Abstract
Heterotrimeric G proteins are essential mediators of intracellular signaling of G protein–coupled receptors. The Gq/11 subfamily consists of Gq, G11, G14, and G16 proteins, of which all but G16 are inhibited by the structurally related natural products YM-254890 and FR900359. These inhibitors act by preventing the GDP/GTP exchange, which is necessary for activation of all G proteins. A homologous putative binding site for YM-254890/FR900359 can also be found in members of the other three G protein families, Gs, Gi/o, and G12/13, but none of the published analogs of YM-254890/FR900359 have shown any inhibitory activity for any of these. To explain why the YM-254890/FR900359 scaffold only inhibits Gq/11/14, the present study delineated the molecular selectivity determinants by exchanging amino acid residues in the YM-254890/FR900359–binding site in Gq and Gs. We found that the activity of a Gs mutant with a Gq-like binding site for YM-254890/FR900359 can be inhibited by FR900359, and a minimum of three mutations are necessary to introduce inhibition in Gs. In all, this suggests that although the YM-254890/FR900359 scaffold has proven unsuccessful to derive Gs, Gi/o, and G12/13 inhibitors, the mechanism of inhibition between families of G proteins is conserved, opening up the possibility of targeting by other, novel inhibitor scaffolds. In lack of a selective Gαs inhibitor, FR900359-sensitive Gαs mutants may prove useful in studies where delicate control over Gαs signaling would be of the essence.
Keywords: FR900359, YM-254890, G protein inhibitor, Gs, Gq, G protein, inhibitor, G protein-coupled receptor (GPCR), site-directed mutagenesis, molecular pharmacology
G protein–coupled receptors (GPCRs) comprise the largest family of membrane-bound receptors in the human genome, with ∼800 genes (around half of which are olfactory) (1). GPCRs regulate a range of important physiological functions in humans (e.g. cardiac function (2), vascular tonus (3), immune function (4), metabolism (5), and neurotransmission (6)). Consequently, they have long been important drug targets, and it is estimated that 34% of all drugs approved by the United States Food and Drug Administration target GPCRs (7).
GPCR signaling is mainly mediated by heterotrimeric G protein complexes, a class of intracellular guanine nucleotide–binding proteins comprised of Gα, Gβ, and Gγ subunits, which can be precoupled to the receptor (8–10) before agonist binding. Upon receptor activation, a conformational change causes the Gα subunit to exchange the prebound GDP with GTP, which causes the heterotrimeric Gαβγ complex to dissociate (11, 12). The Gα subunit is the major effector of downstream signaling, but the Gβγ dimer can also elicit downstream effects (13). G protein signaling is eventually attenuated by hydrolysis of GTP to GDP. Regulators of G protein signaling (RGS) accelerate the intrinsic GTPase activity of the Gα subunit, and GTP hydrolysis restores the Gαβγ complex (11). There are 16 Gα subunit–encoding genes in the human genome, and based on their protein sequence and function, they can be divided into four families: Gs, Gi/o, Gq/11, and G12/13 (11, 14).
Because virtually all known GPCRs couple to and elicit effects via one or more of the four families of G proteins, the elucidation of which G protein(s) a GPCR couples to provides essential knowledge in understanding the physiological function of the receptor. Modulators of G protein activity, therefore, constitute very important and powerful tools for studies of the numerous cellular and physiological responses that depend on G protein activity. As an example, identification of the G protein(s) involved in the observed response for a phenotypic GPCR assay, such as Corning Epic dynamic mass redistribution, can be deconvoluted with selective G protein inhibitors (15, 16). A selective inhibitor has been shown to inhibit tumors driven by oncogenic mutations in Gαq and Gα11; thus, G protein inhibitors might also have therapeutic potential (17, 18). Therefore, there is great interest in the development of selective modulators and inhibitors of all four families of G proteins.
The well-known pharmacological tool compounds capable of modulating G protein function are the two bacterial toxins, cholera toxin and pertussis toxin, that were discovered in the early 1980s and found to activate Gs and inhibit Gi/o, respectively (19, 20). Unfortunately, the complexity and size of the bacterial toxins make them poor starting points for derivation of modulators for other G protein families through rational design by conventional medicinal chemistry approaches. However, a new class of natural G protein inhibitors emerged in the early 2000s when several structurally similar depsipeptides were isolated from the soil bacteria Chromobacterium sp. QS3666 and found to selectively inhibit the Gq/11 family with nanomolar potency (21, 22). The most potent depsipeptide, YM-254890, has been shown by biochemical approaches and structural studies to inhibit exchange of GDP with GTP. The inhibition is achieved by YM-254890 binding into a hydrophobic cleft between the Linker 1 and Switch I loops, which connect the GTPase and helical domains of the Gα subunit (23) (Fig. 1A).
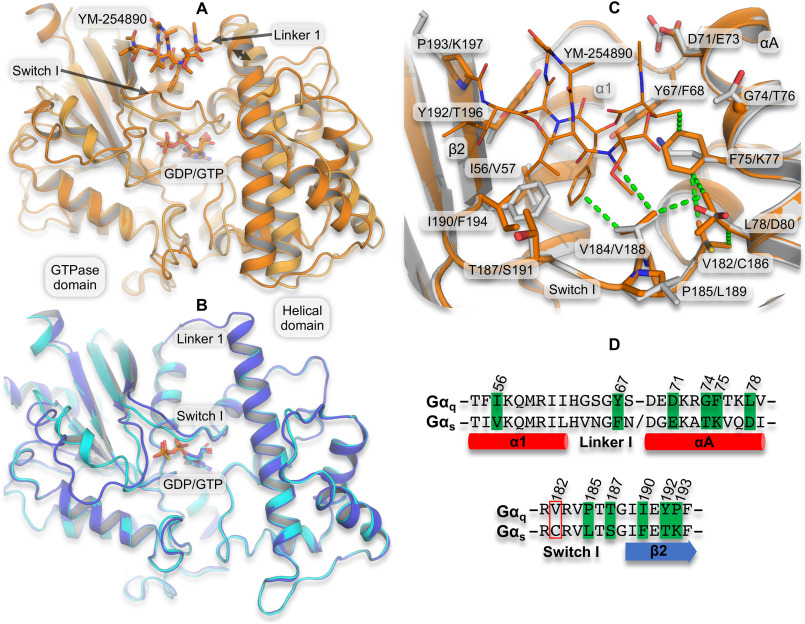
Figure 1.
Structural effect of inhibitor binding and differences in the YM-254890/FR900359 binding site between Gq and Gs. A, superimposition of YM-254890–bound Gαq (dark orange carbon atoms and cartoon representation, YM-254890 and GDP as sticks; PDB code 3AH8) and the GDP/Al4+-activated Gαq without inhibitor (light orange carbon atoms and cartoon representation; PDB code 5DO9) based on the GTPase domain shows a 2.8-Å inhibitor-induced shift in the relative location of the helical domain (double arrow). B, superimposition of the inactive GDP-bound Gs (purple carbon atoms and cartoon; PDB code 6EG8) and the GTPγS-activated Gs (cyan carbon atoms and cartoon; PDB code 1AZT) shows practically identical relative location of the GTPase and helical domains. C, the depsipeptide-binding site in YM-254890-bound Gαq (as in A) superimposed on a homology model of Gαs (gray carbon atoms and cartoon) highlighting (sticks) the binding site residues (within 6 Å and side chains pointing toward YM-254890) that differ between Gαq and Gαs. Val-182/Cys-186 is included, as it is part of a hydrophobic network of amino acids having close vdW contact with each other and with YM-254890 (dotted lines). Labels indicate one-letter amino acid codes and numbers in Gq/Gs. D, sequence alignment of the YM-254890–binding site region in Gαq and Gαs, highlighting differing binding site residues (green). Residues in the red box are not part of the binding site but the hydrophobic network. Sequence numbers are from Gq.
FR900359, which is a natural structural analog of YM-254890, isolated from the leaves of the East Asian plant Ardisia crenata, also inhibits the Gαq, Gα11, and Gα14—but not Gα15/16—members of the Gq/11 family (24). Despite the Gαq/11/14 selectivity, inhibition by FR900359 has been introduced in Gαi1 (25) and Gα16 (26) by mutations, and sequence diversity between the G protein families may thus provide an avenue for development of novel selective inhibitors for these targets.
Other small-molecule inhibitors of G proteins have been discovered, notably the imidazopirazines; BIM-46174 (27) and the more stable BIM-46187 (28). The imidazopirazines are cell-permeable nonselective G protein inhibitors (29) that stabilize the non-guanine nucleotide–bound form of the G proteins (30). Additionally, the anthelmintic drug suramin and its analogs are inhibitors with partial selectivity for Gs, but also act on Gi/o (31, 32), and are not well-suited as tool compounds, due to their lack of cell membrane permeability (29).
Consequently, the FR900359/YM-254890 scaffold has been considered the best lead in the derivatization of selective inhibitors for other G protein families. Because the extent to which the natural plant-derived FR900359 molecule could be modified was very limited (33), a synthetic pathway was needed to fully probe the scaffold's inhibitory potential at other G proteins. We recently reported the first complete de novo total synthesis of FR900359 and YM-254890 along with several novel analogs (34) and have since examined the structure-activity of the YM-254890 scaffold through the synthesis of analogs generated to cover virtually all regions of the chemical structure (35–37). The 34 hitherto published analogs were characterized in assays for the Gq/11, Gi/o, and Gs families of G proteins but were intriguingly found to only possess inhibitory activity for the Gq/11 proteins (34–37).
In the present study, we investigate the molecular determinants for the Gq selectivity of FR900359 by swapping selected Gαs and Gαq residues in the YM-254890–binding site by mutagenesis (23). The inhibitory activity of FR900359 was then assessed on each of these mutant G proteins in HEK293 cells, where the corresponding endogenous G protein family had been knocked out by CRISPR/Cas9 genome editing, providing a background-free system for investigating the effects of the mutations. Herein we report the key amino acid residues responsible for FR900359's selectivity at Gq compared with Gs, as well as the first reported Gs proteins engineered to be amenable to inhibition by FR900359.
Results
YM-254890 putatively stabilizes the inactive Gαq state through a hydrophobic network
To evaluate G protein conformational changes associated with depsipeptide inhibitor binding, we compared the structures of the inactive GDP-bound heterotrimeric YM-254890–bound Gq protein complex (23) and the -activated monomeric Gαq (38). First of all, comparison of the latter with other active state structures (e.g. with phospholipase C) (39) displays very similar conformations (not shown), ruling out influence from the co-crystallized effector, RGS8. Second, superposition of the inactive and active Gq structures based on the structurally conserved parts of the GTPase domain gave a very good fit, showing that this domain is a rigid entity and can be used as a reference domain when comparing Gα conformations (Fig. 1A). Based on this, we observe a rather large difference between backbone atoms in the helices of the helical domain. Thus, relative to the GTP-bound monomeric structure, the GDP– and YM-254890–bound heterotrimeric complex apparently entails a closure of the two Gαq domains moving 2.8 Å closer together as measured across the YM-254890–binding site. The Switch I loop, which is in direct contact with YM-254890 (23) and connects the two domains, also displays different conformations, and both of these changes provide a tighter fit to the depsipeptide inhibitor (Fig. 1A). In the absence of an inactive state Gq complex without YM-254890, we performed a similar comparison of active and inactive state heterotrimeric structures of Gi and Gs to get an impression of how much of this conformational change is caused by binding of YM-254890. A much smaller displacement of the helical domain is observed for Gαi1 (40, 41) (not shown), whereas there is basically no domain movement when comparing the very recent inactive heterotrimeric Gs structure (12) with an active state monomeric Gs structure (42) (Fig. 1B). Crystal structures only show snapshots of the dynamic systems and may not reveal the full picture. However, these differences indicate that binding of YM-254890, and not the presence of GDP or Gβγ, causes the marked conformational change between the GTPase and helical domains plus the switch I loops of the inactive and active Gαq structures. Thus, as the basis for our attempt to introduce FR900359 inhibition in Gs by means of mutagenesis, we hypothesize that FR900359 acts similarly to YM-254890 and that Gαs can undergo a similar conformational change as Gαq.
Focusing on the interactions between Gαq and YM-254890, we identified a small network of hydrophobic residues, Phe-75, Leu-78, Val-182, and Val-184, displaying van der Waals (vdW) contacts to each other and parts of YM-254890 (Fig. 1C). In the Gαq monomeric active structure, this “hydrophobic cluster” is not as tight, which is best illustrated by the shortest distance between Phe-75 and Val-184 being 6.1 Å, whereas it is 3.7 Å in the heterotrimeric YM-254890–bound structure. Thus, the hydrophobic interactions to YM-254890 take part in tethering Val-184 in a position that promotes the Switch I loop conformation and likely also promotes the observed conformational change of the inhibitor-bound structure (23). The structural differences between YM-254890 and FR900359 include an isopropyl instead of the methyl, which is in close contact with residues of the hydrophobic network (Fig. 1C). As these hydrophobic contacts play the largest role for the inhibition by FR900359 (26), this inhibitor was selected for the mutational studies presented in this work.
Three differing binding site positions determine Gq versus Gs FR900359 selectivity
To outline the differences between the established depsipeptide-binding site in Gαq and the putative inhibitor site in Gαs, we constructed a homology model of Gαs in complex with FR900359 (supporting information) and compared the amino acids with side chains that do, or potentially could, interact with YM-254890 and/or FR900359 (within 6 Å). Mapping these positions in a sequence alignment, we found that 11 positions near the inhibitor differ between the two Gα protein subtypes (Fig. 1, C and D). To conduct a pharmacological evaluation of the role of these differing residues in FR900359 inhibition, we employed CRISPR/Cas9 genome-edited HEK293 cell lines lacking Gαq/11 or Gαs genes (referred to as ΔGq/11-HEK293 and ΔGs-HEK293, respectively). These cell lines have been described previously in the literature (see “Experimental procedures”) and provided cellular expression systems to investigate the mutated G proteins without interference from endogenously expressed WT G proteins. We confirmed the lack of G protein activity by observing no response for the endogenously expressed muscarinic M3 (Gαq/11-coupled) and adrenergic β2 (Gαs-coupled) receptors in the ΔGq/11-HEK293 and ΔGs-HEK293 cell lines, respectively (Fig. S1, A and B). However, when transfected with WT Gαq and Gαs, a robust agonist response can be reintroduced (Fig. S1, A and B). These findings were corroborated by Western blotting analysis, wherein no specific band could be observed for the missing G protein in their respective cell line, but similarly G protein–specific bands could be reintroduced by transfection (Fig. S2). Using this setup, we next showed that all G protein mutants generated in this study, both in Gαq and Gαs, resulted in robust agonist-induced responses relative to baseline when activated by the endogenously expressed M3 or β2 receptors and displayed sufficient expression, making them suitable to investigate FR900359 inhibition (Figs. S3 and S4). Additionally, isoproterenol concentration-response curves from β2-adrenergic receptors coupling to WT Gs, Gs-11, and other Gs mutants confirm effective mediation of cAMP production (Fig. S5).
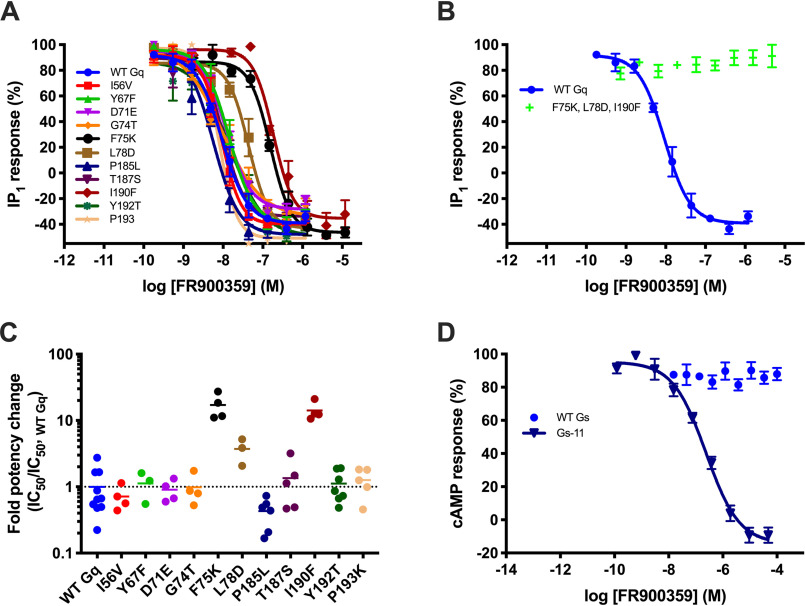
Initially, each of the aforementioned 11 positions in Gαq was individually exchanged with the corresponding amino acid from Gαs to determine which positions were important for binding of FR900359. The single-point mutations of amino acids with limited or no direct contacts to the inhibitor in the Gαq-YM-254890 structure (G74T, P185L, Y192T, and P193K) as well as mutations substituting amino acids able to make similar interactions (I56V, Y67F, D71E, and T187S) showed subtle or no effects on the inhibitory potency of FR900359 on carbachol-induced M3 receptor-mediated IP1 generation (Figs. 1 (C and D) and 2 (A and C) and Table 1). These observations agree with a lack of, or similar, interactions in the FR900359-Gαs model. Only three of the Gαq protein mutations resulted in >3-fold reductions in FR900359 inhibitory potency (F75K, 20.4-fold; L78D, 4.5-fold; I190F, 17.6-fold), and combining these three mutations markedly decreased the potency of FR900359 (Fig. 2B and Table 1).
Figure 2.
Transfer of the Gq-binding site to Gs results in complete inhibition by FR900359. Mutant Gq and Gs proteins were expressed in ΔGq/11-HEK293 and ΔGs-HEK293 cells, respectively, to enable assessment of their potential inhibition by FR900359. After treatment with FR900359, the cells were stimulated with either carbachol (15 μm) or isoproterenol (5 nm), activating the endogenous Gq-coupled muscarinic acetylcholine receptor M3 or the endogenous Gs-coupled β2-adrenergic receptor, respectively. A and B, normalized concentration-inhibition curves of FR900359 on WT Gq and derived Gq mutants. C, -fold change in FR900359 potency for Gq mutants compared with WT. D, normalized concentration-inhibition curves of FR900359 on WT Gs and the Gs-11 mutant (V57I, F68Y, E73D, T76G, K77F, D80L, L189P, S191T, F194I, T196Y, K197P). A, B, and D, each data point represents the mean value for that data point across experiments (n = 3–5). Error bars, S.E. C, each data point represents an independent experiment (n = 3–10), and the bars show the corresponding mean.
Table 1.
Pharmacological data for inhibition by FR900359 on WT Gαq and Gαq mutants
The G proteins were expressed in ΔGq/11-HEK293 cells to enable assessment of their potential inhibition by FR900359. The cells were stimulated with carbachol (15 μm) to activate the endogenous Gq-coupled muscarinic acetylcholine receptor M3 after treatment with FR900359 (associated concentration inhibition curves shown in Fig. 2 (A and B)).
| WT and mutation(s) | FR900359 pIC50 ± S.E. | n | FR900359 − IC50 (nm) (95% confidence interval) |
|---|---|---|---|
| Gq WT | 8.03 ± 0.10 | 10 | 9.31 (5.48–15.8) |
| I56V | 8.10 ± 0.09 | 4 | 7.94 (4.22–15.28) |
| Y67F | 7.91 ± 0.14 | 3 | 12.3 (3.08–49.1) |
| D71E | 7.99 ± 0.08 | 4 | 10.2 (5.69–18.6) |
| G74T | 7.97 ± 0.11 | 4 | 10.7 (4.84–23.6) |
| F75K | 6.72 ± 0.09 | 4 | 190 (97.1–375) |
| L78D | 7.38 ± 0.12 | 3 | 41.7 (12.9–133) |
| P185L | 8.29 ± 0.11 | 5 | 5.12 (2.56–10.2) |
| T187S | 8.03 ± 0.13 | 4 | 9.44 (3.74–23.8) |
| I190F | 6.79 ± 0.06 | 4 | 164 (102–263) |
| Y192T | 7.82 ± 0.10 | 3 | 15.3 (5.86–40.0) |
| P193K | 8.01 ± 0.13 | 3 | 9.89 (2.65–36.9) |
| F75K, L78D, I190F | 3 | >5,000 |
The Gαq-YM-254890 crystal structure suggests a rationale, as both Phe-75 and Leu-78 are part of the hydrophobic network in the binding pocket and Ile-190 as well as Phe-75 are in close contact with the inhibitor (i.e. shortest distances are 3.8 and 3.7 Å, respectively) (Fig. 1C). Introduction of charged amino acids into positions 75 and 78 is predicted from the FR900359-Gαs model to weaken the hydrophobic network (Fig. 1C) and destabilize the inactive GDP-bound state. Combined with changing the shape and nature of the binding pocket, these structural observations can explain the decreased potency caused by the single-point mutations as well as the abolished effect of the triple mutant, where the hydrophobic network is significantly weakened by two simultaneous substitutions of hydrophobic residues for hydrophilic ones. These results underline the importance of the hydrophobic network and suggest that these positions are determinants for the selectivity of FR900359 for Gαq versus Gαs.
Gs is inhibited by FR900359 upon transfer of binding site residues from Gq
In an attempt to introduce a functional depsipeptide-binding site in Gαs, we simultaneously exchanged all 11 differing residue positions in Gαs (Gs-11) with the corresponding Gαq residues. Whereas we observed no inhibition of isoproterenol-induced β2 receptor–mediated cyclic adenosine monophosphate (cAMP) generation by FR900359 at concentrations up to 100 μm for WT Gαs, we observed complete inhibition of the Gs-11 mutant with an IC50 of 231 nm, which is ∼25-fold higher than the IC50 of FR900359 for WT Gαq (Fig. 2D and Table 2). These results indicate that the same conformational change accompanying YM-254890 binding in Gαq is also possible in Gαs with (some of) the introduced mutations likely lowering the energy barrier by enforcing a similar hydrophobic network as observed in Gq (Fig. 1C).
Table 2.
Pharmacological data for FR900359 inhibition of the Gs-11 (V57I, F68Y, E73D, T76G, K77F, D80L, L189P, S191T, F194I, T196Y, and K197P) and derived restored mutants
The G proteins were expressed in ΔGs-HEK293 cells to enable assessment of their potential inhibition by FR900359. The cells were stimulated with isoproterenol (5 nm) to activate the endogenous Gs-coupled β2-adrenergic receptor after treatment with FR900359 (associated concentration inhibition curves shown in Fig. 3 (A and B)).
| Name/Mutation(s) | FR900359 pIC50 ± S.E. | n | FR900359 − IC50 (nm) (95% confidence interval) |
|---|---|---|---|
| Gs-11/V57I, F68Y, E73D, T76G, K77F, D80L, L189P, S191T, F194I, T196Y, K197P | 6.64 ± 0.07 | 5 | 231 (146–363) |
| Val-57–restored/F68Y, E73D, T76G, K77F, D80L, L189P, S191T, F194I, T196Y, K197P | 6.55 ± 0.08 | 3 | 282 (132–604) |
| Phe-68–restored/V57I, E73D, T76G, K77F, D80L, L189P, S191T, F194I, T196Y, K197P | 6.26 ± 0.14 | 3 | 546 (135–2200) |
| Glu-73–restored/V57I, F68Y, T76G, K77F, D80L, L189P, S191T, F194I, T196Y, K197P | 6.85 ± 0.15 | 3 | 143 (33.9–601) |
| Thr-76–restored/V57I, F68Y, E73D, K77F, D80L, L189P, S191T, F194I, T196Y, K197P | 5.94 ± 0.17 | 3 | 1,140 (213–6,080) |
| Lys-77–restored/V57I, F68Y, E73D, T76G, D80L, L189P, S191T, F194I, T196Y, K197P | 7.87 ± 0.20 | 19 | 13.5 (9.29–36.3) |
| 4.84 ± 0.08 | 14,600 (9,840–21,500)a,b | ||
| Asp-80–restored/V57I, F68Y, E73D, T76G, K77F, L189P, S191T, F194I, T196Y, K197P | 5.55 ± 0.15 | 3 | 2,850 (656–12,400) |
| Leu-189–restored/V57I, F68Y, E73D, T76G, K77F, D80L, S191T, F194I, T196Y, K197P | 6.59 ± 0.10 | 3 | 255 (93.3–698) |
| Ser-191–restored/V57I, F68Y, E73D, T76G, K77F, D80L, L189P, F194I, T196Y, K197P | 6.52 ± 0.11 | 7 | 302 (160–570) |
| Phe-194–restored/V57I, F68Y, E73D, T76G, K77F, D80L, L189P, S191T, T196Y, K197P | 4.36 ± 0.07 | 3 | 43,400 (20,800–90,200)b |
| Thr-196–restored/V57I, F68Y, E73D, T76G, K77F, D80L, L189P, S191T, F194I, K197P | 5.05 ± 0.09 | 4 | 8,970 (4,670–17,300)b |
| Lys-197–restored/V57I, F68Y, E73D, T76G, K77F, D80L, L189P, S191T, F194I, T196Y | 5.75 ± 0.07 | 3 | 1,780 (935–3,400) |
a Biphasic inhibition observed.
b Bottom-plateau of concentration-inhibition curve constrained to buffer level to allow curve fitting of low-potency mutant.
Gq and Gs-11 display differential sensitivity to FR900359 inhibition upon swapping of binding site residues
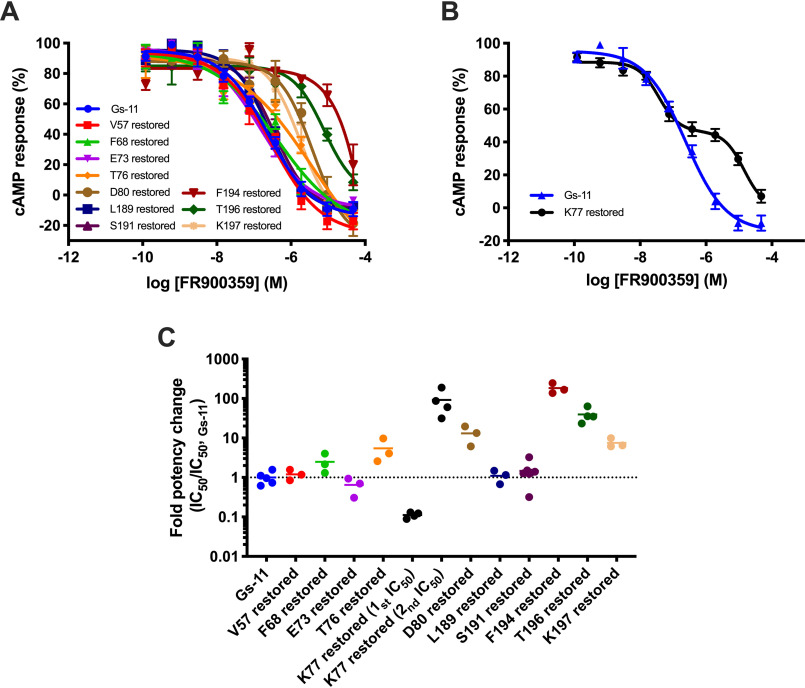
To determine the individual importance of each binding site residue, we prepared Gαs mutants restoring each of the 11 mutations in Gs-11 (above) one at a time. All of the 11 restored Gαs mutants could be inhibited by FR900359 with IC50 values ranging between 13.5 nm and 43.4 μm, and 10 of these showed the expected sigmoidal inhibition curves, although a few were not fully inhibited at the highest tested concentrations (Fig. 3 (A and B) and Table 2). Five of the Gαs-restoring mutations showed subtle or no effect (<3-fold: Val-57–, Phe-68–, Glu-73–, Leu-189–, and Ser-191–restored) compared with Gs-11, which, as for the corresponding Gq mutations (Table 1), can be explained by exchange of similar amino acids and/or having limited interactions to the inhibitor. Restoring Thr-76, Asp-80, and Lys-197 resulted in modestly larger effects than in Gαq, whereas the Lys-77–, Phe-194–, and Thr-196–restored mutants displayed significantly altered potencies or inhibition curves. Surprisingly, restoring the Lys in position 77 (Phe in Gαq) resulted in an atypical biphasic inhibition curve with two phases of equal size but a striking ∼1,000-fold difference in potencies (i.e. 13.5 nm and 14.6 μm) (Fig. 3B and Table 2). This corresponds to 17- and 63-fold higher and lower potencies, respectively, relative to the IC50 of 231 nm of the Gs-11 mutant (Fig. 3C and Table 2). This surprising observation signifies that there may be fundamental differences in the mechanism and/or kinetics of activation and inhibition between the two G protein subtypes.
Figure 3.
The binding site amenable to FR900359 inhibition in Gs introduced by the 11 mutations is sensitive to small changes. To assess their contribution to the inhibitory activity of FR900359, each mutated amino acid comprising the Gs-11 mutant (V57I, F68Y, E73D, T76G, K77F, D80L, L189P, S191T, F194I, T196Y, K197P) was restored in the Gs-11 backbone individually, thereby generating mutants with 10 mutations, referred to by their reintroduced residue (e.g. V57 restored for the mutant without the V57I mutation). The Gs-11 mutant and restored mutants were then expressed in ΔGs-HEK293 cells to enable assessment of their potential inhibition by FR900359. The cells were stimulated with isoproterenol (5 nm) to activate the endogenous Gs-coupled β2-adrenergic receptor after treatment with FR900359. A, normalized concentration-inhibition curves of FR900359 on the restored mutants. Each data point represents the mean value for that data point across experiments (n = 3–7). Error bars, S.E. B, normalized concentration-inhibition curve obtained for FR900359 on the Lys-77–restored mutant showing the biphasic inhibition obtained for this mutant. Each data point represents the mean value across experiments (n = 19). Error bars, S.E. The inhibition curve for the Gs-11 mutant, shown in A, is included for comparison. C, -Fold change in FR900359 potency for the restored mutants compared with the Gs-11 mutant. Each data point represents an independent experiment (n = 3–5) and the bars show the corresponding mean.
A minimum of three mutations in the hydrophobic network or binding site introduce FR900359 inhibition in Gαs
Restoring the individual binding site residues in Gαs revealed which residues are most important for FR900359 activity, but it was pertinent to identify the minimal set of mutations necessary to attain FR900359 inhibition. As an exhaustive combination of single-point mutations was not feasible, we combined the Gαs mutations with the largest effect on inhibition potency (Fig. 3C) and included C186V from the hydrophobic network (Fig. 1, C and D), which was hypothesized to be important for stabilizing the inactive FR900359-binding state. We found that a minimum of three simultaneous mutations in WT Gαs were required to obtain FR900359 inhibition, K77F/D80L/C186V or K77F/F194I/T196Y, although they only resulted in partial inhibition of 42 and 56%, respectively, compared with the full inhibition observed for the Gs-11 mutant (Table 3 and Fig. S6). Full inhibition by FR900359 was observed with a combination of the four restored mutations with the largest effect, K77F/D80L/F194I/T196Y, as well as with two combinations of five mutations (the addition of K197P or C186V; Table 3). Common for these three Gαs mutants is that they display biphasic inhibition curves, as was observed for the Lys-77–restored Gs-11 mutant (Fig. S6C).
Table 3.
Pharmacological data for the inhibition by FR900359 on Gs mutants designed with select high-impact mutations, based on the results of the restored mutants (see Table 2)
The Gs mutants were expressed in ΔGs-HEK293 cells to enable assessment of their potential inhibition by FR900359. The cells were stimulated with isoproterenol (5 nm) to activate the endogenous Gs-coupled β2-adrenergic receptor after treatment with FR900359 (associated concentration-inhibition curves in Fig. S6 (A–C)). The extent of the high-potency inhibition component for partial and biphasic inhibition curves is given in parentheses as a percentage of full inhibition. The Lys-77–restored mutant has been included for comparison.
| Name/Mutation(s) | FR900359 pIC50 ± S.E. | n | FR900359 IC50 (nm) (95% confidence interval) |
|---|---|---|---|
| Gs WT | 3 | >100,000 | |
| T76G, F194I | 3 | >100,000 | |
| T76G, D80L, F194I | 3 | >100,000 | |
| T76G, D80L, F194I, T196Y | 3 | >100,000 | |
| T76G, D80L, F194I, K197P | 3 | >100,000 | |
| T76G, K77F, D80L, F194I | 3 | >100,000 | |
| K77F, D80L, C186V | 7.31 ± 0.14 | 6 | 49.0 (21.2–111) (42%) |
| K77F, D80L, C186V, F194I | 7.01 ± 0.16 | 5 | 97.5 (34.0–279) (47%) |
| K77F, D80L, F194I, K197P | 7.41 ± 0.06 | 3 | 38.9 (22.2–68.2) (49%) |
| K77F, F194I, T196Y | 7.12 ± 0.08 | 8 | 76.0 (50.0–116) (56%) |
| K77F, D80L, F194I, T196Y | 7.17 ± 0.11 | 8 | 67.0 (37.6–119) (48%) |
| 4.77 ± 0.08 | 16,900 (11,100–25,600)a,b | ||
| K77F, D80L, F194I, T196Y, K197P | 7.18 ± 0.09 | 8 | 65.8 (40.6–106) (56%) |
| 5.04 ± 0.01 | 9,162 (8,490–9,890)a,b | ||
| K77F, D80L, C186V, F194I, T196Y | 7.07 ± 0.09 | 7 | 84.5 (50.8–141) (50%) |
| 4.86 ± 0.10 | 11,041 (5,350–19,200)a,b | ||
| Lys-77–restored/V57I, F68Y, E73D, T76G, D80L, L189P, S191T, F194I, T196Y, K197P | 7.87 ± 0.20 | 1 | 13.5 (9.29–36.3) (52%) |
| 4.84 ± 0.08 | 9 | 14,600 (9,840–21,500)a,b |
a Biphasic inhibition observed.
b Bottom-plateau of concentration-inhibition curves constrained to buffer level to allow curve fitting of low potent mutant.
Another interesting observation is that the T76G mutation may have a negative effect on depsipeptide inhibition, as all the Gαs mutants containing this residue replacement are insensitive to FR900359 (Table 3). Our Gαs model indicates that the side-chain methyl on Thr-76 actually can take part in the hydrophobic network, and, although this position contains a Gly in Gαq, the removal of the side chain weakens the hydrophobic network in Gαs resulting in reduced inhibition.
Partial conversion to the Gαq site results in full biphasic or partial inhibition of Gαs
Of the Gαs mutations that do not fully convert the inhibitor site to that of Gαq, four mutants displayed high-potency partial inhibition (IC50 = 38.9–97.5 nm, 42–56%; Fig. S6B). An additional four mutants, including the Gαs Lys-77–restored mutant, showed biphasic inhibition with a 130–1,000-fold difference between the high- and low-potency components (IC50-1 = 13.5–84.5 nm and IC50-2 = 9.2–16.9 μm; Fig. S6C and Table 3).
To ensure that a monophasic curve fit would not describe the putative biphasic inhibition curves better, all FR900359 inhibition curves were subjected to a statistical comparison-of-fits F test in GraphPad Prism (four parameters versus biphasic). Inhibition curves described as biphasic all had significant p values of <0.0001. Interestingly, the high-potency component from all the partial or biphasic inhibition curves represents only 7-fold variation, and the low-potency component from the biphasic curves varies <2-fold (Fig. S6D and Table 3). Additionally, the biphasic curves observed for these mutants were not limited to FR900359, but also observed for YM-254890 (Fig. S7).
Discussion
There is still an unmet need for G protein inhibitors targeting Gαs (and Gα12/13) as pharmacological tools to specifically block signaling from these signaling proteins. The available information from the YM-254890-Gαq structure has so far not been sufficient to clarify requirements for binding and function of similar depsipeptides as potential inhibitors for the other G protein subtypes. However, within the Gq/11 family, we recently introduced both YM-254890 and FR900359 inhibition into Gα16 (26), the only insusceptible subtype within the Gq/11 family (24). For Gα16 only five point mutations were required to introduce Gq-like inhibition due to higher conservation of the depsipeptide-binding pocket. Even though Gαs is one of the most evolutionarily distant subtypes from Gαq (43) and consequently shows larger differences in the depsipeptide-binding site, both in amino acid numbers and properties, the same approach proved successful. Similar to recent results for the Gi family (25), the present study clearly shows that GPCR signaling through Gαs can be inhibited via a site equivalent to the depsipeptide-binding site in the Gq/11 family. The underlying mechanism by stabilizing the inactive state and hampering nucleotide exchange (23, 24) very likely also applies to Gαs. Although not the target of this study, it is tempting to speculate that the same may apply for Gα12/13 due to the similarity in structure and activation mechanism between G protein families (44). However, we show here and in a previous study (26) that the hydrophobic network is important for this inhibition, and a positively charged amino acid (Lys/Arg in Gα12/13) in the position corresponding to Val-184 and Val-188 in this network of Gαq and Gαs, respectively, contradicts this.
Of the 11 differing residue positions among those comprising the inhibitor-binding site in Gαq, we identify three positions that are crucial for Gαq versus Gαs selectivity that abolish FR900359-mediated inhibition when exchanged simultaneously. Conversely, by introducing all 11, or as few as three, Gαq-binding site amino acids into Gαs we clearly show that Gαs can mechanistically be inhibited through the analogue-binding site.
Having introduced the depsipeptide inhibitor–binding site from Gαq into Gαs, we expected similar inhibition profiles and effects of the amino acid exchanges at the same sites. Apparently, FR900359 inhibition of the Gs-11 mutant is much more sensitive to single-point mutations than WT Gαq, which cannot readily be explained. However, in the YM-254890-Gq crystal structure, all of the six residues affecting the Gs-11 mutant the most are part of the hydrophobic network and/or, to a varying degree, show vdW interactions to the inhibitor (Fig. 1C). Moreover, it is common for all six restored mutations that the reintroduced amino acids are more hydrophilic/polar compared with those of the Gαq-binding site (Fig. 1D) and are predicted from the FR900359-Gαs model to weaken the hydrophobic network and the interactions to the inhibitor. The larger effect of reversing hydrophilicity into the Gs-11 mutant clearly demonstrates that the G protein background of the positional single-point mutations plays a significant role, and the hydrophobic nature of the site is more important for the function of FR900359 in the modified Gαs versus the Gαq protein. This is in line with one of the triple mutations that constitute the minimum change to introduce inhibition in Gαs (i.e. the K77F/D80L/C186V mutant), introducing the hydrophobic network from Gαq. In fact, the mutations that introduce FR900359 inhibition in Gαs all increase hydrophobicity of the binding site either with direct vdW interactions to the hydrophobic parts of the inhibitor or as part of the hydrophobic network. Thus, the hydrophilic nature of the binding site and disruption of the hydrophobic network in Gαs provide a poor fit for FR900359, as confirmed by our homology model, and may also destabilize the conformation necessary for binding of YM-254890/FR900359 (Fig. 1A). Additionally, this could be correlated to the different rate constants for deactivation and GTP hydrolysis for the Gαq and Gαs subtypes, which can be heavily affected by, for example, GTPase-activating proteins (45) and, thus, maybe also point mutations.
Whereas introduction of the full Gαq-binding site into Gαs allowed for FR900359 to fully inhibit the agonist response in a normal monophasic fashion, combinations of the 3–5 reverse mutations with the largest effect in Gαs resulted in partial or biphasic inhibition. The phenomenon of biphasic inhibition curves and two different potencies is only observed in mutant receptors, making the relevance for inhibition of WT Gαs speculative. It could, however, indicate a general mechanistic feature of inhibition of Gαs-mediated GPCR signaling through the depsipeptide-binding site, which would be interesting to explore in more detail in future studies by, for example, performing nucleotide exchange assays on purified Gαs mutants to determine whether their biphasic profile is intrinsic to the mutant proteins or depends on association with other proteins, such as the GPCR or Gβγ.
The most straightforward interpretation of a biphasic concentration-response curve is the presence of two different binding sites for the tested compounds with different binding affinities as observed for other receptor targets (46–48). However, because all the mutated positions that introduce and affect the inhibition by FR900359 are located in the same putative binding site, this explanation most likely does not apply to the mutated Gαs proteins. Alternative explanations could be that FR900359 binds to the same binding site in two different states or populations of the Gαs protein (i.e. free versus receptor-precoupled heterotrimeric G protein, free Gα versus Gβγ-complexed Gα, or GDP-bound versus GTP-bound Gα), which due to conformational changes of the binding site could display significantly different binding preferences for the inhibitor. The presence of two different inhibitor conformations separated by a large energy barrier and with different affinities for the same binding site could in theory also result in biphasic curves, but then it should also be the case for Gαq. Irrespective of the reason for the biphasic behavior, the nearly constant level of the high-potency inhibition component for the partial and biphasic inhibition curves (42–56%) indicates a constant ratio between the two states/populations of Gαs, implying that also the apparently monophasic curves with full inhibition could be comprised of two components with indistinguishable potencies. Nevertheless, none of the mentioned hypotheses provide a straightforward explanation for the lack of a consistent pattern in which combinations of Gαs mutations cause partial or mono- or biphasic inhibition.
On the other hand, with respect to our original purpose of investigating the molecular determinants for depsipeptide inhibitor selectivity for Gαq over Gαs, the results are easier to interpret. Due to the apparent difference in susceptibility of inhibition and the number of mutations necessary to introduce inhibition, we assess that it is not feasible to modify the FR900359/YM-254890 scaffold to accommodate the structural changes needed for Gαs activity, which agrees with the lack of Gs activity of the diverse range of FR900359/YM-254890 analogs tested so far (33–37). However, other molecules may be designed to target this site and constitute a path to selective Gs inhibitors. The amino acid replacements between Gαq and Gαs that introduce FR900359 inhibition are predominantly polar to hydrophobic residues with either direct interactions with the inhibitor or as part of a hydrophobic network playing a role in the conformation of the inactive GDP-bound G protein. Thus, a potential selective Gαs inhibitor acting through the investigated depsipeptide-binding site should be able to interact with these hydrophilic residues in the binding site and promote the same conformational change as observed in Gαq.
Beyond Gαs, it should be mentioned that also for the other G protein families, Gαi/o and Gα12/13, none of the published analogs of FR900359/YM-254890 to date have shown any activity (33–37). Similar to Gαs, it was recently shown that the depsipeptide-binding site can be introduced in Gαi1 (25), which again indicates that the site of action and mechanism of inhibition can also be utilized for inhibition of the Gαi/o family. As for the Gα12/13 family, only the structural and functional conservation between the four families of Gα proteins indicate that the mechanism of inhibition could also be conserved. In the absence of a selective inhibitor for Gαs, the FR900359-sensitive Gαs mutants described in the present study may prove useful to researchers in need of an alternative. Expression of these Gαs mutants may be used in experiments in which selective inhibition of, or binding to, Gαs would prove useful (e.g. stabilizing proteins for structural studies or developing disease models where delicate control over Gαs signaling would be of the essence). To this end, it should be noted that the Gαs mutants containing the L189P mutation in the Switch I loop displayed an increased basal activity (Fig. S5B). Although the increased basal activity could be inhibited by FR900359, it might still be preferential for future studies to use the “Leu-189–restored” mutant, as it displayed full monophasic inhibition by FR900359 with WT Gs-like basal activity.
Experimental procedures
Chemicals, reagents, and plasmid constructs
Unless explicitly stated otherwise, all reagents for cell culture were purchased from Thermo Fisher Scientific (Slangerup, Denmark), and all chemicals and reagents used for the assays were bought from Sigma–Aldrich (Copenhagen, Denmark). The FR900359 used in all experiments was isolated from A. crenata leaves as described previously (49).
For experiments involving Gq, the cDNA transcript of the human GNAQ gene (Homo sapiens G protein subunit αq, mRNA, accession no. NM_002072.4) was used as WT and template for further mutations. For Gs, the cDNA transcript of the human GNAS gene (Homo sapiens G protein subunit αs short, mRNA, accession no. AF493898) was used. All cDNAs and derived mutants were purchased from GenScript (Piscataway, NJ, USA) as inserts in a pcDNA3.1(+) vector.
Cell culture and transient transfection of G protein knockout HEK293 cell lines
HEK293A cells with either the Gq or Gs or subfamilies of G proteins knocked out by CRISPR/Cas9 genome editing (ΔGs-HEK293 and ΔGq/11-HEK293, respectively) have been described previously (24, 50). The cells were cultured in Dulbecco's modified Eagle's medium + Glutamax I supplemented with 10% (v/v) fetal bovine serum and 1% (v/v) penicillin-streptomycin (10,000 units/ml) and grown in a tissue culture dish kept at 37 °C in a humidified atmosphere (95% air and 5% CO2).
The cells were transfected using Lipofectamine®2000 with a reverse transfection protocol 48 h prior to experimentation. In brief, for each transfection condition, 2 ml of culture medium containing 500,000 cells was added to each well in a 6-well tissue culture plate. Subsequently, an aliquot of Opti-MEM containing 2.5 µg of G protein cDNA and 2.5 μl of Lipofectamine®2000 was carefully added to the culture media dropwise, prepared according to the manufacturer's protocol.
Functional assay
General
G protein activity was assessed by measuring the generation and accumulation of the intracellular pathway–specific molecules, cAMP and IP1, using assays based on CisBio's homogeneous time-resolved Förster resonance transfer (HTRF) technology. Specifically, the cAMP-Gs dynamic kit (CisBio, Codolet, France) was used to measure Gs activity, whereas the IP-One kit (CisBio) was used to measure Gq activity.
Serial dilutions of FR900359 were prepared from DMSO (Sigma–Aldrich) dissolved stock solutions of either 1 or 20 mm, so that the final DMSO concentration was <1% (v/v) final for every condition. Each concentration in the dilution series was added as 4 μl in technical triplicates to a Greiner Bio-One 384-well small-volume plate (In Vitro, Fredensborg, Denmark) for Gs or a 384-well OptiPlate (PerkinElmer, Skovlunde, Denmark) for Gq.
Gs, cAMP
Subconfluent, transfected ΔGs-HEK293 cells were detached using preheated (37 °C) nonenzymatic cell dissociation solution (Sigma–Aldrich), resuspended in Dulbecco's PBS, and counted on a Countess II automated cell counter (Thermo Fisher Scientific). The cell suspension was centrifuged at 1,200 rpm on a Kubota 2010 (Kubota, Osaka, Japan) and resuspended to 500,000 cells/ml in Assay Buffer (Hanks' balanced salt solution, 20 mm HEPES, pH 7.4, 1 mm CaCl2, 1 mm MgCl2) supplemented with BSA to 0.2% (w/v) and 50 μm 3-isobutyl-1-methylxanthine. From the resultant cell suspension, 5 μl/well (2,500 cells) were added to the Greiner Micro well plate, in which titrated FR900359 was dispensed, and centrifuged at 500 rpm on a Kubota 8100 briefly and incubated for 1 h (37 °C) with an Axygen plate seal (Thermo Fisher Scientific). The cells were then treated with 1 μl/well of either isoproterenol (50 nm (10 × EC80) or 200 nm (10 × EC100)) or Assay Buffer and incubated on a Titramax 100 shaking table (Heidolph, Schwabach, Germany) for 30 min at room temperature. The assay was stopped by cell lysis, and the response was measured by adding 10 μl/well cAMP Detection Solution (cAMP conjugate and lysis buffer (CisBio, Codolet, France) containing 2.5% (v/v) anti-cAMP cryptate conjugate (CisBio) and 2.5% (v/v) cAMP-d2 conjugate (CisBio)), centrifuging the plate at 500 rpm on a Kubota 8100 (Kubota, Osaka, Japan), incubating the plate in darkness at room temperature for 1 h, and reading the plate on an EnVision Multilabel Reader (PerkinElmer Life Science, Waltham, MA, USA) with excitation at 340 nm and measuring emissions at 615 and 665 nm. The resultant fluorescent energy transfer ratios (665/615 nm) were converted to cAMP concentrations by interpolating a cAMP standard curve.
Gq, IP1
Subconfluent, transfected ΔGq/11-HEK293 cells were detached using preheated (37 °C) nonenzymatic cell dissociation solution (Sigma–Aldrich), resuspended in Dulbecco's PBS, and counted on a Countess II automated cell counter. The cell suspension was centrifuged at 1,200 rpm on a Kubota 2010 and resuspended to 2,000,000 cells/ml in Assay Buffer supplemented with BSA to 0.2% (w/v). From the resultant cell suspension, 5 μl/well (10,000 cells), were added to the OptiPlate well plate, in which titrated FR900359 was dispensed, centrifuged at 500 rpm on a Kubota 8100 briefly, and incubated for 1 h (37 °C) with a plate seal. The cells were then treated with 1 μl/well of either carbachol (0.15 mm (10 × EC80) or 1 mm (10 × EC100)) or Assay Buffer, both supplemented with LiCl (Sigma–Aldrich) to achieve a final well concentration of 20 mm and incubated for 1 h (37 °C) with an Axygen plate seal. The assay was stopped by cell lysis, and the response was measured by adding 10 μl/well IP-One Detection Solution (IP1 conjugate and lysis buffer (Cisbio) containing 2.5% (v/v) anti-IP1 cryptate conjugate (Cisbio) and 2.5% (v/v) IP1-d2 conjugate (Cisbio)), centrifuging the plate at 500 rpm on a Kubota 8100, incubating the plate in darkness at room temperature for 1 h, and reading the plate on an EnVision Multilabel Reader with excitation at 340 nm and measuring emissions at 615 and 665 nm. The resultant fluorescent energy transfer ratios (665/615 nm) were converted to IP1 concentrations by interpolating an IP1 standard curve.
Data presentation and analysis
All concentration-inhibition curves were analyzed using the nonlinear regression function, y = bottom + (top − bottom)/(1 + 10^((log IC50 − x) × Hill slope)) found in GraphPad Prism 7.0b. Likewise, statistical analysis (S.D., S.E., and 95% confidence intervals) shown in figures and tables were also calculated using GraphPad Prism 7.0b. For concentration-inhibition curves where a full curve could not be obtained (due to a low FR900359 potency), the bottom of the curve fit was constrained to levels corresponding to buffer; this is noted when applied.
Structural analysis and Gs homology model
Comparison of the YM-254890–bound and unbound structures was performed by superimposing the Gq-YM-254890 structure (PDB code 3AH8 (23)) and a Gq structure without inhibitor (PDB code 5DO9 (38)) in PyMOL (PyMOL Molecular Graphics System, version 2.0.6 (Schrödinger, LLC)) based on the backbone atoms of the GTPase domain, disregarding structurally nonconserved loops (i.e. C, CA, N, and O atoms of residues 39–61, 190–205, 225–235, 247–316, and 325–345) (RMSD = 0.654). The distances between the GTPase and helical domains of Gq in the YM-254890–bound and unbound structures were measured on the CA atoms of residues Phe-75 and Glu-191 (18.4 and 21.2 Å, respectively). Comparison of the inhibitor site in the GDP- and GTPγS-bound Gs structures was performed in a similar fashion based on the backbone atoms of the GTPase domain (i.e. C, CA, N, and O atoms of residues 52–56, 91–95, 196–200, and 208–212) (RMSD = 0.461).
The Gs-FR900359 homology model was constructed using MODELLER (51) (version 9.20), the above-mentioned Gq-YM-254890 crystal structure as template, and the sequence alignment from the CGN-server (44) (RRID:SCR_018960). To adhere as much as possible to the closely related template structure, the “very_fast” keyword was utilized to output the initial model retaining the copied coordinates for conserved residue positions. The YM-254890 inhibitor from the Gq template was included in the Gs model. To obtain G protein structures with FR900359 in the binding site, the propionyl plus isopropyl substituents of FR900359 were manually added to YM-254890 of the Gs model and the Gq structure in PyMOL, ensuring a local minimum “staggered” conformation with the least number of steric clashes.
SDS-PAGE and immunoblotting
Gα protein–deficient HEK293 cells (transfected as described previously with either WT or mutant Gα protein) were pelleted and then resuspended in radioimmune precipitation assay buffer (Sigma) supplemented with cOmplete protease inhibitor mixture (Sigma) and lysed using a probe sonicator for 30 s at 50% impulse. The lysates were then agitated for 1 h at 4 °C and subsequently centrifuged at 15,000 × g for 10 min at 4 °C.
Samples containing an equal amount of protein (as determined by a BCA assay) were prepared from the supernatants by diluting with radioimmune precipitation assay buffer (Sigma) as necessary and by adding 1 m DTT (Sigma) and NuPAGE-LDS 4× (Invitrogen) to a final concentration of 10 and 25%, respectively. Each sample was then exposed to 50 °C on a heating block for 30 s and allowed to cool for 15 min.
The ready protein samples were then separated on a 4–20% Mini-PROTEAN TGX protein gel (Bio-Rad) using electrophoresis (3 A and 200 V for 40 min) and transferred to a polyvinylidene difluoride membrane using a Trans-Blot Turbo transfer system (Bio-Rad) (2.5 A and 25 V for 7 min). The protein-loaded membranes were then blocked for 1 h using 3% BSA TBS-T buffer (0.05% Tween 20). The membrane was then cut in two just below the 70 kDa protein ladder marker to allow for detection of both sample and loading control using different antibodies.
The membrane section containing the Na+/K+ ATPase–loading control was incubated with rabbit monoclonal anti-Na+/K+ ATPase antibody (Abcam, ab76020) diluted 1:10,000 in 1% BSA TBS-T (0.05% Tween 20) overnight at 4 °C. For experiments involving Gq and its mutants, the membrane section containing the G protein was incubated with mouse monoclonal anti-Gq/11/14 antibody (Santa Cruz Biotechnology, Inc., sc-365906) diluted 1:250 in 1% BSA TBS-T (0.05% Tween 20) overnight at 4 °C. Similarly, for experiments with Gs and its mutants, the membrane section was instead incubated with goat polyclonal anti-Gs/olf (Santa Cruz Biotechnology, sc-46975) diluted 1:250 in 5% skim milk powder TBS-T (0.5% Tween 20).
The following day, the membranes were washed three times for 10 min each in TBS-T (0.05% Tween 20) on a rocking table and then incubated for 1 h with different antibodies, depending on which they had previously been incubated with. For membranes with Na+/K+ ATPase, goat polyclonal anti-rabbit HRP-conjugated antibody (Dako, P0448) diluted 1:2,000 in 1% BSA TBS-T (0.05% Tween 20) was used. For membranes with Gq, goat polyclonal anti-mouse HRP–conjugated antibody (Dako, P0447) diluted 1:2,000 in 1% BSA TBS-T (0.05% Tween 20) was used, whereas for Gs, rabbit polyclonal anti-goat HRP–conjugated antibody (Thermo Fisher Scientific, 81-1620) diluted 1:3,000 in 1% BSA TBS-T (0.05% Tween 20) was used instead. The membranes were once again washed three times for 10 min each in TBS-T (0.05% Tween 20) and then briefly incubated with the following chemiluminescent substrates: SuperSignal West Pico (Thermo Fisher Scientific, 34079) for Na+/K+ ATPase and SuperSignal ELISA Femto (Thermo Fisher Scientific, 37075) for Gq and Gs. Resultant luminescence was then measured using a FluorChem HD2 imaging set-up using exposure times ranging from 0.5 to 5 min.
Data availability
All the data described are located within the article and the supporting information.
Supplementary Material
This article contains supporting information.
Author contributions—M. W. B. and M. M. data curation; M. W. B. formal analysis; M. W. B. validation; M. W. B., K. H., M. M., and C. R. U. investigation; M. W. B. and K. H. visualization; M. W. B., K. H., M. M., C. R. U., A. I., J. M. M., and E. K. methodology; M. W. B. and K. H. writing-original draft; M. W. B., K. H., M. M., C. R. U., A. I., J. M. M., G. M. K., E. K., D. E. G., and H. B.-O. writing-review and editing; K. H. software; C. R. U., G. M. K., E. K., D. E. G., and H. B.-O. conceptualization; C. R. U., J. M. M., D. E. G., and H. B.-O. supervision; A. I., G. M. K., and E. K. resources; A. I., C. R. U., D. E. G., E. K., G. M. K., and H. B.-O. funding acquisition; D. E. G. and H. B.-O. project administration.
Funding and additional information—This work was supported by Independent Research Fund Denmark | Medical Sciences 8020-00280B (to D. E. G.); Lundbeck Foundation Grants R163-2013-16327 (to D. E. G.), R178-2014-2060 (to H. B.-O. and D. E. G.), and R171-2014-505 (to C. R. U); Novo Nordisk Foundation Grant NNF18OC0031226; German Research Foundation (DFG) Grants KO 1582/10-1 and KO 1582/10-2 within FOR2372 (to E. K.) as well as KO 902/17-1 and KO 902/17-2 (to G. M. K.); Japan Society for the Promotion of Science (JSPS) KAKENHI Grant 17K08264 (to A. I.); and Japan Agency for Medical Research and Development (AMED) Grants PRIME JP17gm5910013 and LEAP JP17gm0010004 (to A. I.).
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- GPCR
- G protein–coupled receptor
- RGS
- regulator of G protein signaling
- vdW
- van der Waals
- cAMP
- cyclic adenosine monophosphate
- Gs-11
- Gαs mutant exchanging all 11 residue positions of the YM-254890/FR900359–binding site that differ between Gαs and Gαq
- RMSD
- root mean square deviation
- GTPγS
- guanosine 5′-3-O-(thio)triphosphate
- HRP
- horseradish peroxidase.
References
- 1. Alexander S. P., Davenport A. P., Kelly E., Marrion N., Peters J. A., Benson H. E., Faccenda E., Pawson A. J., Sharman J. L., Southan C., Davies J. A., and CGTP Collaborators (2015) The Concise Guide to PHARMACOLOGY 2015/16: G protein-coupled receptors. Br. J. Pharmacol 172, 5744–5869 10.1111/bph.14748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Capote L. A., Mendez Perez R., and Lymperopoulos A. (2015) GPCR signaling and cardiac function. Eur. J. Pharmacol. 763, 143–148 10.1016/j.ejphar.2015.05.019 [DOI] [PubMed] [Google Scholar]
- 3. Ushio-Fukai M. (2009) Vascular signaling through G protein-coupled receptors: new concepts. Curr. Opin. Nephrol. Hypertens. 18, 153–159 10.1097/MNH.0b013e3283252efe [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Li Z., Zhou X., Dai Z., and Zou X. (2012) Classification of G proteins and prediction of GPCRs-G proteins coupling specificity using continuous wavelet transform and information theory. Amino Acids 43, 793–804 10.1007/s00726-011-1133-6 [DOI] [PubMed] [Google Scholar]
- 5. Husted A. S., Trauelsen M., Rudenko O., Hjorth S. A., and Schwartz T. W. (2017) GPCR-mediated signaling of metabolites. Cell Metab. 25, 777–796 10.1016/j.cmet.2017.03.008 [DOI] [PubMed] [Google Scholar]
- 6. Betke K. M., Wells C. A., and Hamm H. E. (2012) GPCR mediated regulation of synaptic transmission. Prog. Neurobiol. 96, 304–321 10.1016/j.pneurobio.2012.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hauser A. S., Attwood M. M., Rask-Andersen M., Schiöth H. B., and Gloriam D. E. (2017) Trends in GPCR drug discovery: new agents, targets and indications. Nat. Rev. Drug Discov. 16, 829–842 10.1038/nrd.2017.178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ferré S. (2015) The GPCR heterotetramer: challenging classical pharmacology. Trends Pharmacol. Sci. 36, 145–152 10.1016/j.tips.2015.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Navarro G., Cordomí A., Casadó-Anguera V., Moreno E., Cai N. S., Cortés A., Canela E. I., Dessauer C. W., Casadó V., Pardo L., Lluís C., and Ferré S. (2018) Evidence for functional pre-coupled complexes of receptor heteromers and adenylyl cyclase. Nat. Commun. 9, 1242 10.1038/s41467-018-03522-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Civciristov S., Ellisdon A. M., Suderman R., Pon C. K., Evans B. A., Kleifeld O., Charlton S. J., Hlavacek W. S., Canals M., and Halls M. L. (2018) Preassembled GPCR signaling complexes mediate distinct cellular responses to ultralow ligand concentrations. Sci Signal. 11, eaan1188 10.1126/scisignal.aan1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Oldham W. M., and Hamm H. E. (2008) Heterotrimeric G protein activation by G-protein-coupled receptors. Nat. Rev. Mol. Cell Biol. 9, 60–71 10.1038/nrm2299 [DOI] [PubMed] [Google Scholar]
- 12. Liu X., Xu X., Hilger D., Aschauer P., Tiemann J. K. S., Du Y., Liu H., Hirata K., Sun X., Guixa-Gonzalez R., Mathiesen J. M., Hildebrand P. W., and Kobilka B. K. (2019) Structural insights into the process of GPCR-G protein complex formation. Cell 177, 1243–1251.e12 10.1016/j.cell.2019.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Smrcka A. V. (2008) G protein βγ subunits: central mediators of G protein-coupled receptor signaling. Cell Mol. Life Sci. 65, 2191–2214 10.1007/s00018-008-8006-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wettschureck N., and Offermanns S. (2005) Mammalian G proteins and their cell type specific functions. Physiol. Rev. 85, 1159–1204 10.1152/physrev.00003.2005 [DOI] [PubMed] [Google Scholar]
- 15. Schröder R., Janssen N., Schmidt J., Kebig A., Merten N., Hennen S., Müller A., Blättermann S., Mohr-Andrä M., Zahn S., Wenzel J., Smith N. J., Gomeza J., Drewke C., Milligan G., et al. (2010) Deconvolution of complex G protein-coupled receptor signaling in live cells using dynamic mass redistribution measurements. Nat. Biotechnol. 28, 943–949 10.1038/nbt.1671 [DOI] [PubMed] [Google Scholar]
- 16. Christensen H. B., Gloriam D. E., Pedersen D. S., Cowland J. B., Borregaard N., and Bräuner-Osborne H. (2017) Applying label-free dynamic mass redistribution assay for studying endogenous FPR1 receptor signalling in human neutrophils. J. Pharmacol. Toxicol. Methods 88, 72–78 10.1016/j.vascn.2017.07.003 [DOI] [PubMed] [Google Scholar]
- 17. Annala S., Feng X., Shridhar N., Eryilmaz F., Patt J., Yang J., Pfeil E. M., Cervantes-Villagrana R. D., Inoue A., Häberlein F., Slodczyk T., Reher R., Kehraus S., Monteleone S., Schrage R., et al. (2019) Direct targeting of Gαq and Gα11 oncoproteins in cancer cells. Sci. Signal. 12, eaau5948 10.1126/scisignal.aau5948 [DOI] [PubMed] [Google Scholar]
- 18. Kostenis E., Pfeil E. M., and Annala S. (2020) Heterotrimeric Gq proteins as therapeutic targets? J. Biol. Chem. 295, 5206–5215 10.1074/jbc.REV119.007061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cassel D., and Pfeuffer T. (1978) Mechanism of cholera toxin action: covalent modification of the guanyl nucleotide-binding protein of the adenylate cyclase system. Proc. Natl. Acad. Sci. U.S.A. 75, 2669–2673 10.1073/pnas.75.6.2669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Katada T., and Ui M. (1982) Direct modification of the membrane adenylate cyclase system by islet-activating protein due to ADP-ribosylation of a membrane protein. Proc. Natl. Acad. Sci. U. S. A. 79, 3129–3133 10.1073/pnas.79.10.3129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Taniguchi M., Nagai K., Arao N., Kawasaki T., Saito T., Moritani Y., Takasaki J., Hayashi K., Fujita S., Suzuki K., and Tsukamoto S. (2003) YM-254890, a novel platelet aggregation inhibitor produced by Chromobacterium sp. QS3666. J. Antibiot. 56, 358–363 10.7164/antibiotics.56.358 [DOI] [PubMed] [Google Scholar]
- 22. Takasaki J., Saito T., Taniguchi M., Kawasaki T., Moritani Y., Hayashi K., and Kobori M. (2004) A novel Gαq/11-selective inhibitor. J. Biol. Chem. 279, 47438–47445 10.1074/jbc.M408846200 [DOI] [PubMed] [Google Scholar]
- 23. Nishimura A., Kitano K., Takasaki J., Taniguchi M., Mizuno N., Tago K., Hakoshima T., and Itoh H. (2010) Structural basis for the specific inhibition of heterotrimeric Gq protein by a small molecule. Proc. Natl. Acad. Sci. U. S. A. 107, 13666–13671 10.1073/pnas.1003553107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schrage R., Schmitz A.-L., Gaffal E., Annala S., Kehraus S., Wenzel D., Büllesbach K. M., Bald T., Inoue A., Shinjo Y., Galandrin S., Shridhar N., Hesse M., Grundmann M., Merten N., et al. (2015) The experimental power of FR900359 to study Gq-regulated biological processes. Nat. Commun. 6, 10156 10.1038/ncomms10156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Onken M. D., Makepeace C. M., Kaltenbronn K. M., Kanai S. M., Todd T. D., Wang S., Broekelmann T. J., Rao P. K., Cooper J. A., and Blumer K. J. (2018) Targeting nucleotide exchange to inhibit constitutively active G protein α subunits in cancer cells. Sci. Signal. 11, eaao6852 10.1126/scisignal.aao6852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Malfacini D., Patt J., Annala S., Harpsøe K., Eryilmaz F., Reher R., Crüsemann M., Hanke W., Zhang H., Tietze D., Gloriam D. E., Bräuner-Osborne H., Strømgaard K., König G. M., Inoue A., et al. (2019) Rational design of a heterotrimeric G protein α subunit with artificial inhibitor sensitivity. J. Biol. Chem. 294, 5747–5758 10.1074/jbc.RA118.007250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Prévost G. P., Lonchampt M. O., Holbeck S., Attoub S., Zaharevitz D., Alley M., Wright J., Brezak M. C., Coulomb H., Savola A., Huchet M., Chaumeron S., Nguyen Q. D., Forgez P., Bruyneel E., et al. (2006) Anticancer activity of BIM-46174, a new inhibitor of the heterotrimeric Gα/Gβγ protein complex. Cancer Res. 66, 9227–9234 10.1158/0008-5472.CAN-05-4205 [DOI] [PubMed] [Google Scholar]
- 28. Ayoub M. A., Damian M., Gespach C., Ferrandis E., Lavergne O., De Wever O., Banères J. L., Pin J. P., and Prévost G. P. (2009) Inhibition of heterotrimeric G protein signaling by a small molecule acting on Gα subunit. J. Biol. Chem. 284, 29136–29145 10.1074/jbc.M109.042333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Smrcka A. V. (2013) Molecular targeting of Gα and Gβγ subunits: a potential approach for cancer therapeutics. Trends Pharmacol. Sci. 34, 290–298 10.1016/j.tips.2013.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schmitz A.-L., Schrage R., Gaffal E., Charpentier T. H., Wiest J., Hiltensperger G., Morschel J., Hennen S., Häußler D., Horn V., Wenzel D., Grundmann M., Büllesbach K. M., Schröder R., Brewitz H. H., et al. (2014) A cell-permeable inhibitor to trap Gαq proteins in the empty pocket conformation. Chem. Biol. 21, 890–902 10.1016/j.chembiol.2014.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Freissmuth M., Boehm S., Beindl W., Nickel P., Ijzerman A. P., Hohenegger M., and Nanoff C. (1996) Suramin analogues as subtype-selective G protein inhibitors. Mol. Pharmacol. 49, 602–611 [PubMed] [Google Scholar]
- 32. Hohenegger M., Waldhoer M., Beindl W., Böing B., Kreimeyer A., Nickel P., Nanoff C., and Freissmuth M. (1998) Gsα-selective G protein antagonists. Proc. Natl. Acad. Sci. U. S. A. 95, 346–351 10.1073/pnas.95.1.346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Reher R., Kühl T., Annala S., Benkel T., Kaufmann D., Nubbemeyer B., Odhiambo J. P., Heimer P., Bäuml C. A., Kehraus S., Crüsemann M., Kostenis E., Tietze D., König G. M., and Imhof D. (2018) Deciphering specificity determinants for FR900359-derived Gqα inhibitors based on computational and structure-activity studies. ChemMedChem 13, 1634–1643 10.1002/cmdc.201800304 [DOI] [PubMed] [Google Scholar]
- 34. Xiong X., Zhang H., Underwood C., Harpsøe K., Gardella T., Wöldike M., Mannstadt M., Gloriam D., Bräuner-Osborne H., and Strømgaard K. (2016) Total synthesis and structure-activity relationship studies of a series of selective G protein inhibitors. Nat. Chem. 8, 1035–1041 10.1038/nchem.2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang H., Xiong X. F., Boesgaard M. W., Underwood C. R., Bräuner-Osborne H., and Strømgaard K. (2017) Structure-activity relationship studies of the cyclic depsipeptide natural product YM-254890, targeting the Gq protein. ChemMedChem 12, 830–834 10.1002/cmdc.201700155 [DOI] [PubMed] [Google Scholar]
- 36. Zhang H., Nielsen A. L., Boesgaard M. W., Harpsøe K., Daly N. L., Xiong X. F., Underwood C. R., Haugaard-Kedström L. M., Bräuner-Osborne H., Gloriam D. E., and Strømgaard K. (2018) Structure-activity relationship and conformational studies of the natural product cyclic depsipeptides YM-254890 and FR900359. Eur. J. Med. Chem. 156, 847–860 10.1016/j.ejmech.2018.07.023 [DOI] [PubMed] [Google Scholar]
- 37. Xiong X. F., Zhang H., Boesgaard M. W., Underwood C. R., Bräuner-Osborne H., and Strømgaard K. (2019) Structure-activity relationship studies of the natural product Gq/11 protein inhibitor YM-254890. ChemMedChem 14, 865–870 10.1002/cmdc.201900018 [DOI] [PubMed] [Google Scholar]
- 38. Taylor V. G., Bommarito P. A., and Tesmer J. J. (2016) Structure of the regulator of G protein signaling 8 (RGS8)-Gαq complex: molecular basis for Gα selectivity. J. Biol. Chem. 291, 5138–5145 10.1074/jbc.M115.712075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lyon A. M., Begley J. A., Manett T. D., and Tesmer J. J. G. (2014) Molecular mechanisms of phospholipase Cβ3 autoinhibition. Structure 22, 1844–1854 10.1016/j.str.2014.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kaya A. I., Lokits A. D., Gilbert J. A., Iverson T. M., Meiler J., and Hamm H. E. (2014) A conserved phenylalanine as a relay between the α5 helix and the GDP binding region of heterotrimeric Gi protein α subunit. J. Biol. Chem. 289, 24475–24487 10.1074/jbc.M114.572875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kaya A. I., Lokits A. D., Gilbert J. A., Iverson T. M., Meiler J., and Hamm H. E. (2016) A conserved hydrophobic core in Gαi1 regulates G protein activation and release from activated receptor. J. Biol. Chem. 291, 19674–19686 10.1074/jbc.M116.745513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sunahara R. K., Tesmer J. J., Gilman A. G., and Sprang S. R. (1997) Crystal structure of the adenylyl cyclase activator Gsα. Science 278, 1943–1947 10.1126/science.278.5345.1943 [DOI] [PubMed] [Google Scholar]
- 43. Syrovatkina V., Alegre K. O., Dey R., and Huang X. Y. (2016) Regulation, signaling, and physiological functions of G-proteins. J. Mol. Biol. 428, 3850–3868 10.1016/j.jmb.2016.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Flock T., Ravarani C. N. J., Sun D., Venkatakrishnan A. J., Kayikci M., Tate C. G., Veprintsev D. B., and Babu M. M. (2015) Universal allosteric mechanism for Gα activation by GPCRs. Nature 524, 173–179 10.1038/nature14663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ross E. M. (2014) G protein-coupled receptors: multi-turnover GDP/GTP exchange catalysis on heterotrimeric G proteins. Cell. Logist. 4, e29391 10.4161/cl.29391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Harpsøe K., Ahring P. K., Christensen J. K., Jensen M. L., Peters D., and Balle T. (2011) Unraveling the high- and low-sensitivity agonist responses of nicotinic acetylcholine receptors. J. Neurosci. 31, 10759–10766 10.1523/JNEUROSCI.1509-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Busnelli M., Kleinau G., Muttenthaler M., Stoev S., Manning M., Bibic L., Howell L. A., McCormick P. J., Di Lascio S., Braida D., Sala M., Rovati G. E., Bellini T., and Chini B. (2016) Design and characterization of superpotent bivalent ligands targeting oxytocin receptor dimers via a channel-like structure. J. Med. Chem. 59, 7152–7166 10.1021/acs.jmedchem.6b00564 [DOI] [PubMed] [Google Scholar]
- 48. Brea J., Castro M., Giraldo J., López-Giménez J. F., Padín J. F., Quintián F., Cadavid M. I., Vilaró M. T., Mengod G., Berg K. A., Clarke W. P., Vilardaga J. P., Milligan G., and Loza M. I. (2009) Evidence for distinct antagonist-revealed functional states of 5-hydroxytryptamine2A receptor homodimers. Mol. Pharmacol. 75, 1380–1391 10.1124/mol.108.054395 [DOI] [PubMed] [Google Scholar]
- 49. Crüsemann M., Reher R., Schamari I., Brachmann A. O., Ohbayashi T., Kuschak M., Malfacini D., Seidinger A., Pinto-Carbó M., Richarz R., Reuter T., Kehraus S., Hallab A., Attwood M., Schiöth H. B., et al. (2018) Heterologous expression, biosynthetic studies, and ecological function of the selective Gq-signaling inhibitor FR900359. Angew. Chem. Int. Ed. Engl. 57, 836–840 10.1002/anie.201707996 [DOI] [PubMed] [Google Scholar]
- 50. Stallaert W., van der Westhuizen E. T., Schönegge A. M., Plouffe B., Hogue M., Lukashova V., Inoue A., Ishida S., Aoki J., Le Gouill C., and Bouvier M. (2017) Purinergic receptor transactivation by the β2-adrenergic receptor increases intracellular Ca2+ in nonexcitable cells. Mol. Pharmacol. 91, 533–544 10.1124/mol.116.106419 [DOI] [PubMed] [Google Scholar]
- 51. Webb B., and Sali A. (2016) Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinformatics 54, 5.6.1–5.6.37 10.1002/cpbi.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data described are located within the article and the supporting information.