-ribosylation at DNA lesions and suppresses XRCC1-mediated SSBR. Similarly, because other BRCT domain–containing proteins also recognize ADPR, prolonged ADPR at DNA lesions by AI26 treatment may trap other BRCT domain–containing DNA damage repair factors. Similarly, because the PIN domain of EXO1 is another ADPR-binding motif, AI26 treatment prolongs ADPR at DNA lesions and traps EXO1 as well. In addition, other ADPR-binding partners may also act as XRCC1 and EXO1 and be trapped at DNA lesions when cells are treated with AI26. It has been shown that major serine ADP-ribosylation targets include nucleosomal histones (18). It is possible that AI26 treatment traps these ADPR-binding partners on histones in the vicinity of DNA lesions.
This trapping mechanism also triggers hypersensitivities of tumor cells to AI26. Because AI26 treatment abolishes ARH3-mediated DNA damage repair, it increases the repair stress on tumor cells that originally have repair defects. This synthetic lethality mechanism selectively kills tumor cells with repair defects such as BRCA1 or BRCA2 mutant cells. Thus, ARH3 is a novel targeting for personalized chemotherapy for cancer patients. Here, we have shown that AI26 is a potent lead compound for the suppression of ARH3, and we expect that its derivatives can be used for cancer therapy in future.
Experimental procedures
Virtual screening
To identify the potent small-molecule inhibitors of ARH3, we employed our in-house developed LiVS pipeline to perform virtual ligand screening from the NCI Developmental Therapeutics Program library with 260,000 compounds. LiVS pipeline integrates our in-house developed methods for drug discovery in silico. It is a multiple-stage (taking the three high-throughput virtual screening (HTVS)/standard precision (SP)/extra precision (XP) mode of Schrödinger Glide docking software in series), multiple-CPU (using parallel computing to speed up docking calculations), and full-coverage (calculating the docking of every compound) program that can screen millions of compounds within weeks. First, the catalytic pocket of ARH3 (PDB code 5ZQY) was selected as the binding site for virtual screening, which was also confirmed as the best druggable site by using our in-house developed druggable site prediction method. Then the HTVS mode (fast, but less accurate) of Glide software was used to initially screen the whole NCI library on 30 computer cores in parallel. The top 10,000 compounds were selected and docked again using the Glide SP mode. Later, the top 1,000 compounds were further picked to dock in XP mode. They were also analyzed and filtered by Lipinski's rule of five (44), HTS frequent hitter (PAINS) (45), protein reactive chemicals (ALARM) (46), and maximized the molecule diversity by using our in-house developed universe diversity score (to measure library diversity, which is independent of library size). Finally, a total of 71 candidate compounds were prepared on the basis of their docking score, modeling analysis, and availability from NCI.
Cell culture
293T, U2OS, HCC1937, HCC1937+BRCA1, PEO-1, and PEO-4 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% FBS and incubated with 5% CO2 at 37 °C.
Protein purification and in vitro PAR digestion assay
Both full-length ARH3 and the F143A mutant were expressed as N-terminal GST tag recombinant proteins in Escherichia coli BL21(DE3) cells and were purified according to the protocols described previously (32). Briefly, all proteins were purified by GSH-Sepharose 4B column, Source 15Q, and Superdex 200 Increase column successively. The three purification columns were purchased from GE Healthcare. According to previous studies, PAR was synthesized from a biochemical assay using the recombinant PARP1 and NAD+ as the synthetase and substrate, respectively (47).
For the in vitro ARH3 inhibition assay, the full-length ARH3 (1 μm) was incubated with 10 μm PAR substrate in the presence of small-molecule compound or DMSO in PBS supplemented with 5 mm MgCl2. After incubation for 30 min at room temperature, the reaction was stopped by heating the samples at 95 °C for 10 min. Samples (2 μl) from each reaction were dotted onto nitrocellulose membranes and then cross-linked at 60 °C for 30 min. The membrane was blocked with 5% milk. The blocked membrane was examined with anti-PAR mAb at 4 °C.
To determine the enzyme kinetic parameters, 0.1 μm proteins were incubated with 0.6–19.2 μm PAR substrate in the abovementioned buffer at room temperature for 0, 20, 40, 60, 90, and 120 s. The reduction of PAR was detected by dot-blotting assay with anti-PAR antibody. Initial reaction rate at the indicated concentration of PAR was measured by fitting the linear portion of the reaction progress curve. Km and Vmax were calculated by Michaelis–Menten equation in the GraphPad Prism 7 software.
Thermal shift assay
To carry out the thermal shift assays, 5 μl of fresh 200× SYPRO orange dye solution was mixed with 45 μl of 5 μm recombinant proteins in the Tris-HCl buffer. The thermal melting curves were measured using a real-time PCR (Applied Biosystems) melt curve program with a ramp rate of 0.5 °C and a temperature range from 25 to 60 °C. A mixture solution only containing the appropriate amount of buffer and SYPRO orange dye was used as the control sample.
ITC assay
ITC was used to calculate the binding affinity between the WT ARH3 (or the F143A mutant) and AI26 at 25 °C using MicroCal PEAK-ITC. The AI26 (100–200 μm) and the recombinant protein (10–20 μm) were loaded in the titration syringe and sample cell, respectively. The titration protocol contains 0.4 μl of preinjection and sequential 19 × 2-μl injections at 200-s intervals.
Target selectivity assays
For TARG1 and MacroD1 inhibition assays, the auto–ADP-ribosylated PARP10 was used as the substrate and prepared according to the previously published methods (24). The 10 μm substrate was incubated with 0.5 μm enzymes with the indicated concentration of AI26 in a 15-μl reaction system for 1 h at 37 °C. Then the reaction mixture was heated for 10 min at 95 °C. The auto–ADP-ribosylated PARP10 was examined by dot-blotting assay with anti-ADPR antibody. For ARH1 inhibition assay, the mouse brain membrane fraction (300 μg) was incubated with cholera toxin (100 μg) in the presence of 10 μm 32P-labeled NAD+. Approximately 5 μg of labeled substrate, 0.5 μm of recombinant ARH1 protein, and AI26 with the indicated concentration were incubated together in PBS with additional 10 mm MgCl2. After incubation for 1 h at room temperature and heating at 95 °C for 10 min, 2-μl samples were dotted onto nitrocellulose membranes.
Liquid chromatography–MS assay
The identity of AI26 was validated by LC–MS. The measurement was carried out with the combined use of the quadrupole TOF accurate mass spectrometer and the HPLC Agilent 1200 nanoflow system. As the key parameter, the mass tolerance was configured to ±5 ppm mass accuracy.
Immunofluorescence
The cells were treated with microirradiation for the analysis of PARylation and recovered in fresh medium at 37 °C. Treated cells were fixed in 5% paraformaldehyde for 15 min at room temperature and permeabilized using 0.5% Triton X-100 (Sigma–Aldrich) for 15 min at room temperature. After washing with PBS buffer, the coverslips was incubated with the primary antibody overnight at 4 °C and followed by washing with PBS three times. In detail, the primary antibody is at 1:200 dilutions in PBS supplemented with 8% goat serum (Sigma–Aldrich). Incubation with the secondary antibody (Sigma–Aldrich) was carried out at room temperature at 1:500 dilutions in 8% goat serum for 1 h in the dark. 4′,6-Diamidino-2-phenylindole (Sigma–Aldrich) was used to counterstain nuclei for 10 min at room temperature in the dark. Coverslips were mounted in VECTASHIELD (Vector Laboratories, Peterborough, UK). The results were analyzed using a fluorescence microscope.
Comet assays
After incubation, the cells were collected and resuspended using ice-cold PBS. 1 × 105/ml cells were mixed with 1% low-melt agarose at 37 °C at a ratio of 1:3 (v/v) and immediately pipetted onto frosted glass slides. To perform neutral comet assay, the glass slides were placed overnight in the neutral lysis buffer (2% sarkosyl, 0.5 m EDTA, and 0.5 mg/ml proteinase K, pH 8.0) at 37 °C in the dark and then washed twice using the rinse buffer consisting of 90 mm Tris-HCl, pH 8.5, 90 mm boric acid, and 2 mm EDTA for 30 min. Electrophoresis was carried out at 20 V for 25 min (0.6 V/cm), and then the slides were stained in phosphatidylinositol (PI, 2.5 µg/ml) for 20 min in the dark. To perform alkaline comet assay, the glass slides were placed in the alkaline lysis buffer (2.5 m NaCl, 100 mm EDTA, 10 mm Tris-HCl, 1% sarkosyl, and 1% Triton X-100, pH 10.0) for 30 min at 4 °C, washed three times in cold distilled water, and placed in fresh prepared alkaline electrophoresis solution (300 mm NaOH and 1 mm EDTA, pH 13.0) for 10 min at 4 °C. The slides were treated with electrophoresis at 20 V (1 V/cm) and 300 mA for 20 min. The slides were then neutralized to pH 7.5 in 0.4 mm Tris-HCl buffer and stained 20 min with PI (2.5 µg/ml) in the dark. The images were viewed under a fluorescence microscope and further analyzed by OpenComet.
Laser microirradiation and imaging of cells
U2OS cells were plated on glass-bottomed culture dishes (NEST Biotechnology) and transfected with the GFP-XRCC1 plasmid, and then these transfected cells were pretreated with or without 10 μm AI26 at 37 °C for 1 h before laser microirradiation. Laser microirradiation was carried out using an IX 71 microscope (Olympus) combined with the MicroPoint laser illumination and ablation system (Photonic Instruments Inc.). The cell exposure time to the laser beam was ∼3.5 ns, and the corresponding pulse energy was 150 mJ at 10 Hz. The same microscope was used to take the images, which were analyzed with the cellSens software (Olympus). The GFP fluorescence located at the laser line was then converted into a numerical value using ImageJ software. Normalized fluorescent curves from 50 cells from three independent experiments were averaged. The error bars represent S.D.
KillerRed activation
The KillerRed was activated using the method described previously (48). Briefly, U2OS tet response element cells carrying the pBROAD3/tetR-KR plasmid were used for activation. These transfected cells were pretreated without or with 10 μm AI26 for 1 h and then exposed to Sylvania 15-Watt cool white fluorescent light bulb for 10 min (height to light is 15 cm). After recovery for ∼1 or 10 min, immunofluorescence staining was carried out to examine the samples with anti-KillerRed antibody (Evrogen), anti-ADPR antibody (purified by our laboratory, or anti-XRCC1 antibody (GeneTex). Images were acquired using a fluorescence microscope and analyzed by ImageJ software.
Clonogenic assay
The cells were seeded in 6-well plates at a density of 1000 cells/well and then treated with AI26 at the indicated concentration. To perform synergistic efficacy analysis, the chemical compounds was added into each well with the indicated concentrations. The cells were treated with AI26, doxorubicin, or camptothecin alone or combination treatments as indicated. After 14 days of incubation, the viable cells were fixed using methanol and then stained using crystal violet. The number of colonies (>50 cells for each colony) was measured. To measure the sensitivity of ARH3-deficient cells to AI26, the cells were seeded into six-well plates (∼1000 cells/well), then pretreated with AI26 for 2 h, and then stimulated with different concentrations of MMS (0.125, 0.25, and 0.5 mm) for 30 min. After a 14-day culture, the viable cells were fixed by methanol and stained with crystal violet. The number of colonies (>50 cells for each colony) was calculated.
Stable cell lines construction
All the knockdown sequences were constructed into the Plko.1 vector. The efficient sequence was screened by subsequent Western blotting. The knockdown sequences for ARH3 were designed and shown as follows: sense, 5´-CCGGGAAGCCTTGTACTACACAGATCTCGAGATCTGTGTAGTACAAGGCTTCTTTTTG-3´; and antisense, 5´-TGTACTACACAGATCTCGAGATCTGTGTAGTACAAGGCTATCTGTGTAGTACAAGGCTTC-3′.
The cells were co-transfected with the ARH3 knockdown plasmids and two packaging plasmids, psPAX2 and pMD2.G. After being transfected for 6 h, the cells were changed to fresh medium. Once transfected for 48 h, the replication-defective virus was harvested, filtered using a 0.45-μm sterile filter membrane, and used to infect host cells with 10 μg/ml Polybrene. The WT cells and infected cells were treated with 1 μg/ml puromycin for a period of time. The stable cell lines could be constructed until the WT cells were killed by puromycin. Finally, the ARH3 expression level was detected using Western blotting.
Data availability
All data are contained within the article and its Supporting Information.
Supplementary Material
Acknowledgments
We thank Dr. Hua Chen for invaluable discussions.
This article contains supporting information.
Author contributions—X. L., R. X., L. L. Y., X. Yang, A. K. S., and C. W. data curation; X. L. and X. Yu writing-original draft; A. K. S. and X. Yu writing-review and editing; H. L. software; C. W. and X. Yu formal analysis; C. W. and X. Yu investigation; X. Yu supervision; X. Yu funding acquisition; X. Yu methodology; X. Yu project administration; S.-H. C. perform experiment.
Funding and additional information—This work was supported by National Natural Science Foundation of China Grants 81874160 and 31670812, Foundation of Hebei Educational Committee Grant ZD2020183, Ministry of Education ChunHui Project Grant 5, Natural Science Foundation of Hebei Province Grant C2018201171, and funds from the Hebei Province Foundation for Returned Overseas Chinese Scholars and from Westlake University. The Drug Discovery & Structural Biology Core facility of City of Hope is supported by National Cancer Institute Grant P30CA033572. A. K. S. was partially supported by the Tower Cancer Research Foundation and by National Cancer Institute Grant CA233664. S.-H. C. was partially supported by National Cancer Institute Grants CA130899 and CA240392 and by the Ovarian Cancer Research Fund. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- PARP
- poly(ADP-ribose) polymerase
- PAR
- poly(ADP-ribose)
- PARylation
- poly(ADP-ribosyl)ation
- MARylation
- mono(ADP-ribosyl)ation
- ADPR
- ADP-ribose
- PDB
- Protein Data Bank
- NCI
- National Cancer Institute
- ITC
- isothermal titration calorimetry
- SSBR
- single-strand break repair
- DSBR
- double-strand break repair
- MMS
- methyl methanesulfonate
- HTVS
- high-throughput virtual screening
- SP
- standard precision
- XP
- extra precision
- BRCT
- BRCA1 C-terminal
- PI
- phosphatidylinositol.
References
- 1. Rouleau M., Patel A., Hendzel M. J., Kaufmann S. H., and Poirier G. G. (2010) PARP inhibition: PARP1 and beyond. Nat. Rev. Cancer 10, 293–301 10.1038/nrc2812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gupte R., Liu Z., and Kraus W. L. (2017) PARPs and ADP-ribosylation: recent advances linking molecular functions to biological outcomes. Genes Dev. 31, 101–126 10.1101/gad.291518.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hassa P. O., and Hottiger M. O. (2008) The diverse biological roles of mammalian PARPS, a small but powerful family of poly-ADP-ribose polymerases. Front. Biosci. 13, 3046–3082 10.2741/2909 [DOI] [PubMed] [Google Scholar]
- 4. Aguiar R. C. T., Takeyama K., He C. Y., Kreinbrink K., and Shipp M. A. (2005) B-aggressive lymphoma family proteins have unique domains that modulate transcription and exhibit poly(ADP-ribose) polymerase activity. J. Biol. Chem. 280, 33756–33765 10.1074/jbc.M505408200 [DOI] [PubMed] [Google Scholar]
- 5. Vyas S., Chesarone-Cataldo M., Todorova T., Huang Y. H., and Chang P. (2013) A systematic analysis of the PARP protein family identifies new functions critical for cell physiology. Nat. Commun. 4, 2240 10.1038/ncomms3240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vyas S., Matic I., Uchima L., Rood J., Zaja R., Hay R. T., Ahel I., and Chang P. (2014) Family-wide analysis of poly(ADP-ribose) polymerase activity. Nat. Commun. 5, 4426 10.1038/ncomms5426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liu C., Vyas A., Kassab M. A., Singh A. K., and Yu X. (2017) The role of poly ADP-ribosylation in the first wave of DNA damage response. Nucleic Acids Res. 45, 8129–8141 10.1093/nar/gkx565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Poirier G. G., de Murcia G., Jongstra-Bilen J., Niedergang C., and Mandel P. (1982) Poly(ADP-ribosyl)ation of polynucleosomes causes relaxation of chromatin structure. Proc. Natl. Acad. Sci. U.S.A. 79, 3423–3427 10.1073/pnas.79.11.3423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Leung A. K. (2014) Poly(ADP-ribose): an organizer of cellular architecture. J. Cell Biol. 205, 613–619 10.1083/jcb.201402114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Teloni F., and Altmeyer M. (2016) Readers of poly(ADP-ribose): designed to be fit for purpose. Nucleic Acids Res. 44, 993–1006 10.1093/nar/gkv1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hottiger M. O. (2015) Nuclear ADP-ribosylation and its role in chromatin plasticity, cell differentiation, and epigenetics. Annu. Rev. Biochem. 84, 227–263 10.1146/annurev-biochem-060614-034506 [DOI] [PubMed] [Google Scholar]
- 12. Zhang Y., Wang J., Ding M., and Yu Y. (2013) Site-specific characterization of the Asp- and Glu-ADP-ribosylated proteome. Nat. Methods 10, 981–984 10.1038/nmeth.2603 [DOI] [PubMed] [Google Scholar]
- 13. Zhen Y., Zhang Y., and Yu Y. (2017) A cell-line–specific atlas of PARP-mediated protein Asp/Glu-ADP-ribosylation in breast cancer. Cell Rep. 21, 2326–2337 10.1016/j.celrep.2017.10.106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Martello R., Leutert M., Jungmichel S., Bilan V., Larsen S. C., Young C., Hottiger M. O., and Nielsen M. L. (2016) Proteome-wide identification of the endogenous ADP-ribosylome of mammalian cells and tissue. Nat. Commun. 7, 12917 10.1038/ncomms12917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Daniels C. M., Ong S. E., and Leung A. K. (2015) The promise of proteomics for the study of ADP-ribosylation. Mol. Cell 58, 911–924 10.1016/j.molcel.2015.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Leutert M., Menzel S., Braren R., Rissiek B., Hopp A. K., Nowak K., Bisceglie L., Gehrig P., Li H., Zolkiewska A., Koch-Nolte F., and Hottiger M. O. (2018) Proteomic characterization of the heart and skeletal muscle reveals widespread arginine ADP-ribosylation by the ARTC1 ectoenzyme. Cell Rep. 24, 1916–1929.e5 10.1016/j.celrep.2018.07.048 [DOI] [PubMed] [Google Scholar]
- 17. Bonfiglio J. J., Fontana P., Zhang Q., Colby T., Gibbs-Seymour I., Atanassov I., Bartlett E., Zaja R., Ahel I., and Matic I. (2017) Serine ADP-ribosylation depends on HPF1. Mol. Cell 65, 932–940.e6 10.1016/j.molcel.2017.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Leidecker O., Bonfiglio J. J., Colby T., Zhang Q., Atanassov I., Zaja R., Palazzo L., Stockum A., Ahel I., and Matic I. (2016) Serine is a new target residue for endogenous ADP-ribosylation on histones. Nat. Chem. Biol. 12, 998–1000 10.1038/nchembio.2180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Palazzo L., Leidecker O., Prokhorova E., Dauben H., Matic I., and Ahel I. (2018) Serine is the major residue for ADP-ribosylation upon DNA damage. eLife 7, e34334 10.7554/eLife.34334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fontana P., Bonfiglio J. J., Palazzo L., Bartlett E., Matic I., and Ahel I. (2017) Serine ADP-ribosylation reversal by the hydrolase ARH3. eLife 6, e28533 10.7554/eLife.28533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Abplanalp J., Leutert M., Frugier E., Nowak K., Feurer R., Kato J., Kistemaker H. V. A., Filippov D. V., Moss J., Caflisch A., and Hottiger M. O. (2017) Proteomic analyses identify ARH3 as a serine mono-ADP-ribosylhydrolase. Nat. Commun. 8, 2055 10.1038/s41467-017-02253-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mashimo M., Kato J., and Moss J. (2014) Structure and function of the ARH family of ADP-ribosyl-acceptor hydrolases. DNA Repair 23, 88–94 10.1016/j.dnarep.2014.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kassab M. A., and Yu X. (2019) The role of dePARylation in DNA damage repair and cancer suppression. DNA Repair 76, 20–29 10.1016/j.dnarep.2019.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen S. H., and Yu X. (2019) Targeting dePARylation selectively suppresses DNA repair-defective and PARP inhibitor-resistant malignancies. Sci. Adv. 5, eaav4340 10.1126/sciadv.aav4340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Min W., and Wang Z. Q. (2009) Poly (ADP-ribose) glycohydrolase (PARG) and its therapeutic potential. Front. Biosci. 14, 1619–1626 10.2741/3329 [DOI] [PubMed] [Google Scholar]
- 26. Kaufmann T., Grishkovskaya I., Polyansky A. A., Kostrhon S., Kukolj E., Olek K. M., Herbert S., Beltzung E., Mechtler K., Peterbauer T., Gotzmann J., Zhang L. J., Hartl M., Zagrovic B., Elsayad K., et al. (2017) A novel non-canonical PIP-box mediates PARG interaction with PCNA. Nucleic Acids Res. 45, 9741–9759 10.1093/nar/gkx604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mortusewicz O., Fouquerel E., Amé J. C., Leonhardt H., and Schreiber V. (2011) PARG is recruited to DNA damage sites through poly(ADP-ribose)- and PCNA-dependent mechanisms. Nucleic Acids Res. 39, 5045–5056 10.1093/nar/gkr099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shirai H., Poetsch A. R., Gunji A., Maeda D., Fujimori H., Fujihara H., Yoshida T., Ogino H., and Masutani M. (2013) PARG dysfunction enhances DNA double strand break formation in S-phase after alkylation DNA damage and augments different cell death pathways. Cell Death Dis. 4, e656 10.1038/cddis.2013.133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lin K. Y., and Kraus W. L. (2017) PARP inhibitors for cancer therapy. Cell 169, 183 10.1016/j.cell.2017.03.034 [DOI] [PubMed] [Google Scholar]
- 30. Bryant H. E., Schultz N., Thomas H. D., Parker K. M., Flower D., Lopez E., Kyle S., Meuth M., Curtin N. J., and Helleday T. (2005) Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 434, 913–917 10.1038/nature03443 [DOI] [PubMed] [Google Scholar]
- 31. Farmer H., McCabe N., Lord C. J., Tutt A. N. J., Johnson D. A., Richardson T. B., Santarosa M., Dillon K. J., Hickson I., Knights C., Martin N. M. B., Jackson S. P., Smith G. C. M., and Ashworth A. (2005) Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434, 917–921 10.1038/nature03445 [DOI] [PubMed] [Google Scholar]
- 32. Wang M., Yuan Z., Xie R., Ma Y., Liu X., and Yu X. (2018) Structure–function analyses reveal the mechanism of the ARH3-dependent hydrolysis of ADP-ribosylation. J. Biol. Chem. 293, 14470–14480 10.1074/jbc.RA118.004284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pourfarjam Y., Ventura J., Kurinov I., Cho A., Moss J., and Kim I. K. (2018) Structure of human ADP-ribosyl-acceptor hydrolase 3 bound to ADP-ribose reveals a conformational switch that enables specific substrate recognition. J. Biol. Chem. 293, 12350–12359 10.1074/jbc.RA118.003586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rosenthal F., Feijs K. L., Frugier E., Bonalli M., Forst A. H., Imhof R., Winkler H. C., Fischer D., Caflisch A., Hassa P. O., Lüscher B., and Hottiger M. O. (2013) Macrodomain-containing proteins are new mono-ADP-ribosylhydrolases. Nat. Struct. Mol. Biol. 20, 502–507 10.1038/nsmb.2521 [DOI] [PubMed] [Google Scholar]
- 35. Breslin C., Hornyak P., Ridley A., Rulten S. L., Hanzlikova H., Oliver A. W., and Caldecott K. W. (2015) The XRCC1 phosphate-binding pocket binds poly (ADP-ribose) and is required for XRCC1 function. Nucleic Acids Res. 43, 6934–6944 10.1093/nar/gkv623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ray Chaudhuri A., and Nussenzweig A. (2017) The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat. Rev. Mol. Cell Biol. 18, 610–621 10.1038/nrm.2017.53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cheruiyot A., Paudyal S. C., Kim I. K., Sparks M., Ellenberger T., Piwnica-Worms H., and You Z. (2015) Poly(ADP-ribose)-binding promotes Exo1 damage recruitment and suppresses its nuclease activities. DNA Repair 35, 106–115 10.1016/j.dnarep.2015.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang F., Shi J., Chen S. H., Bian C., and Yu X. (2015) The PIN domain of EXO1 recognizes poly(ADP-ribose) in DNA damage response. Nucleic Acids Res. 43, 10782–10794 10.1093/nar/gkv939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pessetto Z. Y., Yan Y., Bessho T., and Natarajan A. (2012) Inhibition of BRCT(BRCA1)–phosphoprotein interaction enhances the cytotoxic effect of olaparib in breast cancer cells: a proof of concept study for synthetic lethal therapeutic option. Breast Cancer Res. Treat. 134, 511–517 10.1007/s10549-012-2079-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Moss J., Jacobson M. K., and Stanley S. J. (1985) Reversibility of arginine-specific mono(ADP-ribosyl)ation: identification in erythrocytes of an ADP-ribose-l-arginine cleavage enzyme. Proc. Natl. Acad. Sci. U.S.A. 82, 5603–5607 10.1073/pnas.82.17.5603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mueller-Dieckmann C., Kernstock S., Lisurek M., von Kries J. P., Haag F., Weiss M. S., and Koch-Nolte F. (2006) The structure of human ADP-ribosylhydrolase 3 (ARH3) provides insights into the reversibility of protein ADP-ribosylation. Proc. Natl. Acad. Sci. U.S.A. 103, 15026–15031 10.1073/pnas.0606762103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Oka S., Kato J., and Moss J. (2006) Identification and characterization of a mammalian 39-kDa poly(ADP-ribose) glycohydrolase. J. Biol. Chem. 281, 705–713 10.1074/jbc.M510290200 [DOI] [PubMed] [Google Scholar]
- 43. Li M., Lu L. Y., Yang C. Y., Wang S., and Yu X. (2013) The FHA and BRCT domains recognize ADP-ribosylation during DNA damage response. Genes Dev. 27, 1752–1768 10.1101/gad.226357.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lipinski C. A. (2004) Lead- and drug-like compounds: the rule-of-five revolution. Drug Discov. Today. Technol. 1, 337–341 10.1016/j.ddtec.2004.11.007 [DOI] [PubMed] [Google Scholar]
- 45. Baell J. B., and Holloway G. A. (2010) New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J. Med. Chem. 53, 2719–2740 10.1021/jm901137j [DOI] [PubMed] [Google Scholar]
- 46. Huth J. R., Mendoza R., Olejniczak E. T., Johnson R. W., Cothron D. A., Liu Y., Lerner C. G., Chen J., and Hajduk P. J. (2005) ALARM NMR: a rapid and robust experimental method to detect reactive false positives in biochemical screens. J. Am. Chem. Soc. 127, 217–224 10.1021/ja0455547 [DOI] [PubMed] [Google Scholar]
- 47. Langelier M. F., Planck J. L., Roy S., and Pascal J. M. (2012) Structural basis for DNA damage-dependent poly(ADP-ribosyl)ation by human PARP-1. Science 336, 728–732 10.1126/science.1216338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lan L., Nakajima S., Wei L. Z., Sun L. X., Hsieh C. L., Sobol R. W., Bruchez M., Van Houten B., Yasui A., and Levine A. S. (2014) Novel method for site-specific induction of oxidative DNA damage reveals differences in recruitment of repair proteins to heterochromatin and euchromatin. Nucleic Acids Res. 42, 2330–2345 10.1093/nar/gkt1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the article and its Supporting Information.